Abstract
We report a gene discovery system for poplar trees based on gene and enhancer traps. Gene and enhancer trap vectors carrying the β-glucuronidase (GUS) reporter gene were inserted into the poplar genome via Agrobacterium tumefaciens transformation, where they reveal the expression pattern of genes at or near the insertion sites. Because GUS expression phenotypes are dominant and are scored in primary transformants, this system does not require rounds of sexual recombination, a typical barrier to developmental genetic studies in trees. Gene and enhancer trap lines defining genes expressed during primary and secondary vascular development were identified and characterized. Collectively, the vascular gene expression patterns revealed that approximately 40% of genes expressed in leaves were expressed exclusively in the veins, indicating that a large set of genes is required for vascular development and function. Also, significant overlap was found between the sets of genes responsible for development and function of secondary vascular tissues of stems and primary vascular tissues in other organs of the plant, likely reflecting the common evolutionary origin of these tissues. Chromosomal DNA flanking insertion sites was amplified by thermal asymmetric interlaced PCR and sequenced and used to identify insertion sites by reference to the nascent Populus trichocarpa genome sequence. Extension of the system was demonstrated through isolation of full-length cDNAs for five genes of interest, including a new class of vascular-expressed gene tagged by enhancer trap line cET-1-pop1-145. Poplar gene and enhancer traps provide a new resource that allows plant biologists to directly reference the poplar genome sequence and identify novel genes of interest in forest biology.
INTRODUCTION
Forest trees are of great environmental and economic importance and also display remarkable developmental traits. For example, long-term perennial growth produces both the largest organism (giant sequoia [Sequoia giganteum]) and the longest-lived organism (bristle cone pine [Pinus longaeva]). The genes and mechanisms regulating developmental traits in trees remain largely uncharacterized, however, because classical developmental genetic approaches are not suitable for forest trees. In addition to large size, forest trees have long generation times and suffer from inbreeding depression. Commonly employed strategies such as loss-of-function mutagenesis that require rounds of sexual recombination or self-pollination are thus impractical.
Advances in genetic technologies present new strategies for gene discovery and characterization in trees, and members of the genus Populus (including poplars, aspens, and cottonwoods) are model angiosperms for molecular genetic studies (Bradshaw et al., 2001). Poplars offer the advantages of clonal propagation and a highly efficient Agrobacterium tumefaciens-based transformation system. Transformation with recombinant DNA constructs can be used to approximate classical developmental genetic approaches because recombinant DNA constructs can produce dominant phenotypes that eliminate the need to perform close matings or successive rounds of sexual reproduction. For example, RNA interference produces a dominant phenotype that can be assayed in primary transformants and approximates loss-of-function alleles. Clonal propagation facilitates the replication of selected transformants without the complication of segregation.
The P. trichocarpa genome is currently being sequenced, necessitating new strategies for gene discovery and characterization that directly reference this resource. In Arabidopsis, powerful strategies for gene discovery relate the insertion site of a recombinant DNA construct to phenotypes in plants. In the simplest strategy, random insertion of T-DNA has been used to produce plants harboring individual gene knockouts (e.g. Krysan et al., 1999; Sessions et al., 2002). Each T-DNA insertion provides a molecular tag, facilitating sequencing of the genomic DNA flanking the insertion. The exact insertion site is then determined by reference to the genome sequence, obviating the need for genetic mapping and associated sexual reproduction. Insertions in individual genes are thus indexed with mutant plant phenotypes and together provide a dynamic resource serving the scientific community. For poplars, an attractive strategy is to devise forward genetic screens using more complex recombinant DNAs that create dominant phenotypes. For example, activation tagging involves the random insertion of a strong promoter element that can result in overexpression of genes near the insertion site, and resulting developmental phenotypes are dominant (Kardailsky et al., 1999). This approach has been successfully established in poplars, and activation tagging of a poplar gene encoding a gibberellin catabolism-associated protein (PtaGA2ox1) proves the feasibility of this approach (Busov et al., 2003). Also in poplar, a promoter trap T-DNA carrying a luciferase marker gene was used to identify genes expressed in wood-forming tissues dissected from growing stems, including a gene encoding a ribosomal protein (Johansson et al., 2003).
Gene and enhancer trapping are alternative methods for insertion-based gene discovery that both reference genome sequence data and produce a dominant expression phenotype (Springer, 2000). In general, gene trap vectors carry a reporter gene lacking a functional promoter, while enhancer trap vectors carry a minimal promoter element preceding a reporter gene. In each case, the reporter gene is expressed in a fashion that mimics the normal expression pattern of a gene at the insertion site, as has been shown for Arabidopsis gene and enhancer trap lines (e.g. Springer et al., 1995; Gu et al., 1998; Pruitt et al., 2000). The genomic region flanking the insertion site is amplified via PCR and sequenced, and alignment of the flanking sequence with the genome sequence allows immediate mapping of insertions (Sundaresan et al., 1995). Insertion sites in genes can thus be indexed with expression phenotypes. The same collection of gene or enhancer trap lines can be screened for genes expressed in specific cell or tissue types, expressed at precise times in development or in response to experimental treatments or protein product subcellular localization (Groover et al., 2003). Gene and enhancer traps expressed at precise points in development or in response to experimental treatments can be used as convenient molecular markers. In the case of species amenable to inbreeding, individuals homozygous for gene or enhancer trap insertions can also be assayed for mutant phenotypes in addition to expression phenotypes.
We report identification of genes expressed during primary and secondary vascular development using a system for gene and enhancer trapping in poplars. Using gene and enhancer trap marker lines, we describe the percentage of genes expressed in different vascular tissues of poplar and highlight the common occurrence of genes expressed in both primary and secondary vascular tissue or expressed in vascular tissues of more than one organ. Insertion sites were determined by reference to the poplar genome sequence, and full-length cDNAs were cloned for genes of interest.
RESULTS
Establishing a Collection of Poplar Gene and Enhancer Trap Lines
Our primary goal was to establish systems for insertion-based gene discovery for poplar trees. Towards that goal, a gene trap T-DNA vector was derived from modified Ds transposable element gene trap vector (“Materials and Methods”) used to create a large collection of Arabidopsis gene trap lines at Cold Spring Harbor Laboratory (Springer et al., 1995; Sundaresan et al., 1995; http://genetrap.cshl.org/). As shown in Figure 1A, the gene trap T-DNA vector carries the nptII gene at the 3′ end, which confers kanamycin resistance to transformed plants. The β-glucuronidase (GUS) reporter gene is positioned at the 5′ end of the T-DNA. GUS is preceded by splice acceptor sites, which in turn are preceded by a short plant intron and splice donor sites at the 5′ end of the T-DNA. The gene trap thus acts as both an intron and an exon trap. If inserted in an exon of an expressed gene, the splice donors at the 5′ end of the vector work in conjunction with the splice acceptors preceding GUS, resulting in splicing of vector intron and production of a GUS fusion transcript and protein. In the case of insertion into an intron, the endogenous splice donor works in conjunction with the vector splice acceptors to produce a GUS fusion transcript and protein. In both cases, GUS expression mirrors the normal expression pattern of the interrupted gene.
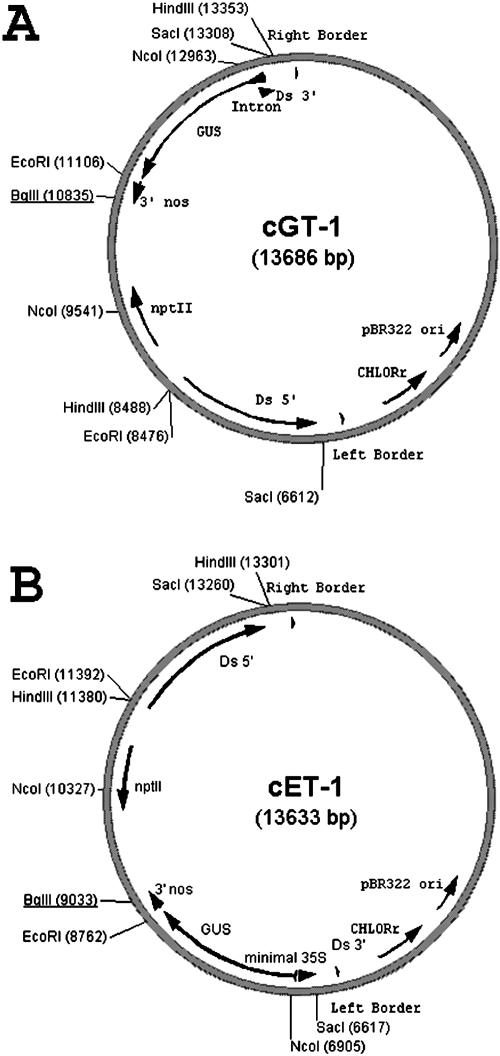
Figure 1.
Poplar gene and enhancer trap constructs. Constructs were assembled as described in “Materials and Methods”. For each construct, the left- and right-hand border sequences define the portion of T-DNA transferred into plant by A. tumefaciens transformation. A, The gene trap (cGT-1) contains the GUS reporter gene adjacent to the right border and is preceded by splice acceptor sites and a short plant intron. GUS expression in plants results from insertion into the transcribed portion of a chromosomal gene, when GUS is in the same orientation as the interrupted gene for expression. A fusion transcript is produced consisting of GUS and upstream exons of the interrupted gene, which results in translation of a corresponding GUS fusion protein. B, The enhancer trap (cET-1) contains the GUS reporter adjacent to the left border and is preceded by a minimal 35S promoter. The minimal promoter is not itself sufficient to drive GUS expression but is activated when inserted in close proximity to transcriptional enhancers (e.g. promoter) of an expressed chromosomal gene. For both constructs, bacterial selection is 100 μg/mL spectinomycin; plant selection is 50 μg/mL kanamycin.
An enhancer trap T-DNA vector (Fig. 1B) was similarly derived from an enhancer trap Ds element (“Materials and Methods”). In the enhancer trap, the GUS gene is preceded by a minimal promoter containing a ribosome entry site but by itself is not sufficient to drive GUS expression. If the enhancer trap inserts near an expressed gene, however, transcriptional enhancers can activate the minimal promoter to drive GUS expression. GUS expression reflects the expression of a gene at or near the insertion site, but no GUS fusion transcript is produced.
A total of 708 gene trap and 674 enhancer trap plant lines were established. The gene and enhancer trap vectors were introduced into poplar clone INRA 717-IB4 (P. tremula x P. alba) as T-DNA by A. tumefaciens-mediated transformation of leaf disc and stem explants (“Materials and Methods”). A single kanamycin-resistant shoot was selected from each explant culture to ensure that all gene and enhancer trap plants harbor a unique insertion event. Approximately 38% of explants yielded a gene or enhancer trap plant 6 cm in height within 5 months from the time of transformation. Southern-blot analysis indicated that gene and enhancer trap plants carried an average of 1.6 (se 0.13, n = 45) insertion events (“Materials and Methods”). Thus, the complication of multiple insertions is limited in this system.
Expression Patterns of Trapped Genes
GUS staining of gene and enhancer trap plantlets (“Materials and Methods”) revealed a variety of gene expression patterns, as shown in Figure 2 and summarized in Table I. Samples from stems, leaves, and shoot apices were tested for GUS histological staining under low stringency conditions (“Materials and Methods”). Eight percent of gene trap lines expressed GUS in some part of the plant. More than 68% of enhancer trap lines expressed GUS, but 44% of these lines showed only faint expression in leaf axils. As staining was not observed in untransformed controls, axil staining likely indicates the enhancer trap minimal promoter is predisposed to expression in the axil. As illustrated by cGT-1-pop1-708 in Figure 2A, a low percentage of lines have apparent ubiquitous GUS expression. More commonly, the expression pattern of tagged genes indicates discrete expression patterns, as revealed at high precision by GUS histological staining. For example, expression in the leaves of cGT-1-pop1-496 is restricted to the hydathodes (Fig. 2B). Expression patterns were found that defined anatomical features such as leaf traces (Fig. 2, C and D). Enhancer trap line cET-1-pop1-164 stains the adaxial side of petioles and represents an organ polarity marker. In stems, all of the cell and tissue types are amenable to GUS reporter analysis, including the interfascicular region (e.g. cET-1-pop1-103; Fig. 2F), the ground tissues (e.g. cET-1-pop1-333; Fig. 2G), the pith (e.g. cET-1-pop1-329; Fig. 2H), and vascular tissues (see below).
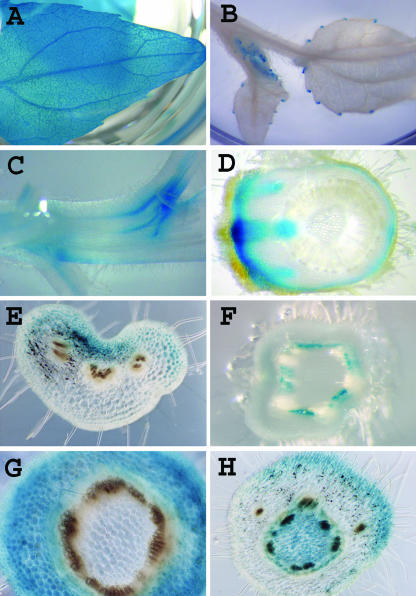
Figure 2.
Gene expression patterns revealed by GUS reporter histological staining resulting from gene or enhancer trap insertion. Most genes are expressed in a discrete subset of tissues or cell types, and expression can be assayed in a variety of organs including roots (not shown), leaves, and stems. Gene trap line cGT-1-pop1-708 (A) is an exception, and the tagged gene is expressed uniformly. Examples are shown of expression patterns that would not be readily revealed by other current gene expression technologies (e.g. microarrays). Gene trap line cGT-1-pop1-496 (B) is expressed in the hydathodes of the leaf. Lines cET-1-pop1-234 (whole mount staining of stem; [C]) and cET-1-pop1-13 (cross-section through node; [D]) are expressed in the vascular leaf traces. Line cET-1-pop1-164 (cross-section through petiole; [E]) includes expression on the adaxial side of the petiole. The different tissue and living cell types of the stem can be assayed for gene expression, including the interfascicular zone stained by cET-1-pop1-103 (F), the ground tissues in cET-1-pop1-333 (G), and the pith in cET-1-pop1-329 (H).
Table I.
GUS expression patterns for poplar gene and enhancer trap lines
| GUS Expression Assay | Enhancer Traps
|
Gene Traps
|
||
|---|---|---|---|---|
| No. Lines | Percent Lines | No. Lines | Percent Lines | |
| Assayed for GUS expression | 674 | 100 | 708 | 100 |
| Expressed GUS | 455 | 68a | 55 | 8b |
| Leaf assayed for GUS expression | 662 | 98a | 701 | 99b |
| Leaf assayed and GUS expressed | 239 | 35c | 42 | 6d |
| Leaf assayed and GUS vascular expressed | 99 | 15c | 17 | 2d |
| Stem assayed for GUS expression | 641 | 95a | 375 | 52b |
| Stem assayed and GUS expressed | 392 | 58c | 37 | 5d |
| Stem assayed and GUS vascular expressed | 33 | 5c | 8 | 1d |
| Stem assayed and GUS axil expressed | 201 | 30c | 10 | 1d |
Expressed as percentage of total enhancer trap lines.
Expressed as percentage of total gene trap lines.
Expressed as total of enhancer trap lines assayed in leaf or stem.
Expressed as total of gene trap lines assayed in leaf or stem.
A secondary goal of the work described here was to identify novel markers and genes expressed during vascular development and wood formation. We reasoned that, while some gene and enhancer trap lines showed clear vascular expression under the low stringency staining conditions used in the initial screen, additional more strongly expressed vascular genes might display poorly localized GUS staining patterns because of diffusion of the GUS reaction product. Gene or enhancer trap plants that had poorly localized, strong GUS staining were thus retested under high stringency staining to identify additional vascular vascular-expressed lines (“Materials and Methods”). As summarized in Table I, 41% of enhancer trap lines with leaf expression were expressed exclusively in veins. Similarly, 40% of gene trap lines with leaf expression were expressed exclusively in veins. This result indicates a large number of genes is required for vascular development and to support the functions of the multiple cell types found in vascular tissues. In the stem, 13% of enhancer traps and 22% of gene traps expressed in the stem were expressed exclusively in the xylem, phloem, or vascular cambium. The larger percentage of genes expressed in the veins of leaves versus stems is partly explained by the exclusive association of collenchyma tissue with the major veins of leaves, whereas collenchyma is found in the ground tissue of the stem (and would not have been scored as vascular in this study).
The relationship of the genetic mechanisms regulating the development and function of the vasculature in different parts of the plant is largely unknown. One possibility is that a unique set of genes and mechanisms regulates vascular development and functions in each organ. The gene expression patterns in stems versus leaves were thus compared to determine if most genes regulating vascular development and function are expressed only in one organ or in multiple organs. As summarized in Table II and illustrated in Figure 3, 30% of enhancer trap lines and 7% of gene trap lines assayed for staining in both leaves and stem showed staining in either leaf veins or stem. Of these lines, 38% of enhancer traps and 41% of gene traps were expressed in both leaf veins and stem. Similarly, screening of over 2,000 Arabidopsis gene trap lines revealed that, of 54 lines showing expression in leaf veins, 70% were also expressed in the vasculature of the root, hypocotyl, or inflorescence stem (A. Groover and R. Martienssen, unpublished data). These observations suggest significant overlap of the genes and mechanisms regulating development and function of the vasculature in different organs of the plant.
Table II.
GUS expression patterns in stem and leaf veins
| GUS Expression Assay | Enhancer Traps
|
Gene Traps
|
||
|---|---|---|---|---|
| Total Lines | Percent Lines | Total Lines | Percent Lines | |
| Assayed GUS expression in both stem and leaf | 639 | 94a | 375 | 53b |
| GUS expressed in leaf veins or stem | 190 | 30c | 27 | 7d |
| GUS expressed in both leaf veins and stem | 73 | 11c | 11 | 3d |
| GUS expressed in leaf veins but not in stem | 26 | 4c | 6 | 2d |
| GUS expressed in stem but not leaf veins | 91 | 14c | 10 | 3d |
Expressed as percent total enhancer trap lines.
Expressed as percentage of total gene trap lines.
Expressed as percentage of enhancer traps assayed in both leaf and stem.
Expressed as percentage of gene traps assayed in both leaf and stem.
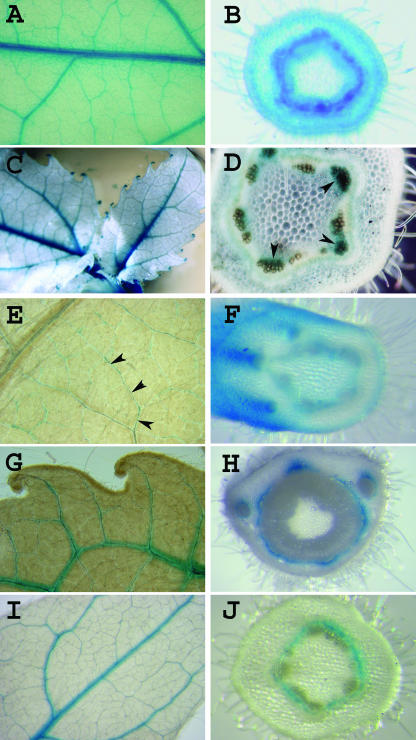
Figure 3.
Vascular expression patterns in leaves and stems of enhancer and gene trap plants. Expression in vascular tissues is often associated with expression in more than one organ. Shown are GUS stained whole mount leaves (A, C, E, G, and I) and stem cross-sections (B, D, F, H, and J) from lines cET-1-pop1-55 (A and B), cGT-1-pop1-18 (C and D), cET-1-pop1-18 (E and F), cET-1-pop1-465 (G and H), and cET-1-pop1-371 (I and J), all of which show vascular expression in both leaves and stems. Staining of vascular bundles in the stem of cGT-1-pop1-18 is indicated by arrowheads (D). Staining of minor veins in cET-1-pop1-18 is indicated for a single vein by arrowheads (E).
Genes Tagged by Gene and Enhancer Trap Insertions
The chromosomal region flanking individual gene or enhancer trap insertion sites was successfully amplified using thermal asymmetric interlaced (TAIL) PCR for 34 lines (“Materials and Methods”). TAIL PCR products were sequenced and used in nucleotide BLAST searches against the 3× draft poplar genome sequence (http://genome.jgi-psf.org/poplar0/poplar0.home.html) that was available at that time. As expected, polymorphisms exist between the TAIL sequences from the P. tremula x P. alba clone used for gene and enhancer trap transformation and the P. trichocarpa clone used for genome sequencing. However, as summarized in Table III , the TAIL products all had strong similarity to a sequenced P. trichocarpa genomic clone. In some cases, overlapping clones were available to make an extended contig of the insertion region (data not shown). Insertion sites were confirmed for all insertions in 12 lines tested by PCR amplification of genomic DNA from each line using one locus-specific PCR primer in conjunction with a primer annealing in the gene or enhancer trap vector (“Materials and Methods”).
Table III.
Insertion sites of gene and enhancer trap T-DNAs in the poplar genome, and similarity of tagged genes to annotated Arabidopsis genes
| Plant Line IDa | TAIL Primerb | GenBank Accession | JGI Poplar Clonec | JGI BLASTE Value |
|---|---|---|---|---|
| cET-1-pop1-009-1 | Ds3 | AY428447 | VAO90278.g2 | 2.00E−23 |
| cET-1-pop1-009-2 | Ds5 | AY428448 | VAO90278.g2 | 2.00E−65 |
| cET-1-pop1-018 | Ds5 | AY428449 | XXI762884.y1 | E−179 |
| cET-1-pop1-028 | Ds3 | AY428450 | XXI671413.g1 | 8.00E−14 |
| cET-1-pop1-031 | Ds3 | AY428451 | XXI699820.g1 | 3.00E−38 |
| cET-1-pop1-045 | Ds3 | AY428452 | XXI171777.b1 | 1.00E−08 |
| cET-1-pop1-049 | Ds5 | AY428453 | XXI191745.y1 | E−132 |
| cET-1-pop1-053 | Ds3 | AY428454 | XXI918518.b2 | 3.00E−54 |
| cET-1-pop1-064 | Ds3 | AY428455 | XXI772438.b1 | 0.00E00 |
| cET-1-pop1-066 | Ds3 | AY428456 | XXI351291.g1 | 2.00E−49 |
| cET-1-pop1-067 | Ds5 | AY428458 | VAO145265.b2 | 7.00E−56 |
| cET-1-pop1-076 | Ds3 | AY428459 | TRE152057.b2 | E−177 |
| cET-1-pop1-076 | Ds5 | AY428460 | XXI832726.b1 | 4.00E−48 |
| cET-1-pop1-103 | Ds5 | AY428461 | XXI914440.y1 | 2.00E−34 |
| cET-1-pop1-108 | Ds3 | AY428462 | TRE19677.y2 | 1.00E−59 |
| cET-1-pop1-145 | Ds5 | AY428463 | XXI682888.g1 | 0.00E00 |
| cET-1-pop1-160 | Ds3 | AY428464 | XXI144847.g1 | 3.00E−04 |
| cET-1-pop1-173 | Ds3 | AY428465 | XXI830746.g1 | 1.00E−10 |
| cET-1-pop1-174 | Ds3 | AY428466 | XXI580558.g1 | E−101 |
| cET-1-pop1-329 | Ds3 | AY428467 | XXI583770.b1 | 7.00E−65 |
| cET-1-pop1-371 | Ds3 | AY428468 | XXI272684.g1 | 0.00E00 |
| cET-1-pop1-409 | Ds3 | AY428470 | XXI569762.b1 | E−136 |
| cET-1-pop1-465 | Ds3 | AY428471 | XXI793081.x1 | E−109 |
| cET-1-pop1-506 | Ds3 | AY428472 | XXI716994.g1 | 0.00E00 |
| cET-1-pop1-540 | Ds3 | AY428473 | XXI362111.b1 | E−137 |
| cET-1-pop1-579 | Ds3 | AY428474 | XXI3267.x1 | E−178 |
| cET-1-pop1-765 | Ds3 | AY428475 | XXI577429.g1 | E−160 |
| cET-1-pop1-777 | Ds3 | AY428476 | XWN136790.x1 | E−123 |
| cGT-1-pop1-018 | Ds3 | AY428477 | XXI154086.b1 | E−106 |
| cGT-1-pop1-048 | Ds5 | AY428478 | XXI743977.b1 | 2.00E−25 |
| cGT-1-pop1-100 | Ds5 | AY428479 | XXI356514.b1 | 0.00E00 |
| cGT-1-pop1-151-1 | Ds5 | AY428480 | XXI441445.x1 | E−114 |
| cGT-1-pop1-253 | Ds3 | AY428481 | XXI742394.g1 | 1.00E−86 |
| cGT-1-pop1-342 | Ds3 | AY428482 | XXI682242.g1 | 2.00E−09 |
| cGT-1-pop1-500 | Ds3 | AY428483 | XWN13513.x1 | E−148 |
| cGT-1-pop1-541 | Ds3 | AY428484 | XXI849877.g1 | 5.00E−34 |
| TAIR BLASTX Cloned | TAIR BLAST E Value | TAIR Annotation | PCR Verified | Organ Staining Pattern |
|---|---|---|---|---|
| Not significant | ND | Leaf/Stem | ||
| Not significant | ND | Leaf/Stem | ||
| At4g35740.1 | 2.00E−17 | DNA helicase TPS1 | Yes | Leaf/Stem/Axil |
| At5g19330.1 | 4.00E−09 | Similar to VAC8 Protein-vacuolar transport. | ND | Hydathode/Axil |
| Not significant | ND | Leaf/Stem | ||
| Not significant | ND | Leaf/Stem | ||
| Not significant | ND | Leaf/Stem | ||
| Not significant | ND | Axil | ||
| Not significant | ND | Leaf/Stem | ||
| Not significant | ND | Leaf/Axil | ||
| Not significant | Yes | Leaf/Stem | ||
| At1g22540.1 | 4.00E−25 | Oligopeptide Transporter | Yes | Axil |
| Not significant | ND | Axil | ||
| Not significant | ND | Leaf/Stem | ||
| Not significant | ND | Leaf/Stem | ||
| At1g80940 | 2.00E−18 | Expressed protein: full-length cDNA: Ceres:37300 | Yes | Leaf/Stem |
| Not significant | Yes | Leaf/Axil | ||
| Not significant | ND | Axil | ||
| Not significant | ND | Leaf/Axil | ||
| Not significant | ND | Leaf/Stem | ||
| At1g45228.1 | E−109 | Similar to RING zinc finger protein | Yes | Leaf/Stem |
| At3g51950.1 | 5.00E−29 | Polyadenylate-Binding Protein | Yes | Leaf/Stem |
| At3g53680.1 | 8.00E−14 | PHD finger transcription factor | Yes | Leaf/Stem |
| At1g63850.1 | 3.00E−58 | Myb DNA Binding | Yes | Leaf/Stem |
| Not significant | ND | Leaf/Stem | ||
| Not significant | ND | Leaf/Stem | ||
| Not significant | ND | Leaf/Stem | ||
| At2g01024.1 | 6.00E−18 | Retroelement | ND | Leaf/Stem |
| Not significant | Yes | Leaf/Stem | ||
| Not significant | ND | Stem | ||
| At3g61880.1 | 2.00E−59 | Cytochrome p450 | Yes | Leaf |
| Not significant | Yes | Leaf/Stem | ||
| Not significant | ND | Leaf/Stem | ||
| Not significant | ND | Leaf | ||
| Not significant | ND | Stem | ||
| Not significant | ND | Leaf/Stem |
cET identifies enhancer trap lines; cGT indicates gene trap lines.
Indicates the orientation of sequencing relative to the Ds transposon borders internal to the T-DNA.
Result from BLASTN at the Joint Genome Institute (http://genome.jgi-psf.org/poplar0/poplar0.home.html).
From The Arabidopsis Information Resource.
ND, Not determined.
The P. trichocarpa genomic sequence reads were not assembled or annotated at the time of this analysis. To determine if gene and enhancer trap insertion sites interrupted protein coding sequence, poplar TAIL DNA sequences were used in BLASTX searches against the Arabidopsis protein database (http://www.arabidopsis.org). For 10 lines, translated TAIL flanking sequence contained significant similarity to one or more Arabidopsis proteins (Table III). The genes identified are diverse in both their expression patterns and protein product (Table III). This suggests that the gene and enhancer traps are not biased toward specific classes of genes or by expression levels.
Full-length cDNAs were cloned for five poplar genes of interest, tagged in cET-1-pop1-18 (encoding a DNA helicase; GenBank accession no. AY442255), cET-1-pop1-145 (encoding a novel protein; GenBank accession no. AY442256), cET-1-pop1-409 (contains a putative RNA recognition motif; GenBank accession no. AY442257), cET-1-pop1-465 (encoding a putative PHD finger transcription factor; GenBank accession no. AY442258), and cET-1-pop1-506 (encoding a putative Myb transcription factor; GenBank accession no. AY442259). For each gene, TAIL PCR sequence was aligned with the gene's putative Arabidopsis ortholog to establish the most likely gene structure and location of conserved exons. Primers were designed to anneal within exon sequence that would amplify the 5′ and 3′ ends of each poplar cDNA using 5′ and 3′ RACE (“Materials and Methods”). Full-length cDNAs were assembled from the sequences of the RACE products, and each contig confirmed using gene-specific primers that amplified each entire cDNA. The cDNA sequences are currently being used to evaluate gene function, by making recombinant DNA constructs to overexpress and down-regulate by RNA interference each gene (A. Groover, J. Fontana, and G. Dupper, unpublished data).
The gene tagged by cET-1-pop1-145 represents a previously undescribed gene class. The full-length poplar cDNA cloned using RACE (GenBank accession no. AY442256) includes 133 bp of 5′ untranslated region, an open reading frame of 666 bp, and 321 bp of 3′ untranslated region and polyadenylation. A BLASTP search against the protein database at GenBank identified a single, highly similar protein in Arabidopsis (accession no. AAF14682.1). Based on additional searches of the Arabidopsis protein database, the protein is apparently encoded by a unigene (only one such gene is present in Arabidopsis). Although similar proteins were not found in the GenBank protein database, a putative ortholog was found in rice (Oryza sativa; accession no. AK102581.1) by using the poplar protein to search translated GenBank DNA sequences using tBLASTn. As shown in Figure 4A, the Arabidopsis, poplar, and rice proteins share high similarity across their lengths. Although well conserved across species, there is no published description of these genes.
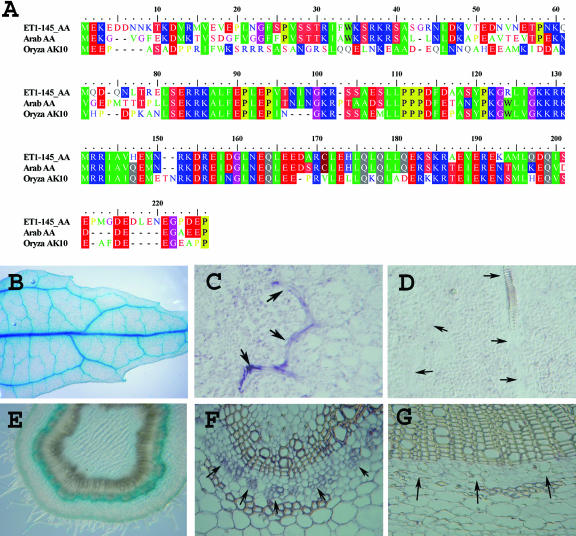
Figure 4.
Comparison of translated cDNA from the gene tagged by cET-pop1-145 to related genes in Arabidopsis and rice, and comparison of cET-pop1-145 GUS expression to expression of the endogenous gene revealed by in situ hybridization. A full-length cDNA was cloned for the gene tagged by cET-pop1-145 (“Materials and Methods”). A, The translated cDNA protein sequence shows high similarity to a single protein product in Arabidopsis and a translated cDNA from rice. B, The enhancer trap line cET-pop1-145 shows expression in the veins of leaves. C, In situ hybridization with an antisense probe recognizing the cET-pop1-145 tagged chromosomal gene labels the veins (arrows) in paradermal leaf sections. D, Hybridization with a control sense probe does not label the veins (position of veins indicated by arrows). E, Enhancer trap line cET-pop1-145 expression in the vasculature of a stem undergoing secondary growth. F, In situ hybridization with an antisense probe recognizing the cET-pop1-145 tagged chromosomal gene labels the phloem (arrows) in cross-section of a stem. G, Hybridization with a control sense probe does not label the stem (arrows indicate corresponding area that was labeled in F).
In situ hybridization was used to confirm that the cET-1-pop1-145 GUS reporter expression pattern faithfully reported the tagged gene's expression. The cET-1-pop1-145 plants express the GUS reporter in leaf veins (Fig. 4B). As shown in Figure 4C, paradermal sections of leaves hybridized with an antisense probe from the tagged gene label an identical pattern of expression in the veins. Control sense probes do not label the leaf veins (Fig. 4D). Similarly, cET-1-pop1-145 plants express the GUS reporter in the vasculature of stems. As shown in Figure 4F, hybridization of stem sections with an antisense probe from the tagged gene labels the phloem, while control sense probes do not result in labeling (Fig. 4G). Thus, the enhancer trap expression faithfully reflects the tagged gene expression pattern in this line, as has been shown for Arabidopsis gene and enhancer traps (e.g. Springer et al., 1995; Gu et al., 1998; Pruitt et al., 2000).
DISCUSSION
Genomic sequence from P. trichocarpa is enabling new approaches for the study of forest tree biology. Developing forward genetic screens that effectively reference the genome sequence is prerequisite to making major gains in forest tree biology similar to those realized for model annual plants. We describe such a screen here, based on dominant expression phenotypes revealed by gene and enhancer traps. The system relies on an efficient transformation system paired with random insertion of recombinant DNA vectors that produce dominant expression phenotypes when inserted into genes. The molecular tagging of the insertion site by the vector facilitates immediate mapping of the insertion through homology to the genomic sequence. The general strategy of producing dominant phenotypes with recombinant DNAs circumvents the need for repeated rounds of sexual reproduction, which has traditionally limited developmental genetic approaches in trees. In comparison to loss-of-function mutagenesis or activation tagging, gene trapping targets a larger class of genes. Loss-of-function or overexpression alleles do not reveal obvious phenotypes for many genes (Bouche and Bouchez, 2001), whereas most expressed genes can be detected through gene or enhancer trap expression phenotypes. This point is illustrated by our identification of a new class of vascular expressed gene, tagged by enhancer trap line cET-1-pop1-145.
The anatomy and biochemistry of wood formation has been well described for various tree species, but the genes and molecular mechanism controlling the underlying developmental processes remain largely unknown. Vascular development is difficult to address using classical mutation genetics because phenotypes resulting from disruption of genes regulating vascular development are expected to result in highly pleiotropic or lethal phenotypes (Nelson and Dengler, 1997). Gene and enhancer traps are well suited for discovery of vascular regulatory genes, as they report gene expression in heterozygous individuals and do not rely on production of an informative mutant phenotype.
Dividing stem cells in the vascular cambium support secondary growth, including wood formation, and results in radial thickening of stems. Important genes regulating secondary growth and wood formation have been tacitly assumed to be expressed exclusively in wood-forming tissues. Because of the ease with which expression can be assayed, the poplar gene and enhancer trap lines described herein allowed precise survey of vascular gene expression throughout the plant. Our results indicate that many genes expressed during secondary growth and wood formation are also expressed in vascular tissues in other parts of the plant, suggesting that differential or subtractive screens that exclude genes expressed in leaves or other organs discard genes that could play a fundamental role in wood formation. Indeed, wood formation has many features shared with the development of primary vascular tissues (e.g. vascular tissues in elongating stems and leaf venation), including the differentiation of the same cell types and patterning of tissues. Although the exact relationship is unknown, provascular cells in primary plant tissues and cells within apical meristems share many characters with vascular cambia, including the ability to divide in an undifferentiated state. A reasonable expectation is that, during the evolution of vascular plants, wood formation arose as a variation of primary vascular development. That secondary growth has arisen independently multiple times during evolution raises the expectation that key aspects of wood formation are regulated by genes and mechanisms regulating primary vascular development.
The percentage of poplar gene trap insertion lines with an expression phenotype reflects gene density because the gene trap must insert into the transcribed portion of a gene in the same reading orientation frame as the native gene in order for expression to occur. For poplar, the genome size has been estimated at 450 to 550 megabases, approximately 5 times the size of the Arabidopsis genome. We observed 8% of gene trap lines with an expression phenotype in poplar versus 23% in Arabidopsis (Groover et al., 2003). This is roughly consistent with the estimated genome size for poplar, assuming similar gene density and a slightly higher average number of gene trap insertions for poplar versus Arabidopsis. In contrast to gene traps, enhancer traps are less precise gene-finding tools, carrying a minimal promoter that can be activated by genes near the insertion site, and do not require insertion into the transcribed portion of a gene in order to be expressed. A greater percentage of enhancer trap insertions result in expression patterns and are thus a richer source of marker lines.
In practical terms, the percentage of poplar gene and enhancer trap lines with an expression phenotype is high enough that, in conjunction with the efficient transformation system, gene trapping is a reasonable approach for gene discovery in poplars. This is in contrast to many other tree species, most notably conifers, which have very large genomes, presumed lower gene density, and lower efficiency of transformation. To make insertion-based gene discovery feasible for such species, recombinant DNA vectors could be designed that produce selection-based phenotypes (e.g. antibiotic resistance) when inserted into a native gene.
MATERIALS AND METHODS
Plant Cultivation and Transformation
Hybrid aspen clone INRA 717-IB4 (Populus tremula x P. alba) was propagated and transformed using the protocol of Han et al. (2000). Briefly, leaf disc and stem segment explants were cocultivated with Agrobacterium tumefaciens strain GV3101 containing either the enhancer or gene trap T-DNA vector prior to induction of callus growth. After induction of shoot growth, one or more shoots from each explant were tested for ability to root in a medium containing kanamycin. A single kanamycin-resistant shoot was selected from each explant to ensure that every line established was derived from a unique insertion event.
Gene and Enhancer Trap Constructs
Enhancer and gene trap T-DNA vectors were derived from vectors originally developed at Cold Spring Harbor (Springer et al., 1995; Sundaresan et al., 1995). The binary vector pCambia 1200 (accession no. AF234292.1) was modified by digestion with XhoI and EcoRI to remove the 1.1-kb T-DNA region conferring hygromycin resistance. The ends of the vector were blunted with Klenow and religated to create p1200<35S::HYG>. The SacI fragment of enhancer trap vector pWS31 (accession no. AF433042.1) was cloned into the SacI site of p1200<35S::HYG> to create cET-1 (Fig. 1A). The SacI fragment of gene trap vector pWS32 (accession no. AF433043.1) was cloned into the SacI site of p1200<35S::HYG> to create cGT-1 (Fig. 1B).
DNA and RNA Manipulations
Poplar genomic DNA was isolated from shoot apices using the DNeasy Plant Miniprep (Qiagen, Valencia, CA), and RNA was isolated using the RNeasy Plant Miniprep (Qiagen) using the manufacturer's protocol. Southern blotting, digoxigenin (DIG) probe labeling, hybridization, and detection were performed with a DIG High Prime DNA Labeling and Detection II kit (Roche, Basel) using the manufacturer's protocol. TAIL PCR was performed as described previously (http://genetrap.cshl.org) using primers Ds3-1 (5′-ACC CGA CCG GAT CGT ATC GGT-3′), Ds3-2 (5′-CGA TTA CCG TAT TTA TCC CGT TC-3′), Ds3-4 (5′-CCG TCC CGC AAG TTA AAT ATG-3′), Ds5-1 (5′-ACG GTC GGG AAA CTA GCT CTA C-3′), Ds5-2 (5′-CCG TTT TGT ATA TCC CGT TTC CGT-3′), Ds5-4 (5′-TAC GAT AAC GGT CGG TAC GG-3′), and AD1 (5′-NTC GAS TWT SGW GTT-3′). The cDNA synthesis and RACE were performed using a SMART RACE cDNA amplification kit (CLONTECH, Palo Alto, CA). Sequences were assembled using Bioedit (Hall, 1999).
In Situ Hybridization
Tissue was fixed in 10% formalin, 5% acetic acid, and 50% ethanol, embedded in Paraplast Xtra (Fisher, Loughborough, Leicestershire, UK), sectioned to 7 μm, and probed with DIG-labeled RNA probes using the Barton lab protocol (http://carnegiedpb.stanford.edu/research/barton/in_situ_protocol.html). The gene tagged by ET1-pop1-145 was PCR-amplified using forward and reverse primers (5′-CCA AGG ATG TGA GGA TGG TTG AAG TTG A-3′ and 5′-TCC CCA ACA AAC CTA CAA AA-3′) and cloned into a TOPO TA vector (Invitrogen, Carlsbad, CA) for in vitro transcription.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes. We are currently establishing additional poplar gene and enhancer trap plants that will be available to screen upon request. Screening results are available on CD-ROM as an Access database.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY442255–AY442259, AAF14682.1, AK102581.1, AF234292.1, AF433042.1, and AF433043.1.
Acknowledgments
We thank Annie Delfino-Mix for greenhouse cultivation of poplar trees and Bethany Heynen for assistance in tissue culture and greenhouse propagation of poplar trees.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.034330.
References
- Bouche N, Bouchez D (2001) Arabidopsis gene knockout: phenotypes wanted. Curr Opin Plant Biol 4: 111–117 [DOI] [PubMed] [Google Scholar]
- Bradshaw HD, Ceulemans R, Davis J, Stettler R (2001) Emerging model systems in plant biology: poplar (Populus) as a model forest tree. J Plant Growth Regul 19: 306–313 [Google Scholar]
- Busov V, Meilan R, Pearce D, Ma C, Rood S, Strauss S (2003) Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from poplar that regulates tree stature. Plant Physiol 132: 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groover A, Fontana J, Arroyo J, Yordan C, McCombie W, Martienssen R (2003) Secretion trap tagging of secreted and membrane-spanning proteins using Arabidopsis gene traps. Plant Physiol 132: 698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q, Ferrandiz C, Yanofsky M, Martienssen R (1998) The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125: 1509–1517 [DOI] [PubMed] [Google Scholar]
- Hall T (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95–98. [Google Scholar]
- Han KH, Meilan R, Ma C, Strauss SH (2000) An Agrobacterium tumefaciens transformation protocol effective on a variety of cottonwood hybrids (genus Populus). Plant Cell Rep 19: 315–320 [DOI] [PubMed] [Google Scholar]
- Johansson AM, Wang C, Stenberg A, Hertzberg M, Little C, Olsson A (2003) Characterization of a PttRPS18 promoter active in the vascular cambium region of hybrid aspen. Plant Mol Biol 52: 317–329 [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N, Dengler N (1997) Leaf vascular pattern formation. Plant Cell 9: 1121–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt R, Vielle-Calzada J, Ploense S, Grossniklaus U, Lolle S (2000) FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc Natl Acad Sci USA 97: 1311–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer P (2000) Gene traps: Tools for plant development and genomics. Plant Cell 12: 1007–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer P, McCombie W, Sundaresan V, Martienssen R (1995) Gene trap tagging of Prolifera, an essential MCM2-3-5 like gene in Arabidopsis. Science 268: 877–880 [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones J, Dean C, Ma H, Martienssen R (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]