Abstract
Although largely involved in innate and adaptive immunity, NF-κB plays an important role in vertebrate development. In chicks, the inactivation of the NF-κΒ pathway induces functional alterations of the apical ectodermal ridge, which mediates limb outgrowth. In mice, the complete absence of NF-κB activity leads to prenatal death and neural tube defects. Here, we report the cloning and characterization of NF-κΒ/IκB proteins in zebra fish. Despite being ubiquitously expressed among the embryonic tissues, NF-κΒ/IκB members present distinct patterns of gene expression during the early zebra fish development. Biochemical assays indicate that zebra fish NF-κΒ proteins are able to bind consensus DNA-binding (κB) sites and inhibitory IκBα proteins from mammals. We show that zebra fish IκBαs are degraded in a time-dependent manner after induction of transduced murine embryo fibroblasts (MEFs) and that these proteins are able to rescue NF-κΒ activity in IκBα−/− MEFs. Expression of a dominant-negative form of the murine IκBα (mIκBαM), which is able to block NF-κΒ in zebra fish cells, interferes with the notochord differentiation, generating no tail (ntl)-like embryos. This phenotype can be rescued by coinjection of the T-box gene ntl (Brachyury homologue), which is typically required for the formation of posterior mesoderm and axial development, suggesting that ntl lies downstream of NF-κΒ. We further show that ntl and Brachyury promoter regions contain functional κB sites and NF-κΒ can directly modulate ntl expression. Our study illustrates the conservation and compatibility of NF-κΒ/IκB proteins among vertebrates and the importance of NF-κΒ pathway in mesoderm formation during early embryogenesis.
The NF-κΒ signaling pathway plays a crucial role in physiological events such as inflammation, immune response, apoptosis, cell growth, and differentiation (9, 18, 24, 43). NF-κΒ transcriptional factors are found in the cytoplasm as heterodimers, associated with IκB proteins that block their nuclear localization domains, thereby preventing the translocation of NF-κΒ to the nucleus (50). More than 150 different stimuli, including bacterial lipopolysaccharides (LPS), proinflammatory cytokines (tumor necrosis factor alpha [TNF-α] and interleukin-1 [IL-1]), hormones, and mitogenic agents, are able to promote NF-κΒ activation (29). After stimulation, IκB proteins are phosphorylated and ubiquitinated, resulting in their degradation by the proteasome. Subsequently, NF-κΒ factors translocate to the nucleus, where they induce the transcription of κΒ DNA-containing genes.
NF-κΒ transcriptional factors are conserved from insects to humans. In Drosophila melanogaster, NF-κΒ factors (Dorsal, Dif, and Relish) are responsible for regulating several biological functions such as (i) dorsoventral embryonic polarity specification, (ii) humoral immunity, (iii) hemopoiesis, and (iv) muscle development (10). The protein Dorsal regulates zygotic genes involved with the subdivision of the embryonic axis in the mesoderm, as well as adjacent and dorsal neuroectoderm.
Developmental studies in chick have shown that NF-κΒ factors are important for limb budding. Inactivation of these factors results in a reduction of limb size and functional alterations of the apical ectodermal ridge, which mediates proper limb outgrowth (7, 17). Expression of genes involved with limb outgrowth (Fgf-8, Shh, and Twist) is attenuated due to the lack of NF-κΒ factors in the nucleus. This suggests the existence of a common signal transduction cascade involved in the developmental process, mediated by Dorsal in Drosophila and its homologue NF-κΒ in vertebrates (48).
Members of the NF-κΒ family have also been shown to be involved with organogenesis and endoderm progression. Mice lacking p65 or p50 subunits exhibit hepatic degeneration and abnormalities of immune and hemopoietic systems, respectively (4, 12, 41). Inactivation of IKK2 (IKKβ) or NEMO (IKKγ), subunits of the IκB kinase (IKK) complex responsible for IκB phosphorylation and NF-κΒ activation, reveals a phenotype similar to that of p65 (RelA)-deficient mice (23, 36).
Despite the advances described in chicks and mice, the role of NF-κΒ in early embryonic processes, such as evolution of the germ layers and morphogenesis, remains uncharacterized. The search for biological models that could decipher the complex processes of early embryogenesis led to the identification of the teleost Danio rerio (zebra fish) as a model system (13). We document here the cloning and functional characterization of NF-κΒ/ΙκΒ members in zebra fish. We demonstrate that zebra fish NF-κΒ/ΙκΒ proteins can be functionally substituted by their mammalian counterparts. Blocking NF-κΒ pathway by overexpressing a dominant-negative form of the inhibitory protein ΙκΒα affects notochord development in zebra fish embryos. Our results show that NF-κΒ proteins might be required for mesoderm differentiation by regulating the T-box gene no tail (ntl), a Brachyury orthologue essential for the morphogenesis of the dorsal mesoderm in zebra fish (1, 14, 40).
MATERIALS AND METHODS
Molecular cloning of zebra fish NF-κB/IκB family members.
The NF-κB/IκB family members were isolated from zebra fish cDNA by RACE (rapid amplification of cDNA ends)-PCR (SMART RACE cDNA amplification kit; Clontech) or standard PCR procedures (Advantage 2 PCR kit; Clontech) by using degenerate primers. RACE-PCR primers were designed based on available expressed sequence tags with high similarity to NF-κB/IκB homologues (basic local alignment search tool [BLAST] analysis).
Molecular cloning of zebra fish ntl promoter.
The ntl promoter region was isolated by standard PCR procedures from zebra fish genomic DNA, after BLAST analysis of ntl mRNA (GenBank no. NM_131162) at the Pre-ensembl database for the zebra fish genome (http://pre.ensembl.org/Danio_rerio/).
Cell cultures.
Human embryonic kidney 293T cells and wild-type and IκBα−/− mouse embryo fibroblasts (MEFs) (20) were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (HyClone) in an atmosphere of 10% CO2 at 37°C.
Zebra fish fibroblast-like cells (ZF4; ATCC CRL-2050) were originally established from 1-day-old zebra fish embryos (8). ZF4 cells were maintained in a 1:1 mixture of Ham F-12 medium and Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum in an atmosphere of 5% CO2 at 28°C.
DNA and lentiviral constructs.
Hemagglutinin (HA)-tagged zebra fish c-rel (HA-zc-rel) cDNA was generated by PCR and subcloned in pcDNA3 plasmid (Invitrogen) to perform in vitro transfections. For cRNA synthesis, dominant-negative form mIκBαM and EGFP cDNAs were in pCS2(−) plasmid. For coinjection assays, ntl cDNA was isolated by PCR and also subcloned into pCS2(−). Cytomegalovirus-driven lentiviral vectors containing the wild-type mIκBα or mIκBαM coding region were kindly donated by Masahito Ikawa (Salk Institute). Myc-tagged iκbαa and iκbαb cDNAs were generated by PCR and subcloned in HIV-based self-inactivating lentiviral vector pCSC-SP-PW, kindly donated by Robert Marr (Salk Institute). Vesicular stomatitis virus G envelope protein-pseudotyped lentiviruses were prepared as described previously (31).
RNA analysis.
Total RNA was extracted from 100 to 200 staged zebra fish embryos or cell cultures (ZF4) by TRIzol solution (Gibco-BRL/Life Technologies) as indicated. After isolation, 20 μg of total RNA was loaded per lane on a formaldehyde-1% agarose gel and blotted onto Hybond-N+ membrane (Amersham Pharmacia Biotech). Northern hybridizations were carried out in 50% formamide-1 M NaCl-1% sodium dodecyl sulfate-10% dextran sulfate at 42°C with DNA probes produced by MegaPrime DNA labeling system (Amersham) in the presence of [α-32P]dCTP. Probes complementary to NF-κΒ-related or ntl mRNAs corresponding to the full-length sequences were isolated by reverse transcription-PCR as described previously. Equal loading of RNA was verified by visualization of 28S rRNA by ethidium bromide staining. The complete coding sequence for ef1α cDNA (elongation factor 1α, 1.4 kb) was amplified and used as a probe to verify the integrity of the RNA in each sample. Membranes were washed four to five times at high stringency in 2× SSC-1% sodium dodecyl sulfate at 65°C.
Whole-mount in situ hybridization and immunohistochemistry.
In situ hybridization was performed as described previously (16) with the following alterations. (i) Embryos were kept in hybridization solution (60% formamide; 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], pH 6.0; 0.1% Tween 20; 50 μg of heparin/ml; 500 μg of torula RNA/ml) for at least 1 h at 70°C, and then fresh hybridization solution containing 20 to 100 ng of digoxigenin-labeled riboprobe preheated at 70°C was added. Hybridizations with the riboprobes specific to c-rel (sense and antisense), ntl, shh, hoxb1b, and fgf-8 were performed at 70°C for 12 to 16 h. (ii) After equilibration in PBT, embryos were washed with antibody blocking solution (5% heat-inactivated sheep serum in phosphate-buffered saline pluse Tween 20) for at least 1 h on an orbital shaker at room temperature and then replaced with fresh antibody blocking solution supplemented with 0.1 U of anti-digoxigenin-AP Fab fragments (Roche)/ml. Incubation was performed at 4°C for 12 to 16 h.
Immunohistochemistry in zebra fish embryos was done as described previously for antibody staining (38) with rabbit anti-mouse IκΒα antibody (C-21; Santa Cruz) and goat anti-rabbit Alexa Fluor 488 (Molecular Probes).
Protein analysis.
MEF or 293T cells with at least 90% of confluence were either not treated or treated with 20 ng of hTNFα (Calbiochem)/ml, 20 ng of hIL-α (Calbiochem)/ml, or 20 to 40 μg of LPS (L-2654; Sigma)/ml at the indicated time points. After treatment, cells were washed with cold phosphate-buffered saline, and cytoplasmic and nuclear extracts were prepared. Western blot analysis and electrophoretic mobility shift assays (EMSA) were performed as previously described (25, 47).
For EMSA with ZF4 cells, total protein extracts were obtained with Totex solution (20 mM HEPES [pH 7.9], 350 mM NaCl, 20% glycerol, 1% NP-40, 1 mM MgCl2, 0.5 mM EDTA, 0.1 mM EGTA, 0.5 mM dithiothreitol, and protease inhibitors). Supernatants from centrifugation at 13,000 × g for 15 min were collected and used for EMSA. The NF-κB and Oct-1 probes were as described previously (25, 47). The end-labeled probes (T4 kinase) were incubated with Totex extracts for 30 min at room temperature. Complexes were separated by electrophoresis on a 5% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA. Dried gels were subjected to phosphorimager analysis.
For immunoprecipitation (IP) assays, ZF4 and 293T cells with at least 90% confluence were either untreated or treated with 20 to 40 μg of lipopolysaccharide (LPS)/ml for 40 min and 20 ng of hTNFα/ml for 30 min, respectively. After protein extraction, samples were equilibrated with IP buffer (20 mM Tris [pH 8.0], 250 mM NaCl, 0.05% NP-40, 3 mM EDTA, 3 mM EGTA). Supernatants were collected and incubated with 2 μl of monoclonal anti-HA tag antibody (a gift from Tony Hunter, Salk Institute) and 20 μl of protein A-Sepharose CL-4B (Amersham) in 1 ml of IP buffer for 12 to 16 h at 4°C. The immunoprecipitates were then washed three times with IP buffer and analyzed by Western blot.
Zebra fish microinjection.
Synthetic capped cRNAs were prepared by using Megascript kit (Ambion) supplemented with cap analog. The ntl, mIκBα, or mIκBαM cRNA (150 ng/μl, 1 nl per embryo) or respective pCS2 constructs (50 ng/μl, 1 nl per embryo) were injected into one-cell stage embryos. Morpholinos for ntl (GACTTGAGGCAGGCATATTTCCGAT), p100 (CATCCATCCTTAGTGCTCCAGCCAT), p65 (CCCACTGGTGAAACATTCCGTCCAT), and a five-mispair p65 control (CCGACTGCTGAAAGATTCGGTCGAT) were obtained from Gene Tools, LLC. Morpholinos were solubilized in 1× Danieau buffer [58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, 5.0 mM HEPES; pH 7.6] and injected into one-cell stage embryos at final concentrations of 0.25 to 0.5 mM (2.3 to 4.5 ng of morpholino per embryo). Embryos were kept in embryo buffer (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) at 28°C for further analysis.
RESULTS
Isolation and expression pattern of NF-κΒ during zebra fish development.
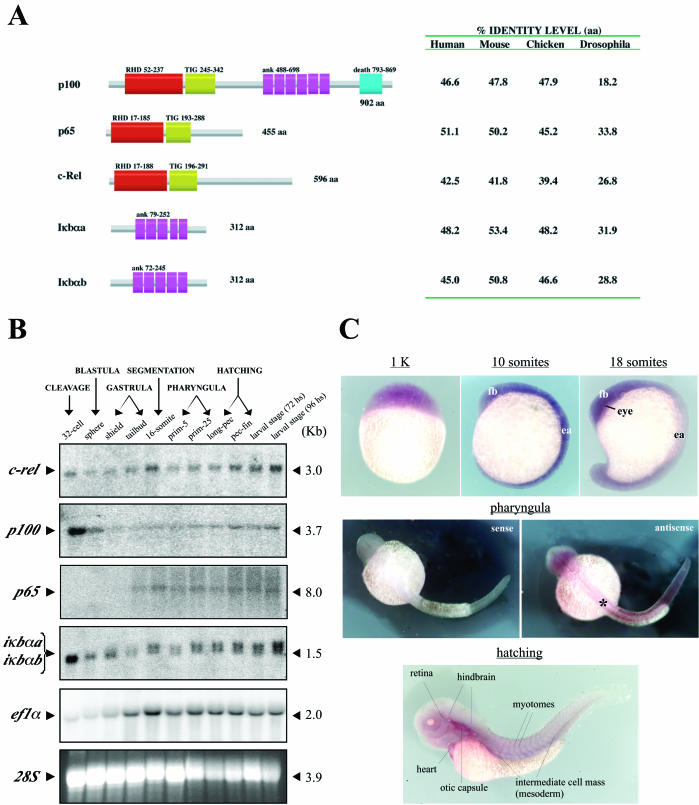
Based on the RACE-PCR approach and amplification by degenerate primers, we isolated the coding cDNA sequences for three NF-κΒ molecules (p65, c-rel, and p100) and two IκBα orthologues (iκbαa and iκbαb) from zebra fish (Fig. 1A). The related proteins contain specific domains conserved in NF-κΒ members from other vertebrates, with an identity level of 39 to 53% (Fig. 1A).
FIG. 1.
NF-κΒ/ΙκΒ members in zebra fish. (A) Diagrammatic structure of NF-κΒ/ΙκΒ proteins in zebra fish. Protein sequence with domain annotation by Pfam analysis (http://pfam.wustl.edu) and amino acid content for p100, p65, c-Rel, Iκbαa, and Iκbαb (GenBank numbers AY163837 to AY163841) are shown. Identity levels were determined by CLUSTAL W analysis, based on protein sequences from human, mouse, chick, and Drosophila orthologues. In comparisons with Drosophila, zebra fish proteins were compared to Relish, Dorsal, Dif, and Cactus, respectively. Annotated domains: RHD, Rel homology domain; TIG, immunoglobulin-like folds; ank, ankyrin repeats; death, death domain. Approximate numbers of amino acids in the domains are shown. (B) Temporal expression of NF-κΒ/ΙκΒ members in zebra fish. A total of 20 μg of total RNA isolated from staged embryos were analyzed by Northern blot with radiolabeled probes specific to c-rel, p100, p65, iκbα, and ef1α (integrity control). The approximate lengths of related mRNAs are indicated. Visualization of 28S rRNA by ethidium bromide was performed as a loading control. (C) Spatial expression of NF-κΒ during zebra fish development. In situ hybridization was performed with the full-length c-Rel digoxigenin antisense probe, containing the highly conserved Rel homology domain motif, which probably recognizes all of the members of the NF-κΒ family. Developmental stages are indicated with respective embryos in lateral view (except for pharyngula period, in dorsal view). A representative staining with sense probe (negative control) is shown at the pharyngula period (32 hpf). The notochord region is indicated (asterisk). fb, forebrain; ea, embryonic axis.
Gene expression analysis of NF-κΒ members during zebra fish development (Fig. 1B) indicated three different patterns: (i) ubiquitous expression at all embryonic stages (c-rel); (ii) higher level of transcripts at earlier stages (maternal origin) (p100), and (iii) activation of transcription during the gastrulation period (embryonic origin) (p65). Expression of the IκBα homologues follows the pattern of p100, suggesting some restrictive control of NF-κΒ activity during cleavage (occurrence of embryonic polarization) and early blastula (blastoderm formation) stages. In situ hybridization analysis of whole zebra fish embryos shows that NF-κΒ is, in general, ubiquitously expressed (Fig. 1C). Similar results have been observed in Xenopus laevis (32, 46). In early somitogenesis, NF-κΒ is expressed in anterior neural tissue and along the embryonic axis (Fig. 1C, 10-somite stage). During the pharyngula period, NF-κΒ is expressed in the notochord region and spinal cord (Fig. 1C). After the embryos hatch, NF-κΒ expression is more distinct in the heart, otic capsule, hindbrain region, muscle tissue (myotomes), and reminiscent mesoderm (Fig. 1C).
Conservation of the NF-κΒ pathway in zebra fish cells.
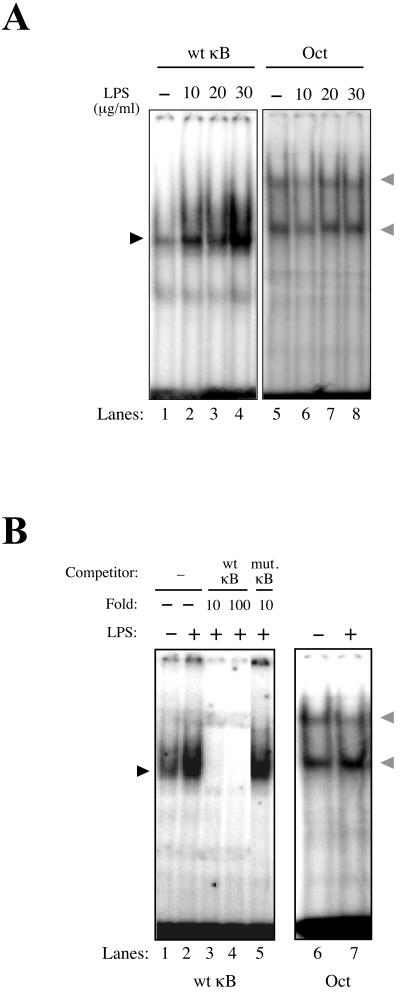
EMSAs were performed to verify whether zebra fish NF-κΒ molecules recognize cognate DNA-binding (κΒ) sites from mammals. In this context, we established zebra fish cell cultures (8) (ZF4; ATCC) to perform detailed biochemical analysis in the zebra fish model. We detected a dose-dependent binding to the NF-κΒ consensus site by using extracts from zebra fish cells (ZF4) after treatment with increasing amounts of LPS (Fig. 2A, lanes 1 to 4) or UV light (data not shown). No increase of LPS-induced DNA-binding activity could be detected with the control Oct-1 oligonucleotide probe (Fig. 2A, lanes 5 to 8), suggesting that LPS can selectively induce NF-κB DNA-binding activity in zebra fish cells. Nonquantitative competition assays with unlabeled consensus (Fig. 2B, lanes 3 to 4) or unlabeled single nucleotide-mutated κB probes (Fig. 2B, lane 5) demonstrated that zebra fish NF-κΒ proteins specifically bind to its cognate decameric sequence. No increase of DNA-binding activity could be detected with Oct-1 probe (Fig. 2B, lanes 6 to 7), reiterating the specificity of NF-κB DNA binding in zebra fish cells.
FIG. 2.
NF-κΒ activation in zebra fish cells. (A) LPS specifically activates NF-κΒ in the zebra fish cell line ZF4. Total protein extracts (60 μg) from ZF4 cells induced with increasing doses of LPS (10 to 30 μg/ml for 30 min) were used for EMSA, with radiolabeled consensus-κΒ (wt κB, 5′-AGTTGAGGGGACTTTCCCAGGC-3′) or control Oct-1 probes (Oct, 5′-TGTCGAATGCAAATCACTAGAA-3′). (B) Total protein extracts (60 μg) from ZF4 cells were used for EMSA with radiolabeled consensus-κΒ (wt κB) probe. Competition assay with nonlabeled consensus (wt κB) or single-mutated κΒ (mut. κB, 5′-AGTTGAGGGCACTTTCCCAGGC-3′) probes was performed after induction of ZF4 cells with 36 μg of LPS/ml for 40 min by using a concentration 10 or 100 times greater compared to the radiolabeled probe. The κΒ site is underlined, and the mutation is indicated in boldface. As a control, nuclear extracts of ZF4 cells not treated (−) or treated (+) with LPS were also used for EMSA with Oct-1 probe (Oct). The NF-κΒ and Oct-DNA complexes are indicated (dark and gray arrowheads, respectively).
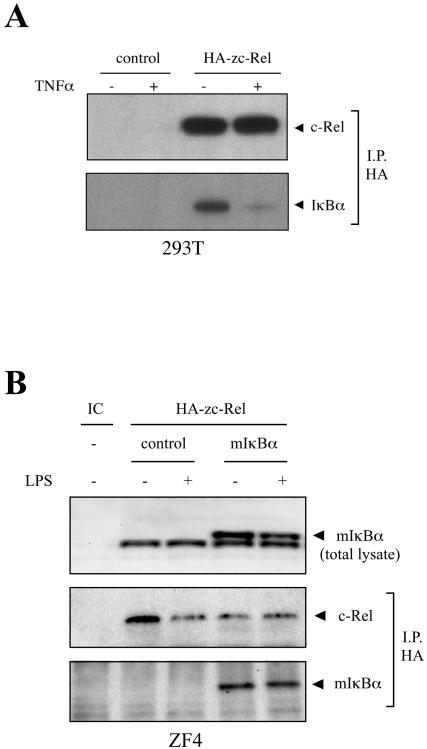
To evaluate the functional conservation of NF-κB proteins from zebra fish with that of other species, the interaction of mammalian IκBα with zebra fish c-Rel was analyzed. Co-IP assays were performed by using protein extracts derived from human (293T) or zebra fish (ZF4) cell lines expressing c-Rel proteins. Transient expression of HA-tagged zebra fish c-rel (HA-zc-rel) in 293T cell lines, followed by IP, showed that endogenous IκBα associates with zebra fish c-Rel (Fig. 3A). Furthermore, in the presence of TNF-α, the level of associated IκBα decreased, due to stimulus-dependent degradation of IκBα (Fig. 3A). With ZF4 cells constitutively expressing mouse IκBα (mIκBα), transfected with zebra fish HA-zc-rel, the same association was observed after IP of HA-zc-Rel (Fig. 3B, bottom panel). In the presence of LPS, decreased levels of mIκBα protein in the total cell extract can be observed by Western blot (Fig. 3B, top panel) that are not evident after IP (Fig. 3B, bottom panel).
FIG. 3.
Zebra fish NF-κΒ proteins (c-Rel) interact with mammalian IκBαs. (A) Zebra fish c-Rel interacts with endogenous IκBα in human cells. 293 T cells from 10-cm plates, transfected with 2.0 μg of EGFP (control) or HA-zc-rel plasmids and either treated (+) or not treated (−) with 20 ng of hTNFα/ml for 30 min, were lysed and immunoprecipitated (I.P.) with HA antibody. IP complexes were eluted and used for immunoblotting with HA or IκBα antibodies. (B) Zebra fish c-Rel interacts with mouse IκBα in zebra fish cells. ZF4 cells (10-cm plates) constitutively expressing EGFP (control) or mouse IκBα (mIκBα) were transfected with 2.0 μg of HA-zc-rel plasmid and treated (+) or not treated (−) with 20 μg of LPS/ml for 40 min. The levels of mIκBα protein were evaluated by Western blotting (50 μg of total protein) with IκBα antibody (top panel). IP procedures were as described for panel A. Nontransfected wild-type ZF4 cells were used as internal control (IC).
Zebra fish IκBαs function like murine IκBα (mIκBα) in mammalian cells.
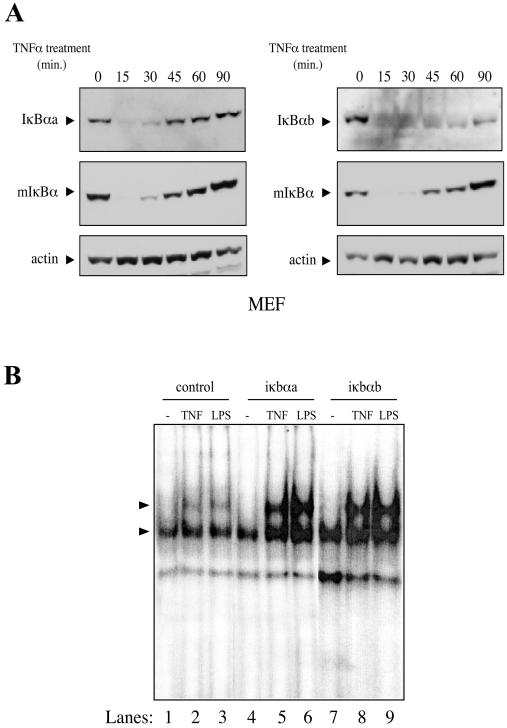
MEFs were transduced with recombinant lentiviruses expressing both zebra fish iκbα homologues to evaluate whether these molecules function like the murine IκBα (mIκBα) in TNF-α-mediated NF-κB activation. TNF-α was able to induce degradation of both zebra fish proteins in a time-dependent manner (Fig. 4A), reiterating the functional compatibility between zebra fish and mammalian NF-κB pathways. It is interesting that after induction with TNF-α, the resynthesis of IκBαb protein takes longer compared to IκBαa (90 min versus 45 min). To further verify whether mIκBα activity could be substituted by its zebra fish homologues, we performed EMSAs on protein extracts derived from knockout IκBα−/− MEFs (20) constitutively expressing each zebra fish iκbα homologue (Fig. 4B). Given the role of IκBα in recruitment of the NF-κB/IκBα complex to the IKK complex, IκBα−/− MEFs have been reported to activate NF-κΒ with a brief delay compared to wild-type cells (37). In contrast to findings with wild-type cells, 15 min of TNF-α or 30 min of LPS treatment is insufficient to fully activate NF-κΒ in IκBα−/− cells (37) (Fig. 4B, lanes 2 and 3). Reconstitution of IκBα−/− MEFs with either zebra fish IκBαa or IκBαb was able to restore rapid induction in response to TNF-α (Fig. 4B, lanes 5 and 8) or LPS (Fig. 4B, lanes 6 and 9). Taken together, these results strengthen the idea that zebra fish IκBαs are functionally identical to mammalian IκBα.
FIG. 4.
Zebra fish IκBαs are functionally identical to mIκBα. (A) Zebra fish IκBαs are degraded after induction with TNF-α in mammalian cells. MEFs constitutively expressing iκbαa-Myc or iκbαb-Myc were treated with 20 ng of hTNFα/ml at different time points and immunoblotted with c-Myc, IκBα, or actin antibodies. (B) Zebra fish IκBαs are able to rescue early NF-κΒ responsiveness in IκBα−/− MEFs. MEFs constitutively expressing EGFP (control), iκbαa-Myc, or iκbαb-Myc were treated with 20 ng of hTNFα/ml for 15 min or 40 μg of LPS/ml for 30 min. Nuclear protein extracts (10 μg) were used for EMSA with radiolabeled consensus κΒ probe. The NF-κΒ-DNA complexes are indicated (arrowheads).
Dominant-negative form of mIκBα (mIκBαM) blocks NF-κΒ activity in zebra fish cells.
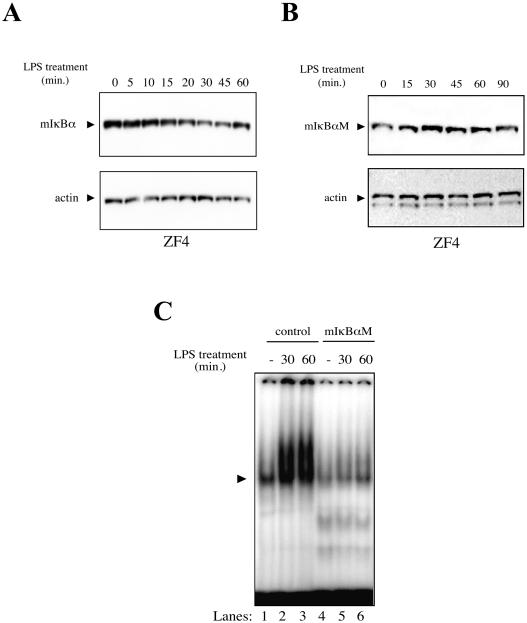
Based on the observation that zebra fish cells are efficiently transduced by lentiviral vectors (R. G. Correa and I. M. Verma, unpublished results), we used this approach to express transgenes in ZF4 cells. The mIκBα protein expressed in ZF4 cells shows time-dependent degradation upon treatment with LPS (Fig. 5A), suggesting that NF-κΒ machinery in zebra fish is able to recognize and degrade the mammalian homologue. In contrast, the dominant-negative form of mIκBα (mIκBαM), which lacks the inducible (Ser 32 and Ser 36) and constitutive phosphorylation sites in the PEST domain (49), was not degraded upon treatment with LPS in ZF4 cells (Fig. 5B). Indeed, EMSAs showed no κB binding activity after LPS induction in ZF4 cells constitutively expressing mIκBαM (Fig. 5C, lanes 4 to 6) compared to control EGFP cells (Fig. 5C, lanes 1 to 3). These results suggest that mutations at the phosphorylation sites in the destruction motif of mIκBα, which are conserved in the zebra fish homologues, can block NF-κΒ activity in zebra fish cells.
FIG. 5.
Dominant-negative form of mIκBα blocks NF-κΒ in zebra fish cells. (A) Mouse IκBα is degraded in a time-dependent manner with LPS in zebra fish cells. ZF4 cells constitutively expressing mIκBα were treated with 40 μg of LPS/ml at different time points, lysed, and analyzed by immunoblotting with monoclonal IκBα or actin antibodies. (B) Mutations at phosphorylation sites of mIκBα block its NF-κΒ mediated degradation in zebra fish cells. The experiment was performed as described for panel A. (C) mIκBαM-expressing ZF4 cells fail to activate NF-κΒ after induction with LPS. Total protein extracts (30 μg) were used for EMSA with radiolabeled consensus κΒ probe. Induction was performed with 40 μg of LPS/ml for 30 or 60 min. EGFP-expressing ZF4 cells were used as a control. The NF-κΒ-DNA complex is indicated (arrowhead).
Induction of no tail-like phenotype by mIκBαM overexpression.
To analyze the role of NF-κΒ pathway in zebra fish development, both DNA and cRNA for wild-type mIκBα and the dominant-negative mIκBαM were microinjected into one-cell stage embryos. Injection of mIκBαM cRNA (150 pg per embryo) resulted in about 40% of embryos with no tail (ntl)-like phenotype. The mIκBαM-expressing embryos were dorsalized, with normal head and eyes and short body length due to posterior deficiency involving smaller or absent tail and trunk distortion (Fig. 6A). At higher doses of injection, embryos fail to gastrulate normally and cyclopia is observed. The mRNA-injected embryos expressing wild-type mIκBα revealed the same phenotype but at a lower frequency (18%, n = 195) compared to mIκBαM (43%, n = 218), since the overexpression of even wild-type IκBα is capable of blocking NF-κΒ activity (5, 6, 19, 45). In contrast, overexpression of c-rel or p65 (75 to 150 pg of cRNA per embryo) did not cause any expansion of the anteroposterior axis and/or duplication of a secondary axis as possibly predicted (data not shown). Protein expression induced by injected cRNA or plasmid DNA was confirmed by Western blot analysis (data not shown).
FIG. 6.
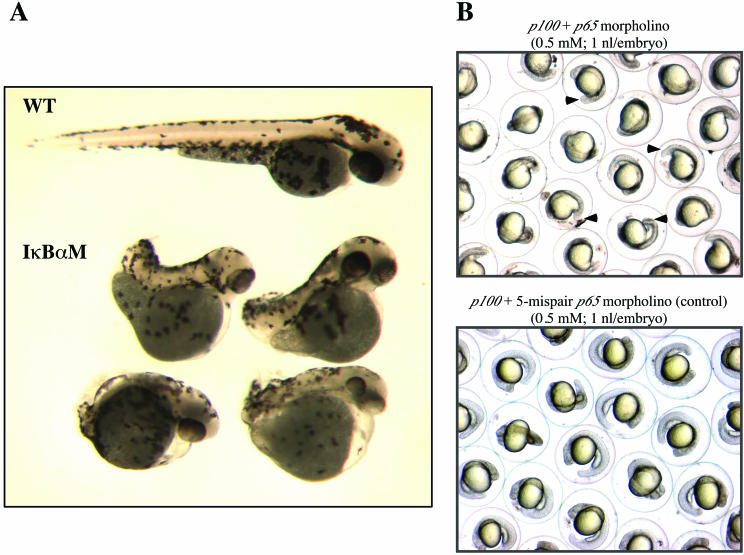
Blocking NF-κΒ in zebra fish embryos disturbs mesoderm development. (A) Wild-type (WT) and mIκBαM cRNA-injected (IκBαM) embryos at 72 hpf (larval stage). Mutated embryos are dorsalized, with partial or complete lack of tail formation and irregularities along the anteroposterior axis. (B) Morpholino-injected embryos at 24 hpf (pharyngula period). Coinjections were performed with 0.5 mM concentrations of each morpholino (4.5 ng of each morpholino per embryo). The knockdown effect induced by p65 morpholino disturbs posterior body morphogenesis, generating embryos with trunk and tail shortening (top panel). Arrowheads indicate lack of the caudal region in some embryos. In parallel, a five-mispair p65 morpholino was injected as a control (bottom panel).
To better correlate the observed mIκBαM phenotype with blockage of NF-κΒ activity in vivo, we designed morpholinos (26) for two zebra fish NF-κΒ genes, p100 and p65, and performed single- and double-knockdown experiments, with 2.3 to 4.5 ng of each morpholino. Single injections of p100 morpholino did not cause any phenotypic changes in the early patterning and/or anteroposterior axis formation (Fig. 6B, bottom panel). Single or double injections of p65 morpholino, along with p100 morpholino, caused incomplete formation of the anteroposterior axis (lack of caudal region) (87%, n = 482) and, in some cases, localized cell death in the hindbrain region (Fig. 6B, top panel). At higher doses of injection, embryos had a delay during early development (arrest in late somitogenesis) and died before 48 hpf. As a control, we performed the injection of a five-mispair p65 control morpholino (2%, n = 346) (Fig. 6B, bottom panel). The phenotype obtained by p65 knockdown by using morpholino technology provides strong evidence that blocking NF-κΒ disturbs mesoderm development in zebra fish.
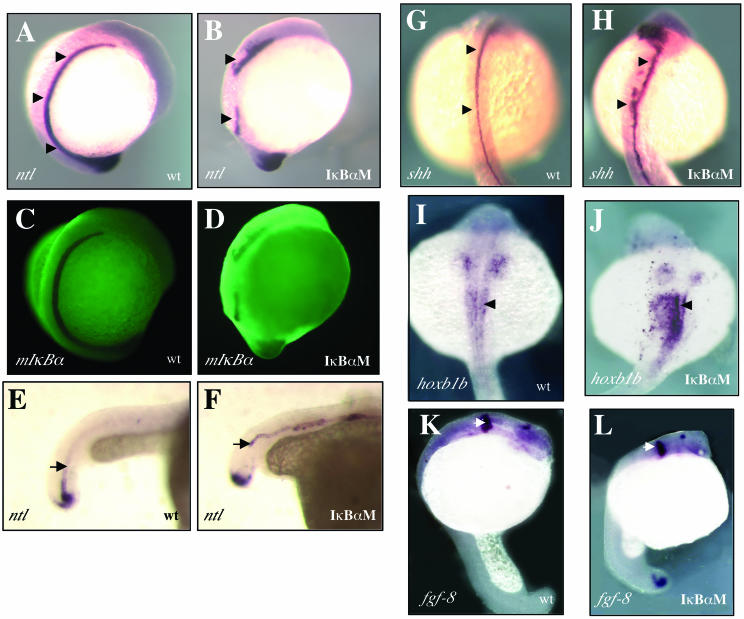
In situ hybridization analysis, with ntl as a marker, indicated a lack of differentiated notochord in mIκBαM expressing embryos, with a greater reduction of ntl expression during somitogenesis (Fig. 7A and B) and dramatic changes in its spatial distribution during the pharyngula period (Fig. 7E and F). No representative changes of ntl levels were observed at the beginning of gastrulation (data not shown). Using sonic hedgehog (shh) as a marker, irregularities in the medial floor plate, such as broadening (1, 27), were also detected (Fig. 7G and H), possibly due to the interruption of ventral cell fates originally promoted by the notochord (28, 30, 44), as typically observed in ntl mutants (1). Interestingly, the anterior spinal cord (hoxb1b as marker) fails to develop axially (Fig. 7I and J) but the central nervous system patterning is apparently unaffected, as judged by the integrity of the midbrain-hindbrain boundary with fgf-8 as a marker (Fig. 7K and L).
FIG. 7.
NF-κΒ activity in vivo is important for notochord differentiation. Whole-mount in situ hybridization was performed with the specific markers ntl (A to F), shh (G and H), hoxb1b (I and J), and fgf-8 (K and L). (A and B) A pronounced reduction of ntl expression is observed in mIκBαM-expressing embryos at the 14-somite stage (lateral view, arrows), which is directly involved in the interruption of mesoderm maturation. (C and D) Fluorescent-antibody staining for mIκBαM protein (indicated as mIκBα) was performed in parallel as a control. (E and F) At 24 hpf, ntl is irregularly expressed and indicates a failure of notochord closure within the tailbud (caudal-lateral view, arrows). As a consequence, the medial floor plate develops irregularly (dorsal view [G and H], arrowheads). Also, the presumptive spinal cord does not develop axially on the anterior-most region (I and J, arrowheads), but the midbrain-hindbrain boundary (K and L, arrowheads) appears to be normal. In situ markers are indicated on the left side of each panel. wt, wild-type embryo; IκBαM, mIκBαM-expressing embryo.
NF-κΒ mediates notochord differentiation by controlling ntl expression.
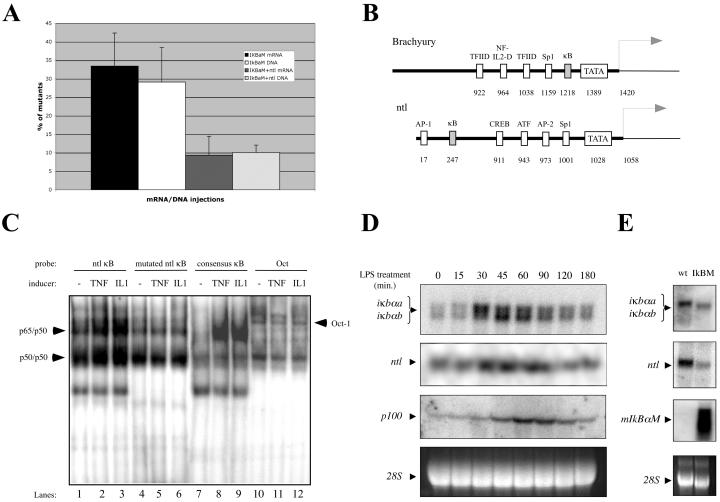
The striking similarity between the mIκBαM and ntl phenotypes (27) led us to evaluate whether reduction of ntl expression by blocking NF-κΒ underlies the observed phenotype of mIκBαM-expressing embryos. Coinjection of DNA or cRNA expressing mIκBαM and ntl in zebra fish embryos resulted in a 65 to 73% rescue of the mutated phenotype compared to injection of mIκBαM alone (10% versus 29% of total embryos after DNA injections and 9% versus 33% after cRNA injections, respectively) (Fig. 8A). This result suggests that the mIκBαM phenotype may be primarily due to reduction in the expression of ntl. To examine if loss of ntl expression has any synergistic effect on the mIκBαM phenotype, we also coinjected 4.5 ng of ntl morpholino (26) with 150 pg of mIκBαM or EGFP cRNAs in zebra fish embryos, and no enhanced phenotype was observed (data not shown), suggesting that NF-κΒ and ntl could be related in a linear way.
FIG. 8.
ntl expression is positively regulated by NF-κΒ. (A) Ectopic expression of ntl is able to rescue mIκBαM (IκBM) phenotype. Graph shows the number of mIκBαM-expressing embryos obtained after injection of 150 ng of cRNA/μl or 50 ng of pCS2 plasmids/μl expressing mIκBαM alone (IκBαM) or with ntl (IκBaM+ntl). The results were plotted based on three independent experiments (150 to 200 embryos per experiment). Rescue effect was considered when embryos were phenotypically indistinguishable from wild-types at 30 hpf (pharyngula period). (B) Promoter analysis of Brachyury (Xenopus) and ntl (zebra fish) genes indicates the presence of κB sites. Significant regulatory sites and their respective positions are shown. Putative initiation of transcription is also indicated (arrows). (C) NF-κΒ dimers interact specifically with the κΒ site in the ntl promoter region. MEF nuclear extract (10 μg) were used for EMSA with radiolabeled oligonucleotides derived from the ntl promoter region. Wild-type (ntl κB, 5′-GTGACGATCTGTGTGAATTTCCTTAATCCAGCATGACT-3′) or mutant (mutated ntl κB, 5′-GTGACGATCTGTGTAAATTTCCTTAATCCAGCATGACT-3′) κΒ-containing region, which differ by a single mutation, were used as probes. The κΒ site is underlined and the mutation is indicated in boldface. Induction was performed with 20 ng of TNF-α/ml for 15 min or 20 ng of IL-1/ml for 20 min. Consensus-κΒ and Oct-1 probes were used as positive and loading controls, respectively. Expected shifts for NF-κΒ dimers (p65/p50 and p50/p50) and Oct-1 are indicated (arrowheads). (D) Induction of putative NF-κΒ target genes by LPS in zebra fish cells. A total of 20 μg of total RNA isolated from ZF4 cells, noninduced or induced with 40 μg of LPS/ml at different time points, were analyzed by Northern blotting with probes specific to iκbα, ntl, and p100. (E) Downregulation on the iκbα and ntl expression in vivo mediated by mIκBαM. A total of 20 μg of total RNA isolated from wild-type (wt) or mIκBαM (IκBM)-expressing embryos (30 hpf) were analyzed as described for panel D by using specific probes for iκbα, ntl, and mIκBαM (positive control).
To verify whether ntl is directly regulated by NF-κΒ, we analyzed the interaction of NF-κΒ proteins to the promoter region of ntl homologues. For this analysis, we cloned 1.1 kb of genomic sequence related to the ntl promoter region (GenBank no. AY288069). Comparison of the ntl promoter to the Xenopus Brachyury (Xbra) promoter region (GenBank no. AF007123) revealed that, despite the relative divergence among the regulatory transcription sites, both promoters have predicted κB sites (Fig. 8B). Using a 38-bp probe derived from the κB-containing portion (5′-GTGAATTTCC-3′) of ntl promoter for EMSA analysis, a mobility shift was observed after induction of MEFs with TNF-α or IL-1 (Fig. 8C, lanes 1 to 3), as typically observed with the consensus κΒ probe (Fig. 8C, lanes 7 to 9). However, no induction of binding was observed when a single mutation was inserted in the κB site (5′-GTAAATTTCC-3′) in the context of the ntl promoter (Fig. 8C, lanes 4 to 6), suggesting that NF-κΒ molecules interact specifically with the ntl promoter region.
To further evaluate whether ntl is a target gene of NF-κΒ, we performed some Northern blot analysis. Treatment of ZF4 cells with LPS induced the mRNA levels of ntl and also of classical NF-κΒ target genes, such as p100 and IκBα, in a time-dependent manner (Fig. 8D). The strong correlation between the temporal expression of ntl and the classical NF-κΒ target genes, in response to LPS, suggests that ntl is a direct target of NF-κΒ. In addition, mIκBαM-expressing embryos (30 hpf) showed a decrease in the RNA levels for endogenous IκBα and ntl compared to wild-type embryos (Fig. 8E). These data support the idea that ntl can be positively regulated at the transcriptional level by NF-κΒ in zebra fish.
DISCUSSION
Zebra fish NF-κΒ proteins are functionally related to their mammalian counterparts.
Rel/NF-κΒ family members are transcriptional factors that regulate the expression of a large number of target genes involved with physiological processes such as immune response, programmed cell death (apoptosis), inflammation, and progression of the cell cycle in different organisms (9, 18, 24, 43). Here we describe the cloning and characterization of NF-κΒ/ΙκΒ members in zebra fish. We demonstrate that the NF-κΒ pathway is functionally conserved in zebra fish and cross-compatible with the mammalian orthologues, suggesting that the role of NF-κΒ signaling pathway is preserved in early vertebrates.
The sequence analysis of NF-κΒ/ΙκΒ proteins in zebra fish indicates a significant level of conservation with mammals (Fig. 1A), despite the fact that these two species segregated from their common ancestor about 450 million years ago. Interestingly, zebra fish NF-κΒ/IκB members are expressed in distinct temporal patterns during early embryogenesis (Fig. 1B), suggesting that each molecule could be possibly involved in specific developmental events, such as embryonic polarization and morphogenesis. Indeed, at later embryonic stages, NF-κΒ is expressed in the notochord region (Fig. 1C, pharyngula period), suggesting a role during mesoderm development.
In our studies, we used a cell line (ZF4) derived from 24-h-old zebra fish embryos (8) as a tool to biochemically characterize the NF-κΒ pathway. We were able to observe that known NF-κΒ inducers in mammalian cells, such as LPS and UV light, were efficient in activating the NF-κΒ pathway in ZF4 cells (Fig. 2 and data not shown). We also observed that zebra fish NF-κΒ proteins can physically interact with mammalian IκBα protein (Fig. 3), providing evidence that zebra fish NF-κΒ molecules are functionally compatible with their mammalian counterparts.
To express mammalian IκBα proteins in zebra fish cells, we used lentivirus-mediated gene delivery. By using this approach, we detected degradation of mIκBα in a time-dependent manner in response to LPS in ZF4 cells (Fig. 5A). Concomitantly, the zebra fish IκBα orthologues, IκBαa and IκBαb, are also degraded after stimuli in transduced MEFs (Fig. 4A). Furthermore, both IκBαa and IκBαb proteins can restore early inducibility of IκBα−/− MEFs (Fig. 4B) in response to TNF-α or LPS. Since most of the human and mouse genes have two paralogues in the zebra fish genome (34) and the divergence of duplicated genes in zebra fish possibly affects expression patterns more than protein activity (33), it is reasonable to conclude that zebra fish iκbαs are duplicated genes functionally identical to mIκBα.
A dominant-negative form of the mIκBα (mIκBαΜ) that lacks the inducible and constitutive phosphorylation sites has already been clearly shown to block NF-κB activity in higher vertebrates (17, 49). In zebra fish cells, the expression of mIκBαΜ was also able to interrupt NF-κΒ activity, since no protein degradation was detected in response to LPS (Fig. 5B) and mIκBαM-transduced cells presented no DNA binding after induction (Fig. 5C). These results validate the use of the murine dominant-negative form of IκBα gene (mIκBαM) to block NF-κB activity in the zebra fish model and, consequently, to investigate whether the NF-κB pathway could be crucial for zebra fish development.
NF-κΒ regulates notochord differentiation in zebra fish.
NF-κB proteins have been shown to be involved in vertebrate and invertebrate development. The complete absence of NF-κB activity in mice leads to prenatal death at day 12 due to neural tube defects (22). In birds, the loss or decrease of NF-κB activity leads to limb deformities (17). In zebra fish, we now show that blockage of NF-κB activity by overexpressing the dominant-negative form of the murine IκBα gene (mIκBαM) leads to embryonic dorsalization and notochord deformities (Fig. 6 and 7) in a fashion similar to that of ntl embryos (27, 40).
The ntl mutant embryos lack a differentiated notochord and the caudal region of their bodies (40). The ntl (no tail) phenotype is characterized by mutations in the zebra fish homologue of the T-box transcription factor Brachyury (ntl), whose expression is required for the formation of the posterior mesoderm and for axial development in both mouse and zebra fish embryos (14, 15, 40). The expression of ntl gene is transient in cells of the germ ring but stable in cells of the presumptive notochord during early embryogenesis (39). Expression of notochord markers, such as twist, sonic hedgehog (shh), and axial, is disturbed in the axial mesoderm of ntl embryos (27). In our experiments, with ntl itself and with shh as in situ markers, we were able to detect lack of notochord differentiation and irregularities in the medial floor plate in mIκBαM-expressing embryos (Fig. 7) as typically observed in ntl embryos (1).
Overexpression of the dominant-negative form of the mIκBα (mIκBαM), which is able to block NF-κΒ activity in zebra fish cells (Fig. 5), affects mesoderm maturation in zebra fish embryos (Fig. 6A). Also, knocking down p65 expression by using morpholino technology (26) revealed a similar effect on the posterior body morphogenesis (Fig. 6B), which provides strong evidence that blocking NF-κΒ disturbs mesoderm development in zebra fish.
Shimada et al. (42) isolated two members of NF-κΒ family (As-rel1 and As-rel2) from the protochordate Halocynthia roretzi (ascidian) and verified that As-rel1 overexpression can interfere with notochord formation, resulting in embryos with shortened tails and a reduced number of notochord cells. In contrast, UV-ventralized Xenopus embryos can be rescued by Drosophila Spätze/Toll proteins, whose effect is inhibited by the coexpression of a dominant Cactus (IκΒ protein) variant (2). Overexpression of the Cactus variant alone caused a slight but reproducible dorsalizing effect in UV-ventralized embryos by unknown reasons (2). Additional studies in Xenopus have shown that cRNA injection of a dominant-negative p65 variant is able to block Xbra expression by inhibiting its autoregulation by embryonic fibroblast growth factor (FGF), which affects mesoderm induction (3). The proposed model suggests that NF-κΒ activates FGF signaling downstream of Xbra (3). We present strong evidence here that, in zebra fish, ntl can be directly regulated by NF-κΒ in a putative FGF-independent manner.
In the present study, we show that activation of NF-κΒ, like that of the members of the transforming growth factor β (35) and FGF (11) families, can regulate ntl expression (Fig. 7 and 8). More specifically, our results showing that (i) coexpression of ntl cRNA is able to rescue the mIκBαM phenotype (Fig. 8A), (ii) Brachyury and ntl promoter regions contain κΒ sites (Fig. 8B), (iii) ntl κΒ sites are able to bind NF-κΒ molecules (Fig. 8C), (iv) ntl is upregulated after LPS treatment (NF-κΒ inducer) in zebra fish cells (Fig. 8D), and (v) ntl is downregulated in mIκBαM-expressing embryos (Fig. 7A to F and 8E) support the idea that ntl is positively regulated by NF-κΒ during notochord development and that the lack of differentiated notochord in mIκBαM-expressing embryos is due to a spatial decrease in ntl expression levels along the developing notochordal tissue. We hypothesize that, during vertebrate embryogenesis, the inductive activity of transcription factors such as NF-κΒ could be counterbalanced by the presence of repressor molecules, such as Sip1 (Smad-interacting protein 1) and homeobox proteins (21), to restrict expression of ntl to the mesoderm and, consequently, promote notochord differentiation.
Understanding the upstream signals that activate NF-κΒ in notochord development will be an important issue to address. Further analysis of cooperation between NF-κΒ and other transcription factors will provide clues to the complexity of the circuitry involved in this modulation. We believe that deciphering the specific role of each NF-κΒ/ΙκΒ protein in zebra fish development will offer important insights into understanding the development of tissues and organogenesis in vertebrates.
Acknowledgments
We thank members of the Verma and Belmonte laboratories for helpful discussions, R. Marr and M. Ikawa for sharing lentiviral preparations and constructs, C. Rodriguez-Esteban for helping with the zebra fish photos, S. L. Amacher for providing shh construct, and M. C. Sogayar and M. D. Weitzman for additional suggestions on the manuscript. R.G.C. is supported by the NIH.
V.T. is supported by the Leukemia and Lymphoma Society. The J.C.I.-B. laboratory is supported by grants from the March of Dimes, HSPO, and the G. Harold and Leila Y. Mathers Charitable Foundation. I.M.V. is an American Cancer Society Professor of Molecular Biology and is supported in part by grants from the National Institutes of Health; the Larry L. Hillblom Foundation, Inc.; the Lebensfeld Foundation; the Wayne and Gladys Valley Foundation; and the H. N. and Frances C. Berger Foundation.
REFERENCES
- 1.Amacher, S. L., B. W. Draper, B. R. Summers, and C. B. Kimmel. 2002. The zebrafish T-box genes no tail and spadetail are required for development of trunk and tail mesoderm and medial floor plate. Development 129:3311-3323. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, N. J., H. Steinbeisser, C. Prothmann, R. DeLotto, and R. A. Rupp. 1998. Conserved Spatzle/Toll signaling in dorsoventral patterning of Xenopus embryos. Mech. Dev. 71:99-105. [DOI] [PubMed] [Google Scholar]
- 3.Beck, C. W., D. J. Sutherland, and H. R. Woodland. 1998. Involvement of NF-κB associated proteins in FGF-mediated mesoderm induction. Int. J. Dev. Biol. 42:67-77. [PubMed] [Google Scholar]
- 4.Beg, A. A., W. C. Sha, R. T. Bronson, S. Ghosh, and D. Baltimore. 1995. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature 376:167-170. [DOI] [PubMed] [Google Scholar]
- 5.Bondeson, J., F. Brennan, B. Foxwell, and M. Feldmann. 2000. Effective adenoviral transfer of IκBα into human fibroblasts and chondrosarcoma cells reveals that the induction of matrix metalloproteinases and proinflammatory cytokines is nuclear factor-κB dependent. J. Rheumatol. 27:2078-2089. [PubMed] [Google Scholar]
- 6.Bondeson, J., B. Foxwell, F. Brennan, and M. Feldmann. 1999. Defining therapeutic targets by using adenovirus: blocking NF-κB inhibits both inflammatory and destructive mechanisms in rheumatoid synovium but spares anti-inflammatory mediators. Proc. Natl. Acad. Sci. USA 96:5668-5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushdid, P. B., D. M. Brantley, F. E. Yull, G. L. Blaeuer, L. H. Hoffman, L. Niswander, and L. D. Kerr. 1998. Inhibition of NF-κB activity results in disruption of the apical ectodermal ridge and aberrant limb morphogenesis. Nature 392:615-618. [DOI] [PubMed] [Google Scholar]
- 8.Driever, W., and Z. Rangini. 1993. Characterization of a cell line derived from zebrafish (Brachydanio rerio) embryos. In Vitro Cell Dev. Biol. Anim. 29A:749-754. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-κB puzzle. Cell 109(Suppl.):S81-S96. [DOI] [PubMed] [Google Scholar]
- 10.Govind, S. 1999. Control of development and immunity by rel transcription factors in Drosophila. Oncogene 18:6875-6887. [DOI] [PubMed] [Google Scholar]
- 11.Griffin, K., R. Patient, and N. Holder. 1995. Analysis of FGF function in normal and no tail zebrafish embryos reveals separate mechanisms for formation of the trunk and the tail. Development 121:2983-2994. [DOI] [PubMed] [Google Scholar]
- 12.Grossmann, M., D. Metcalf, J. Merryfull, A. Beg, D. Baltimore, and S. Gerondakis. 1999. The combined absence of the transcription factors Rel and RelA leads to multiple hemopoietic cell defects. Proc. Natl. Acad. Sci. USA 96:11848-11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grunwald, D. J., and J. S. Eisen. 2002. Headwaters of the zebrafish: emergence of a new model vertebrate. Nat. Rev. Genet. 3:717-724. [DOI] [PubMed] [Google Scholar]
- 14.Halpern, M. E., R. K. Ho, C. Walker, and C. B. Kimmel. 1993. Induction of muscle pioneers and floor plate is distinguished by the zebrafish no tail mutation. Cell 75:99-111. [PubMed] [Google Scholar]
- 15.Herrmann, B. G., S. Labeit, A. Poustka, T. R. King, and H. Lehrach. 1990. Cloning of the T gene required in mesoderm formation in the mouse. Nature 343:617-622. [DOI] [PubMed] [Google Scholar]
- 16.Jowett, T. 1999. Analysis of protein and gene expression. Methods Cell Biol. 59:63-85. [DOI] [PubMed] [Google Scholar]
- 17.Kanegae, Y., A. T. Tavares, J. C. Izpisua Belmonte, and I. M. Verma. 1998. Role of Rel/NF-κB transcription factors during the outgrowth of the vertebrate limb. Nature 392:611-614. [DOI] [PubMed] [Google Scholar]
- 18.Karin, M., Y. Cao, F. R. Greten, and Z. W. Li. 2002. NF-κB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer 2:301-310. [DOI] [PubMed] [Google Scholar]
- 19.Kiriakidis, S., E. Andreakos, C. Monaco, B. Foxwell, M. Feldmann, and E. Paleolog. 2003. VEGF expression in human macrophages is NF-κB-dependent: studies using adenoviruses expressing the endogenous NF-κB inhibitor IκBβ and a kinase-defective form of the IκB kinase 2. J. Cell Sci. 116:665-674. [DOI] [PubMed] [Google Scholar]
- 20.Klement, J. F., N. R. Rice, B. D. Car, S. J. Abbondanzo, G. D. Powers, P. H. Bhatt, C. H. Chen, C. A. Rosen, and C. L. Stewart. 1996. IκBα deficiency results in a sustained NF-κB response and severe widespread dermatitis in mice. Mol. Cell. Biol. 16:2341-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lerchner, W., B. V. Latinkic, J. E. Remacle, D. Huylebroeck, and J. C. Smith. 2000. Region-specific activation of the Xenopus brachyury promoter involves active repression in ectoderm and endoderm: a study using transgenic frog embryos. Development 127:2729-2739. [DOI] [PubMed] [Google Scholar]
- 22.Li, Q., G. Estepa, S. Memet, A. Israel, and I. M. Verma. 2000. Complete lack of NF-κB activity in IKK1 and IKK2 double-deficient mice: additional defect in neurulation. Genes Dev. 14:1729-1733. [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Q., D. Van Antwerp, F. Mercurio, K. F. Lee, and I. M. Verma. 1999. Severe liver degeneration in mice lacking the IκB kinase 2 gene. Science 284:321-325. [DOI] [PubMed] [Google Scholar]
- 24.Li, Q., and I. M. Verma. 2002. NF-κB regulation in the immune system. Nat. Rev. Immunol. 2:725-734. [DOI] [PubMed] [Google Scholar]
- 25.Miyamoto, S., M. Maki, M. J. Schmitt, M. Hatanaka, and I. M. Verma. 1994. Tumor necrosis factor alpha-induced phosphorylation of IκBα is a signal for its degradation but not dissociation from NF-κB. Proc. Natl. Acad. Sci. USA 91:12740-12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nasevicius, A., and S. C. Ekker. 2000. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26:216-220. [DOI] [PubMed] [Google Scholar]
- 27.Odenthal, J., P. Haffter, E. Vogelsang, M. Brand, F. J. van Eeden, M. Furutani-Seiki, M. Granato, M. Hammerschmidt, C. P. Heisenberg, Y. J. Jiang, D. A. Kane, R. N. Kelsh, M. C. Mullins, R. M. Warga, M. L. Allende, E. S. Weinberg, and C. Nusslein-Volhard. 1996. Mutations affecting the formation of the notochord in the zebrafish, Danio rerio. Development 123:103-115. [DOI] [PubMed] [Google Scholar]
- 28.Odenthal, J., F. J. van Eeden, P. Haffter, P. W. Ingham, and C. Nusslein-Volhard. 2000. Two distinct cell populations in the floor plate of the zebrafish are induced by different pathways. Dev. Biol. 219:350-363. [DOI] [PubMed] [Google Scholar]
- 29.Pahl, H. L. 1999. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18:6853-6866. [DOI] [PubMed] [Google Scholar]
- 30.Patten, I., and M. Placzek. 2002. Opponent activities of Shh and BMP signaling during floor plate induction in vivo. Curr. Biol. 12:47-52. [DOI] [PubMed] [Google Scholar]
- 31.Pfeifer, A., and I. M. Verma. 2001. Gene therapy: promises and problems. Annu. Rev. Genomics Hum. Genet. 2:177-211. [DOI] [PubMed] [Google Scholar]
- 32.Richardson, J. C., A. M. Garcia Estrabot, and H. R. Woodland. 1994. XrelA, a Xenopus maternal and zygotic homologue of the p65 subunit of NF-κB: characterisation of transcriptional properties in the developing embryo and identification of a negative interference mutant. Mech. Dev. 45:173-189. [DOI] [PubMed] [Google Scholar]
- 33.Robinson-Rechavi, M., and V. Laudet. 2001. Evolutionary rates of duplicate genes in fish and mammals. Mol. Biol. Evol. 18:681-683. [DOI] [PubMed] [Google Scholar]
- 34.Robinson-Rechavi, M., O. Marchand, H. Escriva, P. L. Bardet, D. Zelus, S. Hughes, and V. Laudet. 2001. Euteleost fish genomes are characterized by expansion of gene families. Genome Res. 11:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodaway, A., H. Takeda, S. Koshida, J. Broadbent, B. Price, J. C. Smith, R. Patient, and N. Holder. 1999. Induction of the mesendoderm in the zebrafish germ ring by yolk cell-derived TGF-β family signals and discrimination of mesoderm and endoderm by FGF. Development 126:3067-3078. [DOI] [PubMed] [Google Scholar]
- 36.Rudolph, D., W. C. Yeh, A. Wakeham, B. Rudolph, D. Nallainathan, J. Potter, A. J. Elia, and T. W. Mak. 2000. Severe liver degeneration and lack of NF-κB activation in NEMO/IKKγ-deficient mice. Genes Dev. 14:854-862. [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt, C., B. Peng, Z. Li, G. M. Sclabas, S. Fujioka, J. Niu, M. Schmidt-Supprian, D. B. Evans, J. L. Abbruzzese, and P. J. Chiao. 2003. Mechanisms of proinflammatory cytokine-induced biphasic NF-κB activation. Mol. Cell 12:1287-1300. [DOI] [PubMed] [Google Scholar]
- 38.Schulte-Merker, S. 2002. Looking at embryos, p. 39-58. In C. Nusslein-Volhard (ed.), Zebrafish: a practical approach, vol. 261. Oxford University Press, Inc., New York, N.Y. [Google Scholar]
- 39.Schulte-Merker, S., R. K. Ho, B. G. Herrmann, and C. Nusslein-Volhard. 1992. The protein product of the zebrafish homologue of the mouse T gene is expressed in nuclei of the germ ring and the notochord of the early embryo. Development 116:1021-1032. [DOI] [PubMed] [Google Scholar]
- 40.Schulte-Merker, S., F. J. van Eeden, M. E. Halpern, C. B. Kimmel, and C. Nusslein-Volhard. 1994. No tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development 120:1009-1015. [DOI] [PubMed] [Google Scholar]
- 41.Sha, W. C., H. C. Liou, E. I. Tuomanen, and D. Baltimore. 1995. Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell 80:321-330. [DOI] [PubMed] [Google Scholar]
- 42.Shimada, M., N. Satoh, and H. Yokosawa. 2001. Involvement of Rel/NF-κB in regulation of ascidian notochord formation. Dev. Growth Differ. 43:145-154. [DOI] [PubMed] [Google Scholar]
- 43.Silverman, N., and T. Maniatis. 2001. NF-κB signaling pathways in mammalian and insect innate immunity. Genes Dev. 15:2321-2342. [DOI] [PubMed] [Google Scholar]
- 44.Smith, J. C. 1995. Mesoderm-inducing factors and mesodermal patterning. Curr. Opin. Cell Biol. 7:856-861. [DOI] [PubMed] [Google Scholar]
- 45.Squadrito, F., B. Deodato, G. Squadrito, P. Seminara, M. Passaniti, F. S. Venuti, M. Giacca, L. Minutoli, E. B. Adamo, M. Bellomo, R. Marini, M. Galeano, H. Marini, and D. Altavilla. 2003. Gene transfer of IκBα limits infarct size in a mouse model of myocardial ischemia-reperfusion injury. Lab. Investig. 83:1097-1104. [DOI] [PubMed]
- 46.Suzuki, K., J. Tsuchida, T. Yamamoto, and J. Inoue. 1998. Identification and expression of the Xenopus homolog of mammalian p100-NFκB2. Gene 206:1-9. [DOI] [PubMed] [Google Scholar]
- 47.Tergaonkar, V., M. Pando, O. Vafa, G. Wahl, and I. Verma. 2002. p53 stabilization is decreased upon NFκB activation: a role for NFκB in acquisition of resistance to chemotherapy. Cancer Cell 1:493-503. [DOI] [PubMed] [Google Scholar]
- 48.Tickle, C. 1998. Worlds in common through NF-κB. Nature 392:547-549. [DOI] [PubMed] [Google Scholar]
- 49.Van Antwerp, D. J., S. J. Martin, T. Kafri, D. R. Green, and I. M. Verma. 1996. Suppression of TNF-alpha-induced apoptosis by NF-κB. Science 274:787-789. [DOI] [PubMed] [Google Scholar]
- 50.Verma, I. M., J. K. Stevenson, E. M. Schwarz, D. Van Antwerp, and S. Miyamoto. 1995. Rel/NF-κB/I κB family: intimate tales of association and dissociation. Genes Dev. 9:2723-2735. [DOI] [PubMed] [Google Scholar]