Abstract
The Saccharomyces cerevisiae Srs2 protein is involved in DNA repair and recombination. In order to gain better insight into the roles of Srs2, we performed a screen to identify mutations that are synthetically lethal with an srs2 deletion. One of them is a mutated allele of the ULP1 gene that encodes a protease specifically cleaving Smt3-protein conjugates. This allele, ulp1-I615N, is responsible for an accumulation of Smt3-conjugated proteins. The mutant is unable to grow at 37°C. At permissive temperatures, it still shows severe growth defects together with a strong hyperrecombination phenotype and is impaired in meiosis. Genetic interactions between ulp1 and mutations that affect different repair pathways indicated that the RAD51-dependent homologous recombination mechanism, but not excision resynthesis, translesion synthesis, or nonhomologous end-joining processes, is required for the viability of the mutant. Thus, both Srs2, believed to negatively control homologous recombination, and the process of recombination per se are essential for the viability of the ulp1 mutant. Upon replication, mutant cells accumulate single-stranded DNA interruptions. These structures are believed to generate different recombination intermediates. Some of them are fixed by recombination, and others require Srs2 to be reversed and fixed by an alternate pathway.
Mutations in the Saccharomyces cerevisiae SRS2 gene were isolated in different genetic screens, namely suppression of the trimethoprim sensitivity of rad6 and rad18 mutants (29), suppression of the DNA damage sensitivity in rad18 cells (2), and hyperrecombination (3). The Srs2 protein shares homologies with the bacterial UvrD, Rep (2), and PcrA helicases. As predicted by the sequence, Srs2 was shown in vitro to have 3′-5′ DNA helicase activity (50).
Genetic studies showed that a number of deleterious phenotypes associated with srs2 null alleles are suppressed by deletions of the RAD51, RAD52, RAD55, and RAD57 genes, which are all involved in the formation of Rad51 nucleofilaments on single-stranded DNA (ssDNA) (1, 9, 26). These suppressed phenotypes include UV sensitivity and synthetic lethality with rad54 (17, 47, 52) and sgs1 (14) mutations, which both (most likely) affect later recombination steps (4, 13, 38). Links between Srs2 and homologous recombination were also revealed by the fact that srs2 null alleles suppress the γ-ray and methyl methanesulfonate (MMS) sensitivities conferred by several leaky alleles of RAD52 and RAD51 but have no suppressor effects on the corresponding deletions (9, 25, 42, 52). These phenotypes led to the proposal that Srs2 could destabilize recombination intermediates poisoned to some extent by the leaky recombination proteins (9). In the absence of Srs2, the leakiness of the proteins would allow efficient recombinational repair to occur. In support of this interpretation, Srs2 was recently shown to disrupt in vitro Rad51 nucleofilaments on ssDNA (27, 66). This activity might well account for the srs2 phenotypes cited above, which can be explained by the abnormal persistence of inappropriate recombination intermediates.
Several data indicate that Srs2 is active during replication. In the mitotic cycle, the expression of the gene is high during the S and G2 phases (17). Furthermore, Srs2 is phosphorylated during the S phase in response to checkpoint activation by DNA damages (36). Finally, Srs2 interacts with Pol32, a subunit of polymerase δ (23). This, together with the genetic interactions between srs2 and rad51 and the physical interaction between Srs2 and Rad51 proteins (27), might indicate that one role of Srs2 is to control recombination intermediates arising during replication and eventually to reverse them.
To find new clues on the role(s) of Srs2 in DNA metabolism, we performed a screen for mutations that are synthetically lethal with an srs2 deletion. Mutations in several genes were isolated. For most of them, the synthetic lethality with an srs2Δ mutation was suppressed by preventing homologous recombination by a RAD51 deletion. However, two mutants behaved differently in that synthetic lethality was not suppressed by a recombinational defect. One of these bore the ulp1-I615N mutation, a recessive temperature-sensitive allele of the essential ULP1 gene. Ulp1 is a protease specific for both the processing of the precursor of the ubiquitin-like Smt3 protein (an ortholog of the mammalian SUMO-1 protein) into its mature form and the cleavage of Smt3 from its conjugates. It is required for cell cycle progression, in which it plays a role in the G2/M cell cycle transition (32, 33). Several homologs of Ulp1 have now been found in different species, including Schizosaccharomyces pombe (63) and Drosophila melanogaster (31). Key roles of sumoylation are indicated by the fact that mammalian SUMO-1 is involved in a broad range of cellular processes, such as p53 and c-jun transcriptional activation, signal transduction, inflammatory and immune responses, and the control of cell growth (reviewed in reference 40). Sumoylation plays a role in proper protein localization, organizing and targeting promyelocytic leukemia nuclear bodies, and protein stabilization and has been proposed to be involved in apoptosis (reviewed in reference 53). SUMO conjugates of histone H4, which may mediate gene silencing, have also been reported (57). Numerous reports have shown close relations between SUMO-1 and genome integrity maintenance (reviewed in reference 46). Indeed, some human proteins involved in genome stability, e.g., the DNA topoisomerases I and II and the DNA helicase of the RecQ family involved in the Werner syndrome (Wrn), are sumoylated after DNA-damaging treatments (24). The Bloom syndrome helicase (Blm), which also belongs to the RecQ helicase family, interacts with the SUMO-1-conjugating enzyme Ubc9 and with Rad51 (see reference 19 for a review). A link between sumoylation and recombination has also been suggested by interactions of SUMO-1 or Ubc9 with Rad51 or Rad52 (54, 55). Finally, sumoylation, i.e., Smt3 conjugation, is believed to play a role during the S phase of the S. cerevisiae cell cycle, since PCNA gets either mono-ubiquitinated or sumoylated. The ubiquitin-Smt3 couple possibly acts like a regulatory switch that directs PCNA for alternative functions (21).
We describe here the phenotypes conferred by the ulp1-I615N mutation screened as synthetically lethal with an srs2 deletion. The mutant is thermosensitive, and at a permissive temperature, it presents severe growth defects, moderate UV sensitivity, a strong mitotic hyperrecombination phenotype, an impairment in meiosis, and an accumulation of Smt3 conjugates. The mutation was found to be synthetically lethal not only with srs2 but also with mutations in genes involved in the RAD51-dependent homologous recombination pathway (rad51, rad52, rad54, rad55, rad50, and mre11). These results, together with those of other experiments described below, led us to postulate that the ulp1 mutation affects replication rather than recombination, resulting in the accumulation of recombinogenic intermediates that require both homologous recombination and Srs2 to be correctly processed. We show that this mutant does indeed accumulate single-stranded interruptions in newly replicated DNA.
MATERIALS AND METHODS
Media.
Complete glucose (YPD), sporulation, and synthetic minimal media were prepared as described by Sherman et al. (56). YPD8 is YPD medium containing 8% instead of 2% glucose. Galactose complete medium (YPGal) was prepared in the same way as YPD, but with 2% galactose instead of 2% glucose. Presporulation and sporulation media were prepared according to the method of Resnick et al. (49). 5-Fluoroorotic acid was used for the selection of Ura− cells at a concentration of 1 mg/ml. G418-YPD plates contained 200 μg of the drug/ml. MMS was used at a final concentration of 0.0175% (vol/vol) in YPD plates. Canavanine was added to suitably supplemented minimal medium at a concentration of 60 μg/ml. Carbendazim (Sigma) was added to liquid YPD medium at a final concentration of 100 μg/ml from a 1% (wt/vol) stock solution in dimethyl sulfoxide. For DNA labeling experiments, YPGal liquid medium was supplemented with either thymidine at a final concentration of 100 μM or bromodeoxyuridine (BrdU) at a final concentration of 200 μM, and for chase experiments, thymidine was used at a final concentration of 0.4 to 1 mM. When used in YPGal plates, thymidine was used at a 1 mM final concentration.
Yeast strains.
The MH yeast strains used for this study (Table 1) are a series of isogenic strains constructed starting from one spore derived from a cross between the FF18733 and GF1906 strains.
TABLE 1.
Strains used for this studya
| Strain | Genotype | Source |
|---|---|---|
| FF18733 | MATahis7-2 leu2-3,112 lys1-1 trp1-289 ura3-52 | F. Fabre |
| GF1906 | MATα ade2Δ ade3Δ leu2-3,112 lys2-1 trp1-289 ura3-52 | G. Faye |
| MH1004 | MATα ade2Δ ade3Δ leu2-3,112 lys2-1 trp1-289 ura3-52 | This study |
| MH1006 | MATa ade2Δ ade3Δ leu2-3,112 lys1::kanMX4 trp1-289 ura3-52 | This study |
| MH983 | MATa srs2::LEU2 ade2Δ ade3Δ leu2-3,112 lys1::kanMX4 trp1-289 ura3-52 | This study |
| MH921 | MATα srs2::LEU2 ade2Δ ade3Δ leu2-3,112 lys2-1 trp1-289 ura3-52 | This study |
| B392 | MATα srs2::LEU2 ulp1-I615N ade2Δ ade3Δ leu2-3,112 lys2-1 trp1-289 ura3-52 + YCp50-SRS2+-ADE3+ | This study |
| FF181079 | rad51::ura3− isogenic to FF18733 | F. Fabre |
| MH1018 | MATa ulp1-I615N ade2Δ ade3Δ leu2-3,112 lys1::kanMX4 trp1-289 ura3-52 | This study |
| MH1024 | MATα ulp1-I615N ade2Δ ade3Δ leu2-3,112 lys2-1 trp1-289 ura3-52 | This study |
| MH1080 | MATaulp1-I615N arg4-RV ade2Δ ade3Δ leu2-3,112 lys1::kanMX4 trp1-289 ura3-52 | This study |
| MH1085 | MATα ulp1-I615N arg4-BglII ade2Δ ade3Δ leu2-3,112 lys2-1 trp1-289 ura3-52 | This study |
| MH1088 | MH1085 × MH1080 | This study |
| FF18744 | srs2::LEU2+ isogenic to FF18733 | F. Fabre |
| FF18958 | rad51::URA3+ isogenic to FF18733 | F. Fabre |
| FF18742 | rad52::URA3+ isogenic to FF18733 | F. Fabre |
| FF18973 | rad54::LEU2+ isogenic to FF18733 | F. Fabre |
| FF181461 | rad55::URA3+ isogenic to FF18733 | F. Fabre |
| FF18964 | rad50::URA3+ isogenic to FF18733 | F. Fabre |
| FF181613 | mre11::URA3+ isogenic to FF18733 | F. Fabre |
| BY4742 | MATα yku70::kanMX4 leu2-Δ0 ura3-Δ0 lys2-Δ0 his3-Δ1 | Euroscarf |
| FPII006-01D | MATa rad59::kanMX4 ura3-52 trp1-Δ63 his3-Δ200 | Euroscarf |
| FF18739 | rad18::LEU2+ isogenic to FF18733 | F. Fabre |
| FF18220 | rad6::URA3+ isogenic to FF18733 | F. Fabre |
| FF181655 | rad14::LEU2+ isogenic to FF18733 | F. Fabre |
| Y301 | MATa rad53 (sad1-1) leu2-3,112 ura3-1 trp1-1 ade2-1 his3,115 can1-100 | S. Elledge |
| YLVE4 | MATacdc21::kanMX4 leu2-3,112::pRS305-GAL1-10-hENT1 trp1Δ2::pRS304-GAL1-10-DmdNK ade2-1 his3-11,15 ura3-52 can1-100 | L. Vernis |
| MH 1367 | MATa ulp1-I615N cdc21::kanMX4 leu2-3,112::pRS305-GAL1-10-hENT1 trp1Δ2::pRS304-GAL1-10-DmdNK ade2-1 his3-11,15 lys2-1 ura3-52 | This study |
All strains (with the exceptions of BY4742, FPII006-01D, Y301, YLVE4, and MH1367) were derived from two isogenic series, FF18733 and GF1906.
To test recombination, we constructed yeast strain MH1088 as follows. Plasmid pSG19 containing the arg4-RV allele and the URA3+ gene was integrated at the ARG4 locus of strain MH1024 after SnaBI linearization in the arg4-RV allele. Clones that had “popped out” the URA3+ plasmid were then selected on 5-fluoroorotic acid- and arginine-containing medium. A stable Ura− Arg− subclone was selected as the MH1080 strain. Strain MH1085 was constructed similarly, by using the URA3+ pSG21 plasmid containing the arg4-BglII allele linearized by NheI to direct integration at the ARG4 locus of strain MH1018. Strains MH1080 and MH1085 were crossed to obtain the MH1088 strain, which is heteroallelic for arg4-RV and arg4-BglII, as verified by a papillation test on synthetic medium lacking arginine.
For BrdU labeling experiments, we constructed the MH1367 strain bearing the ulp1-I615N mutation in a genetic background that allows efficient thymidine incorporation into DNA. It was derived from a temperature-sensitive G418R Leu+ Trp+ spore found among the meiotic progeny of diploids constructed by crossing the MH1024 mutant with one W303 derivative, the YLVE4 strain, described by Vernis et al. (67). The YLVE4 strain bears the cdc21::kanMX4 deletion, which inhibits de novo dTTP synthesis, and single integrated copies of the pGAL-hENT1-LEU2+ and pGAL-DmdNK-TRP1+ constructs, bearing, under the control of the GAL1-10 promoter, one of the human genes of a nucleoside transporter and the D. melanogaster gene of deoxyribonucleoside kinase, respectively (67).
Isolation of synthetically lethal mutants.
We used a previously described colored colony sectoring assay (6). Two isogenic srs2Δ ade2Δ ade3Δ strains (MH921 and MH983 [MATa and MATα, respectively] [Table 1]) were transformed with a derivative of plasmid YCp50 (URA3+ selection marker) containing the SRS2 and ADE3 genes (YCp50-SRS2+-ADE3+). On YPD8 medium, the ability to segregate away the plasmid gives rise to red-and-white sectored colonies and white colonies. Red is associated with the presence of the plasmid (Ade2 cells), and white is associated with the loss of the plasmid (Ade2 Ade3 cells). UV mutagenesis was performed with a dose of 30 J/m2, leaving about 50% survivors. We screened for full red colonies, which are presumably indicative of lethality upon plasmid loss. The synthetic lethality was further ascertained by the absence of sectored or white colonies through three subsequent subcloning steps. We ultimately verified that white segregants could be obtained upon plasmid exchange with a YCp50-ura3Δ-TRP1+-SRS2+ plasmid derivative with a deletion of the URA3 marker and containing the TRP1 and SRS2 genes but not the ADE3 gene and that no plasmid exchange could be obtained with YCp50-ura3Δ-TRP1+ lacking the SRS2 gene.
Plasmids.
The YCp50-SRS2+-ADE3+ plasmid was constructed by ligation of a 3,600-bp SalI-NruI fragment containing the ADE3 gene from the p41 plasmid (kindly provided by G. Faye) into the p14H plasmid (YCp50 with a 5,573-bp HindIII-SalI SRS2 insert) (2) digested with SalI and NruI.
The YCp50-ura3Δ-TRP1+ and YCp50-ura3Δ-TRP1+-SRS2+ plasmids were obtained, respectively, from YCp50 and p14H: the 1,565-bp NruI-SmaI fragment containing the URA3 gene was deleted and the 1,453-bp EcoRI fragment from the ARS1-based YRp7 plasmid was subcloned into the unique EcoRI site of the resulting plasmids.
The plasmid pTZ18-rad51::KanMX4 used to disrupt the RAD51 gene was constructed as follows: a 1,500-bp HincII-EcoRV fragment isolated from the pFA6a-KanMX4 plasmid (a Eurofan plasmid for functional analysis [68]) was introduced into the pTZ18-RAD51 plasmid digested with StuI and NruI. A 4,100-bp AgeI-NheI fragment was used to transform yeast strains.
The genomic library used to retrieve the ULP1+-containing insert was kindly provided by G. Faye and was constructed in a TRP1+ plasmid derived from YRp7.
ULP1+-bearing plasmids were constructed as follows: a 2,617-bp BstBI-NspI fragment containing the ULP1+ gene was purified from a cloned plasmid that complements both the ulp1 thermosensitivity and the synthetic lethality between ulp1 and srs2Δ in the B392 mutant. This fragment was ligated into the YRp7 multicopy vector, the YCp50 and YCp50-ura3Δ-TRP1+ centromeric vectors, and the URA3+ YIp5 integrative vector after digestion with ClaI and SphI.
The pRS304-GAL-RAD52+ plasmid was constructed as follows. The GAL1-10 promoter was amplified by PCR and subcloned into pRS304 at EcoRI and BamHI sites. The RAD52 coding sequence was amplified by a PCR using the oligonucleotides ATAAGAATGCGGCCGCATGGCGTTTTTAAGCTATTTTGCC (NotI site in bold) and CCCGAGCTCTCAAGTAGGCTTGCGTGCATGCAGG (SacI site in bold). The PCR product was restricted with NotI and SacI and subcloned into pRS304-GAL at NotI and SacI sites downstream of the GAL promoter. Digestion by BstXI was done to direct the integration of the pRS304-GAL-RAD52+ plasmid into the chromosomal LEU2 locus.
RAD51 was cloned under the control of the GAL1-10 promoter into the multicopy URA3+ vector pYEDP1/8-2 (11). Digestion by EcoRV was used to direct the integration of this GAL1-10-RAD51-containing plasmid into the chromosomal URA3 locus.
Plasmids pSG19 and pSG21 were kindly provided by S. Gangloff.
Irradiation.
UV irradiation (254 nm) was done on plated cells at a dose rate of 2 to 5 J/m2/s, depending upon the strain under study. γ-Rays were from a 137Cs source and were applied at a dose rate of 0.943 Gy/s to cell suspensions in H2O before plating. We used exponentially growing cells taken from overnight cultures, except for assays to measure the UV sensitivity of haploids, for which stationary-phase cells were treated. To enrich populations of G2 cells, we treated exponential-phase cultures with carbendazim (100 μg/ml) before UV irradiation, for 4 h in the case of ulp1 rad18 double mutants and for 2.5 h for rad18 simple mutants. All of the results presented here correspond to the means of at least two independent experiments.
Determination of recombination rates.
Recombination rates were calculated by the method of the median (30). They were measured in ulp1 homozygous diploids (MH1088) in the presence or absence of either the YCp50-ura3Δ-TRP1+-ULP1+ or YRp7-ULP1+ plasmid. Cells were grown on solid supplemented minimal medium containing arginine and selective for the plasmid when necessary. Thirteen independent samples were used for each strain. Each sample corresponded to a pool of two clones taken after 3 days of incubation at 28°C and suspended in 1 ml of H2O. After hemocytometer counting, we withdrew 10 μl to make appropriate dilutions before plating them onto supplemented minimal arginine-containing medium in order to estimate the number of viable cells. The remaining cell suspension was plated on two supplemented minimal medium plates lacking arginine. Each experiment was done at least in triplicate.
Western immunoblotting.
Yeast protein extracts were prepared from trichloroacetic acid-treated cells, and equal amounts of proteins in 1× Laemmli buffer were loaded onto sodium dodecyl sulfate-10% polyacrylamide gels and run according to standard procedures. After electrophoresis, proteins were transferred onto Hybond ECL nitrocellulose membranes (Amersham Pharmacia Biotech). Smt3-conjugated proteins were detected in 45-μg protein samples by using an anti-Smt3 antibody kindly provided by M. Hochstrasser. Rad51 and Rad52 were detected in 25-μg protein samples by using anti-Rad51 and anti-Rad52 antibodies (Santa Cruz Biotechnology) according to the manufacturer's instructions.
BrdU labeling, chasing, and detection of single-strand breaks in growing cells.
Exponential-phase cells (2 × 106 cells/ml) of strains YLVE4 and MH1367 from overnight cultures in 500 ml of YPGal plus thymidine were collected, washed three times in YPGal devoid of thymidine, and resuspended in the same volume of fresh YPGal supplemented with BrdU at a final concentration of 200 μM. They were incubated for one generation time at 28°C for BrdU labeling (2.1 h for YLVE4 and 3.8 h for MH1367) before sodium azide was added to a final concentration of 0.1% to one-half of the culture. Cells were collected and washed once in H2O, and pellets were frozen in liquid nitrogen before nucleic acid purification by phenol extraction. BrdU was chased from the other half of the culture by washing the cells three times in YPGal and resuspending them in YPGal supplemented with 400 μM or 1 mM thymidine. Cells were further incubated for another doubling time at 28°C. Sodium azide was then added to a final concentration of 0.1%, and cell pellets were collected and treated as described above for nucleic acid purification.
The DNA samples analyzed corresponded to 2 × 107 unchased cells and about twice as many chased ones. One microgram of a λ HindIII digest was deposed in each gel, and after electrophoresis, the corresponding track was moved apart and stained with ethidium bromide for molecular weight calibration. For analyses under denaturing conditions, electrophoresis was performed with 0.6% alkaline agarose gels (50 mM NaCl, 5 mM EDTA) previously equilibrated in migration buffer (30 mM NaOH, 2 mM EDTA) and run at 0.66 V/cm overnight. The DNAs from the samples were transferred by Southern blotting from the gels to Hybond N+ nitrocellulose membranes and revealed by classical Western blotting methods using a 10−3 dilution of an anti-BrdU antibody (Becton Dickinson). For analyses under neutral conditions, samples were run in 0.8% agarose gels that were then treated as described above.
RESULTS
Isolation and properties of the synthetically lethal ulp1-I615N srs2Δ mutant.
To screen synthetically lethal mutations in the presence of an srs2Δ mutant, we used a previously described colored colony sectoring assay (6) starting with two mutagenized srs2Δ strains of opposite mating types (Table 1) containing the URA3+ selectable YCp50-SRS2+-ADE3+ plasmid (see Materials and Methods). Of 68,000 colonies examined at 24°C, we isolated 70 synthetically lethal mutants. All were due to recessive mutations, which allowed us to classify them into at least 13 complementation groups.
Since the deleterious phenotypes of srs2 mutants were suppressed by deletions of the RAD51 gene, we asked whether the synthetic lethality observed for the different mutants was due to abortive recombination events. For this purpose, we performed experiments to delete RAD51 by transformation with a rad51::KanMX4 fragment of one representative mutant from each of the 13 complementation groups. G418-resistant clones that lost the RAD51 function were recovered for 11 strains, as indicated by a complementation test for MMS resistance with a rad51Δ tester strain (FF181079). In contrast, none of the G418-resistant clones obtained for the other two strains corresponded to rad51 deletions. This result strongly suggested that the mutations isolated as synthetically lethal with the srs2 deletion were also synthetically lethal with rad51::kanMX4 for those two strains. Therefore, they would not affect late recombination steps, but rather events occurring prior to recombination. One of these mutations, corresponding to the B392 mutant, is described here, and indeed its synthetic lethality with a rad51Δ mutant was confirmed by further tests, as described below.
The mutant strain B392 exhibited thermosensitive growth at 37°C and was the only representative of its complementation group. A genetic analysis after crossing with the wild-type strain MH1006 showed that B392 contains a single new recessive mutation that is genetically independent from the srs2Δ mutation. B392 cells therefore have the genotype “x srs2Δ,” where x represents the new thermosensitive allele, and contain the plasmid pYCp50-SRS2+-ADE3+.
To clone the corresponding gene, we screened an S. cerevisiae genomic TRP1 library (see Materials and Methods) for sequences able to confer thermoresistance to the B392 mutant at 37°C. Of the 12,000 transformants tested, 1 grew at 37°C. Moreover, the synthetic lethality between the srs2 deletion and the second mutation was abolished in this transformant, as indicated by its viability after the loss of the SRS2+ plasmid.
The complementing plasmid was isolated, amplified in Escherichia coli, and analyzed. It contained an insert of 6,861 bp, corresponding to a genomic sequence extending from nucleotides 509040 to 515900 on chromosome XVI (Saccharomyces Genome Database). This region encompasses two partial and two complete open reading frames, including the ULP1 gene. A deletion analysis indicated that the ULP1 gene was the only open reading frame responsible for thermoresistance and the abolition of synthetic lethality (data not shown).
In order to check that ULP1 was indeed the wild-type copy of the synthetically lethal x mutation and not an extragenic suppressor that would map elsewhere in the genome, we integrated an ULP1 gene flanked by the URA3 marker at the ULP1 locus in a wild-type strain, which resulted in duplicate ULP1 genes separated by URA3. This strain was crossed with a derivative of B392 in which the original plasmid had been replaced by a TRP1+-SRS2+-carrying plasmid. Seventeen tetrads derived from this diploid were analyzed. In all cases, the Ura3+ phenotype had segregated away from the temperature-sensitive phenotype conferred by the synthetically lethal mutation. This result indicates that the mutation lies in, or at least very close to, the ULP1 gene.
The sequence of the mutant allele revealed a single point mutation, a T replaced with an A, at position 1843 from the ATG, leading to the replacement of isoleucine 615 with asparagine in the Ulp1 protein. The allele will be referred to hereafter as ulp1-I615N.
From the tetrads derived from the B392 × MH1006 (SRS2+ ULP1+) strain, we isolated MATa and MATα monosporic clones containing the single ulp1-I615N mutation at the origin of the MH1018 and MH1024 strains, respectively.
The ulp1-I615N mutation confers defects in vegetative growth and during meiosis.
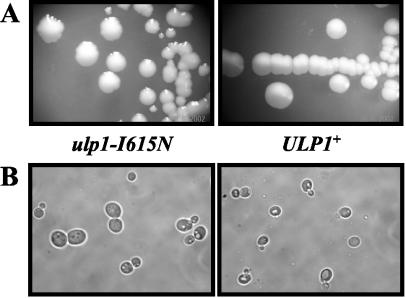
ulp1-I615N mutant cells do not grow at 37 or 14°C, and at permissive temperatures (24 to 28°C) they have severe growth defects. The doubling time for haploids is 3.5 h at 24°C compared to 1.8 h for wild-type cells. The colony morphology is also affected, exhibiting an irregular shape and various sizes (Fig. 1A). Cells are also variable in size, being one to three times larger for the mutant than for its wild-type counterpart (Fig. 1B). Large budded cells with a single nucleus are overrepresented, which is indicative of an impairment in the G2/M transition. Stationary phase is reached at about a two times lower cell density than that for the wild type. Cell death is also increased in the mutant population compared to the wild type: the plating efficiency is about 30% for a growing population and 40 to 50% for stationary phase.
FIG. 1.
The ulp1-I615N mutant shows growth defects at a permissive temperature. (A) Colony morphology of ulp1-I615N mutants (MH1024) and ULP1+ cells (MH1004) after 5 days of incubation at 24°C on YPD plates. (B) Live phase-contrast images of ulp1-I615N and ULP1+ cells in exponential-phase cultures growing at 24°C in YPD.
Meiosis is also impaired in homozygous ulp1-I615N diploids. Under optimal standard conditions for meiosis (24 h in presporulation medium and 48 h in sporulation medium [49]) at 24°C, tetrad frequencies fluctuated from one experiment to another between 0.3 and 5% and those of dyads fluctuated between 0 and 7% for ulp1 homozygotes. In contrast, ULP1+ homozygotes gave rise to 42 to 70% tetrads and 1 to 7% dyads. The spore viability, as determined after the dissection of 12 tetrads, was decreased compared to the wild type. Of 48 spores derived from the mutant diploid, 13 developed into a colony, compared to 35 of 48 for diploid mutant cells complemented with YCp50-ULP1+. These data demonstrate that the ulp1-I615N mutation affects both the sporulation efficiency and spore viability. The spore lethality is likely due to a meiotic defect leading to genetic anomalies that are responsible for cell death. Indeed, ulp1-I615N monosporic clones were recovered at nearly normal frequencies if they originated from meiosis of ulp1/ULP1+ heterozygous diploids (see control line in Table 2).
TABLE 2.
Genetic interactions of the ulp1-I615N mutation with mutated genes (x) of the repair and recombination pathways
| Diploid heterozygous for ulp1-I615N and another mutation (no. of tetrads analyzed) | No. of monosporic clones
|
Nature of genetic interaction with ulp1 | |||
|---|---|---|---|---|---|
| Wild type | ulp1 single mutant | x single mutant | ulp1 x double mutant | ||
| Control: ulp1 × wild type (32) | 64 | 58 | |||
| Mutants specifically deficient in homologous recombination | |||||
| ulp1 × srs2Δ (48) | 44 | 45 | 39 | 1 | Colethal |
| ulp1 × rad51Δ (76) | 66 | 78 | 76 | 2 | Colethal |
| ulp1 × rad52Δ (48) | 46 | 41 | 38 | 3 | Colethal |
| ulp1 × rad54Δ (58) | 57 | 44 | 45 | 1 | Colethal |
| ulp1 × rad55Δ (21) | 10 | 23 | 21 | 1 | Colethal |
| Mutants deficient in both homologous recombination and NHEJ | |||||
| ulp1 × rad50Δ (59) | 59 | 43 | 48 | 0 | Colethal |
| ulp1 × mre11Δ (12) | 10 | 13 | 12 | 0 | Colethal |
| Mutant specifically deficient in NHEJ: ulp1 × yku70Δ (18) | 12 | 20 | 18 | 12 | Viable |
| Mutant mainly deficient in SSA (and homologous recombination): ulp1 × rad59Δ (32) | 28 | 25 | 23 | 27 | Viable |
| Mutants deficient in control of translesional bypass by ubiquitination | |||||
| ulp1 × rad18Δ (64) | 57 | 51 | 48 | 51 | Viable |
| ulp1 × rad6Δ (37) | 39 | 29 | 33 | 28 | Viable |
| Mutant deficient in excision repair: ulp1 × rad14Δ (22) | 21 | 13 | 18 | 21 | Viable |
| Mutant deficient in checkpoint entry: ulp1 × rad53/sad1-1 (31) | 26 | 24 | 32 | 21 | Viable |
| Mutant deficient in replication and repair: ulp1 × rad27Δ (17) | 17 | 16 | 14 | 0 | Colethal |
From these observations, it appears that the ulp1-I615N mutation affects both vegetative growth (abnormal cell cycle and increased cell death) and the meiotic process (sporulation efficiency and spore viability).
The ulp1-I615N mutation leads to accumulation of Smt3 conjugates.
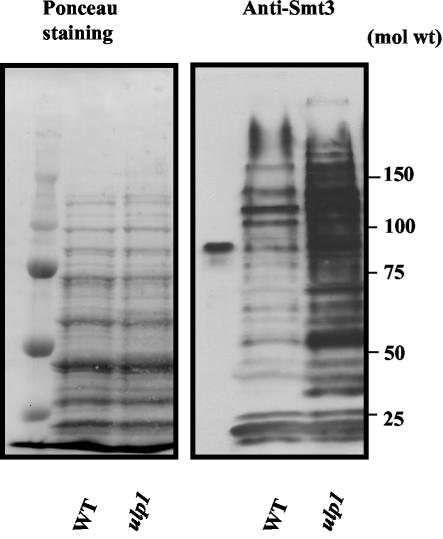
Because Ulp1 catalyzes not only the deconjugation of Smt3 but also the maturation of the Smt3 precursor (32), ulp1 mutations could have led to either an excess of or a reduction in the amount of sumoylated forms of proteins. In order to know which was the effect of the ulp1-I615N mutation, we compared the status of the proteins extracted from mutant and wild-type cells during exponential growth at a permissive temperature. Western blots using an anti-Smt3 antibody revealed that sumoylated proteins were present in a very large excess in the ulp1-I615N mutant compared to those in wild-type cells (Fig. 2). The phenotypes described here are thus most likely related to an excess of sumoylated forms of some Smt3 targets.
FIG. 2.
The ulp1-I615N mutant accumulates Smt3 protein conjugates. The images show Ponceau red staining of proteins after transfer onto a nitrocellulose membrane (left) and immunodetection of Smt3 conjugates with an anti-Smt3 antibody (right). Extracts (45 μg) were obtained from cells growing exponentially at 28°C. WT, wild type.
The ulp1-I615N mutation confers sensitivity to UV but not to γ-rays.
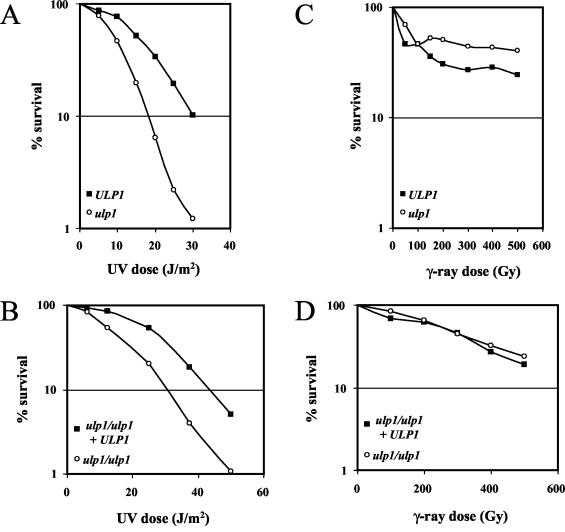
That the ulp1-I615N mutation confers growth defects and is synthetically lethal with an srs2 deletion at a permissive temperature raises the possibility that ULP1, like SRS2, is involved in DNA repair. We therefore asked whether ulp1-I615N confers sensitivity to the lethal effects of UV and γ-irradiation. The response of haploids was compared to that of the corresponding wild-type cells. For diploids, we compared the response of ulp1-I615N homozygotes to that of the same cells complemented with a multicopy plasmid expressing ULP1+. These diploids will be referred to as Ulp1− and Ulp1+, respectively.
The haploid ulp1 mutant was moderately UV sensitive (Fig. 3A) compared to the wild type. The dose reduction factor at a 10% survival rate was 1.6. Similarly, Ulp1− diploids were more sensitive than Ulp1+ cells to UV irradiation (dose reduction factor at a 10% survival rate of 1.4) (Fig. 3B). Diploids have long been known to be more UV resistant than haploids, which is an effect related to homologous recombination. A comparison of haploid and diploid responses in wild-type or mutant cells (Fig. 3A and B) indicated that the diploid effect is not significantly affected by the ulp1 mutation.
FIG. 3.
The ulp1-I615N mutant is slightly sensitive to UV but not to γ-rays. (A and B) Survival curves of ulp1-I615N (MH1024) and ULP1+ (MH1004) haploids after UV (A) or γ (B) irradiation. (C and D) Survival curves of homozygous ulp1-I615N (MH1088) diploids after UV (C) or γ (D) irradiation in the presence or absence of an ULP1+-bearing plasmid.
On the other hand, no sensitivity to γ-rays was observed in haploids or homozygous ulp1 diploids relative to ULP1+ cells: in haploids, the resistant subpopulation corresponding to cells in S/G2 was larger for the mutant than for the wild type (Fig. 3C), as was expected because of the presence of a larger fraction of doublets with a single nucleus in the growing cultures, and in diploids, no difference in survival curves was found for ulp1 and wild-type cells (Fig. 3D). These results indicate that the ulp1 mutation does not affect the recombinational repair of double strand breaks (DSBs), whether it involves sister chromatids (haploids) or homologous chromosomes (diploids).
The ulp1-I615N mutation confers a spontaneous and UV-induced hyperrecombination phenotype.
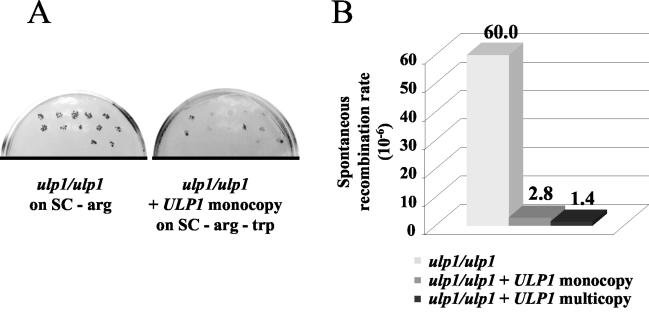
In order to determine whether recombination was affected by the ulp1-I615N mutation, we constructed heteroallelic arg4-RV/arg4-BglII diploids that were homozygous for ulp1 and either complemented or not by the ULP1+ plasmid. Intragenic recombinants are essentially generated by gene conversion in wild-type diploids. We observed that spontaneous recombinants were much more frequent for the mutant than for the Ulp1+ diploids, based on the frequencies of Arg+ papillae after replica plating of colonies on a medium lacking arginine (Fig. 4A). To determine the spontaneous recombination rates, we performed fluctuation tests according to the method of Lea and Coulson (30; also see Materials and Methods). We found the recombination rate to be about 20 times higher in the mutant diploids than in the Ulp1+ diploids (Fig. 4B). Thus, intragenic homologous recombination is highly stimulated by the mutation.
FIG. 4.
Hyperrecombination phenotype of the ulp1-I615N mutant. (A) Papillation test of ulp1 homozygous diploids (MH1088) that were heteroallelic for ARG4 on synthetic medium lacking arginine. Clones with (right) or without (left) an ULP1+ copy were grown on arginine-containing medium before being replica plated onto medium lacking arginine (left) or onto medium lacking arginine and tryptophan to maintain the ULP1-containing plasmid (right). (B) The ulp1 homozygous diploids have a spontaneous hyperrecombination phenotype. Spontaneous recombination rates were measured at the ARG4 locus in the presence or absence of one or several ULP1+ copies.
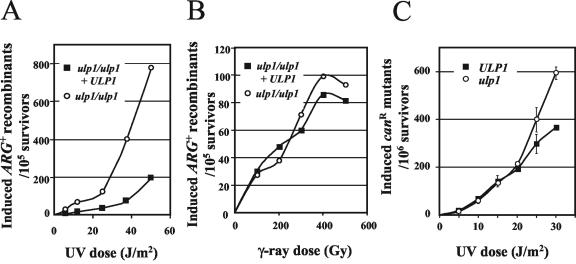
We then asked if radiation-induced recombination was also stimulated by the ulp1-I615N mutation. The frequencies of UV-induced Arg+ recombinants among the survivors were found to be increased in Ulp1− diploids compared to Ulp1+ diploids (dose modification factor = 2) (Fig. 5A). After γ-ray irradiation, the induction of recombinants was similar in the mutant and Ulp1+ diploids (Fig. 5B).
FIG. 5.
Radiation-induced recombination and UV-induced mutagenesis in ulp1-I615N mutants. (A and B) Induced ARG+ recombinant frequency in homozygous ulp1-I615N (MH1088) diploids that were heteroallelic for ARG4 after UV (A) or γ (B) irradiation in the presence or absence of an ULP1+-bearing plasmid. (C) UV-induced canR mutants in ulp1-I615N (MH1024) and ULP1+ (MH1004) haploids. Error bars show standard errors of the means from independent experiments.
We also compared UV-induced mutagenesis in mutant and wild-type haploids when they were treated during the stationary phase of growth. Similar dose-effect relationships for the induction of canR mutants (forward mutations) were found, at least in the low-dose range (Fig. 5C), indicating that mutagenic repair is not significantly affected by the ulp1-I615N mutation in G1 cells.
Altogether, these results indicate that the ulp1-I615N cells (i) accumulate recombination events during growth, (ii) are not affected in γ-ray-induced recombination and DSB repair, and (iii) are sensitive to some extent to the lethal effect of UV, a phenotype linked to increased UV-induced recombination.
RAD51-dependent homologous recombination is essential in the ulp1-I615N mutant.
As mentioned above, we were unable to recover transformants with a deletion of RAD51 in the synthetically lethal mutant B392, indicating that the ulp1-I615N and rad51 mutations themselves are likely to be synthetically lethal. This observation, together with the spontaneous hyperrecombination phenotype of ulp1 cells, raised the possibility that the whole process of homologous recombination is required for the viability of this ulp1 mutant. We next tested for synthetic lethality between the ulp1-I615N mutation and mutations in different genes governing homologous recombination. The ulp1-I615N strain was crossed with single mutants with a deletion of either RAD51, RAD52, RAD54, RAD55, RAD50, or MRE11. The diploids were sporulated and tetrad analysis was performed. The genotypes of the monosporic clones were determined (Table 2). Tetrad analysis allowed us to deduce the presumed genotypes of the missing clones. In all cases, a large majority of the ulp1 rad double mutant spores did not develop into a colony. The rare viable double mutant spores were not analyzed further and may contain some kind of suppressor. The results are clear enough, however, to conclude that all six tested rad mutations affecting the RAD51-dependent homologous recombination process are synthetically lethal with the ulp1 mutation, and therefore that this process is essential for the viability of ulp1 cells. By the same type of analysis, we confirmed the synthetic lethality of the srs2 and ulp1-I615N mutations (Table 2).
All types of homologous recombination events are dependent on RAD52, but a subclass of them do not require the RAD51 pathway and are mediated through pathways involving RAD59 (5). These types of events include the repair of DSBs by break-induced replication (58) and by single-strand annealing (SSA) (61). We therefore studied the genetic interaction of the rad59Δ mutation with ulp1 by a tetrad analysis of double heterozygous diploids. No synthetically lethal interaction was found (Table 2).
DSBs can also be repaired by nonhomologous end joining (NHEJ) (43). RAD50 and MRE11 are involved not only in homologous recombination but also in NHEJ. Thus, the synthetic lethality between rad50 and ulp1 or between mre11 and ulp1 could be due to the impairment of either one or both of the mechanisms. To determine which is true, we studied the genetic interaction between ulp1 and a yku70Δ mutation which specifically blocks NHEJ (7, 8). No synthetic lethality was observed (Table 2), indicating that NHEJ is not required for ulp1-I615N cell viability and that the interaction between ulp1 and rad50 or between ulp1 and mre11 relates to defects in homologous recombination. Thus, among the recombination processes, only the RAD51-dependent mechanisms are required for the viability of ulp1-I615N cells.
We then assayed the genetic interactions of the ulp1-I615N mutant with mutations of key genes involved in other processes participating in DNA repair and genome stability, i.e., genes controlling translesional bypass (rad6Δ and rad18Δ), excision repair (rad14Δ), and checkpoint control (rad53/sad1-1). In each case, the double mutants were viable (Table 2). This set of results indicates that among the repair mechanisms, only RAD51-dependent homologous recombination is essential for the ulp1-I615N mutant to survive.
We also questioned if the ulp1-I615N mutation genetically interacts with the deletion of the RAD27 gene, which encodes an endonuclease (Rad27/FEN1) involved in replication (37, 59), HO-induced switching of the MAT locus (22), and the processing of DNA strand breaks generated at abasic sites (70). Synthetic lethality was observed (Table 2), but whether it relates more specifically to one of the different RAD27 functions remains to be elucidated.
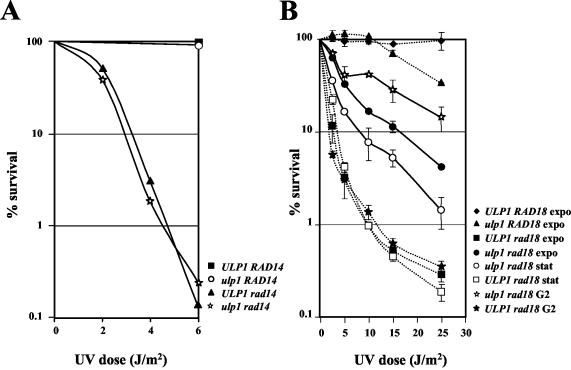
The ulp1-I615N mutation partially suppresses the UV sensitivity associated with a rad18Δ mutation.
Because ulp1 is sensitive to UV, we asked whether ulp1 and rad14 and/or ulp1 and rad18 present genetic interactions with respect to UV repair. The rad14 and rad18 mutations prevent nucleotide excision repair and translesion synthesis, respectively. The ulp1-I615N rad14Δ double mutant responds in the same way as the rad14Δ single mutant (Fig. 6A). However, due to the lack of UV sensitivity of ulp1 cells in a dose range that permits the study of the highly UV-sensitive rad14Δ cells, no conclusions could be drawn about a possible epistatic or additive interaction. A similar experiment was performed with the ulp1-I615N rad18Δ double mutant and revealed an unexpected interaction (Fig. 6B): the survival curve is biphasic, indicating the presence of two subpopulations with different UV sensitivities. Therefore, ulp1 acts as a suppressor of the rad18 sensitivity of haploids, but only in a subpopulation of cells. Moreover, this subpopulation is larger when cells are irradiated in the exponential phase than in stationary phase, suggesting that the resistant subpopulation is comprised of S/G2 cells, which are able to repair by sister chromatid recombination. That the suppression relates to S/G2 cells was further ascertained by a significant increase in the UV-resistant fraction when cell populations were enriched in G2 by preincubation in carbendazim, an inhibitor of spindle microtubule polymerization. Under these conditions, about 70% of the cells were doublets with a single nucleus, which is indicative of a G2 arrest at the time of irradiation. It should be noted that varying the proportion of G1, S, and G2 cells at the time of irradiation (exponential-phase or stationary-phase cells, with cultures enriched to 75 to 80% G2 cells by a carbendazim pretreatment) had only a minor, if any, effect on the sensitivity of rad18 simple mutants (Fig. 6B). This confirms that the biphasic survival curves of the double mutant resulted from a suppression of the UV sensitivity of S/G2 rad18 cells by the ulp1-I615N mutation. Since ulp1 cells were not viable in the absence of recombination, a direct test of the involvement of recombinational repair in ulp1 rad18 cells is precluded a priori. However, we tried to construct ulp1 rad18 rad51 or ulp1 rad18 rad52 triple mutants because of a previous report showing that the synthetic lethality between a mutation of the polymerase δ gene (pol3-13) and rad51 or rad52 is suppressed by a rad18 deletion (15). The situation with ulp1 is different, as we found no viable ulp1 rad18 rad51 or ulp1 rad18 rad52 cells among the meiotic progeny of the triple heterozygous diploids (data not shown). Although we cannot demonstrate it, the most likely explanation for the UV resistance conferred by ulp1 to rad18 cells is a channeling of repair into a recombinational pathway which is successful in at least a fraction of the S/G2 population of cells.
FIG. 6.
Interaction analysis of ulp1-I615N mutation with rad14Δ (A) or rad18Δ (B) mutation in response to UV irradiation. For both panels, UV was applied to cells from each of the four monosporic clones of a tetratype that originated from meiosis of the double heterozygotes described in Table 2. expo, exponentially growing cells; stat, stationary-phase cells; G2, populations enriched for G2 cells by a carbendazim preincubation. Error bars show standard errors of the means from independent experiments.
RAD51 and RAD52 are not deregulated in the mutant and their overexpression does not improve the mutant condition.
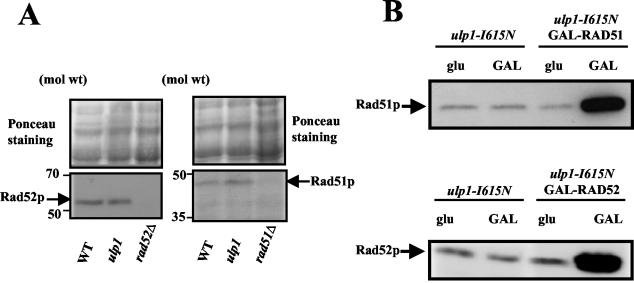
Because the ulp1 mutant has a strong spontaneous hyperrecombination phenotype, we examined the possibility that it correlates with an upregulation of homologous recombination genes. We compared Rad51 and Rad52 levels in mutant and wild-type cells during exponential growth. A Western blot analysis revealed that both protein levels were very comparable in both types of cells (Fig. 7A), indicating that Rad51 and Rad52 levels are not noticeably deregulated in the ulp1-I615N mutant.
FIG. 7.
Rad51 and Rad52 analysis and expression in ulp1-I615N haploids. (A) Rad51 and Rad52 levels are not noticeably affected in the ulp1 mutant. The images show Ponceau red staining of proteins after transfer onto a nitrocellulose membrane and immunodetection of Rad51 and Rad52 proteins with anti-Rad51 and anti-Rad52 antibodies. (B) Overexpression of Rad51 and Rad52 under control of the GAL1-10 promoter in the ulp1 mutant. Immunodetection of the Rad51 and Rad52 proteins was done with anti-Rad51 and anti-Rad52 antibodies.
The essential role of homologous recombination in ulp1 cells could be related to a saturation of recombination proteins in this genetic context. We therefore asked whether artificially up-regulating recombination protein levels could improve the mutant condition. Indeed, the increased expression of homologous recombination genes has been reported to improve mutant phenotypes in several cases: the simultaneous overexpression of Rad51 and Rad52 suppresses the X-ray sensitivity of rad55 and rad57 mutants (16), the overexpression of Rad54 suppresses the UV and MMS sensitivities of rad51 mutants (10), and the temperature-sensitive lethal phenotype of an mgs1 rad18 double mutant is suppressed by RAD52 overexpression (20). We overexpressed RAD51 and RAD52 under the control of the GAL1-10 promoter integrated as a single copy into the chromosome (see Material and Methods). Overexpression was verified by Western blotting (Fig. 7B). We checked that both the GAL-RAD51 and GAL-RAD52 copies were fully functional, as they suppress the MMS sensitivities of rad51Δ and rad52Δ strains, respectively (data not shown). Neither RAD51 nor RAD52 overexpression was found to improve the mutant condition, as shown by comparisons on YPGal medium of the responses of ulp1-I615N cells harboring (test samples) or not harboring (control samples) the GAL1-RAD51 or GAL1-RAD52 fusion: we found no increase in plating efficiencies upon overexpression; control and test samples did not differ in thermosensitivity, as assayed by drop tests at 24, 32, 35, and 37°C; they gave similar UV survival curves; and the synthetic lethality conferred by the ulp1-I615N and srs2Δ mutations was not rescued upon overexpression, as judged from the color sectoring assays used to screen synthetically lethal mutations (data not shown). Therefore, these results suggest that the high spontaneous rate of recombination and the high incidence of spontaneous death in ulp1-I615N mutants reflect a high level of recombinogenic structures rather than an altered recombinational metabolism.
The ulp1-I615N mutant accumulates single-stranded interruptions in newly synthesized DNA.
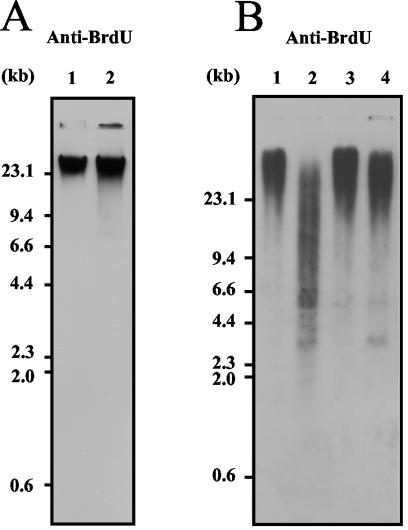
Recombinogenic structures relate to discontinuities in DNA, either DSBs or single-strand breaks. In order to determine if such structures are found in abnormal amounts in ulp1-I615N cells, we examined nucleic acid extracts from exponentially growing mutant and wild-type cells run in neutral and alkaline agarose gels. Ethidium bromide staining did not reveal any noticeable change in migration associated with the ulp1 mutation under neutral electrophoresis conditions, and therefore there was no evidence of DNA DSBs (data not shown). In contrast, after electrophoresis under alkaline conditions, whereas ssDNA from the wild type was concentrated in the high-molecular-weight area of the gel, the DNA extracted from ulp1 cells appeared as a smear (data not shown). Since about 70% of mutant cells from a growing culture did not develop into colonies, we asked whether this smear reflected a physiological consequence of the mutation or if it was due to secondary effects related to cell death. To answer this question, we used a genetic system developed by Vernis et al. (67) which offers the possibility of specifically labeling nascent DNA with BrdU. We therefore labeled the DNAs of living cells over the course of one generation and compared the sizes of newly synthesized DNAs in mutant and control cells by agarose gel electrophoresis followed by Southwestern blotting and immunodetection with an anti-BrdU antibody. Again, no marked difference was seen between the two types of cells when DNAs were separated under neutral conditions (Fig. 8A). In contrast, significant amounts of low-molecular-weight ssDNA fragments were found in the mutant extracts after electrophoresis under denaturing conditions, as evidenced by a smear extending to a position of about 1.9 kb (Fig. 8B, lane 2). Such a smear was not observed for control cells (Fig. 8B, lane 1) unless the extracts were overloaded (10×) (data not shown). This experiment therefore reveals the presence of an abnormally large amount of single-stranded interruptions in the BrdU-labeled DNA of ulp1 cells. When cells were allowed to replicate further in the absence of BrdU and in the presence of an excess of thymidine (chase), it appeared that nearly all of the incorporated BrdU had been chased into the high-molecular-weight fraction (Fig. 8B, lane 4), indicating that single-stranded interruptions had been repaired and did not occur in mutant cells once the DNA was replicated. From these data, it is clear that the ulp1-I615N mutation leads to an abnormally large amount of single-strand DNA interruptions that are specifically associated with DNA replication. This could reflect an increase in the S-phase duration, possibly due to an impairment in Okazaki fragment maturation. We did not detect ssDNA molecules of the size of Okazaki fragments, which are about 500 bp in length (41), in these experiments performed at a permissive temperature. We therefore studied the effect of the ulp1-I615N mutation after labeling at the restrictive temperature of 36.5°C. The newly replicated DNA fragments, even after 5 h of incubation, had sizes extending progressively down to about 700 bp (data not shown), compared to 1.9 kb after labeling at the permissive temperature. Thus, we do not know if the ssDNA accumulation relates to a partial defect in Okazaki fragment maturation, to a decrease in the DNA polymerization rate, or both.
FIG. 8.
Analysis of electrophoretic patterns of BrdU-labeled DNA in ulp1-I615N mutant. (A) Immunodetection of BrdU-labeled DNAs separated under neutral conditions. DNAs were prepared from ULP1+ (YLVE4; lane 1) and ulp1-I615N (MH1367; lane 2) cells after one generation with BrdU. DNAs were revealed by using an anti-BrdU antibody. (B) Immunodetection of BrdU-labeled DNAs separated under alkaline conditions. DNAs were prepared from ULP1+ (YLVE4; lanes 1 and 3) and ulp1-I615N (MH1367; lane 2 and 4) cells. Lanes 1 and 2, DNAs extracted after one generation with BrdU; lanes 3 and 4, DNAs extracted after one generation with BrdU followed by one generation of chasing. DNAs were revealed by using an anti-BrdU antibody.
We conclude from this set of experiments that, as inferred from genetic data, recombinogenic structures accumulate in the mutant during replication. Those specific structures are not DNA DSBs but single-strand breaks in newly synthesized DNA.
DISCUSSION
In a screen for synthetically lethal mutations with an srs2 deletion, we found a mutated ulp1 allele (ulp1-I615N) coding for a protein with a replacement of Ile by Asn at position 615. The ULP1 gene of S. cerevisiae was previously identified and characterized as an essential cysteine protease involved in the maturation and deconjugation from proteins of Smt3, a ubiquitin-like protein ortholog of mammalian SUMO-1 (32). It is known that the conserved C-terminal domain extending from residue 432 to residue 621, also called the Ulp domain, displays full proteolytic activity in deconjugating Smt3/SUMO protein conjugates and in processing the Smt3/SUMO precursor to its mature form (45). The same authors showed evidence of the presence of six α helixes in the C-terminal fold from residues 432 to 621, and Ile615 lies within the last helix of the crystal structure. Rasmol structure viewing software (Bernstein + Sons) was used to visualize the Ulp1-Smt3 structure (PDB no. 1EUV). It indicated that seven amino acids are in the close vicinity of Ile615 (<5 Å): they are Phe456, Met428, Ile435, Phe440, Leu614, Leu616, and Asp618. Asp618 possesses an acidic residue but is orientated towards the external part of the helix. All six other amino acids are hydrophobic, indicating that Ile615 belongs to the hydrophobic core of the protein. Moreover, Ile615 is relatively distant from the Ulp1-Smt3 interface. The replacement of Ile615 by Asn could then affect the folding of the Ulp1 hydrophobic core, as Asn is a polar but small amino acid. Most likely, it does not directly affect the catalytic activity of the protease, but rather the global stability of the protein. We inferred a decrease in the Ulp1 protease function from the fact that Smt3-conjugated proteins were overrepresented in the mutant relative to the wild type (Fig. 2). Thus, the properties of this mutant are likely related to an excess of sumoylated forms of the protein(s).
We showed that the ulp1-I615N mutation has a strong hyperrecombination effect (Fig. 4). Links between recombination and this mutation are also evidenced in this study by the synthetic lethality of ulp1 with mutations in homologous recombination genes and in SRS2 (Table 2). Thus, in ulp1 cells, recombination appears to be simultaneously essential and also deleterious, since Srs2, in some cases at least, was shown to prevent the persistence of otherwise toxic recombination intermediates (9, 13, 27, 42, 52, 66). Protein interactions also support the existence of a relationship between Srs2, homologous recombination, and sumoylation: Srs2 interacts not only with Rad51 (27) but also with Smt3 and Ubc9 (65), the Smt3 E2-conjugating protein. Rad51, in turn, interacts with Ubc9 (65). Links between sumoylation and recombination were also reported for mammalian cells: for human cells, two-hybrid assays revealed that the Rad51 or Rad52 protein interacts with Ubc9 and SUMO-1 (54, 55). Further studies demonstrated the coimmunoprecipitation of SUMO-1 and Rad51 in Chinese hamster cells but failed to evidence covalently modified forms of the Rad51 protein (34). The same authors reported that the overexpression of SUMO-1 results in a reduction in DSB-induced homologous recombination and in cellular resistance to ionizing radiation.
Two possibilities could a priori explain the hyperrecombination effect of the ulp1-I615N mutation. Either this mutation affects the activity of repair genes, leading directly or not to an increase in recombination, or it affects a function that normally avoids the formation of recombinogenic structures and those accumulate in the mutant. We found no evidence showing that the recombination process per se is affected in ulp1-I615N cells, but we demonstrated, by BrdU labeling and chase experiments, the transient presence of frequent single-stranded interruptions in the newly replicated DNA, even in cells growing at a permissive temperature (Fig. 8). No DSBs were detected. The sizes of the ssDNA fragments were larger than those predicted for Okazaki fragments. Thus, the interruptions may be due to abnormal Okazaki fragment maturation and/or increased elongation rates that are or are not related to transient replication arrests. In any case, this replicative defect could well be responsible for the different phenotypes conferred by the mutation, as discussed below.
The accumulation of single-strand breaks during replication explains not only the hyperrecombination phenotype, but also the delay or arrest in the S/G2 mitotic phase associated with low plating efficiencies. This is in accordance with the essential role of Ulp1 in the G2/M transition (32). Meiotic defects, revealed by low sporulation efficiencies and a decreased spore viability, could reasonably result from defective meiotic replication, although we did not further studied meiotic cells.
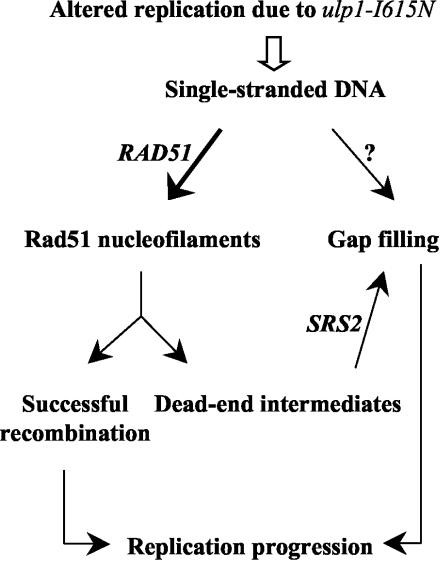
The viability of ulp1-I615N cells depends on Srs2 activities and on homologous recombination. This indicates that, in the mutant, nicks and likely single-stranded gaps accumulated in the newly replicated strands are, at least in part, metabolized through recombination. A possible model showing the repair pathways of these structures is diagrammed in Fig. 9. The requirement of Srs2 is interpreted to mean that Rad51 binds at least a subclass of single-strand gaps, leading to recombination but also to dead-end intermediates that are toxic if not dismantled by Srs2. Such a scheme was previously proposed to explain the negative interaction between sgs1 and srs2 (14). However, in contrast to the case with sgs1 cells, which are viable in the absence of homologous recombination, the ulp1 cells require a functional RAD51-dependent homologous recombination process for viability (Table 2). Therefore, depending upon the context of its initiation, RAD51-dependent recombination is potentially toxic or essential for repair in the ulp1 mutant. It could be that the accumulation of ssDNA regions in the newly replicated DNA results in some cases in the formation of DSBs that require recombination to be repaired. The BrdU technique used here is not sensitive enough to detect a low DSB level. Alternatively, due to the ulp1 mutation, single-stranded structures of a particular class could be repairable only by recombination. These structures could be gaps adjacent to the replication fork on the lagging strand, which cannot be filled by DNA synthesis due to the lack of a 3′ priming end (for a review, see reference 39).
FIG. 9.
Model accounting for the essential roles of both SRS2 and homologous recombination genes in ulp1-I615N cells. The ulp1-I615N mutation is responsible for an altered replication yielding single-stranded gaps in nascent DNA. Recombination is initiated, for at least a fraction of these gaps, by the formation of Rad51 nucleofilaments. A subclass of them is processed by recombination and the others are processed by Srs2, which allows gap filling by DNA synthesis. In the absence of Srs2, toxic recombination intermediates lead to cell death.
Because Smt3 has numerous targets, the possibility that the ulp1-I615N mutation also leads to a modification of recombination activities remains open. However, as stated above, our results strongly support the view that homologous recombination metabolism is not impaired in ulp1 cells. The mutant has a strong spontaneous hyperrecombination phenotype (Fig. 4) and shows an increase in UV-induced intragenic recombinants (Fig. 5A). The γ-ray responses (survival and recombination) are not affected by the ulp1 mutation (Fig. 3C, 3D, and 5B), indicating that DSB repair is normal. Moreover, Rad51 and Rad52 proteins appear to be present in similar amounts in mutant and wild-type cells (Fig. 7A), and overexpression of these proteins (Fig. 7B) did not rescue the mutant phenotype. Altogether, these results indicate that recombination is not impaired, but rather that the effects of the ulp1-I615N mutation on recombination relate to the abnormal incidence of recombinogenic structures.
We did not know what Smt3 target is responsible for the replicative defect in ulp1 cells. A candidate was PCNA, which is known to be involved in DNA replication and several DNA repair processes (for a review, see reference 69). In yeast, PCNA can be modified at the same lysine residue by either a Rad6/Rad18-mediated ligation of a single ubiquitin molecule or sumoylation. These modifications differentially affect responses to DNA damage: the mono-ubiquitinated form of PCNA is required for damage-induced translesion synthesis, while the sumoylated form is involved in the replication of undamaged templates (21, 60). It is then conceivable that, even in untreated cells, an impairment in PCNA modifications due to the ulp1-I615N mutation results in replication defects. After UV irradiation, we observed a genetic interaction between rad18 and ulp1-I615N, which also suggests that the regulation of the ubiquitination/sumoylation modifications plays an important role in DNA repair. The ulp1-I615N mutation suppresses the UV sensitivity of a subpopulation of rad18Δ haploids (Fig. 6B). We believe that the resistant ulp1 rad18 cells are S/G2 cells, which are able to repair by recombination between sister chromatids. Indeed, the size of the resistant subpopulation is larger in G2 cell-enriched cultures than in exponentially growing cells; the latter, in turn, have a larger fraction of resistant cells than cells in stationary phase, in which a large proportion of cells are in G1. A direct test of recombination involvement is precluded by the synthetically lethal interactions between the ulp1-I615N mutation and mutations in recombination genes. However, the coexpression of MATa and MATα, known to favor recombinational repair (28, 44, 51), leads to a similar suppression of the rad18 sensitivity (18), also supporting the hypothesis that the resistance of a subpopulation is due to recombinational repair. Thus, the UV responses strongly suggest that in the rad18 context, the ulp1-I615N mutation channels the repair of otherwise lethal structures into a recombinational process. Whether this channeling relates to PCNA modifications, and consequently to the interplay between polymerases, is an attractive hypothesis that remains to be demonstrated.
It is striking that the viability of several mutants that accumulate small ssDNA fragments during replication depends, like that of the ulp1-I615N mutant, on homologous recombination. This is the case for different PCNA mutants (pol30) (41) and rad27 mutants (62, 64). Rad27 (FEN1) is an endonuclease involved in the processing of Okazaki fragments (37, 59) whose activity is stimulated via binding to PCNA (35). Rad27 is also involved in base excision repair, as shown in rad27 mutants by the accumulation of DNA single-strand breaks generated by endonucleolytic cleavage of abasic sites (70). Like ulp1 cells, the rad27 mutant displays a hyperrecombination phenotype (64) and shows an increased sensitivity to UV light, but not to ionizing radiation (48). Furthermore, the rad27 mutant is also synthetically lethal with the srs2 deletion (12, 26). These similarities support the view that the properties of both ulp1 and rad27 mutants relate to single-stranded interruptions or gaps in newly replicated DNA. Different hypotheses could explain the synthetic lethality conferred by the ulp1-I615N and rad27Δ mutations reported here. It could result from the addition of replicative defects, since both single mutants are viable but sick. In that case, the two mutations could affect the same function, such as the maturation of Okazaki fragments, or different functions, such as base excision repair and the maturation of Okazaki fragments. Alternatively, it could be that abnormally sumoylated proteins during replication lead to ssDNA structures that require Rad27 to be fixed.
In conclusion, these results show that the regulation of sumoylation plays an important role in DNA replication and that in the ulp1-I615N mutant, the formation of single-stranded gaps in the newly replicated DNA is likely due to an abnormal persistence of a sumoylated protein(s). The generation of these structures could by itself explain the mutant phenotypes. The binding of recombination proteins at these gaps would result in some cases in recombinational repair, accounting for the hyperrecombination phenotype. In other cases, it would result in dead-end recombination intermediates that require Srs2 to be dismantled and repaired by an alternative pathway(s).
Acknowledgments
This work was supported by grants from the Centre National de la Recherche Scientifique (UMR2027 CNRS/I.C and UMR217 CEA/CNRS), the Institut National de la Santé et de la Recherche Médicale (INSERM), the Association pour la Recherche sur le Cancer (ARC), the Institut Curie and the Commissariat à l'Energie Atomique (CEA). Christine Soustelle was supported by a postdoctoral fellowship from CEA.
Thanks are due to Serge Urbach for his expert advice about immunodetection, to Patrick Hughes for his comments and corrections on the manuscript, to Serge Gangloff for the pSG plasmids, to Mark Hochstasser for the Smt3 antibody, and to Natalie Declerck for help in using Rasmol. We also thank Gérard Faye and Giuseppe Baldacci for helpful discussions about this work.
REFERENCES
- 1.Aboussekhra, A., R. Chanet, A. Adjiri, and F. Fabre. 1992. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol. 12:3224-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aboussekhra, A., R. Chanet, Z. Zgaga, C. Cassier-Chauvat, M. Heude, and F. Fabre. 1989. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 17:7211-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilera, A., and H. L. Klein. 1988. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics 119:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexeev, A., A. Mazin, and S. C. Kowalczykowski. 2003. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 10:182-186. [DOI] [PubMed] [Google Scholar]
- 5.Bai, Y., and L. S. Symington. 1996. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10:2025-2037. [DOI] [PubMed] [Google Scholar]
- 6.Bender, A., and J. R. Pringle. 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulton, S. J., and S. P. Jackson. 1996. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double- strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15:5093-5103. [PMC free article] [PubMed] [Google Scholar]
- 9.Chanet, R., M. Heude, A. Adjiri, L. Maloisel, and F. Fabre. 1996. Semidominant mutations in the yeast Rad51 protein and their relationships with the Srs2 helicase. Mol. Cell. Biol. 16:4782-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clever, B., H. Interthal, J. Schmuckli-Maurer, J. King, M. Sigrist, and W. D. Heyer. 1997. Recombinational repair in yeast: functional interactions between Rad51 and Rad54 proteins. EMBO J. 16:2535-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullin, C., and D. Pompon. 1988. Synthesis of functional mouse cytochromes P-450 P1 and chimeric P-450 P3-1 in the yeast Saccharomyces cerevisiae. Gene 65:203-217. [DOI] [PubMed] [Google Scholar]
- 12.Debrauwere, H., S. Loeillet, W. Lin, J. Lopes, and A. Nicolas. 2001. Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl. Acad. Sci. USA 98:8263-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabre, F., A. Chan, W. D. Heyer, and S. Gangloff. 2002. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 99:16887-16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25:192-194. [DOI] [PubMed] [Google Scholar]
- 15.Giot, L., R. Chanet, M. Simon, C. Facca, and G. Faye. 1997. Involvement of the yeast DNA polymerase delta in DNA repair in vivo. Genetics 146:1239-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hays, S. L., A. A. Firmenich, and P. Berg. 1995. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl. Acad. Sci. USA 92:6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heude, M., R. Chanet, and F. Fabre. 1995. Regulation of the Saccharomyces cerevisiae Srs2 helicase during the mitotic cell cycle, meiosis and after irradiation. Mol. Gen. Genet. 248:59-68. [DOI] [PubMed] [Google Scholar]
- 18.Heude, M., and F. Fabre. 1993. a/α-control of DNA repair in the yeast Saccharomyces cerevisiae: genetic and physiological aspects. Genetics 133:489-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hickson, I. D. 2003. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3:169-178. [DOI] [PubMed] [Google Scholar]
- 20.Hishida, T., T. Ohno, H. Iwasaki, and H. Shinagawa. 2002. Saccharomyces cerevisiae MGS1 is essential in strains deficient in the RAD6-dependent DNA damage tolerance pathway. EMBO J. 21:2019-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135-141. [DOI] [PubMed] [Google Scholar]
- 22.Holmes, A. M., and J. E. Haber. 1999. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell 96:415-424. [DOI] [PubMed] [Google Scholar]
- 23.Huang, M. E., A. de Calignon, A. Nicolas, and F. Galibert. 2000. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase delta, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr. Genet. 38:178-187. [DOI] [PubMed] [Google Scholar]
- 24.Kawabe, Y., M. Seki, T. Seki, W. S. Wang, O. Imamura, Y. Furuichi, H. Saitoh, and T. Enomoto. 2000. Covalent modification of the Werner's syndrome gene product with the ubiquitin-related protein, SUMO-1. J. Biol. Chem. 275:20963-20966. [DOI] [PubMed] [Google Scholar]
- 25.Kaytor, M. D., M. Nguyen, and D. M. Livingston. 1995. The complexity of the interaction between RAD52 and SRS2. Genetics 140:1441-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein, H. L. 2001. Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Δ with other DNA repair genes in Saccharomyces cerevisiae. Genetics 157:557-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy, H. Klein, T. Ellenberger, and P. Sung. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305-309. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski, W. 1960. Inaktivierungsversuche mit hefestÂmmen verschiedenen ploidiegrades. I. Aufbau homozygoten stämme und dosiseffektkürven für ionisierende strahlen, UV und organische peroxyde. Z. Naturforsch. 1960:495-506. [Google Scholar]
- 29.Lawrence, C. W., and R. B. Christensen. 1979. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J. Bacteriol. 139:866-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lea, D. E., and C. A. Coulson. 1948. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49:264-284. [DOI] [PubMed] [Google Scholar]
- 31.Lehembre, F., P. Badenhorst, S. Muller, A. Travers, F. Schweisguth, and A. Dejean. 2000. Covalent modification of the transcriptional repressor tramtrack by the ubiquitin-related protein Smt3 in Drosophila flies. Mol. Cell. Biol. 20:1072-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, S. J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398:246-251. [DOI] [PubMed] [Google Scholar]
- 33.Li, S. J., and M. Hochstrasser. 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20:2367-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, W., B. Hesabi, A. Babbo, C. Pacione, J. Liu, D. J. Chen, J. A. Nickoloff, and Z. Shen. 2000. Regulation of double-strand break-induced mammalian homologous recombination by UBL1, a RAD51-interacting protein. Nucleic Acids Res. 28:1145-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, X., J. Li, J. Harrington, M. R. Lieber, and P. M. Burgers. 1995. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J. Biol. Chem. 270:22109-22112. [DOI] [PubMed] [Google Scholar]
- 36.Liberi, G., I. Chiolo, A. Pellicioli, M. Lopes, P. Plevani, M. Muzi-Falconi, and M. Foiani. 2000. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 19:5027-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lieber, M. R. 1997. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays 19:233-240. [DOI] [PubMed] [Google Scholar]
- 38.Mazin, A. V., A. A. Alexeev, and S. C. Kowalczykowski. 2003. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 278:14029-14036. [DOI] [PubMed] [Google Scholar]
- 39.McGlynn, P., and R. G. Lloyd. 2002. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell. Biol. 3:859-870. [DOI] [PubMed] [Google Scholar]
- 40.Melchior, F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 41.Merrill, B. J., and C. Holm. 1998. The RAD52 recombinational repair pathway is essential in pol30 (PCNA) mutants that accumulate small single-stranded DNA fragments during DNA synthesis. Genetics 148:611-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milne, G. T., T. Ho, and D. T. Weaver. 1995. Modulation of Saccharomyces cerevisiae DNA double-strand break repair by SRS2 and RAD51. Genetics 139:1189-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore, J. K., and J. E. Haber. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mortimer, R. K. 1958. Radiobiological and genetic studies on a polyploid series (haploid to hexaploid) of Saccharomyces cerevisiae. Radiat. Res. 9:312-326. [PubMed] [Google Scholar]
- 45.Mossessova, E., and C. D. Lima. 2000. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5:865-876. [DOI] [PubMed] [Google Scholar]
- 46.Muller, S., C. Hoege, G. Pyrowolakis, and S. Jentsch. 2001. SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell. Biol. 2:202-210. [DOI] [PubMed] [Google Scholar]
- 47.Palladino, F., and H. L. Klein. 1992. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics 132:23-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reagan, M. S., C. Pittenger, W. Siede, and E. C. Friedberg. 1995. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 177:364-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Resnick, M. A., S. Stasiewicz, and J. C. Game. 1983. Meiotic DNA metabolism in wild-type and excision-deficient yeast following UV exposure. Genetics 104:583-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rong, L., and H. L. Klein. 1993. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 268:1252-1259. [PubMed] [Google Scholar]
- 51.Saeki, T., I. Machida, and S. Nakai. 1980. Genetic control of diploid recovery after gamma-irradiation in the yeast Saccharomyces cerevisiae. Mutat. Res. 73:251-265. [DOI] [PubMed] [Google Scholar]
- 52.Schild, D. 1995. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics 140:115-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seeler, J. S., and A. Dejean. 2001. SUMO: of branched proteins and nuclear bodies. Oncogene 20:7243-7249. [DOI] [PubMed] [Google Scholar]
- 54.Shen, Z., P. E. Pardington-Purtymun, J. C. Comeaux, R. K. Moyzis, and D. J. Chen. 1996. Associations of UBE2I with RAD52, UBL1, p53, and RAD51 proteins in a yeast two-hybrid system. Genomics 37:183-186. [DOI] [PubMed] [Google Scholar]
- 55.Shen, Z., P. E. Pardington-Purtymun, J. C. Comeaux, R. K. Moyzis, and D. J. Chen. 1996. UBL1, a human ubiquitin-like protein associating with human RAD51/RAD52 proteins. Genomics 36:271-279. [DOI] [PubMed] [Google Scholar]
- 56.Sherman, F., G. R. Fink, and C. W. Lawrence. 1974. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 57.Shiio, Y., and R. N. Eisenman. 2003. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA 100:13225-13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Signon, L., A. Malkova, M. L. Naylor, H. Klein, and J. E. Haber. 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 21:2048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sommers, C. H., E. J. Miller, B. Dujon, S. Prakash, and L. Prakash. 1995. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′-to-3′ exonuclease required for lagging strand DNA synthesis in reconstituted systems. J. Biol. Chem. 270:4193-4196. [DOI] [PubMed] [Google Scholar]
- 60.Stelter, P., and H. D. Ulrich. 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425:188-191. [DOI] [PubMed] [Google Scholar]
- 61.Sugawara, N., G. Ira, and J. E. Haber. 2000. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Symington, L. S. 1998. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 26:5589-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor, D. L., J. C. Ho, A. Oliver, and F. Z. Watts. 2002. Cell-cycle-dependent localisation of Ulp1, a Schizosaccharomyces pombe Pmt3 (SUMO)-specific protease. J. Cell Sci. 115:1113-1122. [DOI] [PubMed] [Google Scholar]
- 64.Tishkoff, D. X., N. Filosi, G. M. Gaida, and R. D. Kolodner. 1997. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88:253-263. [DOI] [PubMed] [Google Scholar]
- 65.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 66.Veaute, X., J. Jeusset, C. Soustelle, S. C. Kowalczykowski, E. Le Cam, and F. Fabre. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423:309-312. [DOI] [PubMed] [Google Scholar]
- 67.Vernis, L., J. Piskur, and J. F. Diffley. 2003. Reconstitution of an efficient thymidine salvage pathway in Saccharomyces cerevisiae. Nucleic Acids Res. 31:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 69.Warbrick, E. 2000. The puzzle of PCNA's many partners. Bioessays 22:997-1006. [DOI] [PubMed] [Google Scholar]
- 70.Wu, X., and Z. Wang. 1999. Relationships between yeast Rad27 and Apn1 in response to apurinic/apyrimidinic (AP) sites in DNA. Nucleic Acids Res. 27:956-962. [DOI] [PMC free article] [PubMed] [Google Scholar]