Abstract
Pigment epithelium-derived factor (PEDF), a member of the serine protease inhibitor superfamily, has potent anti-metastatic effects in cutaneous melanoma through its direct actions on endothelial and melanoma cells. Here we show that PEDF expression positively correlates with microphthalmia-associated transcription factor (MITF) in melanoma cell lines and human samples. High PEDF and MITF expression is characteristic of low aggressive melanomas classified according to molecular and pathological criteria, whereas both factors are decreased in senescent melanocytes and naevi. Importantly, MITF silencing down-regulates PEDF expression in melanoma cell lines and primary melanocytes, suggesting that the correlation in the expression reflects a causal relationship. In agreement, analysis of Chromatin immunoprecipitation coupled to high throughput sequencing (ChIP-seq) data sets revealed three MITF binding regions within the first intron of SERPINF1, and reporter assays demonstrated that the binding of MITF to these regions is sufficient to drive transcription. Finally, we demonstrate that exogenous PEDF expression efficiently halts in vitro migration and invasion, as well as in vivo dissemination of melanoma cells induced by MITF silencing. In summary, these results identify PEDF as a novel transcriptional target of MITF and support a relevant functional role for the MITF-PEDF axis in the biology of melanoma.
Keywords: Abbreviations; PEDF, pigment epithelium-derived factor; MITF, microphthalmia-associated transcription factor; RGP, radial growth phase of melanoma; VGP, vertical growth phase of melanoma; CM, cutaneous metastasis of melanoma; VM, visceral metastasis of melanoma; BRAF, v-raf murine sarcoma viral oncogene homolog B; NRAS, neuroblastoma RAS viral (v-ras) oncogene homolog; OIS, oncogene induced senescence; hnRNA, heterogeneous nuclear RNA
Introduction
Pigment epithelium-derived factor (PEDF), a member of the serine protease inhibitor (SERPIN) superfamily, is a 50 kDa secreted glycoprotein that displays multiple biological activities relevant for cancer biology [1], [2]. PEDF was first described as a pro-differentiation and survival factor in neuronal cells [3] and later was identified as the most potent endogenous inhibitor of angiogenesis in the eye [4]. Evidence accumulated over the last decade consolidated its role as a general anti-angiogenic factor in solid tumors [1], [2]. Additionally, PEDF impinges on proliferation, survival, migration and differentiation of a broad spectrum of cancer cell types [1], [2], [5].
We have previously reported that PEDF has potent anti-tumor and anti-metastatic activities in mouse models of melanoma as a consequence of its combined actions: 1) in the tumor microvasculature; hindering melanoma neovascularization through direct actions on endothelial cells and indirect actions modulating the angiogenic profile of melanoma cells; 2) in the melanoma cells; restricting their survival, migratory and invasive capabilities [5], [6]. Using a combination of gene expression profiling analysis and functional assays we have also demonstrated that loss of PEDF expression enables melanoma cells to acquire a migratory and invasive phenotype and consequently it contributes to the metastatic spread of melanoma [7].
Despite the demonstrated relevance of PEDF in melanoma biology, the mechanisms underlying the variations of PEDF levels during malignant progression of melanoma are still largely unknown. In this regard we have recently shown that hypoxia, a condition frequently found in solid tumors and associated to malignant progression, negatively regulates PEDF expression in melanocytes and melanoma cells [8].
Here, we aimed to investigate the regulatory mechanisms controlling PEDF expression in the context of melanoma malignization and melanocyte senescence.
Microphthalmia-associated transcription factor (MITF) has been established as a master regulator of melanocyte and melanoma biology [9], [10], [11]. MITF functions as a rheostat to determine different biological responses in melanocytic cells depending on the level of MITF expression and its post-translational regulation [12], [13]. Very low MITF levels lead to G1 arrested cells with invasive and stem-like properties [12], [14]. Moderate levels of MITF are required for melanocytic cell proliferation [12]. By contrast, elevated MITF levels drive the expression of lineage specific differentiation genes like TYR [9]. In the context of melanoma progression, decreased MITF levels lead to acquisition of invasive properties; whereas high MITF levels favor less aggressive melanoma cells likely more sensitive to therapeutic strategies [15].
The striking similarities in the multiple functional effects described for PEDF and MITF in melanocytic cells prompted us to investigate whether a regulatory and functional link between MITF and PEDF is operative in melanocytic cells.
Herein, we describe that the expression of PEDF and MITF positively correlates and varies with the pathological and molecular staging of melanoma and in the context of melanocyte senescence. Using a combination of approaches we demonstrate that MITF is a direct positive regulator of PEDF expression in melanocytic cells. And finally, we identify PEDF as a novel functional mediator of MITF in the control of melanoma cell migration, invasion and metastatic dissemination.
Materials and Methods
Cell Lines and Cell Culture
Human melanoma cell line 501mel was provided by C. Bertolotto (Institute de Génétique et de Biologie Moléculaire et Cellulair, Illkirch, France) and cultured as described [16]. WM278, WM164, WM88 and 1205Lu melanoma cell lines were provided by M. Herlyn (The Wistar Institute, Philadelphia, PA, USA) and cultured as described previously [17]. M000921, M080306 and M010308 melanoma cell lines were provided by K. Hoek (University Hospital of Zürich, Zürich, Switzerland) and cultured as described [18]. MaMel69, MaMel82 and MaMel26a1 melanoma cell lines provided by D. Schadendorf (Skin Cancer Unit of the Dermatology Department, University Hospital, University Duisburg-Essen, Germany) were cultured in RPMI (Gibco, Carlsbad, CA, USA) medium supplemented with 10% FBS. HEK293T cell line was cultured in DMEM (Gibco) medium supplemented with 10% FBS. Primary human melanocytes (NHEM) were obtained from Lonza (Basel, Switzweland) and cultured in MBM-4 medium with MGM-4 supplements (Lonza).
Melanoma cell lines were classified as weakly or highly aggressive according to the molecular profiling analysis by K. Hoek and collaborators [19]. Molecular profiling classification was validated by functional assays in representative cell lines from the weakly aggressive and highly aggressive cohorts [7], [18].
Western Blot
Whole-cell extracts were prepared by lysing the cells in 125 mM Tris-HCl pH 7.5 and 2% SDS buffer containing protease and phosphatase inhibitors (10 μg/ml leupeptin; 10 μg/ml aprotinin; 1mM sodium orthovanadate; 1 mM PMSF (all from Sigma, St Louis, MO, USA)). Conditioned medium was concentrated as described [20]. Concentrated or direct conditioned medium and whole-cell extracts were separated by SDS-PAGE, transferred to PVDF membranes and incubated with appropriate antibodies. Specific primary antibodies and dilutions are listed in Supplementary Table 1.
RNA Extraction and Quantitative PCR
Total RNA was extracted and purified with the RNeasy Mini Kit (Quiagen, Hilden, Germany) and retrotranscribed to cDNA using high-capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA). For hnRNA extraction, samples were pretreated with DNAse I (Quiagen). See Supplementary Methods for TaqMan probes and oligonucleotides used. The quantitative PCR reaction was performed in an ABI Prism 7900 HT (Applied Biosystems).
Microarray Data Analysis
Published DNA microarray data sets comprising series of melanoma cell lines were downloaded from public database (GEO accession numbers: Mannheim GSE4843; Philadelphia GSE4841) [19] and analyzed as described in [7].
Lentivirus Production and Transduction of Cell Lines
Lentiviruses were produced and tittered as previously described [7]. For transduction of target cells, lentiviruses were used at multiplicity of infection (MOI) of 5-100 depending on the cell line, for 5-8 h, resulting in more than 95% transduced (GFP-positive) cells.
RNA Interference and Over-Expression Strategies
MITF silencing was carried out using the lentiviral vector pGIPz containing shRNAmir sequence V2LHS_257541 from Open Biosystems (Thermo Fisher Scientific, Huntsville, AL, USA). Two additional shRNAmir sequences were used: V2LHS_259964 and V2LHS_76565, from Open Biosystems. Non-silencing shRNAmir sequence (shNS), with no homology to known mammalian genes was used as control, cloned in a pGIPz vector (Open Biosystems). In order to carry out the MITF silencing in 501mel melanoma cell line over-expressing PEDF, parental cells were transduced with control lentiviral vector pRRL-GFP (GFP) or lentiviral vector expressing PEDF, pRRL-PEDF (PEDF) [7], at 10 MOI. Seventy-two hours later, these melanoma cell lines were infected with shNS or shMITF lentiviral vectors at 100 MOI. For MITF over-expression we generated lentiviral constructs in the pCDH-CMV-MCS-EF1-copGFP (kindly provided by H. Rizos [21]) encoding human MITF cDNA as indicated in Supplementary Methods.
Oncogene Induced Senescence (OIS)
OIS in melanocytes was carried out by transducing the lentiviral plasmids indicated below at 5 MOI. pCDH-BRAFV600E lentiviral plasmid [21] was provided by H. Rizos. We also generated lentiviral constructs in the pRRL.CMV.EGFP.wpre.SIN vector encoding the full-length HRASG12V cDNA as described in Supplementary Methods.
Migration and Invasion Assays
Migration and invasion assays were performed seeding 120.000 cells in modified Boyden chambers with polycarbonate filters (6.5 mm diameter, 8.0 μm pore size) (Corning Incorporated, Corning, NY, USA) coated with 0.5 % gelatin (migration assay) [6] or 15 μg growth factor reduced Matrigel (BD Biosciences, Bedford, MA, USA) (invasion assay) [7] diluted in 100 μl serum-free medium, and air dried overnight. Thirty or sixty μg/ml of concentrated conditioned medium (CM) from NIH-3T3 cells was used as chemoattractant. After incubation for 16-24 h (migration assays) or 48 h (invasion assays), non-migrated and non-invaded cells were wiped off using a cotton swab, and the filters were stained with Diff Quik (Dade Behring, Newark, DE, USA). Migrated or invaded cells were counted in 10 fields of maximum migration or invasion under a light microscope at 40x magnification.
Zebrafish Tumor Cell Dissemination Model
The metastatic ability of the tumor cell lines derived from 501mel cells in the zebrafish metastasis model was assayed as described by P. Rouhi and collaborators [22], [23]. Briefly, 501mel-GFP-shNS, 501mel-PEDF-shNS, 501mel-GFP-shMITF and 501mel-PEDF-shMITF cells were labeled in vitro with DiI dye and approximately 100-500 cells were injected per embryo in the perivitelline space of 2 days old transgenic Tg (fli1a:EGFP) zebrafish embryos. Z-stack fluorescent micrographs were taken 3 days after melanoma cell implantation and distally disseminated tumor cells were counted. All experiments were performed in accordance with ethical permissions granted by the North Stockholm Experimental Animal Ethical Committee, Sweden.
Proliferation Assay
Melanocytes S-phase entry was analyzed by the incorporation of 5-ethynyl-2-deoxyuridine (EdU) at 20 μM for 6 h using the Click-iT EdU Imaging Kit (Invitrogen, Carlsbad, CA, USA) as indicated by the manufacturer. EdU-positive cells were counted in 10 different fields at 20x magnification.
Immunofluorescence
Primary antibodies and dilutions are listed in Supplementary Table 1. Primary melanocytes were seeded on glass coverslips in 24-well plates at 2.104 cells per well. Cells were washed in PBS twice and fixed with 4 % paraformaldehyde/PBS for 15 min at room temperature. Cells were rinsed three times with PBS, then permeabilized with 0.5 % Triton X-100/PBS-0.1 M glycine for 10 min, then rinsed and blocked with 3 % BSA/PBS for 1 h. Detection of H3K9Me was performed incubating cells with rabbit polyclonal antibody to H3K9Me for 16 h at 4ºC, then washed and incubated with Alexa Fluor 546-conjugated secondary IgG (Invitrogen) and DAPI (4’,6-diamidino-2-phhenylin-dole) for nuclei detection. Ten fields at 40x magnification were counted. Immunofluorescence in human samples for PEDF and MITF expression was performed on formalin-fixed paraffin-embedded 3 μm sections from tissue microarrays (TMAs) corresponding to naevus (n = 15), RGP melanoma (n = 13), VGP melanoma (n = 19), cutaneous metastases (n = 32) and visceral metastases (n = 17), retrieved from Hospital 12 de Octubre Biobank. After deparaffinization, heat antigen retrieval was performed in EDTA buffer, pH 9.0 using DakoLink platform. Sections were immersed in TBS 5% BSA for 10 min to block non-specific binding and then, incubated with a rabbit polyclonal antibody to PEDF and a mouse monoclonal antibody against MITF. PEDF and MITF were detected using appropriate Alexa Fluor 658 and 488 conjugated anti-rabbit and anti-mouse IgG antibodies (Invitrogen). Sections were incubated with DAPI to visualize cell nuclei. All incubations were performed at room temperature (RT) using a Dako Autostainer. Fluorescence signals were evaluated blindly by a trained pathologist using a Nuance FX Multispectral Imaging System (Cri Caliper Life Sciences), which compensates for unevenness in illumination and background, and then flat fielded and filtered after image deconvolution the spectral data. TIFF images were created. A semiquantitative HistoScore (Hscore) was calculated for nuclear MITF and cytoplasmic PEDF expression in target cells considering the percentage of melanocytic cells showing low, medium, or high fluorescence intensity. The final score was determined after applying a weighting factor to each estimate: Hscore = (low %) x 1 + (medium %) x 2 + (high %) x 3. The results ranged from 0 to 300. For group comparisons, low, intermediate and high expression categories were defined establishing a threshold at Hscores of 100 and 200. Additionally, we estimated the percentage of area of the lesion positive for PEDF or MITF in each sample. Extension of co-localization was determined for each sample as percentage of MITF-positive cells showing PEDF expression. Percentage of biopsies displaying different degrees of co-localization (< 25%, 25%-75% and > 75%) was estimated in naevus and pathological stages of human melanoma.
ChIP-Sequencing
To identify MITF binding sites in the SERPINF1 locus, we analyzed publicly available [24] and unpublished ChIP-seq data provided by I. Davidson. Both ChIP-seq experiments were performed on chromatin from native 501mel cells according to standard protocols as previously described [24], [25], [26]. Peak detection was performed using the MACS software (http://liulab.dfci. harvard.edu/MACS/; [27]) under settings in which the HA–ChIP from untagged 501mel cells was used as a negative control. Only peaks with an associated p-value below 10- 5 were considered significant. The False Discovery Rate for the MITF binding sites in the SERPINF1 locus was below 8%. To generate the figures, the coordinates of the MITF binding sites and peak maximum were uploaded into the UCSC genome browser (http://genome.ucsc.edu/) as custom tracks in bed format [28].
Reporter Assays
Reporter assays were performed using WM278 and MaMel26a1 melanoma cell lines and HEK293T, all of them with no basal expression of MITF. Cells were seeded on 24-well plates (6.104 cells/well) 16 h (HEK293T) or 48 h (WM278 and MaMel26a1) prior to transfection. A mixture containing 0.6 μg of MITF expression vector (pCDH-HA-MITF) or its control (pCDH-GFP), 0.15 μg of the indicated reporter construct (described in Supplementary Methods) or empty plasmid and 0.03 μg of a plasmid encoding for renilla luciferase under the control of a minimal promoter (prolactin-) [29] was used for transfection with Fugene (Promega). Analysis of firefly and renilla luciferase was performed using the Dual Luciferase Reporter System (Promega) in a Lumat LB9507 luminometer (Berthold Technologies, Bad Wildbad, Germany). The luciferase activity was normalized to renilla activity (constitutive expression). The results are represented as the average of the fold induction obtained from three independent experiments in each cell line and error bars represent the standard deviation.
Statistical Analysis
Statistical analysis was performed using GraphPad Instat (GraphPad Software, San Diego, CA, USA). P-values < 0.05 were considered as significant.
Results
MITF and PEDF Expression Positively Correlate During Human Melanoma Progression and Melanocyte Senescence
Previously, we used gene expression profiling analysis of large series of melanoma cell lines to demonstrate that PEDF expression varied with the molecular staging of melanoma cells, with high levels of PEDF being characteristic of the weakly aggressive cohort [7]. Here we extended this analysis to search for factors that positively correlate with PEDF and that could therefore be candidate regulators implicated in maintaining high PEDF levels in melanocytes and weakly aggressive melanoma cells. In this analysis we found that expression of PEDF positively correlated with MITF in two large melanoma sets (Figure 1B and E). Figure 1A and D show that the cohort of weakly aggressive melanomas is characterized by high levels of PEDF and MITF, whereas the expression of both factors is significantly decreased in the highly aggressive cohort (Figure 1C and F).
Figure 1.
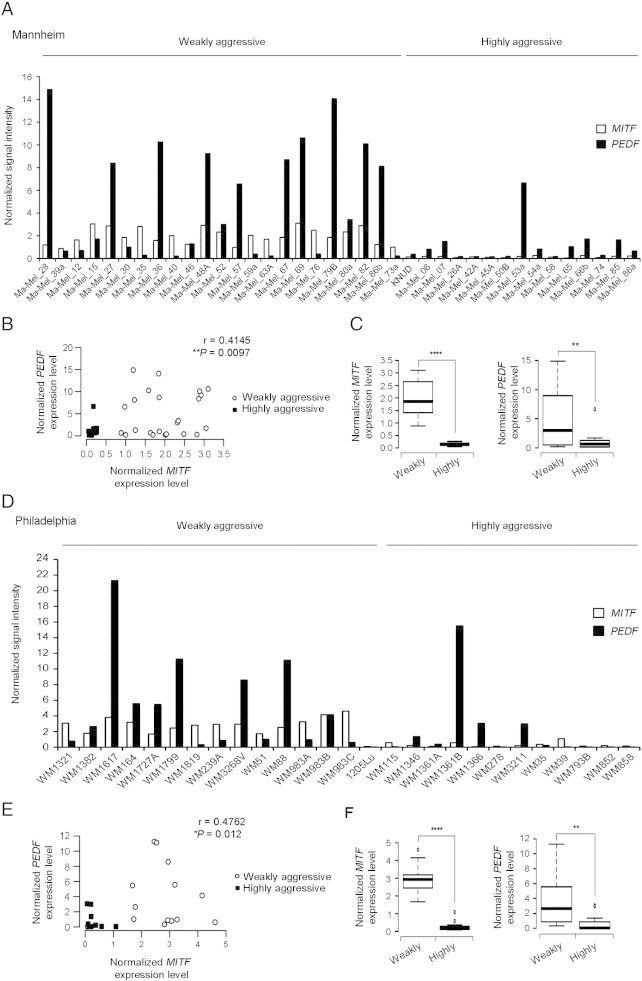
MITF and PEDF expression in weakly aggressive and highly aggressive melanoma cell lines. (A) Analysis of MITF and PEDF expression using microarray gene expression data of Mannheim series of melanoma cell lines. Cell lines were classified by their gene signature into weakly and highly aggressive cohorts. Normalized MITF (empty bars) and PEDF (filled bars) signal intensity are plotted across the sample set. (B) Scattergram showing the positive correlation between MITF and PEDF levels in each cohort of the Mannheim series. Statistical significance was determined by Pearson’s test (r = 0.4145; **P < 0.01). (C) Box-plot of MITF (****P < 0.0001) (left) and PEDF (**P < 0.01) (right) expression in weakly and highly aggressive melanoma cell lines of the Mannheim series. Statistical significance was determined by Welch’s test. Boxes in the plots include values in the 25-75% interval; internal lanes represent the median; circles represent outliers. (D) Analysis of MITF and PEDF expression using microarray gene expression data of Philadelphia series of melanoma cell lines. (E) Scattergram showing the positive correlation between MITF and PEDF levels in each cohort of the Philadelphia series. Statistical significance was determined by Spearman’s test (r = 0.4762; *P < 0.05). (F) Box-plot of MITF (Mann-Whitney’s test; ****P < 0.0001) (left) and PEDF (Welch’s test; **P < 0.01) (right) expression in weakly and highly aggressive melanoma cell lines.
We next evaluated whether both factors also positively correlate in human biopsies of different pathological stages of melanoma by performing double immunoflourescence in tissue microarrays (TMAs) of naevus (n = 15), radial growth phase (RGP) melanoma (n = 13), vertical growth phase (VGP) melanoma (n = 19), cutaneous metastases (CM) of melanoma (n = 32) and visceral metastases (VM) of melanoma (n = 17) (Figure 2). As expected, we detected nuclear MITF expression and cytoplasmatic PEDF expression (Figure 2A). PEDF and MITF expression was variable in primary melanomas and metastases, ranging from moderate to high (Figure 2A-B). However, homogeneous and low expression of both factors was detected in naevus; and high expression for either PEDF or MITF was not detected in any naevus sample (Figure 2A-B). Expression of PEDF and MITF was significantly increased in primary melanoma (RGP or VGP melanoma) compared to naevus (Figure 2C). Also a significant decrease in PEDF and MITF expression was observed in VGP melanoma, cutaneous and visceral metastases compared to RGP melanoma (Figure 2C). Furthermore, a positive correlation of both factors was confirmed comparing biopsies of the different pathological stages of human melanoma progression (Figure 2D).
Figure 2.
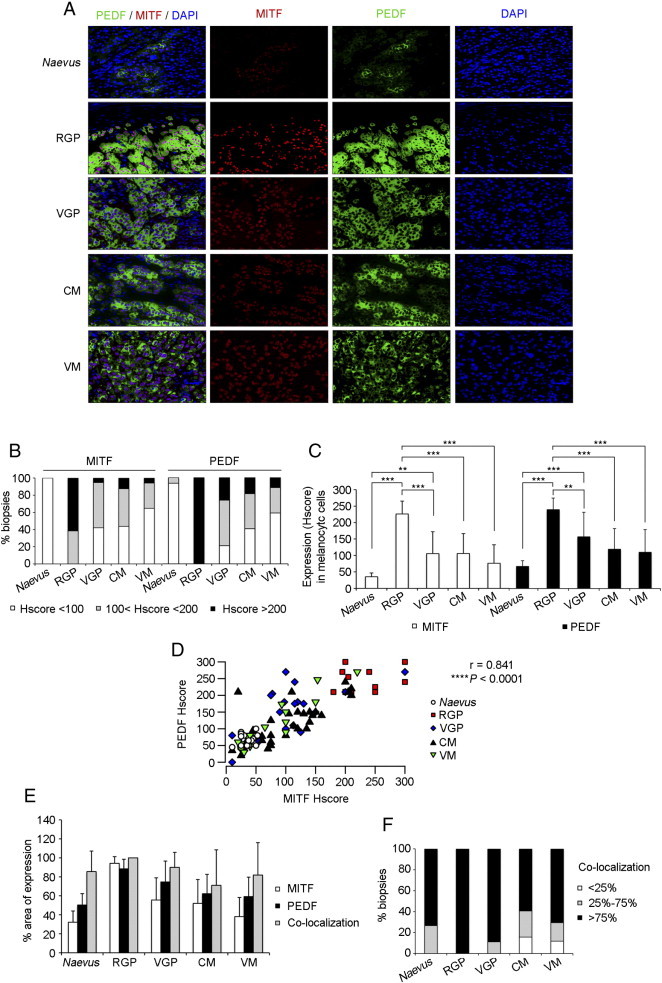
PEDF and MITF expression in human melanoma samples. (A) Representative images (40x magnification) corresponding to double immunofluorescence of PEDF (green) and MITF (red) expression in tissue microarrays (TMAs) from naevus, radial growth phase (RGP) melanoma, vertical growth phase (VGP) melanoma, cutaneous metastasis (CM) of melanoma and visceral metastasis (VM) of melanoma. PEDF was detected in the cytoplasm and MITF in the nucleus of melanocytic cells. (B) Percentage of cases with low, intermediate and high Hscore for PEDF and MITF in naevus, RGP, VGP, CM and VM of melanoma. (C) Quantification of MITF (empty bars) and PEDF (filled bars) expression expressed as Hscore in naevus, RGP, VGP, CM and VM of melanoma. Bars represent average +/- standard deviation (SD), and statistical significance was determined by one-way ANOVA using Tukey-Kramer post-test (**P < 0.01; ***P < 0.001). (D) Scattergram showing the positive correlation between MITF and PEDF expression (Hscore) in human samples. Statistical significance was determined by Pearson’s test (r = 0.841; ****P < 0.0001). (E) Percentage of area of expression of MITF (empty bars), PEDF (black bars) and co-localization (grey bars) in the collection of TMAs comprising naevus and pathological stages of melanoma progression. Bars represent average +/- SD. (F) Percentage of biopsies displaying different degrees of co-localization (< 25%, 25%-75% and > 75%) of PEDF and MITF in naevus and pathological stages of melanoma progression.
These results provide solid evidence for positive correlation between PEDF and MITF levels and co-regulation of their expression according to molecular and pathological staging of melanoma.
Additionally, we analyzed by double immunofluorescence PEDF and MITF co-localization within melanocityc cells in samples of human naevus and melanoma (Figure 2A and E-F). In naevus, where a mean of 50.3% of the total area of the sample expressed PEDF and 32.2% MITF, we observed that in average 85.5% of MITF-positive cells also expressed PEDF (Figure 2E). Analysis of distribution of the samples according to the degree of co-localization showed that PEDF and MITF co-localized in the majority of the samples (Figure 2F). Similar results were also found in primary melanoma and metastases (Figure 2E-F).
In agreement with our results, it has been reported that MITF expression is decreased in benign naevi [12], [30], [31]; which are considered senescent lesions composed by non-proliferating melanocytes in response to oncogenic mutations on BRAF or NRAS [32], [33], [34]. However, the association of PEDF down-regulation with senescence was unknown and therefore we directly addressed whether PEDF was regulated during oncogene-induced senescence (OIS) in primary cultures of melanocytes.
Induction of senescence in primary melanocytes was evaluated assessing characteristic morphological changes, presence of heterochromatin foci and proliferation arrest [35] upon lentiviral transduction of HRASG12V or BRAFV600E (Figure 3A-D). Human primary melanocytes expressed high levels of PEDF and MITF that decreased after induction of senescence by HRASG12V or BRAFV600E (Figure 3E-G). Down-regulation of both intracellular and secreted PEDF, as well as MITF protein was detected by western blot (Figure 3E). p16INK4A was used as a marker of senescence in this analysis [36] (Figure 3E). The changes in protein caused by OIS in primary melanocytes were paralleled by decreased PEDF and MITF mRNA levels as shown by quantitative RT-PCR (Figure 3F-G).
Figure 3.
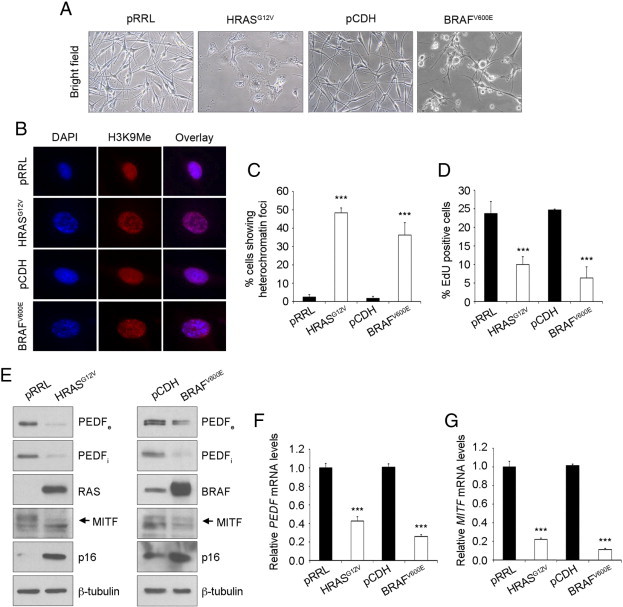
Oncogene induced senescence promotes PEDF and MITF down-regulation in primary melanocytes. (A) Phase-contrast images (20x magnification) showing the morphology of primary melanocytes (NHEM) transduced with control (pRRL or pCDH) and pRRL-HRASG12V (HRASG12V) and pCDH-BRAFV600E (BRAFV600E) lentivirus. (B) Fluorescence images (40x magnification) showing chromatin condensation (DAPI) and H3K9Me marks at day 6 post-infection of control or HRASG12V/BRAFV600E-expressing melanocytes. (C) Percentage of cells showing heterochromatin foci. Bars represent average +/- standard deviation (SD). (D) Proliferation assay by 5-ethyl-2-deoxyuridine (EdU) incorporation at day 6 post-infection of control and HRASG12V/BRAFV600E-expressing melanocytes. Bars represent average +/- SD. (E) Western blot analysis of extracellular PEDF (PEDFe) protein levels in concentrated conditioned medium (CM), intracellular PEDF (PEDFi), RAS, BRAF, MITF, and p16 protein levels in whole-cell extracts from control and HRASG12V/BRAFV600E-expressing melanocytes. β-tubulin was used as loading control. (F-G) Quantitative RT-PCR analysis of PEDF (F) and MITF (G) mRNA levels in control and HRASG12V/BRAFV600E-expressing melanocytes. PEDF and MITF mRNA levels are shown relative to control melanocytes (pRRL or pCDH) after normalization to 18s rRNA levels. Bars represent average +/- SD. Data shown are from a representative experiment that was confirmed in three independent experiments, and statistical significance was determined by one-way ANOVA using Tukey-Kramer post-test (***P < 0.001).
These results, together with the data from naevus, confirm that PEDF and MITF expression are regulated with the same trend during melanocyte senescence, both in vitro and in vivo.
MITF Regulates PEDF Expression in Melanoma Cell Lines and Primary Melanocytes
To directly address the existence of a regulatory link between MITF and PEDF, we silenced MITF by lentiviral transduction of a shRNAmir specific for MITF in the following weakly aggressive melanoma cell lines expressing high PEDF and MITF levels: 501mel (Figure 4), WM164, M000921 and WM88 (Supplementary Figure 1). Interference of MITF led to a significant down-regulation of PEDF mRNA (Figure 4B and Supplementary Figure 1B) and protein (Figure 4C and Supplementary Figure 1C). The extent of PEDF down-regulation by MITF interference was similar to the decrease in tyrosinase (TYR), a well known MITF target (Figure 4B and Supplementary Figure 1B). PEDF down-regulation by MITF interference was confirmed with two additional shRNAmir sequences specific to MITF (data not shown).
Figure 4.
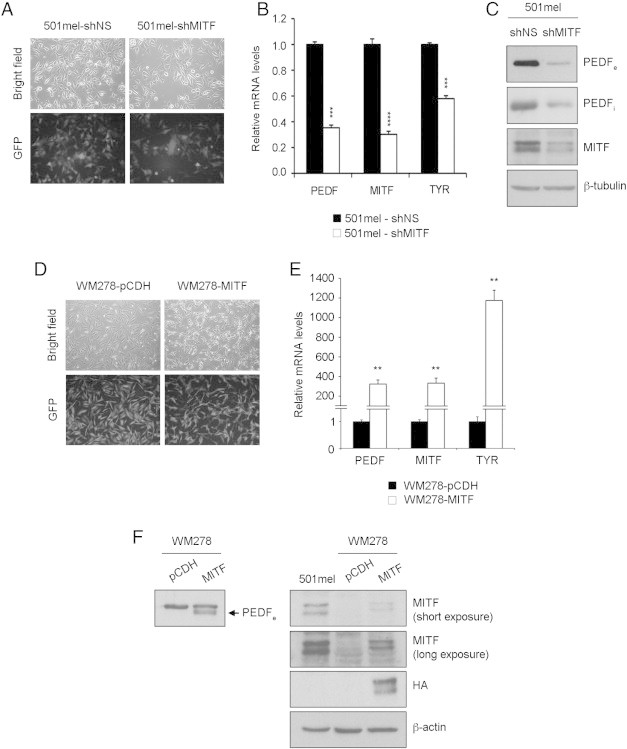
MITF regulates PEDF expression in melanoma cell lines. (A) Transduction efficiency of 501mel melanoma cell line after infection with non-silencing (shNS) or shRNAmir to MITF (shMITF) lentivirus at multiplicity of infection (MOI) of 100. Fluorescence images (10x magnification) show more than 90% GFP-positive cells. (B) Quantitative RT-PCR of PEDF, MITF and TYR mRNA levels in 501mel-shNS and 501mel-shMITF melanoma cell lines. PEDF, MITF and TYR mRNA levels are shown relative to control shNS cells after normalization to GAPDH mRNA levels. Bars represent average +/- standard deviation (SD), and statistical significance was determined by Student’s test (***, P < 0.001; ****P < 0.0001). (C) Western blot analysis of extracellular PEDF (PEDFe) protein levels in concentrated conditioned medium (CM), intracellular PEDF (PEDFi) and MITF protein levels in whole-cell extracts from 501mel-shNS and 501mel-shMITF melanoma cell lines. β–tubulin was used as loading control. (D) Transduction efficiency of WM278 melanoma cell line after infection with control (pCDH) and pCDH-HA-MITF (MITF) lentivirus at 100 MOI. Fluorescence images (10x magnification) show more than 90% GFP-positive cells. (E) Quantitative RT-PCR of PEDF, MITF and TYR mRNA levels in WM278-pCDH and WM278-MITF melanoma cell lines. PEDF, MITF and TYR mRNA levels are shown relative to control pCDH cells after normalization to 18s rRNA levels. Bars represent average +/- SD, and statistical significance was determined by Student’s test (**P < 0.01). (F) Western blot analysis of PEDFe protein levels (PEDF band indicated by an arrow) in direct conditioned medium (left), MITF and HA-tag protein levels (right) in whole-cell extracts from WM278-pCDH, WM278-MITF and 501mel melanoma cell lines. β–actin was used as loading control.
Primary melanocytes express high levels of PEDF, similar to levels in the weakly aggressive melanoma cell lines. We also found that MITF interference decreased PEDF levels in primary human melanocytes (Supplementary Figure 2).
Conversely, MITF over-expression by lentiviral transduction of pCDH-HA-MITF in the WM278 melanoma cell line (which does not express significant levels of MITF and PEDF mRNA) resulted in increased mRNA and protein PEDF levels (PEDF band in WM278 cell line is indicated by an arrow) (Figure 4D-F).
PEDF is a Direct Target of MITF in Human Melanoma Cells
We next tested the hypothesis that PEDF is a direct transcriptional target of MITF. Changes in gene expression due to increased transcription rate are expected to be accompanied by increased heterogeneous nuclear RNA (hnRNA), whereas changes due to increased half-life of mRNA should not have an impact on hnRNA levels. Therefore, we compared levels of hnRNA in a small collection of melanoma cell lines representative of the weakly and highly aggressive cohorts. As expected, mRNA levels of PEDF, MITF and the MITF-target gene TYR were significantly higher in the weakly aggressive cohort than in the highly aggressive cohort (Figure 5A-B). PEDF and MITF hnRNA levels were significantly decreased in the highly aggressive melanoma cell lines compared to the weakly aggressive lines (Figure 5C-D), consistent with transcriptional regulation. In agreement with the proposed role for MITF in the transcriptional regulation of TYR, we found the expected trend in hnRNA and mRNA, although it did not reach statistical significance due to the high TYR hnRNA levels in one sample (MaMel69) (Figure 5C-D).
Figure 5.
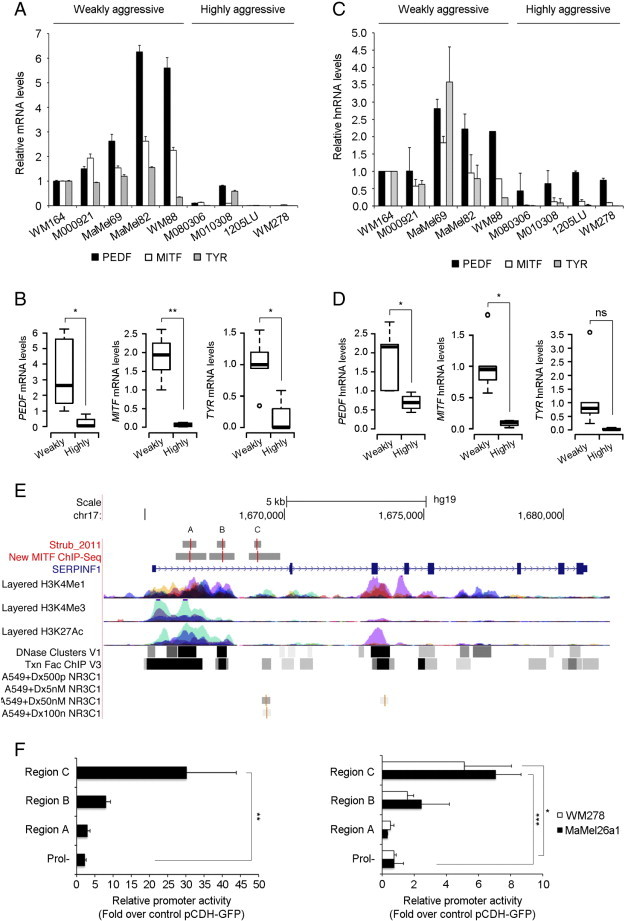
MITF is a direct transcriptional regulator of PEDF expression in melanoma cell lines. (A) Quantitative RT-PCR analysis of PEDF, MITF and TYR mRNA in a set of melanoma cell lines representative of weakly and highly aggressive cohorts. mRNA levels are shown relative to WM164 melanoma cell line after normalization to GAPDH mRNA levels. Bars represent average +/- standard deviation (SD). (B) Box-plot of PEDF (Welch’s test; *P < 0.05) (left), MITF (Welch’s test; **P < 0.01) (middle) and TYR (Student’s test; *P < 0.05) (right) mRNA levels in weakly and highly aggressive melanoma cell lines. Boxes in the plots include values in the 25-75% interval; internal lanes represent the median, circles represent outliers. (C) Quantitative RT-PCR analysis of PEDF, MITF and TYR heterogeneous nuclear RNA (hnRNA) in a set of melanoma cell lines representative of weakly and highly aggressive cohorts. hnRNA levels are shown relative to WM164 melanoma cell line. Bars represent average +/- SD. (D) Box-plot of PEDF (Student’s test; *P < 0.05) (left), MITF (Welch’s test; *P < 0.05) (middle) and TYR (Student’s test; ns) (right) hnRNA levels in weakly and highly aggressive melanoma cell lines. (E) Schematic diagram depicting the human (hg19 assembly) genomic region containing the SERPINF1 gene obtained from UCSC genomic browser (http://genome.ucsc.edu/). The image includes two tracks, “Strub_2011” and “New MITF ChIP-Seq”, showing MITF binding sites determined by ChIP-seq in two independent experiments ([24] and I. Davidson unpublished data). Binding regions are named as A, B and C accordingly to their proximity to ATG origin in first exon. These tracks represent the binding regions as grey boxes, and peaks summits as a vertical red line within boxes. The remaining tracks show information from the UCSC databases (from top to bottom): genes from curated databases (RefSeq), mono- and trimethylated H3K4 histone marks (H3K4Me1 and H3K4Me3) and acetylated H3K27 histone mark (H3K27Ac), DNase hypersensitive regions (DNase clusters), regions bound by transcription factors experimentally assessed by ChIP-seq (Txn Factor ChIP) and regions bound by the glucocorticoid receptor (NR3C1) in cells treated with 500 pM-100 nM dexamethasone (Dx500pM NR3C1, Dx5nM NR3C1, Dx50nM NR3C1 and Dx100nM NR3C1). Note that no peaks are detected at lower doses of dexamethasone (500 pM and 5 nM). The histone marks, DNase HS regions and Txn Factor cluster tracks derive from the comparison across several cell types (see UCSC browser information ofr details). (F) Reporter assays in HEK293T (left), WM278 and MaMel26a1 melanoma cell lines (right). Cell lines were transfected with control (pCDH-GFP) or MITF expression vector (pCDH-HA-MITF) and reporter constructs containing the indicated regions upstream of the firefly luciferase gene. The graphs show the corrected luciferase activity values of each construct and represented as fold change over the activity obtained in cells transfected with control vector (pCDH-GFP). Bars represent the average of values obtained in three independent experiments +/- SD. Statistical significance was determined by one-way ANOVA using Tukey-Kramer post-test (*P < 0.05; **P < 0.01; ***P < 0.001).
To directly address the transcriptional regulation of PEDF by MITF, we analyzed the data from a recently published MITF ChIP-Seq (chromatin immunoprecipitation followed by high-throughput sequencing) study [24] to identify MITF binding sites in the SERPINF1 locus. Three peaks of MITF binding were observed within the first intron of the SERPINF1 locus (Figure 5E, “Strub2011” track). The binding of MITF to these three regions was confirmed by the analysis of the unpublished data from an independent ChIP-Seq experiment (Figure 5E, “New_MITF_ChIP-Seq” track). Importantly, these MITF binding sites lie within regions probably involved in the regulation of the expression of this locus as indicated by their sensitivity to DNase I (“DNase Clusters” track), presence of experimentally determined binding sites for multiple transcription factors (“Txn Factor ChIP” track), pattern of H3K27 acetylation (“Layered H3K27Ac” track) and pattern of H3K4 methylation (“Layered H3K4Me1” and “Layered H3K4Me3” tracks).
Finally, we performed reporter assays in HEK293T and two aggressive melanoma cell lines (WM278 and MaMel26a1), which do not express detectable levels of MITF and PEDF, to analyze the functionality of the three binding regions identified by ChIP-Seq. All three regions had the ability to increase the basal transcription of the luciferase reporter gene under the control of a minimal promoter (data not shown). However, only regions B and C, responded to exogenous MITF (Figure 5F). Although, both regions B and C increased the transcription of the reporter gene in response to MITF in HEK293T and the two melanoma cell lines tested, the differences reached statistical significance only in the case of region C (Figure 5F).
Altogether these results demonstrate that MITF regulates PEDF transcription through direct binding to regulatory regions in the first intron of SERPINF1 gene encoding for PEDF.
PEDF Mediates MITF Effects on Migration and Invasion of Melanoma Cells
Regulation of the migratory and invasive potential of melanoma cells is a fundamental driver of melanoma progression. Therefore, we explored a possible functional connection between MITF and PEDF in the control of melanoma cell migration and invasion. We previously demonstrated that PEDF silencing consistently increases migration and invasion capacity of melanocytes and melanoma cells [7]. An analogous increase of the migratory ability of melanoma cells was consistently observed with MITF silencing [12], [37] in several melanoma cell lines (Figure 6A and Supplementary Figure 3), pointing to a possible functional role for the MITF-PEDF axis.
Figure 6.
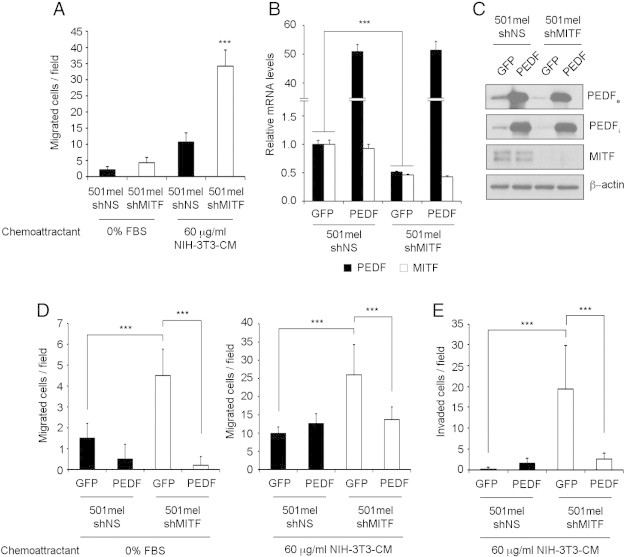
Exogenous PEDF expression blocks activation of migration and invasion induced by MITF silencing. (A) Migration assay of 501mel-shNS and 501mel-shMITF melanoma cell lines toward 0% FBS and 60 μg/ml of concentrated conditioned medium (CM) from NIH-3T3 cells for 24 h. Bars represent average +/- standard deviation (SD), and statistical significance was determined by one-way ANOVA using Tukey-Kramer post-test (***P < 0.001). (B) Quantitative RT-PCR of PEDF and MITF mRNA levels in 501mel-GFP-shNS, 501mel-PEDF-shNS, 501mel-GFP-shMITF and 501mel-PEDF-shMITF melanoma cell lines. PEDF and MITF mRNA levels are shown relative to control GFP-shNS cells after normalization to GAPDH mRNA levels. Bars represent average +/- SD, and statistical significance was determined by Student’s test (***P < 0,001). (C) Western blot analysis of extracellular PEDF (PEDFe) protein levels in direct conditioned medium, intracellular PEDF (PEDFi) and MITF protein levels in whole-cell extracts from 501mel-GFP-shNS, 501mel-PEDF-shNS, 501mel-GFP-shMITF and 501mel-PEDF-shMITF melanoma cell lines. β–actin was used as loading control. (D) Migration assay of 501mel-GFP-shNS, 501mel-PEDF-shNS, 501mel-GFP-shMITF and 501mel-PEDF-shMITF melanoma cell lines toward 0% FBS (left) and 60 μg/ml CM from NIH-3T3 (right) for 24 h. (E) Invasion assay of 501mel-GFP-shNS, 501mel-PEDF-shNS, 501mel-GFP-shMITF and 501mel-PEDF-shMITF melanoma cell lines toward 60 μg/ml CM from NIH-3T3 for 48 h. In migration and invasion assays bars represent average +/- SD, and statistical significance was determined by one-way ANOVA using Tukey-Kramer post-test (***P < 0.001). Shown data are a representative experiment that was confirmed in three independent experiments.
We next studied whether exogenous PEDF expression could bypass the increased migration and invasion induced by MITF silencing. To this end we transduced 501mel cells with lentivirus-PEDF followed by MITF silencing by lentiviral transduction of shRNAmir specific to MITF (Figure 6B-C). Figure 6D-E show that exogenous PEDF expression efficiently blocks activation of migration and invasion induced by MITF interference in 501mel cells, indicating that the effect of MITF in migration and invasion of melanoma cells is mediated, at least in part, by the regulation of PEDF.
To further strengthen the functional connection between MITF and PEDF and the relevance of the MITF-PEDF axis in the biology of melanoma progression we have used the zebrafish metastasis model. Figure 7 shows that silencing of MITF (501mel-GFP-shMITF cells) increases the number of disseminated cells, while simultaneous expression of exogenous PEDF (501mel-PEDF-shMITF cells) efficiently counteracts this functional effect.
Figure 7.
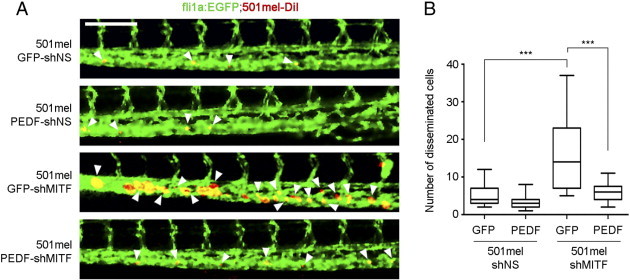
Sustained expression of PEDF halts in vivo dissemination of melanoma cells induced by MITF silencing. (A) Fluorescent micrographs of the posterior tail showing distal dissemination of tumor cells (red) and their close association with the blood vessels of the embryo (green) 3 days after melanoma cell implantation. Size bar indicates 100 micrometers. White arrowheads indicate disseminated tumor cells. (B) The box-plots represent the number of distally disseminated tumor cells per fish (n = 21-25 fish/condition), 3 days after melanoma cell implantation. Statistical significance was determined by one-way ANOVA using Tukey-Kramer post-test (***P < 0.001).
Discussion
PEDF has emerged over the last decade as a dual action factor, with a potent capacity to simultaneously modify the tumor microenvironment and the behavior of the tumor cells [1], [2], [5]. PEDF is a potent angiostatic factor for melanoma [6], [38] and also an effective molecular barrier to restrict the migratory and invasive capacity of melanoma cells [7], [39]. Despite its biological relevance in melanoma, mechanisms underlying variations in PEDF expression during the malignant progression of melanoma are largely unknown. Recently, we have described that microenvironment-derived signals like hypoxia negatively regulate the expression of PEDF in melanocytic cells [8]. Previously identified PEDF regulators in other cell types are the members of the p53 family, p63 and p73 in colorectal cancer cells [40], androgens in prostate cancer cells [41], 17β-estradiol in ovarian cancer cells [42] and retinoic acid and dexamethasone in several cell types [43], [44].
Here, we identify MITF as a major transcriptional regulator of PEDF in the context of melanoma malignization and melanocyte senescence.
Using the information from high-throughput microarray studies available at GEO data bases [7], here we identify a positive correlation of PEDF and MITF expression in melanoma cell lines classified according to molecular profiling criteria. PEDF and MITF are differentially expressed in the cohorts defined by molecular profiling and high levels of both factors are characteristic of the weakly aggressive cohort. Therefore, these analyses support that high expression of both factors restricts the metastatic potential of melanoma cells, which has been directly demonstrated in mouse models of melanoma [7], [12], [15], [39]. Here, we further support this conclusion by showing a positive correlation of PEDF and MITF with the pathological staging of human melanoma.
A novel context in which we have found a link between PEDF and MITF expression is melanocyte senescence. Induction of senescence in melanocytes by forskolin treatment is associated with decreased MITF expression [45], [46]. Moreover, MITF silencing induces senescence in melanoma cells [16], [24]. Here we describe for the first time that oncogene-induced senescence in primary melanocytes down-regulates MITF and PEDF expression. In support of the physiological relevance of this result, benign naevi; which are considered senescent lesions [32], [33], consistently showed decreased levels of both PEDF and MITF compared to primary melanomas. The anti-oxidant properties of PEDF [47], [48], [49], [50] could be playing a role in avoidance of melanocyte senescence. In this regard, it has been recently reported that PEDF delays cellular senescence of human mesenchymal stem cells by reducing oxidative stress [51]. Additionally, PEDF could have an impact on the senescent secretome [36], [52]. In support of this, we have previously demonstrated that exogenous PEDF down-regulates the pro-senescent cytokine IL8 in melanoma cells [53].
Oncogenic mutations in BRAF and NRAS are present in naevi as well as in melanomas; suggesting that suppressive mechanisms of senescence may also be contributing to maintain high PEDF expression in weakly aggressive melanomas, in contrast to low PEDF levels characteristic of senescent melanocytes in naevi. As BRAF and NRAS mutations are mutually exclusive in melanoma, we also analyzed whether they could underlie variations in PEDF expression. Statistical analysis showed no significant association of PEDF expression levels with BRAF or NRAS mutational status (P = 0.55).
Our finding of positive correlation and co-localization of PEDF and MITF in human melanoma samples, prompted us to formally address whether MITF could be a regulator of PEDF in the context of melanoma. Here we show that MITF silencing down-regulates PEDF expression in several melanoma cell lines and primary melanocytes. Conversely, MITF over-expression increases PEDF levels in the highly aggressive WM278 melanoma cell line. Subsequently, by analysis of published and unpublished ChIP-seq data we identify three MITF binding sites located in the first intron of SERPINF1. Importantly, by means of reporter assays, we demonstrate that at least two of these sites are functional in HEK293T and melanoma cells and respond to MITF increasing transcription. Moreover, analysis of publicly available information from the ENCODE project indicates that MITF binding regions overlap with chromatin features associated to transcriptional regulation, further supporting our experimental data. Interestingly, region C overlaps with H3K4me1 chromatin mark and the binding site of the transcription factor NR3C1, the endogenous glucocorticoid receptor (Figure 5E, NR3C1 tracks). Binding of NC3R1 to this region could explain the reported regulation of SERPINF1 by dexamethasone [43], [44]; suggesting that region C is a functional regulatory element of the SERPINF1 locus. Together these results demonstrate that MITF is a positive regulator of PEDF in melanocytic cells.
Finally, we demonstrate a functional connection between MITF and PEDF in the context of melanoma cell migration, invasion and in vivo metastatic dissemination. We show that increased migration and invasion caused by MITF silencing could be efficiently halted by exogenous PEDF expression in 501mel cells, resulting also in a significant decreased dissemination in the zebrafish metastasis model. Recently published results in retinal pigment epithelial cells also demonstrate that silencing of PEDF functionally compensates the inhibition of migration and invasion caused by MITF over-expression [54]. Mechanisms involved in inhibition of migration and invasion by MITF and PEDF are poorly defined. Importantly, we have recently demonstrated that PEDF abrogates the two main modes of melanoma invasion: amoeboid and mesenchymal; through simultaneous inhibition of RhoA and MT1-MMP triggered by the 34-mer epitope through binding to PNPLA2 receptor [39]. Both PEDF and MITF affect the balance of Rho and Rac to inhibit melanoma cell migration [12], [39], although they impinge on melanoma invasion through inhibition of different protease activities (MITF inhibits MMP2, whereas PEDF blocks MT1-MMP) [12], [39].
According to the phenotype switching model of melanoma progression, metastasis is driven by the reversible reprogramming of melanoma cells between proliferative and invasive phenotypes [12], [18], [19]. MITF and PEDF were independently characterized as important regulators of this switch [12], [18]. Here, we identify a regulatory and functional connection linking MITF and PEDF as relevant brakes to melanoma malignization.
Supplementary data.
Supplementary Methods
Acknowledgments
Supported by Grants from Ministerio de Ciencia y Competitividad of Spain: SAF-2010-19256 (BJ); SAF-2011-24225 (LP); SAF-2012-32117 (IP); FIS 11/02568 (JLRP); RD09/0076/0101; PT13/0010/0012 (FR); PI12/01552 (FR) and LiU-Cancer, Svenska Sällskapet för Medicinsk Forskning, Åke Wibergs Stiftelse and Goesta Fraenkels Stifelse (LJ). E.R-F. is the recipient of a postdoctoral fellowship from “Fundación Científica de la Asociación Española Contra el Cáncer”. We thank M. Soengas for her advice in senescence experiments in melanocytes.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2014.06.001.
Footnotes
Conflict of interests
The authors declare no conflict of interests.
The following are the supplementary data related to this article.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2014.06.001.
References
- 1.Craword SE, Fitchev P, Veliceasa D, Volpert OV. The many facets of PEDF in drug discovery and disease: a diamond in the rough or split personality disorder? Expert Opin Drug Discov. 2013;8:769–792. doi: 10.1517/17460441.2013.794781. [DOI] [PubMed] [Google Scholar]
- 2.Becerra SP, Notario V. The effects of PEDF on cancer biology: mechanisms of action and therapeutic potential. Nat Rev Cancer. 2013;13:258–271. doi: 10.1038/nrc3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tombran-Tink J, Barnstable CJ. PEDF: a multifaceted neurotrophic factor. Nat Rev Neurosci. 2003;4:628–636. doi: 10.1038/nrn1176. [DOI] [PubMed] [Google Scholar]
- 4.Dawson DW, Volpert OV, Gillis P, Crawford SE, Xu H, Benedict W, Bouck NP. Pigment epithelium-derived factor: a potent inhibitor of angiogenesis. Science. 1999;285:245–248. doi: 10.1126/science.285.5425.245. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Garcia NI, Volpert OV, Jimenez B. Pigment epithelium-derived factor as a multifunctional antitumor factor. J Mol Med. 2007;85:15–22. doi: 10.1007/s00109-006-0111-z. [DOI] [PubMed] [Google Scholar]
- 6.Garcia M, Fernandez-Garcia NI, Rivas V, Carretero M, Escamez MJ, Gonzalez-Martin A, Medrano EE, Volpert O, Jorcano JL, Jimenez B. Inhibition of xenografted human melanoma growth and prevention of metastasis development by dual antiangiogenic/antitumor activities of pigment epithelium-derived factor. Cancer Res. 2004;64:5632–5642. doi: 10.1158/0008-5472.CAN-04-0230. [DOI] [PubMed] [Google Scholar]
- 7.Orgaz JL, Ladhani O, Hoek KS, Fernandez-Barral A, Mihic D, Aguilera O, Seftor EA, Bernad A, Rodriguez-Peralto JL, Hendrix MJ. Loss of pigment epithelium-derived factor enables migration, invasion and metastatic spread of human melanoma. Oncogene. 2009;28:4147–4161. doi: 10.1038/onc.2009.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Barral A, Orgaz JL, Gomez V, del Peso L, Calzada MJ, Jimenez B. Hypoxia negatively regulates antimetastatic PEDF in melanoma cells by a hypoxia inducible factor-independent, autophagy dependent mechanism. PLoS One. 2012;7:e32989. doi: 10.1371/journal.pone.0032989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheli Y, Ohanna M, Ballotti R, Bertolotto C. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res. 2010;23:27–40. doi: 10.1111/j.1755-148X.2009.00653.x. [DOI] [PubMed] [Google Scholar]
- 10.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Widlund HR, Fisher DE. Microphthalamia-associated transcription factor: a critical regulator of pigment cell development and survival. Oncogene. 2003;22:3035–3041. doi: 10.1038/sj.onc.1206443. [DOI] [PubMed] [Google Scholar]
- 12.Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20:3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goding CR. Commentary. A picture of Mitf in melanoma immortality. Oncogene. 2011;30:2304–2306. doi: 10.1038/onc.2010.641. [DOI] [PubMed] [Google Scholar]
- 14.Cheli Y, Giuliano S, Botton T, Rocchi S, Hofman V, Hofman P, Bahadoran P, Bertolotto C, Ballotti R. Mitf is the key molecular switch between mouse or human melanoma initiating cells and their differentiated progeny. Oncogene. 2011;30:2307–2318. doi: 10.1038/onc.2010.598. [DOI] [PubMed] [Google Scholar]
- 15.Saez-Ayala M, Montenegro MF, Sanchez-Del-Campo L, Fernandez-Perez MP, Chazarra S, Freter R, Middleton M, Pinero-Madrona A, Cabezas-Herrera J, Goding CR. Directed phenotype switching as an effective antimelanoma strategy. Cancer Cell. 2013;24:105–119. doi: 10.1016/j.ccr.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 16.Giuliano S, Cheli Y, Ohanna M, Bonet C, Beuret L, Bille K, Loubat A, Hofman V, Hofman P, Ponzio G. Microphthalmia-associated transcription factor controls the DNA damage response and a lineage-specific senescence program in melanomas. Cancer Res. 2010;70:3813–3822. doi: 10.1158/0008-5472.CAN-09-2913. [DOI] [PubMed] [Google Scholar]
- 17.Li G, Schaider H, Satyamoorthy K, Hanakawa Y, Hashimoto K, Herlyn M. Downregulation of E-cadherin and Desmoglein 1 by autocrine hepatocyte growth factor during melanoma development. Oncogene. 2001;20:8125–8135. doi: 10.1038/sj.onc.1205034. [DOI] [PubMed] [Google Scholar]
- 18.Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L, Hemmi S, Dummer R. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68:650–656. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- 19.Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, Weber BL, Nathanson KL, Phillips DJ, Herlyn M. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Garcia NI, Palmer HG, Garcia M, Gonzalez-Martin A, del Rio M, Barettino D, Volpert O, Munoz A, Jimenez B. 1alpha,25-Dihydroxyvitamin D3 regulates the expression of Id1 and Id2 genes and the angiogenic phenotype of human colon carcinoma cells. Oncogene. 2005;24:6533–6544. doi: 10.1038/sj.onc.1208801. [DOI] [PubMed] [Google Scholar]
- 21.Scurr LL, Pupo GM, Becker TM, Lai K, Schrama D, Haferkamp S, Irvine M, Scolyer RA, Mann GJ, Becker JC. IGFBP7 is not required for B-RAF-induced melanocyte senescence. Cell. 2010;141:717–727. doi: 10.1016/j.cell.2010.04.021. [DOI] [PubMed] [Google Scholar]
- 22.Rouhi P, Jensen LD, Cao Z, Hosaka K, Lanne T, Wahlberg E, Steffensen JF, Cao Y. Hypoxia-induced metastasis model in embryonic zebrafish. Nat Protoc. 2010;5:1911–1918. doi: 10.1038/nprot.2010.150. [DOI] [PubMed] [Google Scholar]
- 23.Rouhi P, Lee SL, Cao Z, Hedlund EM, Jensen LD, Cao Y. Pathological angiogenesis facilitates tumor cell dissemination and metastasis. Cell Cycle. 2010;9:913–917. doi: 10.4161/cc.9.5.10853. [DOI] [PubMed] [Google Scholar]
- 24.Strub T, Giuliano S, Ye T, Bonet C, Keime C, Kobi D, Le Gras S, Cormont M, Ballotti R, Bertolotto C. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene. 2011;30:2319–2332. doi: 10.1038/onc.2010.612. [DOI] [PubMed] [Google Scholar]
- 25.Delacroix L, Moutier E, Altobelli G, Legras S, Poch O, Choukrallah MA, Bertin I, Jost B, Davidson I. Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol Cell Biol. 2010;30:231–244. doi: 10.1128/MCB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobi D, Steunou AL, Dembele D, Legras S, Larue L, Nieto L, Davidson I. Genome-wide analysis of POU3F2/BRN2 promoter occupancy in human melanoma cells reveals Kitl as a novel regulated target gene. Pigment Cell Melanoma Res. 2010;23:404–418. doi: 10.1111/j.1755-148X.2010.00697.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krebs A, Frontini M, Tora L. GPAT: retrieval of genomic annotation from large genomic position datasets. BMC Bioinformatics. 2008;9:533. doi: 10.1186/1471-2105-9-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rincon M, Flavell RA. AP-1 transcriptional activity requires both T-cell receptor-mediated and co-stimulatory signals in primary T lymphocytes. EMBO J. 1994;13:4370–4381. doi: 10.1002/j.1460-2075.1994.tb06757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 31.Maldonado JL, Timmerman L, Fridlyand J, Bastian BC. Mechanisms of cell-cycle arrest in Spitz nevi with constitutive activation of the MAP-kinase pathway. Am J Pathol. 2004;164:1783–1787. doi: 10.1016/S0002-9440(10)63736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett DC. Human melanocyte senescence and melanoma susceptibility genes. Oncogene. 2003;22:3063–3069. doi: 10.1038/sj.onc.1206446. [DOI] [PubMed] [Google Scholar]
- 33.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, Moses TY, Hostetter G, Wagner U, Kakareka J. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 34.Miller AJ, Mihm MC., Jr. Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 35.Haferkamp S, Rizos H. Oncogene-induced senescence pathways in melanocytes. Cell Cycle. 2010;9:4778–4779. doi: 10.4161/cc.9.24.14248. [DOI] [PubMed] [Google Scholar]
- 36.Bansal R, Nikiforov MA. Pathways of oncogene-induced senescence in human melanocytic cells. Cell Cycle. 2010;9:2782–2788. doi: 10.4161/cc.9.14.12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheli Y, Giuliano S, Fenouille N, Allegra M, Hofman V, Hofman P, Bahadoran P, Lacour JP, Tartare-Deckert S, Bertolotto C. Hypoxia and MITF control metastatic behaviour in mouse and human melanoma cells. Oncogene. 2012;31:2461–2470. doi: 10.1038/onc.2011.425. [DOI] [PubMed] [Google Scholar]
- 38.Abe R, Shimizu T, Yamagishi S, Shibaki A, Amano S, Inagaki Y, Watanabe H, Sugawara H, Nakamura H, Takeuchi M. Overexpression of pigment epithelium-derived factor decreases angiogenesis and inhibits the growth of human malignant melanoma cells in vivo. Am J Pathol. 2004;164:1225–1232. doi: 10.1016/s0002-9440(10)63210-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ladhani O, Sanchez-Martinez C, Orgaz JL, Jimenez B, Volpert OV. Pigment epithelium-derived factor blocks tumor extravasation by suppressing amoeboid morphology and mesenchymal proteolysis. Neoplasia. 2011;13:633–642. doi: 10.1593/neo.11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasaki Y, Naishiro Y, Oshima Y, Imai K, Nakamura Y, Tokino T. Identification of pigment epithelium-derived factor as a direct target of the p53 family member genes. Oncogene. 2005;24:5131–5136. doi: 10.1038/sj.onc.1208695. [DOI] [PubMed] [Google Scholar]
- 41.Doll JA, Stellmach VM, Bouck NP, Bergh AR, Lee C, Abramson LP, Cornwell ML, Pins MR, Borensztajn J, Crawford SE. Pigment epithelium-derived factor regulates the vasculature and mass of the prostate and pancreas. Nat Med. 2003;9:774–780. doi: 10.1038/nm870. [DOI] [PubMed] [Google Scholar]
- 42.Cheung LW, Au SC, Cheung AN, Ngan HY, Tombran-Tink J, Auersperg N, Wong AS. Pigment epithelium-derived factor is estrogen sensitive and inhibits the growth of human ovarian cancer and ovarian surface epithelial cells. Endocrinology. 2006;147:4179–4191. doi: 10.1210/en.2006-0168. [DOI] [PubMed] [Google Scholar]
- 43.Tombran-Tink J, Lara N, Apricio SE, Potluri P, Gee S, Ma JX, Chader G, Barnstable CJ. Retinoic acid and dexamethasone regulate the expression of PEDF in retinal and endothelial cells. Exp Eye Res. 2004;78:945–955. doi: 10.1016/j.exer.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 44.Uchida H, Hayashi H, Kuroki M, Uno K, Yamada H, Yamashita Y, Tombran-Tink J, Oshima K. Vitamin A up-regulates the expression of thrombospondin-1 and pigment epithelium-derived factor in retinal pigment epithelial cells. Exp Eye Res. 2005;80:23–30. doi: 10.1016/j.exer.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 45.Schwahn DJ, Xu W, Herrin AB, Bales ES, Medrano EE. Tyrosine levels regulate the melanogenic response to alpha-melanocyte-stimulating hormone in human melanocytes: implications for pigmentation and proliferation. Pigment Cell Res. 2001;14:32–39. doi: 10.1034/j.1600-0749.2001.140106.x. [DOI] [PubMed] [Google Scholar]
- 46.Schwahn DJ, Timchenko NA, Shibahara S, Medrano EE. Dynamic regulation of the human dopachrome tautomerase promoter by MITF, ER-alpha and chromatin remodelers during proliferation and senescence of human melanocytes. Pigment Cell Res. 2005;18:203–213. doi: 10.1111/j.1600-0749.2005.00229.x. [DOI] [PubMed] [Google Scholar]
- 47.Cao W, Tombran-Tink J, Chen W, Mrazek D, Elias R, McGinnis JF. Pigment epithelium-derived factor protects cultured retinal neurons against hydrogen peroxide-induced cell death. J Neurosci Res. 1999;57:789–800. [PubMed] [Google Scholar]
- 48.Yamagishi S, Inagaki Y, Amano S, Okamoto T, Takeuchi M, Makita Z. Pigment epithelium-derived factor protects cultured retinal pericytes from advanced glycation end product-induced injury through its antioxidative properties. Biochem Biophys Res Commun. 2002;296:877–882. doi: 10.1016/s0006-291x(02)00940-3. [DOI] [PubMed] [Google Scholar]
- 49.Yamagishi S, Inagaki Y, Nakamura K, Abe R, Shimizu T, Yoshimura A, Imaizumi T. Pigment epithelium-derived factor inhibits TNF-alpha-induced interleukin-6 expression in endothelial cells by suppressing NADPH oxidase-mediated reactive oxygen species generation. J Mol Cell Cardiol. 2004;37:497–506. doi: 10.1016/j.yjmcc.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura K, Yamagishi S, Matsui T, Yoshida T, Takenaka K, Jinnouchi Y, Yoshida Y, Ueda S, Adachi H, Imaizumi T. Pigment epithelium-derived factor inhibits neointimal hyperplasia after vascular injury by blocking NADPH oxidase-mediated reactive oxygen species generation. Am J Pathol. 2007;170:2159–2170. doi: 10.2353/ajpath.2007.060838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao Y, Yang T, Gu C, Yi D. Pigment epithelium-derived factor delays cellular senescence of human mesenchymal stem cells in vitro by reducing oxidative stress. Cell Biol Int. 2013;37:305–313. doi: 10.1002/cbin.10041. [DOI] [PubMed] [Google Scholar]
- 52.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 53.Orgaz JL, Benguria A, Sanchez-Martinez C, Ladhani O, Volpert OV, Jimenez B. Changes in the gene expression profile of A375 human melanoma cells induced by overexpression of multifunctional pigment epithelium-derived factor. Melanoma Res. 2011;21:285–297. doi: 10.1097/CMR.0b013e32834495c3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma X, Pan L, Jin X, Dai X, Li H, Wen B, Chen Y, Ma A, Qu J, Hou L. Microphthalmia-associated transcription factor acts through PEDF to regulate RPE cell migration. Exp Cell Res. 2012;318:251–261. doi: 10.1016/j.yexcr.2011.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods


