Abstract
TATA binding protein (TBP) is a central transcription factor used by all three cellular RNA polymerases. Changes in the levels of TBP have been shown to have selective effects on gene activity. Overexpression of TBP has been recently shown to contribute to cellular transformation, and elevated levels of TBP occur in a clinically significant proportion of human colon tumors relative to matched normal tissue. To understand the mechanisms by which TBP is regulated, we have analyzed whether activation of the epidermal growth factor receptor (EGFR), a membrane-bound tyrosine receptor kinase that is activated in a large number of human cancers, can serve to regulate cellular TBP. We show that treatment of mouse epidermal cells with EGF produces an increase in TBP levels, which can be blocked with an EGFR-specific inhibitor. In contrast, TBP levels remain unchanged after EGF treatment of EGFR null cells. EGF-mediated increases in TBP are regulated at the transcriptional level, as transient expression of the human TBP promoter is induced with EGF. This regulatory event is dependent upon the downstream activation of Ras and requires the activation of p38, JNK, and ERK mitogen-activated protein kinases. The consequence of elevated TBP on gene expression was further determined. Transcription by RNA polymerase (Pol) I and III was induced by EGF. Directly overexpressing TBP also stimulated transcription from these promoters. Thus, we have identified a new and important target of EGFR signaling, TBP, that contributes to EGF-mediated stimulation of RNA Pol I- and III-dependent gene activity. Since the cellular levels of the products of these genes, tRNAs and rRNAs, determine the translational capacity of cells, this event may be an important contributor to the transforming function of EGF.
The TATA binding protein (TBP) is an essential transcription component used by all three cellular RNA polymerases (Pols). TBP associates with distinct sets of proteins (TBP-associated factors) to form at least three complexes, SL1, TFIID, and TFIIIB, that are required for transcription initiation by RNA Pol I, Pol II, and Pol III, respectively (for review, see reference 10). While TBP is central to these transcription processes, evidence indicates that regulation of this protein can have profound effects on gene expression. Depending on the cell type, TBP can be limiting for the transcription of RNA Pol I- and III-dependent genes, and increasing cellular TBP levels have been shown to enhance transcription of these genes in both insect and mammalian cells (24, 28). In contrast, RNA Pol II-dependent genes are differentially affected by alterations in TBP levels (3, 15, 20). The type of promoter and locations of regulatory elements have been shown to influence the responsiveness of a given gene to TBP increases. Thus, changes in TBP levels have a profound, but selective, impact on gene expression.
The phenotypic consequences that result from changes in TBP levels are beginning to be realized. Heterozygous disruption of TBP in chicken B cells was found to produce abnormalities in cell size and delays in cell cycle progression (25). Recently, when cells from the immortalized but nontumorigenic cell line rat 1A were programmed to express increased amounts of TBP, no change in proliferation rate was observed, yet these cells acquired anchorage-independent growth properties and became tumorigenic in athymic mice (11). Mutant TBP proteins, which cannot participate in RNA Pol II-dependent transcription or bind to consensus TATA sequences, were further used to assess their transforming capabilities. The expression of these mutant TBPs failed to transform cells, indicating that TBP-mediated transformation requires changes in RNA Pol II-dependent transcription and the ability of TBP to bind to consensus TATA-containing promoters (11). Thus, modest changes in TBP expression give rise to significant phenotypic consequences, underlining the importance of understanding the mechanisms by which this central transcription factor is regulated.
Our previous studies have shown that TBP levels can be increased by the activation of specific cellular signaling pathways. The activation of protein kinase C by the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (9), expression of a constitutively activated form of Ras (26), and the activation of signaling pathways produced by expression of the hepatitis B virus X protein (28) have all been shown to produce increases in TBP. This increase is produced at the level of transcription, where Ras-mediated induction of the human TBP promoter involves the downstream effectors Raf and RalGDS, both of which require the activation of mitogen-activated protein kinase (MAPK) kinase (MEK) (12). These studies suggest that TBP levels may be regulated by specific cytokines and growth factors that activate Ras signaling cascades.
To begin to delineate the specific growth factors and receptors that serve to regulate TBP, we have examined the potential role for the epidermal growth factor receptor (EGFR) in stimulating TBP expression. The EGFR, which is a tyrosine kinase, mediates many cellular responses in normal biological processes and in pathological states (for review, see references 2 and 13). Phosphorylation of EGFR, in response to ligand binding, provides docking sites for Src-homology 2-containing and phosphotyrosine binding domain-containing proteins. Upon activation of EGFR, enzymes such as Src and phospholipase Cγ, or adaptor proteins such as SHC, are recruited to the membrane and become activated by phosphorylation, linking EGFR to downstream signaling events. An important and well-studied consequence of EGFR activation involves recruitment of the Grb2-Sos complex, which results in the activation of Ras and MAPK pathways. The pathways triggered by this and other EGFR-mediated responses can lead to a variety of integrated biological responses, including proliferation, differentiation, and apoptosis. As enhanced EGFR activity correlates with the development of cancer and other proliferative diseases, it is important to assess the downstream targets of this response that mediate these processes.
Our results demonstrate that EGF enhances TBP expression through the activation of EGFR. EGF-mediated increases in TBP occur, at least in part, through enhanced transcription of the TBP promoter. Induction of the TBP promoter by EGF requires the activation of Ras, as well as the activation of all three classes of MAPKs. The analysis of TBP promoter mutants revealed that a binding site for an Ets transcription factor is required for this response. We further determined the potential consequence of EGF-mediated induction of TBP on gene activity. EGF induces both RNA Pol I- and Pol III-dependent gene activity. Directly increasing expression of TBP, alone, results in enhanced transcription from these promoters. Together, our results reveal that the central transcription factor, TBP, can be regulated through EGFR signaling. Increases in TBP mediated by EGF serve to coregulate transcription of genes that synthesize rRNAs and tRNAs. Thus, it is likely that this event contributes to the growth potential of cells where EGFR is upregulated.
MATERIALS AND METHODS
Plasmids and reagents.
The DNA constructs used were constructed as previously reported and were as follows: human TBP promoter luciferase reporter plasmids p−4500/+66hTBP, p−736/+66, p−736/+66etsmut, p−84/+66hTBP, p−84/−1hTBP, and p−84/−1hTBPetsmut (8); RasAla15 (27) and the tRNAArg gene, pArg-maxi (5); and human rRNA gene reporter (29). EGF was from Sigma-Aldrich; U0126 was from Promega; and AG 1478, SB 202190, SP 600125, and G418 were from Calbiochem. Eagle minimal essential cell culture medium (MEM) was obtained from Cellgro, and OPTI-MEM I was from Invitrogen.
Transfection assays.
For transient-transfection assays, 105 mouse epidermal (JB6) or Huh-7 cells were seeded in 100-mm or 60-mm dishes. Fifteen hours later, cells were transfected using 1 μl of Lipofectin/μg of DNA (USC Liver Tissue Culture Core Facility). Serum-free medium was added to each dish with Lipofectin-DNA complexes, and cells were further incubated at 37°C for 4 to 5 h. The medium was changed and the cells were incubated overnight before harvesting.
For the preparation of total cell lysates from transfected cells for determining TBP promoter activity, the cells were rinsed with Dulbecco’s phosphate-buffered saline, scraped from the plates, and collected by centrifugation. Cell pellets were resuspended in Promega reporter lysis buffer. Cell suspensions were lysed by incubation on ice for 10 min, followed by freezing and thawing. Cell lysates were centrifuged for 20 min at 10,000 × g at 4°C, and the supernatant was collected for protein and luciferase activity measurements. Protein concentrations of the resultant lysates were measured by the Bradford method using the Bio-Rad protein assay reagent. Lysates prepared from transfected cells were analyzed for luciferase activity using a luminometer and the Promega luciferase assay system as described by the manufacturer. Resultant luciferase activities were normalized to the amount of protein in each lysate. For all transient transfections with promoter-luciferase reporter constructs, the fold change in promoter activity was calculated by determining the level of luciferase specific activity in the presence of the empty vector or in the absence of EGF and setting this value at 1 for each independent experiment. Differences in TBP promoter activity are expressed as the mean ± standard error. At least three independent experiments were conducted for each determination.
The stable JB6 cell line expressing the tRNAArg reporter gene was constructed by cotransfecting 5 μg of DNA of the pArg-maxi gene plasmid and 1 μg of pDCR vector plasmid containing a G418 resistance gene. The cells were incubated at 37°C for 2 days, and selective medium containing 200 μg of G418/ml was then added to propagate the cells.
Immunoblot analysis.
JB6 cells were grown to 80% confluency in 5% fetal bovine serum (FBS) in MEM and then serum deprived using 0.1% FBS in MEM for 3 h. Cells were then incubated with EGF for 30 to 60 min at 37°C. Cells were washed with phosphate-buffered saline twice. Lysis buffer (62.5 mM Tris-HCl [pH 6.8], 0.1% Triton X-100, 50 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) was then added at 1 ml of buffer per ml of cell pellet. Cells were incubated on ice for 15 to 20 min and harvested. The cells were sonicated and centrifuged to collect the cell lysates. Protein concentrations were determined by the Bradford method. Lysates (50 μg of protein) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis. Membranes were probed with either rabbit polyclonal anti-human TBP antibody (Upstate Biotechnology), phospho-ERK, phospho-p38 (Cell Signaling), phospho-JNK (Santa Cruz), or mouse monoclonal anti-β-actin (Boehringer Mannheim), as indicated. Hybond-P membrane was used for protein transfer. Bound primary antibody was visualized using horseradish peroxidase-conjugated secondary antibody (Vector Laboratories) and enhanced chemiluminescence reagents (Amersham).
RNase protection assay.
JB6 cells were transfected with the pArg-maxi gene for 48 h and starved in 0.1% FBS-MEM for 3 h. Cells were treated with EGF at 37°C with 5% CO2 for another 30 min. RNase protection assays were carried out as described in the protocol of Ambion, Inc. Briefly, cells were harvested and cell pellets were resuspended with 1 ml of TRIzol reagent (Invitrogen) and incubated at room temperature for 10 min. Chloroform was added (200 μl), and the mixture was incubated at room temperature for 3 min. Suspensions were centrifuged at 18,000 × g for 20 min at 4°C.
The supernatant was mixed with an equal volume of isopropanol, incubated on dry ice for 15 min, and centrifuged at 14,000 rpm for 20 min at 4°C. The pellet was rinsed with 70% ethanol and resuspended in 20 μl of water. For the preparation of probe, the pArg-maxi gene plasmid was digested with XbaI, and labeled RNA probe was synthesized using the MAXIscript T7 kit (Ambion) with [32P]CTP. RNA (1 μg) was hybridized with the labeled RNA probe and incubated at 42°C overnight. The samples were digested by RNase A/T1 at 37°C for 30 min and precipitated. The samples were incubated on dry ice for 15 min and centrifuged at 14,000 rpm for another 15 min. The pellets were resuspended in gel loading buffer (Ambion) and separated on 8% acrylamide-8 M urea denaturing gels. The bands were visualized by autoradiography, and the RNA products were quantified by a phosphorimager.
In vitro transcription assay.
Cell extracts were prepared as previously described (5). The transcription reaction final mixture contained 20 μg of nuclear extract protein, 0.8 μg of tRNAArg gene template, 20 mM HEPES (pH 7.9), 5 mM MgCl2, 3 mM dithiothreitol, 100 mM KCl, 10% glycerol, 0.5 mM each ATP, CTP, and UTP, and 0.1 mM [α-32P]GTP (6 Ci/mmol) in a final reaction volume of 60 μl. Reactions were incubated for 1 h at room temperature and stopped by the addition of 0.1% SDS and 400 μg of proteinase K/ml. After 15 min at 37°C, RNAs were purified by phenol extraction and ethanol precipitation and analyzed by electrophoresis on 8 M urea-8% polyacrylamide gels. Transcription products were visualized by autoradiography, and the resultant bands were quantified by densitometry using a phosphorimager. The values given in the text for the relative levels of transcription stimulation were based on at least three independent experiments in each case.
Primer extension assay.
The primer, 5′-AGAGTTGAGAGGGTACGTACG-3′, was labeled following the protocol of Zhai and Comai (29). Briefly, the oligonucleotides (120 pmol; USC Microchemical Core Facility) were labeled by T4 nucleotide kinase (Promega) in the presence of [γ-32P]ATP (50 mCi/ml; ICN) at 37°C for 45 min. The reaction was terminated by incubation at 65°C for 10 min. The labeled primer was purified using a column (Amersham) following the manufacturer's instruction.
The primer extension assay was performed as described previously (29). Briefly, RNA was extracted using TRIzol (Invitrogen) 48 h after transfection. RNA (5 μg) was hybridized with the 32P-labeled primers (550,000 cpm) at 42°C for 90 min followed by reverse transcription. DNA was synthesized using avian myeloblastosis virus reverse transcriptase (Promega) for 1 h in the presence of actinomycin D (200 mg/ml; Sigma). The reaction was terminated by incubation at 65°C for 10 min, and the RNA was digested by RNase A (2 μg/ml; Ambion) at 37°C for 15 min followed by phenol-chloroform (24:1) extraction. The newly synthesized DNA was precipitated with 100% ethanol. The DNA was visualized by autoradiography after PAGE with 8 M urea.
RESULTS
Activation of EGFR increases cellular levels of TBP by enhancing TBP promoter activity.
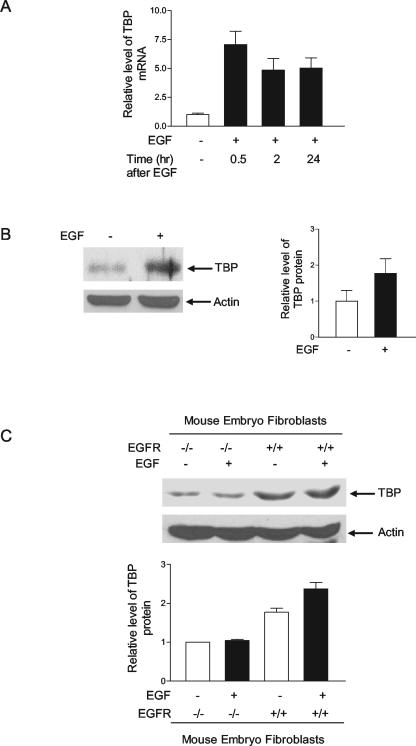
To investigate whether the activation of EGFR regulates TBP expression, mouse epidermal JB6 cells were treated with the EGFR ligand, EGF, for various periods of time. Reverse transcription-PCR analysis was performed to determine the relative levels of TBP mRNA compared to the levels of actin mRNA. As shown in Fig. 1A, treatment of the cells with EGF produced a substantial increase in TBP mRNA after 30 min of EGF treatment, and this response was sustained for at least 24 h. Furthermore, immunoblot analysis showed that TBP levels were similarly increased upon EGF treatment (Fig. 1B). To further determine whether this event specifically required EGFR, matched mouse embryo fibroblast cells that normally express EGFR were compared to cells that lack EGFR. Immunoblot analysis revealed that the endogenous TBP levels were higher in the noninduced EGFR+/+ cells compared to the EGFR−/− cells (Fig. 1C). EGF treatment of these cells resulted in a discernible increase in TBP in the EGFR+/+ cells, but no effect on TBP expression was observed upon EGF treatment of the EGFR−/− cells. These results indicate that activation of EGFR signaling serves to regulate cellular TBP levels.
FIG. 1.
EGF enhances cellular levels of TBP through the activation of EGFR. (A) EGF increases TBP mRNA levels in JB6 cells. Cells were incubated in 0.1% FBS-MEM with 50 ng of EGF/ml for different times as shown. Total RNA was extracted, and reverse transcription-PCR was performed as previously described (11) using primers for TBP and β-actin. The change in TBP mRNA was calculated relative to the levels of β-actin. (B) EGF increases TBP levels in JB6 cells. Cells were incubated with 50 ng of EGF/ml for 30 min as described for panel A. A 100-μg aliquot of protein from the resulting cell lysates was resolved by SDS-PAGE, and TBP levels were determined by immunoblot analysis with antibodies against TBP, as described in Materials and Methods. The same membrane was stripped and reprobed with antibodies against β-actin (left panel). TBP and β-actin levels were quantified by densitometry, and the fold change in TBP was normalized to β-actin (right panel). The data shown are the result of three independent experiments. (C) EGFR and its activation increase cellular TBP levels. Mouse embryo fibroblast cells, EGFR+/+ and EGFR−/−, were treated with 50 ng of EGF/ml for 30 min, and cell lysates were subjected to immunoblot analysis as described in Materials and Methods. The values shown are the means ± standard errors of the means of at least four independent experiments.
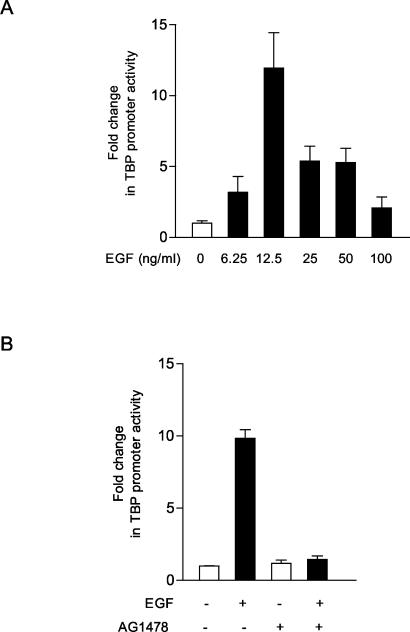
Since an increase in TBP mRNA was observed upon EGF stimulation, we analyzed whether this increase in TBP was mediated at a transcriptional level. A 4.5-kb genomic fragment containing the human TBP promoter linked to a luciferase reporter (−4500/+66hTBP) (8) was transiently expressed in JB6 cells. EGF treatment of these cells resulted in a dose-dependent increase in TBP promoter activity (Fig. 2A). The increase in TBP promoter activity by EGF was dependent on EGFR, as pretreatment of the cells with the EGFR-specific inhibitor AG 1478 served to block EGF-mediated TBP promoter induction (Fig. 2B). Thus, these results indicate that the EGF-induced increase in TBP expression is, at least in part, mediated through activation of TBP promoter activity. The response is highly dose dependent, further indicating that the mechanism for induction of the TBP promoter by EGF is complex.
FIG. 2.
EGF stimulation induces TBP promoter activity. (A) EGF induces TBP promoter activity in a dose-dependent manner. JB6 cells were transiently transfected with 5 μg of the human TBP promoter-luciferase construct p−4500/+66hTBP-luc. Twenty-four hours after transfection, cells were incubated in 0.1% FBS-MEM with various concentrations of EGF overnight, as indicated. Protein lysates were prepared, and luciferase activity measurements were made as described in Materials and Methods. The change in TBP promoter activity was calculated relative to that with no EGF treatment. The results shown are based on at least three independent experiments. (B) The EGFR inhibitor AG 1478 blocks TBP promoter activity stimulated by EGF. Twenty-four hours after the cells were transfected with p−4500/+66hTBP-luc, the cells were pretreated with or without AG 1478 (0.5 μM) for 1 h and then incubated with or without EGF (12.5 ng/ml). The values shown are the means ± standard errors of the means of at least four independent experiments.
EGF-induced stimulation of TBP promoter activity requires the activation of Ras and ERK, p38, and JNK MAPKs.
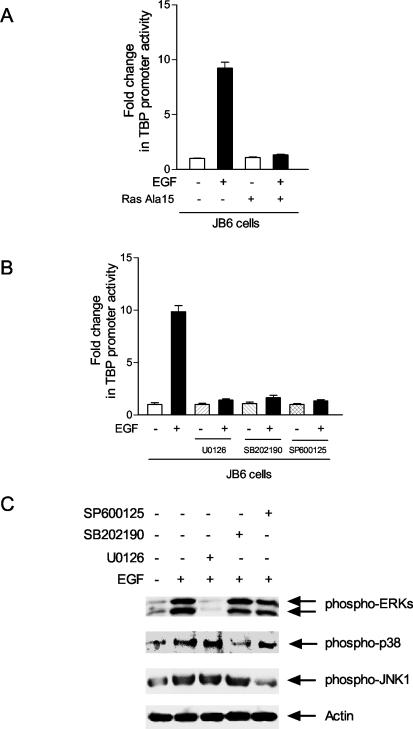
The activation of EGFR by EGF elicits a large cascade of signal transduction events. To define the specific pathways that contribute to the EGF-mediated increase in TBP expression observed, the requirement for Ras activation was first examined. JB6 cells were transiently cotransfected with the TBP promoter-reporter together with a vector that expresses a dominant negative form of Ras, Ras-Ala15. While expression of Ras-Ala15 had no effect on TBP promoter activity in untreated JB6 cells, the expression of Ras-Ala15 eliminated EGF-mediated induction of the promoter (Fig. 3A).
FIG. 3.
EGF-induced stimulation of TBP promoter activity requires the activation of Ras and the three classes of MAPKs. (A) Expression of a dominant negative mutant Ras (RasAla15) inhibits EGF induction of TBP promoter activity. JB6 cells were transiently cotransfected with the TBP promoter construct p−4500/+66hTBP-luc (as described in the legend for Fig. 2A) and 2 μg of the RasA15 expression plasmid or empty vector. The change in TBP promoter activity was determined relative to that with no EGF treatment and in the absence of RasA15 expression. (B) Inhibitors of p38, JNK, and ERK1/2 MAPKs each block EGF stimulation of TBP promoter activity. Cells transfected with p−4500/+66hTBP-luc, as described in the legend for Fig. 2A, were pretreated with U0126 (10 μM), SB202190 (1 μM), or SP600125 (2.5 μM) for 1 h and then incubated with EGF (12.5 ng/ml) for 30 min. The values shown are the means ± the standard errors of the means of at least four independent experiments. (C) Kinase inhibitors are selective for blocking the activation of each of the MAPKs. Cells were pretreated with inhibitors as described for panel B and then incubated with or without EGF (12.5 ng/ml) for 30 min as indicated. Immunoblot analysis of the resultant cell lysates was carried out using specific anti-phospho-MAPKs, as indicated.
The activation of Ras by EGF has been shown to lead to the activation of a variety of MAPKs. These MAPKs fall into three main classes, which include ERK1/2, p38, and JNK (for review, see reference 14). To determine the downstream Ras-activated signaling events that contributed to EGF promoter induction, two approaches were used to assess the potential involvement of the MAPKs. First, selective inhibitors were used to block the activation of the three classes of MAPKs. U0126 is a specific inhibitor for the MAPK kinase (MEK) that inhibits downstream ERK1/2 activation (6), SB202190 is a specific inhibitor for p38 kinases (22), and SP600125 is a specific inhibitor for JNKs (1). Treatment of the JB6 cells with each of the inhibitors had no effect on TBP promoter activity in the absence of EGF (Fig. 3B). However, cotreatment of the cells with EGF and each of the MAPK inhibitors significantly reduced EGF-dependent TBP promoter induction. To ensure that these inhibitors were acting selectively on their MAPK targets, immunoblot analysis was used to determine the amounts of the phosphorylated activated forms of ERK, p38, and JNK upon inhibitor treatment. As shown in Fig. 3C, under the conditions used, each inhibitor selectively inhibited its target without altering the activation of the other MAPKs.
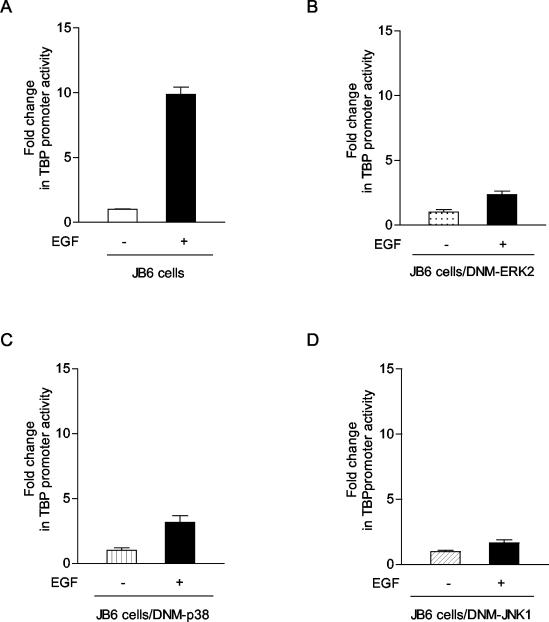
To further ascertain the requirement for each of the MAPKs in EGF-mediated TBP promoter induction, JB6-derived cell lines were used that stably express dominant negative forms of each of the three MAPKs (30). These cell lines have been previously shown to selectively block the activation of each of the MAPKs. The parental JB6 cell line exhibited an approximate 10-fold increase in TBP promoter activity upon EGF treatment (Fig. 4A). However, when the TBP promoter was transiently expressed in the JB6-derived cell lines containing one of the dominant negative forms of ERK2, p38, or JNK, the enhancement of TBP promoter activity observed upon EGF treatment was greatly diminished (Fig. 4B to D). Together, these results support that activation of Ras and all three of the MAPKs are required for induction of the TBP promoter by EGF.
FIG. 4.
Expression of dominant negative forms of ERK2, p38, or JNK1 MAPKs each inhibits EGF induction of TBP promoter activity. Stable cell lines were derived from parental JB6 cells using dominant negative mutants (DNM) of ERK2, p38, or JNK1 (30). Each cell line was transiently transfected with p−4500/+66hTBP-luc (described in the legend for Fig. 2A). The change in TBP promoter activity with EGF treatment is shown. The values shown are the means ± standard errors of the means of at least four independent experiments.
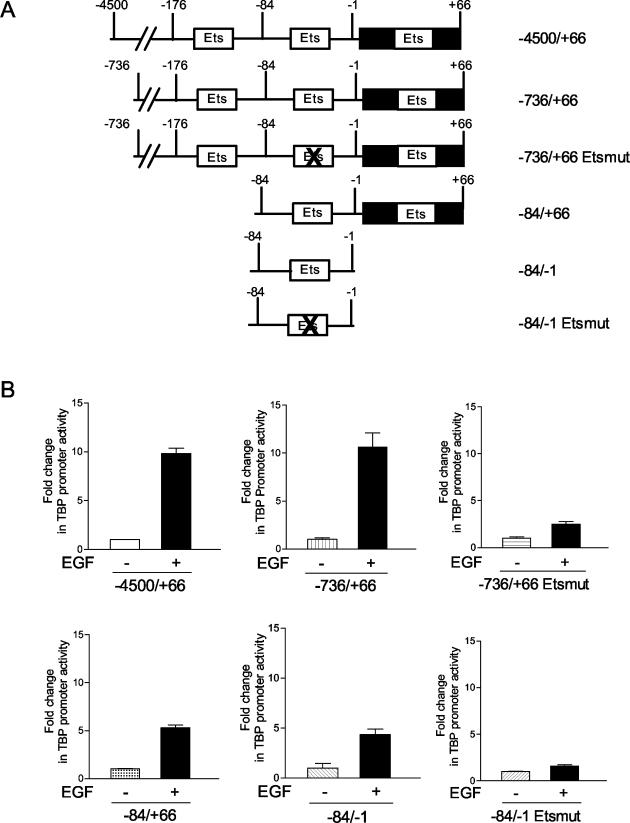
The results above support the idea that the activation of the three classes of MAPKs, together, is required for induction of TBP promoter activity. It is possible that the individual MAPK signaling processes converge on a specific transcription component target(s) that acts in concert to induce the TBP promoter. If this is the case, the EGF-mediated TBP transcription induction should be confined to a relatively small region of the promoter. We therefore determined which regions within the human TBP promoter were required for EGF-mediated induction. Three deletion mutants were examined and compared to the −4500/+66 TBP promoter construct. EGF inducibility of the −736/+66 promoter construct was indistinguishable from that of the −4500/+66 promoter. Analysis of the −84/+66 and −84/−1 TBP promoter constructs, which resulted in the removal of consensus Ets binding sites, displayed an approximate twofold reduction on the ability of the promoter to be induced by EGF, compared to that with the −4500/+66 promoter construct (Fig. 5). Further mutation of the Ets binding site within the −84/−1 fragment abolished EGF-mediated inducibility (Fig. 5). However, mutation at this site within the context of the −84/−1 fragment also resulted in a 20-fold reduction in basal promoter activity (data not shown). In contrast, mutation at this Ets site in the context of the −736/+66 promoter did not alter basal promoter activity (data not shown), yet its ability to be induced upon EGF stimulation was dramatically reduced (Fig. 5). Thus, this Ets binding site between −84/−1 plays a crucial role in EGF-mediated stimulation of the TBP promoter.
FIG. 5.
Analysis of sequences required for EGF induction of the human TBP promoter. JB6 cells were transiently transfected with one of the following hTBP promoter constructs: p−4500/+66, p−736/+66, p−736/+66 etsmut, p−84/+66, p−84/−1, and p−84/−1 etsmut as indicated. Cells in 0.1% FBS-MEM were treated with EGF (12.5 ng/ml) overnight, lysates were prepared, and the resultant luciferase activities were measured as described in Materials and Methods. The values shown are the means ± standard errors of the means of at least four independent experiments.
An EGF-mediated increase in TBP induces RNA Pol I- and Pol III-dependent gene transcription.
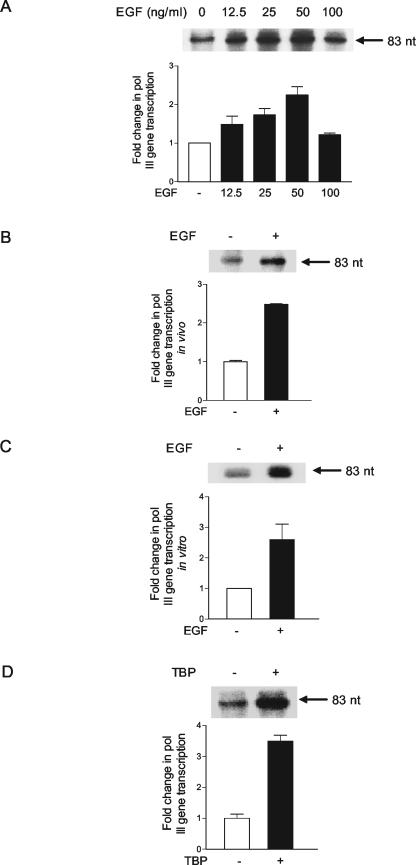
The above results revealed that EGF stimulates increased expression of TBP. In order to understand how this regulatory event might impact cellular gene activity, we examined whether EGF, and the resultant enhanced expression of TBP, was capable of inducing RNA Pol I- and RNA Pol III-dependent gene activity. JB6 cells were transiently transfected with a tRNAArg gene reporter plasmid to monitor RNA Pol III-dependent gene activity. As shown in Fig. 6A, a dose-dependent increase in RNA Pol III-dependent gene activity was observed upon EGF treatment. A JB6-derived cell line that contains the stably integrated tRNAArg gene displayed similar results whereby an approximate 2.5-fold increase in transcription was observed upon EGF treatment (Fig. 6B). To determine whether this regulatory event could also be recapitulated in vitro, extracts were prepared from cells that were incubated in the presence or absence of EGF. The use of several different extract preparations and in vitro transcription assays revealed that the EGF-treated cell extracts displayed a reproducible increase in tRNA gene transcription (Fig. 6C). These results indicate that EGF activates RNA Pol III-dependent gene activity. The ability to reproduce this regulatory event in vitro further indicates that it is the components of the transcription machinery that are targeted in this response. To further determine if EGF-mediated increases in TBP levels could contribute to the induction of tRNA gene activity, JB6 cells were transiently transfected with the tRNAArg gene together with an expression plasmid for TBP. Immunoblot analysis revealed that transfection of the TBP expression plasmid resulted in a less-than-twofold increase in TBP levels (data not shown). However, this increase in TBP produced an approximate 3.5-fold enhancement of tRNA gene activity (Fig. 6D). Together, these results support that EGF stimulates RNA Pol III-dependent gene activity, and at least one EGF-mediated event that drives this process is an increase in cellular TBP levels.
FIG. 6.
EGF or the overexpression of TBP induces RNA Pol III-dependent gene transcription. (A) Transient expression of a tRNA gene is increased by EGF stimulation of JB6 cells. Cells (106) were transfected with 10 μg of the tRNA gene reporter-containing plasmid, pArg-maxi. The cells were treated with increasing amounts of EGF as indicated for 30 min. RNA was extracted, and RNase protection assays were carried out with equal amounts (1 μg) of RNA and a 32P-labeled antisense riboprobe as described in Materials and Methods. The resultant labeled RNAs were separated by electrophoresis and visualized by autoradiography. An example of an autoradiogram is shown in the upper panel, and quantification of three independent experiments is shown in the lower panel. (B) Induction of a stably integrated tRNA gene in JB6 cells by EGF. The stable cell line containing the tRNAArg reporter gene, pArg-maxi, was treated with EGF (50 ng/ml), and an RNase protection assay was carried out as described in Materials and Methods. (C) EGF induction of RNA Pol III-dependent gene transcription can be reproduced in vitro. Nuclear extracts were preparedfrom JB6 cells that had been treated with or without 50 ng of EGF/ml for 30 min, and transcription assays were carried out using 0.4 μg of the tRNAArg gene as the template and 30 μg of nuclear extract. (D) Enhanced expression of TBP in JB6 cells induces transient expression of a tRNA gene. JB6 cells were transiently transfected with 10 μg of pArg-maxi together with 10 μg of an expression vector containing hemagglutinin (HA) epitope-tagged human TBP cDNA or empty vector. RNA was isolated from the cells, and RNase protection assays were carried out as described in Materials and Methods. A resultant autoradiograph is shown in the left panel. Values shown are the means ± standard errors of the means of at least three independent determinations. Cell lysates were prepared from a portion of transfected cells, and immunoblot analysis was carried out as described in Materials and Methods to detect the HA-tagged TBP product (designated HA-TBP) and the endogenous TBP (designated TBP).
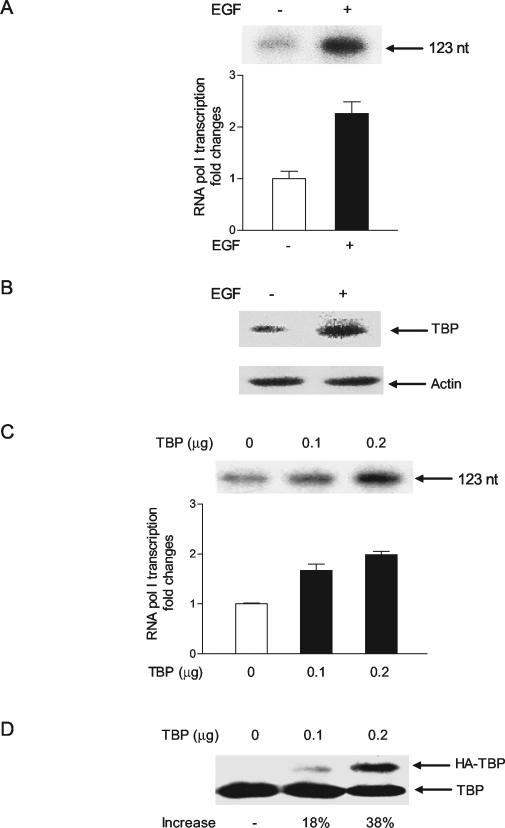
To determine whether EGF could similarly mediate an increase in RNA Pol I-dependent gene activity, the human hepatoma Huh-7 cells were transfected with a human rRNA promoter. EGF treatment of the cells produced an approximate twofold increase in RNA Pol I-dependent promoter activity (Fig. 7A). Immunoblot analysis revealed that EGF treatment of the Huh-7 cells produced an increase in TBP amounts (Fig. 7B), similar to that observed for the JB6 cells (Fig. 1B). Directly increasing TBP expression in these cells also produced a dose-dependent increase in rRNA promoter activity (Fig. 7C). Immunoblot analysis revealed that an approximate 40% increase in TBP expression was capable of producing a twofold increase in RNA Pol I-dependent promoter activity. Thus, RNA Pol I-dependent promoter activity can be stimulated by EGF, and the increases in TBP resulting from this event contribute to the enhanced gene activity observed.
FIG. 7.
EGF stimulation or increased expression of TBP induces RNA Pol I-dependent gene transcription. (A) EGF induces the transcription of an RNA Pol I-dependent promoter in Huh-7 cells. Cells (105) were transiently transfected with 1 μg of a human RNA Pol I-dependent promoter-reporter plasmid as described in Materials and Methods. Twenty-four hours after transfection a portion of the cells was treated with EGF (50 ng/ml) for 30 min. A primer extension assay was used to determine the amount of transcript produced, as described in Materials and Methods. An example of an autoradiogram and quantification of three independent experiments are shown (left panel). The values shown are the means ± standard errors of the means. (B) EFG stimulation of Huh-7 cells produces an increase in TBP concentration. Immunoblot analysis was performed using lysates derived from cells treated with EGF (50 ng/ml) for 30 min (+) or without treatment (−) using antibodies against TBP or actin, as indicated. (C) RNA Pol I-dependent transcription is induced by enhanced expression of TBP. Huh-7 cells were cotransfected with the human RNA Pol I reporter plasmid and different amounts of an hemagglutinin (HA)-tagged TBP expression plasmid as indicated. Primer extension assays were used to determine the amount of transcript produced, and an example of an autoradiogram and quantification of three independent experiments are shown at the top. (D) TBP concentrations are increased by transient transfection of a TBP expression plasmid. Huh-7 cells were cotransfected with the human RNA Pol I reporter plasmid and different amounts of an HA-tagged TBP expression plasmid as indicated. Immunoblot analysis was used to detect the ectopically expressed HA-TBP and endogenous TBP in the transfected cells.
DISCUSSION
Our previous work has shown that ectopic expression of an oncogenic form of Ras is capable of increasing TBP expression, suggesting that receptor tyrosine kinases, which normally mediate Ras signaling, could serve to regulate TBP. Our present studies demonstrate that EGF induces TBP expression in several cell types. This response is rapid, and changes in both TBP mRNA and protein levels can be seen as early as 30 min after exposure of cells to EGF. Analysis of TBP expression in mouse embryo fibroblast EGFR−/− cells revealed that the basal level of TBP is reduced in these cells compared to that in EGFR+/+ cells and that EGFR expression is required for EGF-mediated induction of TBP. These results indicate that at least one member of the EGFR family is capable of regulating cellular TBP amounts. The increase in TBP is mediated, at least in part, at a transcriptional level.
EGFR-induced signaling has been shown to activate ERK1/2 (19) as well as p38 and JNK (16). Using both specific chemical inhibitors as well as dominant negative mutant proteins to selectively block each class of MAPKs, our results indicate that EGF-mediated TBP promoter induction requires the activation of all three classes of MAPKs. This is consistent with our group's previous studies that showed that induction of TBP promoter activity by expression of a constitutively activated form of Ras can be blocked by an ERK1/2 inhibitor (12). Our present studies support that the other two families of MAPKs, p38 and JNK, which are known to be stress inducible, can also regulate TBP expression. Further analysis of the DNA sequences required for promoter induction revealed that an Ets site within 84 bp flanking the gene is essential for EGF stimulation. The Ets family of transcription components has been shown to be direct targets of the MAPKs (17). Interestingly, at least one member, Elk-1, is phosphorylated and activated by all three MAPK cascades (18, 23). Thus, it is possible that EGF-mediated induction of these MAPKs converges on a specific Ets protein that serves to regulate both basal TBP expression as well as its induction in response to specific growth factors.
Activating mutations as well as the overexpression of EGFR have been found to be frequently associated with human malignancies, and therapeutic strategies to inhibit EGFR signaling result in antitumor effects (2). Thus, a considerable effort is being made to develop pharmacological agents that inhibit EGFR dysfunction in human cancer. As EGFR transduces a complex array of signaling cascades, elucidating the targets of this response is essential for understanding its role in promoting oncogenesis. Our studies reveal a new target regulated through EGFR signaling. As changes in the cellular amounts of TBP have been shown to produce specific alterations in transcription (3, 15, 20), the question arises as to whether the EGFR-mediated regulation of TBP levels contributes to its ability to promote oncogenic transformation. Our recent results indicate that this may indeed be the case. There are several lines of evidence supporting that directly modulating TBP levels contributes to the transformation state of cells. Overexpression of TBP in rat1A cells was found to promote anchorage-independent growth properties and the formation of tumors in athymic mice (11). Inhibiting the ability of Ras to enhance expression of TBP in NIH 3T3 cells inhibited Ras transforming function. Initial analysis of human colon epithelium, comparing TBP expression in tumor and normal cells, revealed that increases in TBP mRNA are observed in a significant proportion of human colon tumors (11). Thus, it is likely that the EGFR-mediated signaling events that serve to increase TBP contribute to its oncogenic effects. Importantly, modest (less-than-twofold) changes in TBP levels are sufficient for producing these phenotypic alterations (11). This is consistent with our present studies, whereby EGF treatment of JB6, Huh-7, and mouse embryo fibroblast cells produced small changes in TBP levels, yet these changes were sufficient to drive enhanced transcription of RNA Pol I- and III-dependent promoters. Consistent with these results, heterozygous disruption of a TBP allele in chicken DT cells, which decreases TBP concentrations by 50%, produced abnormalities in cell size and delayed mitosis (25).
We have examined how the activation of EGFR and the resultant increase in TBP levels may alter transcription. We considered that RNA Pol I- and Pol III-dependent transcription could potentially be regulated through EGF induction. As the levels of the major products of these genes, rRNAs and tRNAs, determine the translational capacity of cells, changes in RNA Pol I- and III-dependent transcription have been correlated to the proliferative state of cells. Enhanced activity of EGFR leads to increases in cell growth and proliferation, suggesting that enhanced synthesis of rRNAs and tRNAs could contribute to this response. Our results show that both RNA Pol I- and Pol III-dependent transcription are induced upon EGF stimulation. Further comparison of the level of EGF-mediated induction of a stably integrated tRNA gene to that of a transiently expressed tRNA gene showed that there was no discernible difference. In addition, comparing the transcriptional capacities of extracts prepared from cells incubated with or without EGF revealed an increase in tRNA gene transcription upon EGF stimulation. Together, these results suggest that alterations in chromatin structure are not required for mediating transcription induction and that the transcription components are the targets of this response.
Our previous studies have shown that TBP can be limiting for RNA Pol I- and Pol III-dependent transcription. To determine if the EGF-mediated increase in TBP was contributing to the enhanced RNA Pol I and Pol III gene activity observed, TBP was overexpressed in cells. Our results revealed that this increase in TBP, alone, was capable of stimulating transcription. These results support that one mechanism by which EGF mediates induction of RNA Pol I- and Pol III-dependent transcription is through increases in TBP. As TBP is the one transcription component so far shared by both transcription systems, modulation of TBP levels would serve as a mechanism for the coregulation of RNA Pol I- and Pol III-dependent genes. In this manner, the cell could coordinate the levels of the large rRNAs and 5S RNA needed for ribosome biosynthesis. While we find that TBP is limiting for the transcription of rRNA and tRNA gene promoters in the JB6 and Huh-7 cell lines used, the extent to which enhanced TBP expression contributes to EGF-mediated induction of these genes will likely be cell type dependent. For example, our laboratory has recently shown that RNA Pol III-dependent transcription is not affected by increasing TBP levels in the lung carcinoma H1299 cell line, which does not express functional p53 (4). However, when these cells are programmed to express p53 and the levels of TBP are increased, RNA Pol III-dependent transcription is selectively augmented. Thus, the genetic background of the cell may determine how EGF-mediated increases in TBP affect cellular gene expression.
Given the pleiotropic nature of EGFR-mediated signaling, it would seem likely that other proteins in the RNA Pol I and Pol III transcription complexes, in addition to TBP, might be targeted in this response. Consistent with this idea, ERK activation was previously shown to increase rRNA gene transcription, and ERK was shown to directly phosphorylate the RNA Pol I-specific transcription factor, UBF (21). In contrast, another study showed that stimulation of rRNA synthesis by growth factors was shown to require ERK activation and phosphorylation of the TIF-1A component by ERK and RSK, while no change in UBF phosphorylation state was observed (31). Recently, serum stimulation and resultant activation of ERK was shown to induce RNA Pol III-dependent transcription (7). In this case, the RNA Pol III-specific transcription factor, BRF1, was found to be phosphorylated by ERK. Together, these studies support that the activation of ERK results in the induction of both RNA Pol I- and Pol III-dependent transcription. While it is possible that the cell signaling events and downstream targets within the RNA Pol I and Pol III transcription components could be dependent upon the cell type or specific conditions used, these results collectively support that alterations in more than one transcription factor likely contribute to the changes in gene expression observed. Our present study identifies an additional target, TBP, whose increase in EGF-stimulated cells, alone, can contribute to enhanced RNA Pol I and Pol III transcription. These results further suggest that the activation of other classes of MAPKs, p38 and JNK, can also contribute to enhanced RNA Pol I- and Pol III-dependent transcription.
Our results support that EGF can coregulate an increase in RNA Pol I- and Pol III-dependent transcription through its ability to increase TBP levels. Previous studies have shown that alterations in TBP amounts differentially affect the transcription of RNA Pol II-dependent promoters and that the composition and location of regulatory elements will influence the extent to which the transcription of a particular gene may be changed (3, 15, 20). It is possible that EGF-mediated changes in TBP levels also contribute to qualitative and quantitative changes in RNA Pol II-dependent transcription. In this manner, modulation of TBP levels by EGFR signaling could be used to coregulate critical protein-encoding genes also linked to cell growth and proliferation.
Acknowledgments
We thank Sandra Johnson and our colleagues at the USC Norris Comprehensive Cancer Center for many helpful discussions. We thank Diane Hawley for the TBP promoter constructs and Z. Dong for providing the JB6 cell lines.
This work was supported in part by NIH grant CA74138 to D. L. Johnson and by the USC Cancer Center support grant 5P30CA14089.
REFERENCES
- 1.Bennett, B. L., D. T. Sasaki, B. W. Murray, E. C. O'Leary, S. T. Sakata, W. Xu, J. C. Leisten, A. Motiwala, S. Pierce, Y. Satoh, S. S. Bhagwat, A. M. Manning, and D. W. Anderson. 1998. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA 98:13681-13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blume-Jensen, P., and T. Hunter. 2001. Oncogenic kinase signaling. Nature 411:355-365. [DOI] [PubMed] [Google Scholar]
- 3.Colgan, J., and J. L. Manley. 1992. TFIID can be rate limiting in vivo for TATA-containing but not TATA-lacking, RNA polymerase II promoters. Genes Dev. 6:304-315. [DOI] [PubMed] [Google Scholar]
- 4.Crighton, D., A. Woiwode, C. Zheng, N. Mandavia, J. P. Morton, L. J. Warnock, J. Milner, R. J. White, and D. L. Johnson. 2003. p53 represses RNA polymerase III transcription by targeting the TATA binding protein and inhibiting promoter occupancy by TFIIIB. EMBO J. 22:2810-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dingermann, T. S., S. Sharp, B. Appel, D. DeFranco, S. Mount, R. Heiermann, O. Pongs, and D. Söll. 1981. Transcription of cloned tRNA and 5S RNA genes in a Drosophila cell free extract. Nucleic Acids Res. 9:3907-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Favata, M. G., K. Y. Horiuchi, E. J. Manos, A. J. Daulerio, D. A. Stradley, W. S. Feeser, D. E. Van Dyk, W. J. Pitts, R. A. Earl, F. Hobbs, R. A. Copeland, R. L. Magolda, P. A. Scherle, and J. M. Trzaskos. 1998. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 273:18623-18632. [DOI] [PubMed] [Google Scholar]
- 7.Felton-Edkins, Z. A., J. A. Fairley, E. L. Graham, I. M. Johnston, R. J. White, and P. H. Scott. 2003. The mitogen-activated protein (MAP) kinase ERK induces tRNA synthesis by phosphorylating TFIIIB. EMBO J. 22:2422-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foulds, C. E., and D. K. Hawley. 1997. Analysis of the human TATA binding protein promoter and identification of an Ets site critical for activity. Nucleic Acids Res. 25:2485-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garber, M., A. Vilalta, and D. L. Johnson. 1994. Induction of Drosophila RNA polymerase III gene expression by the phorbol ester, TPA, is mediated by TFIIIB. Mol. Cell. Biol. 14:339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez, N. 1993. TBP, a universal eukaryotic transcription factor? Genes Dev. 7:1291-1308. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, S. S., L. Dubeau, M. Kawalek, A. Dervan, A. Schonthal, C. V. Dang, and D. L. Johnson. 2003. Overexpression of the TATA-binding protein contributes to oncogenesis. Mol. Cell. Biol. 23:3043-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, S. A. S., N. Mandavia, H.-D. Wang, and D. L. Johnson. 2000. Transcriptional regulation of the human TATA-binding protein by Ras cellular signaling. Mol. Cell. Biol. 20:5000-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jorissen, R. N., F. Walker, N. Pouliot, T. P. J. Garrett, C. W. Ward, and A. W. Burgess. 2003. Epidermal growth factor receptor: mechanisms of activation and signaling. Exp. Cell Res. 284:31-53. [DOI] [PubMed] [Google Scholar]
- 14.Liebmann, C. 2001. Regulation of MAP kinase activity by peptide receptor signaling pathway: paradigms of multiplicity. Cell. Signal. 13:777-785. [DOI] [PubMed] [Google Scholar]
- 15.Majello, B., G. Napolitano, P. De Luca, and L. Lania. 1998. Recruitment of human TBP selectively activates RNA polymerase II TATA-dependent promoters. J. Biol. Chem. 273:16509-16516. [DOI] [PubMed] [Google Scholar]
- 16.Minden, A., and M. Karin. 1997. Regulation and function of the JNK subgroup of MAP kinases. Biochim. Biophys. Acta 24:F85-F104. [DOI] [PubMed] [Google Scholar]
- 17.Oikawa, T., and T. Yamada. 2003. Molecular biology of the Ets family of transcription factors. Gene 303:11-34. [DOI] [PubMed] [Google Scholar]
- 18.Price, M. A., F. H. Cruzalegui, and R. Treisman. 1996. The p38 and ERK MAP kinase pathways cooperate to activate ternary complex factors and c-fos transcription in response to UV light. EMBO J. 15:6552-6563. [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson, M. J., and M. H. Cobb. 1997. Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 9:180-186. [DOI] [PubMed] [Google Scholar]
- 20.Sadovsky, Y., P. Webb, G. Lopez, J. D. Baxter, P. M. Fitzpatrick, E. Gizang-Ginsberg, V. Cavailles, M. Parker, and P. J. Kushner. 1995. Transcriptional activators differ in their responses to overexpression of TATA-box-binding protein. Mol. Cell. Biol. 15:1554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stefanovsky, V. Y., G. Pettetier, R. Hannan, T. Gagnon-Kugler, L. I. Rothblum, and T. Moss. 2001. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell 8:1063-1073. [DOI] [PubMed] [Google Scholar]
- 22.Tan, Y., J. Rouse, A. H. Zhang, S. Cariati, P. Cohen, and M. J. Comb. 1996. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 15:4629-4642. [PMC free article] [PubMed] [Google Scholar]
- 23.Treisman, R. 1996. Regulation of transcription by MAP kinase cascades. Curr. Opin. Cell Biol. 8:205-215. [DOI] [PubMed] [Google Scholar]
- 24.Trivedi, A., A. Vilalta, S. Gopalan, and D. L. Johnson. 1996. TATA-binding protein is limiting for both TATA-containing and TATA-lacking RNA polymerase III promoters in Drosophila cells. Mol. Cell. Biol. 16:6909-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Um, M., J. Yamauchi, S. Kato, and J. L. Manley. 2001. Heterozygous disruption of the TATA-binding protein gene in DT40 cells reduces cdc25B phosphatase expression and delayed mitosis. Mol. Cell. Biol. 21:2435-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, H.-D., A. Trivedi, and D. L. Johnson. 1997. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signaling pathway. Mol. Cell. Biol. 17:6838-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, H.-D., A. Trivedi, and D. L. Johnson. 1998. Regulation of RNA polymerase I-dependent promoters by the hepatitis B virus X protein, activated Ras, and the TATA-binding protein. Mol. Cell. Biol. 18:7086-7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, H.-D., C.-H. Yuh, C. V. Dang, and D. L. Johnson. 1995. The hepatitis B virus increases the cellular level of TATA-binding protein which mediates transactivation of RNA polymerase III genes. Mol. Cell. Biol. 15:6720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhai, W., and L. Comai. 2000. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell. Biol. 20:5930-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, Y. G., S. P. Zhong, Z. M. Dong, N. Y. Chen, A. M. Bode, W. Y. Ma, and Z. Dong. 2001. UVA induces Ser381 phosphorylation of p90RSK/MAPKAP-K1 via ERK and JNK pathways. J. Biol. Chem. 276:14572-14580. [DOI] [PubMed] [Google Scholar]
- 31.Zhao, J., X. Yuan, M. Frödin, and I. Grummt. 2003. ERK-dependent phosphorylation of the transcription initiation factor TIF-1A is required for RNA polymerase I transcription and cell growth. Mol. Cell 11:405-413. [DOI] [PubMed] [Google Scholar]