Abstract
Reticulon 3 (RTN3) has previously been shown to interact with BACE1 and negatively regulate BACE1 activity. To what extent RTN3 deficiency affects BACE1 activity is an intriguing question. In this study, we aimed to address this by generating RTN3-null mice. Mice with complete deficiency of RTN3 grow normally and have no obviously discernible phenotypes. Morphological analyses of RTN3-null mice showed no significant alterations in cellular structure, although RTN3 is recognized as a protein contributing to the shaping of tubular endoplasmic reticulum. Biochemical analysis revealed that RTN3 deficiency increased protein levels of BACE1. This elevation of BACE1 levels correlated with enhanced processing of amyloid precursor protein at the β-secretase site. We also demonstrated that RTN3 deficiency in Alzheimer's mouse models facilitates amyloid deposition, further supporting an in vivo role of RTN3 in the regulation of BACE1 activity. Since it has been shown that RTN3 monomer is reduced in brains of Alzheimer's patients, our results suggest that long-lasting reduction of RTN3 levels has adverse effects on BACE1 activity and may contribute to Alzheimer's pathogenesis.
Keywords: Alzheimer's disease, APP, BACE1, neuritic plaques, Nogo, reticulon
Introduction
The reticulon (RTN) protein family is distinguished by the presence of C-terminal reticulon homolog domain (RHD; Oertle et al., 2003; Yan et al., 2006; Yang and Strittmatter, 2007). In human and rodents, this protein family consists of four RTN members (RTN1–4); each member has multiple spliced isoforms divergent through variable N-terminal sequences. RTN proteins are highly conserved and are present in plants, yeasts, vertebrates, and mammals (Oertle et al., 2003; Klinger et al., 2004; Nziengui et al., 2007). Genetic studies using a Saccharomyces cerevisiae model suggest that the prototypical function of RTN protein is likely to shape the tubular ER structure (Zurek et al., 2011). RTN proteins typically have the ω-shape membrane topology in which both N- and C-terminal domains face the cytosolic side, and the RHD appears to determine this topology (Voeltz et al., 2006; He et al., 2007).
The function of RTNs is evolving in multicell organisms and its cellular location is apparently not restricted to the ER compartment in mammals. Neuronal RTN3 is naturally localized in axons and growth cones (Hu et al., 2007; Deng et al., 2013) and RTN4, also known as Nogo, is found on the cell surface (Fournier et al., 2001; Dodd et al., 2005). A growing body of evidence suggests important roles of these proteins in various tissues. For example, RTN4 (Nogo), particularly Nogo-A, has been shown to play a role in inhibition of neurite outgrowth (Strittmatter, 2002; Schwab, 2004). Nogo-A levels are markedly elevated in hippocampi of patients with temporal lobe epilepsy, in brains and muscles of patients with ALS, and in schizophrenic patients (Teng and Tang, 2005; Yang and Strittmatter, 2007). Nogo-B, on the other hand, regulates ICAM-1-mediated leukocyte transmigration and acute inflammation (Di Lorenzo et al., 2011) and appears to be necessary for proper vascular remodeling (Acevedo et al., 2004).
Our interest in this family of proteins is due to the finding that RTNs interact with BACE1 and negatively modulate its processing of APP at the β-secretase site (He et al., 2004; Murayama et al., 2006; Wojcik et al., 2007). We have focused our studies on RTN3, which is richly expressed in neurons. Transgenic mice overexpressing RTN3 in neurons have reduced amyloid deposition in Alzheimer's mouse models (Shi et al., 2009b, 2013; Araki et al., 2013). However, it is not clear whether mice deficient in RTN3 have any significant effects on BACE1 activity.
To address this question, we generated mice with RTN3 deficiency by a standard gene-targeting approach. RTN3-null mice exhibit no obvious abnormality in growth or fertility. RTN3-null mice were used for biochemical and functional characterizations to evaluate the potential effect of RTN3 deficiency on protein expression of selected targets and on formation of cellular structures such as tubular ER. We also bred RTN3-null mice with an Alzheimer's mouse model that overexpresses neuronal mutant APP and presenilin-1 and compared amyloid deposition with or without RTN3 deficiency. Our results show for the first time that RTN3 deficiency elevates protein levels of BACE1 and enhances amyloid deposition.
Materials and Methods
Mouse strains, cell lines, and biochemistry.
RTN3-null mice were generated by targeted deletion of exons 4–9 as illustrated in the Results section. Genotyping of mice was conducted by either PCR or Southern blotting, or both if necessary. The PCR primers for genotyping are as follows: RTN3KO491-F, 5′-ACACTCTACCTGCACTGGACTT-3′; RTN3KO491-R, 5′-GCCAGAGGCCACTTGTGTAG-3′; mRTN3WT336F, 5′-GGTCATCAGCCACATGAAGA-3′; and mRTN3WT336R, 5′-GCCAGGACTGCACAGAGAA-3′.
Tg-APPsw/PSEN1ΔE9 mice (Tg-PA) were purchased from The Jackson Laboratory (stock #004462). RTN3−/−/APPsw/PSEN1ΔE9 mice (R3KOPA mice) were generated by crossing RTN3-null mice with APPsw/PSEN1ΔE9 mice. All mice in the study were maintained and used according to the protocols approved by the Institutional Animal Care and Use Committee at the Cleveland Clinic Foundation.
Antibodies against β-amyloid peptide (6E10; catalog #NE1003-100UL, RRID:AB_564201), calnexin (C4731, RRID: AB_476845), β-actin (A5441, RRID: AB_476744), and APP C terminus (A8717, RRID: AB_258409) were purchased from Sigma-Aldrich. All RTN antibodies were generated in the Yan lab. Antibodies R454 and R458 recognize the N terminus and C terminus of RTN3, respectively; R461 is an antibody specific to RTN4 C terminus and recognizes all RTN4-A, RTN4-B and RTN4-C isoforms (He et al., 2004). RTN1 and RTN2 antibodies were recently generated in the Yan lab and react with the C terminus of RTN1 or RTN2. Alexa Fluor 488- and 568-labeled secondary antibodies were purchased from Invitrogen. DAB (D5905) was purchased from Sigma-Aldrich. Avidin/Biotin Complex Elite Kits were purchased from Vector Laboratories. Complete protease inhibitor tablets were purchased from Roche Biosciences. Four to 12% Bis-Tris NuPAGE gels were purchased from Invitrogen.
Immunohistochemistry and immunoconfocal microscopy.
For Aβ plaque examination, 16 brain sagittal sections (160 μm apart) per mouse were immunolabeled. Aβ deposition was detected with primary antibody 6E10 (1:1000 dilution) and secondary antibody anti-mouse IgG conjugated with Alexa Fluor 568 (1:2000 dilution) as described previously (Shi et al., 2009b). Briefly, half brains of female mice (180 d old) were fixed with 4% paraformaldehyde and O.C.T. embedding and were sagittally sectioned at a thickness of 16 μm. For immunohistochemical staining, fixed brain sagittal sections were initially reacted with anti-Aβ monoclonal antibody 6E10 and subsequently detected by the complex of HRP-conjugated secondary antibody and DAB staining. For confocal experiments, either Alexa Fluor 488- or Alexa Fluor 568-labeled secondary anti-IgG antibody was used for detecting fluorescent signals. Images were examined and captured with a Leica confocal microscope. Aβ plaques in 16 sections per mouse brain were examined by an investigator blinded to treatment conditions. Group means were calculated based on counts of individual animals.
Quantification of Aβ peptides using ELISA.
Insoluble Aβ1–40 and Aβ1–42 were extracted from aggregated Aβ1–40 and Aβ1–42 in brain tissues by the guanidine hydrochloride method. The levels of Aβ1–40 and Aβ1–42 in the left cerebral cortex and hippocampus were separately analyzed using sandwich ELISA according to lab standards as previously described (He et al., 2004).
Western blot analysis of brain proteins.
For Western blotting, snap-frozen brain sections were homogenized on ice in 1% CHAPS extraction buffer containing complete protease inhibitors (Roche Bioscience) and 0.1 mm Na3VO4 for inhibiting phosphatase. The homogenates were rotated for 30 min at 4°C to ensure extraction of membrane proteins. After centrifugation at 15,000 × g for 120 min, supernatants were collected and protein concentration was measured with the BCA protein assay reagent (Pierce). Equal amounts of lysate proteins were resolved on 4–12% Bis-Tris NuPAGE gels, followed by standard Western blotting with antibodies specified above. Chemiluminescent signals were scanned and integrated density values were calculated with a chemiluminescent imaging system (Alpha Innotech).
Results
Generation of mice deficient in RTN3 and analyses of RTN3-null phenotypes
Previous studies showed that overexpression of RTN3 negatively modulates BACE1 activity (He et al., 2004; Kume et al., 2009; Shi et al., 2009a;2013; Araki et al., 2013). To determine whether RTN3 deficiency affects amyloid deposition, we generated RTN3-null mice. The mouse rtn3 gene is 56 kb long and contains 9 exons, with the highly conserved RHD domain encoded by exons 4–9 (Fig. 1A). To ensure disruption of RTN3 function, we designed a construct that removed exons 4–9 through homologous recombination. We predicted that expression of the C-terminally truncated RTN3 is unlikely to be functional, as the RHD domain is required for proper folding and membrane anchoring.
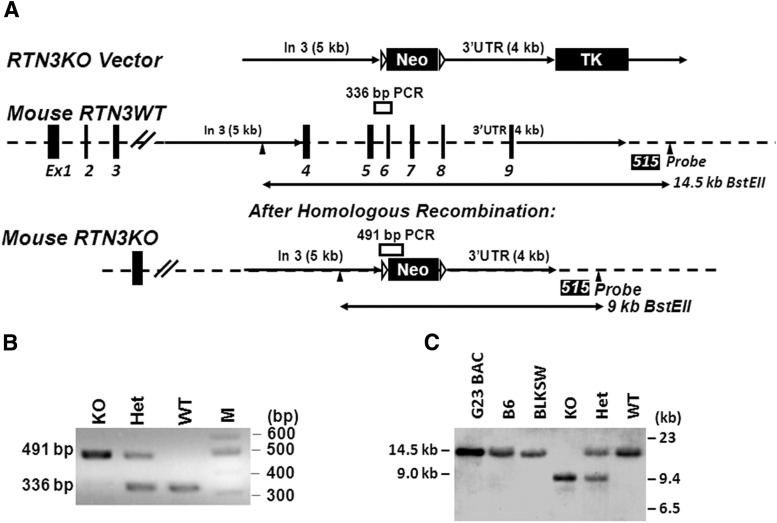
Figure 1.
Generation of RTN3-null mice. A, Schematic illustration of rtn3 gene organization and targeted deletion. The targeting RTN3KO vector was generated by insertion of 4 and 5 kb fragments as marked. The position of PCR genotyping and the probe (515) for Southern blot analysis with BstEII digestion is also specified. B, C, Examples of PCR-genotyping results and Southern blot confirmation of RTN3 deficiency (KO), RTN3 heterozygous (Het), and wild type (WT) are shown. Genomic DNA was prepared from mouse G23 BAC clone, mouse tails from C56BL/6 (B6), and BLKSW strains. All control genomic DNA produced the expected fragment of 14.5 kDa after Best EII digestion. RTN3KO mice produced the 9 kb DNA fragment as expected.
The targeting vector, named the RTN3KO vector, contained a 5′-side insertion of an ∼5 kb fragment, released after restriction enzyme digestion of a BAC DNA (Fig. 1A). The 4 kb fragment on the 3′-side downstream of exon 9 was generated by PCR amplification and inserted between the Neo and TK (thymidine kinase) cassettes. The fidelity of targeting sequences was confirmed by both restriction enzyme digestion analyses and DNA sequencing (data not shown). The validated targeting vector was microinjected into 129 SvEvTac blastocysts according to a standard knock-out strategy. We confirmed properly targeted deletion of the RTN3 gene in founder mice by PCR genotyping and Southern blot analyses. We showed that RTN3-null mice produced a 336 bp PCR fragment as expected while WT mice yielded a larger fragment of 491 bp (Fig. 1B). On Southern blots, genomic DNA from WT mice, after the BstEII digestion, produced a fragment of 14.5 kb while RTN3-null genomic DNA produced a fragment of 9 kb (Fig. 1C). Validated RTN3-null mice were maintained in the congenic C57BL/6 background by routine breeding strategies.
In the RTN family, only RTN4/Nogo knock-out mice were previously produced by three independent groups, and RTN4 KO mice are viable and normal in growth (Kim et al., 2003; Simonen et al., 2003; Zheng et al., 2003). We observed no obvious gross abnormalities among RTN3−/−, RTN3+/−, or RTN3+/+ mice at any age, similar to results seen in RTN4-null mice. The expected normal Mendelian inheritance was seen in the deletion of either one or both alleles of the rtn3 gene, excluding any developmental disadvantage. Overall body weight during various mouse growth stages as well as weights of various tissues such as the brain, heart, liver, pancreas, kidney, thymus, testis, and ovary showed no overt difference among various genotypes. Histopathological examination of various tissues, particularly brain tissues, revealed no readily discernible abnormalities (data not shown). Hence, RTN3-null mice appear similar to RTN4-null mice, which show no obvious growth defects.
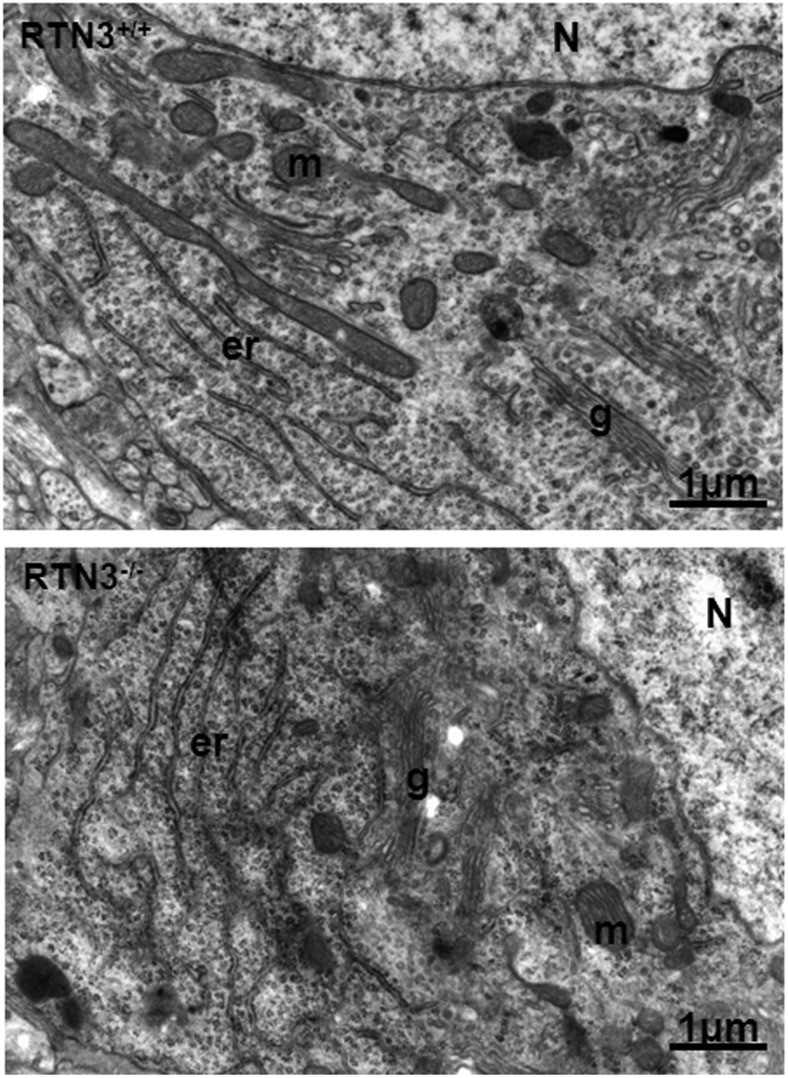
RTNs are considered to be important in shaping tubular ER structure and RTN3 is indeed predominantly localized in the tubular ER (Deng et al., 2013). We examined the ultrastructure of the ER in multiple RTN3-null mice by electron microscopy. It was clear that the loss of RTN3 alone had no effect on the ER tubular structure in our extensive examinations of various cell types (Fig. 2). Consistent with this observation, yeast cells with deletion of all RTN isoforms have normal tubular ER structure (Voeltz et al., 2006). Our results are in line with a prior finding that the effect of RTN on the formation of ER tubular structure is dispensable (Voeltz et al., 2006). Hence, deletion of RTN3 alone did not significantly disrupt the morphological structure of cells or neurons.
Figure 2.
Comparison of neuronal ultrastructure of mouse brains. RTN3-null mice and their WT littermates (2 months old) were fixed for examination by electron microscopy. ER structure is specified with “er,” and no obvious differences were observed between these two different genotype samples. N, nucleus; m, mitochondria; g, Golgi apparatus. Scale bar, 1 μm.
Analysis of RTN3 expression in mice
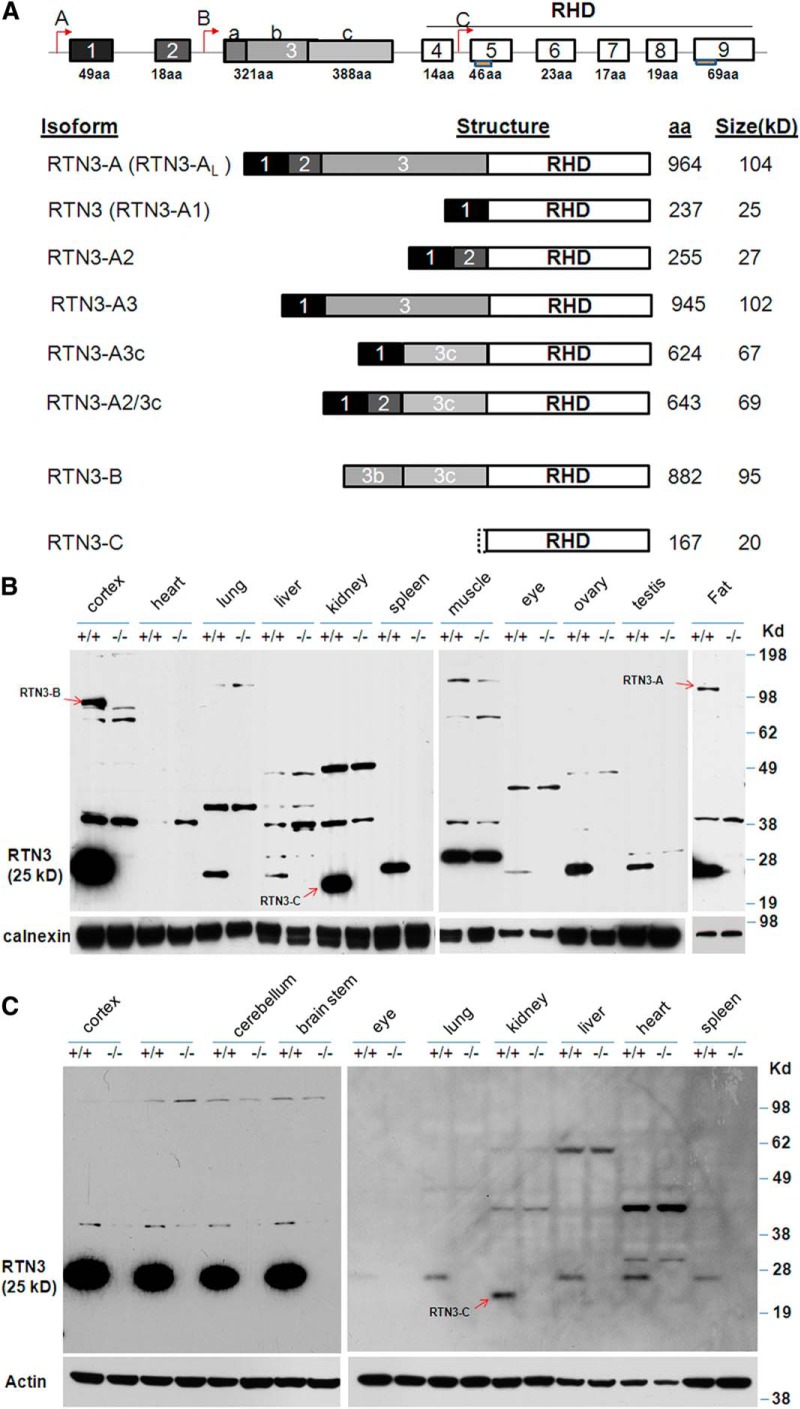
The initial report of RTN3 indicated the presence of only one RTN3 transcript, which encodes 236 aa in human and 237 aa in mice (Moreira et al., 1999). Further in silico searching of EST databases, analyses of the rtn3 genomic structure, and RT-PCR products identified additional splice variants (Cai et al., 2005; Di Scala et al., 2005). Based on published results and our own database analyses, we outlined the possible transcripts derived from mouse rtn3 gene in Figure 3. The nomenclature of each transcript is based on the published data regarding other RTN family members (Oertle and Schwab, 2003; Yan et al., 2006). Two additional translational initiation sites in the rtn3 gene are potentially present and are specified in Figure 3A. We assigned the translated product from the largest RTN3 transcript as “RTN3-A or RTN3-AL,” which starts with the first transcription site and contains all exons; the smallest “RTN3-C” is from RTN3 mRNA transcribed from the “C” initiation site and lacks exons 1–4.
Figure 3.
Expression of RTN3 variants in mouse tissues. A, Extensive in silico searches of the mouse EST database identified multiple spliced transcripts, which are illustrated and specified with a nomenclature consistent with other reticulon members. The transcriptional initiation sites are specified by arrows, and each exon is in a box. RTN3-A1, which contains exon 1 and the common exons 4–9, is the form that is commonly referred to as RTN3. Two transmembrane domains are specified with a boxed line under each relevant exon. B, Tissues were dissected from adult WT and RTN3-null littermates (6 months old in this case) and used for preparing protein lysates. On Western blots, antibody R458 recognizes the C-terminal 16 aa and is expected to recognize all RTN3 variants. Identified RTN3 variants other than RTN3-A1 are marked with arrows. Calnexin antibody was used for load control specification. C, P5 mice were used to prepare protein lysates for evaluation of developmental regulation of RTN3 expression. RTN3 (or RTN3-A1) is a predominant isoform. RTN3-C was only identified in the kidney. Antibody to actin was used to verify sample loading.
The RTN3-null mice allowed us to confirm the presence of various RTN3 products using antibodies specific to RTN3 peptide sequences. In adult mice (6 month olds in this example), the band migrating at ∼25 kDa was broadly expressed and should be translated from the RTN3-A1 transcript (containing exons 1 and 4–9; Fig. 3B). This product is often simply referred as RTN3 due to its much broader and abundant expression. The level of RTN3 in brains was the highest and was comparably weaker in peripheral tissues such as lung, liver, eye, ovary, and testis. We used our RTN3-null mice to confirm the absence of RTN3 in heart and skeletal muscle at this age; cross-reacted bands were present in both WT and RTN3 KO mice and therefore were nonspecific (Fig. 3B).
Expression of RTN3-AL, the expected largest transcript, was readily detected in fat tissues but not in other normal tissues (Fig. 3B). The band migrating near 95 kDa in WT but not RTN3 KO adult brain correlated with a product translated from the transcript RTN3-B; more sensitive RT-PCR experiments have previously identified RTN3-B in the mouse cortex, hippocampus, and cerebellum (Di Scala et al., 2005). The products from RTN3-A2/3c were barely detectable in brain samples and appeared to be ∼100-fold weaker than RTN3 (data not shown). In spinal cord, RTN3-A2, which contains exons 1, 2, and 4–9, was detectable and migrated expectedly near 27 kDa (data not shown). We failed to detect RTN3-A3 and RTN3-A3c transcripts, as such splicing events might be extremely rare under normal conditions. Overall, the brain produces more RTN3 variants than other tissues and its protein levels correlated with rtn3 gene alleles (data not shown).
To determine whether the expression of RTN3 is under transcriptional control during mouse development, we also examined samples from mice at different ages. In P5 animals, the 25 kDa RTN3 was again predominantly expressed in brain. Intriguingly, we detected RTN3 in P5 heart at a level comparable to other peripheral tissues (Fig. 3C), indicating that its expression in the heart is developmentally controlled and is switched off in the adult. In contrast, RTN3-B, which was detectable in adult brains, was not expressed in neonatal brains (Fig. 3C). RTN3-C was normally expressed in both adult and neonatal mouse kidney, suggesting the selective use of this alternative transcriptional initiation site. Together, our results show that the rtn3 gene produces different transcripts through various splicing events; all three transcriptional initiation sites are used and are tissue specific. Whether each RTN3 isoform has a specific function remains to be determined.
RTN3 deficiency increases BACE1 levels
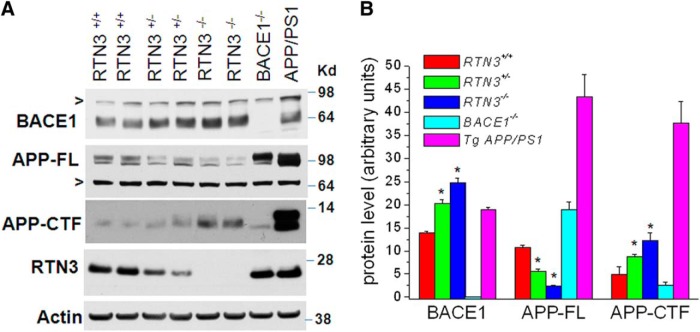
To determine whether RTN3 deficiency affects BACE1 activity, we first examined the expression of BACE1 in brain lysates. Protein levels of BACE1 were markedly increased in 6-month-old mouse brains with homozygous deletion of rtn3 and elevated BACE1 was also seen in RTN3 hemizygous mice (Fig. 4A). This observation was unexpected and was therefore repeatedly verified and quantified (Fig. 4B). In addition, the elevated BACE1 level was also detected in neonatal and young adult RTN3-null brains compared with their WT littermates (data not shown). RT-PCR analysis indicated that levels of BACE1 mRNA were not significantly altered among RTN3−/−, RTN3+/−, and RTN3+/+ mice (data not shown), suggesting that the elevation of BACE1 levels was due to a post-translational event.
Figure 4.
BACE1 levels are elevated in RTN3-null mice. A, Brain lysates were prepared from the indicated genotype and analyzed by Western blotting. BACE1-null mice and Tg-APPswe/PSEN1ΔE9 bitransgenic mice were used for verifying BACE1 expression and APP processing. Antibody R458 was used to verify expression of RTN3 in each mouse. BACE1 levels were elevated in heterozygous RTN3 mice, and this was more pronounced in RTN3-null mice. The nonspecifically reacted band is shown in “>”. Full-length APP and its processing products were detected using antibody A8717. B, Five independent experiments were performed to verify the elevation of BACE1 in RTN3-null mice (6-month-old mice, n = 5 experiments and 2 mice in each experiment, *p < 0.01, ANOVA test).
Increased BACE1 levels correlated with a reduction in full-length APP, consistent with increased processing of APP by elevated BACE1 levels (Fig. 4A). In WT mice, BACE1-cleaved APP fragments (APP-C99 and APP-C89) and α-secretase-cleaved APP fragment (APP-C83) were not readily separated due to low levels of endogenous APP. Transgenic mice produced high levels of APP-C99 due to overexpressing Swedish APP (Fig. 4A, control lane APP/PS1). Total C-terminal fragments of APP, which include APP-C83, APP-C89, and APP-C99, were higher in RTN3-null mice compared with WT control littermates and BACE1-null controls, which is consistent with the fact that elevated BACE1 favors processing of WT APP at the β′ site to generate APP-C89 (R. Wang et al., 1996; Cai et al., 2001; Luo et al., 2001; Huse et al., 2002; Liu et al., 2002; Qahwash et al., 2004), which migrates closer to C83 (α-secretase-cleaved product).
RTN3 deficiency increases amyloid deposition
To determine whether elevated BACE1 levels in RTN3-null mice affect amyloid deposition, we crossed RTN3-null mice with Tg-APPswe/PSEN1ΔE9 bitransgenic mice (Tg-PA mice) to generate RTN3−/−/APPswe/PSEN1ΔE9 compound mice (R3KOPA mice). Tg-PA mice express both the familial mutant PS1 with a deletion of exon 9 and the Swedish mutant APP under the control of a prion promoter, and this line of mice begins to develop amyloid deposition at the age of 5 months old (Borchelt et al., 1997). We examined amyloid deposition in female Tg-PA and R3KOPA mice at the age of 6 months (exactly 180 d). To minimize potential gender effects on Aβ production and amyloid deposition, we collected female mice for both biochemical and immunohistochemical examinations of Aβ generation and amyloid plaque density.
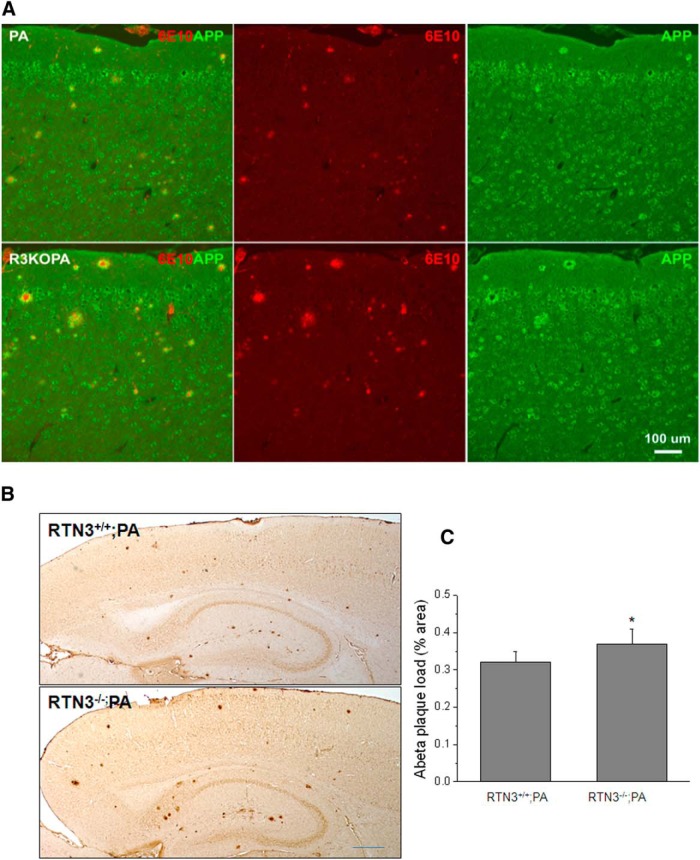
As demonstrated in Figure 5A, RTN3 deficiency did not affect general expression of APP transgene or its localization in neurons and dystrophic neurites. The size of amyloid plaques, detected by monoclonal antibody 6E10 that specifically reacts with Aβ N-terminal region, was visibly larger in R3KOPA mouse brains compared with Tg-PA brains. For further quantitative analysis of amyloid deposition, we conducted DAB staining of mouse brain sections with antibody 6E10 and used ImageJ software to quantify the area occupied by the condensed Aβ plaques. Our results show that the condensed Aβ plaque occupied 0.322 ± 0.0314% of the examined forebrain in Tg-PA mice, and this occupied area was increased to 0.374 ± 0.0414% in R3KOPA mice (increased by ∼16.1%; Fig. 5B,C; p < 0.05, n = 5, paired t test).
Figure 5.
Production of Aβ was increased in RTN3-null AD mice. A, Confocal experiments were performed to compare amyloid deposition in Tg-APPswe/PSEN1ΔE9 (PA) versus RTN3−/−/Tg-APPswe/PSEN1ΔE9 (R3KOPA) mouse brains. The size of amyloid plaques was often larger in R3KOPA mice when compared with PA mice. Scale bar, 100 μm. B, C, Brain sagittal sections were DAB stained with 6E10 antibody and 16 sagittal sections (160 μm apart) from each genotype were examined. The Aβ plaque load was determined by percentage of the area occupied by condensed deposits in the total examined area (*p < 0.05, n = 5, paired t test). Scale bar, 600 μm.
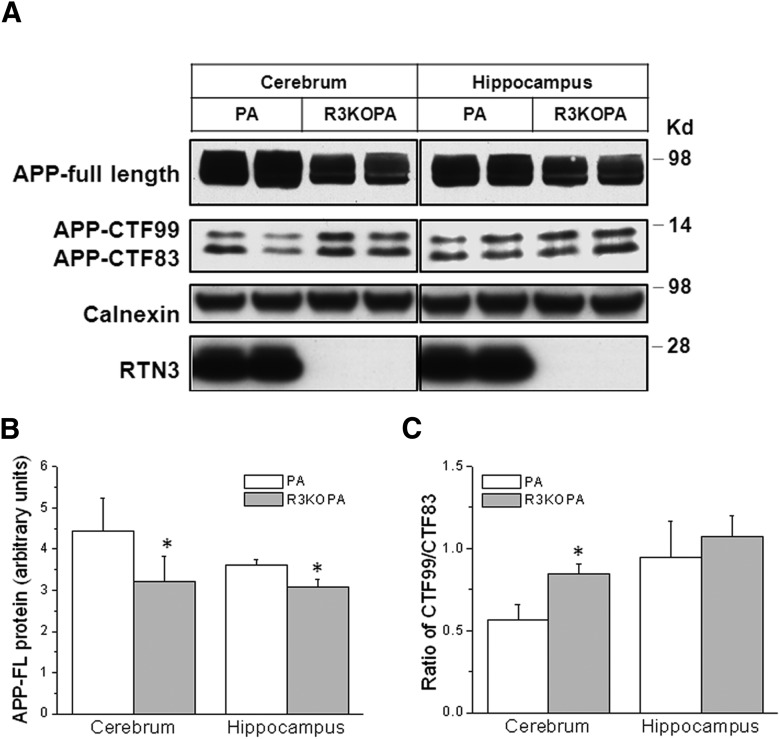
We also examined the effect of RTN3 deficiency on APP processing in the brains of R3KOPA mice by Western blotting. The levels of high molecular weight APP species, which include full-length APP and large secreted N-terminal APP fragments after α- or β-secretase cleavage, were reduced in both cortex and hippocampi of R3KOPA mice compared with PA controls (Fig. 6A,B). APP-C-terminal fragments were better separated in transgenic mice than in WT mice, because Swedish mutant APP is a favored substrate of BACE1, and much less C89 was produced. The levels of BACE1-cleaved CTF99 were elevated in cortical regions but less in hippocampus (Fig. 6A). Further quantitative measurement of CTF99 levels compared with CTF83 also suggested higher BACE1 activity in brains (Fig. 6C; 0.567 ± 0.089 in PA cortex vs 0.846 ± 0.061 in R3KOPA cortex; 0.943 ± 0.223 in PA hippocampus vs 1.073 ± 0.126 in R3KOPA hippocampus; n = 6 in total; *p < 0.05; Student's t test).
Figure 6.
Increased APP processing in R3KOPA mouse brains. A, Cortical and hippocampal protein lysates from 6-month-old PA and R3KOPA mice were examined by Western blotting. Antibody A8717 was used to detect APP and its BACE1-cleaved APP C-terminal fragment CTF99 and α-secretase-cleaved fragment CTF83. RTN3 deficiency was verified by antibody R458. Calnexin antibody was used to verify protein loading. B, The relative protein levels of full-length APP were normalized by comparing levels of calnexin and are shown in bar graphs. C, Relative BACE1 activity was manifested by the relative ratio of CTF99 to CTF83; higher BACE1 activity tends to produce more APP-C99 (n = 3 experiments, 2 mice in each experiment, *p < 0.05, ANOVA test).
We also extracted total Aβ from the specified mouse hippocampal samples by the guanidine hydrochloride method to measure Aβ1–40 and Aβ1–42 levels by sandwich ELISA. We found a small increase of both Aβ1–40 (40.3 ± 3.97 in Tg-PA vs 45.6 ± 3.71 pmol/g in R3KOPA) and Aβ1–42 levels (477.4 ± 40.38 in Tg-PA vs 536.9 ± 44.68 pmol/g in R3KOPA; p < 0.05, n = 5). Together, our immunohistochemical and biochemical results showed that RTN3 deficiency facilitated deposition of Aβ in brains of R3KOPA mice compared with Tg-PA mice.
Discussion
BACE1, a critical enzyme for generating β-amyloid peptides (Aβ), is being actively pursued as a therapeutic target for Alzheimer's disease (Vassar et al., 2009; Yan and Vassar, 2014). In additional to the chemical inhibition of BACE1, the regulation of BACE1 activity by cellular factors is of great interest for exploring alternative methods of inhibition. Increased expression of RTN3 has been shown to reduce generation of amyloid peptides via interacting with Alzheimer's BACE1 and to decrease Aβ generation (He et al., 2004; Murayama et al., 2006; Wojcik et al., 2007; Shi et al., 2009b, 2013; Araki et al., 2013); overexpressed RTN3 has also been linked to the development of hippocampal dystrophic neurites (Hu et al., 2007; Shi et al., 2009a), another pathological feature in AD brains. The critical remaining question is whether RTN3 deficiency has any effect on Aβ generation. In this study, we have used RTN3-null mice to address this question. We demonstrated that RTN3 deficiency increases expression of BACE1 and therefore enhances Aβ generation, further supporting an in vivo role of RTN3 in regulating BACE1 activity.
By using RTN3-null mice, we can ascertain the expression of RTN3 in various tissues by excluding nonspecifically reacted proteins on Western blots. This knowledge is important because different RTN3 variants might have specific biological functions in specific tissues or cells. Consistent with this concept, RTN4-B and RTN4-C variants appear to play specific roles in different tissues (Acevedo et al., 2004; Schanda et al., 2011; Tashiro et al., 2013). We have shown that RTN3 (also called RTN3-A1), similar to its binding partner BACE1, is strongly expressed in brain, mainly in various types of neurons, and is sparsely detected in resting astrocytes or microglia. However, significantly elevated expression of RTN3 was observed in hyperactive astrocytes (data not shown), suggesting a regulated event of gene expression. Future investigations should determine why RTN3 is expressed when astrocytes are activated. To our surprise, despite the presence of possible various splicing events in the mouse genome, we have identified only a few RTN3 alternatively spliced transcripts during normal mouse growth. Among brains and peripheral tissues that we have examined, RTN3-AL was weakly detected in adult but not neonatal mouse brains (Fig. 3). The reason for expressing this large RTN3 transcript is still unclear, as RTN3 (or called RTN3-A1) is a highly expressed RTN3 variant. RTN3-A2 was mainly detected in spinal cord samples but was hardly present in other brain regions. Alternative use of the transcriptional “B” site occurs in the normal adult brain as RTN3-B was found in adult but not neonatal brains (Fig. 3). Alternative use of transcriptional “C” site, which produces RTN3-C, was only found in the kidney. Apparently, expression of RTN3 variants occurs via multiple transcriptional regulations under certain conditions. It is expected that each RTN3 variant has its own unique biological function and this RTN3-null mouse model will be useful for revealing these individual functions.
While the specific biological functions of RTN3 variants remain poorly understood, genetic and biochemical studies suggest that RTN proteins are largely localized in the ER and that their general functions are likely to mediate the shaping of tubular ER (Voeltz et al., 2006). To determine whether RTN3 deficiency affects ER structure, we conducted ultrastructural examinations of RTN3-null mice by EM. Compared with their WT littermates, RTN3-null neurons exhibited no obvious alterations in the distinctive architecture of the ER (Fig. 2). More specifically, morphological ER structures of RTN3-null neurons or other tissues were not altered. Our data therefore suggest that RTN3 is dispensable for the formation of tubular ER. Since RTN3 family members, i.e., RTN1, are also expressed by neurons, these RTNs may potentially compensate for RTN3 deficiency and preserve the tubular ER structure in RTN3-null mice. Such a possibility will be further investigated when mice null for additional RTN members are available.
We have previously demonstrated that levels of RTN3 monomer are reduced in brains of Alzheimer's patients and that RTN3 is colocalized with BACE1 in various subcellular compartments of neurons (Hu et al., 2007; Kume et al., 2009; Heath et al., 2010). In RTN3-null mice, we unexpectedly observed that BACE1 protein levels were elevated (Fig. 4). This increase appears in different ages of RTN3-null mouse brains compared with their age-matched WT littermates, and more obvious in older than younger RTN3-null mice (data not shown). Consistently, we noticed that elevated BACE1 produced more APP-C89 in mice that express endogenous APP (Fig. 4). In brains of Alzheimer's patients, BACE1 protein levels are elevated (H. Wang et al., 2013). Likely, a reduction of RTN3 monomer in AD brains partially contributes to this BACE1 elevation. BACE1 is normally degraded via the ubiquitin-lysosome pathway (Qing et al., 2004; Tesco et al., 2007; Kang et al., 2010) and RTN3 overexpression reduces axonal localization of BACE1 (Deng et al., 2013). Our future studies will test hypotheses regarding whether RTN3 deficiency increases BACE1 levels in axons and reduces its degradation due to enriched ubiquitin-lysosome in soma and reduced ubiquitin-lysosome in axons.
In Tg-APPswe/PSEN1ΔE9 bitransgenic mice with RTN3 deficiency, amyloid deposition was increased (Fig. 5). However, this increase was not dramatic. Several possibilities may account for this modest effect. For example, this AD mouse model expresses Swedish mutant APP, which is preferentially processed by BACE1 due to the mutation in the β-secretase site. Small increases in BACE1 levels might have a weak effect on cleavage of Swedish mutant APP. In addition, BACE1 activity has already increased slightly in areas surrounding amyloid plaques in Alzheimer's mouse models (Kandalepas et al., 2013). The higher BACE1 levels in general reduce the likelihood of a large impact of small subsequent elevations in BACE1. On the other hand, RTN3 deficiency leads to a small elevation in RTN1 expression in RTN3-null mouse brains, and RTN1 is naturally expressed by many neurons (data not shown). Small elevations in RTN1 levels may somewhat counter the RTN3 deficiency. Although the effect was small, there is a clear tendency of RTN3 deficiency to facilitate BACE1 processing of its APP substrate and to regulate Aβ deposition. Alternatively, it is perhaps better to test whether ectopic expression of RTN3 via AAV-mediated gene delivery will reduce BACE1 activity and decrease Aβ generation.
RTN3, as a membrane integral protein, has been shown to affect various cellular functions. For example, RTN3 has been shown to mediate monocyte recruitment to the arterial wall and their transformation into macrophages in the pathogenesis of atherosclerosis (Chen et al., 2012) and to induce ER overload response (Kuang et al., 2005). Higher levels of RTN3 are found in astrocytoma tumor cells but not in glia cells of noncancerous brain tissues (Huang et al., 2004). Hence, the RTN3-null mouse model will also be a powerful tool for revealing the role of RTN3 in neurodegeneration and other human diseases.
Footnotes
This work is partially supported by National Institutes of Health grants (AG025493 and NS074256) and an award from Alzheimer's Association (NPSPAD-10-174543) to R.Y. We thank Drs. Qinyuan Fan and Hailong Hou for their discussions during the study and Dr. Chris Nelson for critical reading of this manuscript.
The authors declare no competing financial interests.
References
- Acevedo L, Yu J, Erdjument-Bromage H, Miao RQ, Kim JE, Fulton D, Tempst P, Strittmatter SM, Sessa WC. A new role for Nogo as a regulator of vascular remodeling. Nat Med. 2004;10:382–388. doi: 10.1038/nm1020. [DOI] [PubMed] [Google Scholar]
- Araki W, Oda A, Motoki K, Hattori K, Itoh M, Yuasa S, Konishi Y, Shin RW, Tamaoka A, Ogino K. Reduction of beta-amyloid accumulation by reticulon 3 in transgenic mice. Curr Alzheimer Res. 2013;10:135–142. doi: 10.2174/1567205011310020003. [DOI] [PubMed] [Google Scholar]
- Borchelt DR, Ratovitski T, van Lare J, Lee MK, Gonzales V, Jenkins NA, Copeland NG, Price DL, Sisodia SS. Accelerated amyloid deposition in the brains of transgenic mice coexpressing mutant presenilin 1 and amyloid precursor proteins. Neuron. 1997;19:939–945. doi: 10.1016/S0896-6273(00)80974-5. [DOI] [PubMed] [Google Scholar]
- Cai H, Wang Y, McCarthy D, Wen H, Borchelt DR, Price DL, Wong PC. BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat Neurosci. 2001;4:233–234. doi: 10.1038/85064. [DOI] [PubMed] [Google Scholar]
- Cai Y, Saiyin H, Lin Q, Zhang P, Tang L, Pan X, Yu L. Identification of a new RTN3 transcript, RTN3–A1, and its distribution in adult mouse brain. Brain Res Mol Brain Res. 2005;138:236–243. doi: 10.1016/j.molbrainres.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Chen Y, Xiang R, Zhao S. The potential role of RTN3 in monocyte recruitment and atherosclerosis. Mol Cell Biochem. 2012;361:67–70. doi: 10.1007/s11010-011-1089-6. [DOI] [PubMed] [Google Scholar]
- Deng M, He W, Tan Y, Han H, Hu X, Xia K, Zhang Z, Yan R. Increased expression of reticulon 3 in neurons leads to reduced axonal transport of beta site amyloid precursor protein-cleaving enzyme 1. J Biol Chem. 2013;288:30236–30245. doi: 10.1074/jbc.M113.480079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo A, Manes TD, Davalos A, Wright PL, Sessa WC. Endothelial reticulon-4B (Nogo-B) regulates ICAM-1-mediated leukocyte transmigration and acute inflammation. Blood. 2011;117:2284–2295. doi: 10.1182/blood-2010-04-281956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Scala F, Dupuis L, Gaiddon C, De Tapia M, Jokic N, Gonzalez de Aguilar JL, Raul JS, Ludes B, Loeffler JP. Tissue specificity and regulation of the N-terminal diversity of reticulon 3. Biochem J. 2005;385:125–134. doi: 10.1042/BJ20040458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd DA, Niederoest B, Bloechlinger S, Dupuis L, Loeffler JP, Schwab ME. Nogo-A, -B, and -C are found on the cell surface and interact together in many different cell types. J Biol Chem. 2005;280:12494–12502. doi: 10.1074/jbc.M411827200. [DOI] [PubMed] [Google Scholar]
- Fournier AE, GrandPre T, Strittmatter SM. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature. 2001;409:341–346. doi: 10.1038/35053072. [DOI] [PubMed] [Google Scholar]
- Heath JE, Siedlak SL, Zhu X, Lee HG, Thakur A, Yan R, Perry G, Smith MA, Castellani RJ. Widespread distribution of reticulon-3 in various neurodegenerative diseases. Neuropathology. 2010;30:574–579. doi: 10.1111/j.1440-1789.2010.01107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Lu Y, Qahwash I, Hu XY, Chang A, Yan R. Reticulon family members modulate BACE1 activity and amyloid-beta peptide generation. Nat Med. 2004;10:959–965. doi: 10.1038/nm1088. [DOI] [PubMed] [Google Scholar]
- He W, Shi Q, Hu X, Yan R. The membrane topology of RTN3 and its effect on binding of RTN3 to BACE1. J Biol Chem. 2007;282:29144–29151. doi: 10.1074/jbc.M704181200. [DOI] [PubMed] [Google Scholar]
- Hu X, Shi Q, Zhou X, He W, Yi H, Yin X, Gearing M, Levey A, Yan R. Transgenic mice overexpressing reticulon 3 develop neuritic abnormalities. EMBO J. 2007;26:2755–2767. doi: 10.1038/sj.emboj.7601707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Yang H, Zhou Y, Liu J, Yin B, Peng X, Qiang B, Yuan J. Overexpression of human reticulon 3 (hRTN3) in astrocytoma. Clin Neuropathol. 2004;23:1–7. [PubMed] [Google Scholar]
- Huse JT, Liu K, Pijak DS, Carlin D, Lee VM, Doms RW. Beta-secretase processing in the trans-Golgi network preferentially generates truncated amyloid species that accumulate in Alzheimer's disease brain. J Biol Chem. 2002;277:16278–16284. doi: 10.1074/jbc.M111141200. [DOI] [PubMed] [Google Scholar]
- Kandalepas PC, Sadleir KR, Eimer WA, Zhao J, Nicholson DA, Vassar R. The Alzheimer's beta-secretase BACE1 localizes to normal presynaptic terminals and to dystrophic presynaptic terminals surrounding amyloid plaques. Acta Neuropathol. 2013;126:329–352. doi: 10.1007/s00401-013-1152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang EL, Cameron AN, Piazza F, Walker KR, Tesco G. Ubiquitin regulates GGA3-mediated degradation of BACE1. J Biol Chem. 2010;285:24108–24119. doi: 10.1074/jbc.M109.092742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Li S, GrandPré T, Qiu D, Strittmatter SM. Axon regeneration in young adult mice lacking Nogo-A/B. Neuron. 2003;38:187–199. doi: 10.1016/S0896-6273(03)00147-8. [DOI] [PubMed] [Google Scholar]
- Klinger M, Diekmann H, Heinz D, Hirsch C, Hannbeck von Hanwehr S, Petrausch B, Oertle T, Schwab ME, Stuermer CA. Identification of two NOGO/RTN4 genes and analysis of Nogo-A expression in Xenopus laevis. Mol Cell Neurosci. 2004;25:205–216. doi: 10.1016/j.mcn.2003.09.021. [DOI] [PubMed] [Google Scholar]
- Kuang E, Wan Q, Li X, Xu H, Liu Q, Qi Y. ER Ca(2+) depletion triggers apoptotic signals for endoplasmic reticulum (ER) overload response induced by overexpressed reticulon 3 (RTN3/HAP) J Cell Physiol. 2005;204:549–559. doi: 10.1002/jcp.20340. [DOI] [PubMed] [Google Scholar]
- Kume H, Konishi Y, Murayama KS, Kametani F, Araki W. Expression of reticulon 3 in Alzheimer's disease brain. Neuropathol Appl Neurobiol. 2009;35:178–188. doi: 10.1111/j.1365-2990.2008.00974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Doms RW, Lee VM. Glu11 site cleavage and N-terminally truncated A beta production upon BACE overexpression. Biochemistry. 2002;41:3128–3136. doi: 10.1021/bi015800g. [DOI] [PubMed] [Google Scholar]
- Luo Y, Bolon B, Kahn S, Bennett BD, Babu-Khan S, Denis P, Fan W, Kha H, Zhang J, Gong Y, Martin L, Louis JC, Yan Q, Richards WG, Citron M, Vassar R. Mice deficient in BACE1, the Alzheimer's beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat Neurosci. 2001;4:231–232. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- Moreira EF, Jaworski CJ, Rodriguez IR. Cloning of a novel member of the reticulon gene family (RTN3): gene structure and chromosomal localization to 11q13. Genomics. 1999;58:73–81. doi: 10.1006/geno.1999.5807. [DOI] [PubMed] [Google Scholar]
- Murayama KS, Kametani F, Saito S, Kume H, Akiyama H, Araki W. Reticulons RTN3 and RTN4-B/C interact with BACE1 and inhibit its ability to produce amyloid beta-protein. Eur J Neurosci. 2006;24:1237–1244. doi: 10.1111/j.1460-9568.2006.05005.x. [DOI] [PubMed] [Google Scholar]
- Nziengui H, Bouhidel K, Pillon D, Der C, Marty F, Schoefs B. Reticulon-like proteins in Arabidopsis thaliana: structural organization and ER localization. FEBS Lett. 2007;581:3356–3362. doi: 10.1016/j.febslet.2007.06.032. [DOI] [PubMed] [Google Scholar]
- Oertle T, Schwab ME. Nogo and its paRTNers. Trends Cell Biol. 2003;13:187–194. doi: 10.1016/S0962-8924(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Oertle T, Klinger M, Stuermer CA, Schwab ME. A reticular rhapsody: phylogenic evolution and nomenclature of the RTN/Nogo gene family. FASEB J. 2003;17:1238–1247. doi: 10.1096/fj.02-1166hyp. [DOI] [PubMed] [Google Scholar]
- Qahwash I, He W, Tomasselli A, Kletzien RF, Yan R. Processing amyloid precursor protein at the beta-site requires proper orientation to be accessed by BACE1. J Biol Chem. 2004;279:39010–39016. doi: 10.1074/jbc.M407101200. [DOI] [PubMed] [Google Scholar]
- Qing H, Zhou W, Christensen MA, Sun X, Tong Y, Song W. Degradation of BACE by the ubiquitin-proteasome pathway. FASEB J. 2004;18:1571–1573. doi: 10.1096/fj.04-1994fje. [DOI] [PubMed] [Google Scholar]
- Schanda K, Hermann M, Stefanova N, Gredler V, Bandtlow C, Reindl M. Nogo-B is associated with cytoskeletal structures in human monocyte-derived macrophages. BMC Res Notes. 2011;4:6. doi: 10.1186/1756-0500-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab ME. Nogo and axon regeneration. Curr Opin Neurobiol. 2004;14:118–124. doi: 10.1016/j.conb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Shi Q, Hu X, Prior M, Yan R. The occurrence of aging-dependent reticulon 3 immunoreactive dystrophic neurites decreases cognitive function. J Neurosci. 2009a;29:5108–5115. doi: 10.1523/JNEUROSCI.5887-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Prior M, He W, Tang X, Hu X, Yan R. Reduced amyloid deposition in mice overexpressing RTN3 is adversely affected by preformed dystrophic neurites. J Neurosci. 2009b;29:9163–9173. doi: 10.1523/JNEUROSCI.5741-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Prior M, Zhou X, Tang X, He W, Hu X, Yan R. Preventing formation of reticulon 3 immunoreactive dystrophic neurites improves cognitive function in mice. J Neurosci. 2013;33:3059–3066. doi: 10.1523/JNEUROSCI.2445-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonen M, Pedersen V, Weinmann O, Schnell L, Buss A, Ledermann B, Christ F, Sansig G, van der Putten H, Schwab ME. Systemic deletion of the myelin-associated outgrowth inhibitor Nogo-A improves regenerative and plastic responses after spinal cord injury. Neuron. 2003;38:201–211. doi: 10.1016/S0896-6273(03)00226-5. [DOI] [PubMed] [Google Scholar]
- Strittmatter SM. Modulation of axonal regeneration in neurodegenerative disease: focus on Nogo. J Mol Neurosci. 2002;19:117–121. doi: 10.1007/s12031-002-0021-7. [DOI] [PubMed] [Google Scholar]
- Tashiro K, Satoh A, Utsumi T, Chung C, Iwakiri Y. Absence of Nogo-B (Reticulon 4B) Facilitates Hepatic Stellate Cell Apoptosis and Diminishes Hepatic Fibrosis in Mice. Am J Pathol. 2013;182:786–795. doi: 10.1016/j.ajpath.2012.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng FY, Tang BL. Nogo signaling and nonphysical injury-induced nervous system pathology. J Neurosci Res. 2005;79:273–278. doi: 10.1002/jnr.20361. [DOI] [PubMed] [Google Scholar]
- Tesco G, Koh YH, Kang EL, Cameron AN, Das S, Sena-Esteves M, Hiltunen M, Yang SH, Zhong Z, Shen Y, Simpkins JW, Tanzi RE. Depletion of GGA3 stabilizes BACE and enhances beta-secretase activity. Neuron. 2007;54:721–737. doi: 10.1016/j.neuron.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Kovacs DM, Yan R, Wong PC. The {beta}-secretase enzyme BACE in health and Alzheimer's disease: regulation, cell biology, function, and therapeutic potential. J Neurosci. 2009;29:12787–12794. doi: 10.1523/JNEUROSCI.3657-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Wang H, Li R, Shen Y. beta-Secretase: its biology as a therapeutic target in diseases. Trends Pharmacol Sci. 2013;34:215–225. doi: 10.1016/j.tips.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Sweeney D, Gandy SE, Sisodia SS. The profile of soluble amyloid beta protein in cultured cell media. Detection and quantification of amyloid beta protein and variants by immunoprecipitation-mass spectrometry. J Biol Chem. 1996;271:31894–31902. doi: 10.1074/jbc.271.50.31894. [DOI] [PubMed] [Google Scholar]
- Wojcik S, Engel WK, Yan R, McFerrin J, Askanas V. NOGO is increased and binds to BACE1 in sporadic inclusion-body myositis and in AbetaPP-overexpressing cultured human muscle fibers. Acta Neuropathol. 2007;114:517–526. doi: 10.1007/s00401-007-0281-y. [DOI] [PubMed] [Google Scholar]
- Yan R, Vassar R. Targeting the beta secretase BACE1 for Alzheimer's disease therapy. Lancet Neurol. 2014;13:319–329. doi: 10.1016/S1474-4422(13)70276-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Shi Q, Hu X, Zhou X. Reticulon proteins: emerging players in neurodegenerative diseases. Cell Mol Life Sci. 2006;63:877–889. doi: 10.1007/s00018-005-5338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YS, Strittmatter SM. The reticulons: a family of proteins with diverse functions. Genome Biol. 2007;8:234. doi: 10.1186/gb-2007-8-12-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Ho C, Li S, Keirstead H, Steward O, Tessier-Lavigne M. Lack of enhanced spinal regeneration in Nogo-deficient mice. Neuron. 2003;38:213–224. doi: 10.1016/S0896-6273(03)00225-3. [DOI] [PubMed] [Google Scholar]
- Zurek N, Sparks L, Voeltz G. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic. 2011;12:28–41. doi: 10.1111/j.1600-0854.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]