Abstract
Protein inhibitor of activated STAT Y (PIASy) is the shortest member of the PIAS family and has been reported to modulate the transcriptional activities of STAT1, lymphoid enhancer factor 1 (LEF-1), and the androgen receptor. PIAS proteins have also been identified as E3 ligases for the small ubiquitin-like modifier (SUMO) proteins. PIASy in particular has been reported to mediate SUMO-2/3 modification of LEF-1, sequestering it into nuclear bodies, and SUMO-1 ligation to c-Myb, modulating its transcriptional activation properties. We have cloned murine Piasy and a splice variant which omits exon 6, containing the nuclear retention PINIT motif. Cell culture studies indicate that both the full length and the splice variant are localized in the nucleus but differentially enhance SUMO ligation. To further understand the functions of PIASy, we have generated PIASy-deficient mice. Surprisingly, Piasy−/− mice appear phenotypically normal. Activation of STAT1 is not significantly perturbed in Piasy−/− cells, and sumoylation patterns for SUMO-1 or SUMO-3 modification are similar when comparing tissues and embryonic fibroblasts from wild-type and knockout mice. Our study demonstrates that at steady state, PIASy is either dispensable or compensated for by other PIAS family members or by other mechanisms when deleted.
Protein inhibitor of activated STAT (PIAS) proteins were named for their abilities to inhibit DNA binding and transcription activation by the STAT (signal transducer and activator of transcription) family of transcription factors (1, 16). They have since been shown to have broader transcriptional regulation properties beyond that of STAT inhibition (reviewed in reference 30). The mammalian family consists of at least five members, PIAS1, PIAS3, PIASxα, PIASxβ, and PIASy (PIASxα and xβ being splice variants) (31). PIAS proteins contain a zinc ring finger domain (1), an N-terminal LXXLL coregulator motif (15), a C-terminal acidic domain involved in binding TIF2 (9), and a recently identified PINIT motif involved in nuclear retention (4).
In addition to their roles in regulating transcription, PIAS proteins have also been identified as E3 ligases for the small ubiquitin-like modifier proteins 1 and 2/3 (SUMO-1 and SUMO-2/3) (11). SUMO proteins are structurally similar to ubiquitin (18) but, in contrast to ubiquitin, do not normally target proteins for degradation. SUMO addition to lysine residues of target proteins proceeds in a fashion similar to that of ubiquitination and has been extensively studied in Saccharomyces cerevisiae (3, 10, 19, 30). Although sumoylation is evolutionarily conserved from yeast to humans, it is not known what effects the loss of function mutations in mammalian systems would produce. Siz1 and Siz2 (SAP and Miz1 domains) have been identified as SUMO E3 ligating enzymes in yeast (11) and share homology to the zinc ring finger domains of PIAS proteins. Siz1 and Siz2 double mutants grow poorly and also display a marked decrease in SUMO modification but do not totally abolish it (11).
In mammalian systems, functions for SUMO modification include roles in subnuclear structure formation, modulation of protein-protein interactions, transcriptional control, and stabilization of proteins by blocking of ubiquitination sites (13, 18). The roles of PIAS proteins in sumoylating transcription factors such as p53, c-Jun, androgen receptor, c-Myb, and lymphoid enhancer factor 1 (LEF-1) have been reported recently (2, 22, 26, 28). In the case of LEF-1, PIASy represses LEF-1-mediated gene activation and acts as an E3 ligase for addition of SUMO-2/3 moieties to LEF-1. However, repression of LEF-1-mediated gene activation by PIASy was not lost if the consensus lysine sumoylation residues on LEF-1 were mutated to arginine. PIASy was also reported to enhance SUMO-1 modification of c-Myb, leading to a reduction in its transcriptional activation properties (2). Conversely, sumoylation of p53 leads to an increase in its transcriptional activation activity (5, 25), and this modification is mediated through either the direct or tightly associated E3 ligase activity of PIAS1 (12). An independent report showed that PIASy is able to repress p53-mediated activation of target genes with no effect on apoptosis (21).
PIASy is the shortest member of the PIAS family and has been characterized as a specific inhibitor of STAT1 but by a mechanism other than inhibition of STAT DNA binding as described for PIAS1 and PIAS3 (15). Similar to PIAS1 and PIASxα, PIASy is also able to repress androgen receptor-mediated gene activation. How this repression occurs is not yet known for PIASy, but for PIAS1 and PIASxα, it is at least in part related to SUMO E3 ligase activity (6, 22). During mouse embryonic development, PIASy, also described as PIASγ, is expressed in the limb buds, neuroepithelium, and inner root sheath of hair follicles (32).
To study the biological function of PIASy, we have cloned the murine Piasy cDNA along with a naturally occurring splice variant lacking a region that contains the PINIT motif and demonstrated that both are localized in the nucleus and that overexpression of each is capable of differential enhancement of SUMO ligation. Furthermore, to examine the in vivo physiological functions of PIASy, we generated PIASy-deficient mice. Characterization of the mutant mice indicates that PIASy is not essential for embryogenesis or adult life at a steady state and that, despite enhanced SUMO ligation in overexpression studies, there is no absolute requirement for PIASy for addition of SUMO-1 or SUMO-3 to target proteins.
MATERIALS AND METHODS
Cloning of mouse PIASy cDNAs.
Degenerate PCR primers, designed to amplify a region near the conserved zinc ring finger domain of the PIAS gene family, were chosen from a multiple alignment of PIAS human and mouse genes: forward primer, 5′-GCATCGATGA(A/T/G/C)AC(A/C)AG(T/C)TG(T/C)CC(T/A)CA(A/G)GA-3′; reverse primer, 5′-CGTCGACGC(T/C)(T/G)(T/G)(T/C)TT(A/G)TC(A/G)CA(G/C)AC(A/G)GG(A/G)CA-3′.
Spleen mononuclear cells were isolated from mice, and total RNA was prepared by using Trizol reagent (Gibco-BRL). After DNase treatment, 2 μg of RNA was reverse transcribed to first-strand cDNA by using oligo(dT) primers by avian myeloblastosis virus reverse transcriptase (RT) under conditions recommended by the manufacturer (Gibco-BRL). PCRs were simultaneously performed at various annealing temperatures using a Robocycler gradient thermal cycler. Thermal cycling parameters were as follows: one cycle at 94°C for 3 min; 40 cycles at 94°C for 1 min, 47 to 58°C for 1 min, and 72°C for 1.5 min; and one cycle at 72°C for 10 min. PCR products were T/A cloned into pCRTopo2.1 (Invitrogen), sequenced, and identified by BLAST search. Full-length mouse PIASy was cloned by using the PCR product as a probe to screen a Uni-ZAP XR mouse embryonic day 10.5 cDNA library (Stratagene) per manufacturer's protocol and was verified by DNA sequencing. PIASyE6− was cloned by utilizing flanking restriction enzyme sites in the full-length cDNA. Full-length PIASy and PIASyE6− were cloned in frame into a 5′ Myc-tagged mammalian expression vector (pCMV-Tag3; Stratagene).
PIASy and PIASyE6− subcellular localization.
The cDNAs for mouse PIASy and PIASyE6− were fused in frame to the green fluorescent protein (GFP) expression fusion construct pEGFP-C1 (Clontech). NIH 3T3 cells were grown on coverslips in six-well dishes and transfected with either pEGFP-PIASy, pEGFP-PIASyE6−, or pEGFP. Cells were grown in Dulbecco's modified Eagle medium plus 10% fetal calf serum (FCS) (Gibco-BRL), and transfections were performed with Effectene reagent (QIAGEN, Valencia, Calif.) per the manufacturer's protocol. Forty-eight hours posttransfection, the cells were washed and fixed in 4% para-formaldehyde for 10 min. Coverslips were placed on glass microscope slides with mounting media containing 4′,6′-diamidino-2-phenylindole (DAPI; Vector Labs) and allowed to dry. Samples were visualized by using an Olympus BX60 microscope with a ×100 oil immersion objective under UV illumination.
E3 SUMO ligase assay.
The mouse SUMO-2 sequence was acquired from GenBank (accession no. AF063847) and used to clone the mouse SUMO-2 cDNA into an N-terminal Flag-tagged expression vector (pCMV-Tag2; Stratagene). The Flag-SUMO-1 expression construct was a generous gift from Ke Shuai. NIH 3T3 cells were transfected (Effectene; QIAGEN) with 1 to 2 μg of Flag-tagged SUMO-1 and SUMO-2 along with Myc epitope-tagged PIASy or PIASyE6− expression vectors. Twenty-four to 48 h posttransfection, cells were harvested in radioimmunoprecipitation assay buffer (1× phosphate-buffered saline, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) containing protease inhibitor cocktail (Complete; Roche) and N-ethylmaleamide at a concentration of 10 mM followed by Western blot analysis as described below. Flag antibody (Stratagene) was used at a concentration of 1:10,000 for 1 to 2 h and Myc antibody (Cell Signaling Technology) at a concentration of 1:1,000.
Gene targeting of Piasy.
The bacterial artificial chromosome (BAC) containing the Piasy gene was isolated from a 129S6/SvEvTac strain mouse genomic BAC library and used to clone 3-kb 5′ and 1.4-kb 3′ targeting arms flanking the NeoR gene. The diphtheria toxin gene was cloned upstream of the 5′ targeting arm for negative selection. Fifty micrograms of targeting vector was linearized by NotI digestion and electroporated into 2 × 107 embryonic stem (ES) cells. Twenty-four hours later, ES cells were subjected to selection by 400 μg of G418 (Roche)/ml for 8 to 10 days. Surviving ES cell colonies were individually picked and checked for homologous recombination by genomic Southern blotting. ES cell clones positive for homologous recombination were expanded and injected into C57BL/6 blastocysts to generate chimeric mice as previously described (34). Male chimeras were backcrossed to C57BL/6 females for germ line transmission of the Piasy knockout allele.
Genotyping of Piasy wild-type and knockout alleles.
Genotyping of ES cell clones was performed by genomic Southern blotting. Digestion of genomic DNA with EcoRI was performed overnight, followed by electrophoresis on a 0.7% agarose gel. DNA was transferred to nitrocellulose membranes (Hybond; Amersham) and probed with a [α-32P]CTP-labeled 5′ genomic probe flanking the 5′ targeting arm, which identifies an 11.4-kb wild-type allele and a 7.2-kb knockout allele.
RT-PCR of embryonic fibroblast RNA.
Timed matings were set up between heterozygous animals. At E13.5, embryos were dissected from the mother, individually trypsinized, and expanded on gelatinized tissue culture dishes in Dulbecco's modified Eagle medium (Gibco-BRL) plus 10% FCS. Total RNA was isolated from confluent 10-cm tissue culture dishes (RNeasy; QIAGEN) and subjected to RT-PCR with primers specific for the murine Piasy cDNA. The forward primer lies in exon 2 (5′-GTATCAGACCTGCAGATGCTGC-3′), while the reverse primer lies in exon 11 (5′-CACCAGGCCTTTCTGGAACG-3′). β-Actin primers were used as a control. Thermal cycling parameters were as follows: one cycle at 94°C for 3 min; 40 cycles at 94°C for 1 min, 62°C for 1 min, and 72°C for 1.5 min; and one cycle at 72°C for 10 min.
Western blots.
Mice were sacrificed and various tissues were dissected. Tissues were placed in radioimmunoprecipitation assay buffer containing protease inhibitor and 10 mM N-ethylmaleamide and homogenized on ice using a RotorStator Power Gen 125 (Fisher Scientific). Homogenized samples were passed through a Qiashredder column (QIAGEN) for 2 min at 14,000 rpm. Phenylmethylsulfonyl fluoride was added to a concentration of 100 μg/ml, and lysates were placed on ice for 30 to 60 min. Forty micrograms of protein per lane was loaded for Western blotting. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose. Blots were blocked in Tris-buffered saline with 0.1% Tween 20 (TBST) containing 5% nonfat dry milk for 1 to 2 h at room temperature. Primary antibodies were incubated overnight at 4°C and washed three to five times for 10 min each in TBST. (PIASy antibody was supplied by Ke Shuai, SUMO-1 and SUMO-3 antibody were from Zymed, phosphotyrosine-STAT1 and STAT1 were from Cell Signaling Technology, and LEF-1 and c-Myb antibodies were from Santa Cruz Biotechnology.) Horseradish peroxidase-conjugated immunoglobulin secondary antibodies (Amersham Life Sciences) were added to TBST plus 5% milk and incubated on blots for 1 to 2 h followed by 3 to 5 10-min washes in TBST. Western blots were incubated with LumiGLO ECL reagent (Cell Signaling Technology), exposed to X-ray film, and developed. For stress treatment of mouse embryonic fibroblasts (MEFs), 107 cells were trypsinized and incubated at 43°C for 10 min for heat shock or with 7% ethanol at 37°C for 10 min for ethanol treatment.
Tissue sections.
Mice were euthanized and tissues dissected for sectioning. Tissues were collected in 10% formalin and fixed overnight at room temperature. Fixed tissues were sent to Impath Histological Services (Marina del Rey, Calif.) for paraffin embedding and sectioning. Sections were mounted on slides and stained with hematoxylin and eosin.
IFN-γ stimulation.
For Western blotting, primary splenocytes were suspended in Iscove's modified Dulbecco's medium plus 0.5% FCS plus 100 ng of gamma interferon (IFN-γ)/ml for 20 min, washed, and then resuspended in Iscove's modified Dulbecco's medium plus 0.5% FCS and incubated at 37°C for 0, 15, and 30 min before preparation of protein lysates for Western blot analysis with STAT1 and phospho-STAT1 (Tyr 701) antibodies (Cell Signaling Technology). For Northern analysis, splenocytes were treated with 100 ng of IFN-γ/ml for the indicated time points before preparation of total RNA (RNeasy; QIAGEN).
Northern blots.
Approximately 10 μg of total RNA from tissues was electrophoresed on a 1.0% formaldehyde agarose gel at 100 V for 3 h. RNA was transferred to nitrocellulose overnight in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) by capillary action. Blots were UV cross-linked and baked at 80°C for 2 to 3 h before hybridization with [α-32P]CTP-labeled probes and exposure to phosphorimager cassettes. For IFN-γ stimulations, quantitations were performed using ImageQuant software with care taken to stay within the linear response range.
PML subnuclear localization.
MEF cells were grown on coverslips in six-well dishes and transfected with Flag-PML or empty Flag expression vectors. Forty-eight hours posttransfection, the cells were washed and fixed in 4% para-formaldehyde for 10 min, permeabilized in 0.2% Triton X-100 for 5 min, and quenched in 0.1% sodium borohydride. Cells were blocked in 10% goat serum followed by Flag antibody incubation at 1:1,000 overnight at 4°C. Cells were washed 3 times in TBS, followed by Cy3-labeled goat anti-mouse secondary incubation for 1 h, and then they were washed three times. Coverslips were mounted in media containing DAPI (Vector Labs) and allowed to dry.
Mo-MULV replication assay.
One microgram of plasmid encoding Moloney murine leukemia virus (Mo-MuLV) was transfected into mouse NIH 3T3 cells by using Effectene (QIAGEN). Cells were split 1:8 every 3 to 4 days for 2 weeks and then allowed to grow to confluency. Supernatant was collected and filtered through a 0.22-μm-pore-size filter and frozen at −80°C. For infection, cells were split to approximately 30% confluency, followed by addition of 1 ml of virus stock the next day. After 1 h of incubation, medium was added back to normal levels and cells were allowed to grow to confluency. Cells were split 1:8 every 3 to 4 days for 2 weeks to maintain cells in a log phase of growth. Viral RNA was purified from media of infected cells by using a viral RNA kit (Roche) and subjected to RT-PCR (One Step; Invitrogen) for the presence of Gag RNA; the primers are described in reference 14. Thermal cycling parameters were as follows: one cycle at 50°C for 30 min; one cycle at 94°C for 2 min; 20 to 30 cycles at 94°C for 15 s, 60°C for 30 s, and 72°C for 1 min; and one cycle at 72°C for 10 min.
RESULTS
Mouse PIASy and PIASyE6− are localized in the nucleus.
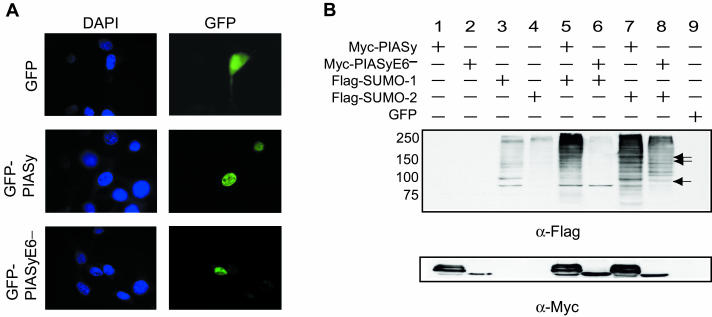
To study the function of PIASy, we cloned the mouse Piasy cDNA from mouse spleen mononuclear cells by RT-PCR. In addition to the full-length cDNA, a short form missing an internal region coding for amino acids 218 to 260 was also cloned (PIASyE6−). Subsequent sequence analysis indicated that this short form was missing only exon 6, indicating a splice variant. Contained in exon 6 is the coding region for the recently identified PINIT motif involved in nuclear retention of PIAS3 and conserved in all mammalian PIAS proteins (4) (Fig. 1). To determine the subcellular localization of full-length mouse PIASy and PIASyE6−, we generated PIASy-GFP and PIASyE6−-GFP fusion constructs and expressed them in NIH 3T3 cells. Expression of GFP alone showed green fluorescence throughout the cell. However, green fluorescence was seen exclusively in the nucleus when fused to either full-length PIASy or PIASyE6− (Fig. 2A). These data indicated that, similar to human PIASy (15), full-length mouse PIASy is also localized in the nucleus, and that a lack of the PINIT motif did not alter subcellular localization in an overexpression setting.
FIG. 1.
Primary amino acid alignment of full-length mouse PIASy (top) and PIASyE6− (bottom). Exon 6 codes for 43 amino acids consisting of residues 218 to 260. The PINIT motif is boxed and is missing in PIASyE6−. Alignment was performed by the Clustal method.
FIG. 2.
Mouse PIASy and PIASyE6− are nuclear localized and differentially enhance SUMO ligation. (A) Transient transfections into NIH 3T3 cells of GFP, GFP-PIASy, or GFP-PIASyE6− expression constructs. Cells were visualized with a ×100 objective under UV illumination. The same field is shown in each row. First column, DAPI stain for nuclei; second column, green fluorescence. (B) Western blots of NIH 3T3 cells transiently transfected with expression constructs designated by the grid above the blots. Shown is duplicate Western blotting with anti-Flag antibody in the top panel and anti-Myc antibody in the bottom panel. Molecular weights are indicated to the left of the top panel. Arrows indicate differences in the pattern of SUMO-2 ligation by PIASy and PIASyE6− in lane 7 versus 8.
PIASy and PIASyE6− enhance sumoylation differentially.
In order to assess the abilities of mouse PIASy and PIASyE6− to act as SUMO E3 ligases, Myc-tagged expression constructs for each were generated and coexpressed along with either Flag-tagged SUMO-1 or SUMO-2 constructs (Fig. 2B). When SUMO-1 or SUMO-2 is expressed alone, only a modest signal is seen in the high-molecular-weight range by Flag Western blots representing SUMO-modified substrates (Fig. 2B, lanes 3 and 4). When full-length PIASy is coexpressed with either SUMO-1 or SUMO-2, an enhancement in the high-molecular-weight SUMO-1 and SUMO-2 signal is seen (Fig. 2B, lanes 5 and 7). Interestingly, when PIASyE6− is coexpressed with SUMO-1 or SUMO-2, enhancement of SUMO-2, but not SUMO-1, ligation is observed (Fig. 2B, lanes 6 and 8). Furthermore, the pattern of SUMO-2 ligation to substrates is different than that observed when full-length PIASy is coexpressed (compare lanes 7 and 8). These data are consistent with the reported abilities of PIAS proteins to act as SUMO E3 ligases and suggest that the region coded by exon 6 is involved in both substrate specificity of SUMO-2 substrates and the ability of PIASy to ligate either SUMO-1 or SUMO-2 moieties to target proteins.
Gene targeting of the Piasy gene.
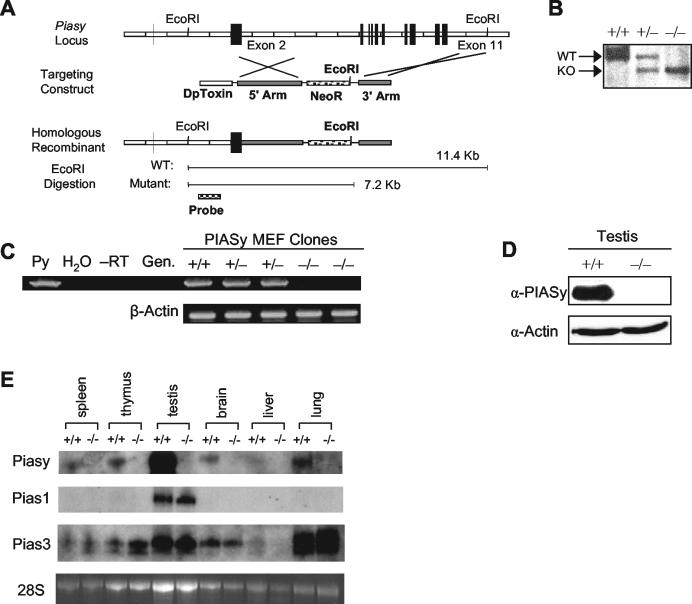
In order to determine the in vivo functions of PIASy, we took a loss of function approach by generating a Piasy-deficient mouse strain. The mouse Piasy gene is localized on chromosome 10 and is organized into 11 exons spanning a 14.7-kb region (Fig. 3A). Genetic deletion of the Piasy gene in ES cells was achieved by electroporating a targeting construct designed to delete exons 3 through 11 (Fig. 3A). This strategy replaced the majority of the coding region, including the zinc ring finger domain with the neomycin resistance gene. Homologous recombination was confirmed by genomic Southern blot analysis.
FIG. 3.
Gene targeting of the Piasy gene. (A) Gene targeting diagram of the Piasy locus. The targeting construct deletes exon 3 to 11. Homologous recombination introduces a new EcoRI site shown in bold. Digestion of genomic DNA with EcoRI and probing with the indicated 5′ probe yields a 11.4-kb wild-type (WT) band and a 7.2-kb mutant band. DpToxin, diphtheria toxin. (B) Genotyping by EcoRI-digested genomic Southern blotting. The wild-type band is 11.4 kb, and the knockout (KO) band is 7.2 kb. (C) RT-PCR of MEFs isolated from heterozygous intercrosses shows no expression in null clones. Genotypes are labeled above each lane. Templates: lane 1, PIASy cDNA as positive-control template (Py); lane 2, water template (H2O); lane 3, no reverse transcriptase in cDNA preparation (−RT); lane 4, wild-type tail DNA template (Gen.); lanes 5 to 9, MEF RNA. Bottom panel, β-actin primers used as control for RNA. (D) Western blot of Piasy+/+ and Piasy−/− testes. Top panel blotted with PIASy antibody, bottom panel blotted with antiactin antibody. (E) Northern analysis of paired tissues from Piasy+/+ and Piasy−/− animals. The PIAS-specific probes used are indicated on the left. Bottom panel, ethidium bromide-stained 28S rRNA indicating similar levels of RNA present in each set of tissues examined.
Piasy+/− ES cells were injected into C57BL/6 blastocysts and implanted into pseudopregnant recipients. Germ line transmission was achieved by backcrossing male chimeras to C57BL/6 females, and subsequent heterozygous intercrosses produced Piasy−/− offspring identified by genomic Southern blotting (Fig. 3B). To confirm complete deletion of Piasy, MEFs were isolated from E13.5 +/+, +/−, and −/− embryos and used for RT-PCR analysis with primers specific for the Piasy mRNA (Fig. 3C). An amplified band of the correct 1.5-kb size was seen in both wild-type and heterozygous samples but not Piasy−/− cells. PIASy cDNA was used here as a template for positive control, while water, RNA without reverse transcription, and genomic DNA all returned the expected negative results. β-Actin primers were used to verify the presence of cDNA in the PIASy knockout samples. Loss of PIASy expression was further confirmed by Western blot analysis of Piasy+/+ and Piasy−/− testes (Fig. 3D). Furthermore, Northern analysis for PIAS1 and PIAS3 expression in paired Piasy+/+ and Piasy−/− primary tissues showed no significant increase in expression of either PIAS1 or PIAS3 (Fig. 3E, second and third panels, respectively). Taken together, these results indicate that we have successfully deleted the Piasy gene.
PIASy is not essential for mouse embryonic development.
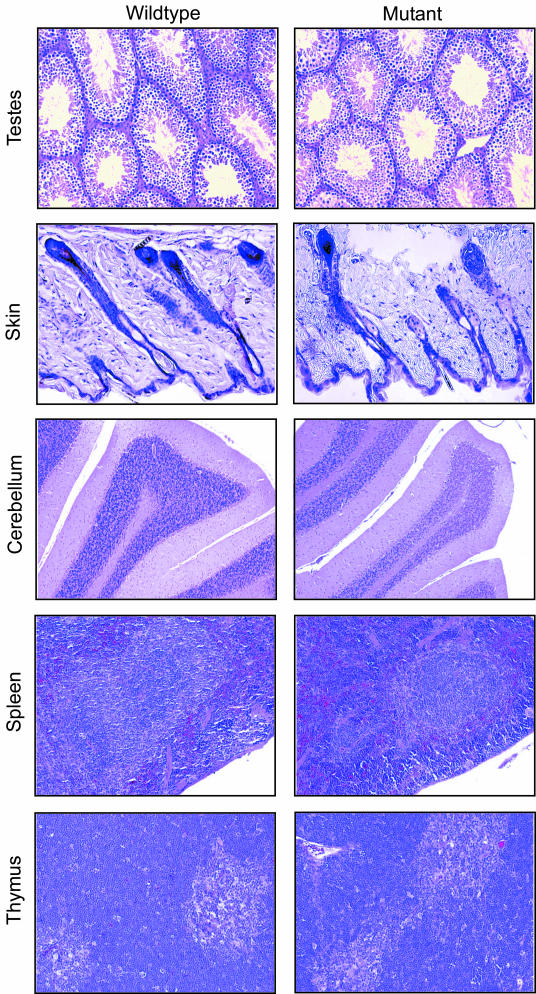
Intercrosses of heterozygous animals produced viable Piasy−/− offspring on a mixed 129SvJ-C57BL/6 as well as a pure 129SvJ genetic background, indicating that PIASy is not essential for embryonic development and viability of the adult animal. The frequency of their production is lower than the expected 1:2:1 (+/+:+/−:−/−) Mendelian distribution on a mixed 129 × C57BL/6 genetic background. However, pure strain 129 animals displayed a genotype distribution approximating Mendelian prediction (Table 1). Piasy+/+ and Piasy−/− animals did not show any differences upon examination of gross anatomy. Organ weights of knockouts and their wild-type littermates at either 6 weeks or 6 months of age were similar (data not shown). Histological analysis showed no obvious defects in the skin and brain even though PIASy expression is reported in the inner root sheath of hair follicles and developing neurons (Fig. 4) (32). Since PIASy is highly expressed in human testis (6), we tested whether deletion of PIASy would lead to male sterility. Histological analysis demonstrated that mature sperm is present in the testes of both Piasy+/+ and Piasy−/− animals (Fig. 4). Furthermore, when Piasy−/− males were mated to Piasy−/− females, they produced litters of Piasy−/− mice and raised them to weaning age, indicating that despite the loss of Piasy expression both males and females are still fertile.
TABLE 1.
Genotype frequencies from Piasy+/− intercrosses on mixed 129 × C57 and pure 129 genetic backgrounds
| Strain | Genotype frequency (no./%)
|
Total | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| 129 × C57 | 59/35 | 82/48 | 28/17 | 169 |
| Pure 129 | 18/23 | 42/55 | 17/22 | 77 |
FIG. 4.
Hematoxylin and eosin-stained tissue sections of Piasy+/+ and Piasy−/− animals. Left column shows tissue from Piasy+/+ mice, and right column shows tissue from Piasy−/− mice. Tissues were indicated to the left. Magnification: testes, ×20; skin, ×20; cerebellum, ×10; spleen, ×20; thymus, ×20. No histological abnormalities were identified.
STAT activation in PIASy null cells.
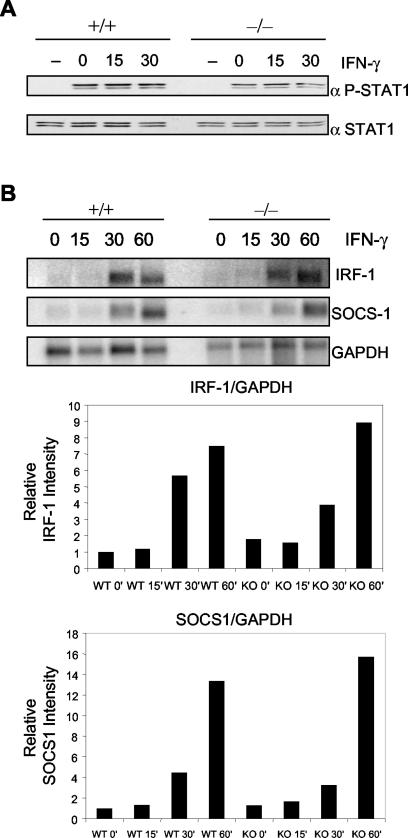
In order to check for deficits in STAT1 tyrosine phosphorylation and subsequent gene activation, we stimulated Piasy+/+ and Piasy−/− primary splenocytes with IFN-γ. Wild-type and knockout cells responded similarly as measured by STAT-1 phosphorylation after a 20-min pulse of IFN-γ followed by incubation in low-serum media from 0 to 30 min (Fig. 5A). Since PIASy has been shown to serve as a corepressor for STAT1 (15), we examined STAT1-mediated gene activation of IRF-1 and SOCS-1 by Northern analysis in primary splenocytes. IFN-γ treatment of Piasy−/− primary splenocytes showed a slight delay in induction of both IRF-1 and SOCS-1 at 30 min of treatment compared to that of the wild-type control. However, by 60 min both genes were induced to slightly higher levels in the null splenocytes (Fig. 5B).
FIG. 5.
No major differences in STAT1 activation in Piasy−/− primary splenocytes. (A) Western blots for STAT1 tyrosine phosphorylation upon IFN-γ treatment in +/+ and −/− splenocytes. Top panel, blotted for phospho-STAT1; bottom panel, blotted for STAT1. Cells were untreated or pulsed with IFN-γ (20 min) and then washed and incubated for the indicated time in minutes before lysis. (B) Northern analysis of IFN-γ-induced gene induction of IRF-1 and SOCS-1 from Piasy+/+ and Piasy−/− splenocytes over 60 min with quantitative bar graphs normalized to the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) levels below. The time of stimulation in minutes is indicated across the top. The probes used are indicated to the right of Northern blots. WT, wild type; KO, knockout.
Sumoylation in PIASy knockouts.
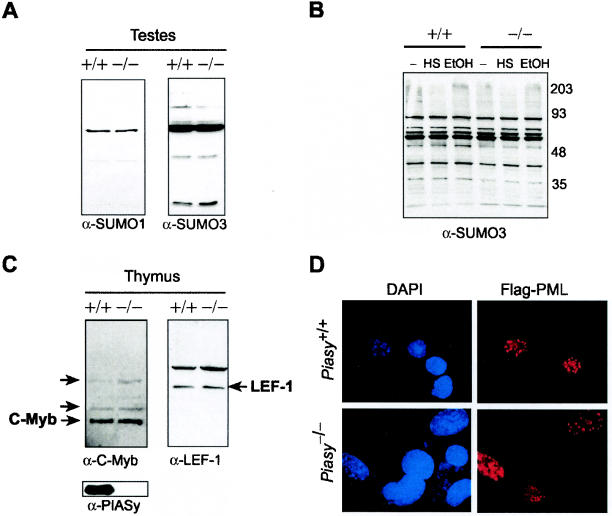
To assess whether or not sumoylation is impaired in PIASy knockouts, the patterns of SUMO-1- and SUMO-3-modified proteins in the testes were compared by Western blotting due to its high expression of PIASy (Fig. 6A). No differences were observed in the patterns of either SUMO-1 or SUMO-3 modification in testes lysates. Saitoh and Hinchey reported that modification of proteins in the high-molecular-weight range with SUMO-2/3 moieties occurs following cellular stress such as heat shock or ethanol treatment (27). Therefore, we tested both heat shock and ethanol treatment on Piasy+/+ and Piasy−/− fibroblasts but did not find any difference in SUMO-3 modification when using a commercially available antibody (Fig. 6B). Thus, deletion of Piasy in MEF cells has no significant effects on overall sumoylation patterns.
FIG. 6.
No difference in SUMO-1 or SUMO-3 modification patterns between Piasy+/+ and Piasy−/− cells. (A) Western blots for SUMO-1 and SUMO-3 from wild-type and mutant testis lysates. (B) Western blot of Piasy+/+ and Piasy−/− MEF cells untreated (−), heat shocked (HS), or ethanol treated (EtOH) and blotted for SUMO-3. (C) Thymus Western blots for c-Myb (left panel) and LEF-1 (right panel). +/+, lysate in the first lane; −/−, lysate in the second lane. The expected mobility of each protein is labeled. Arrows on the c-Myb blot identify the SUMO-1-modified bands reported by Dahle et al. (2). The arrow in the right panel points to the expected mobility of LEF-1 in SDS-PAGE. (D) Immunofluorescence of Flag-PML nuclear bodies in Piasy+/+ and Piasy−/− MEF cells. The same field is shown across each row at ×100 magnification.
We then turned our attention to several specific sumoylated proteins reported previously in the literature. Since transcription factors c-Myb and LEF-1 are found to be sumoylated by PIASy, we examined their sumoylation status in wild-type and knockout thymus. As shown in Fig. 6C, there was no apparent difference in the mobility of c-Myb or LEF-1 when comparing Piasy+/+ and Piasy−/− thymus. Similar results were also seen when using lysates prepared from bone marrows (data not shown). The higher-molecular-weight bands representing the SUMO-1-modified c-Myb described by Dahle et al. (2) are indicated in Fig 6C, as are the bands indicating the expected mobility of LEF-1 in SDS-PAGE. The upper bands may represent a modified form of LEF-1 or nonspecific binding of the antibody. If the former is true, there remains no difference between the presence and absence of PIASy. Western blotting with PIASy antibody shows that PIASy is indeed expressed at a high level in the wild-type thymus but is absent in the knockout (Fig. 6C, bottom panel). Taken together, there do not appear to be any obvious differences in these proteins reported to be SUMO-1 or SUMO-3 modified between Piasy+/+ and Piasy−/− animals.
PML nuclear bodies have been reported to contain a multitude of proteins, including SP100, p53, BRCA1, and Daxx (for an extensive list, see reference 20). Despite the abundance of distinct proteins present, only loss of PML has been shown to lead to disassociation of the nuclear body. PML itself can be SUMO-1 modified, and its sumoylation has been suggested to be critical for normal nuclear body formation and interaction with Daxx (8). Lysine-to-arginine mutation of the SUMO acceptor sites of PML produced aberrant nuclear aggregates when transfected into PML−/− cells, while wild-type PML restored speckled nuclear body structures (35). Coupled with the fact that PIASy can direct LEF-1 to nuclear body structures (26), we decided to determine if PML nuclear body formation is dependent on PIASy. To this end, we transfected Flag-tagged PML (generously provided by P. P. Pandolfi) into Piasy+/+ and Piasy−/− MEF cells and performed immunofluorescence using anti-Flag antibody to visualize PML nuclear bodies. Nuclear bodies in a speckled pattern were seen in both wild-type and knockout MEF cells transfected with the Flag-PML construct (Fig. 6D), indicating that PIASy is not essential for nuclear body formation.
Mo-MuLV infection and replication do not require PIASy.
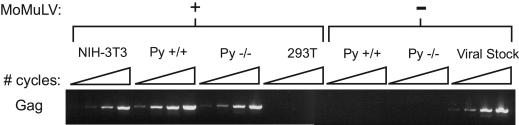
A number of viral proteins have recently been shown to associate with the sumoylation machinery or to be covalently modified by SUMO residues. Sumoylation is therefore likely to be important for some aspects of the viral life cycle (reviewed in reference 33). For example, the bovine papillomavirus E1 protein was reported to be SUMO-1 modified (23). This modification was subsequently shown to be required for nuclear accumulation of the virus, and mutation of the modified lysine residue resulted in a loss of replication capacity correlating with an inability to target to the nucleus (24). While direct roles for PIAS proteins in viral life cycles have not been previously shown, PIASy has been suggested to play an essential role in the life cycle of Mo-MuLV by participating in SUMO-1 modification of viral capsid protein (S. P. Goff, personal communication). To test this hypothesis, we exposed mouse NIH 3T3, Piasy+/+, and Piasy−/− MEFs and human 293T cells to live Mo-MuLV and assayed viral replication competence at 2 weeks postinfection. RT-PCR was performed for the presence of viral Gag mRNA harvested from the media alone. As shown in Fig. 7, viral Gag mRNA could be detected from NIH 3T3, Piasy+/+, and Piasy−/− MEF cells with as few as 20 PCR amplification cycles but not from human 293T cells, indicating that cells lacking PIASy can indeed support Mo-MuLV infection and replication. Media from cells not exposed to virus did not show amplification of viral Gag mRNA. These data indicate that PIASy is not essential for Mo-MuLV infection and replication.
FIG. 7.
Mo-MuLV is able to infect and replicate in PIASy−/− fibroblasts. Analysis of viral replication in cells exposed and unexposed to live Mo-MuLV by semiquantitative RT-PCR for viral Gag transcript. NIH 3T3 cells are mouse cells, while 293T cells are human. Media from unexposed cells show no Gag transcript. Gag transcript was amplified from viral stock used in initial infection as a positive control (last lanes). For each cell type infected, samples were subjected to increasing numbers of thermal cycles: 20, 23, 27, and 30.
DISCUSSION
While splice variants do exist for the PIAS family (PIAS3/PIAS3L, PIASxα/PIASxβ), our cloning of murine PIASyE6− through a degenerate PCR strategy identifies the existence of a previously unreported mRNA splice variation. For PIAS3, mutagenesis of the PINIT motif to PSDST resulted in a redistribution of the PIAS3 protein into both cytoplasmic and nuclear compartments (4). Noting that the recently identified PINIT motif lies in exon 6 of PIASy, our original prediction was that PIASyE6− would also be redistributed to both cytoplasmic and nuclear compartments. However, our data indicated that PIASyE6− is still exclusively nuclear localized, suggesting that the PINIT motif in PIASy is not as critically involved in nuclear retention as it is for PIAS3. It should also be noted that PIASy contains a conservative substitution of leucine for isoleucine in the fourth position of the motif, PINLT. Whether our result for PIASyE6− subcellular localization reflects other differences between PIAS3 and PIASy or a lack of the entire exon remains to be determined.
The results from our sumoylation assays indicate that PIASy can indeed behave as a SUMO E3 ligase. Furthermore, PIASyE6− can also act as a SUMO-2 E3 ligase but displays no activity for SUMO-1 ligation. This result raises the possibility that exon 6 of PIASy is crucial for directing SUMO-1 ligation to target proteins but is not critical for overall E3 ligase activity because PIASyE6− is still able to enhance SUMO-2 ligation. Therefore, for PIASy, recognition of which SUMO moiety to ligate to target proteins at least in part depends on the presence or absence of exon 6. Moreover, because a difference is seen in the pattern of SUMO-2 ligation to target proteins between full-length PIASy and PIASyE6−, exon 6 may also direct specificity of target substrates as to which SUMO moiety to utilize. Identifying the substrates that are differentially modified will be a key to understanding PIASy versus PIASyE6− functions.
In order to further understand the function of PIASy, we have taken a genetic loss of function approach. Piasy−/− animals appear to be normal for the various parameters examined in this study. Both male and female Piasy−/− mice are fertile and appear normal. On a mixed 129 × C57 genetic background, the frequency of producing heterozygous offspring from heterozygous intercrosses is skewed higher in the wild-type animals than in the knockout animals by a ratio of 2:1. This is likely to be a function of the mixed genetic background, as the frequencies observed on a 129 pure background are close to Mendelian distribution (Table 1).
Since PIAS proteins were identified as inhibitors of the JAK/STAT pathway, we examined whether or not there are deficits in cytokine signaling. Loss of PIASy did not affect STAT1 activation and tyrosine phosphorylation in primary splenocytes as shown in Fig. 5A. Because PIASy has previously been shown to be a corepressor of STAT1, we further examined STAT1-mediated gene activation by Northern analysis of IFN-γ-stimulated splenocytes over the course of 60 min (Fig. 5B). A slight delay in the kinetics of activation of both IRF-1 and SOCS-1 between 15 and 30 min was observed with levels of expression being induced in Piasy−/− slightly higher than in Piasy+/+ splenocytes at 60 min. The repressive role of PIASy on STAT1 does not explain the modest delay in kinetics of STAT1 activation we observed, and more detailed studies will be required to examine this unexpected effect.
The results from our investigation of sumoylation defects caused by loss of PIASy, either globally or on specific proteins, are more or less negative, implying that PIASy SUMO E3 ligase activity is dispensable in the mouse at steady state. We also studied sumoylation status under stress conditions and found no appreciable difference when Piasy+/+ and Piasy−/− MEFs were treated at 43°C for 10 min (heat shock) or 7% ethanol for 20 min. One interpretation is that PIASy is not involved in sumoylation following these particular cellular stresses or the responses may be cell type specific (MEFs versus COS-7 cells). However, the lack of increased sumoylation even in wild-type cells may indicate differences in the proteins recognized by the antibody used here and the antibody used by Saitoh and Hinchey (27).
A wealth of evidence has identified PIAS proteins as SUMO E3 ligases for a number of targets. Although both LEF-1 and c-Myb have been reported to be sumoylated by PIASy (2, 26), we were not able to show any differences in their mobilities through SDS-PAGE in a PIASy null setting. Immunoprecipitation of endogenous sumoylated c-Myb or LEF-1 from wild-type or knockout primary tissues was not detected and likely underscores the fact that the wealth of data on specific sumoylated proteins in the literature relies on in vitro and overexpression systems. Similarly, no difference could be detected on PML nuclear localization when Flag-tagged PML was transfected into Piasy+/+ and Piasy−/− MEFs. This result, however, cannot preclude that PIASy is able to sumoylate the aforementioned proteins, but it indicates that PIASy is not the sole SUMO E3 ligase capable of acting on these particular substrates in vivo.
Sumoylation in the context of the immune system and viral life cycle is an interesting and growing area of research. However, at least for Mo-MuLV infection, our semiquantitative RT-PCR data demonstrate that PIASy is not essential. Nevertheless, PIASy may still have a role, as the replication assay performed here cannot exclude the possibility that Mo-MuLV replication is partially impaired in PIASy null cells. It is possible that compensation by other family members, other types of E3 ligases, or SUMO E2 activity alone is sufficient to provide the necessary Mo-MuLV capsid SUMO modification.
To summarize, our overexpression studies of mouse PIASy and PIASyE6− indicate that subcellular localization is not altered between the two forms despite the lack of the PINIT motif in the short form. However, their respective abilities to utilize SUMO-1 or SUMO-2 and to modify at least some target substrates differ depending on the presence or absence of exon 6 and represent another method for fine-tuning PIAS function.
Despite SUMO ligase activity in an overexpression setting, PIASy is not critical in an in vivo loss of function setting. We have not observed any significant deficits in STAT1-mediated gene activation or sumoylation defects in Piasy−/− fibroblasts and animals. Recently, PIASy has been reported to play a role in modulating transforming growth factor β signaling by acting on SMAD proteins (7, 17). Examination of transforming growth factor β signaling in a PIASy null setting will be examined in the future to determine if any deficits exist in this pathway. The lack of an apparent phenotype may be due to compensation by other PIAS family members even though we do not observe any upregulation of PIAS1 or PIAS3. Therefore, generation of compound PIAS null mice will be a necessary future goal to understand loss of function for this family of genes.
Acknowledgments
We thank members of our laboratory and Ke Shuai for providing insightful discussion and antibodies to PIASy. We thank Jack Lenz for providing the Mo-MuLV plasmid and Pier Paolo Pandolfi for the Flag-PML construct.
K.A.W. is partially supported by USPHS National Research Service Award GM07185 and the Dr. Norman Sprague, Jr., fellowship program from UCLA. R.K. was supported by the National Institutes of Health National Research Service Award Institutional Training Grant T32 HD07549. H.W. is a V Foundation scholar and an assistant investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Chung, C. D., J. Liao, B. Liu, X. Rao, P. Jay, P. Berta, and K. Shuai. 1997. Specific inhibition of Stat3 signal transduction by PIAS3. Science 278:1803-1805. [DOI] [PubMed] [Google Scholar]
- 2.Dahle, O., T. O. Andersen, O. Nordgard, V. Matre, G. Del Sal, and O. S. Gabrielsen. 2003. Transactivation properties of c-Myb are critically dependent on two SUMO-1 acceptor sites that are conjugated in a PIASy enhanced manner. Eur. J. Biochem. 270:1338-1348. [DOI] [PubMed] [Google Scholar]
- 3.Dohmen, R. J., R. Stappen, J. P. McGrath, H. Forrova, J. Kolarov, A. Goffeau, and A. Varshavsky. 1995. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J. Biol. Chem. 270:18099-18109. [DOI] [PubMed] [Google Scholar]
- 4.Duval, D., G. Duval, C. Kedinger, O. Poch, and H. Boeuf. 2003. The ′PINIT' motif, of a newly identified conserved domain of the PIAS protein family, is essential for nuclear retention of PIAS3L. FEBS Lett. 554:111-118. [DOI] [PubMed] [Google Scholar]
- 5.Gostissa, M., A. Hengstermann, V. Fogal, P. Sandy, S. E. Schwarz, M. Scheffner, and G. Del Sal. 1999. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 18:6462-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gross, M., B. Liu, J. Tan, F. S. French, M. Carey, and K. Shuai. 2001. Distinct effects of PIAS proteins on androgen-mediated gene activation in prostate cancer cells. Oncogene 20:3880-3887. [DOI] [PubMed] [Google Scholar]
- 7.Imoto, S., K. Sugiyama, R. Muromoto, N. Sato, T. Yamamoto, and T. Matsuda. 2003. Regulation of transforming growth factor-beta signaling by protein inhibitor of activated STAT, PIASy through Smad3. J. Biol. Chem. 278:34253-34258. [DOI] [PubMed] [Google Scholar]
- 8.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jimenez-Lara, A. M., M. J. Heine, and H. Gronemeyer. 2002. PIAS3 (protein inhibitor of activated STAT-3) modulates the transcriptional activation mediated by the nuclear receptor coactivator TIF2. FEBS Lett. 526:142-146. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, E. S., and G. Blobel. 1997. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272:26799-26802. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, E. S., and A. A. Gupta. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106:735-744. [DOI] [PubMed] [Google Scholar]
- 12.Kahyo, T., T. Nishida, and H. Yasuda. 2001. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8:713-718. [DOI] [PubMed] [Google Scholar]
- 13.Kim, K. I., S. H. Baek, and C. H. Chung. 2002. Versatile protein tag, SUMO: its enzymology and biological function. J. Cell. Physiol. 191:257-268. [DOI] [PubMed] [Google Scholar]
- 14.Lai, L., H. Liu, X. Wu, and J. C. Kappes. 2001. Moloney murine leukemia virus integrase protein augments viral DNA synthesis in infected cells. J. Virol. 75:11365-11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, B., M. Gross, J. ten Hoeve, and K. Shuai. 2001. A transcriptional corepressor of Stat1 with an essential LXXLL signature motif. Proc. Natl. Acad. Sci. USA 98:3203-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, B., J. Liao, X. Rao, S. A. Kushner, C. D. Chung, D. D. Chang, and K. Shuai. 1998. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. USA 95:10626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long, J., I. Matsuura, D. He, G. Wang, K. Shuai, and F. Liu. 2003. Repression of Smad transcriptional activity by PIASy, an inhibitor of activated STAT. Proc. Natl. Acad. Sci. USA 100:9791-9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melchior, F. 2000. SUMO-nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 19.Meluh, P. B., and D. Koshland. 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 6:793-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234-7242. [DOI] [PubMed] [Google Scholar]
- 21.Nelson, V., G. E. Davis, and S. A. Maxwell. 2001. A putative protein inhibitor of activated STAT (PIASy) interacts with p53 and inhibits p53-mediated transactivation but not apoptosis. Apoptosis 6:221-234. [DOI] [PubMed] [Google Scholar]
- 22.Nishida, T., and H. Yasuda. 2002. PIAS1 and PIASxα function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J. Biol. Chem. 277:41311-41317. [DOI] [PubMed] [Google Scholar]
- 23.Rangasamy, D., and V. G. Wilson. 2000. Bovine papillomavirus E1 protein is sumoylated by the host cell Ubc9 protein. J. Biol. Chem. 275:30487-30495. [DOI] [PubMed] [Google Scholar]
- 24.Rangasamy, D., K. Woytek, S. A. Khan, and V. G. Wilson. 2000. SUMO-1 modification of bovine papillomavirus E1 protein is required for intranuclear accumulation. J. Biol. Chem. 275:37999-38004. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez, M. S., J. M. Desterro, S. Lain, C. A. Midgley, D. P. Lane, and R. T. Hay. 1999. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 18:6455-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitoh, H., and J. Hinchey. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252-6258. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt, D., and S. Muller. 2002. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. USA 99:2872-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt, D., and S. Muller. 2003. PIAS/SUMO: new partners in transcriptional regulation. Cell. Mol. Life Sci. 60:2561-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seufert, W., B. Futcher, and S. Jentsch. 1995. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature 373:78-81. [DOI] [PubMed] [Google Scholar]
- 31.Shuai, K. 2000. Modulation of STAT signaling by STAT-interacting proteins. Oncogene 19:2638-2644. [DOI] [PubMed] [Google Scholar]
- 32.Sturm, S., M. Koch, and F. A. White. 2000. Cloning and analysis of a murine PIAS family member, PIASgamma, in developing skin and neurons. J. Mol. Neurosci. 14:107-121. [DOI] [PubMed] [Google Scholar]
- 33.Wilson, V. G., and D. Rangasamy. 2001. Viral interaction with the host cell sumoylation system. Virus Res. 81:17-27. [DOI] [PubMed] [Google Scholar]
- 34.Wu, H., X. Liu, R. Jaenisch, and H. F. Lodish. 1995. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell 83:59-67. [DOI] [PubMed] [Google Scholar]
- 35.Zhong, S., S. Muller, S. Ronchetti, P. S. Freemont, A. Dejean, and P. P. Pandolfi. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748-2752. [PubMed] [Google Scholar]