Abstract
Reprogramming technologies have emerged as a promising approach for future regenerative medicine. Here we report on the establishment of a novel methodology allowing for the conversion of human fibroblasts into Hematopoietic Progenitor-like Cells (HPC) with macrophage differentiation potential. SOX2 overexpression in human fibroblasts, a gene found to be upregulated during hematopoietic reconstitution in mice, induced the rapid appearance of CD34+ cells with a concomitant upregulation of mesoderm-related markers. Profiling of Cord Blood hematopoietic progenitor cell populations identified miR-125b as a factor facilitating commitment of SOX2-generated CD34+ cells to immature hematopoietic-like progenitor cells with grafting potential. Further differentiation towards the monocytic lineage resulted in the appearance of CD14+ cells with functional phagocytic capacity. In vivo transplantation of SOX2/miR-125b-generated CD34+ cells facilitated the maturation of the engrafted cells towards CD45+ cells and ultimately the monocytic/macrophage lineage. Altogether, our results indicate that strategies combining lineage conversion and further lineage specification by in vivo or in vitro approaches could help to circumvent long-standing obstacles for the reprogramming of human cells into hematopoietic cells with clinical potential.
Keywords: Monocytes, macrophages, reprogramming, blood, miRNAs, Transdifferentiation
Introduction
The discovery of induced pluripotent stem cell (iPSC) technologies in 2006 [1] opened the door for the future application of autologous stem-cell therapies in the clinic. Recent reports have pointed out potential caveats associated with the use of iPSCs [2] and boosted research on additional reprogramming strategies [3]. In this regard, lineage conversion methodologies have emerged as a complementary approach to iPSC technology [4, 5]. Two distinct lineage conversion strategies have been described based on the differential use of transcription factors (TFs). Direct lineage conversion relies on the overexpression of TFs directly regulating the expression of genes defining target cell identity [6-8]. In contrast, indirect lineage conversion has been described to rely on the overexpression of pluripotency-related factors leading to the generation of partially dedifferentiated intermediate cellular states and thus, requiring further signals for lineage specification [3, 9-11].
In terms of potential clinical applications, the hematopoietic system represents one of the most suitable tissues for stem cell-based therapies as it can be relatively easily reconstituted upon bone marrow or umbilical cord blood cell transplantation. However, and even though much effort has focused on the derivation of hematopoietic cells from iPSCs, their grafting and differentiation potential remains limited. Two different possibilities might be contributing to these limitations: first, iPSC-differentiated hematopoietic cells might represent an immature hematopoietic cellular state as described for other differentiated lineages [12]; second, intrinsic differences in the murine niche might be limiting the engraftment of human cells. Interestingly, two recent reports have explored the possibility of using lineage conversion strategies for tackling the issues associated to the use of iPSCs. On the one hand, Szabo et al. described the direct conversion of human fibroblasts into CD45+ positive cells with grafting potential, albeit grafting was limited to the site of injection [13]. More recently, Pereira et al. demonstrated the possibility of establishing a hemogenic gene expression program in vitro, though in vivo grafting and differentiation potential were not analyzed [14]. Noticeably, both reports demonstrated that the processes of conversion occurred in a direct fashion in which the TFs utilized for inducing the process were able to directly bind to hematopoietic gene promoters.
Here we report on the conversion of human fibroblasts into functional macrophage-like progenitor cells by using a novel strategy combining TF- and micro-RNA-based partial reprogramming alongside further in situ maturation steps accomplished upon in vivo transplantation into the hematopoietic niche.
Materials and methods
Reagents and antibodies
The following antibodies were used for flow cytometry, cell sorting and immunostainings respectively: mouse anti-human CD34-APC (Miltenyi and BD biosciences), CD34-PE (Miltenyi), mouse anti-human CD45-FITC (Miltenyi), CD45-APC (BD biosciences), CD45-VB (Miltenyi), mouse anti-human HLA ABC-VB (BD biosciences), CD31-FITC (BD biosciences), CD31-PE (BD biosciences), CD14-APC (Miltenyi), CD15-FITC (Miltenyi), mouse anti-human CD-133-APC (Miltenyi), mouse anti-human CD38-PE (BD biosciences), mouse anti-human CD90-FITC (BD biosciences), HLA-PE (BD biosciences), mouse APC isotype control (BD biosciences), mouse FITC isotype control (BD biosciences), mouse PE isotype control and mouse Vioblue isotype control (BD biosciences), OCT4 (C-10, SantaCruz, sc-5279, 1:100), LAMP-2 (1:50, Abcam), α-tubulin mouse IgG (1:500; Sigma), Anti mouse IgG-Cy3 (1:200; Jackson), Anti rabbit-IgG- Dylight 649 (1:200; Jackson), Alexa fluor 488-, 594- or 647-conjugated anti-rabbit or anti-mouse or anti-goat secondary antibodies (1/500, Jackson Immunoresearch), DAPI (5 mg ml−1) (1:2000; Invitrogen).
For ChIP assay experiment, the following antibodies were used: anti-rabbit-IgGs (sc-2027 Santa Cruz Biotechnology), and rabbit anti-SOX2 (ab59776; Abcam). A total of 5 μg of each antibody was used for ChIP experiments.
Cell culture
Neonatal human foreskin fibroblasts were purchased from ATCC (HFF-1). Additionally, two primary fibroblast lines were derived from human skin biopsies obtained with informed consent from patients. All three lines were negative for hematopoietic markers including CD34 and CD45. Fibroblasts were cultured in DMEM containing 10% FBS, 2 mM GlutaMAX (Invitrogen), 50 U/ml penicillin and 50 mg/ml streptomycin (Invitrogen). All cell lines were maintained in an incubator (37°C, 5% CO2) with media changes every second day.
Constructs, retroviral production and lentiviral production
Lentiviral constructs for the overexpression of miRNA species, 10a, 155, 126, 125a-5p and scramble-GFP were purchased from System Biosciences, while miR-125b was purchased from Genecopoeia. Retroviral constructs for the reprogramming factors used in this study - i.e. Oct4, SOX2 - were purchased from addgene. Moloney-based retroviral vectors (pMX) were co-transfected with packaging plasmids (gag-pol and VSVg) in HEK-293T cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Lentiviral vectors were co-transfected with packaging plasmids (pMDL, Rev and VSVg) in HEK-293T cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For retroviruses, viral supernatants were collected after 48 hours and stored at 4°C for being used the next days (up to 7 days). For lentiviruses, viral supernatants were collected after 48 hours and an ultracentrifugation step was performed (19,400rpm for 2 hr). Then, lentiviral particles were concentrated in PBS (500X) and stored at −80°C until further use.
Retroviral transductions
1.105 HFFs were transduced with pMX retroviruses expressing empty-Orange (Ø), Oct4-Orange, Oct4, SOX2-Orange or SOX2 as follow. Transduction was performed by centrifuging the cells in the presence of the respective viruses (MOI 0.75) and polybrene (4ug/ml) at 1800rpm for 45minutes at room temperature. The same procedure was repeated an additional time after allowing the cells to rest overnight. One day post-transduction, media was changed and cells were kept during 6 days in the presence of “dedifferentiation media” (DMEM/F12 (Invitrogen) supplemented with 20% Knockout Serum Replacement (Invitrogen), 1 mM L-glutamine, 0.1 mM non-essential amino acids, 55 M 2-β-mercaptoethanol and 10 ng/ml bFGF (peprotech)).
Lentiviral transductions with microRNAs and media switch
Fibroblasts were infected with lentiviral particles previously produced, either to test the overexpression of a specific miRNA or to test the differentiation towards blood progenitors. Regarding the latter, and after different tests, we decided to use a protocol of transduction in which fibroblasts were first co-transduced with both SOX2 and miR-125b (or the respective controls). Upon co-transduction, cells were maintained in dedifferentiation media during 6 days and afterwards switched to the media described for the expansion of CB-HPCs (see below) for another 8 days with media changes every other day.
Purification of total RNA, miRNA and RT- PCR (mRNA and miRNA)
Samples used for mRNA or miRNA expression analysis were resuspended in Trizol reagent and total RNA (including miRNAs) was extracted using the miRNA easy Qiagen kit (#217004) following the manufacturer's instructions. The Cloned AMV First-Strand cDNA Synthesis Kit (#12328, Invitrogen) was used to obtain cDNA to quantify gene expression. For miRNA analysis, the NCode Express SYBR GreenER (Life Technologies) kit was used. miRNA primers were all from the NCODE database (life technologies). All quantitative PCRs were performed in Applied Biosciences thermocyclers. Primers used throughout the manuscript are listed in Supplementary Tables 1 and 2.
ChIP Assay
SOX2 chromatin occupancy was measured in fibroblasts after 7 days of transduction with SOX2 and compared against uninfected fibroblasts. 3*105 cells were fixed using 1% PFA, treated with glycine 0.1M and resuspended in nuclei lysis buffer (SDS 1%, EDTA 100mM, Tris-HCl 50mM pH7.5). Chromatin samples were sonicated with a Branson Digital Sonifier to generate DNA fragments from 200 to 1000 bp, diluted with ChIP RIPA Buffer (Tris-HCl 0.1mM pH7.5, EDTA 1mM, EGTA 0.5mM, Triton X-100 1%, SDS 0.1%, Deoxycolic acid 0.1%, NaCl 140mM) and incubated with Dynabeads Protein A (Invitrogen) coupled to the specific antibody. After overnight incubation, the immunocomplexes were eluted in ChIP elution buffer (Tris-HCl 20mM pH7.5, EDTA 5mM, NaCl 140mM, SDS 1%, proteinase K 40μg/ml) and the crosslinking was reverted overnight at 65°C. DNA was recovered by phenol/chloroform extraction and quantified by quantitative PCR using SYBR green on an ABI Prism 7300 system. Specific primers used for the ChIP experiment were reported and validated elsewhere [13] (see supplementary table 1). Data obtained by realtime PCR for each specific antibody were normalized to IgG control and plotted as percentage of input (% input ChIP − % input IgG).
Cord blood hematopoietic progenitor isolation
Human Cord Blood bags were obtained from Banc de Sang i Teixits, Hospital Duran i Reynals, Barcelona. Mononuclear cells (MNC) were isolated from CB using Lympholyte-H (Cederlane, Ontario, CA) by density gradient centrifugation. CD34+ cells were positively selected using Mini-Macs immunomagnetic separation system (Miltenyi Biotec, Bergisch Gladbach, Germany). The CD34+ fraction was immunophenotyped by FACS, for the CD45, CD38, CD133, CD90 antibodies.
Expansion of Hematopoietic stem/progenitor cells
30,000/well (24-well plates) sorted CD34+ cells were co-cultured on Bone Marrow Mesenchymal Stem Cells (LONZA) previously plated at a density of 50,000 cells/cm2. Cells were cultured in 1 ml of IMDM supplemented with Human Serum (1%), Human albumin (0,4%, griffols), insulin-transferrin-selenium 1X (GIBCO), Flt3, TPO and SCF (50 ng/ml, Peprotech). 0.5 ml medium/well were added every 4 days.
Hematopoietic colony-forming assays
Hematopoietic clonogenic assays were performed in 35-mm low adherent plastic dishes (Stem Cell Technologies, Vancouver, BC, Canada) using 1.1 ml/dish of methylcellulose semisolid medium (MethoCult H4434 classic, Stem Cell Technologies) according to the manufacturer's instructions. Briefly, enriched CD34+ cells were sorted and immediately plated at various densities: 1.5 × 103/ml, 3 × 103/ml and 6 × 103/ml. All assays were performed in duplicate. Colony-forming units (CFU) and Burst-forming units (BFU) were identified after 14 days of incubation according to their colony morphology as erythroid (CFU-E and BFU-E), granulocyte, erythroid, macrophage, megakaryocyte (CFU-GEMM), granulocyte-macrophage (CFU-GM), and macrophage (CFU-M).
Myeloid differentiation and phagocytosis assays
14 days after initiating the conversion process (see retroviral transductions and lentiviral transductions with microRNAs and media switch), CD45+ cells were sorted from HFF infected with SOX2 + miR125b or scr-miR. CD45+ sorted cells were further cultured during 6 days with RPMI supplemented with IL-4 and M-CSF (10μg/ml. After myeloid induction, 5*104 cells were plated with 1*107 solid Fluoresbrite 0.5-μm BB carboxylate microshperes (Polysciences), previously coated with human serum 50%. After 90 minutes of incubation cells were washed twice with cold PBS and bead uptake was analyzed by flow cytometry. Experiments were performed in technical duplicates with 3 biological replicates. Fibroblasts undifferentiated or partially converted after 7 days of SOX2 induction were used as negative controls and monocytes from human cord blood samples were used as a positive control. After phagocytosis experiment cells were recovered, fixed with PFA 4% and processed for immunostaining.
Immunocytochemistry and fluorescence Microscopy
Briefly, cells were washed thrice with PBS and fixed using 4% PFA in 1X PBS. After fixation, cells were blocked and permeabilized for 1 hour at 37° C with 5% BSA/5% appropriate serum/1X PBS in the presence of 0.1% Triton X100. Subsequently, cells were incubated with the indicated primary antibody either for 1hr at room temperature or overnight at 4° C. The cells were then washed thrice with 1X PBS and incubated for 1 hour at 37° C with the respective secondary antibodies and 20 minutes with DAPI. Cells were washed thrice with 1X PBS before analysis. Sections were analyzed by using an Olympus 1X51 upright microscope equipped with epifluorescence and TRITC, FITC, and DAPI filters. Confocal image acquisition was performed using a Zeiss LSM 780 laser-scanning microscope (Carl Zeiss Jena, Germany) with 20X, 40X or 63X immersion objectives.
Flow cytometry analysis
Cells were harvested, washed once with PBS and further incubated with the corresponding antibodies in the presence of FACS blocking buffer (1×PBS/10%FCS) for 1 hour on ice in the absence of light. After incubation, cells were washed thrice with 1 ml of FACS blocking buffer and resuspended in a total volume of 200 μl prior to analysis. A minimum of 10,000 cells in the living population were analyzed by using a BD LSRII flow cytometry machine equipped with 5 different lasers and the BD FACSDiva software. Percentages are presented after subtracting isotype background and referring to the total living population analyzed based on FSC/SSC gating and the use of Propidium Iodide and/or 7-AAD staining. Results are representative of at least two independent experiments with a minimum of two technical replicates per experiment.
Cell sorting
CD34+ cells were stained as described above and sorted by using a BDAria II and/or MoFlow FACS sorter (BD Biosystems). Alternatively, CD34+ cells were enriched using anti-CD34 conjugated magnetic beads (Miltenyi biotec) according to the manufacturer's instructions with slight modifications. Briefly, up to 109 cells were incubated with constant mixing at 4° C with 100 μl of the corresponding magnetic beads in the presence of 100 μl of Fc-blocking solution in a total volume of 500 μl FACS blocking buffer. After 1 hour, cells were sorted by two consecutive rounds of column separation in order to increase purity by applying MACS separation magnets. Shortly, cells were passed through the first MS separation column allowing binding of labeled cells. Non-labeled cells were washed thoroughly with 3 ml FACS blocking buffer prior to elution of the labeled fraction. Eluted labeled cells were then subjected to a second purification step as described above.
In vivo transplantation experiments
For each condition indicated in the manuscript (converted CD34+/CD45- cells; CD34+/CD45- obtained from CB samples; Fibroblasts + scr-miR or Fibroblasts), 1.105 cells were injected in the bone marrow of 6-8 weeks old NSG female mice previously irradiated (2,75 Cy). Mice were treated with antibiotics and monitored for the next 8 weeks. Subsequently, mice were sacrificed and samples of peripheral blood, spleen, lymph nodes, backbone and bone marrow from both legs were recovered and treated in order to obtain single cells for further analysis. Cells were immunophenotyped by flow cytometry and fluorescence microscopy as described.
Analysis of endogenous murine regenerative responses
6-8 week old NSG, C57BL/6, Oct4-GFP and Sox2-GFP (the Jackson laboratory) female mice were irradiated with 2,25 Cy. Mice were treated with antibiotics and bone marrow aspirates were collected at day 3 post-irradiation for RNA analysis. After depletion of red blood cells, two different cell populations were sorted out including MSCs and HSCs. MSCs were identified as positive for CD29, CD73 and CD44, and HSC for CD34, C-Kit and Sca-1. After cell sorting, cells were immediately processed to recover total RNA. mRNA levels of specific genes were measured by qRT-PCR and compared with the expression levels respectively found in non-irradiated NSG mice (n=4)
Collection of Mouse Samples
Samples from peripheral blood, ipsilateral as well as contralateral femoral bone marrow, spleen and spine of the sacrificed mice were culture in X-VIVO 10 medium (Lonza) during 1 day. The presence of GFP cells was analyzed by fluorescence microscopy and cells were recovered in trizol for further RT-PCR and PCR analysis
Detection of human samples by PCR
The DNA from the total sample of each organ was extracted by trizol extraction (Tri Reagent- MRC) following the manufacturer's instructions. Briefly, 5 to 50 ng of DNA were used to perform a PCR specific for the alpha satellite of the human chromosome 17.
Analysis of GFP cells in bone marrow samples
Transversal sections of spinal bone, cryopreserved and fixed with PFA (4%), were obtained from each mice in order to detect GFP-human positive cells. Each slice was stained with Masson's trichrome staining to detect the bone marrow and the subsequent slice was immunostained with anti-GFP- alexa 488 antibodies.
Ex-vivo culture
CD34/45 cells were collected and resuspended in X-VIVO 10 medium. Cells were cultured in methylcellulose for colony forming experiments. After 14 days, single cells were recovered and subjected to RT-PCR and/or immunofluorescence phenotyping.
Statistical evaluation
Statistical analyses of all endpoints were performed by using unpaired Student t and Mann-Whitney tests (one-tailed, 95% confidence intervals) using Graphpad 5.0 (Prism). All data are presented as mean ± standard deviation (s.d.) and represent a minimum of two independent experiments with at least three technical replicates.
Results
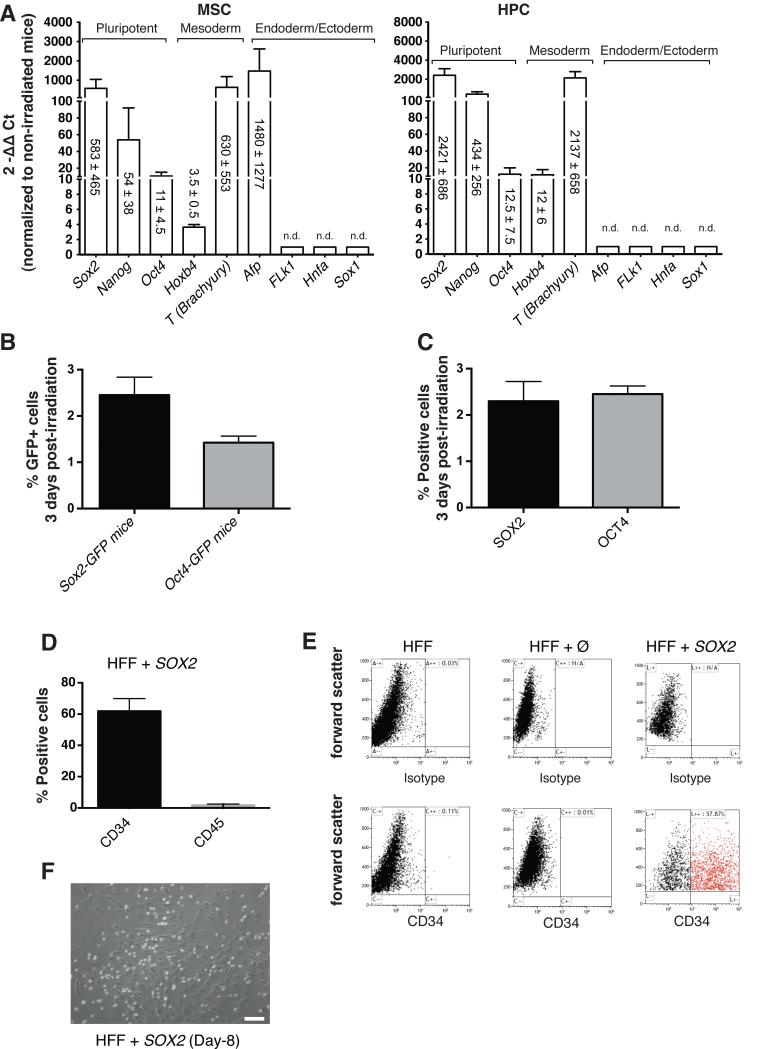
In order to establish a novel lineage conversion strategy for the generation of human blood progenitor cells, we first investigated whether endogenous reconstitution of the murine hematopoietic system involved the upregulation of pluripotency-related genes. Upon sublethal irradiation, endogenous reconstitution was allowed to proceed for a period of 3 days. Subsequently, the expression levels of pluripotent and lineage-specific TFs was evaluated in two different stem/progenitor cell populations residing within the bone marrow: mesenchymal stem cells (MSCs) and hematopoietic progenitor cells (HPCs). Murine MSCs (CD29+, CD73+, CD44+), and HPCs (CD34+, c-Kit+, Sca-1+) were sorted out and RNA levels compared to the basal gene expression levels observed in non-irradiated mice (Fig. 1A). We observed the rapid upregulation of Sox2 as well as other pluripotency-related TFs such as Nanog and Oct4, albeit to a lower extent as compared to Sox2, in both MSCs and HPCs (Fig. 1A). Next, we sought to investigate the expression of SOX2 and OCT4 by flow cytometry (Fig. 1B,C). We made use of knock-in wild-type animals in where GFP expression is driven by Sox2 and Oct4 promoters respectively. Our results demonstrated that a small fraction of the cells in the marrow expressed SOX2 (~2.5%) and OCT4 (~1.5%) upon irradiation (Fig. 1B). Then, we further validated our results by performing intracellular stainings for SOX2 and OCT4 in wild-type animals lacking any kind of reporter. Similar to the use of knock-in animals, intracellular flow cytometry staining demonstrated the appearance of a small population of cells expressing SOX2 and OCT4 (Fig. 1C). Together, these results elaborate on previous observations highlighting the role of Sox2 during regeneration [15] and suggest a potential role for pluripotency-related TFs during the early regenerative responses of the hematopoietic system in vivo.
Figure 1. SOX2 overexpression upon mouse total body irradiation identifies SOX2 as a potent TF for the conversion of fibroblasts into CD34+ cells.
A) 3 days after sublethal irradiation, murine mesenchymal stem cells (MSCs, left panel) and hematopoietic progenitor cells (HPCs, right panel) were sorted out for further gene expression analysis and compared with non-irradiated samples. mRNA expression of bone marrow aspirates demonstrated strong upregulation of Sox2 as well as other pluripotency-related markers during the endogenous murine regenerative response. n.d. stands for not detectable. B) Percentage of GFP+ cells detected in bone marrow aspirates from either Sox2-GFP or Oct4-GFP mice, three days after sublethal irradiation. C) Percentage of SOX2+ and OCT4+ cells detected in bone marrow aspirates from wild type mice, three days after sublethal irradiation. Non-irradiated control mice, for each mouse strain used in B and C, were respectively analyzed to confirm the absence of positive cells. D) Percentage of CD34+ cells obtained 8 days after SOX2 transduction in human foreskin fibroblasts (HFF). E) Representative flow cytometry plots showing the absence of CD34+ cells in HFF and HFF infected with an empty vector (Ø) whereas a CD34+ cell population can be clearly identified in SOX2-infected HFF at 8 days post-transduction. F) Representative bright field picture of SOX2-infected fibroblasts showing the appearance of small and round-shape cells resembling hematopoietic-related cells. Of note, fibroblast-like cells remained attached to the plate. Data are represented as mean +/− s.d., n=>3. Scale bar: 100μm (F).
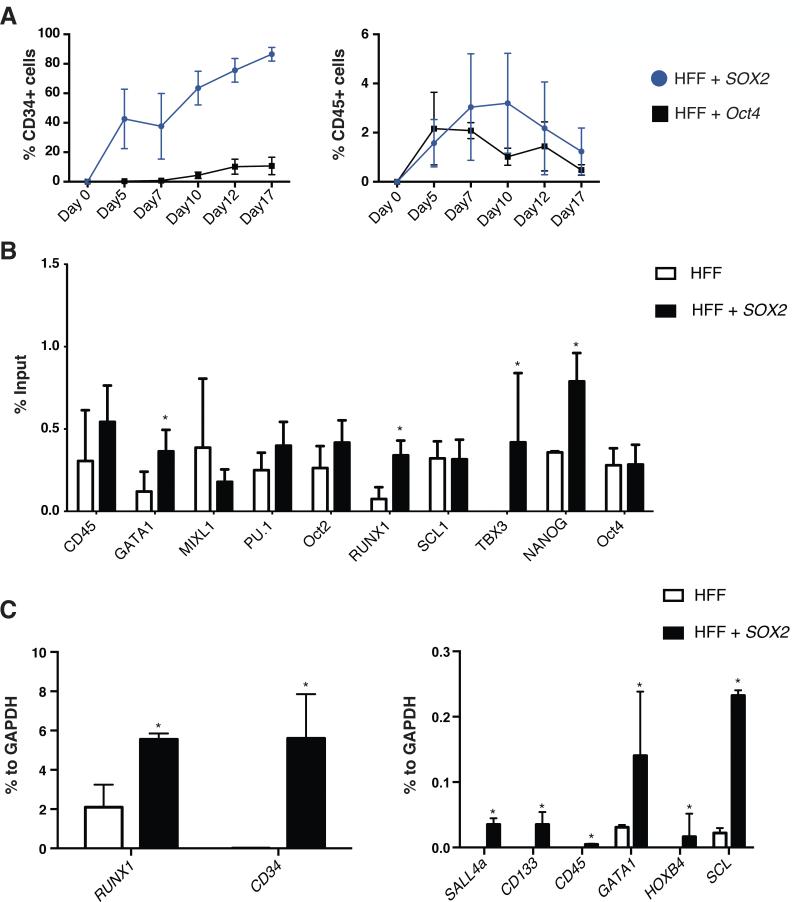
We next wondered whether overexpression of pluripotency factors might facilitate the generation of plastic cell intermediates [3, 9, 11, 16] that could be driven towards a hematopoietic fate upon exposure to specific hematopoietic signals. We decided to focus on SOX2 for several reasons: a) we have previously reported that SOX2 overexpression led to the appearance of CD34+ cells [9] unable to differentiate to terminal lineages unless other pluripotent factors were additionally used; b) Sox2 demonstrated the highest upregulation among all different pluripotent genes analyzed during murine BM reconstitution in HPCs. As expected, SOX2 overexpression led to the efficient conversion of human fibroblasts into CD34+ cells whereas CD45+ cells were marginally observed (Fig. 1D,E). We routinely transduced 1.105 human fibroblasts at a defined MOI of 0.75 with transduction efficiencies reaching 70-80% and, due to the proliferative nature of the transduced cells, generally obtained ~1.25×105 CD34+ positive cells 7 days after. SOX2-converted cells rapidly detached from the culture surface and, albeit some remaining fibroblast-like cells could be observed, small floating viable cells with round morphology accounted for the majority of cells in culture (Fig.1F). Oct4 has been reported to trigger a process of human cell conversion into CD45+ hematopoietic cells by direct mechanisms involving binding to hematopoietic gene promoters for driving transcription [13]. Therefore, in order to investigate the differences between Oct4- and SOX2-driven conversion processes, we first performed time-course analyses monitoring CD34 and CD45 expression. As expected, Oct4 overexpression resulted in lower amounts of CD34+ cells as compared to SOX2 (Fig. 2A) in line with the results reported by Szabo et al. [13]. For the purpose of defining whether SOX2 was driving human cell conversion by a direct mechanism similar to as described for Oct4, we performed Chromatin Immunoprecipitation (ChIP experiments) including a set of genes bound by OCT4 during HPC conversion [13]. Contrary to OCT4 [13], SOX2 was found to bind to genes typical of mesoderm and early hematopoietic specification (RUNX1, TBX3) (Fig. 2B). In addition, we observed a slight yet statistically significant upregulation of early hematopoietic markers at the RNA level, including RUNX1, SCL (TAL1), SALL4 and HOXB4 after SOX2 overexpression (Fig. 2C). Together, our results demonstrate that the sole overexpression of SOX2 suffices to induce an early hematopoietic-like phenotype.
Figure 2. SOX2 primes human fibroblasts towards a mesoderm/hematopoietic fate.
A) Kinetic of expression for CD34 and CD45 markers in HFF infected with SOX2 or Oct4. B) mRNA expression profiling of the converted CD34+ cells (SOX2-infected HFF) showing upregulation of mesoderm and early hematopoietic-related markers when compared to the original cell population. C) ChIP assay shows SOX2 binding to the regulatory regions of pluripotency and hematopoietic genes. Levels were determined by quantitative PCR and are expressed as the input percentage. Data are represented as mean +/− s.d. *p<0.05, =>3.
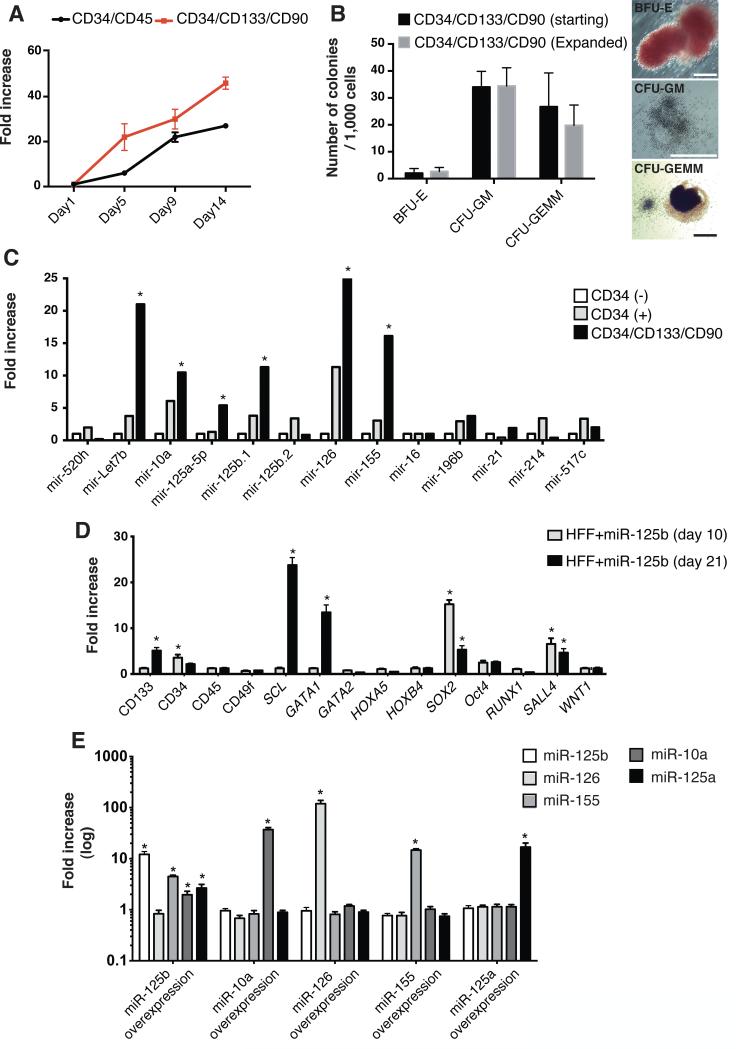
Accordingly, we next sought to identify specific factors contributing to hematopoietic commitment. We focused on miRNAs, as they have emerged as important players during development [17], reprogramming [4, 18, 19] and regenerative processes [20, 21]. We decided to use Cord Blood HPCs (CB-HPCs) because of their availability and their reported uses in hematopoietic transplantation experiments. Due to the limited ex vivo expansion potential of CB-HPCs, we first devised and validated a protocol allowing for CB-HPC expansion (Fig. 3A-B). Next we analyzed the expression of a set of miRNAs previously identified as differentially regulated in CD34+ populations derived from cord blood, peripheral blood and bone marrow [22-25]. Our results highlighted 6 miRNAs specifically upregulated in the CD34+/CD133+/CD90+ expanded population, Let7b, miR 10a, 125a-5p, 125b.1, 126 and 155 (Fig. 3C). miR-125b has been reported to be a major regulator of hematopoiesis in mice, a strong enhancer of engraftment upon transplantation [23, 26, 27] as well as to play a major role in myeloid and lymphoid commitment and associated malignancies [25]. Therefore, we first focused our attention on the effects of miR-125b overexpression. One day after transduction with miR-125b alone we observed a surviving population of ~ 75% of the cells by flow cytometry. Twenty-one days after overexpression, due to the proliferative nature of the cells, the living population reached ~95% with a ~4-fold expansion in the total number of cells. miR-125b resulted in the significant upregulation of key regulators of hematopoiesis including SCL (TAL1), GATA1, and SALL4 (Fig. 3D) [28, 29]. Most interestingly, miR-125b overexpression in human fibroblasts resulted in the endogenous upregulation of three other miRNAs identified in CB-HPCs ((miR-155, -10a and -125a (Fig. 3E)). Overexpression of miR-10a, -155, -125a-5p and -126, however, did not lead to the endogenous upregulation of additional miRNAs. Thus, suggesting miR-125b as the most suitable target for hematopoietic specification. We additionally discarded Let7b, due to its relatively ubiquitous expression among the different blood lineages. Together, our results highlighted a role for miR-125b in hematopoietic specification and indicated a preponderant and/or upstream role for miR-125b in controlling the expression of other miRNAs identified in CB-HPCs.
Figure 3. miR-125b expression is upregulated in human hematopoietic progenitor cells.
A) Expansion of endogenous Cord Blood hematopoietic progenitor populations by defined media conditions (starting number of cells: 30,000 at day 0). B) Hematopoietic Colony Forming (CFU) assays demonstrating maintenance of differentiation potential upon Cord Blood expansion. On the right, representative pictures of differentiated colonies. C) miRNA expression profile in the different Cord Blood populations as compared to the CD34-/CD45+ fraction. D) mRNA profiling of human fibroblasts overexpressing miR-125b demonstrates upregulation of pro-hematopoietic markers as well as a positive feedback loop leading to SOX2 upregulation. E) miRNA profiling of human fibroblasts individually overexpressing each of the miRNAs identified in CD34+/CD133+/CD90+ Cord Blood populations. Data are represented as mean +/− s.d. *p<0.05, =>3. Scale bars: 200μm (B).
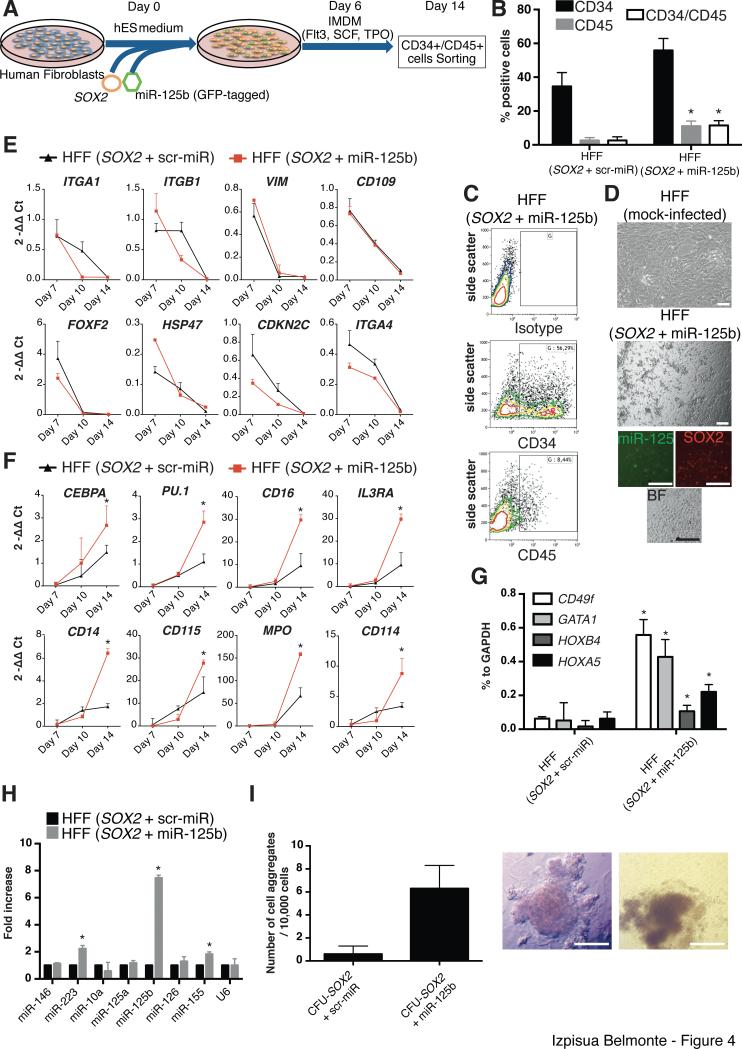
Next, we evaluated the role that SOX2 and miR-125b could play in combination with each other (Fig. 4A). Overexpression of miR-125b alone did not result in the expression of neither CD34 nor CD45 and could therefore not be used for the sorting experiments explained below (Fig. S1). Overexpression of miR-125b and SOX2 resulted in significantly higher percentages of CD45+ cells (Fig. 4B,C), lost of typical fibroblast morphology and detachment from the culture dish (Fig. 4D). qPCR analysis demonstrated the rapid downregulation of fibroblasts-related markers upon SOX2 overexpression independently of miR-125b co-overexpression (Fig. 4E). Accompanying erasure of initial cell identity, we observed the significant upregulation of hematopoietic markers related to myeloid lineages (Fig. 4F) only when both SOX2 and miR-125b were overexpressed together. In support of these results, sorting and RNA profiling of the generated CD34+/CD45+ cells obtained upon SOX2 + miR-125b overexpression demonstrated the significant upregulation of CD49f [30], HOXA5, HOXB4 and GATA1 (Fig. 4G). Previous reports have highlighted the role of PU.1 (SPI1) in driving the conversion of somatic cells to macrophages [31-33]. Therefore, we analyzed PU.1 expression upon SOX2 and miR-125b transduction. Interestingly, our data demonstrated a downregulation of PU.1 in the initial culture conditions followed by an upregulation when SOX2 + miR-125b-derived cells were exposed to cytokine containing media (Figs. 4F and S2). Furthermore, these CD34+/CD45+ cells demonstrated higher expression levels of miRNAs 155 and 223, both typically expressed in myeloid progenitor cells [34, 35] (Fig. 4H). Side by side comparison of cells overexpressing SOX2 with and without miR-125b highlighted that miR-125b contributed to cell self-renewal as exemplified by an increased number of colonies in clonal assays (Fig. 4I). Last, considering that CD34 can also be expressed in vascular progenitor cells [9], we next analyzed its expression at the protein level. Flow cytometry analyses demonstrated the lack of CD31 expression upon conversion (data not shown). Taken together, our results indicate that SOX2 sufficed for the priming of human fibroblasts towards an immature hematopoietic-like phenotype, as best exemplified by the appearance of CD34+ cells and the upregulation of early hematopoietic markers, whereas miR-125b allowed for further maturation into CD34+/CD45+ cells presenting a gene expression signature resembling early myeloid cells.
Figure 4. miR-125b facilitates human fibroblast conversion towards an hematopoietic-like fate.
A) Schematic representation of the conversion process. B) Flow Cytometry analysis of CD34 and CD45 after conversion of human Foreskin Fibroblasts (HFFs) overexpressing SOX2 with miR-125b or the respective scrambled control. C) Representative flow cytometry plots showing the emergence of CD34+ cells in HFFs infected with SOX2 and miR-125b, at day 14 after initiation of the conversion process. D) Representative bright field pictures and immunofluorescence of HFF overexpressing SOX2 with miR-125b, at day 14, showing the appearance of cell clusters displaying a hematopoietic-like morphology. E,F) Dynamic gene expression profile of SOX2-infected HFFs with miR-125b or a scrambled control demonstrating downregulation of fibroblasts-related genes (E) and upregulation of hematopoietic-related genes (F). Of note, the upregulation of hematopoietic genes was significantly higher when HFFs were co-infected with SOX2 and miR-125b (F). G) mRNA expression profiling of the converted CD34+/CD45+ cells (from the respective groups) demonstrating the acquisition of a hematopoietic-like signature. H) miRNA expression profiling of the converted CD34+/CD45+ cells. I) Clonal assays demonstrating enhanced self-renewal properties in the converted cells. Data are represented as mean +/− s.d. *p<0.05, n=>3. Scale bars: 100μm (D), 200 μm (I).
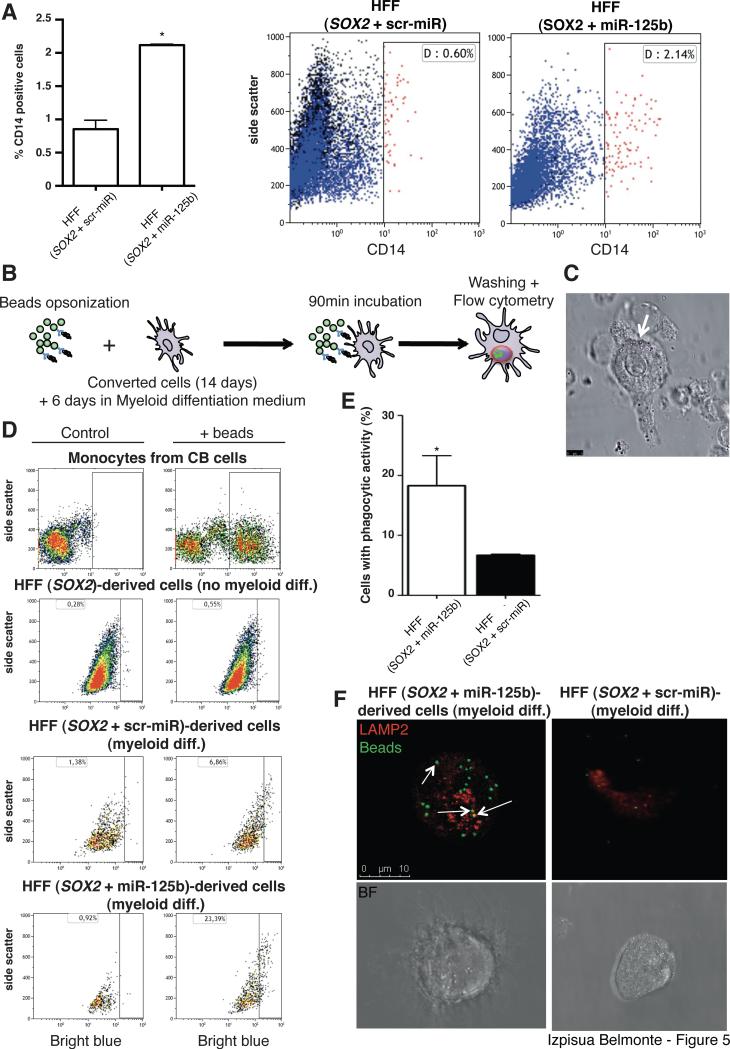
We next evaluated the ability of the obtained cells to differentiate into functional macrophages. After 6 days of differentiation we observed the appearance of CD14+ monocyte-like cells. The percentage of CD14+ cells was significantly increased when SOX2 was combined with miR-125b (Fig. 5A). To evaluate functionality of the generated cells, we tested phagocytic capacity by incubating the converted cells in the presence of beads opsonized with human serum (Fig. 5B, C). Comparative analyses confirmed the monocytic/macrophage-like nature of the differentiated cells (Fig. 5D). Though overall phagocytosis was higher when monocytes were derived from CB-HPCs, an expected result considering the purity of this control cell population, overexpression of miR-125b resulted in enhanced phagocytosis when compared to cells infected with a scrambled miRNA control (scr-miR) (Fig. 5D,E). Lastly, we decided to confirm that bead engulfment was due to phagocytosis and not to unspecific endocytosis processes. Immunofluorescence analyses confirmed that only CD45+/CD14+ cells obtained by SOX2+miR-125b overexpression internalized the beads in compartments expressing LAMP-2, a marker specifically involved in phagosome maturation (Fig. 5F). On the contrary, SOX2 + scr-miR converted fibroblasts displayed a highly reduced capacity for beads internalization (Fig. 5F). Taken together, our results indicated that reprogramming intermediates obtained upon SOX2 and miR-125b overexpression could be specifically differentiated toward macrophage-like cells with phagocytic activity.
Figure 5. Differentiation of fibroblast-derived CD34+ cells results in the acquisition of functional macrophage-like properties.
A) Differentiation of Fibroblast-derived CD34+ cells for 6 days leads to the appearance of CD14+ monocytic-like cells as assessed by Flow Cytometry. B) Experimental setup for the testing of phagocytic capacity in the converted fibroblasts. C) Representative picture of macrophage-like cells obtained after differentiation and incubation with opsonized beads. White arrow indicates the presence of internalized beads. D) Representative Flow Cytometry dot plots illustrating phagocytic properties in the converted fibroblasts (HFF). Cord Blood (CB) cells were used as positive controls. E) Flow Cytometry quantification demonstrated significantly higher phagocytic capacity in SOX2+miR-125b converted fibroblasts. F) Representative picture of a single fibroblast converted with SOX2+miR125b (left panels) or SOX2+scr-miR (right panels) after monocytic differentiation. Arrows indicate co-localization of the internalized beads with LAMP-2. Data are represented as mean +/− s.d. *p<0.05, n=3. Scale bars: 5μm (C), 10 μm (F).
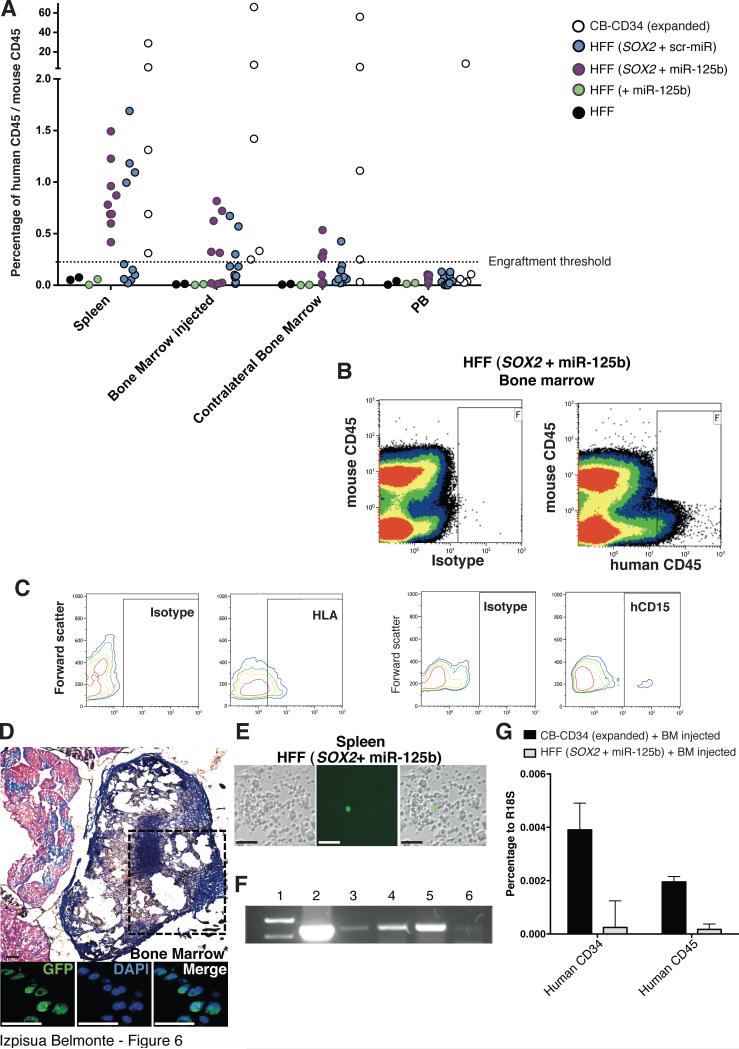
We next wondered whether the in vivo niche could suffice for the specification of the converted human cells in the absence of in vitro differentiation procedures. To test this hypothesis, CD34+/CD45- cells derived from fibroblasts overexpressing SOX2 + scr-miR or SOX2 + miR-125b were FACS sorted after 7 days of differentiation, and injected intrafemorally into sublethally irradiated NSG mice. Mice were routinely monitored and samples from the peripheral blood, injected femur, non-injected femur, backbone, lymph nodes and spleen obtained after 8 weeks. Importantly, we only sorted and analyzed human CD45+/murine CD45- cells to avoid the presence of chimeric events as a consequence of cell fusion processes that might occur in vivo. Interestingly, CD45+ human cells (HLA positive) were detected not only in the injected site but also in the spleen and contralateral bone marrow in a similar fashion to that observed upon CBHPC transplantation (Fig. 6A-C). Noticeably, the appearance of human CD45+ cells upon CD34+CD45- cell transplantation suggested that mere circulation was not responsible for the localization of human cells in distant sites and indicated that maturation and migration of the injected cells occurred as a result of exposure to the in vivo niche (Fig. 6A,B). In support of this conclusion CD45+ human cells were not detected in animals injected with any of the control groups (HFF and HFF+miR-125b). The human origin of cells harvested from each tissue was further confirmed by the presence of GFP positive cells in the bone marrow and ex-vivo cultures (Figs. 6D, E), by PCR specific for the chromosome 17 (Fig. 6F) as well as by RT-PCR for the expression of human CD34 and CD45 (Fig. 6G). Overexpression of miR-125b resulted in greater engraftment as compared to cells overexpressing SOX2 + scr-miR (Fig. 6A). Indeed, and contrary to animals injected with SOX2 + scr-miR converted cells, all animals receiving cells generated by SOX2 + miR-125b demonstrated the presence of human CD45+ cells in at least one of the different sites analyzed. These results confirmed that miR-125b was important to further commit SOX2-converted cells towards a more mature progenitor state with enhanced engraftment capacities. Most noticeably, SOX2 and miR-125b converted CD34+ cells were able to mature in vivo as indicated by the expression of CD45 and CD15 (Figure 6C). RNA and flow cytometry analyses for CD3 (T-cells) and CD19 (B-cells) suggested the lack of differentiation towards lymphoid lineages in vitro or in vivo (data not shown). Together, our results demonstrate that the combination of SOX2 and miR-125b allows for the conversion of human fibroblasts towards monocytes/macrophages with engraftment capacity as well as facilitates further maturation, migration and repopulation of distant hematopoietic compartments.
Figure 6. miR-125b facilitates engraftment into the murine niche.
A) Sorted CD34+CD45- cells were injected in sublethally irradiated NSG mice, and mice were sacrificed after 8 weeks. Representative plots indicating positive engraftment, migration and in vivo maturation, as exemplified by progression towards a CD45+ phenotype, only when SOX2 was combined with miR-125b prior injection (lower than 0,1%). B,C) Representative FACS analyses of cells recovered from the injected bone marrow indicating the presence of CD45 (B) HLA, and CD15 (C) positive human cells. D) The bones of injected mice were recovered, fixed and stained to detect the presence of GFP positive cells (corresponding to converted human cells expressing the miR-125b-gfp construct). Upper panel shows a representative transversal section after Masson's trichrome staining. Lower panels demonstrating the presence of GFP positive cells in the bone marrow of the vertebral column of the injected animals. E) GFP positive human cells were found ex-vivo upon recovery of cells from the spleen of mice injected with CD34+ cells obtained after conversion of HFFs with SOX2 and miR-125b. F) PCR of the human chromosome 17 α-satellite sequence (1, Ladder. 2, Bone marrow (BM) from mice injected with Cord Blood. 3-6, Cells from a mouse injected with CD34+ cells from (HFF+SOX2+miR-125b): 3, Contralateral BM. 4, Injected BM. 5, Spleen. 6, Peripheral blood.). G) mRNA expression levels of human CD34 and CD45 in cells recovered from the transplanted animals. Data are represented as mean +/− s.d. n=>3. Scale bars: 50μm (D), 100 μm (E).
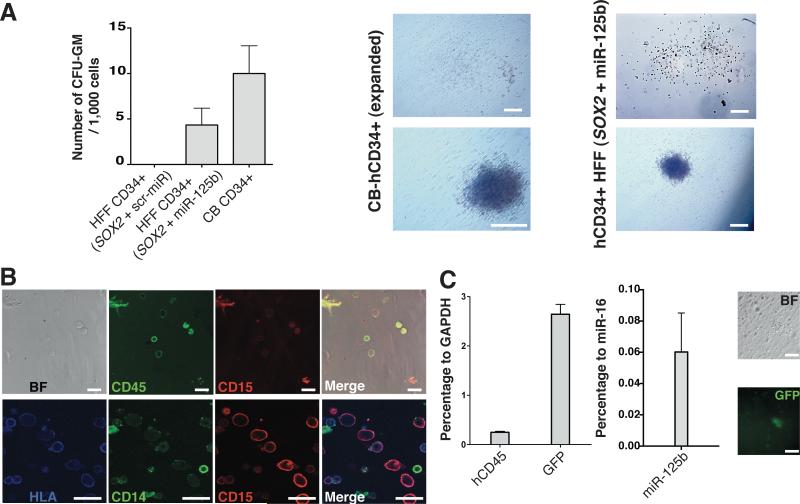
To further confirm whether maturation was occurring in vivo, we next decided to perform secondary differentiation assays to evaluate differentiation potential as well as the presence of hematopoietic progenitor populations derived from the injected Fibroblast-derived CD34+ fibroblasts. In vivo differentiated CD34+/CD45+ were sorted out from the different tissues and subjected to CFU assays. Contrary to SOX2+miR-125b converted fibroblasts prior to in vivo maturation, in vivo maturated sorted cells gave rise to well-defined hematopoietic colonies resembling those observed upon Cord Blood differentiation (Fig. 7A). Importantly, only cells generated by SOX2 and miR-125b overexpression gave rise to Granulocyte-macrophages colonies, in line with the gene expression signature of the generated cells prior to injection (Fig. 7A). Furthermore, cells recovered from the colonies were positively immunostained for HLA, CD45, CD15 and CD14, corroborating their human origin and monocytic identity while still expressing GFP and miR-125b (Figs. 7B,C). Interestingly, the lack of colony formation when other conditions were analyzed demonstrated specificity for SOX2 and miR-125b converted cells and ruled out contamination with murine HPCs. Together, these results further suggest that CD34+ cells generated by SOX2 and miR-125b overexpression possess an immature phenotype and that further maturation is accomplished as a result of the in vivo niche.
Figure 7. miR-125b facilitates in vivo maturation towards a monocyte-like fate.
A) Human CD34+CD45+ cells were sorted out from the engrafted animals and cultured in methylcellulose to assess the ability to give rise to human CFU. On the left, average number of CFU-GM observed after 14 days of culture of CD34+/45+ cells recovered from mice injected with CD34+CD45- human cells. On the right, representative images of the differentiated colonies ex-vivo demonstrating the formation of myeloid Granulocyte Monocyte-Colonies (CFU-GM). B) Single cells derived from the colonies were immunostained for HLA as well as the human hematopoietic marker CD45, and the monocyte/macrophage markers CD14 and CD15. C) The levels of human CD45, GFP and miR-125b were assessed by RT-PCR in single cells dissociated from the exvivo colonies (left panel). Right panels, representative images of a GFP-expressing colony. Data are represented as mean +/− s.d. *p<0.05, n=>3. Scale bars: 200μm (A), 20 μm (B), 100μm (C).
Discussion
In summary, here we report on a novel methodology for the conversion of human fibroblasts towards macrophage-like progenitor cells with the ability to engraft into murine recipients. Our results complement previous studies where direct lineage conversion strategies were reported suitable to obtain HPC, either by the overexpression of Oct4 [13] or of a specific cocktail of transcription factors [14]. However, in vivo grafting potential was only analyzed when Oct4 was used as a driver of the conversion process [13]. Additionally, numerous previous reports have demonstrated the possibility of reprogramming different somatic lineages to macrophages. Among them, the most notorious work has been performed by the Graf laboratory including the in vitro and even the in vivo conversion of several different murine hematopoietic lineages to a macrophage fate including B-cells [31], T-cells [32], the conversion of murine fibroblasts to functional macrophages upon PU.1 and C/EBPalpha/beta overexpression [33] and even the conversion of human leukemic cells to non-malignant macrophages [36]. Contrary to these strategies, our results highlight the suitability of lineage conversion processes based on the use of pluripotency factors for the generation of defined human cell types. However, one interesting observation was the fact that SOX2 and miR-125b overexpression results in the concomitant endogenous upregulation of PU.1 and C/EBP alpha, both genes described to induce the conversion of somatic cell lineages to macrophages upon overexpression [31-33], when exposed to hematopoietic cytokines. Thus, opening the possibility of certain overlapping conversion mechanisms triggered, in this case, by SOX2 and miR-125b. Contrary to previous reports leading to a more direct conversion of different cell types to functional macrophages, our strategy required coupling a pre-hematopoietic cellular state, induced by the sole overexpression of SOX2 and miR-125b, to robust differentiation conditions, including exposure of committed cells to the in vivo niche, for the completion of the lineage conversion process. Most importantly, in vivo transplantation of the converted human cells resulted in further differentiation of the injected cells towards a macrophage fate into the murine niche. The fact that the Yamanaka factors do not seem to determine a specific cell identity, as compared to processes of direct lineage conversion in which the overexpressing molecules are most generally defining target cell identity is better understood in the context of three recent publications [16, 37, 38]. Transcription factors driving the reprogramming of somatic cells to iPSCs act as “pioneer” TFs opening the chromatin structure and further facilitating and driving the binding of TFs defining cell identity to specific promoters. These observations are well in agreement with the observations by Polo et al. describing that chromatin remodeling processes take place in the very early stages of reprogramming whereas final cell identity is achieved later on once cells are poised to become iPSCs [16]. Our results indicate that SOX2 alone is able to induce an intermediate state and subsequent lineage specification could be accomplished upon miR-125b overexpression, probably due to the synergistic action of its reported downstream targets Lin28, p53, CDX2, STAT3, ETS1, MCL1, and IL-6 [39-41]. Of note while unable to bind hematopoietic gene promoters targeted by Oct4[13], SOX2 overexpression sufficed for the upregulation of genes related to mesodermal lineages and hematopoietic specification. Thus, opening the possibility of SOX2 acting in a direct or indirect manner similar to what has been described for the generation of Neural Progenitor Cells (NPCs) [42, 43]. Most strikingly, SOX2 and Oct4 have been reported to control the expression of ectodermal and mesendodermal genes respectively [44]. Therefore, making the use of SOX2 for the conversion of fibroblasts to hematopoietic lineages is somewhat counterintuitive. Yet, and along the same line, a recent report has found that Oct4 can be conversely utilized for the transdifferentiation of fibroblasts to NPCs [45]. Collectively, these observations, together with the reprogramming of somatic cells to iPSCs by lineage specifiers [46, 47], highlight the need of further investigations and add over the multiple pleiotropic roles that so-called “pluripotent factors” play in defining cell identity.
Conclusion
In here we describe a methodology that allows for the indirect lineage conversion of human fibroblasts into Hematopoietic Progenitor-like Cells (HPC) with macrophage differentiation potential. Our approach complements other reprogramming strategies for the lineage conversion of human somatic cells into human hematopoietic cells with in vivo reconstitution potential. Future studies unveiling the signals required for lymphoid lineage specification alongside the development of strategies for enhancing engraftment of hematopoietic cells generated by lineage conversion approaches might contribute to the development of novel therapeutics with clinical implications bypassing the risks associated to the use of pluripotent stem cells.
Supplementary Material
Acknowledgments
We thank M. Schwarz for administrative support. J.P. was partially supported by a Juan de la Cierva postdoctoral fellowship. Y.X. and L.K. were partially supported by a postdoctoral training grant from the California Institute for Regenerative Medicine (CIRM). E.N. was partially supported by a F.M. Kirby Foundation postdoctoral fellowship. I.S.M. was partially supported by a Nomis Foundation postdoctoral fellowship. Work in the laboratory of J.C.I.B. was supported by grants from NIH (5U01HL107442), CIRM, the G. Harold and Leila Y. Mathers Charitable Foundation and The Leona M. and Harry B. Helmsley Charitable Trust 2012PG-MED002.
Footnotes
Author contributions:
Julian Pulecio: Conception and design, Collection of Data, Data analysis and interpretation, Manuscript writing.
Emmanuel Nivet: Conception and design, Collection of Data, Data analysis and interpretation, Manuscript writing.
Ignacio Sancho-Martinez: Conception and design, Collection of Data, Data analysis and interpretation, Manuscript writing.
Marianna Vitaloni: Conception and design, Collection of Data,
Guillermo Guenechea: Collection of Data
Leo Kurian: Collection of Data
Ilir Dubova: Collection of Data
Yun Xia: Collection of Data
Juan Bueren: Data analysis and interpretation
Leopoldo Laricchia-Robbio: Conception and design, Data analysis and interpretation
Juan Carlos Izpisua Belmonte: Conception and design, Data analysis and interpretation, Manuscript writing, Final approval of manuscript.
Competing interests:
The authors declare no competing financial interests.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Panopoulos AD, Ruiz S, Izpisua Belmonte JC. iPSCs: induced back to controversy. Cell Stem Cell. 2011;8:347–348. doi: 10.1016/j.stem.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Sancho-Martinez I, Baek SH, Izpisua Belmonte JC. Lineage conversion methodologies meet the reprogramming toolbox. Nat Cell Biol. 2012;14:892–899. doi: 10.1038/ncb2567. [DOI] [PubMed] [Google Scholar]
- 4.Anokye-Danso F, Trivedi CM, Juhr D, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vierbuchen T, Wernig M. Direct lineage conversions: unnatural but useful? Nat Biotechnol. 2011;29:892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vierbuchen T, Ostermeier A, Pang ZP, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ieda M, Fu JD, Delgado-Olguin P, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 9.Kurian L, Sancho-Martinez I, Nivet E, et al. Conversion of human fibroblasts to angioblast-like progenitor cells. Nat Methods. 2013;10:77–83. doi: 10.1038/nmeth.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efe JA, Hilcove S, Kim J, et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 11.Ladewig J, Koch P, Brustle O. Leveling Waddington: the emergence of direct programming and the loss of cell fate hierarchies. Nat Rev Mol Cell Biol. 2013;14:225–236. doi: 10.1038/nrm3543. [DOI] [PubMed] [Google Scholar]
- 12.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szabo E, Rampalli S, Risueno RM, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 14.Pereira CF, Chang B, Qiu J, et al. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell. 2013;13:205–218. doi: 10.1016/j.stem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnold K, Sarkar A, Yram MA, et al. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polo JM, Anderssen E, Walsh RM, et al. A Molecular Roadmap of Reprogramming Somatic Cells into iPS Cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanyam D, Lamouille S, Judson RL, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoo AS, Sun AX, Li L, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams AH, Valdez G, Moresi V, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eulalio A, Mano M, Dal Ferro M, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 22.Georgantas RW, 3rd, Hildreth R, Morisot S, et al. CD34+ hematopoietic stemprogenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao R, Sun J, Zhang L, et al. MicroRNAs play a role in the development of human hematopoietic stem cells. Journal of cellular biochemistry. 2008;104:805–817. doi: 10.1002/jcb.21668. [DOI] [PubMed] [Google Scholar]
- 24.Merkerova M, Vasikova A, Belickova M, et al. MicroRNA expression profiles in umbilical cord blood cell lineages. Stem Cells Dev. 2010;19:17–26. doi: 10.1089/scd.2009.0071. [DOI] [PubMed] [Google Scholar]
- 25.O'Connell RM, Chaudhuri AA, Rao DS, et al. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci U S A. 2010;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaudhuri AA, So AY, Mehta A, et al. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28A. Proc Natl Acad Sci U S A. 2012;109:4233–4238. doi: 10.1073/pnas.1200677109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ooi AG, Sahoo D, Adorno M, et al. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21505–21510. doi: 10.1073/pnas.1016218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bresnick EH, Katsumura KR, Lee HY, et al. Master regulatory GATA transcription factors: mechanistic principles and emerging links to hematologic malignancies. Nucleic Acids Res. 2012;40:5819–5831. doi: 10.1093/nar/gks281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ichikawa M, Asai T, Chiba S, et al. Runx1/AML-1 ranks as a master regulator of adult hematopoiesis. Cell cycle (Georgetown, Tex. 2004;3:722–724. [PubMed] [Google Scholar]
- 30.Notta F, Doulatov S, Laurenti E, et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- 31.Xie H, Ye M, Feng R, et al. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 32.Laiosa CV, Stadtfeld M, Xie H, et al. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 33.Feng R, Desbordes SC, Xie H, et al. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Connell RM, Zhao JL, Rao DS. MicroRNA function in myeloid biology. Blood. 2011;118:2960–2969. doi: 10.1182/blood-2011-03-291971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El Gazzar M, McCall CE. MicroRNAs regulatory networks in myeloid lineage development and differentiation: regulators of the regulators. Immunol Cell Biol. 2012;90:587–593. doi: 10.1038/icb.2011.74. [DOI] [PubMed] [Google Scholar]
- 36.Rapino F, Robles EF, Richter-Larrea JA, et al. C/EBPalpha induces highly efficient macrophage transdifferentiation of B lymphoma and leukemia cell lines and impairs their tumorigenicity. Cell Rep. 2013;3:1153–1163. doi: 10.1016/j.celrep.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar M, Lu Z, Takwi AA, et al. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene. 2011;30:843–853. doi: 10.1038/onc.2010.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le MT, Teh C, Shyh-Chang N, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun YM, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 2013;6:6. doi: 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ring KL, Tong LM, Balestra ME, et al. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11:100–109. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thier M, Worsdorfer P, Lakes YB, et al. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell. 2012;10:473–479. doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 44.Loh KM, Lim B. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell. 2011;8:363–369. doi: 10.1016/j.stem.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Mitchell RR, Szabo E, Benoit YD, et al. Activation of Neural Cell Fate Programs Toward Direct Conversion of Adult Human Fibroblasts into Tri-potent Neural Progenitors Using OCT-4. Stem Cells Dev. 2014 doi: 10.1089/scd.2014.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montserrat N, Nivet E, Sancho-Martinez I, et al. Reprogramming of human fibroblasts to pluripotency with lineage specifiers. Cell Stem Cell. 2013;13:341–350. doi: 10.1016/j.stem.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 47.Shu J, Wu C, Wu Y, et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–975. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.