Abstract
HLA class I molecules and killer cell immunoglobulin-like receptors (KIR) form a diverse system of ligands and receptors that individualize human immune systems in ways that improve the survival of individuals and populations. Human settlement of Oceania by island-hopping East and Southeast Asian migrants started ~3,500 years ago. Subsequently, New Zealand was reached ~750 years ago by ancestral Māori. To examine how this history impacted KIR and HLA diversity, and their functional interaction, we defined at high resolution the allelic and haplotype diversity of the 13 expressed KIR genes in 49 Māori and 34 Polynesians. Eighty KIR variants, including four ‘new’ alleles, were defined; as were 35 centromeric and 22 telomeric KIR region haplotypes, which combine to give >50 full-length KIR haplotypes. Two new and divergent variant KIR form part of a telomeric KIR haplotype, which appears derived from Papua New Guinea and was probably obtained by the Asian migrants en route to Polynesia. Māori and Polynesian KIR are very similar, but differ significantly from African, European, Japanese and Amerindian KIR. Māori and Polynesians have high KIR haplotype diversity with corresponding allotype diversity being maintained throughout the KIR locus. Within the population each individual has a unique combination of HLA class I and KIR. Characterizing Māori and Polynesians is a paucity of HLA-B allotypes recognized by KIR. Compensating for this deficiency are high frequencies (>50%) of HLA-A allotypes recognized by KIR. These HLA-A allotypes are ones that modern humans likely acquired from archaic humans at a much earlier time.
Keywords: KIR, NK cells, HLA class I, immune diversity, Māori, Polynesian
Introduction
Natural killer (NK) cells are essential components of the human immune system that provide defense against infection and cancer (Cooper et al. 2009; Lanier 2005). They also contribute to placentation and the success of reproduction (Parham and Moffett 2013). Education of NK cells and activation of their effector functions are controlled by the integration of signals generated through a variety of cell-surface receptors (Long et al. 2013; Narni-Mancinelli et al. 2013). Several of these NK cell receptors are specific for HLA class I ligands (Moretta et al. 2014). Of particular interest here is the diverse family of killer-cell immunoglobulin-like receptors (KIR) that recognize four mutually exclusive epitopes of the highly polymorphic HLA-A, -B and -C molecules. These epitopes comprise the A3/11 epitope carried by HLA-A*03 and HLA-A*11 allotypes, the Bw4 epitope carried by around one third of HLA-A and -B allotypes, the C2 epitope carried by the subset of HLA-C allotypes having lysine at position 80, and the C1 epitope carried by the complementary subset of HLA-C allotypes that have asparagine at position 80 (Dohring et al. 1996; Gumperz et al. 1995; Winter and Long 1997). In addition, the unusual and geographically restricted HLA-B*46 and -B*73 allotypes also carry the C1 epitope (Moesta et al. 2008).
The KIR are encoded by a close-packed gene family that is part of the leukocyte receptor complex (LRC) on chromosome 19 (Wilson et al. 2000). There are two distinctive components to the genetic diversity of the KIR locus. One is variability in KIR gene content, the other is allelic polymorphism of the KIR genes. These two dimensions of variability synergize to diversify KIR haplotypes (Shilling et al. 2002), which although numerous divide into two broad and functionally distinctive groups called KIR A and B (Uhrberg et al. 1997). More than 200 studies of KIR gene content in various human populations and clinical cohorts (Bashirova et al. 2006; Hollenbach et al. 2012; Norman et al. 2001) have associated combinatorial diversity of KIR and HLA with a range of diseases, including infections, autoimmune disorders, and pregnancy syndromes, as well as the outcome of transplantation (Hiby et al. 2004; Khakoo and Carrington 2006; Leung et al. 2004; Martin and Carrington 2013; Venstrom et al. 2012; Zambello et al. 2014).
Whereas many studies describe KIR gene content diversity in anthropologically well-defined human populations (Hollenbach et al. 2012; Norman et al. 2001; Parham 2005), relatively few reports describe their allelic diversity. These studies examined Japanese (Yawata et al. 2006), Yucpa South Amerindians (Gendzekhadze et al. 2009), Ga-Adangbe West Africans (Norman et al. 2013) and Europeans (Middleton et al. 2007; Vierra-Green et al. 2012). Nonetheless, these studies demonstrate that the allelic and haplotypic diversity of the KIR genes varies significantly between human populations. Importantly, they are complemented by functional studies showing how allotypic KIR variation can modulate the interaction of KIR with HLA class I and thereby influence NK cell function (Ahlenstiel et al. 2008; Bari et al. 2009; Frazier et al. 2013; Hilton et al. 2012; O’Connor et al. 2014; Thananchai et al. 2007). To extend high-resolution analysis of the combinatorial diversity of KIR and HLA alleles to the distinctive populations of the Remote Oceanic region of the South Pacific, we report here on Māori from New Zealand and Polynesians from Samoa, Tokelau and the Cook Islands.
Colonized approximately 750 years ago by Māori ancestors, New Zealand (Aotearoa) was one of the last places on earth to be populated by humans (Wilmshurst et al. 2011). Remote Oceania was settled quickly (Friedlaender et al. 2008), through successive migration and expansion to colonize each newly discovered island (Bellwood et al. 2011). Along the migration route from Northwest to South and East Oceania is a gradient of genetic diversity with Māori having the lowest diversity (Chambers 2013; Kayser et al. 2006; Whyte et al. 2005). The migrants from Eastern Asia to the South Pacific experience some limited genetic exchange with people of Near Oceania (e.g. Papua New Guinea), which had then been populated for ~40,000 years (Chambers 2013; Friedlaender et al. 2008). In the ancestry of Remote Oceanians there was also sex-biased gene flow, involving females that originated in Taiwan or Near Oceania and males from Island Southeast Asia (Duggan et al. 2014; Kayser et al. 2006; Soares et al. 2011; Wollstein et al. 2010). During the 3,500 years since the settlement of Remote Oceania began (Burley et al. 2012; Hurles et al. 2003) gene flow between Remote Oceanic population groups preserved their close relationships relative to other population groups (Edinur et al. 2013; Friedlaender et al. 2008; Wollstein et al. 2010). But adding further complexity, were genetic influences from Western Europeans that began in the nineteenth century (Underhill et al. 2001).
The HLA allele frequencies of Māori and Polynesian populations reflect this ancestry, with common alleles and haplotypes being also common in the populations of Taiwan, Island Southeast Asia or Near Oceania (Gonzalez-Galarza et al. 2011; Kostyu et al. 1984; Tracey and Carter 2006). Rare alleles and haplotypes tend to stem from modern or historical admixture, such as those shared by Europeans and New Zealand Māori or by Amerindians and Easter Islanders (Edinur et al. 2012; Roberts et al. 2013; Thorsby 2012). A previous study (Velickovic et al. 2006) analyzed the gene content of the KIR locus in four Polynesian populations: Cook Islands, Samoa, Tokelau and Tonga. These populations exhibited similar KIR-gene frequencies and extent of gene-content variation. Individual populations had 19–29 different KIR genotypes, with a total of 46 KIR genotypes being observed. The goal of our study was to determine KIR and HLA class I allele variation in Māori and Polynesian populations, how it compares to that in African, European, Asian and Amerindian populations, and its potential for diversifying the functional interactions of KIR with HLA class I.
Materials and Methods
Study populations
We studied the KIR of 49 Māori (from New Zealand) and 34 Polynesians (from either Cook Islands, Samoa or Tokelau), cohorts that were previously analyzed for HLA class I (Edinur et al. 2012). All these individuals were unrelated. They self-identified as having four Māori or four Polynesian grandparents and had no known admixture with any other ethnic group in their family history. These indicators are likely to accurately reflect their ancestry, because these people have a strong respect for genealogical heritage (known as whakapapa by the Māori). The individuals studied were all volunteer blood donors at the Wellington Blood Transfusion Service and were resident in Wellington at the time of blood donation. Genomic DNA samples made from peripheral blood cells were drawn from the Victoria University of Wellington DNA Bank. Ethical approval for this study was obtained from the New Zealand Central Region Ethics Committee, the Victoria University of Wellington Human Ethics Committee, and the Stanford University Administrative Panels on Laboratory Care and Human Subjects in Medical Research.
PCR amplifications
Individual exons of the KIR genes were amplified by PCR and subject to Sanger sequencing, followed by cloning and/or pyrosequencing. The KIR genes were analyzed in nine groups; KIR3DL3, 2DL23/S2, 2DL5, 2DS3/5, 2DL1/S1, 2DL4, 3DL1/S1, 2DS4 and 3DL2, as described previously (Norman et al. 2013). Each of 39 reactions was performed separately in a 12μl mixture containing PCR buffer (Invitrogen, Carlsbad, CA), 2.5mM MgCl2, 0.2mM mix of dNTPs (Sigma-Aldrich, St. Louis, MO), 1μM of each respective oligonucleotide primer (as described in (Norman et al. 2013)), 1U of recombinant Taq DNA polymerase (Invitrogen Carlsbad CA) and 50–200ng of DNA template. Amplification was performed using a Veriti 96 well thermocycler (Applied Biosystems, Foster City, CA) set to 9600 emulation mode, with a 3 minute initial heat activation step at 94°C, 10 cycles consisting of denaturing at 94°C for 10 seconds, annealing at 65°C for 60 seconds, and another 20 cycles consisting of a denaturing step of 94°C for 10 seconds, annealing at 61°C for 50 seconds and elongation at 72°C for 30 seconds. The PCR products obtained from this step were used as a template for a nested PCR. The reaction mixture consisted of PCR Buffer containing 15mM MgCl2 (Qiagen, Valencia, CA), 0.2mM dNTPs (Sigma-Aldrich, St. Louis, MO), 1μM of each oligonucleotide primers as described (Norman et al. 2013)), 1U of HotStarTaq Plus DNA Polymerase (Qiagen, Valencia, CA). Amplification was performed using the same conditions as the first round of PCR, but with 30 cycles in the final step (45 total). The PCR products from this step were then genotyped by pyrosequencing.
Sample preparation for pyrosequencing
A 5μl aliquot of biotinylated PCR product was incubated with 4μl Sepharose beads (Amersham Biosciences, Piscataway, NJ) in a 100μl final volume at room temperature with agitation at 1100rpm for 15 minutes. To remove unattached PCR products, the beads were washed with 70% ethanol, DNA denatured with 0.2M NaOH, then washed again with 10mM Tris-Acetate (pH 7.6) and suspended in 12μl of annealing buffer (20mM Tris-Acetate (pH 7.6), 2mM Mg-Acetate) containing 30μM sequencing primers. To anneal the primer and template, the mixture was incubated at 75°C for 2 minutes followed by slow (10–15 minutes) cooling to room temperature. Pyrosequencing was then performed using PSQ 96 Gold reagents, and a PSQ HS 96A instrument (Qiagen, Valencia, CA).
Sanger sequencing
Standard DNA sequencing reactions were performed in forward and reverse directions using BigDye Terminator v3.1 and analyzed using an ABI-3730 sequencer (ABI, Foster City CA). When required, PCR products were cloned using Topo-pcr2.1 vector (Invitrogen, Carlsbad, CA) and sequenced using M13 and internal primers. Four newly-discovered KIR alleles were validated as recommended by the curators of the Immuno Polymorphism Database (IPD) (Robinson et al. 2013). For each individual in which a ‘new allele’ was detected, five or more clones for the allele were sequenced. The allele sequences were submitted to the IPD database (http://www.ebi.ac.uk/ipd/kir/). Their names and accession numbers (in parentheses) are KIR3DL1*080 (KF941346), 3DL1*086 (KM026529), 2DL4*028 (KF941350), and 2DL3*00110 (KJ013515).
KIR and HLA class I genotyping
Confirmation of KIR gene presence/absence was performed using polymerase chain reaction-sequence specific oligonucleotides (PCR-SSO) and Luminex technology (LABType SSO KIR, One Lambda, Canoga Park, CA). The HLA class I genotypes were as described previously for all the Māori and 26 of the Polynesians (Edinur et al. 2013). The other eight of the 34 Polynesians, were genotyped for HLA class I using the LABType SSO method.
Population genetics
Allele frequencies were determined by direct counting. Their fit to Hardy-Weinberg equilibrium proportions was examined, and established, using an exact test. Expected heterozygosity was calculated from the sum of the square of the frequencies (He = 1- SSF). Differences in the allele-frequency distributions between populations were compared using 2 x n contingency tables and a chi-square test, with only common alleles (>5%) being examined in order to eliminate any bias due to rare alleles. Centromeric and telomeric allele-level KIR haplotypes were determined for each individual based on patterns of commonly-segregating alleles at adjacent KIR genes. Full-length allelic level KIR haplotype frequencies were estimated using the Expectation-Maximization algorithm of Haplo Stats implemented in the R programming language (R Development Core Team 2008). Gene-content haplotypes were compiled from these allele-level haplotypes. Individuals having duplicated KIR genes were identified from pyrosequencing of component alleles and their gene copy number confirmed using real-time PCR (Jiang et al. 2012). In such cases the additional copy of the duplicated gene was not included in the allele frequency calculations. Individuals appearing homozygous for combinations of KIR2DL4 and 3DL1/S1 alleles were also tested for gene copy number using real-time PCR to determine if they carried a deletion KIR haplotype that lacks the KIR2DL4 and KIR3DL1/S1 genes. Comparisons were made between Māori, Polynesians and the other populations for which KIR alleles and haplotypes have been determined at high-resolution: Yucpa Amerindians from Venezuela: (Gendzekhadze et al. 2009), Ga-Adangbe sub-Saharan Africans from Ghana: (Norman et al. 2013), Europeans from Northern Ireland (Middleton et al. 2007) and the USA (Vierra-Green et al. 2012), and Japanese (Yawata et al. 2006). European HLA frequencies are from the “USA (Eur)” population (N=564) of (Meyer et al. 2007).
Pairwise mismatch analysis
For separate centromeric or telomeric KIR haplotypes, the sequences of the composite alleles were concatenated and mismatch distributions were calculated with p-dist, using Mega 6 (Tamura et al. 2011) set to pairwise deletion. The haplotypes having two copies of KIR2DL4 and KIR3DL1, observed in one Māori and one Polynesian, were not included in the analysis.
Results
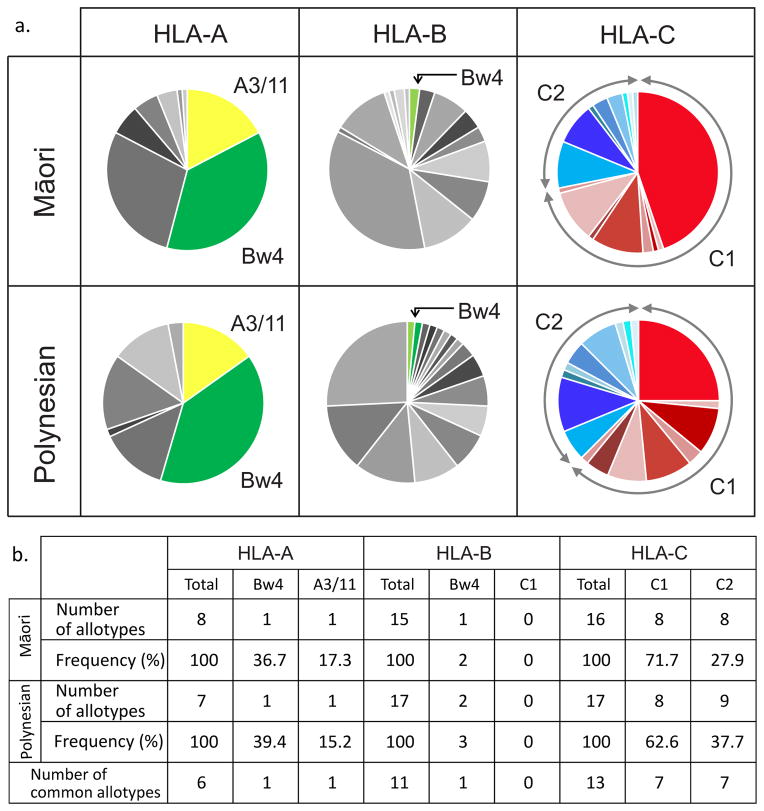
High-resolution allele-level HLA-A, -B and -C typing of cohorts of unrelated New Zealand Māori and Polynesian individuals shows that all three loci exhibit substantial polymorphism and that the spectrum of alleles in the two populations is similar ((Edinur et al. 2013) and Fig. 1a). Of the 50 HLA-A, B, C alleles detected, 30 are held in common and none of the alleles present in only one of the populations has a frequency greater than 5% (Supplementary Fig. 1). In contrast, shared, high-frequency alleles in the Māori are A*02:06 (29%), A*11:01 (17%), A*24:02 (37%), B*55:02 (36%) and C*01:02 (45%), and these are also common alleles in the Polynesians. HLA class I allotypes bearing the four, mutually exclusive, epitopes recognised by KIR are present in both populations. Whereas all HLA-C allotypes carry either the C1 or the C2 epitope, and 54% of the HLA-A allotypes carry either the Bw4 or the A3/11 epitope, only 2–3% of the HLA-B allotypes carry an epitope recognized by KIR (Fig. 1b). In effect, HLA-B in the Māori and Polynesian populations is rarely functioning as a ligand for KIR and thus appears dedicated to presenting peptide antigens to the T-cell receptors of CD8 T cells. With this knowledge of HLA class I in hand, we determined the coding region sequences for the alleles for all 13 functional KIR genes in the Māori and Polynesian cohorts.
Figure 1. Māori and Polynesian KIR ligands are provided by HLA-A and HLA-C but not by HLA-B.
a. The allele-frequency spectra of HLA class I. Each segment of the pie corresponds to a distinct allotype, which are matched in shade between the two populations: (yellow) – allotype has A3/11 epitope, (green) – Bw4 epitope, (red) – C1 epitope, (blue) – C2 epitope, (gray) indicates the allotype does not carry a KIR ligand. The allele names and frequencies are given in Supplementary Figure 1 and (Edinur et al. 2013).
b. The number of HLA-A, -B or -C allotypes present within each category of KIR ligand, and their frequencies in the Māori and Polynesian populations.
Māori and Polynesians have eighty familiar KIR alleles and four ‘new’ KIR alleles
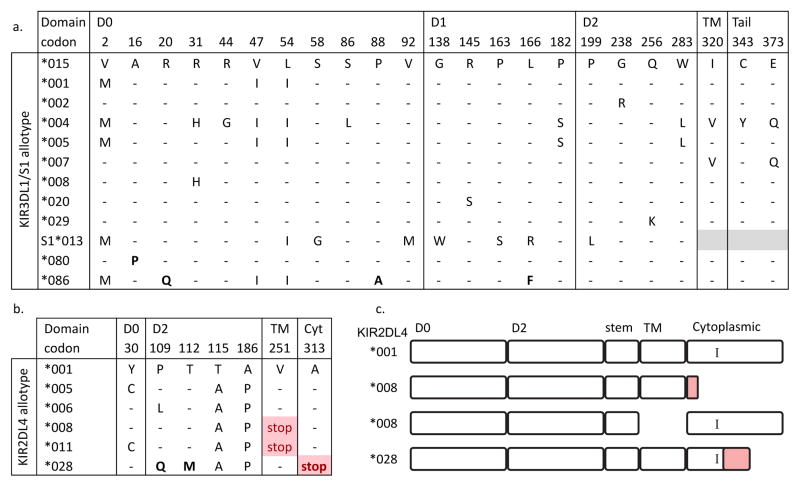
A total of 84 KIR alleles encoding 66 allotypes were defined in the Māori and Polynesian populations (Supplementary Fig. 2). Of these, 71 alleles encoding 49 allotypes were present in both populations. Among the 84 Māori and Polynesian KIR alleles, four alleles (KIR2DL3*00110, 2DL4*028, 3DL1*080 and 3DL1*086) were previously unknown and appear to be specific to these Remote Oceanic populations. Two of the new alleles, KIR2DL3*00110 and KIRDL1*080, differ from other Māori and Polynesian alleles by single nucleotide substitutions, whereas KIR3DL1*086 and KIRDL4*028 differ by multiple dispersed substitutions. KIR2DL3*00110 differs from KIR2DL3*00101 by a synonymous substitution and is present in Māori and Polynesians. KIR3DL1*080 is unique to the Māori and differs from 3DL1*015 by a non-synonymous substitution that replaces alanine at position 16 in the D0 domain with proline (Fig. 2a). This substitution has potential to affect functional recognition of the Bw4 epitope.
Figure 2. Defining characteristics of the newly-discovered KIR3DL1 and KIR2DL4 allotypes.
a. The amino acid differences that distinguish the KIR3DL1/S1 allotypes found in Māori and Polynesians. 3DL1*015 is used as a reference and the two newly-discovered allotypes (3DL1*080 and *086) are shown at the bottom with their unique substitutions in bold. 3DL1*086 also has a synonymous (g -> a) substitution at nucleotide 1288.
b. The amino acid differences that distinguish the KIR2DL4 allotypes present in the Māori and Polynesians. Only those residues that differ from the reference allotype (2DL4*001) are shown. The newly-discovered substitutions are shown in bold. Red indicates the stop codons introduced by preceding frame-shift mutations.
c. Shows the proteins that are predicted to be made from the 2DL4*028 allele compared to 2DL4*001 (full length) and 2DL4*008, of which the latter may form a truncated (upper) or secreted protein (lower) (Goodridge et al. 2007). Red indicates the altered polypeptide sequence due to the frame-shift. I - shows the position of the ITIM motif.
New alleles KIR3DL1*086 and KIR2DL4*028 are present both in Māori and Polynesians, where they are present in the same subsets of individuals. This distribution suggests KIR3DL1*086 and KIR2DL4*028 segregate on the same KIR haplotypes. KIR3DL1*086 differs from other KIR3DL1 alleles by four nucleotide substitutions. One substitution, in exon 9, is synonymous, where the other three substitutions are non-synonymous and change the residues at positions 20 and 88 in the D0 domain and position 166 in the D1 domain (Fig. 2a). Of the three amino-acid substitutions, alanine 88 and phenylalanine 166 are present in none of the 81 KIR3DL1/S1 allotypes previously defined, and glutamine 20 is present only in KIR3DL1*027, which was detected in just one African individual from Zimbabwe (Norman et al. 2007). Residues 20 and 166 of KIR3DL1/S1 have been subject to positive diversifying selection in the hominid lineage (Norman et al. 2007). Residue 166 is part of the F loop of the D1 domain that contacts Bw4-bearing HLA molecules and is the only residue to contact the bound peptide directly and affect the ligand-receptor interaction (Vivian et al. 2011). The replacement of leucine by phenylalanine in KIR3DL1*086 could have the effect of making this allotype recognize Bw4-bearing HLA molecules that carry a different set of bound peptides.
KIR2DL4*028 differs from other Māori and Polynesian KIR2DL4 by four nucleotide changes. These comprise coding changes at positions 109 and 112 in the D1 domain and a 2bp deletion in exon 9 (Fig. 2b). None of these changes has been identified in the 27 KIR2DL4 allotypes described to date (Goodridge et al. 2007; Hou et al. 2011; Robinson et al. 2013). The glutamine residue at position 109 introduces a third alternative amino acid at a known polymorphic site (Fig. 2b), whereas the 2bp deletion causes a frame-shift starting at codon 287, causing premature termination at residue 312 and producing a polypeptide with a truncated cytoplasmic domain (Fig. 2c). Although the single immunoreceptor tyrosine-based inhibitory motif (ITIM) of 2DL4 is retained by 2DL4*028, it is unknown if the protein remains functional as a membrane-bound receptor. The loss of 65 amino acids from the cytoplasmic tail could give 2DL4*028 a similar phenotype to the previously defined ‘2DL4-9a’ alleles (Goodridge et al. 2007). These alleles, for example KIR2DL4*008, have a single base-pair deletion in exon 7 that changes the reading frame, causes premature termination at codon 251 in the transmembrane region (Fig. 2c) and produces a soluble protein that is not expressed at the cell surface (Goodridge et al. 2007; Kikuchi-Maki et al. 2003).
Māori and Polynesians have similar KIR allele frequencies that differ from those of other populations
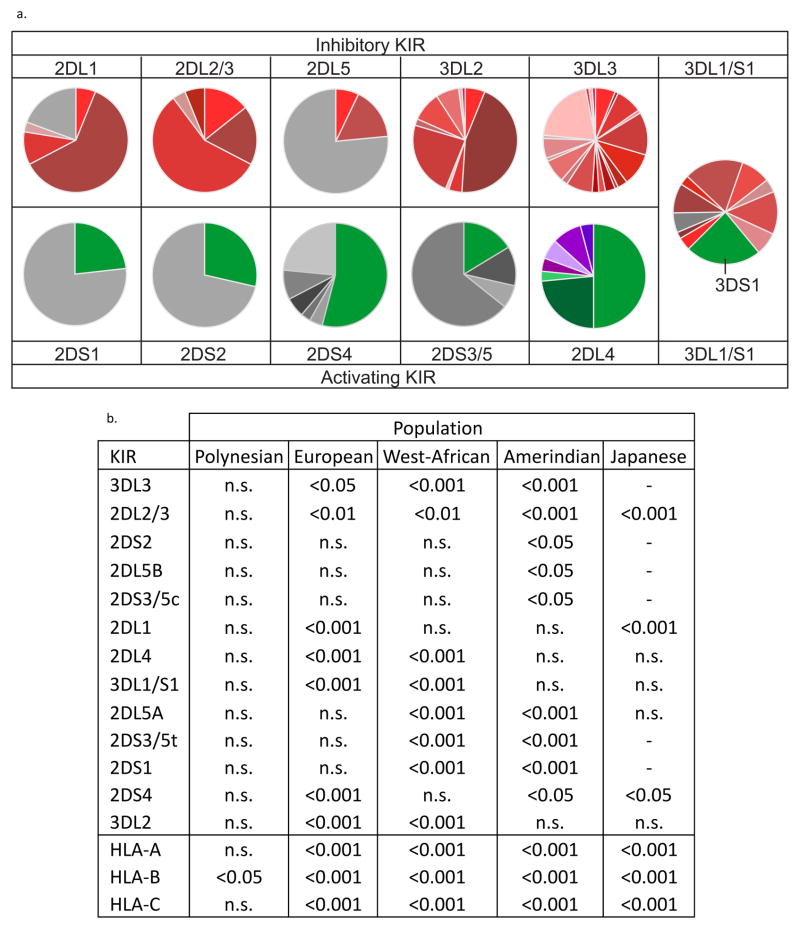
Comparison of the Māori KIR genes shows the inhibitory KIR are more polymorphic than the activating KIR (Fig. 3a). Inhibitory KIR have between 5 (KIR2DL1) and 21 (KIR3DL3) allotypes, whereas the presence and absence of a functional gene is the principal source of activating KIR variation. Exemplifying this distinction is the KIR3DL1/S1 gene, which encodes both inhibitory (KIR3DL1) and activating (KIR3DS1) receptors. Whereas, KIR3DS1 is represented by a single allotype, KIR3DL1 is highly polymorphic. Distinguishing KIR2DL4 is its specificity for HLA-G, a class I MHC molecule expressed only by trophoblast cells, implicating KIR2DL4 in the reproductive function of NK cells (Rajagopalan and Long 2012). KIR2DL4, which although retains inhibitory signalling potential (Yusa et al. 2002), is the one activating receptor (Kikuchi-Maki et al. 2003) with considerable polymorphism (Fig. 3a). In exhibiting these distinctive differences between inhibitory and activating receptors, the Māori and Polynesians are similar to the other populations for which KIR alleles and haplotypes have been described at high resolution (Gendzekhadze et al. 2009; Norman et al. 2013; Vierra-Green et al. 2012; Yawata et al. 2006). The exception is KIR2DS5, which is polymorphic in sub-Saharan Africans (Hou et al. 2009; Norman et al. 2013).
Figure 3. Comparison of Māori and Polynesian KIR allele frequencies with those of other populations.
a. The allele-frequency spectra for Māori KIR. Shades of (red) represent inhibitory KIR, (green) - activating KIR, (purple) - truncated allotypes of KIR2DL4. The allele names and frequencies are given in Supplementary Figure 2.
b. Shown are the results of contingency table comparisons of allele-frequency spectra for KIR (upper) and HLA class I (lower). The populations shown are compared with the Māori; p values are the α level of significance following Bonferroni correction. (n.s.) – not significant. (−) – no data available from population.
In comparing KIR alleles and their frequencies, the Polynesians are so similar to the Māori that no statistically significant difference could be detected between the two groups for any KIR (Fig. 3b). In contrast, when Māori or Polynesians were compared to European, West African, and Amerindian populations, highly significant differences in allele frequencies were detected for a majority of the KIR genes, as was similarly observed for HLA-A, B and C (Fig. 3b). In general, the differences are larger for genes encoding inhibitory KIR than activating KIR. Thus, KIR3DL3 and 2DL2/3 display the highest differences and 2DS2, 2DL5B and 2DS3/5c display the least differences (Fig. 3b). The Japanese, who reside geographically closer to Remote Oceania than the other populations, have KIR allele frequencies that are more similar to those of Māori and Polynesians, with only three genes, centromeric KIR2DL2/3 and KIR2DL1, and telomeric KIR2DS4, showing statistically significant difference. A common feature of the telomeric KIR region of Māori, Polynesians and Japanese is an unusually low frequency of KIR2DL4 alleles encoding truncated KIR2DL4 allotypes. Such alleles are at 20% frequency in Japanese (Yawata et al. 2006) and ~25% in Māori and Polynesians, including a contribution of 4% from the new KIR2DL4*028 allotype (Supplementary Fig. 2). In contrast, the West-Africans and the Amerindians have allele frequencies for truncated forms of KIR2DL4 that are 40% (Norman et al. 2013) and 57% (Gendzekhadze et al. 2009), respectively. Although 2DL4*028 is not present in the Japanese population studied, the overall similarity between Japanese, Māori and Polynesians suggests a common ancestry of telomeric KIR segments.
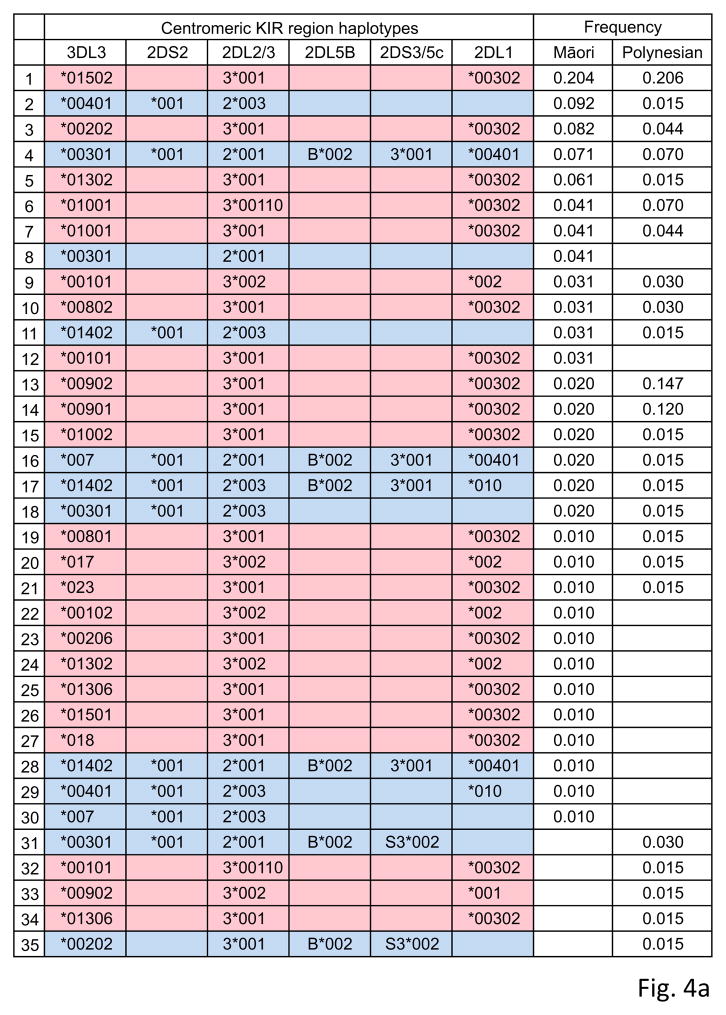
KIR haplotype diversity in Māori and Polynesians
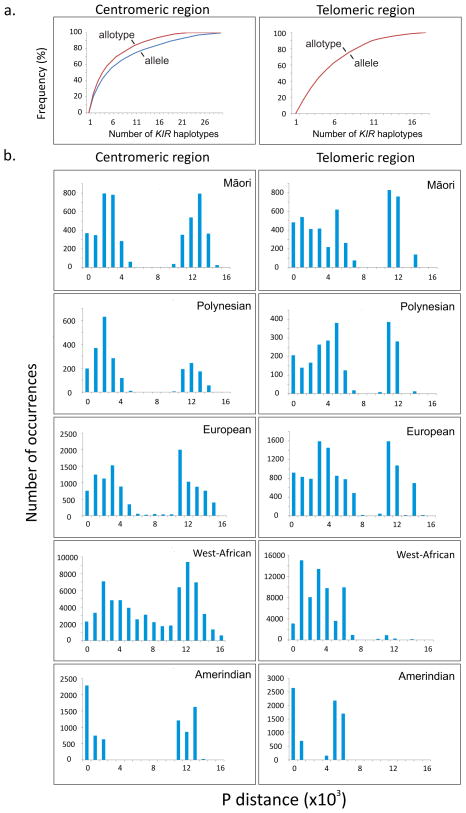
From the patterns of linkage disequilibrium (LD) within the centromeric and telomeric regions of the KIR haplotype (Middleton et al. 2007; Norman et al. 2013; Vierra-Green et al. 2012) we inferred the centromeric and telomeric haplotypes for each Māori and Polynesian individual and the haplotype frequencies for the two populations. We observed a total of 35 centromeric and 22 telomeric haplotypes, the common ones being present in both populations (76–91% cumulative frequency; Fig. 4). The number of different centromeric and telomeric KIR haplotypes did not change when non-expressed and synonymous KIR variants were excluded from the analysis (Fig. 5a). Pairwise comparison of Māori and Polynesian haplotype sequences gives a striking bimodal mismatch distribution for both the centromeric and telomeric regions (Fig. 5b). Underlying the bimodal distribution is the sequence divergence of the KIR A and B haplotypes and their presence at comparable frequencies. The bimodal distribution is a strong indicator that haplotypic diversity is maintained by balancing selection throughout the KIR locus in these populations. Comparable bimodal distributions are seen for the European, West-African and Amerindian KIR haplotypes, with the exception of the telomeric region of the West-African KIR locus which has a paucity of telomeric B haplotype regions. This deficiency could have arisen from disease-specific selection (Norman et al. 2007). Although having a striking bimodal distribution, the Amerindian KIR haplotypes have considerably less sequence diversity than the other populations, consistent with their overall low genetic diversity and the extended migration that was required for modern humans to reach the Americas. In contrast the Māori and Polynesians have comparable KIR diversity to the European population, indicating that their more recent history of migration did not cause extensive loss of genetic diversity.
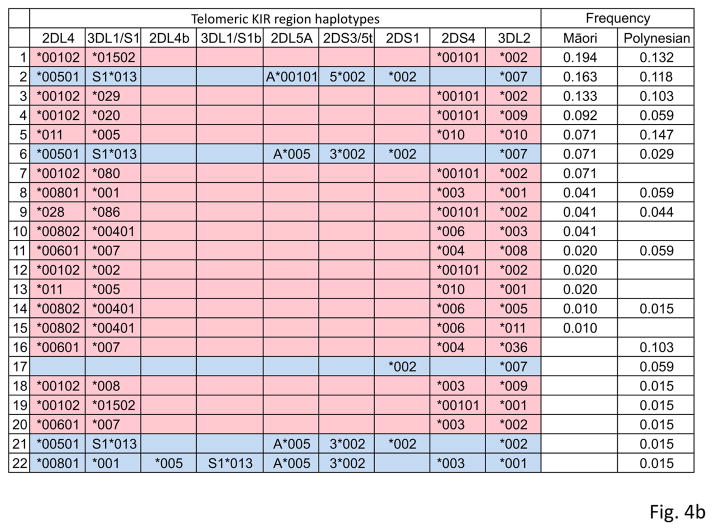
Figure 4. Allelic Diversity of Māori and Polynesian KIR haplotypes.
The centromeric (panel a) and telomeric (panel b) KIR haplotypes observed in the Māori and Polynesians. (Red) indicates KIR A and (blue) KIR B motifs. Haplotype frequencies are shown at the right. Individuals with duplicated segments (designated 2DL4b-3DL1/S1b) were identified from pyrosequencing of component alleles and those with deleted 2DL4-3DL1/S1 segments using real-time PCR (Jiang et al. 2012).
Figure 5. Bimodal KIR diversity in centromeric and telomeric regions of Māori and Polynesian KIR haplotypes.
a. Cumulative frequency plots of KIR/KIR haplotype frequencies. (red) indicates allele-level haplotypes (blue) represent allotype-level haplotypes (i.e. considering only substitutions that alter the sequence or number of expressed KIR); the telomeric region the plots are identical.
b. Shows histograms of mismatch distributions of centromeric (left) and telomeric region (right) haplotypes determined by percentage difference (p-distance).
With a frequency of 20%, the haplotype encoding KIR3DL3*015, 2DL3*001 and 2DL1*003 is the most common centromeric KIR haplotype in Māori and Polynesians, but it is rare in other populations or absent (Supplementary Fig. 3). The commonest telomeric haplotype (which encodes KIR2DL4*001, 3DL1*015, 2DS4*001 and 3DL2*001), is also present at a frequency of ~20% (Supplementary Fig. 3). This haplotype is also common in Amerindians and Japanese, but rare in West-African and European populations (Supplementary Fig. 3). Telomeric KIR haplotypes with deleted or duplicated 2DL4-3DL1/S1 segments were also detected at low frequency (Fig. 4). These recombinant haplotypes are known to be at low to intermediate frequency in Europeans, Asians and Sub-Saharan Africans (Gomez-Lozano et al. 2005; Norman et al. 2013; Williams et al. 2003).
An unusual KIR haplotype shared by Māori, Polynesians and a Papua New Guinean
As predicted from the concordance of KIR2DL4*028 and 3DL1*086 in the Māori and Polynesians, these two new alleles segregate together on a single telomeric KIR A haplotype segment, which also contains KIR2DS4*001 and 3DL2*002 (haplotype 9 in Fig. 4b). Because KIR2DL4*028 and 3DL1*086 differ from other Māori and Polynesian alleles by several dispersed nucleotide substitutions we hypothesized that this haplotype was acquired through admixture with another population group. Given the history of the Māori and Polynesian populations, one candidate source for this unusual haplotype was Papua New Guinea. To investigate this possibility we studied the telomeric KIR genes of an individual from Papua New Guinea whose genome had been sequenced (Meyer et al. 2012). Using a novel bioinformatics approach (Kidd et al. 2014), we extracted the sequence reads that are specific to the telomeric KIR segment and mapped them to their corresponding genes. With this analysis we identified both of the telomeric KIR haplotypes in the Papua New Guinean individual. One haplotype was identical to 2DL4*028-3DL1*086 containing haplotype of the Māori and Polynesians. This identity indicates that this unusual haplotype was acquired from a Near Oceanic individual. That studying a single Papua New Guinean revealed this haplotype suggests it has substantial frequency in the population of Papua New Guinea. The second telomeric KIR haplotype in the genome of the Papua New Guinean contains the 2DL4*005, 3DS1*01301, 2DL5A*001, 2DS5*002, 2DS1*002 and 3DL2*007 alleles. This is the most frequent B haplotype worldwide, being common to all populations outside of Africa (Supplementary Fig. 3) and a good candidate for having been introduced into modern humans by introgression from archaic humans (Abi-Rached et al. 2011).
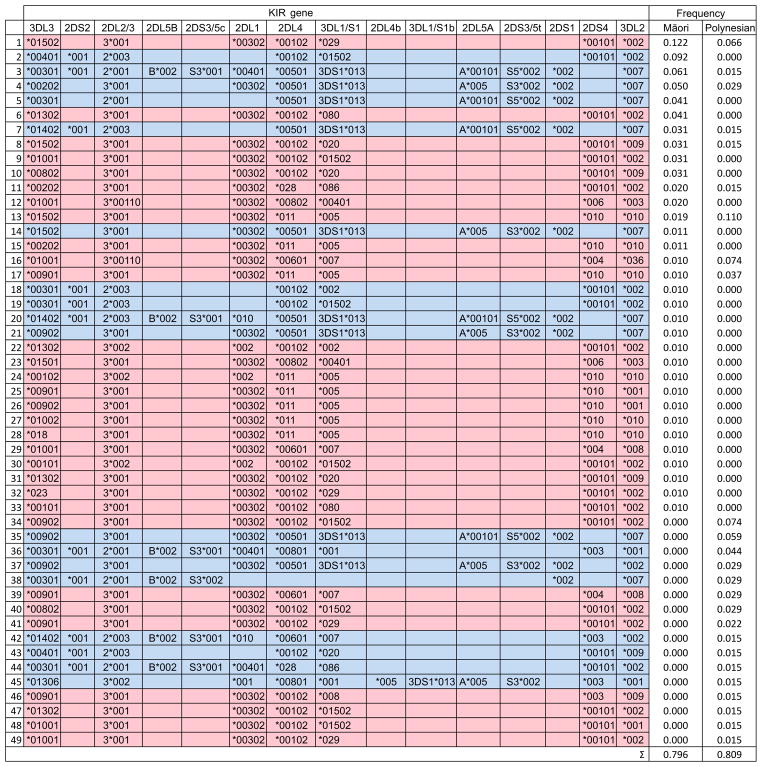
Determination of complete Māori and Polynesian KIR haplotypes from centromeric and telomeric haplotypes
The fact that meiotic recombination shuffles centromeric and telomeric segments to diversify KIR haplotypes (Bashirova et al. 2006; Wilson et al. 2000), complicated the determination of which combinations of centromeric and telomeric KIR make up the complete haplotypes for each Māori and Polynesian individual. To address this question we estimated population frequencies of full-length KIR haplotypes using the expectation-maximization (EM) algorithm. This approach enabled us to define 80% of the Māori and Polynesian KIR haplotypes. Of these 49 haplotypes, 23 haplotypes are present in more than one individual and 26 were observed only once (Fig. 6). On the basis of high-resolution KIR genotyping we have demonstrated that the Māori have a minimum of 35 different KIR haplotypes and for the Polynesians there are minimally 25 KIR haplotypes. Among these haplotypes is allelic diversity of 31 KIR A haplotypes and 18 KIR B haplotypes (Fig. 6). In terms of gene content, the canonical KIR A haplotype is the most common haplotype (estimated frequency of 60% in Māori and 68% in Polynesian), as is true for nearly all populations examined to date (Hollenbach et al. 2013; Parham 2005). The second most frequent gene-content haplotype observed in the Māori (10.2%) and Polynesians (13.3%) is a B haplotype that possesses the centromeric A motif (Fig. 6). This KIR B haplotype, also the most common B haplotype in other non-African populations (Hollenbach et al. 2012), has only five variants distinguished by allele differences in Māori and Polynesians (Fig. 6). The two divergent new KIR, 2DL4*028 and 3DL1*086, are carried on one A (haplotype 11 in Fig. 6) and one B (haplotype 44 in Fig. 6) haplotype. Although there was no significant difference between Māori and Polynesians in the frequency spectra of the KIR haplotypes (possibly due to small sample sizes) only nine of the 49 full-length haplotypes are shared by the two populations (~35% combined frequency: Fig. 6). This finding suggests the Māori and Polynesians share many low-frequency haplotypes that have yet to be discovered.
Figure 6. Complete Māori and Polynesians KIR haplotypes.
Frequencies of full-length KIR haplotypes estimated using the EM algorithm. Only haplotypes with an estimated frequency equal to, or above, a single observation are shown (~20% of the haplotypes from each population are not accounted for). (Red) indicates KIR A and (blue) KIR B haplotypes.
HLA-A KIR ligands compensate for the paucity of HLA-B KIR ligands in the Māori
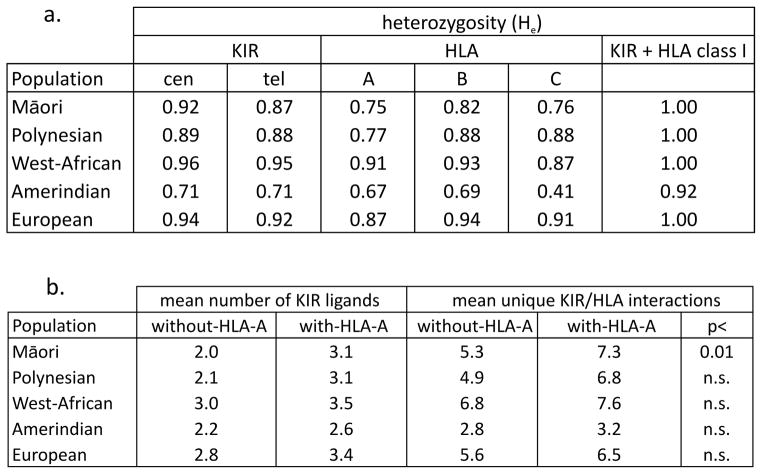
Consistent with results from genome-wide comparisons (Chambers 2013; Friedlaender et al. 2008), Māori and Polynesians have greater KIR diversity than Amerindians, but somewhat less than sub-Saharan Africans and Europeans (Fig. 7a). Overall, however, the level of immunogenetic diversity is such that each Māori and Polynesian individual studied has a unique KIR/HLA class I compound genotype, implying a unique repertoire of NK-cell interactions. We previously observed such individualization by KIR/HLA class I in the highly heterozygous West African population (Norman et al. 2013). Comparison with the other populations studied at high resolution shows that Europeans similarly achieve immune individuality. Moreover, >90% of the Yucpa Amerindians have unique receptor/ligand genotypes (He=0.92: Fig. 7a) although they have relatively few HLA and KIR alleles and haplotypes (Gendzekhadze et al. 2009). To examine how this genetic diversity could impact NK cell function, we investigated pairwise diversity of HLA class I ligands and their cognate KIR.
Figure 7. HLA-A has restored KIR/ligand heterozygosity in Māori and Polynesians.
a. Shown in the two columns at the left are the heterozygosity values (He) for the centromeric (cen) and telomeric (tel) KIR allotypes in the Māori and Polynesians compared to the West-African, Amerindian and European populations. The central three columns give the heterozygosity for HLA-A, -B and -C. The column on the right gives the heterozygosity values for the combination of KIR and HLA class I.
b. The two columns on the left shows the mean number of KIR ligands per individual, with and without the inclusion of HLA-A. The next two columns show the mean number of unique KIR and HLA class I epitope interactions per individual, with and without the inclusion of HLA-A. The column on the right gives the statistical significance as measured using the Wilcoxon test (implemented in R) with p values corrected for the number of populations.
In Māori and Polynesians, there is a high frequency of HLA-A and a low frequency of HLA-B molecules that are KIR ligands, features not seen in West-African, European and Amerindian populations. Thus in Māori and Polynesians, ~54% of HLA-A carry either the A3/11 or Bw4 epitope, whereas only 2–3% of HLA-B carry Bw4 (Fig. 1). The contribution of HLA-A has the effect of increasing the mean number of KIR ligands per individual from 2.0 to 3.1, a value higher than for Amerindians but below the values for West-Africans and Europeans (Fig. 7b). Based on the reported interactions of KIR with the C1, C2, Bw4 and A3/11 epitopes of HLA class I (Graef et al. 2009; Hilton et al. 2012; Lanier 2005; Liu et al. 2014; Moesta et al. 2008; Moretta et al. 2014), for each Māori and Polynesian individual, we determined the number of functional interactions between their HLA-A, -B and -C molecules (Edinur et al. 2013) and their cell-surface expressed KIR (Supplementary Fig. 1). The Māori and Polynesians have, respectively, a mean of 7.3 and 6.8 unique ligand-receptor interactions per individual (Fig. 7b), values that are higher than the 6.5 mean in Europeans but lower than the 7.6 mean of West Africans (Norman et al. 2013). The impact of HLA-A on increasing the number of functional interactions is of statistical significance for the Māori but not for the other populations (Fig. 7b). The lack of significance for the Polynesians could be due to small sample size, which is not the case for the other populations. These results suggest that the paucity of KIR ligands contributed by Māori and Polynesian HLA-B is compensated by the increased contribution of KIR ligands by HLA-A.
Discussion
The first analysis of KIR variation in the populations of Remote Oceania was performed at the low resolution of KIR gene content and examined the Polynesians of the Cook Islands, Samoa, Tokelau and Tonga (Velickovic et al. 2006). Considerable KIR gene content diversity was observed, which was similar in the four populations. Here we focused on high-resolution, allele-level KIR analysis of the New Zealand Māori, for which this is the first study of KIR variation. We also performed high-resolution KIR analysis of a mixed Polynesian cohort for comparison with the Māori. We show that the high level of KIR diversity in Polynesians (Velickovic et al. 2006) is further enhanced by allelic variation within the multiple gene content haplotypes. We show that the Māori have similar KIR diversity to Polynesians both in quantity and quality.
The Māori samples were carefully selected to represent their ancestry because even recent admixture can perturb the allele frequency spectra that are characteristic of these small populations (Edinur et al. 2013; Roberts et al. 2013). Analysis of the 13 expressed KIR genes identified a total of 80 allelic variants in the Māori, of which 54 encode distinct allotypes. Similar numbers -- 76 alleles encoding 51 allotypes -- were identified in Polynesians. Although these numbers are half those present in West-Africans or Europeans (Middleton et al. 2007; Norman et al. 2013; Vierra-Green et al. 2012), high diversity is achieved in both Māori and Polynesians because of their relatively balanced allele frequencies (Fig. 5). Despite repeated cycles of population contraction and expansion during the rapid colonization of the South Pacific islands, the KIR locus of the survivors retained very high diversity. The same phenomenon is seen in Yucpa South Amerindians, who maximize KIR diversity with yet fewer haplotypes (Gendzekhadze et al. 2009). These finding indicate the need for human populations to retain a critical level of NK cell receptor variation. The human NK system has dual roles in immunity and reproduction that are potentially competing. That the risk of pre-eclampsia is significantly elevated in Māori women (Anderson et al. 2012), suggests an imbalance between the two roles persists in these populations. This imbalance may be due to the relatively high frequencies of KIR A haplotypes, which are associated with preeclampsia in homozygous women who carry a foetus expressing a C2 motif (Hiby et al. 2004).
Ligands for KIR are four epitopes of the polymorphic HLA-A, -B and -C molecules. In the Remote Oceanic populations we studied there is a paucity of HLA-B allotypes that express KIR ligands; less than 3% of Māori and Polynesian HLA-B have the Bw4 epitope and none have the C1 epitope (Fig. 7). In apparent compensation for this loss, the Remote Oceanic populations have acquired high frequencies (>50%) of HLA-A allotypes having the A3/11 or Bw4 epitopes recognized by KIR (Fig. 7). The A3/11 epitope carried by HLA-A*11 is recognized by KIR2DS2, 2DS4 and 3DL2 (Dohring et al. 1996; Graef et al. 2009; Liu et al. 2014) and the Bw4 epitope carried by HLA-A*24 is recognized by KIR3DL1 (Thananchai et al. 2007). The HLA-A*11 and -A*24 alleles are common throughout Southeast Asia and Oceania and there is evidence that they were introduced into the modern human population through admixture with archaic humans, such as Denisovans, and then rose to high frequency through adaptive introgression (Abi-Rached et al. 2011). Thus, a strong candidate for the mechanism that drove the adaptive introgression of HLA-A*11 and HLA-A*24 is their capacity to provide the Bw4 and A3/11 epitopes that either restored or replaced KIR ligands that had been lost by the small migrating populations of modern humans.
Estimates of remaining ancestry proportions place the genetic influence of Near Oceanians on modern day Remote Oceanians at 15–30% (Kimura et al. 2008; Wollstein et al. 2010). Analyzing the frequency spectra of HLA haplotypes, which characterize human populations (Fernandez Vina et al. 2012), has also identified this genetic influence on Māori and Polynesians (Edinur et al. 2013; Edinur et al. 2012). Because knowledge of KIR allele diversity is restricted to very few populations, relative ancestry proportions cannot yet be calculated from KIR data. Relating to this question, however, was our discovery of two divergent KIR alleles (KIR2DL4*028 and KIR3DL1*086) that are in complete LD and segregate in the Māori and Polynesian population on a single telomeric KIR haplotype that is also shared with a Papua New Guinean individual. The likely source of this haplotype was Near Oceania, because 3DL1*086 was not discovered in exploratory surveys for ‘new’ KIR3DL1 alleles that encompassed several East and Southeast Asian population groups (Norman et al. 2007; Tao et al. 2014; Yawata et al. 2006). Furthermore, finding the 2DL4*028-3DL1*086 haplotype in the single Papua New Guinean individual we studied suggests it is at high frequency in the Papua New Guinean population. In turn, Near Oceanian populations have the highest proportion of archaic-human genetic ancestry of any modern population group (Meyer et al. 2012). Thus, the divergent sequence and geographic distribution of the 2DL4*028-3DL1*086 haplotype are consistent with it having been given to modern humans outside of Africa by archaic humans.
Supplementary Material
Shown in the two columns on the right of each panel are the frequencies of HLA-C (panel a) HLA-A (panel b) and HLA-B (panel c) allotypes in the Māori and Polynesians. In the central columns are shown the KIR that can recognize each allotype.
Shown are the KIR alleles and their frequencies in the Māori and Polynesian populations. KIR allotypes that are known or predicted to be unexpressed at the cell surface are given in red. Alleles of the centomeric KIR genes are given in panel a; alleles of the telomeric KIR genes are given in panel b.
In the depiction on the left are shown the Māori and Polynesian centromeric (panel a) and telomeric (panel b) KIR haplotypes with only variation that alters protein sequence or expression being taken into account. Boxes corresponding to KIR A haplotypes are colored red and KIR B haplotypes are colored blue. In the columns on the right the frequency of each haplotype is compared to that in Amerindian, European and sub-Saharan African populations that have also been studied at high resolution. (Gendzekhadze et al. 2009; Norman et al. 2013; Vierra-Green et al. 2012). The Japanese population (Yawata et al. 2006) was not fully typed for centromeric KIR, so only the telomeric segment is shown.
Acknowledgments
We thank Jyothi Jayaraman for technical assistance, Eric Long for advice and Derek Middleton for use of genotype data. This study was supported by U.S. National Institutes of Health grants AI17892 (PP, PJN, NNG) and GM109030 (JAH, PJN, PP), the Medical Research Council of the UK (JAT) and by the Victoria University of Wellington, NZ and Ministry of Higher Education, Malaysia.
References
- Abi-Rached L, et al. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science (New York, NY) 2011;334:89–94. doi: 10.1126/science.1209202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlenstiel G, Martin MP, Gao X, Carrington M, Rehermann B. Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. The Journal of clinical investigationthe. 2008;118:1017–1026. doi: 10.1172/jci32400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson NH, Sadler LC, Stewart AW, Fyfe EM, McCowan LM. Ethnicity, body mass index and risk of pre-eclampsia in a multiethnic New Zealand population. The Australian & New Zealand journal of obstetrics & gynaecology. 2012;52:552–558. doi: 10.1111/j.1479-828X.2012.01475.x. [DOI] [PubMed] [Google Scholar]
- Bari R, et al. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood. 2009;114:5182–5190. doi: 10.1182/blood-2009-07-231977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annual review of genomics and human genetics. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- Bellwood P, Chambers G, Ross M, Hung H-C. Are ‘cultures’ inherited? Multidisciplinary perspectives on the origins of Austronesian-speaking peoples prior to 1000 BC. In: Roberts B, Vander Linden M, editors. Investigating Archaeological Cultures: Material Culture, Variability and Transmission. Springer; Berlin: 2011. pp. 321–354. [Google Scholar]
- Burley D, Weisler MI, Zhao JX. High precision u/th dating of first Polynesian settlement. PloS one. 2012;7:e48769. doi: 10.1371/journal.pone.0048769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers GK. Genetics and the Origins of the Polynesians. John Wiley & Sons, Ltd; Chichester: 2013. [DOI] [Google Scholar]
- Cooper MA, Colonna M, Yokoyama WM. Hidden talents of natural killers: NK cells in innate and adaptive immunity. EMBO reports. 2009;10:1103–1110. doi: 10.1038/embor.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohring C, Scheidegger D, Samaridis J, Cella M, Colonna M. A human killer inhibitory receptor specific for HLA-A1,2. J Immunol. 1996;156:3098–3101. [PubMed] [Google Scholar]
- Duggan AT, et al. Maternal History of Oceania from Complete mtDNA Genomes: Contrasting Ancient Diversity with Recent Homogenization Due to the Austronesian Expansion. American journal of human genetics. 2014;94:721–733. doi: 10.1016/j.ajhg.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinur HA, et al. HLA and MICA polymorphism in Polynesians and New Zealand Maori: implications for ancestry and health. Human immunology. 2013;74:1119–1129. doi: 10.1016/j.humimm.2013.06.011. [DOI] [PubMed] [Google Scholar]
- Edinur HA, Dunn PP, Hammond L, Selwyn C, Velickovic ZM, Lea RA, Chambers GK. Using HLA loci to inform ancestry and health in Polynesian and Maori populations. Tissue antigens. 2012;80:509–522. doi: 10.1111/tan.12026. [DOI] [PubMed] [Google Scholar]
- Fernandez Vina MA, et al. Tracking human migrations by the analysis of the distribution of HLA alleles, lineages and haplotypes in closed and open populations. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2012;367:820–829. doi: 10.1098/rstb.2011.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier WR, Steiner N, Hou L, Dakshanamurthy S, Hurley CK. Allelic variation in KIR2DL3 generates a KIR2DL2-like receptor with increased binding to its HLA-C ligand. J Immunol. 2013;190:6198–6208. doi: 10.4049/jimmunol.1300464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlaender JS, et al. The genetic structure of Pacific Islanders. PLoS genetics. 2008;4:e19. doi: 10.1371/journal.pgen.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendzekhadze K, Norman PJ, Abi-Rached L, Graef T, Moesta AK, Layrisse Z, Parham P. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18692–18697. doi: 10.1073/pnas.0906051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Lozano N, Estefania E, Williams F, Halfpenny I, Middleton D, Solis R, Vilches C. The silent KIR3DP1 gene (CD158c) is transcribed and might encode a secreted receptor in a minority of humans, in whom the KIR3DP1, KIR2DL4 and KIR3DL1/KIR3DS1 genes are duplicated. European journal of immunology. 2005;35:16–24. doi: 10.1002/eji.200425493. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic acids research. 2011;39:D913–919. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge JP, et al. Three common alleles of KIR2DL4 (CD158d) encode constitutively expressed, inducible and secreted receptors in NK cells. European journal of immunology. 2007;37:199–211. doi: 10.1002/eji.200636316. [DOI] [PubMed] [Google Scholar]
- Graef T, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. The Journal of experimental medicine. 2009;206:2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. The Journal of experimental medicine. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiby SE, Walker JJ, O’Shaughnessy KM, Redman CW, Carrington M, Trowsdale J, Moffett A. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. The Journal of experimental medicine. 2004;200:957–965. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton HG, et al. Mutation at positively selected positions in the binding site for HLA-C shows that KIR2DL1 is a more refined but less adaptable NK cell receptor than KIR2DL3. J Immunol. 2012;189:1418–1430. doi: 10.4049/jimmunol.1100431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbach JA, et al. 16(th) IHIW: population global distribution of killer immunoglobulin-like receptor (KIR) and ligands. International journal of immunogenetics. 2013;40:39–45. doi: 10.1111/iji.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbach JA, Nocedal I, Ladner MB, Single RM, Trachtenberg EA. Killer cell immunoglobulin-like receptor (KIR) gene content variation in the HGDP-CEPH populations. Immunogenetics. 2012;64:719–737. doi: 10.1007/s00251-012-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Chen M, Jiang B, Kariyawasam K, Ng J, Hurley CK. In contrast to other stimulatory natural killer cell immunoglobulin-like receptor loci, several KIR2DS5 alleles predominate in African Americans. Human immunology. 2009;70:733–737. doi: 10.1016/j.humimm.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Jiang B, Chen M, Ng J, Hurley CK. The characteristics of allelic polymorphism in killer-immunoglobulin-like receptor framework genes in African Americans. Immunogenetics. 2011;63:549–559. doi: 10.1007/s00251-011-0536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles ME, Maund E, Nicholson J, Bosch E, Renfrew C, Sykes BC, Jobling MA. Native American Y chromosomes in Polynesia: the genetic impact of the Polynesian slave trade. American journal of human genetics. 2003;72:1282–1287. doi: 10.1086/374827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, et al. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome research. 2012;22:1845–1854. doi: 10.1101/gr.137976.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, et al. Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Molecular biology and evolution. 2006;23:2234–2244. doi: 10.1093/molbev/msl093. [DOI] [PubMed] [Google Scholar]
- Khakoo SI, Carrington M. KIR and disease: a model system or system of models? Immunological reviews. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- Kidd JM, et al. Exome capture from saliva produces high quality genomic and metagenomic data. BMC genomics. 2014;15:262. doi: 10.1186/1471-2164-15-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi-Maki A, Yusa S, Catina TL, Campbell KS. KIR2DL4 is an IL-2-regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN-gamma production. J Immunol. 2003;171:3415–3425. doi: 10.4049/jimmunol.171.7.3415. [DOI] [PubMed] [Google Scholar]
- Kimura R, et al. Gene flow and natural selection in oceanic human populations inferred from genome-wide SNP typing. Molecular biology and evolution. 2008;25:1750–1761. doi: 10.1093/molbev/msn128. [DOI] [PubMed] [Google Scholar]
- Kostyu D, et al. HLA in two islands of French Polynesia. Tissue antigens. 1984;23:217–228. doi: 10.1111/j.1399-0039.1984.tb00036.x. [DOI] [PubMed] [Google Scholar]
- Lanier LL. NK cell recognition. Annual review of immunology. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- Leung W, et al. Determinants of antileukemia effects of allogeneic NK cells. J Immunol. 2004;172:644–650. doi: 10.4049/jimmunol.172.1.644. [DOI] [PubMed] [Google Scholar]
- Liu J, Xiao Z, Ko HL, Shen M, Ren EC. Activating killer cell immunoglobulin-like receptor 2DS2 binds to HLA-A*11. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2662–2667. doi: 10.1073/pnas.1322052111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annual review of immunology. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MP, Carrington M. Immunogenetics of HIV disease. Immunological reviews. 2013;254:245–264. doi: 10.1111/imr.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, et al. In: Hansen JA, editor. Single Locus Polymorphism of Classical HLA Genes; Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference; Seattle, WA: IHWG Press; 2007. pp. 653–704. [Google Scholar]
- Meyer M, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science (New York, NY) 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton D, Meenagh A, Gourraud PA. KIR haplotype content at the allele level in 77 Northern Irish families. Immunogenetics. 2007;59:145–158. doi: 10.1007/s00251-006-0181-7. [DOI] [PubMed] [Google Scholar]
- Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- Moretta L, et al. Human NK Cells: From Surface Receptors to the Therapy of Leukemias and Solid. Tumors Frontiers in immunology. 2014;5:87. doi: 10.3389/fimmu.2014.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narni-Mancinelli E, Ugolini S, Vivier E. Tuning the threshold of natural killer cell responses. Current opinion in immunology. 2013;25:53–58. doi: 10.1016/j.coi.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Norman PJ, et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nature genetics. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- Norman PJ, et al. Co-evolution of human leukocyte antigen (HLA) class I ligands with killer-cell immunoglobulin-like receptors (KIR) in a genetically diverse population of sub-Saharan Africans. PLoS genetics. 2013;9:e1003938. doi: 10.1371/journal.pgen.1003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman PJ, Stephens HA, Verity DH, Chandanayingyong D, Vaughan RW. Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics. 2001;52:195–205. doi: 10.1007/s002510000281. [DOI] [PubMed] [Google Scholar]
- O’Connor GM, et al. Mutational and structural analysis of KIR3DL1 reveals a lineage-defining allotypic dimorphism that impacts both HLA and peptide sensitivity. J Immunol. 2014;192:2875–2884. doi: 10.4049/jimmunol.1303142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P. MHC class I molecules and KIRs in human history, health and survival. Nature reviews Immunology. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nature reviews Immunology. 2013;13:133–144. doi: 10.1038/nri3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. A language and environment for statistical computing. Vienna, Austria: 2008. [Google Scholar]
- Rajagopalan S, Long EO. Cellular senescence induced by CD158d reprograms natural killer cells to promote vascular remodeling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20596–20601. doi: 10.1073/pnas.1208248109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RL, et al. Prevalence of HLA-B27 in the New Zealand population: effect of age and ethnicity. Arthritis research & therapy. 2013;15:R158. doi: 10.1186/ar4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Halliwell JA, McWilliam H, Lopez R, Marsh SG. IPD--the Immuno Polymorphism Database. Nucleic acids research. 2013;41:D1234–1240. doi: 10.1093/nar/gks1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, Parham P. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168:2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- Soares P, et al. Ancient voyaging and Polynesian origins. American journal of human genetics. 2011;88:239–247. doi: 10.1016/j.ajhg.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao SD, He YM, Ying YL, He J, Zhu FM, Lv HJ. KIR3DL1 genetic diversity and phenotypic variation in the Chinese Han population. Genes and immunity. 2014;15:8–15. doi: 10.1038/gene.2013.55. [DOI] [PubMed] [Google Scholar]
- Thananchai H, et al. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- Thorsby E. The Polynesian gene pool: an early contribution by Amerindians to Easter Island. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2012;367:812–819. doi: 10.1098/rstb.2011.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey MC, Carter JM. Class II HLA allele polymorphism: DRB1, DQB1 and DPB1 alleles and haplotypes in the New Zealand Maori population. Tissue antigens. 2006;68:297–302. doi: 10.1111/j.1399-0039.2006.00671.x. [DOI] [PubMed] [Google Scholar]
- Uhrberg M, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Marzuki S, Oefner PJ, Cavalli-Sforza LL, Chambers GK. Maori origins, Y-chromosome haplotypes and implications for human history in the Pacific. Human mutation. 2001;17:271–280. doi: 10.1002/humu.23. [DOI] [PubMed] [Google Scholar]
- Velickovic M, Velickovic Z, Dunckley H. Diversity of killer cell immunoglobulin-like receptor genes in Pacific Islands populations. Immunogenetics. 2006;58:523–532. doi: 10.1007/s00251-006-0124-3. [DOI] [PubMed] [Google Scholar]
- Venstrom JM, et al. HLA-C-dependent prevention of leukemia relapse by donor activating KIR2DS1. The New England journal of medicine. 2012;367:805–816. doi: 10.1056/NEJMoa1200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierra-Green C, et al. Allele-level haplotype frequencies and pairwise linkage disequilibrium for 14 KIR loci in 506 European-American individuals. PloS one. 2012;7:e47491. doi: 10.1371/journal.pone.0047491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian JP, et al. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature. 2011;479:401–405. doi: 10.1038/nature10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte AL, Marshall SJ, Chambers GK. Human evolution in Polynesia. Human biology. 2005;77:157–177. doi: 10.1353/hub.2005.0045. [DOI] [PubMed] [Google Scholar]
- Williams F, Maxwell LD, Halfpenny IA, Meenagh A, Sleator C, Curran MD, Middleton D. Multiple copies of KIR 3DL/S1 and KIR 2DL4 genes identified in a number of individuals. Human immunology. 2003;64:729–732. doi: 10.1016/s0198-8859(03)00089-2. [DOI] [PubMed] [Google Scholar]
- Wilmshurst JM, Hunt TL, Lipo CP, Anderson AJ. High-precision radiocarbon dating shows recent and rapid initial human colonization of East Polynesia. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1815–1820. doi: 10.1073/pnas.1015876108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MJ, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. 1997;158:4026–4028. [PubMed] [Google Scholar]
- Wollstein A, et al. Demographic history of Oceania inferred from genome-wide data. Current biology: CB. 2010;20:1983–1992. doi: 10.1016/j.cub.2010.10.040. [DOI] [PubMed] [Google Scholar]
- Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. The Journal of experimental medicine. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa S, Catina TL, Campbell KS. SHP-1- and phosphotyrosine-independent inhibitory signaling by a killer cell Ig-like receptor cytoplasmic domain in human NK cells. J Immunol. 2002;168:5047–5057. doi: 10.4049/jimmunol.168.10.5047. [DOI] [PubMed] [Google Scholar]
- Zambello R, Teramo A, Barila G, Gattazzo C, Semenzato G. Activating KIRs in Chronic Lymphoproliferative Disorder of NK Cells: Protection from Viruses and Disease Induction? Frontiers in immunology. 2014;5:72. doi: 10.3389/fimmu.2014.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shown in the two columns on the right of each panel are the frequencies of HLA-C (panel a) HLA-A (panel b) and HLA-B (panel c) allotypes in the Māori and Polynesians. In the central columns are shown the KIR that can recognize each allotype.
Shown are the KIR alleles and their frequencies in the Māori and Polynesian populations. KIR allotypes that are known or predicted to be unexpressed at the cell surface are given in red. Alleles of the centomeric KIR genes are given in panel a; alleles of the telomeric KIR genes are given in panel b.
In the depiction on the left are shown the Māori and Polynesian centromeric (panel a) and telomeric (panel b) KIR haplotypes with only variation that alters protein sequence or expression being taken into account. Boxes corresponding to KIR A haplotypes are colored red and KIR B haplotypes are colored blue. In the columns on the right the frequency of each haplotype is compared to that in Amerindian, European and sub-Saharan African populations that have also been studied at high resolution. (Gendzekhadze et al. 2009; Norman et al. 2013; Vierra-Green et al. 2012). The Japanese population (Yawata et al. 2006) was not fully typed for centromeric KIR, so only the telomeric segment is shown.