Abstract
Transcriptional activation of eukaryotic genes depends on the precise and ordered recruitment of activators, chromatin modifiers/remodelers, coactivators, and general transcription factors to the promoters of target genes. Using the human matrix metalloproteinase 9 (MMP-9) gene as a model system, we investigated the sequential assembly and dynamic formation of transcription complexes on a human promoter under the influence of mitogen signaling. We find that, coincident with activation of the MMP-9 gene, activators, chromatin remodeling complexes, and coactivators are recruited to the preassembled MMP-9 promoter in a stepwise and coordinated order, which is dependent on activation of MEK-1/extracellular signal-regulated kinase and NF-κB signaling pathways. Conversely, corepressor complexes are released from the MMP-9 promoter after transcriptional activation. Histone modifications shift from repressive to permissive modifications concurrent with activation of the MMP-9 gene. Chromatin remodeling induced by Brg-1 is required for MMP-9 gene transcription, which is concomitant with initiation of transcription. Therefore, coordination of cell signaling, chromatin remodeling, histone modifications, and stepwise recruitment of transcription regulators is critical to precisely regulate MMP-9 gene transcription in a temporally and spatially dependent manner. Given the important role of MMP-9 in both normal development and pathological conditions, understanding MMP-9 gene regulation is of great relevance.
Gene transcription in eukaryotic cells is controlled by protein complexes, including general and tissue-specific transcription factors, coregulators, chromatin-remodeling complexes, and complexes responsible for signal-specific histone modifications (26). As eukaryotic DNA is packaged into chromatin, generally a repressive structure for transcriptional activation, transcription in the context of chromatin requires remodeling processes to reconfigure the chromatin, so that activators, coactivators, and general transcription factors (GTFs) have access to promoters of target genes (12). Chromatin remodeling is dependent on either ATP-dependent chromatin-remodeling-complex-induced structural modifications of nucleosomes or histone acetyltransferase- (HAT) and histone methyltransferase-mediated covalent modifications of the N-terminal tails of core histones (12). The SWI/SNF chromatin-remodeling complex can alter chromatin structure by either shifting nucleosomes along the DNA or twisting DNA to modulate the nucleosome structure (42). Brg-1 and Brm are two ATPase subunits of the SWI/SNF complex. Recruitment of the SWI/SNF complex to target promoters requires protein-protein interactions through Brg-1 and other transcription regulators, as Brg-1 does not recognize sequence-specific DNA (21).
The basic unit of chromatin is the nucleosome, a protein and DNA complex formed by 147 bp of DNA wrapped around the histone octamer (12). The N-terminal tails of core histones have several basic amino acid residues that are subject to modifications such as acetylation, methylation, phosphorylation, and ubiquitination (6). Histone acetylation requires the activities of HATs, including GCN5, p/CAF, CBP/p300, and the p160 family. Acetylation of lysine residues by HATs has a critical role in relaxing the compact structure of nucleosomes (6). Histone deacetylases (HDACs) reverse the acetylation of histones (6). HDAC-1 and HDAC-2 interact with the corepressor Sin3A to suppress gene transcription, while HDAC-3 and HDAC-4 associate with NcoR and SMRT corepressor complexes to inhibit transcription (20). Methylation of arginine and lysine residues in the N-terminal tail of H3 or H4 has important regulatory effects on gene transcription (23). Methylation of H3-K9 is linked to gene silencing, DNA methylation, and heterochromatin formation (24). Methylation of H3-K9 has also been shown to be involved in regulation of gene transcription in euchromatin (33). Methylation of H3-K4, H4-R3, H3-R26, and H3-R17 is associated with nuclear receptor-induced transcriptional activation (5, 27).
GTFs are important for recruiting RNA polymerase II (Pol II) to target promoters and subsequent initiation of transcription (17, 26). Mediator complexes such as TRAP100 and TRAP250 are critical for interactions between activators and GTFs (17, 26). Pol II controls the synthesis of mRNA in eukaryotic cells. The C-terminal domain (CTD) is composed of 52 tandem repeats of heptapeptide YSPTSPS that are subject to phosphorylation. The two major phosphorylation sites of Pol II are Ser2 and Ser5; phosphorylation at these sites results in two forms of Pol II (hypophosphorylated IIa and hyperphosphorylated IIo) (28). In the process of transcription initiation, the predominant phosphorylation of Pol II is on Ser5, while in transcription elongation, the major form is Ser2 phosphorylation (9).
Using the human matrix metalloproteinase 9 (MMP-9) gene as a model system, we investigated the sequential assembly and dynamic formation of transcription complexes on the MMP-9 promoter induced by mitogen signaling. MMPs are a family of structurally related zinc-dependent endopeptidases capable of degrading extracellular matrix components. MMPs also regulate the availability and function of cytokines and growth factors through sequence-specific cleavage (43). MMP-9 has important roles in normal growth and development, stem cell differentiation, and pathological conditions such as tumor invasion and angiogenesis (40). Transcriptional regulation is the rate-limiting step in MMP-9 synthesis. cis-acting elements found in the ∼2.2-kb human MMP-9 promoter include AP-1, NF-κB, Sp1, and Ets-1 binding sites. Maximal induction of MMP-9 gene expression requires all these elements, although the proximal AP-1 site is critical for MMP-9 gene transcription (36). Our results demonstrate that coordination of cell signaling (extracellular signal-regulated kinase [ERK] and NF-κB pathways), chromatin remodeling, histone modifications, and the stepwise recruitment of transcription regulators is critical to the regulation of MMP-9 gene expression.
MATERIALS AND METHODS
Materials and reagents.
Phorbol 12-myristate 12-acetate (PMA) and MTA [5′-deoxy-(5′-methylthio) adenosine] were purchased from Sigma (St. Louis, Mo.). Micrococcal nuclease (MNase) was purchased from Worthington Biochemicals (Lakewood, N.J.). Antibodies against AcH3, AcH4, pS10AcK14-H3, MeR26-H3, MeR3-H4, MeK9-H3, MeK4-H3, CARM1, and trichostatin A (TSA) were purchased from Upstate Biotechnology (Lake Placid, N.Y.). The antibody against p65 was purchased from Abcam (Cambridge, United Kingdom). Antibodies against Ser5-P-Pol II-CTD and Ser2-P-Pol II-CTD were purchased from Covance (Princeton, N.J.). All other antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.).
Cell lines.
HeLa cells were maintained in Dulbecco's modified Eagle's medium with 2 mM l-glutamine, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 10% fetal bovine serum.
Gelatin substrate gel zymography.
Zymography was performed as described previously (31). In brief, HeLa cells were treated with PMA for 36 h. Supernatants were collected and concentrated and then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis in 8% polyacrylamide gels that were copolymerized with 1 to 2 mg of gelatin/ml. The gels were developed and quantified as described previously (31).
Total RNA isolation and RNase protection assay (RPA).
Experiments were performed and quantified as previously described (31). Twenty micrograms of total RNA was hybridized with human MMP-9 (50 × 103 cpm) and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; 25 × 103 cpm) riboprobes at 42°C overnight. The hybridized mixture was then treated with RNase A/T1 (1:200) at room temperature for 1 h and analyzed by 5% denaturing (8 M urea) polyacrylamide gel electrophoresis. Values for MMP-9 mRNA expression were normalized to GAPDH mRNA levels for each experimental condition.
Preparation of nuclei.
Isolation of nuclei from HeLa cells was carried out as described previously (2). Briefly, cells were spun at 300 × g for 5 min at 4°C and washed twice with ice-cold phosphate-buffered saline. Pelleted cells were resuspended in 10 mM Tris-HCl (pH 7.4)-10 mM NaCl-3 mM MgCl2-0.5% (vol/vol) Nonidet P-40. After 5 min of incubation on ice, the lysate was spun at 500 × g for 10 min at 4°C, and then the nuclei were resuspended in MNase digestion buffer.
Nuclear run-on.
HeLa cells were serum starved for 12 h before stimulation by PMA. Nuclei were purified as described above. Nuclear run-on transcription was carried out in the presence of 300 μCi of [α-32P]UTP. After digestion with DNase I and protease K, RNA was extracted by Trizol (Invitrogen). Hybridization was performed on a nylon membrane dotted with MMP-9 cDNA, GAPDH cDNA, and the control vector. Transcription intensity was determined by normalizing the intensity of MMP-9 to that of GAPDH.
Mapping of nucleosomes by MNase.
Nuclei were isolated from HeLa cells as described above. Then, 2 × 107 nuclei in 200 μl of MNase digestion buffer (30 mM Tris-HCl [pH 8.3], 150 mM KCl, 10 mM CaCl2, 5 mM MgCl2, 20% glycerol, 0.05 mM EDTA) were incubated with increasing amounts of MNase at 30°C for 10 min. The digestion reaction was stopped by the addition of 0.5 mM EDTA. Genomic DNA was purified as described previously (2).
Southern blotting.
Twenty micrograms of control DNA or DNA isolated from MNase-digested nuclei was first digested by appropriate restriction enzymes and then separated in a 1.5% (vol/vol) agarose gel and transferred to a Hybond N+ membrane (Amersham-Pharmacia, Piscataway, N.J.). The UV-cross-linked blot was hybridized with random-primer-labeled probes.
Restriction enzyme hypersensitivity analysis.
Nuclei were isolated from HeLa cells as described above. The purified nuclei were digested with 5 U of EcoRI and EcoRV per μg of DNA for 20 min at 25°C. Genomic DNA was then purified and subjected to Southern blotting as described above.
LM-PCR.
Ligation-mediated PCR (LM-PCR) was performed as previously described (13). MNase-digested DNA (1 μg) was kinased and ligated with the unidirectional linker. The amplification PCR included 22 PCR cycles with upper and lower primers. The labeling PCR was performed for 5 cycles with end-labeled upper and lower primers. The products were fractioned on a 6% denaturing sequence gel and exposed to the PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
ChIP and chromatin reimmunoprecipitation (Re-ChIP).
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (1, 37). Nuclei from cross-linked cells were resuspended in Tris-EDTA buffer and sonicated. The soluble chromatin was adjusted into radioimmunoprecipitation assay (RIPA) buffer (0.1% sodium dodecyl sulfate, 1% Triton X-100, 0.1% sodium deoxycholate, 140 mM NaCl) and precleared. Immunoprecipitation was performed with 2 to 5 μg of appropriate antibodies, and the immune complexes were absorbed with protein A beads (Upstate Biotechnology) or protein A/G beads (Pierce) blocked with bovine serum albumin and salmon sperm DNA. For the ChIP assay for Ser2 and Ser5 phosphorylation of the Pol II CTD, protein L beads (Pierce) were used. Immunoprecipitated DNA was amplified by a primer pair corresponding to a 269-bp fragment (−67 to −336) from the human MMP-9 promoter and subjected to semiquantitative PCR. The PCR products were resolved in 1.5% agarose gels in 1× TAE electrophoresis buffer, and the gels were stained with ethidium bromide. In some experiments, PCR was performed for 25 to 28 cycles in the presence of 2.5 μCi of [α-32P]dCTP and the products were fractionated in 4% polyacrylamide gels. The dried gels were exposed to the PhosphorImager. Densitometry was used to quantify the PCR results, and all results were normalized by the respective input values. For reimmunoprecipitation, the chromatin complex was eluted from beads with 10 mM dithiothreitol at 37°C, diluted with RIPA buffer, and then immunoprecipitated with specific antibodies.
Transient transfection and luciferase assays.
Luciferase reporter plasmids containing 670 bp of the human MMP-9 promoter and serial deletion constructs were obtained from D. Boyd (M. D. Anderson Cancer Center, Houston, Tex.) (15). Chloramphenicol acetyltransferase (CAT) reporter plasmids driven by site-directed mutant versions of the MMP-9 promoter were obtained from A. R. Mackay (University of L'Aquila, L'Aquila, Italy) (11). Transient transfection was performed as previously described (31) with Lipofectamine (Life Technologies, Grand Island, N.Y.). Cells were also transfected with a promoterless vector control (pGL3-basic) and pRL-null vector (Promega). Cell extracts were assayed in triplicate with the Dual-Luciferase reporter assay system (Promega). For the site-directed mutation experiments, cell extracts were assayed in triplicate with the CAT enzyme-linked immunosorbent assay kit (Roche Diagnostics Corporation). The luciferase or CAT activity from the vector control was arbitrarily set at 1 for calculation of induction.
RESULTS
AP-1, NF-κB, and Sp1 binding sites are indispensable for PMA-induced MMP-9 gene transcription in HeLa cells.
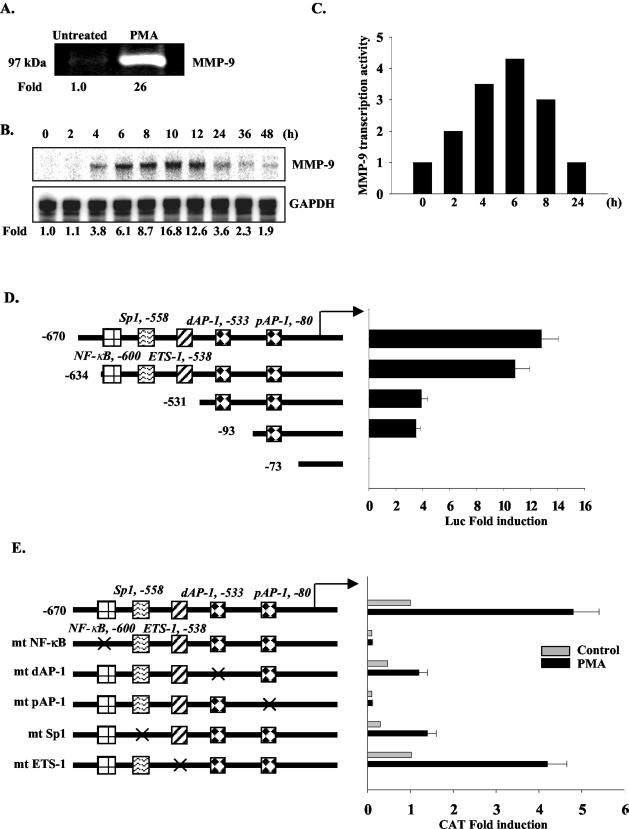
PMA was utilized to stimulate MMP-9 gene expression in HeLa cells. PMA-induced MMP-9 protein expression was determined by zymography assay (Fig. 1A). MMP-9 mRNA expression was examined at different time points after PMA stimulation. MMP-9 mRNA was first detected 4 h after addition of PMA, peaked at 10 h (16.8-fold induction), and then declined over time (Fig. 1B). These results were confirmed by reverse transcription-PCR analysis (data not shown). Nuclear run-on analysis was performed to examine the transcription rate of the MMP-9 gene. MMP-9 gene transcription in response to PMA was detectable at 2 h, peaked at 6 h, and then declined over time (Fig. 1C). To define the cis-acting elements critical for MMP-9 induction in HeLa cells, serial deletion and site-directed mutation constructs of the human MMP-9 promoter were tested. Deletion of the −670 to −634 region led to a small reduction in PMA-induced MMP-9 promoter activity, while deletion of the −634 to −531 region substantially reduced MMP-9 promoter activity. Deletion to −73 bp of the MMP-9 promoter abolished PMA-induced promoter activity (Fig. 1D). Mutation of the NF-κB element, the distal and proximal AP-1 sites (dAP-1 and pAP-1), and the Sp1 site inhibited PMA-induced MMP-9 promoter activity, while mutation of the Ets-1 site had only a modest effect (Fig. 1E). These results indicate that NF-κB, the distal and proximal AP-1 sites, and the Sp1 site are indispensable for PMA-induced MMP-9 gene transcription in HeLa cells.
FIG. 1.
NF-κB, AP-1, and Sp1 binding sites are indispensable for PMA-induced MMP-9 gene transcription in HeLa cells. (A) Serum-starved HeLa cells were treated without or with PMA (50 ng/ml) for 36 h, and supernatants were collected and subjected to zymography. Induction is shown. (B) Kinetics of MMP-9 mRNA levels after PMA stimulation for up to 48 h were detected by RPA. Induction, representative of four experiments, is shown. (C) Nuclear run-on analysis of MMP-9 gene transcription in HeLa cells treated with PMA for up to 24 h. The normalized value of the MMP-9 transcript was graphed. Data are representative of two experiments. (D) Schematic diagram of the −670 human MMP-9 promoter and serial deletion constructs. Serial deletion constructs of the MMP-9 promoter linked to the luciferase reporter were transiently transfected into HeLa cells and stimulated with PMA for 12 h, and then luciferase activity was analyzed. Data are means ± standard deviations (SD) from three experiments. (E) Site-directed mutant constructs of the MMP-9 promoter were transiently transfected into HeLa cells and stimulated with PMA for 12 h, and then CAT activity was examined. Data are means ± SD from four experiments.
The human MMP-9 promoter is wrapped into a regular array of nucleosomes.
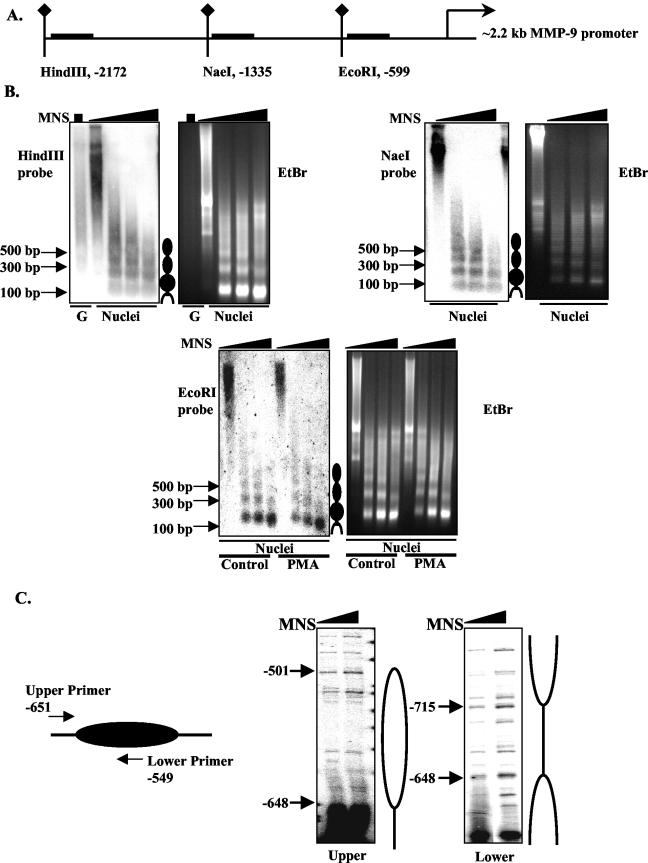
To determine the chromatin structure of the MMP-9 gene, limited MNase digestion in combination with Southern blotting was utilized to map nucleosomes on the MMP-9 promoter. Nuclei were isolated from HeLa cells and then digested with increasing amounts of MNase. DNA from MNase-digested nuclei was digested with HindIII, NaeI, or EcoRI, and the presence of nucleosomes was examined with probes generated from the immediate downstream sequences of the three restriction sites (Fig. 2A). Regularly positioned nucleosomes were found in the 2.2-kb human MMP-9 promoter, as shown by the ladder pattern (∼180 bp apart) obtained from Southern blotting (Fig. 2B). Purified genomic DNA (lane G) subjected directly to MNase digestion did not produce this ladder pattern, confirming that the ladder pattern induced by the MNase digestion digest is due to nucleosome protection (Fig. 2B). PMA stimulation did not lead to the disappearance or translocation of the MNase-protected bands; however, the intensity of MNase-protected bands from PMA-treated nuclei was less than that from untreated nuclei, suggesting the nucleosomes in the promoter region are relaxed during transcriptional activation (Fig. 2B). The boundary of the nucleosome covering the −634 to −531 region of the MMP-9 promoter was mapped by LM-PCR with the primers shown (Fig. 2C). With the upper primer, the results indicate that the nucleosome located in the −634 to −531 region starts at −648 and ends at −501 (Fig. 2C). With the lower primer, the boundary of the nucleosome located in the −634 to −531 region was confirmed to start at −648 (Fig. 2C). Our results indicate that the human MMP-9 promoter is packaged into an ordered chromatin structure and that the nucleosome located between −501 and −648 covers the NF-κB, dAP-1, and Sp1 elements.
FIG. 2.
The human MMP-9 promoter is wrapped into a regular array of nucleosomes. (A) Schematic diagram of the 2.2-kb human MMP-9 promoter, with the positions of the HindIII, NaeI, and EcoRI restriction sites indicated. The horizontal bars indicate the probes used for Southern blotting. (B) Nuclei from unstimulated HeLa cells were digested with increasing amounts of MNase (MNS; 0 to 50 U). The purified DNA was subsequently digested by HindIII, NaeI, or EcoRI. DNA fragments were analyzed by Southern blotting with the corresponding probes shown above. Genomic DNA purified from PMA-treated nuclei was also subjected to Southern blotting with the HindIII probe. Ethidium bromide (EtBr)-stained agarose gels of the respective digested genomic DNA are also shown. (C) Schematic diagram shows the position of labeling primers that were used to map the boundary of the nucleosome. The boundaries of the nucleosome covering the AP-1 and NF-κB sites were detected by LM-PCR with the upper and lower primers. As for the upper primer, sites hypersensitive to MNase digestion were found upstream of nucleotide −648 and downstream of nucleotide −501. For the lower primer, hypersensitive sites were located between nucleotides −648 and −715, thus defining the linker region of nucleosome 4.
Chromatin remodeling is required for transcription of the MMP-9 gene.
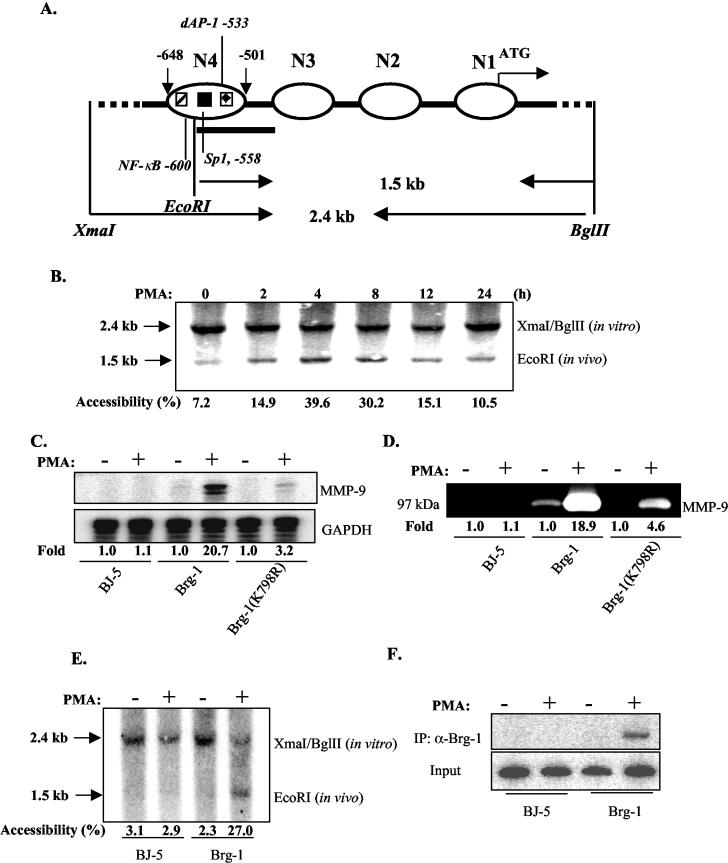
Chromatin undergoes structural reconfiguration or remodeling to facilitate transcription complex formation. Restriction enzyme hypersensitivity analysis was utilized to examine chromatin remodeling in relation to MMP-9 transcription. We focused on the N4 nucleosome that covers the NF-κB, dAP-1, and Sp1 elements (Fig. 3A). Nuclei purified from unstimulated and PMA-stimulated HeLa cells were digested with EcoRI, and purified DNA was further digested with XmaI/BglII and then blotted with the probe at the EcoRI site (Fig. 3A). There is only modest accessibility for EcoRI to nucleosome N4 in unstimulated HeLa cells, as the 1.5-kb band resulting from in vivo digestion with EcoRI is very weak (Fig. 3B). After PMA stimulation, accessibility for EcoRI increases, peaking at 4 h and then decreasing over the 24-h time course, suggesting that PMA stimulation resulted in chromatin remodeling on the MMP-9 promoter.
FIG. 3.
Chromatin remodeling is required for transcription of the MMP-9 gene. (A) Schematic diagram of nucleosomes located in the −670 region of the human MMP-9 promoter. The positions of the four nucleosomes spanning the −670 promoter region are shown. (B) HeLa cells were treated without or with PMA for up to 24 h, and nuclei were purified and then digested by EcoRI. The extracted genomic DNA was then digested by XmaI and BglII. DNA fragments were analyzed by Southern blotting with the probe downstream of the EcoRI site. Accessibility was defined as the ratio (percentage) of in vivo digestion normalized against the sums of in vivo and in vitro digestion. (C) SW-13 cells were transfected with BJ-5, Brg-1, and Brg-1(K798R). Transfected cells were treated with PMA for 12 h, and expression levels of MMP-9 mRNA were examined by RPA. Induction is shown. (D) Supernatants from the transfected cells from panel C were subjected to zymography, and induction is shown. (E) SW-13 cells were transfected with BJ-5 and Brg-1 for 36 h, and then cells were treated without and with PMA for 4 h. Purified nuclei were subjected to an EcoRI accessibility assay. Percentages of accessibility are shown. (F) SW-13 cells were transfected with BJ-5 and Brg-1 for 36 h, and then cells were treated without and with PMA for 4 h. Soluble chromatin was purified from formaldehyde-cross-linked cells. Immunoprecipitation was performed with the anti-Brg-1 antibody, and precipitated genomic fragments were subjected to PCR analysis for the presence of the MMP-9 promoter sequence.
Brg-1 is the ATPase subunit of the SWI/SNF complex, and Brg-1-deficient SW-13 cells are deficient in chromatin remodeling (29). Therefore, SW-13 cells were used to examine the requirement for chromatin remodeling for MMP-9 gene expression. SW-13 cells transfected with the control vector BJ-5 do not express MMP-9 mRNA or protein upon PMA stimulation; however, reconstitution with Brg-1 restored the ability of the cells to express MMP-9 upon PMA stimulation (Fig. 3C and D). Reconstitution with the ATPase-null mutant Brg-1 protein [Brg-1(K798R)] had only a modest effect in restoring responsiveness to PMA stimulation (Fig. 3C and D). These results indicate that Brg-1 is essential for MMP-9 gene transcription. The role of Brg-1 in the chromatin remodeling of the MMP-9 promoter was next examined (Fig. 3E). Chromatin remodeling, as assessed by detection of the 1.5-kb band generated from in vivo digestion with EcoRI, was absent in SW-13 cells, either in the absence or presence of PMA. However, reconstitution of Brg-1 restored PMA-induced chromatin remodeling on the N4 nucleosome of the MMP-9 promoter (Fig. 3E). Results from a ChIP assay indicated that reconstitution of Brg-1 in SW-13 cells led to recruitment of Brg-1 to the MMP-9 promoter upon PMA stimulation (Fig. 3F). Thus, chromatin remodeling mediated by Brg-1 is important for MMP-9 gene transcription.
MEK-1/ERK and NF-κB signaling pathways are involved in PMA-induced MMP-9 gene transcription.
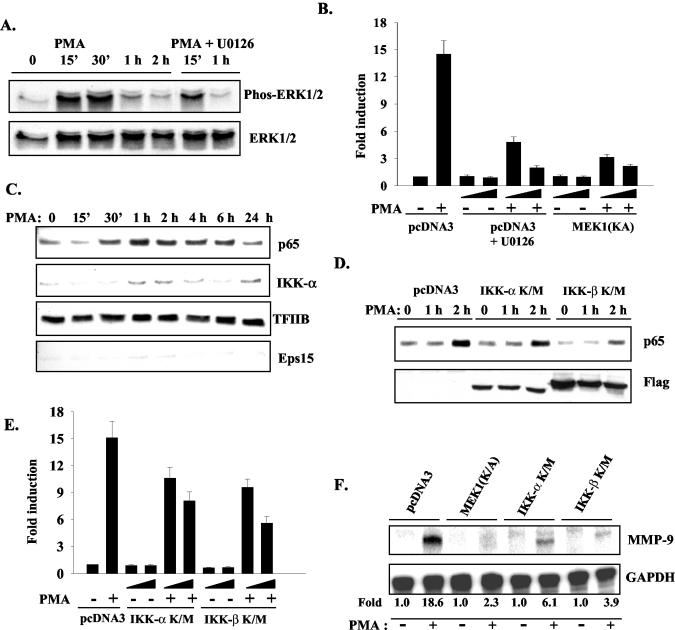
PMA induces target gene expression through numerous signaling pathways, including mitogen-activated protein kinase (MAPK), NF-κB, and phosphatidylinositol 3-kinase pathways in a variety of cell types (16). To investigate the signaling pathways critical for PMA induction of MMP-9 gene expression in HeLa cells, the MAPK pathway was first evaluated. When pharmacological inhibitors for ERK1 and 2 (ERK1/2), c-Jun N-terminal kinase, and the p38 pathways were used, it was found that only the ERK1/2 pathway was involved (data not shown). PMA treatment leads to the phosphorylation of ERK1/2, which was blocked by the MEK-1 inhibitor U0126 (Fig. 4A). In addition, transfection of a dominant negative (DN) MEK-1(K/A) construct decreased phosphorylation of ERK1/2 induced by PMA (data not shown). Both U0126 and MEK-1(K/A) abolished PMA-induced MMP-9 promoter activity in a dose-dependent manner (Fig. 4B).
FIG. 4.
MEK-1/ERK and NF-κB signaling pathways are involved in PMA-induced MMP-9 gene transcription. (A) HeLa cells were treated with PMA, or PMA plus the MEK-1 inhibitor U0126 (20 μM) for up to 2 h. Total cell lysates were subjected to immunoblotting with antibodies against phosphorylated ERK1/2 and total ERK1/2. (B) The MMP-9 promoter construct (0.2 μg) was transiently transfected into HeLa cells with pcDNA3 or increasing amounts of MEK-1(K/A) (200 or 500 ng). pcDNA3 was used to normalize the amount of DNA in each condition. After 12 h of recovery, transfected cells were treated with serum-free medium or PMA [for MEK-1(K/A) transfected cells] or serum-free medium without or with PMA plus U0126 (10 or 20 μM; for pcDNA3 transfected cells) for 12 h. Luciferase activity was determined. Data are means ± standard deviations (SD) from three experiments. (C) HeLa cells were treated with PMA for 24 h, and then nuclear extracts were subjected to immunoblotting with antibodies against p65 and IKK-α. Expression levels of TFIIB were used as nuclear protein loading control, and Eps15 was used as a cytoplasmic marker. (D) HeLa cells were transfected with pcDNA3, IKK-α (K/M), and IKK-β (K/M) for 48 h, and the transfected cells were treated with PMA for up to 2 h. Nuclear extracts from transfected cells were immunoblotted for p65, and total cell lysates were blotted for the Flag tag of the IKK mutant constructs. (E) The MMP-9 promoter construct (0.2 μg) was transfected into HeLa cells with increasing amounts of pcDNA3, IKK-α (K/M), and IKK-β (K/M). Transfected cells were treated without or with PMA for 12 h, and luciferase activity was determined. Data are means ± SD from three experiments. (F) HeLa cells were transfected with pcDNA3, MEK-1(K/A), IKK-α (K/M), or IKK-β (K/M). The transfected cells were treated without or with PMA for 10 h, and MMP-9 mRNA levels were determined by RPA. Induction is shown.
The involvement of the NF-κB pathway in MMP-9 gene transcription was also investigated. HeLa cells were treated with PMA for up to 24 h, and nuclear translocation of p65 was determined. PMA treatment leads to nuclear accumulation of p65, peaking at 1 to 2 h after stimulation (Fig. 4C). Interestingly, nuclear accumulation of IκB kinase α (IKK-α) occurred with the same kinetics as those for p65 (Fig. 4C). Use a DN construct of IKK-β (IKK-β K/M) resulted in inhibition of the nuclear translocation of p65, while a DN construct of IKK-α (IKK-α K/M) had only a modest effect on p65 nuclear accumulation (Fig. 4D). However, both the IKK-α and IKK-β DN constructs partially inhibited PMA-induced MMP-9 promoter activity (Fig. 4E), suggesting that IKK-α enhances MMP-9 expression independent of its effect on the nuclear accumulation of p65. The MEK-1(K/A), IKK-α K/M, and IKK-β K/M constructs inhibited PMA-induced endogenous MMP-9 gene expression, as revealed by RPA (Fig. 4F). Therefore, the MEK-1/ERK and NF-κB pathways are critical for PMA-induced MMP-9 gene expression.
Dynamic recruitment of activators and modifications of histones in transcriptional activation of the MMP-9 gene.
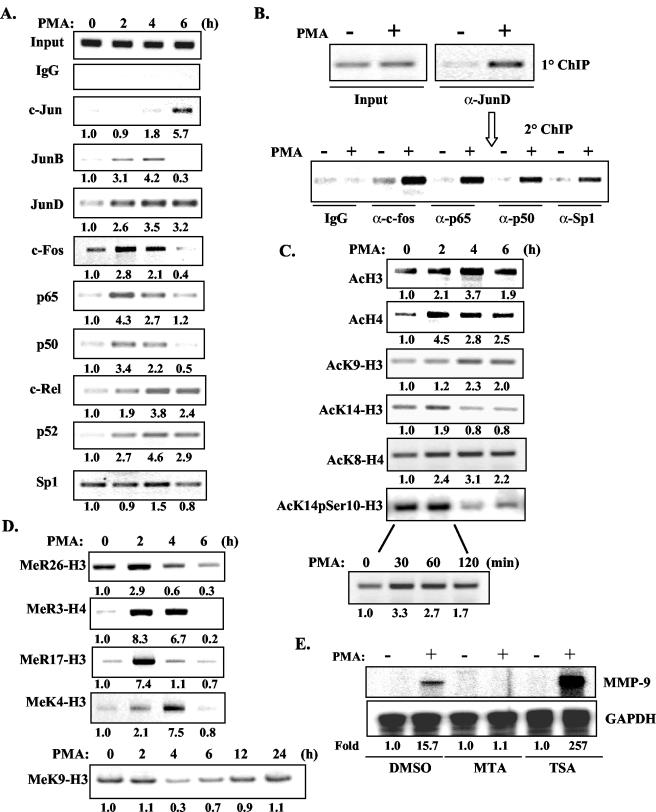
Although previous studies suggested that NF-κB, Sp1, and AP-1 factors play critical roles in MMP-9 gene transcription, it is still unknown what transcription factors are recruited to the endogenous MMP-9 promoter and, more importantly, how this is coupled to MMP-9 gene transcription. To monitor transcription factor binding in vivo, ChIP assays were performed with antibodies against c-Jun, JunB, JunD, c-Fos, p65, p50, c-Rel, p52, Sp1, and normal rabbit immunoglobulin G (as a negative control). PCR analysis of the positive control (input) indicated that the soluble chromatin samples obtained from each time point had equal amounts of chromatin fragments containing the MMP-9 promoter (Fig. 5A). In untreated cells, the major AP-1 subunits bound to the MMP-9 promoter were JunD and c-Fos. Increased binding of JunB, JunD, and c-Fos was observed 2 to 4 h after addition of PMA. The binding of JunB and c-Fos returned to basal levels after 6 h, while the binding level of JunD was relatively stable over the 6-h time course. Occupancy of c-Jun on the MMP-9 promoter was observed relatively late, at 6 h (Fig. 5A). Regarding NF-κB subunits, p65, p50, c-Rel, and p52 were weakly associated with the MMP-9 promoter in untreated cells. Strong binding of p50 and p65 was observed at 2 to 4 h after PMA stimulation, while c-Rel and p52 were recruited at later time points (4 to 6 h) (Fig. 5A). The earliest time point at which enhanced binding of p65, p50, c-Fos, and JunD was observed on the MMP-9 promoter was 30 min after PMA stimulation (data not shown). The binding of Sp1 throughout the 6-h time course was relatively stable (Fig. 5A). Nuclear translocation of AP-1, NF-κB, and Sp1 transcription factors induced by PMA was examined by immunoblotting; recruitment of these transcription factors to the MMP-9 promoter correlated temporally with the kinetics of nuclear translocation (data not shown).
FIG. 5.
Dynamic recruitment of activators and modification of histones in transcriptional activation of the MMP-9 gene. (A) HeLa cells were treated with PMA for up to 6 h, and the cells were cross-linked with formaldehyde. The soluble chromatin was subjected to immunoprecipitation with antibodies against the AP-1, NF-κB, and Sp1 transcription factors. The basal level was set as 1.0, and induction upon PMA treatment was compared to that. IgG, immunoglobulin G. (B) ChIP was performed on HeLa cells treated with PMA for 2 h with an antibody against JunD. Then, soluble chromatin was eluted from the beads and subjected to a second round of ChIP with the antibodies shown (c-Fos, p65, p50, and Sp1). (C) ChIP was performed with antibodies against histone acetylation and phosphorylation. (D) ChIP was performed with antibodies against histone methylation. (E) HeLa cells were pretreated with MTA (1 mM) and TSA (50 ng/ml) for 30 min, and then PMA was added for an additional 10 h. Total RNA was subjected to RPA analysis of MMP-9 mRNA levels. Induction is shown. DMSO, dimethyl sulfoxide.
Re-ChIP assays were performed to examine what transcription factors localized simultaneously on the MMP-9 promoter with JunD. HeLa cells were treated with PMA for 2 h, and then soluble chromatin was immunoprecipitated with the anti-JunD antibody. The eluted chromatin was then diluted and reimmunoprecipitated with antibodies against c-Fos, p65, p50, and Sp1. c-Fos, p65, p50, and Sp1 were present in the chromatin immunoprecipitated by the antibody against JunD (Fig. 5B), demonstrating that these transcription factors are simultaneously present on the MMP-9 promoter after PMA treatment.
In addition to having functional roles in gene transcriptional regulation, covalent histone modifications are indicators of the recruitment of histone modifier complexes, such as HATs, HDACs, and histone methyltransferases, to promoters (19). To study histone modifications during PMA-induced MMP-9 gene transcription, ChIP assays were performed. Enhanced acetylation of H3 and H4 was observed 2 to 4 h after PMA stimulation (Fig. 5C), which corresponds to the onset of PMA-induced MMP-9 mRNA synthesis. Enhanced acetylation of H3 and H4 after PMA stimulation was observed as early as 30 min (data not shown). In addition, enhanced acetylation of K9-H3 was observed 4 to 6 h after PMA stimulation, K14-H3 acetylation was modestly enhanced 2 h after PMA treatment, and K8-H4 acetylation was observed 2 to 6 h after PMA treatment. Phosphorylation of Ser10 and acetylation of lysine 14 of H3 were slightly increased after 2 h of PMA stimulation and then diminished at 4 to 6 h. A more detailed kinetic analysis of 0 to 120 min of PMA stimulation demonstrated enhanced modification at 30 to 60 min, suggesting that the AcK14pSer10-H3 modification occurs prior to other modifications (Fig. 5C). We also found that MeR26-H3, MeR3-H4, and MeR17-H3 are involved in PMA-induced transcriptional activation of the MMP-9 gene (Fig. 5D). Furthermore, the ChIP assay for MeK4-H3 demonstrated that PMA-induced methylation of lysine 4 in H3 is optimal at 4 h and then diminishes over time (Fig. 5D). However, methylation of K9-H3, generally a suppressive modification, demonstrated a different kinetic pattern. MeK9-H3 was decreased upon PMA stimulation, reaching its lowest levels at 4 to 6 h, and then was restored to basal levels at 12 to 24 h (Fig. 5D). To confirm the functional role of histone modifications in MMP-9 gene transcription, HeLa cells were pretreated with MTA, a specific protein methyltransferase inhibitor, or TSA, an HDAC inhibitor, for 30 min and then PMA induction of MMP-9 mRNA expression was evaluated. MTA suppressed PMA-induced MMP-9 mRNA levels, while TSA substantially enhanced MMP-9 mRNA expression (Fig. 5E).
Recruitment of general transcription machinery, coactivators, and chromatin-remodeling complexes to the MMP-9 promoter.
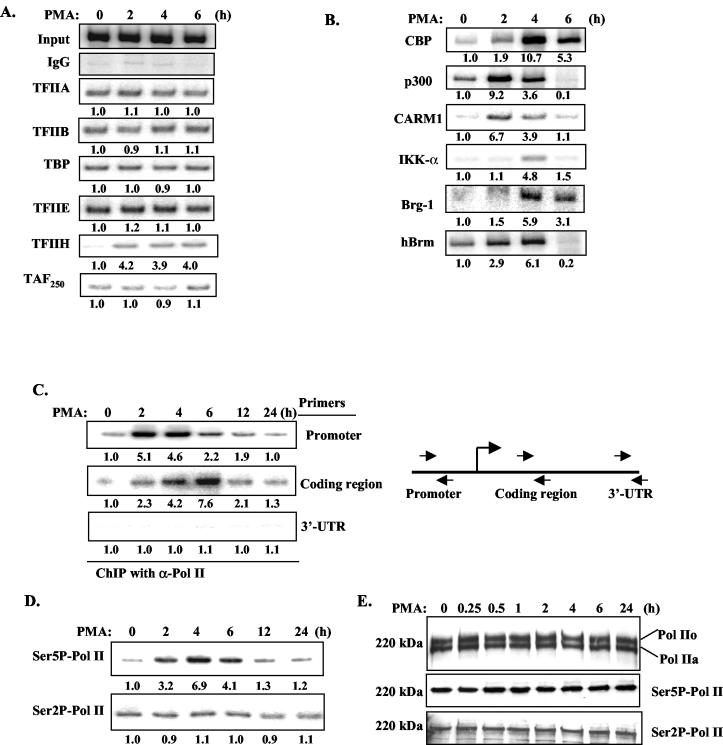
To examine the recruitment of general transcriptional machinery over the time course of PMA-induced MMP-9 gene activation, ChIP assays were performed. The binding of TFIIA, TFIIB, TATA binding protein, TFIIE, and TAF250 was relatively stable over the 0- to 6-h time course, while recruitment of TFIIH increased 2 h after PMA stimulation (Fig. 6A). Coactivators were recruited in a PMA-inducible manner (Fig. 6B). The peak of p300 and CARM1 recruitment was at 2 h, while, for CBP, maximal binding occurred at 4 h. Binding of IKK-α on the MMP-9 promoter was observed 4 h after PMA stimulation (Fig. 6B). However, recruitment of IKK-β, MEK-1, MEKK-1, or ERK1/2 on the MMP-9 promoter was not detected at any time points (data not shown). The recruitment of chromatin-remodeling complexes, as represented by the binding of Brg-1 and Brm to the MMP-9 promoter, occurred 2 to 4 h after PMA stimulation (Fig. 6B). The levels of GTFs and coactivators were also examined by immunoblotting; protein levels were stable over the 24-h time course of PMA stimulation (data not shown).
FIG. 6.
Recruitment of general transcription machinery, coactivators, and chromatin-remodeling complexes to the MMP-9 promoter. (A) ChIP was performed with antibodies against general transcription factors in HeLa cells treated with PMA for up to 6 h. The basal level was set as 1.0, and induction upon PMA treatment was compared to that. IgG, immunoglobulin G. (B) ChIP was performed with antibodies against CBP, p300, CARM1, IKK-α, Brg-1, and Brm. (C) DNA purified from ChIP with anti-Pol II antibodies was subjected to PCR analysis with primer pairs corresponding to the promoter, coding region, and 3′ UTR of the MMP-9 gene. (D) ChIP was performed with antibodies against Ser5 and Ser2 phosphorylation of Pol II CTD. (E) Nuclear extracts from HeLa cells treated with PMA for up to 24 h were immunoblotted with antibodies against total Pol II and Ser2 or Ser5 phosphorylation of Pol II CTD.
ChIP assays were also performed to examine the kinetics of Pol II recruitment. The binding of Pol II to the MMP-9 promoter increased between 2 and 4 h and diminished afterward (Fig. 6C). As the average size of chromatin fragments is ∼500 to 1,000 bp (data not shown), PCR was also performed with primers specific for the coding region (+2630 to +2842 of the human MMP-9 genomic sequence) and 3′ untranslated region (UTR; +7533 to +7853 of the human MMP-9 genomic sequence) of the MMP-9 gene, which are more than 3 kb apart. Maximal binding of Pol II at the coding region was at 4 to 6 h after PMA stimulation, which was ∼2 h later than the peak of Pol II at the promoter region; however, the binding of Pol II at the 3′ UTR was not detected (Fig. 6C). Therefore, these data suggest the transition of Pol II over the entire genomic locus of the MMP-9 gene during transcriptional activation. Antibodies against phospho-Ser2 and -Ser5 of the Pol II CTD were used to examine changes in phosphorylation during MMP-9 gene transcription by ChIP assay. Ser5 phosphorylation of Pol II CTD increased 2 h after PMA stimulation, reached maximal levels at 4 h, and then diminished over time (Fig. 6D). Ser2 phosphorylation was relatively stable over the entire time course (Fig. 6D). Data obtained from immunoblotting illustrated that total Pol II levels were relatively stable over the 24-h course of PMA stimulation, as were the levels of Pol II CTD Ser2 phosphorylation and Ser5 phosphorylation after PMA stimulation (Fig. 6E).
Dynamic occupancy of the MMP-9 promoter by corepressor complexes during MMP-9 gene activation.
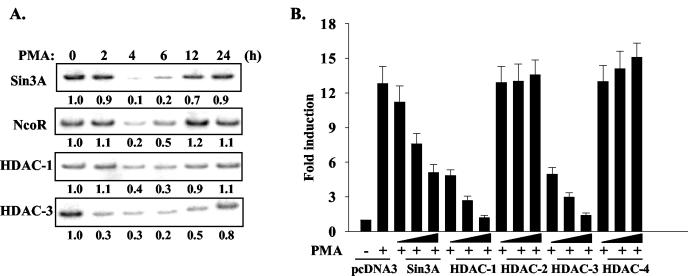
Recruitment of corepressor complexes on the MMP-9 promoter was examined by ChIP assays. The corepressor complexes Sin3A/HDAC-1 and NcoR/HDAC-3 occupied the MMP-9 promoter in unstimulated HeLa cells (Fig. 7A). Upon PMA stimulation, Sin3A/HDAC-1 and NcoR/HDAC-3 were removed from the MMP-9 promoter at 4 to 6 h and then restored at 12 to 24 h (Fig. 7A). To evaluate the functional significance of Sin3A and HDACs on MMP-9 gene transcription, increasing amounts of these corepressor constructs were transfected with the MMP-9 promoter reporter construct. Sin3A, HDAC-1, and HDAC-3 suppressed PMA-induced MMP-9 promoter activity in a dose-dependent manner, while HDAC-2 and HDAC-4 had no effect (Fig. 7B). Therefore, the Sin3A/HDAC-1 and NcoR/HDAC-3 corepressor complexes function to suppress MMP-9 gene transcription and are removed from the promoter after activation of the MMP-9 gene.
FIG. 7.
Functional involvement and dynamic occupancy of the MMP-9 promoter by corepressor and HDAC complexes in transcription of the MMP-9 gene. (A) HeLa cells were treated with PMA for up to 24 h. ChIP was performed with antibodies against Sin3A, NcoR, HDAC-1, and HDAC-3. The basal level was set as 1.0, and induction upon PMA treatment was compared to that. (B) The MMP-9 promoter construct (0.2 μg) was transiently transfected into HeLa cells with increasing amounts of Sin3A, HDAC-1, HDAC-2, HDAC-3, and HDAC-4 expression vectors. pcDNA3 was used to normalize the amount of DNA in each condition. After 12 h of recovery, transfected cells were treated with serum-free medium or PMA for 12 h. Luciferase activity was determined from the cell lysates. Data are means ± standard deviations from three experiments.
DISCUSSION
We investigated chromatin remodeling and dynamic formation of transcriptional complexes during PMA-induced activation of the MMP-9 gene. Activators, chromatin-remodeling complexes, and coactivators were recruited to the preassembled MMP-9 promoter in vivo in a stepwise and coordinated order, which was dependent on activation of the MEK-1/ERK and NF-κB signaling pathways. In contrast, corepressor complexes were released from the MMP-9 promoter after transcriptional activation. Histone modifications shifted from repressive to permissive modifications concurrent with activation of the MMP-9 gene. Chromatin remodeling induced by Brg-1 was required for MMP-9 gene transcription and was concomitant with initiation of transcription. Thus, coordination of cell signaling, chromatin remodeling, histone modifications, and stepwise recruitment of transcription regulators is critical to regulate MMP-9 gene expression.
Dynamics of chromatin remodeling and transcription complex formation during MMP-9 gene transcription.
Chromatin structure analysis illustrated that the endogenous MMP-9 promoter is packaged into regularly phased nucleosomes and, more importantly, that the chromatin structure of the MMP-9 promoter is remodeled in coordination with the activation of MMP-9 gene transcription. ChIP assays demonstrated that recruitment of Brg-1 and Brm peaked 4 h after PMA stimulation, the time point at which maximal relaxation of chromatin on the MMP-9 promoter occurs. In addition, Brg-1-mediated chromatin remodeling was functionally required for MMP-9 gene transcription. Therefore, chromatin remodeling is a critical step involved in transcriptional activation of the MMP-9 gene.
The composition of AP-1 and NF-κB factors on the MMP-9 promoter dynamically changed over the course of MMP-9 induction. JunD, c-Fos, p65, and p50 associated with the MMP-9 promoter in unstimulated cells. After PMA treatment, the binding of JunD and c-Fos increased and c-Jun appeared at a later time (6 h) on the promoter. Recruitment of p65/p50 increased between 2 and 4 h, while at 4 to 6 h the NF-κB composition changed to c-Rel/p52. The Re-ChIP assays illustrated that JunD, c-Fos, p65, p50, and Sp1 were simultaneously associated with the MMP-9 promoter 2 h after PMA stimulation, suggesting the assembly of an enhanceosome induced by PMA. Previous studies proposed that AP-1 factors can act as either activators or repressors, depending on the composition of subunits and partner proteins in the transcription complex (7). In addition, the composition of NF-κB dimers constantly changes during lipopolysaccharide-induced gene transcription (35). These findings suggest that dynamic changes in the composition of AP-1 and NF-κB dimers constitute an important regulatory mechanism to fine tune transcription of the MMP-9 gene.
Our results illustrate that general transcription machinery, chromatin-remodeling complexes, coactivators, and corepressors display distinct patterns of recruitment. GTFs, except TFIIH and Pol II, are present on the MMP-9 promoter in untreated cells, and PMA stimulation does not alter binding, suggesting that the MMP-9 promoter is preassembled. This has been proposed as a mechanism for a prompt response to an inductive signal (38, 39). The recruitment kinetics of Pol II and TFIIH and the phosphorylation of Ser5 Pol II CTD correlated with transcriptional activation of the MMP-9 gene, suggesting that Pol II, TFIIH, and Ser5 phosphorylation are the components of the general transcription machinery recruited to the MMP-9 promoter upon initiation of transcription. Interestingly, we found that Ser5 phosphorylation in the promoter region of the human MMP-9 gene is more closely correlated with transcriptional activation than that of Ser2. This may be caused by the distribution of CTD phosphorylation during transcription, since Ser5 phosphorylation is more concentrated in the promoter region, while Ser2 phosphorylation is evenly distributed along the promoter and coding regions (9, 22). It has also been shown that gamma interferon-activated CIITA (major histocompatibility complex class II transactivator) gene transcription is more dependent on Ser5 phosphorylation than on Ser2 phosphorylation (39). CBP and p300, the two structural and functionally related HATs, displayed differential recruitment kinetics. Several groups have proposed that CBP and p300 may play distinct roles in transcription activation (14). Considering our data and results from other groups (37), we propose that p300 may be involved in transcription initiation and that CBP is important for transcription and disassembly of transcription complexes by acetylating other proteins. Indeed, the maximal levels of phosphorylation of Ser5 Pol II CTD, which is a marker of transcription initiation, occur at the same time point (4 h) at which CBP displays maximal association on the MMP-9 promoter. Our results also demonstrate that IKK-α translocates into the nucleus and binds the MMP-9 promoter with the same kinetics as that of CBP; thus IKK-α may function to facilitate recruitment of CBP (3, 45). Our data also demonstrated that CARM1 is involved in MMP-9 gene transcription activation. CARM1 is recruited to the MMP-9 promoter concurrent with CBP and p300. Therefore, in addition to inducing methylation of R17-H3, CARM1 may methylate arginine residues in the KIX domains of CBP and p300 and subsequently enhance transcription of the MMP-9 gene by enhancing the affinity of activators and coactivators (44). Indeed, CARM1 can activate MMP-9 promoter activity in synergy with CBP and p300 (data not shown).
In contrast to activation forces, the corepressor complexes, including Sin3A/HDAC-1 and NcoR/HDAC-3, associate with the MMP-9 promoter with kinetics distinct from those for activators and coactivators. This suggests that transcriptional activation of the MMP-9 gene requires simultaneous release of corepressor complexes and recruitment of activators and coactivators. The switching of corepressors and coactivators may be regulated by the MAPK pathway, as the MEK-1 kinase pathway has been shown to inhibit the activities of NcoR and SMRT and, at the same time, enhance nuclear export of corepressor complexes (4, 18).
Histone code of MMP-9 gene activation.
In addition to acetylation of H3 and H4 and arginine methylation of R17-H3, R26-H3, and R3-H4, we found that lysine methylation of K4-H3 and K9-H3 and phosphorylation of S10-H3 are also involved in transcriptional activation of the MMP-9 gene. We observed relatively high levels of Ser10 phosphorylation and K14 acetylation of H3 on the MMP-9 promoter prior to PMA stimulation. It has been shown that elevated levels of H3 phosphorylation may be responsible for relaxed chromatin structure and aberrant gene expression in transformed cells (8, 41). In addition, Ser10-H3 phosphorylation is functionally linked to Gcn5-mediated acetylation of AcK14-H3 (10, 30). Therefore, constitutive and induced K14 acetylation and phosphorylation of Ser10 of H3 may contribute to the prompt response of the MMP-9 gene to an exogenous stimulus such as PMA. Concurrent with transcription complex formation, histone modifications that correlate with gene activation increase, following the recruitment kinetics of activators, coactivators, and chromatin-remodeling complexes. However, for suppressive modifications such as methylation of K9-H3, levels decrease upon transcription activation and then are restored. In agreement with our findings, Saccani and Natoli have shown that changes of K9-H3 methylation strongly correlate with Pol II recruitment and release in inducible inflammatory genes (34). In addition, it has been reported that c-Jun/c-Fos-induced chromatin-remodeling activity increased 10-fold on an acetylated nucleosome template, suggesting that there is an inherent connection among chromatin remodeling, regulator recruitment, and histone modifications (32). Thus, histone modifications may not only be the “phenotypes” of histone modification complex recruitment, but also may play important functional roles in modulating transcriptional complex formation. It has been suggested that chromatin-remodeling complexes may serve as “translators” to remodel nucleosomes corresponding to the instruction of the histone code (19).
Coordinated and stepwise recruitment of transcription complexes on a preassembled MMP-9 promoter.
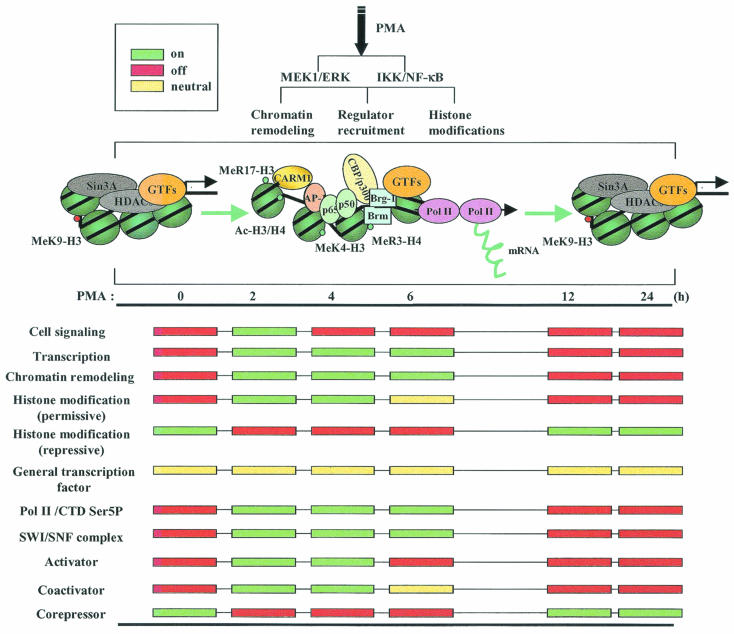
Previous studies indicate that the time course of gene expression and ordered recruitment is accomplished by gene-specific transcription programs (1, 37, 38). Two models are proposed to explain the possible mechanisms of eukaryotic transcription regulation (25). The first is the stepwise-assembly model, which suggests that activators are first targeted to the promoter and then chromatin remodeling and histone modifications by SWI/SNF and coactivators allow the recruitment of Pol II, GTFs, and mediator complexes and finally trigger mRNA synthesis by promoter clearance (25). The second model suggests a preassembly process for transcription activation, which proposes that transcription complexes are formed as a complex including all the major components of transcription, such as GTFs, and some of the coactivators, but without Pol II, TATA binding protein, or the chromatin-remodeling complex (25). Our results suggest that transcription activation of the MMP-9 gene depends on the coordinated and stepwise recruitment of transcription complexes on the preassembled MMP-9 promoter. In the basal state, the MMP-9 promoter is occupied by GTFs and corepressor complexes. The major histone modification is methylation of K9-H3, which defines the suppressive chromatin structure on the MMP-9 promoter. Induction of the MMP-9 gene triggers activation of the ERK and NF-κB pathways, inducing nuclear translocation of transcription factors and subsequent recruitment to the MMP-9 promoter. Concurrently, corepressor complexes are released from the promoter and coactivator and chromatin-remodeling complexes are recruited. Concurrent with regulator recruitment, the chromatin is relaxed by the SWI/SNF complex, resulting in the binding of Pol II and other coactivators such as CBP. Histone modifications switch from repressive modifications to inductive modifications. Subsequently, mRNA is synthesized through Pol II elongation following the MMP-9 coding sequence after initiation of Ser5 phosphorylation at Pol II CTD. Finally, coincident with methylation of K9-H3 and restoration of corepressor complexes, coactivator and chromatin-remodeling complexes are removed from the MMP-9 promoter, which correlates with attenuation of MMP-9 gene expression (Fig. 8). Therefore, cell-specific factors and signaling pathways regulate transcriptional activation of the human MMP-9 gene by modulating its gene-specific transcriptional program.
FIG. 8.
Transcription program of the human MMP-9 gene. The sequential recruitment of activators, coactivators, chromatin remodeling complexes, and general transcription machinery to the preassembled MMP-9 promoter results in transcriptional activation of the human MMP-9 gene. See text for details.
Acknowledgments
This work was supported by Public Health Service grants CA-97247 from the National Cancer Institute and NS-39954 from the National Institute of Neurological Disorders and Stroke to E.N.B.
We thank B. Bernstein, D. Boyd, G. R. Crabtree, J. E. Kudlow, F. T. Lin, A. R. Mackay, H. Nakano, M. G. Rosenfeld, M. R. Stallcup, E. Seto, and K. Zhao for providing valuable reagents.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Albert, T., J. Wells, J. O. Funk, A. Pullner, E. E. Raschke, G. Stelzer, M. Meisterernst, P. J. Farnham, and D. Eick. 2001. The chromatin structure of the dual c-myc promoter P1/P2 is regulated by separate elements. J. Biol. Chem. 276:20482-20490. [DOI] [PubMed] [Google Scholar]
- 3.Anest, V., J. L. Hanson, P. C. Cogswell, K. A. Steinbrecher, B. D. Strahl, and A. S. Baldwin. 2003. A nucleosomal function for IκB kinase-α in NF-κB-dependent gene expression. Nature 423:659-663. [DOI] [PubMed] [Google Scholar]
- 4.Baek, S. H., K. A. Ohgi, D. W. Rose, E. H. Koo, C. K. Glass, and M. G. Rosenfeld. 2002. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-κB and beta-amyloid precursor protein. Cell 110:55-67. [DOI] [PubMed] [Google Scholar]
- 5.Bauer, U. M., S. Daujat, S. J. Nielsen, K. Nightingale, and T. Kouzarides. 2002. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 3:39-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein, L. R., and S. E. Walker. 1999. Tumor promotion resistant cells are deficient in AP-1 DNA binding, JunD DNA binding and JunD expression and form different AP-1-DNA complexes than promotion sensitive cells. Biochim. Biophys. Acta 1489:263-280. [DOI] [PubMed] [Google Scholar]
- 8.Chadee, D. N., M. J. Hendzel, C. P. Tylipski, C. D. Allis, D. P. Bazett-Jones, J. A. Wright, and J. R. Davie. 1999. Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J. Biol. Chem. 274:24914-24920. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, C., and P. A. Sharp. 2003. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and gamma-actin genes. Mol. Cell. Biol. 23:1961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, P., K. G. Tanner, W. L. Cheung, P. Sassone-Corsi, J. M. Denu, and C. D. Allis. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5:905-915. [DOI] [PubMed] [Google Scholar]
- 11.Farina, A. R., A. Tacconelli, A. Vacca, M. Maroder, A. Gulino, and A. R. Mackay. 1999. Transcriptional up-regulation of matrix metalloproteinase-9 expression during spontaneous epithelial to neuroblast phenotype conversion by SK-N-SH neuroblastoma cells, involved in enhanced invasivity, depends upon GT-box and nuclear factor κB elements. Cell Growth Differ. 10:353-367. [PubMed] [Google Scholar]
- 12.Fry, C. J., and C. L. Peterson. 2002. Transcription. Unlocking the gates to gene expression. Science 295:1847-1848. [DOI] [PubMed] [Google Scholar]
- 13.Garrity, P. A., and B. J. Wold. 1992. Effects of different DNA polymerases in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc. Natl. Acad. Sci. USA 89:1021-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 15.Gum, R., E. Lengyel, J. Juarez, J. H. Chen, H. Sato, M. Seiki, and D. Boyd. 1996. Stimulation of 92-kDa gelatinase B promoter activity by ras is mitogen-activated protein kinase kinase 1-independent and requires multiple transcription factor binding sites including closely spaced PEA3/ets and AP-1 sequences. J. Biol. Chem. 271:10672-10680. [DOI] [PubMed] [Google Scholar]
- 16.Hazzalin, C. A., and L. C. Mahadevan. 2002. MAPK-regulated transcription: a continuously variable gene switch? Nat. Rev. Mol. Cell Biol. 3:30-40. [DOI] [PubMed] [Google Scholar]
- 17.Hochheimer, A., and R. Tjian. 2003. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 17:1309-1320. [DOI] [PubMed] [Google Scholar]
- 18.Hong, S. H., and M. L. Privalsky. 2000. The SMRT corepressor is regulated by a MEK-1 kinase pathway: inhibition of corepressor function is associated with SMRT phosphorylation and nuclear export. Mol. Cell. Biol. 20:6612-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 20.Jepsen, K., and M. G. Rosenfeld. 2002. Biological roles and mechanistic actions of co-repressor complexes. J. Cell Sci. 115:689-698. [DOI] [PubMed] [Google Scholar]
- 21.Kadam, S., G. S. McAlpine, M. L. Phelan, R. E. Kingston, K. A. Jones, and B. M. Emerson. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 14:2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12:198-209. [DOI] [PubMed] [Google Scholar]
- 24.Lachner, M., and T. Jenuwein. 2002. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14:286-298. [DOI] [PubMed] [Google Scholar]
- 25.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 26.Levine, M., and R. Tjian. 2003. Transcription regulation and animal diversity. Nature 424:147-151. [DOI] [PubMed] [Google Scholar]
- 27.Li, J., Q. Lin, H. G. Yoon, Z. Q. Huang, B. D. Strahl, C. D. Allis, and J. Wong. 2002. Involvement of histone methylation and phosphorylation in regulation of transcription by thyroid hormone receptor. Mol. Cell. Biol. 22:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, P. S., A. Tremeau-Bravard, and M. E. Dahmus. 2003. The repetitive C-terminal domain of RNA polymerase II: multiple conformational states drive the transcription cycle. Chem. Rec. 3:235-245. [DOI] [PubMed] [Google Scholar]
- 29.Liu, R., H. Liu, X. Chen, M. Kirby, P. O. Brown, and K. J. Zhao. 2001. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106:309-318. [DOI] [PubMed] [Google Scholar]
- 30.Lo, W. S., R. C. Trievel, J. R. Rojas, L. Duggan, J. Y. Hsu, C. D. Allis, R. Marmorstein, and S. L. Berger. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5:917-926. [DOI] [PubMed] [Google Scholar]
- 31.Ma, Z., H. Qin, and E. N. Benveniste. 2001. Transcriptional suppression of matrix metalloproteinase-9 gene expression by IFN-γ and IFN-β: critical role of STAT-1α. J. Immunol. 167:5150-5159. [DOI] [PubMed] [Google Scholar]
- 32.Ng, K. W., P. Ridgway, D. R. Cohen, and D. J. Tremethick. 1997. The binding of a Fos/Jun heterodimer can completely disrupt the structure of a nucleosome. EMBO J. 16:2072-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 34.Saccani, S., and G. Natoli. 2002. Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev. 16:2219-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saccani, S., S. Pantano, and G. Natoli. 2003. Modulation of NF-κB activity by exchange of dimers. Mol. Cell 11:1563-1574. [DOI] [PubMed] [Google Scholar]
- 36.Sato, H., and M. Seiki. 1993. Regulatory mechanism of 92 kDa type IV collagenase gene expression which is associated with invasiveness of tumor cells. Oncogene 8:395-405. [PubMed] [Google Scholar]
- 37.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 38.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 39.Spilianakis, C., A. Kretsovali, T. Agalioti, T. Makatounakis, D. Thanos, and J. Papamatheakis. 2003. CIITA regulates transcription onset via Ser5-phosphorylation of RNA Pol II. EMBO J. 22:5125-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sternlicht, M. D., and Z. Werb. 2001. How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol. 17:463-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strelkov, I. S., and J. R. Davie. 2002. Ser-10 phosphorylation of histone H3 and immediate early gene expression in oncogene-transformed mouse fibroblasts. Cancer Res. 62:75-78. [PubMed] [Google Scholar]
- 42.Sudarsanam, P., and F. Winston. 2000. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 16:345-351. [DOI] [PubMed] [Google Scholar]
- 43.Vu, T. H., and Z. Werb. 2000. Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev. 14:2123-2133. [DOI] [PubMed] [Google Scholar]
- 44.Xu, W., H. Chen, K. Du, H. Asahara, M. Tini, B. M. Emerson, M. Montminy, and R. M. Evans. 2001. A transcriptional switch mediated by cofactor methylation. Science 294:2507-2511. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto, Y., U. N. Verma, S. Prajapati, Y. T. Kwak, and R. B. Gaynor. 2003. Histone H3 phosphorylation by IKK-α is critical for cytokine-induced gene expression. Nature 423:655-659. [DOI] [PubMed] [Google Scholar]