Abstract
Haploinsufficiency of the NSD1 gene is a hallmark of Sotos syndrome, and rearrangements of this gene by translocation can cause acute myeloid leukemia. The NSD1 gene product is a SET-domain histone lysine methyltransferase that has previously been shown to interact with nuclear receptors. We describe here a novel NSD1-interacting protein, Nizp1, that contains a SCAN box, a KRAB-A domain, and four consensus C2H2-type zinc fingers preceded by a unique finger derivative, referred to herein as the C2HR motif. The C2HR motif functions to mediate protein-protein interaction with the cysteine-rich (C5HCH) domain of NSD1 in a Zn(II)-dependent fashion, and when tethered to RNA polymerase II promoters, represses transcription in an NSD1-dependent manner. Mutations of the cysteine or histidine residues in the C2HR motif abolish the interaction of Nizp1 with NSD1 and compromise the ability of Nizp1 to repress transcription. Interestingly, converting the C2HR motif into a canonical C2H2 zinc finger has a similar effect. Thus, Nizp1 contains a novel type of zinc finger motif that functions as a docking site for NSD1 and is more than just a degenerate evolutionary remnant of a C2H2 motif.
Sotos syndrome is a neurological disorder characterized by overgrowth from the prenatal stage through childhood, with advanced bone age, a dysmorphic face with macrocephaly and a pointed chin, mental retardation, and a possible susceptibility to tumors (32). The most prevalent cause of Sotos syndrome is haploinsufficiency caused by point mutations and microdeletions within the NSD1 gene at 5q35 (21, 27). NSD1 mutations have also been identified in cases of Weaver syndrome, an overgrowth disorder closely related to Sotos syndrome (11). Moreover, in a recurrent translocation [t(5;11)(q35;p15.5)] associated with de novo childhood acute myeloid leukemia, NSD1 was found fused to the NUP98 nucleoporin gene (17). In mammals, there are two highly related NSD1-like (NSD) genes, namely, NSD2 (also referred to as WHSC1 [40] and MMSET [8]) and NSD3 (4), both of which have been implicated in cancer. NSD2 maps to 4p16.3 in the Wolf-Hirschhorn syndrome critical region (40) and was found to be disrupted by t(4;14) translocations causing lymphoid multiple myeloma (8). NSD3 maps to a region that is frequently rearranged in several tumor cell lines and primary breast carcinomas (4). This region was also found fused to the NUP98 gene in cases of acute myeloid leukemia associated with the t(8;11)(p11.2;p15) translocation (37).
It was recently shown that mice devoid of NSD1 are defective in early embryogenesis (34). Homozygous null mutants display a high incidence of apoptotic cell death in embryonic ectodermal cells as early as embryonic day 6.5 (E6.5) and fail to complete gastrulation (34). These embryonic defects clearly indicate that NSD1 plays crucial roles in early postimplantation development and also demonstrate that, at least during early embryogenesis, the members of the NSD family, though structurally related, exert distinct, nonredundant functions. The molecular mechanisms by which these proteins act are still largely unknown, although a role in transcriptional regulation and modulation of the chromatin structure has been suggested (reference 34 and references therein).
NSD1, along with the other members of the family, contains a SET [Su(var)3-9, Enhancer of zeste, Trithorax] domain (4, 16, 40). The SET domain was originally identified in Drosophila melanogaster proteins and was later found in a variety of eukaryotic chromatin-associated proteins that function as histone lysine methyltransferases (HMTases) (reviewed in reference 22). It was recently reported that NSD1 has a catalytically active SET domain that specifically methylates recombinant histone H3 at lysine 36 (H3-K36) and histone H4 at lysine 20 (H4-K20) (34). HMTase activities specific to each of these lysine residues have been described for yeasts, Drosophila, and mammals (12, 30, 42) and have been shown to be associated with transcriptionally inactive chromatin. Thus, the molecular mechanisms through which NSD1 carries out its function during embryogenesis may relate in part to the transcriptional silencing of developmentally regulated genes.
In addition to the SET domain, all three NSD proteins contain five plant homeodomain (PHD) motifs (4, 16, 40). The PHD motif is a conserved C4HC3 zinc finger domain that binds to phosphoinositides (14) and predominantly occurs in proteins that function at the chromatin level (1). The biological importance of this domain is underscored by its involvement in the pathogenesis of several human disorders, including cancer (13, 15, 18). In the NSD proteins, the PHDI to PHDIV motifs are adjacent to each other and are located N-terminal to the SET domain, whereas the PHDV motif is C-terminal to this domain (Fig. 1A). Adjacent to the C terminus of PHDV, an NSD-specific cysteine-rich domain of unknown function was previously identified and was designated a C5HCH motif according to the sequence arrangement of the conserved cysteine and histidine residues (4). Finally, NSD proteins also have a PWWP (proline-tryptophan-tryptophan-proline) domain (41) located between the PHD finger cluster (PHDI to PHDIV) and the SET domain. This domain, originally described for the hepatoma-derived growth factor-related protein, is also present in various nuclear proteins involved in cell growth and differentiation (see reference 41 and references therein).
FIG. 1.
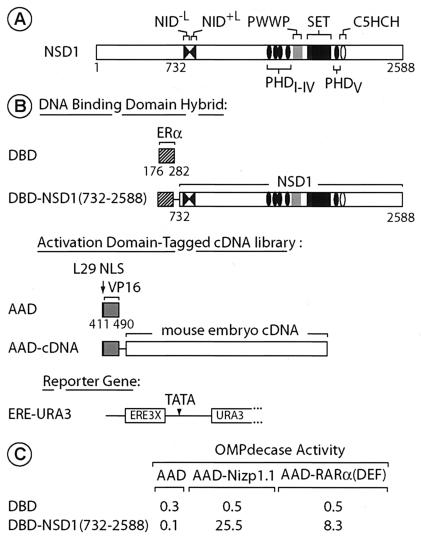
Isolation of Nizp1.1 cDNA. (A) Schematic representation of the domain organization of NSD1. The structural and functional domains are indicated. Numbers refer to amino acid positions (16). (B) Schematic representation of the ERα DBD, unfused or fused to the C-terminal region of NSD1 (amino acids 732 to 2588), which corresponds to the bait. The VP16 AAD-tagged mouse embryo cDNA expression library is represented below. The AAD tag also includes the nuclear localization signal (NLS) of the yeast ribosomal protein L29. The URA3 reporter gene, which is regulated by three EREs (ERE3X) in the yeast reporter strain PL3, is shown at the bottom. (C) Two-hybrid interaction between Nizp1.1 and NSD1(732-2588). PL3 transformants expressing the indicated DBD and AAD fusion proteins were grown in liquid medium containing uracil. Extracts were prepared and assayed for OMPdecase activity, which is expressed in millimoles of substrate per minute per milligram of protein. Values (±10%) are the averages from three independent transformants.
Initially identified in a screen for proteins interacting with the ligand binding domain (LBD) of retinoic acid receptor (RAR) (16), NSD1 was subsequently shown to differ from the other two NSDs in that it contains two N-terminally located nuclear receptor interaction domains, NID−L and NID+L (4, 16) (Fig. 1A). NID−L binds the unliganded LBDs of RARs and thyroid hormone receptors (TRs) in a ligand-inhibited manner, whereas NID+L binds the LBDs of RARs, TRs, retinoid X receptors, and estrogen receptors (ERs) in a ligand-dependent manner (16). Although the functional relevance of these interactions has not yet been established, they provide additional evidence for NSD1 playing a role in gene-specific regulation.
Considering the importance of NSD1 in controlling transcriptional regulation and development, we looked for additional molecules that would bind to NSD1 and possibly function as upstream targets or regulators of NSD1. Using NSD1 as bait in a yeast two-hybrid screen, we identified a novel NSD1-interacting zinc finger protein, designated Nizp1. Nizp1 contains multiple C2H2 zinc finger motifs and a novel C2HR zinc finger derivative that acts as a necessary and sufficient NSD1 interaction and repression domain.
MATERIALS AND METHODS
Plasmids.
Details on individual plasmid constructs, which were all verified by sequencing, are available upon request. For yeast two-hybrid assays, DNA binding domain (DBD) and acidic activation domain (AAD) fusion proteins were expressed from the multicopy plasmids pBL1 and pASV3, respectively (24). These plasmids express their inserts under the control of the phosphoglycerate kinase promoter. pBL1 contains a HIS3 marker and directs the synthesis of epitope (F region of human ER)-tagged ERα DBD fusion proteins. pASV3 contains a LEU2 marker and a cassette expressing a nuclear VP16 AAD (24). For in vitro binding assays, the indicated cDNAs were fused to glutathione S-transferase (GST) in the pGEX-2TK vector (Pharmacia Biosciences). The His-tagged Nizp1(C2HR) construct was obtained by cloning of the cDNA encoding amino acids 397 to 434 of Nizp1 into the pRSET plasmid (Invitrogen). For transfection studies in mammalian cells, the GAL4(1-147) chimera was constructed in pG4MpolyII (28). For the expression of VP16-tagged proteins in mammalian cells, the VP16 AAD (amino acids 411 to 490) was fused to the simian virus 40 T-antigen nuclear localization signal in front of the polylinker sequence of a pSG5-based expression vector named pNL1. pNL1-VP16 was then used to construct various fusions of Nizp1 and NSD1. The chimeric protein ER(C)-VP16, which is made up of amino acids 176 to 280 of ERα and amino acids 411 to 490 of VP16, has been described previously (28). For the expression of FLAG epitope-tagged proteins, the indicated cDNAs were fused in frame into a modified version of the puromycin resistance-encoding vector pSG5puro named pSG5puroFLAG (29). The 17M2-ERE-G-SEAP reporter, bearing two GAL4 DNA binding sites (UAS and 17M2) and an estrogen response element (ERE) in front of a β-globin (G) promoter-SEAP (secreted alkaline phosphatase) fusion, was obtained from the 17M2-ERE-G-CAT reporter plasmid (28), in which the bacterial chloramphenicol acetyltransferase coding sequence was replaced with the SEAP sequence. For the expression of GFP-Nizp1, the entire coding sequence of Nizp1 was cloned in frame into the pEGFP-C2 vector (Clontech).
cDNA library screening.
A VP16-tagged cDNA library derived from 9.5- to 12.5-day-postcoitum mouse embryonic poly(A)+ RNAs in the LEU2 plasmid pASV3 (16) was introduced by lithium acetate transformation into the Saccharomyces cerevisiae L40 reporter strain [MATa trp1-901 leu2-3,112 his3-Δ200 ade2 LYS2::(lexAo)4-HIS3 URA3::(lexAo)8-lacZ] expressing LexA-NSD1(732-2588) from a derivative of the TRP1 pBTM116 vector. Approximately 6 × 106 yeast transformants were selected on Trp− Leu− plates and replated at a multiplicity of infection of ∼10 onto Trp− Leu− His− plates containing 2 mM 3-aminotriazole. After 3 to 5 days, clones were isolated and retested for β-galactosidase activity. Library plasmids from the positive isolates were recovered into Escherichia coli HB101 (leuB mutant) and introduced (i) into L40 expressing either LexA, LexA-NSD1(732-2588), or LexA-lamin as a control for specificity and (ii) into the yeast reporter strain PL3 (MATα ura3-Δ1 his3-Δ200 leu2-Δ1 trp1::3ERE-URA3) expressing NSD1(732-2588) fused to the ERα DBD [DBD-NSD1(732-2588)] (Fig. 1B) or an unfused DBD as a control. A cDNA that scored positive in both yeast two-hybrid systems was designated NSD1-interacting zinc finger protein 1 clone 1 (Nizp1.1). A cDNA encoding the DEF region of RARα was also identified in the screen, in agreement with previously reported results (16). The 1,755-bp full-length coding sequence of Nizp1 was obtained by PCR amplification with specific primers that had appropriate restriction enzyme cutting sites, with an EST clone (GenBank accession no. BF165312) including Nizp1 nucleotides 116 to 3531 used as a template (see Fig. 2A).
FIG. 2.
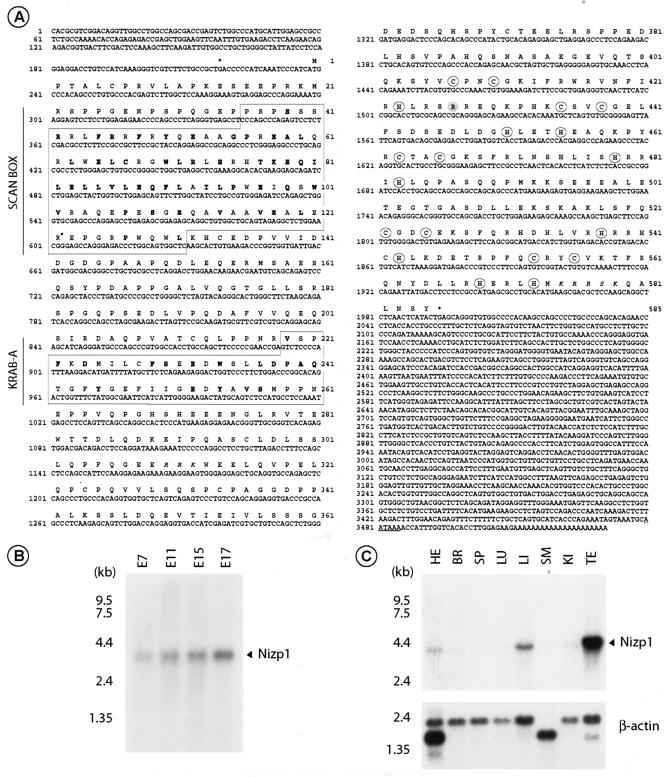
Characterization of mouse Nizp1 cDNA. (A) Nucleotide and deduced amino acid sequences of mouse cDNA encoding Nizp1. The 3,531 nucleotides of Nizp1 cDNA, the putative translation start site (position +1), the 583-amino-acid ORF, and 5′-flanking termination codons (marked by asterisks) are shown. Amino acid residues matching the consensus sequences in the SCAN box (amino acids 36 to 131 [boxed]) and the KRAB A domain (amino acids 219 to 259 [boxed]) are shown in bold. Cysteine (C) and histidine (H) residues belonging to the C2HR motif and C2H2-type zinc fingers are circled. Italicized sequences denote the putative nuclear localization signals. (B and C) Northern blot analyses of Nizp1. RNA blots containing poly(A)+ RNAs from the indicated mouse embryo developmental stages (B) and various adult mouse tissues, including the heart (HE), brain (BR), spleen (SP), lung (LU), liver (LI), skeletal muscle (SM), kidney (KI), and testis (TE) (C), were hybridized with a Nizp1-specific cDNA fragment (nucleotides +752 to +1923, corresponding to the library-derived cDNA insert, Nizp1.1). A single mRNA species of 3.9 kb was seen, as indicated by an arrowhead. In panel C, hybridization to a β-actin probe is shown as a control for mRNA integrity and loading. The positions of RNA size markers are shown on the left.
Northern blot analysis.
Northern blots containing poly(A)+ RNAs from mouse embryos at various developmental stages and from various adult mouse tissues (Clontech Laboratories Inc.) were hybridized with the library-derived Nizp1.1 cDNA clone encoding amino acids 251 to 562 of Nizp1 (nucleotides 751 to 1923) or a β-actin cDNA fragment (control) labeled with [α-32P]dCTP by random priming according to the manufacturer's instructions.
Electrophoretic mobility shift assays.
A 145-bp DNA fragment that was isolated from P19 genomic DNA and cloned into pBluescript (GenBank accession no. AT155467) (35) was end labeled with [α-32P]dCTP by using Klenow DNA polymerase and was then incubated with 50 ng of purified GST-Nizp1 fusion proteins in a buffer containing 25 mM Tris-HCl (pH 7.9), 25 mM NaCl, 2 mM MgCl2, 20% glycerol, 5 mM dithiothreitol, and 0.1% Tween 20 for 15 min at room temperature. For competition experiments, 1.0 μg of nonspecific DNA [poly(dI-dC) · poly(dI-dC)] was incubated with the reaction. Samples were loaded into a native 8% Tris-glycine gel and electrophoresed at room temperature. The gels were then dried and exposed to autoradiography.
Yeast two-hybrid interaction assays.
S. cerevisiae PL3 transformants were grown exponentially for five generations in a selective medium supplemented with uracil, and cell extracts were assayed for orotidine 5′-monophosphate decarboxylase (OMPdecase) activity as described previously (24).
Mammalian transfection and reporter assays.
Cells were transiently transfected in six-well dishes by using Superfect (QIAGEN) with various combinations of expression and reporter plasmid DNAs. Forty-eight hours after transfection, the medium was assayed for SEAP activity as described previously (31), and the cells were harvested and assayed for β-galactosidase activity as described previously (28).
Cell line establishment, nuclear extract preparation, and immunoprecipitation.
Mouse N2A neuroblastoma cells (5 × 105 cells per 10-cm-diameter plate) were transfected with 10 μg of an expression vector encoding GFP-Nizp1. Cells were selected with 1 mg of Geneticin (GIBCO BRL)/ml added to the growth medium 48 h after transfection over a period of 10 days with regular medium changes. Several drug-resistant clones were picked and tested for the expression of GFP-Nizp1 by fluorescence analysis. A GFP-Nizp1-positive cell line was propagated and retransfected with a selection plasmid encoding either unfused FLAG (pSG5puroFLAG) or FLAG-tagged NSD1. Puromycin (10 μg/ml) was added 48 h after transfection, and stable puromycin-resistant clones were isolated after 10 days of selection. Cells were harvested by use of a rubber policeman into ice-cold 10 mM phosphate-buffered saline, pH 7.2, washed three times in phosphate-buffered saline, and resuspended in nucleus isolation buffer (NIB; 15 mM Tris-HCl [pH 7.5], 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 1 mM dithiothreitol, 250 mM sucrose, and a protease inhibitor cocktail). An equal volume of NIB containing 0.6% NP-40 was added, and the suspension was gently mixed and incubated on ice for 5 min. Nuclei were pelleted by centrifugation at 2,000 × g for 5 min at 4°C and washed with NIB before a final suspension in nucleus extraction buffer (NEB; 25 mM Tris-HCl [pH 8.0], 10% glycerol, 0.2% NP-40, and a protease inhibitor cocktail) containing 500 mM NaCl. After incubation on ice for 15 min, the samples were briefly vortexed and centrifuged at 15,000 × g for 30 min at 4°C. The resulting nuclear extracts were mixed with an equal volume of NEB and incubated with 50 μl of a specific antibody coupled to protein G-Sepharose beads for 12 h at 4°C. The immunoprecipitates were washed five times in NEB containing 250 mM NaCl, and the final pellets were resuspended in 50 μl of 2× Laemmli sodium dodecyl sulfate (SDS) buffer (100 mM Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 2% β-mercaptoethanol, 0.002% bromophenol blue) and analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting as previously described (28).
In vitro binding assays.
GST pull-down assays were performed as previously described (28). When required, 10 mM 1,10-phenanthroline (final concentration) was added to the binding buffer (BB). The 1,10-phenanthroline was purchased from Sigma and was added from a 0.5 M stock in ethanol; as a negative control, an equal amount of ethanol was added to BB. Varying the NaCl and detergent concentrations in BB revealed that the NSD1-Nizp1 interaction was stable in 100 to 900 mM NaCl and 0.1 to 2% NP-40.
Nucleotide sequence accession number.
The DDBJ/EMBL/GenBank accession number for the Nizp1 sequence reported in this paper is AY303242.
RESULTS
Identification of Nizp1, a novel NSD1-interacting protein with several conserved domains.
The yeast two-hybrid system was used to isolate mouse cDNAs encoding proteins that interact with NSD1. Since the N-terminal region of NSD1 (amino acids 1 to 731) contains an efficient transcriptional activation domain which can function in yeast (16), the region of the C-terminal 1,857 amino acids of NSD1 [NSD1(732-2588)] (Fig. 1A) fused to the LexA protein (amino acids 1 to 202) was used as bait to screen a library of mouse embryonic cDNAs fused to the DNA sequence encoding the AAD of the VP16 protein in the yeast reporter strain L40 (see Materials and Methods). Isolated cDNAs were classified as positive when they were retested in another version of the two-hybrid system using the DBD of ERα fused to NSD1 [DBD-NSD1(732-2588)] (Fig. 1B) or an unfused ERα DBD as a control in the yeast reporter strain PL3, which contains a URA3 reporter gene driven by three EREs (ERE-URA3; Fig. 1B) (24). Among the positive clones, one fused cDNA (designated AAD-Nizp1.1 for AAD-NSD1-interacting zinc finger protein 1 clone 1; Fig. 1C) showed an absolute requirement for coexpressed DBD-NSD1(732-2588) to transactivate the URA3 reporter. When the OMPdecase activity of the URA3 gene product was measured, the coexpression of DBD-NSD1(732-2588) with AAD-Nizp1.1 resulted in a >250-fold increase (Fig. 1C). This increase was similar to that resulting from the interaction of NSD1 and RARα [Fig. 1C, DBD-NSD1(732-2588) and AAD-RARα(DEF)].
Sequencing of the Nizp1.1 cDNA clone revealed a novel 1,173-bp open reading frame (ORF) (nucleotides 751 to 1923 in Fig. 2A) fused in frame over its entire length with the VP16 AAD. BLAST searches of the GenBank database revealed identities with several overlapping mouse expressed sequence tags that represented more 5′ and 3′ regions of the cDNA. We determined the complete sequences of two overlapping cDNA clones from which expressed sequence tags G919060 (Image clone 4947290) and BF165312 (Image clone 4019493) were derived and found a 3,531-bp cDNA (hereafter called Nizp1 cDNA) encompassing an ORF of 1,755 bp flanked by a 5′ untranslated region of 237 bp and a 3′ untranslated region of 1,539 bp and containing a consensus polyadenylation signal (AATAAA) at nucleotides 3480 to 3485 (Fig. 2A). A Northern blot analysis of poly(A)+ RNAs from mouse embryos at various developmental stages (days E7 to E17) revealed a single 3.9-kb mRNA species (Fig. 2B) which was also detected in most adult mouse tissues tested, with the highest levels occurring in the testes and moderate levels occurring in the heart, liver, and kidneys (Fig. 2C). After a longer exposure, the transcript was also detected at very low levels in the brain, spleen, and lungs, but not in skeletal muscle (data not shown). BLAST searches of the mouse and human genome sequences showed the presence of a single Nizp1 locus on chromosomes 11B2 and 1q44, respectively.
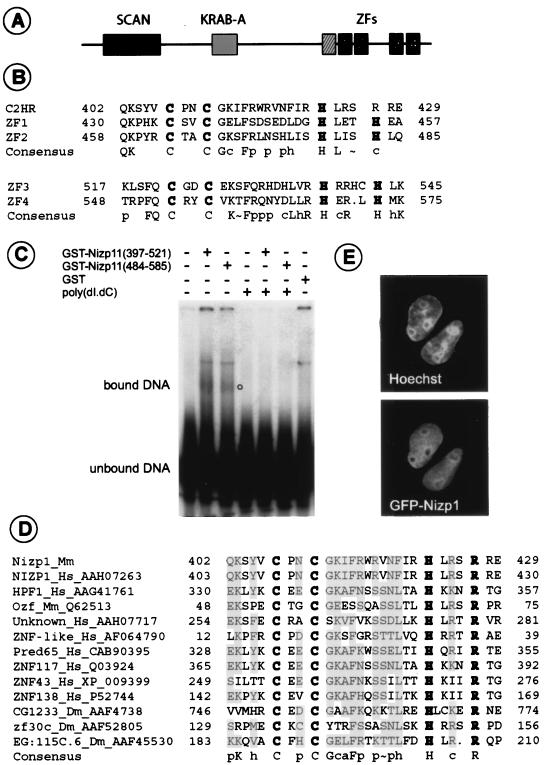
The putative protein translated from the coding sequence of Nizp1 (Fig. 2A) consists of 585 amino acids, with a calculated molecular mass of 66.5 kDa. A protein database analysis revealed several conserved sequence motifs that are found in transcription factors (Fig. 2A and 3A). The N-terminal region of Nizp1 (amino acids 36 to 131) contains a SCAN box, or leucine-rich region (LeR) (Fig. 2A). This motif was previously identified in a number of C2H2 zinc finger proteins and was shown to function as an oligomerization domain mediating self-association or association with other proteins bearing SCAN domains (9, 38). A Krüppel-associated box-A (KRAB-A) subdomain was identified in Nizp1 between amino acid residues 219 and 259 (Fig. 2A). The KRAB domain is a conserved module that is only found in association with genes containing C2H2 zinc finger motifs (9) and may be comprised of A and B subdomains, or alternatively, only A subdomains (reference 25 and references therein). The KRAB-A subdomain has been shown to act as a potent transcriptional repressor element when it is targeted to DNA (26; also see Discussion). At the C terminus, Nizp1 contains four putative zinc fingers (ZF1 to ZF4) that occur in two clusters (Fig. 2A and 3A) and have moderate homology with each other (Fig. 3B). They all conform well to the C2H2 zinc finger consensus CX2-4CX3Fx5LX2HX3-4H (where “X” is any amino acid) (19) and display DNA binding activity in vitro (Fig. 3C). They are preceded by a degenerate copy of this domain which has an Arg residue in place of the last His residue (Fig. 2A and 3B). This finger derivative, herein referred to as the C2HR motif, was also identified in the putative human Nizp1 counterpart (GenBank accession no. AAHO7263) (Fig. 3D) as well as in several (un)characterized zinc finger proteins from mammals and Drosophila (Fig. 3D).
FIG. 3.
Structural features, DNA binding activity, and nuclear localization of Nizp1. (A) Schematic representation of conserved domains of Nizp1. Black boxes indicate conventional C2H2-type zinc finger motifs (ZF1 to ZF4), while the striped box denotes a degenerate C2HR motif. (B) Alignment of the non-C2H2 and C2H2 fingers of Nizp1. Based on sequence homology, these putative fingers were classified into two subgroups. The consensus sequence for each subgroup is indicated according to the following convention: a, aliphatic residues (A, I, L, and V); c, charged residues (D, E, K, R, and H); h, hydrophobic residues (L, I, V, M, F, Y, and W); p, polar residues (C, D, E, H, K, N, Q, R, S, and T); ∼, S or T. Conserved cysteine (C) and histidine (H) residues are shown in bold. (C) DNA binding analysis of Nizp1 zinc finger clusters. Purified GST-zinc finger fusion proteins bearing residues 397 to 521 and 484 to 585 of Nizp1 and unfused GST were incubated with a radioactively labeled 145-bp DNA fragment. For competition experiments, 1.0 μg of poly(dI-dC) was used. Complexes were resolved by native polyacrylamide gel electrophoresis. The empty circle denotes the position of complexes. (D) Alignment of the C2HR motifs. Sequences are listed according to genus name, species name, and the corresponding GenBank accession number. Mm, Mus musculus; Hs, Homo sapiens; Dm, Drosophila melanogaster. The shading indicates 70% consensus. (E) Nuclear localization of GFP-Nizp1 in transfected P19 EC cells. The transfected cells were counterstained with the blue fluorescent DNA-binding dye Hoechst 33342 immediately before analysis by fluorescence microscopy. Images were collected from a single cell by using filter sets that selectively discriminate GFP and Hoechst 33342 fluorescence.
To examine the subcellular localization of Nizp1, we fused the entire coding sequence of Nizp1 3′ to sequences encoding green fluorescent protein (GFP). The fusion protein (GFP-Nizp1) was introduced into P19 embryonal carcinoma (P19 EC) cells by transient transfection, and its localization was assessed by fluorescence microscopy. Cells transfected with GFP-Nizp1 showed a nuclear fluorescence pattern, with exclusion from nucleoli (Fig. 3E). Under similar conditions, control cells expressing GFP alone produced a diffuse distribution of fluorescence in both the cytoplasm and the nucleus (data not shown).
Nizp1 coimmunoprecipitates with NSD1.
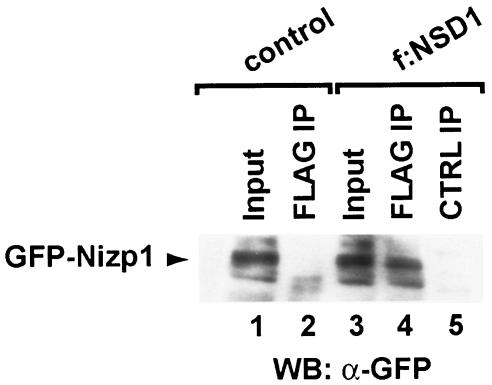
The identification of Nizp1.1 as an NSD1 interaction partner in yeast led us to investigate whether the two full-length proteins were also capable of interaction in mammalian cells. Two mouse Neuro-2A (N2A)-derived cell lines that stably expressed GFP-Nizp1 alone (control) or in combination with FLAG-tagged full-length NSD1 (f:NSD1) were generated (see Materials and Methods). Nuclear extracts from these cell lines were immunoprecipitated with the anti-FLAG antibody M2, and the immunoprecipitates were probed for the presence of the GFP-Nizp1 protein (Fig. 4). Coimmunoprecipitation of GFP-Nizp1 was clearly detected in the f:NSD1 immunoprecipitate (Fig. 4, lane 4), but not in control immunoprecipitates (lane 2). Also, no signal from GFP-Nizp1 was detected in f:NSD1 immunoprecipitations when an irrelevant antibody was used (lane 5). These results indicate that full-length Nizp1 and NSD1 can interact with one another in nuclear extracts of Neuro-2A cells.
FIG. 4.
Nizp1 coimmunoprecipitates with NSD1. Nuclear extracts from Neuro-2A (N2A) cells expressing GFP-Nizp1 alone (control) or in combination with FLAG-NSD1 (f:NSD1) were analyzed by Western blotting either directly (input) or after immunoprecipitation with the M2 anti-FLAG antibody (FLAG IP) or with an irrelevant antibody (anti-SNF5; CTRL IP). A Western blot of the immunoprecipitates probed with an anti-GFP monoclonal antibody is shown. Lanes 1 and 3 (input) correspond to 1/20 the amount of nuclear extract used for immunoprecipitation. The arrowhead shows the position of the GFP-Nizp1 protein.
Nizp1 interacts with the cysteine-rich (C5HCH) domain of NSD1.
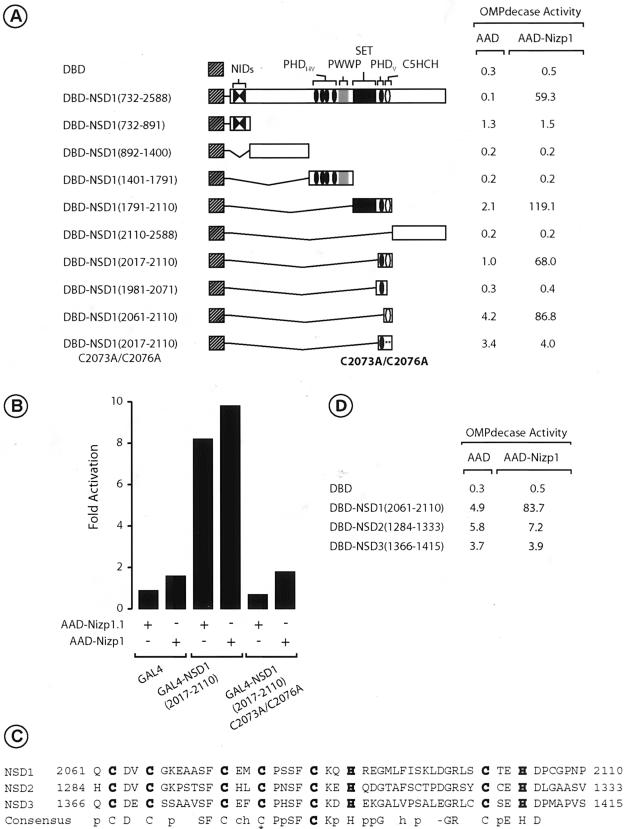
To identify the Nizp1-interacting domain of NSD1, we performed a deletion analysis with the yeast two-hybrid system. Various deletion mutants targeting key functional and structural domains of NSD1 (Fig. 5A) were fused to the ERα DBD and tested for the ability to interact with full-length Nizp1 fused to the VP16 activation domain in the yeast strain PL3. A mutant lacking N-terminal residues 1 to 2016, including NID−L and NID+L, PHD fingers PHDI to PHDIV, the PWWP motif, and the SET HMTase domain, as well as C-terminal residues 2111 to 2588 interacted with Nizp1 [Fig. 5A, DBD-NSD1(2017 to 2110)]. This region of NSD1 contains the PHD finger PHDV (positions 2018 to 2060) and the C5HCH motif (positions 2062 to 2103). Further deletions defined the minimal region required for the interaction as a 50-amino-acid region (positions 2061 to 2110) containing only the C5HCH motif (Fig. 5A). Mutations in this motif that changed cysteine residues 2073 and 2076 to alanine eliminated the interaction [Fig. 5A, DBD-NSD1(2017-2110)C2073A/C2076A] and provided additional evidence that the C5HCH motif of NSD1 is capable of interacting with Nizp1 in yeast.
FIG. 5.
Nizp1 interacts with C5HCH motif of NSD1. (A) Mapping of Nizp1-interacting domain in NSD1. Plasmids expressing individual regions of NSD1 fused to the ERα DBD (as indicated) were introduced into PL3 together with either the unfused VP16 AAD or an AAD fusion containing full-length Nizp1. Transformants were grown in liquid medium containing uracil. Extracts were prepared and assayed for OMPdecase activity, which is expressed as millimoles of substrate per minute per milligram of protein. Values (±10%) are the averages from three independent transformants. (B) The NSD1 C5HCH motif interacts with Nizp1 in mammalian cells. HT1080 cells were transiently cotransfected with 1 μg of the 17M2-ERE-G-SEAP reporter, 1 μg of pCH110 (expressing β-galactosidase), and 1 μg of expression vectors coding for the indicated GAL4 and AAD chimeras. Forty-eight hours after transfection, the secreted alkaline phosphatase activity was measured. The data are expressed relative to the SEAP activity measured in the presence of the unfused AAD expression vector. All values (±15%) represent the averages of two independent transfections performed in triplicate after normalization to the internal control β-galactosidase activity of pCH110. (C) Alignment of the C5HCH motifs. Numbers refer to amino acid positions in the corresponding protein. Cysteine and histidine residues are shown in bold. A consensus sequence is indicated below the alignment using the convention described for Fig. 3B. The asterisk indicates the position of the Weaver syndrome missense mutation C2183S in the C5HCH motif of NSD1 (11). (D) The NSD2 and NSD3 C5HCH motifs have no Nizp1 binding capability. PL3 transformants expressing the indicated DBD and AAD fusions were treated as described for panel A. Note that in all transformants the expression of the DBD and AAD fusion proteins was confirmed by Western blotting using the antibodies F3 against the F region of ERα and 2GV4 against VP16, respectively (data not shown).
We confirmed this result in mammalian cells by using a mammalian two-hybrid assay. Residues 2017 to 2110 of NSD1, comprising the C5HCH motif, were expressed as a fusion protein with the GAL4 DBD [GAL4-NSD1(2017-2110)], which was tested for its interaction with VP16 AAD fusion proteins containing the library-derived Nizp1.1 cDNA sequence (amino acids 173 to 562; AAD-Nizp1.1) or the entire coding sequence of Nizp1 (AAD-Nizp1). These fusion proteins were transfected into human HT1080 cells together with a GAL4-responsive SEAP reporter gene, and SEAP production was assayed. The cotransfection of GAL4-NSD1(2017-2110) with either AAD-Nizp1.1 or AAD-Nizp1 significantly activated the reporter (Fig. 5B). In keeping with the yeast two-hybrid data, no increase in reporter gene activity was observed with a GAL4 fusion protein containing the mutated NSD1 C5HCH motif [Fig. 5B, GAL4-NSD1(2017-2110)C2073A/C2076A].
Since NSD2 and NSD3 also contain C-terminal C5HCH motifs (4) (Fig. 5C), we investigated whether Nizp1 could interact with these motifs. The corresponding regions were fused to the ERα DBD and tested for their interaction with AAD-Nizp1 in yeast. No significant increase in reporter gene activity above the AAD background was observed (Fig. 5D), indicating that in contrast to the case for the NSD1 C5HCH motif, the NSD2 and NSD3 C5HCH motifs did not interact with Nizp1. These motifs share about 45% amino acid sequence identity (Fig. 5C). Thus, Nizp1 may recognize particular amino acid residues within the C5HCH motif of NSD1 which are not conserved in the NSD2 and NSD3 C5HCH sequences.
Nizp1 interacts with NSD1 through the C2HR motif.
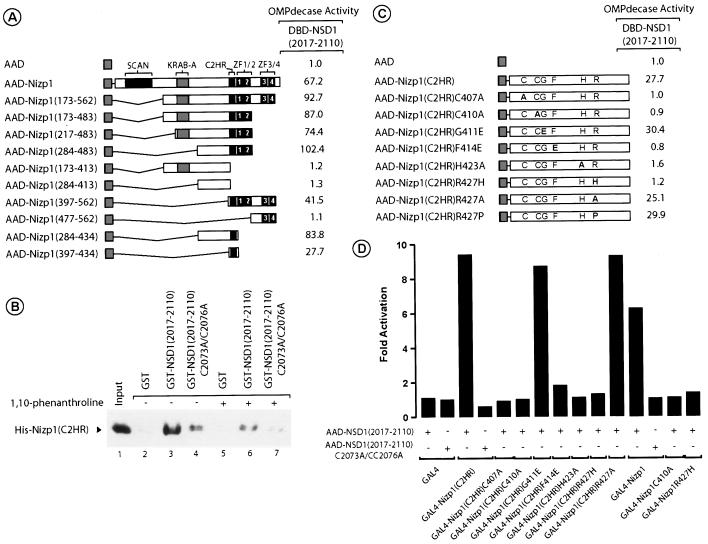
To determine which sequences within Nizp1 are responsible for the interaction between Nizp1 and NSD1, we introduced several deletions into Nizp1 and fused these mutants to the VP16 AAD. The resulting AAD-Nizp1 fusion proteins were assayed for their interactions with DBD-NSD1(2017-2110) in the yeast reporter strain PL3 (Fig. 6A). As indicated by the reporter gene activity, this deletion analysis showed that an NSD1-interacting domain is present between residues 397 and 434 and is centered around the C2HR motif (amino acids 402 to 429; Fig. 3B) [Fig. 6A, AAD-Nizp1(397-434)].
FIG. 6.
The C2HR motif of Nizp1 acts as a necessary and sufficient NSD1-interacting domain. (A) Mapping of the NSD1-interacting domain of Nizp1. Plasmids expressing individual regions of Nizp1 fused to the VP16 AAD were introduced into PL3 together with DBD-NSD1(2017-2110). Transformants were grown in liquid medium containing uracil. Extracts were prepared and assayed for OMPdecase activity, which is expressed as millimoles of substrate per minute per milligram of protein. Values (±10%) are the averages from three independent transformants. The expression of the various AAD fusion proteins was confirmed by Western blotting (data not shown). (B) The Nizp1 C2HR motif binds directly to the C5HCH motif of NSD1 in a metal-dependent manner. Purified His-Nizp1(C2HR) (residues 397 to 434) was incubated in a batch assay with control GST (lanes 2 and 5), GST-NSD1(2017-2110) (lanes 3 and 6), or GST-NSD1(2017-2110)C2037A/C2076A (lanes 4 and 7) bound to glutathione-Sepharose beads in the absence (−) or presence (+) of 10 mM 1,10-phenanthroline. Bound Nizp1 was detected by Western blotting with an anti-histidine tag antibody. Lane 1 shows 1/10 of the amount of input His-Nizp1, the position of which is indicated by an arrow. (C) Effects of C2HR point mutations on NSD1 interaction in yeast. PL3 transformants expressing the indicated DBD and AAD fusion proteins were treated as described for panel A. (D) The integrity of the C2HR motif in Nizp1 is essential for its interaction with NSD1 in mammalian cells. HT1080 cells were transiently cotransfected with 1 μg of the 17M2-ERE-G-SEAP reporter, 1 μg of pCH110 (expressing β-galactosidase), and 1 μg of expression vectors coding for the indicated GAL4 and AAD chimeras. Forty-eight hours after transfection, the secreted alkaline phosphatase activity was measured. The data are expressed relative to the SEAP activity measured in the presence of the unfused AAD expression vector. All values (±15%) represent the averages of two independent triplicate transfections after normalization to the internal control β-galactosidase activity of pCH110.
The direct binding of the C2HR motif of Nizp1 to NSD1 was studied in vitro by using purified recombinant proteins. A purified E. coli-expressed histidine-tagged Nizp1 fusion protein bearing residues 397 to 434 [referred to hereafter and in Fig. 6B as His-Nizp1(C2HR)] was assessed for its ability to interact with GST-NSD1(2017-2110) and GST-NSD1(2017-2110)C2073A/C2076A immobilized on glutathione-Sepharose beads. Western blotting indicated that His-Nizp1(C2HR) was efficiently retained on GST-NSD1(2017-2110) beads (Fig. 6B, lane 3). In contrast, we detected very little binding to GST-NSD1(2017-2110)C2073A/C2076A (Fig. 6B, lane 4) and no interaction with GST alone (Fig. 6B, lane 2). Thus, the C2HR motif of Nizp1 can interact with the C5HCH motif of NSD1 both in yeast and in vitro. To determine whether this direct interaction was metal dependent, we performed GST pull-down assays in the presence of 1,10-phenanthroline (a zinc chelator). As shown in Fig. 6B, the amount of His-Nizp1(C2HR) retained on GST-NSD1(2017-2110) beads was severely reduced in the presence of 10 mM 1,10-phenanthroline, thus indicating that the C2HR-C5HCH interaction required metal ions.
To determine the amino acid sequence regions within the C2HR motif that are necessary for the NSD1 interaction, we created a series of C2HR derivatives with point mutations in the most highly conserved residues by in vitro site-directed mutagenesis (Fig. 6C). Each mutant was tested for its ability to interact with NSD1(2017-2110) in yeast. Unlike the case for the wild-type C2HR motif, no interaction was detected with the mutants in which the putative zinc-coordinating residues C407, C410, and H423 were replaced with alanine residues (Fig. 6C), indicating that these residues may contribute to the formation of a zinc finger structure that is critical for the NSD1 interaction (see Discussion). Similarly, an F414E mutant in which the conserved aromatic residue at position 414 was replaced with an aspartate residue failed to interact with NSD1, whereas a glycine replacement immediately downstream of the second cysteine residue yielded a G411E mutant that was not affected in its ability to interact with NSD1 (Fig. 6C). We also mutated the R427 arginine residue to histidine in an R427H mutant to mimic the C2H2 configuration of a classical zinc finger. This mutant was unable to interact with NSD1 (Fig. 6C), suggesting that the unusual C2HR configuration is also critical for the interaction. For a further investigation of the importance of the R427 residue for the interaction, this residue was also replaced with an alanine or a proline residue. The resulting C2HA and C2HP motifs both interacted with NSD1 with the same efficiency as C2HR (Fig. 6C), further supporting the view that the non-C2H2 finger configuration is an essential feature of this particular protein interaction domain.
We confirmed these results in mammalian cells (Fig. 6D). Wild-type and mutated C2HR derivatives were fused to the GAL4 DBD, and the resulting GAL4 fusion proteins were tested for interactions with AAD-NSD1(2017-2110) or NSD1(2017-2110)C2073A/C2076A in transiently cotransfected HT1080 cells. As observed in yeast cells, the mutants C407A, C410A, H423A, F414E, and R427H were defective in mediating the NSD1 interaction, whereas the mutants G411E and R427A displayed wild-type activities (Fig. 6D). To definitively demonstrate that the C2HR motif is actually functional in Nizp1, we introduced the deleterious mutations C410A and R427H into full-length Nizp1 (Fig. 6D). In contrast to wild-type Nizp1, Nizp1C410A and Nizp1R427H showed no interaction with NSD1 (Fig. 6D). Taken together, these results indicate that the C2HR motif of Nizp1 acts as a necessary and sufficient NSD1-interacting domain.
The Nizp1 C2HR motif exerts transcriptional repression through NSD1 recruitment.
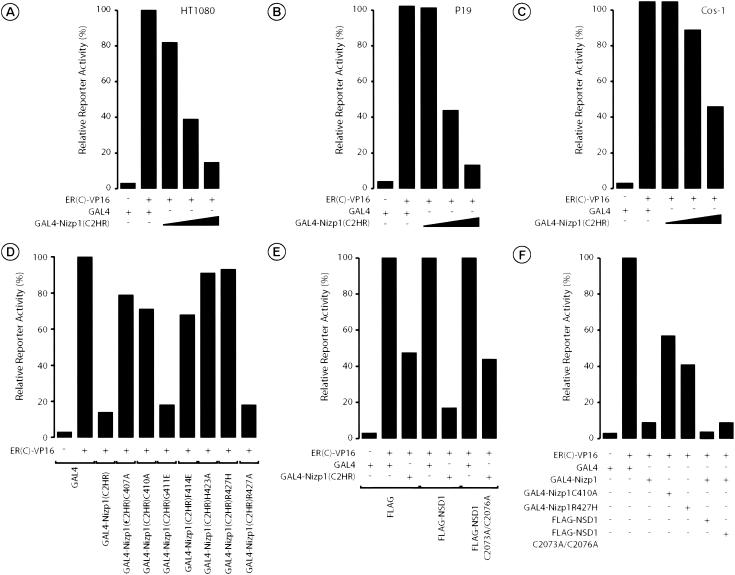
Having established that the C2HR motif functions to mediate the interaction with NSD1 and that NSD1 contains several transcriptional repression domains (16, 34), we set out to determine if the region of Nizp1 harboring this novel protein-protein interaction motif can confer a transcriptional repression function on a heterologous DBD. HT1080 cells were transiently transfected with the GAL4 and ER reporter 17M2-ERE-G-SEAP in the presence of the chimeric transactivator ER(C)-VP16 (see Materials and Methods). As expected, ER(C)-VP16 stimulated transcription of the reporter gene (Fig. 7A). Interestingly, transcriptional activation was significantly repressed by GAL4-Nizp1(C2HR) in a dose-dependent manner (Fig. 7A). This GAL4 DBD fusion protein did not affect transcription from a reporter gene lacking the GAL4 binding sites (data not shown), indicating that the repression by Nizp1(C2HR) is dependent on site-specific DNA binding. Such a specific repression was also observed in transfected P19 EC and Cos-1 cells (Fig. 7B and C).
FIG. 7.
The Nizp1 C2HR motif is a novel transcriptional repression domain acting through interaction with the NSD1 corepressor. (A to C) The Nizp1 C2HR motif represses activated transcription in a dose-dependent manner when targeted to the promoter. The 17M2-ERE-G-SEAP (1 μg) reporter and 1 μg of the pCH110-β-gal plasmid (as an internal control) were cotransfected into HT1080 (A), P19 EC (B), or Cos-1 (C) cells with the indicated pSG5-based vectors expressing the activator ER(C)-VP16 (50 ng) and the unfused GAL4 DBD (1 μg) or residues 397 to 434 of Nizp1, comprising the C2HR motif [GAL4-Nizp1(C2HR)] (0.04, 0.2, or 1 μg). SEAP activities are expressed relative to controls of ER(C)-VP16 and the unfused GAL4 (taken as 100%). Values (±15%) represent the averages of two independent triplicate transfections after normalization to β-galactosidase activities. (D) The repressive activity of the Nizp1 C2HR motif correlates with its ability to interact with NSD1. Cotransfection assays were conducted in HT1080 cells with the indicated GAL4-Nizp1(C2HR) mutants (1 μg) as described for panel A. The expression of the GAL4 fusion proteins was confirmed by Western blotting using the antibody 2GV3 against the GAL4 DBD (data not shown). (E) Enhancement of C2HR-mediated repression by exogenous NSD1. Cos-1 cells were transfected with the 17M2-ERE-G-SEAP reporter plasmid (1 μg), 1 μg of the pCH110-β-gal plasmid, 50 ng of ER(C)-VP16, and 1 μg of a GAL4 expression vector encoding GAL4 or GAL4-Nizp1(C2HR), together with 5 μg of unfused FLAG, FLAG-tagged wild-type NSD1, or FLAG-NSD1C2073A/C2076A. SEAP activities are expressed relative to those measured in the presence of the unfused GAL4 expression vector (taken as 100%). Values (±15%) represent the averages of two independent triplicate transfections after normalization to β-galactosidase activities. (F) The full repressing activity of Nizp1 requires the NSD1 interaction. Cotransfection assays were conducted in HT1080 cells with the indicated wild-type and mutated GAL4-Nizp1 chimeras (1 μg), together with 5 μg of unfused FLAG, FLAG-tagged wild-type NSD1, or FLAG-NSD1C2073A/C2076A, as described for panel A. SEAP activities are expressed as described above. The expression of the GAL4 fusion proteins was confirmed by Western blotting using the antibody 2GV3 against the GAL4 DBD (data not shown).
We next examined the repressive activities of C2HR mutants which were impaired in the ability to interact with NSD1. The interaction-deficient mutations C407A, C410A, F414E, H423A, and R427H significantly inhibited the ability of GAL4-Nizp1(C2HR) to repress transcription, whereas the silent mutations G411E and R427A had no effect (Fig. 7D). These results provide strong evidence for a tight correlation between the efficiency of the interaction of mutated C2HR motifs with NSD1 and their ability to repress transcription, indicating that this interaction may be involved in transcriptional repression.
To further investigate the functional consequences of the C2HR-NSD1 interaction, we assayed the repressive activity of the Nizp1 C2HR motif alone or in combination with overexpressed NSD1 in transiently transfected Cos-1 cells. As shown in Fig. 7E, NSD1 enhanced the repression by GAL4-Nizp1(C2HR) up to threefold. In control experiments, the coexpression of NSD1 did not suppress ER(C)-VP16-stimulated transcription in the presence of the GAL4 DBD alone (Fig. 7E). Moreover, an NSD1 mutant protein, NSD1C2073A/C2076A, bearing mutations that abolished the interaction with the C2HR motif, did not enhance GAL4-C2HR-mediated repression (Fig. 7E).
The above data indicate that NSD1 may be a C2HR-interacting corepressor. To evaluate the role of the C2HR-NSD1 interaction in the context of full-length Nizp1, we tested the effects of the interaction-deficient mutations C410A and R427H on the repression activity of the GAL4-Nizp1 protein (Fig. 7F). Upon transient transfection in HT1080 cells, GAL4-Nizp1 efficiently repressed ER(C)-VP16-mediated activation of the reporter gene (Fig. 7F). The mutations C410A and R427H significantly reduced, but did not fully relieve, this repression (Fig. 7F, compare GAL4-Nizp1 with GAL4-Nizp1C410A and GA4L-Nizp1R427H), while a modest (2.3-fold) but reproducible increase in the repression activity of GAL4-Nizp1 was observed by overexpressing NSD1, but not NSD1C2073A/C2076A (Fig. 7F). Taken together, these results strongly suggest that NSD1 is involved in the repression mediated by full-length Nizp1 but that other mechanisms of repression may also operate.
DISCUSSION
Nizp1 belongs to SCAN-KRAB subfamily of C2H2-type zinc finger proteins.
C2H2-type zinc finger proteins represent one of the largest and most diverse superfamilies of nucleic acid binding proteins in eukaryotes. It has been estimated that over 500 genes in the human genome encode proteins with zinc finger domains of the C2H2 type (23). These zinc finger proteins are commonly composed of tandem arrays of two or more zinc fingers, and they have been classified into various subgroups on the basis of the presence of other conserved sequence elements (9, 23). One of these subgroups consists of proteins predicted to have a SCAN-KRAB-(C2H2)x domain organization, in which the KRAB repression domain is complete, or more frequently, consists of only the A domain (9). Members of this subgroup are encoded by vertebrate-specific genes (23), and among these are genes important in development, gene regulation, and diseases, such as the hypoalphalipoproteinemia susceptibility gene, ZNF202, which encodes a transcriptional repressor that binds to elements found in genes involved in lipid metabolism (44); ZNF215, a Beckwith-Wiedemann syndrome-associated gene (3); and ZNF213, which is linked with familial Mediterranean fever (7). In the present study, we have identified and characterized a novel mouse SCAN-KRAB-C2H2 zinc finger gene that has been named Nizp1 and found to be expressed in many tissues. The corresponding human NIZP1 gene was mapped to chromosome 1q44, a locus associated with several human diseases and neurological disorders (see reference 10 and references therein).
Nizp1 encodes a 585-amino-acid protein which localizes to the nucleus. Along with an N-terminal SCAN box associated with a KRAB A domain, Nizp1 contains a C-terminal C2HR motif followed by four classical C2H2-type zinc fingers that are capable of binding to DNA directly. Using transient transfection and reporter assays, we have shown that Nizp1 can confer a transcriptional repression function to a heterologous DNA binding domain. Taken together, these results suggest that Nizp1 functions in the nucleus as a DNA-binding transcriptional repressor.
The C2HR motif as a novel NSD1 recruitment and repression motif.
Although the recruitment of corepressors is a widely used mechanism of transcriptional repression, the signals and molecular determinants for corepressor-repressor interactions remain largely unknown. In some instances, these interactions have been shown to be mediated by short sequence motifs. Thus, the Drosophila transcriptional repressors Dorsal and Hairy recruit the Groucho corepressor via a WPRW motif (6), and fusion of this motif to the GAL4 DBD results in a chimera that can repress transcription in a Groucho-dependent manner. Another peptide motif involved in corepressor recruitment is the PLDLS motif, which is critical for mediating an interaction of the E1A oncogene product with CtBP (see reference 43 and references therein). We report here the discovery of a novel transcriptional repression domain that maps to the C2HR motif of Nizp1 and offer a number of lines of evidence indicating that NSD1 is a C2HR corepressor: (i) NSD1 interacts with the C2HR domain both in vivo and in vitro, (ii) mutations in the C2HR motif that abolish repression concomitantly abolish the interaction with NSD1, and (iii) the overexpression of NSD1 enhances C2HR-mediated repression in a manner dependent on the integrity of the domain in NSD1 that binds the C2HR motif.
The metal dependence of the C2HR-NSD1 interaction led us to explore the possibility that the C2HR motif, first identified in silico as a degenerate C2H2-type zinc finger, may in fact represent a true zinc finger. We therefore examined the effects of alterations at the cysteine and histidine residues predicted to coordinate zinc and found that these residues are crucial for binding to NSD1. We also investigated the functional roles of other amino acids within the C2HR sequence which may constitute putative fourth zinc-coordinating residues, such as the glutamate and histidine residues at positions 429 and 433, respectively (Fig. 2A). However, the replacement of these amino acids by an Ala residue, E429A and H433A, did not affect the ability of Nizp11(397-434) to interact with NSD1 and to repress transcription on its own (data not shown). Taken together, our data are consistent with and extend those of previous reports in which three zinc chelators within CCHX zinc finger derivatives have been demonstrated to be sufficient to bind zinc and maintain the finger in an active form (reference 39 and references therein). Finally, to investigate the importance of the unusual C2HR configuration for the interaction, we mutagenized Arg residue 427 into a His residue and found that the mutated motif has no NSD1 binding activity, indicating that the C2HR (as opposed to C2H2) motif is a prerequisite for the NSD1 interaction.
Through a deletion analysis, we mapped the C2HR-interacting domain of NSD1 to its C-terminal C5HCH motif, which resembles the PHD finger (1, 16). The PHD finger is a conserved C4HC3 zinc binding motif that binds two zinc atoms in a cross-brace fashion (33). Supporting the notion that the C5HCH motif of NSD1 may also have zinc binding activity, we found that mutations of the putative zinc-coordinating residues, the cysteine residues at positions 2073 and C2073, prevent NSD1 from interacting with—and corepressing the silencing activity of—the Nizp1 C2HR motif. Thus, the C5HCH-C2HR interaction reported here may represent a finger-finger interaction. Further structural studies are clearly required to clarify this issue.
Similar to NSD1, human and mouse NSD2 and NSD3 possess C-terminal C5HCH motifs (4). However, these motifs failed to interact with Nizp1. This result reinforces the specificity and selectivity of NSD1 recruitment by the Nizp1 C2HR motif and raises the possibility that different C5HCH motifs may bind distinct C2HR motifs. In contrast to the mammalian NSD family members, the only NSD ortholog in flies, Drosophila NSD, has no C5HCH motif (4), indicating that this motif may have evolved late in evolution. Interestingly, this parallels the species specificity of the SCAN-KRAB-(C2H2)x subfamily of zinc finger proteins, which are restricted to vertebrates (9, 23), and suggests that the C5HCH-C2HR interaction may play a role in a vertebrate-specific function. It is noteworthy in this regard that a missense mutation in the C5HCH motif of NSD1 (C2183S) has recently been described for a Weaver outgrowth syndrome patient (11). This mutation affects the fifth conserved cysteine residue within the motif, possibly disrupting the secondary structure of the domain (Fig. 5C), and points to the biological importance of the C5HCH motif in controlling cell proliferation during mammalian development.
Transcriptional repression by Nizp1.
It was previously shown that NSD1 has an HMTase domain that specifically methylates histone H3 at lysine 36 (H3-K36) and histone H4 at lysine 20 (H4-K20) (34). Although the functional consequences of these modifications remain to be determined, HMTase activities specific to each of these lysine residues that are associated with transcriptionally inactive chromatin have been described for yeast, Drosophila, and mammals (12, 30, 42). Thus, based on these findings and our present data, it is tempting to speculate that Nipzp1 is a DNA-binding transcriptional repressor whose function is to target the H3-K36 and H4-K20 HMTase activities of NSD1 to specific promoters in vivo.
The recruitment of NSD1, however, may not be sufficient to account for all of the Nizp1 repressor properties. Indeed, we found that in full-length Nizp1, the C2HR mutations preventing NSD1 interaction severely reduced but did not abolish the Nizp1 repression activity, suggesting that there is an additional repression domain(s) within the protein. One possible candidate for a second transcriptional repression domain is the KRAB A domain. This highly conserved domain has been shown to silence transcription through the recruitment of the corepressor protein TIF1β/KAP-1 (reference 2 and references therein). Interestingly, TIF1β/KAP-1, like NSD1, acts on the chromatin template, but the underlying mechanism implicates other chromatin-modifying activities. TIF1β/KAP-1 has been reported to mediate silencing through histone deacetylation, H3-K9 methylation, and direct binding to heterochromatin-associated proteins (reviewed in reference 5). Thus, one attractive possibility for Nizp1 is that it utilizes several chromatin-modifying activities, leading to various changes in chromatin structure and organization to silence transcription. Such a mechanism is not unprecedented, since the presence of several repressor activities in the same transcriptional factor acting synergistically or independently, depending on the cellular target and/or the target promoter, has already been described for numerous transcriptional repressors (reviewed in references 20 and 36), indicating that this accumulation of multiple domains of similar function within a single protein may be a common mechanism that contributes to the fine-tuning of gene expression.
Acknowledgments
We are grateful to S. Vicaire for DNA sequencing, F. Ruffenach and A. Staub for oligonucleotide synthesis, the cell culture staff for their help, and our colleagues of the Department of Physiological Genetics and Nuclear Signaling for helpful discussions.
This work was supported by the Carlsberg Foundation, the Danish Cancer Society, the Novo Nordisk Foundation, the Danish Research Councils, the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, l'Hôpital Universitaire de Strasbourg (H.U.S.), the Association pour la Recherche sur le Cancer, the Collèege de France, the Fondation pour la Recherche Médicale (FRM), and Bristol-Myers-Squibb.
REFERENCES
- 1.Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. [DOI] [PubMed] [Google Scholar]
- 2.Abrink, M., J. A. Ortiz, C. Mark, C. Sanchez, C. Looman, L. Hellman, P. Chambon, and R. Losson. 2001. Conserved interaction between distinct Kruppel-associated box domains and the transcriptional intermediary factor 1β. Proc. Natl. Acad. Sci. USA 98:1422-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alders, M., A. Ryan, M. Hodges, J. Bliek, A. P. Feinberg, O. Privitera, A. Westerveld, P. F. Little, and M. Mannens. 2000. Disruption of a novel imprinted zinc-finger gene, ZNF215, in Beckwith-Wiedemann syndrome. Am. J. Hum. Genet. 66:473-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angrand, P. O., F. Apiou, A. F. Stewart, B. Dutrillaux, R. Losson, and P. Chambon. 2001. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics 74:79-88. [DOI] [PubMed] [Google Scholar]
- 5.Cammas, F., and R. Losson. 2002. TIF1β in heterochromatin-mediated silencing. Recent Res. Dev. Mol. Cell. Biol. 3:561-603. [Google Scholar]
- 6.Chen, G., and A. J. Courey. 2000. Groucho/TLE family proteins and transcriptional repression. Gene 249:1-16. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X., M. Hamon, Z. Deng, M. Centola, R. Sood, K. Taylor, D. L. Kastner, and N. Fischel-Ghodsian. 1999. Identification and characterization of a zinc finger gene (ZNF213) from 16p13.3. Biochim. Biophys. Acta 1444:218-230. [DOI] [PubMed] [Google Scholar]
- 8.Chesi, M., E. Nardini, R. S. Lim, K. D. Smith, W. M. Kuehl, and P. L. Bergsagel. 1998. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 92:3025-3034. [PubMed] [Google Scholar]
- 9.Collins, T., J. R. Stone, and A. J. Williams. 2001. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 21:3609-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuisset, L., J. P. Drenth, J. M. Berthelot, A. Meyrier, G. Vaudour, R. A. Watts, D. G. Scott, A. Nicholls, S. Pavek, C. Vasseur, J. S. Beckmann, M. Delpech, and G. Grateau. 1999. Genetic linkage of the Muckle-Wells syndrome to chromosome 1q44. Am. J. Hum. Genet. 65:1054-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douglas, J., S. Hanks, I. K. Temple, S. Davies, A. Murray, M. Upadhyaya, S. Tomkins, H. E. Hughes, T. R. Cole, and N. Rahman. 2003. NSD1 mutations are the major cause of Sotos syndrome and occur in some cases of Weaver syndrome but are rare in other overgrowth phenotypes. Am. J. Hum. Genet. 72:132-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang, J., Q. Feng, C. S. Ketel, H. Wang, R. Cao, L. Xia, H. Erdjument-Bromage, P. Tempst, J. A. Simon, and Y. Zhang. 2002. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 12:1086-1099. [DOI] [PubMed] [Google Scholar]
- 13.Gibbons, R. J., S. Bachoo, D. J. Picketts, S. Aftimos, B. Asenbauer, J. Bergoffen, S. A. Berry, N. Dahl, A. Fryer, K. Keppler, K. Kurosawa, M. L. Levin, M. Masuno, G. Neri, M. E. Pierpont, S. F. Slaney, and D. R. Higgs. 1997. Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat. Genet. 17:146-148. [DOI] [PubMed] [Google Scholar]
- 14.Gozani, O., P. Karuman, D. R. Jones, D. Ivanov, J. Cha, A. A. Lugovskoy, C. L. Baird, H. Zhu, S. J. Field, S. L. Lessnick, J. Villasenor, B. Mehrotra, J. Chen, V. R. Rao, J. S. Brugge, C. G. Ferguson, B. Payrastre, D. G. Myszka, L. C. Cantley, G. Wagner, N. Divecha, G. D. Prestwich, and J. Yuan. 2003. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 114:99-111. [DOI] [PubMed] [Google Scholar]
- 15.Gunduz, M., M. Ouchida, K. Fukushima, H. Hanafusa, T. Etani, S. Nishioka, K. Nishizaki, and K. Shimizu. 2000. Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res. 60:3143-3146. [PubMed] [Google Scholar]
- 16.Huang, N., E. vom Baur, J.-M. Garnier, T. Lerouge, J.-L. Vonesch, Y. Lutz, P. Chambon, and R. Losson. 1998. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J. 17:3398-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaju, R. J., C. Fidler, O. A. Haas, A. J. Strickson, F. Watkins, K. Clark, N. C. Cross, J. F. Cheng, P. D. Aplan, L. Kearney, J. Boultwood, and J. S. Wainscoat. 2001. A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood 98:1264-1267. [DOI] [PubMed] [Google Scholar]
- 18.Kalkhoven, E., J. H. Roelfsema, H. Teunissen, A. den Boer, Y. Ariyurek, A. Zantema, M. H. Breuning, R. C. Hennekam, and D. J. Peters. 2003. Loss of CBP acetyltransferase activity by PHD finger mutations in Rubinstein-Taybi syndrome. Hum. Mol. Genet. 12:441-450. [DOI] [PubMed] [Google Scholar]
- 19.Klug, A., and J. W. Schwabe. 1995. Protein motif 5. Zinc fingers. FASEB J. 9:597-604. [PubMed] [Google Scholar]
- 20.Kobayashi, M., R. E. Goldstein, M. Fujioka, Z. Paroush, and J. B. Jaynes. 2001. Groucho augments the repression of multiple Even skipped target genes in establishing parasegment boundaries. Development 128:1805-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurotaki, N., K. Imaizumi, N. Harada, M. Masuno, T. Kondoh, T. Nagai, H. Ohashi, K. Naritomi, M. Tsukahara, Y. Makita, T. Sugimoto, T. Sonoda, T. Hasegawa, Y. Chinen, H.-A. Tomita, A. Kinoshita, T. Mizuguchi, K.-I. Yoshiura, T. Ohta, T. Kishino, Y. Fukushima, N. Niikawa, and N. Matsumoto. 2002. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet. 30:365-366. [DOI] [PubMed] [Google Scholar]
- 22.Lachner, M., and T. Jenuwein. 2002. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14:286-298. [DOI] [PubMed] [Google Scholar]
- 23.Lander, E. S., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 24.Le Douarin, B., P. Pierrat, E. vom Baur, P. Chambon, and R. Losson. 1995. A new version of the two-hybrid assay for detection of protein-protein interactions. Nucleic Acids Res. 23:876-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Looman, C., M. Abrink, C. Mark, and L. Hellman. 2002. KRAB zinc finger proteins: an analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Mol. Biol. Evol. 19:2118-2130. [DOI] [PubMed] [Google Scholar]
- 26.Margolin, J. F., J. R. Friedman, W. K. Meyer, H. Vissing, H. J. Thiesen, and F. J. Rauscher. 1994. Krüppel-associated boxes are potent transcriptional repression domains. Proc. Natl. Acad. Sci. USA 91:4509-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagai, T., N. Matsumoto, N. Kurotaki, N. Harada, N. Niikawa, T. Ogata, K. Imaizumi, K. Kurosawa, T. Kondoh, H. Ohashi, M. Tsukahara, Y. Makita, T. Sugimoto, T. Sonoda, T. Yokoyama, K. Uetake, S. Sakazume, Y. Fukushima, and K. Naritomi. 2003. Sotos syndrome and haploinsufficiency of NSD1: clinical features of intragenic mutations and submicroscopic deletions. J. Med. Genet. 40:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen, A. L., J. A. Ortiz, J. You, M. Oulad-Abdelghani, R. Khechumian, A. Gansmuller, P. Chambon, and R. Losson. 1999. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 18:6385-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen, A. L., M. Oulad-Abdelghani, J. A. Ortiz, E. Remboutsika, P. Chambon, and R. Losson. 2001. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell 7:729-739. [DOI] [PubMed] [Google Scholar]
- 30.Nishioka, K., J. C. Rice, K. Sarma, H. Erdjument-Bromage, J. Werner, Y. Wang, S. Chuikov, P. Valenzuela, P. Tempst, R. Steward, J. T. Lis, C. D. Allis, and D. Reinberg. 2002. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 9:1201-1213. [DOI] [PubMed] [Google Scholar]
- 31.Nordentoft, I., and P. Jorgensen. 2003. The acetyltransferase 60 kDa trans-acting regulatory protein of HIV type 1-interacting protein (Tip60) interacts with the translocation E26 transforming-specific leukaemia gene (TEL) and functions as a transcriptional co-repressor. Biochem. J. 374:165-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Opitz, J. M., D. W. Weaver, and J. F. Reynolds. 1998. The syndromes of Sotos and Weaver: reports and review. Am. J. Med. Genet. 79:294-304. [DOI] [PubMed] [Google Scholar]
- 33.Pascual, J., M. Martinez-Yamout, H. J. Dyson, and P. E. Wright. 2000. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. J. Mol. Biol. 304:723-729. [DOI] [PubMed] [Google Scholar]
- 34.Rayasam, G. V., O. Wendling, P. O. Angrand, M. Mark, K. Niederreither, L. Song, T. Lerouge, G. L. Hager, P. Chambon, and R. Losson. 2003. NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J. 16:3153-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Remboutsika, E., K. Yamamoto, M. Harbers, and M. Schmutz. 2002. The bromodomain mediates transcriptional intermediary factor 1α-nucleosome interactions. J. Biol. Chem. 277:50318-50325. [DOI] [PubMed] [Google Scholar]
- 36.Roberts, S. G. E. 2000. Mechanisms of action of transcription activation and repression domains. Cell. Mol. Life Sci. 57:1149-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosati, R., R. La Starza, A. Veronese, A. Aventin, C. Schwienbacher, T. Vallespi, M. Negrini, M. F. Martelli, and C. Mecucci. 2002. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15). Blood 15:3857-3860. [DOI] [PubMed] [Google Scholar]
- 38.Sander, T. L., K. F. Stringer, J. L. Maki, P. Szauter, J. R. Stone, and T. Collins. 2003. The SCAN domain defines a large family of zinc finger transcription factors. Gene 310:29-38. [DOI] [PubMed] [Google Scholar]
- 39.Simpson, R. J., E. D. Cram, R. Czolij, J. M. Matthews, M. Crossley, and J. P. Mackay. 2003. CCHX zinc finger derivatives retain the ability to bind Zn(II) and mediate protein-DNA interactions. J. Biol. Chem. 278:28011-28018. [DOI] [PubMed] [Google Scholar]
- 40.Stec, I., T. J. Wright, G. J. van Ommen, P. A. de Boer, A. van Haeringen, A. F. Moorman, M. R. Altherr, and J. T. den Dunnen. 1998. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum. Mol. Genet. 7:1071-1082. [DOI] [PubMed] [Google Scholar]
- 41.Stec, I., S. B. Nagl, G. J. van Ommen, and J. T. den Dunnen. 2000. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 473:1-5. [DOI] [PubMed] [Google Scholar]
- 42.Strahl, B. D., P. A. Grant, S. D. Briggs, Z. W. Sun, J. R. Bone, J. A. Caldwell, S. Mollah, R. G. Cook, J. Shabanowitz, D. F. Hunt, and C. D. Allis. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner, J., and M. Crossley. 2001. The CtBP family: enigmatic and enzymatic transcriptional co-repressors. Bioessays 23:683-690. [DOI] [PubMed] [Google Scholar]
- 44.Wagner, S., M. A. Hess, P. Ormonde-Hanson, J. Malandro, H. Hu, M. Chen, R. Kehrer, M. Frodsham, C. Schumacher, M. Beluch, C. Honer, M. Skolnick, D. Ballinger, and B. R. Bowen. 2000. A broad role for the zinc finger protein ZNF202 in human lipid metabolism. J. Biol. Chem. 275:15685-15690. [DOI] [PubMed] [Google Scholar]