Abstract
The HEXIM1 protein inhibits the kinase activity of P-TEFb (CDK9/cyclin T) to suppress RNA polymerase II transcriptional elongation in a process that specifically requires the 7SK snRNA, which mediates the interaction of HEXIM1 with P-TEFb. In an attempt to define the sequence requirements for HEXIM1 to interact with 7SK and inactivate P-TEFb, we have identified the first 18 amino acids within the previously described nuclear localization signal (NLS) of HEXIM1 as both necessary and sufficient for binding to 7SK in vivo and in vitro. This 7SK-binding motif was essential for HEXIM1's inhibitory action, as the HEXIM1 mutants with this motif replaced with a foreign NLS failed to interact with 7SK and P-TEFb and hence were unable to inactivate P-TEFb. The 7SK-binding motif alone, however, was not sufficient to inhibit P-TEFb. A region C-terminal to this motif was also required for HEXIM1 to associate with P-TEFb and suppress P-TEFb's kinase and transcriptional activities. The 7SK-binding motif in HEXIM1 contains clusters of positively charged residues reminiscent of the arginine-rich RNA-binding motif found in a wide variety of proteins. Part of it is highly homologous to the TAR RNA-binding motif in the human immunodeficiency virus type 1 (HIV-1) Tat protein, which was able to restore the 7SK-binding ability of a HEXIM1 NLS substitution mutant. We propose that a similar RNA-protein recognition mechanism may exist to regulate the formation of both the Tat-TAR-P-TEFb and the HEXIM1-7SK-P-TEFb ternary complexes, which may help convert the inactive HEXIM1/7SK-bound P-TEFb into an active one for Tat-activated and TAR-dependent HIV-1 transcription.
During transcription by RNA polymerase II (Pol II), phosphorylation of the carboxy-terminal domain (CTD) of the largest subunit of Pol II by the positive transcriptional elongation factor b (P-TEFb) is crucial for the transition from the abortive to the productive phase of transcriptional elongation, leading to the generation of full-length RNA transcripts (for reviews, see references 7 and 14). Active P-TEFb is composed of CDK9 and its regulatory subunit, cyclin T1 (CycT1) (14). Unlike other CDKs and their cyclin partners whose functions are closely related to the cell cycle regulation, P-TEFb is constitutively expressed and, hence, its protein level varies little throughout the cell cycle (5). Not only is P-TEFb essential for the expression of most protein-encoding genes (2, 16), but also it is indispensable for the replication of human immunodeficiency virus type 1 (HIV-1), since it is a specific host cellular cofactor for the viral Tat protein (7, 14). P-TEFb is recruited by Tat to the HIV-1 long terminal repeat (LTR) through the formation of a stable ternary complex consisting of P-TEFb, Tat, and the TAR RNA stem-loop structure located at the 5′ end of the nascent viral transcript (14). Once recruited, P-TEFb phosphorylates the Pol II CTD and stimulates the production of full-length HIV-1 transcripts.
Not every P-TEFb complex in the cell can function as a transcription factor. P-TEFb has been shown to lose its ability to phosphorylate the CTD and stimulate transcriptional elongation when associated with 7SK RNA (11, 20), a 330-nucleotide small nuclear RNA (snRNA) conserved in higher eukaryotes (10, 18, 24). Our recent in vitro reconstitution studies indicated that the association with 7SK is necessary but not sufficient to inactivate P-TEFb (21). Rather, a protein factor called HEXIM1, which has been identified as the third protein component of the 7SK-P-TEFb snRNP formed in vivo (9, 21), potently and specifically inhibits the kinase and transcriptional activities of P-TEFb in a 7SK-dependent manner (21). HEXIM1 has previously been identified as a nuclear protein whose expression is rapidly induced in cells treated with hexamethylene bis-acetamide (12), a potent inducer of cell differentiation (8). The 7SK-dependent inhibition of P-TEFb by HEXIM1 can be explained by the observation that 7SK plays a scaffolding role in mediating the interaction between HEXIM1 and P-TEFb, enabling HEXIM1 to exert its inhibitory effect on P-TEFb (9, 21).
Roughly half of nuclear P-TEFb in HeLa cells is sequestered in an inactive state within the 7SK snRNP. However, this population of P-TEFb can be rapidly dissociated from 7SK and HEXIM1 upon treatment of cells with certain stress-inducing agents (9, 11, 20, 21), some of which have been shown to stimulate the CTD phosphorylation and HIV-1 transcription at low dosages (1, 17). In addition, hypertrophic signals have been shown to activate P-TEFb in cardiac myocytes by disrupting the 7SK snRNP, leading to an accumulation of active, 7SK/HEXIM1-free P-TEFb in the cell. This in turn causes the stimulation of CTD phosphorylation as well as a global increase in RNA and protein contents, which eventually promote cardiac hypertrophy (15). Hence, the abilities of these stress- or hypertrophy-inducing agents to release 7SK/HEXIM1 and activate P-TEFb provide a mechanistic explanation for their effects on transcriptional activation.
Although the signaling pathway(s) leading to the stress- or hypertrophy-induced disruption of the 7SK snRNP is currently unknown, reversible phosphorylation of CDK9 is likely to play an important role in governing the formation and disruption of the snRNP. We have recently shown that phosphorylation of CDK9, on possibly the conserved Thr186 in the T-loop, is important for the 7SK-P-TEFb interaction (3). Mutation of only Thr186, but not the other known phosphorylated residues in CDK9, abolishes the 7SK-P-TEFb interaction both in vivo and in vitro.
It is important to further characterize the various interactions within the 7SK-HEXIM1-P-TEFb snRNP, which ultimately control the nuclear level of active P-TEFb for general and disease-related transcription. In this study, we focused on the newly discovered HEXIM1 protein and investigated the sequence requirements for HEXIM1 to bind 7SK and P-TEFb and to inactivate P-TEFb. We have identified an 18-amino-acid (aa) arginine-rich 7SK-binding motif within the nuclear localization signal (NLS) of HEXIM1 that was essential for HEXIM1's inhibitory effect on transcription through a 7SK-mediated interaction with P-TEFb. However, the 7SK-binding motif alone was incapable of inactivating P-TEFb, and the region of HEXIM1 C-terminal to this motif was essential for this function. Interestingly, the arginine-rich 7SK-binding motif in HEXIM1 is highly homologous to the TAR RNA-binding domain of the HIV-1 Tat protein. We speculate that this conserved arginine-rich RNA-binding motif has evolved to facilitate the specific interactions of P-TEFb with HEXIM1 and Tat via the 7SK and TAR RNA, respectively.
MATERIALS AND METHODS
cDNA constructs.
Wild-type HEXIM1, ΔN, and HEXIM1-ΔNLS cloned into pFlag-CMV2 were a generous gift from H. Tanaka from the University of Tokyo, Tokyo, Japan. The pFlag-CMV2 plasmids containing the cDNAs for HXM1-NLSSV40, HXM1-NLSTat, ΔN-NLSSV40, HXM1(150-158A), HXM1(159-167A), and HXM1(168-177A) were generated by a standard PCR protocol using custom-designed primers. For the generation of NLSHXM1-green fluorescent protein (GFP) and NLSSV40-GFP constructs, the sequences encoding the Flag-tagged NLS of HEXIM1 or simian virus 40 (SV40) T antigen were joined to the 5′ end of GFP by PCR and cloned into pcDNA3 (Invitrogen) using the HindIII and XhoI sites.
Affinity purification of HEXIM1 and its associated factors for Northern and Western analyses.
Flag-tagged wild-type and mutant HEXIM1 proteins as well as the NLS-GFP fusion proteins were expressed from cDNA constructs in HeLa cells transfected with the Lipofectamine Plus reagent (Invitrogen). Nuclear extract (NE) was prepared from the transfected cells 48 h later. The HEXIM1 complexes were affinity purified from NE by incubating at 4°C for 2 h with anti-Flag agarose beads (Sigma), followed by extensive washes with buffer D (20 mM HEPES-KOH [pH 7.9], 15% glycerol, 0.2 mM EDTA, 0.2% NP-40, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride) containing 0.3 M KCl (D0.3 M) and elution with the Flag peptide dissolved in D0.1 M. The eluted materials were analyzed by Western blotting with anti-Flag (M2; Sigma), anti-CDK9, and anti-CycT1 antibodies (Santa Cruz Biotechnology) and Northern hybridization using the 32P-labeled full-length 7SK antisense RNA as the probe.
In vitro transcription assay.
Flag-tagged wild-type and mutant HEXIM1 proteins used in the transcription reactions were affinity purified by anti-Flag immunoprecipitation from NE of transiently transfected 293T cells. After extensive washing with the high-salt buffer D containing 1.0 M KCl (D1.0 M) and then D0.05 M, the immobilized Flag-HEXIM1 proteins were eluted with 0.5 mg of Flag peptide/ml dissolved in D0.05 M. Prior to the transcription reactions, HeLa NE in D0.1 M was preincubated at 30°C for 20 min with the affinity-purified wild-type or mutant Flag-HEXIM1 proteins, in the presence of 0.5 mM ATP and 5 mM MgCl2. This step caused the phosphorylation of the endogenous 7SK/HEXIM1-free P-TEFb, enabling its subsequent binding to the exogenously added HEXIM1 to form the HEXIM1-7SK-P-TEFb complex (3). The in vitro transcription reactions containing two HIV-1 DNA templates, pHIV+TAR-G400 and pHIVΔTAR-G100, were performed essentially as described previously (22).
Immunofluorescence staining.
Cells transfected with various HEXIM1 cDNA constructs were grown in Lab-Tek II chamber slides (Nalge Nunc International) to ∼70% confluence and fixed with 4% formaldehyde in phosphate-buffered saline (PBS) at room temperature for 20 min. The rest of the procedure was performed on ice. Cells were permeabilized with ice-cold 0.1% Triton X-100 in PBS for 5 min and then washed in PBS, followed by incubation with 10% goat serum in PBS for 15 min to block nonspecific binding sites. Mouse anti-Flag antibody was added to a final concentration of 1 μg/ml. After incubation for 1 h, cells were washed extensively in PBS and then incubated for 30 min with goat anti-mouse immunoglobulin G conjugated to Alexa Fluor 488 at a 1:2,000 dilution (Molecular Probes).
Gel mobility shift assay.
Flag-tagged NLSHXM1-GFP and NLSSV40-GFP proteins were immunoprecipitated from NE of transfected HeLa cells. Proteins immobilized on anti-Flag beads were incubated with RNase A and washed stringently with the high-salt buffer D1.0 M to strip away any associated RNA and proteins. The Flag peptide-eluted proteins were then incubated with the 32P-labeled in vitro-transcribed sense- or antisense 7SK RNA in D0.05 M plus 1 mg of bovine serum albumin/ml, and 0.7 mg of yeast tRNA/ml. To supershift the complex, the anti-Flag monoclonal antibody was added at 2.4 μg/reaction mixture. The RNA-protein complexes were resolved in a 4% nondenaturing polyacrylamide gel in 0.5× Tris-glycine electrophoresis buffer at room temperature.
Luciferase assay.
HeLa cells grown in six-well cluster dishes were cotransfected with 100 ng of a luciferase reporter construct driven by the HIV-1 LTR (21) and with increasing concentrations of plasmids expressing either wild-type HEXIM1, NLS-replaced or -deleted HEXIM1 mutants, or NLS-GFP fusion proteins. The total amount of plasmids was kept constant for each transfection by adjusting with the empty vector. Luciferase activity was measured 48 h later.
In vitro kinase assay.
In vitro kinase reactions to detect the phosphorylation of GST-CTD by P-TEFb and to analyze the effect of wild-type and mutant HEXIM1 proteins on P-TEFb's kinase activity were performed as described previously (21).
RESULTS
Targeting HEXIM1 to the nucleus through a heterologous NLS domain.
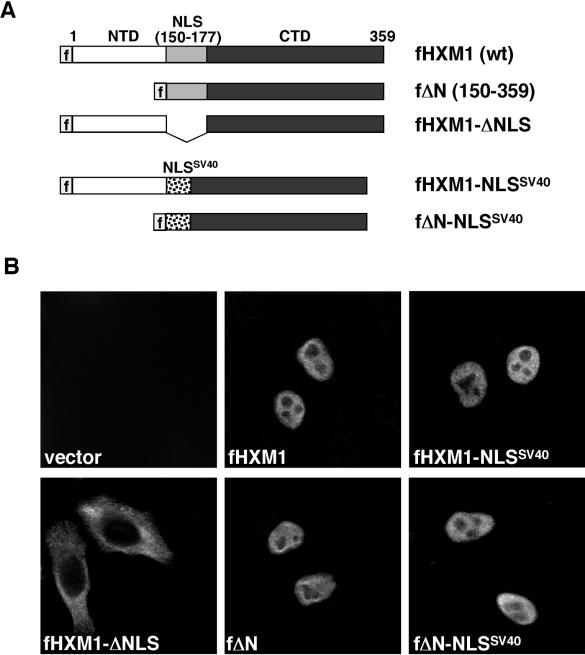
The full-length HEXIM1 protein, consisting of 359 aa (Fig. 1A), can be divided into three regions: an N-terminal proline-rich region (aa 1 to 149), a central region (aa 150 to 177) containing an unusually long bipartite NLS (12), and a C-terminal acidic region (aa 178 to 359) enriched in Asp and Glu residues. We have previously shown that two HEXIM1 truncation mutants, lacking either the N- or C-terminal region but still retaining the central NLS domain, have similar affinities for 7SK when compared to the wild-type HEXIM1 (21). Since the only region common to these two truncated mutants is the NLS, this domain could potentially contain both an NLS as well as a 7SK-binding motif.
FIG. 1.
HEXIM1 can be targeted to the nucleus through a heterologous NLS domain. (A) Domain structure of the Flag-tagged wild-type HEXIM1 (fHXM1) containing the NTD and CTD and a central NLS. Also shown are fΔN, a truncated HEXIM1 missing the NTD; fHXM1-ΔNLS, a HEXIM1 mutant missing the central NLS; fHXM1-NLSSV40, a mutant with the native NLS replaced with the NLS of the SV40 T antigen; and finally fΔN-NLSSV40, containing the SV40 NLS in place of the native NLS in fΔN. (B) The wild-type and mutant HEXIM1 proteins described in panel A were expressed from transfected plasmids in HeLa cells, and their subcellular localizations were examined by anti-Flag immunofluorescence staining.
To test whether the 28-aa NLS of HEXIM1 contains a 7SK-binding motif, it was replaced by the 10-aa prototype NLS (PKKKRKVEDP) derived from the SV40 large T antigen (6), resulting in the generation of HXM1-NLSSV40 (Fig. 1A). Next, the subcellular localization of the Flag-tagged wild-type HEXIM1 (fHXM1) and fHXM1-NLSSV40 in transiently transfected HeLa cells was examined by anti-Flag immunofluorescence staining. Both proteins were similarly located in the nucleus (Fig. 1B), indicating that the SV40 NLS was able to functionally replace the native HEXIM1 NLS as a nuclear targeting signal. Like the transfected fHXM1, the endogenous HEXIM1 was also stained as a nuclear protein (data not shown). As a control, a HEXIM1 mutant lacking the central NLS region (Fig. 1A, fHXM1-ΔNLS) was excluded from the nucleus (Fig. 1B), as previously reported (12).
We have previously shown that the N-terminally truncated HEXIM1 mutant ΔN (aa 150 to 359) efficiently interacted with 7SK and P-TEFb, causing the inhibition of P-TEFb's kinase and transcriptional activities (21). This finding implicates the C-terminal region of HEXIM1 as the business end of the molecule required for targeting and inactivating P-TEFb. Like fHXM1 and fHXM1-NLSSV40, both fΔN and fΔN-NLSSV40, in which the 28-aa NLS of HEXIM1 has been replaced with the SV40 NLS (Fig. 1A), were found to localize to the nucleus of the transfected HeLa cells (Fig. 1B).
The NLS domain of HEXIM1 is necessary for HEXIM1 to bind to 7SK and P-TEFb.
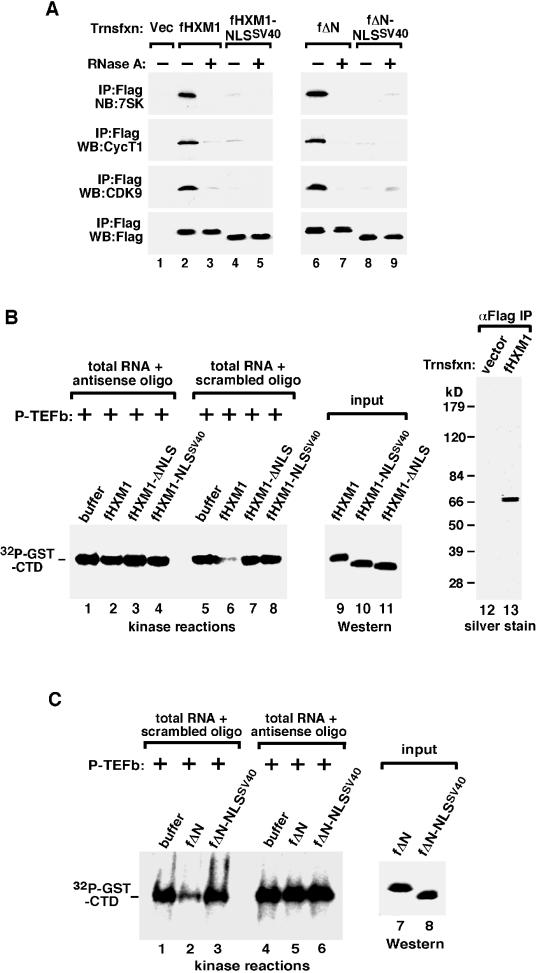
Given that both fHXM1-NLSSV40 and fΔN-NLSSV40 were properly targeted to the nucleus, we asked whether these two proteins could interact with 7SK and P-TEFb like their wild-type counterparts. Flag-tagged wild-type and mutant HEXIM1 proteins and their associated factors were affinity purified from the NE of transfected HeLa cells and analyzed by Western and Northern blotting. As expected, all the known components of the 7SK snRNP (7SK, CDK9, and CycT1) were associated with both fHXM1 and fΔN (Fig. 2A, lanes 2 and 6). Consistent with a previously demonstrated structural role of 7SK in bridging the HEXIM1-P-TEFb interaction (21), degradation of 7SK by RNase A during immunoprecipitation disrupted the associations with P-TEFb by both fHXM1 and fΔN (lanes 3 and 7). In contrast to these two proteins containing the native HEXIM1 NLS domain, fHXM1-NLSSV40 and fΔN-NLSSV40 interacted with very little 7SK and P-TEFb, either in the presence or absence of RNase A (Fig. 2A, lanes 4, 5, 8, and 9). These results suggest that although the NLS of the SV40 large T antigen was able to direct the nuclear localization of the backbone HEXIM1 protein, the SV40 NLS-substituted HEXIM1 mutants apparently lost the ability to form the 7SK snRNP. Thus, the native NLS of HEXIM1 does not act merely as a nuclear targeting signal but may also be required for 7SK binding, which in turn mediates the interaction between HEXIM1 and P-TEFb.
FIG. 2.
The native NLS domain is required for HEXIM1 to bind to 7SK and P-TEFb and inhibit P-TEFb's kinase activity. (A) The NLS-substituted HEXIM1 mutants fail to bind to 7SK and P-TEFb in vivo. Anti-Flag immunoprecipitation (IP) was performed in NE of HeLa cells transfected with either an empty vector (lane 1) or constructs expressing the indicated Flag-tagged HEXIM1 proteins (lanes 2 to 9). The peptide-eluted immune complexes were analyzed by Western blotting (WB) for the presence of CDK9, CycT1, and HEXIM1 and by Northern blotting (NB) for 7SK. (B) The native NLS is required for HEXIM1 to inhibit the P-TEFb kinase in a 7SK-dependent manner. Kinase reaction mixtures contained the affinity-purified wild-type or mutant HEXIM1, P-TEFb, GST-CTD, and the indicated RNA fractions. Total RNA was extracted from HeLa NE, which had been subjected to deoxyoligonucleotide-directed RNase H cleavage in the presence of 7SK antisense or scrambled oligonucleotide. Lanes 1 to 8, phosphorylated GST-CTD as detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography. Lanes 9 to 11, relative amounts of HEXIM1 proteins used in the kinase reactions as determined by anti-Flag Western blotting. Lanes 12 and 13, purity of wild-type fHXM1 affinity purified from transfected cells as indicated by silver staining. (C) The HEXIM1 NLS is required for fΔN to inhibit P-TEFb kinase. Kinase reactions containing the indicated HEXIM1 mutants were performed exactly as for panel B.
The NLS-substituted HEXIM1 mutants fail to inhibit P-TEFb's kinase activity.
Next, in vitro kinase reactions with P-TEFb as the kinase, GST-CTD as the substrate, and protein-free total nuclear RNA as the source of 7SK were performed to examine the abilities of wild-type and mutant HEXIM1 to inhibit P-TEFb (21). Affinity purified from NE of transfected 293T cells, the immobilized fHXM1 proteins were stripped off their associated factors by washing with the high-salt buffer D1.0 M (containing 1.0 M KCl) (21) prior to elution with the Flag peptide. The purity was confirmed by silver staining analysis (Fig. 2B, lanes 12 and 13, and data not shown). In kinase reactions, wild-type fHXM1 and fΔN inhibited P-TEFb's phosphorylation of GST-CTD in a process that specifically required the presence of intact 7SK in the reactions (Fig. 2B, lanes 2 and 6, and C, lanes 2 and 5). No inhibition was detected when 7SK in the total RNA fraction was first destroyed by the 7SK-antisense deoxyoligonucleotide-mediated RNase H cleavage (21). As for fHXM1-NLSSV40 and fΔN-NLSSV40, consistent with their failure to bind to 7SK and P-TEFb, neither was able to inhibit P-TEFb whether or not 7SK was present in the reaction mixture (Fig. 2B, lanes 4 and 8, and C, lanes 3 and 6). Because fHXM1-ΔNLS was mostly cytoplasmic (Fig. 1B) and unable to bind to 7SK/P-TEFb (data not shown), it also failed to inhibit the P-TEFb kinase activity (Fig. 2B, lanes 3 and 7).
The NLS domain is required for HEXIM1 to inhibit reporter gene expression in vivo.
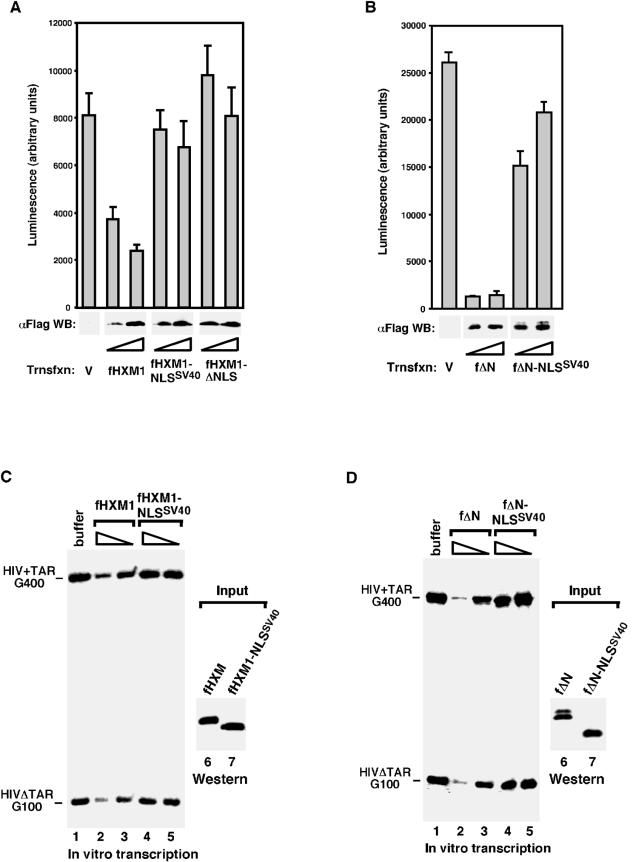
We have previously shown that when transiently expressed in HeLa cells, both wild-type and ΔN HEXIM1 efficiently inhibited the expression of the cotransfected luciferase reporter gene driven by a variety of Pol II promoters (21). To test whether the central NLS domain of HEXIM1 is required for this inhibition, we compared fHXM1 with fHXM1-NLSSV40 and fΔN with fΔN-NLSSV40 for their abilities to suppress the expression of the luciferase gene driven by the HIV-1 LTR. Consistent with their abilities to inhibit the P-TEFb kinase in vitro, luciferase gene expression was significantly inhibited by both fHXM1 and fΔN (Fig. 3A and B). In contrast, no or only very weak inhibition was observed with fHXM1-NLSSV40 or fΔN-NLSSV40. As expected, fHXM1-ΔNLS, which lacks the central NLS domain, failed to inhibit the luciferase gene expression.
FIG. 3.
The native NLS domain is required for HEXIM to inhibit transcription in vivo and in vitro. (A and B) The HEXIM1 NLS is required for both fHXM1 and fΔN to inhibit reporter gene transcription in vivo. HeLa cells were transfected with an HIV-1 LTR-luciferase reporter construct together with either an empty vector (V) or the indicated HEXIM1-expressing plasmid (at a twofold increment). The total amount of transfected DNA was kept constant by adjusting with the empty vector. Top panels: luciferase activities measured 48 h later; data from three independent experiments are shown. Bottom panels: levels of the Flag-tagged HEXIM1 proteins in transfected cell lysate determined by anti-Flag Western blotting. (C and D) The NLS-substituted HEXIM1 mutants fail to inhibit transcription in vitro. Flag-tagged wild-type and mutant HEXIM1 proteins were affinity purified from NE of transfected HeLa cells, stripped off their associated factors, and added into transcription reaction mixtures containing standard HeLa NE and two transcription templates, HIV+TAR-G400 and HIVΔTAR-G100. RNA fragments transcribed from two G-less cassettes inserted, respectively, into the two templates at a position ∼1 kb downstream of the HIV-1 promoter were indicated. The amounts of HEXIM1 proteins used in the reaction mixtures were examined by Western blotting in lanes 6 and 7.
NLS-substituted HEXIM1 mutants fail to inhibit transcription in vitro.
Having demonstrated that the transfected fHXM1 and fΔN could suppress the reporter gene expression in vivo, we next performed in vitro transcriptional studies to seek a direct confirmation of the ability of these two proteins to inhibit transcription and the requirement for their NLS domain in this process (Fig. 3C and D). Flag-tagged HEXIM1 proteins were affinity purified from NE of transfected cells, stripped off their associated factors with high salt (Fig. 2B and data not shown), and then added into transcription reaction mixtures containing standard HeLa NE and two HIV-1 DNA templates, pHIV+TAR-G400 and pHIVΔTAR-G100 (23). Again, both fHXM1 and fΔN, but not fHXM1-NLSSV40 or fΔN-NLSSV40, were able to directly inhibit HIV-1 promoter-distal transcription (i.e., transcriptional elongation) of two G-less cassettes (G400 and G100) that were inserted, respectively, into the two templates at ∼1 kb downstream of the start site (Fig. 3C and D).
The isolated NLS domain of HEXIM1 binds strongly to 7SK but poorly to P-TEFb in vivo.
The data presented so far are consistent with the idea that the central positively charged domain in HEXIM1 functions as not only as an NLS but also as a 7SK-binding domain required for HEXIM1 to target and inactivate P-TEFb through the mediation of 7SK. However, a trivial explanation also exists, which suggests that the replacement of the native HEXIM1 NLS with a foreign sequence may cause a conformational change in the overall protein structure, leading to a loss of HEXIM1's 7SK-binding ability. To rule out this latter possibility, it was important to demonstrate directly that the 28-aa HEXIM1 NLS domain indeed harbors a 7SK-binding motif.
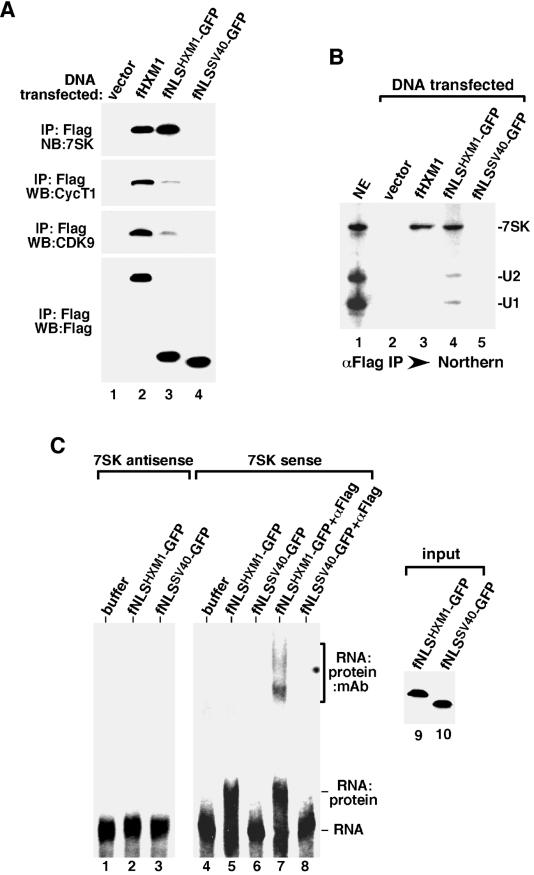
To this goal, we fused the Flag-tagged NLS domains of HEXIM1 and SV40 T antigen to the N terminus of GFP, resulting in the generation of two fusion proteins, fNLSHXM1-GFP and fNLSSV40-GFP, respectively. When tested for their abilities to bind to 7SK/P-TEFb in transiently transfected HeLa NE, 7SK was found to coimmunoprecipitate with fNLSHXM1-GFP (Fig. 4A, lane 3) but not fNLSSV40-GFP (lane 4). Compared to wild-type fHXM1, fNLSHXM1-GFP bound to slightly more 7SK (lanes 2 and 3), suggesting that a fully functional 7SK-binding motif is present within the 28-aa NLS domain of HEXIM1. However, unlike fHXM1, NLSHXM1-GFP coimmunoprecipitated only trace amounts of CycT1 and CDK9 (Fig. 4A), indicating that the HEXIM1 NLS alone was not sufficient to recruit the same amount of P-TEFb that was found in the full-length HEXIM1/7SK complex. Because the HEXIM1 ΔN mutant is fully able to bind to P-TEFb (21) (Fig. 2A), a region from aa 178 to 359, which is outside of the NLS but within the C-terminal half of HEXIM1, is likely to contribute to the recruitment of P-TEFb through additional protein-protein interactions with P-TEFb.
FIG. 4.
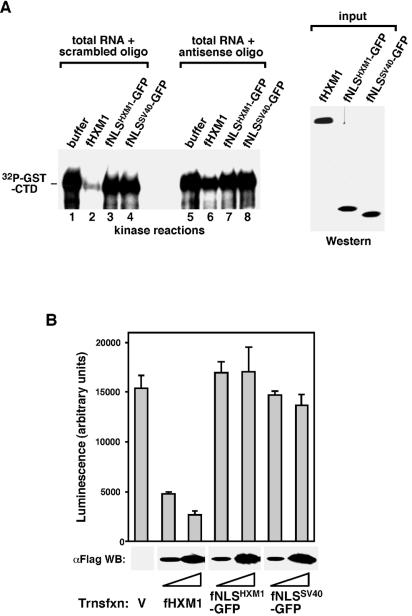
Direct and selective binding of the isolated NLS domain of HEXIM1 to 7SK in vivo and in vitro. (A) Efficient in vivo binding of the isolated NLS of HEXIM1 to 7SK but not P-TEFb. Anti-Flag immunoprecipitation (IP) was performed in NE of HeLa cells transfected with either an empty vector or vectors expressing the indicated Flag-tagged proteins. The peptide-eluted immune complexes were examined by Western blotting (WB) with antibodies against CDK9, CycT1, and Flag and Northern blotting (NB) with antisense 7SK RNA as the probe. (B) The isolated NLS of HEXIM1 binds 7SK with slightly reduced selectivity. Results shown are from Northern analyses of the levels of 7SK, U1, and U2 snRNAs present in HeLa NE and the various immune complexes (αFlag IP) obtained in panel A. (C) Direct binding of the isolated HEXIM1 NLS to 7SK in the absence of other factors. The indicated GFP fusion proteins were affinity purified from transfected cells, stripped off their associated factors, and analyzed in a gel mobility shift assay for their abilities to interact with the antisense (lanes 1 to 3) and sense (lanes 4 to 8) 7SK RNA. Anti-Flag monoclonal antibody was also present in the reaction mixtures in lanes 7 and 8. The amounts of GFP fusion proteins used in the reactions were examined by Western blotting in lanes 9 and 10.
The isolated HEXIM1 NLS binds 7SK with slightly reduced specificity.
Given that the 28-aa HEXIM1 NLS alone was fully capable of interacting with 7SK, we examined the specificity of this interaction by analyzing whether other abundant snRNA species (e.g., U1 and U2 snRNAs) besides 7SK could also coimmunoprecipitate with fNLSHXM1-GFP in transfected HeLa NE. Northern analyses in Fig. 4B revealed that the immunoprecipitates from cells transfected with either an empty vector (lane 2) or a construct producing fNLSSV40-GFP (lane 5) did not contain any 7SK, U1, or U2. Wild-type fHXM1, on the other hand, interacted with only 7SK but not U1 and U2, as expected (lane 3). Interestingly, although fNLSHXM1-GFP associated with a similar amount of 7SK as did fHXM1, it also coprecipitated small amounts of U1 and U2 in NE (lane 4), indicating that the specificity of the isolated HEXIM1 NLS for 7SK was somewhat reduced compared to that of the full-length protein. Quantification of the amount of snRNAs coimmunoprecipitated with fNLSHXM1-GFP indicated that, compared to the three RNA species present in NE (lane 1), 7SK was 8 times more likely than U1 and U2 to bind to fNLSHXM1-GFP. Thus, although the isolated HEXIM1 NLS showed a slightly reduced specificity for 7SK, it was still much higher than for the other snRNA species tested.
The isolated HEXIM1 NLS binds 7SK directly in the absence of other factors.
To rule out the possible involvement of another unknown factor(s) in mediating the in vivo interaction between fNLSHXM1-GFP and 7SK, a gel mobility shift assay was performed to determine whether there was a direct interaction between these two molecules in vitro. fNLSHXM1-GFP and fNLSSV40-GFP proteins were immunoprecipitated from the transfected HeLa NE, treated with RNase A and high salt (0.8 M KCl) to remove any associated factors (3), and eluted off the beads with the Flag peptide. The two proteins, affinity purified to single polypeptides as determined by silver staining analysis (data not shown), were subsequently incubated with 32P-labeled, in vitro-transcribed sense or antisense 7SK RNA. The RNA-protein complexes were then resolved in a nondenaturing polyacrylamide gel. As shown in Fig. 4C, incubation with fNLSHXM1-GFP but not fNLSSV40-GFP caused a shift in the mobility of only the sense 7SK RNA. The fNLSHXM1-GFP-7SK complex could be supershifted with the anti-Flag monoclonal antibody, which reacted with Flag-tagged fNLSHXM1-GFP. In agreement with the above-described high specificity of the isolated HEXIM1 NLS for 7SK, fNLSHXM1-GFP did not bind to the antisense 7SK RNA (Fig. 4C). Taken together, these results confirmed that the NLS of HEXIM1 indeed contains a bona fide 7SK-binding motif.
The HEXIM1 NLS alone is unable to inhibit P-TEFb kinase and Pol II transcription.
Although it was fully active in binding 7SK, the isolated HEXIM1 NLS interacted poorly with P-TEFb when fused to GFP (Fig. 4A). Reflecting this inability to target P-TEFb, fNLSHXM1-GFP failed to inhibit P-TEFb's kinase activity whether or not 7SK was present in the reaction mixture (Fig. 5A). Furthermore, unlike wild-type fHXM1, neither fNLSHXM1-GFP nor fNLSSV40-GFP produced from a transfected plasmid was able to inhibit the expression of a cotransfected luciferase reporter gene driven by the HIV-1 LTR (Fig. 5B). Thus, although the isolated HEXIM1 NLS bound to 7SK, this binding alone did not inactivate P-TEFb. Because the N-terminally deleted fΔN was fully active in binding to and suppressing P-TEFb (Fig. 2 and 3), a region outside of the NLS but within the C-terminal half of HEXIM1 likely cooperated with the 7SK-binding motif and was required for HEXIM1 to target and inhibit P-TEFb.
FIG. 5.
The HEXIM1 NLS alone fails to inhibit P-TEFb kinase and Pol II transcription. (A) The isolated HEXIM1 NLS cannot inhibit the P-TEFb kinase. Kinase reactions containing P-TEFb as the kinase, GST-CTD as the substrate, and the total RNA fraction treated by RNase H cleavage in the presence of 7SK-antisense or scrambled oligonucleotide were performed as for Fig. 2B. Buffer alone, fHXM1, or the indicated affinity-purified GFP fusion proteins were added into the reaction mixtures to inhibit the P-TEFb kinase. (B) The isolated NLS of HEXIM1 does not suppress the luciferase gene expression driven by the HIV-1 LTR. HeLa cells were cotransfected with the HIV-1 LTR-luciferase reporter construct and the indicated expression plasmids as for Fig. 3A. Top panel: luciferase activity was measured 48 h later, and data from three independent experiments are shown. Bottom panel: anti-Flag Western analysis of the levels of the transfected Flag-tagged proteins in whole-cell lysate.
The first 18 residues of the HEXIM1 NLS are important for 7SK binding.
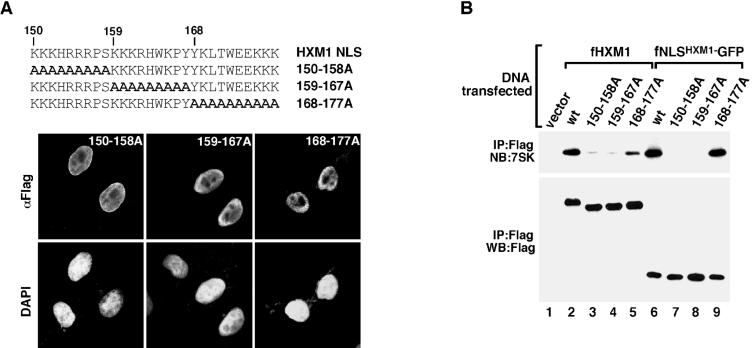
To further define the residues within the HEXIM1 NLS that are important for 7SK binding, we divided this 28-aa domain into three sections of roughly equal sizes and mutated all residues within each section to Ala (Fig. 6A). The alanine-substitution mutants generated in either the full-length HEXIM1 (Fig. 6A) or the NLS-GFP (data not shown) background were all localized to the nucleus as revealed by fluorescence microscopy, indicating that there was functional redundancy for nuclear targeting among the residues within the HEXIM1 NLS. The abilities of these alanine-substitution mutants to interact with 7SK were then assessed by anti-Flag immunoprecipitation from NE of transfected HeLa cells. Substitution of aa 150 to 158 and aa 159 to 167 in either the HEXIM1 (Fig. 6B, lanes 3 and 4) or the NLS-GFP (lanes 7 and 8) background significantly weakened or completely abolished 7SK binding. In contrast, mutation of aa 168 to 177 into Ala in HEXIM1 only moderately reduced its binding to 7SK (Fig. 6B, lane 5), and the same mutation introduced into the NLS-GFP background had no detectable effect on 7SK binding (lane 9). Thus, while aa 150 to 167 within the HEXIM1 NLS were mostly indispensable for 7SK binding, aa 168 to 177 had a relatively minor role in this process.
FIG. 6.
The first 18 residues of the HEXIM1 NLS are required for 7SK binding. (A) Functional redundancy for nuclear targeting among the residues within the HEXIM1 NLS. Top: amino acid sequences of the NLS domains of wild-type and mutant HEXIM1 with the indicated alanine substitutions within the NLS. Bottom: the wild-type and mutant HEXIM1 proteins were expressed from transfected plasmids in HeLa cells, and their subcellular localizations were examined by anti-Flag immunofluorescence staining. Nuclei of both transfected and untransfected cells were revealed by 4′,6′-diamidino-2-phenylindole staining and used as a reference. (B) The first 18 residues of HEXIM1 NLS are important for 7SK binding in vivo. HeLa cells were transfected with plasmids expressing the indicated fHXM1 or fNLSHXM1-GFP fusion proteins containing either wild-type or the various alanine-substituted NLS sequences described for panel A. Anti-Flag immune complexes derived from NE of transfected cells were analyzed by Western and Northern blotting as indicated.
A duplicated arginine-rich TAR-binding motif of HIV-1 Tat can substitute for the HEXIM1 7SK-binding motif.
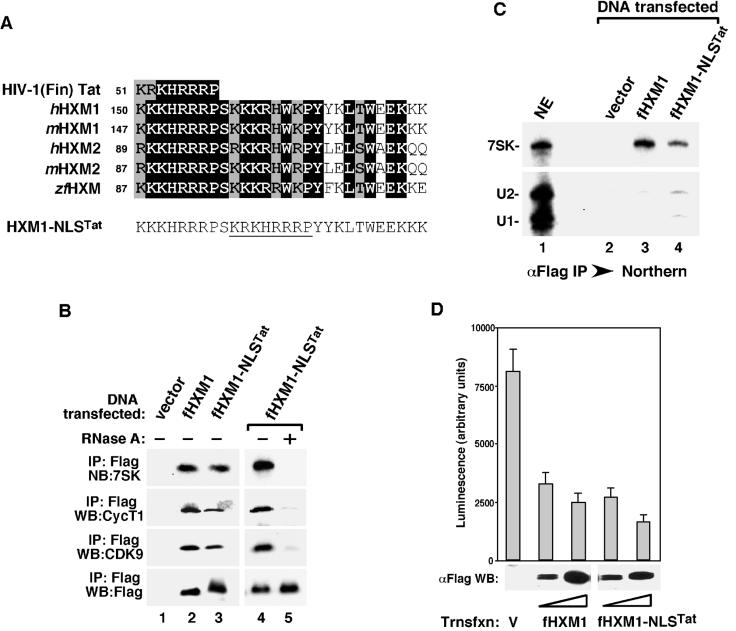
Consistent with the observation that aa 150 to 167 of the HEXIM1 NLS are important for 7SK binding, sequence alignment among the human and mouse HEXIM1 and HEXIM2 (unpublished data) as well as the zebrafish HEXIM proteins has revealed that the highest amino acid identity or similarity within the 28-aa NLS domain is indeed concentrated near the N-terminal two-thirds of this region (Fig. 7A). These 18 residues displayed an intriguing pattern of two consecutive segments linked by a conserved serine, with each segment consisting of six to seven basic residues followed by a proline (KKKHRRRP and KKKRHWKP) (Fig. 7A). Comprehensive BLAST analysis revealed that the first segment (aa 150 to 157) was remarkably similar to the arginine-rich TAR RNA-binding motif of the HIV-1 Tat protein, particularly that of the Finland strain (Fig. 7A). There is only one conserved amino acid change (Lys versus Arg) in the entire eight-residue segment shared by the two proteins. However, this Tat-like eight-residue segment was not sufficient to mediate HEXIM1-7SK binding, as illustrated by the failure to bind 7SK by both the full-length HEXIM1 and the NLS-GFP fusion protein containing wild-type sequences in this segment but Ala substitutions in the second segment of the 7SK-binding motif of HEXIM1 (Fig. 6B, 150 to 158A mutants).
FIG. 7.
A duplicated arginine-rich TAR-binding motif of HIV-1 Tat can substitute for the NLS of HEXIM1 for 7SK binding. (A) Sequence alignment of the arginine-rich TAR-binding motif of Tat (HIV-1 Finland strain AAC29057) with the NLS derived from human and mouse HEXIM1 (hHXM1 and mHXM1) and their homologues hHXM2 (AK056946), mHXM2 (BC026458), and the zebra fish (Danio rerio) zfHXM (BG307670). Reverse type indicates amino acid identity, and shaded boxes indicate similarity. The numbers refer to the positions of the first residue of the segments within the respective proteins. The NLS region of a HEXIM1 mutant (HXM1-NLSTat) containing the arginine-rich TAR-binding motif of Tat near the center (underlined) is also shown. (B) Substitution of the middle one-third of the HEXIM1 NLS with the arginine-rich TAR-binding motif of Tat restores 7SK binding. Anti-Flag immunoprecipitates were isolated from NE of HeLa cells transfected with either an empty vector or constructs expressing the indicated Flag-tagged HEXIM1 proteins and analyzed as described in the legend for Fig. 2A. Lanes 4 and 5 contain fHXM1-NLSTat complex immunoprecipitated in the presence (+) or absence (−) of RNase A. (C) fHXM1-NLSTat binds 7SK with slightly decreased specificity. Shown are Northern analyses of the levels of 7SK, U1, and U2 snRNAs present in HeLa NE and the various immune complexes (αFlag IP) obtained in panel B. (D) Overexpressed fHXM1-NLSTat suppresses the luciferase gene expression driven by the HIV-1 LTR. HeLa cells were cotransfected with the HIV-1 LTR-luciferase reporter construct and the indicated HEXIM1 expression plasmids. Luciferase activity was measured and analyzed as for Fig. 3A.
Since the second segment of HEXIM1's bipartite 7SK-binding motif is also positively charged and shows partial resemblance to the TAR-binding motif of Tat, we decided to replace this segment with the arginine-rich Tat sequences. The fHXM1-NLSTat mutant thus created contained essentially a duplicated TAR-binding motif of Tat separated by a conserved Ser (Fig. 7A). Remarkably, this substitution mostly restored the ability of HEXIM1 to bind to 7SK and P-TEFb, as demonstrated by the detection of 7SK, CDK9, and CycT1 associated with the immunoprecipitated fHXM1-NLSTat (Fig. 7B). Like the situation involving wild-type fHXM1, degradation of 7SK by RNase A prior to immunoprecipitation also disrupted the binding of fHXM1-NLSTat to P-TEFb (lanes 4 and 5), again revealing an RNA-dependent interaction between the two. However, compared to wild-type fHXM1, fHXM1-NLSTat appeared to bind to 7SK with slightly decreased specificity, as suggested by its coprecipitation with small amounts of U1 and U2 snRNAs in transfected cell NE (Fig. 7C). Quantitative analysis of the amounts of 7SK, U1, and U2 in both the immunoprecipitates and NE indicated that fHXM1-NLSTat was at least 10 times more likely to interact with 7SK than with U1 and U2, demonstrating its fairly selective if not entirely exclusive binding to 7SK. Importantly, like fHXM1, the ability of fHXM1-NLSTat to target P-TEFb through a 7SK-mediated interaction resulted in an efficient inhibition of HIV-1 LTR-driven luciferase gene expression in vivo (Fig. 7D). Together, these data reinforce the idea that a bipartite HIV-1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA Pol II transcription through a 7SK-mediated interaction with and inactivation of P-TEFb.
DISCUSSION
HEXIM1 and its homologues constitute a unique class of cyclin-dependent kinase inhibitors that require an RNA cofactor, 7SK, to mediate their interactions with and inactivation of the target kinase, P-TEFb. 7SK has been shown to interact with P-TEFb containing the phosphorylated T-loop in CDK9, and the different phosphorylation states control the formation and disruption of the 7SK snRNP in response to various physiological stimuli (3). In contrast, little is known about the molecular determinants in HEXIM1 that are key for interacting with 7SK and suppressing P-TEFb. Here, we demonstrate that a previously described NLS located near the center of HEXIM1 also functions as a 7SK-binding domain. While the region N-terminal to this domain plays a regulatory role in modulating the interaction between HEXIM1 and 7SK (21), the C-terminal region is essential for HEXIM1 to bind to and inhibit P-TEFb.
This important role of the HEXIM1 C-terminal region has been implicated by several observations. First, this region has been shown to interact directly with CycT1 in a Saccharomyces cerevisiae two-hybrid assay (9). Consistent with this, the C-terminal-deleted HEXIM1 binds significantly less P-TEFb in vivo and thus has markedly reduced ability to inactivate P-TEFb when compared to the full-length protein (21). Finally, because of the lack of the C-terminal region that is key for forming the complete 7SK snRNP, the isolated 7SK-binding domain of HEXIM1 interacted efficiently with 7SK but poorly with P-TEFb (Fig. 4A) and was therefore unable to inactivate P-TEFb (Fig. 5). Together, these data support a model that the 7SK-binding domain and the C-terminal P-TEFb-targeting region are both required for HEXIM1 to inhibit P-TEFb in a 7SK-dependent fashion. When the HEXIM1 NLS was replaced with a foreign NLS, the mutant HEXIM1 was properly directed to the nucleus but failed to bind to 7SK and therefore could not interact with P-TEFb. This in turn resulted in the inability of the mutant to inhibit P-TEFb's kinase and transcriptional activities (Fig. 2 and 3). These data reaffirm the notions that the 7SK snRNA plays a scaffolding role in mediating the HEXIM1-P-TEFb interaction and that P-TEFb is the functional target of HEXIM1/7SK's inhibitory action on transcription.
We have pinpointed the first 18 aa (aa 150 to 167) within the HEXIM1 NLS domain as residues constituting the 7SK-binding motif. When fused to GFP, they are sufficient for 7SK binding. Of those 18 residues, there are 14 invariable and 4 highly conserved ones, as revealed by sequence alignment of the human, mouse, and zebrafish HEXIM1 homologues or paralogues (Fig. 7A). This coincides with the fact that the 7SK snRNA sequences are also highly conserved throughout evolution (e.g., human and mouse 7SK sequences are 99% identical). The high degree of conservation of both the amino acids and nucleotides involved in the 7SK-HEXIM1 interaction indicates an important and highly conserved function of this interaction in controlling P-TEFb activity and RNA Pol II transcription across diverse animal species.
The 7SK-binding motif in HEIXM1 displays an interesting pattern of two consecutive segments connected by a conserved serine, with each segment consisting of six to seven positively charged residues followed by a proline (KKKHRRRP and KKKRHWKP) (Fig. 7A). While the first segment aligned almost perfectly with the arginine-rich TAR RNA-binding motif (KRKHRRRP) found in the HIV-1 Tat protein (Fig. 7A), the second segment also showed partial resemblance to the TAR-binding motif of Tat.
Despite the seemingly haphazard arrangement of clusters of positively charged residues within the 7SK-binding motif in HEXIM1, this motif nevertheless confers stringent specificity for 7SK binding when present within the HEXIM1 backbone (Fig. 4B). This observation could be explained by the structural insights obtained through the characterization of the folds of RNA-bound arginine-rich peptides and the architecture of their peptide-binding RNA pockets in viral and phage systems (for a review, see reference13). Studies of these model peptide-RNA complexes reveal diverse strategies of recognition based on structural transitions and adaptations involving both the RNA molecules and the peptides. Induced RNA structures include noncanonical elements, such as mismatches, base triplets, and looped-out bases, which sculpture the RNA major groove to create specific peptide-binding pockets and surfaces. On the other hand, although the arginine-rich peptides are related at the level of their primary amino acid sequences, different peptides can fold as an isolated alpha-helix, beta-hairpin, or helix-bend-helix, depending on the induced architectures of their RNA targets (13). Regarding the HEXIM1-7SK interaction, future structural studies will reveal to what extent adaptive transitions are involved in defining the specificity of this interaction and what specific role each segment of the bipartite 7SK-binding motif may play in contributing to this process.
It is important to note that when the isolated 7SK-binding motif of HEXIM1 is fused to GFP, the specificity for 7SK is somewhat reduced (Fig. 4B). This observation suggests that other parts of HEXIM1 or the incorporation of P-TEFb into the snRNP are likely to provide additional determinants to further enhance the specificity for 7SK binding. In support of a key role of P-TEFb in this process, it has been shown that P-TEFb can directly and specifically bind 7SK in vitro (3). In fact, both the conserved Thr186 in the CDK9 T-loop and a CycT1 region between aa 255 and 333 are specifically required for the binding (3).
The ability of P-TEFb to enhance the RNA-binding specificity of another protein is not without a precedent. In fact, binding of the HIV-1 Tat to the TAR stem-loop structure in vitro is notoriously not very specific. Although the binding depends on the specific sequences in the stem and a 3-nucleotide bulge region of the TAR RNA, mutations in the apical loop that destroy Tat-activated HIV-1 transcription do not affect the Tat-TAR interaction in vitro (7). The inclusion of P-TEFb into the complex, however, dramatically changes the situation. The ability of CycT1 to interact with both Tat and the TAR loop region enhances the specificity of the Tat-TAR interaction and confers dependence on specific TAR loop sequences for Tat-TAR-P-TEFb complex formation (4, 19). Given the observations that P-TEFb can specifically bind to 7SK in vitro (3) and that a direct interaction between HEXIM1 and CycT1 has been observed in a yeast two-hybrid assay (9), it is likely that a similar mechanism involving multiple protein-RNA and protein-protein interactions may be used by P-TEFb to enhance the specificity of the HEXIM1-7SK interaction. These observations, together with the discovery of a similar arginine-rich RNA-binding motif in both Tat and HEXIM1, strongly suggest that a similar architectural plan may exist to form both the Tat-TAR-P-TEFb and the HEXIM1-7SK-P-TEFb ternary complexes. This hypothesis, while yet to be confirmed, raises an intriguing possibility that the formation of the Tat-TAR-P-TEFb complex in HIV-infected cells may out-compete and preclude the formation of the inactive HEXIM1-7SK-P-TEFb complex, leading to the activation of HIV-1 transcription.
Acknowledgments
We thank K. Luo for helpful suggestions.
This work was supported by grants from the National Institutes of Health (AI-41757) and American Cancer Society (RSG-01-171-01-MBC) to Q.Z., a postdoctoral fellowship (F03-B-204) from the Universitywide AIDS Research Program to J.H.N.Y., and a fellowship from the Berkeley Scholar Program to R.C., a visiting scholar from Xiamen University, People’s Republic of China.
REFERENCES
- 1.Casse, C., F. Giannoni, V. T. Nguyen, M.-F. Dubois, and O. Bensaude. 1999. The transcriptional inhibitors, actinomycin D and alpha-amanitin, activate the HIV-1 promoter and favor phosphorylation of the RNA polymerase II C-terminal domain. J. Biol. Chem. 274:16097-16106. [DOI] [PubMed] [Google Scholar]
- 2.Chao, S.-H., and D. H. Price. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276:31793-31799. [DOI] [PubMed] [Google Scholar]
- 3.Chen, R., Z. Yang, and Q. Zhou. 2004. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J. Biol. Chem. 279:4153-4160. [DOI] [PubMed] [Google Scholar]
- 4.Garber, M. E., P. Wei, and K. A. Jones. 1998. HIV-1 Tat interacts with cyclin T1 to direct the P-TEFb CTD kinase complex to TAR RNA. Cold Spring Harb. Symp. Quant. Biol. 63:371-380. [DOI] [PubMed] [Google Scholar]
- 5.Garriga, J., S. Bhattacharya, J. Calbo, R. M. Marshall, M. Truongcao, D. S. Haines, and X. Grana. 2003. CDK9 is constitutively expressed throughout the cell cycle, and its steady-state expression is independent of SKP2. Mol. Cell. Biol. 23:5165-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldfarb, D. S., J. Gariepy, G. Schoolnik, and R. D. Kornberg. 1986. Synthetic peptides as nuclear localization signals. Nature 322:641-644. [DOI] [PubMed] [Google Scholar]
- 7.Jones, K. A. 1997. Taking a new TAK on tat transactivation. Genes Dev. 11:2593-2599. [DOI] [PubMed] [Google Scholar]
- 8.Marks, P. A., V. M. Richon, H. Kiyokawa, and R. A. Rifkind. 1994. Inducing differentiation of transformed cells with hybrid polar compounds: a cell cycle-dependent process. Proc. Natl. Acad. Sci. USA 91:10251-10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michels, A. A., V. T. Nguyen, A. Fraldi, V. Labas, M. Edwards, F. Bonnet, L. Lania, and O. Bensaude. 2003. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 23:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy, S., F. Altruda, E. Ullu, M. Tripodi, L. Silengo, and M. Melli. 1984. DNA sequences complementary to human 7 SK RNA show structural similarities to the short mobile elements of the mammalian genome. J. Mol. Biol. 177:575-590. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 12.Ouchida, R., M. Kusuhara, N. Shimizu, T. Hisada, Y. Makino, C. Morimoto, H. Handa, F. Ohsuzu, and H. Tanaka. 2003. Suppression of NF-κB-dependent gene expression by a hexamethylene bisacetamide-inducible protein HEXIM1 in human vascular smooth muscle cells. Genes Cells 8:95-107. [DOI] [PubMed] [Google Scholar]
- 13.Patel, D. J. 1999. Adaptive recognition in RNA complexes with peptides and protein modules. Curr. Opin. Struct. Biol. 9:74-87. [DOI] [PubMed] [Google Scholar]
- 14.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sano, M., M. Abdellatif, H. Oh, M. Xie, L. Bagella, A. Giordano, L. H. Michael, F. J. DeMayo, and M. D. Schneider. 2002. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 8:1310-1317. [DOI] [PubMed] [Google Scholar]
- 16.Shim, E. Y., A. K. Walker, Y. Shi, and T. K. Blackwell. 2002. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 16:2135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valerie, K., A. Delers, C. Bruck, C. Thiriart, H. Rosenberg, C. Debouck, and M. Rosenberg. 1988. Activation of human immunodeficiency virus type 1 by DNA damage in human cells. Nature 333:78-81. [DOI] [PubMed] [Google Scholar]
- 18.Wassarman, D. A., and J. A. Steitz. 1991. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol. Cell. Biol. 11:3432-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 20.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317-322. [DOI] [PubMed] [Google Scholar]
- 21.Yik, J. H., R. Chen, R. Nishimura, J. L. Jennings, A. J. Link, and Q. Zhou. 2003. Inhibition of P-TEFb (CDK9/cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12:971-982. [DOI] [PubMed] [Google Scholar]
- 22.Zhou, Q., D. Chen, E. Pierstorff, and K. Luo. 1998. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 17:3681-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou, Q., and P. A. Sharp. 1995. Novel mechanism and factor for regulation by HIV-1 Tat. EMBO J. 14:321-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zieve, G., and S. Penman. 1976. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell 8:19-31. [DOI] [PubMed] [Google Scholar]