Abstract
The retinoblastoma (RB) tumor suppressor is a critical negative regulator of cellular proliferation. Repression of E2F-dependent transcription has been implicated as the mechanism through which RB inhibits cell cycle progression. However, recent data have suggested that the direct interaction of RB with replication factors or sites of DNA synthesis may contribute to its ability to inhibit S phase. Here we show that RB does not exert a cis-acting effect on DNA replication. Furthermore, the localization of RB was distinct from replication foci in proliferating cells. While RB activation strongly attenuated the RNA levels of multiple replication factors, their protein expression was not diminished coincident with cell cycle arrest. During the first 24 h of RB activation, components of the prereplication complex, initiation factors, and the clamp loader complex (replication factor C) remained tethered to chromatin. In contrast, the association of PCNA and downstream components of the processive replication machinery was specifically disrupted. This signaling from RB occurred in a manner dependent on E2F-mediated transcriptional repression. Following long-term activation of RB, we observed the attenuation of multiple replication factors, the complete cessation of DNA synthesis, and impaired replicative capacity in vitro. Therefore, functional distinctions exist between the “chronic” RB-mediated arrest state and the “acute” arrest state. Strikingly, attenuation of RB activity reversed both acute and chronic replication blocks. Thus, continued RB action is required for the maintenance of two kinetically and functionally distinct modes of replication inhibition.
The retinoblastoma (RB) tumor suppressor is a critical negative regulator of cellular proliferation that is targeted at high frequency in human cancer (5, 30, 51, 61, 72, 73). While RB has principally been considered as a regulator of G1 phase, the importance of RB in governing DNA synthesis has become increasingly clear (14, 33, 35, 41, 52, 58, 60). Currently, there are two mechanisms through which RB has been postulated to inhibit replication. In the first, DNA synthesis may be influenced by the direct interaction of RB with components of the replication process. For example, recent data have suggested a direct role for RB/E2F in regulating origin function during Drosophila chorion gene amplification (10). Furthermore, under certain conditions RB has been localized to sites of DNA replication, although this remains controversial (16, 32). Additionally, RB has been demonstrated to directly interact with the DNA replication factors MCM7, DNA polymerase α, and replication factor C (RFC) p140 subunit (24, 55, 64, 67). The importance of these interactions during cellular DNA replication has yet to be determined. However, these studies collectively suggest that RB could function in cis to directly inhibit DNA replication.
In the second model, RB-mediated S-phase inhibition may be attributed to the active repression of requisite DNA replication factor expression. RB is known to bind to the E2F family of transcription factors and antagonize their function (19). Active transcriptional repression by RB is mediated through the simultaneous recruitment of cofactors such as histone deacetylases and SWI/SNF chromatin remodeling enzymes (26). Active RB has been demonstrated to elicit repression of E2F-regulated S-phase genes, such as MCMs, DNA polymerase subunits, and deoxynucleoside triphosphate synthetic enzymes (43). Additionally, E2F-dependent transcriptional repression of preRC genes has been recently shown to regulate DNA synthesis in Drosophila (13). Thus, it has been alternatively postulated that the role of RB in DNA replication control is dependent on the transcriptional regulation of S-phase genes.
There are numerous potential targets for RB, as strict regulation and coordination of S-phase progression is achieved through several precise steps (6, 31, 65). Origins of replication are marked by the stable chromatin association of a heterohexameric origin recognition complex (ORC1-6) (6). During mitotic exit, the MCM complex (MCM2-7) is loaded onto chromatin by Cdc6 and Cdt1, forming a functional prereplication complex (preRC) (17, 44, 53). The establishment of preRCs has been termed “replication licensing,” as it confers the ability of DNA synthesis to initiate at a given origin (7). Throughout S phase, sequential initiation of replication at licensed origins occurs in a tightly coordinated manner (23). During initiation of replication, the heterotrimeric protein complex RPA binds to exposed single-stranded DNA as origin firing occurs and de novo synthesis begins by the action of the DNA polymerase α-primase complex (6, 31, 70). Processive replication requires the activity of replication factor C (RFC), the clamp loader complex responsible for the recruitment of PCNA (47, 70). Subsequent bidirectional replication fork progression requires DNA polymerases (δ and/or ɛ) and the recruitment of additional factors (e.g., DNA ligase I) by interaction with PCNA (70). The regulation of these events is thought to be catalyzed by the combined activities of Cdc7 and cyclin-dependent kinase 2 (CDK2) complexes, although their essential substrates have yet to be clearly defined (6, 71).
Although CDK2, cyclin E, and cyclin A are E2F-regulated genes, RB-mediated arrest does not dramatically affect the expression or activity of CDK2/cyclin E (2, 33). However, recent studies have questioned the absolute requirement of cyclin E and CDK2 in cellular proliferation (22, 54, 68). Consistent with this finding, cells arrested by active RB alleles exhibit significant loss of cyclin A expression (2, 36, 41). Importantly, the RB-mediated repression of cyclin A-associated kinase activity leads to the disruption of PCNA association with chromatin (60). However, the proximal target in this pathway was not identified. Here, we sought to more rigorously determine the effect of RB activation on the DNA replication machinery. We report that the program of cell cycle arrest elicited by RB occurs in two temporally and functionally distinct manners. First, RB can block DNA replication acutely and transiently by specifically targeting the recruitment and/or maintenance of critical elongation factors to chromatin. Second, chronic RB activation mediates the gradual downregulation of requisite replication factors, including both licensing factors and components of the processive replication machinery. Importantly, both the acute and long-term effects require functional RB and are not permanent arrest states. These data suggest that RB can inhibit DNA replication via two kinetically distinct, reversible pathways—transient regulation of the elongation phase and a more stable arrest state under conditions of persistent RB activation.
MATERIALS AND METHODS
Cell culture, plasmids, transfection, and flow cytometry.
Floxed Rb mice (RbF19/F19) were sacrificed by CO2 anesthetization followed by cervical dislocation. Fibroblasts were isolated form the peritoneal fascia by excision, mincing, and dissociation with 0.2-μg/ml collagenase (type I; Sigma) supplemented with 100 U of DNase I (Roche) at 37°C for 40 min with constant agitation. After washing in phosphate-buffered saline, dissociated tissue was incubated in 0.25% trypsin (Gibco) at 37°C for 20 min with constant agitation. Isolated cells were washed twice and plated in tissue culture dishes. Murine adult fibroblasts (MAFs) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum, 100-U/ml penicillin-streptomycin, and 2 mM glutamine at 37°C in 5% CO2. Rat-16, A2-4, and A5-1 cell lines were cultured as previously described (43). A5C1 cells harboring inducible expression of hemagglutinin (HA)-tagged RbΔcdk were cultured as described previously (41). The pTRE2-RbΔcdk plasmid was constructed by subcloning the HindIII/XbaI fragment encoding full-length RbΔcdk from pcDNA3.1 into the HindIII/NheI sites of pTRE2 (Clontech). The BL-1 cell line was generated by cotransfection of pTRE2-RbΔcdk and pTK-Hyg into Rat-16 cells and subsequent selection in 250-μg/ml hygromycin B. The U24-4 cell line was generated by cotransfection of pTRE-PSM-RB.7LP and pBabe-puro into the parental Tet-Off U24 cell line and subsequent selection in medium containing 2.5-μg/ml puromycin. Cells were synchronized in S phase by the addition of aphidicolin (APH) at a final concentration of 5 μg/ml for 24 h. Transfections were performed with FuGENE 6 (Roche) according to the manufacturer's protocol. A5-1 cells were cotransfected with green fluorescent protein (GFP)-RPA34, GFP-PCNA, and GFP-DNA ligase I as described previously (12, 37, 63). Stable cell lines were generated by selection with 2.5-μg/ml puromycin (Calbiochem), and clones were established by limiting dilution. The H2B-GFP, E2F1, E2F-DB, and PSM-RB expression plasmids have been previously described (33, 79). Rat-1 cells were transfected at an H2B-GFP/PSM-RB/E2F plasmid ratio of 1:6:6. Clinical grade cis-diamminedichloroplatinum II (cisplatin [CDDP]; Bristol Oncology) was added to cell culture medium as indicated. Flow cytometry was performed as described previously (33).
PCR-mediated analysis of recombination.
Confirmation of the conditional knockout of the Rb gene was determined by utilizing genomic DNA harvested from GFP- and Cre recombinase (GFP-Cre)-encoding adenovirus strain-infected MAFs. Genomic DNA was used in PCRs with primers as previously described (42). The PCR conditions were as follows: 95°C for 5 min; followed by 35 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 1 min; followed by final extension at 72°C for 7 min.
Adenoviral infections.
For the conditional knockout of Rb, MAFs were infected with adenovirus encoding either GFP alone or GFP and at a ratio of approximately 2 × 107 virus particles per 10-cm-diameter dish. Actual infection efficiency was 90 to 95% as determined by GFP fluorescence. A2-4 cells were infected with either GFP- or E2F2-encoding adenovirus as previously described (43). Rat-1 cells were infected with GFP or p16ink4a as previously described (43).
Biochemical fractionation and Western blotting.
Preparation of cell lysates, chromatin-bound pellet fractions, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blotting were carried out as previously described (2, 60). PSM-RB was detected with 851 antibody. HA-RBΔcdk was detected with HA probe antibody (Y-11; Santa Cruz). Cyclin A (C-19), CDK2 (M2), CDK4 (H-22), MCM7 (141.2), PCNA (PC10), ribonucleotide reductase (RNR) R2 subunit (I-15), β-tubulin (D-10), and lamin B (M-20) antibodies were from Santa Cruz. MCM5 (M14020) and DNA polymerase δ (D73020) antibodies were from Transduction Laboratories. Antibodies against RFCp37 (J. Hurwitz), HsDbf4/ASK (H. Masai), RFCp140 and cdc6 (B. Stillman), MCM2 (I. Todorov), vimentin (W. Ip), and RPA34 (M. Wold) were kind gifts.
Xenopus in vitro replication assays.
Xenopus egg extracts were prepared as previously described (75). Preparation of intact nuclei using digitonin was carried out as previously described (75). Nuclei were resuspended at a concentration of 10,000/μl of extract. DNA replication efficiency was determined by quantifying the amount (in cpm) of trichloroacetic acid (TCA)-precipitable [α-32P]dATP or [α-32P]dCTP (75). Purified Xenopus geminin-DEL (a nondegradable form of geminin) protein was purified as described previously (53) and added at a final concentration of 80 nM to extracts as indicated.
Live cell imaging and photobleaching.
A5-1 cells stably expressing GFP-PCNA were seeded on 25-mm-diameter coverslips. For imaging, coverslips were maintained at 37°C in live cell imaging chambers (Atto) in a water-jacketed stage incubator. Photobleaching was performed on a Zeiss LSM510 laser scanning confocal unit mounted on a Zeiss axiovert inverted microscope equipped with a C-Apochromat ×63 1.4 NA objective. A 2.9- by 2.9-μm area of the nucleus was photobleached for the indicated lengths of time with 100% transmission of 488-nm light from an argon laser running at 6.25 W. For fluorescence recovery after photobleaching (FRAP) analysis, fluorescent intensity values of the bleached area and of a distal unbleached area of the nucleus of equal size were measured every 50 ms for the indicated lengths of time following photobleaching. These values were compared to produce a relative fluorescent intensity to normalize for prebleach intensity. The data presented were collected from 24 nuclei per condition from two independent experiments.
BrdU incorporation and immunofluorescence microscopy.
Bromodeoxyuridine (BrdU) incorporation and immunofluorescence microscopy were performed as described previously (33, 34). BrdU labeling was carried out for 30 min (A5C1 cells), 8 h (MAFs), or 1 h (A2-4 cells) prior to formaldehyde fixation and immunodetection. A5C1 cells cultured in the absence of Dox for 24 h were fixed in 3.7% formaldehyde, permabilized with 0.3% Triton X-100 in phosphate-buffered saline, and immunostained with HA antibody. RB immunostaining was performed as previously described (3). Prior to MCM7 and PCNA staining (MAFs), cells were fixed in ice-cold methanol for 5 min. Extraction of soluble PCNA (A2-4, BL-1, U24-4, and A5C1) was performed as previously described (60).
RESULTS
Active RB alleles inhibit S-phase progression.
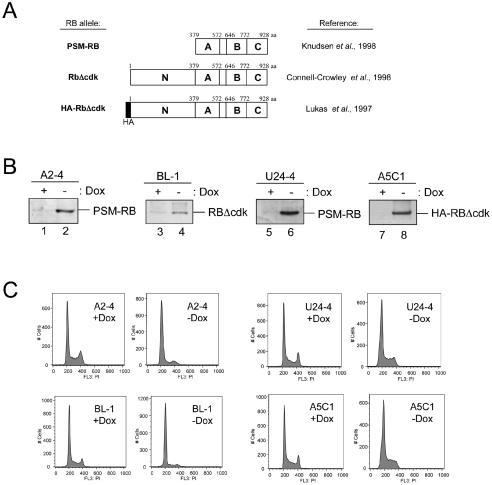
To investigate the action of RB on replication control, we utilized several cell lines that inducibly express alleles of RB that are resistant to the robust CDK activity in S-phase cells. Specifically, we employed a phosphorylation-site mutant allele of RB (PSM-RB; amino acids 379 to 928), RbΔcdk (amino acids 1 to 928), and HA-tagged RbΔcdk (amino acids 1 to 928) alleles that potently induce cell cycle arrest (Fig. 1A) (15, 33, 39, 41). These alleles vary in terms of the specific phosphorylation sites mutated, the species of origin, and the presence of an N-terminal domain. While the N terminus is dispensable for RB-mediated transcriptional repression and tumor suppression (78), it has been speculated to contribute to the inhibition of replication through RB (64). Thus, the comparison between PSM-RB and the other alleles enables determination of the functional role of the N terminus. Similarly, the PSM-RB and HA-RbΔcdk proteins are partially phosphorylated in cells, yet maintain the capacity to constitutively repress E2F activity (33, 39). In contrast, the RbΔcdk allele is completely resistant to phosphorylation (15). This distinction is important, since it has been postulated that low-level phosphorylation may be required to activate RB or alternatively that the partial phosphorylation of PSM-RB and HA-RbΔcdk could disrupt specific functions (20, 27). As such, the comparison of the behavior between these alleles will enable us to assess the potential role of partial phosphorylation or the N terminus of RB in the control of DNA replication. As shown in Fig. 1B, we utilized rat fibroblast cell lines that express either PSM-RB (A2-4 cell line) or RbΔcdk (BL-1 cell line) upon removal of the tetracycline analog doxycycline (Dox) from the media (15, 60). Similarly, accumulation of PSM-RB (U24-4 cell line) or HA-RbΔcdk (A5C1 cell line) was detected in human osteosarcoma (U2OS) cell lines (41). As expected, expression of the CDK-refractory RB alleles prevented the progression of cells into G2/M as determined by both flow cytometry and BrdU incorporation (Fig. 1C) (data not shown). Thus, these cell lines represent appropriate models to study RB-mediated inhibition of S-phase progression.
FIG. 1.
Cell cycle inhibition by multiple active RB alleles. (A) Diagram of active RB alles used in this study, with corresponding references. The N-terminal (N), C-terminal (C), and A and B pocket regions are identified. aa, amino acids. (B) A2-4, BL-1, U24-4, or A5C1 cell lines were cultured in the presence (+Dox) or absence (−Dox) of Dox for 24 h. Cells were harvested, and equal amounts of total protein were resolved by SDS-PAGE and immunoblotted for RB. (C) A2-4, BL-1, U24-4, or A5C1 cell lines were cultured in the presence or absence of Dox for 24 h. Cells were then fixed, stained with propidium iodide, and processed for flow cytometry. Histograms represent 10,000 gated events.
Active RB inhibits S phase by indirect mechanisms.
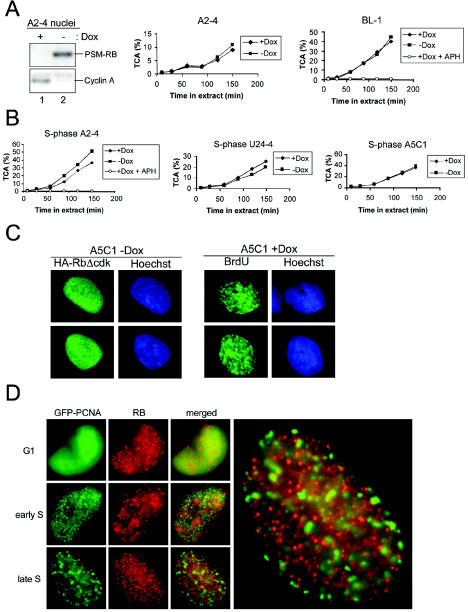
Initially, we investigated whether RB acts in a manner consistent with direct interaction versus transcriptional repression to inhibit S-phase progression. RB has been demonstrated to interact with greater than 100 proteins (46). While E2F transcription factors are considered the key targets of RB action in cell cycle control (19), RB has also been demonstrated to directly interact with certain replication factors. Thus, direct interaction of active RB with MCM7, DNA polymerase α, or RFCp140 may contribute to the S-phase inhibitory function of RB (55, 64, 67). Furthermore, evidence from Drosophila has suggested that RB/E2F complexes may exert influence on replication origin firing (10). To address whether active RB alleles might directly inhibit DNA replication, we utilized an established in vitro replication system (75). In the presence of Xenopus egg extract, the DNA synthesis that occurs is subject to in vivo cell cycle controls. In this assay, nuclei are prepared in a manner that protects the integrity of the nuclear envelope. These nuclei are then resuspended in Xenopus egg extract in the presence of radiolabeled dATP or dCTP and an energy regenerating system. The amount of DNA synthesis is determined by the incorporation of radiolabeled deoxyribonucleotide into TCA-precipitable material. As shown in Fig. 2A, nuclei from asynchronous or RB-arrested cultures were incubated in Xenopus egg extract and DNA replication efficiency was monitored. As a control, the presence of PSM-RB and the downregulation of protein levels of cyclin A (a major target of RB-mediated transcriptional repression) in the preparations of nuclei were verified by immunoblotting (Fig. 2A, left panel). Nuclei from A2-4 (middle panel) or BL-1 (right panel) cells were competent for replication irrespective of the presence of active RB alleles (Fig. 2A). In contrast, addition of the DNA polymerase inhibitor APH completely blocked replication (Fig. 2A, right panel). In order to confirm these observations specifically in S-phase cells, cultures were synchronized with APH prior to the induction of active RB (60). Consistently, in vitro replication in nuclei from S-phase cells was not affected by the presence of active RB alleles (Fig. 2B). These results demonstrate that RB-mediated arrest can be overcome by the supply of exogenous factors in vitro, indicating that the active RB alleles do not act in cis to inhibit replicative function. Importantly, the presence or absence of the N-terminal portion of the active RB alleles had no observable effect on replication efficiency. Thus, RB-arrested nuclei can be functionally complemented by the provision of Xenopus egg extract.
FIG. 2.
Active RB alleles do not inhibit in vitro replication or localize to replication sites. (A) The indicated cell lines were incubated in the presence (+Dox) or absence (−Dox) of Dox for 24 h. Intact nuclei were utilized for immunoblotting to detect PSM-RB and cyclin A (left panel) or resuspended in Xenopus egg extract in the presence of [α-32P]dATP or [α-32P]dCTP, and samples were removed at the indicated time points (middle and right panels). Relative DNA replication efficiency was determined by measuring the percentage of acid-precipitable cpm (TCA).(B) Nuclei from indicated cell lines were utilized as in panel A, except cultures were first synchronized in S phase by using APH. To verify the DNA polymerase-dependent incorporation of [α-32P]dATP or [α-32P]dCTP, APH was added to Xenopus egg extract as indicated. (C) A5C1 cells cultured in the presence of Dox (right panel) or in the absence of Dox for 24 h (left panel). Cells cultured in the presence of Dox were pulse-labeled with BrdU for 30 min, fixed, and stained for BrdU. Cells cultured in the absence of Dox were fixed, and immunostaining for HA-RbΔcdk was performed. (D) U2OS cells stably expressing GFP-PCNA were fixed and immunostained for endogenous RB. Representative photomicrographs of G1- and S-phase cells are shown.
Interactions of RB with discrete nuclear regions has been the subject of ongoing controversy. For example, immunofluorescent detection of the interaction of RB with replication proteins (e.g., MCMs) or the presence of RB at replication foci has been a disputed issue (16, 32). In order to examine the localization of active RB in arrested cells, we compared sites of BrdU incorporation in cycling cells and staining for HA-RbΔcdk. In our hands, immunofluorescent detection of sites of BrdU incorporation in proliferating A5C1 cells revealed distinct patterns when compared to detection of the HA-RbΔcdk allele in arrested cells (Fig. 2C, right and left panels). Thus, the inhibitory RB allele does not localize to discrete foci that resemble active replication sites. In order to visualize the nuclear distribution of endogenous RB relative to replication factories, we exploited a GFP-tagged PCNA protein known to localize to sites of active DNA replication (37, 63). In G1 cells, GFP-PCNA is diffuse and endogenous RB shows a relatively punctate distribution throughout the nucleoplasm (Fig. 2D, top panel set). This result is consistent with the previously published distribution of endogenous RB (16). In S-phase cells, GFP-PCNA is concentrated in subnuclear foci demarcating active sites of DNA replication (Fig. 2D, middle and bottom panel sets). High-resolution imaging of S-phase cells demonstrates that the endogenous RB does not colocalize with GFP-PCNA (Fig. 2D, right panel). Collectively, these experiments argue against a direct effect of active RB functioning in cis to inhibit S phase.
RB-mediated transcriptional repression of DNA replication machinery.
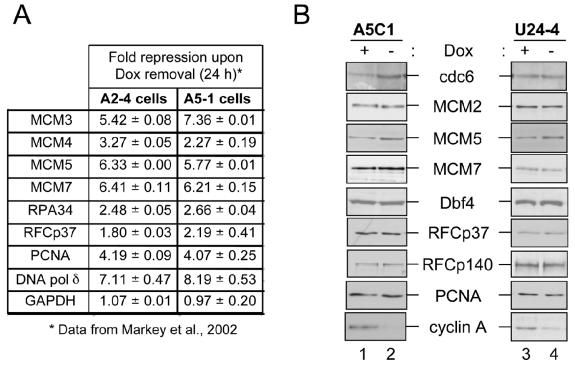
The principal cellular target of RB is typically considered to be the E2F family of transcription factors (19, 26). E2F proteins regulate the transcription of numerous genes required for S-phase and G2/M progression and a myriad of additional cellular processes (11, 29, 48, 76). To address the possible role of transcriptional repression during the RB-induced DNA replication block, the levels of several potential target genes were assessed. Microarray data obtained from PSM-RB-inducible cell lines were analyzed to determine whether active RB signaling might deplete requisite DNA replication factors (43). As illustrated in Fig. 3A, RNA levels of a large number of replication proteins were significantly repressed upon the induction of PSM-RB in both A2-4 and A5-1 cells. These results suggest that RB/E2F-mediated transcriptional repression might regulate preRC formation, initiation or elongation by limiting available protein levels. Since the majority of antibodies against replication components were more reactive with human proteins, we initially utilized the U2OS-based A5C1 and U24-4 cell lines. Surprisingly, assessment of total protein amounts of various preRC components (cdc6 and MCMs), an initiation factor (Dbf4), and elongation factors (RFC subunits p37 and p140, PCNA, and DNA polymerase δ) revealed no significant attenuation (Fig. 3B). In contrast, the expression of cyclin A is notably lost following 24 h of active RB induction. These findings suggested that transcriptional repression of replication components is not responsible for the rapid induction of cell cycle arrest.
FIG. 3.
RB-mediated repression of DNA replication target genes. (A) A2-4 or A5-1 cells were incubated in the presence (+Dox) or absence (−Dox) of Dox for 24 h. RNA was harvested and utilized for microarray analysis. Shown are the averages of two independent experiments comparing the relative RNA levels from the −Dox condition to the +Dox condition. These data are adapted from Markey et al. (43). (B) A5C1 and U24-4 cells were cultured in the presence (lanes 1 and 3) or absence (lanes 2 and 4) of Dox for 24 h. Equal total protein amounts were resolved by SDS-PAGE and immunoblotted for the indicated proteins.
Acute S-phase arrest by RB targets replication elongation factors.
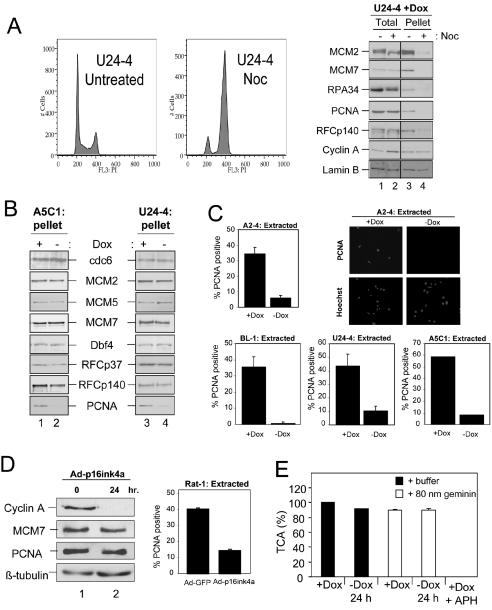
Due to the relatively rapid cessation of DNA synthesis following RB induction, we hypothesized that rather than targeting the expression of critical replication factors, their activity and/or function might be affected. To address this possibility, we next analyzed the stable chromatin association of specific candidate proteins during RB-mediated cell cycle arrest (18, 44, 53). To ensure that chromatin association of these putative targets was dependent on cell cycle position and not artifactual, U24-4 cells were initially synchronized in G2/M by using nocodazole (53) (Fig. 4A, right and left panels). Cells accumulated with 4 N DNA content following nocodazole treatment (Fig. 4A, left panel). In certain instances, protein modifications (as detected by mobility shift) could be detected in the total protein fraction (e.g., MCM2 and RPA34) but no substantial alterations in protein levels were observed (Fig. 4A, right panel). Examination of the chromatin-bound pellet fraction, however, revealed that G2/M synchronization led to dissociation of replication factors from chromatin, thereby validating the technique as a means to assess RB action on the replication fork (Fig. 4A, right panel). To do so, we analyzed the presence of the same replication proteins in the chromatin-bound fraction during RB-mediated arrest in both A5C1 and U24-4 cells (Fig. 4B). Upon the detailed examination of potential targets, we observed that preRC components, initiation factors, and even the clamp loader complex, RFC, remained associated with chromatin in the presence of active RB. However, tethering of PCNA was specifically disrupted in both cell lines, indicating that PCNA is the focal point of RB action (60).
FIG. 4.
Active RB alleles do not disrupt RFC tethering, but PCNA activity is compromised. (A) U24-4 cells were cultured either untreated or in the presence of 50-ng/ml nocodazole (Noc) for 16 h. Samples were harvested for flow cytometric analysis (left panel). Total (lanes 1 and 2) or chromatin-bound (pellet, lanes 3 and 4) fractions from untreated (lanes 1 and 3) or nocodazole-treated (lanes 2 and 4) cultures were resolved by SDS-PAGE and immunoblotted for the indicated proteins (right panel). Lamin B was utilized as a loading control. (B) A5C1 and U24-4 cells were cultured as in panel A, and equal amounts of chromatin-bound fractions (pellet) were resolved by SDS-PAGE and immunoblotted for the indicated proteins. (C) A2-4, BL-1, U24-4, and A5C1 cells were cultured in the presence (+Dox) or absence (−Dox) of Dox for 24 h. Cells were extracted in situ with CSK buffer, and PCNA was detected by immunofluorescence. Nuclei were counterstained with Hoechst stain, and the percentage of PCNA-positive cells was determined. The values shown are means ± standard deviation. At least 200 cells were counted per experiment, and representative photomicrographs are shown. (D, left panel) Asynchronously proliferating Rat-1 cells were infected with recombinant adenovirus expressing p16ink4a. Cells were harvested at 0 (lane 1) or 24 (lane 2) h postinfection. Equal total protein levels were resolved by SDS-PAGE, and the indicated proteins were detected by immunoblotting. (Right panel) Rat-1 cells were infected with GFP- or p16ink4a-encoding adenoviruses and subjected to in situ extraction with CSK buffer at 24 h postinfection. Cells were subsequently fixed, and PCNA was detected by immunofluorescence. The values shown are means ± standard deviation. At least 200 cells were counted per experiment. (E) Intact nuclei from A2-4 cells cultured in the presence or absence of Dox were resuspended in Xenopus egg extract in the presence of [α-32P]dATP that was supplemented with either buffer (filled bars) or 80 nM geminin-DEL (open bars). Following a 150-min reaction, the relative DNA replication efficiency was determined by measuring the relative percent of acid-precipitable cpm with Dox and buffer set to 100%. The values shown are means ± standard deviation.
To verify that PCNA was a common target in RB signaling, we evaluated this event in different cell types harboring distinct RB alleles and in response to the activation of the endogenous RB. As shown in Fig. 4C, following 24 h of Dox removal, PCNA tethering was disrupted irrespective of the specific active RB allele or U2OS versus Rat-1 cells. These results combined with the preceding analyses strongly argue that all of the RB alleles utilized act in the same manner to inhibit DNA replication. The p16ink4a CDK inhibitor is known to elicit an RB-dependent cell cycle arrest (40, 61). To study the action of p16ink4a, we utilized a recombinant adenovirus expressing human p16ink4a to trigger the dephosphorylation of endogenous RB and cell cycle arrest. Analyses of target gene expression 24 h postinfection indicated that p16ink4a, like the active RB alleles, failed to influence the protein levels of MCM7 or PCNA, yet dramatically attenuated the levels of cyclin A (Fig. 4D). Additionally, PCNA chromatin tethering in the presence of adenoviral p16ink4a expression was similarly compromised (Fig. 4D). Thus, activation of endogenous RB via p16ink4a disrupts PCNA activity and the effect of RB activation is common to all cells studied.
Together, these data argue that active RB functions to specifically target PCNA and not earlier stages of replication. To determine if there was a functional action of RB on the preRC, we exploited the replication inhibitior geminin. The geminin protein binds to Cdt1 and inhibits the recruitment of nontethered MCM proteins to form the preRC (7). Therefore, resistance to recombinant nondegradable geminin (geminin-DEL) in an in vitro replication assay demonstrates that nuclei have functional preRC(s) (53). Consistent with the chromatin tethering data indicating the presence of a preRC, the addition of recombinant geminin failed to inhibit in vitro replication of nuclei that had been arrested through the action of RB for 24 h. Thus, RB specifically inhibits PCNA activity while maintaining functional preRC(s).
Action of RB on PCNA is dependent on transcriptional repression.
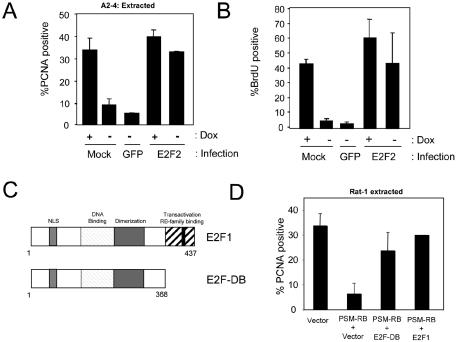
To determine whether RB-mediated disruption of PCNA activity could be overcome by stimulating E2F activity, the ectopic expression of E2F2 was utilized. We have previously observed that overexpression of E2F2 effectively alleviates RB-mediated repression of an E2F reporter and endogenous targets, such as cyclin A (43). As shown in Fig. 5A, mock-infected or GFP-infected A2-4 cells cultured in the absence of Dox for 24 h (−Dox) failed to retain PCNA on chromatin when compared to mock-infected control (+Dox). In contrast, the introduction of E2F2 via adenovirus infection stimulated an increase in chromatin-associated PCNA. In order to correlate the tethering of PCNA with S-phase progression, we next analyzed the ability of ectopic E2F2 expression to stimulate DNA synthesis (Fig. 5B). The induction of PSM-RB expression potently inhibited S phase as determined by BrdU incorporation. As predicted, GFP-encoding adenovirus failed to rescue the DNA replication inhibition. In contrast, the introduction of E2F2 completely restored S-phase progression in cells expressing PSM-RB. Therefore, these studies indicate that an E2F-regulated event specifically targets PCNA loading. To delineate the specific role of E2F-mediated transcriptional repression in this event as opposed to sequestration of RB via the ectopically expressed E2F, we employed an allele of E2F-1 (E2F-DB) that lacks the RB binding domain and transcriptional activation domains (Fig. 5C). This allele has been previously shown to disrupt RB-mediated transcriptional repression and cell cycle inhibition by displacing endogenous E2F assembled complexes from target promoters (57, 79). Consistent with these prior data, we found that E2F-DB disrupted RB-mediated transcriptional repression in Rat-1 cells (not shown). Additionally, we found that the E2F-DB allele efficiently restored PCNA tethering in the presence of PSM-RB (Fig. 5D). Therefore, these data indicate that RB/E2F-dependent transcriptional repression is responsible for attenuating PCNA activity.
FIG. 5.
E2F participates in the RB-mediated regulation of PCNA. (A) A2-4 cells were incubated in the presence (+Dox) or absence (−Dox) of Dox for a total of 24 h. Cells were either mock infected or infected with GFP- or E2F2-encoding adenovirus for the final 16 h. Cells were then extracted in situ and stained for PCNA. The percentage of PCNA-positive nuclei is shown as the mean ± standard deviation. More than 200 nuclei per condition were counted from two independent experiments. (B) Asynchronously proliferating A2-4 cells were cultured and infected as in panel A. BrdU incorporation was determined by indirect immunofluorescence following a 1-h pulse-labeling. The percentage of BrdU-positive nuclei is shown as the mean ± standard deviation. (C) Schematic representation of E2F1 and the E2F-DB alleles utilized. (D) Rat-1 cells were cotransfected with an H2B-GFP expression plasmid (to mark transfected cells) and the indicated plasmids. Thirty hours posttransfection, cells were subjected to in situ extraction, fixed, and stained for PCNA. The percentage of transfected (H2B-GFP positive) nuclei that were PCNA positive is shown as the mean ± standard deviation.
Active RB disrupts focal accumulation of elongation factors.
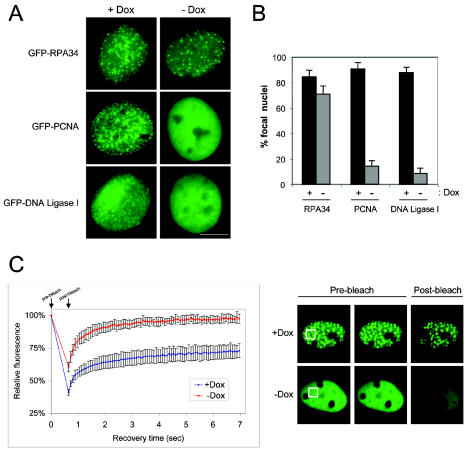
As the effect of RB on DNA replication factors had only been documented by indirect immunofluorescence or biochemical assay, we sought to directly monitor the regulation of these replication factors in living cells. In this manner, the effect of RB on replication protein dynamics could be observed. For this analysis, GFP-fusion proteins with genes encoding the RPA p34 subunit (RPA34), PCNA, and DNA ligase I were utilized. Each of these proteins has been extensively characterized, and their colocalization at foci of active DNA synthesis has been demonstrated (12, 37, 63). Expression plasmids encoding the GFP fusions were stably integrated into a PSM-RB inducible cell line to examine their dynamic behavior. Initially, S-phase-synchronized cells were cultured in the presence or absence of Dox for 12 h, imaging was performed by fluorescence microscopy, and the percentage of cells with focal distribution of GFP fluorescence was determined (Fig. 6A). Consistent with the results obtained by biochemical fractionation, GFP-RPA34 foci were observed in cells irrespective of PSM-RB expression, indicative of unperturbed association with chromatin (Fig. 6A and B). In contrast, GFP-PCNA appeared diffuse in the presence of active RB, indicating that PCNA is displaced from or prevented from associating with replication sites (Fig. 6A and B). PCNA itself is responsible for the recruitment of additional accessory factors, such as DNA ligase I, to elongating replication forks (45). Strikingly, the focal pattern of GFP-DNA ligase I observed in S-phase cells was also perturbed during RB-mediated arrest (Fig. 6A and B). In order to determine the effect of active RB on the dynamics of these proteins, nuclear photobleaching experiments were performed. FRAP can be utilized to determine the immobile versus mobile fraction of a fluorescent protein (38). GFP-PCNA-expressing cells indicated that the focal PCNA at active replication sites is relatively immobile (Fig. 6C, +Dox). In contrast, the diffuse distribution of GFP-PCNA observed in PSM-RB-expressing cells coincided with high mobility (Fig. 6C, −Dox). Together, these findings indicate that active RB blocks the stable recruitment of PCNA to replication forks, leading to freely diffusible PCNA, and additionally leads to the mobilization of DNA ligase I.
FIG. 6.
FRAP analysis reveals the mobilization of GFP-PCNA and downstream factors by active RB. (A) Cells harboring inducible expression of PSM-RB were transfected with GFP-RPA34, GFP-PCNA, or GFP-DNA ligase I and pBabe-puro, and stable clones were isolated by puromycin selection. Cells were synchronized in early S phase by culture in medium containing 2-μg/ml APH for 12 h. Synchronized cells were then cultured in the presence or absence of Dox for 12 h and subsequently released from APH block for 30 min prior to fixation and imaging. Bar, 5 μm. (B) The percentage of cells with focal GFP fluorescence as in panel A was determined. The values shown are means ± standard deviation. (C) The cell line stably expressing GFP-PCNA was cultured as in panel A and then utilized for FRAP analysis. Shown are the fluorescence recovery curves following 0.42 s of photobleaching (left panel) and representative images (right panel).
Chronic RB activation invokes replicative exit in S phase.
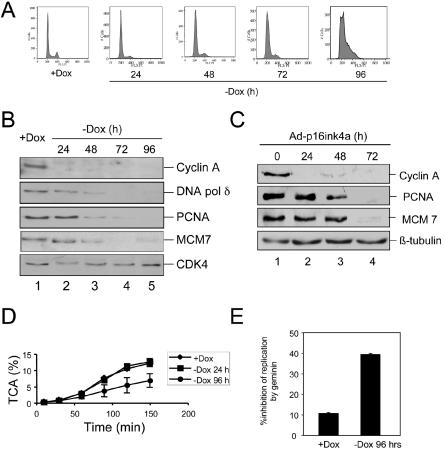
Although the acute arrest program elicited by RB specifically targeted the chromatin binding activity of PCNA and downstream factors, the significance of RB-mediated transcriptional repression of DNA replication factors had yet to be determined. To assess whether RB attenuated the protein levels of known E2F targets involved in DNA replication, kinetic analyses were performed following induction of active RB. As determined by flow cytometric analysis, cells entered but could not complete S phase after 96 h of Dox removal (Fig. 7A). This observation was consistent with previous data demonstrating the failure of RB to impose a sustained G1 arrest and with nonprocessive replication occurring in the presence of active RB (41, 60). To investigate whether RB significantly attenuated critical DNA replication factors at these later time points, protein levels were analyzed by immunoblotting. Interestingly, the levels of all target proteins examined—MCMs, RPA34, DBF4, RFCp37, and RFCp140—were appreciably attenuated 72 to 96 h after active RB induction (Fig. 7B) (data not shown). Similarly, when endogenous RB was activated through the expression of p16ink4a, we observed a delayed attenuation of MCM7 and PCNA expression (Fig. 7C). Since the acute replication block could be overcome by the addition of Xenopus egg extract, we next sought to determine whether arrested nuclei at the 96-h time point could support DNA replication in vitro (Fig. 7D). When nuclei from the chronic arrest state (−Dox for 96 h) were incubated with egg extract, they were significantly impaired for DNA replication as compared to asynchronous controls or acutely arrested cells (−Dox for 24 h). The ability of these nuclei to support replication in the absence of MCMs and other factors is likely due to compensation by the exogenous Xenopus proteins. To specifically probe the activity of endogenous preRC complexes, recombinant geminin-DEL was added to the reaction (Fig. 7E). While geminin-DEL addition had a minimal effect on control (+Dox), under the conditions of chronic arrest geminin-DEL addition further inhibited replication by an additional 40% (Fig. 7E). Similar inhibition of replication was observed when geminin-DEL was expressed in chronically arrested cells (not shown). Thus, chronic activation of RB results in significant repression of critical replication factors and reduced replicative capacity, albeit with delayed kinetics relative to cyclin A expression and PCNA inhibition.
FIG. 7.
Chronic RB activation leads to S-phase exit. (A) A2-4 cells were cultured in the presence of Dox (+Dox) or Dox was removed and samples were collected at the indicated time points. Cells were fixed in ethanol and processed for flow cytometry. (B) Cells cultured as in panel A were harvested at the time indicated, and equal total protein was resolved by SDS-PAGE. Immunoblotting was performed to detect the indicated proteins. (C) Rat-1 cells were infected with recombinant adenovirus encoding p16ink4a, and cells were harvested at the indicated time points postinfection. Total protein was resolved by SDS-PAGE, and the indicated proteins were detected by immunoblotting. (D) A2-4 cells were cultured in the presence (+Dox) or absence (−Dox) of Dox for 96 h. Intact nuclei were prepared and incubated in Xenopus egg extract in the presence of [α-32P]dCTP. Relative DNA replication efficiency was determined by measuring the percent of acid-precipitable cpm (TCA). (E) A2-4 cells were cultured in the presence or absence of Dox for 96 h. Intact nuclei were incubated in Xenopus egg extract in the presence of [α-32P]dCTP and 80 nM geminin-DEL for 150 min. Relative DNA replication efficiency was determined by measuring the percentage of acid-precipitable cpm (TCA). The percentage of inhibition of replication by the inclusion of geminin-DEL is shown as the mean ± standard deviation.
Endogenous RB mediates both acute and chronic replication checkpoints.
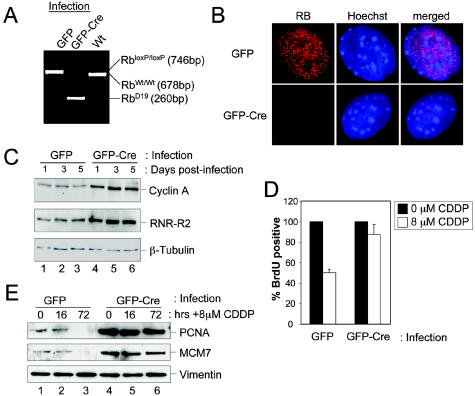
To further determine the action of endogenous RB in the induction of these kinetically distinct replication arrest states, we utilized MAFs with floxed Rb alleles (RbloxP/loxP) (42, 69). Infection of isolated MAFs with adenovirus encoding GFP and Cre recombinase (GFP-Cre) resulted in excision of exon 19 of the RbloxP allele, as shown by genomic PCR (Fig. 8A). GFP infection served as a negative control (Fig. 8A). Analyses of RB protein expression following GFP-Cre infection revealed a rapid loss of RB protein, which could be readily observed by immunoblotting (9; data not shown). Since it has been speculated that RB may localize differently in primary and immortal/tumor cells, we also analyzed RB localization via immunostaining (32). As shown in Fig. 8B, we observed that RB exhibited a focal staining pattern throughout the nucleoplasm that was similar to that observed in U2OS cells. This staining was specifically lost in the Cre-infected cultures (Fig. 8B). As a consequence of RB loss, deregulation of established RB/E2F targets was observed (Fig. 8C). For example, relative levels of cyclin A and RNR R2 subunit (RNR-R2) increase upon loss of RB expression. To verify that RB was required to invoke an acute cell cycle arrest comparable to that observed in inducible systems, asynchronously proliferating control (GFP) or Rb knockout (GFP-Cre) cells were treated with 8 μM cisplatin (CDDP) for 16 h. DNA damage induced by CDDP has been previously shown to activate endogenous RB, leading to the transcriptional repression of E2F-regulated target genes and ultimately cell cycle arrest (28, 35, 62). The ability of CDDP-treated MAFs to traverse the cell cycle was monitored by BrdU incorporation (Fig. 8D). While control cells exhibited an acute checkpoint response to CDDP treatment, RB-deficient cells continued to incorporate BrdU unchecked. To investigate the effects of CDDP on levels of replication proteins, GFP (control)- and Cre-infected MAFs were cultured following exposure to 8 μM CDDP for the indicated time (Fig. 8E). Consistent with acute arrest mediated by active RB alleles, protein levels of RB/E2F-regulated DNA replication factors (MCM7 and PCNA) were unchanged after 16 h. However, these proteins were not detectable 72 h post-CDDP treatment. This delayed response is RB-dependent, as no attenuation of MCM7 and PCNA was observed in the Cre-infected cells (Fig. 8E). These data indicate that endogenous RB triggers both acute and chronic S-phase arrest programs in response to genotoxic stress.
FIG. 8.
Endogenous RB mediates both acute and chronic arrest states. (A) MAFs with a wild-type Rb gene (RbWt/Wt) or MAFs with loxP sites flanking exon 19 of the Rb gene (RbloxP/loxP) were cultured. RbloxP/loxP MAFs were infected with GFP (control)- or GFP-Cre-encoding adenovirus. Genomic DNA was isolated 72 h postinfection and utilized for PCR. Recombination/excision of exon 19 is detectable by the amplification of a novel PCR product (RbD19). (B) MAFs of the RbloxP/loxP genotype were infected with GFP- or GFP-Cre-encoding adenoviruses and cultured on glass coverslips. Cells were fixed, and endogenous RB protein was detected by immunostaining. (C) MAFs of the RbloxP/loxP genotype were infected with GFP (lanes 1 to 3)- or GFP-Cre (lanes 4 to 6)-encoding adenovirus. Cells were harvested either 1, 3, or 5 days postinfection, and equal total protein levels were resolved by SDS-PAGE and immunoblotted for the proteins indicated. (D) MAFs of the RbloxP/loxP genotype were infected with GFP- or GFP-Cre-encoding adenovirus. Seventy-two hours postinfection, cells were either untreated (0 μM) or treated with 8 μM cisplatin (CDDP) for 16 h. Subsequently the cells were pulse-labeled with BrdU, and BrdU incorporation was then monitored by immunofluorescence microscopy. The values shown are means ± standard deviation. Experiments were performed twice with greater than 200 cells counted per experiment. (E) MAFs of the RbloxP/loxP genotype infected with GFP- and Cre-encoding adenovirus were treated with 8 μM CDDP for 0 or 16 h. Cells were then cultured for an additional 56 h in drug-free medium. At the indicated times post-CDDP addition, cells were harvested and equal total protein amounts were resolved by SDS-PAGE. The indicated proteins were detected by immunoblotting.
Active RB-mediated arrest programs are reversible.
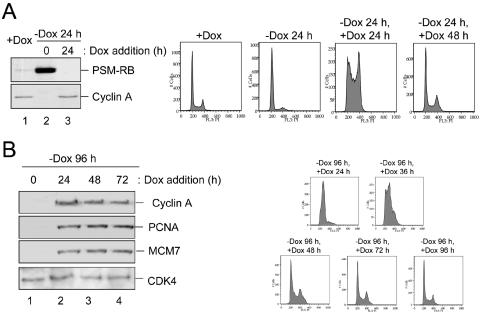
To date, the majority of replicative exit pathways studied (e.g., hydroxyurea or DNA damage induced) represent terminal cell cycle end points akin to senescence (8, 59). To determine whether the temporally distinct S-phase inhibitory mechanisms enacted by RB were permissive for the resumption of DNA replication, the reversibility of each arrest state was evaluated. Although this has not been previously determined, it could be envisioned that acute RB-mediated arrest represents a transient replication pausing mechanism to allow for the repair of damaged DNA. Readdition of Dox to the media of acutely arrested cells led to the rapid loss of active RB expression (Fig. 9A, left panel). Following the attenuation of active RB, cyclin A levels were restored. As shown in Fig. 9A (right panel), cells complete S phase in a relatively synchronous fashion, following cessation of active RB expression and reaccumulation of cyclin A. These results agree with the idea that acute RB arrest did not irreversibly disrupt preRC assembly or replicative competence; rather, the progression of replicating forks was impeded.
FIG. 9.
Acute and chronic arrest states can be reversed by loss of active RB signaling. (A, left panel) A2-4 cells were cultured in the presence of Dox (+Dox, lane 1) or the absence of Dox (−Dox, lane 2). Cells were cultured in the absence of Dox for 24 h, and then Dox was added back to the media for 24 h (−Dox 24 h, +Dox 24 h, lane 3). Equal total protein amounts were resolved by SDS-PAGE, and RB and cyclin A were detected by immunoblotting. (Right panel) A2-4 cells were additionally processed as indicated for flow cytometry. (B, left panel) A2-4 cells were cultured in the absence of Dox for 96 h, and then Dox was added back to the medium for the indicated time points. Equal total protein was resolved by SDS-PAGE, and immunoblotting was performed to detect the indicated proteins. (Right panel) Samples were also utilized for flow cytometry.
Since chronic RB activation led to the dramatic attenuation of preRC components and elongation factors, we hypothesized that these cells might fail to reenter the cell cycle upon the attenuation of RB expression. Additionally, preRC assembly is normally inhibited in cells with S-phase DNA content (7). As previously shown, cells cultured for 96 h in medium lacking Dox exhibit an S-phase DNA content (Fig. 7A). At this point, Dox was added to the medium, thereby attenuating expression of the Tet-regulated RB allele (not shown). Alleviation of repression by RB resulted in the accumulation of target protein levels (Fig. 9B, left panel). To determine whether the availability of replication factors was sufficient to enable cell cycle progression, cell cycle position was monitored by flow cytometry (Fig. 9B, right panel). Surprisingly, these cells were able to resume S-phase progression following the derepression of RB/E2F targets. Furthermore, these cells exhibited no increase in DNA content above 4 N, suggesting that their genome was accurately duplicated and followed by a normal mitotic division. Long-term analyses of these cells indicated that cell cycle progression and proliferation ensued following the cessation of active RB signaling (data not shown).
Acute loss of endogenous RB permits S-phase progression after checkpoint induction.
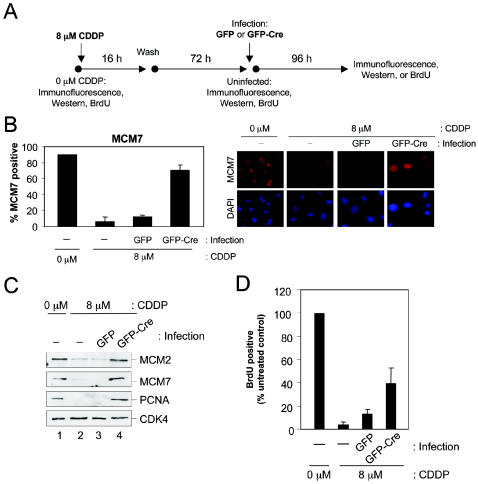
These observations suggested that RB-mediated transcriptional repression may be essential for maintaining an S-phase exit state. This possibility was addressed in the context of DNA damage-induced RB activation. To establish the requirement of endogenous RB for the prevention of aberrant S-phase reentry following DNA damage, we employed the protocol illustrated in Fig. 10A. Chronic RB activation by CDDP treatment was followed by GFP or GFP-Cre infection and cells were subsequently analyzed for protein expression and BrdU incorporation. As shown in Fig. 10B, immunofluorescent detection revealed that loss of RB led to derepression of MCM7, as compared to GFP-infected controls (Fig. 10B). PCNA dererepression was similarly detected by immunofluorescence (not shown). These observations were further confirmed by immunoblotting for MCM2, MCM7, and PCNA (Fig. 10C). These findings support the notion that RB is essential for long-term suppression of E2F targets, similar to senescence (50). The deregulation of MCM and PCNA expression did not reveal whether the fibroblasts had resumed S phase. To directly determine the replicative capacity of these cells, BrdU incorporation was monitored following the adenoviral infection of chronically arrested MAFs. Importantly, the observed accumulation of MCM7 and PCNA coincided with DNA replication as monitored by BrdU incorporation in the RB knockout cells (Fig. 10D). These data indicate that endogenous RB is required to maintain S-phase arrest and prevent the replication of damaged DNA even following chronic exposure and the downregulation of replicative factors.
FIG. 10.
RB ablation reverses the DNA damage-induced chronic arrest state. (A) Schematic diagram illustrating the protocol utilized for analysis of chronic arrest reversal in RB conditional knockout cells. (B) MAFs of the RbloxP/loxP genotype were cultured as depicted in panel A, and MCM7 was detected by immunofluorescence at the indicated time points. (Left panel) percentage of cells with MCM7-positive staining was determined. The data shown (mean ± standard deviation) are from two independent experiments with at least 150 cells counted per experiment. (Right panel) Representative photomicrographs of MCM7 immunostaining. Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI). (C) MAFs of the RbloxP/loxP genotype were cultured as depicted in panel A. Equal total protein was resolved by SDS-PAGE, and the indicated proteins were detected by immunoblotting. (D) MAFs of the RbloxP/loxP genotype were cultured as depicted in panel A. At the indicated time points, BrdU incorporation was monitored by immunofluorescent staining. The percentage of BrdU-positive cells was determined relative to untreated control. The data shown (mean ± standard deviation) are from two independent experiments with at least 150 cells counted per experiment.
DISCUSSION
Understanding how RB suppresses proliferation and, conversely, how loss of RB deregulates cell cycle control is critical for elucidating the tumor suppressive functions of RB. Here, we demonstrate a role for RB in the mediation of two kinetically distinct S-phase inhibitory mechanisms. Both pathways lead to the inhibition of DNA synthesis differing only with regard to the status of the replication machinery—acute disruption of the elongation factor recruitment versus a more sustained attenuation of requisite factors. Importantly, the loss of RB compromises the appropriate replicative inhibition elicited by DNA damage and even enables those cells that have undergone replicative exit to resume cell cycle progression.
Recent studies have suggested that RB utilizes the direct association with components of the replication machinery to inhibit DNA synthesis. This mechanism was first supported by the finding that RB can interact with MCM7 through its N-terminal domain (64). Subsequent data have suggested that RB and E2F can interact with replication foci (32). Additionally, in Drosophila it has been documented that the influence of RB on replication control occurs through a nontranscriptional mechanism (10). Lastly, it has been recently shown that the RB protein can be detected at sites of replication initiation during the inhibition of replication following DNA damage (4). Based on these studies, it would be predicted that RB could function in cis to inhibit DNA replication potentially dependent on the N terminus of RB (24, 25). We failed to observe replicative inhibition by RB in in vitro replication assays with both N-terminal-truncated and full-length RB alleles, although these alleles clearly inhibited DNA replication in cell culture. This contrasts with the effect of direct inhibition of DNA synthesis as occurs through the use of APH or the addition of a Ran mutant incapable of hydrolyzing GTP (77). These results indicate that the cellular inhibition of DNA replication does not involve a cis-acting effect of RB on the replication machinery. In keeping with a trans-acting effect of RB, we failed to detect a localization of RB with sites of replication. This was evident both with endogenous RB and with the active RB alleles. These results are consistent with a recent study demonstrating that RB does not colocalize with replication foci or MCM complexes in mammalian cells (16).
In addition to direct effects on the replication machinery, it has been postulated that downregulation of E2F target genes represents the critical means through which RB impacts DNA replication (19, 26). Specifically, microarray studies by numerous laboratories have documented MCMs, DNA polymerase subunits, and several other replication components as targets of E2F control (29, 48). As such, it would be predicted that these targets are downregulated during RB-mediated arrest. However, we fail to observe downregulation of protein levels of any replication proteins concurrent with the rapid arrest induced by RB. Additionally, analysis of protein activities documented that multiple replication factors in the preRC and initiation complex were functional or chromatin tethered when cells are arrested by RB. These results indicate that RB does not preclude numerous steps associated with replication but does specifically perturb the activity of PCNA. We have previously found that PCNA is a target of active RB signaling through cyclin A (60) but further define that the effect of RB is highly specific to the loading of PCNA and does not affect RFC directly upstream of PCNA. These results suggest that the retention of replication factors during a short-term response to RB serves a purpose. One of the physiological signals known to induce RB-dependent S-phase inhibition is DNA damage (28, 35). The continued presence of RPA and PCNA would be required for the repair of genetic lesions prior to S-phase completion (56, 74). Consistent with this prediction, we observed the relocalization of GFP-RPA34 and GFP-PCNA to sites of CDDP-induced damage in living cells arrested by RB (not shown) (60). Since PCNA represents a later stage in the replication process, the RB-mediated replication pause could be readily reversed following the repair of damage. In fact, the specific disruption of replication complexes by RB may represent a means to free PCNA from replicative roles to repair.
The results observed with the rapid response to RB activation questions the involvement of E2F/RB-mediated control of replication factors. Based on recent work in Drosophila, it is clear that levels of MCM proteins play critical roles in the regulation of replication by the E2F/RB signaling axis (13). We find that RB does, in fact, target the expression of a large number of replication factors at the protein level, but this occurs with delayed kinetics. Such a finding is consistent with the relatively long half-life that has been documented for MCM3 (49). Under the conditions of chronic RB-mediated arrest, cells arrest with an S-phase DNA content but lack the expression of replication factors as is typically only observed in quiescent cells. This molecular state is obviously not common in cell cycle control but has been observed following prolonged replication arrest and, as we show here, occurs in cells with severely damaged DNA. In addition, the existence of analogous kinetically distinct arrest states mediated by RB has been recently demonstrated in the context of the G2/M checkpoint (21). In quiescent cells, resynthesis and assembly of preRC complexes are believed to allow for the subsequent entry into the cell cycle (66). However, those cells chronically arrested with hydroxyurea fail to reenter the cycle, and the prolonged replicative block following DNA damage has been associated with an irreversible senescence program (8). As such, this suggests that temporally delayed block elicited by RB represents a means to permanently retreat from the cell cycle and prevent replication from occurring following catastrophic DNA damage or other replicative insults.
The action of RB in mediating long-term replicative exit has clear physiological relevance. Severe DNA damage leads to blocks in replication to prevent mutations, and this program can induce a senescence-like state (59). Consistent with chronic RB activation playing a similar role, RB-arrested cells exhibit a senescent morphology and stain positively for senescence-associated β-galactosidase (1; Williams et al., submitted for publication). In the case of CDDP damage and RB-mediated arrest, cells are present in S phase concurrent with the exit from the cell cycle and the downregulation of replication factors. What happens to replication structures or forks under these conditions is unknown, but both long-term CDDP damage and PSM-RB-mediated arrests are reversible following the attenuation of RB activity. This finding demonstrates that replicative exit programs are not necessarily irreversible but are dependent on RB for their maintenance. RB clearly controls the expression of multiple replication factors, and when this control is lost, the factors are resynthesized. Importantly, they are not only synthesized but function to mediate DNA replication. How these factors assemble and position the replication machinery to prevent rereplication under these conditions is not clear and the subject of ongoing study. Irrespective, these results indicate that RB plays a critical role in the maintenance of cell cycle exit and that loss of RB represents a critical means through which arrested cells can reenter the cell cycle.
Acknowledgments
We thank Karen Knudsen for critical reading of the manuscript and all members of the Knudsen laboratories for helpful comments. We thank Jiri Bartek for the A5C1 cell line. We are grateful to J. Wade Harper for providing RbΔcdk plasmid, Bruce Stillman for cdc6 and RFCp140 antibody, Jerard Hurwitz for RFCp37 antibody, Hisao Masai for HsDbf4 (ASK) antibody, Marc Wold for RPA antibody, Wallace Ip for vimentin antibody, and Liang Zhu for providing U24 cells. Gustavo Leone and James DeGregori kindly provided GFP, GFP-Cre, and E2F2 adenoviruses. We also thank Sandy Schwemberger and George Babcock for expert flow cytometric analyses.
S.P.A. was supported by the Albert J. Ryan Foundation and a U.C. Distinguished Graduate Fellowship. C.N.M. is supported by NCI training grant T32 CA 59268. D.M.G. is supported by NIH grant GM-57233-01. E.S.K. is supported by NCI grant CA-106471.
REFERENCES
- 1.Alexander, K., and P. W. Hinds. 2001. Requirement for p27KIP1 in retinoblastoma protein-mediated senescence. Mol. Cell. Biol. 21:3616-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angus, S. P., A. F. Fribourg, M. P. Markey, S. L. Williams, H. F. Horn, J. DeGregori, T. F. Kowalik, K. Fukasawa, and E. S. Knudsen. 2002. Active RB elicits late G1/S inhibition. Exp. Cell Res. 276:201-213. [DOI] [PubMed] [Google Scholar]
- 3.Angus, S. P., D. A. Solomon, L. Kuschel, R. F. Hennigan, and E. S. Knudsen. 2003. Retinoblastoma tumor suppressor: analyses of dynamic behavior in living cells reveal multiple modes of regulation. Mol. Cell. Biol. 23:8172-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avni, D., H. Yang, F. Martelli, F. Hofmann, W. M. ElShamy, S. Ganesan, R. Scully, and D. M. Livingston. 2003. Active localization of the retinoblastoma protein in chromatin and its response to S phase DNA damage. Mol. Cell 12:735-746. [DOI] [PubMed] [Google Scholar]
- 5.Bartek, J., J. Bartkova, and J. Lukas. 1997. The retinoblastoma protein pathway in cell cycle control and cancer. Exp. Cell Res. 237:1-6. [DOI] [PubMed] [Google Scholar]
- 6.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 7.Blow, J. J., and B. Hodgson. 2002. Replication licensing—defining the proliferative state? Trends Cell Biol. 12:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borel, F., F. B. Lacroix, and R. L. Margolis. 2002. Prolonged arrest of mammalian cells at the G1/S boundary results in permanent S phase stasis. J. Cell Sci. 115:2829-2838. [DOI] [PubMed] [Google Scholar]
- 9.Bosco, E. E., C. N. Mayhew, R. F. Hennigan, J. Sage, T. Jacks, and E. S. Knudsen. 2004. RB signaling prevents replication-dependent DNA double-strand breaks following genotoxic insult. Nucleic Acids Res. 32:25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosco, G., W. Du, and T. L. Orr-Weaver. 2001. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 3:289-295. [DOI] [PubMed] [Google Scholar]
- 11.Cam, H., and B. D. Dynlacht. 2003. Emerging roles for E2F: beyond the G1/S transition and DNA replication. Cancer Cell 3:311-316. [DOI] [PubMed] [Google Scholar]
- 12.Cardoso, M. C., C. Joseph, H. P. Rahn, R. Reusch, B. Nadal-Ginard, and H. Leonhardt. 1997. Mapping and use of a sequence that targets DNA ligase I to sites of DNA replication in vivo. J. Cell Biol. 139:579-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cayirlioglu, P., W. O. Ward, S. C. Silver Key, and R. J. Duronio. 2003. Transcriptional repressor functions of Drosophila E2F1 and E2F2 cooperate to inhibit genomic DNA synthesis in ovarian follicle cells. Mol. Cell. Biol. 23:2123-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chew, Y. P., M. Ellis, S. Wilkie, and S. Mittnacht. 1998. pRB phosphorylation mutants reveal role of pRB in regulating S phase completion by a mechanism independent of E2F. Oncogene 17:2177-2186. [DOI] [PubMed] [Google Scholar]
- 15.Connell-Crowley, L., S. J. Elledge, and J. W. Harper. 1998. G1 cyclin-dependent kinases are sufficient to initiate DNA synthesis in quiescent human fibroblasts. Curr. Biol. 8:65-68. [DOI] [PubMed] [Google Scholar]
- 16.Dimitrova, D. S., and R. Berezney. 2002. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J. Cell Sci. 115:4037-4051. [DOI] [PubMed] [Google Scholar]
- 17.Dimitrova, D. S., T. A. Prokhorova, J. J. Blow, I. T. Todorov, and D. M. Gilbert. 2002. Mammalian nuclei become licensed for DNA replication during late telophase. J. Cell Sci. 115:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimitrova, D. S., I. T. Todorov, T. Melendy, and D. M. Gilbert. 1999. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 146:709-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 20.Ezhevsky, S. A., A. Ho, M. Becker-Hapak, P. K. Davis, and S. F. Dowdy. 2001. Differential regulation of retinoblastoma tumor suppressor protein by G1 cyclin-dependent kinase complexes in vivo. Mol. Cell. Biol. 21:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flatt, P. M., L. J. Tang, C. D. Scatena, S. T. Szak, and J. A. Pietenpol. 2000. p53 regulation of G2 checkpoint is retinoblastoma protein dependent. Mol. Cell. Biol. 20:4210-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geng, Y., Q. Yu, E. Sicinska, M. Das, J. E. Schneider, S. Bhattacharya, W. M. Rideout, R. T. Bronson, H. Gardner, and P. Sicinski. 2003. Cyclin E ablation in the mouse. Cell 114:431-443. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gladden, A. B., and J. A. Diehl. 2003. The cyclin D1-dependent kinase associates with the pre-replication complex and modulates RB.MCM7 binding. J. Biol. Chem. 278:9754-9760. [DOI] [PubMed] [Google Scholar]
- 25.Goodrich, D. W. 2003. How the other half lives, the amino-terminal domain of the retinoblastoma tumor suppressor protein. J. Cell Physiol. 197:169-180. [DOI] [PubMed] [Google Scholar]
- 26.Harbour, J. W., and D. C. Dean. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14:2393-2409. [DOI] [PubMed] [Google Scholar]
- 27.Harbour, J. W., R. X. Luo, A. Dei Santi, A. A. Postigo, and D. C. Dean. 1999. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 98:859-869. [DOI] [PubMed] [Google Scholar]
- 28.Harrington, E. A., J. L. Bruce, E. Harlow, and N. Dyson. 1998. pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc. Natl. Acad. Sci. USA 95:11945-11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishida, S., E. Huang, H. Zuzan, R. Spang, G. Leone, M. West, and J. R. Nevins. 2001. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21:4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaelin, W. G., Jr. 1997. Alterations in G1/S cell-cycle control contributing to carcinogenesis. Ann. N. Y. Acad. Sci. 833:29-33. [DOI] [PubMed] [Google Scholar]
- 31.Kelly, T. J., and G. W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69:829-880. [DOI] [PubMed] [Google Scholar]
- 32.Kennedy, B. K., D. A. Barbie, M. Classon, N. Dyson, and E. Harlow. 2000. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 14:2855-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knudsen, E. S., C. Buckmaster, T. T. Chen, J. R. Feramisco, and J. Y. Wang. 1998. Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev. 12:2278-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knudsen, K. E., K. C. Arden, and W. K. Cavenee. 1998. Multiple G1 regulatory elements control the androgen-dependent proliferation of prostatic carcinoma cells. J. Biol. Chem. 273:20213-20222. [DOI] [PubMed] [Google Scholar]
- 35.Knudsen, K. E., D. Booth, S. Naderi, Z. Sever-Chroneos, A. F. Fribourg, I. C. Hunton, J. R. Feramisco, J. Y. J. Wang, and E. S. Knudsen. 2000. RB-dependent S-phase response to DNA damage. Mol. Cell. Biol. 20:7751-7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knudsen, K. E., A. F. Fribourg, M. W. Strobeck, J. M. Blanchard, and E. S. Knudsen. 1999. Cyclin A is a functional target of retinoblastoma tumor suppressor protein-mediated cell cycle arrest. J. Biol. Chem. 274:27632-27641. [DOI] [PubMed] [Google Scholar]
- 37.Leonhardt, H., H. P. Rahn, P. Weinzierl, A. Sporbert, T. Cremer, D. Zink, and M. C. Cardoso. 2000. Dynamics of DNA replication factories in living cells. J. Cell Biol. 149:271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lippincott-Schwartz, J., E. Snapp, and A. Kenworthy. 2001. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2:444-456. [DOI] [PubMed] [Google Scholar]
- 39.Lukas, J., T. Herzinger, K. Hansen, M. C. Moroni, D. Resnitzky, K. Helin, S. I. Reed, and J. Bartek. 1997. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes Dev. 11:1479-1492. [DOI] [PubMed] [Google Scholar]
- 40.Lukas, J., D. Parry, L. Aagaard, D. J. Mann, J. Bartkova, M. Strauss, G. Peters, and J. Bartek. 1995. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature 375:503-506. [DOI] [PubMed] [Google Scholar]
- 41.Lukas, J., C. S. Sorensen, C. Lukas, E. Santoni-Rugiu, and J. Bartek. 1999. p16INK4a, but not constitutively active pRb, can impose a sustained G1 arrest: molecular mechanisms and implications for oncogenesis. Oncogene 18:3930-3935. [DOI] [PubMed] [Google Scholar]
- 42.Marino, S., M. Vooijs, H. van Der Gulden, J. Jonkers, and A. Berns. 2000. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14:994-1004. [PMC free article] [PubMed] [Google Scholar]
- 43.Markey, M. P., S. P. Angus, M. W. Strobeck, S. L. Williams, R. W. Gunawardena, B. J. Aronow, and E. S. Knudsen. 2002. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res. 62:6587-6597. [PubMed] [Google Scholar]
- 44.Méndez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montecucco, A., R. Rossi, D. S. Levin, R. Gary, M. S. Park, T. A. Motycka, G. Ciarrocchi, A. Villa, G. Biamonti, and A. E. Tomkinson. 1998. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 17:3786-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris, E. J., and N. J. Dyson. 2001. Retinoblastoma protein partners. Adv. Cancer Res. 82:1-54. [DOI] [PubMed] [Google Scholar]
- 47.Mossi, R., and U. Hubscher. 1998. Clamping down on clamps and clamp loaders—the eukaryotic replication factor C. Eur. J. Biochem. 254:209-216. [PubMed] [Google Scholar]
- 48.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Musahl, C., H. P. Holthoff, R. Lesch, and R. Knippers. 1998. Stability of the replicative Mcm3 protein in proliferating and differentiating human cells. Exp. Cell Res. 241:260-264. [DOI] [PubMed] [Google Scholar]
- 50.Narita, M., S. Nunez, E. Heard, A. W. Lin, S. A. Hearn, D. L. Spector, G. J. Hannon, and S. W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113:703-716. [DOI] [PubMed] [Google Scholar]
- 51.Nevins, J. R. 2001. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10:699-703. [DOI] [PubMed] [Google Scholar]
- 52.Niculescu, A. B., III, X. Chen, M. Smeets, L. Hengst, C. Prives, and S. I. Reed. 1998. Effects of p21Cip1/Waf1 at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol. Cell. Biol. 18:629-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okuno, Y., A. J. McNairn, N. den Elzen, J. Pines, and D. M. Gilbert. 2001. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 20:4263-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ortega, S., I. Prieto, J. Odajima, A. Martin, P. Dubus, R. Sotillo, J. L. Barbero, M. Malumbres, and M. Barbacid. 2003. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat. Genet. 35:25-31. [DOI] [PubMed] [Google Scholar]
- 55.Pennaneach, V., I. Salles-Passador, A. Munshi, H. Brickner, K. Regazzoni, F. Dick, N. Dyson, T. T. Chen, J. Y. Wang, R. Fotedar, and A. Fotedar. 2001. The large subunit of replication factor C promotes cell survival after DNA damage in an LxCxE motif- and Rb-dependent manner. Mol. Cell 7:715-727. [DOI] [PubMed] [Google Scholar]
- 56.Prosperi, E. 1997. Multiple roles of the proliferating cell nuclear antigen: DNA replication, repair and cell cycle control. Prog. Cell Cycle Res. 3:193-210. [DOI] [PubMed] [Google Scholar]
- 57.Rowland, B. D., S. G. Denissov, S. Douma, H. G. Stunnenberg, R. Bernards, and D. S. Peeper. 2002. E2F transcriptional repressor complexes are critical downstream targets of p19(ARF)/p53-induced proliferative arrest. Cancer Cell 2:55-65. [DOI] [PubMed] [Google Scholar]
- 58.Saudan, P., J. Vlach, and P. Beard. 2000. Inhibition of S-phase progression by adeno-associated virus Rep78 protein is mediated by hypophosphorylated pRb. EMBO J. 19:4351-4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmitt, C. A., J. S. Fridman, M. Yang, S. Lee, E. Baranov, R. M. Hoffman, and S. W. Lowe. 2002. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109:335-346. [DOI] [PubMed] [Google Scholar]
- 60.Sever-Chroneos, Z., S. P. Angus, A. F. Fribourg, H. Wan, I. Todorov, K. E. Knudsen, and E. S. Knudsen. 2001. Retinoblastoma tumor suppressor protein signals through inhibition of cyclin-dependent kinase 2 activity to disrupt PCNA function in S phase. Mol. Cell. Biol. 21:4032-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherr, C. J., and F. McCormick. 2002. The RB and p53 pathways in cancer. Cancer Cell 2:103-112. [DOI] [PubMed] [Google Scholar]
- 62.Spitkovsky, D., A. Schulze, B. Boye, and P. Jansen-Durr. 1997. Down-regulation of cyclin A gene expression upon genotoxic stress correlates with reduced binding of free E2F to the promoter. Cell Growth Differ. 8:699-710. [PubMed] [Google Scholar]
- 63.Sporbert, A., A. Gahl, R. Ankerhold, H. Leonhardt, and M. C. Cardoso. 2002. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell 10:1355-1365. [DOI] [PubMed] [Google Scholar]
- 64.Sterner, J. M., S. Dew-Knight, C. Musahl, S. Kornbluth, and J. M. Horowitz. 1998. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol. Cell. Biol. 18:2748-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stillman, B. 1996. Cell cycle control of DNA replication. Science 274:1659-1664. [DOI] [PubMed] [Google Scholar]
- 66.Sun, W., M. Hola, K. Pedley, S. Tada, J. J. Blow, I. T. Todorov, S. E. Kearsey, and R. F. Brooks. 2000. The replication capacity of intact mammalian nuclei in Xenopus egg extracts declines with quiescence, but the residual DNA synthesis is independent of Xenopus MCM proteins. J. Cell Sci. 113:683-695. [DOI] [PubMed] [Google Scholar]
- 67.Takemura, M., T. Kitagawa, S. Izuta, J. Wasa, A. Takai, T. Akiyama, and S. Yoshida. 1997. Phosphorylated retinoblastoma protein stimulates DNA polymerase alpha. Oncogene 15:2483-2492. [DOI] [PubMed] [Google Scholar]
- 68.Tetsu, O., and F. McCormick. 2003. Proliferation of cancer cells despite CDK2 inhibition. Cancer Cell 3:233-245. [DOI] [PubMed] [Google Scholar]
- 69.Vooijs, M., H. te Riele, M. van der Valk, and A. Berns. 2002. Tumor formation in mice with somatic inactivation of the retinoblastoma gene in interphotoreceptor retinol binding protein-expressing cells. Oncogene 21:4635-4645. [DOI] [PubMed] [Google Scholar]
- 70.Waga, S., and B. Stillman. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721-751. [DOI] [PubMed] [Google Scholar]
- 71.Walter, J. C. 2000. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J. Biol. Chem. 275:39773-39778. [DOI] [PubMed] [Google Scholar]
- 72.Wang, J. Y., E. S. Knudsen, and P. J. Welch. 1994. The retinoblastoma tumor suppressor protein. Adv. Cancer Res. 64:25-85. [DOI] [PubMed] [Google Scholar]
- 73.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 74.Wold, M. S. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66:61-92. [DOI] [PubMed] [Google Scholar]
- 75.Wu, J. R., G. Yu, and D. M. Gilbert. 1997. Origin-specific initiation of mammalian nuclear DNA replication in a Xenopus cell-free system. Methods 13:313-324. [DOI] [PubMed] [Google Scholar]
- 76.Wu, L., C. Timmers, B. Maiti, H. I. Saavedra, L. Sang, G. T. Chong, F. Nuckolls, P. Giangrande, F. A. Wright, S. J. Field, M. E. Greenberg, S. Orkin, J. R. Nevins, M. L. Robinson, and G. Leone. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414:457-462. [DOI] [PubMed] [Google Scholar]
- 77.Yamaguchi, R., and J. Newport. 2003. A role for Ran-GTP and Crm1 in blocking re-replication. Cell 113:115-125. [DOI] [PubMed] [Google Scholar]
- 78.Yang, H., B. O. Williams, P. W. Hinds, T. S. Shih, T. Jacks, R. T. Bronson, and D. M. Livingston. 2002. Tumor suppression by a severely truncated species of retinoblastoma protein. Mol. Cell. Biol. 22:3103-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang, H. S., A. A. Postigo, and D. C. Dean. 1999. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFbeta, and contact inhibition. Cell 97:53-61. [DOI] [PubMed] [Google Scholar]