In spite of differences among human oncogenic DNA viruses, there are some common characteristics. The viral genome persists in host cells and expresses latent gene products, and the strategy of cell transformation is generally the same: tumor viruses target cell signaling pathways. The goal of oncogenic viruses is obvious: to protract cell cycle progression and protect cells from apoptosis, thus perpetuating the virus genome. Therefore, the common cellular targets for tumor virus oncoproteins are the most important transcriptional factors involved in oncogenesis, such as c-myc, NF-κB, AP-1, p53, and others (62). Because there are innumerable cellular pathways that can regulate the transcriptional machinery of the cell, there are many opportunities for tumor viruses to dysregulate them to the benefit of the virus. It would not be surprising if any cell signaling pathway implicated in oncogenesis could be corrupted by oncogenic viruses.
For several years, the Wnt signaling pathway has been the object of intense attention in diverse biological areas. The classic Wnt pathway was initially characterized by its role in development; later came the realization that dysregulation of this signal transduction pathway plays an important role in human tumorigenesis, especially in carcinomas (7, 8, 14, 33, 47, 69). A central effector of the Wnt pathway is β-catenin, a multifunctional protein that was first known as a component of the cadherin cell adhesion complex (7, 9, 16, 33). Normally, the level of “free” β-catenin in the cytoplasm is tightly regulated by rapid degradation through the ubiquitin-proteasome system (1, 49, 52). According to the most accepted model, β-catenin is phosphorylated by glycogen synthase kinase (GSK)-3β as a part of the adenomatous polyposis coli protein (APC)/Axin complex and is subsequently subjected to ubiquitination and proteasomal degradation (45, 59). Wnt signaling interrupts the action of GSK-3β through an unknown mechanism, and thus β-catenin is no longer degraded. Accumulated β-catenin translocates into the nucleus, where it forms a complex with the T-cell factor (TCF)/Lef transcription factor and transactivates the expression of Wnt targets such as c-myc, cyclin D1, and others (7, 66).
Recently, a quite different general way to dysregulate this pathway, namely, through the actions of oncogenic viruses, has been recognized. Several groups have reported that human tumor viruses and their products are involved in the activation of the β-catenin/TCF pathway (25, 30, 32, 60, 71). Different viruses appear to use different mechanisms to activate β-catenin. The latency-associated nuclear antigen protein of Kaposi's sarcoma-associated herpesvirus dysregulates the GSK-3β/APC/Axin phosphorylation complex by binding to GSK-3β (30). The large T-antigen of the human polyomavirus JC virus binds directly to β-catenin and activates it through an unclear mechanism (25, 32). The latent membrane protein 2A of Epstein-Barr virus (EBV) activates β-catenin through phosphatidylinositol 3-kinase/AKT activation, leading to phosphorylation and inactivation of GSK-3β in epithelial cells (60), a pathway that was previously implicated in β-catenin activation (23, 31, 35, 72). Finally, in B lymphocytes latently infected with EBV, β-catenin is stabilized, cytoplasmic β-catenin is associated with active deubiquitinating enzymes (DUBs), and the β-catenin/TCF pathway is activated in type III latency but not in type I (71).
Ubiquitin-proteasome-mediated degradation of β-catenin is the key mechanism for the regulation of β-catenin levels in cells. In general, the degradation of proteins via ubiquitin-proteasome machinery involves three components: ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3) (19, 39). Phosphorylation of β-catenin is essential for the ubiquitination and degradation of β-catenin, which are mediated by the β-TrCP ubiquitin ligase in combination with the Skp1-Cullin1 ubiquitin-conjugating complex (29, 49, 87). β-TrCP1 is a member of the F box- and WD40 repeat-containing family of proteins that recognizes β-catenin as a substrate for ubiquitination only when it is phosphorylated at both serine residues in the conserved DSGXXS motif (49, 87, 88). Targeted disruption of the β-TrCP1 gene leads to impaired degradation and thus accumulation and nuclear translocation of β-catenin (61).
Recently, a new, phosphorylation-independent pathway of β-catenin degradation has been uncovered. In this case a distinct ubiquitin ligase complex, Siah-SIP-Skp1-Ebi, promotes the degradation of β-catenin through a mechanism independent of GSK-3β-mediated phosphorylation (54, 55). Interestingly, the Siah gene is a p53-inducible gene, and Siah mediates p53-inducible degradation of β-catenin (54, 55). Since each of the DNA tumor viruses encodes a protein that can interact with p53 (62) and as a result can inactivate p53 function, this may be another way for viruses to dysregulate the β-catenin/TCF pathway.
It is beginning to emerge that tumor viruses modulate the ubiquitin-proteasome machinery of host cells for their needs, mostly on the ubiquitin ligase (E3) level. In some cases the components of E3 can be encoded by the virus; in other cases viruses preempt host ubiquitin ligases, redirecting them to a substrate that normally would not be recognized (6).
Herpes simplex virus-infected cell protein 0 (ICP0) is a unique example of a viral ubiquitin ligase protein with two independent E3 sites. The N terminus of ICP0 contains a RING finger domain that is found in the largest known class of E3 ubiquitin ligases. The RING domain of ICP0 can induce the accumulation of polyubiquitin chains in the presence of the E2 conjugating enzymes UbcH5a and UbcH6 in vitro (12) and is required for the ubiquitin-dependent degradation of ICP0 substrates in vivo (27, 34). The targets of the RING domain of ICP0 include promyelocytic leukemia antigen and Sp100, constituents of nuclear structures known as ND10s (17, 28, 65). The C-terminal region of ICP0 contains a different ubiquitin ligase domain that does not have a RING finger but that binds the E2 enzyme UbcH3 (80). UbcH3 is the major E2 enzyme in the E1/E2/E3 complex that promotes ubiquitination and degradation of cyclin D1 (90). Together with the evidence that ICP0 can stabilize cyclin D1 without binding to it (56), these results lead to the attractive hypothesis that the C-terminal domain of ICP0 acts as a pseudo-E3 ligase, competitively inhibiting the proteasomal degradation of cyclin D1 (6).
Other examples of tumor virus-encoded ubiquitin ligases are the two membrane-bound proteins of Kaposi's sarcoma-associated herpesvirus, MIR1 and MIR2 (21). Both MIR1 and MIR2 can ubiquitinate major histocompatibility complex class I chains, which leads not to proteasomal but to endolysosomal degradation (20, 21).
In contrast, the strategies of human papillomavirus (HPV) and adenovirus for co-opting the host ubiquitin-proteasome system are quite different and have an important common target—the p53 tumor suppressor protein (6). The HPV E6 protein forms a complex with the cellular ubiquitin ligase E6AP, which leads to the ubiquitination and degradation of p53 in HPV-infected cells (74, 77, 78). E6AP contains the C-terminal ubiquitin ligase HECT domain that has been shown to interact with a number of E2 conjugating enzymes, including UbcH5, UbcH6, UbcH7, and UbcH8 (43, 44, 70). This is an interesting situation because the virus product prefers to alter endogenous substrate specificity; normally, p53 is a target for MDM2 ubiquitin ligase (38, 42), which belongs to a different, RING finger-containing, class of ubiquitin ligases (81). Although other targets for E6AP besides p53 have been found (51, 63), the biological role of E6AP in uninfected cells is still unclear. Furthermore, the ability of HPV to modulate cell protein turnover by proteasomal degradation is not restricted to E6. The HPV E7 protein is involved in ubiquitin-dependent degradation of pRb (13, 37), but the ligase partner of E7 is unknown.
In adenovirus-infected cells the level of p53 is reduced, and two adenovirus products, E1B 55K and E4orf6, have been shown to be necessary for the decrease of p53 transcriptional activity (24, 46, 58, 73). Later it became clear that these adenovirus products are involved in forming a specific ubiquitin ligase complex responsible for p53 ubiquitination and proteasomal degradation. The assembly of E1B and E4orf6 into a multiprotein complex containing elongins B and C, Cullin5, and Rbx1 is required for E1B 55K/E4orf6-dependent p53 ubiquitination and proteasomal degradation (36, 67). It is not clear which E2 conjugating enzymes are important for the ubiquitin ligase activity of this complex. UbcH5 was used in experiments in vitro, but in vivo assays detected UbcH3 in the complex with E4orf6 (67).
As a conclusion, it is pretty clear that the targeting of cellular proteins to proteasomal degradation through ubiquitination by way of viral or cellular ubiquitin ligases is an important aspect of infection and cell transformation by tumor viruses.
Deubiquitination, as a process counteracting ubiquitination, has recently received increasing attention. Since the first time deubiquitinating activity—cleavage of ubiquitin from histone H2A—was described (2), a huge number of DUBs have been identified. At least four distinct families of DUBs are now known. Well-defined classes include ubiquitin-processing proteases (UBP) and ubiquitin carboxyl-terminal hydrolases (UCH) (18, 22). Recently, two new groups have been characterized: the Jab1/Csn5 and MPN domain-containing proteins (the JAMM group of hydrolases) (82, 83, 89) and a family of cysteine proteases that contains an ovarian tumor domain (OTU) (5, 26). More details regarding the structure of DUBs and the mechanism of their function can be found in several excellent reviews (48, 85, 86). For years, the common understanding of the functional role of DUBs was limited to removing monoubiquitin from proteins such as histones or recycling ubiquitin from polyubiquitinated peptides after proteasomal degradation. In the last several years, DUBs have also been implicated in such fundamentally important biological processes as cell growth, differentiation, oncogenesis, development, and regulation of chromosome structure (86), but knowledge of the specific biologic roles of DUBs has been lacking.
For a long time, the existence of a large number of tissue-specific DUBs sharing little sequence similarity suggested the possibility that this class of proteins had a largely unexplored substrate specificity (18, 22, 84). It is becoming clearer that a number of proteins regulating cellular mechanisms for homeostasis in all eukaryotes are controlled not only by phosphorylation and dephosphorylation but also by ubiquitination and deubiquitination. The extraordinary importance of DUBs was shown recently: the herpesvirus-associated ubiquitin-specific protease (HAUSP) can remove ubiquitin from the p53 tumor suppressor protein and rescue it from degradation, leading to p53-mediated cell growth suppression (53), and the tumor suppressor CYLD (which is mutated in individuals with familial cylindromatosis [10]) turns out to be a DUB that negatively regulates NF-κB signaling (15, 50, 68, 79).
While the ability of tumor viruses to manipulate the ubiquitination system is now appreciated, the question of whether viral manipulation of the opposite wing of these linked systems—deubiquitination—holds true remains largely unstudied. If oncogenic viruses target signaling pathways in a ubiquitin-dependent manner and DUBs are also important regulators of these pathways, it would be logical to suggest that tumor viruses should affect cellular deubiquitinating processes as well. Consistent with this hypothesis, ICP0 binds HAUSP (34), a DUB that is important for p53 stabilization (53), thus adding another route by which a viral product may inactivate p53. Interestingly, EBV nuclear antigen 1 (EBNA1) also interacts with HAUSP, and this interaction influences EBNA1 transcriptional activity (40, 41).
Recently, two cytokine-inducible DUBs—DUB-1 and DUB-2—have been described. These hematopoiesis-specific genes with unclear function are rapidly induced after cytokine stimulation (3). Interleukin-2-inducible DUB-2 is constitutively expressed in human T-lymphotropic virus 1 (HTLV-1)-transformed cells (57). This DUB prolongs cytokine-induced activation of signal transducers and activators of transcription and suppresses apoptosis following cytokine withdrawal (57). Since interleukin-2 is constitutively expressed in HTLV-1-infected cells, this may be an example of an oncogenic virus regulating DUB expression indirectly, through the activation of another gene.
The idea that a human-oncogenic virus might direct synthesis of its own DUB is an intriguing scenario that has been examined recently. Adenovirus infection increases deubiquitinating activity in infected cells via the adenovirus proteinase Avp, which can function as a DUB in vitro and in vivo (4). Compared with classical DUBs, Avp seems to act as an enzyme of low specificity, which suggests that this viral DUB might deubiquitinate different viral and cellular ubiquitinated substrates, none of which is as yet identified (4).
The activity of DUBs in complex samples, such as mammalian cell extracts, has been difficult to examine. A new chemistry-based proteomics approach has allowed the detection of active DUBs with specific, active-site-directed probes against this group of enzymes (11). Recently it was shown that EBV immortalization of B cells results in the activation of a set of DUBs (64). The authors found that in normal B cells the level of DUB activity is low in general, which is interesting in itself and somewhat surprising. A relatively similar set of active DUBs was detected in both EBV-immortalized B cells and in Burkitt's lymphoma (BL) cell lines, with higher UCH-L1 or UCH-L3 activity in BL cell lines (64). Using the same probe for active DUBs, we have also observed a DUB in two different EBV-immortalized B-cell lines with the molecular weight of UCH-L1 as a major specific band (unpublished data). Also reported was that EBV infection of B cells induced activation of HAUSP, which was detected in both EBV-positive and -negative BL cell lines (64). Since HAUSP fulfills an important step in the stabilization and activation of p53, these results raise the questions of why either the tumor virus in EBV-immortalized cells or c-myc translocation in BL cells activates this DUB and what the possible functional consequence of this somewhat counterintuitive activation of p53 in transformed cells might be.
Interestingly, the activity of the FAM DUB, which had previously been implicated in β-catenin stabilization (75), is increased in both EBV-immortalized and BL cells (64). This observation suggests that β-catenin may be stabilized in both types of cell lines. But according to our results, β-catenin is not stabilized in several BL cell lines, including intensively studied EBV-positive BL lines (71) and the EBV-negative BL line DG75 (unpublished data). Since we observed stabilized β-catenin in another EBV-negative BL cell line, BJAB (unpublished data), these contrary observations may indicate the existence of genetic or cell signaling variations in BL cell lines. Of course, it should be noted that β-catenin is not the only target of FAM; AF-6 (also called afadin), an actin-binding multidomain protein which is involved in Ras signaling, is also subject to FAM-dependent stabilization (76). Our preliminary data suggest that cytoplasmic β-catenin in EBV-infected cells forms a complex with a DUB with a molecular weight of ∼34,000, the size of UCH-L3 (unpublished data), but a biological function for this association is unknown. Another interesting observation is that most DUBs identified in EBV-infected cells become detectable after a certain period of time (more than a month), suggesting that expression (or increase of activity) of the enzymes is independent of the direct expression of viral latency products. It raises the speculation that EBV might induce DUBs in an indirect manner, probably through activation of intermediate messengers. In any event, all of this evidence supports the suggestion that tumor viruses may utilize deubiquitinating systems as well as ubiquitination to support their needs, but our knowledge in this area is very limited, and obviously much more research is needed in the future.
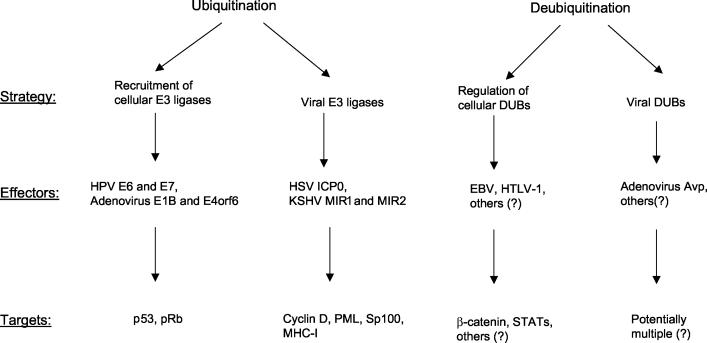
It has become clear that a number of proteins regulating cellular mechanisms for homeostasis in all eukaryotes may be controlled by both ubiquitination and deubiquitination and that oncogenic viruses play a certain role in the dysregulation of cell signaling pathways that intervene in this system. In normal cells the balance between the two processes is likely determined by a dynamic equilibrium and is highly regulated. Tumor viruses may affect ubiquitination directly, by using their own ubiquitinating enzymes, or indirectly, by use of endogenous cellular components of the ubiquitin system. The recent studies of a DUB encoded by an oncogenic virus (4), as well as the suggestion that a tumor virus can regulate a cell signaling pathway through deubiquitination (71), raise the same possibility for the deubiquitinating system, and investigations have begun in this new area of viral functionality (Fig. 1). The identification of the mechanisms whereby tumor viruses regulate the host ubiquitinating-deubiquitinating systems, including the viral products involved in this regulation, are central aspects of continuing studies. The roles of the ubiquitinating and deubiquitinating machinery in the oncogenic potential of human tumor viruses seem to be important not only for viral oncogenesis but probably also for the understanding of viral function in general.
FIG. 1.
Human oncogenic DNA viruses use ubiquitination-deubiquitination systems to dysregulate cell signaling pathways. The strategies for both processes involve recruitment of either cellular or viral enzymes by viral products, in several cases by identified effectors that act on specific cellular targets.
ADDENDUM IN PROOF
Since acceptance of this paper, Li et al. (M. Li, C. L. Brooks, N. Kon, and W. Gu, Mol. Cell 13:879-886, 2004) and Cummings et al. (J. M. Cummings, C. Rago, M. Kohli, K. W. Kinzler, C. Lengauer, and B. Vogelstein, Nature 428:486, 2004) reported that HAUSP can increase p53 proteasomal degradation through deubiquitination of MDM2. These data might help to explain why EBV infection induces HAUSP activation.
REFERENCES
- 1.Aberle, H., A. Bauer, J. Stappert, A. Kispert, and R. Kemler. 1997. β-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 16:3797-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, M. W., N. R. Ballal, I. L. Goldknopf, and H. Busch. 1981. Protein A24 lyase activity in nucleoli of thioacetamide-treated rat liver releases histone 2A and ubiquitin from conjugated protein A24. Biochemistry 20:1100-1104. [DOI] [PubMed] [Google Scholar]
- 3.Baek, K. H. 2002. Lymphocyte-specific murine deubiquitinating enzymes induced by cytokines. Am. J. Hematol. 71:340-345. [DOI] [PubMed] [Google Scholar]
- 4.Balakirev, M. Y., M. Jaquinod, A. L. Haas, and J. Chroboczek. 2002. Deubiquitinating function of adenovirus proteinase. J. Virol. 76:6323-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balakirev, M. Y., S. O. Tcherniuk, M. Jaquinod, and J. Chroboczek. 2003. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 4:517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banks, L., D. Pim, and M. Thomas. 2003. Viruses and the 26S proteasome: hacking into destruction. Trends Biochem. Sci. 28:452-459. [DOI] [PubMed] [Google Scholar]
- 7.Barker, N., and H. Clevers. 2000. Catenins, Wnt signaling and cancer. Bioessays 22:961-965. [DOI] [PubMed] [Google Scholar]
- 8.Barker, N., P. J. Morin, and H. Clevers. 2000. The Yin-Yang of TCF/beta-catenin signaling. Adv. Cancer Res. 77:1-24. [DOI] [PubMed] [Google Scholar]
- 9.Bienz, M., and H. Clevers. 2003. Armadillo/beta-catenin signals in the nucleus—proof beyond a reasonable doubt? Nat. Cell Biol. 5:179-182. [DOI] [PubMed] [Google Scholar]
- 10.Bignell, G. R., W. Warren, S. Seal, M. Takahashi, E. Rapley, R. Barfoot, H. Green, C. Brown, P. J. Biggs, S. R. Lakhani, C. Jones, J. Hansen, E. Blair, B. Hofmann, R. Siebert, G. Turner, D. G. Evans, C. Schrander-Stumpel, F. A. Beemer, A. van den Ouweland, D. Halley, B. Delpech, M. G. Cleveland, I. Leigh, J. Leisti, S. Rasmussen, et al. 2000. Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Genet. 25:160-165. [DOI] [PubMed] [Google Scholar]
- 11.Borodovsky, A., H. Ovaa, N. Kolli, T. Gan-Erdene, K. D. Wilkinson, H. L. Ploegh, and B. M. Kessler. 2002. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 9:1149-1159. [DOI] [PubMed] [Google Scholar]
- 12.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyer, S. N., D. E. Wazer, and V. Band. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56:4620-4624. [PubMed] [Google Scholar]
- 14.Brantjes, H., N. Barker, J. van Es, and H. Clevers. 2002. TCF: Lady Justice casting the final verdict on the outcome of Wnt signalling. Biol. Chem. 383:255-261. [DOI] [PubMed] [Google Scholar]
- 15.Brummelkamp, T. R., S. M. Nijman, A. M. Dirac, and R. Bernards. 2003. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 424:797-801. [DOI] [PubMed] [Google Scholar]
- 16.Bullions, L. C., and A. J. Levine. 1998. The role of beta-catenin in cell adhesion, signal transduction, and cancer. Curr. Opin. Oncol. 10:81-87. [DOI] [PubMed] [Google Scholar]
- 17.Chelbi-Alix, M. K., and H. de The. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935-941. [DOI] [PubMed] [Google Scholar]
- 18.Chung, C. H., and S. H. Baek. 1999. Deubiquitinating enzymes: their diversity and emerging roles. Biochem. Biophys. Res. Commun. 266:633-640. [DOI] [PubMed] [Google Scholar]
- 19.Ciechanover, A. 1998. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17:7151-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coscoy, L., and D. Ganem. 2003. PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol. 13:7-12. [DOI] [PubMed] [Google Scholar]
- 21.Coscoy, L., D. J. Sanchez, and D. Ganem. 2001. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 155:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Andrea, A., and D. Pellman. 1998. Deubiquitinating enzymes: a new class of biological regulators. Crit. Rev. Biochem. Mol. Biol. 33:337-352. [DOI] [PubMed] [Google Scholar]
- 23.Desbois-Mouthon, C., A. Cadoret, M. J. Blivet-Van Eggelpoel, F. Bertrand, G. Cherqui, C. Perret, and J. Capeau. 2001. Insulin and IGF-1 stimulate the β-catenin pathway through two signalling cascades involving GSK-3β inhibition and Ras activation. Oncogene 20:252-259. [DOI] [PubMed] [Google Scholar]
- 24.Dobner, T., N. Horikoshi, S. Rubenwolf, and T. Shenk. 1996. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science 272:1470-1473. [DOI] [PubMed] [Google Scholar]
- 25.Enam, S., L. Del Valle, C. Lara, D. D. Gan, C. Ortiz-Hidalgo, J. P. Palazzo, and K. Khalili. 2002. Association of human polyomavirus JCV with colon cancer: evidence for interaction of viral T-antigen and beta-catenin. Cancer Res. 62:7093-7101. [PubMed] [Google Scholar]
- 26.Evans, P. C., T. S. Smith, M. J. Lai, M. G. Williams, D. F. Burke, K. Heyninck, M. M. Kreike, R. Beyaert, T. L. Blundell, and P. J. Kilshaw. 2003. A novel type of deubiquitinating enzyme. J. Biol. Chem. 278:23180-23186. [DOI] [PubMed] [Google Scholar]
- 27.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761-770. [DOI] [PubMed] [Google Scholar]
- 28.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuchs, S. Y., A. Chen, Y. Xiong, Z. Q. Pan, and Z. Ronai. 1999. HOS, a human homolog of Slimb, forms an SCF complex with Skp1 and Cullin1 and targets the phosphorylation-dependent degradation of IkappaB and beta-catenin. Oncogene 18:2039-2046. [DOI] [PubMed] [Google Scholar]
- 30.Fujimuro, M., F. Y. Wu, C. ApRhys, H. Kajumbula, D. B. Young, G. S. Hayward, and S. D. Hayward. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300-306. [DOI] [PubMed] [Google Scholar]
- 31.Fukumoto, S., C. M. Hsieh, K. Maemura, M. D. Layne, S. F. Yet, K. H. Lee, T. Matsui, A. Rosenzweig, W. G. Taylor, J. S. Rubin, M. A. Perrella, and M. E. Lee. 2001. Akt participation in the Wnt signaling pathway through Dishevelled. J. Biol. Chem. 276:17479-17483. [DOI] [PubMed] [Google Scholar]
- 32.Gan, D. D., and K. Khalili. 2004. Interaction between JCV large T-antigen and beta-catenin. Oncogene 23:483-490. [DOI] [PubMed] [Google Scholar]
- 33.Giles, R. H., J. H. van Es, and H. Clevers. 2003. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys. Acta. 1653:1-24. [DOI] [PubMed] [Google Scholar]
- 34.Hagglund, R., and B. Roizman. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haq, S., A. Michael, M. Andreucci, K. Bhattacharya, P. Dotto, B. Walters, J. Woodgett, H. Kilter, and T. Force. 2003. Stabilization of beta-catenin by a Wnt-independent mechanism regulates cardiomyocyte growth. Proc. Natl. Acad. Sci. USA 100:4610-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harada, J. N., A. Shevchenko, A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helt, A. M., and D. A. Galloway. 2003. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis 24:159-169. [DOI] [PubMed] [Google Scholar]
- 38.Hengstermann, A., L. K. Linares, A. Ciechanover, N. J. Whitaker, and M. Scheffner. 2001. Complete switch from Mdm2 to human papillomavirus E6-mediated degradation of p53 in cervical cancer cells. Proc. Natl. Acad. Sci. USA 98:1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 40.Holowaty, M. N., Y. Sheng, T. Nguyen, C. Arrowsmith, and L. Frappier. 2003. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. J. Biol. Chem. 278:47753-47761. [DOI] [PubMed] [Google Scholar]
- 41.Holowaty, M. N., M. Zeghouf, H. Wu, J. Tellam, V. Athanasopoulos, J. Greenblatt, and L. Frappier. 2003. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J. Biol. Chem. 278:29987-29994. [DOI] [PubMed] [Google Scholar]
- 42.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 43.Huang, L., E. Kinnucan, G. Wang, S. Beaudenon, P. M. Howley, J. M. Huibregtse, and N. P. Pavletich. 1999. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science 286:1321-1326. [DOI] [PubMed] [Google Scholar]
- 44.Huibregtse, J. M., M. Scheffner, S. Beaudenon, and P. M. Howley. 1995. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 92:2563-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jope, R. S., and G. N. Bijur. 2002. Mood stabilizers, glycogen synthase kinase-3beta and cell survival. Mol. Psychiatry 7(Suppl. 1):S35-S45. [DOI] [PubMed] [Google Scholar]
- 46.Kao, C. C., P. R. Yew, and A. J. Berk. 1990. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology 179:806-814. [DOI] [PubMed] [Google Scholar]
- 47.Kikuchi, A. 2000. Regulation of beta-catenin signaling in the Wnt pathway. Biochem. Biophys. Res. Commun. 268:243-248. [DOI] [PubMed] [Google Scholar]
- 48.Kim, J. H., K. C. Park, S. S. Chung, O. Bang, and C. H. Chung. 2003. Deubiquitinating enzymes as cellular regulators. J. Biochem. (Tokyo) 134:9-18. [DOI] [PubMed] [Google Scholar]
- 49.Kitagawa, M., S. Hatakeyama, M. Shirane, M. Matsumoto, N. Ishida, K. Hattori, I. Nakamichi, A. Kikuchi, and K. Nakayama. 1999. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. EMBO J. 18:2401-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kovalenko, A., C. Chable-Bessia, G. Cantarella, A. Israel, D. Wallach, and G. Courtois. 2003. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature 424:801-805. [DOI] [PubMed] [Google Scholar]
- 51.Kumar, S., A. L. Talis, and P. M. Howley. 1999. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J. Biol. Chem. 274:18785-18792. [DOI] [PubMed] [Google Scholar]
- 52.Latres, E., D. S. Chiaur, and M. Pagano. 1999. The human F box protein beta-Trcp associates with the Cul1/Skp1 complex and regulates the stability of beta-catenin. Oncogene 18:849-854. [DOI] [PubMed] [Google Scholar]
- 53.Li, M., D. Chen, A. Shiloh, J. Luo, A. Y. Nikolaev, J. Qin, and W. Gu. 2002. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416:648-653. [DOI] [PubMed] [Google Scholar]
- 54.Liu, J., J. Stevens, C. A. Rote, H. J. Yost, Y. Hu, K. L. Neufeld, R. L. White, and N. Matsunami. 2001. Siah-1 mediates a novel beta-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol. Cell 7:927-936. [DOI] [PubMed] [Google Scholar]
- 55.Matsuzawa, S. I., and J. C. Reed. 2001. Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol. Cell 7:915-926. [DOI] [PubMed] [Google Scholar]
- 56.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223-1233. [DOI] [PubMed] [Google Scholar]
- 57.Migone, T. S., M. Humbert, A. Rascle, D. Sanden, A. D'Andrea, and J. A. Johnston. 2001. The deubiquitinating enzyme DUB-2 prolongs cytokine-induced signal transducers and activators of transcription activation and suppresses apoptosis following cytokine withdrawal. Blood 98:1935-1941. [DOI] [PubMed] [Google Scholar]
- 58.Moore, M., N. Horikoshi, and T. Shenk. 1996. Oncogenic potential of the adenovirus E4orf6 protein. Proc. Natl. Acad. Sci. USA 93:11295-11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morin, P. J. 1999. β-Catenin signaling and cancer. Bioessays 21:1021-1030. [DOI] [PubMed] [Google Scholar]
- 60.Morrison, J. A., A. J. Klingelhutz, and N. Raab-Traub. 2003. Epstein-Barr virus latent membrane protein 2A activates β-catenin signaling in epithelial cells. J. Virol. 77:12276-12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakayama, K., S. Hatakeyama, S. Maruyama, A. Kikuchi, K. Onoe, R. A. Good, and K. I. Nakayama. 2003. Impaired degradation of inhibitory subunit of NF-kappa B (I kappa B) and beta-catenin as a result of targeted disruption of the beta-TrCP1 gene. Proc. Natl. Acad. Sci. USA 100:8752-8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nevins, J. 2001. Cell transformation by viruses, p. 245-284. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 63.Oda, H., S. Kumar, and P. M. Howley. 1999. Regulation of the Src family tyrosine kinase Blk through E6AP-mediated ubiquitination. Proc. Natl. Acad. Sci. USA 96:9557-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ovaa, H., B. M. Kessler, U. Rolen, P. J. Galardy, H. L. Ploegh, and M. G. Masucci. 2004. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc. Natl. Acad. Sci. USA 101:2253-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peifer, M., and P. Polakis. 2000. Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science 287:1606-1609. [DOI] [PubMed] [Google Scholar]
- 67.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Regamey, A., D. Hohl, J. W. Liu, T. Roger, P. Kogerman, R. Toftgard, and M. Huber. 2003. The tumor suppressor CYLD interacts with TRIP and regulates negatively nuclear factor kappaB activation by tumor necrosis factor. J. Exp. Med. 198:1959-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roose, J., and H. Clevers. 1999. TCF transcription factors: molecular switches in carcinogenesis. Biochim. Biophys. Acta 1424:M23-M37. [DOI] [PubMed] [Google Scholar]
- 70.Scheffner, M., U. Nuber, and J. M. Huibregtse. 1995. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 373:81-83. [DOI] [PubMed] [Google Scholar]
- 71.Shackelford, J., C. Maier, and J. S. Pagano. 2003. Epstein-Barr virus activates beta-catenin in type III latently infected B lymphocyte lines: association with deubiquitinating enzymes. Proc. Natl. Acad. Sci. USA 100:15572-15576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharma, M., W. W. Chuang, and Z. Sun. 2002. Phosphatidylinositol 3-kinase/Akt stimulates androgen pathway through GSK3beta inhibition and nuclear beta-catenin accumulation. J. Biol. Chem. 277:30935-30941. [DOI] [PubMed] [Google Scholar]
- 73.Steegenga, W. T., N. Riteco, A. G. Jochemsen, F. J. Fallaux, and J. L. Bos. 1998. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene 16:349-357. [DOI] [PubMed] [Google Scholar]
- 74.Talis, A. L., J. M. Huibregtse, and P. M. Howley. 1998. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J. Biol. Chem. 273:6439-6445. [DOI] [PubMed] [Google Scholar]
- 75.Taya, S., T. Yamamoto, M. Kanai-Azuma, S. A. Wood, and K. Kaibuchi. 1999. The deubiquitinating enzyme Fam interacts with and stabilizes beta-catenin. Genes Cells 4:757-767. [DOI] [PubMed] [Google Scholar]
- 76.Taya, S., T. Yamamoto, K. Kano, Y. Kawano, A. Iwamatsu, T. Tsuchiya, K. Tanaka, M. Kanai-Azuma, S. A. Wood, J. S. Mattick, and K. Kaibuchi. 1998. The Ras target AF-6 is a substrate of the fam deubiquitinating enzyme. J. Cell Biol. 142:1053-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thomas, M., and L. Banks. 1998. Inhibition of Bak-induced apoptosis by HPV-18 E6. Oncogene 17:2943-2954. [DOI] [PubMed] [Google Scholar]
- 78.Thomas, M., D. Pim, and L. Banks. 1999. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene 18:7690-7700. [DOI] [PubMed] [Google Scholar]
- 79.Trompouki, E., E. Hatzivassiliou, T. Tsichritzis, H. Farmer, A. Ashworth, and G. Mosialos. 2003. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 424:793-796. [DOI] [PubMed] [Google Scholar]
- 80.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 98:8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vargas, D. A., S. Takahashi, and Z. Ronai. 2003. Mdm2: a regulator of cell growth and death. Adv. Cancer Res. 89:1-34. [DOI] [PubMed] [Google Scholar]
- 82.Verma, R., L. Aravind, R. Oania, W. H. McDonald, J. R. Yates III, E. V. Koonin, and R. J. Deshaies. 2002. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298:611-615. [DOI] [PubMed] [Google Scholar]
- 83.Verma, R., S. Chen, R. Feldman, D. Schieltz, J. Yates, J. Dohmen, and R. J. Deshaies. 2000. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell. 11:3425-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilkinson, K. D. 1997. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 11:1245-1256. [DOI] [PubMed] [Google Scholar]
- 85.Wilkinson, K. D. 2000. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin. Cell Dev. Biol. 11:141-148. [DOI] [PubMed] [Google Scholar]
- 86.Wing, S. S. 2003. Deubiquitinating enzymes—the importance of driving in reverse along the ubiquitin-proteasome pathway. Int. J. Biochem. Cell Biol. 35:590-605. [DOI] [PubMed] [Google Scholar]
- 87.Winston, J. T., P. Strack, P. Beer-Romero, C. Y. Chu, S. J. Elledge, and J. W. Harper. 1999. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 13:270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu, G., G. Xu, B. A. Schulman, P. D. Jeffrey, J. W. Harper, and N. P. Pavletich. 2003. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol. Cell 11:1445-1456. [DOI] [PubMed] [Google Scholar]
- 89.Yao, T., and R. E. Cohen. 2002. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419:403-407. [DOI] [PubMed] [Google Scholar]
- 90.Yu, Z. K., J. L. Gervais, and H. Zhang. 1998. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc. Natl. Acad. Sci. USA 95:11324-11329. [DOI] [PMC free article] [PubMed] [Google Scholar]