Abstract
GATA transcription factors are a group of DNA binding proteins broadly distributed in eukaryotes. The GATA factors DNA binding domain is a class IV zinc finger motif in the form CX2CX17–20CX2C followed by a basic region. In plants, GATA DNA motifs have been implicated in light-dependent and nitrate-dependent control of transcription. Herein, we show that the Arabidopsis and the rice (Oryza sativa) genomes present 29 and 28 loci, respectively, that encode for putative GATA factors. A phylogenetic analysis of the 57 GATA factors encoding genes, as well as the study of their intron-exon structure, indicates the existence of seven subfamilies of GATA genes. Some of these subfamilies are represented in both species but others are exclusive for one of them. In addition to the GATA zinc finger motif, polypeptides of the different subfamilies are characterized by the presence of additional domains such as an acidic domain, a CCT (CONSTANS, CO-like, and TOC1) domain, or a transposase-like domain also found in FAR1 and FHY3. Subfamily VI comprises genes that encode putative bi-zinc finger polypeptides, also found in metazoan and fungi, and a tri-zinc finger protein which has not been previously reported in eukaryotes. The phylogeny of the GATA zinc finger motif, excluding flanking regions, evidenced the existence of four classes of GATA zinc fingers, three of them containing 18 residues in the zinc finger loop and one containing a 20-residue loop. Our results support multiple models of evolution of the GATA gene family in plants including gene duplication and exon shuffling.
GATA factors are a class of transcriptional regulators present in fungi, metazoans, and plants that normally recognize the consensus sequence WGATAR (W = T or A; R = G or A; Lowry and Atchley, 2000). The DNA binding domain of GATA factors is constituted by a type IV zinc finger in the form CX2CX17–20CX2C followed by a highly basic region. In animals, where GATA factors have been shown to play critical roles in development, differentiation, and control of cell proliferation, the GATA DNA binding domain adopts the form CX2CX17CX2C (Patient and McGhee, 2002). Vertebrate and many invertebrate GATA proteins present two of these zinc fingers, where only the C-terminal finger (C-finger) is involved in DNA binding. The N-terminal zinc finger (N-finger) can modulate the binding of the C-finger to specific GATA sites (Trainor et al., 2000), bind DNA with different specificity (Pedone et al., 1997; Newton et al., 2001), or mediate the interaction with transcription cofactors of the Friend of GATA (FOG) family (Tsang et al., 1997; Fox et al., 1998). The majority of the fungal GATA factors contain a single zinc finger domain and mostly fall into two different categories: those with 17-residue loops (CX2CX17CX2C; also called zinc finger type IVa) and those with 18-residue loops (CX2CX18CX2C; also called zinc finger type IVb; Teakle and Gilmartin, 1998). Nineteen- and 20-residue zinc finger loops are also found, albeit rarely, in fungi. For example, ASH1 is a Saccharomyces cerevisiae GATA factor with 20 residues in the zinc finger loop that binds to the promoter of the HO nuclease gene (Maxon and Herskowitz, 2001). Fungal GATA factors have been shown to be involved in diverse functions such as nitrogen control, siderophore biosynthesis, light-regulated photomorphogenesis, circadian regulation, and mating-type switching (for review, see Scazzocchio, 2000). The structures of two GATA binding domains have been solved by NMR: the chicken GATA1 (cGATA1) C-finger (Omichinski et al., 1993) and the Aspergillus nidulans AreA single zinc finger (Starich et al., 1998). The domain is formed by two anti-parallel β-sheets followed by an α-helix and a nonstructured basic tail. Side chains of the zinc finger make hydrophobic contacts in the major groove of the DNA whereas the carboxy-terminal basic tail contacts the phosphate backbone in the minor groove in the case of the cGATA1 structure and in the major groove in the case of the A. nidulans structure.
Evidence of the existence of GATA factors in plants came first with the identification of GATA motifs in the regulatory regions of light and circadian clock responsive genes (for review, see Terzaghi and Cashmore, 1995; Argüello-Astorga and Herrera-Estrella, 1998). In vitro electrophoretic mobility shift assays and DNase I footprinting experiments carried out with plant nuclear extracts also demonstrated the existence of constitutive and light-regulated GATA DNA binding activities strongly suggesting a role for plant GATA factors in light-mediated regulation (Lam and Chua, 1989; Schindler and Cashmore, 1990; Borello et al., 1993). On the other hand, in vivo footprinting experiments also demonstrated the existence of GATA DNA motifs in the promoter of the spinach (Spinacia oleracea) nitrite reductase gene that were differentially protected depending on the availability of ammonium (Rastogi et al., 1997). The gene for the first GATA factor identified in plants (NTL1) was isolated from tobacco (Nicotiana tabacum) by PCR as a plant homolog of the Neurospora crassa GATA factor NIT2 (Daniel-Vedele and Caboche, 1993). In this fungus, NIT2 activates the expression of genes for nitrogen metabolic enzymes during ammonium deprivation. Whether or not NTL1 is related to nitrogen control in plants has not been reported. Teakle et al. have recently demonstrated that four different but highly related Arabidopsis GATA factors (AtGATA1 to AtGATA4) are able to bind GATA and GAT motifs previously defined as targets for nuclear GATA binding proteins (Teakle et al., 2002). All the characterized GATA factors in plants present one single zinc finger domain with 18 residues in the zinc finger loop. Interestingly, the in vivo function of the plant GATA factors remains very poorly defined. As a first step toward understanding the role of GATA proteins in plants we set out to conduct a genome-wide survey of GATA related sequences in Arabidopsis and rice. Our analysis demonstrates the existence of 29 different loci encoding putative GATA factors in Arabidopsis and 28 in rice. Phylogenetic and gene structure analysis shows the existence of four different subfamilies of GATA factor genes in Arabidopsis, all of them encoding proteins with one single zinc finger. Rice and Arabidopsis share three subfamilies of GATA factor genes that most likely evolved before the divergence of dicots and monocots, but several different groups of GATA factors are exclusive for rice. Interestingly, the rice genome presents two genes encoding two-zinc finger GATA factors and one gene encoding a protein with three GATA-like zinc fingers. This finding implies a revision of the current knowledge about the GATA family in plants.
RESULTS
The GATA Gene Family in Arabidopsis
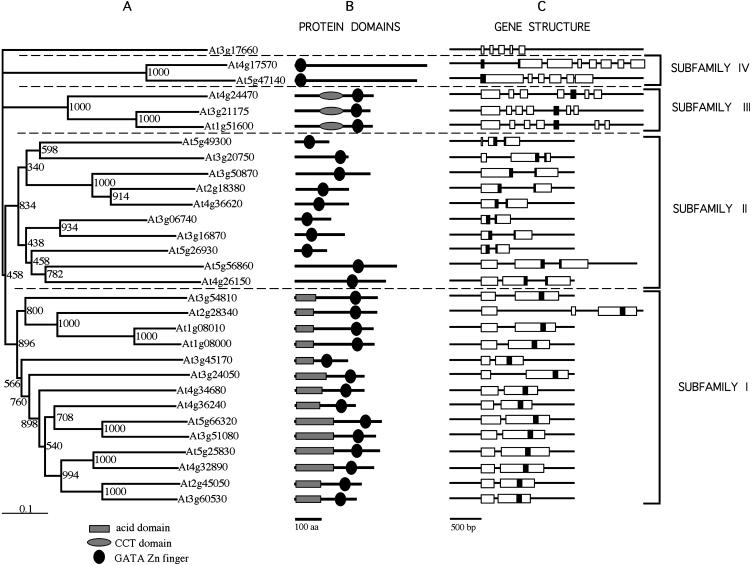
BLAST searches in available Arabidopsis databases using the Arabidopsis GATA1, the cGATA1, and the A. nidulans AreA full-length protein sequences showed the existence of 29 different Arabidopsis loci encoding proteins containing GATA-like zinc fingers (Table I). All the deduced protein sequences present only one zinc finger domain. Twenty-six of the amino acid sequences contain zinc finger motifs with 18 residues in the zinc finger loop. The other three GATA proteins (encoded by At4g24470, At3g21175, and At1g51600) exhibit zinc fingers with 20-residue loops. To determine the relationships between the different members of the GATA family in Arabidopsis we performed an alignment of the 29 full-length GATA proteins. Phylogenetic trees were generated with the Neighbor-Joining method (Saitou and Nei, 1987) using the At3g17660 sequence as outgroup. The At3g17660 gene encodes a zinc finger protein distantly related to the GATA family. The polypeptide phylogeny evidenced the existence of four well-resolved subfamilies of genes. Figure 1 shows the phylogenetic tree of polypeptide sequences together with their domains organization as well as the intron-exon organization of the corresponding genes. Subfamily I is formed by 14 genes with two exons (with one exception, At2g28340) where the 3′ last exon encodes the complete zinc finger motif and the carboxy-terminal basic region. All proteins encoded by these genes exhibit a single zinc finger with 18 residues in the zinc finger loop (CX2CX18CX2C) and an acidic amino-terminal domain with pI below 4. The function of this acidic domain is unknown, but acid regions are typical transactivation domains found in many different families of transcription factors (Schwechheimer et al., 1998). Subfamily II is constituted by 10 genes with two or three exons. In all genes the DNA sequence encoding the zinc finger has been split between the two last exons (Fig. 1). Subfamily II GATA factors also exhibit 18 residues in the zinc finger loop. The GATA factor encoded by the At3g20750 gene presents four residues between the first and the second Cys residues of the zinc finger (CTNMMC). A similar irregularity has been found in the Caenorhabditis elegans GATA factor END-1, which maintains its ability to recognize GATA DNA motives (Shoichet et al., 2000). Different subgroups within subfamilies I and II can be inferred from the tree; however they were supported by low bootstrapping values.
Table I.
Arabidopsis GATA genes
| AGI Name | Other Names | Amino Acid Number | Protein ID Code | REFSEQ Accession | Number of EST |
|---|---|---|---|---|---|
| Subfamily I | |||||
| At1g08000 | 308 | NP_172278 | NM_100674.2 | 6 | |
| At1g08010 | 303 | NP_172279 | NM_100675.2 | 7 | |
| At2g28340 | 315 | NP_180401 | NM_128393.1 | 0 | |
| At2g45050 | GATA-2a | 264 | NP_182031 | NM_130069.2 | 12 |
| At3g24050 | GATA-1a | 274 | NP_189047 | NM_113310.2 | 13 |
| At3g45170 | 204 | NP_190103 | NM_114386.1 | 0 | |
| At3g51080 | 312 | NP_190677 | NM_114968.2 | 2 | |
| At3g54810 | 322 | NP_850704 | NM_180373.1 | 13 | |
| At3g60530 | GATA-4a | 240 | NP_191612 | NM_115917.2 | 7 |
| At4g32890 | 308 | NP_195015 | NM_119442.2 | 10 | |
| At4g34680 | GATA-3a | 269 | NP_195194 | NM_119634.2 | 4 |
| At4g36240 | 238 | NP_195347 | NM_119792.2 | 2 | |
| At5g25830 | 331 | NP_197955 | NM_122484.1 | 4 | |
| At5g66320 | 339 | NP_201433 | NM_126030.2 | 8 | |
| Subfamily II | |||||
| At2g18380 | 207 | NP_179429 | NM_127395.1 | 0 | |
| At3g06740 | 149 | NP_566290 | NM_111554.2 | 5 | |
| At3g16870 | 190 | NP_188312 | NM_112563.2 | 5 | |
| At3g20750 | 208 | NP_188711 | NM_112966.1 | 0 | |
| At3g50870 | 294 | NP_566939 | NM_114947.2 | 3 | |
| At4g26150 | 352 | NP_194345 | NM_118748.2 | 1 | |
| At4g36620 | 211 | NP_195380.1 | NM_119825.1 | 0 | |
| At5g26930 | 120 | NP_198045 | NM_122575.1 | 1 | |
| At5g49300 | 139 | NP_199741 | NM_124307.1 | 0 | |
| At5g56860 | 398 | NP_200497 | NM_125069.2 | 3 | |
| Subfamily III | |||||
| At1g51600 | 302 | NP_564593 | NM_104038.1 | 3 | |
| At4g24470 | ZIMb | 309 | NP_849435 | NM_179104.1 | 6 |
| At3g21175 | 295 | NP_850618 | NM_180287.1 | 9 | |
| Subfamily IV | |||||
| At4g17570 | 510 | NP_193491 | NM_117864.2 | 3 | |
| At5g47140 | 470 | NP_199525 | NM_124085.2 | 0 |
Figure 1.
Phylogenetic analysis of Arabidopsis GATA genes. A, Neighbor-Joining tree of full-length amino acid sequences from Arabidopsis GATA genes. Bootstrap values from 1,000 replicates are shown. The scale bar corresponds to 0.1 estimated amino acid substitutions per site. B, Protein domain organization of the corresponding polypeptides. C, Exon-intron structure of the corresponding genes. Position of the nucleotide sequence that codifies for the GATA zinc finger is depicted in black.
Subfamily III is formed by three genes (At1g51600, At4g24470, and At3g21175) that encode GATA factors with 20 residues in the zinc finger loop (CX2CX20CX2C). These three genes are constituted by seven exons where the zinc finger is encoded by the fifth exon (Fig. 1). In addition, the three proteins of this subfamily present another conserved domain in the middle region of the protein. BLAST searches of the plant databases demonstrated that this domain is also present in the flowering time controller protein CONSTANS (CO), in other CO-like proteins (Robson et al., 2001; Griffiths et al., 2003), as well as in TOC1 (timing of cab expression1) and related pseudo response regulators (Strayer et al., 2000). This domain, called CCT (CO, COL, and TOC1), was identified for the first time in CO and it is thought to mediate protein-protein interactions. An alignment of the CCT domain of CO, TOC1, the three Arabidopsis GATA proteins of the subfamily III, and other rice ortologues is shown in Figure 2.
Figure 2.
Alignment of the CCT domains from Arabidopsis and rice GATA factors. The CCT domains of Arabidopsis TOC1 (AF272039) and CO (X94937) are also included. Identical residues in at least 8 of the 10 sequences are shaded in back.
Finally, subfamily IV is formed by two closely related genes with a nonhomogeneous intron-exon composition and is characterized by the presence of a CX2CX18CX2C zinc finger domain at the amino-terminal end of the protein. No other known domains were found in the rest of the protein (Fig. 1).
Analysis of the Arabidopsis expressed sequence tag (EST) databases indicated that partial or complete cDNA sequences have been reported for 22 of the 29 GATA genes (Table I). The total number of ESTs found for a given cDNA provides an indication of the expression level of the corresponding gene. Interestingly, the analysis of 127 ESTs evidenced that GATA genes of the subfamily I are (with some exceptions) much more represented than genes from subfamily II. Thus, 69% of the ESTs correspond to cDNAs of subfamily I genes (14 genes) while 15% and 15.5% of the ESTs correspond to cDNAs of subfamily II genes (10 genes) and III (3 genes), respectively. Finally, 3 ESTs were found for one of the genes of subfamily IV, At4g17570, but not for At5g47140.
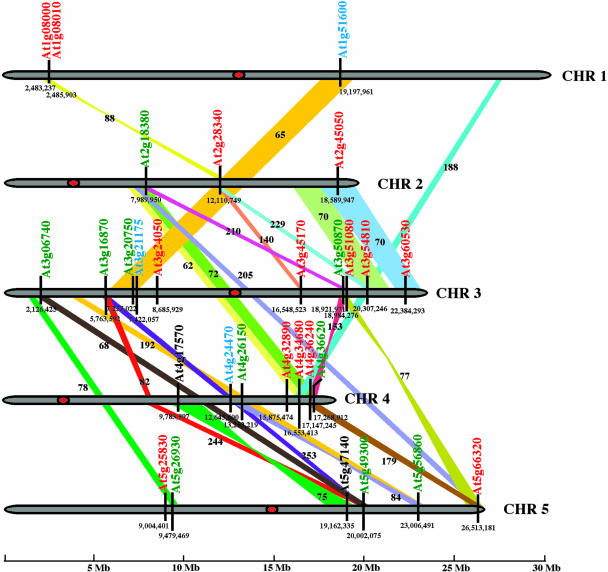
The high number of members of the Arabidopsis GATA factors family contrasts with the relatively small size of the gene family in metazoan and fungi (6–11 members), raising the question of how the expansion of this family occurred in the plant lineage. The topology of the phylogenetic tree shown in Figure 1 suggests in some cases a clear paralogous pattern of gene divergence, i.e. evolution by gene duplication. To further investigate this question we analyzed the location of GATA genes in the Arabidopsis chromosomes. Twenty-five of the 29 GATA genes are found in previously identified chromosomal duplications (Simillion et al., 2002; Fig. 3). In some of the duplication events one of the duplicated GATA genes has been lost; however, about 70% of the duplicated GATA genes have been retained. This is notably more than the 28% of gene preservation after duplications found by Simillion et al. in their whole genome analysis. In most of the cases, the paralogous relationship deduced of the duplication events is supported by the phylogeny of Figure 1. The following pair of genes are contained in duplications that occurred about 75 ± 22 million of years ago (age estimation according to Simillion et al., 2002), and therefore are close paralogous GATA genes in Arabidopsis: At1g51600 and At3g21175; At2g18380 and At4g36620; At2g45050 and At3g60530; At3g51080 and At5g66320; At4g17570 and At5g47140; and At4g26150 and At5g56860. At3g28340 is closely related to a pair of genes disposed in tandem in chromosome 1, suggesting that a first duplication gave rise to At2g28340 in chromosome 2 and the ancestor of At1g08000 and At1g08010 in chromosome 1. Subsequently, this putative gene underwent a tandem duplication event that generated At1g08000 and At1g08010. In some cases the evolutive history of some clades shown in Figure 1 can be explained in detail. For example, according to Simillion et al., a duplication occurred about 210 ± 70 million years ago between chromosomes 2 and 3 involving both At3g50870 and the ancestor of At2g18380 and At4g36620. Then a second duplication occurred between chromosomes 2 and 4, about 72 ± 20 million years ago, that originated At2g18380 and At4g36620 (Figs. 1 and 3). Similarly, the genes At3g51080, At5g66320, and At4g36240 are also the consequence of two different events of segment duplication. The genes At5g25830 and At4g32890 also appear closely related paralogues in Figure 1; however, they are not positioned in previously defined duplicated segments. These two genes could be placed in a very small duplicated chromosomal segment not previously identified, or they could have suffered a complex evolutionary history.
Figure 3.
Chromosomal positions of Arabidopsis GATA genes. Subfamily I gene are depicted in red, subfamily II genes are shown in green, subfamily III genes are shown in blue, and subfamily IV genes are shown in black. Colored bands connect corresponding duplicated segments that contain GATA genes. Numbers next to the bands indicate estimated age (in millions of years) of the duplication according to Simillion et al., 2002. Centromers are marked in red. Numbers below the genes correspond to the nucleotide chromosomal coordinates of the gene. The scale is in megabases.
The GATA Gene Family in Oryza sativa
BLAST searches in several rice databases using Arabidopsis full-length GATA protein sequences from the different subfamilies, as well as sequences from the cGATA1 and the A. nidulans AreA proteins, identified 28 different rice loci encoding proteins containing GATA-like zinc fingers (Table II). A first inspection of the amino acid sequences suggested a higher complexity and variety of GATA genes in rice in comparison to Arabidopsis. Twenty-five of the protein sequences contain only one zinc finger. Two of the sequences (OsGATA25 and OsGATA26) present two GATA-type zinc fingers, and one deduced sequence (OsGATA24) presents three GATA-type zinc fingers. In addition, OsGATA24 also contains one-half of a fourth GATA related zinc finger. While two-zinc finger GATA factors are well known in animals and some fungi, there is no previous evidence of the existence of two-GATA zinc finger proteins in plants. Furthermore, to our knowledge, proteins containing three or four GATA-type zinc fingers have never been reported in eukaryotes.
Table II.
Rice GATA genes
| Protein Name | Locus IDa | BAC or ESTb | Amino Acid Number | Number of Exons | Number of EST | Chromosome |
|---|---|---|---|---|---|---|
| Subfamily I | ||||||
| OsGATA1 | 5840.t00010 | P0483D07 | 386 | 2 | 1 | 1 |
| OsGATA2 | 4221.t00003 | P0519A12 | 387 | 3 | 2 | 2 |
| OsGATA3 | AAK98698 | OSJNBa0049O12 | 418 | 2 | 30 | 2 |
| OsGATA4 | AAO17352 | OJ1172F09 | 219 | 2 | 0 | 3 |
| OsGATA5 | 8341.t00009 | OSJNBa0011L07 | 390 | 3 | 2 | 4 |
| AK060574 | ||||||
| OsGATA6 | BAC05593 | OSJNBa0014K08 | 387 | 2 | 2 | 5 |
| OsGATA7 | AAP54978 | OSJNBa0010C11 | 387 | 2 | 3 | 10 |
| Subfamily II | ||||||
| OsGATA8 | BAB93299 | B1045F02 | 131 | 3 | 5 | 1 |
| OsGATA9 | BAB91742 | P0671D01 | 242 | 2 | 0 | 1 |
| OsGATA10 | BAC57629 | P0020E09 | 140 | 3 | 0 | 1 |
| OsGATA11 | 6135.t00004 | B1131G07 | 353 | 3 | 1 | 2 |
| AK099607 | ||||||
| OsGATA12 | AAP46233 | OSJNBb0027B08 | 136 | 3 | 6 | 3 |
| OsGATA13 | 3000.t00009 | P0431G05 | 155 | 3 | 7 | 5 |
| AA752076 | ||||||
| OsGATA14 | 4755.t00021 | OJ1781H11 | 250 | 2 | 0 | 5 |
| OsGATA15 | 2985.t00001 | OJ1126B10 | 279 | 2 | 2 | 5 |
| AK101287 | ||||||
| OsGATA16 | 3760.t00004 | OSJNBa0006A22 | 348 | 4 | 9 | 6 |
| AK069851, AK068715 | ||||||
| Subfamily III | ||||||
| OsGATA17 | 4709.t00014 | P0479D12 | 328 | 7 | 1 | 2 |
| OsGATA18 | AAK14414 | OSJNBb0072E24 | 319 | 8 | 2 | 3 |
| AK105025 | ||||||
| OsGATA19 | 3894.t00015 | OSJNBa0027J18 | 308 | 9 | 2 | 3 |
| OsGATA20 | 3408.t00024 | P0468G03 | 332 | 8 | 2 | 6 |
| OsGATA21 | 7344.t00013 | OSJNBb0077M24 | 303 | 6 | 0 | 11 |
| B1056G08 | ||||||
| Subfamily V | ||||||
| OsGATA22 | AAN65443 | OSJNBb0076N15 | 778 | 7 | 2 | 3 |
| OsGATA23 | 2129.t00011 | OJ1505_A06 | 786 | 6 | 2 | 7 |
| Subfamily VI | ||||||
| OsGATA24 | AAP54112 | OSJNBb0011A08 | 528 | 3 | 0 | 10 |
| OsGATA25 | 7071.t00014 | OJ1014_F06 | 309 | 2 | 7 | 12 |
| AK101421 | ||||||
| OsGATA26 | 6475.t00002 | OJ2007_H06 | 415 | 1 | 2 | 12 |
| AK068306 | ||||||
| Subfamily VII | ||||||
| OsGATA27 | AAM22716 | OJ1004C08.22 | 271 | 1 | 0 | 3 |
| OsGATA28 | 5259.t00014 | OSJNBb0009F15 | 431 | 1 | 0 | 11 |
Locus GenBank or TIGR accessions.
BAC or EST accessions. Two accessions, one for a BAC and one for a EST are supplied for the same locus when exist discrepancy between the ORF sequence proposed in the databases and the cDNA sequence.
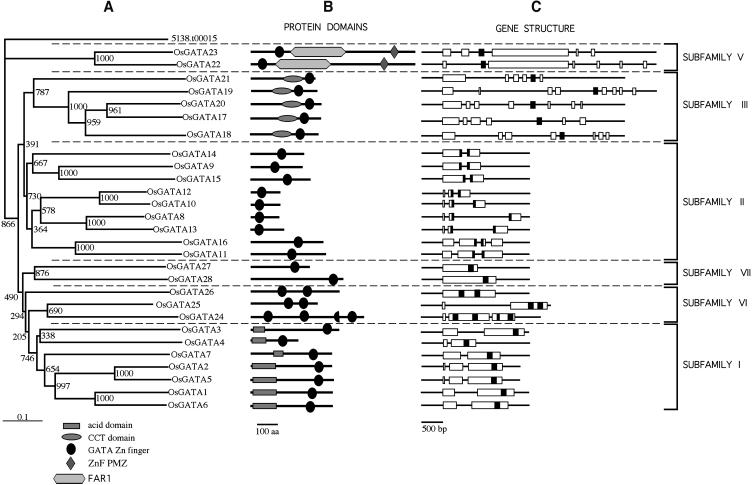
To determine the relationships among the different members of the GATA family in rice, we performed a phylogenetic analysis of the 28 full-length GATA protein sequences. After alignment, phylogenetic trees were also generated with the Neighbor-Joining method using the 5138.t00015 (TIGR locus accession) sequence as outgroup. This rice locus encodes a CX2CX17CX2C zinc finger protein distantly related to the GATA type zinc finger. Figure 4 shows the phylogenetic tree of polypeptide sequences together with the domain and the intron-exon organization of the corresponding deduced proteins and genes, respectively. The relationship between the Arabidopsis and the rice GATA proteins was investigated by generating an alignment of the 57 identified GATA protein sequences followed by the construction of a Neighbor-Joining phylogenetic tree (data not shown). The phylogeny of the rice sequences evidenced the existence of several subfamilies of GATA factors. The combined phylogeny demonstrated that subfamilies I, II, and III from Arabidopsis are also present in rice. Sequences with similar features to that of Arabidopsis subfamily IV are absent in rice. However, new subfamilies were found exclusively in rice.
Figure 4.
Phylogenetic analysis of rice GATA genes. A, Neighbor-Joining tree of full-length amino acid sequences from rice GATA genes. Bootstrap values from 1,000 replicates are shown. The scale bar corresponds to 0.1 estimated amino acid substitutions per site. B, Protein domain organization of the corresponding polypeptides. C, Exon-intron structure of the corresponding genes. Position of the nucleotide sequence that codifies for the GATA zinc finger is depicted in black.
The rice subfamily I is constituted by 7 genes with two or three exons where the 3′last exon encodes the complete zinc finger motif and the carboxy-terminal basic region (Fig. 4). As in Arabidopsis, the proteins encoded by these genes present an 18-residue zinc finger loop and an amino-terminal acidic domain. The combined phylogeny between the Arabidopsis and the rice sequences allowed us to propose putative ortholog groups of genes. For example At5g25830, At4g32890, At2g45050, and At3g60530 (subfamily I, Fig. 1) are closely related genes in Arabidopsis. The OsGATA1 and OsGATA6 genes appear more related to these genes than to other members of the rice subfamily I, suggesting that ancient plants that existed before the monocot/dicot divergence already presented two or more GATA factors of subfamily I. Subfamily II is constituted by 9 genes. Gene structure is also conserved between Arabidopsis and rice in this family. Thus, these genes present two or three exons (except OsGATA16 that has four exons) and in all cases the DNA sequence encoding the zinc finger has been split between two exons. All these proteins also present 18 residues in the zinc finger loop.
In rice, subfamily III is constituted by 6 genes that have between 6 and 9 exons. As its Arabidopsis counterparts, the zinc finger, but not the carboxy-terminal basic region, is encoded in the fifth exon. This is a small exon of around 100 bp which encodes the 28 amino acids of the zinc finger motif almost exactly. Similar to Arabidopsis, proteins encoded by subfamily III genes in rice are characterized by the presence of 20 residues in the zinc finger loop (CX2CX20CX2C) and a CCT domain (see alignment in Fig. 2). Subfamily V is constituted by a group of sequences found only in rice. The two genes that form this subfamily (OsGATA22 and OsGATA23) encode large proteins with a CX2CX20CX2C zinc finger in the amino terminal part of the protein. Searches of additional domains using the SMART (Letunic et al., 2002) and Pfam (Bateman et al., 2002) databases indicated that both proteins present a FAR1 domain (pfam03101) and a PMZ domain (plant mutator transposase zinc finger, smart00575). Both domains are found in a number of transposases of the MULE family of transposons (Lisch et al., 2001). Interestingly, both domains are also found in FAR1 and FHY3, two proteins involved in the phytochrome A signal transduction pathway (Hudson et al., 2003). Full-length or partial cDNAs for OsGATA22 and OsGATA23 have been identified, supporting the authenticity and the expression of these genes. Subfamily VI is not well supported phylogenetically, but all the genes grouped in this subfamily encode GATA factors with more than one zinc finger. In the phylogenetic tree of Figure 4, these sequences appear distantly related to sequences from subfamily I. However, these clades arise deep within the tree and their association with subfamily I and even the phylogenetic relationship between them is supported by low bootstrap values. The putative proteins OsGATA25 and OsGATA26 present two zinc fingers. The N-finger of OsGATA25 has an atypical configuration with 16 residues in the zinc finger loop. The C-finger presents the standard CX2CX18CX2C motif. OsGATA25 is encoded by two exons, being both zinc fingers encoded by the second exon. The other bi-finger protein, OsGATA26, does not seem to be closely related to OsGATA25, given the lack of conservation of the flanking regions, beside the zinc finger motif. The distance between the zinc fingers is also not conserved, being 55 and 102 amino acids in OsGATA25 and OsGATA26 sequences, respectively. Finally, both zinc fingers of OsGATA26 present the standard CX2CX18CX2C motif. The protein encoded by OsGATA24 contains three zinc fingers in the form CX2CX18CX2C. In addition, a sequence that clearly aligns with a half GATA motif is found after the second zinc finger (CRHCGSTETPLWR), which may be the remains of an ancestral entire zinc finger (see Fig. 5). OsGATA24 has three exons, where the first and the second zinc fingers are encoded in the second exon, and the third zinc finger together with the half zinc finger are encoded in the last exon. Full-length cDNAs are available for OsGATA25 and OsGATA26 but not for OsGATA24 (Table II).
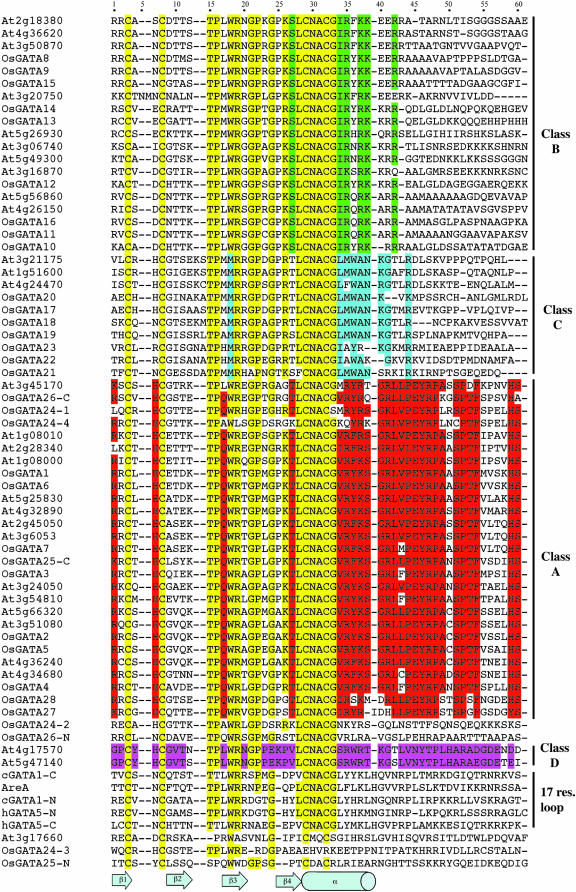
Figure 5.
Amino acid sequence alignment of Arabidopsis and rice GATA-like zinc finger domains. We aligned the 55-amino acid region of the cGATA1 sequence (residues 162–216) containing all sites that physically interact with DNA to the corresponding regions of other GATA domains. When two zinc fingers are present in the same polypeptide, the N-finger is denoted by -N and the C-finger is denoted by -C. In the case of OsGATA24 with four fingers, the different domains are numbered from the amino- to the carboxy terminus. Five nonplant zinc fingers are also included: cGATA1-N, cGATA1-C, hGATA5-N, hGATA-C, and AreA. Residues conserved in all GATA motifs or in most of the plant GATA domains are highlighted in yellow. Residues specifically conserved in Class A, B, C, or D, zinc fingers are highlighted in red, green, blue or pink, respectively. Conservative changes were defined as those that have a value higher than +2 in the BLOSUM62 scoring matrix (that means that the following amino acid changes were considered as conservatives: E-D, R-K, L-I, V-I, Y-F, Y-H, and Y-W). The bottom part of the figure shows the secondary structure elements corresponding to the indicated amino acids in the structure of the cGATA1C-finger domain for reference (Omichinski et al., 1993).
Finally, the phylogeny shown in Figure 4 groups two sequences (OsGATA27 and OsGATA28) in a well-supported branch. Deduced sequences from both genes present a single zinc finger in the form CX2CX18CX2C. In contrast to genes of other subfamilies, OsGATA27 and OsGATA28 have only one exon. In the combined phylogeny between the Arabidopsis and the rice sequences, OsGATA27 and OsGATA28 appear as an independent clade not related to any sequence from Arabidopsis (not shown). We have grouped these two genes in the subfamily VII.
Four Different GATA Zinc Finger Motifs in Plants
Next, we wanted to examine in detail the relationships between the zinc fingers of the GATA factors from different subfamilies. Structural studies have demonstrated that the chicken GATA1 (cGATA1) DNA binding domain makes specific contacts with DNA in a region of about 55 residues (from amino acid −2 to residue +53 with respect to the first Cys; Omichinski et al., 1993). We aligned the corresponding amino acid sequences from each of the Arabidopsis and rice GATA factors (Fig. 5). The bottom part of Figure 5 also shows the elements of the secondary structure of the cGATA1 C-finger as reference. For those proteins containing more than one zinc finger, separated operational taxonomic units, one for each zinc finger and basic adjacent region, were analyzed (denoted by -N or -C or by numbers in the case of the four domains of OsGATA24). In addition, the sequences of five zinc fingers from different eukaryotes have been included in the alignments: the N- and the C-finger of cGATA1 (cGATA1-N and cGATA1-C, respectively, in Fig. 5), the N- and the C-finger of human GATA5 (hGATA5-N and hGATA5-C, respectively, in Fig. 5), and the fungal AreA domain, all containing 17 residues in the zinc finger loop. A first inspection of the peptide alignment indicates that besides the four Cys, other residues of the zinc finger are generally conserved in animals, fungi, and plant sequences. Thr-15, Arg-19, Gly-24, and the amino acids around the second pair of Cys residues (L/VCNACG) are conserved in almost all the sequences, and some of them seem to be involved in maintaining the structural integrity of the zinc finger (Omichinski et al., 1993). In contrast, other residues are specific for the plant sequences, such as Pro-16, Gly-21, and Pro-22. The only sequences that do not match this consensus are OsGATA25-N, OsGATA24-2, OsGATA24-3, and OsGATA24-4, all belonging to multi-zinc finger proteins, suggesting the possibility that these domains may not be involved in DNA binding.
Figure 6 shows the phylogenetic tree constructed using the amino acid sequence alignment shown in Figure 5. Five well-supported clusters of sequences can be observed. In addition, some other branches, originating very deeply in the tree, are only distantly related to the four major groups. These clades correspond again to domains from multi-zinc finger proteins. One of the clusters contains all the CX2CX17CX2C motifs (none of them from plants). Another group comprises 27 sequences, including zinc fingers from Arabidopsis and rice GATA factors classified in subfamilies I, VI, and VII. We have named this clade Class A of plant zinc finger domains. These zinc fingers contain an 18-residue loop and are characterized by the presence of Gln and Thr in the seventh and seventeenth positions of the zinc finger loop (Gln-17 and Thr-27 in alignment of Fig. 5) in most of the sequences. All the members of this class also present high sequence conservation in the α-helix and the unstructured amino-terminal region. Class B comprises 19 plant sequences also with 18 residues in the zinc finger loop. All these zinc fingers correspond to GATA factors encoded by subfamily II genes. These domains are characterized by the presence of a Ser residue in position 29 and a conserved IRX(R/K) K sequence in position 34 to 38, which corresponds to the carboxy-terminal part of the α-helix of the cGATA1-C. Class C consist of 10 zinc fingers and all of them present 20 residues in the zinc finger loop. GATA factors from subfamilies III and V contain this type of zinc finger motif. The alignment of Figure 5 suggests that Class C domains evolved from CX2CX18CX2C zinc fingers by the insertion of two amino acids between the fourth and the fifth residues of the loop. Furthermore, these proteins present Met at position 18 instead of Trp. This Trp residue is absolutely conserved in all the CX2CX17CX2C and the CX2CX18CX2C eukaryotic GATA motifs, and it appears to be involved in maintaining the structural integrity in the metal binding region (Omichinski et al., 1993). Finally, the fourth class of plant GATA motifs, Class D, is characterized by the presence of Glu in the position 24 instead of the universally conserved Gly residue. In this case, high conservation is found all over the domain. This type of zinc finger is present in the Arabidopsis polypeptides encoded by the two genes that form subfamily IV.
Figure 6.
Phylogenetic tree of the amino acid sequences of Arabidopsis and rice GATA-like zinc finger domains. The tree was inferred by the Neighbor-Joining method from the alignment shown in Figure 5. Deduced sequence of the At3g17660 gene was used as outgroup. The scale bar corresponds to 0.1 estimated amino acid substitutions per site.
Next we wanted to examine the relationship between the plant GATA factors and the other eukaryotic GATA factors. To this end, we carried out BLAST searches of the general GenBank databases using sequences of the different subfamilies of the plant GATA factors. The first interesting result was that sequence similarity between plant GATA factors and other eukaryotic GATA factors is exclusively restricted to the DNA binding domain (zinc finger and the contiguous basic region). Then, we performed an alignment of all the Arabidopsis and rice GATA zinc finger amino acid sequences (excluding the very divergent OsGATA25-N, OsGATA26-N, OsGATA24-3, and OsGATA24-4) together with other 24 GATA zinc finger sequences from different eukaryotic origins. We have included sequences containing 17, 18, 19, or 20 amino acids in the zinc finger loop. The outcoming alignment was used to construct a phylogenetic tree (Fig. 7). The topology of the tree indicates that all the fungal and metazoan CX2CX17CX2C fingers form a well-supported clade. In addition, high bootstrapping values also support the plant Class A, B, C, and D zinc finger lineages. All these lineages originate very deeply in the tree, suggesting that ancient plants may have had members of these groups. In agreement with this, we have found sequences encoding type A, B, and C zinc fingers in the genomes and EST collections of several other angiosperms. The relationship between the fungal and the plant GATA zinc fingers is uncertain given the low support of the clades that comprise fungal and plant sequences. Interestingly, with the exception of ASH1, all the plant and fungi GATA zinc fingers with 18-, 19-, and 20-residue loops contain the conserved residues Gly-21 and Pro-22, which are not present in the CX2CX17CX2C fingers. Furthermore, all 18-residue loop zinc fingers seem to be derived from 17-residue loops by insertion of one amino acid around position 15 of the loop. Nineteen- and 20-residue loop zinc fingers also present this insertion and, therefore, seem to derivate from 18-residue zinc fingers. These data support the common origin of the fungal and plant zinc fingers and suggest a monophyletic origin for all the GATA zinc finger domains with more than 17-residue loops.
Figure 7.
Phylogenetic tree of GATA-like zinc finger domains from plant, and representative metazoan and fungi, proteins. After alignment of 87 GATA zinc finger amino acid sequences from Arabidopsis, rice, and other eukaryotes, a Neighbor-Joining tree was constructed, using the deduced sequence of the Arabidopsis At3g17660 gene as an outgroup. The triangles represent the clades comprising all Class A, B, C, and D sequences. Names of the proteins are followed by the taxa name and the number of residues in the zinc finger loop. The scale bar corresponds to 0.1 estimated amino acid substitutions per site.
DISCUSSION
Evolution and Divergence of Genes Encoding GATA Factors
Previous studies on the evolution and diversity of the GATA family of transcription factors in eukaryotes maintained that plant GATA factors possess only one zinc finger in the form CX2CX18CX2C (Teakle and Gilmartin, 1998; Lowry and Atchley, 2000). However, our study reveals that the family of GATA factors in plants is much more varied and complex. Up to seven different subfamilies of GATA genes can be defined based on their phylogenetic relationships. Further support for this classification is gained by comparison of their exon-intron structure, where the structure is conserved among the members within each subfamily. Phylogenetic analysis of the GATA DNA binding domain allows us to propose the existence of four different classes of GATA zinc fingers in plants that show important differences in the number and the type of residues in the zinc finger loop, as well as differences in the adjacent basic region. Different gene subfamilies encode proteins that present different classes of zinc fingers or the same class of zinc finger but different flanking domains. Genes from subfamilies I, VI, and VII encode GATA factors with zinc finger Class A, subfamily II genes encode proteins with a zinc finger Class B, subfamily III and V genes encode proteins with a zinc finger Class C, and finally, subfamily IV genes encode proteins with a zinc finger Class D. Genes from subfamilies I, II, and III are present both in Arabidopsis and rice, indicating that these subfamilies appeared before the divergence between monocot and dicot. The other subfamilies are not present in both species, opening the possibility that they have evolved after the divergence between monocot and dicot or that some subfamilies have been lost in one or the other species analyzed. How have these different subfamilies evolved? The topology of the phylogenetic trees suggests that evolution within each subfamily has proceeded mainly by gene duplication. This is supported by the analysis of the duplicated segments of the Arabidopsis genome (Fig. 3). However, the low level of similarity, apart from the zinc finger, between the different subfamilies argues against common ancestry for the flanking sequences, suggesting that the different subfamilies have appeared by modular evolution via shuffling of exons encoding the zinc finger domains. For example, subfamilies III and V present a zinc finger Class C (CX2CX20CX2C). This zinc finger is encoded by a small exon of about 100 bp both in genes of the subfamilies III (exon five) and V. Since subfamily III seems to be an ancient subfamily present in monocot and dicot, one possibility is that subfamily V has evolved later by a rearrangement that introduced the fifth exon of a subfamily III gene into a gene that encoded for a protein of the FAR1 family.
Despite the evolutionary history of the plant GATA gene family, an interesting aspect is the high number of GATA factors encoded by plant genomes in contrast to the relatively low number of these transcription factors found in other eukaryotes. For example, 6 GATA encoding genes are found in humans, 8 in Drosophila melanogaster, 10 in C. elegans, 11 in S. cerevisiae, and 4 in Schizosaccharomyces pombe. The reason for the expansion of this gene family in plants remains obscure but contrasts with the small number of demonstrated functions of these transcription factors in Arabidopsis. On the other hand, the high number of members of the family suggests a high functional redundancy, which may explain the low success of classical genetic strategies in the elucidation of the function of GATA factors in plants.
Most of the animal GATA factors present two zinc fingers. While the C-finger is involved in DNA binding, several different functions have been attributed to the N-finger (see introduction). Two genes encoding GATA factors with two zinc fingers are found in the rice genome. Interestingly, in both cases the C-finger belongs to the Class A while the N-finger shows variations with respect to the consensus. This suggests that, as in animals, the N-finger may be involved in other functions different to DNA binding, whereas the C-finger is probably responsible for the DNA binding activity. Plant bi-zinc finger GATA factors do not seem to be closely related to animal or fungi bi-zinc finger GATA factors. This is supported by the fact that both zinc fingers of animals and fungus GATA factors present 17 residues in the zinc finger loop while rice bi-finger proteins have 18 residues in the zinc finger loops (except the OsGATA25-N that have 16). Thus, it is more likely that rice bi-finger proteins appeared by the tandem duplication of a CX2CX18CX2C zinc finger, followed by the divergence of the N-finger to adapt to their specific new function. Lowry and Atchley have suggested that the GAF2, SREP, SREA, URBS1, and SRE fungi bi-zinc finger GATA factors could have raised independently of the metazoan bi-zinc finger GATA factors (Lowry and Atchley, 2000). Interestingly, the areA300 mutant of A. nidulans is a tandem duplication that creates a protein with two fingers separated by 114 amino acids (Caddick and Arst, 1990). Therefore, the acquisition of a second zinc finger by GATA factors might have occurred independently several times in the evolution, and it is the consequence of evolutive convergence. Furthermore, OsGATA25 and OsGATA26 genes are unrelated sequences besides the zinc finger domain, which results in deep origin of their respective clades in the phylogenetic tree of Figure 4. In addition, distance between both zinc fingers is not conserved, being 55 and 102 amino acids in OsGATA25 and OsGATA26 sequences, respectively. All these data suggest that OsGATA25 and OsGATA26 may be the result of two independent tandem duplications.
The OsGATA24 gene encodes a putative protein with three complete zinc fingers and a region that clearly resembles a half zinc finger. To our knowledge, this is the first time that the existence of a protein with three GATA-like zinc fingers is reported in eukaryotes (Lowry and Atchley, 2000). Although with some modifications compared to the consensus, the three OsGATA24 complete zinc fingers (OsGATA24–1, OsGATA24–2, and OsGATA24–4) are related to the Class A. The origin of OsGATA24 is uncertain. One obvious possibility is the tandem duplication of a bi-zinc finger gene. However, the very different distance between the first couple of fingers (146 amino acids) and the second couple of fingers (36 amino acids) suggests a more complex evolutionary history.
What DNA Motifs Bind the GATA Factors?
Most of the sequence conservation within each class of plant GATA zinc fingers was found in the regions that correspond to the α-helix of the cGATA1 C-finger (Fig. 5). In the structure of the cGATA1 C-finger these residues contact DNA in the major groove. This suggests that each zinc finger group may have some different DNA binding site specificities. Furthermore, the four GATA factors analyzed by Teakle et al. belong to the subfamily I; however, they show different binding specificities, indicating that not all GATA factors from the same subfamily bind to the same DNA sequence motif (Teakle et al., 2002). Most of the animal CX2CX17CX2C GATA C-fingers bind the consensus sequence WGATAR (W = T or A; R = G or A). However, the single zinc finger of the A. nidulans AreA protein, which clusters together with the animal GATA C-fingers (Lowry and Atchley, 2000; Fig. 7), is able to efficiently bind in vivo and in vitro to CGATAR sites (Ravagnani et al., 1997; Starich et al., 1998). Interestingly, the Leu to Val mutation at position 7 of the zinc finger loop results in preference for TGATAG sites over (A/C)GATAG sites. This residue is strictly conserved among all the characterized GATA factors having17-residue zinc finger loops, including both N- andC-fingers. Omichinski et al. have shown that this Leu residue contributes substantially to specific DNA binding with three different hydrophobic interactions (Omichinski et al., 1993). Plant GATA zinc fingers present 18- or 20-residue loops and therefore the role of the residue in position 7 in DNA binding may be different from that in 17-residue zinc finger loops. Nevertheless, position 7 of the zinc finger loop is Leu or Met in Class B and D plant GATA domains. Ravagnani et al. also show that substitution of Leu by Met did not change dramatically DNA binding specificity in the AreA protein (Ravagnani et al., 1997). Class A plant GATA domains present Gln in the seventh position of the zinc finger loop. Recently, Teakle et al. showed that the polypeptides encoded by At2g45050, At3g24050, At3g60530, and At4g34680 genes (AtGATA1 to AtGATA4), all containing a class A zinc finger, can bind some GATA and GAT domains but a detailed study of DNA binding specificity has not been carried out (Teakle et al., 2002). There is no amino acid conservation in the seventh position of the 20-residue loops of Class C zinc fingers. Interestingly, the only GATA factor with 20 residues in the zinc finger loop that has been carefully investigated is the S. cerevisiae ASH1 protein, which recognizes a YTGAT motif (Maxon and Herskowitz, 2001). Finally, the N. crassa proteins WC1 (White Collar-1) and WC2 (White Collar-2), whose single zinc fingers contain 18-residue loops and cluster with the plant GATA zinc fingers in the phylogenetic tree of Figure 7, bind the consensus sequence GATN (Froehlich et al., 2002). Taken together, this indicates that a considerable number of DNA motifs, including motifs different to GATA, could also be considered as potential targets of plant GATA factors.
Functions of GATA Factors in Arabidopsis and Rice
GATA DNA motifs have been mostly implicated in light-dependent gene regulation in plants. I-boxes, originally defined as GATAA sequences, and other GATA-related motifs have been found in many light-regulated genes such as the RBCS, CAB (chlorophyll A/B binding protein), and GAP (glyceraldydyde-3-phosphate dehydrogenase) genes (Castresana et al., 1987; Giuliano et al., 1988; Gilmartin et al., 1990; Jeong and Shih, 2003). Deletion of some of these elements strongly reduces promoter activity. These motifs are often associated with other light-dependent cis-regulatory elements including G-boxes (Argüello-Astorga and Herrera-Estrella, 1998). Puente et al. demonstrated that combinations of some of these motifs, but not the individual elements alone, may confer light-inducible expression to a reporter gene, independently of the basal promoter context (Puente et al., 1996). Furthermore, GATA-related motifs are also found in constitutive promoters such as the cauliflower mosaic virus 35S. Several GATA-binding activities from nuclear extracts have been characterized by gel mobility shift experiments such as 3AF1, GAF-1, and ASF-2(Lam and Chua, 1989; Gilmartin et al., 1990). The proteins responsible for these biding activities have not been identified. None of the recently characterized AtGATA1 to AtGATA4 proteins seems to have the same specificity as previously identified nuclear GATA-motifs binding activities (Teakle et al., 2002). However, these proteins are able to bind either in vitro or ex vivo (in S. cerevisiae introduced constructs) to GAF-1 and ASF2 DNA sites. Therefore, while several lines of evidence strongly suggest a role of GATA factors in light-mediated transcriptional regulation, there are not conclusive data that demonstrate this implication.
The fact that some GATA factors present domains also found in light signal transduction proteins could be related to its putative roles in light signaling. For example, GATA proteins encoded by genes of the subfamily III present a CCT domain. This domain is found in two proteins involved in light signaling: TOC1 and CO. TOC1 seems to be an important part of the circadian oscillator, controlling positively the level of LHY/CCA1 (for review, see Hayama and Coupland, 2003). TOC1 belongs to a novel family of response regulators, all of them characterized by the presence of a CCT domain (Strayer et al., 2000). CO acts between the circadian clock and genes controlling meristem identity, and therefore it has an essential role in regulating flowering time by photoperiod (Mouradov et al., 2002; Hayama and Coupland, 2003). A number of CONSTANS-like genes with unknown functions also contain a CCT domain. The role of the CCT domain is unclear (Robson et al., 2001).
Rice GATA factors encoded by subfamily V genes contain two domains also found in FAR1 and FHY3 proteins (Fig. 4). far1 and fhy3 mutants display a phenotype of reduced inhibition of hypocotyl elongation in far-red light, suggesting that they are involved in the phytochrome A signaling pathway. It has been recently reported that both proteins are related to transposases of type II MuDR family transposons (Hudson et al., 2003). Hudson et al. also have shown that, when fused to a Gal4 DNA binding domain, FAR1 can activate transcription of reporter genes containing Gal4 cis regulatory elements. This defines a new class of transcriptional regulators with transposase homology. Interestingly, OsGATA22 and OsGATA23 are naturally occurring fusions between a GATA DNA binding domain and a FAR1-like protein. Whether these domains (CCT and FAR1) have a specific role in light signaling or have a more general role in transcription is presently unknown.
Our genomic analysis shows the complexity and the potential interest of the GATA family of transcription factors in Arabidopsis and rice. Since direct genetics has not been successful in elucidating the role of these proteins in plant transcriptional regulation, reverse genetic approaches will probably be required. For those studies it will be potentially interesting to know the paralogous and ortologous relationships established in our study. It would also be interesting to establish the DNA binding specificities displayed by the different classes of plant GATA zinc fingers as well as to investigate the role of accompanying domains in light signaling and transcription.
MATERIALS AND METHODS
Sequence Selection, Gene Structure, and Localization on Chromosomes
To collect all Arabidopsis proteins containing GATA-like zinc fingers BLAST searches of the Arabidopsis genome were conducted at two different addresses: the National Center for Biological Information (NCBI; http://www.ncbi.nlm.nih.gov/BLAST/Genome/ara.html) and the Arabidopsis Information Resources (TAIR; http://www.arabidopsis.org/wublast/index2.html). BLAST searches were carried out using the amino acid sequence of several GATA factors from different origins (chicken GATA1, Aspergillus nidulans AreA, and Arabidopsis AtGATA1 and Neurospora crassa WC1 proteins). All sequences with an E-value below 4 × 10−4 were selected for further analysis. Arabidopsis nucleotide and proteins sequences as well as information regarding the gene structure was obtained from the Munich Information Center for Protein Sequences Database (MIPS, MATDB; http://mips.gsf.de/proj/thal/db). Arabidopsis EST sequences were searched in the TIGR Gene Indices at TIGR (http://tigrblast.tigr.org/tgi/), as well as in the GenBank EST collection at the TAIR BLAST 2.0 page (http://www.arabidopsis.org/Blast/), using the deduced nucleotide sequence of each Arabidopsis GATA gene. Deduced amino acid and cDNA sequences were compared, when possible, with those of the corresponding EST records.
Arabidopsis gene positions on chromosomes were determined using SeqViewer (http://arabidopsis.org/servlets/sv). Gene duplications and their presence on duplicated segments were investigated using the MIPS Redundancy Viewer (http://mips.gsf.de/proj/thal/db/gv/rv/) and the Simillion database (Simillion et al., 2002; http://www.psb.rug.ac.be/bioinformatics/simillion_pnas02).
To identify GATA transcription factor sequences in rice (Oryza sativa ssp. japonica and Oryza sativa ssp. indica), we searched four different databases using the BLAST program and derivatives: (1) sequences for japonica were obtained from the Rice Annotated Protein Database at The Institute for Genome Research (http://tigrblast.tigr.org/euk-blast/index.cgi?project=osa1); (2) genomic sequences for japonica and indica were also obtained from the rice BLAST page at the NCBI (http://www.ncbi.nlm.nih.gov/BLAST/Genome/PlantBlast.shtml?7); (3) rice EST sequences were searched in the TIGR Gene Indices at TIGR (http://tigrblast.tigr.org/tgi/); and (4) in the Knowledge-based Oryza Molecular biological Encyclopedia (http://cdna01.dna.affrc.go.jp/cDNA/Wblast.html) at the National Institute of Agrobiological Sciences. All sequences with an E-value below 4 × 10−4 were selected for further analysis. Nucleotide, amino acid sequences, gene structure, and chromosomal positions were obtained from the same databases mentioned before. Amino acid and cDNA sequences were corrected using the EST sequence information when discrepancy was found.
Sequence Annotation, Alignment, and Phylogenetic Analysis
Conserved structural or functional domains of all amino acid sequences were annotated according to SMART (Letunic et al., 2002) and Pfam (Bateman et al., 2002) databases.
Multiple alignments of amino acid sequences were performed using ClustalW (Thompson et al., 1994) or ClustalX (Thompson et al., 1997) and manually corrected. The weighing matrix used was BLOSUM62. Alignments will be provided upon request. Phylogenetic trees were constructed by the Neighbor-Joining method (Saitou and Nei, 1987) using Clustal or PAUP programs (Swofford, 1998), and 1,000 bootstrap replicates were performed.
Note Added in Proof
Analysis of the updated databases has revealed the existence of an additional rice GATA factor encoding gene not previously included in our work. We have named the gene OsGATA20 (EST GenBank accession AK070729). Inclusion of the deduced amino acid sequence in our previously generated alignments and phylogenetic trees indicates that OsGATA29 is the only rice GATA gene of subfamily IV. This implies that subfamily IV is present both in Arabidopsis and rice, suggesting that this subfamily appears also before the divergence between monocot and dicot, as previously commented for subfamilies I, II, and III.
Acknowledgments
We thank Marika Lindahl, Gabriel Gutierrez, and Jose L. Crespo for critical reading of the manuscript.
This work was supported by Ministerio de Ciencia y Tecnología (grant nos. BMC2002–03198 and BMC2001–2635) and by Junta de Andalucía (group CV1–802).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.037788.
References
- Argüello-Astorga G, Herrera-Estrella L (1998) Evolution of light-regulated plant promoters. Annu Rev Plant Physiol Plant Mol Biol 49: 525–555 [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL (2002) The Pfam protein families database. Nucleic Acids Res 30: 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borello U, Ceccarelli E, Giuliano G (1993) Constitutive, light-responsive and circadian clock-responsive factors compete for the different l box elements in plant light-regulated promoters. Plant J 4: 611–619 [DOI] [PubMed] [Google Scholar]
- Caddick MX, Arst HN, Jr. (1990) Nitrogen regulation in Aspergillus: are two fingers better than one? Gene 95: 123–127 [DOI] [PubMed] [Google Scholar]
- Castresana C, Staneloni RJ, Malik VS, Cashmore AR (1987) Molecular characterization of two clusters of genes encoding the Type I CAB polypeptides of PSII in Nicotiana plumbaginifolia. Plant Mol Biol 10: 117–126 [DOI] [PubMed] [Google Scholar]
- Daniel-Vedele F, Caboche M (1993) A tobacco cDNA clone encoding a GATA-1 zinc finger protein homologous to regulators of nitrogen metabolism in fungi. Mol Gen Genet 240: 365–373 [DOI] [PubMed] [Google Scholar]
- Fox AH, Kowalski K, King GF, Mackay JP, Crossley M (1998) Key residues characteristic of GATA N-fingers are recognized by FOG. J Biol Chem 273: 33595–33603 [DOI] [PubMed] [Google Scholar]
- Froehlich AC, Liu Y, Loros JJ, Dunlap JC (2002) White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297: 815–819 [DOI] [PubMed] [Google Scholar]
- Gilmartin PM, Sarokin L, Memelink J, Chua NH (1990) Molecular light switches for plant genes. Plant Cell 2: 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR (1988) An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA 85: 7089–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131: 1855–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Coupland G (2003) Shedding light on the circadian clock and the photoperiodic control of flowering. Curr Opin Plant Biol 6: 13–19 [DOI] [PubMed] [Google Scholar]
- Hudson ME, Lisch DR, Quail PH (2003) The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J 34: 453–471 [DOI] [PubMed] [Google Scholar]
- Jeong MJ, Shih MC (2003) Interaction of a GATA factor with cis-acting elements involved in light regulation of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase in Arabidopsis. Biochem Biophys Res Commun 300: 555–562 [DOI] [PubMed] [Google Scholar]
- Lam E, Chua NH (1989) ASF-2: a factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in Cab promoters. Plant Cell 1: 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res 30: 242–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch DR, Freeling M, Langham RJ, Choy MY (2001) Mutator transposase is widespread in the grasses. Plant Physiol 125: 1293–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry JA, Atchley WR (2000) Molecular evolution of the GATA family of transcription factors: conservation within the DNA-binding Domain. J Mol Evol 50: 103–115 [DOI] [PubMed] [Google Scholar]
- Maxon ME, Herskowitz I (2001) Ash1p is a site-specific DNA-binding protein that actively represses transcription. Proc Natl Acad Sci USA 98: 1495–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell Suppl 14: S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishii A, Takemura M, Fujita H, Shikata M, Yokota A, Kohchi T (2000) Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during the reproductive phase in Arabidopsis thaliana. Biosci Biotechnol Biochem 64: 1402–1409 [DOI] [PubMed] [Google Scholar]
- Newton A, Mackay J, Crossley M (2001) The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J Biol Chem 276: 35794–35801 [DOI] [PubMed] [Google Scholar]
- Omichinski JG, Clore GM, Schaad O, Felsenfeld G, Trainor C, Appella E, Stahl SJ, Gronenborn AM (1993) NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science 261: 438–446 [DOI] [PubMed] [Google Scholar]
- Patient RK, McGhee JD (2002) The GATA family (vertebrates and invertebrates). Curr Opin Genet Dev 12: 416–422 [DOI] [PubMed] [Google Scholar]
- Pedone PV, Omichinski JG, Nony P, Trainor C, Gronenborn AM, Clore GM, Felsenfeld G (1997) The N-terminal fingers of chicken GATA-2 and GATA-3 are independent sequence-specific DNA binding domains. EMBO J 16: 2874–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente P, Wei N, Deng XW (1996) Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J 15: 3732–3743 [PMC free article] [PubMed] [Google Scholar]
- Rastogi R, Bate NJ, Sivasankar S, Rothstein SJ (1997) Footprinting of the spinach nitrite reductase gene promoter reveals the preservation of nitrate regulatory elements between fungi and higher plants. Plant Mol Biol 34: 465–476 [DOI] [PubMed] [Google Scholar]
- Ravagnani A, Gorfinkiel L, Langdon T, Diallinas G, Adjadj E, Demais S, Gorton D, Arst HN Jr, Scazzocchio C (1997) Subtle hydrophobic interactions between the seventh residue of the zinc finger loop and the first base of an HGATAR sequence determine promoter-specific recognition by the Aspergillus nidulans GATA factor AreA. EMBO J 16: 3974–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F, Costa MM, Hepworth SR, Vizir I, Pineiro M, Reeves PH, Putterill J, Coupland G (2001) Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J 28: 619–631 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Scazzocchio C (2000) The fungal GATA factors. Curr Opin Microbiol 3: 126–131 [DOI] [PubMed] [Google Scholar]
- Schindler U, Cashmore AR (1990) Photoregulated gene expression may involve ubiquitous DNA binding proteins. EMBO J 9: 3415–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Smith C, Bevan MW (1998) The activities of acidic and glutamine-rich transcriptional activation domains in plant cells: design of modular transcription factors for high-level expression. Plant Mol Biol 36: 195–204 [DOI] [PubMed] [Google Scholar]
- Shoichet SA, Malik TH, Rothman JH, Shivdasani RA (2000) Action of the Caenorhabditis elegans GATA factor END-1 in Xenopus suggests that similar mechanisms initiate endoderm development in ecdysozoa and vertebrates. Proc Natl Acad Sci USA 97: 4076–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 13627–13632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich MR, Wikstrom M, Arst HN Jr, Clore GM, Gronenborn AM (1998) The solution structure of a fungal AREA protein-DNA complex: an alternative binding mode for the basic carboxyl tail of GATA factors. J Mol Biol 277: 605–620 [DOI] [PubMed] [Google Scholar]
- Starich MR, Wikstrom M, Schumacher S, Arst HN Jr, Gronenborn AM, Clore GM (1998) The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity. J Mol Biol 277: 621–634 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Swofford DL (1998) PAUP Phylogenetic Analysis Using Parsimony (and Other Methods), Version 4. Sinauer Associates, Sunderland, MA
- Teakle GR, Gilmartin PM (1998) Two forms of type IV zinc-finger motif and their kingdom-specific distribution between the flora, fauna and fungi. Trends Biochem Sci 23: 100–102 [DOI] [PubMed] [Google Scholar]
- Teakle GR, Manfield IW, Graham JF, Gilmartin PM (2002) Arabidopsis thaliana GATA factors: organisation, expression and DNA-binding characteristics. Plant Mol Biol 50: 43–57 [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR (1995) Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol 46: 445–474 [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor CD, Ghirlando R, Simpson MA (2000) GATA zinc finger interactions modulate DNA binding and transactivation. J Biol Chem 275: 28157–28166 [DOI] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH (1997) FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90: 109–119 [DOI] [PubMed] [Google Scholar]