Abstract
Recent genome wide association studies (GWAS) have implicated bridging integrator 1 (BIN1) as a late-onset Alzheimer’s disease (AD) susceptibility gene. There are at least 15 different known isoforms of BIN1, with many being expressed in the brain including the longest isoform (iso1), which is brain-specific and localizes to axon initial segments and nodes of Ranvier. It is currently unknown what role BIN1 plays in AD. We analyzed BIN1 protein expression from a large number (N = 71) of AD cases and controls from five different brain regions [hippocampus, inferior parietal (IP) cortex, inferior temporal (IT) cortex, frontal cortex (BA9), and superior and middle temporal gyri (SMTG)]. We found that the amount of the largest isoform of BIN1 was significantly reduced in the AD brain compared to age-matched controls, and smaller BIN1 isoforms were significantly increased. Further, BIN1 was significantly correlated with the amount of neurofibrillary tangle (NFT) pathology but not with either diffuse or neuritic plaques, or with the amount of amyloid-β peptide. BIN1 is known to be abnormally expressed in another human disease, myotonic dystrophy, which also features prominent NFT pathology. These data suggest that BIN1 is likely involved in AD as a modulator of NFT pathology, and that this role may extend to other human diseases that feature tau pathology.
Keywords: Alzheimer’s disease, myotonic dystrophy, tau, amyloid-β peptide, cellular nucleic acid binding protein, ZNF9
Introduction
Alzheimer’s disease (AD), the most common form of late life dementia, is characterized by two hallmark neuropathological lesions: senile plaques, composed primarily of the amyloid-β (Aβ) peptide, and neurofibrillary tangles (NFT), containing hyperphosphorylated tau protein [1]. Numerous mutations in the amyloid precursor protein (APP) gene and the presenilin 1 and 2 (PSEN1 and PSEN2, respectively) genes, which are involved in Aβ generation, are responsible for familial, early-onset AD. However, familial AD cases are rare, comprising only a small percentage of the total number. The cause of the majority of sporadic, late-onset AD cases is not known, and large scale efforts have been undertaken to discover additional genetic factors that might influence disease risk. The best understood genetic risk factor for AD is the apolipoprotein E (APOE) gene, although recent genome wide association studies (GWAS) have identified several additional genes that may contribute [2-4].
Bridging Integrator 1 (BIN1; sometimes called amphiphysin 2, or AMPH2) was originally discovered as a MYC-interacting protein and possible tumor suppressor [5]. Functional deletions of the BIN1 gene are found in many types of cancers including primary breast carcinomas [5] and metastatic prostate cancers [6]. There are at least 15 different isoforms of BIN1, with many being expressed in the brain including the longest isoform (iso1), which is brain-specific and localizes to axon initial segments and nodes of Ranvier [7]. In the central nervous system, BIN1 interacts with proteins such as clatherin, dynamin, amphiphysin 1, synaptojanin, and endophilin, and may function as an adapter protein that regulates synaptic vesicle endocytosis and cytoskeletal dynamics [8, 9]. The largest isoform encodes a predicted 95 kDa protein that is believed to be expressed exclusively in neurons. A number of smaller ubiquitously expressed BIN1 isoforms of unknown function are also present in the brain, indicating various roles for the protein depending on splicing patterns.
Recently, BIN1 has been implicated as the second most important late-onset Alzheimer’s disease susceptibility genetic locus after APOE [2-4]. The exact role that BIN1 plays in AD is currently unknown. However, two recent papers have provided some useful insights, indicating that BIN1 has little impact on APP processing or Aβ production [10], but may be a direct modulator of tau pathology [11]. Interestingly, BIN1 is also abnormally expressed in both forms of myotonic dystrophy, types I (DM1) and II (DM2), and is related to the muscle weakness phenotype [12]. Tau [13-15] and NFT pathology [16, 17] are also prominent features of both forms of myotonic dystrophy, as is neurodegeneration [18]. The expression of BIN1 in healthy and diseased brain has not been examined in much detail. In this brief report, we present evidence that the expression of BIN1 isoforms are altered in Alzheimer’s disease brain in multiple brain regions, and that this is related to NFT pathology. Interestingly, in a small preliminary study, we also discovered that the ZNF9 protein (also called the cellular nucleic acid binding protein, or CNBP [19]) was also correlated with BIN1 expression in the human brain. An expansion in the first intron of ZNF9 causes DM2. These data suggest that BIN1 is likely involved in AD as a modulator of tangle pathology, and that this role may extend to other human diseases that feature tau pathology.
Materials and Methods
Human Subjects and Tissue
Human tissue collection and handling conformed to Public Health Service (PHS) and University of Kentucky Institutional Review Board (IRB) guidelines, including written informed consent from all participants. Study of pathology specimens are covered under exemption four of the PHS guidelines and do not require a specific protocol number; all samples were coded and anonymized. Samples (N = 71) were obtained from the low post-mortem interval (PMI) tissue repository of the Alzheimer’s Disease Center at the University of Kentucky, Sanders-Brown Center on Aging. Individual case characteristics and their detailed neuropathology (including how tangles are counted) have been extensively documented elsewhere [20-22], and this set comprised both controls [n = 19, 5 M / 14 F; Age (years), 85.2 ± 11.3; PMI (hours), 3.1 ± 0.7; Brain Weight (g), 1136 ± 149; Braak (stage), 1.1 ± 1.0] and probable AD cases [n = 52, 19 M / 33 F; Age (years), 85.9 ± 7.8; PMI (hours), 2.9 ± 0.9; Brain Weight (g), 1120 ± 114; Braak (stage), 4.6 ± 1.8]. A smaller set (N = 6) was obtained from the University of Maryland Brain and Tissue Bank, including three cases of myotonic dystrophy (2 × DM1, 1 × DM2). One AD case used for immunohistochemical characterization studies was provided by the University of California Irvine Alzheimer’s Disease Research Center. Details of the tissue collection procedures and consensus diagnosis have been described previously [20-23]. Overall, the cases were broadly similar for age (83.4 ± 11.8 years), and post-mortem interval (3.7 ± 3.2 hours).
Frozen samples were homogenized using a PowerMax Advanced Homogenizing System 200 (VWR, Batavia, IL) in five volumes (wet w/v) of phosphate-buffered saline supplemented with a complete protease inhibitor cocktail with EDTA (PIC; Amresco, Solon, OH). Whole tissue homogenate was centrifuged at 2000 × g for 15 minutes to pellet insoluble material, followed by an additional spin at 20,800 × g for 30 minutes at 4 °C. Pelleted material was sequentially extracted in an equal volume of 2% SDS (w/v, with PIC) followed by 70% (v/v) formic acid (FA). In each case, the pellet was extracted by brief sonication (10 × 0.5-second microtip pulses at 20% power (100 W); Fisher sonic dismembrator, model 500, Fisher Scientific, Pittsburgh, PA) followed by centrifugation to pellet insoluble material (detergent-soluble fraction: 20,800 × g for 30 minutes at 14 °C; FA fraction: 20,800 × g for 1 hour at 4 °C). Protein content was determined by bicinchoninic acid (BCA) assay (Pierce Biotechnology, Rockford, IL). Neuropathologic data (neuritic plaques, NP; diffuse plaques, DP; neurofibrillary tangles, NFT) were provided by the Sanders-Brown COA neuropathology core. Amyloid-β peptide (for total Aβ, Aβ40, and Aβ42) immunoassay procedures and antibodies have been recently described elsewhere [20-22].
Tissue Culture
Human neuroglioma cells (H4; ATCC, Manassas, VA) were cultured in Opti-MEM supplemented with 10% FBS and 1% Pen-Strep at 37°C under 5% CO2. Cells were transfected with FuGene® HD transfection reagent (Promega, Madison, WI) and harvested 24-72 hours later. Mammalian expression vectors used were pcDNA3-BIN1(-10) (iso9; gift from Dr. George Prendergast) and pCMV6-XL5 (BIN1-1 [iso1] and empty vector; Origene, Rockville, MD). Iso1 and iso9 were the principal isoforms identified in the human brain (Figure 1), based on their relative molecular weights. Cells were lysed in 1X RIPA buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 1.0% Triton X-100, and 0.5% sodium deoxycholate) with PIC.
Figure 1. Identification of BIN1 Isoforms.

(A) The largest isoform (iso1) is found in neurons and nerve terminals, whereas the smaller isoform (iso9) is relatively generic, and is expressed ubiquitously. The major differences between these two forms are insertions encoded by variants of exons 6 and 12. (B) Comparison with H4 cells overexpressing different isoforms of BIN1 indicated that antibody 99D detects the largest full-length isoform 1 (~80 kDa; iso1) in human brain along with at least two smaller isoforms (~60kDa), one of which is likely isoform 9 (iso9; V = empty vector), based on migration at the similar relative molecular weights.
SDS-PAGE and Immunoblots
Total protein was measured by BCA assay (Pierce). Equal amounts of protein were separated using a range of SDS-PAGE Criterion (Bio-Rad, Hercules, CA) or Nu-PAGE (Invitrogen, Grand Island, NY) gels, electrically transferred to 0.45 μM nitrocellulose membranes, and blocked overnight with a solution of 1% bovine serum albumin (Calbiochem / EMD Millipore, Billerica, MA) and 2% Block Ace (AbD Serotec, Raleigh, NC) in PBS. Some human samples (PBS fraction) were directly spotted onto membranes using a MINIFOLD I spot blot system (Whatman, Piscataway, NJ). Commercially available primary antibodies used were rabbit monoclonal anti-BIN1 (Epitomics, Burlingame, CA), mouse monoclonal anti-BIN1 (99D; Sigma-Aldrich, St. Louis, MO), rabbit polyclonal anti-GAPDH (Abcam, Cambridge, MA; this antibody was HRP-conjugated and did not require a secondary antibody for detection), mouse monoclonal anti-tau (Tau46; Cell Signaling, Boston, MA), rabbit monoclonal anti-BACE1 (5606, Cell Signaling), rabbit monoclonal anti-BACE1 (D10E5; Cell Signaling), and mouse monoclonal anti-β-Actin (AC15; Sigma-Aldrich). We have previously reported the specificity of our in-house rabbit anti-ZNF9 antibody [24]. HRP-conjugated secondary antibodies and enhanced chemiluminescent detection reagents were used (Pierce), and densitometry data were obtained using Scion Image.
Immunohistochemistry
For histology experiments, human frontal cortex tissue of an individual diagnosed with Alzheimer’s disease was used (provided by the UC Irvine ADRC). Sections were cut at 50 μm and kept in 1X PBS with 0.02% sodium azide for long-term storage at 4°C. Immunohistochemical methods have been published previously [25] but briefly, sections were double-stained for BIN1 (99D; 1:3000) and PHF-1 (pSer396/Ser404; kindly provided by Dr. Peter Davies, 1:1000); both antibodies were diluted in TrisB (0.1 M Tris, 0.85% NaCl, 0.1% Triton X-100, 2.0% bovine serum albumin, pH 7.4 – 7.6). Following an overnight incubation at room temperature in the first primary antibody (PHF-1), the tissue was incubated in biotinylated secondary antibody (Vector Laboratories, Burlingame, CA). After several PBS washes, sections were incubated for one hour in an avidin-biotin complex (ABC; Vector Laboratories) and detection was performed using 3′-diaminobenzidine and hydrogen peroxide (DAB; Vector Labs). Since both antibodies used were from a mouse host, tissue was incubated in 37% formaldehyde at 37°C between stains. Following the overnight incubation at room temperature in the second primary antibody (BIN1), the tissue underwent the same protocol for both biotinylated secondary antibody and ABC (Vector Labs). However, detection was instead performed using SG substrate (Vector Labs).
Data Analysis
Data were analyzed using SPSS® (SPSS Inc., Chicago, IL). Simple group comparisons were made using either Student’s t-test, or the Mann Whitney U-test, where appropriate. Group data were analyzed by a general linear model (GLM) analysis of variance, covarying for age and postmortem interval when necessary, and post hoc comparisons were performed using Dunnett’s or Tukey’s test. Correlations were determined using either Pearson’s r or Spearman’s π, where appropriate. For multiple comparisons, inflation of type I error rate was controlled by the Holm-Bonferroni method [26].
Results
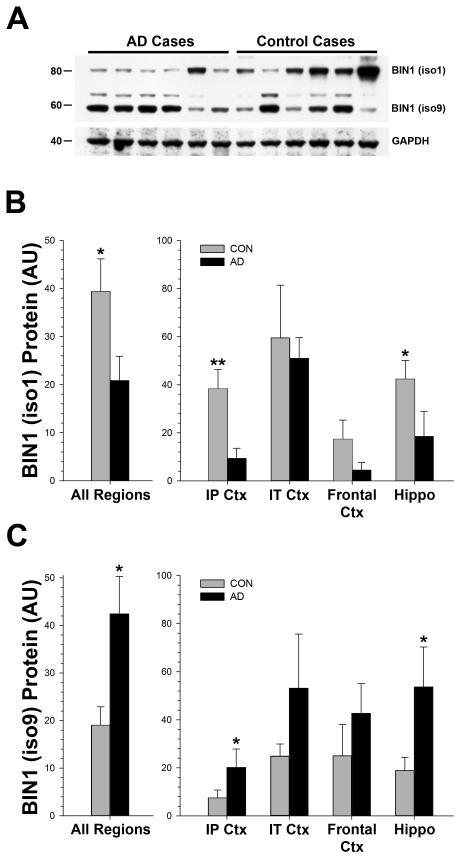
Although the overall amount of BIN1 showed a modest trend (p<0.10) towards increased levels in the AD brain across all regions analyzed, there were striking differences in the two most prominent bands, iso1 and iso9 (Figure 2). There was a consistent increase in the amount of the smaller iso9 (F[1,40] = 7.12, p < 0.02), and a corresponding decrease in the amount of the larger iso1 (F[1,40] = 6.25, p < 0.02). Similar results were obtained using a different antibody, Epitomics anti-BIN1 (Figure 3), although this antibody did not detect the larger iso1 band. Using this second antibody, we performed a densitometric analysis of all cases and brain regions, and found a similar increase in BIN1 in the AD cases (F[1,189] = 16.71, p < 0.0001). There was also a significant negative correlation between the isoform 1 and 9 bands within subjects (99D; π = −0.271, p < 0.04), and between these bands using different antibodies (99D and the Epitomics anti-BIN1 antibody; π = −0.382, p < 0.005)(not shown). These data suggest that there is a significant shift of BIN1 expression in the AD brain towards smaller isoforms of the protein.
Figure 2. Small M.W. Isoforms of BIN1 Increase in Alzheimer’s Disease Brain.
(A) Representative Western blot showing that smaller BIN1 isoforms (~40-60 kDa; antibody 99D) were increased in AD cases compared to controls (2% SDS extracts from frontal cortex, area BA9, are shown); GAPDH levels are shown to confirm equal gel loading. (B) The larger BIN1 isoform (iso1) was significantly decreased in AD cases across brain regions analyzed individually, and when all regions were combined and treated as a single variable in the analysis. (C) In contrast, the smallest isoform (iso9) showed a corresponding increase. Mann-Whitney U-Test; * = p<0.05, ** = p<0.01; error bars represent the standard error of the mean (s.e.m.).
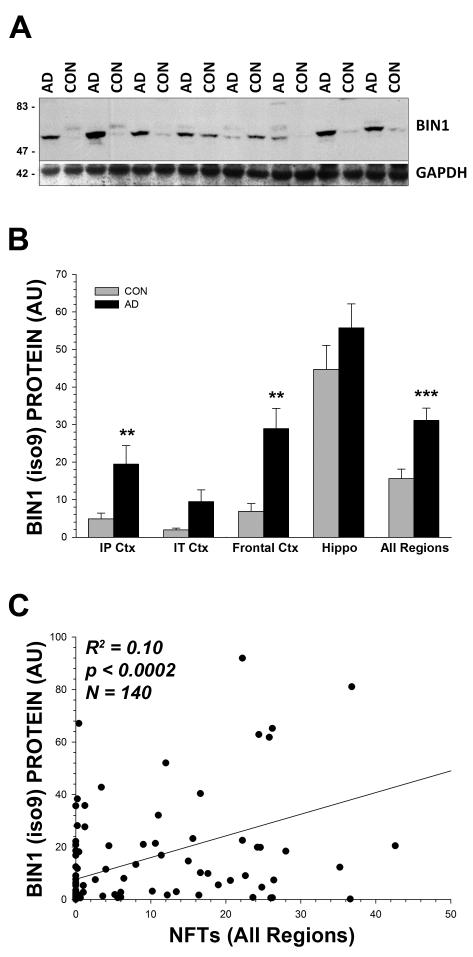
Figure 3. Confirmation of Increase in Smaller Isoforms of BIN1 and Relationship with AD Neuropathology.
(A) Representative Western blot using a different antibody (Epitomics anti-BIN1; 2% SDS extracts from frontal cortex, area BA9, are shown); GAPDH levels are shown to confirm equal gel loading. Interestingly, the Epitomics antibody was considerably less effective at detecting the higher molecular weight isoform of BIN1 (iso1), indicating that the epitope for this antibody may be modified in the human brain. (B) Densitometric analysis confirms that the smaller isoforms of BIN1 were increased across brain regions in the AD cases using this antibody (similar to Figure 2, above; the difference in the SMTG was also significant, p<0.05; not shown). (C) The smaller BIN1 isoform was significantly correlated with the number of NFTs in the brain over a large number of cases (sections were taken from the same brain region as the tissue sample used for Western blotting). Mann-Whitney U-Test; * = p<0.05, ** = p<0.01, *** = p < 0.001; error bars represent the standard error of the mean (s.e.m.).
We next wanted to examine the relationship between BIN1 expression and standard indices of AD neuropathology. The amount of BIN1 (as determined by Epitomics anti-BIN1 spot blot) was not significantly correlated with either Aβ40 or Aβ42 in any extractable fraction (PBS, SDS, or FA; mean p value = 0.57; not shown). Consistent with these data, BIN1 was also not correlated with either diffuse (π = −0.084, p < 0.4) or neuritic (π = −0.077, p < 0.4) plaques. However, the smaller BIN1 isoform (iso9) was significantly and positively correlated with the number of NFTs (π = 0.393, p < 0.0001; R2 = 0.10, p < 0.0002; Figure 3C). BIN1 was ubiquitously distributed throughout the brain, and was strongly colocalized with PHF1 positive neurons, as expected based on the NFT correlation (Supplementary Figure 1).
BIN1 is known to be abnormally expressed in another human disease, myotonic dystrophy (DM), which also features prominent NFT pathology [12-15]. We obtained a small number of cases of DM (types 1 and 2) and performed a preliminary study (Supplementary Figure 2) to determine if the relationship between NFTs and BIN1 was specific to AD or a more general phenomenon. As expected, we found differences in tau expression in samples of frontal cortex from DM brains (obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland). BIN1 expression trended towards an increase, but in these cases this was the higher molecular weight iso1. Interestingly, we also found an increase in the amount of the β-secretase enzyme BACE1, which may show general increases in neurodegenerative disease [20]. Expression of the cellular nucleic acid binding protein (CNBP), encoded by the ZNF9 gene, was also increased. A noncoding RNA expansion in this gene is known to cause type II myotonic dystrophy [27], but expression may be abnormal in both DM1 and DM2 [28], suggesting that it plays a more general role in the disease process. The amount of BIN1 was highly correlated with both tau (R2 = 0.69, p < 0.05) and CNBP (R2 = 0.53, p = 0.05) in this set; CNBP was also strongly correlated with BACE1 expression (R2 = 0.71, p < 0.02) (not shown). CNBP may have a broad role in protein translation [24, 29-31], leading us to look at the relationship between this protein and BIN1 in a subset of our AD cases (Supplementary Figure 2C). We found that CNBP expression was highly correlated with BIN1 in the AD brain (R2 = 0.65, p < 0.001).
Discussion
Recent interest in BIN1 has risen as a consequence of large, genome wide association studies linking the BIN1 gene to the risk of late-onset AD [2-4]. Although we currently know little about the biological mechanism that underlies the linkage, recent studies have connected BIN1 with the modulation of tau function [11] and with the progression of AD [32]. A recent study found a modest increase in plasma BIN1 in AD cases versus controls, and a significant decrease with disease severity, as measured by a negative correlation with MMSE scores [33]. However, there appears to be no association between tau-related CSF biomarkers and several known single nucleotide polymorphisms in BIN1 [34], and Tg2576 mice show no significant change in BIN1 protein expression [35]. Our data confirm that a connection with tau exists, and suggest that BIN1 is a likely mediator of tau expression and tangle pathology in the AD brain. We also demonstrated a tentative connection between tau and BIN1 expression in myotonic dystrophy, a neuromuscular disorder that also features NFT pathology [16, 17], and that this connection may be mediated by a DM-related protein, CNBP. How BIN1 may directly affect tau expression and, ultimately, NFT formation is unknown. The major biological role of BIN1 is in the process of receptor mediated endocytosis [36, 37], and it is possible that dysfunction in this system is responsible for the observed effects on tau. Future studies, especially in cell culture and animal models which coexpress BIN1 and tau, will be required to elucidate this interaction.
Supplementary Material
Acknowledgements
The authors thank the University of Pennsylvania viral vector core for AAV plasmids, Dr. George C. Prendergast at the Lankenau Institute for Medical Research (Wynnewood, PA) for the BIN1 plasmids, and Dr. Erin L. Abner for assistance with collating the human case data for analysis. This work was supported by the National Institutes of Health (NIH) (http://www.nih.gov) grants NS058382, NS083692, GM103486, and AG005119. Tissue was provided by the Alzheimer’s Disease Center (AG028383) at the University of Kentucky (http://www.mc.uky.edu/coa), and the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland (http://medschool.umaryland.edu/btbank) (N01-HD-9-0011). One autopsy case was provided by the UC Irvine ADRC (P50AG16573). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- [1].Niedowicz DM, Nelson PT, Murphy MP. Alzheimer’s disease: pathological mechanisms and recent insights. Curr Neuropharmacol. 2011;9:674–684. doi: 10.2174/157015911798376181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- [4].Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EM, Ramirez-Lorca R, Debette S, Longstreth WT, Jr., Janssens AC, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MM. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA: the journal of the American Medical Association. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sakamuro D, Elliott KJ, Wechsler-Reya R, Prendergast GC. BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nat Genet. 1996;14:69–77. doi: 10.1038/ng0996-69. [DOI] [PubMed] [Google Scholar]
- [6].Cher ML, Bova GS, Moore DH, Small EJ, Carroll PR, Pin SS, Epstein JI, Isaacs WB, Jensen RH. Genetic alterations in untreated metastases and androgen-independent prostate cancer detected by comparative genomic hybridization and allelotyping. Cancer Res. 1996;56:3091–3102. [PubMed] [Google Scholar]
- [7].Ren G, Vajjhala P, Lee JS, Winsor B, Munn AL. The BAR domain proteins: molding membranes in fission, fusion, and phagy. Microbiol Mol Biol Rev. 2006;70:37–120. doi: 10.1128/MMBR.70.1.37-120.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta. 2006;1761:897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- [9].Dowling JJ, Gibbs EM, Feldman EL. Membrane traffic and muscle: lessons from human disease. Traffic. 2008;9:1035–1043. doi: 10.1111/j.1600-0854.2008.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Glennon EB, Whitehouse IJ, Miners JS, Kehoe PG, Love S, Kellett KA, Hooper NM. BIN1 Is Decreased in Sporadic but Not Familial Alzheimer’s Disease or in Aging. PLoS One. 2013;8:e78806. doi: 10.1371/journal.pone.0078806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chapuis J, Hansmannel F, Gistelinck M, Mounier A, Van Cauwenberghe C, Kolen KV, Geller F, Sottejeau Y, Harold D, Dourlen P, Grenier-Boley B, Kamatani Y, Delepine B, Demiautte F, Zelenika D, Zommer N, Hamdane M, Bellenguez C, Dartigues JF, Hauw JJ, Letronne F, Ayral AM, Sleegers K, Schellens A, Broeck LV, Engelborghs S, De Deyn PP, Vandenberghe R, O’Donovan M, Owen M, Epelbaum J, Mercken M, Karran E, Bantscheff M, Drewes G, Joberty G, Campion D, Octave JN, Berr C, Lathrop M, Callaerts P, Mann D, Williams J, Buee L, Dewachter I, Van Broeckhoven C, Amouyel P, Moechars D, Dermaut B, Lambert JC. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry. 2013 doi: 10.1038/mp.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fugier C, Klein AF, Hammer C, Vassilopoulos S, Ivarsson Y, Toussaint A, Tosch V, Vignaud A, Ferry A, Messaddeq N, Kokunai Y, Tsuburaya R, de la Grange P, Dembele D, Francois V, Precigout G, Boulade-Ladame C, Hummel MC, de Munain AL, Sergeant N, Laquerriere A, Thibault C, Deryckere F, Auboeuf D, Garcia L, Zimmermann P, Udd B, Schoser B, Takahashi MP, Nishino I, Bassez G, Laporte J, Furling D, Charlet-Berguerand N. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med. 2011;17:720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- [13].Wang Y, Wang J, Gao L, Lafyatis R, Stamm S, Andreadis A. Tau exons 2 and 10, which are misregulated in neurodegenerative diseases, are partly regulated by silencers which bind a SRp30c.SRp55 complex that either recruits or antagonizes htra2beta1. J Biol Chem. 2005;280:14230–14239. doi: 10.1074/jbc.M413846200. [DOI] [PubMed] [Google Scholar]
- [14].Leroy O, Wang J, Maurage CA, Parent M, Cooper T, Buee L, Sergeant N, Andreadis A, Caillet-Boudin ML. Brain-specific change in alternative splicing of Tau exon 6 in myotonic dystrophy type 1. Biochim Biophys Acta. 2006;1762:460–467. doi: 10.1016/j.bbadis.2005.12.003. [DOI] [PubMed] [Google Scholar]
- [15].Dhaenens CM, Tran H, Frandemiche ML, Carpentier C, Schraen-Maschke S, Sistiaga A, Goicoechea M, Eddarkaoui S, Van Brussels E, Obriot H, Labudeck A, Gevaert MH, Fernandez-Gomez F, Charlet-Berguerand N, Deramecourt V, Maurage CA, Buee L, de Munain AL, Sablonniere B, Caillet-Boudin ML, Sergeant N. Mis-splicing of Tau exon 10 in myotonic dystrophy type 1 is reproduced by overexpression of CELF2 but not by MBNL1 silencing. Biochim Biophys Acta. 2011;1812:732–742. doi: 10.1016/j.bbadis.2011.03.010. [DOI] [PubMed] [Google Scholar]
- [16].Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- [17].Maurage CA, Udd B, Ruchoux MM, Vermersch P, Kalimo H, Krahe R, Delacourte A, Sergeant N. Similar brain tau pathology in DM2/PROMM and DM1/Steinert disease. Neurology. 2005;65:1636–1638. doi: 10.1212/01.wnl.0000184585.93864.4e. [DOI] [PubMed] [Google Scholar]
- [18].Minnerop M, Weber B, Schoene-Bake JC, Roeske S, Mirbach S, Anspach C, Schneider-Gold C, Betz RC, Helmstaedter C, Tittgemeyer M, Klockgether T, Kornblum C. The brain in myotonic dystrophy 1 and 2: evidence for a predominant white matter disease. Brain. 2011;134:3530–3546. doi: 10.1093/brain/awr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lusis AJ, Rajavashisth TB, Klisak I, Heinzmann C, Mohandas T, Sparkes RS. Mapping of the gene for CNBP, a finger protein, to human chromosome 3q13.3-q24. Genomics. 1990;8:411–414. doi: 10.1016/0888-7543(90)90303-c. [DOI] [PubMed] [Google Scholar]
- [20].Holler CJ, Webb RL, Laux AL, Beckett TL, Niedowicz DM, Ahmed RR, Liu Y, Simmons CR, Dowling AL, Spinelli A, Khurgel M, Estus S, Head E, Hersh LB, Murphy MP. BACE2 Expression Increases in Human Neurodegenerative Disease. The American journal of pathology. 2012;180:337–350. doi: 10.1016/j.ajpath.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Beckett TL, Webb RL, Niedowicz DM, Holler CJ, Matveev S, Baig I, LeVine H, 3rd, Keller JN, Murphy MP. Postmortem Pittsburgh Compound B (PiB) binding increases with Alzheimer’s disease progression. J Alzheimers Dis. 2012;32:127–138. doi: 10.3233/JAD-2012-120655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Niedowicz DM, Beckett TL, Matveev S, Weidner AM, Baig I, Kryscio RJ, Mendiondo MS, LeVine H, 3rd, Keller JN, Murphy MP. Pittsburgh compound B and the postmortem diagnosis of Alzheimer disease. Ann Neurol. 2012;72:564–570. doi: 10.1002/ana.23633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Murphy MP, Beckett TL, Ding Q, Patel E, Markesbery WR, St Clair DK, LeVine H, 3rd, Keller JN. Abeta solubility and deposition during AD progression and in APPxPS-1 knock-in mice. Neurobiol Dis. 2007;27:301–311. doi: 10.1016/j.nbd.2007.06.002. [DOI] [PubMed] [Google Scholar]
- [24].Niedowicz DM, Beckett TL, Holler CJ, Weidner AM, Murphy MP. APP(DeltaNL695) expression in murine tissue downregulates CNBP expression. Neurosci. Lett. 2010;482:57–61. doi: 10.1016/j.neulet.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Saing T, Dick M, Nelson PT, Kim RC, Cribbs DH, Head E. Frontal cortex neuropathology in dementia pugilistica. J Neurotrauma. 2012;29:1054–1070. doi: 10.1089/neu.2011.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Holm S. A Simple Sequentially Rejective Multiple Test Procedure. Scand J Statist. 1979;6:65–70. [Google Scholar]
- [27].Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, NAylor SL, Day JW, Ranum LPW. Mytotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- [28].Jones K, Jin B, Iakova P, Huichalaf C, Sarkar P, Schneider-Gold C, Schoser B, Meola G, Shyu AB, Timchenko N, Timchenko L. RNA Foci, CUGBP1, and ZNF9 are the primary targets of the mutant CUG and CCUG repeats expanded in myotonic dystrophies type 1 and type 2. Am J Pathol. 2011;179:2475–2489. doi: 10.1016/j.ajpath.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sammons MA, Antons AK, Bendjennat M, Udd B, Krahe R, Link AJ. ZNF9 activation of IRES-mediated translation of the human ODC mRNA is decreased in myotonic dystrophy type 2. PloS one. 2010;5:e9301. doi: 10.1371/journal.pone.0009301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Huichalaf C, Schoser B, Schneider-Gold C, Jin B, Sarkar P, Timchenko L. Reduction of the rate of protein translation in patients with myotonic dystrophy 2. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2009;29:9042–9049. doi: 10.1523/JNEUROSCI.1983-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gerbasi VR, Link AJ. The myotonic dystrophy type 2 protein ZNF9 is part of an ITAF complex that promotes cap-independent translation. Molecular & cellular proteomics: MCP. 2007;6:1049–1058. doi: 10.1074/mcp.M600384-MCP200. [DOI] [PubMed] [Google Scholar]
- [32].Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PLoS One. 2012;7:e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sun L, Tan MS, Hu N, Yu JT, Tan L. Exploring the Value of Plasma BIN1 as a Potential Biomarker for Alzheimer’s Disease. J Alzheimers Dis. 2013 doi: 10.3233/JAD-130392. [DOI] [PubMed] [Google Scholar]
- [34].Kauwe JS, Cruchaga C, Karch CM, Sadler B, Lee M, Mayo K, Latu W, Su’a M, Fagan AM, Holtzman DM, Morris JC, Alzheimer’s Disease Neuroimaging I. Goate AM. Fine mapping of genetic variants in BIN1, CLU, CR1 and PICALM for association with cerebrospinal fluid biomarkers for Alzheimer’s disease. PLoS One. 2011;6:e15918. doi: 10.1371/journal.pone.0015918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Thomas RS, Lelos MJ, Good MA, Kidd EJ. Clathrin-mediated endocytic proteins are upregulated in the cortex of the Tg2576 mouse model of Alzheimer’s disease-like amyloid pathology. Biochem Biophys Res Commun. 2011;415:656–661. doi: 10.1016/j.bbrc.2011.10.131. [DOI] [PubMed] [Google Scholar]
- [36].Pant S, Sharma M, Patel K, Caplan S, Carr CM, Grant BD. AMPH-1/Amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat Cell Biol. 2009;11:1399–1410. doi: 10.1038/ncb1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Meunier B, Quaranta M, Daviet L, Hatzoglou A, Leprince C. The membrane-tubulating potential of amphiphysin 2/BIN1 is dependent on the microtubule-binding cytoplasmic linker protein 170 (CLIP-170) Eur J Cell Biol. 2009;88:91–102. doi: 10.1016/j.ejcb.2008.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.