Abstract
Overexpression or amplification of ACTR (also named AIB1, RAC3, p/CIP, TRAM-1, and SRC-3), a member of the p160 family of coactivators for nuclear hormone receptors, has been frequently detected in multiple types of human tumors, including breast cancer. However, its role in cancer cell proliferation and the underlying mechanism are unclear. Here, we show that overexpression of ACTR not only enhances estrogen-stimulated cell proliferation but also, more strikingly, completely negates the cell cycle arrest effect by tamoxifen and pure antiestrogens. Unexpectedly, we found that ACTR directly interacts, through its N-terminal domain, with E2F1 and is recruited to E2F target gene promoters. Elevation of ACTR in quiescent cells strongly stimulates the transcription of a subset of E2F-responsive genes that are associated with the G1/S transition. We also demonstrated, by adenovirus vector-mediated RNA interference, that ACTR is required for E2F1-mediated gene expression and the proliferation of estrogen receptor (ER)-negative breast cancer cells. Moreover, the ability of elevated ACTR to promote estrogen-independent cell proliferation depends on the function of E2F1 and the association between ACTR and E2F1, but not ER. Thus, our results reveal an essential role of ACTR in control of breast cancer cell proliferation and implicate the ACTR-E2F1 pathway as a novel mechanism in antiestrogen resistance.
ACTR (also named AIB1, RAC3, and TRAM1; named SRC-3 and p/CIP in the mouse) is a member of the p160/SRC coactivator family (17, 20, 30, 32). Like other p160 coactivators, ACTR/AIB1 was identified as a nuclear cofactor that associates with hormone-bound nuclear receptors and mediates the transcriptional activation function of the receptors (2, 7, 25, 44, 45). Structural and functional studies revealed that the p160s contain functional domains for histone acetyltransferase activity and interactions with ligand-bound receptors, the coregulator proteins CBP and p300, PCAF, and arginine methyltransferase CARM1. Evidence from biochemical studies suggests that the association with the above-mentioned cofactors is important for the p160s to mediate the transactivation by nuclear receptors (15, 42). It is generally conceived that the p160s are recruited to hormone-responsive genes through their interaction with activated receptors and then nucleate the assembly of a coactivator complex, which in turn remodels chromatin through histone modifications and facilitates RNA polymerase II transcription. The role of ACTR/AIB1 as well as the other p160s in animal development and physiology has been explored by genetic approaches (18, 35, 51, 56, 57). Similar to the other p160 knockouts, p/CIP/SRC-3 knockout mice exhibited abnormal development and reduced function of their reproductive systems. Unlike the SRC-1 knockout mice, however, animals with the SRC-3/p/CIP gene deleted displayed significant somatic growth retardation and a reduced capacity of growth factor-stimulated cell proliferation, although the underlying molecular mechanism is unclear (51, 56).
The growth of estrogen receptor (ER)-positive human breast cancers is generally dependent on stimulation by estrogens (1). Experimental evidence indicates that ER mediates the mitogenic effect of estrogen primarily by up-regulating transcription of cyclin D1 (37, 41). Hyperphosphorylation of pRb by increased cyclin D1-Cdk activities leads to accelerated G1 progression. Antiestrogens such as tamoxifen are effective in antagonizing the growth of ER-positive breast cancer by arresting cells in G0/G1. However, resistance to such endocrine therapy often evolves after prolonged treatment. Several potential mechanisms have been proposed to account for the development of antiestrogen resistance (1). Overexpression of ERBB2 and/or deregulation of other kinase-mediated signaling could allow ER to be phosphorylated and activated independent of hormones, which could enhance ER's ability to interact with the coactivators and activate transcription. Down-regulation of transcriptional corepressors may inhibit the transcriptional repression activity of antiestrogen-bound ER. These molecular events would culminate in increased expression and activity of cyclin D1-Cdks. On the other hand, subgroups of ER-positive and -negative breast cancers do not contain high levels of cyclin D1; instead, they overexpress cyclin E (22, 26), suggesting that alternative mechanisms contribute to cell cycle progression in breast cancer.
The identification of ACTR/AIB1 as a gene amplified (in less than 10%) and/or overexpressed in over 30% of breast cancers provided the first genetic link of aberrant ACTR/AIB1 function to tumorigenesis (2). The fact that ACTR/AIB1 is a coactivator for nuclear receptors, including ER, and the initial analysis showing a correlation between ACTR/AIB1 amplification and positive ER status in breast cancers support the assertion that deregulated ACTR/AIB1 accelerates breast cancer cell proliferation by amplifying the mitogenic effect of estrogen through potentiating the transactivation activity of ER. However, subsequent studies with unselected breast cancer samples found that overexpression of ACTR/AIB1 does not correlate with ER status, but instead correlates with abnormal levels of HER-2/neu/ERBB2 (4). Further investigations found ACTR/AIB1 amplification and/or overexpression in other malignancies, including gastric, pancreatic, and hepatocellular carcinomas (19, 39, 50). It is likely that aberrant ACTR/AIB1 function in cancer may target other nuclear receptors and/or cellular factors, in addition to ER.
We report here that ACTR is required for both estrogen-stimulated and ER-independent breast cancer cell proliferation and that ACTR overexpression renders ER-positive cells completely resistant to antiestrogens. We provide several lines of evidence that ACTR can function as a coactivator for one of the key cell cycle regulators, E2F1, which underlies the change of gene expression and cell proliferation we observed in breast cancer cells with ACTR overexpression. Therefore, our results suggest a novel mode of ACTR/AIB1 function in breast cancer and directly link ACTR/AIB1 to cell cycle control mechanisms.
MATERIALS AND METHODS
RNAi adenovirus vectors and adenovirus vectors for gene expression.
For RNA interference (RNAi) vector construction, the human H1 gene promoter sequence was inserted into the pShuttle plasmid from the Adeasy system (21). Oligodeoxynucleotides containing coding sequences for small interfering RNA (siRNA) were inserted downstream of the H1 promoter (5). For ACTR expression, the original pShuttle-CMV was modified for improved expression. ACTR cDNAs were inserted into the modified vector as described before (27). The resulting pShuttle constructs were used to generate recombinant adenoviruses, following the protocols described previously (21). Viruses were purified by centrifugation in a CsCl step gradient. Viral titers were determined using 293 cells by end point cytopathic effect assay with adeno-GFP as reference.
Cell cultures, stable transfectants, and reporter gene assay.
All cells were obtained from the American Type Culture Collection and maintained in the recommended media with necessary supplements (Gibco). For generation of sublines of T-47D cells ectopically expressing ACTR, T-47D cells were transfected with pcD-HCMV-ACTR-HA and pcD-HCMV empty vector using Fugene (Roche). Transfected cells were selected in medium containing G418 (700 μg/ml). Individual clones were analyzed for ectopic ACTR expression using antihemagglutinin (α-HA; F-7; Santa Cruz) and ACTR (Transduction Labs) antibodies. Reporter gene assays with promoter-luc were performed by transfecting T-47D cells with firefly luciferase reporter plasmids pGL3-Cdk2 promoter (3.5 kb) or cyclin E promoter (1.2 kb), pRL-SV40 Renilla luciferase plasmid (Promega), and pcD-HCMV-ACTR-HA and pCMV-E2F1. Assays with 3x E2F-tk-luc and 3x ERE-tk-luc were performed by cotransfection in 293T cells with pCMV-E2F1 and pSh-HCMV-ACTR-HA or its mutant forms using Lipofectamine. Assays with pSh-Ri-ACTR, pSh-Ri-ERa, or pSh-Ri-GFP were performed by cotransfection with 3x E2F-tk-luc and pCMV-E2F1. Forty-eight hours after transfection, cells were harvested for Western and luciferase assays using a dual luciferase assay kit (Promega). All reporter gene assays were performed in triplicate, with the entire experiment repeated at least twice.
Cell proliferation and fluorescence-activated cell sorter (FACS) analysis of cell cycle.
T-47D and MDA-MB 361 cells were deprived of hormone by maintaining them in medium supplemented with 5% hormone-depleted fetal bovine serum (FBS) for the indicated times before infection with the adenovirus vectors. ER-negative cells were treated in the same manner as ER-positive ones, except they were maintained in medium supplemented with 2% hormone-depleted FBS during the proliferation assay. Cell proliferation was measured every 2 days by either a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay (Sigma) or by cell counting of coded samples in triplicate. Proliferation assays in the presence of antagonists 4(OH)-tamoxifen (Sigma) and ICI 182,780 (herein referred to as ICI; Tocris) were conducted in similar fashion, except cells were hormone deprived for at least 24 h and then treated with 10−8 M antagonist for 36 h prior to infection with adenovirus vectors. Cells were refed with fresh medium containing specified ligands every 2 days. For cell cycle analysis by flow cytometry, cells were hormone deprived and treated with ICI for 36 h before infection with either adeno-ACTR or adeno-GFP. At different times after infection, cells were labeled with 10 μM 5-bromo-2′-deoxyuridine (BrdU; Roche). An additional 10 h later, cells were harvested by trypsinization and washed twice with phosphate-buffered saline. Cells were fixed with chilled 70% ethanol and permeabilized in 2 N HCl and 0.1% Triton X-100. Following neutralization with 0.1 M sodium tetraborate (Sigma), cells were incubated with fluorescein isothiocyanate-conjugated α-BrdU antibody (Roche) for 30 min in the presence of 0.1% bovine serum albumin and 0.5% Tween 20. Cells were washed twice and resuspended in phosphate-buffered saline containing 5 μg of propidium iodide (Sigma)/ml. Flow cytometry analysis was performed on a FACScan (Coulter). Data were collected from 20,000 cells per sample.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described previously (27) with the following modifications. T-47D cells grown to 70% confluence were hormone deprived, treated with 10−8 M ICI, and infected with adenovirus vectors as described above for flow cytometry analysis. After the indicated times, cells were fixed in 1% formaldehyde for 8 min. Cells were then washed and collected for sonication. The crude chromatin solution was diluted and incubated with specific antibodies overnight at 4°C. PCR was performed using 5 μl of purified ChIP DNA for 28 cycles with promoter-specific primers. Primer sequences are listed in the Appendix.
Western blotting and quantitative RT-PCR analysis.
Whole-cell lysates were prepared from cells treated and harvested at the indicated times in buffer containing 50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 10% glycerol, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail. Western blotting analysis of proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using the following specific antibodies: α-ACTR, α-TIF2, α-cyclin A, α-cyclin B, and α-Cdk4 (Transduction Labs); α-Cdk2 (M2), α-Cdc2 (17) α-cyclin D1 (H295), α-cyclin D2 (C17), α-cyclin E (HE12), α-SRC-1 (M341), α-p107 (C18), α-p130 (C20), α-p21 (M19), α-p27 (M197), α-E2F1 (KH95), α-E2F3 (C18), and α-E2F4 (C20; Santa Cruz); α-E2F2 (BD), α-Rb (IF8), and α-ERα (Ab-15; Neomarkers); and α-β-actin (Sigma). For quantitative reverse transcriptase PCR (RT-PCR) analysis of gene expression, total RNA was isolated with TRIzol reagent. Three micrograms of total RNA was used for a reverse transcription reaction with Moloney murine leukemia virus RT and oligo(dT)18 primers; cDNAs were diluted 10-fold, and a 5-μl dilution was used for the PCRs. Gene sequences were amplified in the presence of SYBR Green fluorophore and detected using the iCycler real-time PCR equipment (Bio-Rad). Fluorescent values after each elongation step were collected along with a melting curve analysis at the end of the PCR. Fold difference (N) was calculated using the equation N = E(CT of ACTR) − (CT of GFP), between cells infected with adeno-ACTR and adeno-GFP, or as N = E(CT of E2) − (CT of ICI), between cells treated with estrogen and ICI. E is the PCR efficiency of the specific primer pair used and was determined by the standard curve [E =10−(1/slope of standard curve)]. CT is the threshold cycle, which is defined as the cycle number at which the fluorescence signal generated by the PCR product is first detected. The primer sequences are listed in the Appendix.
Coimmunoprecipitation and GST pull-down.
For immunoprecipitation, T-47D cells infected with either adeno-ACTR or adeno-GFP and 293T cells transfected with the expression constructs were lysed in 150 mM NaCl, 0.5% NP-40, 2 mM EDTA, 10% glycerol, and 50 mM Tris-HCl (pH 8.0). Cell lysates were sonicated, and cell debris was removed by centrifugation. The lysates were precleared by incubation with protein A- or G-Sepharose beads (Zymed) for 1 h at 4°C and then immunoprecipitated with α-E2F1 (KH95), α-E2F4 (A-20; Santa Cruz), α-ACTR (8), or control α-mouse immunoglobulin G (IgG) monoclonal antibody, followed by incubation with protein A or G beads. After extensive washing, the precipitates were analyzed by Western blotting with α-ACTR (Transduction Labs), α-E2F1, or α-Flag antibodies. For glutathione S-transferase (GST) pull-down, ACTR proteins either purified as described previously (8) or produced in the presence of [35S]methionine were incubated with an equal amount of bead-bound GST fusion proteins at 4°C for 30 min in a binding buffer containing 20 mM HEPES (pH 7.9), 150 mM KCl, 1 mM EDTA, 4 mM MgCl2, 1 mM dithiothreitol, 0.02% NP-40, 10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride, and 2 mg of bovine serum albumin/ml. The beads were washed three times with the binding buffer and resuspended in SDS-PAGE sample buffer. ACTR proteins retained on the beads were separated by SDS-PAGE and visualized by Western blotting or autoradiography.
RESULTS
ACTR is required for both E2-stimulated and E2/ER-independent breast cancer cell growth.
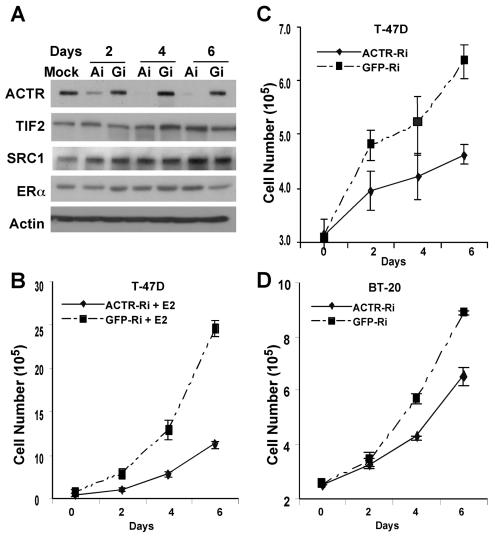
We first examined the role of ACTR in the proliferation of estrogen-responsive breast cancer cells. To this end, we constructed adenoviral vectors to mediate the expression of siRNA for specific silencing of ACTR expression. As shown in Fig. 1A, the ACTR protein level in T-47D cells was dramatically reduced 2 days after the cells were treated with ACTR RNAi (Ai) adenovirus vector, while no significant effect was observed with the green fluorescent protein (GFP) RNAi (Gi) vector. Importantly, the expression of other p160 coactivators SRC-1 and TIF2/GRIP, as well as ERα, was not significantly affected (compare Ai with Gi lanes). To examine the effect of ACTR depletion on estrogen-stimulated cell proliferation, we treated ACTR-depleted T-47D cells with 10−8 M 17β-estradiol (E2). The results in Fig. 1B indicate that depletion of ACTR dramatically inhibited E2-stimulated cell proliferation. This remarkable effect prompted us to examine whether hormone-independent growth was also affected by ACTR depletion. As expected, T-47D cells proliferated slowly without hormone stimulation (Fig. 1C; note that cell numbers were only doubled after 6 days). Strikingly, in the same experiment as in Fig. 1B, depletion of ACTR further reduced the slow growth rate of T-47D cells in the absence of E2 (Fig. 1C). We also depleted ACTR in ER-negative cells. Results in Fig. 1D demonstrate that high levels of ACTR are critical for hormone-independent proliferation of BT-20 cells. Taken together, these results revealed that, in addition to mediating estrogen induction of cell proliferation, ACTR plays an important role in breast cancer cell proliferation that is independent of estrogen and ER.
FIG. 1.
Depletion of ACTR by adenovirus-mediated RNAi inhibits the proliferation of both ER-positive and -negative breast cancer cells. Cells growing in six-well plates were hormone deprived prior to infection with an equal amount of adeno-ACTR-RNAi (labeled ACTR-Ri or Ai) or adeno-GFP-RNAi (labeled GFP-Ri or Gi) at an estimated multiplicity of infection of 50. Different days after infection, cells were harvested for Western analysis (A) or cell proliferation assay (B to D). For estrogen (E2)-stimulated proliferation analyses (as in panel B), cells were treated with 10−8 M 17β-estradiol (E2) when infected. Cells were refed with fresh medium containing E2 every 2 days or harvested and counted in triplicate. For hormone-independent cell proliferation, cells were maintained in hormone-deprived medium. Cells mock infected showed no difference in cell growth from cells infected with adeno-GFP-Ri.
Elevation of ACTR expression promotes hormone-independent cell growth and overcomes antiestrogen-mediated cell cycle arrest.
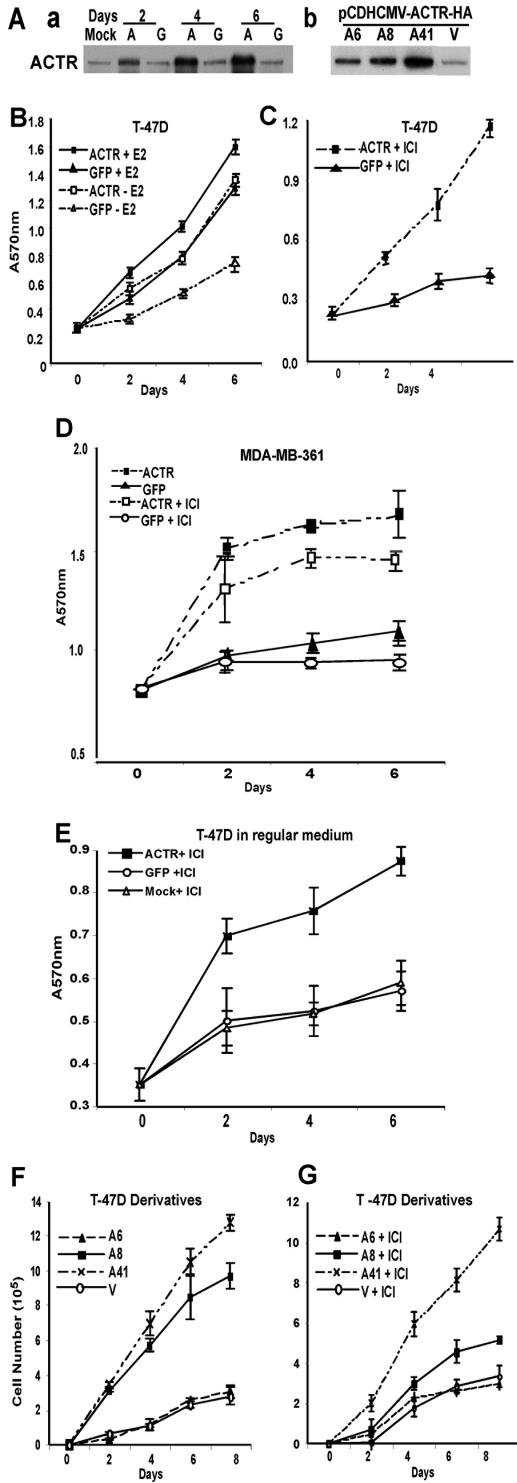
To directly address the consequences of ACTR overexpression on cell proliferation, we ectopically expressed ACTR using adenovirus vectors in a number of human breast cancer cells with low to moderate levels of endogenous ACTR protein (Fig. 2A, panel a, and data not shown). The ectopic expression of ACTR was controlled at levels comparable to those in MCF-7 cells and tumors where ACTR is amplified and overexpressed (34) and did not affect the expression of other coactivators and ER (unpublished data). Cell proliferation was measured by MTT assay at different days after the viral vector treatment. As expected, increased ACTR expression significantly enhanced E2-stimulated cell proliferation (Fig. 2B, compare ACTR+E2 and GFP+E2). Remarkably, overexpression of ACTR increased hormone-independent cell proliferation to a rate that was equivalent to E2-stimulated proliferation (compare ACTR-E2 and GFP+E2). Antiestrogens such as ICI effectively inhibit E2-stimulated cell proliferation by disruption of ER-coactivator interactions and induction of ER proteolysis (10, 12, 47, 53). Strikingly, when T-47D cells were rendered quiescent by treatment with 4(OH)-tamoxifen or the pure antiestrogen ICI, elevation of ACTR effectively promoted their proliferation, even in the continuous presence of the antiestrogens (Fig. 2C and unpublished data). Similar activities of elevated ACTR to stimulate hormone-independent cell proliferation and/or overcome the growth suppression by antiestrogens were observed in other E2-responsive cells, such as MDA-MB-361 (Fig. 2D) and ZR-75-1 (data not shown), or when cells were treated with ICI in regular growth medium (Fig. 2E).
FIG.2.
Overexpression of ACTR stimulates hormone-independent cell proliferation and confers antiestrogen resistance. (A) Ectopic expression of ACTR in T-47D cells by the adenovirus vector (a) or by stable transfection (b). ACTR protein levels in T-47D cells infected with adeno-ACTR (labeled with an A) or adeno-GFP (labeled with a G) or mock infected (Mock) at different days after infection (a), or levels in ACTR stable transfectants of T-47D cells (A6, A8, and A41) and an empty vector transfectant (labeled with a V) (b) were determined by Western analysis of whole-cell lysates from cells grown in hormone-depleted medium. (B) T-47D cells were hormone deprived for 60 h before infection by adenovirus vectors for ACTR or GFP with expression at an estimated multiplicity of infection of 100 and treated with 10−8 M E2 (+E2) or the solvent (-E2). Different days after infection, cell proliferation was measured by the colorimetric MTT assay in six replicates. (C) T-47D cells were hormone deprived for 24 h and treated with 10−8 M ICI for 36 h prior to adenovirus vector infection. Cells were then maintained in medium supplemented with 10−8 M ICI. Cell proliferation was monitored as described for panel B. (D) MDA-MB 361 cells were treated and analyzed as for panels B (except no E2) and C. (E) T-47D cells grown in regular medium (RPMI plus 10% FBS) were treated with 10−7 M ICI for 36 h prior to adenoviral infection. Cell proliferation was measured as described in the legend for panel B. (F and G) The same number of T-47D stable transfectant cells was plated. Different days after plating, cell proliferation was measured by cell enumeration in triplicates in the absence of hormone (F) or in the presence of 10−8 M ICI (G).
These effects are unlikely to be caused by the viral vectors, since cells treated with a viral vector expressing GFP and cells mock treated had no difference in proliferation (unpublished data). Nevertheless, to completely rule out the possibility that viral vector itself might contribute to the effect of ACTR on cell growth, we generated several T-47D derivatives that showed different levels of ectopic ACTR expression (Fig. 2A, panel b, and unpublished data). Significantly, the elevated levels of ACTR in these stable cell lines correlated well with their activity to stimulate hormone-independent cell proliferation (Fig. 2F) and their ability to overcome the growth suppressive effect of antiestrogens ICI and 4(OH)-tamoxifen (Fig. 2G and data not shown). These results strongly suggest that overexpression of ACTR not only facilitates estrogen-stimulated cell proliferation but also, more significantly, promotes hormone-independent cell proliferation and renders estrogen-responsive breast cancer cells resistant to antiestrogens.
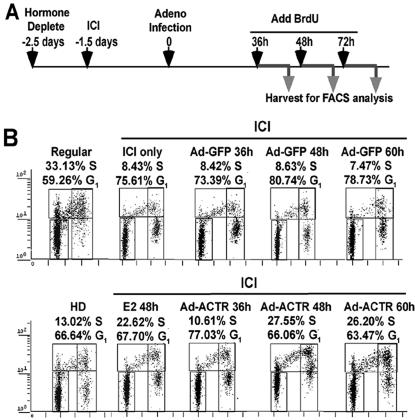
Given that elevation of ACTR overcomes the cell cycle arrest effect of antiestrogens, we postulated that the mechanism of ACTR promotion of cell proliferation may entail its direct involvement in cell cycle progression. We thus examined the effect of ACTR overexpression on cell cycle distributions. As illustrated in Fig. 3A, T-47D cells were first hormone deprived, then treated with ICI and adenovirus vectors, and pulse-labeled for 10 h with BrdU before they were harvested for cell cycle analysis by FACS. In agreement with previous reports that antiestrogens inhibit ER-positive cell growth by causing cell cycle arrest (36, 52), we found that treating T-47D cells with ICI inhibited S-phase entry by arresting cells primarily in G1 (Fig. 3B, ICI only). Expression of GFP did not change their cell cycle distribution. In contrast, 48 to 58 h after ACTR adeno-vector treatment, a marked increase of cell population in S phase was observed (from 8.6% in Ad-GFP to 27.5% in Ad-ACTR [Fig. 3B]). Concomitantly, the number of cells in G1 decreased (from 80.7 to 66.0%). Significantly, the number of BrdU-positive cells among ACTR-overexpressing cells was even higher than that in cells treated with E2 (27.5 versus 22.6%), in keeping with the strong effect of elevated ACTR on cell proliferation. These results indicate that elevated levels of ACTR in ER-positive breast cancer cells overcome the antiestrogen-mediated growth arrest by induction of cell cycle reentry.
FIG. 3.
Elevation of ACTR promotes cell cycle progression in antiestrogen-treated quiescent cells. (A) Schematics of the time course for treatment of T-47D cells for cell cycle analysis by FACS (see Materials and Methods for details). (B) Two-dimensional FACS distribution of T-47D cells treated as above, grown in regular medium (Regular), or hormone deprived for 60 h (HD), or treated with 10−7 M E2 (E2 48 h), and then labeled with BrdU for 10 h prior to harvesting and staining with propidium iodide. The horizontal axis in the dot plots reflects DNA content, and the vertical axis shows DNA synthesis. The populations of cells in G1 and S phases are indicated. Cells mock infected showed no difference in cell cycle distribution from cells infected with adeno-GFP (data not shown).
Elevated ACTR executes a gene expression program distinct from that of E2-ER by selective induction of E2F target genes.
Because ACTR functions as a transcription cofactor, we reasoned that the effect of ACTR on cell cycle progression might be due to its ability to modulate the expression of cell cycle regulatory genes. In order to identify gene expressions directly affected by elevated levels of ACTR, we first rendered T-47D cells quiescent with ICI and then treated them with adeno-vectors as illustrated in Fig. 3A. For comparison, we also included cells treated with E2 or ICI. We first performed Western analysis of the key cell cycle regulatory proteins (Fig. 4A). Significant levels of ectopic ACTR expression could be detected 24 h after adeno-vector treatment. In accordance with previous findings, ICI treatment dramatically reduced the expression of ERα in T-47D cells, which could be restored by E2. Ectopic expression of ACTR or GFP, however, did not restore the expression of ER, indicating that ACTR-stimulated cell proliferation is not due to an up-regulation of ER expression. Importantly, the expressions of E2F1, E2F3, cyclin E, cyclin A, and Cdk2 were markedly increased within 36 h after ACTR adeno-vector treatment. Notably, the increased expression of E2F1 could be readily detected at 24 h, thus closely correlating with the ectopic expression of ACTR. Furthermore, a reproducible but less dramatic increase was also seen with cyclin B and the Rb-related protein p107. Interestingly, while E2 strongly stimulated cyclin D1 expression, it did not markedly affect E2F1 expression, as seen with ACTR. On the contrary, overexpression of ACTR did not have any effect on the expression of cyclin D1. (Fig. 4A and unpublished data). Finally, overexpression of ACTR appeared not to affect the expression of p53, Erb2, and Akt (data not shown). These results suggest that, in ER-positive cells, elevated levels of ACTR induce the expression of key cell cycle regulators that are different from those induced by E2.
FIG.4.
Overexpression of ACTR selectively induces the expression of E2F1-responsive genes, but not ER target genes. (A) The expression of cell cycle regulatory proteins in T-47D cells was analyzed by Western blotting. Cells were treated with ICI and infected with adeno-ACTR (labeled with an A) or adeno-GFP (labeled with a G) as described in the legend for Fig. 3A, and at different hours after infection cells were harvested for Western blotting. Cells were refed every 2 days with fresh medium containing 10−8 M ICI. T-47D cells treated with 10−7 M E2 for 48 h or continuously treated with ICI were included for comparison. (B, panels a to g) Quantitative RT-PCR analysis of E2F-responsive gene expression in T-47D cells that were treated and/or infected with adeno-ACTR and adeno-GFP as described for panel A. Details of the procedures and calculation of difference are described in Materials and Methods. (h) RT-PCR analysis (28 cycles at a Tm of 55°C) of E2F1 target genes that did not show any significant changes of expression due to elevated levels of ACTR. E2F1 is shown for comparison.
To determine whether the detected changes occur at the mRNA level, we performed quantitative analysis of gene transcripts in cells treated with ICI and adenovirus vectors, as in Fig. 4A. Indeed, the increase of E2F1, cyclin E, Cdk2, and cyclin A proteins in cells with ACTR overexpression correlated well with the increase of their RNA transcripts (Fig. 4B, panels a to c, and data not shown). The apparent attenuation at 48 h after adeno-vector treatment may reflect the fact that cells exit G1 phase at that time point. Consistent with the Western results, cyclin D1 transcription was strongly induced by E2, but it was not influenced by the overexpression of ACTR (Fig. 4B, panel d).
As essentially all of the genes with expression induced by ACTR, including cyclin E, cyclin A, cyclin B, Cdk2, p107, and E2F1 itself, are well characterized as targets of transcriptional regulation by E2F1 (13, 33, 38, 48), we examined whether other E2F1-responsive genes may also be subject to ACTR regulation. Results in Fig. 4B, panels e to g, demonstrate that Cdc25A, a cell cycle regulator, Cdc6, a component of DNA replication machinery, and dihydrofolate reductase, an enzyme involved in nucleotide biosynthesis, were all up-regulated by elevated levels of ACTR. In contrast, the expression of other E2F1 target genes, including Cdc2, Apaf1, and caspases 7, 8, and 9, was not significantly altered in cells with a high level of ACTR (panel h and data not shown). Taking together, these results indicate that ACTR-induced promotion of cell cycle progression correlates closely with its selective induction of E2F1-responsive genes that are critical for G1/S transition.
ACTR is recruited to E2F1 target genes.
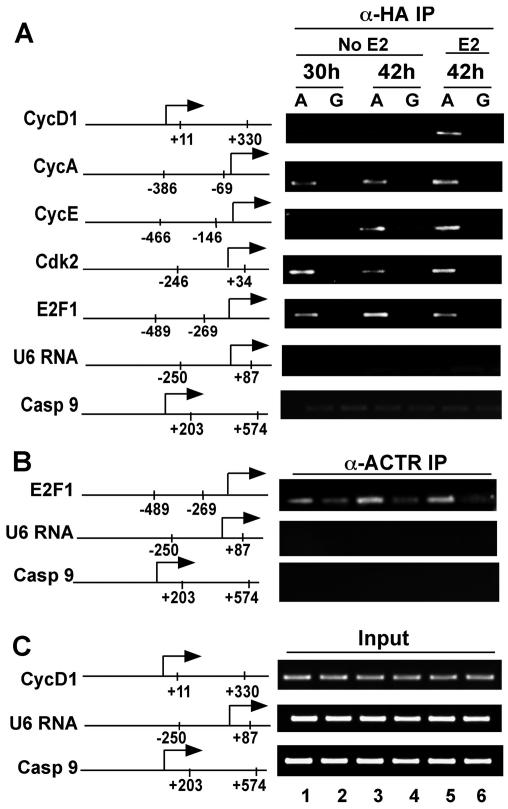
Having demonstrated that overexpression of ACTR enhances the expression of a subset of E2F1-responsive genes, we asked if ACTR is directly involved in E2F1-mediated control of their transcription. Thus, we examined whether ACTR is recruited to the target gene promoters by ChIP assay. Results in Fig. 5A show that, in keeping with the E2-dependent role of ACTR in control of cyclin D1 expression, ectopically expressed ACTR, detected by an α-HA antibody, was recruited to the cyclin D1 promoter only in the presence of E2 (lane 5). In contrast, the occupancy of ACTR at the E2F1 target gene promoters, including Cdk2, cyclin E, cyclin A, and E2F1 itself, was readily detected both in the presence and absence of hormone. No ChIP signals were observed in cells treated with GFP-adeno-vector. To determine whether the endogenous ACTR occupies E2F gene promoters, we performed a ChIP assay with an α-ACTR antibody. Indeed, a low amount of endogenous ACTR was recruited to the E2F1 target gene independent of E2 (Fig. 5B, lanes 2 and 4). Ectopic expression of ACTR markedly increased the overall ACTR occupancy (compare lanes 1, 3, and 5 with lanes 2 and 4). Importantly, promoter sequences from genes that are not subject to ACTR regulation, such as caspase 9 and U6, were not detected in the immunoprecipitates (Fig. 5A and B), indicating that the observed ACTR chromatin association is gene specific and not due to a nonspecific activity of the antibodies used. These results provide a direct link of ACTR to the transcriptional control of E2F1 target genes and suggest that ACTR may act as a coactivator for E2F1.
FIG. 5.
ACTR is selectively recruited to the promoters of cell cycle genes. ChIP analysis of ACTR occupancy on E2F-responsive genes in T-47D cells infected with adeno-ACTR-HA (labeled with an A) and adeno-GFP (labeled with a G), or treated with 10−7M E2, after cells were hormone deprived and treated with ICI. Cells were harvested 30 and 42 h after infection. The ChIP assay was performed using α-HA (A) or α-ACTR (B) antibodies. (C) Fractions (5%) of total soluble chromatin preparations made from cells treated for panels A and B were taken before immunoprecipitation and used as input; the DNA was isolated in the same way as for the ChIP assay. Usually 1/10 of the DNA isolated from the input and the ChIP samples was used as template for generation of the PCR products presented. Regions of genomic sequence analyzed by PCR are indicated on the schematic of each gene promoter.
ACTR, via one of its N-terminal domains, associates specifically with E2F1.
Although p160 coactivators, including ACTR, were initially identified as nuclear hormone receptor cofactors by virtue of their direct interactions with the receptors, recent studies have begun to address the possible role of p160 coactivators in other biological pathways mediated by nonreceptor transcription factors (3, 9, 23, 24, 40, 54, 55). The direct involvement of ACTR in regulation of E2F1-responsive genes implies that ACTR may associate with E2F1. To test this, we first performed coimmunoprecipitation assays to detect the ACTR-E2F1 complex in T-47D cells. Cells were treated with ICI and adeno-vectors as in the experiment shown in Fig. 3. Cell lysates were incubated with α-E2F1 or α-IgG monoclonal antibodies, and the presence of ACTR in immunoprecipitates was analyzed by Western blotting with an α-ACTR antibody. Results in Fig. 6A show that a fraction of endogenous ACTR was coprecipitated with E2F1 (top panel, lane 3) and that elevation of the ACTR level increased the efficiency of ACTR-E2F1 complex formation (lane 4). We also performed reciprocal coimmunoprecipitation assays with HEK-293T cells. ACTR was expressed by transient transfection together with E2F1 or Flag-E2F4. Protein complexes associated with α-ACTR antibody were then analyzed by Western blotting using α-E2F1 or α-Flag antibodies. In line with the results using α-E2F1 antibody for coimmunoprecipitation, we found that the α-ACTR antibody effectively coimmunoprecipitated E2F1 (Fig. 6B, lane 3). Interestingly, ACTR appeared not to coimmunoprecipitate with E2F4, the other member of the E2F family (Fig. 6B, lanes 4 and 5).
FIG. 6.
ACTR interacts directly with E2F1 in vivo and in vitro through the N-terminal regions of ACTR and E2F1. (A) ACTR-E2F1 complex in T-47D cells was analyzed by coimmunoprecipitation assay with cells treated with ICI and infected with adeno-ACTR (labeled with an A) or adeno-GFP (labeled with a G). Two days after infection, cell lysates were prepared and immunoprecipitated with α-E2F1 or α-mouse IgG. The presence of ACTR and E2F1 in the immunoprecipitates was detected by Western blotting using α-ACTR and α-E2F1 antibodies. Input was equivalent to 20% of the cell lysate used in the IP. (B) 293T cells were cotransfected with expression plasmids for ACTR and E2F1 or Flag-E2F4 as indicated. Whole-cell lysates were immunoprecipitated with α-ACTR or α-E2F4, followed by Western analysis with α-ACTR, α-E2F1, or α-Flag antibodies. The input represents 20% of the cell lysates. (C) In vitro direct interaction between ACTR and E2F1 was analyzed by GST pull-down assay using purified proteins. Purified Flag-ACTR proteins were incubated with either GST, GST-E2F1, or GST-E2F4 immobilized on glutathione beads. After extensive washing, proteins retained on beads were separated by SDS-PAGE and detected by Western analysis using α-Flag antibody. Input represents 30% of Flag-ACTR used in the assay. (D and E) A GST pull-down assay was also performed with in vitro-translated and 35S-labeled full-length or fragments of ACTR proteins and GST or GST-E2F1. Labeled proteins retained on the beads after washing were analyzed by SDS-PAGE and autoradiography, together with 10% of the translated products used in the incubation. Note that the Gal4 DNA-binding domain was fused to small ACTR fragments to enhance their in vitro translation signals. (F) GST fusions of full-length and fragments of E2F1 were incubated with in vitro-translated N-terminal fragments of ACTR. The left panel shows Coomassie stain of GST and the GST-E2F proteins used.
Next, we examined whether ACTR directly interacts with E2F1 by performing GST pull-down experiments using purified ACTR and GST-E2F proteins. Purified Flag-ACTR was incubated with either GST, GST-E2F1, or GST-E2F4 immobilized on glutathione beads. After extensive washing, proteins retained on beads were separated by SDS-PAGE and detected by Western analysis using the α-Flag antibody. As demonstrated in Fig. 6C, full-length ACTR interacted specifically with E2F1, but not with E2F4 or GST alone. To examine the domains in ACTR that are required for its interaction with E2F1, we performed similar experiments using in vitro-translated ACTR proteins. Results shown in Fig. 6D indicate that the N terminus (amino acids [aa] 1 to 459) of ACTR interacted strongly with E2F1, whereas the central and C-terminal regions of ACTR, which are contained in the ACTR-A38 variant with domains for interactions with nuclear receptors (RID), CBP/p300 (CID), CARM1, and histone acetyltransferase activity, did not show strong interactions with E2F1. Interestingly, a C-terminal deletion of ACTR, leaving only the putative bHLH and PAS domains (aa 1 to 343) intact, completely abolished the interaction, suggesting that the region immediately downstream of the PAS domains is critical for mediating ACTR-E2F1 interaction. Consistent with this notion, a small ACTR fragment of about 110 aa (aa 343 to 454) from a region between the PAS and the RID domains displayed a strong interaction with E2F1 (Fig. 6D, bottom portion), while an internal deletion of the same region diminished the interaction (Fig. 6E, bottom portion); we thus have named this region as the E2F1 interaction domain, or EID. In contrast, fragments (aa 264 to 329 and aa 395 to 621) either upstream or downstream of the EID showed no detectable activity to associate with E2F1 (Fig. 6D). Furthermore, deletion of the bHLH and/or the PAS domains in the full-length ACTR proteins or the N terminus (aa 1 to 459) did not show any negative effect on their interaction with E2F1 (Fig. 6E and F, right panels). Domain mapping on E2F1 demonstrated that ACTR proteins interacted primarily with the N-terminal region (aa 1 to 284) of E2F1, which contains the N terminus and part of the DNA binding and dimerization domains (Fig. 6F). Collectively, these results suggest that ACTR interacts directly with the N-terminal region of E2F1 through a previously undefined domain in the N terminus of ACTR.
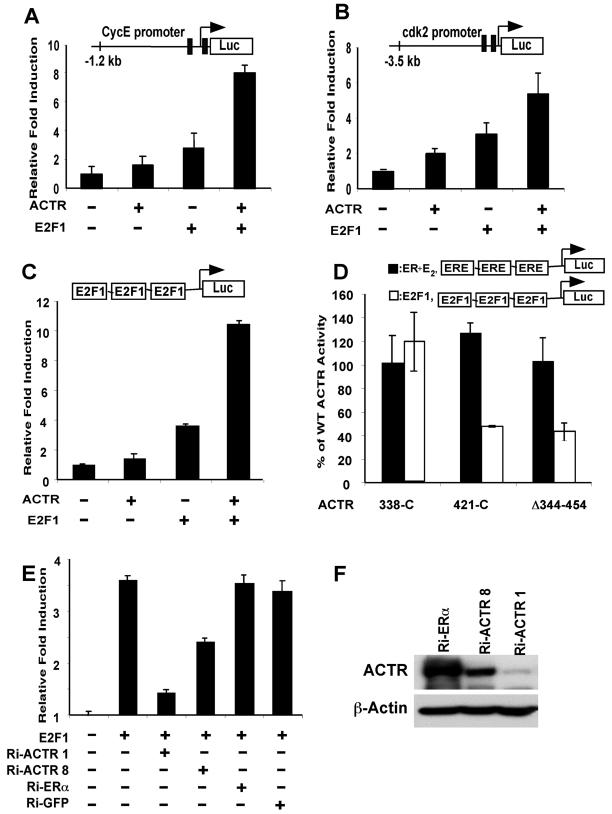
ACTR is required for transactivation by E2F1.
To further establish that ACTR functions as an E2F1 coactivator, we first asked whether ACTR stimulates E2F1-responsive promoters in reporter gene assays (Fig. 7A and B). T-47D cells were transfected with the luciferase gene directly linked to cyclin E or cdk2 promoter sequences. Expression of ACTR slightly enhanced the reporter gene activity, presumably in part through its interaction with the endogenous E2F1. Importantly, simultaneous expression of ACTR and E2F1 significantly increased E2F1-mediated induction of the two promoter activities. To rule out the possibility that ACTR targets transcription factors other than E2F1 on the promoters, we carried out the transfection assay with three E2F1 binding sites linked to a tk-TATA-luciferase reporter. Similar to the effect observed on the E2F1 target gene promoters, cotransfection of ACTR markedly augmented E2F1-mediated transactivation through the defined E2F1 sites (Fig. 7C). More importantly, ACTR deletion mutants, such as ACTRΔ344-454, which specifically impairs its interaction with E2F1, displayed diminished activities to stimulate E2F1 transactivation (Fig. 7D). In contrast, the same mutants possessed the full activity of wild-type ACTR to potentiate ER-mediated induction of ERE-reporter activity. They also showed equivalent expression levels in transfected cells (data not shown). These results not only demonstrate a direct enhancement of E2F1 transactivation by ACTR but also reveal a structural requirement for ACTR to act as a coactivator for E2F1, which is distinct from that for ER.
FIG. 7.
ACTR enhances the transactivation by E2F1. (A and B) Activity of ACTR to enhance the E2F1-responsive promoter function was measured in T-47D cells using human cyclin E and cdk2 promoters linked to the luciferase gene. Cells were transiently transfected with the reporters and ACTR or E2F1 expression constructs as indicated. Induction represents the normalized luciferase activity in cells transfected with ACTR and/or E2F1 expression plasmids divided by the activity in cells transfected with the reporter alone. (C) The ability of ACTR to enhance E2F1 transactivation was analyzed using 3x E2F- tk-luc, a reporter with three E2F1 binding sites linked to luciferase gene. 293T cells were transfected with the reporter and plasmids expressing ACTR and/or E2F1. (D) The activity of wild-type and mutant ACTR to enhance E2F1 transactivation was compared with their activity on E2-stimulated ER transactivation. 293T cells were transfected with either 3x E2F-tk-luc or 3x ERE-tk-luc reporter, along with expression plasmids for E2F1 or ERα, and wild-type ACTR or its mutant forms (N-terminal deletions 338-C and 421-C and an internal deletion, Δ344-454). Percentage of wild-type ACTR activity was calculated by dividing the normalized luciferase activity of mutant ACTR with the normalized wild-type ACTR activity. The ERE reporter was analyzed from cells treated with 10−7 M E2. Note that ACTR (wild type) increased E2-stimulated transactivation by ER about fourfold. (E) The requirement for ACTR in E2F1 transactivation was examined in a reporter assay as for panel C, with ACTR expression silenced by vector-mediated RNAi. 293T cells were cotransfected with 3x E2F-tk-luc and an equal amount of vectors expressing sh-RNAi sequences targeting ACTR, ERα, or GFP as indicated. (F) 293T cells were cotransfected with pcD-HCMV-ACTR and the pSh-RNAi vectors as indicated. Cell lysates were prepared 3 days after transfection for Western analysis.
To more rigorously test the notion that ACTR can act as an E2F1 coactivator, we examined the effect of ACTR depletion on the transactivation activity of E2F1. As demonstrated in Fig. 7E, cotransfection of an ACTR RNAi vector, Ri-ACTR1, mediating the expression of siRNAs specifically against ACTR, dramatically reduced E2F1 induction of reporter activity, while no significant effect was observed with the ER and GFP-RNAi vectors. Consistent with a weaker potency to silence ACTR expression (Fig. 7F), the other ACTR RNAi vector, Ri-ACTR8, was less effective in suppression of E2F1 transactivation. These results, together with the data described above, strongly suggest that ACTR is indeed an E2F1 coactivator and is required for E2F1-mediated transcriptional activation of gene expression.
ACTR-induced hormone-independent breast cancer cell proliferation requires its association with E2F1 but not ER.
The results described above imply in many ways that induction of E2F1-responsive gene expression and cell growth by elevated ACTR may not involve ER. For instance, depletion of ACTR in ER-negative cells decreases their hormone-independent proliferation (Fig. 1). More strikingly, overexpression of ACTR in ER-positive cells promotes cell cycle progression in the presence of the pure ER antagonist ICI (Fig. 3). Antiestrogens such as ICI not only decrease cellular ER protein levels, they also effectively disrupt both hormone-dependent and -independent interactions between ERs (both ERα and β) and the p160 coactivators (10, 47), therefore making it unlikely for ERs to contribute to the growth-promoting activity of ACTR in the presence of antiestrogens. Nevertheless, to further address the role of ACTR association with E2F1 and ER in mediating E2-independent cell proliferation, we used several mutant forms of ACTR that are selectively defective in interactions with E2F1 and ER (Fig. 8A). The p160 coactivators interact with nuclear receptors primarily through the three LXXLL motifs in their receptor interaction domain (20). The ACTR-AAA mutant protein has the three LXXLL motifs changed to LXXAA, which abolishes ACTR association with the ER ligand-binding domain (6, 8, 11, 28, 31). When expressed in T-47D cells, the ACTR-AAA protein showed a strong growth-promoting activity, equivalent to that of ACTR wild type, suggesting that the growth-promoting function of ACTR is not due to an enhanced association between ACTR and the ligand-binding domain of ER. However, eliminating the ACTR association with E2F1 by an N-terminal truncation, as in ACTR-A38 (containing aa 421C), largely abolished the function of ACTR to promote hormone-independent proliferation (Fig. 8B). In contrast, expression of other N-terminal-truncated ACTRs (D98 and D338) that retain the ACTR-E2F1 interaction domain (EID) induced a hormone-independent cell proliferation with the same efficacy as the wild-type ACTR. Importantly, all the ACTR variants were expressed at a similar level as in the wild type (Fig. 8A) and possessed the full activity of wild-type ACTR to function as a nuclear receptor coactivator, as shown in Fig. 7D and the previous study (7).
FIG. 8.
Stimulation of hormone-independent cell proliferation by ACTR requires ACTR interaction with E2F1, but not ER. (A and B) T-47D cells were infected with adenovirus expressing wild-type and mutant forms of ACTR, as depicted. The expression levels of different ACTR proteins in the cells 2 days after infection were determined by Western blotting using α-HA antibody. Cell proliferation in the absence of hormone was monitored by MTT assay 4 days after infection as described in the legend for Fig. 2B, and the percentage of wild-type ACTR activity was determined by dividing the proliferation induced by the mutant ACTR with that of wild-type ACTR. (C and D) ER-negative cells (HCC-1806 and HCC-1937) infected with adeno-ACTR or adeno-GFP were monitored for cell proliferation by MTT assay. (E) T-47D and its derivative, A41 cells, hormone deprived for 2 days, were infected with an equal titer of the RNAi adenoviruses for E2F1 or GFP, as for Fig. 1A. Cells were then replenished with hormone-depleted medium either with (T-47D) or without (A41) 10−7 M E2. Cell proliferation was determined at 6 days after infection, with the values from Ri-GFP adenovirus-infected cells set as 100 arbitrarily. Cells were also collected at day 6 postinfection for Western analysis using α-E2F1 and α-β-actin. Note that no significant effect of GFP-RNAi vector on cell proliferation was observed when compared to mock infection. (F) Potential pathways for elevated levels of ACTR to stimulate breast cancer cell proliferation.
Finally, to further establish the role of E2F1 and ER in ACTR-induced hormone-independent breast cancer cell proliferation, we first overexpressed ACTR in a number of ER-negative human breast cancer cell lines (HCC1806, HCC1937, and BT-20) that do not express ERs or respond to estrogen for growth (unpublished results). Results in Fig. 8C and D (and data not shown) demonstrated that overexpression of ACTR markedly accelerated the proliferation of these ER-negative cells in the absence of hormone. We then examined the effect of E2F1 depletion on the proliferation of T-47D cells, and our newly established T-47D derivative A41, which ectopically expresses ACTR, showed robust E2-independent proliferation (Fig. 2A, panel b, and F) and an elevated level of E2F1 (Fig. 8E, compare lane 4 with lane 2). Both types of cells were treated with an equal titer of the E2F1 RNAi or the control GFP RNAi adenovirus vectors. E2-stimulated proliferation of T-47D cells, and hormone-independent proliferation of A41 cells as well as their E2F1 protein levels were determined 6 days after the RNAi vector treatment. As shown in Fig. 8E, lane 1, although E2F1 expression was effectively silenced in E2-treated T-47D cells, the effect on cell proliferation was modest (about 30% decrease when compared to the control cells), suggesting that factors other than E2F1 play a major role in ER-mediated cell proliferation. In contrast, a less-efficient knocking down of E2F1 in the ACTR-overexpressing A41 cells (compare E2F1 levels in lane 3 with lane 1) resulted in a dramatic inhibition of hormone-independent proliferation (about 80% decrease), indicating that E2F1 is a key mediator of ACTR-induced cell proliferation in the absence of E2. The results above, together with our findings described in Fig. 1 and 2, provide strong evidence that the ability of ACTR to promote hormone-independent breast cancer cell proliferation does not requires its association with ER, but instead involves its interaction with E2F1.
DISCUSSION
Apart from its established role in mediating nuclear receptor function in hormone signaling, the p160 coactivator ACTR/AIB1 has been strongly implicated in cancer, due to the frequent detection of its amplification or overexpression in several types of human tumors, particularly breast cancer. A presumed and not fully tested mechanism for the coactivator action has been that elevated levels of ACTR/AIB1 promote breast cancer cell proliferation through ER. Here, we provide strong evidence that reveals an unexpected pathway for ACTR to promote hormone-independent cell proliferation as well as antiestrogen resistance.
To understand the role of ACTR/AIB1 in breast cancer, we examined the function of ACTR/AIB1 in control of cell proliferation and gene expression using a number of cell culture models. In line with its role as a coactivator for ER, we found that depletion of ACTR/AIB1 inhibited E2-stimulated cell proliferation. Remarkably, however, we observed that overexpression of ACTR/AIB1 in ER-positive cells resulted in strong enhancement of hormone-independent cell proliferation. More strikingly, elevation of the coactivator expression completely negated the growth-suppressive effect of antiestrogens ICI or tamoxifen and allowed the cells to proliferate robustly in the presence of these antiestrogens. Furthermore, we found that in ER-negative cells, depletion of ACTR/AIB1 suppressed their proliferation, whereas its elevation elicited the opposite effect. Unexpectedly, we found that ACTR/AIB1 can associate with the key cell cycle regulatory protein E2F1, and elevated levels of ACTR/AIB1 strongly stimulate the expression of progrowth E2F-responsive genes. Together, these results support the hypothesis that ACTR/AIB1 plays an important role in control of cell proliferation by both E2-ER-dependent and -independent pathways and that the aberrant function of ACTR/AIB1 in breast cancer, as a result of gene amplification and/or overexpression, can bypass the E2-ER-dependent process through its function as an E2F1 coactivator, therefore helping tumor cells acquire antiestrogen resistance.
Based on previous findings and the results from this study, we propose that ACTR/AIB1 can function in control of breast cancer cell proliferation in at least three different modes. First, ACTR/AIB1 can function primarily as a coactivator for E2-bound ER, thereby mediating estrogen-dependent breast cancer cell growth (Fig. 8F, left side), which likely exists in the majority of ER-positive primary breast cancers. Alternatively, ACTR/AIB1 associates with ER independently of estrogen, which is stimulated by growth factor-mediated phosphorylation of ER and/or ACTR/AIB1 (16, 46). Conceivably, this latter ACTR/AIB1 action mode can manifest itself in breast cancers where HER2/neu and/or mitogen-activated protein kinase mediate sustained progrowth signaling. The third mode is the aberrant function of ACTR/AIB1 to potentiate E2F-responsive gene expression by acting as an E2F1 coactivator which, as our results suggest, could operate in both ER-negative and ER-positive breast cancers (Fig. 8F, right side). In support of our prediction, recent studies revealed that overexpression of ACTR/AIB1 in breast cancer did not correlate with positive ER status (4). In fact, amplification and/or overexpression of ACTR/AIB1 has been detected in a broad spectrum of malignancies with high frequencies (19, 39, 50), where steroid hormones such as estrogens are unlikely to play a critical role in tumor growth. Therefore, our results and the findings from clinical studies strongly support the notion that aberrant ACTR may function in the regulatory circuitries of cell proliferation that are independent of ER.
Distinct from the action of E2-ER, the ACTR-E2F1 mode activates E2F1 itself, cyclin E, and the other E2F-responsive genes, but not cyclin D1. Although overexpression of cyclin D1 is clearly linked to breast cancer, both E2F1 and cyclin E have recently been recognized as potential proliferative markers of the disease and closely correlated with poor outcomes (22, 26, 58). Furthermore, overexpression of cyclin E in E2-responsive cells could effectively overcome the growth arrest effect by antiestrogens (14). Therefore, deregulated cyclin E and E2F1 functions could represent an important alternative mechanism to derail the control of cell proliferation in breast cancer. However, whether elevated levels and/or function of ACTR triggers the alternative mechanism and leads to a hormone-independent tumor growth is likely determined by cellular context. Future studies aiming at defining the ACTR-E2F1-cyclin E axis will help determine the significance of the ACTR-E2F1 pathway in breast cancer progression.
The similarity of gene expression profiles generated by elevated ACTR and E2F1 led to our investigation of whether ACTR functions as an E2F1 coactivator. In this study, we presented multiple lines of evidence to support our conclusion. First, we found that ACTR is selectively recruited to the promoters of E2F1 responsive genes that are activated by ACTR, indicating a direct involvement of ACTR in transcriptional control of a subset of E2F1 target genes. We then demonstrated that ACTR interacts with E2F1 directly in vitro and in vivo and that ACTR is required for E2F1-mediated transactivation. Significantly, our domain mapping study localized the E2F1 interaction to an N-terminal domain of ACTR that is distinct from the receptor interaction domains. Moreover, we found that loss of E2F1 interaction specifically abolishes the ability of ACTR to enhance E2F1 transcription and promote hormone-independent cell proliferation. Interestingly, several nuclear proteins have recently been shown to interact with different members of the p160s through their bHLH-PAS domains and/or adjacent regions (3, 9, 55). Although our results indicate that the bHLH-PAS domains are not required for ACTR to promote cell proliferation, we cannot rule out that the N terminus may play some regulatory role in this regard. Recently, we found that the other p160 members SRC-1 and TIF2/GRIP1 may also interact with E2F1 with varying strengths, and they share some but not all the progrowth functions of ACTR we have observed in both normal and malignant human cells (unpublished data). These results, together with the report that overexpression of SRC-3/ACTR in a prostate cancer cell line affects primarily cell growth (59), underscore the functional complexity of the p160 gene family in control of cellular function.
In our attempt to explore the potential mechanism how elevation of ACTR in quiescent cells activates E2F1-responsive genes, we found that overexpression of ACTR altered the promoter occupancies by different E2F family members (unpublished results). The E2Fs can be divided into subgroups with opposing activities to control transcription of cell cycle genes (48). In cells reentering the cell cycle by serum stimulation, decreased promoter occupancies of repressing E2Fs followed by increased occupancies of activating E2Fs have been observed (43). Serum stimulation is believed to cause hyperphosphorylation of Rb and related pocket proteins by Cdks, which in turn results in the relocalization of freed repressive E2Fs to the cytoplasm and subsequent recruitment of activating E2Fs. We speculate that an increased expression of ACTR expands the free pool of the activator ACTR-E2F complexes in quiescent cells and enhances the potential of the complex to be recruited to the E2F-responsive promoters, which in turn displaces the repressive E2F complex. The ACTR-E2F complex could activate transcription through its associated chromatin-modifying activities and the function to facilitate the recruitment of RNA polymerase II-containing complexes, as recently demonstrated (27). It is worth noting that CBP and PCAF were shown to interact with E2F1 and modulate its transactivation (29, 49). It would be of interest to determine how ACTR, p300/CBP, and PCAF may relate to each other in control of the transcriptional output of E2F transcription factors and cell proliferation.
Acknowledgments
We are grateful to Joseph Nevins, Pat Nakatani, Joan Massague, and Xinbin Chen for reagents. We thank Hsing-Jien Kung, Ron Wisdom, and Marty Privalsky for insightful discussion and critical reading of the manuscript. We are indebted to Ai-Hong Ma, Dan Robinson, Michelle Ellis-Hutchings, and Zhenyu Zhong for technical help.
This work was supported in part by a grant from the National Institutes of Health (DK60019) and from the California Breast Cancer Research Program (7KB-0140). M.C.L. is supported by an NIH Molecular and Cellular Biology training grant.
Appendix
The primers listed in Table A1 were used in the quantitative RT-PCR and the analysis of genomic DNA sequences enriched in the ChIP.
TABLE A1.
Primers used in this study
| Primer | Forward sequence | Reverse sequence |
|---|---|---|
| RT-PCR primers | ||
| ACTR | GATTAGGAGAAAACTTGGATCC | TGTTTCCGTCTCGATTCACCA |
| Actin | GAGAAAATCTGGCACCACACC | ATACCCCTCGTAGATGGGCAC |
| Cyclin A | CCCCCAGAAGTAGCAGAGTTTGTG | GCTTTGTCCCGTGACTGTGTAGAG |
| Cyclin D1 | TCCTGTGCTGCGAAGTGGAAAC | AAATCGTGCGGGGTCATTGC |
| Cyclin E | ATACAGACCCACAGAGACAG | TGCCATCCACAGAAATACTT |
| Cdk2 | TTTGCTGAGATGGTGACTCGC | CACTGGAGGAGGGGTGAGATTAG |
| E2F1 | CGCATCTATGACATCACCAACG | GAAAGTTCTCCGAAGAGTCCACG |
| Cdc2 | CCAAGTATTTCTTCAGATCCATGG | GCACTTGGCTTCAAAGCTG |
| Cdc6 | AAAGAGAATGGTCCCCCTCACTC | AGTTTTTCCAGTTCCAGGAGCAC |
| Cdc25A | TGAAGAATGAGGAGGAGACCCC | CTGATGTTTCCCAGCAACTGTATG |
| DHFR | CTTCCCAGATTCAAGCGATTCTC | TCAAGCAACCATCATCCCTCAC |
| Apaf1 | GTCATCATCTTCTCTTAGCCACTGG | TTGCTGCCACCATTATCCTTG |
| Caspase 9 | GGTTTTGTTTCCTGGAGGGACC | TGCTAAGAGCCTGTCTGTCACTGG |
| ChlP primers | ||
| Cyclin A | GAAAACGGAGAATCGGAGATACTG | ACCCAAAGGAAACTGAGCAGGG |
| Cdk2 | GATGGAACGCAGTATACCTCTC | AAAGCAGGTACTTGGGAAGAGTG |
| CyclinE | GCCGCCTGTCCATTCATCC | GGTCCTGTGGAGCCTGTAGCC |
| E2F1 | GCAAGTTGAGGATGGAAGAGGTG | TGGGGACACGGGAACATAGG |
| Caspase 9 | GCTCTTCCTTTGTTCATCTCCTGC | ACGCCGCAACTTCTCACAGTC |
| U6 RNA | GAGGGCCTATTTCCCATGATTC | GAATTTGCGTGTCATCCTTGC |
| Cdc2 | TGAGGTAGAAACAAAGCACAGCG | TCCCAGCATTGGCACAGTTC |
REFERENCES
- 1.Ali, S., and R. C. Coombes. 2002. Endocrine-responsive breast cancer and strategies for combating resistance. Nat. Rev. Cancer 2:101-112. [DOI] [PubMed] [Google Scholar]
- 2.Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965-968. [DOI] [PubMed] [Google Scholar]
- 3.Belandia, B., and M. G. Parker. 2000. Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J. Biol. Chem. 275:30801-30805. [DOI] [PubMed] [Google Scholar]
- 4.Bouras, T., M. C. Southey, and D. J. Venter. 2001. Overexpression of the steroid receptor coactivator AIB1 in breast cancer correlates with the absence of estrogen and progesterone receptors and positivity for p53 and HER2/neu. Cancer Res. 61:903-907. [PubMed] [Google Scholar]
- 5.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 6.Chang, C., J. D. Norris, H. Gron, L. A. Paige, P. T. Hamilton, D. J. Kenan, D. Fowlkes, and D. P. McDonnell. 1999. Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors alpha and beta. Mol. Cell. Biol. 19:8226-8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98:675-686. [DOI] [PubMed] [Google Scholar]
- 9.Chen, S. L., D. H. Dowhan, B. M. Hosking, and G. E. Muscat. 2000. The steroid receptor coactivator, GRIP-1, is necessary for MEF-2C-dependent gene expression and skeletal muscle differentiation. Genes Dev. 14:1209-1228. [PMC free article] [PubMed] [Google Scholar]
- 10.Cheskis, B. J., N. J. McKenna, C. W. Wong, J. Wong, B. Komm, C. R. Lyttle, and B. W. O'Malley. 2003. Hierarchical affinities and a bipartite interaction model for estrogen receptor isoforms and full-length steroid receptor coactivator (SRC/p160) family members. J. Biol. Chem. 278:13271-13277. [DOI] [PubMed] [Google Scholar]
- 11.Darimont, B. D., R. L. Wagner, J. W. Apriletti, M. R. Stallcup, P. J. Kushner, J. D. Baxter, R. J. Fletterick, and K. R. Yamamoto. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12:3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dauvois, S., P. S. Danielian, R. White, and M. G. Parker. 1992. Antiestrogen ICI 164,384 reduces cellular estrogen receptor content by increasing its turnover. Proc. Natl. Acad. Sci. USA 89:4037-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeGregori, J., T. Kowalik, and J. R. Nevins. 1995. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol. Cell. Biol. 15:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhillon, N. K., and M. Mudryj. 2002. Ectopic expression of cyclin E in estrogen responsive cells abrogates antiestrogen mediated growth arrest. Oncogene 21:4626-4634. [DOI] [PubMed] [Google Scholar]
- 15.Dilworth, F. J., and P. Chambon. 2001. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene 20:3047-3054. [DOI] [PubMed] [Google Scholar]
- 16.Font de Mora, J., and M. Brown. 2000. AIB1 is a conduit for kinase-mediated growth factor signaling to the estrogen receptor. Mol. Cell. Biol. 20:5041-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamble, M. J., and L. P. Freedman. 2002. A coactivator code for transcription. Trends Biochem. Sci. 27:165-167. [DOI] [PubMed] [Google Scholar]
- 18.Gehin, M., M. Mark, C. Dennefeld, A. Dierich, H. Gronemeyer, and P. Chambon. 2002. The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and p/CIP. Mol. Cell. Biol. 22:5923-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghadimi, B. M., E. Schrock, R. L. Walker, D. Wangsa, A. Jauho, P. S. Meltzer, and T. Ried. 1999. Specific chromosomal aberrations and amplification of the AIB1 nuclear receptor coactivator gene in pancreatic carcinomas. Am. J. Pathol. 154:525-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 21.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keyomarsi, K., S. L. Tucker, T. A. Buchholz, M. Callister, Y. Ding, G. N. Hortobagyi, I. Bedrosian, C. Knickerbocker, W. Toyofuku, M. Lowe, T. W. Herliczek, and S. S. Bacus. 2002. Cyclin E and survival in patients with breast cancer. N. Engl. J. Med. 347:1566-1575. [DOI] [PubMed] [Google Scholar]
- 23.Korzus, E., J. Torchia, D. W. Rose, L. Xu, R. Kurokawa, E. M. McInerney, T. M. Mullen, C. K. Glass, and M. G. Rosenfeld. 1998. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science 279:703-707. [DOI] [PubMed] [Google Scholar]
- 24.Lazaro, J. B., P. J. Bailey, and A. B. Lassar. 2002. Cyclin D-cdk4 activity modulates the subnuclear localization and interaction of MEF2 with SRC-family coactivators during skeletal muscle differentiation. Genes Dev. 16:1792-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, H., P. J. Gomes, and J. D. Chen. 1997. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc. Natl. Acad. Sci. USA 94:8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loden, M., M. Stighall, N. H. Nielsen, G. Roos, S. O. Emdin, H. Ostlund, and G. Landberg. 2002. The cyclin D1 high and cyclin E high subgroups of breast cancer: separate pathways in tumorigenesis based on pattern of genetic aberrations and inactivation of the pRb node. Oncogene 21:4680-4690. [DOI] [PubMed] [Google Scholar]
- 27.Louie, M. C., H. Q. Yang, A. H. Ma, W. Xu, J. X. Zou, H. J. Kung, and H. W. Chen. 2003. Androgen-induced recruitment of RNA polymerase II to a nuclear receptor-p160 coactivator complex. Proc. Natl. Acad. Sci. USA 100:2226-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mak, H. Y., S. Hoare, P. M. Henttu, and M. G. Parker. 1999. Molecular determinants of the estrogen receptor-coactivator interface. Mol. Cell. Biol. 19:3895-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Balbas, M. A., U. M. Bauer, S. J. Nielsen, A. Brehm, and T. Kouzarides. 2000. Regulation of E2F1 activity by acetylation. EMBO J. 19:662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonnell, D. P., and J. D. Norris. 2002. Connections and regulation of the human estrogen receptor. Science 296:1642-1644. [DOI] [PubMed] [Google Scholar]
- 31.McInerney, E. M., D. W. Rose, S. E. Flynn, S. Westin, T. M. Mullen, A. Krones, J. Inostroza, J. Torchia, R. T. Nolte, N. Assa-Munt, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 33.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osborne, C. K., V. Bardou, T. A. Hopp, G. C. Chamness, S. G. Hilsenbeck, S. A. Fuqua, J. Wong, D. C. Allred, G. M. Clark, and R. Schiff. 2003. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J. Natl. Cancer Inst. 95:353-361. [DOI] [PubMed] [Google Scholar]
- 35.Picard, F., M. Gehin, J. Annicotte, S. Rocchi, M. F. Champy, B. W. O'Malley, P. Chambon, and J. Auwerx. 2002. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell 111:931-941. [DOI] [PubMed] [Google Scholar]
- 36.Planas-Silva, M. D., and R. A. Weinberg. 1997. Estrogen-dependent cyclin E-cdk2 activation through p21 redistribution. Mol. Cell. Biol. 17:4059-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prall, O. W., B. Sarcevic, E. A. Musgrove, C. K. Watts, and R. L. Sutherland. 1997. Estrogen-induced activation of Cdk4 and Cdk2 during G1-S phase progression is accompanied by increased cyclin D1 expression and decreased cyclin-dependent kinase inhibitor association with cyclin E-Cdk2. J. Biol. Chem. 272:10882-10894. [DOI] [PubMed] [Google Scholar]
- 38.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 16:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakakura, C., A. Hagiwara, R. Yasuoka, Y. Fujita, M. Nakanishi, K. Masuda, A. Kimura, Y. Nakamura, J. Inazawa, T. Abe, and H. Yamagishi. 2000. Amplification and over-expression of the AIB1 nuclear receptor co- activator gene in primary gastric cancers. Int. J. Cancer 89:217-223. [PubMed] [Google Scholar]
- 40.Sheppard, K. A., D. W. Rose, Z. K. Haque, R. Kurokawa, E. McInerney, S. Westin, D. Thanos, M. G. Rosenfeld, C. K. Glass, and T. Collins. 1999. Transcriptional activation by NF-κB requires multiple coactivators. Mol. Cell. Biol. 19:6367-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sicinski, P., J. L. Donaher, S. B. Parker, T. Li, A. Fazeli, H. Gardner, S. Z. Haslam, R. T. Bronson, S. J. Elledge, and R. A. Weinberg. 1995. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell 82:621-630. [DOI] [PubMed] [Google Scholar]
- 42.Stallcup, M. R. 2001. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene 20:3014-3020. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 44.Takeshita, A., G. R. Cardona, N. Koibuchi, C. S. Suen, and W. W. Chin. 1997. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J. Biol. Chem. 272:27629-27634. [DOI] [PubMed] [Google Scholar]
- 45.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear- receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 46.Tremblay, A., G. B. Tremblay, F. Labrie, and V. Giguere. 1999. Ligand-independent recruitment of SRC-1 to estrogen receptor beta through phosphorylation of activation function AF-1. Mol. Cell 3:513-519. [DOI] [PubMed] [Google Scholar]
- 47.Tremblay, G. B., A. Tremblay, F. Labrie, and V. Giguere. 1998. Ligand-independent activation of the estrogen receptors alpha and beta by mutations of a conserved tyrosine can be abolished by antiestrogens. Cancer Res. 58:877-881. [PubMed] [Google Scholar]
- 48.Trimarchi, J. M., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3:11-20. [DOI] [PubMed] [Google Scholar]
- 49.Trouche, D., A. Cook, and T. Kouzarides. 1996. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res. 24:4139-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, Y., M. C. Wu, J. S. Sham, W. Zhang, W. Q. Wu, and X. Y. Guan. 2002. Prognostic significance of c-myc and AIB1 amplification in hepatocellular carcinoma. A broad survey using high-throughput tissue microarray. Cancer 95:2346-2352. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Z., D. W. Rose, O. Hermanson, F. Liu, T. Herman, W. Wu, D. Szeto, A. Gleiberman, A. Krones, K. Pratt, R. Rosenfeld, C. K. Glass, and M. G. Rosenfeld. 2000. Regulation of somatic growth by the p160 coactivator p/CIP. Proc. Natl. Acad. Sci. USA 97:13549-13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watts, C. K., A. Brady, B. Sarcevic, A. deFazio, E. A. Musgrove, and R. L. Sutherland. 1995. Antiestrogen inhibition of cell cycle progression in breast cancer cells in associated with inhibition of cyclin-dependent kinase activity and decreased retinoblastoma protein phosphorylation. Mol. Endocrinol. 9:1804-1813. [DOI] [PubMed] [Google Scholar]
- 53.Wijayaratne, A. L., and D. P. McDonnell. 2001. The human estrogen receptor-alpha is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J. Biol. Chem. 276:35684-35692. [DOI] [PubMed] [Google Scholar]
- 54.Wu, R. C., J. Qin, Y. Hashimoto, J. Wong, J. Xu, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 2002. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) coactivator activity by I kappa B kinase. Mol. Cell. Biol. 22:3549-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, X., H. Li, and J. D. Chen. 2001. The human homologue of the yeast DNA repair and TFIIH regulator MMS19 is an AF-1-specific coactivator of estrogen receptor. J. Biol. Chem. 276:23962-23968. [DOI] [PubMed] [Google Scholar]
- 56.Xu, J., L. Liao, G. Ning, H. Yoshida-Komiya, C. Deng, and B. W. O'Malley. 2000. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc. Natl. Acad. Sci. USA 97:6379-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu, J., Y. Qiu, F. J. DeMayo, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1998. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science 279:1922-1925. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, S. Y., S. C. Liu, L. F. Al-Saleem, D. Holloran, J. Babb, X. Guo, and A. J. Klein-Szanto. 2000. E2F-1: a proliferative marker of breast neoplasia. Cancer Epidemiol. Biomarkers Prev. 9:395-401. [PubMed] [Google Scholar]
- 59.Zhou, G., Y. Hashimoto, I. Kwak, S. Y. Tsai, and M. J. Tsai. 2003. Role of the steroid receptor coactivator SRC-3 in cell growth. Mol. Cell. Biol. 23:7742-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]