Abstract
Stk11, commonly known as Lkb1, is a tumor suppressor that regulates cellular energy metabolism and stem cell function. Satellite cells are skeletal muscle resident stem cells that maintain postnatal muscle growth and repair. Here, we used MyoDCre/Lkb1flox/flox mice (called MyoD-Lkb1) to delete Lkb1 in embryonic myogenic progenitors and their descendant satellite cells and myofibers. The MyoD-Lkb1 mice exhibit a severe myopathy characterized by central nucleated myofibers, reduced mobility, growth retardation and premature death. Although tamoxifen-induced postnatal deletion of Lkb1 in satellite cells using Pax7CreER mice bypasses the developmental defects and early death, Lkb1 null satellite cells lose their regenerative capacity cell-autonomously. Strikingly, Lkb1 null satellite cells fail to maintain quiescence in non-injured resting muscles and exhibit accelerated proliferation but reduced differentiation kinetics. At the molecular level, Lkb1 limits satellite cell proliferation through the canonical AMPK/mTOR pathway, but facilitates differentiation through phosphorylation of GSK-3β, a key component of the WNT signaling pathway. Together, these results establish a central role of Lkb1 in muscle stem cell homeostasis, muscle development and regeneration.
Keywords: Liver kinase b1 (Lkb1), Serine/threonine kinase 11 (Stk11), Myogenesis, Stem cells
Introduction
The skeletal muscle comprises ~40% of the body mass in adults and plays important roles in whole body energy metabolism, insulin sensitivity and motility. Adult skeletal muscles have a remarkable capacity to regenerate after damage due to the contribution of a population of muscle resident stem cells called satellite cells [1-3]. Lineage tracing and transplantation studies have demonstrated that satellite cells can undergo efficient self-renewal and myogenic differentiation in vivo, two defining features of stem cells [4-6]. In adult resting muscles, satellite cells are predominantly quiescent, heterogeneous, and reside between the basal lamina and plasma membrane of myofibers [7, 8]. Upon muscle injury, quiescent satellite cells are activated to proliferate, differentiate and fuse into multinuclear myofibers, resulting in repair of the damaged muscle [9]. The cell fate status of satellite cells can be distinguished based on the expression pattern of the myogenic transcription factors Pax7, MyoD and MyoG. In this regard, Pax7+/MyoD–, Pax7+/MyoD+ and Pax7–/MyoD+/MyoG+ cells represent self-renewal, proliferation and differentiation progenies, respectively [10-12]. Various signaling pathways and factors, such as Notch [13-17], Wnt [18, 19], interleukin-6 [20], and insulin-like growth factor 1 [21] have been reported to regulate the cell fate choices of satellite cells. Emerging evidence indicates that metabolic status regulates satellite cell quiescence [22-24]. However, it is unknown what signaling governs the metabolic state of satellite cells and how such signaling regulates satellite cell function.
Serine/threonine protein kinase 11 (Stk11), commonly known as liver kinase B1 (Lkb1), was originally identified as a tumor suppressor protein mutated in Peutz-Jeghers syndrome [25, 26]. Subsequent studies have shown that Lkb1 plays an important role in various cellular processes including cancer initiation and progression [27-29], cellular polarity [30-32], cell adhesion [33], cell death [34], and cell energy metabolism [35]. In Drosophila, mutation of Lkb1 disrupts normal spindle orientation and abrogates the asymmetric division of neuroblasts [36]. In C. elegans, Lkb1 (PAR-4) complex mediates caspase-independent elimination of cells destined to programed cell death during development [37]. Loss of Lkb1 in pancreatic β-cells alters their morphology, increases cell size and insulin production, and enhances glucose tolerance of the mice [32, 38]. In the thymus, Lkb1 is necessary for the differentiation of thymocytes and production of mature T lymphocytes [39]. Similarly, Lkb1 promotes cell cycle progression of immature chondrocytes, and loss of Lkb1 leads to expansion of immature chondrocytes and formation of tumors [40]. In the bone marrow, Lkb1 is emerging as a critical factor in hematopoietic stem cell biology involved in maintaining stem cell survival, quiescence and metabolic homeostasis, and regulating cell cycle progression and energy metabolism [35, 41-43]. In human embryonic stem cells, Lkb1 inhibition upregulates pluripotent genes and downregulates differentiation genes [44]. Many of the evolutionary conserved functions of Lkb1 that have been observed in various animal models and tissue types are mediated through the AMP-activated protein kinase (AMPK) nexus. Lkb1 phosphorylates and activates the AMPK family of proteins which consequently regulate the mammalian target of rapamycin (mTOR) pathway [41, 45-47].
Previous reports have shown that Lkb1 global knockout mice are embryonic lethal and die at E8.5-11, indicating that Lkb1 is essential for embryonic development [48]. Studies of adult mice have shown that skeletal/cardiac muscle-specific deletion of Lkb1 driven by MCKCre affected exercise performance, insulin sensitivity, glucose uptake, and lipid oxidation [49-51], but the knockout mice appeared to be healthy without any obvious muscle atrophy until 30-50 weeks of age [50]. Since MCK-Cre is only activated in differentiated myofibers and mature muscles, the role of Lkb1 in myogenic progenitor cells, muscle development and postnatal muscle regeneration was not examined. In this study, we used MyoDCre knockin mice to specifically delete Lkb1 in all muscle progenitor cells and their descendent skeletal muscles. Deletion of Lkb1 in the MyoD lineage cells results in defective myogenesis and a severe myopathy leading to premature death. The Lkb1 null satellite cells spontaneously activate and exhibit accelerated proliferation but reduced differentiation, and are unable to regenerate injured muscles. Furthermore, we demonstrate that Lkb1 acts through two separate pathways to regulate the proliferation and differentiation of satellite cells.
Material and methods
Animals
All procedures involving mice were guided by Purdue University Animal Care and Use Committee. Mice were housed in the animal facility with free access to standard rodent chow and water. All the mouse strains were obtained from Jackson Laboratory (Bar Harbor, ME) under these stock numbers: #014140 (MyoDCre), #012476 (Pax7CreER), #014143 (Lkb1flox/flox). PCR genotyping was done using protocols described by the supplier.
Muscle myoblast isolation and culture
Primary myoblasts were isolated using type I collagenase and dispase B digestion [52]. Briefly, the hind limb skeletal muscles from the WT and Lkb1-KO mice were collected, minced and digested. The digestions were stopped with F-10 Ham's medium containing 20% FBS and centrifuged at 450 × g for 5 minutes. Then the cells were seeded on collagen-coated dishes and cultured in growth medium containing F-10 Ham's medium, with 20% fetal bovine serum (FBS), 4 ng/mL basic fibroblast growth factor, and 1% penicillin–streptomycin at 37 °C with 5% CO2. The medium was changed every 2 days.
Cell growth rate and differentiation assay
Cell growth rate was determined as previously described [13]. Primary myoblasts were seeded in 6-well plates (1×104 cells per well) and cultured under standard myoblast condition with or without drug treatment. The cells were harvested at different times and counted by using a hemocytometer. For differentiation assays, MyoD-Lkb1 and WT myoblasts were seeded onto 6-well plates (1×105 cells per well) and induced with 2% horse serum for 3 days. Then cultures were collected for RNA isolation or immunostained for myosin heavy chain (MF20) as described previously [53]. The fusion and differentiation indexes were determined as described previously [54]. Briefly, the fusion index was determined as the percentage of the nuclear number in myotubes (cells with two or more nuclei) and the differentiation index was calculated as the ratio of the nuclei within myotubes and MF20+ mononucleated cells. In addition, the percentage of undifferentiated cells was determined as reserve index [55].
Single myofiber isolation and culture
Single myofibers were isolated from the extensor digitorum longus (EDL) muscles after digestion with collagenase A (Sigma) and trituration as previously described [13]. Suspended fibers were cultured in horse serum-coated plates (60 mm) in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT), 2% chicken embryo extract (Accurate Chemical, Westbury, NY), and 1% penicillin-streptomycin for 3 days. Freshly isolated fibers and cultured fibers were then fixed in 4% PFA and stained for Pax7, MyoD, and Lkb1.
Total RNA extraction, cDNA synthesis and real-time PCR
Total RNA extraction, cDNA synthesis and real-time PCR were performed as described [52, 56]. Briefly, total RNA was extracted from cells using Trizol Reagent according to the manufacturer's instructions. RNA was treated with RNase-free DNase l to remove contaminating genomic DNA. The purity and concentration of total RNA were measured by a spectrophotometer (Nanodrop 3000, Thermo Fisher) at 260 nm and 280 nm. Ratios of absorption (260/280 nm) of all samples were between 1.8 and 2.0. Then 5 μg of total RNA were reversed transcribed using random primers and MMLV reverse transcriptase. Real-time PCR was carried out with a Roche Lightcycler 480 PCR System using SYBR Green Master Mix and gene-specific primers. The 2-ΔΔCT method was used to analyze the relative changes in gene expression normalized against 18S rRNA as internal control.
Protein Extraction and Western Blot Analysis
Protein extraction and western blot analysis were conducted as previously described [52]. Briefly, total protein was isolated from cells or tissues using RIPA buffer containing 50mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% NP-40, 0.5% sodium Deoxycholate and 0.1% SDS. Protein concentrations were determined using the Pierce BCA Protein Assay Reagent (Pierce Biotechnology). Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore Corporation) and after blocking in 5% fat-free milk for 1 hour at RT, the filters were incubated with first antibodies in 5% milk overnight at 4°C. The antibodies used for this work were: Desimin, pS6, S6, phospho-GSK3β (Ser9), β-catenin (Cell Signaling), GSK3β (BD Bioscience), Pax7, MyoG (F5D) (Developmental Studies Hybridoma Bank), rabbit anti -Ki67 and active Caspase 3 (Abcam), mouse anti-Ki67, laminin, MyoD, pAMPK, AMPK, and GAPDH (Santa Cruz Biotechnology). Secondary antibodies (anti-rabbit IgG or anti-mouse IgG, Jackson ImmunoResearch) were diluted 8,000-fold. Immunodetection was performed using enhanced chemiluminescence (ECL) with Western blotting substrate (Pierce Biotechnology) and detected with a Gel Logic 2200 imaging system (Carestream).
Muscle injury and regeneration
To induce muscle regeneration, mice were anesthetized using a ketamine-xylazine cocktail and cardiotoxin (CTX) was injected (50μl of 10μM solution, Sigma) into TA muscle [13]. Muscles were then harvested at different time points post injection to assess the completion of regeneration and repair.
Hematoxylin-eosin (HE), Masson's Trichrome and immuno- staining
Whole muscle tissues from the WT and Lkb1-KO mice were dissected and frozen immediately in OCT compound. Frozen muscles were cross sectioned (10 μm) using a Leica CM1850 cryostat. For H&E staining, the sections were stained in haematoxylin (30 min), rinsed in running tap water and stained in eosin (1 min). For Masson's staining, the sections were fixed in 4% PFA at RT for 1hr and re-fixed in Bouin's solution at RT overnight. Then the sections were stained as previously described [57]. The sections were mounted and the images were captured with a Nikon D90 digital camera installed on a Nikon (Diaphot) inverted microscope. Immunostaining was performed as previously described [13]. Briefly, muscle sections were fixed (4% PFA), incubated in blocking buffer (5% goat serum, 2% BSA, 0.2% triton X-100 and 0.1% sodium azide in PBS, 1 h) followed by incubation with the primary antibodies diluted in blocking buffer overnight. After washing with PBS, the samples were incubated with secondary antibodies and Hoechst for 45 minutes at room temperature. Fluorescent images were captured as single channel grayscale images using a Leica DM 6000B fluorescent microscope with a 20X objective (NA 0.70) and assembled in Photoshop. Images for WT and conditional knockout samples were captured using identical parameters and both control and cKO images were adjusted identically in Photoshop.
Data Analysis
All experimental data are presented as mean ± SEM. Comparisons were made by unpaired two-tailed Student's t-tests or one-way ANOVA, as appropriate. Effects were considered significant at P < 0.05.
Results
Myogenic lineage-specific deletion of Lkb1 leads to defects in muscle development and growth
We first examined the expression pattern of Lkb1 during muscle development and in postnatal satellite cells. Using antibody staining, we found that Lkb1 is ubiquitously expressed in E10.5 embryos and highly expressed in MyoD+ muscle progenitors from E10.5-E17.5 (Fig. S1A). Realtime PCR results showed that expression level of Lkb1 in somites and hindlimbs (limb buds) increases from E10.5 to E17.5 (Fig. S1B-D). In addition, Lkb1 protein was detectable in quiescent satellite cells and its expression increased in a fraction of myoblasts during proliferation (Fig. S1E, F). These results indicate that Lkb1 is expressed in skeletal muscle progenitors and may play a role in muscle development.
To directly investigate the roles of Lkb1 in myogenic progenitors and skeletal muscle development, we used the Cre-loxP recombination system involving MyoDCre and Lkbflox/flox mice (Fig. S2A). Previous studies have established that MyoDCre specifically marks all embryonic myogenic progenitors that give rise to postnatal satellite cells and myofibers [59]. Thus, in the MyoDCre/ Lkb1flox/flox mice (abbreviated as MyoD-Lkb1) all myogenic lineage cells should contain a deletion of exons 3-6 in the Lkb1 gene, leading to loss of the kinase domain and premature translational termination of the Lkb1 protein (Fig. S2A). Realtime qPCR and Western blot analyses confirmed efficient and specific depletion of Lkb1 in skeletal muscles as well as myoblasts (Fig. S2B-C), but not in non-skeletal muscle tissues including heart, brown adipose tissue (BAT) and white adipose tissue (WAT) (Fig. S2D-E).
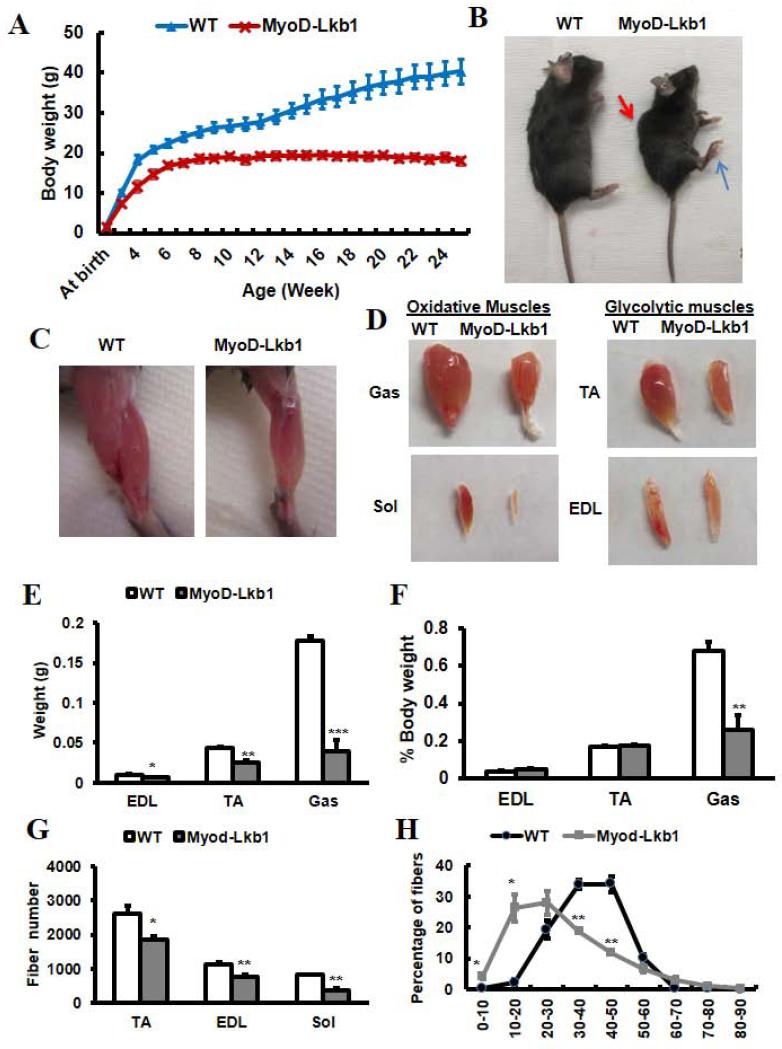
Strikingly, the MyoD-Lkb1 mice were born smaller compared to WT littermates (WT: 1.80 ± 0.04 g; MyoD-Lkb1: 1.55 ± 0.05 g), and failed to thrive during postnatal growth (Fig. 1A). The MyoD-Lkb1 mice reached maximal body weight within 2 months, after which their weight remained the same (Fig. 1A). Even in the first 2 months, the MyoD-Lkb1 mice grew more slowly than their wildtype (WT) littermates (Fig. 1A). At 21-weeks, the MyoD-Lkb1 mice weighed less than half of their WT littermates (Fig. 1A). In addition, the MyoD-Lkb1 mice had a spinal deformity that was evident at 3~4-weeks and obvious kyphosis at 8~10-weeks (Fig. 1B). The MyoD-Lkb1 mice also had abnormal hindlimb postures (Fig. 1B) and severely compromised mobility (Supplementary Movie 1), indicating muscle weakness. Moreover, a portion of MyoD-Lkb1 mice began to die after weaning, at ~45% mortality within 6 months (n = 15). By contrast, the MyoDCre/Lkb1flox/+ mice appeared normal and indistinguishable from WT littermates (Fig. S2F), excluding a haploinsufficiency effect of Lkb1 in myogenesis.
Figure 1.
MyoDCre-mediated deletion of Lkb1 impairs muscle development and growth. (A) Growth curve of the WT (Lkb1flox/flox) and MyoD-Lkb1 (MyoDCre/ Lkb1flox/flox) mice. P < 0.01 for all timepoints measured, n=8 for WT and n=5 for MyoD-Lkb1 mice. (B) Gross morphology of WT and MyoD-Lkb1 mice at 10-wk-old. Kyphosis (red arrow) and abnormal hindlimb postures (blue arrow) were indicated in the MyoD-Lkb1 mouse. (C-D) Representative images of hindlimbs (C) and hindlimb muscles (D) showing reduced muscle mass in the MyoD-Lkb1 mice. (E-F) Absolute (E) and relative to body (F) weight of EDL, TA and Gas muscles at 10-wk-old. (G) Total myofiber numbers in TA, EDL and Sol muscles at 10-wk-old. n=4. (H) Myofiber size distribution of TA muscles at 10-wk-old. n = 4. Error bars represent SEM. * P< 0.05, ** P< 0.01, *** P< 0.001.
We next examined if the reduced body weight in the MyoD-Lkb1 mice is due to a reduction of muscle mass. At 10-week-old when their body weight peaked, the MyoD-Lkb1 mice had smaller hindlimbs (Fig. 1C), and reduced size in various muscles (Fig. 1D), including gastrocnemius (Gas), soleus (Sol), extensor digitorum longus (EDL) and tibialis anterior (TA). The weight of the EDL, TA and Gas were reduced by 30% - 75% in the MyoD-Lkb1 mice (Fig. 1E). Interestingly, when normalized to body weight, the relative weights of EDL and TA, which consist primarily of glycolytic myofibers, were not affected by the Lkb1 deletion (Fig. 1F). However, the normalized weight of the Gas muscle containing both glycolytic and oxidative myofibers was reduced by 60% (Fig. 1F), and the Sol muscle, containing predominantly oxidative (Type I and IIa) myofibers, was almost invisible (Fig. 1D). As control, the relative mass of kidney, lung, liver, heart and spleen were unchanged or slightly increased (Fig. S2G). These results suggest that the decreased body mass of the MyoD-Lkb1 mice was mainly due to reductions in the size of the skeletal muscles.
To determine if the reduced muscle mass observed in the MyoD-Lkb1 mice was due to changes in myofiber number or size, we enumerated the total number of myofibers in TA, EDL and Sol muscles. All MyoD-Lkb1 muscles examined contained a significantly reduced number of myofibers compared to the WT mice (Fig. 1G), suggesting defects in myofiber formation during embryonic development. Consistently, we observed a reduction of differentiated MyoG+ mononuclear myocytes in E10.5 MyoD-Lkb1 embryos, but the abundance of Pax7+ and MyoD+ cells were not affected (Fig. S3). In addition, MyoD-Lkb1 deletion led to a left-shift of the fiber size distribution curve, with most of the myofibers ranging from 10-40 μm in diameter (Fig. 1H). By contrast, WT myofibers predominantly ranged from 20-60 μm in diameter (Fig. 1H). The reduced myofiber size suggests postnatal growth defects. Thus, Lkb1 deletion resulted in both developmental and postnatal growth defects in skeletal muscles.
MyoD-Lkb1 mice develop a severe myopathy
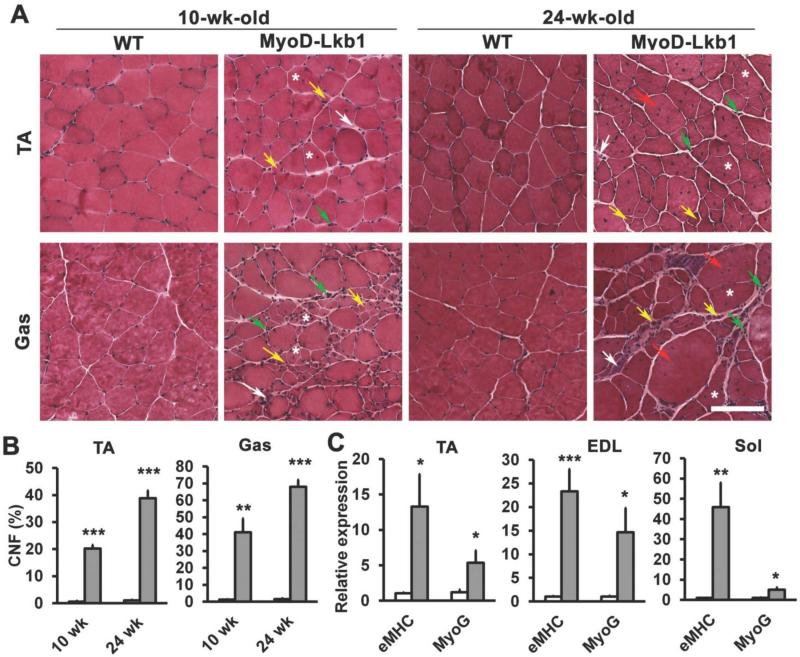
In addition to reduced mass, MyoD-Lkb1 muscles exhibited several striking characteristics of myopathy. These included the presence of numerous centrally nucleated myofibers (CNF) (Fig. 2A, asterisks). The percentage of CNF in the MyoD-Lkb1 TA and Gas muscles was 20% - 40% at 10-weeks, and 40% - 70% at 24-weeks, compared to only 1% - 2% in the WT muscles regardless of age (Fig. 2B). The MyoD-Lkb1 myofibers also exhibited an irregular pattern characterized by increased abundance of very small and very large fibers (Fig. 2A, also refer to the size distribution curve in Fig. 1H). In addition, the MyoD-Lkb1 muscles had higher levels of embryonic myosin heavy chain (eMHC/Myh3) and MyoG mRNA that is indicative of ongoing muscle regeneration (Fig. 2C), and possessed an increased presence of leaky myofibers that were labeled with Evans blue (EB) vital dye (Fig. S4A). Finally, the MyoDLkb1 muscles had elevated inflammatory infiltration manifested by an accumulation of interstitial mononuclear cells (Fig. 2A), an increased number of CD11b+ macrophages (Fig. S4B), and more deposition of interstitial fibrotic tissues (Fig. 2A; Fig. S4C). These results combined with the general kyphosis, reduced mobility and premature death of the mutant mice suggest that the MyoD-Lkb1 deletion results in a severe muscle pathology.
Figure 2.
Lkb1 deficiency results in severe myopathy. (A) Hematoxylin and eosin (H&E) staining of TA and Gas muscle cross sections at 10- and 24-wk-old. Characteristics of muscle pathology are indicated: centronuclear myofibers (asterisks), very small (yellow arrows) and very large (red arrows) myofibers, interstitial mononuclear cells (green arrows), fibrotic tissues (white arrows). (B) Percentage of centrally nucleated fibers (CNF) in TA and Gas muscles. n=4 for 10-wk-old mice, n=5 for 24-wk-old mice. (C) Relative expression of the embryonic muscle myosin heavy chain (eMHC/Myh3) and MyoG in different muscles at 10-wk-old. Error bars represent SEM. * P< 0.05, ** P< 0.01, *** P< 0.001. Scale bars: 100 μm.
Muscle dysfunction is often associated with reduced mitochondria content and fiber type switching. Deletion of Lkb1 reduced the expression of mitochondria-specific genes Pgc1a, Pgc1b, Cox7a1, Cox5b and Cox8b, especially in the oxidative Sol muscles (Fig. S4D-E), and the muscle fiber-type was shifted to a less oxidative state. In particular, Lkb1 deletion led to a shift of type I to IIA fibers in the Sol muscle (Fig. S4F-H), and a shift of type IIA to IIX or IIB fibers in the TA muscle (Fig. S4I). These results are consistent with the observation that Lkb1 cKO led to more robust reduction of oxidative muscles than glycolytic muscles (Fig. 1D). Taken together, the results indicate that the muscle pathology found in Lkb1 deficient mice is characterized by reductions of oxidative capacity and muscle fiber type switching towards the glycolytic phenotype.
Spontaneous activation of MyoD-Lkb1 null satellite cells in resting muscles
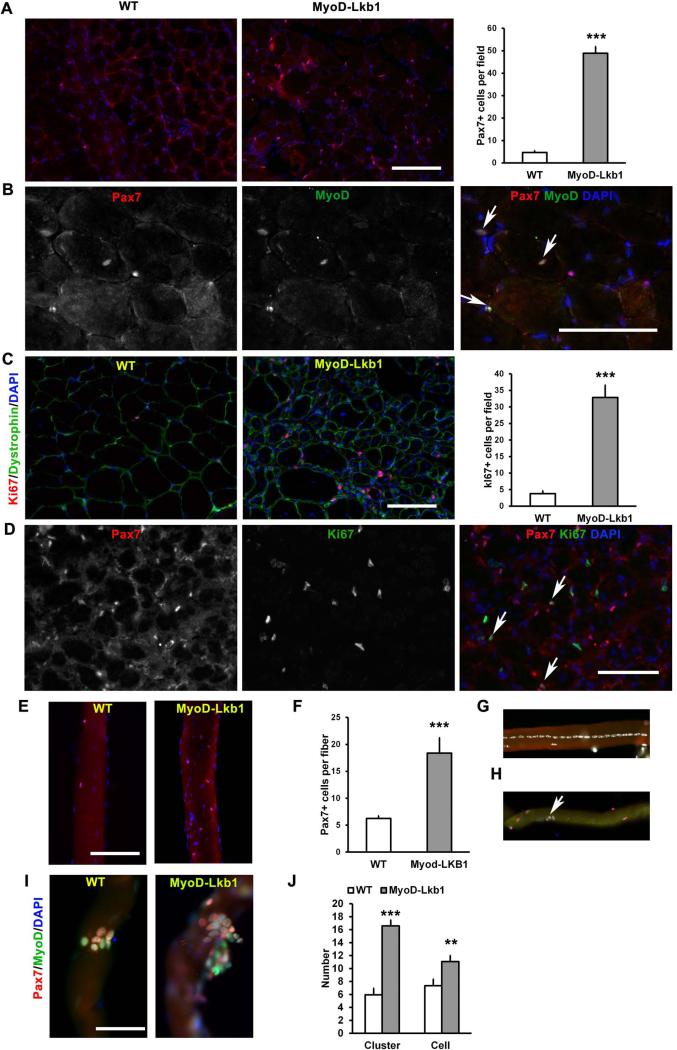
To investigate the function of Lkb1 in muscle stem cells, we examined satellite cells and their descendent myoblasts derived from the MyoD-Lkb1 mice. Notably, there were about 10 times more Pax7+ satellite cells in the resting muscles of MyoD-Lkb1 mice compared to WT mice (Fig. 3A). In addition, a significant portion of the Pax7+ cells co-expressed MyoD in the MyoD-Lkb1 muscles (Fig. 3B). By contrast Pax7+/MyoD+ cells were never found in resting WT muscles at this age (10-wk-old). As quiescent satellite cells are MyoD- and only acquire MyoD expression during activation [10-12], our observation indicates that a portion of satellite cells lose their quiescence and are activated to proliferate in resting muscles of the MyoD-Lkb1 mice. In agreement with this notion, we found that there were ~10 times more Ki67+ proliferating cells in Gas muscles of the MyoD-Lkb1 compared to the WT mice (Fig. 3C), and a significant portion of the Pax7+ cells co-expressed Ki67 in the MyoD-Lkb1 but not WT muscles (Fig. 3D). Together, these results indicate that deletion of Lkb1 spontaneously activates satellite cells, leading to an increased number of satellite cells in resting muscles of adult mice.
Figure 3.
Lkb1 deficient satellite cells are spontaneously activated in resting muscles. (A) Abundance of Pax7+ satellite cells in cross sections of Gastrocnemius muscles from 10-wk-old mice. n = 5. (B) A portion of Lkb1 null satellite cells co-express the activation marker MyoD (Pax7+/MyoD+, arrows) in MyoD-Lkb1 resting muscles at 10-wk-old. (C) Abundance of Ki67+ proliferating cells in 10-wk-old WT and MyoD-Lkb1 muscles. (D) A portion of Pax7+ cells co-expressed Ki67 (Pax7+/Ki67+, arrows) in MyoD-Lkb1 resting muscles at 10-wk-old. (E-F) Abundance of Pax7+ satellite cells on freshly isolated EDL myofibers. n = 5 pairs of mice (~50 myofibers were analyzed per mouse). (G) A representative centronucleated myofiber from resting EDL muscle of MyoD-Lkb1. (H) A portion of Pax7+ cells co-express MyoD in a freshly isolated MyoD-Lkb1 myofiber. (I-J) WT and MyoD-Lkb1myofibers cultured for 72 h showing clusters of myoblasts co-expressing Pax7 and MyoD (I), and quantification of clusters/fiber and cells/cluster (J). n = 3 pairs of mice (~25 myofibers were analyzed per mouse). Error bars represent SEM, ** P< 0.01, *** P< 0.001. Scale bars: 100 μm.
In parallel, we isolated fresh single fibers from the EDL muscles of the MyoD-Lkb1 and WT mice and stained with Pax7 to label satellite cells (Fig. 3E). Consistent with results from the whole muscle cross sectional analysis, there were more Pax7+ cells per fiber in the MyoD-Lkb1 EDL muscles (Fig. 3F). In addition, central nucleated myofibers were readily detected in the MyoD-Lkb1 preparations (Fig. 3G). Furthermore, a portion of Pax7+ satellite cells co-expressed MyoD in freshly isolated EDL fibers (Fig. 3H). The activation of MyoD in the absence of muscle regeneration (evident from the lack of centronuclei) indicates that these satellite cells are spontaneously activated (Fig. 3H). Upon culture, the satellite cells on the MyoD-Lkb1 EDL fibers proliferated faster than the WT satellite cells, indicated by the formation of larger and more clusters of Pax7+/MyoD+ cells (Fig. 3I-J). These results further demonstrate that Lkb1 deficiency activates quiescent satellite cells and promotes their proliferation.
Pax7CreER-mediated postnatal deletion of Lkb1 in satellite cells inhibits muscle regeneration
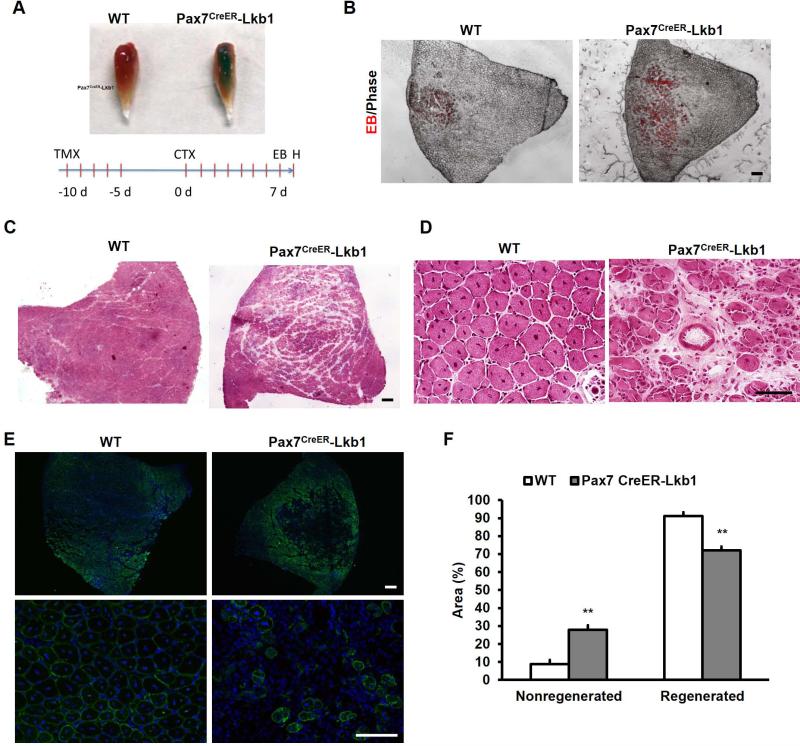
To examine if Lkb1 deletion affects satellite cell function in vivo, we induced muscle regeneration by injecting cardiotoxin (CTX) into TA muscles. We established Pax7CreER/Lkb1flox/flox mice (called Pax7CreER-Lkb1 henceforth) to drive deletion of Lkb1 specifically in postnatal satellite cells in an inducible manner. In the adult Pax7CreER-Lkb1 mice, tamoxifen (TMX) injection activates CreER in Pax7-expressing satellite cells, thereby bypassing the developmental and growth defects observed in the MyoD-Lkb1 mice. After TMX induction, TA muscles of WT and Pax7CreER-Lkb1 mice were injured with CTX, and muscles were examined 8 days after CTX treatment (Fig. 4A). One day prior to harvesting muscle, the mice were intraperitoneally (IP) injected with Evans blue (EB) dye, which accumulates in damaged muscle fibers and facilitates visualization of the non-regenerated myofibers (Fig. 4A). EB signal in the CTX-treated Pax7CreER-Lkb1 muscles was much more profound than in the WT muscles (Fig.4A-B), suggestive of poor regeneration. Consistently, H&E staining showed that the Pax7CreER-Lkb1 muscles were poorly repaired, and contained extensive empty space (Fig. 4C). Closer examination revealed that while the WT muscles were nicely regenerated with uniformly patterned centronuclear myofibers and little interstitial space, the Pax7CreER-Lkb1 muscles contained only a few regenerated fibers with a large number of empty spaces and many degenerated myofibers (Fig. 4D). In addition, while the WT muscles were completely filled with newly regenerated desmin+ myofibers near the center of injury, the Pax7CreER-Lkb1 muscles only had about half desmin+ myofibers (Fig. 4E). On the quantitative basis, a significantly smaller regenerated area but larger non-regenerated area was found in the TA muscles of Pax7CreER-Lkb1 mice compared to the WT mice (Fig. 4F). Together, these results provide compelling evidence that Lkb1 is necessary to maintain the regenerative capacity of satellite cells in vivo.
Figure 4.
Lkb1 deficiency impairs muscle regeneration. (A) Schematic of experimental design and representative images of whole TA muscles at Day 8 post CTX. Blue indicates Evans Blue (EB). (B) Representative TA muscle sections showing EB fluorescence (in Red). (C-D) Low and high magnification images of TA muscle sections stained with H&E. (E) Low and high magnification TA muscle sections stained with Desmin antibody to label newly regenerated myofibers. (F) Quantification of the regenerated and non-regenerated areas of TA muscle sections. Error bars represent SEM, n=5. ** P< 0.01. Scale bars: 100 μm.
Deletion of Lkb1 promotes proliferation but inhibits differentiation of myoblasts
The elevated satellite cell number but reduced muscle regeneration suggests that Lkb1 null satellite cells have imbalanced proliferation/differentiation kinetics. To explore this possibility, we first examined proliferation and differentiation of satellite cells in regenerating muscles of Pax7CreER-Lkb1 mice. At Day 7 after CTX treatment, there were ~60% more Pax7+ cells in the Pax7CreER-Lkb1 compared to the WT muscles (Fig. S5A). Likewise, there were roughly twice as many Ki67+ cells in the Pax7CreER-Lkb1 as in the WT muscles (Fig. S5B). By contrast, the number of MyoG+ differentiating myoblasts was reduced by nearly 3 times in the Pax7CreER-Lkb1muscles (Fig. S5C). Therefore, Lkb1 deletion promotes proliferation and inhibits differentiation of satellite cells in vivo.
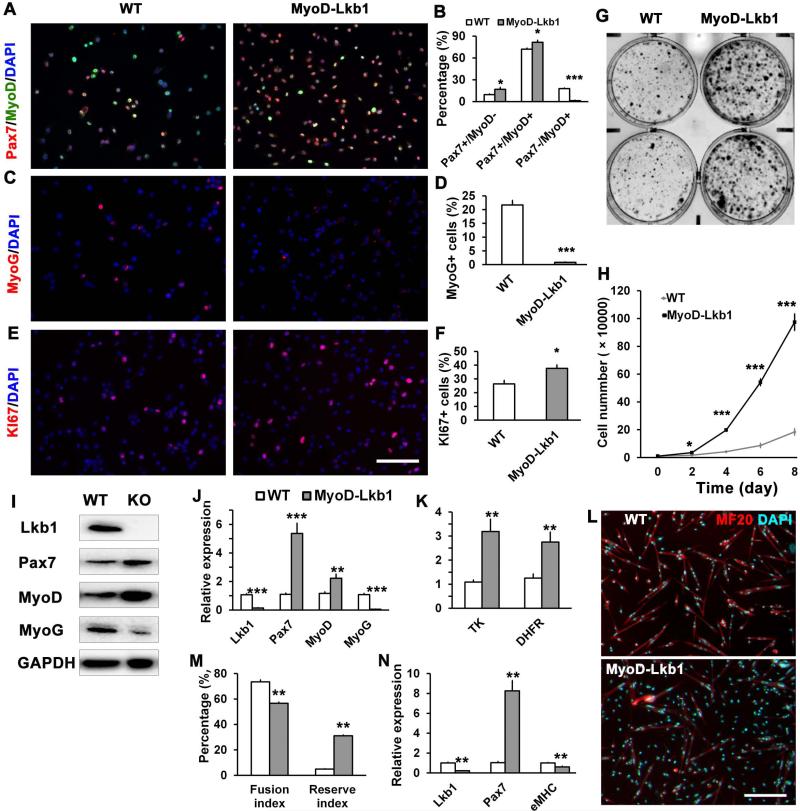
We further examined satellite cell-derived primary myoblasts isolated from MyoD-Lkb1 mice and WT littermates. Previous studies have established that Pax7+/MyoD-, Pax7+/MyoD+ and Pax7-/MyoD+ specifically mark the self-renewing, proliferating, and differentiating myoblasts, respectively [10, 11, 60]. Following this paradigm, we examined relative expression pattern of Pax7 and MyoD based on immunofluorescence (Fig. 5A). Knockout of Lkb1 increased the proportion of self-renewing (Pax7+/MyoD-) and proliferating (Pax7+/MyoD+) myoblasts, but diminished the differentiating (Pax7-/MyoD+) myoblasts (Fig. 5B). Using myogenin (MyoG) as an alternative differentiation marker, we found that Lkb1 null myoblasts had reduced abundance of MyoG+ cells (Fig. 5C-D). We also assessed myoblast proliferation using Ki67 as a marker, and found that there were a higher percentage of Ki67+ cells in the MyoD-Lkb1 compared to WT myoblast cultures (Fig. 5E-F). Analysis of colony formation (Fig. 5G) and cell growth curve (Fig. 5H) further confirmed that MyoD-Lkb1 deletion increased the proliferation and colonization of myoblasts. Western blot analysis confirmed that the protein levels of Pax7 and MyoD were higer, while MyoG was loweer, in the MyoD-Lkb1 than WT myoblasts (Fig. 5I). Moreover, compared WT myoblasts, MyoD-Lkb1 myoblasts expressed higher levels of proliferation (TK and DHFR) and myogenic progenitor (Pax7 and MyoD) marker genes, but lower levels of differentiation gene MyoG (Fig. 5J-K). Upon serum withdrawal induced differentiation, MyoD-Lkb1 myoblasts differentiated and fused less efficiently (Fig. 5L). This is apparent from their lower differentiation and fusion indices but higher percentage of non-differentiated reserve cells (Fig. 5M). Gene expression analysis confirmed that differentiated MyoD-Lkb1 myoblasts expressed higher levels of Pax7 but lower levels of eMHC (Fig. 5N). Similar defects were observed in Pax7CreER-Lkb1 myoblasts (Fig. S6). Collectively, these results suggest that Lkb1 regulate muscle progenitor cell fate through maintaining homeostasis of self-renewal, proliferation and differentiation.
Figure 5.
Lkb1 deletion promotes proliferation but inhibits differentiation of satellite cell derived primary myoblasts. (A-B) Representative images of primary myoblasts from WT and MyoD-Lkb1 mice labeled with Pax7 (red) and MyoD (green), and the percentage of quiescent (Pax7+MyoD-), proliferating (Pax7+MyoD+) and differentiating (Pax7-MyoD+) cells. Nuclei were stained with DAPI (blue). (C-D) Myoblasts stained with differentiation marker MyoG (red) and DAPI; and percentage of MyoG+ cells (D). (E-F) Myoblasts stained with proliferating marker Ki67 (red) and DAPI, the percentage of Ki67+ cells. (G) Colony assay comparing growth of WT and MyoD-Lkb1 myoblasts. (H) Growth curve analysis of the cultured WT and MyoD-Lkb1 myoblasts. (I) Western blot analysis of protein extracts from WT and MyoD-Lkb1 myoblasts. (J-K) mRNA levels of the myogenic (J) and proliferation (K) marker genes in the WT and MyoDLkb1 myoblasts. (L-N) WT and MyoD-Lkb1 myoblasts were differentiated for 3 days. Shown are the myotube morphology (indicated by MF20 staining, Red), fusion index (% nuclei in mytubes), and reserve cell index (% nuclei that are MF20-), and relative mRNA levels of Lkb1, Pax7 and eMHC. Error bars represent SEM, n = 5-7. * P< 0.05, ** P< 0.01, *** P< 0.001. Scale bars: 100 μm.
Lkb1 regulates progenitor cell proliferation through AMPK-mTOR pathway
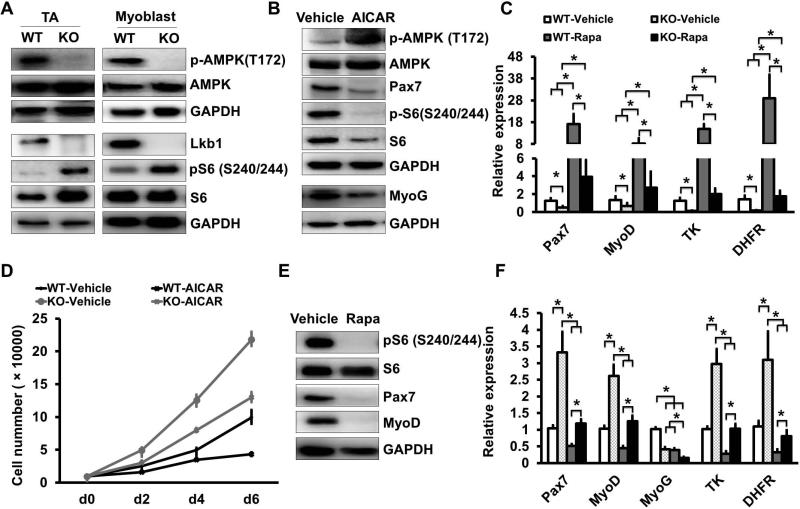
To understand the molecular mechanisms through which Lkb1 regulates muscle stem cells, we examined the canonical AMPK-mTOR pathway downstream of Lkb1 [47]. Lkb1 phosphorylates and activates AMPK, which in turn inhibits the mTOR pathway [35, 41-43]. Indeed, phosphorylated AMPK (pAMPK, T172) levels were lower in both whole muscle and cultured myoblasts of MyoD-Lkb1 than WT mice (Fig. 6A). Consequently, the total and phosphorylated levels of S6 (S240/244), a surrogate downstream target of mTOR, were increased in MyoD-Lkb1 myoblast (Fig. 6A). Next, we examined if AMPK activation can rescue the proliferation of MyoD-Lkb1 myoblasts. The AMPK activator AICAR significantly increased pAMPK level and reduced the levels of Pax7, MyoD, pS6 and total S6 proteins in WT myoblasts (Fig. 6B). Importantly, AICAR treatment rescued the expression of Pax7, MyoD, TK and DHFR in MyoD-Lkb1 myoblasts (Fig. 6C), and restored the growth rate of the Lkb1 deficient myoblasts to similar levels of untreated WT myoblasts (Fig. 6D). We further examined if inhibition of mTOR can similarly rescue the proliferation of MyoD-Lkb1 myoblasts. When WT myoblasts were treated with the mTOR inhibitor rapamycin (Rapa), pS6, Pax7 and MyoD were significantly decreased (Fig. 6E). Consistently, mTORflox/flox myoblast treated with adeno-Cre, or Pax7CreER/mTORflox/flox myoblast treated with 4-OH-TMX had significantly lower levels of Pax7, MyoD, TK and DHFR expression (Fig. S7A-B). Notably, mTOR inhibition restored Pax7, MyoD, TK and DHFR expression in MyoD-Lkb1 myoblasts to similar levels of untreated WT myoblasts (Fig. 6F), and rescued the proliferation of the Lkb1 null myoblasts (Fig. S7C-E). Taken together, these results demonstrate that Lkb1 acts through the AMPK-mTOR pathway to regulate myoblast proliferation.
Figure 6.
Lkb1 regulates myoblast proliferation through AMPK-mTOR pathway. (A) Western blots showing relative levels of phosphorylated AMPK (pAMPK, T172) and pS6 (S240/244) in WT and MyoD-Lkb1 muscles and myoblasts. (B) AMPK activator AICAR (1 mM) treatment increased pAMPK level and reduced the levels of Pax7, MyoD, pS6 and total S6 proteins. (C) AICAR treatment rescued Pax7, MyoD, TK and DHFR expression in MyoD-Lkb1 myoblasts. (D) Growth curve of the WT and MyoD-Lkb1 myoblasts after treated with AICAR. (E) mTOR inhibitor rapamycin (Rapa, 10 nM) treatment decreased pS6, Pax7 and MyoD expression. (F) Rapa rescued expression of Pax7, MyoD, TK and DHFR in MyoD-Lkb1 myoblasts. Error bars represent SEM, n=5. P < 0.05 between bars labeled by different letters (a, b, c and d); * P< 0.05, ** P< 0.01.
Lkb1 phosphorylates GSK-3β at Ser9 to regulate myogenic differentiation
We further examined if the reduced differentiation of Lkb1 null myoblasts is also mediated by the AMPK-mTOR pathway. Contrary to our expectation, activation of AMPK with AICAR reduced MyoG and eMHC expression in WT myoblasts, and failed to rescue MyoG and eMHC expression in MyoD-Lkb1 myoblasts (Fig. S8A-B). Furthermore, inhibition of mTOR with Rapa or by Cre-mediated deletion of mTOR reduced MyoG expression in WT myoblasts (Fig. S8C), and failed to rescue MyoG expression in MyoD-Lkb1 myoblasts (Fig. S8D). These data indicate that Lkb1 regulates MyoG expression and myogenic differentiation through an AMPK-mTOR independent pathway.
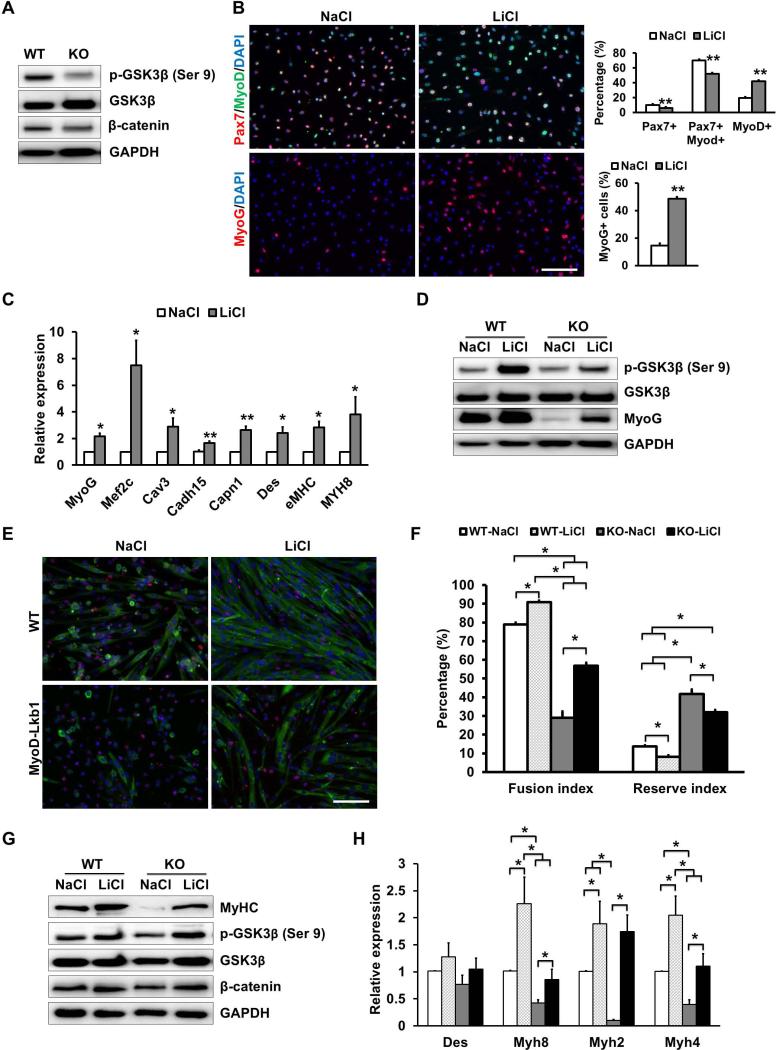
Lkb1 has been shown to phosphorylate GSK-3β at Ser9, leading to its inactivation [61, 62]. As GSK-3β is a key negative regulator of canonical Wnt signaling and myogenesis [19], we asked if Lkb1 regulates myoblast differentiation through GSK-3β. Interestingly, MyoD-Lkb1 myoblasts had significantly reduced levels of pGSK-3β (Ser9) (Fig. 7A), suggesting that the Lkb1 null myoblasts had elevated activation of GSK-3β. Next, we examined if inhibition of GSK3β with LiCl restores the differentiation of Lkb1 null myoblasts. In WT myoblasts, LiCl treatment significantly reduced the proportions of self-renewal (Pax7+) and proliferating cells (Pax7+MyoD+), while increasing the MyoD+ or MyoG+ differentiating cells (Fig. 7B). The mRNA levels of differentiation and fusion related genes (MyoG, Mef2c, Cav3, Cadh15, Capn1, Des, Myh3 and Myh8) were also upregulated by LiCl (Fig. 7C). Importantly, LiCl enhanced the phosphorylation of GSK3β (Ser9) and rescued MyoG expression in MyoD-Lkb1 myoblasts (Fig. 7D). Moreover, inhibition of GSK3β by LiCl promoted myotube formation in both WT and MyoD-Lkb1 myoblasts (Fig. 7E-F). Western blot and realtime PCR results confirmed that inhibition of GSK3β completely rescued the expression of differentiation marker genes in MyoD-Lkb1 myoblasts (Fig. 7G-H). These data demonstrate that Lkb1 regulates MyoG expression and myogenic differentiation through GSK3β.
Figure 7.
Lkb1 acts through GSK3β to regulate myogenic differentiation. (A) Relative protein levels of phosphorylated GSK3β (pGSK3β, Ser9) and β-catenin expression in WT and MyoDLkb1 myoblasts. (B) Morphology and percentage of quiescent (Pax7+MyoD-) and proliferating (Pax7+MyoD+) and differentiating (Pax7-MyoD+ or MyoG+) myoblasts treated with a GSK3β inhibitor (LiCl, 20 mM) or control (NaCl, 20 mM) media. (C) Expression of differentiation and fusion related genes after LiCl treatment. (D) LiCl rescued pGSK3 β and MyoG expression in MyoD-Lkb1 myoblasts. (E) MF20 staining (Red) showing differentiation efficiency of WT and MyoD-Lkb1 myoblasts cultures with or without LiCl. Scale bars: 100 μm. (F) LiCl partially rescued the differentiation efficiency (fusion index) of MyoD-Lkb1 myoblasts. (G-H) LiCl rescued the expression of differentiation marker genes in Lkb1 null myotubes. Error bars represent SEM, n=5. P< 0.05 between bars labeled by different letters (a, b, c and d), * means P< 0.05, ** P< 0.01.
Discussion
Previous studies involving conditional inactivation of Lkb1 in post-mitotic myofibers have provided insights into the role of Lkb1 in mature muscle physiology and metabolism [49, 63-66]. Overall, the reported phenotypes of myofiber-specific Lkb1 inactivation appear to be mild or even beneficial, ranging from compromised motor performance and fatty acid oxidation to improved glucose utilization and insulin sensitivity [49, 51, 63, 65]. However, how Lkb1 regulates muscle stem and progenitor cell function in vivo has been unknown prior to this study.
Our study provides the first evidence for an indispensable role of Lkb1 in skeletal muscle development and muscle stem cell function. We used two different approaches (Cre drivers) to achieve muscle progenitor/stem cell specific deletion of Lkb1. First, MyoDCre was used to drive deletion of Lkb1 in embryonic myogenic lineage cells that give rise to all skeletal muscles and muscle satellite cells [59, 67]. We confirmed the specificity of the MyoDCre driver by demonstrating Lkb1 deletion in skeletal muscles and myoblasts, but not in adipose tissue, heart and several other tissues. Hence, in the MyoD-Lkb1 mice, all myogenic progenitors and their descendant myofibers should have the Lkb1 gene inactivated at around E10.5 when embryonic myogenesis begins [59]. In this regard, the severe myopathy observed in the postnatal MyoDLkb1 mice can be attributed to defects in myogenic stem cells and/or post-mitotic myofibers. Given the relatively mild and late onset of muscle dysfunction that has been reported for the myofiber specific deletion of Lkb1 [50], the severe myopathy in the MyoD-Lkb1 mice that we observed in this study was most likely caused by malfunction of muscle progenitors, supported by reduced number of MyoG+ cells in somites of MyoD-Lkb1 embryos at E10.5. Combined with smaller body weight at birth and reduced fiber number, these results demonstrate that Lkb1 is necessary for proper embryonic myogenesis. We further used Pax7CreER to deplete Lkb1 specifically in Pax7-expressing satellite cells upon TMX induction in the adult. This approach bypassed any developmental defects due to ablation of Lkb1 in embryonic myoblasts and allowed us to dissect the role of Lkb1 in postnatal satellite cells without confounding effects of Lkb1 deletions in myofibers. We found that satellite cells acutely depleted of Lkb1 had aberrant proliferation and differentiation kinetics, and failed to regenerate injured muscles. This observation provides compelling in vivo evidence that Lkb1 is necessary for the normal function of postnatal satellite cells.
The early onset myopathy phenotype observed in the MyoD-Lkb1 mice is quite intriguing. It has been reported that MCK-Cre mediated deletion of Lkb1 results in late onset skeletal muscle atrophy starting from the age of 7-months [50]. The old MCK-Lkb1 mice had about 20% reduction in body weight and compromised motor function, accompanied by reduced activation of mTOR, Pgc1a and mitochondrial protein content [50]. However, as MCK-Cre induces Lkb1 deletion in both skeletal and cardiac muscles, the MCK-Lkb1 mice also had dilated atria [50], suggesting dysfunction of the heart. Therefore, it is unknown whether the skeletal muscle myopathy in the old MCK-Lkb1 is due to secondary effects of heart dysfunction. More recently, Tanner et al used Myog-Cre to induce deletion of Lkb1 in the skeletal (but not cardiac) muscles, however the study did not examine if the mice developed late onset skeletal muscle myopathy [63]. In contrast, we found that MyoDCre mediated deletion of Lkb1 leads to severe myopathy with a high degree of centronuclear fibers, infiltration of CD11b+ macrophages and fibrosis. Notably, the MyoD-Lkb1 mice also developed kyphosis, a sign of severe muscle weakness, accompanied by abnormal posture of the hindlimbs and loss of motility. Importantly, the MyoD-Lkb1 mice were already smaller at birth, and the overt pathological phenotype was evident as early as 2-weeks leading to a greater than 50% reduction in body weight observed in 21-week-old individuals. These studies together suggest that actively cycling muscle progenitors and post-mitotic myofibers have different responses to Lkb1 mutation. The much more severe and earlier onset myopathy in the MyoD-Lkb1 mice suggests that Lkb1 plays more important role in myogenic progenitors and myogenesis than in differentiated/mature myofibers.
We found that Lkb1 deletion also results in muscle fiber type switching towards the glycolytic phenotype. It is well known that PGC1 and other mitochondria biogenesis related genes play important roles in muscle oxidative functions [68, 69]. Previous reports also demonstrate that muscle dysfunction is often linked to mitochondrial dysfunction and aberrant mTOR signal pathway [70, 71], and that MCK-Cre mediated deletion of Lkb1 affects the oxidation functions of skeletal muscles [51, 72]. Consistently, we found that Lkb1 deletion dramatically decreased the expression of mitochondrial genes including Pgc1a, Pgc1b, Cox7a1, Cox5b and Cox8b. Based on previous studies and our current work, we postulate that Lkb1 deletion decreases phosphorylation and activation of AMPK, which subsequently affects mitochondrial and β-oxidation related genes including PGC1a and Acc. Interestingly, inactivation of mTOR or ablation of Raptor, a key component of the mTOR1 complex, has been shown to cause metabolic dysfunction and result in muscle dystrophy [70, 71]. By contrast, we found an increased activation of the mTOR substrate protein pS6 in the MyoD-Lkb1 mice. Moreover, the myopathy was not rescued by rapamycin-mediated inhibition of mTOR (data not shown), indicating that activation of the mTOR pathway is not responsible for the muscle pathology in the MyoD-Lkb1 mice, and other downstream targets of Lkb1 may have mediated the phenotype. For example, the Lkb1 substrate SIK1 has been shown to regulate myogenesis through the class II HDACs [73-75]. Further studies are needed to elucidate the exact mechanism that lead to the myopathy in the MyoD-Lkb1 mice.
Our finding that deletion of Lkb1 promotes the proliferation but inhibits differentiation of muscle stem cells highlights the function of Lkb1 in cell cycle regulation. Previous reports have shown that Lkb1 mutation stimulates the proliferation of β cells [38], peripheral T cells [39], mesenchymal cells [76], epithelial cells [77] and other cell types [78]. The finding that Lkb1 null satellite cells fail to maintain quiescence in uninjured muscle mimics the role of Lkb1 in hematopoietic stem cells (HSC). Lkb1 ablation leads to an immediate loss of HSC quiescence and increased multipotent progenitors but reduced long-term self-renewal HSC [35, 41-43, 45, 46]. Lkb1 is known as a regulator of AMPK and the mTOR pathway [41] and our results demonstrate that Lkb1 regulates the proliferation of muscle stem cells through this pathway. Deletion/inhibition of Lkb1 in mature myofibers also led to a significant reduction of AMPK activation, or AMPKμ2 subunit phosphorylation [49, 63-66]. Consistently, previous reports have shown that deletion of Lkb1 acts through the mTOR pathway to regulate pancreatic β cell proliferation [38]. However, Lkb1 deletion increased epithelial cell proliferation via AMPK dependent and independent pathways [77]. Similarly, Lkb1 regulates HSC quiescence through an AMPK-, mTORC1-, and FoxO-independent mechanism [35, 41-43, 45, 46]. We show here that Lkb1 acts through GSK-3β to regulate myogenic differentiation. As GSK-3β is a key component of WNT signaling pathway, if Lkb1-mediated phosphorylation of GSK-3β alters WNT signaling transduction during myogenesis has yet to be elucidated.
In light of the critical role played by Lkb1 in muscle stem cell proliferation and differentiation, it is not surprising that satellite cell specific ablation of Lkb1 abolishes the regenerative capacity of injured muscles. Although Lkb1 mutation spontaneously activates satellite cells and promotes their proliferation, the activated satellite cells are defective in myogenic differentiation. Specifically, the seemingly increased number of satellite cells in the young MyoD-Lkb1 mice not only fails to promote regeneration, but on the contrary leads to defective regeneration of injured muscles. This observation highlights the importance of temporal control of different signaling pathways during myogenesis. For example, activation of canonical Wnt signaling during the late differentiation phase, but not the early proliferation phase, of myogenesis is essential for proper muscle regeneration [79].
Conclusion
In this study we demonstrate that Lkb1 plays a critical role in myogenesis and muscle stem cell function. Muscle progenitor cell-specific deletion of Lkb1 during development leads to striking muscle pathology and premature postnatal death while adult satellite cell specific deletion of Lkb1 disrupts satellite cell homeostasis and prevents the regeneration of injured muscles. We further discovered that Lkb1 employs two separate mechanisms to regulate satellite cell homeostasis: while Lkb1 regulates proliferation through the classical AMPK/mTOR pathway, it controls muscle differentiation through GSK-3β. These results establish Lkb1 as a novel regulator of myoblast proliferation and differentiation.
Supplementary Material
Acknowledgement
The project was partially supported by funding from NIH (R01AR060652) and an incentive grant from Purdue University Office of Vice President for Research (OVPR) to SK. We thank Jun Wu and Cindy Yang for mouse colony maintenance and technical support; Xiaoqi Liu (Biochemistry, Purdue University) for sharing reagents (antibodies); and Paul Collodi for critical comments.
Footnotes
Author contributions
TS and SK conceived the research; TS, PZ, XL, PB, and FY performed the experiments; TS, PZ, SK analyzed the data; TS, SK wrote the paper.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Sambasivan R, Yao R, Kissenpfennig A, et al. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- 2.Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy MM, Lawson JA, Mathew SJ, et al. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins CA, Olsen I, Zammit PS, et al. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Montarras D, Morgan J, Collins C, et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 6.Kuang S, Kuroda K, Le Grand F, et al. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang YX, Rudnicki MA. Satellite cells, the engines of muscle repair. Nat Rev Mol Cell Biol. 2012;13:127–133. doi: 10.1038/nrm3265. [DOI] [PubMed] [Google Scholar]
- 8.Brack AS, Rando TA. Tissue-specific stem cells: lessons from the skeletal muscle satellite cell. Cell Stem Cell. 2012;10:504–514. doi: 10.1016/j.stem.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuang S, Rudnicki MA. The emerging biology of satellite cells and their therapeutic potential. Trends Mol Med. 2008;14:82–91. doi: 10.1016/j.molmed.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Zammit PS, Golding JP, Nagata Y, et al. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olguin HC, Olwin BB. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev Biol. 2004;275:375–388. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halevy O, Piestun Y, Allouh MZ, et al. Pattern of Pax7 expression during myogenesis in the posthatch chicken establishes a model for satellite cell differentiation and renewal. Dev Dyn. 2004;231:489–502. doi: 10.1002/dvdy.20151. [DOI] [PubMed] [Google Scholar]
- 13.Wen Y, Bi P, Liu W, et al. Constitutive Notch activation upregulates Pax7 and promotes the self-renewal of skeletal muscle satellite cells. Mol Cell Biol. 2012;32:2300–2311. doi: 10.1128/MCB.06753-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuang S, Gillespie MA, Rudnicki MA. Niche regulation of muscle satellite cell self-renewal and differentiation. Cell Stem Cell. 2008;2:22–31. doi: 10.1016/j.stem.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 16.Bjornson CRR, Cheung TH, Liu L, et al. Notch Signaling Is Necessary to Maintain Quiescence in Adult Muscle Stem Cells. Stem Cells. 2012;30:232–242. doi: 10.1002/stem.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mourikis P, Sambasivan R, Castel D, et al. A Critical Requirement for Notch Signaling in Maintenance of the Quiescent Skeletal Muscle Stem Cell State. Stem Cells. 2012;30:243–252. doi: 10.1002/stem.775. [DOI] [PubMed] [Google Scholar]
- 18.Brack AS, Conboy MJ, Roy S, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 19.von Maltzahn J, Chang NC, Bentzinger CF, et al. Wnt signaling in myogenesis. Trends in Cell Biology. 2012;22:602–609. doi: 10.1016/j.tcb.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serrano AL, Baeza-Raja B, Perdiguero E, et al. Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab. 2008;7:33–44. doi: 10.1016/j.cmet.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Musaro A, Giacinti C, Borsellino G, et al. Stem cell-mediated muscle regeneration is enhanced by local isoform of insulin-like growth factor 1. P Natl Acad Sci USA. 2004;101:1206–1210. doi: 10.1073/pnas.0303792101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rocheteau P, Gayraud-Morel B, Siegl-Cachedenier I, et al. A Subpopulation of Adult Skeletal Muscle Stem Cells Retains All Template DNA Strands after Cell Division. Cell. 2012;148:112–125. doi: 10.1016/j.cell.2011.11.049. [DOI] [PubMed] [Google Scholar]
- 23.Montarras D, L'honore A, Buckingham M. Lying low but ready for action: the quiescent muscle satellite cell. Febs J. 2013;280:4036–4050. doi: 10.1111/febs.12372. [DOI] [PubMed] [Google Scholar]
- 24.Cheung TH, Quach NL, Charville GW, et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–U247. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemminki A, Markie D, Tomlinson I, et al. A serine/threonine kinase gene defective in Peutz-Jegheus syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 26.Jenne DE, Reimann H, Nezu J, et al. Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet. 1998;18:38–44. doi: 10.1038/ng0198-38. [DOI] [PubMed] [Google Scholar]
- 27.Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 28.Gao Y, Xiao Q, Ma H, et al. LKB1 inhibits lung cancer progression through lysyl oxidase and extracellular matrix remodeling. Proc Natl Acad Sci U S A. 2010;107:18892–18897. doi: 10.1073/pnas.1004952107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardie DG. The LKB1-AMPK pathway-friend or foe in cancer? Cancer Cell. 2013;23:131–132. doi: 10.1016/j.ccr.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Mirouse V, Swick LL, Kazgan N, et al. LKB1 and AMPK maintain epithelial cell polarity under energetic stress. J Cell Biol. 2007;177:387–392. doi: 10.1083/jcb.200702053. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Amin N, Khan A, St Johnston D, et al. LKB1 regulates polarity remodeling and adherens junction formation in the Drosophila eye. Proc Natl Acad Sci U S A. 2009;106:8941–8946. doi: 10.1073/pnas.0812469106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granot Z, Swisa A, Magenheim J, et al. LKB1 Regulates Pancreatic beta Cell Size, Polarity, and Function. Cell Metab. 2009;10:296–308. doi: 10.1016/j.cmet.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zagorska A, Deak M, Campbell DG, et al. New Roles for the LKB1-NUAK Pathway in Controlling Myosin Phosphatase Complexes and Cell Adhesion. Sci Signal. 2010:3. doi: 10.1126/scisignal.2000616. [DOI] [PubMed] [Google Scholar]
- 34.Karuman P, Gozani O, Odze RD, et al. The Peutz-Jegher gene product LKB1 is a mediator of p53-dependent cell death. Mol Cell. 2001;7:1307–1319. doi: 10.1016/s1097-2765(01)00258-1. [DOI] [PubMed] [Google Scholar]
- 35.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468:653–U669. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonaccorsi S, Mottier V, Giansanti MG, et al. The Drosophila Lkb1 kinase is required for spindle formation and asymmetric neuroblast division. Development. 2007;134:2183–2193. doi: 10.1242/dev.02848. [DOI] [PubMed] [Google Scholar]
- 37.Denning DP, Hatch V, Horvitz HR. Programmed elimination of cells by caspase-independent cell extrusion in C. elegans. Nature. 2012;488:226–230. doi: 10.1038/nature11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu A, Ng ACH, Depatie C, et al. Loss of Lkb1 in Adult beta Cells Increases beta Cell Mass and Enhances Glucose Tolerance in Mice. Cell Metab. 2009;10:285–295. doi: 10.1016/j.cmet.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 39.Tamas P, Macintyre A, Finlay D, et al. LKB1 is essential for the proliferation of T-cell progenitors and mature peripheral T cells. Eur J Immunol. 2010;40:242–253. doi: 10.1002/eji.200939677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai LP, Lilley BN, Sanes JR, et al. Lkb1/Stk11 regulation of mTOR signaling controls the transition of chondrocyte fates and suppresses skeletal tumor formation. P Natl Acad Sci USA. 2013;110:19450–19455. doi: 10.1073/pnas.1309001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krock B, Skuli N, Simon MC. The Tumor Suppressor LKB1 Emerges as a Critical Factor in Hematopoietic Stem Cell Biology. Cell Metab. 2011;13:8–10. doi: 10.1016/j.cmet.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 42.Gurumurthy S, Xie SZ, Alagesan B, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–U675. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gan BY, Hu JA, Jiang S, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–U125. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai D, Chen Y, Wang F, et al. LKB1 controls the pluripotent state of human embryonic stem cells. Cell Reprogram. 2012;14:164–170. doi: 10.1089/cell.2011.0068. [DOI] [PubMed] [Google Scholar]
- 45.Durand EM, Zon LI. STEM CELL The blood balance. Nature. 2010;468:644–645. doi: 10.1038/468644a. [DOI] [PubMed] [Google Scholar]
- 46.David R. STEM CELLS LKB1 maintains the balance. Nat Rev Mol Cell Bio. 2011:12. doi: 10.1038/nrm3032. [DOI] [PubMed] [Google Scholar]
- 47.Lizcano JM, Goransson O, Toth R, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. Embo J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ylikorkala A, Rossi DJ, Korsisaari N, et al. Vascular abnormalities and deregulation of VEGF in Lkb1-deficient mice. Science. 2001;293:1323–1326. doi: 10.1126/science.1062074. [DOI] [PubMed] [Google Scholar]
- 49.Koh HJ, Arnolds DE, Fujii N, et al. Skeletal muscle-selective knockout of LKB1 increases insulin sensitivity, improves, glucose homeostasis, and decreases TRB3. Molecular and Cellular Biology. 2006;26:8217–8227. doi: 10.1128/MCB.00979-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomson DM, Hancock CR, Evanson BG, et al. Skeletal muscle dysfunction in muscle-specific LKB1 knockout mice. J Appl Physiol. 2010;108:1775–1785. doi: 10.1152/japplphysiol.01293.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeppesen J, Maarbjerg SJ, Jordy AB, et al. LKB1 Regulates Lipid Oxidation During Exercise Independently of AMPK. Diabetes. 2013;62:1490–1499. doi: 10.2337/db12-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shan TZ, Liang XR, Bi PP, et al. Myostatin knockout drives browning of white adipose tissue through activating the AMPK-PGC1 alpha-Fndc5 pathway in muscle. Faseb J. 2013;27:1981–1989. doi: 10.1096/fj.12-225755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shan T, Liang X, Bi P, et al. Distinct populations of adipogenic and myogenic Myf5-lineage progenitors in white adipose tissues. J Lipid Res. 2013;54:2214–2224. doi: 10.1194/jlr.M038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bondesen BA, Jones KA, Glasgow WC, et al. Inhibition of myoblast migration by prostacyclin is associated with enhanced cell fusion. Faseb J. 2007;21:3338–3345. doi: 10.1096/fj.06-7070com. [DOI] [PubMed] [Google Scholar]
- 55.Yoshida N, Yoshida S, Koishi K, et al. Cell heterogeneity upon myogenic differentiation: down-regulation of MyoD and Myf-5 generates ‘reserve cells’. J Cell Sci. 1998;111:769–779. doi: 10.1242/jcs.111.6.769. [DOI] [PubMed] [Google Scholar]
- 56.Shan TZ, Liu WY, Kuang SH. Fatty acid binding protein 4 expression marks a population of adipocyte progenitors in white and brown adipose tissues. Faseb J. 2013;27:277–287. doi: 10.1096/fj.12-211516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meinen S, Barzaghi P, Lin S, et al. Linker molecules between laminins and dystroglycan ameliorate laminin-α2–deficient muscular dystrophy at all disease stages. The Journal of cell biology. 2007;176:979–993. doi: 10.1083/jcb.200611152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luukko K, Ylikorkala A, Tiainen M, et al. Expression of LKB1 and PTEN tumor suppressor genes during mouse embryonic development. Mechanisms of development. 1999;83:187–190. doi: 10.1016/s0925-4773(99)00050-7. [DOI] [PubMed] [Google Scholar]
- 59.Kanisicak O, Mendez JJ, Yamamoto S, et al. Progenitors of skeletal muscle satellite cells express the muscle determination gene, MyoD. Dev Biol. 2009;332:131–141. doi: 10.1016/j.ydbio.2009.05.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu W, Wen Y, Bi P, et al. Hypoxia promotes satellite cell self-renewal and enhances the efficiency of myoblast transplantation. Development. 2012;139:2857–2865. doi: 10.1242/dev.079665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ossipova O, Bardeesy N, DePinho RA, et al. LKB1 (XEEK1) regulates Wnt signalling in vertebrate development. Nature Cell Biology. 2003;5:889–894. doi: 10.1038/ncb1048. [DOI] [PubMed] [Google Scholar]
- 62.Asada N, Sanada K. LKB1-Mediated Spatial Control of GSK3 beta and Adenomatous Polyposis Coli Contributes to Centrosomal Forward Movement and Neuronal Migration in the Developing Neocortex. Journal of Neuroscience. 2010;30:8852–8865. doi: 10.1523/JNEUROSCI.6140-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tanner CB, Madsen SR, Hallowell DM, et al. Mitochondrial and performance adaptations to exercise training in mice lacking skeletal muscle LKB1. Am J Physiol Endocrinol Metab. 2013;305:E1018–1029. doi: 10.1152/ajpendo.00227.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miura S, Kai Y, Tadaishi M, et al. Marked phenotypic differences of endurance performance and exercise-induced oxygen consumption between AMPK and LKB1 deficiency in mouse skeletal muscle: changes occurring in the diaphragm. Am J Physiol-Endoc M. 2013;305:E213–E229. doi: 10.1152/ajpendo.00114.2013. [DOI] [PubMed] [Google Scholar]
- 65.Thomson M, Porter BB, Tall JH, et al. Skeletal muscle and heart LKB1 deficiency causes decreased voluntary running and reduced muscle mitochondrial marker enzyme expression in mice. Am J Physiol-Endoc M. 2007;292:E196–E202. doi: 10.1152/ajpendo.00366.2006. [DOI] [PubMed] [Google Scholar]
- 66.Sakamoto K, McCarthy A, Smith D, et al. Deficiency of LKB1 in skeletal muscle prevents AMPK activation and glucose uptake during contraction. Embo J. 2005;24:1810–1820. doi: 10.1038/sj.emboj.7600667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wood WM, Etemad S, Yamamoto M, et al. MyoD-expressing progenitors are essential for skeletal myogenesis and satellite cell development. Dev Biol. 2013;384:114–127. doi: 10.1016/j.ydbio.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin J, Wu H, Tarr PT, et al. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 69.Gerhart - Hines Z, Rodgers JT, Bare O, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC - 1α. The EMBO journal. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Risson V, Mazelin L, Roceri M, et al. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. Journal of Cell Biology. 2009;187:859–874. doi: 10.1083/jcb.200903131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bentzinger CF, Romanino K, Cloetta D, et al. Skeletal Muscle-Specific Ablation of raptor, but Not of rictor, Causes Metabolic Changes and Results in Muscle Dystrophy. Cell Metab. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 72.Thomson DM, Brown JD, Fillmore N, et al. LKB1 and the regulation of malonyl-CoA and fatty acid oxidation in muscle. American Journal of Physiology-Endocrinology and Metabolism. 2007;293:E1572–E1579. doi: 10.1152/ajpendo.00371.2007. [DOI] [PubMed] [Google Scholar]
- 73.Berdeaux R, Goebel N, Banaszynski L, et al. SIK1 is a class IIHDAC kinase that promotes survival of skeletal myocytes. Nat Med. 2007;13:597–603. doi: 10.1038/nm1573. [DOI] [PubMed] [Google Scholar]
- 74.Stewart R, Akhmedov D, Robb C, et al. Regulation of SIK1 abundance and stability is critical for myogenesis. P Natl Acad Sci USA. 2013;110:117–122. doi: 10.1073/pnas.1212676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McKinsey TA, Zhang CL, Lu JR, et al. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Katajisto P, Vaahtomeri K, Ekman N, et al. LKB1 signaling in mesenchymal cells required for suppression of gastrointestinal polyposis. Nat Genet. 2008;40:455–459. doi: 10.1038/ng.98. [DOI] [PubMed] [Google Scholar]
- 77.Lo B, Strasser G, Sagolla M, et al. Lkb1 regulates organogenesis and early oncogenesis along AMPK-dependent and -independent pathways. Journal of Cell Biology. 2012;199:1117–1130. doi: 10.1083/jcb.201208080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hezel AF, Gurumurthy S, Granot Z, et al. Pancreatic Lkb1 deletion leads to acinar polarity defects and cystic neoplasms. Molecular and Cellular Biology. 2008;28:2414–2425. doi: 10.1128/MCB.01621-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brack AS, Conboy IM, Conboy MJ, et al. A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell Stem Cell. 2008;2:50–59. doi: 10.1016/j.stem.2007.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.