Abstract
Several different transcription factors, including estrogen receptor, progesterone receptor, and ETS family members, have been implicated in human breast cancer, indicating that transcription factor-induced alterations in gene expression underlie mammary cell transformation. ESE-1 is an epithelium-specific ETS transcription factor that contains two distinguishing domains, a serine- and aspartic acid-rich (SAR) domain and an AT hook domain. ESE-1 is abundantly expressed in human breast cancer and trans-activates epithelium-specific gene promoters in transient transfection assays. While it has been presumed that ETS factors transform mammary epithelial cells via their nuclear transcriptional functions, here we show (i) that ESE-1 protein is cytoplasmic in human breast cancer cells; (ii) that stably expressed green fluorescent protein-ESE-1 transforms MCF-12A human mammary epithelial cells; and (iii) that the ESE-1 SAR domain, acting in the cytoplasm, is necessary and sufficient to mediate this transformation. Deletion of transcriptional regulatory or nuclear localization domains does not impair ESE-1-mediated transformation, whereas fusing the simian virus 40 T-antigen nuclear localization signal to various ESE-1 constructs, including the SAR domain alone, inhibits their transforming capacity. Finally, we show that the nuclear localization of ESE-1 protein induces apoptosis in nontransformed mammary epithelial cells via a transcription-dependent mechanism. Together, our studies reveal two distinct ESE-1 functions, apoptosis and transformation, where the ESE-1 transcription activation domain contributes to apoptosis and the SAR domain mediates transformation via a novel nonnuclear, nontranscriptional mechanism. These studies not only describe a unique ETS factor transformation mechanism but also establish a new paradigm for cell transformation in general.
ETS transcription factors play crucial roles in several different biological processes, including differentiation and tumorigenesis (36, 41). All ETS factors are characterized by a conserved winged helix-turn-helix DNA binding domain (DBD), the ETS domain, which mediates binding to ets consensus sites in target genes. ETS proteins, which function as activators and/or repressors of gene transcription, can be regulated by both protein-protein interactions and mitogen-activated protein kinase (MAPK)-mediated phosphorylation (36, 41). For example, members of at least six different ETS factor subfamilies are key nuclear effectors of the Ras/Raf/MAPK pathway, serving as direct targets of MAPK phosphorylation and thus playing critical regulatory roles in cell survival and proliferation (36, 41, 50).
Multiple lines of evidence support a causative role for ETS factors in carcinogenesis (19, 36, 41). The founding member of the ETS family, the v-ETS oncogene, causes hematopoetic malignancies in chickens (27). ETS factors are also associated with malignancies in humans. For example, chromosomal translocations in which the amino-terminal region of EWS is fused to the carboxy-terminal ETS domain of Fli-1 are found in 95% of human Ewing's sarcomas (15). Further, a variety of epithelial cancers (e.g., lung, breast, colon, and prostate) are associated with the up-regulation of one or more ETS factors, including ETS-1, ETS-2, ER81, PEA-3, and ESE-1 (19, 36, 41).
ETS factors have also been shown to play tumorigenic roles in breast cancer. Transgenic mice in which one ETS-2 allele has been disrupted are more resistant to breast tumorigenesis than are their wild-type counterparts (31). Further, the transformed phenotypes of both the BT-20 and the MMT breast cancer cell lines are inhibited by the expression of dominant-negative ETS DBD constructs, suggesting that the transforming activity of ETS factors depends on their transcription factor function (14, 40). While the precise causative function of specific ETS factors in human breast cancer has not been fully delineated, the ESE-1 ETS factor has been particularly implicated in breast tumorigenesis (2, 3, 7, 8, 18, 29, 30).
The epithelium-specific ETS (ESE) factor subfamily, which consists of ESE-1 (also known as ESX, Elf-3, Jen, and ERT), ESE-2, ESE-3, and PDEF, consists of epithelium-restricted transcription factors that function in epithelial cell differentiation (19). For example, gene knockout experiments have revealed that ESE-1 plays a key role in the morphogenesis and terminal differentiation of the murine small intestinal epithelium (32). Additionally, ESE-1 expression is induced upon keratinocyte differentiation (34), and ESE-1 regulates the expression of keratinocyte (5, 34), bronchial (38), and retinal (23) epithelial cell gene markers.
ESE-1 is a 46-kDa protein whose transcription factor function has been well documented in transient transfection-reporter gene assays, suggesting that ESE-1 carries out its biological effects by acting as a nuclear transcription factor (2, 3, 9, 18, 19, 29, 32, 34, 54). In addition to an ETS DBD (amino acids [aa] 274 to 354), ESE-1 includes a Pointed domain (aa 60 to 128), a transcription activation domain (TAD; aa 129 to 159), a serine- and aspartic acid-rich (SAR) domain (aa 189 to 229), and an AT hook domain (aa 238 to 259) (Fig. 1A) (1, 6, 7, 34). In a series of nuclear magnetic resonance-based structure-function studies, Asada et al. (2, 3) defined within the ESE-1 TAD a short α-helical motif (137SWIIELLE144) that mediates binding to both TAFII31 of the TATA binding protein complex and DRIP130/Sur2 of the Mediator complex (2, 3, 10). In addition, although these studies were mostly limited to transient transfection-gene reporter assays, their results are all consistent with the conclusion that ESE-1 functions as a nuclear regulator of transcription.
FIG. 1.
Structural organization of the human ESE-1 protein. The ESE-1 protein is depicted from the amino end (N) to the carboxy end (C). The Pointed domain, TAD, SAR domain, AT hook/NLS domain, and ETS domain are shown. Short black lines reveal the positions of NLS1 (aa 238 to 269) and putative NES1 (aa 102 to 112), NES2 (aa 275 to 284), and NLS2 (aa 316 to 323).
Compared to other ETS factors, ESE-1 appears to play an important role in human breast cancer. The ESE-1 gene locus (1q32.1) is amplified in approximately one-half of early human breast tumors, and ESE-1 mRNA overexpression is found in approximately 40% of human ductal carcinomas in situ (DCIS) (7, 47). Further, not only is overexpression of the proto-oncogenic HER-2 receptor positively associated with ESE-1 overexpression (7), but also HER-2-mediated signaling induces ESE-1 gene transcription (30). It was recently found (40a) that stable ESE-1 expression transforms MCF-12A human mammary epithelial cells, imparting to these cells a phenotype characterized by enhanced motility and invasiveness and an epithelial-to-mesenchymal morphological transition. MCF-12A stable transfectants expressing ESE-1 were also found to be capable of anchorage-independent growth in soft agarose and to form tumor-like organelles in three-dimensional Matrigel cultures (40a). In addition, it was demonstrated that MCF-12A cell growth becomes epidermal growth factor independent and that MAPK becomes constitutively phosphorylated when ESE-1 is stably expressed in these cells (40a). Taken together, these data suggest an oncogenic role for ESE-1 in human breast cancer.
Although the nuclear transcriptional function of other ETS factors is critical for their transforming function, here we show an entirely unanticipated cytoplasmic role for ESE-1 protein in cell transformation. Multiple, independently generated pools of MCF-12A cells stably expressing distinct epitope-tagged versions of full-length ESE-1 were able to form colonies in soft agarose, whereas control cell lines showed significantly reduced colony formation. Examination of functional ESE-1 domains by internal deletion analysis revealed that domains associated with transcription factor function or nuclear localization are not required for transformation. In contrast, internal deletion and domain fusion experiments demonstrated that the unique 50-aa ESE-1 SAR domain is necessary and sufficient to induce MCF-12A cell transformation. Consistent with the discovery that nuclear mechanisms do not underlie ESE-1 transforming function, we show that enforced nuclear targeting of stably expressed full-length ESE-1 constructs with internal deletions or only the SAR domain inhibits their transforming properties and that transiently expressed intact ESE-1 protein is nuclear and apoptotic. In further support of this interpretation, we present immunohistochemical (IHC) data showing that endogenous ESE-1 protein is cytoplasmic in primary human breast cancer tissues and in the T-47D breast cancer cell line. Taken together, our studies reveal that ESE-1 protein function depends on subcellular localization and demonstrate that cytoplasmically localized ESE-1 protein mediates breast cell transformation via a novel nonnuclear, nontranscriptional mechanism.
MATERIALS AND METHODS
Mammalian cell cultures.
The MCF-12A, MCF-10A, T-47D, and SK-BR-3 human breast cell lines (American Type Culture Collection, Manassas, Va.) were maintained as described previously (18). For all cell lines, subconfluent cells between passages 5 and 10 were used for all experiments.
IHC analysis.
Slides were deparaffinized in xylene and rehydrated in graded ethanols. Microwave antigen retrieval was performed by boiling slides in 10 mM citrate buffer for 10 min. The following procedures were performed by using a Ventana ES Autostainer at 37°C with 1× APK washes (Ventana Medical Systems, Tucson, Ariz.) between the steps. Endogenous peroxidase was blocked with 3% H2O2 for 4 min. Slides were incubated with primary antibody (1:20) for 32 min. The signal was enhanced by using an amplification kit (Ventana Medical Systems). Endogenous avidin and biotin were blocked and slides were visualized with biotinylated goat secondary antibody (anti-mouse immunoglobulin G [IgG] or IgM and anti-rabbit IgG) and peroxidase-labeled streptavidin followed by the chromogen diaminobenzidine (DAB), substrate, and copper enhancer (Ventana Medical Systems). Slides were counterstained with dilute Gill's no. 2 hematoxylin for 4 min, dehydrated in graded ethanols, and treated with xylene.
Adenovirus vectors.
The expression virus Ad.ESE-1 (adenovirus encoding untagged ESE-1 protein) was constructed by ligating a BglII-cut fragment encompassing the human ESE-1 coding sequence (18) to BglII-cut shuttle plasmid pAdTrack-CMV (Stratagene Inc., La Jolla, Calif.). Bacterial recombination and viral packaging were carried out according to the manufacturer's specifications (Stratagene) to produce Ad.empty (empty control virus) and Ad.ESE-1 stocks. The ESE-1 coding sequence in the adenovirus genome was verified by dideoxy sequencing at the UCHSC Cancer Center DNA Sequencing Core Facility.
GFP-ESE-1 mammalian expression vectors.
An EcoRI ESE-1 cDNA fragment excised from pCR2.1-ESE-1 (18) was ligated to EcoRI-cut plasmid pEGFP-C3 (Invitrogen, Inc., Carlsbad, Calif.), in frame and downstream of the green fluorescent protein (GFP) gene (see Fig. 4A), to produce pEGFP-ESE-1. Internal deletions in GFP-ESE-1 were generated by using two PCRs, one generating the amino-terminal regions and one generating the carboxy-terminal regions flanking each internal deletion. An EcoRI site was engineered at the outside ends of each fragment, and a shared restriction site was included at the deletion junction, allowing in-frame ligation. GFP-ESE-1 fusion plasmids with internal deletions were produced by three-fragment ligation of the EcoRI-cut pEGFP-C3 vector and the two portions of each deletion construct noted above.
FIG. 4.
Stable expression of GFP-ESE-1 transforms MCF-12A cells. (A) RT-PCR strategy. Diagram of GFP-ESE-1 showing the positions of the forward 5′ GFP-specific primer (top arrow) and the reverse 3′ untranslated region-specific primer (bottom arrow) used in RT-PCR assays for GFP-ESE-1 (1,285 bp) and GFP only (169 bp) in stably transfected MCF-12A cell pools. (B) Expression of GFP-ESE-1 and of GFP only in stably transfected MCF-12A cells. The products of RT-PCRs with the primers described above and total RNA purified from each cell population were analyzed by 1% agarose gel electrophoresis. Lane 1 shows a no-template negative RT-PCR control. Two GFP-ESE-1 stable transfectant pools, GFP-ESE-1A (lane 2) and GFP-ESE-1B (lane 3), and two GFP-only stable transfectant pools, GFP A (lane 4) and GFP B (lane 5), were independently generated from MCF-12A cells. Lanes 2 and 3 show the expected GFP-ESE-1-specific 1,285-bp product, and lanes 4 and 5 show the expected GFP-only-specific 169-bp product. Total RNA from each stable transfectant pool was also treated with RNase A (GFP-ESE-1A, GFP-ESE-1B, GFP A, and GFP B in lanes 6 to 9, respectively) prior to RT-PCR. (C) Anchorage-independent colony formation by control MCF-12A and T-47D cells and by stably transfected MCF-12A cell pools. Cells were plated in soft agarose, and typical colonies were photographed 8 days later (magnification, ×40). The 12A and GFP-only panels each show a cluster of three cells (untransfected MCF-12A control cells and MCF-12A cells stably transfected with GFP only, respectively). The T-47D and GFP-ESE-1 panels each show a multicellular colony (T-47D human breast cancer cells and MCF-12A cells stably transfected with GFP-ESE-1, respectively).
The primers used for these constructs were as follows, with bold type indicating BglII (left) and EcoRI (right) restriction sites, italic type indicating the ESE-1 coding sequence, and underlining indicating the ESE-1 start and stop codons: 5′ ESE-1—CGAGATCTCCGGAATTCATATGGCTGCAACCTGTGA, directed against the 5′ end of ESE-1 and used to generate the 5′ segment of all constructs; and 3′ ESE-1—CGAGATCTCCGGAATTCATTCAGTTCCGACTCTGGA, directed against the 3′ end of ESE-1 and used to generate the 3′ segment of all constructs.
The antisense primers used to generate amino-terminal and carboxy-terminal PCR products were as follows, with italic type indicating the ESE-1 coding sequence, bold type indicating restriction sites, and the deletion name and junction-specific restriction site shown in parentheses (A, amino terminal; C, carboxy terminal): 5′-GAATTCACTAGTGCTGGCCTTCTCTG (A-ΔPointed, SpeI), 5′-GAATTCACTAGTAGCTCTTCTGAT (C-ΔPointed, SpeI), 5′-GAATTCGGGCCCGAGGTCTCGCAGCTGGG (A-ΔTAD, ApaI), 5′-GAATTCGGGCCCTTTGACCAG (C-ΔTAD, ApaI), 5′-GAATTCTCTAGAGACGTCAGAGCTGCCAG (A-ΔSAR, XbaI), 5′-GAATTCTCTAGAGACTGCAAGAAGGGGGA (C-ΔSAR, XbaI), 5′-GCGGCGCTCGAGATCGCTGGGGAAGAGCTTG (A-ΔAT/NLS, XhoI), and 5′-GCGGCGCTCGAGGCGCCCAGAGGCACCCAC (C-ΔAT/NLS, XhoI).
pEGFP-SAR was generated by amplifying ESE-1 aa 190 to 239 with the following PCR primer pair, with bold indicating BglII (left) and EcoRI (right) restriction sites and italic type indicating the ESE-1 SAR domain coding sequence: 5′-CGAGATCTCCGGAATTCATTCCCCTGGCAGCTCTGACGTCTC (sense) and 5′-CGAGATCTCCGGAATTCATATCCCCCTTCTTGCAGTC (antisense). The SAR PCR product was ligated to the pEGFP-C3 vector at the EcoRI site to generate a GFP-SAR in-frame fusion (see Fig. 6A). To generate the GFP-NLS-ESE-1, GFP-NLS-ESE-1ΔAT/NLS, and GFP-NLS-SAR constructs, we modified the two different sense PCR primers used to generate the ESE-1, ESE-1ΔAT/NLS, and SAR coding sequences to include an in-frame simian virus 40 (SV40) nuclear localization signal (NLS) sequence. These NLS sense primers were used in the PCR strategy described above to produce the NLS-ESE-1, NLS-ESE-1ΔAT/NLS, and NLS-SAR coding sequences, which were then ligated in-frame to the pEGFP-C3 vector at the EcoRI site. For the NLS-ESE-1 and NLS-ESE-1ΔAT/NLS PCRs, the sense NLS primer used was as follows, with bold roman type indicating an EcoRI site, nonbold italic type indicating the ESE-1 coding sequence, bold italic type indicating the SV40 NLS, and underlining indicating the ESE-1 start codon: 5′-CCGGAATTCATCCAAAAAAGAAGAGAAAGGTAATGGCTGCAACCTGTGA; the sense NLS primer used for the NLS-SAR PCR was as follows (same format as that just described): 5′-CCGGAATTCATCCAAAAAAGAAGAGAAAGGTATCCCCTGGCAGCTCTGAC. For all plasmids, the sequence, orientation, and frame (relative to the upstream GFP coding sequence) of each DNA insert were verified by DNA sequencing.
FIG. 6.
Deletion of the ESE-1 TAD impairs GFP-ESE-1-mediated apoptosis in transiently transfected MCF-12A cells, and the SAR domain, either as GFP-SAR or GFP-NLS-SAR, is insufficient to recapitulate GFP-ESE-1 apoptotic function. (A) Map and amino acid sequence of GFP-SAR and GFP-NLS-SAR. For GFP-SAR, GFP is indicated by a broken green box (not to scale) followed by the in-frame fusion of a 50-aa ESE-1 region (aa 190 to 239) spanning all but the first 2 aa of the SAR domain. The amino acid sequence of the 50-aa region is shown, with the SAR sequence in bold italic type and carboxy-terminal ESE-1-flanking amino acids in plain type. The SV40 NLS sequence and the in-frame insertion site used to generate GFP-NLS-SAR are also shown. (B) Subcellular localization of GFP-SAR and GFP-NLS-SAR in transiently transfected MCF-12A cells. MCF-12A cells transiently transfected with the GFP-SAR or GFP-NLS-SAR expression plasmid were stained with DAPI, and typical cells in each culture were imaged by digital deconvolution UV microscopy (magnification, ×100). The DAPI, GFP, and merge images are as described in the legend to Fig. 3A, and broken white lines represent cell outlines. Panels 1 to 3 show the diffuse cytoplasmic and nuclear localization of GFP-SAR in transiently transfected MCF-12A cells. Panels 4 to 6 shows the exclusive nuclear localization of transiently expressed GFP-SAR that results from the SV40 NLS fusion. (C) Analysis of the roles of the ESE-1 TAD and SAR domain in the apoptotic activity of transiently expressed GFP-ESE-1. MCF-12A cells were transiently transfected with the GFP-only, GFP-ESE-1, GFP-ESE-1ΔTAD, GFP-SAR, or GFP-NLS-SAR expression plasmid. As described in the legend to Fig. 3C, apoptosis-specific annexin V staining in each transfectant population was examined 12 h later. The percentage of green and red cells in GFP-only transient transfectants was normalized to 1, and the fold increase in apoptosis for GFP-ESE-1, GFP-ESE-1ΔTAD, GFP-SAR, and GFP-NLS-SAR transient transfectants is shown. Three separate transfection experiments were performed for each plasmid, with the same amounts of both plasmid DNA and MCF-12A cells in each experiment, and the resulting data are shown as means and standard deviations. Finally, Western blots corresponding to each transient transfectant are shown immediately below the apoptosis bar graph. From top to bottom, panels show GFP-ESE-1 and GFP-ESE-1ΔTAD (anti-GFP Western blot), GFP-SAR and GFP-NLS-SAR (anti-GFP Western blot), GFP only (anti-GFP Western blot), and β-tubulin (anti-β-tubulin Western blot) for each cell population. Whole-cell extracts were prepared from each transfectant population 28 h after transfection, and 50 μg of total protein from each extract was subjected to SDS-PAGE and Western blotting.
RT-PCR.
Reverse transcription (RT)-PCR was performed by using an Omniscript RT kit (Qiagen, Inc., Valencia, Calif.) to reverse transcribe 5 μg of whole-cell RNA, prepared by using an RNA STAT-60 kit (Iso-Tex, Inc., Friendswood, Tex.), from each pool of untransfected cells and stable transfectants. GFP fusion transcripts were examined by using a sense primer specific for a carboxy-terminal portion of GFP and an antisense primer directed against a transcribed but untranslated sequence immediately downstream of the insertion site in plasmid pEGFP-C3 (see Fig. 4A). For RNase-negative controls, 5 μg of each RNA was incubated with 10 μg of DNase-free RNase (5 Prime→3 Prime, Inc., Boulder, Colo.) for 2 h at 37°C prior to RT.
Fluorescence microscopy.
MCF-12A, MCF-10A, T-47D, and SK-BR-3 cells were electroporated as described previously (18) with 30 μg of pEGFP-C3 control plasmid or with 30 μg of pEGFP-ESE-1 fusion construct plasmid. Respective transfectants were plated on sterile 15-mm glass coverslips with complete medium (Ham’s F12-Dulbecco’s modified Eagle medium [Invitrogen, Inc.] containing 100 ng of cholera toxin per ml, 0.5 μg of hydrocortisone per ml, 10 μg of insulin per ml, 20 ng of epidermal growth factor per ml, and 5% horse serum) and were incubated for 12 h at 37°C in 5% CO2 prior to imaging. Alternatively, subconfluent MCF-12A stable transfectants (see below) were cultured on 15-mm glass coverslips. All coverslips were washed three times in 37°C phosphate-buffered saline (PBS), and some were then incubated in PBS containing 300 nM 4′,6′-diamidino-2-phenylindole (DAPI) for 5 min at 37°C. Some transfectants were incubated in PBS containing 300 nM DAPI and 4 μM N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl)-pyridinium dibromide (FM 4-64) for 5 min at 37°C. Stained transfectants were imaged in a live-cell perfusion chamber by using an Olympus IX70 Infinity inverted microscope fitted with an Olympus NA 1.35 ×100 oil immersion objective. Fluorescence imaging was performed by using a 100-W mercury arc lamp with DAPI (excitation, 360 and 340 nm; emission, 457 and 450 nm), fluorescein isothiocyanate (FITC) (excitation, 490 and 420 nm; emission, 528 and 538 nm), or rhodamine-Texas red (excitation, 555 and 528 nm; emission, 617 and 673 nm) filters. Individual images were obtained by using a Quantix cooled charge-coupled device camera (Photometrics, Inc., Huntington Beach, Calif.) controlled by deconvolution software (DeltaVision, Inc., Montreal, Quebec, Canada) on an Indigo 2 R5000 computer (Silicon Graphics, Inc., Mountain View, Calif.).
Apoptosis assays.
MCF-12A, MCF-10A, T-47D, and SK-BR-3 cells were transfected with either the pEGFP-C3 or the pEGFP-ESE-1 plasmid as described above. Additionally, MCF-12A cells were transiently transfected with the GFP-ESE-1ΔTAD, GFP-SAR, or GFP-NLS-SAR plasmid as described above. Transfected cells were initially plated in complete medium, which was replaced with fresh complete medium 8 h later. Cells were stained 28 h posttransfection by using an annexin V-Alexa Fluor 647 conjugate apoptosis detection kit according to the manufacturer's protocol (Molecular Probes, Inc., Eugene, Oreg.). The numbers of green fluorescent (GFP-ESE-1 or GFP only) and red fluorescent (annexin V-Alexa Fluor 647) cells in each transfectant population were analyzed by using a FACSCalibur fluorescence-activated cell sorter (Becton Dickinson, Inc., San Jose, Calif.).
Western blotting.
MCF-12A, MCF-10A, T-47D, and SK-BR-3 cells were transfected as described above. Transfected cells were harvested at 12 to 24 h posttransfection in warmed PBS (37°C) containing 3 mM EDTA, and each cell pellet was lysed in a solution containing 1% sodium dodecyl sulfate (SDS), 10 mM EDTA, and 50 mM Tris. Total protein (50 to 70 μg) from each lysate was subjected to SDS-10% polyacrylamide gel electrophoresis (PAGE) and then was transferred to Immobilon-P membranes (Millipore, Inc., Bedford, Mass.). Membranes were probed by using a polyclonal rabbit anti-GFP IgG antibody (Santa Cruz, Inc., Santa Cruz, Calif.; 1:1,000 dilution) followed by a polyclonal goat anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody (Bio-Rad, Inc., Hercules, Calif.; 1:5,000 dilution). After anti-GFP antibody analysis, each blot was stripped by using a strong reblotting reagent (Chemicon, Inc., Temecula, Calif.) and then was reprobed with a monoclonal mouse anti-β-tubulin IgG antibody (Oncogene Research Products, Inc., Boston, Mass.; 1:1,000 dilution). To image β-tubulin, each blot was incubated with a polyclonal goat anti-mouse HRP-conjugated secondary antibody (Bio-Rad; 1:5,000 dilution).
Stable cell lines.
MCF-12A cells were transfected as described above. The medium in each transfectant culture was replaced 12 h later with complete medium supplemented with G418 (Invitrogen) to 500 μg/ml. Three weeks later, pools of G418-resistant cells were expanded from each transfectant culture in complete medium supplemented with 500 μg of G418/ml (Schedin et al., unpublished). Log-phase stable transfectants between passages 5 and 10 were used for all experiments.
Soft-agarose assays.
Stable transfectants, untransfected MCF-12A cells, or T-47D cells were resuspended in complete medium plus 0.35% agarose at 42°C. The resulting cell suspensions were plated on solidified agarose base layers and incubated at 37°C in 5% CO2 (Schedin et al., unpublished). Ten drops of complete medium (with or without G418) were added to each culture every 3 days to prevent dehydration.
RESULTS
Endogenous ESE-1 protein is restricted to the cytoplasm of human breast cancer cells.
Previous in vitro transient transfection studies demonstrated that plasmid-encoded ESE-1 protein transactivates several different human breast cancer gene promoters, including the HER-2 promoter, via an ets binding site (2, 3, 7, 9, 18, 19, 25, 34, 47). Given these data, we initially tested whether the binding of endogenous ESE-1 protein to the HER-2 genomic promoter might underlie the action of ESE-1 in breast cancer. However, although we were able to optimize an anti-ESE-1 chromatin immunoprecipitation assay, we were unable to detect binding between the endogenous ESE-1 protein and the endogenous HER-2 promoter in ESE-1-positive T-47D human breast cancer cells (data not shown). Possible explanations for this negative finding are that the ESE-1 protein is not responsible for HER-2 gene transcription in vivo and/or that endogenous ESE-1 does not reside in the nucleus.
To examine intracellular ESE-1 protein localization, we used an affinity-purified anti-ESE-1 polyclonal antibody to stain paraffin-embedded MCF-12A and T-47D human breast cell lines and human breast tissues (Fig. 2). Since the nontransformed MCF-12A human mammary epithelial cell line does not express ESE-1, we used MCF-12A cells as negative controls in these studies (note the lack of any DAB signal in Fig. 2A) (18). The anti-ESE-1 antibody robustly stained the cytoplasm but not the nuclei of T-47D cells, as demonstrated by the cytoplasmic restriction of the DAB signal (Fig. 2B). This observation was confirmed in three separate anti-ESE-1 IHC studies with T-47D cells and indicates that endogenous ESE-1 protein is cytoplasmically localized in T-47D cancer cells. Further, this finding explains our inability to detect in vivo ESE-1-HER-2 promoter binding by chromatin immunoprecipitation.
FIG. 2.
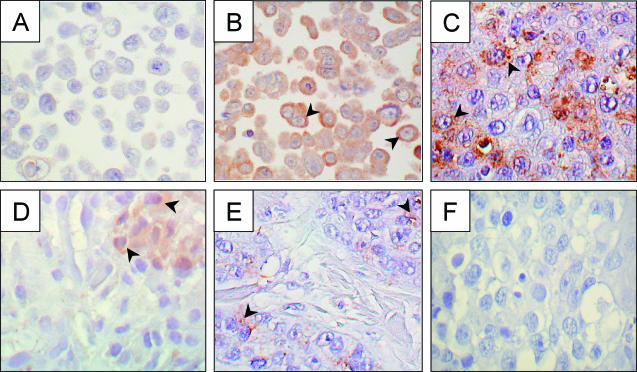
IHC staining of endogenous ESE-1 protein in MCF-12A and T-47D cell lines, in human breast cancer, and in normal breast tissue. Sections from paraffin-embedded MCF-12A (A), T-47D (B), human DCIS (C, D, and F), and normal breast tissue (E) blocks were incubated with an anti-ESE-1 primary antibody (A to E) or with rabbit immune serum (F) and then stained with an HRP-conjugated secondary antibody. Panels C (DCIS), E (normal breast), and F (DCIS) represent adjacent sections from the same paraffin-embedded block, whereas panel D shows anti-ESE-1 antibody staining of a separate DCIS sample. The brown stain represents positive anti-ESE-1 antibody staining, and arrowheads indicate representative stained cells in each sample (B to E). The absence of brown stain (A and F) represents the absence of anti-ESE-1 antibody binding (negative control). Stained sections were photographed at a magnification of ×40.
To test whether the cytoplasmic restriction of anti-ESE-1 staining is also a feature of primary human breast cancer, we performed anti-ESE-1 IHC analysis with a sample of high-grade invasive ductal carcinoma that contained an extensive component of high-grade DCIS (Fig. 2C). Anti-ESE-1 IHC analysis of this cancer tissue showed predominantly cytoplasmic staining, with signal intensity ranging from absent to robust among stained carcinoma cells. Although few epithelial cells in the adjacent normal breast tissue were stained with the anti-ESE-1 antibody, the staining pattern remained cytoplasmic (Fig. 2E). In addition, anti-ESE-1 IHC analysis of a second case of invasive human DCIS revealed a more focal staining pattern, but with a similar cytoplasmic anti-ESE-1 signal (Fig. 2D). In contrast, two cases of poorly differentiated, HER-2 receptor-negative invasive ductal carcinoma were negative for anti-ESE-1 antibody staining (data not shown). Interestingly, samples from both of the ESE-1-positive breast carcinoma cases shown in Fig. 2 also stained positively for the HER-2 receptor (data not shown). Finally, substitution of the anti-ESE-1 antibody with rabbit immune serum inhibited IHC staining in a DCIS sample adjacent to that shown in Fig. 2C and from the same paraffin block (Fig. 2F). This finding, along with the results for the MCF-12A cell negative control (Fig. 2A), demonstrates the specificity of anti-ESE-1 IHC staining. Taken together, the results of these IHC studies, although limited by low sample number, suggest that HER-2 expression and ESE-1 expression are positively correlated in human breast cancer and that endogenous ESE-1 protein is cytoplasmically restricted in both normal and transformed human breast epithelia.
Transiently expressed GFP-ESE-1 is localized in the nucleus and specifically induces apoptosis in nontransformed human breast epithelial cells.
Although our anti-ESE-1 IHC data indicate that endogenous ESE-1 protein is cytoplasmic in human breast cancer cells (Fig. 2), various investigators have shown that ESE-1 activates transcription in transiently transfected cells (7, 9, 18, 34, 54). To determine the subcellular localization of transiently expressed ESE-1 protein, we used GFP fusion and deconvolution UV microscopy to image real-time GFP-ESE-1 in living MCF-12A cells (Fig. 3A). Representative MCF-12A cells were imaged 12 h after transfection with either pEGFP-ESE-1 (Fig. 3A, panels 1 to 3) or pEGFP-C3 (Fig. 3A, panels 4 to 6). All cells were treated with DAPI and FM 4-64, which stain DNA blue and plasma membranes red, respectively (Fig. 3A, panels 1 and 4) (24, 49). The colocalization of green GFP (Fig. 3A, panel 2) and blue DAPI (Fig. 3A, panel 3) fluorescent signals observed here indicates that GFP-ESE-1 is specifically nuclear in transiently transfected MCF-12A cells. In Fig. 3A, panel 2, only two of four cells showed detectable GFP-ESE-1, suggesting that only these two cells were transfected. Typically, we obtained ∼20 to 30% transfection efficiency per ∼106 MCF-12A cells transfected, and multiple replicate studies revealed that 100% of GFP-positive transient transfectants (∼105 cells) expressed GFP-ESE-1 in the nucleus and that ∼5% also expressed some cytoplasmic GFP-ESE-1 (data not shown). In contrast to the results for GFP-ESE-1, 100% of MCF-12A cells transiently transfected with pEGFP-C3 showed diffuse nuclear and cytoplasmic GFP-only localization (Fig. 3A, panels 4 to 6).
FIG. 3.
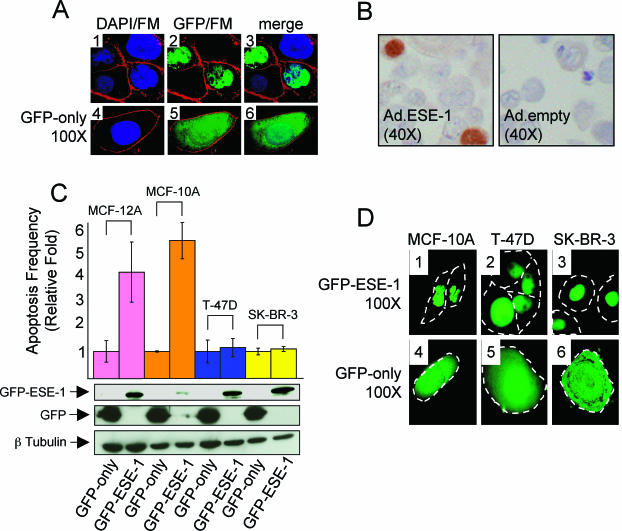
Transiently expressed ESE-1 protein is localized in the nucleus and induces apoptosis in MCF-12A and MCF-10A cells but not in T-47D and SK-BR-3 cells. (A) Fluorescence imaging of transiently expressed GFP-ESE-1 and GFP only in living MCF-12A cells. Representative MCF-12A cells transiently transfected with either the pEGFP-ESE-1 plasmid (panels 1 to 3; four cells shown) or the pEGFP-C3 control plasmid (panels 4 to 6; one cell shown) were imaged by digital deconvolution UV microscopy (magnification, ×100) 12 h after transfection. All transfectants were stained with DAPI or with FM 4-64 prior to microscopy. Panels 1 and 4 show overlays of DAPI (blue) and FM 4-64 (red) fluorescence images. Panels 2 and 5 show overlays of GFP (green) and FM 4-64 (red) fluorescence images. Panels 3 and 6 represent overlays of all three (DAPI, GFP, and FM 4-64) fluorescence signals. Although only four representative cells are shown in the GFP-ESE-1 panels and only one representative cell is shown in the GFP-only panels, a ∼30% transfection efficiency was consistently achieved per 106 cells transfected and ∼105 green fluorescent cells were present following each transfection. Approximately 95% of cells transiently transfected with GFP-ESE-1 showed a nucleus-only GFP-ESE-1 pattern, as shown here, while the other approximately 5% showed a primarily nuclear GFP signal with an associated faint cytoplasmic GFP signal (data not shown). All cells transfected with GFP only showed the pattern displayed in panels 4 to 6. (B) IHC staining of ESE-1 in Ad.ESE-1- and Ad.empty-infected MCF-12A cells. Sections from paraffin-embedded MCF-12A cells infected with either Ad.ESE-1 or Ad.empty were stained with an anti-ESE-1 antibody and then counterstained with an HRP-conjugated secondary antibody. The brown DAB precipitate represents positive anti-ESE-1 antibody staining in the cell nuclei (left panel), and the lack of brown stain (right panel) shows the absence of ESE-1 protein expression in the negative control. Stained sections were photographed at a magnification of ×40. (C) Transiently expressed nuclear GFP-ESE-1 induces apoptosis in MCF-12A and MCF-10A cells but not in T-47D or SK-BR-3 cells. MCF-12A, MCF-10A, T-47D, and SK-BR-3 cells were transfected with either pEGFP-C3 or pEGFP-ESE-1, cultured in complete medium for 28 h, and then stained with an apoptosis-specific annexin V-Alexa Fluor 647 conjugate. FACS analysis was used to quantitate the number of cells positive for both green (GFP-ESE-1 or GFP only) and red (annexin V; apoptosis specific) fluorescence in each transfectant population. The percentage of green and red cells in each GFP-only population was normalized to 1, and the fold increase in apoptotic transient transfectants of MCF-12A, MCF-10A, T-47D, and SK-BR-3 GFP-ESE-1 cells is shown. Three separate transfection experiments were performed for each plasmid, with the same amounts of both plasmid DNA and cells in each experiment, and the resulting data are shown as means and standard deviations. Finally, Western blots corresponding to each transiently transfected cell line are shown immediately below the apoptosis bar graph. From top to bottom, panels show GFP-ESE-1 (anti-GFP Western blot), GFP only (anti-GFP Western blot), and β-tubulin (anti-β-tubulin Western blot) for each cell population. Whole-cell extracts were prepared from each transfectant population 12 h after transfection, and 70 μg of total protein from each extract was used for SDS-PAGE and Western blotting. (D) Fluorescence imaging of transiently expressed GFP-ESE-1 and GFP only in living MCF-10A, T-47D, and SK-BR-3 cells. Representative MCF-10A, T-47D, and SK-BR-3 cells transiently transfected with either the pEGFP-ESE-1 plasmid (panels 1 to 3) or the pEGFP-C3 control plasmid (panels 4 to 6) were imaged for GFP fluorescence by digital deconvolution UV microscopy (magnification, ×100) 12 h after transfection. Broken white lines depict cell outlines. GFP-ESE-1 localized in the nucleus was observed in MCF-10A cells (panel 1, two cells shown), T-47D cells (panel 2, four cells shown), and SK-BR-3 cells (panel 3, three cells shown), whereas GFP-only fluorescence was equally distributed between the nucleus and the cytoplasm in each GFP-only-transfected cell line (panels 4 to 6, one cell shown per transient transfection). Details for transfection efficiencies, numbers of GFP-ESE-1- and GFP-only-positive cells, and subcellular distributions of GFP-ESE-1 and GFP only in each transient transfectant population were as described for panel A.
As a different method of examining ESE-1 protein localization, we infected MCF-12A cells with Ad.ESE-1 or Ad.empty and performed anti-ESE-1 IHC analysis 12 h later (Fig. 3B). The brown nuclear stain shown in two cells in Fig. 3B, left panel, reveals that untagged ESE-1 protein is localized in the nucleus in these cells. As was found for the GFP signal in MCF-12A cells transiently transfected with pEGFP-ESE-1, 100% of Ad.ESE-1-infected, DAB-positive cells showed nuclear DAB localization, and ∼5% also showed some cytoplasmic staining (data not shown). In contrast, Ad.empty-infected MCF-12A cells failed to show staining with the anti-ESE-1 antibody (Fig. 3B, right panel). Because both Ad.ESE-1 and Ad.empty contained an independent GFP-only expression cassette, we used GFP fluorescence to ensure equal adenovirus infection rates prior to each IHC experiment. Taken together, the results of our separate imaging approaches (fluorescence imaging and IHC analysis) show that transiently expressed ESE-1 protein is localized in the nucleus and suggest that this nuclear localization facilitates ESE-1 transcription factor function in transiently transfected cells.
Nevertheless, the nuclear localization of transiently expressed, vector-encoded ESE-1 protein (Fig. 3A and B) and the cytoplasmic localization of endogenous ESE-1 protein (Fig. 2) must be reconciled. Given these two distinct ESE-1 protein localization patterns, we hypothesized that the subcellular localization of transient (nuclear) versus endogenous (cytoplasmic) ESE-1 protein influences mammary epithelial cell growth and/or survival. To test this hypothesis, we performed a time course experiment during which the localization of GFP-ESE-1 was monitored hourly beginning 12 h after transfection. No changes in the absolute number of GFP-ESE-1-positive cells or in the pattern of nuclear GFP-ESE-1 localization (Fig. 3A) were noted over the next 12 h (24 h after transfection). However, at 25 h after transfection, we noted a significant loss of GFP-positive cells, and by 35 h posttransfection, GFP-positive cells were no longer detected. Cell disappearance was limited to transfectants having a nuclear GFP signal, and only GFP-negative cells remained in the population 35 h after transfection. These observations implied that nuclear GFP-ESE-1 induces MCF-12A cell death. To test this hypothesis, we stained MCF-12A cells with a red fluorescent, apoptosis-specific annexin V conjugate 28 h after pEGFP-ESE-1 or pEGFP-C3 transfection and then used fluorescence-activated cell sorting (FACS) to quantitate cells having both GFP fluorescence (green; transfected cells) and annexin V fluorescence (red; apoptotic cells) in each population (48). These experiments showed a 4.4-fold increase in apoptotic GFP-ESE-1-transfected cells over GFP-only-transfected cells (Fig. 3C) and thus suggest that GFP-ESE-1 localized in the nucleus induces apoptosis within 28 h of transfection.
In order to confirm the apoptotic function of nuclear GFP-ESE-1, we performed the same fluorescence imaging and annexin V apoptosis studies with the nontransformed MCF-10A human breast epithelial cell line, which is genetically distinct from the MCF-12A cell line, as well as the fully transformed T-47D and SK-BR-3 human breast cancer cell lines (Fig. 3C and D) (42, 45). With MCF-10A cells, these studies showed a 5.5-fold increase in apoptotic GFP-ESE-1 transfectants over GFP-only controls (Fig. 3C). In contrast, comparisons between pEGFP-ESE-1 and pEGFP-C3 transfections with either T-47D or SK-BR-3 cells (Fig. 3C) revealed that the T-47D and SK-BR-3 breast cancer cell lines are resistant to nuclear GFP-ESE-1-mediated apoptosis. This latter finding suggests that the effect of nuclear GFP-ESE-1 on MCF-12A and MCF-10A cell viability is not simply a result of GFP-ESE-1 overexpression in transiently transfected cells. To confirm this interpretation, we used anti-GFP Western blotting to examine GFP-ESE-1 or GFP-only expression in each transiently transfected cell line (Fig. 3C). The top panel in Fig. 3C shows similar levels of GFP-ESE-1 expression among the MCF-12A, T-47D, and SK-BR-3 cell lines, whereas MCF-10A cells transfected with pEGFP-ESE-1, which showed the most robust GFP-ESE-1-mediated apoptotic response, expressed the least GFP-ESE-1. GFP-only expression levels, in contrast, were similar among all four pEGFP-C3-transfected cell lines (Fig. 3C, middle panel) and, for each cell line, the level of GFP-only expression (GFP-only transfectants) was higher than that of GFP-ESE-1 (GFP-ESE-1 transfectants). The bottom panel in Fig. 3C shows approximately equivalent β-tubulin protein levels in all eight transient tranfectants and served as a loading control. Together, these Western blot data confirm that the differential abilities of nuclear GFP-ESE-1 to induce apoptosis in MCF-12A and MCF-10A cells versus T-47D and SK-BR-3 cells are not due to differences in GFP-ESE-1 expression. Rather, these data indicate that cellular transformation status modulates ESE-1 apoptotic function among these transiently transfected cell lines.
Finally, in order to examine the subcellular localization of GFP-ESE-1 and GFP only in each cell line, we used deconvolution UV microscopy to image real-time GFP fluorescence 12 h after pEGFP-ESE-1 or pEGFP-C3 plasmid transfection (Fig. 3D). Figure 3D shows GFP-ESE-1 localization in transiently transfected MCF-10A (panel 1), T-47D (panel 2), and SK-BR-3 (panel 3) cells. Each transfected cell is outlined and, as in MCF-12A cells (Fig. 3A, panels 2 and 3), the GFP signal is restricted to the nucleus in each cell. In contrast, GFP-only localization is diffusely distributed throughout the cytoplasm and nucleus of each cell transiently transfected with pEGFP-C3 (Fig. 3D, panels 4 to 6). Together, these imaging data confirm nuclear GFP-ESE-1 localization in all four cell lines following transient pEGFP-ESE-1 transfection and therefore indicate that the differences in GFP-ESE-1-mediated apoptosis observed between transiently transfected MCF-12A and MCF-10A versus T-47D and SK-BR-3 cells are not the result of differential GFP-ESE-1 localization.
Stable expression of GFP-ESE-1 mediates anchorage-independent MCF-12A cell growth in soft agarose.
It was recently shown that the stable expression of hemagglutinin (HA) epitope-tagged ESE-1 (HA-ESE-1) results in MCF-12A cell transformation, indicating that stable HA-ESE-1 expression, in contrast to transient GFP-ESE-1 expression, is compatible with cell survival (40a). To determine whether GFP-ESE-1-expressing stable transfectants displayed a similar phenotype, we used G418 selection to generate two independent GFP-ESE-1-expressing MCF-12A stable transfectant pools and two independent GFP-only-expressing MCF-12A stable transfectant pools. All G418-resistant cells in the two GFP-ESE-1 transfectant pools were negative for GFP fluorescence, yet RT-PCR studies designed to distinguish GFP-ESE-1 (1,285 bp) from GFP only (169 bp) (Fig. 4A) revealed the exclusive expression of either the GFP-ESE-1 fusion transcript (Fig. 4B, lanes 2 and 3) or GFP-only mRNA (Fig. 4B, lanes 4 and 5) in respective stable transfectants. Further, DNA sequencing verified in-frame GFP fusion to the mutation-free ESE-1 coding sequence in each GFP-ESE-1 stable transfectant population. RT-PCR-negative controls, including no-template (Fig. 4B, lane 1) and RNase (Fig. 4B, lanes 6 to 9) controls, confirmed the absence of DNA contaminants in each reaction. Finally, both GFP-only-expressing MCF-12A stable transfectant pools showed the same diffuse nuclear and cytoplasmic GFP signals as those seen in transiently transfected cells (Fig. 3A, panels 5 and 6, and data not shown), indicating that the GFP-only distribution was not affected by G418 selection. In contrast, we have consistently been unable to detect GFP-ESE-1 expression in any of our independently generated MCF-12A stable transfectant pools. The inability of deconvolution UV microscopy to identify stably expressed GFP-ESE-1, while RT-PCR clearly revealed stable GFP-ESE-1 transcript expression, indicates that stable GFP-ESE-1 expression in MCF-12A cells is below the level of detection afforded by UV microscopy. This situation may reflect a low level of expression of the GFP-ESE-1 gene in stably transfected MCF-12A cells or may indicate that GFP-ESE-1 is unstable and thus undetectable in the context of stable transfection.
In order to examine the biological relevance of stable GFP-ESE-1 expression in MCF-12A cells, we next tested whether stable GFP-ESE-1 expression could induce a transformation phenotype similar to that mediated by HA-ESE-1 (40a). We assayed for colony formation in soft agarose because (i) such colony formation reflects anchorage-independent cell proliferation and thus represents a cardinal feature of transformation (12, 39) and (ii) the stable expression of HA-ESE-1 induces colony formation in MCF-12A cells (40a). Typical colonies were photographed 8 days after soft-agarose seeding (Fig. 4C; see Fig. 5C for quantitation of colony formation). Control, untransfected MCF-12A cells (Fig. 4C, 12A panel) and GFP-only stable transfectants (Fig. 4C, GFP-only panel) generated small, two- or three-cell clusters, revealing their inability to form colonies within 8 days of seeding. In contrast, both positive control T-47D human breast cancer cells (Fig. 4C, T-47D panel) and GFP-ESE-1-expressing MCF-12A stable transfectants (Fig. 4C, GFP-ESE-1 panel) formed many large, multicellular colonies within 8 days of seeding. These findings suggest that although fluorescence microscopy failed to detect stable GFP-ESE-1 expression, stably transfected GFP-ESE-1 recapitulates the transformation phenotype observed in HA-ESE-1-expressing stable transfectants (40a). Moreover, these data indicate that N-terminal GFP fusion does not interfere with ESE-1 function.
FIG.5.
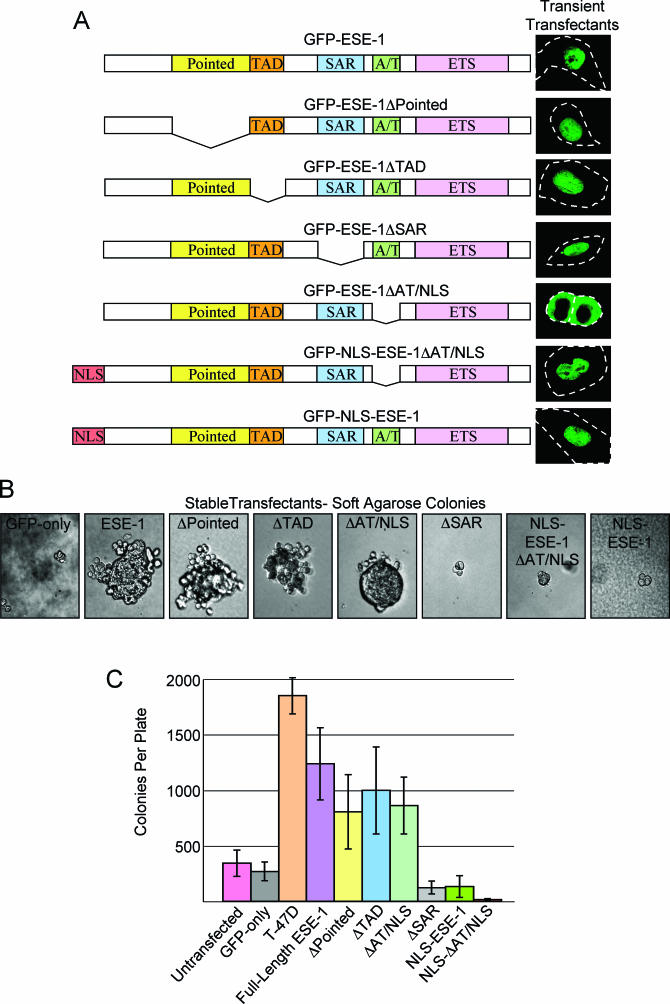
Deletion mapping of the GFP-ESE-1 transforming domain. (A) Structural organization and transiently expressed protein localization for ESE-1 internal deletion mutants and SV40 NLS fusion constructs. For each of the seven constructs shown, the ESE-1 structural domains are depicted in the same color scheme at that used in Fig. 1, and each is fused to GFP at its amino terminus (data not shown). Intact GFP-ESE-1 is depicted at the top, followed by constructs with internal deletions of the Pointed domain (GFP-ESE-1ΔPointed), TAD (GFP-ESE-1ΔTAD), SAR domain (GFP-ESE-1ΔSAR), and AT-hook/NLS domain (GFP-ESE-1ΔAT/NLS). The bottom two constructs contain an in-frame insertion of the SV40 NLS between GFP and the ESE-1ΔAT/NLS deletion (GFP-NLS-ESE-1ΔAT/NLS) and between GFP and the intact ESE-1 sequence (GFP-NLS-ESE-1). A representative image of the corresponding GFP fusion localization in transiently transfected MCF-12A cells 12 h after transfection is shown to the right of each construct. Broken white lines depict cell outlines; except for the GFP-ESE-1ΔAT/NLS construct (two cells shown), one representative cell is shown for each construct. (B) Anchorage-independent growth of MCF-12A cell pools stably transfected with GFP-ESE-1 internal deletion or SV40 NLS fusion constructs. Soft-agarose cultures were prepared and photographed as described in the legend to Fig. 4C (magnification, ×40). A representative colony for each GFP-only and GFP-ESE-1 construct depicted in panel A is shown. Small, two- to four-cell clusters were generated in MCF-12A cell pools stably transfected with GFP-only, GFP-ESE-1ΔSAR, GFP-NLS-ESE-1ΔAT/NLS, and GFP-NLS-ESE-1. Large, multicellular colonies were generated in MCF-12A cell pools stably transfected with intact GFP-ESE-1, GFP-ESE-1ΔPointed, GFP-ESE-1ΔTAD, and GFP-ESE-1ΔAT/NLS. (C) Quantitation of anchorage-independent colony formation. Untransfected MCF-12A and T-47D cells and two independent, stably transfected MCF-12A cell populations for each GFP construct were seeded in triplicate soft-agarose cultures, and all colonies visible by ×6 light microscopy were counted 21 days later. Each bar shows the mean and standard deviation number of total colonies per plate for each cell line or transfectant population.
The 50-aa SAR domain, but not transcription-specific or nuclear targeting domains, is required for ESE-1-mediated transformation.
Having established the soft-agarose colony formation assay as a sensitive marker of GFP-ESE-1-mediated transformation, we next used this assay to map the ESE-1 domain(s) required for transformation. The Pointed, TAD, SAR, AT hook/NLS, and ETS domains (Fig. 1) have been identified by either functional assays or by their similarity to domains in other proteins (1-3, 6, 7, 34). We generated a series of GFP-ESE-1 internal deletion constructs in which regions encompassing the Pointed (ΔPointed), TAD (ΔTAD), SAR (ΔSAR), and AT hook/NLS (ΔAT/NLS) domains were deleted in frame from GFP-ESE-1 (Fig. 5A). Further, to test the role of nuclear localization in ESE-1-mediated transformation, we also fused the strong SV40 large-T-antigen NLS sequence in frame between GFP and ESE-1 in the GFP-ESE-1 (GFP-NLS-ESE-1) and GFP-ESE-1ΔAT/NLS (GFP-NLS-ESE-1ΔAT/NLS) plasmid constructs (53). Each plasmid was used to transiently transfect MCF-12A cells, and transfected cells were imaged by fluorescence microscopy 12 h later (Fig. 5A). Figure 5A shows representative GFP fluorescence localization images for full-length GFP-ESE-1, GFP-ESE-1ΔPointed, GFP-ESE-1ΔTAD, GFP-ESE-1ΔSAR, GFP-ESE-1ΔAT/NLS, GFP-NLS-ESE-1ΔAT/NLS, and GFP-NLS-ESE-1 in transiently transfected MCF-12A cells. Each cell is outlined and, in all but one example, GFP fluorescence is specifically nuclear in transiently transfected MCF-12A cells. In contrast, transiently expressed GFP-ESE-1ΔAT/NLS is almost entirely cytoplasmic in MCF-12A cells. We designed the ΔAT/NLS internal deletion to precisely remove exon 7, since this region contains a putative bipartite NLS that overlaps and extends beyond the AT hook domain (6, 7). Thus, the data shown here confirm that the endogenous ESE-1 NLS is located in this exon 7 region from aa 231 to 268. In addition, comparison of the GFP localization pattern for the transiently expressed GFP-ESE-1ΔAT/NLS construct (cytoplasmic) with that of the transiently expressed GFP-NLS-ESE-1ΔAT/NLS construct (nuclear) reveals that the SV40 NLS fusion restores the nuclear localization of the GFP-ESE-1ΔAT/NLS construct in transiently transfected cells (Fig. 5A). As described previously for full-length GFP-ESE-1, we typically obtained ∼20 to 30% transfection efficiency per ∼106 MCF-12A cells transfected with any of the seven constructs shown in Fig. 5A. Further, multiple replicate studies revealed that 100% of GFP-positive transient transfectants (∼105 cells) expressed GFP-ESE-1, GFP-ESE-1ΔPointed, GFP-ESE-1ΔTAD, GFP-ESE-1ΔSAR, GFP-NLS-ESE-1ΔAT/NLS, and GFP-NLS-ESE-1 in the nucleus and that ∼5% of GFP-positive transient transfectants also expressed some cytoplasmic GFP fusion construct, with the GFP-NLS-ESE-1 construct showing the least cytoplasmic GFP localization (data not shown). Finally, ∼97% of GFP-positive GFP-ESE-1ΔAT/NLS transient transfectants showed exclusively a cytoplasmic GFP signal, and the remaining 3% also showed some nuclear protein localization (data not shown).
Having defined the subcellular localization of each GFP-ESE-1 fusion or deletion construct in transiently transfected MCF-12A cells, we next tested the transforming function of each construct in stably transfected cells. Individual stable pools of MCF-12A cells transfected with the respective GFP-ESE-1 fusion or deletion constructs were established and then assayed for their ability to induce colony formation in soft agarose (Fig. 5B). As in the full-length GFP-ESE-1 stable transfectants described above, GFP fluorescence was not detectable by fluorescence microscopy in any of these stable transfectants. However, as in the full-length GFP-ESE-1 stable transfectants, stable, mutation-free expression of each construct in the respective stable pools was confirmed by RT-PCR and by RT-PCR product sequencing (data not shown). The data in Fig. 5B show that intact GFP-ESE-1 mediates the formation of large, multicellular colonies, whereas GFP only fails to do so, recapitulating the results shown in Fig. 4C. Stable pools of MCF-12A cells expressing GFP-ESE-1ΔPointed, GFP-ESE-1ΔTAD, or GFP-ESE-1ΔAT/NLS all retained the ability to form large, multicellular colonies, similar in size and number to those generated by full-length GFP-ESE-1 (Fig. 5B). Although the colonies shown for GFP-ESE-1ΔPointed and GFP-ESE-1ΔTAD appeared to have a distinct morphology, examination of multiple colonies in each GFP-ESE-1, GFP-ESE-1ΔPointed, GFP-ESE-1ΔTAD, and GFP-ESE-1ΔAT/NLS stable pool revealed a similar colony morphology range. In contrast, deletion of the SAR domain (GFP-ESE-1ΔSAR) resulted in diminished colony formation, and these colonies were indistinguishable in size and number from those observed in GFP-only soft-agarose cultures (Fig. 4C and 5B). These data show that transcription-related and putative NLS domains of ESE-1 (i.e., the Pointed domain, TAD, and NLS) are not required for ESE-1-mediated anchorage-independent growth, whereas the 50-aa acidic SAR domain is critical for ESE-1 transforming activity. A corollary of these conclusions is that the SAR domain, which lacks TAD function (6) and any putative NLS, mediates ESE-1-induced transformation via a cytoplasmic mechanism.
To directly test these interpretations, we next examined the transforming functions of our SV40 NLS fusion constructs, GFP-NLS-ESE-1 and GFP-NLS-ESE-1ΔAT/NLS (Fig. 5A). Although the parental forms of these constructs demonstrated equivalent transforming activities (Fig. 5B, ESE-1 and ΔAT/NLS colonies), the addition of the SV40 NLS, which specifically enforces nuclear localization, completely blocked their transforming capacities (Fig. 5B, NLS-ESE-1 and NLS-ESE-1ΔAT/NLS colonies). These data not only further support our hypothesis that the transcription-related and putative NLS domains of ESE-1 are not required for anchorage-independent growth but also indicate that ESE-1 nuclear localization results in the loss of its transforming ability. Thus, the localization of transiently expressed GFP-ESE-1ΔPointed and ΔTAD (nuclear; Fig. 5A) must differ from that of these same two proteins in stably transfected MCF-12A cells (proteins undetectable by fluorescence microscopy).
In addition to imaging individual colonies, we also quantitated differences in colony formation among soft-agarose cultures (Fig. 5C). For these studies, two independent stable transfectant pools were analyzed for each GFP-ESE-1 construct, and each pool was cultured in triplicate. Triplicate soft-agarose cultures were also prepared for T-47D and untransfected MCF-12A cells. Colonies that were visible under ×6 magnification after 21 days in cultures were counted. Figure 5C shows that negative control untransfected MCF-12A cells and MCF-12A cells expressing GFP only formed ∼359 and ∼277 colonies, respectively. Although no colonies were observed in these negative control cultures at 8 days postseeding (Fig. 4C, 12A and GFP-only panels), the presence of colonies at 21 days indicates that untransfected MCF-12A cells do have some minimal anchorage-independent growth capacity and that this capacity is manifested between 8 and 21 days postseeding (11). In contrast, positive control T-47D cells formed ∼1,838 colonies (Fig. 5C), demonstrating a ∼6-fold increase relative to the results for negative controls. Similarly, GFP-ESE-1 stable transfectants formed ∼1,287 colonies per culture, each GFP-ESE-1ΔPointed stable culture showed ∼810 colonies, GFP-ESE-1ΔTAD stable transfectants formed ∼1,031 colonies per culture, and each GFP-ESE-1ΔAT/NLS stable culture showed ∼895 colonies. While the colony numbers associated with these GFP-ESE-1 constructs were statistically equivalent, they were, on average, ∼3.5-fold higher than the average for the negative control. These results corroborate the colony imaging data presented in Fig. 5B, showing that neither the ESE-1 Pointed, TAD, nor AT hook/NLS domain is required for ESE-1-induced anchorage-independent growth. In contrast, deletion of the SAR domain (GFP-ESE-1ΔSAR) resulted in ∼116 colonies per culture, confirming that the ESE-1 SAR domain is required for ESE-1-mediated colony formation. Similarly, GFP-NLS-ESE-1 stable transfectant pools formed ∼136 colonies per culture, and each GFP-NLS-ESE-1ΔAT/NLS stable culture showed only ∼15 colonies, indicating that the transforming functions of both GFP-ESE-1 and GFP-ESE-1ΔAT/NLS are blocked by the SV40 NLS fusion. Indeed, the significantly lower colony numbers observed for GFP-ESE-1ΔSAR, GFP-NLS-ESE-1, and GFP-NLS-ESE-1ΔAT/NLS relative to those observed for untransfected or GFP-only controls suggest that these GFP-ESE-1 variants may function as dominant-negative effectors with regard to MCF-12A cell growth and/or survival.
The ESE-1 SAR domain is insufficient to mediate ESE-1 apoptotic activity in transiently transfected MCF-12A cells, but in-frame deletion of the ESE-1 TAD impairs ESE-1 apoptotic activity.
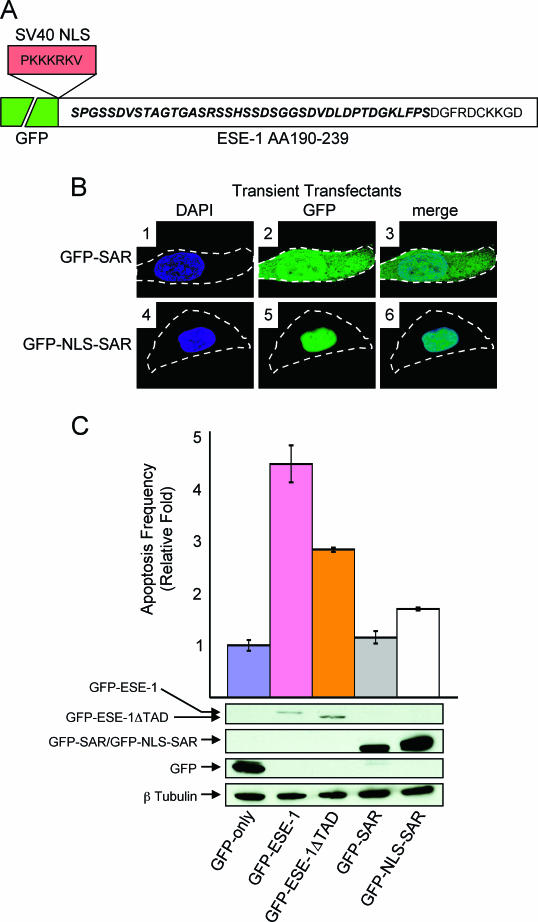
As described previously, full-length GFP-ESE-1 is localized in the nucleus in transiently transfected MCF-12A cells and, in this context, induces apoptosis (Fig. 3C). Having determined that the transcription factor function of ESE-1 is not required for ESE-1-mediated MCF-12A cell transformation (Fig. 5B and C), we next investigated the role of ESE-1 transcription factor function in ESE-1-mediated apoptosis. Further, because our soft-agarose culture data revealed an essential role for the ESE-1 SAR domain in GFP-ESE-1-induced cell transformation (Fig. 5B and C), we generated two additional constructs to specifically test the apoptotic function of the ESE-1 SAR domain. To do this, we fused the isolated ESE-1 SAR domain, spanning ESE-1 aa 190 to 239 and including all but the first 2 aa of the SAR domain, in frame and downstream of GFP (GFP-SAR) (Fig. 6A). In addition, we also fused the SV40 NLS sequence in frame between the GFP and SAR portions of this construct (GFP-NLS-SAR) (Fig. 6A). Figure 6B shows the subcellular localization of GFP-SAR (panels 1 to 3) and GFP-NLS-SAR (panels 4 to 6) in transiently transfected MCF-12A cells. All transiently transfected cells were stained with DAPI prior to fluorescence imaging, as described previously, and each cell is outlined. Although a single representative cell is shown for each construct (Fig. 6B), 100% of GFP-SAR transient transfectants showed diffuse nuclear and cytoplasmic GFP fluorescence (panels 2 and 3), and 100% of GFP-NLS-SAR transient transfectants showed a nuclear GFP signal (panels 5 and 6), with ∼5% also showing some cytoplasmic GFP fluorescence (data not shown). These data indicate that GFP-SAR, having a predicted molecular mass of ∼32 kDa, can diffuse throughout the cell, whereas the SV40 NLS fusion results in the nuclear retention of GFP-NLS-SAR.
Having defined the subcellular localization of GFP-SAR and GFP-NLS-SAR in transiently transfected MCF-12A cells, we next examined the apoptotic functions of these two fusions. Further, to test the role of ESE-1 transcription factor function in apoptosis, we also examined the apoptotic properties of the GFP-ESE-1ΔTAD construct. MCF-12A cells were transiently transfected with the GFP-only (negative control), GFP-ESE-1 (positive control), GFP-ESE-1ΔTAD, GFP-SAR, or GFP-NLS-SAR expression plasmids. At 28 h after transfection, each transfectant population was stained with the apoptosis-specific annexin V conjugate. As shown previously (Fig. 3C), transient expression of GFP-ESE-1 induced a 4.5-fold increase in apoptotic MCF-12A cells compared to the results obtained with the GFP-only control, whereas deletion of the TAD, in GFP-ESE-1ΔTAD, resulted in impaired (2.8-fold) ESE-1 apoptotic function (Fig. 6C). These data reveal that deletion of the ESE-1 TAD impairs ESE-1 apoptotic function and also indicate that ESE-1 transcription factor function contributes to ESE-1-mediated apoptosis. Indeed, we found that our remaining GFP-ESE-1 deletion constructs (GFP-ESE-1ΔPointed, GFP-ESE-1ΔSAR, and GFP-ESE-1ΔAT/NLS) also showed impaired apoptotic function in transiently transfected MCF-12A cells (data not shown). Finally, transient expression of GFP-SAR did not increase MCF-12A cell apoptosis compared to the results obtained with the GFP-only control (Fig. 6C), indicating that the SAR domain by itself is insufficient to recapitulate ESE-1 apoptotic function. Interestingly, enforced nuclear localization of the SAR domain, as GFP-NLS-SAR, did result in a small increase (1.7-fold) in apoptosis in transiently transfected MCF-12A cells. Given that the SAR domain appears to play a functional role in ESE-1-mediated apoptosis, as suggested by the impaired apoptotic function of the GFP-ESE-1ΔSAR construct (data not shown), these data indicate that the optimal contribution of the SAR domain to GFP-ESE-1-mediated apoptosis requires nuclear SAR domain localization.
As described above, we also performed anti-GFP Western blotting with whole-cell extracts prepared from each transient transfectant population (Fig. 6C). Figure 6C shows the relative expression of full-length GFP-ESE-1 and GFP-ESE-1ΔTAD compared to that of GFP-SAR and GFP-NLS-SAR and to that of GFP only in transiently transfected MCF-12A cells. The data reveal that full-length GFP-ESE-1 (∼73 kDa) and GFP-ESE-1ΔTAD (∼70 kDa), which demonstrated the greatest apoptotic function, were expressed at the lowest levels, compared to GFP-SAR, GFP-NLS-SAR, and GFP only. These findings indicate that the high levels of plasmid-encoded protein expression associated with transient transfection were not, in themselves, responsible for the differential apoptotic functions of the constructs tested. Finally, this interpretation is supported by the data shown in Fig. 6C for anti-β tubulin, which served as a loading control; similar β-tubulin protein levels were found in all of the extracts analyzed.
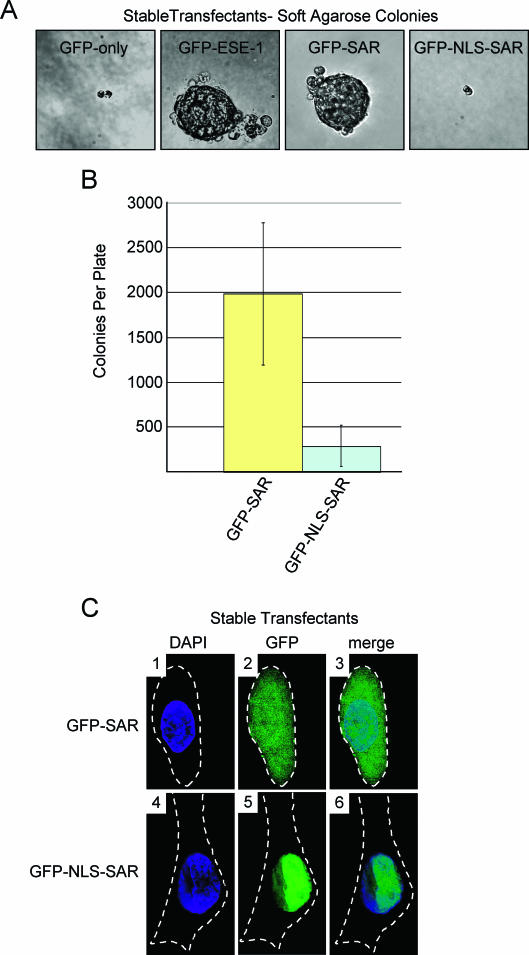
Stable cytoplasmic expression of a 50-aa region spanning the ESE-1 SAR domain is sufficient to induce anchorage-independent proliferation of MCF-12A cells.
Having shown that the ESE-1 SAR domain is required for ESE-1-induced anchorage-independent growth (Fig. 5B and C) but is insufficient to mediate ESE-1 apoptotic function (Fig. 6C), we next tested whether this domain is sufficient to transform MCF-12A cells. Moreover, we also tested whether the subcellular localization of the SAR domain may affect its transforming function. Three independent pools of stably transfected MCF-12A cells were generated for both the GFP-SAR and the GFP-NLS-SAR constructs (Fig. 6A) as detailed in Materials and Methods. Appropriate expression of GFP-SAR and GFP-NLS-SAR mRNAs in each independent GFP-SAR- or GFP-NLS-SAR-expressing MCF-12A stable transfectant pool was confirmed by RT-PCR (data not shown), and DNA sequencing of these RT-PCR products revealed the anticipated, mutation-free fusion sequence in each construct. To examine ESE-1 SAR domain function and the role of the SV40 NLS fusion in anchorage-independent proliferation, we imaged GFP-SAR and GFP-NLS-SAR stable transfectants for soft-agarose colony formation 8 days postseeding. The GFP-only negative control again failed to show any significant colony formation, whereas the GFP-ESE-1-positive control cells formed large, multicellular colonies (Fig. 7A, GFP-only and GFP-ESE-1 panels). Notably, the GFP-SAR stable transfectants also formed large, multicellular colonies 8 days postseeding (Fig. 7A, GFP-SAR panel). However, fusion of the SV40 NLS to the GFP-SAR construct inhibited GFP-SAR-mediated colony formation (Fig. 7A, GFP-NLS-SAR panel). Quantitation of colony formation from three independent stable pools, cultured in triplicate, 21 days postseeding revealed that the GFP-SAR-expressing cells formed ∼1,985 colonies per culture, whereas the GFP-NLS-SAR-expressing cells each yielded ∼288 colonies (Fig. 7B). While the GFP-SAR stable transfectants displayed somewhat variable colony numbers, the average colony number in these cultures was statistically equivalent to that observed for full-length GFP-ESE-1 stable transfectants in soft-agarose cultures (Fig. 7B and Fig. 5C) and was ∼6.9-fold higher than the average colony number in GFP-NLS-SAR cultures. These data demonstrate that the 50-aa region spanning the ESE-1 SAR domain is sufficient to mediate MCF-12A cell transformation and that enforced nuclear localization, induced by the SV40 NLS fusion, blocks SAR domain function in MCF-12A mammary epithelial cell transformation.
FIG. 7.
Quantitation of colony formation and analysis of subcellular localization of of GFP-SAR and GFP-NLS-SAR in stably transfected MCF-12A cell pools. (A) Anchorage-independent growth of MCF-12A cell pools stably transfected with GFP-SAR or GFP-NLS-SAR. Soft-agarose cultures were prepared for each cell line, and representative colonies were photographed as described in the legends to Fig. 4C and 5B (magnification, ×40). GFP-only and GFP-NLS-SAR stable transfectant pools each showed two- to four-cell clusters, and GFP-ESE-1 and GFP-SAR stable transfectant pools each showed large, multicellular colonies. (B) Quantitation of anchorage-independent colony formation in soft-agarose cultures seeded with MCF-12A cell pools stably transfected with GFP-SAR or GFP-NLS-SAR. Three independent MCF-12A populations stably transfected with the GFP-SAR or GFP-NLS-SAR expression plasmid were seeded in triplicate soft-agarose cultures, and colonies were counted as described in the legend to Fig. 5C. Each bar shows the mean and standard deviation number of total colonies per plate for each transfectant population. (C) Subcellular localization of stably expressed GFP-SAR and GFP-NLS-SAR. Pools of MCF-12A cells stably transfected with the GFP-SAR or GFP-NLS-SAR expression plasmid were stained with DAPI, and typical cells in each culture were imaged by digital deconvolution UV microscopy (magnification, ×100). The DAPI, GFP and merge images are as described in the legend to Fig. 3A, and broken white lines represent cell outlines. Panels 1 to 3 show the diffuse cytoplasmic and nuclear localization of stably expressed GFP-SAR. Panels 4 to 6 show the exclusive nuclear localization that results from fusing the SV40 NLS to GFP-SAR.
In an attempt to determine the subcellular localization of GFP-SAR and GFP-NLS-SAR in stable transfectants, we stained each stable pool with DAPI and imaged representative cells by deconvolution UV microscopy. Although GFP-ESE-1-expressing MCF-12A stable transfectants produced the appropriate GFP-ESE-1 mRNA (Fig. 5B), they failed to show a detectable GFP signal, as described above. In contrast, stable expression of both GFP-SAR and GFP-NLS-SAR was detectable by UV microscopy (Fig. 7C), indicating that the full-length ESE-1 protein contains a destabilizing element(s) that is not present in the 50-aa SAR domain and/or that MCF-12A cells tolerate a higher level of expression of the isolated SAR domain. Furthermore, as in transiently transfected MCF-12A cells (Fig. 6B), stably expressed GFP-SAR is diffusely localized to both the cytoplasm and the nucleus, as shown by the nuclear and cytoplasmic GFP signals in individual stable transfectants (Fig. 7C, panels 1 to 3). In contrast, GFP-NLS-SAR is exclusively nuclear in stably transfected MCF-12A cells (Fig. 7C, panels 4 to 6). Having shown that GFP-SAR localizes to both the cytoplasm and the nucleus and promotes anchorage-independent growth, whereas GFP-NLS-SAR is restricted to the nucleus and does not promote anchorage-independent growth, we can conclude that it is the cytoplasmic localization of the 50-aa SAR domain that is responsible for MCF-12A cell transformation.
DISCUSSION
The transforming function of ETS family transcription factors, which include the products of nuclear oncogenes (e.g., v-ETS, Tel-AML1, and EWS-Fli1), generally depends on their ability to dysregulate transcriptional programs involved in cell growth (36, 41). In this study, we identified in ETS transcription factor ESE-1 a unique 50-aa SAR domain (aa 190 to 239) that is necessary and sufficient to transform MCF-12A mammary epithelial cells. Moreover, we reveal that this SAR-mediated transformation occurs via a novel nonnuclear, nontranscriptional mechanism. Specifically, we used anchorage-independent growth in soft agarose to assay the transforming capacity of GFP-ESE-1 and, using loss-of-function internal deletion and gain-of-function domain fusion analyses, we showed that the ESE-1 SAR domain mediates MCF-12A cell transformation from the cytoplasm. While internal deletion of ESE-1 domains that are required for transcription factor function or nuclear localization did not affect ESE-1-induced transformation, fusion of the SV40 large-T-antigen NLS to several ESE-1 constructs, including the SAR domain, uniformly inhibited transforming capacity. Two additional experimental results supported the hypothesis that nuclear ESE-1 localization is irrelevant to its transforming function. First, using IHC analysis, we discovered that the endogenous ESE-1 protein is cytoplasmically localized in human breast cancer tissues, T-47D breast cancer cells, and normal breast epithelium (Fig. 2). Indeed, our data are consistent with a previous report in which the investigators, using two different anti-ESE-1 antibody preparations in two separate IHC procedures, showed that anti-ESE-1 IHC staining is primarily cytoplasmic in human synovial tissues (21). Second, we showed that transiently expressed GFP-ESE-1 is nuclear and specifically induces apoptosis in nontransformed human mammary epithelial cell lines (Fig. 3), whereas fully transformed human breast cell lines are resistant to the apoptotic effect of nuclear GFP-ESE-1. Moreover, it appears that the TAD function of ESE-1 contributes to the apoptotic effect of GFP-ESE-1 (Fig. 6C). Taken together, our studies reveal that ESE-1 has two distinct but separable biological functions, apoptosis and transformation, and that each function is associated with a different ESE-1 subdomain and a distinct subcellular ESE-1 protein localization. These data raise the possibility that nuclear-cytoplasmic shuttling of the ESE-1 protein regulates apoptosis and thus controls cell survival.
Although GFP-ESE-1 was easily detected, was localized in the nucleus, and was proapoptotic in transiently transfected MCF-12A cells, stably expressed GFP-ESE-1 could not be detected by fluorescence microscopy and instead resulted in MCF-12A cell transformation. Our inability to detect GFP-ESE-1 by deconvolution UV microscopy in stably transfected cells could indicate that GFP-ESE-1 was not expressed. However, since we showed that independently generated stable GFP-ESE-1 cell pools were reproducibly transformed and expressed mutation-free GFP-ESE-1 transcripts, we hypothesized that stably expressed GFP-ESE-1 is short-lived, thus resulting in relatively low GFP-ESE-1 levels. Further, we suggest that stably expressed GFP-ESE-1 is cytoplasmic, since its nuclear localization in transiently transfected cells results in apoptosis (as described above). Consistent with our interpretation that cytoplasmic GFP-ESE-1 is expressed at very low levels, previous studies by Niswender et al. (33) showed that the detection of intracellular GFP fusions by fluorescence microscopy requires relatively high GFP fusion concentrations—at least 1 μM. Importantly, we were able to demonstrate by RT-PCR that all of our stable cell pools expressed the appropriate GFP-ESE-1 fusion mRNA and that the GFP-ESE-1 fusion sequence was intact in each sample (Fig. 4B and 5A and data not shown).
The inability to detect stably transfected GFP-ESE-1 in established, transformed MCF-12A stable transfectants could indicate that the transforming effects of ESE-1 result from an initial transitory GFP-ESE-1 function, after which GFP-ESE-1 is no longer required. Arguing against this possibility is our observation that pools of MCF-12A cells stably transfected with either the GFP-SAR or the GFP-NLS-SAR construct showed a detectable GFP fluorescence signal (Fig. 7C), indicating continuous stable expression of GFP-SAR and GFP-NLS-SAR at ≥1 μM. Importantly, GFP-SAR localized to both the nucleus and the cytoplasm of stably transfected cells, and these stable transfectants fully recapitulated the GFP-ESE-1 transformation phenotype (Fig. 7A and B). In contrast, stably expressed GFP-NLS-SAR was restricted to the nucleus and failed to transform MCF-12A cells (Fig. 7A to C). Finally, targeted inhibition of ESE-1 expression in T-47D cells resulted in cell death, indicating that continuous ESE-1 expression is required for breast cancer cell survival (M. Gonzales, J. D. Prescott, and A. Gutierrez-Hartmann, unpublished data). Together, these data support our hypothesis that the continuous expression of cytoplasmically localized GFP-ESE-1 is responsible for MCF-12A cell transformation and indicate that the 50-aa SAR domain is sufficient to mediate this cytoplasmic mechanism.
The studies presented here, combined with previous studies by various investigators (7, 9, 18, 34, 47), suggest that transiently expressed ESE-1 protein has classic ETS nuclear transcription factor function and that the result of ESE-1 transcriptional activity in nontransformed mammary epithelial cells is apoptosis. Specifically, our data show that transiently expressed ESE-1 protein is targeted to the nucleus, independent of the vector delivery method (plasmid transfection or adenovirus infection [Fig. 3A, B, and D]) and of the N-terminal peptide fusion (none [Fig. 3B], GFP [Fig. 3A], or HA [data not shown]). Furthermore, transiently transfected ESE-1 can transactivate various reporter constructs, and reporter assays have been used to localized the ESE-1 TAD (aa 129 to 159) (Fig. 1) (6). Indeed, we show that deletion of the ESE-1 TAD (GFP-ESE-1ΔTAD) impairs GFP-ESE-1-mediated apoptosis, indicating that ESE-1 transactivating function contributes to the apoptotic action of the nuclear ESE-1 protein (Fig. 6C). Preliminary gene array studies have also revealed that transiently expressed ESE-1 can induce proapoptotic genes (e.g., those for NF-κB, Bbc-3, and PLA-2) (Gonzales et al., unpublished). Similarly, several other ETS factors, including ETS-1, ETS-2, and PU.1, have been shown to induce apoptosis via a transcriptional mechanism (43, 51, 52). These data suggest that the nuclear apoptotic function of ESE-1 transcriptional activity may be regulated by mechanisms that restrain the ESE-1 protein in the cytoplasm. Indeed, we have identified two putative nuclear export signals (NESs) in ESE-1 (102LCNCALEELRL112 and 275LWEFIRDILI285) (Fig. 1) that match the LX1-4LX2-4(L/I)X(L/I) NES consensus and that are similar to the functional NESs found in MEK (32ALQKKLEELELDE44), cyclic AMP-dependent protein kinase inhibitor alpha (37LALKLAGLDI46), and human immunodeficiency virus Rev (73LQLPPLERLTL84) (20). These two putative ESE-1 NES motifs, in combination with the NLS defined here, may regulate ESE-1 subcellular localization. Cytoplasmic ESE-1 protein restriction may thus have a normal function in the breast epithelium. Indeed, cytoplasmic localization is a normal mechanism whereby the nuclear functions of the ETS repressors NET, ERF, TEL, and YAN are regulated (17, 26, 28, 44). Additionally, cytoplasmic localization is a normal mechanism for regulating the apoptotic functions of several other transcription factors, including NF-κB and beta-catenin (16, 37). Thus, the data presented here suggest that ESE-1, like other ETS factors, has nuclear transcription factor function and that this function promotes the transcription of proapoptotic genes in mammary epithelial cells.
As noted above, the ESE-1 protein has been assigned several domains based on either homology or functional analysis (Fig. 1). The 23-aa region spanning ESE-1 aa 236 to 259 has been proposed to contain two short AT hook domains that may recognize AT-rich segments in the DNA duplex minor groove and that are similar to the AT hook domain found in the Drosophila tramtrack protein (34). While this putative AT hook region has not been shown to contribute to ESE-1 DNA binding specificity, we showed, by internal deletion analysis, that this region contains a functional NLS (Fig. 5A). Indeed, this region includes a lysine-rich putative bipartite NLS (237KKGDPKHDKRKRGRPRKLSKEYWDCLEGKKSK267) (Fig. 1) (www.prosite.com). Interestingly, previously identified ETS NLS motifs are localized within the ETS DBD; these include the sequences GKRKNKPK (ETS-1), GLRKNKTN (Elk-1), and GERKRKPG (ER71) (4, 13, 22). The ESE-1 ETS domain also appears to contain a putative NLS (Fig. 1), 316GQKKKNSN323, which is also highly conserved in the ESE family members ESE-2 and ESE-3 (25). Although this basic sequence alone is insufficient to mediate nuclear localization (Fig. 5A), it may contribute to nuclear import.
Two additional members of the ESE family, ESE-2 and ESE-3, have been identified on the basis of conservation within their Pointed and ETS domains (25, 35, 46). ESE-2 is primarily a transcription activator, while ESE-3 is localized in the nucleus and appears to repress phorbol ester and Ras/MAPK/ETS signaling (46). Despite their similarities, sequence alignment of the ESE-1, ESE-2, and ESE-3 proteins reveals that the SAR, AT hook, 5′ putative NES, and putative bipartite NLS domains are limited to ESE-1 (25). Thus, ESE-1 appears to be unique in its structural organization, dual DNA binding specificity, bipartite NLS function, cytoplasmic localization, and SAR domain transforming action. These data indicate that ESE-1 has unique cellular functions.
A key discovery in this report is that the ESE-1 SAR domain is a novel and autonomously transforming 50-aa sequence whose function depends on its cytoplasmic localization. This SAR region (Fig. 6A) is highly acidic (pI, ∼4.33) and was originally identified on the basis of its similarity to the C-terminal TAD (aa 370 to 420) of the lymphocyte-restricted HMG-box transcription factor SOX4 (6, 7). However, amino acid sequence analysis indicates that a 15-residue polyserine tract within SOX4 aa 370 to 420 accounts for most of the identity between ESE-1 SAR and SOX4 TAD, suggesting that the homology between these two sequences may simply reflect their similar serine richness. Functional studies have also demonstrated that the SAR region lacks TAD capacity (6). Moreover, additional sequence analysis has indicated that the SAR domain lacks conserved DBD, NLS, or NES motifs, and the absence of such signals is supported by the diffuse nuclear and cytoplasmic localization of GFP-SAR (Fig. 6B and 7C). Indeed, enforced nuclear targeting of GFP-SAR (as GFP-NLS-SAR) reveals that SAR-induced transformation is actually inhibited by nuclear localization (Fig. 7A and B). Therefore, direct regulation of gene transcription is not the basis for SAR-mediated MCF-12A cell transformation. These data raise the intriguing question of which functional motif(s) within the SAR domain underlies its biological functions. Andreoli et al. (1) suggested that the Xenopus and Drosophila segment polarity protein disheveled and the nuclear period clock protein contain amino acid sequences that are similar to those of the ESE-1 SAR domain; however, these authors showed neither the precise amino acid sequences in question nor the degree of identity among the three proteins. Our own computational analysis failed to identify regions of significant similarity between these two proteins and the SAR domain (data not shown). Indeed, all of our analyses to date have indicated not only that the SAR domain is absent from all other ETS factors but that this domain, which we have not found in any other protein, is unique. Finally, our data also reveal that the transforming and apoptotic effects of the SAR domain are separate, with the former requiring cytoplasmic localization and the latter, while modest, requiring nuclear localization (Fig. 6C and 7B and C). In summary, our studies define the SAR domain as a novel transforming peptide sequence whose transforming function is mediated via a cytoplasmic mechanism.
Acknowledgments
We are grateful to David M. Prescott, Heide Ford, and Pepper Schedin for critical review of the manuscript. We thank the members of the Gutierrez-Hartmann laboratory for their helpful comments and suggestions. We also thank Audrey Brumback and Steven Fadul for providing FM 4-64 vital dye and technical assistance with confocal microscopy. Digital deconvolution confocal fluorescence microscopy equipment was provided by the UCHSC Light Microscopy Facility.
DNA sequencing and FACS analysis were provided by Core Facilities of the University of Colorado Comprehensive Cancer Center (supported by grant NIH P30 CA 46934). This work was supported by grants DOD DAMD17-02-1-0352 and DOD DAMD17-00-1-0474 to J.D.P. and by grant DOD DAMD17-00-1-0476 to A.G.-H.
REFERENCES
- 1.Andreoli, J. M., S. I. Jang, E. Chung, C. M. Coticchia, P. M. Steinert, and N. G. Markova. 1997. The expression of a novel, epithelium-specific ets transcription factor is restricted to the most differentiated layers in the epidermis. Nucleic Acids Res. 25:4287-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asada, S., Y. Choi, and M. Uesugi. 2003. A gene-expression inhibitor that targets an alpha-helix-mediated protein interaction. J. Am. Chem. Soc. 125:4992-4993. [DOI] [PubMed] [Google Scholar]
- 3.Asada, S., Y. Choi, M. Yamada, S. C. Wang, M. C. Hung, J. Qin, and M. Uesugi. 2002. External control of Her2 expression and cancer cell growth by targeting a Ras-linked coactivator. Proc. Natl. Acad. Sci. USA 99:12747-12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulukos, K. E., P. Pognonec, B. Rabault, A. Begue, and J. Ghysdael. 1989. Definition of an Ets1 protein domain required for nuclear localization in cells and DNA-binding activity in vitro. Mol. Cell. Biol. 9:5718-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabral, A., D. F. Fischer, W. P. Vermeij, and C. Backendorf. 2003. Distinct functional interactions of human Skn-1 isoforms with Ese-1 during keratinocyte terminal differentiation. J. Biol. Chem. 278:17792-17799. [DOI] [PubMed] [Google Scholar]
- 6.Chang, C. H., G. K. Scott, M. A. Baldwin, and C. C. Benz. 1999. Exon 4-encoded acidic domain in the epithelium-restricted Ets factor, ESX, confers potent transactivating capacity and binds to TATA-binding protein (TBP). Oncogene 18:3682-3695. [DOI] [PubMed] [Google Scholar]
- 7.Chang, C. H., G. K. Scott, W. L. Kuo, X. Xiong, Y. Suzdaltseva, J. W. Park, P. Sayre, K. Erny, C. Collins, J. W. Gray, and C. C. Benz. 1997. ESX: a structurally unique Ets overexpressed early during human breast tumorigenesis. Oncogene 14:1617-1622. [DOI] [PubMed] [Google Scholar]
- 8.Chiang, S. Y., R. W. Burli, C. C. Benz, L. Gawron, G. K. Scott, P. B. Dervan, and T. A. Beerman. 2000. Targeting the ets binding site of the HER2/neu promoter with pyrrole-imidazole polyamides. J. Biol. Chem. 275:24246-24254. [DOI] [PubMed] [Google Scholar]
- 9.Choi, S. G., Y. Yi, Y. S. Kim, M. Kato, J. Chang, H. W. Chung, K. B. Hahm, H. K. Yang, H. H. Rhee, Y. J. Bang, and S. J. Kim. 1998. A novel ets-related transcription factor, ERT/ESX/ESE-1, regulates expression of the transforming growth factor-beta type II receptor. J. Biol. Chem. 273:110-117. [DOI] [PubMed] [Google Scholar]
- 10.Choi, Y., S. Asada, and M. Uesugi. 2000. Divergent hTAFII31-binding motifs hidden in activation domains. J. Biol. Chem. 275:15912-15916. [DOI] [PubMed] [Google Scholar]
- 11.Cifone, M. A. 1982. In vitro growth characteristics associated with benign and metastatic variants of tumor cells. Cancer Metastasis Rev. 1:335-347. [DOI] [PubMed] [Google Scholar]
- 12.Cohen, B. D., C. B. Siegall, S. Bacus, L. Foy, J. M. Green, I. Hellstrom, K. E. Hellstrom, and H. P. Fell. 1998. Role of epidermal growth factor receptor family members in growth and differentiation of breast carcinoma. Biochem. Soc. Symp. 63:199-210. [PubMed] [Google Scholar]
- 13.De Haro, L., and R. Janknecht. 2002. Functional analysis of the transcription factor ER71 and its activation of the matrix metalloproteinase-1 promoter. Nucleic Acids Res. 30:2972-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delannoy-Courdent, A., V. Mattot, V. Fafeur, W. Fauquette, I. Pollet, T. Calmels, C. Vercamer, B. Boilly, B. Vandenbunder, and X. Desbiens. 1998. The expression of an Ets1 transcription factor lacking its activation domain decreases uPA proteolytic activity and cell motility, and impairs normal tubulogenesis and cancerous scattering in mammary epithelial cells. J. Cell Sci. 111:1521-1534. [DOI] [PubMed] [Google Scholar]
- 15.Delattre, O., J. Zucman, B. Plougastel, C. Desmaze, T. Melot, M. Peter, H. Kovar, I. Joubert, P. de Jong, G. Rouleau, et al. 1992. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359:162-165. [DOI] [PubMed] [Google Scholar]
- 16.de Martin, R., J. A. Schmid, and R. Hofer-Warbinek. 1999. The NF-kappaB/Rel family of transcription factors in oncogenic transformation and apoptosis. Mutat. Res. 437:231-243. [DOI] [PubMed] [Google Scholar]
- 17.Ducret, C., S. M. Maira, A. Dierich, and B. Wasylyk. 1999. The net repressor is regulated by nuclear export in response to anisomycin, UV, and heat shock. Mol. Cell. Biol. 19:7076-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckel, K. L., J. J. Tentler, G. J. Cappetta, S. E. Diamond, and A. Gutierrez-Hartmann. 2003. The epithelial-specific ETS transcription factor ESX/ESE-1/Elf-3 modulates breast cancer-associated gene expression. DNA Cell Biol. 22:79-94. [DOI] [PubMed] [Google Scholar]
- 19.Feldman, R. J., V. I. Sementchenko, and D. K. Watson. 2003. The epithelial-specific Ets factors occupy a unique position in defining epithelial proliferation, differentiation and carcinogenesis. Anticancer Res. 23:2125-2131. [PubMed] [Google Scholar]
- 20.Fukuda, M., I. Gotoh, Y. Gotoh, and E. Nishida. 1996. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J. Biol. Chem. 271:20024-20028. [DOI] [PubMed] [Google Scholar]
- 21.Grall, F., X. Gu, L. Tan, J. Y. Cho, M. S. Inan, A. R. Pettit, U. Thamrongsak, B. K. Choy, C. Manning, Y. Akbarali, L. Zerbini, S. Rudders, S. R. Goldring, E. M. Gravallese, P. Oettgen, M. B. Goldring, and T. A. Libermann. 2003. Responses to the proinflammatory cytokines interleukin-1 and tumor necrosis factor alpha in cells derived from rheumatoid synovium and other joint tissues involve nuclear factor kappaB-mediated induction of the Ets transcription factor ESE-1. Arthritis Rheum. 48:1249-1260. [DOI] [PubMed] [Google Scholar]
- 22.Janknecht, R., R. Zinck, W. H. Ernst, and A. Nordheim. 1994. Functional dissection of the transcription factor Elk-1. Oncogene 9:1273-1278. [PubMed] [Google Scholar]
- 23.Jobling, A. I., Z. Fang, D. Koleski, and M. J. Tymms. 2002. Expression of the ETS transcription factor ELF3 in the retinal pigment epithelium. Investig. Ophthalmol. Vis. Sci. 43:3530-3537. [PubMed] [Google Scholar]
- 24.Kapuscinski, J. 1995. DAPI: a DNA-specific fluorescent probe. Biotechnol. Histochem. 70:220-233. [DOI] [PubMed] [Google Scholar]
- 25.Kas, K., E. Finger, F. Grall, X. Gu, Y. Akbarali, J. Boltax, A. Weiss, P. Oettgen, R. Kapeller, and T. A. Libermann. 2000. ESE-3, a novel member of an epithelium-specific ets transcription factor subfamily, demonstrates different target gene specificity from ESE-1. J. Biol. Chem. 275:2986-2998. [DOI] [PubMed] [Google Scholar]
- 26.Le Gallic, L., D. Sgouras, G. Beal, Jr., and G. Mavrothalassitis. 1999. Transcriptional repressor ERF is a Ras/mitogen-activated protein kinase target that regulates cellular proliferation. Mol. Cell. Biol. 19:4121-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leprince, D., A. Gegonne, J. Coll, C. de Taisne, A. Schneeberger, C. Lagrou, and D. Stehelin. 1983. A putative second cell-derived oncogene of the avian leukaemia retrovirus E26. Nature 306:395-397. [DOI] [PubMed] [Google Scholar]
- 28.Lopez, R. G., C. Carron, and J. Ghysdael. 2003. v-SRC specifically regulates the nucleo-cytoplasmic delocalization of the major isoform of TEL (ETV6). J. Biol. Chem. 278:41316-41325. [DOI] [PubMed] [Google Scholar]
- 29.Neve, R., C. H. Chang, G. K. Scott, A. Wong, R. R. Friis, N. E. Hynes, and C. C. Benz. 1998. The epithelium-specific ets transcription factor ESX is associated with mammary gland development and involution. FASEB J. 12:1541-1550. [DOI] [PubMed] [Google Scholar]
- 30.Neve, R. M., B. Ylstra, C. H. Chang, D. G. Albertson, and C. C. Benz. 2002. ErbB2 activation of ESX gene expression. Oncogene 21:3934-3938. [DOI] [PubMed] [Google Scholar]
- 31.Neznanov, N., A. K. Man, H. Yamamoto, C. A. Hauser, R. D. Cardiff, and R. G. Oshima. 1999. A single targeted Ets2 allele restricts development of mammary tumors in transgenic mice. Cancer Res. 59:4242-4246. [PubMed] [Google Scholar]
- 32.Ng, A. Y., P. Waring, S. Ristevski, C. Wang, T. Wilson, M. Pritchard, P. Hertzog, and I. Kola. 2002. Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology 122:1455-1466. [DOI] [PubMed] [Google Scholar]
- 33.Niswender, K. D., S. M. Blackman, L. Rohde, M. A. Magnuson, and D. W. Piston. 1995. Quantitative imaging of green fluorescent protein in cultured cells: comparison of microscopic techniques, use in fusion proteins and detection limits. J. Microsc. 180:109-116. [DOI] [PubMed] [Google Scholar]
- 34.Oettgen, P., R. M. Alani, M. A. Barcinski, L. Brown, Y. Akbarali, J. Boltax, C. Kunsch, K. Munger, and T. A. Libermann. 1997. Isolation and characterization of a novel epithelium-specific transcription factor, ESE-1, a member of the ets family. Mol. Cell. Biol. 17:4419-4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oettgen, P., K. Kas, A. Dube, X. Gu, F. Grall, U. Thamrongsak, Y. Akbarali, E. Finger, J. Boltax, G. Endress, K. Munger, C. Kunsch, and T. A. Libermann. 1999. Characterization of ESE-2, a novel ESE-1-related Ets transcription factor that is restricted to glandular epithelium and differentiated keratinocytes. J. Biol. Chem. 274:29439-29452. [DOI] [PubMed] [Google Scholar]
- 36.Oikawa, T., and T. Yamada. 2003. Molecular biology of the Ets family of transcription factors. Gene 303:11-34. [DOI] [PubMed] [Google Scholar]
- 37.Oren, M. 2003. Decision making by p53: life, death and cancer. Cell Death Differ. 10:431-442. [DOI] [PubMed] [Google Scholar]
- 38.Reddy, S. P., H. Vuong, and P. Adiseshaiah. 2003. Interplay between proximal and distal promoter elements is required for squamous differentiation marker induction in the bronchial epithelium: role for ESE-1, Sp1, and AP-1 proteins. J. Biol. Chem. 278:21378-21387. [DOI] [PubMed] [Google Scholar]
- 39.Rhim, J. S. 2001. Molecular and genetic mechanisms of prostate cancer. Radiat. Res. 155:128-132. [DOI] [PubMed] [Google Scholar]
- 40.Sapi, E., M. B. Flick, S. Rodov, and B. M. Kacinski. 1998. Ets-2 transdominant mutant abolishes anchorage-independent growth and macrophage colony-stimulating factor-stimulated invasion by BT20 breast carcinoma cells. Cancer Res. 58:1027-1033. [PubMed] [Google Scholar]
- 40a.Schedin, P. J., K. L. Eckel-Mahan, S. M. McDaniel, J. D. Prescott, K. S. Brodsky, J. J. Tentler, and A. Gutierrez-Hartmann. 2004. ESX induces transformation and functional epithelial to mesenchymal transition in MCF-12A mammary epithelial cells. Oncogene 9:1766-1779. [DOI] [PubMed] [Google Scholar]
- 41.Sharrocks, A. D. 2001. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell. Biol. 2:827-837. [DOI] [PubMed] [Google Scholar]
- 42.Soule, H. D., T. M. Maloney, S. R. Wolman, W. D. Peterson, Jr., R. Brenz, C. M. McGrath, J. Russo, R. J. Pauley, R. F. Jones, and S. C. Brooks. 1990. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50:6075-6086. [PubMed] [Google Scholar]
- 43.Teruyama, K., M. Abe, T. Nakano, C. Iwasaka-Yagi, S. Takahashi, S. Yamada, and Y. Sato. 2001. Role of transcription factor Ets-1 in the apoptosis of human vascular endothelial cells. J. Cell. Physiol. 188:243-252. [DOI] [PubMed] [Google Scholar]
- 44.Tootle, T. L., P. S. Lee, and I. Rebay. 2003. CRM1-mediated nuclear export and regulated activity of the Receptor Tyrosine Kinase antagonist YAN require specific interactions with MAE. Development 130:845-857. [DOI] [PubMed] [Google Scholar]
- 45.Trempe, G. L. 1976. Human breast cancer in culture. Recent Results Cancer Res. 57:33-41. [DOI] [PubMed] [Google Scholar]
- 46.Tugores, A., J. Le, I. Sorokina, A. J. Snijders, M. Duyao, P. S. Reddy, L. Carlee, M. Ronshaugen, A. Mushegian, T. Watanaskul, S. Chu, A. Buckler, S. Emtage, and M. K. McCormick. 2001. The epithelium-specific ETS protein EHF/ESE-3 is a context-dependent transcriptional repressor downstream of MAPK signaling cascades. J. Biol. Chem. 276:20397-20406. [DOI] [PubMed] [Google Scholar]
- 47.Tymms, M. J., A. Y. Ng, R. S. Thomas, B. C. Schutte, J. Zhou, H. J. Eyre, G. R. Sutherland, A. Seth, M. Rosenberg, T. Papas, C. Debouck, and I. Kola. 1997. A novel epithelial-expressed ETS gene, ELF3: human and murine cDNA sequences, murine genomic organization, human mapping to 1q32.2 and expression in tissues and cancer. Oncogene 15:2449-2462. [DOI] [PubMed] [Google Scholar]
- 48.Vermes, I., C. Haanen, H. Steffens-Nakken, and C. Reutelingsperger. 1995. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immunol. Methods 184:39-51. [DOI] [PubMed] [Google Scholar]
- 49.Vida, T. A., and S. D. Emr. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wasylyk, B., J. Hagman, and A. Gutierrez-Hartmann. 1998. Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem. Sci. 23:213-216. [DOI] [PubMed] [Google Scholar]
- 51.Wolvetang, E. J., T. J. Wilson, E. Sanij, J. Busciglio, T. Hatzistavrou, A. Seth, P. J. Hertzog, and I. Kola. 2003. ETS2 overexpression in transgenic models and in Down syndrome predisposes to apoptosis via the p53 pathway. Hum. Mol. Genet. 12:247-255. [DOI] [PubMed] [Google Scholar]
- 52.Yamada, T., and T. Oikawa. 1997. Induction of apoptosis by overexpression of the PU.1/Spi-1 gene in murine erythroleukemia cells. Leukemia 11(Suppl. 3):480-481. [PubMed] [Google Scholar]
- 53.Yoneda, Y. 1997. How proteins are transported from cytoplasm to the nucleus. J. Biochem. (Tokyo) 121:811-817. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida, N., S. Yoshida, M. Araie, H. Handa, and Y. Nabeshima. 2000. Ets family transcription factor ESE-1 is expressed in corneal epithelial cells and is involved in their differentiation. Mech. Dev. 97:27-34. [DOI] [PubMed] [Google Scholar]