Abstract
Using DNA microarrays, we compared global transcript stability profiles following chemical inhibition of transcription to rpb1-1 (a temperature-sensitive allele of yeast RNA polymerase II). Among the five inhibitors tested, the effects of thiolutin and 1,10-phenanthroline were most similar to rpb1-1. A comparison to various microarray data already in the literature revealed similarity between mRNA stability profiles and the transcriptional response to stresses such as heat shock, consistent with the fact that the general stress response includes a transient shutoff of general mRNA transcription. Genes encoding factors involved in rRNA synthesis and ribosome assembly, which are often observed to be coordinately down-regulated in yeast microarray data, were among the least stable transcripts. We examined the effects of deletions of genes encoding deadenylase components Ccr4p and Pan2p and putative RNA-binding proteins Pub1p and Puf4p on the genome-wide pattern of mRNA stability after inhibition of transcription by chemicals and/or heat stress. This examination showed that Ccr4p, the major yeast mRNA deadenylase, contributes to the degradation of transcripts encoding both ribosomal proteins and rRNA synthesis and ribosome assembly factors and mediates a large part of the transcriptional response to heat stress. Pan2p and Puf4p also contributed to the degradation rate of these mRNAs following transcriptional shutoff, while Pub1p preferentially stabilized transcripts encoding ribosomal proteins. Our results indicate that the abundance of ribosome biogenesis factors is controlled at the level of mRNA stability.
mRNA turnover is an important factor in the regulation of gene expression in eukaryotic cells and complements transcriptional regulation by endowing the cell with the capability to rapidly vary the levels of existing transcripts (7, 27). mRNA half-lives range from 3 min to more than 100 min in Saccharomyces cerevisiae (20, 58) and from ∼15 min to more than 10 h in mammals (27). The details of the eukaryotic mRNA decay pathway are best understood in S. cerevisiae, where many of the major proteins involved have been identified and characterized (53, 60). Deadenylation and decapping are the two most important steps in the mRNA degradation pathway and typically occur sequentially (10, 55, 60), although decapping is not absolutely dependent upon deadenylation (54). Deadenylation is carried out by the Pan2/Pan3 and the Ccr4/Caf1 poly(A) nuclease complexes (54), and decapping is carried out by the decapping enzymes Dcp1 and/or Dcp2, all of which are conserved among eukaryotes (33, 44). A deadenylated and decapped mRNA is degraded by the 5′→3′ exonuclease Rat1 and/or Xrn1 (22) and by a 3′→5′ exonucleolytic protein complex known as the exosome (26).
Transcript stability is traditionally thought to be regulated by specific sequences or structures in the 3′ or 5′ untranslated region (UTR) and by cognate trans-acting factors that recognize, and in some cases may bind to, these elements (40). The identification of cis- and trans-acting factors affecting mRNA stability has led to the general hypothesis that regulators of mRNA stability may represent a functional equivalent of bacterial operons and eukaryotic pathway-specific transcription factors (30, 31). Several of these trans-acting factors constitute a link between mRNA stability and translation (27). For example, at low iron concentrations, the iron regulatory protein IRP-1 (identical to aconitase) binds to the iron-responsive element (IRE) in the 5′ UTR of ferritin mRNA and blocks translation, while IREs in the 3′ UTR of the transferrin receptor control transcript stability (51). Similarly, AU-rich elements (AREs) in the 3′ UTRs of many mammalian protooncogene and chemokine transcripts modulate transcript stability (12, 13), and the binding affinity of proteins in the mammalian ELAV family to AREs correlates with the stabilization or destabilization of these mRNAs (9). However, cis- and trans-acting determinants of relative stability have not been identified for most mRNAs despite the fact that transcripts have characteristic and widely varying half-lives that often correlate with the function of the encoded protein (58). For example, in contrast to textbook examples of mRNA stability determinants such as IREs and AREs, it has been reported that the cis determinants responsible for the relatively short half-lives of yeast ribosomal protein transcripts may reside in the coding sequence (20). Coupled with the fact that cis determinants of mRNA stability may be secondary structures or composite sequence features (rather than short, contiguous motifs), this finding suggests that such cis elements may be difficult to identify and dissect by either computational approaches or conventional techniques in molecular biology.
Genome sequencing has identified dozens of proteins with RNA-interacting motifs (e.g., helicases, nucleases, and RNA-binding proteins), and these potential trans-acting factors represent an alternative starting point for the identification of determinants and modifiers of mRNA stability. To thoroughly test the effect of any experimental perturbation on mRNA stability in trans, a whole-genome approach is needed that can detect overall changes in mRNA half-lives as well as identify whether specific functional classes of transcripts are affected. Two general strategies have been described for using microarrays to study mRNA stability. The first strategy is to measure the relative abundance of transcripts between two conditions, e.g., a wild-type strain and a mutant strain (37). Steady-state measurements, however, fail to capture the actual dynamics of mRNA decay and will reflect both primary and secondary effects. The second strategy is to measure the absolute half-lives of mRNAs over a time course following the inhibition of transcription (8, 18, 45, 58). While this strategy has the advantage that decay rates are measured directly, it requires a separate time course and a separate set of arrays for each sample analyzed, and it does not take advantage of the strength of the two-color (i.e., red and green) microarray system in detecting small changes in relative transcript abundance.
A troublesome caveat of any mRNA decay experiment that involves either genetic or chemical inhibition of transcription is that side effects of these treatments, and possibly the act of shutting off transcription itself, may affect mRNA decay. For example, many studies of mRNA stability in yeast (20, 38, 41, 58) have employed the rpb1-1 allele (42), a temperature-sensitive mutant in the catalytic subunit of RNA polymerase II (Pol II) (2). Shutoff with rpb1-1 requires a temperature shift, which even in a wild-type strain causes a rapid decrease in mRNA levels (21, 38). This decrease appears to be due to a rapid and transient shutoff of general transcription and decay of mRNAs at the natural rate (39). Levels of ribosomal protein transcripts are affected most prominently (21, 32, 38, 58); in addition to having short inherent half-lives, these transcripts comprise roughly 50% of the mRNA of actively growing yeast (38). The transcription inhibitors thiolutin and 1,10-phenanthroline, like heat and many other environmental perturbations, also elicit the stress response transcriptional program at least partially (1, 11, 15).
The experiments we describe here were initiated with the purpose of establishing a general microarray-based method for analyzing the effects of known or potential trans-acting factors on mRNA degradation and/or stabilization in which relative changes in abundance between the mutant and wild type are compared after inhibiting transcription in both cultures in order to identify subsets of transcripts that are preferentially affected. We began by examining the specificity of a panel of transcription inhibitors using DNA microarrays to compare their effects to those of rpb1-1. We observed that the poly(A)+ mRNA decay profile bears a strong resemblance to the heat shock response, consistent with a major role for mRNA decay in determining the heat shock response. We also observed that transcripts encoding ribosome biogenesis factors are among the least stable in the transcription-inhibited yeast cell. We then exemplified the system for the analysis of trans-acting factors by comparing pub1-Δ, ccr4-Δ, puf4-Δ, and pan2-Δ strains to the wild type before and after treatment with transcription inhibitors (and heat shock in the case of ccr4-Δ). This analysis confirmed that Ccr4p is a major mediator of poly(A)+ mRNA stability in yeast and also showed that Ccr4p at least partially mediates the heat shock response. The stability of ribosome biogenesis factors or ribosomal protein-encoding transcripts was affected by all four of the mutants analyzed. This result shows that the down-regulation of mRNAs encoding ribosome biogenesis factors frequently observed in microarray data are due at least in part to their coordinate regulation at the level of transcript stability and that this represents the posttranscriptional functional equivalent of a bacterial operon as proposed by Keene and Tenenbaum (30, 31).
MATERIALS AND METHODS
Strains, yeast culture, RNA extraction, and labeling.
The rpb1-1 strain YF2475 (MATa rpb1Δ187::HIS3 [pRP1-10; rpb1-1 on a LEU2/CEN/ARS plasmid]) was a gift of Dave Jansma (University of Toronto). All other strains were homozygous diploids from the Saccharomyces Genome Deletions collection; the wild-type strain used in all experiments was BY4743, which is the wild-type diploid for the deletions consortium strains (16). Cultures were grown to a final density of ∼107 cells/ml in yeast-peptone-dextrose medium at 30°C with vigorous shaking, with the exception of the rpb1-1 strain, which was grown in yeast-peptone-dextrose medium at 25°C (permissive temperature); transcription was shut off by transferring the culture from the permissive (25°C) to the nonpermissive (37°C) temperature by adding an equivalent volume of medium warmed to 49°C. At the indicated time points, cells were harvested by 2 min of centrifugation at 3,000 rpm in a tabletop centrifuge (Eppendorf 5810) at room temperature and immediately frozen in liquid N2. RNA was extracted by hot phenol followed by ethanol precipitation, and mRNA was purified on oligo(dT) cellulose (New England Biolabs). Two micrograms of poly(A)+ mRNA was reverse transcribed for cDNA synthesis and labeled with Cy3 and Cy5 dyes as described previously (24).
Chemical inhibitors.
All drugs were purchased from Sigma-Aldrich except for thiolutin, which was a gift from Pfizer. The following drugs were used: 1,10-phenanthroline (100 μg/ml in ethanol), 6-azauracil (2 mg/ml), thiolutin (3 μg/ml in dimethyl sulfoxide), ethidium bromide (200 μg/ml), and cordycepin (33 μg/ml).
Microarray manufacture and use.
The Operon Yeast Oligo set, which includes a 70-mer oligonucleotide representing each of 6,300 yeast open reading frames, was diluted to a final concentration of 1 mg/ml in a solution of 50% dimethyl sulfoxide and 0.05% sodium dodecyl sulfate and spotted onto poly-l-lysine slides by using a robotic spotter with 16 pins (Virtek, Toronto, Canada). Blocking, hybridization, and washing procedures were done as described previously (19), with the exceptions that washes were restricted to 20 s each. All microarray hybridizations were performed in duplicate, with fluors reversed on the second array. Slides were scanned on an Axon GenePix 4000 scanner.
Data analysis.
All microarray feature extraction, normalization, clustering, statistical analyses, graphs, and figures were generated with Matlab (Mathworks). Initial spot intensities and ratios for each oligonucleotide were determined by the median of pixels within each spot after local background subtraction. Normalization followed, according to the method of Yang et al. (62), whereby a lowess smoother is applied to the ratios of each experiment over intensity.
Gene ontology (GO) annotations were downloaded from ftp://genome-ftp.stanford.edu/pub/yeast/ (accessed November 20, 2002); each annotation was “propagated upward” along all parental branches of the GO graph.
The Kolmogorov-Smirnov (KS) goodness-of-fit statistic was used to detect, for any functional category of size n, deviations of the mRNA intensity ratio rank distributions from the uniform distribution and is given by the equation D = max|FO − FE|, where FO and FE denote the observed and expected cumulative distribution functions, respectively. The KS test is distribution free and applicable for sample sizes of at least 35 (47). For any GO category, the calculated P value represents the significance levels of the KS test at which the null hypothesis of uniform rank distribution can be rejected.
In the calculation of P values in Fig. 2 and 3, we used the asymptotically normal distribution of the Spearman rank correlation coefficient in the test for independence (17).
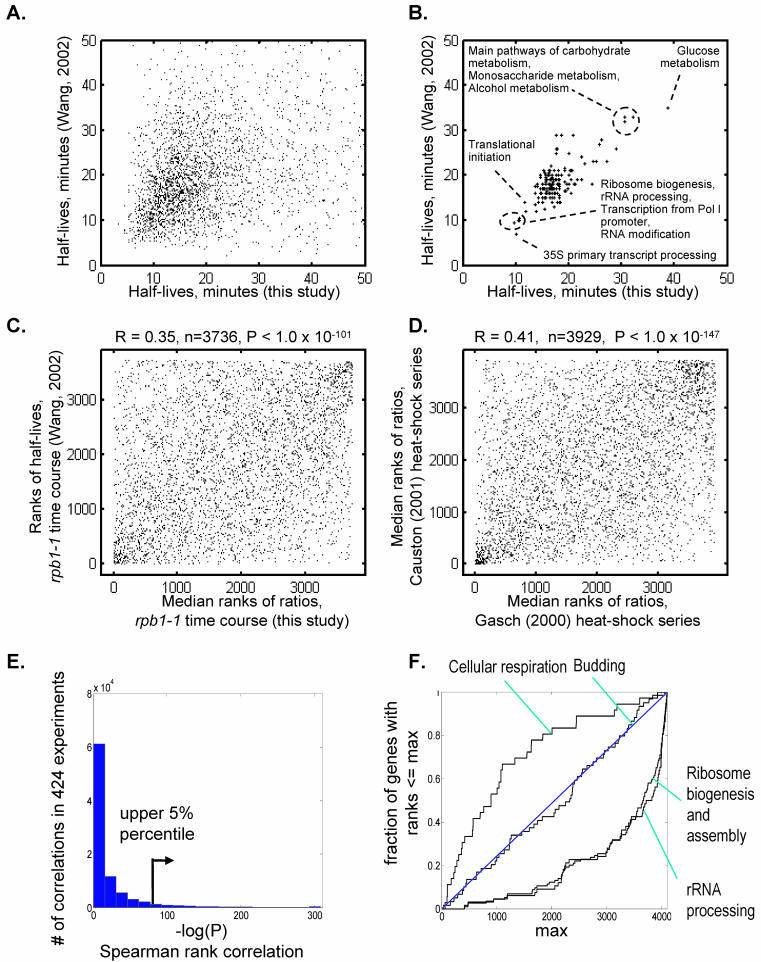
FIG. 2.
Statistical analysis of mRNA stability and comparison to data in the literature. (A) Calculated half-lives for 2,867 genes in our data compared to the calculated half-lives reported by Wang et al. (58). The genes were selected on the basis that (i) their spot intensities at t = 0 in our data are above 50 counts per pixel and (ii) they are present in the 4,686 genes for which Wang et al. reported half-lives. (B) Median mRNA half-lives in both studies for 195 functional categories from the GO Biological Process database that are represented in the set of 2,867 transcripts by 20 or more members. (C) Comparison of mRNA stability ranks from our rpb1-1 data (median ranks for the 5-, 12.5-, and 30-min time points in our two rpb1-1 experiments) and the ranked stabilities reported by Wang et al. (58) using the Spearman rank coefficient over 3,736 genes. The ranks are in increasing order of stability; i.e., genes in the lower left corner are the most stable and those in the upper right corner are the least stable. (D) Analogous comparison of data from Gasch et al. (15) and Causton et al. (11). (E) Histogram of Spearman rank P values among 89,676 possible comparisons among 424 different microarray experiments (61) using the same 3,736 genes as described for panel A. (F) Schematic representation of the KS test. Proceeding from the lowest to the highest rank, the proportion of genes in a given category is compared to the proportion of all genes. Deviation from the diagonal indicates significance (see Materials and Methods). As for panel A, genes with lower rank numbers are more stable.
FIG. 3.
Diagram showing P values of Spearman rank correlations among experiments in this study and the heat shock time course of Gasch et al. (15). A total of 3,736 genes were used to determine the correlations; these are the genes present and considered “good” for our data, the Wang et al. data (58), and the Gasch et al. data (15). Phen., 1,10-phenanthroline; 6-azaU, 6-azauracil; rep, replicate.
Half-lives were calculated for 3,966 mRNAs with the highest spot intensities at t = 0 (median of >50 counts/pixel after background subtraction, where counts scale from 0 to 65,535). For the time points 5, 12.5, and 30 min in the two rpb1-1 experiments, the intensities were normalized to the sum of all spot intensities and reweighted with a factor 2−t/τ_total, where the half-life τ_total of total mRNA was assumed to be 23 min. Ratios between these intensities and the intensities at t = 0 were calculated and averaged over rpb1-1 trials 1 and 2. For each mRNA species i, its average ratios ri(t) for t = 5, 12.5, and 30 min were equated with 2−t/τ_i(t), where τ_i(t) denotes the half-life of mRNA i inferred from time point t; the lifetime for mRNA species i was determined as the average over the three pointwise inferences, τ_i(t).
Data availability.
The following data can be downloaded from http://hugheslab.med.utoronto.ca/Grigull/: all processed data files from all experiments, an assembled ratio file for all experiments, data tables underlying each of the figures, a table of P values from the KS test for all 342 functional categories over the ranks in genes shared by Fig. 1 and the Wang et al. study data (58), and the Spearman P values among all of our experiments and those of the Gasch et al. (15) and Wu et al. (61) studies.
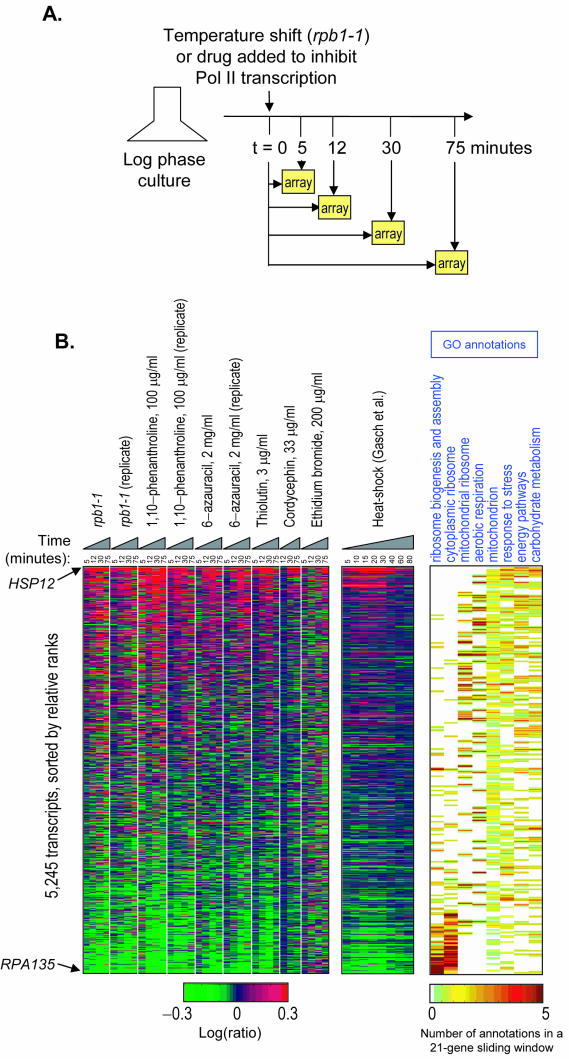
FIG.1.
Comparison of chemical inhibitors of transcription to rpb1-1 and relationship of transcript stabilities to classes of gene functions catalogued by GO (4, 25). (A) Schematic diagram of our experimental strategy to compare genetic and chemical inhibition of transcription using a two-color assay comparing successive time points to the first time point (t = 0). (B) Left panel, log ratio of 5,245 transcripts in nine different time course experiments following the procedure described for panel A. The genes were ordered according to the median rank of each gene among the rpb1-1, 1,10-phenanthroline, and 6-azauracil experiments. Approximately 1,000 yeast genes not meeting minimum intensity and spot quality measures were omitted. Center panel, ratios over a heat shock time course (15) are shown, with the gene order maintained from the microarray data at left. Right panel, density is shown for eight of the GO annotation classes identified as having nonrandom distributions of relative transcript stability, with gene order again maintained from the microarray data at the left.
RESULTS
Analysis of rpb1-1.
We began by analyzing an rpb1-1 strain as a reference for the chemical inhibitors of transcription. Since the rpb1-1 strain is slow growing, even at the permissive temperature, it is likely that it has alterations in steady-state transcript levels in comparison to the wild type, as is typical for mutants with a growth defect (24). Therefore, we used the protocol illustrated in Fig. 1A, in which mRNA from time points following the temperature shift (or application of the inhibitor for experiments below) were each compared to the starting culture with DNA microarrays. In this experimental design, 2 μg of poly(A)+ mRNA is used at each time point to create cDNA. However, because the pool of mRNA is deadenylating over the time point, the actual pool of mRNA in the cell that is selected on oligo(dT) cellulose will decrease and will be enriched in mRNAs with slow deadenylation rates and ongoing low-level transcription. Consequently, the relative ranks of ratios should reflect the relative ranks of deadenylation rates (i.e., stability) because the mRNAs that are deadenylated more slowly than average will be more stable and, hence, their proportion will gradually increase relative to that of the average mRNA, whereas those that are rapidly deadenylated will be less stable and will gradually decrease in proportion to the average. This differs substantially from the protocol used by Wang et al. (58), in which a two-color microarray system and an rpb1-1 strain were also employed but with genomic DNA as the reference sample on the microarrays, rather than mRNA from the starting culture.
To increase signal intensity on microarray spots and to reduce potential cross-hybridization from noncoding RNAs (which make up the vast majority of cellular RNA), we used equal amounts of oligo(dT)-primed, poly(A)+-selected mRNA in each channel of each array. Thus, our analysis was restricted to mRNAs that contain poly(A) tails; subsequent events following deadenylation were not observed. Previous studies have established that deadenylation is the key step that determines mRNA decay and is usually the step that is regulated (for reviews, see references 10 and 55). Consistent with this, Wang et al. (58) showed that the rpb1-1 decay profile is very similar, whether the labeling reaction was primed with oligo(dT) or random primers.
As with most two-color microarray experiments, the data resulting from each of these hybridizations are relative ratios for each gene relative to its abundance in the starting sample and also relative to the average transcript. These data are expressed on a log scale where positive numbers indicate an increase in relative abundance and negative numbers indicate a relative decrease. Figure 1B shows the data for 5,245 genes ranked by their average ratio (which should represent their relative stability) over multiple experiments. As an example, in our first rpb1-1 time course, the log ratios at the 30-min time point (which is close to the estimated average 23-min half-life of yeast mRNAs [58]), the HSP12 transcript has a log ratio of 1.40 (101.4 = 25-fold increased relative to the average transcript), indicating that it is a relatively stable transcript. In contrast, RPA135 (a subunit of Pol I) has a log ratio of −1.22 (16-fold reduced relative to the average transcript), indicating that it is a very unstable transcript. All of the microarray data described herein are available in the online supplemental material at http://hugheslab.med.utoronto.ca/Grigull/, including ratios, spot intensities, and estimates of measurement error.
Although it was not the primary purpose of our study, it is possible to calculate the half-lives of individual poly(A)+ mRNAs from our data by assuming a mass decay rate (in this case, t1/2 = 23 min). The overall distribution of half-lives in our data mirrored the distribution in the Wang et al. study (58) (Fig. 2A), with most transcripts having half-lives between 10 and 20 min but some having much longer half-lives. Disagreements between the two studies may stem from measurement error; there is a threefold higher correlation between the two studies with genes that are highly expressed in our data (R = 0.48 at the 90th percentile of microarray spot intensity) compared to those that are weakly expressed (R = 0.15 at the 10th percentile) (Fig. S1). The two studies were in good agreement, however, when half-lives were averaged for all genes within any given functional category (Fig. 2B). The two studies had similar spreads in half-lives within individual categories (Fig. S2). Assuming that poor measurements tend to increase the spread within a category (i.e., to randomize the measured half-lives), this indicates that neither study is more precise than the other overall.
A more general way of comparing microarray data from different sources is to use relative ranks. The Spearman rank correlation coefficient measures the strength of the associations between two variables (36) and is conceptually similar to the Pearson correlation (which is normally used in microarray data analysis); essentially, the measurements being compared (e.g., microarray ratios or transcript half-lives) are placed on an equivalent scale by conversion to relative ranks (Fig. 2C). Importantly, the likelihood (i.e., P value) that a given Spearman correlation would be obtained by chance is easily calculated, which is not the case for the Pearson correlation. The Spearman correlation is also more robust against outliers (e.g., sporadic errors in microarray data). The Spearman rank correlation between our data and the Wang data (58) is 0.35 and, despite numerous outliers (i.e., points that disagree between the two studies; Fig. 2C, upper left and lower right corners), has a P value of <1.0 × 10−101. As a reference, for 424 different microarray experiments in the literature (Wu et al. [61] and references therein), the P values for the 89,676 possible rank correlations are shown in a histogram in Fig. 2B; 10−101 would be well within the top 5%. These 424 experiments contain time courses of sporulation, mating, and the cell cycle as well as multiple instances of different mutants in the same pathway; hence, a few percent of the correlations are expected to be highly significant.
This analysis also revealed other examples in which replicates of the same microarray experiment by two different labs yielded highly significant correlations but with many disagreements regarding individual genes; for example, Fig. 2D shows a comparison between the heat shock series of Gasch et al. (15) and that of Causton et al. (11), which have a correlation coefficient of only 0.41. In summary, these analyses confirmed that the experimental protocol used here yields relative half-lives that are consistent with previous observations, particularly with regard to groups of functionally related genes, and the fact that there is not absolute agreement on every measurement is typical of comparisons among microarray studies.
Comparison of five chemical inhibitors of transcription to rpb1-1.
We next examined the effects of five known or putative chemical inhibitors of Pol II transcription with the same experimental protocol (Fig. 1A). Thiolutin is a transcription inhibitor that interacts with all three RNA polymerases (52). 1,10-Phenanthroline is a metal chelator that most likely inhibits Pol II by sequestering magnesium (29). 6-Azauracil is a nucleotide base analog that strongly affects intracellular GTP levels and is commonly used to screen for mutants affecting transcript elongation (14, 48). Cordycepin is an adenosine analog that is incorporated into transcripts as if it was a normal base but lacks a 3′ hydroxyl group that can attack the alpha phosphorus on the incoming base. This process leads to chain termination because the RNA polymerase has no proofreading ability (M. Crowley, personal communication). Ethidium bromide is a DNA intercalater that presumably interferes with interaction between RNA polymerase and its DNA template (49). Chemical concentrations (see Materials and Methods) were chosen as the first in a concentration series that prevented growth of overnight cultures (data not shown) in order to minimize secondary effects. To our knowledge, it has not been demonstrated that cordycepin and ethidium bromide are effective inhibitors of Pol II transcription in yeast, although they do inhibit growth. For thiolutin, 1,10-phenanthroline, and 6-azauracil, however, we confirmed by dot blot analysis of total RNA with an oligo(dT) probe that chemical treatment resulted in a reduction in mRNA levels which, like those in rpb1-1 (but unlike those in heat-shocked samples), did not recover after 80 min (data not shown).
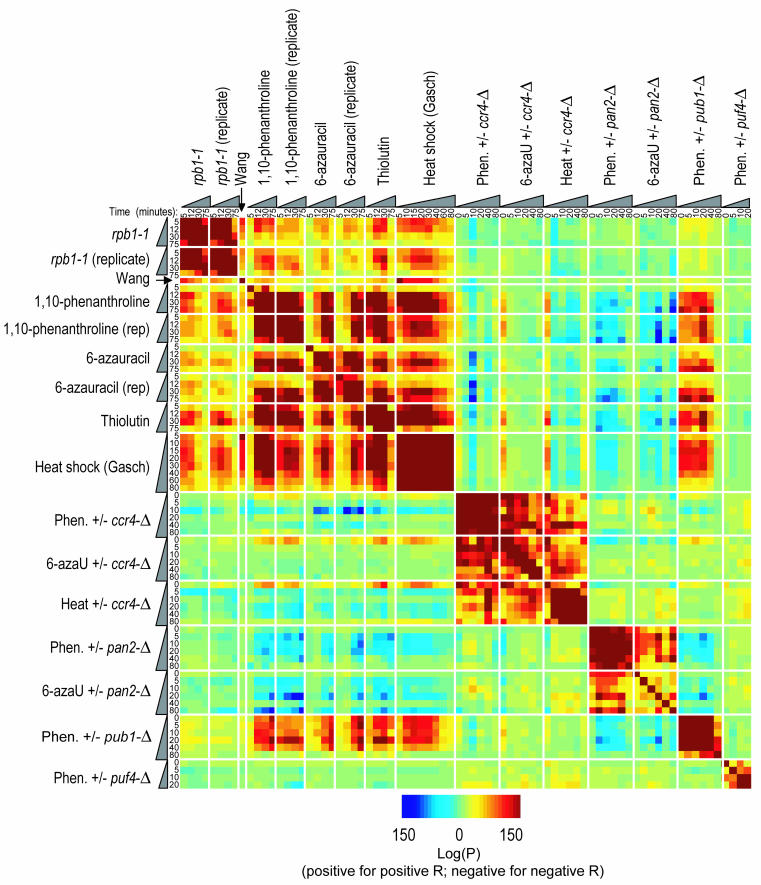
Figure 1B shows the microarray ratios obtained for 5,245 yeast genes over each time point in all nine experiments (rpb1-1, 1,10-phenanthroline, and 6-azauracil were each repeated, and the cordycepin 5-min time point sample was lost due to technical reasons). The P values of the Spearman rank correlation coefficients among all of these experiments are given in the supplemental materials. A positive correlation with the rpb1-1 data was obtained with all of the chemical inhibitors of transcription except ethidium bromide. The highest correlations to rpb1-1 were obtained with 1,10-phenanthroline and thiolutin as well as 6-azauracil to a slightly lesser extent (Fig. 3 and supplemental material). In addition, these chemicals correlated more closely with each other than with rpb1-1 (Fig. 3), suggesting that either (i) the drugs share secondary effects or (ii) rpb1-1 is itself not an ideal model of transcriptional shutoff in a wild-type cell.
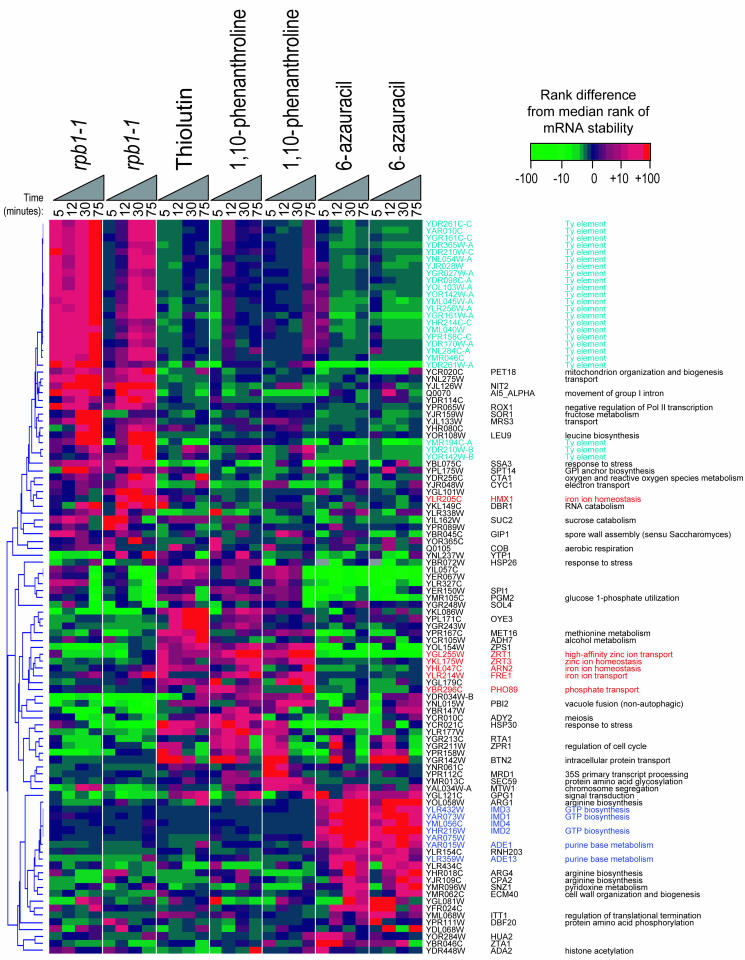
We also examined the characteristic differences between the relative ranks of transcript stabilities obtained with rpb1-1 and different transcription inhibitors in order to further compare them (Fig. 4). We identified 104 genes whose rank positions were skewed in the rpb1-1, thiolutin, 1,10-phenanthroline, and 6-azauracil experiments and grouped them by hierarchical clustering. We then assessed the resulting clusters for a statistically significant enrichment of any functional categories using the hypergeometric P value (46, 50). The rpb1-1 experiments specifically featured relative up-regulation of Ty transcripts (i.e., transposons [Fig. 4, light blue]). This result may be due to the temperature shift (which, among our experiments, was unique to rpb1-1). However, induction of Ty elements was not seen in heat shock (15). We also observed that the rpb1-1 strain grows slowly even at the permissive temperature; hence, rpb1-1 may be dysfunctional at any temperature. 1,10-Phenanthroline caused apparent stabilization of a group of transcripts highly enriched (P < 0.000016) for the functional category “ionic homeostasis” (46) (Fig. 4, highlighted in red). This may be physiologically relevant (i.e., these transcripts may be either stabilized or up-regulated by any residual transcriptional activity in the transcription-inhibited cells) since 1,10-phenanthroline is a metal chelator. Thiolutin had a similar effect, and this raises the possibility that thiolutin may impact metal ions, which to our knowledge has not been previously proposed. 6-Azauracil affected a group of transcripts enriched for genes involved in purine metabolism (P < 3 × 10−7) (Fig. 4, highlighted in dark blue). These transcripts include IMD1, IMD2, IMD3, and IMD4, the four yeast genes that each encode an IMP dehydrogenase, the target of 6-azauracil. The induction of IMD2 in response to 6-azauracil has been previously described and is believed to be regulated at the level of transcription since it is dependent upon transcription elongation factors (48). Hence, the transcriptional shutoff by 6-azauracil is probably incomplete.
FIG. 4.
Differences in relative transcript stabilities among rpb1-1, thiolutin, 1,10-phenanthroline, and 6-azauracil. A total of 104 transcripts were selected that had an increase or decrease in relative rank of at least 30 (relative to the median rank) in at least 2 of the 28 time points shown, with no missing data points. The differences in relative ranks relative to the median are shown in the clustergram.
Relationship between transcriptional shutoff and stress response.
To ask whether the poly(A)+ mRNA decay signature is a major component of other microarray expression profiles in the literature, and also to examine the relationship between our poly(A)+ mRNA decay profiles and previously reported heat and stress responses in the literature, we next compared our poly(A)+ mRNA decay data to over 450 other microarray expression profiles in the literature (11, 15, 61) (Fig. S3 and data not shown) using the P value of the Spearman rank correlation coefficients as described above. Correlations between our poly(A)+ mRNA decay profiles were most similar to responses to environmental stresses such as heat and nutrient deprivation (Fig. 3 and data not shown). This result is consistent with mRNA decay being a major mediator of the heat shock response (39) and with a previous report that 1,10-phenanthroline at least partially induces the stress response (1). Hence, the mRNA decay profile following transcriptional shutoff may be experimentally inseparable from the stress response; likewise, what is learned by studying the mRNA decay profile may also apply to the stress response.
Correlations between gene function and transcript stability following transcription inhibition.
An intriguing aspect of the correspondence between poly(A)+ mRNA decay profiles and those of heat and other stresses is the possibility that stress-induced transcriptional alterations in groups of functionally related transcripts may be mediated at the posttranscriptional level. In particular, Gasch et al. (15) noted the down-regulation of many ribosome biogenesis factors, as have others (61); however, the mechanism controlling this regulation has not to our knowledge been conclusively identified. Wang et al. (58) noted many correlations among the stabilities of transcripts that encode components of protein complexes, although ribosome biogenesis factors, aside from the ribosomal proteins themselves, were not specifically mentioned.
To survey the general correlation between gene function and transcript abundance in different studies, we applied the KS test (47), which estimates the likelihood (i.e., assigns a P value) that the distribution of a subset of ranks is nonrandom (i.e., that the distribution deviates from the expected uniform distribution that would be obtained by assigning ranks at random). Figure 2F illustrates the KS test. The expected distribution, if the genes in a given functional category appear at random among the ranks of stabilities, is a line along the diagonal; this is shown for “budding” which is a randomly selected functional category with a P value of ∼1 (i.e., not significant) by this test. In contrast, mRNAs encoding proteins involved in cellular respiration and glucose metabolism, a subset of which has previously been noted to be controlled at the level of mRNA stability (34), are extremely significant in both our data and the Wang et al. data (Fig. 2B and F), tending to be more stable than would be expected by chance.
We tested the rank distributions of 304 GO categories (4, 25) with 35 or more genes in the category (a rule-of-thumb cutoff intended to avoid sampling error) (47). Among the 304 categories, 63 were significant at P < 0.001 in our data (supplemental materials). Among the most significant categories we observed were ribosome biogenesis factors and ribosome assembly proteins; in both our data and the Wang et al. data (58), these proteins appear to be among the least stable yeast transcripts (Fig. 1B and 2B and F).
Examination of relative transcript stabilities in ccr4-Δ, pan2-Δ, pub1-Δ, and puf4-Δ.
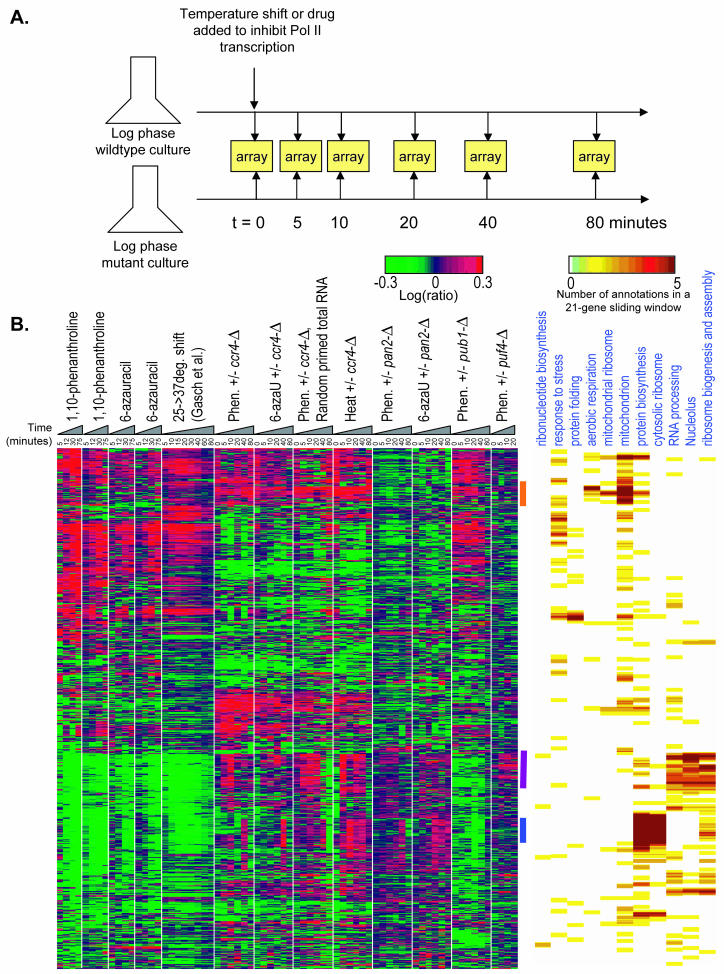
Finally, we applied the protocol outlined in Fig. 5A to compare relative transcript stabilities between mutant and wild-type strains in order to identify potential trans-acting factors that specifically regulate distinct functional categories. The four mutant strains we analyzed were selected on the basis that the mutated genes encode known or potential regulators of transcript stability. Ccr4p and Pan2p are components of the two yeast mRNA deadenylases (54). Pub1p is the yeast homolog of HuR in the ELAV protein family, which binds AREs (9, 56). Puf4p is one of five proteins in yeast that carry the Pumilio domain, an RNA-binding domain that mediates binding to specific mRNAs in at least one other yeast protein (Puf3p), promoting decay (43). Briefly, the wild-type and mutant cultures were grown simultaneously to similar densities; chemical inhibition was initiated in both cultures just after time zero, and mRNA from identical time points in the two cultures was compared in the two channels by using microarrays. The pub1-Δ and puf4-Δ strains were examined only once with 1,10-phenanthroline as the transcriptional inhibitor. The ccr4-Δ and pan2-Δ strains were each examined twice, first by using 1,10-phenanthroline as the transcriptional inhibitor and then by using 6-azauracil. These two chemicals were chosen because they are commercially available and their mechanisms of action are well characterized but different from each other. The microarray data are represented in Fig. 5B and show (i) that the absence of any of these proteins from the cell affects mRNA stability in specific ways and (ii) that this observation is reproducible over more than one transcription inhibitor; very similar results were obtained with 1,10-phenanthroline and 6-azauracil. We also verified that in ccr4-Δ, essentially the same results are obtained by using random-primed total RNA; i.e., the effects we observed for poly(A)+ mRNA, which are presumably due to differences in deadenylation rates between the wild type and the mutant, are reflected in the overall decay (Fig. 5B).
FIG. 5.
Comparison of the specific effects of genetic perturbations, known or suspected to affect transcript stabilities, following a time course after chemical inhibition of transcription. (A) Schematic diagrams of the experimental method. (B) Left panel, clustering analysis of 1,115 genes with ratios of twofold or more in the four or more time points among the experiments shown. Genes that are transposons (Ty elements) were omitted. Right panel, density is shown for 11 GO annotation classes enriched in one or more clusters according to the hypergeometric P value (50). The gene order (vertical axis) is identical from left to right. Phen., 1,10-phenanthroline; 6-azaU, 6-azauracil.
There are three ways that ratios for specific mRNAs can arise in this procedure. The first is as steady-state alterations in transcript abundance that are present before transcription is inhibited; these will include primary and secondary effects as previously noted (37). Steady-state alterations can be identified because they should be present in the first time point and, if they are not a result of altered mRNA stability, their ratios will not change substantially over the time course. The second way ratios can arise is if the perturbation causes an overall effect on mRNA half-lives; this will have the effect of a gradual alteration in ratios that is proportional to mRNA half-life, because transcripts with short half-lives will have a greater proportional “gain” in amount at any given time point. These alterations can be identified because the slopes of the ratios over time should be proportional to the half-lives of the mRNAs over at least the part of the time course where accurate measurements can be obtained. The third way ratios can arise is by an alteration in the degradation rates of specific mRNA classes in the mutant.
Ccr4p exemplifies how we distinguished between these effects. First, the zero time point (i.e., steady state) contains many transcriptional alterations which do not change appreciably over the time course; for example, a group of transcripts encoding predominantly mitochondrial proteins are indicated by an orange bar near the top of Fig. 5B. Second, visual inspection suggests that in the ccr4-Δ mutant, mRNA stability is affected almost independently of the generic stability of the most transcripts following transcriptional inhibition (Fig. 5B; compare the heat shock columns with the 1,10-phenanthroline and 6-azauracil columns). This idea is confirmed by the statistical analysis presented in Fig. 3. The average P value for the rank correlations between ccr4-Δ experiments and transcriptional shutoff experiments is only 10−6.5, which lies in the central 50% of pairwise rank correlations among the Wu et al. reference set (61) (Fig. 2B). A similar result was obtained when we corrected for the alterations in the first time point (i.e., subtracted the ratio for the first time point from all subsequent time points for each gene) or if we examined the ranks of the slopes of the ratios over the time course (data not shown). This makes it unlikely that the effects seen in ccr4-Δ reflect merely a uniform increase of lifetimes (which would lead to apparent up-regulation of unstable transcripts and down-regulation of stable transcripts in our experimental protocol with equal amounts of total mRNAs from the mutant and the wild type for hybridization).
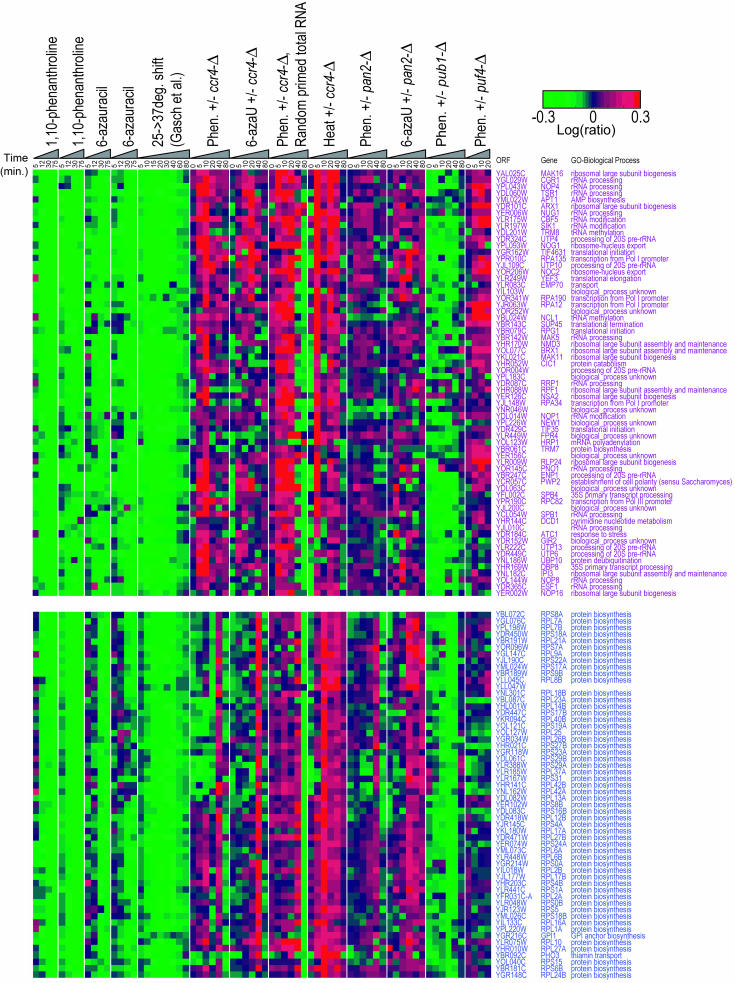
Two classes of transcripts that do appear to be stabilized by ccr4-Δ correspond to those that encode ribosomal proteins and those that encode ribosome biogenesis factors (Fig. 5B, purple and blue bars, respectively). These transcripts are easily identified because they are almost completely unaffected at the zero time point and their ratio gradually increases over time (in late time points, the signal in one or both channels is lost and microarray measurements become unreliable; the effect is more pronounced for ribosome biogenesis factors which have the shortest mRNA half-lives) (Fig. 5B). Figure 6 shows the individual genes for these two groups.
FIG. 6.
Individual transcripts encoding nucleolar proteins (top) and ribosomal proteins (bottom), corresponding to the purple and blue bars shown in Fig. 5B. Phen., 1,10-phenanthroline; 6-azaU, 6-azauracil.
There were striking differences in the classes of transcripts that were affected in the mutants. In the pub1-Δ strain, as in the ccr4-Δ strain, transcripts encoding mitochondrial proteins displayed altered abundance at the zero time point, and there was similarly little change in relative ratios of these transcripts over the time course. In contrast to ccr4-Δ, however, pub1-Δ displayed preferential stabilization of transcripts encoding ribosomal proteins. In pan2-Δ and puf4-Δ, there were few alterations at the zero time point, while the stabilization of transcripts encoding ribosomal proteins and ribosome biogenesis factors was similar to that of ccr4-Δ, but less penetrant. Among the four mutants, the data from pan2-Δ showed the strongest relationship to the poly(A)+ mRNA decay profile (Fig. 3 and 5B), indicating that its effects are most related to simply extending overall mRNA half-lives, consistent with the notion that Pan2p acts constitutively on all mRNAs. This finding also raises the possibility that Ccr4p, which is the major yeast deadenylase (54), has a preference for transcripts encoding ribosome biogenesis factors and ribosomal proteins.
These data show that there is information that can be obtained only by examining the changes in the transcript ratios over the time course, validating the experimental approach shown in Fig. 5A, which was the original aim of this study. None of the effects on transcripts encoding nucleolar or ribosomal proteins are evident from comparisons of the mutant versus the wild type at the zero time point, indicating that the time course following transcriptional shutoff (outlined in Fig. 5A) is necessary to detect the influence of each of these factors on the decay of these transcripts. In fact, the zero time point analysis (i.e., steady-state comparison) may be misleading; one would conclude that pub1-Δ and ccr4-Δ primarily cause alterations in the abundance of a variety of mitochondrial transcripts (Fig. 5B), but these do not appear to be due to an alteration in stability of these transcripts.
Ccr4p is a mediator of the heat shock response.
Since the transcriptional response to heat shock consists largely of a transcriptional shutoff followed by mRNA decay (39), and since Ccr4p appears to preferentially contribute to the degradation of transcripts whose abundance is known to decrease in response to heat shock (15), we reasoned that Ccr4p is likely to be required for this aspect of the heat shock response. To test this, we examined the effects of ccr4-Δ on mRNA levels following a time course after heat shock (Fig. 5B). The results were similar to those obtained by treatment of the ccr4-Δ mutant with 1,10-phenanthroline and 6-azauracil as the inhibitor of transcription; while the stability of most transcripts was largely unaffected, a clear stabilization of transcripts encoding ribosomal proteins and ribosome biogenesis factors was observed. This result shows that Ccr4p is a mediator of the heat shock response.
DISCUSSION
Evidence that diverse biological processes are regulated by controlled transcript decay continues to accumulate in the literature. For example, nocturnin, which is related to the yeast Ccr4p deadenylase, is specifically expressed in the Xenopus laevis retina and controls melatonin levels (5). In contrast to the hundreds of known sequence-specific transcriptional activators, however, relatively few factors have been identified that regulate the stability of different classes of functionally related mRNA transcripts. The substrate range has not been reported for most of the factors already implicated in mRNA stability, even in yeast, an organism for which obtaining deletion mutants is now trivial (16).
Regulation of nucleolar proteins at the level of mRNA stability.
Among the specific functional categories that from our analyses appear to be regulated at the level of mRNA stability are rRNA biogenesis and ribosome assembly. Many of the proteins in these categories are located in the nucleolus. The rRNA biogenesis and ribosome assembly categories overlap with one another but are distinct from the ribosomal proteins. Although yeast ribosome biosynthesis is already known to be regulated at multiple levels (38, 57, 59), it has not to our knowledge been previously reported that yeast nucleolar proteins are regulated at the level of mRNA stability. In fact, the observation that the abundance of these transcripts is largely unchanged from that of the wild type at the zero time point (Fig. 5 and 6) suggests that their transcription rate may be down-regulated in these mutants to compensate for their increased stability. Our data, and also those of Wang et al. (58) (Fig. 2B) indicate that these are among the least stable transcripts in the cell, at least upon the inhibition of transcription. We have identified three potential trans-acting factors that affect the relative degradation rates of these transcripts: Ccr4p, Pan2p, and Puf4p, which all appear to contribute to their degradation. Pub1p, however, appears to be a stabilizing factor for ribosomal protein transcripts, which have previously been noted to have short half-lives upon transcriptional shutoff (20, 58).
Rapid down-regulation of transcripts encoding nucleolar proteins (and also ribosomal proteins) is a feature of many microarray analyses of various perturbations of yeast in the literature (15, 61). Transcriptional coregulation of these genes has previously been attributed to two different promoter elements (PAC and RRPE) shared by many of these genes (50). The inherent instability of these transcripts would explain their rapid decline in abundance upon down-regulation of transcription. However, it is also possible that their stability is itself part of the regulatory mechanism; this possibility is supported by the fact that three of the mutants we analyzed affected the deadenylation rate of these transcripts and also that their deadenylation appears more rapid in response to heat shock than to transcription inhibition, on the basis of the ccr4-Δ data in Fig. 5B. Regardless, the mRNA degradation rate plays an important role in the regulation of this group of transcripts, as we show by analysis of the heat shock response in the presence and absence of ccr4-Δ. Hence, this group of transcripts appears to represent, in at least one sense, the posttranscriptional functional equivalent of a bacterial operon, as proposed by Keene and Tenenbaum (30, 31).
Does transcriptional shutoff itself impact mRNA decay?
Since these experiments were conducted over a time course of inhibition of transcription, one caveat of our results is that they may be relevant only to this condition (e.g., they may correspond to posttranscriptional mechanisms that react to the loss of general transcription). The same argument applies to many studies in the literature that have utilized inhibition of transcription to monitor mRNA decay (20, 38, 41, 58). A previous comparison of mRNA half-lives measured by both in vivo radiolabeling and Northern blotting following transcriptional shutoff suggested that there is a loose but positive correlation between half-lives measured with and without shutoff (20). However, only a small number of genes (11, plus 3 unnamed cDNAs) were compared. It may ultimately be worthwhile to perform such experiments on a genome-wide scale.
An online tool for identifying biologically significant trends in relative ranks data.
The KS test provides a generally applicable method for identifying categories of transcripts that are significantly stabilized or destabilized under any given condition. For the convenience of researchers wishing to confirm our findings or analyze their own data, we have implemented a web-accessible version of the KS test at http://kstest.med.utoronto.ca, in which lists of ranked yeast gene names can be entered and a summary of KS test P values can be retrieved. The KS test is useful for analyzing yeast data of any type that can be sorted, including not only microarray ratios but other types of genomic data such as absolute levels of gene or protein expression, colony sizes, or even screening results.
Is the stability of yeast nucleolar proteins regulated by UTR elements?
A variety of computational tools can also be applied to identify possible cis-acting elements in UTRs of transcripts with shared stability characteristics (3, 23, 28, 35). The potential of such analyses has recently been demonstrated in Arabidopsis thaliana (6). However, we have as yet been unable to identify any statistically significant elements shared by a majority of the 3′ or 5′ UTRs among the different groups of functionally related transcripts described here (Fig. 1B, 4, and 5B).
Global analysis of potential trans-acting mRNA stability factors.
The methods described here (in particular, the procedure shown in Fig. 5A) can be used to determine whether a mutation in any protein known or suspected to be involved in mRNA stability preferentially stabilizes or destabilizes certain classes of transcripts. Despite its effect in up-regulating a small group of transcripts enriched in functions related to ion homeostasis (Fig. 4), 1,10-phenanthroline is the most practical of the five chemicals we tested for applying this method in yeast. It is inexpensive and easily obtained, the mechanism of action is understood, and it appears to work slightly faster than the other chemicals we tested (Fig. 1B). Thiolutin has a very similar effect (Fig. 1B, 3, and 4) and is effective at lower concentrations, but it is not available commercially. To our knowledge, the precise mode of action of thiolutin is unknown, although our results (Fig. 4) suggest that it may act in the same way as 1,10-phenanthroline.
In contrast to microarray analysis of mRNA populations between the mutant and the wild type at steady state, our approach captures a greater number of differential transcripts (Fig. 5B) and affords higher confidence and statistical power through the identification of transcripts whose relative abundance changes consistently across the time course. In comparison to measuring the poly(A)+ decay time courses of wild-type and mutant cultures independently, this method also takes advantage of the ability of the two-color system to highlight relatively small differences in relative transcript abundance. It will be of interest to analyze the multitude of factors known or suspected to affect stability of mRNAs.
Acknowledgments
This work was supported by grants from Genome Canada, NSERC, and the Connaught Foundation (University of Toronto) to T.R.H.
We thank Rick Collins, Jim Ingles, and Alan Cochrane for critical evaluation of the manuscript, Ben Blencowe for helpful advice, Pfizer for the gift of thiolutin, Dave Jansma for the rpb1-1 strain, and Stuart Yang and Naveed Mohammad for technical and computational support.
REFERENCES
- 1.Adams, C. C., and D. S. Gross. 1991. The yeast heat shock response is induced by conversion of cells to spheroplasts and by potent transcriptional inhibitors. J. Bacteriol. 173:7429-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, L. A., J. K. Wong, V. D. Fitzpatrick, M. Moyle, and C. J. Ingles. 1988. The C-terminal domain of the largest subunit of RNA polymerase II of Saccharomyces cerevisiae, Drosophila melanogaster, and mammals: a conserved structure with an essential function. Mol. Cell. Biol. 8:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, J. S. J., and R. Parker. 2000. Computational identification of cis-acting elements affecting post-transcriptional control of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 28:1604-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, et al. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baggs, J. E., and C. B. Green. 2003. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr. Biol. 13:189-198. [DOI] [PubMed] [Google Scholar]
- 6.Bateman, A. 2002. The SGS3 protein involved in PTGS finds a family. BMC Bioinformatics 3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belasco, J. G., and G. Brawerman (ed.). 1993. Control of messenger RNA stability. Academic Press, San Diego, Calif.
- 8.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2001. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99:9697-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaxall, B. C., A. Pende, S. C. Wu, and J. D. Port. 2002. Correlation between intrinsic mRNA stability and the affinity of AUF1 (hnRNP D) and HuR for A+U-rich mRNAs. Mol. Cell. Biochem. 232:1-11. [DOI] [PubMed] [Google Scholar]
- 10.Cao, D., and R. Parker. 2001. Computational modeling of eukaryotic mRNA turnover. RNA 7:1192-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Causton, H. C., B. Ren, S. S. Koh, P. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 13.Chen, C. Y., R. Gherzi, S. E. Ong, E. L. Chan, R. Rajmakers, G. J. Prujin, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451-464. [DOI] [PubMed] [Google Scholar]
- 14.Exinger, F., and F. Lacroute. 1992. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 22:9-11. [DOI] [PubMed] [Google Scholar]
- 15.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 17.Gibbons, J. D., and S. Chakraborti. 1992. Nonparametric statistical inference, 3rd ed. Marcel Dekker Inc., New York, N.Y.
- 18.Gutierrez, R. A., R. M. Ewing, J. M. Cherry, and P. J. Green. 2002. Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl. Acad. Sci. USA 99:11513-11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, et al. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29:548-556. [DOI] [PubMed] [Google Scholar]
- 20.Herrick, D., R. Parker, and A. Jacobson 1990. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2269-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herruer, M. H., W. H. Mager, H. A. Raue, P. Vreken, E. Wilms, and R. J. Planta. 1988. Mild temperature shock affects transcription of yeast ribosomal protein genes as well as the stability of their mRNAs. Nucleic Acids Res. 16:7917-7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu, C. L., and A. Stevens. 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13:4826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu, Y. J. 2002. Prediction of structural motifs in a family of coregulated RNA sequences. Nucleic Acids Res. 30:3886-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 25.Issel-Tarver, L., K. R. Christie, K. Dolinski, R. Andrada, R. Balakrishnan, C. A. Ball, G. Binkley, S. Dong, S. S. Dwight, D. G. Fisk, M. Harris, M. Schroeder, A. Sethuraman, K. Tse, S. Weng, D. Botstein, and J. M. Cherry. 2002. Saccharomyces Genome Database. Methods Enzymol. 350:329-346. [DOI] [PubMed] [Google Scholar]
- 26.Jacobs, J. S., A. R. Anderson, and R. P. Parker. 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17:1497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobson, A., and S. W. Peltz. 1996. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 65:693-773. [DOI] [PubMed] [Google Scholar]
- 28.Jacobson, A. B., and M. Zuker. 1993. Structural analysis by energy dot plot of a large mRNA. J. Mol. Biol. 233:261-269. [DOI] [PubMed] [Google Scholar]
- 29.Johnston, J. R. 1994. Molecular genetics of yeast: a practical approach. Oxford University Press, New York, N.Y.
- 30.Keene, J. D. 2001. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc. Natl. Acad. Sci. USA 98:7018-7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keene, J. D., and S. A. Tenenbaum. 2002. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 9:1161-1167. [DOI] [PubMed] [Google Scholar]
- 32.Kief, D. R., and J. R. Warner. 1981. Hierarchy of elements regulating synthesis of ribosomal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 1:1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LaGrandeur, T. E., and R. Parker. 1998. Isolation and characterization of Dcp1, the yeast mRNA decapping enzyme. EMBO J. 17:1487-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LaGrandeur, T. E., and R. Parker. 1999. The cis acting sequences responsible for the differential decay of the unstable MFA2 and stable PGK1 transcripts in yeast include the context or the translational start codon. RNA 5:420-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le, S. Y., J. H. Chen, D. Konings, and J. V. Maizel, Jr. 2003. Discovering well-ordered folding patterns in nucleotide sequences. Bioinformatics 19:354-361. [DOI] [PubMed] [Google Scholar]
- 36.Lehmann, E. L., and H. J. M. D'Abrera. 1998. Nonparametrics: statistical methods based on ranks, revised ed. Prentice-Hall, Englewood Cliffs, N.J.
- 37.Lelivelt, M. J., and M. R. Culbertson. 1999. Yeast Upf proteins for RNA surveillance affect global expression of the yeast transcriptome. Mol. Cell. Biol. 19:6710-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, B., C. R. Nierras, and J. R. Warner. 1999. Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol. Cell. Biol. 19:5393-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindquist, S. 1981. Regulation of protein synthesis during heat shock. Nature 293:311-314. [DOI] [PubMed] [Google Scholar]
- 40.Mignone, F., C. Gissi, S. Liuni, and G. Pesole. 2002. Untranslated regions of mRNAs. Genome Biol. 3:REVIEWS0004.1-REVIEWS0004.10. [Online.] http://genomebiology.com/2002/3/3/REVIEWS/0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore, P. A., F. A. Sagliocco, R. M. Wood, and A. J. Brown. 1991. Yeast glycolytic mRNAs are differentially regulated. Mol. Cell. Biol. 11:5330-5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nonet, M., C. Scafe, J. Sexton, and R. Young. 1987. Eukaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol. 7:1602-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olivas, W., and R. Parker. 2000. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 19:6602-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker, R., and H. Song. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11:121-127. [DOI] [PubMed] [Google Scholar]
- 45.Raghavan, A., R. L. Ogilvie, C. Reilly, M. L. Abelson, S. Raghavan, J. Vasdevani, M. Krathwol, and P. R. Bohjanen. 2002. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 30:5529-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson, M. D., J. Grigull, N. Mohammad, and T. R. Hughes. 2002. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sachs, L. 1982. Applied statistics. Springer-Verlag, New York, N.Y.
- 48.Shaw, R. J., and D. Reines. Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol. 20:7427-7437. [DOI] [PMC free article] [PubMed]
- 49.Straney, D. C., and D. M. Crothers. 1987. Effect of drug-DNA interactions upon transcription initiation at the lac promoter. Biochemistry 26:1987-1995. [DOI] [PubMed] [Google Scholar]
- 50.Tavazoie, S., J. D. Hughes, M. J. Campbell, R. J. Cho, and G. M. Church. 1999. Systematic determination of genetic network architecture. Nat. Genet. 22:281-285. [DOI] [PubMed] [Google Scholar]
- 51.Theil, E. C. 1993. The IRE (iron regulatory element) family: structures which regulate mRNA translation or stability. Biofactors 4:87-93. [PubMed] [Google Scholar]
- 52.Tipper, D. J. 1973. Inhibition of yeast ribonucleic acid polymerases by thiolutin. J. Bacteriol. 116:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tucker, M., and R. Parker. 2000. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem. 69:571-595. [DOI] [PubMed] [Google Scholar]
- 54.Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis, and R. Parker. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104:377-386. [DOI] [PubMed] [Google Scholar]
- 55.Tucker, M., R. R. Staples, M. A. Valencia-Sanchez, D. Muhlrad, and R. Parker. 2002. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 21:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vasudevan, S., and S. W. Peltz. 2001. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol. Cell 7:1191-1200. [DOI] [PubMed] [Google Scholar]
- 57.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 58.Wang, Y., C. L. Liu, J. D. Storey, R. J. Tibshirani, D. Herschlag, and P. O. Brown. 2002. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA 99:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 60.Wilusz, C. J., M. Wormington, and S. W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2:237-246. [DOI] [PubMed] [Google Scholar]
- 61.Wu, L. F., T. R. Hughes, A. P. Davierwala, M. D. Robinson, R. Stoughton, and S. J. Altschuler. 2002. Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat. Genet. 31:255-265. [DOI] [PubMed] [Google Scholar]
- 62.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The following data can be downloaded from http://hugheslab.med.utoronto.ca/Grigull/: all processed data files from all experiments, an assembled ratio file for all experiments, data tables underlying each of the figures, a table of P values from the KS test for all 342 functional categories over the ranks in genes shared by Fig. 1 and the Wang et al. study data (58), and the Spearman P values among all of our experiments and those of the Gasch et al. (15) and Wu et al. (61) studies.
FIG.1.
Comparison of chemical inhibitors of transcription to rpb1-1 and relationship of transcript stabilities to classes of gene functions catalogued by GO (4, 25). (A) Schematic diagram of our experimental strategy to compare genetic and chemical inhibition of transcription using a two-color assay comparing successive time points to the first time point (t = 0). (B) Left panel, log ratio of 5,245 transcripts in nine different time course experiments following the procedure described for panel A. The genes were ordered according to the median rank of each gene among the rpb1-1, 1,10-phenanthroline, and 6-azauracil experiments. Approximately 1,000 yeast genes not meeting minimum intensity and spot quality measures were omitted. Center panel, ratios over a heat shock time course (15) are shown, with the gene order maintained from the microarray data at left. Right panel, density is shown for eight of the GO annotation classes identified as having nonrandom distributions of relative transcript stability, with gene order again maintained from the microarray data at the left.