Abstract
Gametophytic apomixis is asexual reproduction as a consequence of parthenogenetic development of a chromosomally unreduced egg. The trait leads to the production of embryos with a maternal genotype, i.e. progeny are clones of the maternal plant. The application of the trait in agriculture could be a tremendous tool for crop improvement through conventional and nonconventional breeding methods. Unfortunately, there are no major crops that reproduce by apomixis, and interspecific hybridization with wild relatives has not yet resulted in commercially viable germplasm. Pennisetum squamulatum is an aposporous apomict from which the gene(s) for apomixis has been transferred to sexual pearl millet by backcrossing. Twelve molecular markers that are linked with apomixis coexist in a tight linkage block called the apospory-specific genomic region (ASGR), and several of these markers have been shown to be hemizygous in the polyploid genome of P. squamulatum. High resolution genetic mapping of these markers has not been possible because of low recombination in this region of the genome. We now show the physical arrangement of bacterial artificial chromosomes containing apomixis-linked molecular markers by high resolution fluorescence in situ hybridization on pachytene chromosomes. The size of the ASGR, currently defined as the entire hemizygous region that hybridizes with apomixis-linked bacterial artificial chromosomes, was estimated on pachytene and mitotic chromosomes to be approximately 50 Mbp (a quarter of the chromosome). The ASGR includes highly repetitive sequences from an Opie-2-like retrotransposon family that are particularly abundant in this region of the genome.
Apomixis is one of the most intriguing subjects in plant reproductive biology (Nogler, 1984; Koltunow, 1993; van Dijk and van Damme, 2000; Grimanelli et al., 2001; Ozias-Akins et al., 2003). Two broad modalities of seed reproduction are amphimixis and apomixis, which refer to sexual and asexual reproduction, respectively. Apomixis can be subdivided into two categories, adventitious embryony and gametophytic apomixis, and within gametophytic apomixis, apospory and diplospory. Adventitious embryos arise directly from somatic cells of the ovule. In diplospory, the megaspore mother cell does not undergo meiosis and forms an unreduced embryo sac. In apospory, the megaspore mother cell may undergo meiosis, but the products typically degenerate leaving an unreduced embryo sac that arises from a somatic cell of the ovule.
It has been reported from genetic studies that diplospory is controlled by at least two loci in dandelions (Taraxacum species; Richards, 1973; van Dijk et al., 1999), Erigeron annuus (Noyes and Rieseberg, 2000), and Tripsacum dactyloides (Blakey et al., 2001). On the other hand, apospory is controlled by a single locus, or a tight cluster of loci, in Cenchrus ciliaris (Sherwood et al., 1994), Pennisetum squamulatum (Ozias-Akins et al., 1998), Brachiaria brizantha (Pessino et al., 1997, 1998), and Paspalum simplex (Labombarda et al., 2002). P. squamulatum is one species used extensively to study apomixis. The trait has been transferred to a sexual relative, P. glaucum (pearl millet), by cross and backcross hybridization between P. squamulatum as the paternal and pearl millet as the maternal and recurrent parent (Dujardin and Hanna, 1989).
Apomixis has been genetically mapped in P. squamulatum and shown to be linked with 12 molecular markers assayed as sequence-characterized amplified regions (SCARs; Ozias-Akins et al., 1998). As the 12 SCAR markers show no recombination with the trait, the linkage block has been termed the apospory-specific genomic region (ASGR; Ozias-Akins et al., 1998; Roche et al., 1999). Bacterial artificial chromosome (BAC) libraries from an apomictic hybrid of P. squamulatum and from a related apomict, C. ciliaris, have been constructed (Roche et al., 2002). BAC clones from each library that contain low copy-number SCAR markers have been probed by fluorescence in situ hybridization (FISH) onto mitotic chromosomes of the respective species of origin and were observed to locate to a single chromosome indicating hemizygosity (Goel et al., 2003). Since the spatial resolution of physical cytogenetic mapping is 4.0 to 5.0 Mb on mitotic chromosomes, the physical order of different BACs could not be determined. FISH using pachytene chromosomes, which are less highly condensed than mitotic chromosomes, has been an effective technique for constructing molecular cytogenetic maps in species such as Arabidopsis (Fransz et al., 1996; Jackson et al., 2000; Ziolkowski and Sadowski, 2002), Lycopersicon esculentum (Zhong et al., 1998; Peterson et al., 1999), Medicago truncatula (Kulikova et al., 2001), Sorghum bicolor (Islam-Faridi et al., 2002), and rice (Oryza sativa; Cheng et al., 2001; Ohmido et al., 2001; Zhao et al., 2002).
In this study, we have used FISH on pachytene chromosomes to physically map BAC clones containing SCAR markers that show no genetic recombination. Additional BAC clones that mapped to the ASGR and contain highly repetitive sequences were used to establish tentative boundaries for the ASGR and to estimate the size of the ASGR on pachytene and mitotic chromosomes by image analysis. The results further clarify the structure of the ASGR-carrier chromosome and support previous hypotheses to explain the low level of recombination in this chromosomal region.
RESULTS
Physical Mapping of BACs on Pachytene Chromosomes
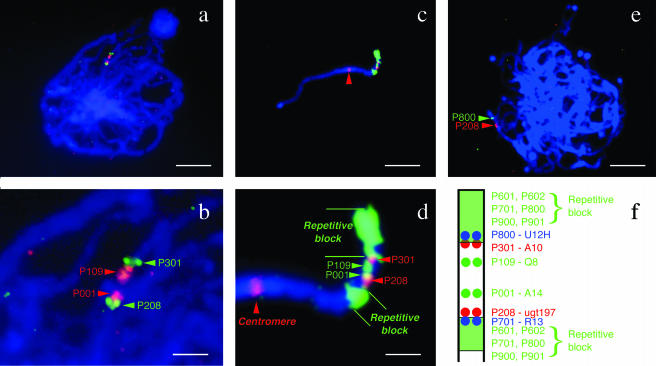
All BAC clones, labeled and hybridized to pachytene chromosomes of BC3, PS24, and PS26, showed a hybridization signal on the distal end of the short arm of a single chromosome. The relative signal positions among multiple BACs could be determined on the same pachytene chromosome by dual-color FISH, probing with two BACs at a time (Fig. 1). The three genotypes tested (accessions PS24, PS26, and backcross BC3) showed an identical arrangement of BACs on the ASGR-carrier chromosome. FISH results defined two groups of BACs. The first group of BACs (P001, P109, P205, P208, and P301) showed discrete signals that could be readily positioned with respect to one another on the pachytene chromosome (Fig. 1, a and b). The signals from hybridization of P205 and P208, that contain the same SCAR marker ugt197, were overlapping. Sometimes, the signal of P208 is separated into two sites, indicating there are duplicated loci. The second group of BACs (P601, P602, P701, P800, P900, and P901) did not demonstrate unique hybridization sites among its members, but strong signal was detected in two large blocks on the ASGR-carrier chromosome that flanked the first group of BACs (Fig. 1, c and d). This observation suggested that the second group of BACs contained similar repetitive sequences that hybridized to the two chromosomal blocks. A discrete hybridization signal could be observed when one of the BACs from the second group (P800) was used as the probe in combination with unlabeled blocking DNA from BAC P602 (Fig. 1e). P800 and P602 were combined because of the low and high frequency, respectively, of Opie-2 like retrotransposon sequences that they contained (Table I). Although the signal of P701 after blocking with P602 was not as clear compared with P800, we were able to determine that it was located close to and on the centromeric side of P208. The repetitive block signal displayed by P900 was eliminated by blocking with P602 and no low-copy signal could be detected. The physical arrangement of the BACs is schematized in Figure 1f.
Figure 1.
High resolution FISH mapping of BACs linked with apomixis on pachytene chromosomes of apomictic backcross (BC3) and P. squamulatum accessions (PS24, PS26). a, Dual-color FISH of pachytene chromosomes in BC3 using low-copy BACs containing apomixis-linked markers. b, 4× magnification of image in a. c, The ASGR-carrier chromosome at pachytene in PS24 hybridized with low- and high-copy BACs containing apomixis-linked markers. Red arrow indicates centromere. d, 4× magnification of image c. e, Dual-color FISH of pachytene chromosome spread from PS26 using labeled BACs P208 and P800 and blocked with DNA from BAC P602. f, Schematic of the BAC order as inferred from multiple FISH experiments. The bars in a, c, e and b, d = 10 μm and 2.5 μm, respectively.
Table I.
Sequences from apomixis-linked BAC clones with significant similarity to retroelements (BLASTx E value < e − 06)
| BAC ID | No. High-Quality Reads | No. Retroelement Hits | No. Opie-2-Like Hits |
|---|---|---|---|
| P602 | 85 | 25 | 20 |
| P708 | 69 | 15 | 9 |
| P800 | 77 | 3 | 3 |
| P900 | 72 | 18 | 4 |
| P002 | 73 | 18 | 0 |
| P102 | 72 | 13 | 0 |
| P201 | 69 | 7 | 0 |
Related Retrotransposon-Like Sequences in the Repetitive BACs
A few of the BAC clones that contained repetitive SCAR markers (P602, P708, P800, and P900) and hybridized to the two chromosomal blocks were shotgun cloned and sequenced at low coverage (approximately 0.5×). In the seven partially sequenced BACs (Table I), between 4% and 30% of the BLAST queries had hits (E-value < e −06) to retrotransposon-like sequences, and of that subset of sequences, 22% to 100% were similar to the Opie-2 family from maize (Zea mays; SanMiguel et al., 1996).
Sequences from BAC clones that contained low-copy SCARs (P002, P102, and P201) also had BLASTx hits to retroelements (10%–30%); however, none of these were of the Opie-2 class (Table I). This result, in addition to the ability to block the repetitive signal from P800 with the BAC that contained the highest frequency of retrotransposon-like sequences (P602), suggests that the two blocks of FISH signal at the ASGR may be due to a single retrotransposon family. To further investigate this possibility, shotgun subclones from P602, ranging in size from 2 to 4 kb, were end sequenced. Five clones that showed similarity to various regions of Opie-2-like sequences from rice (BAB03384; AAN60494) were used individually as FISH probes. Clone AA5 gave a FISH signal similar to, though less intense than P602 (Fig. 2A), and was completely sequenced (Fig. 2B; GenBank AY375366). Out of 2,443 bp, the last 467 nucleotides contain an open reading frame that is 25% identical and 45% similar to the 5′ end of the gag protein of Opie-2 from maize (Fig. 2B). When multiple shotgun clones with similarity to the Opie-2 sequences of rice were mixed together, the FISH signal was very similar to that from P602.
Figure 2.
A, FISH analysis of subclone AA5 from P602. B, Comparison of an open reading frame from subclone AA5 (AY375366) to AAN60494 (rice) and Opie-2 of maize (AAC49501).
DNA Content of the ASGR-Carrier Chromosome
We used a crude method to estimate DNA content in the ASGR-carrier chromosome based on the product of the DNA content of the total genome and the proportion of the total chromosome area occupied by the chromosome of interest. Because meiotic chromosomes are highly condensed at metaphase I, all chromosomes appear to have the same amount of DNA per unit area (based on 4′,6-diamino-phenylindole [DAPI] staining), thus relative chromosome area should be suitable to estimate DNA content in each chromosome. For such an estimate, the chromosome area was measured for each of 26 spreads of PS24 around metaphase I. The area of the ASGR-carrier chromosome, identified by hybridization with one of the repeat-containing BACs, was calculated as 1.92% ± 0.25% of the total diploid genome. This percentage is slightly higher than the average for all chromosomes of P. squamulatum (mean = 1.79%) which was calculated after determining that both PS24 and PS26 contained 56 chromosomes. Extrapolating from our genome size estimate for P. squamulatum (5,150 Mbp/1C for PS26; Roche et al., 2002), the DNA content of the ASGR-carrier chromosome is estimated to be 198 ± 26 Mbp, approximately 50% larger than the entire Arabidopsis genome.
Size of the FISH-Delimited Hemizygous Region
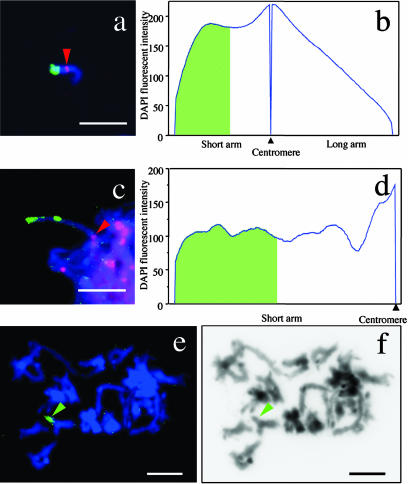
To estimate the size of the hemizygous region of the ASGR-carrier chromosome, P602 and a centromeric probe were hybridized with mitotic chromosome spreads of PS24 and PS26 and pachytene chromosome spreads of PS24 (Fig. 3). The centromeric probe was used to accurately position the centromere for arm-length measurements. In order to estimate the physical size of the hemizygous region, we analyzed at least 30 mitotic chromosomes from each genotype and 11 pachytene chromosomes from one genotype. There was little difference between the results from the two accessions of P. squamulatum or from mitotic and pachytene chromosomes. Quantitative data for characteristics of the ASGR-carrier chromosome are shown in Table II. One of these measurements examines the distribution of DAPI fluorescence intensity, which should roughly reflect the DNA distribution pattern (although results could be somewhat skewed by the A/T bias of DAPI), on the ASGR-carrier chromosome from mitotic and pachytene spreads of PS24 (Fig. 3, b and d). The DNA distribution pattern of mitotic chromosomes in PS26 showed the same pattern as PS24. The length and arm ratio of the mitotic ASGR-carrier chromosome in PS24 and PS26 was 6.91 ± 1.75 μm, 6.84 ± 1.66 μm and 1.33 ± 0.27, 1.34 ± 0.21, respectively. There was no significant difference between PS24 and PS26 in chromosome length (t = 0.24, P = 0.81) or arm ratio (t = 0.15, P = 0.88). This result indicates that the ASGR-carrier chromosome in both PS24 and PS26 is morphologically similar. For the ASGR itself, the percentage of the short arm covered in PS24 and PS26 was 58.24% ± 8.89% and 53.79% ± 8.12%, respectively. From the DNA content of the entire ASGR-carrier chromosome calculated above, the DNA content of the hemizygous region alone was estimated from the percent area under the DNA distribution curve occupied by the ASGR (Fig. 3b) to be 55.67 ± 9.74 Mbp (in PS24) and 50.61 ± 13.42 Mbp (in PS26).
Figure 3.
a, c, and e, The ASGR-carrier chromosome in PS24 hybridized with BAC P602 (green), which contains the highest frequency of Opie-2-like sequences, and a centromeric probe (red, arrowheads). b and d, The DNA distribution pattern based on DAPI fluorescence intensity. The green area indicates the hemizygous region. f, Inverted DAPI image of e. Green arrowhead indicates the hemizygous region. a and b, Mitotic chromosome. c and d, Pachytene chromosome. e and f, chromosomes at diplotene. Bars = 10 μm.
Table II.
Characteristics of the ASGR-carrier chromosome in P. squamulatum
| Mitosis
|
Pachytene
|
|||||
|---|---|---|---|---|---|---|
| PS26
|
PS24
|
PS24
|
||||
| Mean | sd | Mean | sd | Mean | sd | |
| N (number of observations) | 30 | 55 | 11 | |||
| Short Arm Length (μm) | 2.94 | 0.71 | 2.98 | 0.73 | 15.67 | 3.21 |
| Long Arm Length (μm) | 3.90 | 1.05 | 3.93 | 1.13 | - | - |
| Chromosome length (μm) | 6.84 | 1.66 | 6.91 | 1.75 | - | - |
| Arm Ratio (L/S) | 1.34 | 0.21 | 1.33 | 0.27 | - | - |
| DNA Density/Short Arm (Mbp/μm) | 45.45 | 11.17 | 42.97 | 10.11 | 8.03 | 1.52 |
| DNA Density/Long Arm (Mbp/μm) | 19.37 | 6.69 | 20.88 | 5.60 | - | - |
| DNA Density/ASGR (Mbp/μm) | 32.79 | 7.20 | 33.54 | 6.86 | 7.05 | 2.83 |
| DNA Density/Short Arm w/o ASGR | 60.89 | 19.41 | 57.93 | 20.59 | 8.89 | 1.61 |
| DNA content in ASGR (Mbp) | 50.61 | 13.42 | 55.67 | 9.74 | 48.99 | 14.40 |
The entire ASGR-carrier chromosome from pachytene spreads could rarely be traced because of its extended length and the large number of chromosomes in P. squamulatum. Therefore, only the short arm of the ASGR-carrier chromosome was measured (Fig. 3, c and d). The short arm length was 15.67 ± 3.21 μm in pachytene chromosomes, approximately 5 times greater than the length in mitotic chromosomes. The size estimate for the hemizygous region alone was 46.92% ± 6.84% of the short arm. The DNA content of the short arm on meiotic chromosomes is 121.32 ± 10.15 Mbp when calculated from the DNA content of the ASGR-carrier chromosome and the percent area of the short arm under the DNA distribution curve (Fig. 3d). The DNA content of the hemizygous region alone was estimated to be 48.99 ± 14.4 Mbp (Fig. 3d). The estimate of DNA content in the hemizygous region was slightly lower for the pachytene data, probably because of the higher resolution that could be achieved in measuring the boundaries of FISH signals.
To estimate the size of the region flanked by lower copy BACs, the distance between BACs P208 and P301 was measured on five pachytene chromosomes of BC3. The distance was 1.81 ± 0.45 μm, and the DNA content was estimated to be 12.75 ± 3.20 Mbp based on the DNA density of this region of the chromosome in PS24.
Degree of Chromatin Condensation in the ASGR-Carrier Chromosome
We evaluated whether the hemizygous region differs in its degree of chromatin condensation by comparing the DNA density of different regions of the ASGR-carrier chromosome estimated from DAPI-stained mitotic and pachytene chromosomes. The short arm of the mitotic ASGR-carrier chromosome was more highly condensed than the long arm based on DAPI staining intensity (Fig. 3b). The average DNA density on the short arm was twice that of the long arm (39.25 ± 9.23 Mbp/μm versus 19.07 ± 5.12 Mbp/μm). Across the short arm, the hemizygous region had a lower DNA density than the remaining portion of the short arm (30.64 ± 6.26 Mbp/μm versus 52.91 ± 18.80 Mbp/μm), probably because of the highly condensed pericentromeric region. The DNA distribution pattern (Fig. 3b) showed two peaks, the first around the centromere, and a second peak within the hemizygous region. At least a part of the hemizygous region forms a large condensed block at mitotic metaphase. The DNA distribution patterns in PS26 were comparable to those in PS24. In pachytene, the DNA density of the short arm of the ASGR-carrier chromosome showed five peaks (Fig. 3d), with the highest peak around the centromere. The two peaks in the ASGR were comparable in amplitude with a third peak between the hemizygous region and the centromere.
To compare condensation rates among different regions of the ASGR-carrier chromosome, we plotted the relationship between total chromosome length and the relative lengths of three chromosomal segments: the long arm of the ASGR-carrier chromosome, the hemizygous region, and the remaining segment of the short arm. Chromosome length is a function of the mitotic stage; therefore, if there are any differences in the condensation rate for each segment, relative lengths will vary. Linear regression of the percent length of a chromosomal region versus the total chromosome length resulted in the following equations: y = 51.27 + 0.76x (long arm), y = 33.00 − 1.12x (hemizygous region), y = 15.76 + 0.37x (the remaining segment of the short arm). Among these regression equations, only the hemizygous region shows a negative regression coefficient. This result indicates that the hemizygous region has a slower condensation rate than the other segments. Based on these observations, the hemizygous region, with a high DNA density and slow condensation rate during mitosis, displays characteristics that distinguish it from the remainder of the chromosome. Moreover, the extension of this region on pachytene chromosomes was only 5 to 6 times greater than on mitotic chromosomes, which is comparable to the smallest degree of extension previously described (7-fold in rye [Secale cereale] and 40-fold in rice; de Jong et al., 1999).
Pairing of the ASGR-Carrier Chromosome
The ASGR is present on only one chromosome as a hemizygous region, but the ASGR-carrier chromosome does indeed pair at meiosis with another chromosome having no ASGR (Fig. 3, e and f). Although the ASGR-carrier chromosome can form bivalents, it is possible that a portion of the chromosome is asynaptic or desynaptic. To determine if the ASGR does not synapse, we observed the ASGR-carrier chromosome at meiotic stages ranging from pachytene to diakinesis. In at least 80% of the spreads (Table III), the short arm of the ASGR-carrier chromosome appeared to be unpaired or lacking chiasmata (Fig. 3, e and f).
Table III.
Evidence for pairing or chiasmata in the short arm of the ASGR-carrier chromosome
| Diplotene/Diakinesis
|
Pachytene
|
|||||
|---|---|---|---|---|---|---|
| PS24 | PS24 | PS26 | ||||
| Number % | ||||||
| Bivalent | 4 | 7.7 | 2 | 10.5 | 6 | 18.2 |
| Univalent | 48 | 92.3 | 17 | 89.5 | 27 | 81.8 |
| Total | 52 | 100.0 | 19 | 100.0 | 33 | 100.0 |
DISCUSSION
Hemizygosity of small DNA sequences has been prevalently observed in genomic regions associated with apomixis (Ozias-Akins et al., 1998; Labombarda et al., 2002). We recently showed that BACs containing low-copy number SCARs that cosegregate with apomixis (Roche et al., 2002) were localized to a single mitotic chromosome of polyploid P. squamulatum (Goel et al., 2003). We now show that these BAC signals can be separated on pachytene chromosomes, and thus the physical order and approximate distances of the markers contained within them can be estimated. The physical ordering of BAC clones will help confirm walking steps across the locus. Identifying the order of the BAC clones will also help to determine the regions of the ASGR which are required for apomixis via deletion analysis. The discovery of repetitive sequences which flank the relatively low-copy region of the ASGR is informative for structural identification of the ASGR-carrier chromosome, since the repeat sequence organization is a unique identifier of this region of the genome.
An Extensive Hemizygous Region Resides on the ASGR-Carrier Chromosome of P. squamulatum
All 11 ASGR-linked BACs examined thus far have produced a prominent hybridization signal on only one chromosome. While Group 1 BACs gave distinct signals that could be physically ordered, Group 2 BACs displayed a repetitive signal that flanked the low-copy ASGR clones. In one instance, it was possible to determine the chromosomal location of the low-copy sequences in Group 2 BAC P800 by blocking the repeat signal. These collective results reveal that the physical size of the hemizygous region in P. squamulatum is enormous (approximately 50 Mbp or a quarter of the chromosome). The repetitive elements flanking the low-copy region of the ASGR delineate the largest physical proportion of the ASGR (approximately 74%). Why did this large hemizygous region arise on the ASGR-carrier chromosome? One possibility is that this region was derived by introgression of alien chromatin from another plant species. We are testing other apomictic Pennisetum species for their distribution patterns of homologous repetitive sequences, and have evidence that the Opie-2-like retrotransposon is abundant throughout the buffelgrass genome (Y. Akiyama, unpublished data). Perhaps a progenitor such as buffelgrass, with dispersed and abundant Opie-2-like repeats, was involved in a wide hybridization event that led to the speciation of P. squamulatum. Since only one chromosome in P. squamulatum shows a high concentration of such repeats, wide hybridization would have to have been followed by additional backcrosses that would have resulted in elimination/replacement of other chromosomes that contained clusters of these repeats or by large-scale deletion of clusters on other chromosomes. It is perhaps more likely that such retrotransposons would be activated and proliferate upon wide hybridization (Liu and Wendel, 2000), spreading to other genomes, which could imply that a P. squamulatum-like progenitor (with respect to the distribution of the Opie-2-like repeat) was involved in the speciation of buffelgrass.
An equally plausible hypothesis is that the region has arisen from local proliferation of a retrotransposon family. It is known that RIRE7, a gypsy-type retrotransposon in rice, is localized only in centromeric regions interspersed between tandem repeat sequences (TrsD; Kumekawa et al., 2001). In Allium cepa, Ty1-copia group retrotransposons are distributed across the genome but are enriched in the terminal heterochromatic region of chromosomes (Pearce et al., 1996b). Although copia-type retrotransposons in many plant species are often distributed throughout the whole genome (Schmidt et al., 1995; Pearce et al., 1996a; SanMiguel et al., 1996; Suoniemi et al., 1996; Meyers et al., 2001), a cluster distribution also has been reported (Belyayev et al., 2001). Copia-like retrotransposons in cultivated rice showed highly restricted distribution on the molecular-linkage map (Wang et al., 1999). The identification of subclones from P602, which contain Opie-2-like sequences and when alone or in combination produce FISH signals similar to the Group 2 clones, suggests that some if not most of the high-copy region of the ASGR is due to the proliferation of an Opie-2-like retrotransposon. Similar sequences also are present in the pearl millet genome as observed by Southern-blot hybridization (J.A. Conner, unpublished data), but our data suggest that they are not clustered. Additional sequence data from the Opie-2-like retrotransposon family members that are present in the ASGR and related species could allow us to use long terminal repeat sequences to date insertion events and to distinguish between the introgression and local proliferation hypotheses. More recent insertion events would be expected to have a higher sequence similarity between the two long terminal repeats from one element (SanMiguel et al., 1998).
Suppressed Recombination in the ASGR
Polyploid Pennisetum species often do not show strict bivalent pairing at meiosis (Dujardin and Hanna, 1984). Although mostly bivalents were observed in P. squamulatum, univalents, trivalents, quadrivalents, and hexavalents also were present. Our FISH results showed that although the ASGR-carrier chromosome can form a bivalent, the short arm, including the ASGR, usually is asynaptic or desynaptic. Chromosome pairing during meiosis is considered to be exquisitely sensitive to structural change, such as an insertion, inversion, or translocation (Jauhar and Joppa, 1996), and this sensitivity probably is a consequence of the nucleotide sequence and microsynteny of homo(eo)logous chromosomes. Our FISH results revealed hemizygosity over a large region of the ASGR-carrier chromosome, which likely indicates extensive nucleotide sequence divergence over at least one-quarter of the chromosome relative to the pairing partner.
The lack of recombination in apomixis-associated genomic regions has been extensively discussed (Ozias-Akins et al., 1998; Roche et al., 1999, 2001; Labombarda et al., 2002; Goel et al., 2003). Although the location of the ASGR on a univalent B chromosome was initially speculated, chromosome pairing evidence presented here does not support this hypothesis. The extensive hemizygosity that we now show for the ASGR almost certainly would have an impact on chromosome synapsis and/or crossing-over. In addition, the hemizygous region of the ASGR-carrier chromosome remains more highly condensed than the long arm of the same chromosome, and combined with the low recombination observed and the abundance of repetitive sequence, displays multiple characteristics often associated with heterochromatin (Bennetzen, 2000; Henikoff, 2000). In Arabidopsis, heterochromatic regions with low recombination were largely composed of transposable and retrotransposable elements (Cold Spring Harbor Laboratory, 2000). Moreover, it has been reported that chromosomal regions with abundant retrotransposons in maize are relatively inert recombinationally (Fu et al., 2002; Yao et al., 2002).
Whether the apomictic phenotype is encoded by a complex locus or a single gene is a crucial question, but one that will be difficult to address in natural apomicts that have the phenotype associated with a nonrecombining region of the genome. Low recombination is characteristic of complex loci; for example, the S-locus for self incompatibility in Brassica (Boyes et al., 1997; Casselman et al., 2000), and disease resistance loci in lettuce (Lactuca sativa; Meyers et al., 1998), rice (Chauhan et al., 2002), and wheat (Triticum aestivum; Neu et al., 2002). It remains tempting to speculate that the low recombination preserves gene linkages that are required in order to elicit an apomictic phenotype (Ozias-Akins et al., 1998; Roche et al., 2001). Alternatively, it could simply be a consequence of sequence divergence due to the reproductive isolation of the apomictic genome coupled with susceptibility of the now hemizygous region to somatic invasion by retrotransposons (Ozias-Akins et al., 2003). Unfortunately, the large size of the hemizygous region in P. squamulatum and the consequently low level of recombination reduce the feasibility of positional cloning unless other means are used to narrow the region essential for apomixis, e.g. through deletion mutagenesis.
MATERIALS AND METHODS
Plant Materials
Genotypes used in this study included the apomictic species, Pennisetum squamulatum accession numbers PI248534 (PS24) and PI319196 (PS26), and an apomictic backcross (BC3) of pearl millet (P. glaucum) with a trispecific hybrid (Dujardin and Hanna, 1989). For pachytene chromosome preparation, immature panicles of BC3, PS24, and PS26, grown in the field in summer, were collected and fixed in 3:1 ethanol to acetic acid (fixative solution). For mitotic chromosome preparation, root tips of all genotypes were collected from greenhouse-grown plants and pretreated for 3 h by soaking in a saturated solution of α-bromonaphthalene at 4°C prior to fixation.
BAC Clones
The construction of BAC libraries and isolation of ASGR-linked BAC clones (P001, P002, derived by SCAR A14M; P102, P109, SCAR Q8M; P201, P205, P208, SCAR ugt197; P301, SCAR A10H) previously have been reported (Roche et al., 2002). BACs containing SCAR markers with repetitive DNA (P601, 602, SCAR X18R; P701, P708, SCAR R13; P800, SCAR U12H; P900, 901, SCAR W10M) were isolated by PCR screening of pooled BAC DNA (Liu et al., 2000). These BACs were derived from the polyhaploid apomictic line MS228-20 that contains a complete haploid genome of pearl millet and a partial genome of P. squamulatum (Dujardin and Hanna, 1986; Roche et al., 2002). Sample sequencing of selected BACs was carried out from 96 shotgun subclones each.
Mitotic Chromosome Preparation
Mitotic chromosome spreads were prepared by the enzymatic maceration/air-drying (EMA) method (Fukui, 1996). The root tips were macerated with an enzyme mixture (1.33% cellulase Onozuka RS, 0.5% macerozyme R-200, 0.23% pectolyase Y-23, 0.33 mm EDTA, pH 4.2) for 2 h for P. squamulatum. The digested cells in fixative solution were spread over glass slides with forceps; the slides were briefly exposed to steam and dried on a heat block at 85°C (Henegariu et al., 2001).
Pachytene Chromosome Preparation
Pachytene chromosome spreads were prepared according to Ohmido et al. (2001) with modifications. To find anthers in the pachytene stage, one approximately 1.5-mm anther from a floret was squashed with acetocarmine and examined under the microscope. When the correct stage was found, the remaining two anthers were macerated in an enzyme mixture (0.43% cellulase RS, 0.43% pectolyase Y23, 0.43% cytohelicase, 43 mm citrate buffer, pH 4.5; Zhong et al., 1996) on a glass slide in a humid chamber at 37°C for 5 to 6 h. The enzyme solution was removed and the residue on the slides was thinned by water. After removing the water, the digested cells were spread in fixative solution as for mitotic chromosome preparations.
Pretreatment and Hybridization
Pretreatment and hybridization of spreads were carried out according to Henegariu et al. (2001). The spreads were flooded with 500 μL ethanol, covered with a coverslip and gauze moistened with ethanol, then heated on a heat block at 85°C for 2 min. After gradually cooling to room temperature, slides were incubated in 0.005% pepsin/0.01 n HCl at 37°C for 90 s, rinsed in 1× phosphate-buffered saline (PBS), fixed in 1% formaldehyde in 1× PBS containing 50 mm MgCl2, rinsed in 1× PBS, and dehydrated sequentially in 70%, 95%, and 100% ethanol (5 min at room temperature for each step). After air drying, the chromosomes were denatured in 100 μL 70% formamide in 2× SSC at 85°C for 90 s, then rinsed sequentially in 70%, 95%, and 100% ethanol to remove formamide. The hybridization mixture contained approximately 5 μg/mL biotin- or digoxigenin-labeled probe, 5% dextran sulfate, and 50% formamide in 2× SSC. The hybridization mixture was denatured at 85°C for 10 min and cooled on ice, then 10 μL of the denatured hybridization mixture was applied to the denatured slide and covered with a 22 × 30 mm coverslip. The coverslip was sealed with rubber cement and the slide was incubated in a humidified chamber at 37°C for at least 12 h.
Detection of Signals
Probes were detected in dual-color FISH according to Goel et al. (2003). The digoxigenin-labeled probes were detected with fluorescein-conjugated antibody according to the manufacturer's instructions (Roche, Indianapolis). Biotin-labeled probes were detected with texas-red streptavidin (Vector Laboratories, Burlingame, CA). To amplify the biotin signal, biotinylated anti-streptavidin (Vector Laboratories) was applied for a second layer, then texas-red streptavidin was applied again. The slides were mounted in Vectashield (Vector Laboratories) containing 1.5 μg/mL DAPI and observed under an Olympus (Tokyo) BX50 fluorescence microscope.
Rehybridization
Several BACs could be analyzed on the same spread by rehybridization. To remove probe, the slides were incubated in a humidified chamber at 37°C for 30 min to allow easy removal of the coverslip. The slides were then washed twice in 0.2% Tween 20 in 1× PBS at room temperature for 15 min each, once in 1× PBS at room temperature for 5 min, and once in 70% formamide at 70°C for 3 min. The slides were dehydrated in 70%, 95%, and 100% ethanol for 5 min each. After air-drying, the slides were denatured, hybridized, and further processed as for the first hybridization.
Image Analysis
Images were captured by a cooled CCD camera (Sensys Photometrics, Tucson, AZ) through Image Pro ver 4.1 software (Media Cybernetics, Silver Spring, MD). Image analysis was carried out by using Object-Image 2.08 (http://simon.bio.uva.nl/object-image.html) and CHIAS3 (Kato and Fukui, 1998) on a Macintosh Power Book. The macros of CHIAS3 are largely modified for analysis of dual-color FISH.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY375366.
Acknowledgments
We thank Gunawati Gunawan, Evelyn Perry, Anne Bell, and Jacolyn Merriman for technical assistance.
This work was supported by the University of Georgia Experiment Station, USDA National Research Initiative (award no. 99–35300–7691), and by the National Science Foundation (award no. 0115911).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.033969.
References
- Belyayev A, Raskina O, Nevo E (2001) Chromosomal distribution of reverse transcriptase-containing retroelements in two Triticeae species. Chromosome Res 9: 129–136 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL (2000) The many hues of plant heterochromatin. Genome Biol 1: reviews 107.1–107.4 [DOI] [PMC free article] [PubMed]
- Blakey CA, Goldman SL, Dewald CL (2001) Apomixis in Tripsacum: comparative mapping of a multigene phenomenon. Genome 44: 222–230 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Nasrallah ME, Vrebalov J, Nasrallah JB (1997) The self-incompatibility (S) haplotypes of Brassica contain highly divergent and rearranged sequences of ancient origin. Plant Cell 9: 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casselman AL, Vrebalov J, Conner JA, Singhal A, Giovannoni J, Nasrallah ME, Nasrallah JB (2000) Determining the physical limits of the Brassica S locus by recombinational analysis. Plant Cell 12: 23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan RS, Farman ML, Zhang HB, Leong SA (2002) Genetic and physical mapping of a rice blast resistance locus, Pi-CO39(t), that corresponds to the avirulence gene AVR1-CO39 of Magnaporthe grisea. Mol Genet Genomics 267: 603–612 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Presting GG, Buell CR, Wing RA, Jiang J (2001) High-resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics 157: 1749–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cold Spring Harbor Laboratory, Washington University Genome Sequencing Center, PE Biosystems Arabidopsis Sequencing Consortium (2000) The complete sequence of a heterochromatic island from a higher eukaryote. Cell 100: 377–386 [PubMed] [Google Scholar]
- De Jong JH, Fransz P, Zabel P (1999) High resolution FISH in plants: techniques and applications. Trends Plant Sci 4: 258–263 [DOI] [PubMed] [Google Scholar]
- Dujardin M, Hanna WW (1984) Microsporogenesis, reproductive behavior, and fertility in five Pennisetum species. Theor Appl Genet 67: 197–201 [DOI] [PubMed] [Google Scholar]
- Dujardin M, Hanna WW (1986) An apomictic polyhaploid obtained from a pearl millet x Pennisetum squamulatum apomictic interspecific hybrid. Theor Appl Genet 72: 33–36 [DOI] [PubMed] [Google Scholar]
- Dujardin M, Hanna WW (1989) Developing apomictic pearl millet - characterization of a BC3 plant. J Genet Breed 43: 145–151 [Google Scholar]
- Fransz PF, Alonso-Blanco C, Liharska TB, Peeters AJ, Zabel P, De Jong JH (1996) High-resolution physical mapping in Arabidopsis thaliana and tomato by fluorescence in situ hybridization to extended DNA fibres. Plant J 9: 421–430 [DOI] [PubMed] [Google Scholar]
- Fu H, Zheng Z, Dooner HK (2002) Intraspecific violation of genetic colinearity and its implications in maize. Proc Natl Acad Sci USA 99: 1082–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui K (1996) Plant chromosomes at mitosis. In K Fukui, S Nakayama, eds, Plant Chromosomes, Laboratory Methods. CRC Press, Boca Raton, FL, pp 1–17
- Goel S, Chen ZB, Conner JA, Akiyama Y, Hanna WW, Ozias-Akins P (2003) Delineation by FISH of a single hemizygous chromosomal region responsible for aposporous embryo sac formation in Pennisetum squamulatum and Cenchrus ciliaris. Genetics 163: 1069–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimanelli D, Leblanc O, Perotti E, Grossniklaus U (2001) Developmental genetics of gametophytic apomixis. Trends Genet 17: 597–604 [DOI] [PubMed] [Google Scholar]
- Henegariu O, Heerema NA, Wright LL, Bray-Ward P, Ward DC, Vance GH (2001) Improvements in cytogenetic slide preparation: controlled chromosome spreading, chemical aging and gradual denaturing. Cytometry 43: 101–109 [PubMed] [Google Scholar]
- Henikoff S (2000) Heterochromatin function in complex genomes. Biochim Biophys Acta 1470: O1–O8 [DOI] [PubMed] [Google Scholar]
- Islam-Faridi MN, Childs KL, Klein PE, Hodnett G, Menz MA, Klein RR, Rooney WL, Mullet JE, Stelly DM, Price HJ (2002) A molecular cytogenetic map of sorghum chromosome 1. Fluorescence in situ hybridization analysis with mapped bacterial artificial chromosomes. Genetics 161: 345–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SA, Cheng ZK, Wang ML, Goodman HM, Jiang JM (2000) Comparative fluorescence in situ hybridization mapping of a 431-kb Arabidopsis thaliana bacterial artificial chromosome contig reveals the role of chromosomal duplications in the expansion of the Brassica rapa genome. Genetics 156: 833–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauhar PP, Joppa LR (1996) Chromosome pairing as a tool in genome analysis: merits and limitations. In PP Jauhar, ed, Methods of Genome Analysis in Plants. CRC Press, Boca Raton, FL, pp 9–37
- Kato S, Fukui K (1998) Condensation pattern (CP) analysis of plant chromosomes by an improved chromosome image analysing system. CHIAS III. Chromosome Res 6: 473–479 [DOI] [PubMed] [Google Scholar]
- Koltunow A (1993) Apomixis: Embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 5: 1425–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulikova O, Gualtieri G, Geurts R, Kim DJ, Cook D, Huguet T, de Jong JH, Fransz PF, Bisseling T (2001) Integration of the FISH pachytene and genetic maps of Medicago truncatula. Plant J 27: 49–58 [DOI] [PubMed] [Google Scholar]
- Kumekawa N, Ohmido N, Fukui K, Ohtsubo E, Ohtsubo H (2001) A new gypsy-type retrotransposon, RIRE7: preferential insertion into the tandem repeat sequence TrsD in pericentromeric heterochromatin regions of rice chromosomes. Mol Genet Genomics 265: 480–488 [DOI] [PubMed] [Google Scholar]
- Labombarda P, Busti A, Caceres ME, Pupilli F, Arcioni S (2002) An AFLP marker tightly linked to apomixis reveals hemizygosity in a portion of the apomixis-controlling locus in Paspalum simplex. Genome 45: 513–519 [DOI] [PubMed] [Google Scholar]
- Liu B, Wendel JF (2000) Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome 43: 874–880 [PubMed] [Google Scholar]
- Liu YG, Nagaki K, Fujita M, Kawaura K, Uozumi M, Ogihara Y (2000) Development of an efficient maintenance and screening system for large-insert genomic DNA libraries of hexaploid wheat in a transformation-competent artificial chromosome (TAC) vector. Plant J 23: 687–695 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Chin DB, Shen KA, Sivaramakrishnan S, Lavelle DO, Zhang Z, Michelmore RW (1998) The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10: 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Tingey SV, Morgante M (2001) Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res 11: 1660–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu C, Stein N, Keller B (2002) Genetic mapping of the Lr20-Pm1 resistance locus reveals suppressed recombination on chromosome arm 7AL in hexaploid wheat. Genome 45: 737–744 [DOI] [PubMed] [Google Scholar]
- Nogler GA (1984) Gametophytic apomixis. In BM Johri, ed, Embryology of Angiosperms. Springer-Verlag, Berlin, pp 448–452
- Noyes RD, Rieseberg LH (2000) Two independent loci control agamospermy (apomixis) in the triploid flowering plant Erigeron annuus. Genetics 155: 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmido N, Kijima K, Ashikawa I, de Jong JH, Fukui K (2001) Visualization of the terminal structure of rice chromosomes 6 and 12 with multicolor FISH to chromosomes and extended DNA fibers. Plant Mol Biol 47: 413–421 [DOI] [PubMed] [Google Scholar]
- Ozias-Akins P, Akiyama Y, Hanna WW (2003) Molecular characterization of the genomic region linked with apomixis in Pennisetum/Cenchrus. Funct Integr Genomics 3: 94–104 [DOI] [PubMed] [Google Scholar]
- Ozias-Akins P, Roche D, Hanna WW (1998) Tight clustering and hemizygosity of apomixis-linked molecular markers in Pennisetum squamulatum implies genetic control of apospory by a divergent locus that may have no allelic form in sexual genotypes. Proc Natl Acad Sci USA 95: 5127–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce SR, Harrison G, Li D, Heslop-Harrison J, Kumar A, Flavell AJ (1996. a) The ty1-copia group retrotransposons in Vicia species: copy number, sequence heterogeneity and chromosomal localization. Mol Gen Genet 250: 305–315 [DOI] [PubMed] [Google Scholar]
- Pearce SR, Pich U, Harrison G, Flavell AJ, Heslop-Harrison JS, Schubert I, Kumar A (1996. b) The ty1-copia group retrotransposons of Allium-cepa are distributed throughout the chromosomes but are enriched in the terminal heterochromatin. Chromosome Res 4: 357–364 [DOI] [PubMed] [Google Scholar]
- Pessino SC, Evans C, Ortiz JPA, Armstead I, do Valle CB, Hayward MD (1998) A genetic map of the apospory-region in Brachiaria hybrids: identification of two markers closely associated with the trait. Hereditas 128: 153–158 [Google Scholar]
- Pessino SC, Ortiz JPA, Leblanc O, do Valle CB, Evans C, Hayward MD (1997) Identification of a maize linkage group related to apomixis in Brachiaria. Theor Appl Genet 94: 439–444 [Google Scholar]
- Peterson DG, Lapitan NL, Stack SM (1999) Localization of single- and low-copy sequences on tomato synaptonemal complex spreads using fluorescence in situ hybridization (FISH). Genetics 152: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AJ (1973) The origin of Taraxacum agamospecies. Bot J Linn Soc 66: 189–211 [Google Scholar]
- Roche D, Cong P, Chen Z, Hanna WW, Gustine DL, Sherwood RT, Ozias-Akins P (1999) An apospory-specific genomic region is conserved between Buffelgrass (Cenchrus ciliaris L.) and Pennisetum squamulatum Fresen. Plant J 19: 203–208 [DOI] [PubMed] [Google Scholar]
- Roche D, Conner JA, Budiman MA, Frisch D, Wing R, Hanna WW, Ozias-Akins P (2002) Construction of BAC libraries from two apomictic grasses to study the microlinearity of their apospory-specific genomic regions. Theor Appl Genet 104: 804–812 [DOI] [PubMed] [Google Scholar]
- Roche D, Hanna W, Ozias-Akins P (2001) Is supernumerary chromatin involved in gametophytic apomixis of polyploidy plants? Sex Plant Reprod 13: 343–349 [Google Scholar]
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL (1998) The paleontology of intergene retrotransposons of maize. Nat Genet 20: 43–45 [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melakeberhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768 [DOI] [PubMed] [Google Scholar]
- Schmidt T, Kubis S, Heslop-Harrison JS (1995) Analysis and chromosomal localization of retrotransposons in sugar-beet (Beta-vulgaris L): lines and ty1-copia-like elements as major components of the genome. Chromosome Res 3: 335–345 [DOI] [PubMed] [Google Scholar]
- Sherwood RT, Berg CC, Young BA (1994) Inheritance of apospory in buffelgrass. Crop Sci 34: 1490–1494 [Google Scholar]
- Suoniemi A, Anamthawat-Jonsson K, Arna T, Schulman AH (1996) Retrotransposon-bare-1 is a major, dispersed component of the barley (Hordeum-vulgare L) genome. Plant Mol Biol 30: 1321–1329 [DOI] [PubMed] [Google Scholar]
- Van Dijk PJ, Tas IC, Falque M, Bakx-Schotman T (1999) Crosses between sexual and apomictic dandelions (Taraxacum). II. The breakdown of apomixis. Heredity 83: 715–721 [DOI] [PubMed] [Google Scholar]
- Van Dijk P, van Damme J (2000) Apomixis technology and the paradox of sex. Trends Plant Sci 5: 81–84 [DOI] [PubMed] [Google Scholar]
- Wang S, Liu N, Peng K, Zhang Q (1999) The distribution and copy number of copia-like retrotransposons in rice (Oryza sativa L.) and their implications in the organization and evolution of the rice genome. Proc Natl Acad Sci USA 96: 6824–6828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Zhou Q, Li J, Smith H, Yandeau M, Nikolau BJ, Schnable PS (2002) Molecular characterization of meiotic recombination across the 140-kb multigenic a1-sh2 interval of maize. Proc Natl Acad Sci USA 99: 6157–6162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Zhang Y, Cheng ZK, Chen MS, Wang SY, Feng Q, Huang YC, Li Y, Tang YS, Zhou B, et al (2002) A fine physical map of the rice chromosome 4. Genome Res 12: 817–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XB, de Jong JH, Zabel P (1996) Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization (FISH). Chromosome Res 4: 24–28 [DOI] [PubMed] [Google Scholar]
- Zhong XB, Fransz PF, Wennekes-van Eden J, Ramanna MS, van Kammen A, Zabel P, de Jong JH (1998) FISH studies reveal the molecular and chromosomal organization of individual telomere domains in tomato. Plant J 13: 507–517 [DOI] [PubMed] [Google Scholar]
- Ziolkowski PA, Sadowski J (2002) FISH-mapping of rDNAs and Arabidopsis BACs on pachytene complements of selected Brassicas. Genome 45: 189–197 [DOI] [PubMed] [Google Scholar]