Abstract
Because of its central regulatory role, T-cell receptor (TCR) signal transduction is a common target of viruses. We report here the identification of a small signaling protein, ORF5, of the T-lymphotropic tumor virus herpesvirus saimiri (HVS). ORF5 is predicted to contain 89 to 91 amino acids with an amino-terminal myristoylation site and six SH2 binding motifs, showing structural similarity to cellular LAT (linker for activation of T cells). Sequence analysis showed that, despite extensive sequence variation, the myristoylation site and SH2 binding motifs were completely conserved among 13 different ORF5 isolates. Upon TCR stimulation, ORF5 was efficiently tyrosine phosphorylated and subsequently interacted with cellular SH2-containing signaling proteins Lck, Fyn, SLP-76, and p85 through its tyrosine residues. ORF5 expression resulted in the marked augmentation of TCR signal transduction activity, evidenced by increased cellular tyrosine phosphorylation, intracellular calcium mobilization, CD69 surface expression, interleukin-2 production, and activation of the NF-AT, NF-κB, and AP-1 transcription factors. Despite its structural similarity to cellular LAT, however, ORF5 could only partially substitute for LAT function in TCR signal transduction. These results demonstrate that HVS utilizes a novel signaling protein, ORF5, to activate TCR signal transduction. This activation probably facilitates viral gene expression and, thereby, persistent infection.
Upon engagement of the T-cell antigen receptor (TCR), many cellular proteins, including TCR subunits, adaptor proteins, and other effector molecules, are phosphorylated and subsequently are involved in the formation of molecular complexes at the site of contact with the antigen-presenting cells (33, 51, 54) The earliest signaling event following TCR engagement is the sequential activation of the non-receptor protein tyrosine kinases (PTKs) of the Src and Syk families. Activated Src family PTKs, Lck and Fyn, in T cells subsequently phosphorylate tyrosine residues within a consensus sequence termed the immunoreceptor tyrosine-based activation motif in the cytosolic tails of the TCR subunits. The phosphorylated immunoreceptor tyrosine-based activation motifs of TCR in turn recruit the Syk family PTKs ZAP70 and Syk through their Src homology 2 (SH2) domains (33). Together with Lck and Fyn, the ZAP70 and Syk kinases then promote the phosphorylation of many intracellular signaling molecules, including phospholipase C-γ1 (PLC-γ1), Cbl, Vav, LAT (linker for activation of T cells) and SLP-76 (SH2-containing leukocyte protein-76) (12, 25, 41, 45, 46, 53). Phosphorylation of these cellular signaling molecules ultimately induces various cellular events such as cytoskeletal alteration, intracellular Ca++ influx, transcription factor activation, and cytokine/chemokine production (4, 20, 38, 43, 59).
Two adaptor proteins, LAT and SLP-76, have been shown to be critical for the optimal activation of T cells and to function in linking proximal signaling events to distal downstream signaling actions. LAT is a lipid raft-associated protein that has a short transmembrane domain, sites for palmitoylation at the N terminus, and 10 tyrosine-based signaling motifs in its cytoplasmic tail. Upon TCR stimulation, LAT rapidly becomes phosphorylated on its multiple tyrosine residues by ZAP70, and the phosphorylated tyrosine residues of LAT then serve as binding sites for the SH2 domains of PLC-γ1, Grb2, and Gads (20, 61). Because of its important role in TCR signal transduction, a T-cell line devoid of LAT has defects in Ca2+ mobilization, production of IP3, induction of CD69, activation of Ras and interleukin-2 (IL-2) gene expression, and tyrosine phosphorylation of Vav, SLP-76, and PLC-γ1 (20, 61). LAT−/− mice exhibit a complete block in thymocyte development at the pro-T3 stage and defects in mast cell functions (62). SLP-76, on the other hand, is a cytoplasmic adaptor protein consisting of three domains: an N-terminal acidic region containing several tyrosine residues that are also phosphorylated by ZAP70 after TCR engagement (19, 24, 37, 52, 57); a central proline-rich region that binds the SH3 domains of Grb2, Gads, and Lck; and a carboxy-terminal SH2 domain that interacts with the Fyn binding protein FYB/SLAP130 (43, 58). A gene-targeting approach has shown that SLP-76 is also necessary for T-cell development. SLP-76−/− mice lack peripheral T cells and also lack CD4+ CD8+ double-positive thymocytes and CD25− CD44− and CD4− CD8− double-negative thymocytes (9-11, 42-44). SLP-76-deficient cell lines are also defective in TCR-mediated PLC-γ1 phosphorylation, extracellular signal-regulated kinase activation, Ca2+ mobilization, and IL-2 promoter activity (59). This indicates that LAT and SLP-76 are essential for linking TCR/CD3 to downstream signal cascades.
Herpesvirus saimiri (HVS) is a member of the gammaherpesvirus family, which includes human Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus (KSHV). HVS infection is endemic and nonpathogenic in its natural host, squirrel monkeys (Saimiri sciureus) (15). However, HVS infection of other species of New World primates results in rapidly progressing fatal T-cell lymphomas and leukemias (14, 29). Besides its potent oncogenicity in vivo, HVS can immortalize peripheral blood mononuclear cells of human, rhesus monkey, and common marmoset to IL-2-independent growth in vitro (2, 5). Cell lines derived from peripheral blood mononuclear cells of common marmoset by in vitro immortalization with HVS represent a restricted lymphocyte subpopulation that is CD2+, CD8+, CD4−, and CD56+; thus, these cells are derived from suppressor/cytotoxic T lymphocytes (31). Sequence divergence at the left end of the viral genome defines three subgroups (A, B, and C) of HVS that differ with respect to their oncogenic potential. Subgroup A and C strains immortalize common marmoset T lymphocytes to IL-2-independent permanent cell growth in vitro, but none of the subgroup B strains tested had this ability (16, 50). Subgroup C strains are further capable of immortalizing human, rhesus monkey, and rabbit T lymphocytes into continuously proliferating T-cell lines and also of inducing lymphoma in rhesus monkeys and rabbits (2, 5).
Herpesvirus persists in its host through an ability to establish a latent infection and periodically reactivates to produce infectious virus. To do this, lymphotropic gammaherpesviruses, particularly Epstein-Barr virus, KSHV, and HVS, have been shown to deregulate lymphocyte signal transduction (8, 27, 34). Two HVS proteins, ORF14 and Tip, have been previously found to target and alter TCR signal transduction. HVS ORF14 protein, which shows a high homology to the mouse mammalian tumor virus (MMTV) superantigen (vSag), is detected primarily during lytic replication, is N glycosylated, and is secreted into supernatant. Like MMTV vSag, ORF14 is able to induce the proliferation of CD4-positive human T cells through an interaction with major histocompatibility complex (MHC) class II molecules (17). However, unlike MMTV vSag, which activates T cells in a TCR Vβ-dependent manner, ORF14 induces the polyclonal proliferation of T cells independent of the TCR Vβ subtype (17, 32). Mutational analysis has shown that ORF14 is not required for viral replication but is required for in vitro and in vivo viral oncogenesis and persistent infection in animals (32). A second HVS protein, called Tip (tyrosine kinase-interacting protein), is expressed primarily during viral latency. As seen with ORF14, Tip is not required for viral replication but is required for T-cell transformation in culture and for lymphoma induction in primates (17). As a lipid raft resident protein, Tip interacts with the SH3 domain of Lck tyrosine kinase, and this interaction interferes with early events in the TCR signal transduction pathway (3, 27, 35). Furthermore, Tip interacts with a novel cellular endosomal protein, p80, which contains an amino-terminal WD repeat domain and a carboxy-terminal coiled-coil domain (39, 40). Interaction of Tip with p80 facilitates endosomal vesicle formation and subsequent recruitment of Lck and TCR into the endosomes for degradation. Thus, T-lymphotropic HVS has evolved elaborate mechanisms to deregulate TCR signal transduction at multiple points in the viral life cycle. These mechanisms are likely necessary for the establishment and maintenance of persistent infection and for the induction of disease in the infected host.
We report here the identification of a novel viral signaling protein, ORF5, of HVS that contains an amino-terminal myristoylation for membrane attachment and six SH2 binding motifs for signal transduction. Tyrosine-phosphorylated ORF5 effectively interacted with cellular SH2-containing signaling proteins Lck, Fyn, SLP-76, and p85 through its tyrosine residues, and these interactions strongly enhanced TCR signal transduction activity. These results demonstrate a novel viral strategy: HVS harbors several viral proteins that activate or inactivate TCR signal transduction depending on the stage of viral life cycle.
MATERIALS AND METHODS
DNA sequencing.
The region spanning the entire ORF5 gene of HVS subgroup A strain A11 and subgroup C strain C488 was used to design primers for PCR of ORF5 (GenBank accession no. AJ131936): forward 5′ EcoRI CGC GAA TTC TGT TAG ATG TGG ATG CTT TGA GAG C, corresponding to nucleotides (nt) 12583 to 12549 of A11 and nt 12339 to 12285 of C488, and reverse 5′ XhoI CGC CTC GAG CAC AAT TTC TTC ATA AAT ATG, corresponding to nt 11722 to 11742 of C488. PCR products were then cloned into the EcoRI/XhoI sites of pSP72. Sequencing was performed with the M13 forward sequencing primer and an ABI PRISM 377 automatic DNA sequencer.
RNA extraction and reverse transcriptase PCR (RT-PCR).
One microgram of total RNA was used for the synthesis of first-strand cDNA with a kit for 3′ rapid amplification of cDNA ends (Invitrogen, San Diego, Calif.) according to the manufacturer's instructions. Aliquots of cDNA samples were then PCR amplified with a combination of forward ORF5 gene-specific primers and a reverse universal adaptor primer provided with the kit. The amplified PCR products were separated on 2% agarose gels, visualized by ethidium bromide staining, purified with a QiaEx gel purification kit (Qiagen, Chatsworth, Calif.), cloned into pCRII.1 TOPO (Invitrogen), and sequenced with T7 sequencing primer.
Plasmids.
pTracerEF.GFP (Invitrogen) and pEF.IRES.puro were used for transient and stable expression of ORF5, respectively. In addition, the ORF5-coding sequence was cloned into the EcoRI/XhoI sites of pGEX4T-1 (Clontech, Palo Alto, Calif.). The Myc-tagged Lck and Fyn, Flag-tagged SLP-76, and Myc-tagged LAT were kindly provided by Sabine Lang, Gary Koretzky, and Lawrence Samelson. In vitro site-directed mutagenesis was carried out to construct ORF5 tyrosine-to-phenylalanine (YF) and glycine-to-alanine (G2A) mutants. Each ORF5 mutant was completely sequenced to verify the presence of the mutation and the absence of any other changes.
Cell lines and antibodies.
Cultures of 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Jurkat T cells were grown in RPMI 1640 supplemented with 10% fetal bovine serum. Jurkat T E6.1 cells (55), and J.CaM1 (49), JCaM2.5 (21, 22), J14-v-29 (59), and P116 (56) cells, which have deficient expression of Lck, LAT, SLP-76, and ZAP70, respectively, were used in this study. J.CaM1 and P116 cells were purchased from the American Type Culture Collection (Manassas, Va.). J.CaM2.5, J14-v-29, and LAT-reconstituted LAT#3 cells were kindly provided by A. Weiss (University of California, San Francisco). Polyclonal antibodies used in this study were anti-Fyn, anti-ZAP70, anti-PLC-γ1 and anti-LAT (Santa Cruz Biotechnology, Santa Cruz, Calif.). Monoclonal antibodies were anti-CD3 magnetic beads and anti-CD3/CD28 magnetic beads (Dynal, Lake Success, N.Y.); anti-CD3 (PharMingen, San Diego, Calif.); anti-CD3ζ, anti-p85 subunit of phosphatidylinositol 3-kinase (PI3 kinase), and anti-c-Myc (Santa Cruz Biotechnology); anti-Lck (Transduction Laboratories, Lexington, Ky.); anti-V5 (Invitrogen); antiphosphotyrosine antibody 4G10 (Upstate Biotechnology, Lake Placid, N.Y.); and anti-CT-B antibody (Sigma, St. Louis, Mo.).
Transfection and luciferase assay.
Calcium phosphate (Clontech) was used for transient expression in 293T cells. Electroporation at 260 V and 975 μF was used for transient expression in Jurkat T cells. Stable Jurkat T-cell lines expressing ORF5 were selected and maintained in the presence of puromycin (5 μg/ml).
For the luciferase reporter assay, Jurkat T cells were electroporated with 20 μg of luciferase reporter plasmid, 2 μg of β-galactosidase plasmid, and the indicated amounts of pTracerEF.GFP and pTracerEF.GFP-ORF5 to a total amount of 50 μg of DNA per transfection. At 36 h posttransfection, cells were washed twice with phosphate-buffered saline (PBS), lysed, and analyzed by luciferase assay (Promega, Madison, Wis.). Luciferase activity was normalized to β-galactosidase activity and presented as fold induction compared with the control.
Protein purification and mass spectrometry.
To identify ORF5 binding proteins, 10 liters of Jurkat T cells was lysed with lysis buffer (20 mM HEPES [pH 7.4], 100 mM NaCl, 1% NP-40) containing protease and phosphatase inhibitors. After centrifugation, the supernatant was filtered through a 0.45-μm-pore-size syringe filter and precleared by mixing with glutathione S-transferase (GST)-bound glutathione beads twice. Precleared lysates were mixed with glutathione beads containing GST, GST-ORF5 fusion protein, or tyrosine-phosphorylated GST-ORF5 (GST-pORF5) protein for 4 h. Afterwards, the beads were extensively washed with lysis buffer. Proteins bound to the beads were eluted, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and subjected to peptide sequencing at the Harvard Mass Spectrometry Facility.
Metabolic labeling.
At 24 h posttransfection, 293T cells were incubated with Dulbecco's modified Eagle's medium containing 5% dialyzed fetal bovine serum and [3H]myristic acid (100 μCi/ml) overnight. The cells were then washed once with 1× PBS, lysed in ice-cold radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.5], 200 mM NaCl, 1% TritonX-100, 0.5% Na-deoxycholate, 0.1% SDS, and proteinase inhibitors), and immunoprecipitated with an anti-V5 or anti-Myc antibody.
GST pull-down assays.
GST fusion proteins were purified from either the Escherichia coli Top10 or the TKX1 strain, which contains a mammalian elk tyrosine kinase expression vector (Stratagene, La Jolla, Calif.). Jurkat T-cell lysates were incubated with glutathione beads containing GST fusion protein in binding buffer (20 mM HEPES [pH 7.4], 100 mM NaCl, 1% NP-40, and protease inhibitors) at 4°C for 2 h. The glutathione beads were then washed four times with binding buffer, and the proteins associated with the beads were analyzed by SDS-PAGE and subjected to immunoblot assay with a phosphorimager (BAS-1500; Fuji Film Co., Tokyo, Japan).
In vitro kinase assay.
The immune complex kinase mixture was resuspended with 20 μl of kinase buffer (20 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 2 mM dithiothreitol, and 10 μCi of [γ-32P]ATP) and incubated for 20 min at room temperature. The reaction was performed in the absence and presence of the Src substrate enolase (Sigma). The kinase reaction was stopped by adding SDS sample loading buffer, and the mixture was subjected to Nu-PAGE (4 to 12% gradient gel; Invitrogen) in MES (morpholinepropanesulfonic acid) buffer for 40 min. The dried gel was autoradiographed with a Fuji phosphorimager.
Cell stimulation, immunoprecipitation, and immunoblotting.
Jurkat T cells (5 × 106) were stimulated with anti-human CD3 magnetic beads at 37°C for the indicated times. Stimulation was stopped by the addition of an equal volume of ice-cold 2× 1% NP-40 lysis buffer. Cell lysates were then precleared by rocking at 4°C for 1 to 2 h in the presence of protein A/G-agarose (Santa Cruz Biotechnology). The precleared cell lysates were incubated with the relevant antibodies for 2 h, followed by protein A/G-agarose for 2 h. Immunoprecipitates were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Amersham Pharmacia Biotech, Chicago, Ill.). Membranes were reacted with appropriate antibodies in 5% skim milk-PBS-Tween 20 (or 3% bovine serum albumin-PBS-Tween 20 for phosphotyrosine blots). The protein was visualized with chemiluminescent detection reagents (Pierce, Rockford, Ill.) and detected with a Fuji phosphorimager.
Calcium mobilization analysis.
Cells (2 × 106) were loaded with 1 μM indo-1 (Molecular Probes, Eugene, Oreg.) in 200 μl of RPMI complete medium for 30 min at 37°C, washed once with complete medium, resuspended in 1 ml of RPMI complete medium, and then put on ice until analyzed. Baseline calcium levels were established for 1 min prior to the addition of the antibody. Cells were stimulated with 10 μg of anti-human CD3 antibody (PharMingen) per ml. Baseline absolute intracellular calcium levels were determined by using ionophore and EGTA. Data were collected and analyzed on a FACS Vantage instrument (Becton Dickinson, Mountain View, Calif.).
CD69 surface expression.
Jurkat T cells were transfected with pTracerEF.GFP or pTracerEF.GFP-ORF5. At 24 h postelectroporation, Ficoll-Histopaque was used to remove dead cells. The live cells were stimulated with anti-CD3 Dynabeads or phorbol myristate acetate (PMA) (50 ng/ml), or were given medium alone as a control, for an additional 16 h at 37°C and then were fixed with 1% paraformaldehyde. Fluorescence-activated cell sorter analysis was performed with a FACS Scan (Becton Dickinson) with phycoerythrin-conjugated mouse anti-human CD69 antibody (PharMingen).
Immunofluorescence.
Cells were fixed with 2% paraformaldehyde for 15 min, permeabilized with cold acetone for 15 min, blocked with 10% goat serum in PBS for 30 min, and reacted with 1:5,000 dilutions of primary antibodies in PBS for 30 min at room temperature. After incubation, cells were washed extensively with PBS, incubated with 1:100 Alexa 488-conjugated goat anti-mouse antibody (Molecular Probes, Eugene, Oreg.) in PBS for 30 min at room temperature, and washed three times with PBS. Nuclear staining was performed for 1 min with Lo-Pro 3 (Molecular Probes) diluted 1:50,000. Confocal microscopy was performed with a TCS SP laser-scanning microscope (Leica Microsystems, Exton, Pa.) fitted with a 40× Leica objective (PL APO, 1.4NA), using Leica imaging software. Images were collected at a 512- by 512-pixel resolution. The stained cells were optically sectioned in the z axis, and the images in the different channels (photomultiplier tubes) were collected simultaneously. The step size in the z axis varied from 0.2 to 0.5 μm to obtain 30 to 50 slices per imaged file. The images were transferred to a Macintosh G4 computer (Apple Computer, Cupertino, Calif.), and National Institutes of Health Image version 1.61 software was used to render the images.
Measurements of secreted IL-2.
Cells (106) were cultured in 24-well tissue culture plates with or without anti-CD3 and anti-CD28 Dynabeads in 1 ml of RPMI1640 complete medium. The supernatants were harvested 8 h later, and IL-2 production was measured with an enzyme-linked immunosorbent assay kit (PharMingen).
Isolation of lipid rafts.
Lipid rafts were isolated by floatation on discontinuous sucrose gradients (6, 7). Briefly, 293T cells (five 10-cm-diameter dishes) were washed with ice-cold PBS and lysed for 30 min on ice in 1% Triton X-100 in TNEV (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA) containing phosphatase inhibitors and protease inhibitor cocktail (Roche, Indianapolis, Ind.). The lysate was further homogenized with 10 strokes in a Wheaton loose-fitting Dounce homogenizer. Nuclei and cellular debris were pelleted by centrifugation at 900 × g for 10 min. For the discontinuous sucrose gradient, 1 ml of cleared cell lysates was mixed with 1 ml of 85% sucrose in TNEV and transferred to the bottom of a Beckman 14- by 89-mm centrifuge tube. The diluted lysate was overlaid with 6 ml of 35% sucrose in TNEV and finally with 3.5 ml of 5% sucrose in TNEV. The samples were then centrifuged in an SW41 rotor at 200,000 × g for 20 h at 4°C, and 1-ml fractions were collected from the top of the gradient.
RESULTS
Sequence analysis of HVS ORF5 isolates.
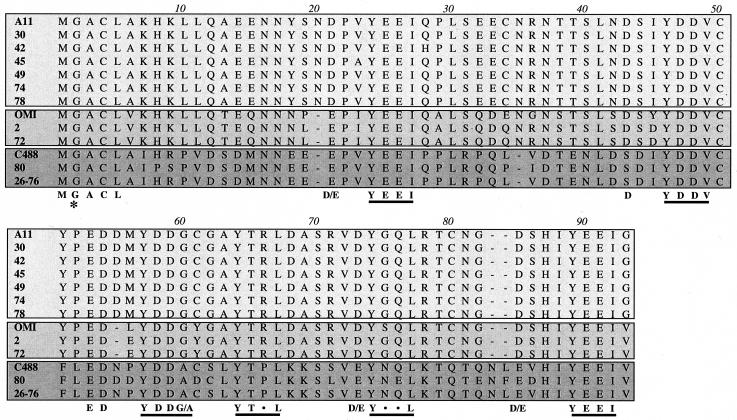
The ORF5 genes of HVS strains A11 and strain C488 were originally predicted to encode 81 and 72 amino acids, respectively (1, 18). However, the extensive sequence variation at the amino terminus suggested potential splicing of the ORF5-coding sequence. To test this hypothesis, we performed RT-PCR with ORF5 gene-specific primers by using the purified mRNA from HVS-infected OMK cells. This showed that HVS strain A11 ORF5 was spliced from nt 12329 to 12255 and that HVS strain C488 ORF5 was spliced from nt 12054 to 11979. As a result, both sequences encoded an ORF5 protein of 89 to 91 amino acids (Fig. 1). We determined the primary amino acid sequences of ORF5 mRNAs from 11 additional HVS subgroup A and C isolates. All ORF5 alleles are predicted to encode 89 to 91 amino acids (Fig. 1).
FIG. 1.
Sequence analysis of ORF5 alleles of HVS. Amino acid sequences of ORF5 proteins from 13 different isolates of HVS were aligned by using the Cluster W program. Sequences are grouped into three clusters: the A11 cluster of HVS subgroup A (light gray), the OMI cluster of subgroup A (gray), and HVS subgroup C (dark gray). The putative myristoylation at the second glycine residue is indicated by asterisks, and the six potential SH2 binding motifs are underlined.
Amino acid substitutions, deletions, or insertions in ORF5 proteins from different isolates were compared with ORF5 from strains A11 and C488. This analysis showed that amino acid sequence identity of ORF5 among subgroup C isolates was 83 to 100% (Table 1). However, the ORF5 alleles of subgroup A isolates were further divided into two separate clusters: the ORF5 alleles of A11, 30, 42, 45, 49, 74, and 78 (the A11 cluster) showed 97 to 100% identity, and the ORF5 alleles of OMI, 2, and 72 (the OMI cluster) showed 93 to 100% identity (Table 1). Interestingly, the amino acid identity of ORF5 between the A11 cluster and OMI cluster was 77 to 80%, whereas the amino acid identity of ORF5 between subgroup A and subgroup C was only 46 to 50% (Table 1). This indicates that the ORF5 sequence is relatively conserved within each subgroup, whereas it shows a significant variation between subgroups.
TABLE 1.
Amino acid identity of ORF5 proteins among HVS isolates
| HVS strain | % Amino acid identity with ORF5 from straina:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A11 | 30 | 42 | 45 | 49 | 74 | 78 | OMI | 2 | 72 | C488 | 80 | 26-76 | |
| A11 | 100 | 100 | 98 | 98 | 100 | 100 | 100 | 78 | 80 | 80 | 50 | 50 | 50 |
| 30 | 100 | 98 | 98 | 100 | 100 | 100 | 78 | 80 | 80 | 50 | 50 | 50 | |
| 42 | 100 | 97 | 98 | 98 | 98 | 77 | 79 | 79 | 50 | 49 | 50 | ||
| 45 | 100 | 98 | 98 | 98 | 78 | 80 | 80 | 49 | 49 | 49 | |||
| 49 | 100 | 100 | 100 | 78 | 80 | 80 | 50 | 50 | 50 | ||||
| 74 | 100 | 100 | 78 | 80 | 80 | 50 | 50 | 50 | |||||
| 78 | 100 | 78 | 80 | 80 | 50 | 50 | 50 | ||||||
| OMI | 100 | 93 | 93 | 46 | 47 | 46 | |||||||
| 2 | 100 | 100 | 47 | 48 | 47 | ||||||||
| 72 | 100 | 47 | 48 | 47 | |||||||||
| C488 | 100 | 83 | 100 | ||||||||||
| 80 | 100 | 83 | |||||||||||
| 26-76 | 100 | ||||||||||||
Groups of boldface numbers, from left to right, indicate the A11 cluster of subgroup A, the OMI cluster of subgroup A, and subgroup C, respectively.
The presence of potential structural motifs was assessed based on primary amino acid sequence. Interestingly, full-length ORF5 contained the putative myristoylation site at the second glycine (G2) residue, which appeared to be strikingly conserved in all isolates (Fig. 1). Furthermore, despite extensive sequence variation throughout the open reading frame, ORF5 proteins of all isolates showed a complete conservation of six YxxI/L/V sequences preceded by negatively charged glutamic acid and/or aspartic acid residues; these motifs most closely match the consensus sequences for Src family kinase SH2 binding (ExxYxxI/L/V) (47, 48). Based on sequence conservation, both the myristoylation and SH2 binding motifs are predicted to have important roles in ORF5 function. Since HVS subgroup C strain 488 has been well characterized (28), the ORF5 protein from this strain was selected for the rest of study.
Amino-terminal myristoylation and lipid raft localization of ORF5.
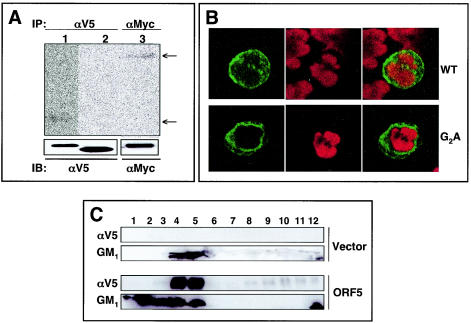
To test whether the G2 residue of ORF5 is the site for the addition of the myristate moiety, we constructed expression vectors containing the carboxy-terminal V5-tagged wild-type (wt) ORF5 and a glycine-to-alanine (G2A) ORF5 mutant. At 24 h posttransfection, 293T cells were metabolically labeled with [3H]myristate and immunoprecipitated with an anti-V5 antibody. The carboxy-terminal Myc-tagged Lck, which contains an amino-terminal myristoylation site, was included as a control. wt ORF5 and Lck were readily labeled with [3H]myristate, whereas ORF5 G2A was not (Fig. 2A). Interestingly, wt ORF5 migrated slightly slower in SDS-PAGE than did the G2A mutant, suggesting that the amino-terminal myristoylation slows the migration of ORF5.
FIG. 2.
Myristoylation and membrane and lipid raft localization of ORF5. (A) Myristoylation of ORF5. 293T cells transiently transfected with the V5-tagged wt ORF5 (lane 1), the ORF5 G2A mutant (lane 2), and Myc-tagged Lck (lane 3) were metabolically labeled with [3H]myristate overnight, lysed in radioimmunoprecipitation assay buffer, and immunoprecipitated (IP) with anti-V5 (αV5) or anti-Myc (αMyc) antibody. Immunoprecipitates were then analyzed by SDS-PAGE and fluorography (top) or by an αV5 and αMyc immunoblot (IB) (bottom). (B) Localization of ORF5. At 36 h postelectroporation with wt ORF5 or ORF5 G2A mutant expression vector, Jurkat T cells were fixed with 2% paraformaldehyde, permeabilized, and reacted with mouse monoclonal anti-V5 antibody followed by Alexa 488-conjugated goat anti-mouse antibody. The cells were then stained with Lo-Pro-3 nuclear dye. Cells were visualized by laser-scanning microscopy, and representative optical sections are presented. (C) ORF5 localization in lipid rafts. 293T cells were transfected with empty vector alone or V5-tagged ORF5 expression vector. At 48 h posttransfection, cells were lysed with 1% Triton X-100 and subjected to discontinuous centrifugation. Each fraction was subjected to immunoblot assay with anti-V5 antibody. HRP-conjugated CT-B (GM1) was used as a control to locate the lipid raft fractions.
Since ORF5 contained the myristoylation at its amino-terminal glycine residue, we compared the localization of wt ORF5 with that of the G2A mutant by using confocal immunofluorescence. At 48 h postelectroporation with wt ORF5 and the G2A mutant expression vector, human Jurkat T cells were fixed, permeabilized, and reacted with an anti-V5 antibody. wt ORF5 was localized primarily at the plasma membrane, whereas the G2A mutant was detected in the cytoplasm, with a dramatic reduction of plasma membrane localization (Fig. 2B). This result indicates that the amino-terminal myristoylation is necessary for the efficient localization of ORF5 at the plasma membrane.
The membrane lipid raft microdomain has been described as a functional platform for signaling of various stimuli and trafficking of signal receptor molecules (6, 7). To further investigate the potential lipid raft localization of ORF5, 293T cells were transfected with empty vector or ORF5 expression vector. At 48 h posttransfection, cells were lysed with 1% Triton X-100 and subjected to discontinuous centrifugation. Fractions 3-4-5, 6-7-8, and 9-10-11 were previously shown to contain the raft region, the interval region, and the soluble region of the gradients, respectively (39). Immunoblot assay showed that ORF5 was present primarily in lipid raft fractions (Fig. 2C). The position and integrity of the lipid rafts in the sucrose gradient were confirmed by the presence of the raft-associated GM1 ganglioside (Fig. 2C). These results collectively indicate that ORF5 is modified by the amino-terminal myristoylation and is constitutively present in lipid rafts.
Interaction of tyrosine-phosphorylated ORF5 with SH2-containing cellular proteins.
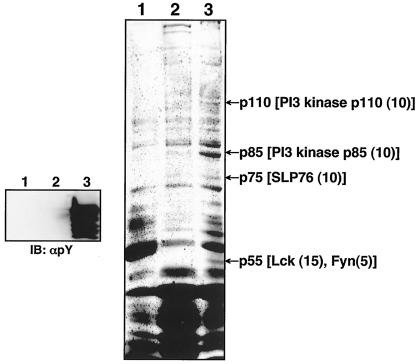
To identify cellular proteins interacting with ORF5, a GST pull down assay was performed with bacterial GST fusion proteins. Unphosphorylated GST-ORF5 was produced from the E. coli Top10 strain, and tyrosine-phosphorylated GST-ORF5 (GST-pORF5) was produced from the E. coli TKX1 strain, which contains the elk tyrosine kinase. The elk tyrosine kinase has broad specificity and efficiently phosphorylates many mammalian proteins in E. coli. We found that over 60% of GST-ORF5 protein purified from E. coli TKX1 was tyrosine phosphorylated by elk kinase (Fig. 3).
FIG. 3.
Identification of cellular proteins interacting with the tyrosine-phosphorylated ORF5. (Left) Glutathione-Sepharose beads containing GST (lane 1), GST-ORF5 (lane 2), or tyrosine-phosphorylated GST-ORF5 (GST-pORF5) (lane 3) were resolved by SDS-PAGE, followed by immunoblotting (IB) with an antiphosphotyrosine antibody (αpY). (Right) Glutathione-Sepharose beads containing GST (lane 1), GST-ORF5 (lane 2), or phosphorylated GST-ORF5 (GST-pORF5) (lane 3) were mixed with lysates of Jurkat T cells. ORF5 binding proteins were resolved by SDS-PAGE and visualized by Coomassie blue staining. Arrows indicate the PI3 kinase catalytic subunit (p110), PI3 kinase regulatory subunit (p85), SLP-76 (p75), Lck, and Fyn (p55). Numbers in parentheses indicate the frequency of peptides of each cellular protein obtained from mass spectrometry.
In an effort to identify cellular proteins interacting with ORF5, we performed bulk purification with over 10 liters of Jurkat T-cell pellets with GST, GST-ORF5, or GST-pORF5 fusion protein. Several cellular proteins specifically interacted with GST-pORF5 protein but not with GST and GST-ORF5 protein (Fig. 3). Mass spectrometry revealed that these cellular proteins were Lck, Fyn, SLP-76, and the PI3 kinase regulatory subunit (p85) and catalytic subunit (p110) (Fig. 3). Interestingly, all of these cellular proteins except p110 contain an SH2 domain. It seems likely that Lck, Fyn, SLP-76, and p85 interact with the phosphorylated ORF5 through their SH2 domains, whereas p110 interacts through its association with the p85 regulatory subunit.
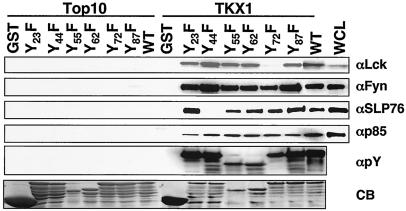
Identification of the tyrosine residues of ORF5 that are responsible for binding to cellular proteins.
To define the tyrosine residues of ORF5 that are responsible for binding to cellular proteins, each tyrosine residue of ORF5 was mutated to phenylalanine to generate Y23F, Y44F, Y55F, Y62F, Y72F, and Y87F. These GST-ORF5 mutant fusion proteins were purified from the Top10 or TKX1 bacterial strain and used for in vitro binding assays as described for Fig. 3. While the tyrosine-phosphorylated fusion proteins containing wt, Y23F, Y44F, Y55F, Y62F, and Y87F bound to Lck at similar levels, the tyrosine-phosphorylated GST-ORF5 Y72F fusion did not do so under the same conditions, indicating that phosphorylated Y72 is the primary site for Lck interaction (Fig. 4). In addition, the tyrosine-phosphorylated GST-ORF5 Y44F fusion was not able to bind to SLP-76, while all other tyrosine-phosphorylated GST-ORF5 fusion proteins efficiently bound to SLP-76, indicating that phosphorylated Y44 is the principal site for SLP-76 interaction (Fig. 4). In contrast, none of the Y-F mutations of ORF5 abolished the interaction with Fyn and p85, suggesting that Fyn and p85 likely bind to multiple tyrosine residues of ORF5 (Fig. 4). Finally, ORF5 binding to these cellular proteins appeared to be specific, because these interactions were detected only with the tyrosine-phosphorylated GST-ORF5 proteins and not with the unphosphorylated GST-ORF5 proteins (Fig. 4). These results indicate that ORF5 efficiently binds to cellular SH2-containing Lck, Fyn, p85, and SLP-76 signaling molecules through its phosphorylated tyrosine residues.
FIG. 4.
Identification of tyrosine residues of ORF5 responsible for binding to cellular proteins. GST and GST-ORF5 mutant fusion proteins purified from either E. coli strain Top10 or TKX1 were mixed with Jurkat T-cell lysates. Polypeptides associated with GST and GST-ORF5 fusion proteins were separated by SDS-PAGE and subjected to immunoblotting with anti-Lck, anti-Fyn, and anti-p85 antibodies. Whole-cell lysate (WCL) obtained from 293T cells transfected with a Flag-tagged SLP-76 expression vector and anti-Flag antibody were used for this assay. WCL was included as a control. GST and GST-ORF5 mutant fusion proteins purified from either E. coli strain Top10 or TKX1 were resolved by SDS-PAGE and subjected to immunoblotting with an antiphosphotyrosine (αpY) antibody and also stained with Coomassie blue (CB).
ORF5 interaction with Src family PTKs.
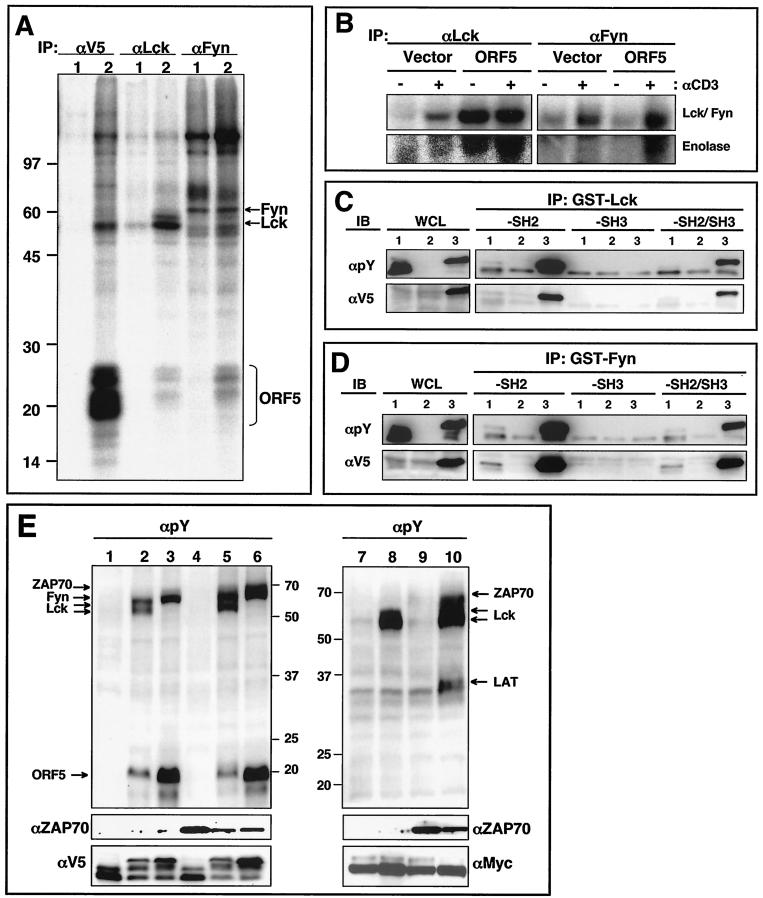
We next examined the interaction of ORF5 with the Src family PTKs Lck and Fyn by performing in vitro immune complex kinase assays. To generate cells stably expressing ORF5, pEF.IRES.puro expression vector containing the V5-tagged ORF5 gene was electroporated into human Jurkat T cells, followed by selection for puromycin resistance. ORF5 protein was readily detected in puromycin-resistant Jurkat-ORF5 cells by immunoblotting with an anti-V5 antibody. Lysates of Jurkat-vector and Jukat-ORF5 cells were used for immunoprecipitation with anti-V5, anti-Lck, or anti-Fyn antibodies, followed by in vitro kinase reaction with [γ-32P]ATP. In vitro kinase reaction with V5 immunoprecipitates revealed that ORF5 protein was readily phosphorylated by associated kinases (Fig. 5A). In addition to the 20- to 25-kDa ORF5 protein, cellular proteins of 55, 85, 110, and 140 kDa were present in the ORF5 immune complex. In vitro kinase reactions with Lck and Fyn immune complexes also showed that both contained 20- to 25-kDa bands that comigrated with ORF5 (Fig. 5A). Furthermore, Lck kinase activity was greatly enhanced in Jurkat-ORF5 cells compared with that in Jurkat-vector cells. To further delineate the ORF5 effect on Lck and Fyn kinase activities, Jurkat-vector and Jurkat-ORF5 cells were unstimulated or stimulated with an anti-CD3 antibody, followed by immunoprecipitation with anti-Lck and anti-Fyn antibodies and in vitro kinase reaction. Enolase was also included in the in vitro kinase reaction as a substrate. The results of autophosphorylation of Lck and Fyn and transphosphorylation of enolase showed that Jurkat-ORF5 cells contained the enhanced kinase activities of Lck and Fyn compared to Jurkat-vector cells in the absence of anti-CD3 stimulation, whereas Lck and Fyn kinase activities were further increased in Jurkat-ORF5 cells upon anti-CD3 stimulation (Fig. 5B). These results suggest that Lck and Fyn interact with and efficiently phosphorylate ORF5 protein in living cells and that Lck and Fyn kinase activities are likely activated by ORF5 interaction.
FIG. 5.
Interaction of ORF5 with Src family kinases. (A) In vitro immune complex kinase reactions. Jurkat-vector (lanes 1) and Jurkat-ORF5 (lanes 2) cell lysates were immunoprecipitated (IP) with anti-V5 (αV5), anti-Lck, and anti-Fyn antibodies. Subsequently, immunoprecipitates were subjected to an in vitro immune complex kinase assay with [γ-32P]ATP. Arrows indicate Fyn, Lck, and ORF5 proteins. Molecular weight markers (in thousands) are indicated at the left. (B) In vitro Lck and Fyn kinase reactions. Jurkat-vector and Jurkat-ORF5 cells were unstimulated (−) or stimulated (+) with an anti-CD3 antibody for 3 min, and cell lysates were then immunoprecipitated with anti-Lck and anti-Fyn antibodies. Subsequently, immunoprecipitates were subjected to an in vitro immune complex kinase assay with [γ-32P]ATP and 5 μg of enolase. (C) In vitro interaction of tyrosine-phosphorylated ORF5 with the SH2 domain of Lck kinase. 293T cells were transfected with the V5-tagged ORF5 expression vector (lanes 1), Myc-tagged Lck expression vector (lanes 2), or V5-tagged ORF5 and Myc-tagged Lck expression vector (lanes 3). Whole-cell lysates (WCL) were used for immunoblotting (IB) with anti-V5 or antiphosphotyrosine (αpY) antibody to show ORF5 expression and its tyrosine phosphorylation. The same WCL were then used for binding assay with Sepharose bead-conjugated Lck-SH2, Lck-SH3, or Lck-SH2/SH3 followed by immunoblotting with either an anti-V5 or antiphosphotyrosine antibody. (D) In vitro interaction of tyrosine-phosphorylated ORF5 with the SH2 domain of Fyn kinase. Samples and procedures were as described for panel C, except for the use of Fyn in place of Lck kinase. (E) Lck and Fyn but not ZAP70 phosphorylate ORF5. 293T cells were transfected with the V5-tagged ORF5, Myc-tagged LAT, Lck, Fyn, and ZAP70 expression vector in various combinations. At 48 h posttransfection, cell lysates were used for immunoblotting with an antiphosphotyrosine antibody. The bottom panels show the expression levels of ZAP70, ORF5, and LAT. Arrows indicate Lck, Fyn, ZAP70, LAT, and ORF5. Lane 1, ORF5; lane 2, Lck plus ORF5; lane 3, Fyn plus ORF5; lane 4, ZAP70 plus ORF5; lane 5, Lck plus ZAP70 plus ORF5; lane 6, Fyn plus ZAP70 plus ORF5; lane 7, LAT; lane 8, Lck plus LAT; lane 9, ZAP70 plus LAT; lane 10, Lck plus ZAP70 plus LAT. Numbers on the left are molecular weights in thousands.
To further demonstrate that ORF5 is a target and interacting protein for Lck and Fyn kinases, 293T cells were transfected with the V5-tagged ORF5 expression vector alone or together with Lck or Fyn expression vector. At 48 h posttransfection, cell lysates were used for immunoblotting with antiphosphotyrosine or anti-V5 antibodies or for in vitro binding assay with GST-Lck-SH2/SH3 or GST-Fyn-SH2/SH3 fusion proteins. Immunoblotting showed that ORF5 was effectively phosphorylated by Lck and Fyn kinases and that the tyrosine-phosphorylated form of ORF5 migrated more slowly in SDS-PAGE than the unphosphorylated form of ORF5 (Fig. 5C and D, lanes 3). In vitro binding assays showed that the SH2 domains of Lck and Fyn strongly bound to tyrosine-phosphorylated ORF5 protein but did not bind to unphosphorylated ORF5 protein (Fig. 5C and D). These results demonstrate that ORF5 is readily phosphorylated by Lck and Fyn in living cells and that the tyrosine-phosphorylated ORF5 in turn interacts with the SH2 domains of Lck and Fyn.
To investigate whether the phosphorylation of ORF5 by Lck and Fyn was specific, 293T cells were cotransfected with the V5-tagged ORF5, Lck, Fyn, and ZAP70 in various combinations. In addition, ORF5 was replaced with the Myc-tagged LAT as a control for ZAP70-mediated tyrosine phosphorylation. At 48 h posttransfection, whole-cell lysates were used for immunoblotting with antiphosphotyrosine, anti-ZAP70, anti-V5, and anti-Myc antibodies. This showed that ORF5 tyrosine phosphorylation was readily detected in cells expressing Lck or Fyn, whereas it was not detected in cells expressing ZAP70 (Fig. 5E, lanes 2, 3, and 4). Furthermore, ZAP70 expression did not enhance the Lck- or Fyn-mediated tyrosine phosphorylation of ORF5 (Fig. 5E, lanes 5 and 6). Finally, the migration of ORF5 was significantly retarded due to its tyrosine phosphorylation in cells expressing Lck or Fyn but not in cells expressing ZAP70 (Fig. 5E). By contrast, the tyrosine phosphorylation of LAT was apparent in cells expressing Lck and ZAP70, suggesting that, as previously shown (61), Lck activates ZAP70, which in turn phosphorylates LAT (Fig. 5E, lane 10). These results indicate that unlike cellular LAT, ORF5 is effectively tyrosine phosphorylated by Lck and Fyn kinases but not by ZAP70 kinase.
Induction of TCR-mediated tyrosine phosphorylation of cellular proteins by ORF5.
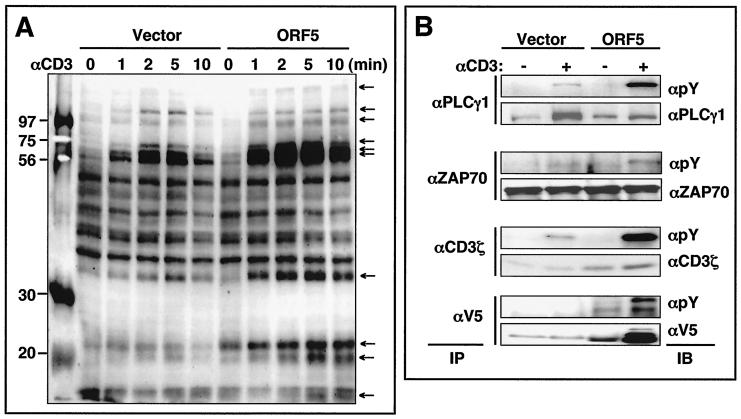
An early signaling event following TCR engagement is the increase in tyrosine phosphorylation of cellular proteins (25). To examine the effect of ORF5 on TCR signal transduction, Jurkat-vector and Jurkat-ORF5 cells were stimulated with an anti-CD3 antibody for various times, and whole-cell lysates were used for immunoblotting with an antiphosphotyrosine antibody. No significant difference in TCR or CD3 surface expression between Jurkat-vector and Jurkat-ORF5 cells was detected (data not shown). However, immunoblotting revealed that while the tyrosine phosphorylation of numerous cellular proteins was rapidly induced in both Jurkat-vector and Jurkat-ORF5 cells upon TCR stimulation, the level of tyrosine phosphorylation was much higher in Jurkat-ORF5 cells (Fig. 6A). Particularly, the levels and rates of tyrosine phosphorylation of specific cellular proteins (15, 19, 22, 32, 53, 55, 70, 85, 110, and 140 kDa) were significantly higher in Jurkat-ORF5 cells than in Jurkat-vector cells. To further demonstrate the increase of tyrosine phosphorylation of cellular proteins in Jurkat-ORF5 cells upon TCR stimulation, PLC-γ1, ZAP70, and CD3ζ proteins were immunoprecipitated with their specific antibodies and reacted with an antiphosphotyrosine antibody. The tyrosine phosphorylation of PLC-γ1, ZAP70, and CD3ζ proteins was considerably higher in Jurkat-ORF5 cells than in Jurkat-vector cells upon TCR stimulation (Fig. 6B). Furthermore, the tyrosine phosphorylation of ORF5 was also detected after TCR stimulation. These results demonstrate that ORF5 expression enhances cellular tyrosine phosphorylation induced by TCR signal transduction.
FIG. 6.
TCR-mediated tyrosine phosphorylation of cellular proteins. (A) Jurkat-vector and Jurkat-ORF5 cells were stimulated with anti-CD3 (αCD3) Dynabeads at 37°C for the indicated times. Equivalent amounts of whole-cell lysates (A) were then separated by SDS-PAGE and immunoblotting with an antiphosphotyrosine antibody. Molecular weight markers (in thousands) are indicated to the left, and the arrows indicate the tyrosine-phosphorylated proteins. (B) The same whole-cell lysates were then used for immunoprecipitation (IP) with anti-PLC-γ1, anti-ZAP70, anti-CD3ζ, and anti-V5 antibodies. After extensive washing, immunoprecipitates were separated by SDS-PAGE and analyzed by immunoblotting (IB) with an antiphosphotyrosine (αpY) antibody or an antibody specific for PLC-γ1, ZAP70, CD3ζ, or V5-ORF5.
Upregulation of TCR signal transduction by ORF5.
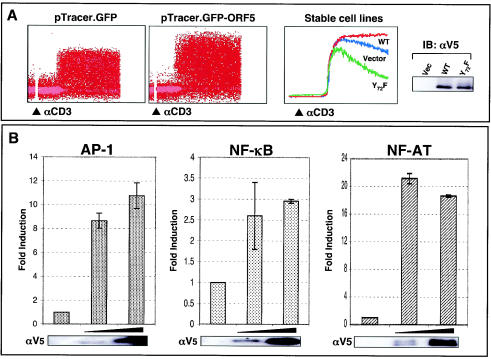
Upon engagement with the MHC complexes on the antigen-presenting cells or target cells, TCR initiates an array of signal transduction events: rapid tyrosine phosphorylation of cellular proteins, increased intracellular free calcium, activation of cellular transcription factor activity, and, ultimately, production of IL-2 (33). To further examine the effect of ORF5 on TCR-induced intracellular free calcium mobilization, Jurkat T cells containing vector or wt ORF5 were treated with an anti-CD3 antibody and monitored by flow cytometry for the level of intracellular free calcium. To demonstrate the specificity of ORF5 activity in TCR signal transduction, Jurkat T cells stably expressing the ORF5 Y72F mutant lacking Lck binding were constructed and included in this assay. Control Jurkat-vector cells exhibited a rapid increase in intracellular calcium concentration immediately after anti-CD3 antibody stimulation, whereas Jurkat T cells transiently or stably expressing ORF5 showed a significantly enhanced level of intracellular calcium under the same conditions (Fig. 7A). In striking contrast, Jurkat T cells stably expressing the ORF5 Y72F mutant showed a reduced level of intracellular calcium compared to control Jurkat-vector and Jurkat-ORF5 cells (Fig. 7A). The ORF5 Y72F mutant was expressed at a level equivalent to that for wt ORF (Fig. 7A, right panel). This indicates that, as seen with tyrosine phosphorylation, TCR-mediated intracellular calcium mobilization is also greatly enhanced by ORF5 and that ORF5 interaction with Lck is necessary for this activity.
FIG. 7.
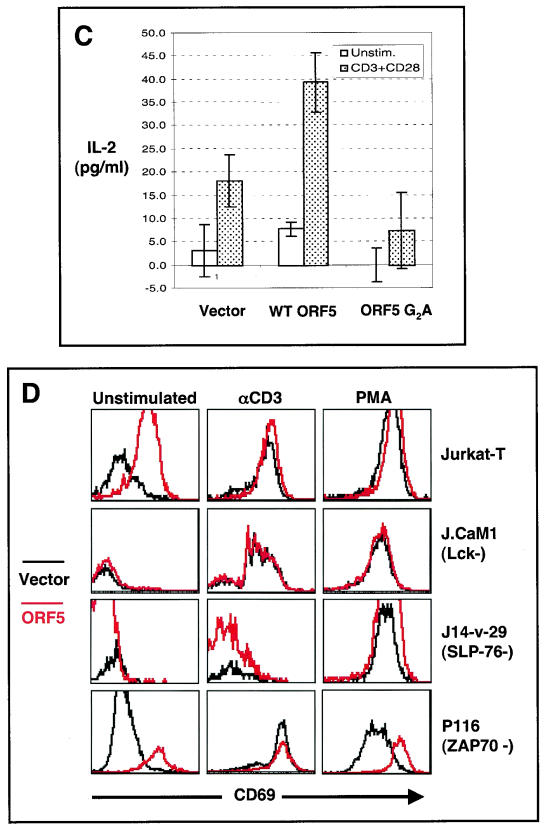
Enhancement of TCR signal transduction by ORF5. (A) Intracellular calcium mobilization. Jurkat T cells electroporated with pTracerEF.GFP or pTracerEF.GFP-ORF5 (left two panels) and stable Jurkat-vector, Jurat-ORF5, or Jurkat-ORF5 Y72F cell lines (middle panel) were stimulated with an anti-CD3 (αCD3) antibody. Calcium mobilization of GFP-positive cell populations was monitored over time by changes in the ratio of violet to blue (405 to 485 nm) fluorescence. Data for calcium mobilization in the left two panels are presented as a histogram of the number of cells with a particular fluorescence ratio (y axis) versus time (x axis). Data in the middle panel are presented as linear lines of the averaged number of responding cells. The breaks in the graphs indicate the time intervals during the addition of CD3 antibody. Data were similar in two independent experiments. wt ORF5 and ORF5 Y72F mutant expressions in Jurkat T cells were determined by immunoblotting (IB) with an anti-V5 antibody (right panel). (B) Activation of AP-1, NF-κB, and NF-AT transcription factor activity by ORF5. Jurkat T cells were electroporated with 20 μg of plasmids containing the luciferase reporter vector containing an AP-1-, NF-κB-, or NF-AT-dependent promoter sequence with increasing amounts of ORF5 expression vector. At 36 h posttransfection, the level of luciferase activity was measured by using cell lysates. Luciferase activity was normalized to β-galactosidase activity for transfection efficiency. Shown is the fold induction obtained with vector, based on the average and standard error from two independent experiments. The same cell lysates were used for immunoblot analysis to confirm the expression of ORF5 (bottom panels). (C) IL-2 production. At 36 h postelectroporation with vector, ORF5 expression vector, or ORF5 G2A mutant expression vector, Jurkat T cells were stimulated with or without anti-CD3-CD28 Dynabeads for an additional 8 h. The supernatants were used to assay IL-2 production by enzyme-linked immunosorbent assay. (D) Surface expression of CD69. At 24 h postelectroporation with pTracerEF.GFP or pTracerEF.GFP-ORF5, Jurkat T, Lck-deficient J.CaM1, SLP76-deficient J14-v-76, or ZAP70-deficient P116 cells were unstimulated or stimulated with either anti-CD3 Dynabeads or PMA (50 ng/ml) for an additional 16 h. GFP-positive cell populations were then gated and examined for CD69 surface expression by flow cytometry.
Furthermore, we examined whether ORF5 expression induced the activation of cellular AP-1, NF-κB, and NF-AT transcription factors. At 36 h postelectroporation with each transcription factor luciferase reporter construct cotransfected with increasing amounts of ORF5 expression vector, lysates of Jurkat T cells were used to measure luciferase activity. AP-1 and NF-AT transcription factor activity was drastically activated by ORF5 expression, whereas NF-κB transcription factor activity was only weakly induced under the same conditions (Fig. 7B).
Finally, the production of IL-2 cytokine and the surface expression of the CD69 early lymphocyte activation marker, which are the consequences of TCR signal transduction (13), were measured in Jurkat T cells electroporated with empty vector or ORF5 expression vector. At 36 h posttransfection, these cells were costimulated with or without anti-CD3 and anti-CD28 antibody for 8 h. The ORF5 G2A mutant was included in this assay as a control. IL-2 production was observed in vector-transfected Jurkat T cells only after CD3-CD28 costimulation (Fig. 7C). In ORF5-expressing cells, IL-2 production was detected in the absence of CD3-CD28 costimulation, whereas it was considerably enhanced in the presence of CD3-CD28 costimulation (Fig. 7C). In contrast, ORF5 G2A-expressing cells without CD3-CD28 costimulation did not produce IL-2 cytokine, whereas with CD3-CD28 costimulation they did produce IL-2, but at a lower level than control Jurkat T cells and Jurkat T cells expressing wt ORF5 (Fig. 7C).
In concert with IL-2 production, ORF5 expression also upregulated the surface expression of CD69 activation antigen in Jurkat T cells regardless of TCR stimulation (Fig. 7D). To further explore the upregulation of CD69 surface expression by ORF5, we expressed ORF5 in Lck-deficient J.CaM1 cells, SLP-76-defient J14-v-29 cells, and ZAP70-deficient P116 cells, which were derived from Jurkat T cells (49, 56, 59). In contrast to its activity in parental Jurkat T cells, ORF5 expression was incapable of inducing the upregulation of CD69 surface expression in Lck-deficient J.CaM1 cells and SLP-76-defient J14-v-29 cells, indicating that Lck and SLP-76 are required for ORF5's ability to induce CD69 surface expression (Fig. 7D). In striking contrast, ORF5 upregulated CD69 surface expression in ZAP70-deficient P116 cells in the absence of TCR stimulation as efficiently as in parental Jurkat T cells (Fig. 7D). Furthermore, PMA stimulation synergistically enhanced the CD69 surface expression induced by ORF5 expression in ZAP70-deficient P116 cells (Fig. 7D). These results collectively demonstrate that ORF5 expression efficiently enhances TCR signal transduction activity, as evidenced by the increase of tyrosine phosphorylation of cellular proteins, intracellular calcium mobilization, CD69 surface expression, IL-2 production, and the activation of NF-AT, NF-κB, and AP-1 transcription factor activity. Furthermore, this activity requires Lck and SLP-76 expression, but not ZAP70 expression.
Partial substitution of ORF5 for LAT function.
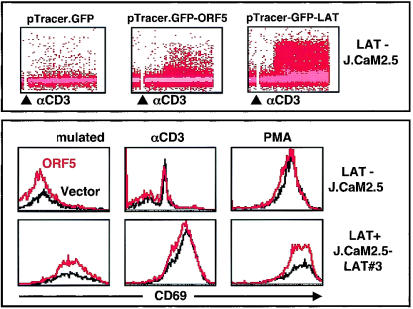
LAT has been shown to be critical for the optimal activation of T cells and to function in linking proximal signaling events to distal downstream signaling actions (20, 61). The LAT-deficient J.CaM2.5 variant of Jurkat T cells fails to elevate intracellular calcium, increase CD69 activation marker, or activate NF-AT and AP-1 transcription factor activity upon TCR stimulation (20). Since ORF5 contains an amino-terminal membrane attachment site and six SH2 binding motifs, showing structural similarity to LAT, we next investigated whether ORF5 was capable of substituting for LAT function in TCR signal transduction. J.CaM2.5 cells were electroporated with pTracerEF.GFP, pTracerEF.GFP-ORF5, or pTracerEF.GFP-LAT expression vector. At 36 h postelectroporation, cells were stimulated with anti-CD3 antibody, and GFP-positive J.CaM2.5 cells were then monitored for intracellular free calcium mobilization and CD69 surface expression by flow cytometry. wt LAT expression efficiently induced the elevation of intracellular calcium and CD69 surface expression upon TCR stimulation (Fig. 8). However, ORF5 expression only partially substituted for LAT function in inducing intracellular calcium mobilization upon CD3 stimulation and did not substitute at all for LAT in the upregulation of CD69 surface expression (Fig. 8). GFP expression resulted in no effect on either intracellular calcium mobilization or CD69 surface expression under the same conditions (Fig. 8). These results indicate that, despite its structural similarity, ORF5 is only capable of partially substituting for LAT function in linking proximal signaling events to the downstream intracellular calcium influx. These results also suggest that ORF5 is likely distinct from LAT in its action in TCR signal transduction.
FIG. 8.
Partial substitution of ORF5 for LAT function in TCR signal transduction. (Top) Intracellular calcium mobilization. J.CaM2.5 LAT-deficient cells electroporated with pTracerEF.GFP, pTracerEF.GFP-ORF5, or pTracerEF.GFP-LAT were stimulated with anti-CD3 (αCD3) Dynabeads, and calcium mobilization in GFP-positive cell populations was then monitored over time. The procedure and data presentation are as described for Fig. 7A. Data were similar in two independent experiments. (Bottom) Surface expression of CD69. At 24 h postelectroporation with pTracerEF.GFP or pTracerEF.GFP-ORF5, J. CaM2.5 cells were unstimulated or stimulated with either anti-CD3 Dynabeads or PMA (50 ng/ml) for an additional 16 h. GFP-positive cell populations were then gated and examined for CD69 surface expression by flow cytometry.
DISCUSSION
In this study, we identified a novel viral signaling adaptor protein, ORF5, encoded by T-lymphotropic HVS. Sequence comparison of ORF5 proteins from 13 HVS isolates revealed that ORF5 contains two conserved motifs: a myristoylation site and six SH2 binding motifs. Protein binding assays showed that tyrosine-phosphorylated ORF5 interacted with cellular SH2-containing proteins Lck, Fyn, SLP-76, and PI3 kinase. The consequence of ORF5 interaction with these cellular signaling molecules was the strong augmentation of TCR signal transduction, evidenced by increased tyrosine phosphorylation, intracellular calcium mobilization, IL-2 production, CD69 surface expression, and activation of cellular transcription factors AP-1, NF-AT, and NF-κB. Thus, these results demonstrate that HVS ORF5 functions as an adaptor to efficiently link proximal signaling events to distal downstream signaling actions.
Previous sequence analysis indicated the presence of a small open reading frame, encoding ORF5, in the HVS subgroup A and C genomes (1, 18). Despite strong conservation of the ORF5 gene between two strains, the ORF5 proteins of subgroup A strain 11 and subgroup C strain 488 showed dramatic sequence variation throughout the coding sequence, particularly in the amino-terminal region. Our RT-PCR analysis demonstrated that the ORF5 genes of both viruses are spliced near the 5′ end of the coding sequence, which leads to the addition of 14 identical amino acids to their amino terminus, providing the amino-terminal myristoylation site for membrane attachment. Sequence comparison of 13 ORF5 alleles showed that ORF5 was relatively conserved within each subgroup but showed a dramatic sequence divergence between subgroups; for example, there was 97 to 100% identity among members of the A11 cluster of subgroup A but only 46 to 50% identity between the A11 cluster and subgroup C. Among more than 80 different open reading frames compared between HVS subgroup A11 and C488 strains, only two other open reading frames have been shown to display considerable sequence divergence (2, 14, 36). These are the STP oncoprotein and the Tip signaling protein. Neither of these proteins is required for viral replication, but they are required for in vitro T-cell immortalization and in vivo lymphoma induction (28). Our study has added ORF5 to this list of divergent viral proteins. Interestingly, all three viral proteins play important roles in the deregulation of cellular signal transduction: STP activates Ras and TRAF signal transduction (26), Tip deregulates Lck signal transduction (28), and ORF5 enhances TCR signal transduction. This unusually high diversity between HVS subgroups reflects some unknown, powerful biological selection process that has been acting preferentially on these viral signaling proteins. This also suggests that, similar to the case for STP and Tip, ORF5 may be dispensable for viral replication but indispensable for viral oncogenesis. This possibility is under active investigation.
The divergence of the ORF5 amino acid sequence raises the intriguing possibility that different ORF5 alleles may have different signaling activities or different cellular targets. However, we found that, despite the dramatic divergence throughout the ORF5-coding sequence, all six SH2 binding motifs were completely conserved among 13 isolates. In addition, HVS strain A11 ORF5 also induced CD69 surface expression and NF-AT and AP-1 transcription factor activity as efficiently as HVS stain C488 ORF5 (data not shown). Thus, despite dramatic amino acid sequence divergence, ORF5 alleles likely have similar activity and cellular targets in TCR signal transduction.
Upon engagement with the MHC complex on antigen-presenting cells or target cells, TCR initiates an array of signal transduction events: rapid tyrosine phosphorylation of cellular proteins, increased intracellular free calcium, and ultimately, activation of cellular transcription factor activity. Numerous studies have demonstrated a critical role of adaptor proteins in TCR signal transduction (12, 23, 25, 41, 45, 46, 53). Particularly, LAT is a prominent adaptor molecule that is essential for linking the proximal signaling events of the TCR pathway to the distal downstream signaling events. Interestingly, ORF5 displays a similarity to LAT; both have an amino-terminal membrane attachment motif and multiple SH2 binding motifs, and both augment TCR signal transduction. Despite this similarity, however, the level of functional substitution of ORF5 for LAT was only minimal. This finding indicates that ORF5 is similar but distinct from LAT. This notion is also supported by our finding that while ZAP70 is the primary kinase to phosphorylate LAT in the TCR signaling pathway (61), Lck and Fyn but not ZAP70 were capable of efficiently phosphorylating ORF5. This suggests that ORF5 enhances TCR signal transduction in a way distinct from that of LAT.
We used in vitro GST pull-down assays with bacterially purified tyrosine-phosphorylated ORF5 to identify cellular interacting proteins. This revealed that tyrosine-phosphorylated ORF5 interacted with cellular SH2-containing Lck, Fyn, SLP-76, and p85. This interaction was specific, because unphosphorylated ORF5 was unable to interact with these cellular proteins. In addition, the precise point mutations containing the replacements of tyrosine residues of ORF5 with phenylalanine resulted in the specific loss of interaction with cellular proteins. Furthermore, the lack of Lck or SLP-76 expression abolished the ability of ORF5 to augment CD69 surface expression, and the ORF5 Y72F mutant, which lacked Lck binding, was incapable of enhancing TCR signal transduction. Finally, when we used the same GST pull-down procedure with bacterially purified tyrosine-phosphorylated KSHV K1 and HVS Tip, which both also contain SH2 binding motifs, we identified completely different repertoires of cellular SH2-containing proteins (unpublished results). These features support the validity and specificity of our procedure to identify cellular proteins interacting with tyrosine-phosphorylated ORF5. However, since this procedure may lead to a biased representation of cellular interacting proteins, we will continue our efforts to identify additional cellular proteins interacting with ORF5 by using a modified yeast two-hybrid screen (30) and/or in vivo binding protein purification.
Herpesviruses have a large genome and thus have the luxury of being able to utilize multiple genes to achieve the desired association with the host. The reason that these viruses retain such genes is not to cause disease in the host but to facilitate their persistence in the host. Since HVS is T lymphotropic, the regulation of TCR may be an essential part of the HVS life cycle that allows it to persist in the host. We have found that at least three HVS proteins directly target TCR signal transduction. ORF14 protein, a homolog of MMTV vSag, is detected primarily during lytic replication and induces the polyclonal proliferation of CD4-positive T cells through an interaction with MHC class II-TCR molecules (60). Tip, which is expressed primarily during viral latency, interacts with Lck tyrosine kinase and p80 endosomal protein and thus interferes with early events in the TCR signal transduction pathway and induces a rapid endocytosis of the TCR complex (39, 40). Finally, ORF5, which is expressed primarily during viral replication cycle but not during the viral latent cycle (unpublished results), interacts with cellular SH2-containing proteins, which results in the strong augmentation of TCR signal transduction. This indicates that T-lymphotropic HVS elaborately regulates TCR signal transduction at multiple points in the viral life cycle and that the positive or negative activity of HVS toward TCR signal transduction also differs, depending on the stage of viral life cycle: ORF5 and ORF14 lytic proteins activate TCR signal transduction to facilitate viral progeny production, whereas Tip latent protein inactivates TCR signal transduction to avoid host immune recognition and to establish and/or maintain viral latency. Future study of the molecular mechanisms of HVS-mediated TCR signal transduction will lead to a better understanding of viral persistence and disease progression and also provide a novel means for investigating the cellular regulatory systems of signal transduction pathways.
Acknowledgments
We especially thank A. Weiss, G. Koretzky, and L. Samelson for providing cell lines and expression vector, S. Gygi for mass spectrometry analysis, M. Connole for flow cytometry analysis, J. Macke for manuscript editing, and members of our laboratory for helpful discussion and comments.
This work was partly supported by Public Health Service grants CA31363, AI38131, and RR00168. P. Feng is a Leukemia and Lymphoma Society Fellow, and J. Jung is a Leukemia and Lymphoma Society Scholar.
REFERENCES
- 1.Albrecht, J. C., J. Nicholas, D. Biller, K. R. Cameron, B. Biesinger, C. Newman, S. Wittmann, M. A. Craxton, H. Coleman, B. Fleckenstein, et al. 1992. Primary structure of the herpesvirus saimiri genome. J. Virol. 66:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biesinger, B., J. J. Trimble, R. C. Desrosiers, and B. Fleckenstein. 1990. The divergence between two oncogenic herpesvirus saimiri strains in a genomic region related to the transforming phenotype. Virology 176:505-514. [DOI] [PubMed] [Google Scholar]
- 3.Biesinger, B., A. Y. Tsygankov, H. Fickenscher, F. Emmrich, B. Fleckenstein, J. B. Bolen, and B. M. Broker. 1995. The product of the herpesvirus saimiri open reading frame 1 (tip) interacts with T cell-specific kinase p56lck in transformed cells. J. Biol. Chem. 270:4729-4734. [DOI] [PubMed] [Google Scholar]
- 4.Burack, W. R., A. M. Cheng, and A. S. Shaw. 2002. Scaffolds, adaptors and linkers of TCR signaling: theory and practice. Curr. Opin. Immunol. 14:312-316. [DOI] [PubMed] [Google Scholar]
- 5.Cabanillas, J. A., R. Cambronero, A. Pacheco-Castro, M. C. Garcia-Rodirguez, J. M. Martin-Fernandez, G. Fontan, and J. R. Regueiro. 2002. Characterization of herpesvirus saimiri-transformed T lymphocytes from common variable immunodeficiency patients. Clin. Exp. Immunol. 127:366-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, P. C., A. Cherukuri, M. Dykstra, S. Malapati, T. Sproul, M. R. Chen, and S. K. Pierce. 2001. Floating the raft hypothesis: the roles of lipid rafts in B cell antigen receptor function. Semin. Immunol. 13:107-114. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, P. C., M. L. Dykstra, R. N. Mitchell, and S. K. Pierce. 1999. A role for lipid rafts in B cell antigen receptor signaling and antigen targeting. J. Exp. Med. 190:1549-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, J. K., B. S. Lee, S. N. Shim, M. Li, and J. U. Jung. 2000. Identification of the novel K15 gene at the rightmost end of the Kaposi's sarcoma-associated herpesvirus genome. J. Virol. 74:436-446. [PMC free article] [PubMed] [Google Scholar]
- 9.Clements, J. L., J. R. Lee, B. Gross, B. Yang, J. D. Olson, A. Sandra, S. P. Watson, S. R. Lentz, and G. A. Koretzky. 1999. Fetal hemorrhage and platelet dysfunction in SLP-76-deficient mice. J. Clin. Investig. 103:19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clements, J. L., S. E. Ross-Barta, L. T. Tygrett, T. J. Waldschmidt, and G. A. Koretzky. 1998. SLP-76 expression is restricted to hemopoietic cells of monocyte, granulocyte, and T lymphocyte lineage and is regulated during T cell maturation and activation. J. Immunol. 161:3880-3889. [PubMed] [Google Scholar]
- 11.Clements, J. L., B. Yang, S. E. Ross-Barta, S. L. Eliason, R. F. Hrstka, R. A. Williamson, and G. A. Koretzky. 1998. Requirement for the leukocyte-specific adapter protein SLP-76 for normal T cell development. Science 281:416-419. [DOI] [PubMed] [Google Scholar]
- 12.Crespo, P., K. E. Schuebel, A. A. Ostrom, J. S. Gutkind, and X. R. Bustelo. 1997. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature 385:169-172. [DOI] [PubMed] [Google Scholar]
- 13.D'Ambrosio, D., D. A. Cantrell, L. Frati, A. Santoni, and R. Testi. 1994. Involvement of p21ras activation in T cell CD69 expression. Eur. J. Immunol. 24:616-620. [DOI] [PubMed] [Google Scholar]
- 14.Desrosiers, R. C., A. Bakker, J. Kamine, L. A. Falk, R. D. Hunt, and N. W. King. 1985. A region of the herpesvirus saimiri genome required for oncogenicity. Science 228:184-187. [DOI] [PubMed] [Google Scholar]
- 15.Desrosiers, R. C., and L. A. Falk. 1982. Herpesvirus saimiri strain variability. J. Virol. 43:352-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desrosiers, R. C., D. P. Silva, L. M. Waldron, and N. L. Letvin. 1986. Nononcogenic deletion mutants of herpesvirus saimiri are defective for in vitro immortalization. J. Virol. 57:701-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duboise, M., J. Guo, S. Czajak, H. Lee, R. Veazey, R. C. Desrosiers, and J. U. Jung. 1998. A role for herpesvirus saimiri orf14 in transformation and persistent infection. J. Virol. 72:6770-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ensser, A., M. Thurau, S. Wittmann, and H. Fickenscher. 2003. The genome of herpesvirus saimiri C488 which is capable of transforming human T cells. Virology 314:471-487. [DOI] [PubMed] [Google Scholar]
- 19.Fang, N., D. G. Motto, S. E. Ross, and G. A. Koretzky. 1996. Tyrosines 113, 128, and 145 of SLP-76 are required for optimal augmentation of NFAT promoter activity. J. Immunol. 157:3769-3773. [PubMed] [Google Scholar]
- 20.Finco, T. S., T. Kadlecek, W. Zhang, L. E. Samelson, and A. Weiss. 1998. LAT is required for TCR-mediated activation of PLCgamma1 and the Ras pathway. Immunity 9:617-626. [DOI] [PubMed] [Google Scholar]
- 21.Goldsmith, M. A., and A. Weiss. 1988. Early signal transduction by the antigen receptor without commitment to T cell activation. Science 240:1029-1031. [DOI] [PubMed] [Google Scholar]
- 22.Goldsmith, M. A., and A. Weiss. 1987. Isolation and characterization of a T-lymphocyte somatic mutant with altered signal transduction by the antigen receptor. Proc. Natl. Acad. Sci. USA 84:6879-6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han, J., B. Das, W. Wei, L. Van Aelst, R. D. Mosteller, R. Khosravi-Far, J. K. Westwick, C. J. Der, and D. Broek. 1997. Lck regulates Vav activation of members of the Rho family of GTPases. Mol. Cell. Biol. 17:1346-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackman, J. K., D. G. Motto, Q. Sun, M. Tanemoto, C. W. Turck, G. A. Peltz, G. A. Koretzky, and P. R. Findell. 1995. Molecular cloning of SLP-76, a 76-kDa tyrosine phosphoprotein associated with Grb2 in T cells. J. Biol. Chem. 270:7029-7032. [DOI] [PubMed] [Google Scholar]
- 25.June, C. H., M. C. Fletcher, J. A. Ledbetter, G. L. Schieven, J. N. Siegel, A. F. Phillips, and L. E. Samelson. 1990. Inhibition of tyrosine phosphorylation prevents T-cell receptor-mediated signal transduction. Proc. Natl. Acad. Sci. USA 87:7722-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung, J. U., and R. C. Desrosiers. 1992. Herpesvirus saimiri oncogene STP-C488 encodes a phosphoprotein. J. Virol. 66:1777-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung, J. U., S. M. Lang, U. Friedrich, T. Jun, T. M. Roberts, R. C. Desrosiers, and B. Biesinger. 1995. Identification of Lck-binding elements in tip of herpesvirus saimiri. J. Biol. Chem. 270:20660-20667. [DOI] [PubMed] [Google Scholar]
- 28.Jung, J. U., J. J. Trimble, N. W. King, B. Biesinger, B. W. Fleckenstein, and R. C. Desrosiers. 1991. Identification of transforming genes of subgroup A and C strains of herpesvirus saimiri. Proc. Natl. Acad. Sci. USA 88:7051-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaschka-Dierich, C., I. Bauer, B. Fleckenstein, and R. C. Desrosiers. 1981. Episomal and nonepisomal herpesvirus DNA in lymphoid tumor cell lines. Haematol. Blood Transfus. 26:197-203. [DOI] [PubMed] [Google Scholar]
- 30.Keegan, K., and J. A. Cooper. 1996. Use of the two hybrid system to detect the association of the protein-tyrosine-phosphatase, SHPTP2, with another SH2-containing protein, Grb7. Oncogene 12:1537-1544. [PubMed] [Google Scholar]
- 31.Kiyotaki, M., R. C. Desrosiers, and N. L. Letvin. 1986. Herpesvirus saimiri strain 11 immortalizes a restricted marmoset T8 lymphocyte subpopulation in vitro. J. Exp. Med. 164:926-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knappe, A., C. Hiller, M. Thurau, S. Wittmann, H. Hofmann, B. Fleckenstein, and H. Fickenscher. 1997. The superantigen-homologous viral immediate-early gene ie14/vsag in herpesvirus saimiri-transformed human T cells. J. Virol. 71:9124-9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, J., and A. Weiss. 2001. T cell receptor signalling. J. Cell Sci. 114:243-244. [DOI] [PubMed] [Google Scholar]
- 34.Longnecker, R. 2000. Epstein-Barr virus latency: LMP2, a regulator or means for Epstein-Barr virus persistence? Adv. Cancer Res. 79:175-200. [DOI] [PubMed] [Google Scholar]
- 35.Lund, T. C., P. C. Prator, M. M. Medveczky, and P. G. Medveczky. 1999. The Lck binding domain of herpesvirus saimiri tip-484 constitutively activates Lck and STAT3 in T cells. J. Virol. 73:1689-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medveczky, M. M., E. Szomolanyi, R. Hesselton, D. DeGrand, P. Geck, and P. G. Medveczky. 1989. Herpesvirus saimiri strains from three DNA subgroups have different oncogenic potentials in New Zealand White rabbits. J. Virol. 63:3601-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musci, M. A., D. G. Motto, S. E. Ross, N. Fang, and G. A. Koretzky. 1997. Three domains of SLP-76 are required for its optimal function in a T cell line. J. Immunol. 159:1639-1647. [PubMed] [Google Scholar]
- 38.Myung, P. S., N. J. Boerthe, and G. A. Koretzky. 2000. Adapter proteins in lymphocyte antigen-receptor signaling. Curr. Opin. Immunol. 12:256-266. [DOI] [PubMed] [Google Scholar]
- 39.Park, J., N. H. Cho, J. K. Choi, P. Feng, J. Choe, and J. U. Jung. 2003. Distinct roles of cellular Lck and p80 proteins in herpesvirus saimiri Tip function on lipid rafts. J. Virol. 77:9041-9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, J., B. S. Lee, J. K. Choi, R. E. Means, J. Choe, and J. U. Jung. 2002. Herpesviral protein targets a cellular WD repeat endosomal protein to downregulate T lymphocyte receptor expression. Immunity 17:221-233. [DOI] [PubMed] [Google Scholar]
- 41.Peterson, E. J., J. L. Clements, N. Fang, and G. A. Koretzky. 1998. Adaptor proteins in lymphocyte antigen-receptor signaling. Curr. Opin. Immunol. 10:337-344. [DOI] [PubMed] [Google Scholar]
- 42.Pivniouk, V., E. Tsitsikov, P. Swinton, G. Rathbun, F. W. Alt, and R. S. Geha. 1998. Impaired viability and profound block in thymocyte development in mice lacking the adaptor protein SLP-76. Cell 94:229-238. [DOI] [PubMed] [Google Scholar]
- 43.Pivniouk, V. I., and R. S. Geha. 2000. The role of SLP-76 and LAT in lymphocyte development. Curr. Opin. Immunol. 12:173-178. [DOI] [PubMed] [Google Scholar]
- 44.Pivniouk, V. I., T. R. Martin, J. M. Lu-Kuo, H. R. Katz, H. C. Oettgen, and R. S. Geha. 1999. SLP-76 deficiency impairs signaling via the high-affinity IgE receptor in mast cells. J. Clin. Investig. 103:1737-1743. [PMC free article] [PubMed] [Google Scholar]
- 45.Samelson, L. E. 1999. Adaptor proteins and T-cell antigen receptor signaling. Prog. Biophys. Mol. Biol. 71:393-403. [DOI] [PubMed] [Google Scholar]
- 46.Secrist, J. P., L. Karnitz, and R. T. Abraham. 1991. T-cell antigen receptor ligation induces tyrosine phosphorylation of phospholipase C-gamma 1. J. Biol. Chem. 266:12135-12139. [PubMed] [Google Scholar]
- 47.Songyang, Z., S. E. Shoelson, M. Chaudhuri, G. Gish, T. Pawson, W. G. Haser, F. King, T. Roberts, S. Ratnofsky, R. J. Lechleider, et al. 1993. SH2 domains recognize specific phosphopeptide sequences. Cell 72:767-778. [DOI] [PubMed] [Google Scholar]
- 48.Songyang, Z., S. E. Shoelson, J. McGlade, P. Olivier, T. Pawson, X. R. Bustelo, M. Barbacid, H. Sabe, H. Hanafusa, T. Yi, et al. 1994. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol. Cell. Biol. 14:2777-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Straus, D. B., and A. Weiss. 1992. Genetic evidence for the involvement of the lck tyrosine kinase in signal transduction through the T cell antigen receptor. Cell 70:585-593. [DOI] [PubMed] [Google Scholar]
- 50.Szomolanyi, E., P. Medveczky, and C. Mulder. 1987. In vitro immortalization of marmoset cells with three subgroups of herpesvirus saimiri. J. Virol. 61:3485-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wange, R. L., and L. E. Samelson. 1996. Complex complexes: signaling at the TCR. Immunity 5:197-205. [DOI] [PubMed] [Google Scholar]
- 52.Wardenburg, J. B., C. Fu, J. K. Jackman, H. Flotow, S. E. Wilkinson, D. H. Williams, R. Johnson, G. Kong, A. C. Chan, and P. R. Findell. 1996. Phosphorylation of SLP-76 by the ZAP-70 protein-tyrosine kinase is required for T-cell receptor function. J. Biol. Chem. 271:19641-19644. [DOI] [PubMed] [Google Scholar]
- 53.Weiss, A., G. Koretzky, R. C. Schatzman, and T. Kadlecek. 1991. Functional activation of the T-cell antigen receptor induces tyrosine phosphorylation of phospholipase C-gamma 1. Proc. Natl. Acad. Sci. USA 88:5484-5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weiss, A., and D. R. Littman. 1994. Signal transduction by lymphocyte antigen receptors. Cell 76:263-274. [DOI] [PubMed] [Google Scholar]
- 55.Weiss, A., R. L. Wiskocil, and J. D. Stobo. 1984. The role of T3 surface molecules in the activation of human T cells: a two-stimulus requirement for IL 2 production reflects events occurring at a pre-translational level. J. Immunol. 133:123-128. [PubMed] [Google Scholar]
- 56.Williams, B. L., K. L. Schreiber, W. Zhang, R. L. Wange, L. E. Samelson, P. J. Leibson, and R. T. Abraham. 1998. Genetic evidence for differential coupling of Syk family kinases to the T-cell receptor: reconstitution studies in a ZAP-70-deficient Jurkat T-cell line. Mol. Cell. Biol. 18:1388-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, J., D. G. Motto, G. A. Koretzky, and A. Weiss. 1996. Vav and SLP-76 interact and functionally cooperate in IL-2 gene activation. Immunity 4:593-602. [DOI] [PubMed] [Google Scholar]
- 58.Wunderlich, L., A. Goher, A. Farago, J. Downward, and L. Buday. 1999. Requirement of multiple SH3 domains of Nck for ligand binding. Cell Signal. 11:253-262. [DOI] [PubMed] [Google Scholar]
- 59.Yablonski, D., M. R. Kuhne, T. Kadlecek, and A. Weiss. 1998. Uncoupling of nonreceptor tyrosine kinases from PLC-gamma1 in an SLP-76-deficient T cell. Science 281:413-416. [DOI] [PubMed] [Google Scholar]
- 60.Yao, Z., E. Maraskovsky, M. K. Spriggs, J. I. Cohen, R. J. Armitage, and M. R. Alderson. 1996. Herpesvirus saimiri open reading frame 14, a protein encoded by T lymphotropic herpesvirus, binds to MHC class II molecules and stimulates T cell proliferation. J. Immunol. 156:3260-3266. [PubMed] [Google Scholar]
- 61.Zhang, W., J. Sloan-Lancaster, J. Kitchen, R. P. Trible, and L. E. Samelson. 1998. LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell 92:83-92. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, W., C. L. Sommers, D. N. Burshtyn, C. C. Stebbins, J. B. DeJarnette, R. P. Trible, A. Grinberg, H. C. Tsay, H. M. Jacobs, C. M. Kessler, E. O. Long, P. E. Love, and L. E. Samelson. 1999. Essential role of LAT in T cell development. Immunity 10:323-332. [DOI] [PubMed] [Google Scholar]