Summary
To test the safety and activity of 5-aza-2’-deoxycytidine (decitabine) in patients with relapsed/refractory acute lymphocytic leukaemia (ALL), we conducted a phase 1 study with two parts: administering decitabine alone or in combination with Hyper-CVAD (fractionated cyclophosphamide, vincristine, doxorubicin and dexamethasone alternating with high-dose methotrexate and cytarabine). Patients participated in either part of the study or in both parts sequentially. In the initial part, decitabine was administered intravenously at doses of 10–120 mg/m2/day for 5 days every other week in cycles of 28 days. In the combination part, patients were treated on the first five days of Hyper-CVAD with intravenous decitabine at 5–60 mg/m2/day. A total of 39 patients received treatment in the study: 14 in the first part only, 16 sequentially in both parts and 9 in the second part only. Decitabine was tolerated at all doses administered, and grade 3 or 4 toxic effects included non-life-threatening hepatotoxicity and hyperglycaemia. Induction of DNA hypomethylation was observed at doses of decitabine up to 80 mg/m2. Some patients who had previously progressed on Hyper-CVAD alone achieved a complete response when decitabine was added. Decitabine alone or given with Hyper-CVAD is safe and has clinical activity in patients with advanced ALL.
Keywords: Precursor Cell Lymphoblastic Leukaemia-Lymphoma, Clinical Trial, DNA Methylation, decitabine
Introduction
Aberrant DNA methylation is a common feature of human malignancies, including leukaemia (Florean, et al 2011). Two hypomethylating agents, 5-azacitidine and 5-aza-2’-deoxycytadine (decitabine), are approved for patients with myelodysplastic syndrome (MDS) and are used in acute myeloid leukaemia (AML) of the elderly (Keating 2009, Lubbert and Minden 2005, Saba and Wijermans 2005). Because of the frequency of DNA methylation alterations in other human malignancies, these agents are under broad investigation (Estécio and Issa 2011, Ren, et al 2011).
Acute lymphocytic leukaemia (ALL) in adults is frequently characterized by relapse from residual disease and poor prognosis. A hypermethylated genome appears to be critical in the pathogenesis and relapse of lymphoid malignancies, just as it is in myeloid malignancies (Garcia-Manero, et al 2009, Klimek and Tallman 2011). For example, detection of residual methylation in treated ALL patients corresponds with relapse (Narayan, et al 2011). Existing clinical reports have suggested that hypomethylating agents may have a role in treating lymphoid leukaemia (Issa, et al 2004, Paulson, et al 2011, Willemze, et al 1993, Yánez, et al 2009). However, few prior studies have investigated hypomethylating agents in ALL specifically (Garcia-Manero, et al 2002, Garcia-Manero, et al 2003, Garcia-Manero, et al 2009, Hoshino, et al 2007, Klimek and Tallman 2011, Narayan, et al 2011, Wong, et al 2012). Pre-clinical in vitro studies have indicated that restoration of epigenetically silenced genes, a phenomenon observed with decitabine, results in cell death in ALL-derived cell lines (Kuang, et al 2007). Decitabine has also been given in combination with cytotoxic chemotherapy to patients with solid tumours who have previously been treated with chemotherapy alone (Appleton, et al 2007), and pre-clinical studies have suggested decitabine sensitizes leukaemia cells to cytarabine by hypomethylation (Qin, et al 2007). Based on this information, we hypothesized that decitabine alone or in combination with cytotoxic chemotherapy would prove beneficial to patients with relapsed, refractory ALL.
We conducted a two-part phase 1 study to investigate the safety, pharmacodynamics and clinical activity of decitabine alone and in combination with Hyper-CVAD (fractionated cyclophosphamide, vincristine, doxorubicin and dexamethasone alternating with high-dose methotrexate and cytarabine), a commonly used cytotoxic chemotherapeutic regimen in ALL (Kantarjian, et al 2000). Patients participated in either part alone, or in both parts sequentially. Patients who had previously been treated with Hyper-CVAD were allowed to participate in the second part, where decitabine was given with Hyper-CVAD. Our study design allowed us to test dose escalation of decitabine alone or in combination with Hyper-CVAD and the demethylation effects of various decitabine doses in ALL. It also provided a limited understanding of the sensitivity of advanced ALL to decitabine. Our analysis revealed that decitabine was tolerated at all doses tested and that it induced DNA hypomethylation up to doses of 80 mg/m2. Responses were observed in patients on decitabine alone and on decitabine in combination with HyperCVAD.
Methods
Study group eligibility
Patients of any age and performance status with documented relapsed or refractory ALL were eligible for this study. Other eligibility included total bilirubin less than 51.3 mmol/l, liver function tests less than 5 times the upper limit of normal, and a creatinine level less than 265.2 mmol/l. Any prior therapy or any number of prior therapies (full lines of treatment) were acceptable, even for the second part using decitabine with Hyper-CVAD. Patients must have completed prior chemotherapy at least 1 week before entering this study, and must have recovered from the toxic effects of such therapy. Nursing and pregnant patients were excluded, as were patients with uncontrolled active illnesses, such as infection. Approval for the study was obtained from the University of Texas MD Anderson Cancer Center Institutional Review Board. All patients gave written informed consent in accordance with the Declaration of Helsinki following institutional guidelines. This trial was registered at www.clinicaltrials.gov as NCT00349596.
Treatment
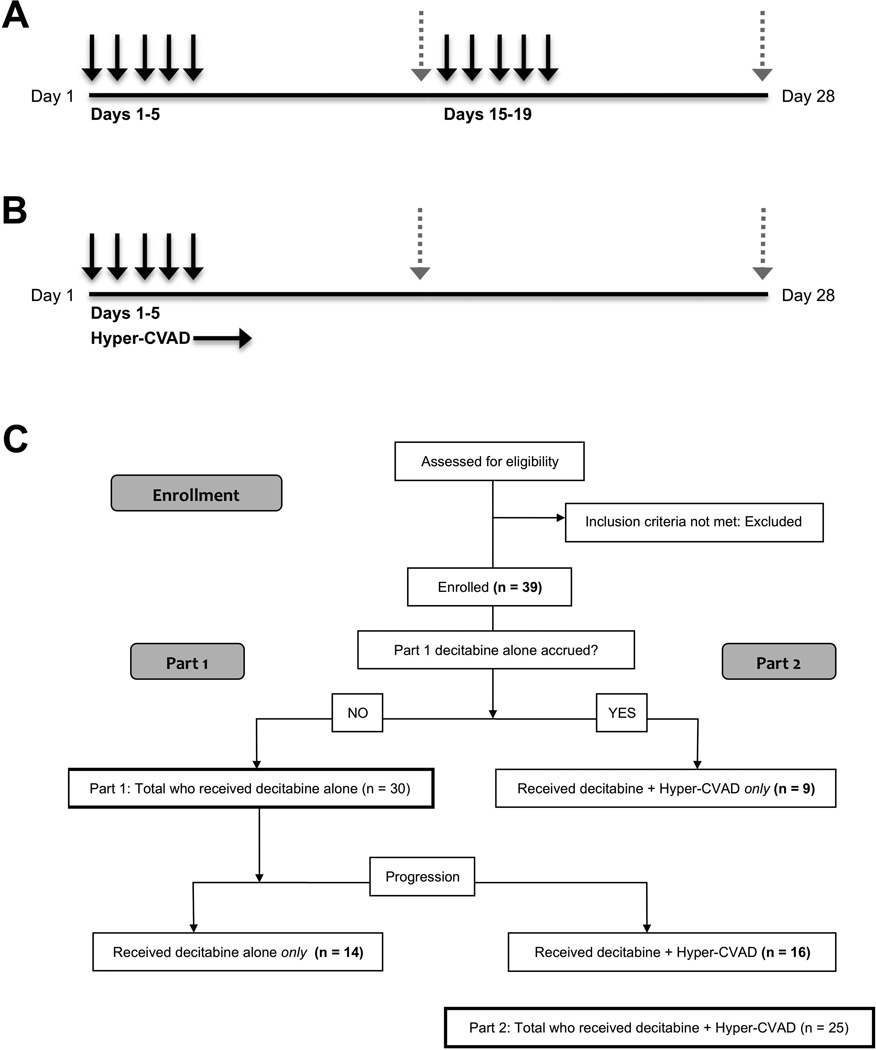
Our phase 1 investigation consisted of two sequential parts. In the first part, decitabine alone was administered. If a patient progressed after receiving decitabine alone, there was the option of participation in the second part of the study, where decitabine was used in conjunction with Hyper-CVAD. Once accrual for the first part was complete (n = 30), patients were enrolled in the second part of the study directly. In the initial part, decitabine was given IV daily over 1 h for 5 days every other week on a 28-day course (Fig 1A). The following doses were studied: 10, 20, 40, 60, 80, 100, and 120 mg/m2 per day (cumulative doses of 100, 200, 400, 600, 800, 1000, and 1200 mg/m2, respectively, per course). In the second part of the study, decitabine was administered on days 1–5 of Hyper-CVAD (Fig 1B). Hyper-CVAD was administered as previously described (Kantarjian, et al 2000). The following doses of decitabine were studied: 5, 10, 15, 20, 40, and 60 mg/m2 (cumulative doses of 50, 100, 150, 200, 400, and 600 mg/m2, respectively, per course). An extension cohort was treated in both parts, using optimal dosage (determined for each part by considering dosages with the most responses, fewest and least severe toxic effects, and most significant global DNA demethylation).
Fig 1.
Dosing schedule and patient numbers for both parts of the decitabine in this acute lymphoblastic leukaemia phase 1 trial. A–B) One complete 28-day course is shown for both studies. Black arrows represent dosing of decitabine and dashed grey arrows represent bone marrow aspiration for disease assessment on day 14 and 28. A) For the first part of the study, decitabine was administered alone IV daily for 5 days every other week (days 1–5 and days 15–19). B) In the second part, decitabine was administered IV daily for 5 days, starting on day 1 of each cycle of Hyper-CVAD (fractionated cyclophosphamide, vincristine, doxorubicin and dexamethasone alternating with high-dose methotrexate and cytarabine). C) Flow chart showing accrual and numbers of patients enrolled on each part of the trial including those receiving only decitabine, those who were on both parts, and those who received only decitabine+HyperCVAD.
Toxicity assessment and dose escalation
Toxicities were graded using the National Cancer Institute Common Toxicity Criteria for Adverse Events (NCI CTCAE), version 3 (http://ctep.cancer.gov/protocolDevelopment/electronic_applications/docs/ctcaev3.pdf). Patients were allowed to receive supportive care measures, including steroids, antibiotics, antiemetics and growth factors, as clinically indicated and according to institutional guidelines. A traditional 3+3 trial design was used for all dosage increases (Le Tourneau, et al 2009, Storer 1989). If 2 or more patients had a grade 3 or higher non-haematological toxic effect, the dose of decitabine was considered toxic. The maximum tolerated dose (MTD) was defined as the dose immediately below the one producing a dose-limiting toxic effect (DLT) in 2 or more patients enrolled at that dosage level. The MTD was based on toxic effects experienced during the first course of therapy. The second part of the study was designed to evaluate the safety of the combination of decitabine and Hyper-CVAD. Accrual and dose escalation were performed in the same fashion as for the first part of the study. Patients were allowed to receive supportive care measures, including steroids, antibiotics, antiemetics and growth factors, as clinically indicated and according to institutional guidelines.
Response criteria and statistical methods
Response assessments were carried out with bone marrow aspiration on day 14 (±7 days), day 28 (±7 days) and thereafter as indicated to document response. A complete response (CR) was defined by the disappearance of all signs and symptoms related to disease, in addition to normalization of peripheral blood counts defined by an absolute neutrophil count (ANC) of 1 × 109/l or higher, platelet count of 100 × 109/l or higher, and 5% or less bone marrow blasts. A CR without the recovery of platelets (CRp) was defined as bone marrow with 5% blasts or less and normalization of ANC to 1 × 109/l or higher, but without platelet recovery (platelet count less than 100 × 109/l). Complete marrow response (mCR) was defined as bone marrow blasts less than 5% but without recovery of peripheral blood counts (ANC less than 1 × 109/l and platelet count less than 100 × 109/l). Haematological improvement was defined as at least 50% decrease in bone marrow blasts. Stable disease was defined as no increase in the percentage of marrow blasts (within 10% of initial percentage) after the first course of therapy and no increase in total number of white blood cells and circulating blasts (within 10% of initial percentage). Remission duration was calculated from the date of first response until relapse, and survival was calculated from the date of therapy initiation until death from any cause. Patients were removed from the study for progressive disease. Kaplan-Meier curves were used to evaluate survival and relapse-free survival, and comparisons between curves were performed using a log rank test.
Isolation of human mononuclear cells
Human mononuclear cells were used to analyse DNA methylation. Peripheral blood from patients was drawn into tubes containing heparin at a concentration of 30 U/ML. Mononuclear cells were separated using Ficoll-Paque PLUS gradient centrifugation (Amersham Biosciences AB, Uppsala, Sweden). The monocyte-enriched cell fraction was collected and then washed twice with calcium- and magnesium-free phosphate-buffered saline. After centrifugation, the phosphate-buffered saline was removed and the samples frozen at −80°C.
Analysis of DNA methylation
To analyse the changes in DNA methylation with treatment, we investigated the dynamics of long interspersed nuclear element (LINE) methylation (which corresponds to the level of global methylation). Techniques for DNA extraction, modification, and polymerase chain reaction (PCR) techniques have been described previously (Garcia-Manero, et al 2006, Shen, et al 2003). To determine the degree of methylation, we used a bisulfite pyrosequencing assay that has been described previously (Garcia-Manero, et al 2006, Issa, et al 2005). We analysed logarithms of gene methylation levels. The genomic methylation patterns of specific groups (such as cohorts of various dosage levels or responders vs. non-responders) were compared by adjusting for the baseline methylation of each particular gene tested. We analysed the LINE methylation of decitabine-treated patients on days 0, 2, 5, 14, 16, 19 and 26 of treatment.
Results
Study group
A total of 39 patients received treatment in the study, and included 14 patients who participated in the first part of the study only (decitabine alone), 16 patients who participated in both parts of the study sequentially (decitabine alone until progression, followed by decitabine + Hyper-CVAD), and 9 patients who participated in the second part only (decitabine + Hyper-CVAD). In total, 30 patients were treated using decitabine alone, and 25 were treated using decitabine + Hyper-CVAD (Fig 1C). Table I shows patients’ clinical and treatment characteristics. The median age of all patients treated was 33 years (range, 4–67 years) and all patients had received at least one prior therapy (range, 1–7 therapies; median, 3 therapies). Most patients (32 patients; 82%) had B-cell ALL and the remaining 7 patients (18%) had T-cell ALL.
Table I.
Patient characteristics (n = 39).
| Characteristic | Total |
|---|---|
| Patients, n | 39 |
| Age, years; median (range) | 33 (4–67) |
| Gender, n (%) | |
| Female | 14 (36) |
| Male | 25 (64) |
| Prior therapies, n; median (range) | 3 (1–7) |
| Primary resistant, n (%) | 9 (23) |
| ≥ 2 prior therapies, n (%) | 30 (77) |
| Prior transplant | 7 (18) |
| CR1 | |
| Duration, weeks: median (range) | 40 (0–424) |
| No response / primary resistant, n (%) | 8 (21) |
| Duration of CR1 < 52 weeks, n (%) | 22 (56) |
| Duration of CR1 ≥ 52 weeks, n (%) | 17 (44) |
| Cytogenetics, n (%) | |
| Diploid | 7 (18) |
| Ph+ | 4 (10) |
| −5/−7 | 6 (15) |
| 11q deletion | 1 (3) |
| Other | 21 (54) |
| Phenotype, n (%) | |
| B-cell | 32 (82) |
| T-cell | 7 (18) |
| WBC, × 109/l; median (range) | 5.3 (0–131.9) |
| Creatinine, mmol/l; median (range) | 70.7 (26.5–159.1) |
| Bilirubin, mmol/l; median (range) | 8.6 (3.4–32.5) |
n, number; CR1, first complete remission; Ph+, Philadelphia chromosome-positive; −5/−7, deletion of chromosome 5 and/or 7; 11q deletion, deletion of chromosome 11q; WBC, white blood cell count.
Decitabine dose escalation and toxicities
We assessed 7 dose levels of decitabine (10, 20, 40, 60, 80, 100, 120 mg/m2) for the decitabine alone part of the study, and a total of 6 dose levels of decitabine (5, 10, 15, 20, 40, 60 mg/m2) for the decitabine + Hyper-CVAD part. The number of patients at each dose level for each part of the study who experienced any therapy-related toxicity grade 1–4 is shown in Table II. No DLT was observed in any patient during either part, and all dosages were generally well-tolerated. Therefore, the MTD was not reached, as the maximum dose of decitabine in both parts of the study was tolerated. An optimal dose was determined and used in extension phases for both parts. The optimal dose was determined based on responses at each dose level, toxic effects observed at each dose level and the effect of each dose on DNA methylation. At 60 mg/m2 for the decitabine alone study, there was one responder, no grade 3 or 4 toxic effects and maximal hypomethylation (see DNA methylation analysis). At 40 mg/m2 for the decitabine + Hyper-CVAD study, one patient had CR and no patients had grade 3 or 4 toxic effects. These dosages were selected for the extension cohorts. The most frequent grade 3 or 4 toxic effects associated with decitabine + Hyper-CVAD therapy were hepatic dysfunction (10 patients) and hyperglycaemia (3 patients). Additionally, one patient each had renal dysfunction, diarrhoea, vomiting and anorexia; no other grade 3 or 4 toxic effects were experienced.
Table II.
Frequency of non-haematological drug-related toxicities during course 1 for both parts of the trial (n = 39).
| Decitabine dose (mg/m2) |
Patients enrolled (n) |
Toxicities (n) |
|||
|---|---|---|---|---|---|
| Grade 1 | Grade 2 | Grade 3 | Grade 4 | ||
| Decitabine alone | |||||
| 10 | 4 | 15 | 1 | 0 | 0 |
| 20 | 3 | 6 | 1 | 1 | 0 |
| 40 | 3 | 2 | 2 | 1 | 0 |
| 60 | 3 | 5 | 2 | 0 | 0 |
| 80 | 3 | 3 | 1 | 2 | 1 |
| 100 | 3 | 3 | 2 | 1 | 0 |
| 120 | 4 | 5 | 2 | 0 | 0 |
| 60 (expansion) | 7 | 9 | 4 | 1 | 0 |
| Decitabine + Hyper-CVAD | |||||
| 5 | 3 | 6 | 7 | 1 | 0 |
| 10 | 3 | 4 | 4 | 2 | 0 |
| 15 | 3 | 7 | 9 | 1 | 1 |
| 20 | 3 | 5 | 3 | 2 | 0 |
| 40 | 3 | 5 | 2 | 0 | 0 |
| 60 | 3 | 5 | 7 | 3 | 0 |
| 40 (expansion) | 7 | 9 | 4 | 5 | 2 |
| Toxicity (Decitabine alone / Decitabine + HyperCVAD) | |||||
| Nausea | 9 / 10 | 0 / 7 | 0 / 0 | 0 / 0 | |
| Vomiting | 3 / 8 | 1 / 1 | 0 / 1 | 0 / 0 | |
| Diarrhoea | 4 / 2 | 4 / 1 | 0 / 1 | 0 / 0 | |
| Constipation | 2 / 1 | 0 / 0 | 0 / 0 | 0 / 0 | |
| Anorexia | 1 / 1 | 0 / 0 | 0 / 1 | 0 / 0 | |
| Altered taste | 1 / 0 | 0 / 0 | 0 / 0 | 0 / 0 | |
| Lip numbness | 1 / 0 | 0 / 0 | 0 / 0 | 0 / 0 | |
| Ileus | 0 / 0 | 0 / 1 | 0 / 0 | 0 / 0 | |
| Mucositis | 3 / 4 | 2 / 3 | 0 / 0 | 0 / 0 | |
| Hyperglycaemia | 2 / 2 | 3 / 11 | 1 / 2 | 1 / 2 | |
| Increase in ALT | 6 / 2 | 1 / 3 | 2 / 2 | 0 / 0 | |
| Increase in AST | 3 / 1 | 1 / 0 | 1 / 1 | 0 / 1 | |
| Increase in alkaline phosphatase | 3 / 6 | 0 / 0 | 0 / 2 | 0 / 0 | |
| Increase in total bilirubin | 3 / 3 | 1 / 4 | 0 / 3 | 0 / 1 | |
| Increase in lipase/amylase | 0 / 0 | 0 / 0 | 0 / 1 | 0 / 0 | |
| Increase in creatinine | 0 / 1 | 0 / 1 | 0 / 0 | 0 / 0 | |
| Acute renal failure | 0 / 0 | 0 / 0 | 0 / 1 | 0 / 0 | |
| Haematuria | 0 / 0 | 0 / 1 | 0 / 0 | 0 / 0 | |
| Fatigue | 1 / 0 | 1 / 0 | 0 / 0 | 0 / 0 | |
| Oedema | 3 / 0 | 0 / 0 | 1 / 0 | 0 / 0 | |
| Flushing | 1 / 0 | 0 / 0 | 0 / 0 | 0 / 0 | |
| Dizziness | 1 / 0 | 0 / 0 | 0 / 0 | 0 / 0 | |
| Itching | 1 / 0 | 0 / 0 | 0 / 0 | 0 / 0 | |
| Headache | 0 / 0 | 0 / 3 | 0 / 0 | 0 / 0 | |
n, number; Hyper-CVAD, fractionated cyclophosphamide, vincristine, doxorubicin and dexamethasone alternating with high-dose methotrexate and cytarabine; ALT, alanine transaminase; AST, aspartate transaminase
DNA methylation analysis
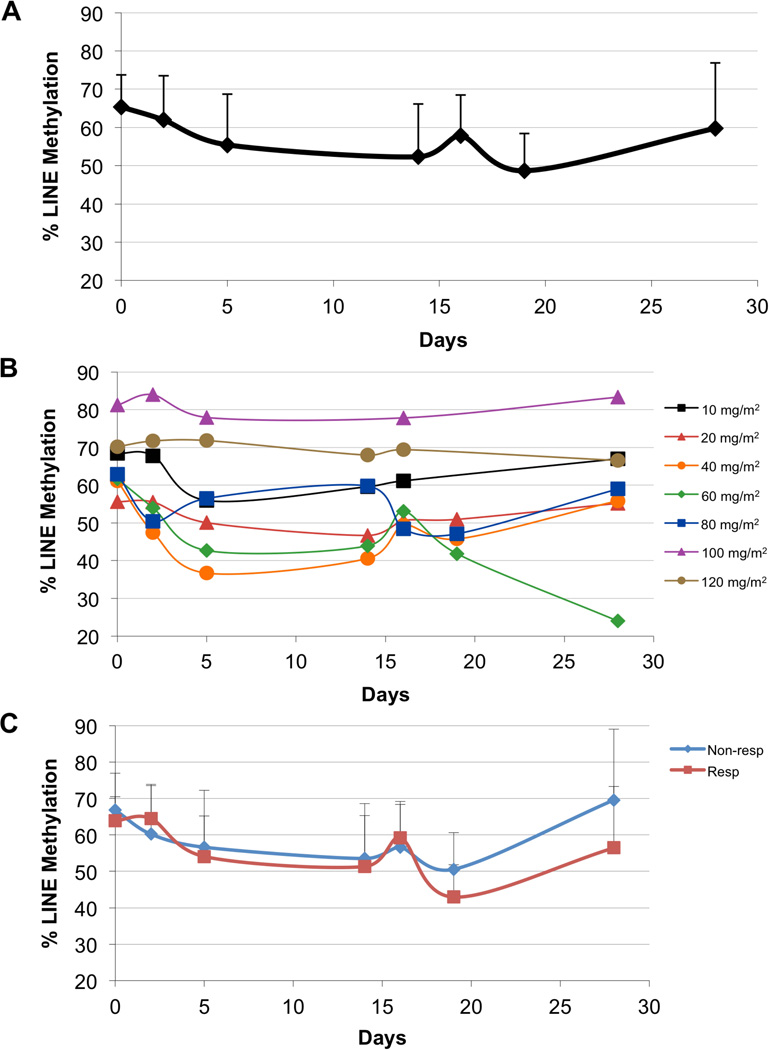
We investigated the kinetics of DNA methylation in patients’ mononuclear cells whilst receiving treatment with decitabine alone during this study. Global DNA methylation was assessed using the LINE pyrosequencing assay (Fig 2A). Global methylation gradually decreased from day 0 to day 14, which corresponded with the administration of decitabine on days 1–5. There was a return to a more hypermethylated state by day 16, with a decrease again by day 19, which corresponded to the second administration of decitabine. By day 26, the average methylation increased to near baseline.
Fig 2.
Dynamics of global DNA methylation with decitabine therapy. Global DNA methylation was assessed with long interspersed nuclear element (LINE) methylation, where a bisulphite pyrosequencing assay was performed on patients’ mononuclear cells on days 0, 2, 5, 14, 16, 19 and 26 of treatment. A) Average methylation of all patients tested. B) Methylation of patients by dosage of decitabine. C) Methylation of responders (Resp) vs. non-responders (Non-resp).
We also evaluated the effects of the decitabine dosage on changes in global methylation (Fig 2B). For many dosages, methylation gradually decreased in the first 14 days of therapy, increased by day 16, and decreased again during the second treatment. Specifically, a dose of 60 mg/m2 yielded a maximal level of global demethylation. Most notably, at the dosage of 60 mg/m2, LINE methylation continued to decrease from day 16 to day 28, whereas for most other dosages, methylation increased after a second nadir around day 19. Doses of 100 and 120 mg/m2 did not induce global hypomethylation to the extent that doses of 10–80 mg/m2 did. Finally, we also plotted the LINE methylation of responders (CRp and mCR) versus non-responders (Fig 2C). The patterns of demethylation were similar; however, responders had a non-significantly larger decrease in methylation at the end of cycle 1 and were noted to have larger fluctuations in global methylation with treatment.
Clinical activity
The responses observed in both parts of the study are shown in Table III. The overall response rate (including CR, CRp and mCR) was 21% in the decitabine alone part and 56% in the decitabine + Hyper-CVAD part. In the decitabine alone part, no patient achieved CR, 1 patient (3%) had CRp (dose level 80 mg/m2) and 5 patients (17%) had mCR. In the decitabine + Hyper-CVAD part, 6 patients (24%) achieved CR, 1 patient (4%) had CRp and 7 patients (28%) had mCR. Table III also shows the number of courses needed to yield treatment response at each dosage. One patient achieved CR only after 3 courses of therapy with decitabine + Hyper-CVAD. This patient also received one course of decitabine alone with no response (NR) prior to participating in the decitabine + Hyper-CVAD part of the study. Two patients who achieved CR on the second part of the study subsequently received haematopoietic stem cell transplants.
Table III.
Clinical responses in both parts of the trial.
| Decitabine dose (mg/m2) |
n | CR (n) | CRp (n) | mCR (n) | OR (%) | Courses to response (n) |
|---|---|---|---|---|---|---|
| Decitabine alone (n = 30) | ||||||
| 10 | 4 | 0 | 0 | 1 | 33 | 2 |
| 20 | 3 | 0 | 0 | 1 | 33 | 1 |
| 40 | 3 | 0 | 0 | 0 | 0 | -- |
| 60 | 3 | 0 | 0 | 1 | 33 | 2 |
| 80 | 3 | 0 | 1 | 0 | 33 | 2 |
| 100 | 3 | 0 | 0 | 0 | 0 | -- |
| 120 | 4 | 0 | 0 | 2 | 50 | 1, 2 |
| 60 (expansion) | 7 | 0 | 0 | 0 | 0 | -- |
| Overall | 30 | 0 | 1 | 5 | 21 | 1–2 |
| Decitabine + Hyper-CVAD (n = 25) | ||||||
| 5 | 3 | 2 | 0 | 0 | 66 | 3,1 |
| 10 | 3 | 1 | 0 | 1 | 66 | 1,1 |
| 15 | 3 | 0 | 0 | 1 | 33 | 1 |
| 20 | 3 | 2 | 0 | 1 | 75 | 1, 2, 1 |
| 40 | 3 | 1 | 0 | 0 | 33 | 1 |
| 60 | 3 | 0 | 0 | 2 | 66 | 1,1 |
| 40 (expansion) | 7 | 0 | 1 | 2 | 50 | 1, 1, 1, 1 |
| Overall | 25 | 6 | 1 | 7 | 56 | 1–3 |
n, number; CR, complete response; CRp, complete response without recovery of platelets; mCR, complete marrow response; OR, overall response; Hyper-CVAD, fractionated cyclophosphamide, vincristine, doxorubicin and dexamethasone alternating with high-dose methotrexate and cytarabine.
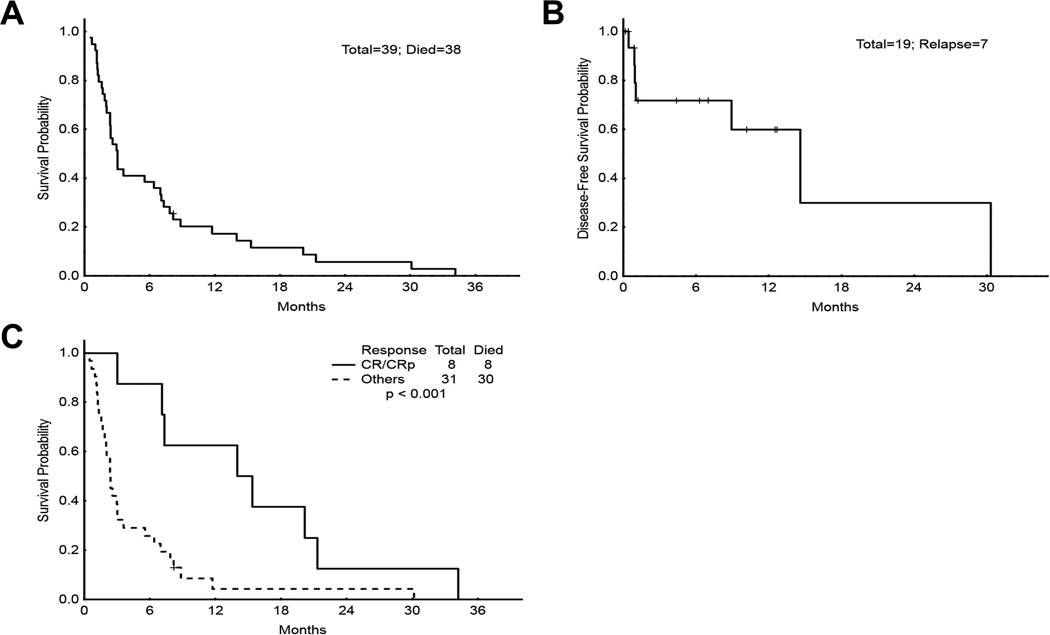
Median survival among all patients was 3.0 months (range, 0.5–34.2 months, Fig 3A). Median disease-free survival among patients who had any response (CR, CRp, or mCR) was 14.6 months (range, 0.0–30.3 months, Fig 3B). We compared the survival rates of responders (CR/CRp only) with those of non-responders for both parts (Fig 3C). Responders to decitabine or decitabine + Hyper-CVAD had a statistically significant improved survival over non-responders (P< 0.001), with a median survival of 15.3 months (range, 3.0–34.2 months) versus 2.4 months (range, 0.5–30.1 months).
Fig 3.
Survival analyses. A) Overall survival for patients in both parts of the trial (n = 39). B) Disease-free survival for CR, CRp, and mCR patients in both parts (n = 19). C) Survival of responders versus non-responders in both parts. Median survival for CR/CRp group was 15.3 months (range, 3.0–34.2 months), and median survival for all others was 2.4 months (range, 0.5–30.1 months). CR, complete response; CRp, complete response without recovery of platelets; mCR, complete marrow response.
Subset analyses of CR/CRp patients and patients previously receiving HyperCVAD
We evaluated the characteristics of the 8 patients that achieved a CR or CRp in both parts of the study (Table IV). One patient who responded to decitabine alone (CRp) had T-cell ALL. Of the 8 patients who achieved CR or CRp, 5 patients had received Hyper-CVAD alone before enrolling in the current trial. Of those 5 patients, 2 (40%) had achieved CR on Hyper-CVAD prior to relapse, while 3 (60%) had disease that was refractory to Hyper-CVAD before the addition of decitabine on protocol. Outcomes of other patients who had previously received Hyper-CVAD–based therapies without decitabine are reviewed in Table V. In addition to the 5 patients who achieved a CR or CRp on trial, there were 8 patients who achieved a mCR (4 patients on decitabine and 4 patients on decitabine + Hyper-CVAD). Out of 7 patients who had NR to previous Hyper-CVAD treatment, one patient each had CR, CRp and mCR on trial. Out of 19 patients who had previously responded and then progressed after Hyper-CVAD, a total of 10 patients had CR, CRp or mCR on study. Individual patients’ responses to all known prior therapies are included in the supporting information (Table SI).
Table IV.
Characteristics of patients who achieved CR/CRp as best response in either part of the trial (n = 8).
| Characteristic | n |
|---|---|
| T-ALL Phenotype | 1 |
| Cytogenetics | |
| Diploid | 2 |
| Ph+ | 0 |
| −5/−7 | 2 |
| Other | 4 |
| Prior therapies, median (range) | 2 (1–4) |
| Prior decitabine | 4 |
| Prior response to decitabine | 1 mCR |
| Prior Hyper-CVAD | 5 |
| Prior response to Hyper-CVAD | 2 CR, 3 NR |
n, number; T-ALL, T-cell acute lymphoblastic leukaemia; Ph+, Philadelphia chromosome-positive; −5/−7, deletion of chromosome 5 and/or 7; CR, complete response; CRp, complete response without recovery of platelets; mCR, complete marrow response; Hyper-CVAD, fractionated cyclophosphamide, vincristine, doxorubicin and dexamethasone alternating with high-dose methotrexate and cytarabine.
Table V.
Responses among patients who had previously received Hyper-CVAD alone or with other therapy, such as rituximab, tyrosine kinase inhibitor or L-asparaginase (n = 26). NR includes disease refractory to treatment.
| Responses | CR | CRp | mCR | NR | Total |
|---|---|---|---|---|---|
| Prior response on Hyper-CVAD | 19 | 0 | 0 | 7 | 26 |
| Best response on trial | 3 | 2 | 8 | 13 | 26 |
| Response on Decitabine alone | 0 | 1 | 4 | 10 | 15 |
| Response on Decitabine + Hyper-CVAD | 3 | 1 | 4 | 7 | 15 |
| Prior CR on Hyper-CVAD | 2 | 1 | 7 | 9 | 19 |
| Prior NR on Hyper-CVAD | 1 | 1 | 1 | 4 | 7 |
Hyper-CVAD, fractionated cyclophosphamide, vincristine, doxorubicin and dexamethasone alternating with high-dose methotrexate and cytarabine; CR, complete response; CRp, complete response without recovery of platelets; mCR, complete marrow response; NR, no response.
Discussion
We report one of the first trials to study the safety and clinical activity of decitabine in ALL in a significant cohort of patients. We have demonstrated that decitabine is safe and well tolerated at all doses tested, both alone and in combination with Hyper-CVAD–based chemotherapy. We have also shown that decitabine induces global hypomethylation in ALL patients at doses as high as 80 mg/m2. Finally, we noted that decitabine has clinical activity in relapsed or refractory ALL, particularly when given in combination with Hyper-CVAD. Overall response rate was 21% in the decitabine alone group and 56% in the decitabine plus Hyper-CVAD group. A significant number of patients achieved meaningful responses with this approach to therapy. Two patients who participated in the decitabine plus Hyper-CVAD part of the study achieved a CR and were able to undergo haematopoietic stem cell transplantion. Other patients who achieved CR did not undergo transplant for other reasons, including (most commonly) short duration of CR. Responses were achieved in both children and adults, and no conclusion can be made about the relative effectiveness of decitabine in younger versus older ALL patients.
We sought to understand the pharmacodynamics of decitabine with respect to its demethylation activity in ALL. Decitabine is a DNA methyltransferase inhibitor that causes dose-dependent demethylation of genomic DNA as well as histone modification in a specific pattern (Jabbour, et al 2008). The doses of decitabine used in this study were determined partly based on its pharmacology as well as previous experience with its use as a hypomethylating agent in MDS, AML and chronic myeloid leukaemia (Momparler 2005). Based on our current understanding of its hypomethylating behaviour at particular doses, decitabine probably acts through the restoration and reactivation of epigenetically silenced genes that inhibit leukaemic growth (Baylin 2005). Its clinical effectiveness appears to depend on repeating cycles of low-dose administration, so that the drug may be incorporated into the DNA of proliferating leukaemic cells (Jones and Taylor 1980). We demonstrated demethylation activity of decitabine at doses up to 80 mg/m2, and a dosage of 60 mg/m2 was most effective for demethylation of mononuclear cells from patients tested in our study.
Doses of decitabine used in this study for patients with ALL were higher than those used in MDS. In this study, the higher dosages in the range studied appeared to generate superior results in our patients. The higher dosages may reflect disease-specific, genomic-level changes that must take place for the drug to exert its effect in ALL patients. There are a number of potential reasons for disease-dependent effective dosage differences. First, different drug concentrations may exert effects on different groups of genes critical in the pathogenesis of lymphoid versus myeloid malignancies. Second, results from different dosages may reflect differences in the number of tumour cells, total tumour mass or the kinetics of leukaemic cell proliferation in lymphoid versus myeloid malignancies. Lastly, decitabine is often administered on a 10-day schedule for AML, on either days 1–5 and 8–12 or days 1–10 of a 28-day cycle (Blum, et al 2010, Issa, et al 2004). In the present decitabine-alone study, decitabine was administered on a 5-day schedule every other week (days 1–5 and days 15–19 of a 28-day cycle). Dosing schedule may prove important when considering the incorporation of decitabine in the context of each type of malignancy treated.
Three patients who had NR to prior Hyper-CVAD–based therapies achieved a response in this trial with the addition of decitabine and a total of 13 responders (3 CR, 2 CRp and 8 mCR) had previously undergone Hyper-CVAD–based therapies and subsequently had disease progression. Based on the current study, there is no evidence that decitabine restores sensitivity to Hyper-CVAD. Our results do, however, suggest that decitabine plus Hyper-CVAD may serve as a viable option for patients with refractory or relapsed ALL who have disease progression after Hyper-CVAD alone.
The idea that key epigenetic events are critical in ALL pathogenesis is supported by recent work demonstrating that methylation signatures of genetically distinct adult B-cell ALL samples correlate to specific genetic dysregulation (Geng, et al 2012). Additionally, there is growing evidence that ALL leukaemic cells utilize stem and progenitor cellular and molecular pathways in the initiation and progression of disease (Pui, et al 2004). This aberrant subpopulation may be responsible for evading treatment and relapse (Reya, et al 2001). The NOTCH pathway, for example, is involved in cell-fate decisions in the lymphoid branch of the developmental haematopoietic tree, and is also highly dysregulated in some ALL (Grabher, et al 2006). Recently, it was shown that the NOTCH-HES pathway is epigentically dysregulated in B-ALL through hypermethlyation (Kuang, et al 2013). Additionally, several genes shown to be methylated in ALL are involved in WNT signalling, a known stem cell pathway regulator (Clevers 2006, Garcia-Manero, et al 2009). As haematopoietic maturation relies on proper methylation and demethylation of critical developmental genes, these pathways may also be dysregulated in leukaemic cells (Cedar and Bergman 2011). Decitabine may be important in the elimination of these stem-like cells.
Although there were fewer patients who responded in the decitabine alone group, this treatment yielded fewer side effects, and may be an option for ALL patients who cannot tolerate intensive therapies. Certain subsets of ALL patients, determined by molecular profiling, may benefit from decitabine therapy, however these were not identified in the current study. Future work will focus on identifying those patients who will most benefit from decitabine treatment, and whether combination with other agents, such as histone deacetylase inhibitors, will be beneficial in ALL (Kalac, et al 2011). While the precise mechanism of this treatment is investigated, additional studies are warranted using decitabine alone, or in combination with Hyper-CVAD, as an option for patients with relapsed, refractory ALL.
Supplementary Material
Acknowledgements
We are grateful to personnel at The University of Texas MD Anderson Cancer Center for support, technical assistance, data analysis, and care of patients who participated in this study. This research is supported by the following grants: National Cancer Institute, USA (5R21CA126457); Leukemia & Lymphoma Society, USA (6173-07); The University of Texas MD Anderson Cancer Center Support Grant (CA016672).
Footnotes
Competing interests: the authors have no competing interests.
Authorship and Disclosures
Contribution: C.B.B. analysed data and wrote the manuscript; D.A.T. designed the research, performed research and contributed essential research tools; H.Y., M.K., S.P. performed research, analysed data; F.R., M.E.R., S.O., A.R.K.F., G.B., S.D., E.J. contributed essential research tools; H.K. designed the research and contributed essential research tools. G.G.-M. designed the research, performed research, analysed data, funded the studies, wrote the manuscript and contributed essential research tools. The authors have no competing interests.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Table SI. Individual patient characteristics, prior therapies/responses, treatment courses on protocol and responses.
References
- Appleton K, Mackay HJ, Judson I, Plumb JA, McCormick C, Strathdee G, Lee C, Barrett S, Reade S, Jadayel D, Tang A, Bellenger K, Mackay L, Setanoians A, Schätzlein A, Twelves C, Kaye SB, Brown R. Phase I and pharmacodynamic trial of the DNA methyltransferase inhibitor decitabine and carboplatin in solid tumors. Journal of Clinical Oncology. 2007;25:4603–4609. doi: 10.1200/JCO.2007.10.8688. [DOI] [PubMed] [Google Scholar]
- Baylin SB. DNA methylation and gene silencing in cancer. Nature Clinical Practice Oncology. 2005;(2 Suppl 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- Blum W, Garzon R, Klisovic RB, Schwind S, Walker A, Geyer S, Liu S, Havelange V, Becker H, Schaaf L, Mickle J, Devine H, Kefauver C, Devine SM, Chan KK, Heerema NA, Bloomfield CD, Grever MR, Byrd JC, Villalona-Calero M, Croce CM, Marcucci G. Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7473–7478. doi: 10.1073/pnas.1002650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Epigenetics of haematopoietic cell development. Nature reviews Immunology. 2011;11:478–488. doi: 10.1038/nri2991. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Estécio MRH, Issa J-PJ. Dissecting DNA hypermethylation in cancer. FEBS letters. 2011;585:2078–2086. doi: 10.1016/j.febslet.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florean C, Schnekenburger M, Grandjenette C, Dicato M, Diederich M. Epigenomics of leukemia: from mechanisms to therapeutic applications. Epigenomics. 2011;3:581–609. doi: 10.2217/epi.11.73. [DOI] [PubMed] [Google Scholar]
- Garcia-Manero G, Daniel J, Smith TL, Kornblau SM, Lee M-S, Kantarjian HM, Issa J-PJ. DNA methylation of multiple promoter-associated CpG islands in adult acute lymphocytic leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2002;8:2217–2224. [PubMed] [Google Scholar]
- Garcia-Manero G, Jeha S, Daniel J, Williamson J, Albitar M, Kantarjian HM, Issa J-PJ. Aberrant DNA methylation in pediatric patients with acute lymphocytic leukemia. Cancer. 2003;97:695–702. doi: 10.1002/cncr.11090. [DOI] [PubMed] [Google Scholar]
- Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, Yang H, Rosner G, Verstovsek S, Rytting M, Wierda WG, Ravandi F, Koller C, Xiao L, Faderl S, Estrov Z, Cortes J, O’Brien S, Estey E, Bueso-Ramos C, Fiorentino J, Jabbour E, Issa J-P. Phase 1/2 study of the combination of 5-aza-2’-deoxycytidine with valproic acid in patients with leukemia. Blood. 2006;108:3271–3279. doi: 10.1182/blood-2006-03-009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Manero G, Yang H, Kuang S-Q, O’Brien S, Thomas D, Kantarjian H. Epigenetics of acute lymphocytic leukemia. Seminars in Hematology. 2009;46:24–32. doi: 10.1053/j.seminhematol.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Brennan S, Milne TA, Chen W-Y, Li Y, Hurtz C, Kweon S-M, Zickl L, Shojaee S, Neuberg D, Huang C, Biswas D, Xin Y, Racevskis J, Ketterling RP, Luger SM, Lazarus H, Tallman MS, Rowe JM, Litzow MR, Guzman ML, Allis CD, Roeder RG, Müschen M, Paietta E, Elemento O, Melnick AM. Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia. Cancer discovery. 2012;2:1004–1023. doi: 10.1158/2159-8290.CD-12-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nature Reviews Cancer. 2006;6:347–359. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- Hoshino K, Quintás-Cardama A, Yang H, Sanchez-Gonzalez B, Garcia-Manero G. Aberrant DNA methylation of the Src kinase Hck, but not of Lyn, in Philadelphia chromosome negative acute lymphocytic leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 2007;21:906–911. doi: 10.1038/sj.leu.2404615. [DOI] [PubMed] [Google Scholar]
- Issa J-PJ, Garcia-Manero G, Giles FJ, Mannari R, Thomas D, Faderl S, Bayar E, Lyons J, Rosenfeld CS, Cortes J, Kantarjian HM. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2’-deoxycytidine (decitabine) in hematopoietic malignancies. Blood. 2004;103:1635–1640. doi: 10.1182/blood-2003-03-0687. [DOI] [PubMed] [Google Scholar]
- Issa J-PJ, Gharibyan V, Cortes J, Jelinek J, Morris G, Verstovsek S, Talpaz M, Garcia-Manero G, Kantarjian HM. Phase II study of low-dose decitabine in patients with chronic myelogenous leukemia resistant to imatinib mesylate. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:3948–3956. doi: 10.1200/JCO.2005.11.981. [DOI] [PubMed] [Google Scholar]
- Jabbour E, Issa J-P, Garcia-Manero G, Kantarjian H. Evolution of decitabine development: accomplishments, ongoing investigations, and future strategies. Cancer. 2008;112:2341–2351. doi: 10.1002/cncr.23463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Kalac M, Scotto L, Marchi E, Amengual J, Seshan VE, Bhagat G, Ulahannan N, Leshchenko VV, Temkin AM, Parekh S, Tycko B, O'Connor OA. HDAC inhibitors and decitabine are highly synergistic and associated with unique gene-expression and epigenetic profiles in models of DLBCL. Blood. 2011;118:5506–5516. doi: 10.1182/blood-2011-02-336891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian HM, O’Brien S, Smith TL, Cortes J, Giles FJ, Beran M, Pierce S, Huh Y, Andreeff M, Koller C, Ha CS, Keating MJ, Murphy S, Freireich EJ. Results of treatment with hyper-CVAD, a dose-intensive regimen, in adult acute lymphocytic leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2000;18:547–561. doi: 10.1200/JCO.2000.18.3.547. [DOI] [PubMed] [Google Scholar]
- Keating GM. Azacitidine: a review of its use in higher-risk myelodysplastic syndromes/acute myeloid leukaemia. Drugs. 2009;69:2501–2518. doi: 10.2165/11202840-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Klimek VM, Tallman MS. Hypomethylating agents in acute lymphoblastic leukemia: untapped potential? Leukemia & Lymphoma. 2011;52:7–8. doi: 10.3109/10428194.2010.524330. [DOI] [PubMed] [Google Scholar]
- Kuang S-Q, Ling X, Sanchez-Gonzalez B, Yang H, Andreeff M, Garcia-Manero G. Differential tumor suppressor properties and transforming growth factor-beta responsiveness of p57KIP2 in leukemia cells with aberrant p57KIP2 promoter DNA methylation. Oncogene. 2007;26:1439–1448. doi: 10.1038/sj.onc.1209907. [DOI] [PubMed] [Google Scholar]
- Kuang S-Q, Fang Z, Zweidler-McKay PA, Yang H, Wei Y, Gonzalez-Cervantes EA, Boumber Y, Garcia-Manero G. Epigenetic inactivation of Notch-Hes pathway in human B-cell acute lymphoblastic leukemia. PLoS ONE. 2013;8:e61807. doi: 10.1371/journal.pone.0061807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tourneau C, Lee JJ, Siu LL. Dose escalation methods in phase I cancer clinical trials. Journal of the National Cancer Institute. 2009;101:708–720. doi: 10.1093/jnci/djp079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubbert M, Minden M. Decitabine in acute myeloid leukemia. Seminars in Hematology. 2005;42:S38–S42. doi: 10.1053/j.seminhematol.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Momparler RL. Pharmacology of 5-Aza-2’-deoxycytidine (decitabine) Seminars in Hematology. 2005;42:S9–S16. doi: 10.1053/j.seminhematol.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Narayan G, Freddy AJ, Xie D, Liyanage H, Clark L, Kisselev S, Un Kang J, Nandula SV, McGuinn C, Subramaniyam S, Alobeid B, Satwani P, Savage D, Bhagat G, Murty VV. Promoter methylation-mediated inactivation of PCDH10 in acute lymphoblastic leukemia contributes to chemotherapy resistance. Genes, chromosomes & cancer. 2011;50:1043–1053. doi: 10.1002/gcc.20922. [DOI] [PubMed] [Google Scholar]
- Paulson K, Kumar R, Ahsanuddin A, Seftel MD. Azacytidine as a novel agent in the treatment of acute lymphoblastic leukemia. Leukemia & Lymphoma. 2011;52:134–136. doi: 10.3109/10428194.2010.512965. [DOI] [PubMed] [Google Scholar]
- Pui C-H, Relling MV, Downing JR. Acute lymphoblastic leukemia. New England Journal of Medicine. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- Qin T, Youssef EM, Jelinek J, Chen R, Yang AS, Garcia-Manero G, Issa J-PJ. Effect of cytarabine and decitabine in combination in human leukemic cell lines. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:4225–4232. doi: 10.1158/1078-0432.CCR-06-2762. [DOI] [PubMed] [Google Scholar]
- Ren J, Singh BN, Huang Q, Li Z, Gao Y, Mishra P, Hwa YL, Li J, Dowdy SC, Jiang S-W. DNA hypermethylation as a chemotherapy target. Cellular signalling. 2011;23:1082–1093. doi: 10.1016/j.cellsig.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Saba HI, Wijermans PW. Decitabine in myelodysplastic syndromes. Seminars in Hematology. 2005;42:S23–S31. doi: 10.1053/j.seminhematol.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Shen L, Toyota M, Kondo Y, Obata T, Daniel S, Pierce S, Imai K, Kantarjian HM, Issa J-PJ, Garcia-Manero G. Aberrant DNA methylation of p57KIP2 identifies a cell-cycle regulatory pathway with prognostic impact in adult acute lymphocytic leukemia. Blood. 2003;101:4131–4136. doi: 10.1182/blood-2002-08-2466. [DOI] [PubMed] [Google Scholar]
- Storer BE. Design and analysis of phase I clinical trials. Biometrics. 1989;45:925–937. [PubMed] [Google Scholar]
- Willemze R, Archimbaud E, Muus P. Preliminary results with 5-aza-2’-deoxycytidine (DAC)-containing chemotherapy in patients with relapsed or refractory acute leukemia. The EORTC Leukemia Cooperative Group. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 1993;(7 Suppl 1):49–50. [PubMed] [Google Scholar]
- Wong NC, Ashley D, Chatterton Z, Parkinson-Bates M, Ng HK, Halemba MS, Kowalczyk A, Bedo J, Wang Q, Bell K, Algar E, Craig JM, Saffery R. A distinct DNA methylation signature defines pediatric pre-B cell acute lymphoblastic leukemia. Epigenetics : official journal of the DNA Methylation Society. 2012:7. doi: 10.4161/epi.20193. [DOI] [PubMed] [Google Scholar]
- Yánez L, Bermúdez A, Richard C, Bureo E, Iriondo A. Successful induction therapy with decitabine in refractory childhood acute lymphoblastic leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 2009;23:1342–1343. doi: 10.1038/leu.2009.58. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.