Abstract
The Smoothened receptor (SMO) mediates signal transduction in the hedgehog pathway, which is implicated in normal development and carcinogenesis. SMO antagonists can suppress the growth of some tumors; however, mutations at SMO have been found to abolish their anti-tumor effects, a phenomenon known as chemoresistance. Here we report three crystal structures of human SMO bound to the antagonists SANT1 and Anta XV, and the agonist, SAG1.5, at 2.6–2.8Å resolution. The long and narrow cavity in the transmembrane domain of SMO harbors multiple ligand binding sites, where SANT1 binds at a deeper site as compared with other ligands. Distinct interactions at D4736.55 elucidated the structural basis for the differential effects of chemoresistance mutations on SMO antagonists. The agonist SAG1.5 induces a conformational rearrangement of the binding pocket residues, which could contribute to SMO activation. Collectively, these studies reveal the structural basis for the modulation of SMO by small molecules.
Introduction
The hedgehog (Hh) signal transduction network plays essential roles in the maintenance of normal embryonic development and postnatal tissue integrity in many eukaryotes ranging from Drosophila to humans 1,2. Aberrant activation of the Hh signaling pathway apparently both promotes carcinogenesis–particularly in basal cell carcinomas and medulloblastomas–and supports the tumor microenvironment in many other cancers 3. The smoothened receptor (SMO) belongs to the Class Frizzled (or Class F) receptors, which is part of the G protein-coupled receptor (GPCR) superfamily. SMO is normally negatively regulated by a catalytic amount of the 12-transmembrane domain protein Patched 4. In the vertebrate canonical Hh signaling pathway, the binding of the Hh signaling proteins to Patched can induce the translocation of SMO to primary cilium, thereby inducing the processing of GLI transcription factors into their active forms, which subsequently undergo nuclear translocation and activate GLI targeted genes 2. It has been proposed that Patched acts as a transporter and controls SMO activity by controlling the availability of small molecule lipid modulators of SMO 4. Although the identity of the endogenous small molecule modulator of SMO is unknown, a number of exogenous small molecules that modulate SMO activity have been discovered 5. Notably, the naturally occurring teratogen cyclopamine, which was the first selective SMO ligand, inhibits Hh signaling presumably via SMO antagonism by targeting to its 7-transmembrane (7TM) domain6. Given the importance of inhibiting Hh signaling pathways in various cancers, small molecules that target SMO are under intensive development and several lead compounds are currently in clinical trials, including Vismodegib (GDC-0449, Supplementary Figure 1) which was approved by the U.S. Food and Drug Administration (FDA) in 2012 for treating basal cell carcinoma 7,8.
Despite the tremendous progress in explicating SMO pharmacology and the recent success in obtaining the first human SMO structure 9, a molecular understanding of the structural basis for small molecule recognition of SMO remains elusive. This is not only important for understanding the structure-activity relationships (SAR) of chemically distinct SMO ligands, but also for providing a mechanistic understanding of chemoresistant mutations. For example, the D4736.55H (superscripts indicate residue numbering using the Ballesteros-Weinstein nomenclature for class F receptors 9,10) mutation in human SMO makes antagonists such as GDC-0449 unable to inhibit the receptor 11; while several other compounds are insensitive to this drug resistance mutation 12–14. Delineating the structural basis for the differential effects of mutations on diverse ligands would provide a rational platform for the development of drugs to counteract emerging drug resistance effects.
Additionally, the modulation of SMO by small molecules reveals complicated effects on ligand efficacy. For example, SAG (3-chloro-N-[4-(methylamino)cyclohexyl]-N-[3-(pyridin-4-yl)benzyl]benzo[b]thiophene-2-carboxamide) 15,16 and derivatives including SAG1.5 (3-chloro-4,7-difluoro-N-[trans-4-(methylamino)cyclohexyl]-N-[[3-(4-pyridinyl)phenyl]methyl]-1-benzothiophene-2-carboxamide) 16 (Supplementary Figure 1) act as potent agonists, inducing cilia translocation of SMO and GLI transcription factor activation. Cyclopamine and SANT1 (N-[(1E)-(3,5-dimethyl-1-phenyl-1H-pyrazol-4-yl)methylidene]-4-(phenylmethyl)-1-piperazinamine) 15 (Supplementary Figure 1), for instance, both inhibit GLI transcription factor activation, while SANT1 inhibits and cyclopamine induces cilia translocation of SMO 17. In addition, molecules such as cyclopamine and GDC-0449, which were initially discovered as potent antagonists for the canonical Hh signaling pathway, act as agonists to induce the glucose uptake response and drive Warburg-like metabolism in fat and muscle cells via a transcription factor independent pathway 18, perhaps accounting for the unwanted side effects of weight loss and muscle cramping observed in humans. Clearly, determining the binding modes of different ligands to SMO represents an essential first step for correlating a ligand’s chemotype with its pattern of functional modulation, thereby illuminating the pharmacology and biology of SMO.
In this study, we report three crystal structures of the human SMO 7TM domain bound to SMO small molecule antagonists SANT1, Anta XV (2-(6-(4-(4-benzylphthalazin-1-yl)piperazin-1-yl)pyridin-3-yl)propan-2-ol) 19 and agonist SAG1.5 (Supplementary Figure 1). Structure-guided mutagenesis and competition binding analysis probed the specific effects of key amino acid residues in these structures. A further examination of these structures alongside the structures of the human SMO in complex with LY2940680 9 and cyclopamine 20 provides a comprehensive framework for understanding the structural basis of molecular recognition and modulation at SMO by small molecules.
Results
Crystallization and structures of SMO-ligand complexes
To investigate the structural basis of activation and inhibition of SMO, we solved the structure of SMO in complex with ligands of different functional properties including the antagonists SANT1 and Anta XV, and the potent agonist SAG1.5. Using the previously reported SMO construct 9 with the N-terminal cysteine-rich domain (CRD, residues 1–189) replaced by a thermostabilized apocytochrome b562RIL (BRIL), and a truncated C-terminus at Q555 (BRIL-ΔCRD-SMO-ΔC), we obtained crystals of SMO bound to SANT1, Anta XV and SAG1.5. The structure of the SMO_SANT1 complex was solved at 2.8 Å resolution in a C2221 space group (Table 1). However, initial crystals of SMO with Anta XV and SAG1.5 diffracted to only 3.5 Å and 8 Å, respectively. In order to improve the resolution of the Anta XV and SAG1.5 bound SMO complexes, we applied another fusion strategy by replacing intracellular loop 3 (ICL3, from P434 to K440) with BRIL (ΔCRD-SMO-BRIL(ICL3)-ΔC). Using this construct, the structure of the SMO_Anta XV complex was solved in a P212121 space group at 2.6 Å resolution; the structure of the SMO_SAG1.5 complex was solved in a C2 space group with resolution cut-off at 2.9Å, 2.5 Å and 3.3 Å along a*, b* and c* axes, respectively, due to the anisotropic nature of the diffraction (Table 1). In addition, we have solved the SMO_cyclopamine complex structure (PDB id: 4O9R) using a newly developed technique of serial femtosecond crystallography with LCP grown crystals (LCP-SFX) at an X-ray free electron laser source. Albeit at a lower resolution, this structure unambiguously located the binding site of cyclopamine 20. The crystal packing analysis (Supplementary Figure 2) reveals that the SMO_SANT1, SMO_Anta XV and SMO_cyclopamine complexes were crystallized as monomers, while the SMO_SAG1.5 complex was crystallized as a dimer, similar to the previously reported SMO_LY2940680 complex 9 (PDB id: 4JKV), with a dimer interface involving helices IV and V.
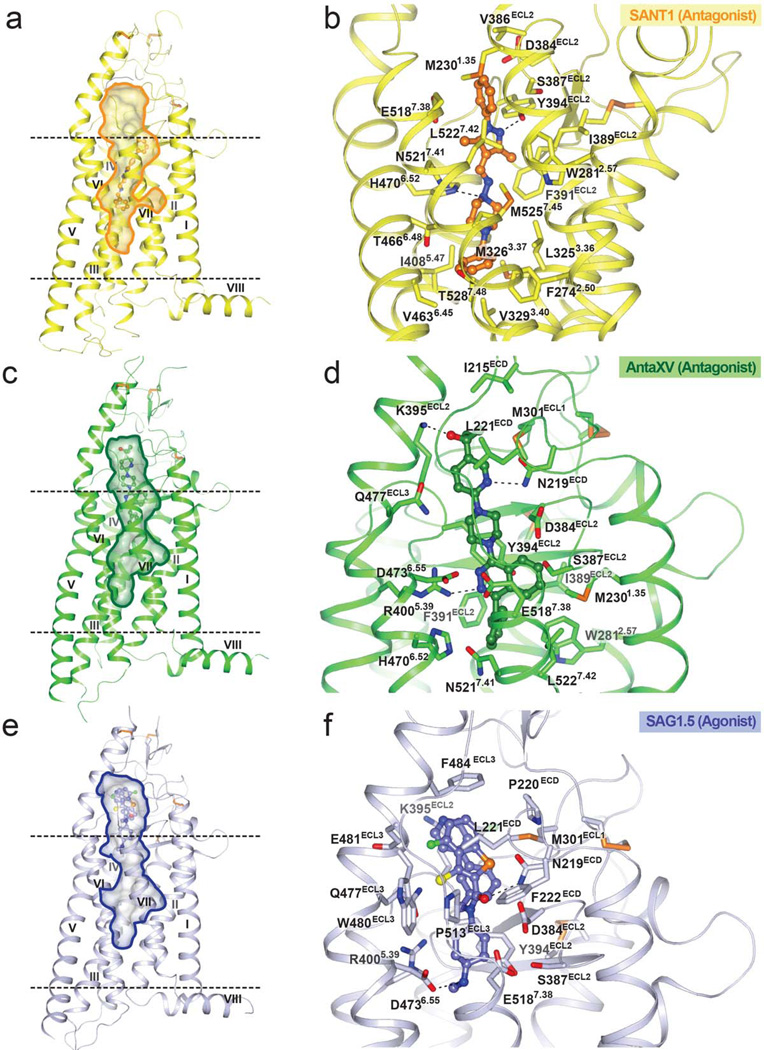
Along with the SMO_LY2940680 and SMO_cyclopamine complex structures, the newly solved structures show that the long and narrow cavity formed by the extracellular domain (ECD) linker domain, extracellular loops and the 7TM bundle provides multiple binding sites for small molecule ligands (Fig. 1 and Supplementary Figure 3). Similar to LY2940680 and cyclopamine, the binding sites of Anta XV and SAG1.5 are formed mostly by residues from the ECD linker domain and extracellular loops (Fig. 1d,f). Additionally, Anta XV, SAG1.5 and LY2940680 form hydrogen bonds with the N219ECD side chain, which provides an important anchor for ligands that bind close to the entrance of the ligand binding cavity. In contrast, SANT1 binds at a unique site in the cavity, which extends much deeper toward the center of the 7TM bundle and is formed mainly by residues from all the transmembrane α-helices, with the exception of helix IV (Fig. 1b).
Figure 1. Overall structures of SMO receptor bound to different ligands and the ligand binding pockets.
(a), (c) and (e) Overall structures of SMO receptor in complex with SANT1, Anta XV and SAG1.5, respectively. The lipid bilayer is shown in dashed lines. The ligand binding cavity is shown in surface presentation. SMO_SANT1 structure (yellow); SMO_Anta XV structure (green); SMO_SAG1.5 structure (light blue) are shown in ribbon presentation. (b), (d) and (f) The structures of the ligand binding pockets for SANT1 (orange carbon), Anta XV (green carbon) and SAG1.5 (light blue carbon) are shown. Polar interactions between the receptor and the ligands are shown as dashed lines and interacting residues are labeled.
The deep binding site of SANT1
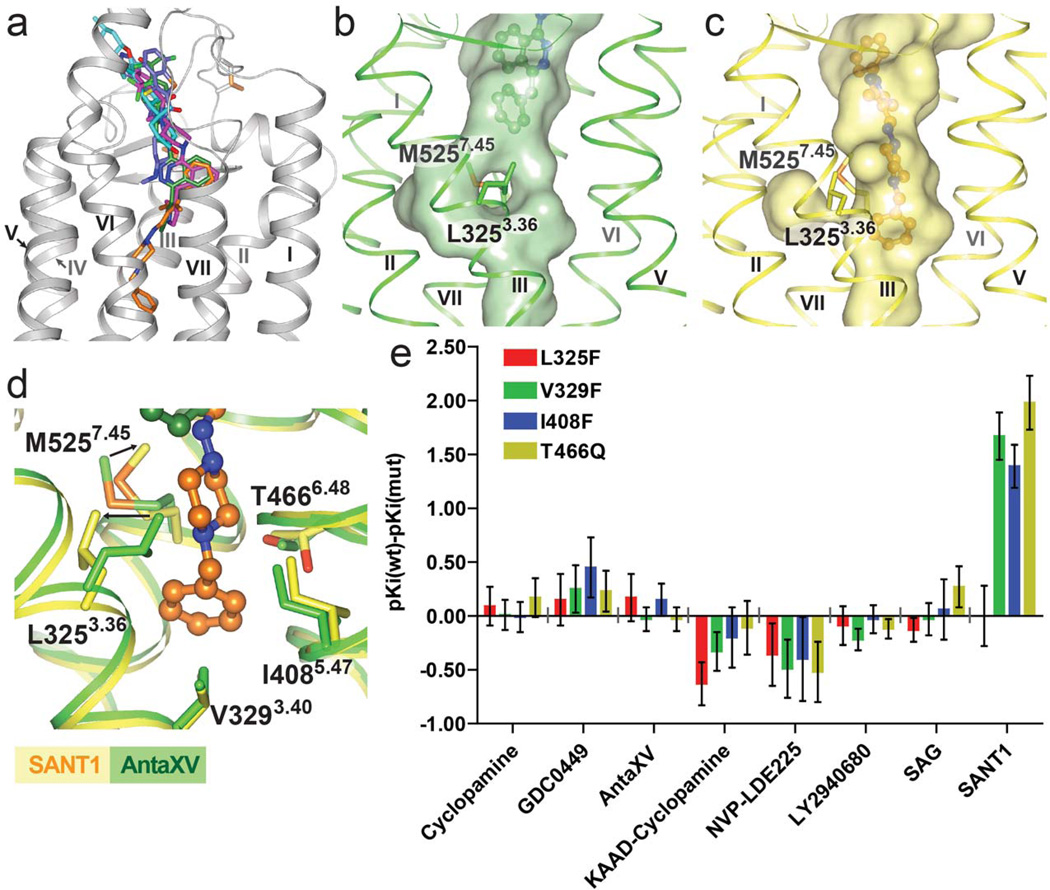
The binding site of SANT1 is unusually deep in the 7TM helical bundle, with the ligand extending at least 9 Å deeper into the cavity compared to LY2940680 (Fig. 2a). The only extracellular loop that interacts with SANT1 is extracellular loop 2 (ECL2), which is positioned inside the helical bundle and forms non-polar contacts and a hydrogen bond from Y394ECL2 to the phenyl ring and the pyrazole ring of SANT1, respectively (Fig. 1b). In the helical bundle, this binding pocket is very narrow, providing a snug fit for the long and linear structure of SANT1. The side chain of H4706.52 forms a hydrogen bond to a nitrogen in the piperazine ring (Fig. 1b). Superposition of the SMO_SANT1 and the SMO_SAG1.5 structures (Fig. 2a) reveals only a minimal overlay between SANT1 and SAG1.5, thereby providing a structural explanation for previous observations that SANT1 allosterically modulates the binding of SAG 21, which is a close derivative of SAG1.5 (Supplementary Figure 1). In the SMO_SANT1 complex, the compound induces a slight expansion in the deep part of the cavity, as compared to the other ligands that do not reach this sub-pocket (Fig. 2b,c). This change in the pocket is manifested mainly in a conformational rearrangement of the side chains, while minimally affecting the protein backbone. The side chain of L3253.36 protrudes into the central cavity when this deep pocket is not occupied, but rotates out from the cavity making space for the binding of SANT1 (Fig. 2d). Additionally, the side chain of M5257.45 moves towards SANT1 to make contact with the piperazine moiety of the ligand. To further investigate the binding pocket for SANT1, we introduced bulky side chain mutations, L3253.36F, V3293.40F, I4085.47F and T4666.48Q, in the bottom part of the pocket (Fig. 2d), aiming to block this binding site without disturbing the overall receptor conformation. While the binding of the radioligand 3H-cyclopamine remains unchanged in agreement with the SMO_cyclopamine structure, competition radioligand binding experiments reveals that three of these bulky mutations, V3293.40F, I4085.47F and T4666.48Q, substantially reduce the binding of SANT1 (Fig. 2e and Supplementary Table 1). Interestingly, the L3253.36F mutation has no effect on SANT1 binding, consistent with the modeling of the L3253.36F mutation which shows that the SMO_SANT1 complex can easily accommodate a phenyl side chain in the rotamer state observed for L3253.36 in the SANT1 bound structure (Fig. 2d). Binding of the other tested ligands was not substantially affected by any of these bulky side chain mutations (Fig. 2e), corroborating the unique nature of the deep binding site for SANT1 among the other SMO ligands studied.
Figure 2. The deep binding pocket of SANT1.
(a) Localization of different ligands in the binding pocket: SANT1 (orange), Anta XV (green), SAG1.5 (light blue), LY2940680 (magenta), and cyclopamine (cyan). (b) and (c) The shapes of the deep binding pockets in the structures of SMO_Anta XV (green) and SMO_SANT1 (yellow), respectively. (d) Superposition of the deep binding pockets in the structures of SMO_Anta XV and SMO_SANT1. The side chain movements induced by SANT1 binding are shown by arrows. (e) ΔpKi values (pKi wild type – pKi mutants) of the designed mutants for different SMO ligands in 3H-cyclopamine competition experiments. Shown in the graph are mean ± SEM of 3–4 independent experiments, see also Supplementary Table 1.
The binding mode of cyclopamine and KAAD-cyclopamine
The naturally occurring steroidal jerveratrum alkaloid cyclopamine was the first small molecule found to bind to and inhibit Hh signaling via SMO. Cyclopamine binds close to the extracellular entrance, and the secondary amine of its 3-methyl-piperidine group points outside of the pocket, while the 3,β-hydroxyl is buried deep in the pocket (Supplementary Figure 4a) 20. This binding pose is consistent with previous SAR studies of cyclopamine derivatives, which showed that adducts to the 3,β-hydroxyl dramatically reduced activity, while the secondary amine permits addition of bulky groups via long aliphatic linkers 22. One such molecule is KAAD-cyclopamine (3-keto-N-(aminoethyl-aminocaproyl-dihydrocinnamoyl)cyclopamine) (Supplementary Figure 1), derived by attaching a long chain substitution to the secondary amine of cyclopamine. Our data shows that the binding of KAAD-cyclopamine to SMO is not impacted by mutations blocking the deep binding pocket (Fig. 2e), consistent with a predicted binding mode where the long chain substitution extends out of the 7TM cavity through the extracellular ligand entrance (Supplementary Figure 4b). The long aliphatic linker protruding out of the pocket has also been shown to enable attachments as large as a fluorescent BODIPY moiety to cyclopamine 6.
Differential binding modes of Anta XV and LY2940680
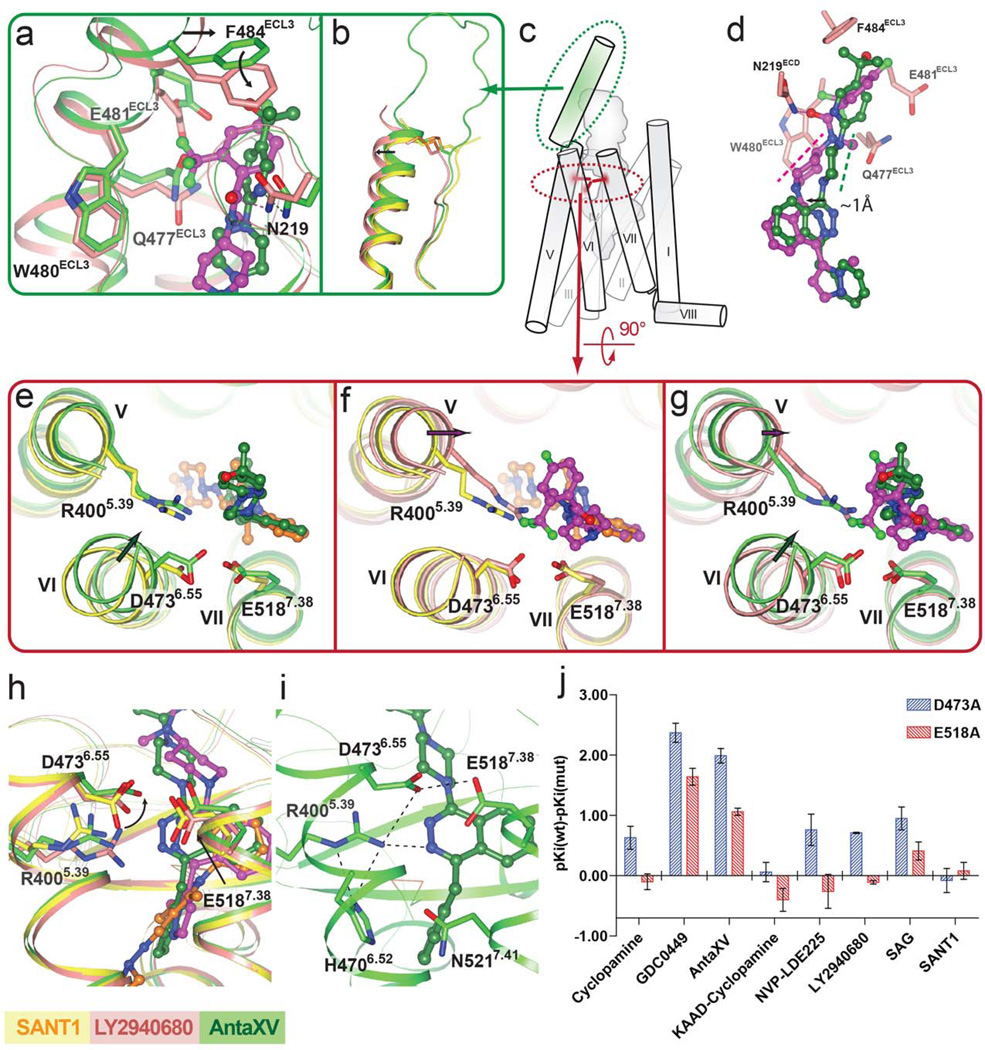
Although Anta XV and LY2940680 share a phthalazine ring core and a similar overall shape, different substitutions on the phthalazine ring result in distinct binding modes of these ligands. In the previously solved SMO_LY2940680 structure 9, the 4-fluoro-2-trifluoromethylphenyl moiety of LY2940680 forms extensive interactions with residues from ECL3, including Q477, W480, E481 and F484, which stacks to the phenyl ring of LY2940680 through a π-π interaction (Fig. 3a). In contrast, Anta XV does not have contacts with ECL3, and the side chain residue conformations of ECL3 in the SMO_Anta XV structure are more similar to those in the structure of SMO_SANT1, which also lacks ECL3 contacts (Supplementary Figure 5). The differential interaction of LY2940680 with ECL3 apparently results in an overall shift of the α-helical portion of the ECL3 (Fig. 3b,c).
Figure 3. D4736.55 plays different roles in the recognition of Anta XV and LY2940680.
(a)–(i) The receptor structures in SMO_Anta XV, SMO_SANT1 and SMO_LY2940680 (PDB id: 4JKV, molecule A) complexes are shown in green, yellow and salmon, respectively. Ligand structures of Anta XV, SANT1 and LY2940680 are shown in green, orange and magenta carbons, respectively. (a) Superposition of SMO_LY2940680 and SMO_Anta XV structures near ECL3. The conformational change of F484ECL3 induced by LY2940680 binding is shown by an arrow. See also Supplementary Figure 4 for comparison with SMO_SANT1 structure. (b) Superposition of the SMO_SANT1, SMO_Anta XV and SMO_LY2940680 structures reveals a different conformation of ECL3 in LY2940680 bound structure (shown by arrow). (c) Localization of ECL3 (green) and polar residue cluster (R4005.39, D4736.55 and E5187.38, red) in SMO. (d) Superposition of the binding poses of LY2940680 and Anta XV reveals distinct orientations (dashed lines parallel to the equatorial bonds of the six-membered chair conformation rings) of the six membered rings connected to the phthalazine ring. The shift of the phthalazine of LY2940680 compared to that of Anta XV is shown by arrow. (e) Superposition of SMO_Anta XV and SMO_SANT1 structures. (f) Superposition of SMO_LY2940680 and SMO_SANT1 structures. (g) Superposition of SMO_LY2940680 and SMO_Anta XV structures. Ligand induced shifts of helices are shown by arrows. (h) Superposition of the ligand binding pocket residues of SMO_SANT1, SMO_Anta XV and SMO_LY2940680 structures. Ligand interaction induced conformation change at D4736.55 is indicated by arrow. (i) Polar residue cluster in the ligand binding pocket of the SMO_Anta XV structure. Polar interactions are shown as dashed lines. (j) ΔpKi values (pKi wild type– pKi mutants) of the D4736.55A and E5187.38A mutants for different SMO receptor ligands in 3H-cyclopamine competition experiments. Shown in the graph are mean ± SEM of 3–4 independent experiments, see also Supplementary Table 2.
Together with the extensive interactions between the 4-fluoro-2-trifluoromethylphenyl group of LY2940680 and ECL3, the hydrogen bond between N219 and the carbonyl group of LY2940680 potentially defines the orientation of piperidine ring substituent on the phthalazine core, which is distinct from the orientation of the piperazine of Anta XV (Fig. 3d). In this orientation, the phthalazine core of LY2940680 adopts an axial position in the chair conformation of the six-membered piperidine ring, which was estimated to be slightly (ΔE~0.72 kcal/mol) 23 suboptimal compared to the equatorial position. In contrast, on the piperazine ring of Anta XV, the phthalazine core sits in the equatorial position; this difference in ring conformation is accompanied by a 1 Å shift of the phthalazine ring between the two ligands (Fig. 3d). The shift is also associated with a shift at the guanidinium group of R4005.39, which forms hydrogen bond to the phthalazine core of Anta XV and LY2940680, as well as the extracellular end of helix V where R4005.39 locates (Fig. 3e–g).
This difference in the positions of the phthalazine core in Anta XV and LY2940680 may explain their distinctive interactions with D4736.55, a key residue in a polar interaction cluster within the ligand binding cavity (Figs. 3c,i). Firstly, the side chain of D4736.55 makes a direct interaction with Anta XV (3.3 Å) whereas its interaction with LY2940680 is much weaker as revealed by a longer distance (4.0 Å in molecule A, 4.3 Å in molecule B). Secondly, the interaction between D4736.55 and Anta XV is accompanied by an inward shift of the extracellular tip of helix VI, as compared to SMO_SANT1 and SMO_LY2940680 structures where their respective ligands do not interact with D4736.55 (Fig. 3e–g). The difference in this interaction is also reflected in different rotamer states of the carboxylate group of D4736.55 (Fig. 3h). All these structural data point to a more important role for D4736.55 in Anta XV recognition compared to LY2940680.
Mutation of D4736.55 into histidine has been identified as a cause of chemoresistance for GDC-0449, while LY2940680 has been reported to be unaffected by this mutation 24. To investigate the role of D4736.55 in the binding of different ligands, we performed 3H-cyclopamine competition binding assays on a D4736.55A mutant, as well as an E5187.38A mutant that could impact the conformation of D4736.55. The natural drug-resistance mutation D4736.55H and other mutations that could impact the conformation of D4736.55, such as R4005.39A, H4706.52A, and N5217.41A (Fig. 3h), unfortunately could not be assessed in the radioligand competition assay, as they abolished the binding of radioligand 3H-cyclopamine. Among the mutations that retained binding of the radioligand, the D4736.55A and E5187.38A mutations impaired the binding of GDC-0449 and Anta XV, with D4736.55A responsible for a more than 100 fold affinity drop for both ligands. In contrast, the effect of D4736.55A mutation on LY2940680 was modest (<7 fold), consistent with the lack of such a direct interaction in the SMO_LY2940680 structure (Fig. 3j and Supplementary Table 2). Moreover, these mutations have no effect on the binding of SANT1, consistent with the fact that in the SMO_SANT1 structure D4736.55 plays little role in ligand binding. These results thus provide a direct structural explanation for the differential effect of ligand binding pocket mutations on ligands with distinct binding poses.
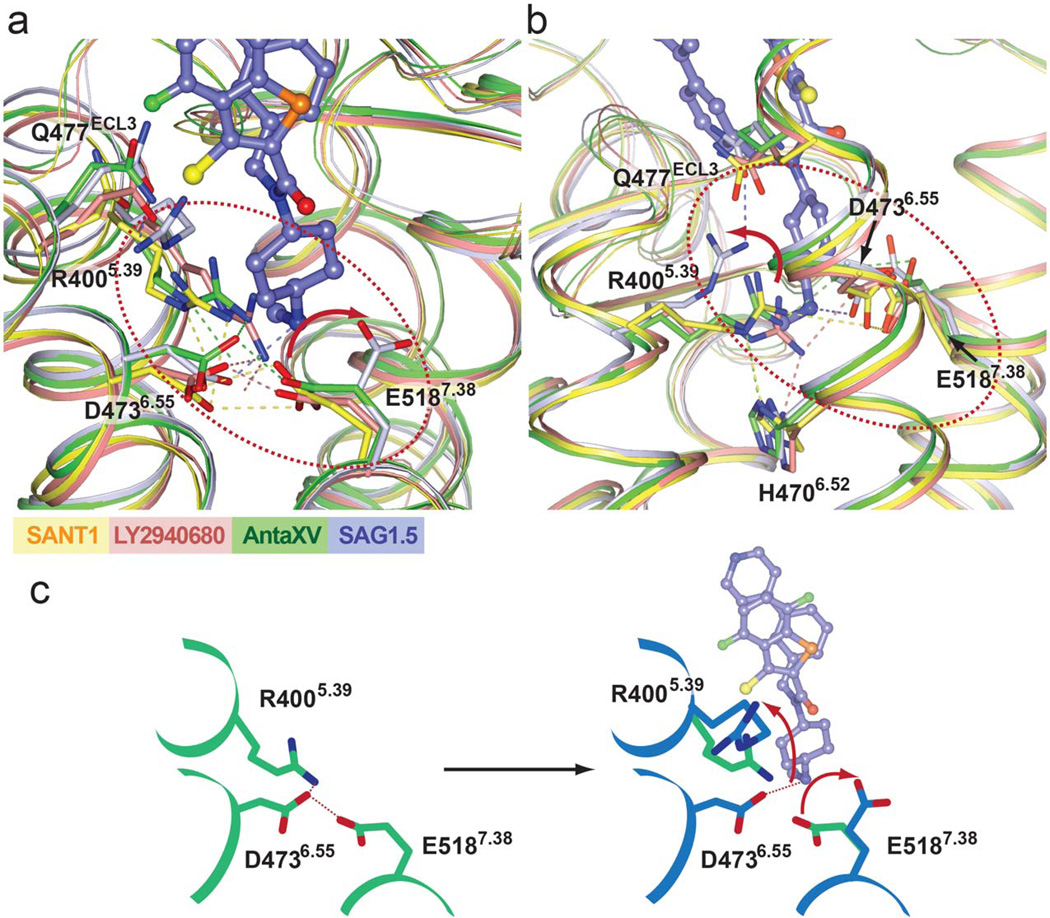
Remodeling of the SMO binding pocket by agonist SAG1.5
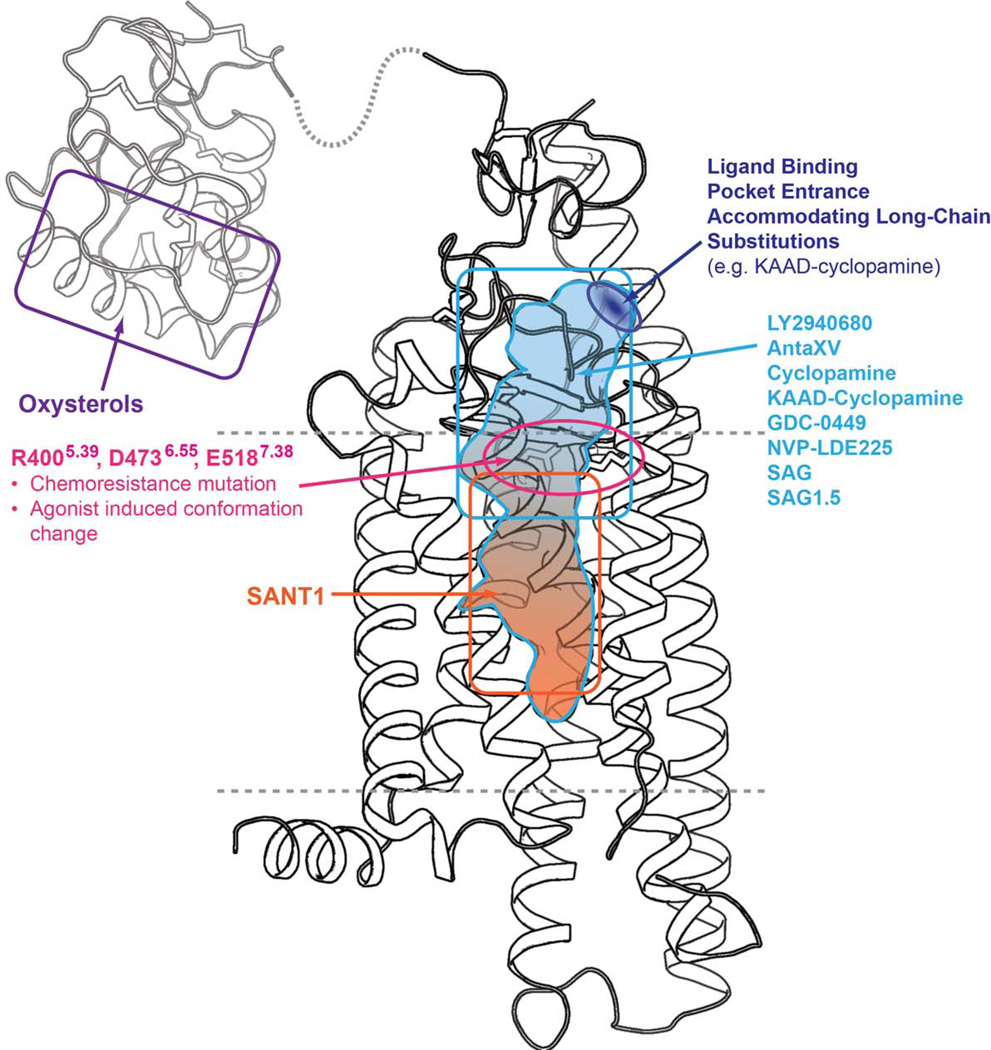
When bound to SMO, the two aromatic rings of SAG1.5 pack against each other, forming a bulky system that fits tightly into the pocket surrounded by the ECD linker domain and extracellular loops close to the extracellular entrance (Fig. 1f). This clamping of the bulky aromatic groups of SAG1.5 by the extracellular loop structures points the cyclohexane deeper into the 7TM bundle, aligning the positively charged methyl amino group in the vicinity of polar residue cluster, R4005.39, D4736.55 and E5187.38 (Figs. 1f and 4), which are the only residues from the seven helixes that interact with the agonist. In the antagonist bound structures, the D4736.55 and E5187.38 side chains point towards each other and interact directly, or in the case of the SMO_LY2940680 structure, through water mediated interactions. In the agonist bound structure, the positively charged amino group of SAG1.5 forms an ionic interaction with D4736.55, while the carboxyl group of E5187.38 moves away and no longer interacts with D4736.55 (Fig. 4a). In the antagonist bound structures, R4005.39 is anchored by polar interactions with D4736.55, H4706.52 and/or the ligands. In the agonist bound structure, the guanidine group of R4005.39 moves up to form a hydrogen bond with the side chain of Q477ECL3 (Fig. 4b). This remodeling at the residues R4005.39, D4736.55 and E5187.38 (Fig. 4c) is the most pronounced change in the ligand binding pocket, differentiating the SMO_SAG1.5 structure from the antagonist-bound structures. In addition, subtle modifications at the amino group of the SAG scaffold have been shown to change the functional property of the ligand, i.e. from agonist to antagonist 25, indicating that this amino group plays a critical role in the SMO modulation by SAG1.5. While previous studies showed that oxysterol binding at the N terminus of SMO can induce activation 26–28, the SAG1.5 bound SMO structure reveals an alternative site at the 7TM domain that is responsible for the small molecule induced activation of the receptor (Fig. 5).
Figure 4. Agonist SAG1.5 induced conformational changes in the ligand binding pocket.
(a) Superposition of the ligand binding pocket residues of different structures of SMO. The receptor structure and side chain carbons are shown for SMO_Anta XV (green), SMO_SANT1 (yellow), SMO_LY2940680 (salmon; PDB id: 4JKV, molecule A), and SMO_SAG1.5 (light blue) structures. The ligand SAG1.5 is shown as light blue carbons. The hydrogen bond interactions between side chains are show as dashed lines in the corresponding color of each structure. (b) A different view of the superposition of the ligand binding pocket residues. H4706.52 which interacts with R4005.39 in antagonist bound structures is shown. Red arrows (in (a) and (b)) are shown to indicate the conformational changes of E5187.38 and R4005.39 induced by SAG1.5 binding. (c) Schematic presentation of agonist induced conformation change: green is antagonist bound state, while blue is agonist bound state.
Figure 5. Map of the structural basis for the modulation of SMO by small molecules.
The N terminal CRD domain is shown using the CRD of zebrafish SMO (PDB id: 4C79). The boundaries of the membrane bilayer are shown as dashed lines.
On the intracellular side, the most pronounced difference between agonist and antagonist bound structures is observed at Y262ICL1, H3614.46 and W3654.50 that are conserved among class F receptors 9. In all the antagonist bound structures, we observed a hydrogen bonding network among these residues. In the agonist bound structure, the hydrogen bond between Y262ICL1 and H3614.46 is broken, which is associated with an inward movement of P263ICL1, which is also conserved among class F receptors 9, to make contact with W5357.55 (Supplementary Figure 6). Mutation of W5357.55 into leucine has been shown to make the SMO receptor constitutively active leading to carcinogenesis 29. In addition, W5357.55 is located at the intracellular end of helix VII, immediately adjacent to helix VIII (Supplementary Figure 6), which packs against helix I, parallel to the membrane layer and plays an important role during the activation of SMO since mutation at the residues W545 and R546 located in helix VIII impairs cilia translocation 30. These conformational changes observed at Y262ICL1, H3614.46, W3654.50 and P263ICL1 could impact W5357.55, and are conceivably important for SMO activation. We note however that the change of the hydrogen bond state could result from the slightly lower pH in the crystallization condition of the agonist bound receptor. Thus, the involvement of this conformational change during SMO activation needs to be investigated in future studies. With the exception of this local conformational change, the agonist SAG1.5 bound SMO structure does not show any large scale movements in helices VI and VII, which are the hallmark of class A GPCR activation 31. This may be due to the stark differences in signaling mechanisms between the two GPCR classes, or the possibility that in the absence of downstream effectors SAG1.5 induces only part of the activation-related conformational changes in SMO or other factors (Discussion).
Discussion
The newly solved structures of SMO bound to SANT1, Anta XV, SAG1.5, along with the LY2940680 and cyclopamine bound structures, reveal a variety of distinctive poses for structurally diverse small molecule ligands in the long and narrow cavity defined by the 7TM helices, ECD and extracellular loops of SMO (Fig. 5). The antagonist SANT1, for instance, binds very deep in the pocket, whereas the other ligands studied remain closer to the extracellular entrance, demonstrating that the entire long and narrow pocket can be targeted by small molecule ligands. Meanwhile, the extracellular entrance can provide a path for accommodating the attachment of reporter moieties or other bulky groups to the ligands through a long chain flexible linker such as that of KAAD-cyclopamine.
Even when the binding sites largely overlap, as for instance with LY2940680 and Anta XV, minor differences in recognition modes result in a discrete response to binding pocket mutations. LY2940680, for example, has been reported to bind the D4736.55H mutant that provides chemoresistance to the approved cancer drug GDC-0449 24. This biological phenomenon accords nicely with our structural analysis, which shows that LY2940680 forms weak interactions with D4736.55. In contrast, the binding of Anta XV requires a more substantial involvement of D4736.55, as revealed by the structural and mutagenesis data, despite its similar scaffold with LY2940680. The most striking difference between the binding mode of LY2940680 and Anta XV is a strong interaction between LY2940680 and ECL3. We hypothesize that this interaction restrains the orientation of LY2940680, which leads to a shift of the phthalazine core of LY2940680 away from D4736.55 compared to Anta XV, weakening the role of D4736.55 in ligand binding. This reduced involvement of D4736.55 in binding might also underlie the effect of chemical modifications on existing compounds which could overcome the drug resistant mutation D4736.55H as shown in studies with derivatives of GDC-0449 12 and Anta XV 14. The long and continuous cavity straddling the ECL region and 7TM domain of SMO provides a variety of binding sub-sites suitable for ligand interaction. Taken together, this structural information should facilitate the development of compounds, which bypass this chemoresistant mutation or have a larger contacting surface so that disruption of a local structure, as chemoresistance mutations do, could not abolish the ligand binding.
We observed remodeling of the polar interaction network between R4005.39, D4736.55 and E5187.38 in the binding pocket induced by SAG1.5; such remodeling could conceivably serve as an activation trigger. However, due to the lack of knowledge regarding the immediate biochemical events downstream of SMO 7TM domain activation, a mechanistic connection between the ligand-triggered activation at the extracellular side and the intracellular coupling with downstream proteins remains to be established. SMO has shown in some studies an ability to activate G proteins 32 which presumably requires an opening of the intracellular crevice for G protein binding 33 as has been established in class A GPCRs. Some conformationally selective ligands are capable of converting class A GPCRs into a partially activated state where the change in the ligand binding pocket propagates into the intracellular side of the receptors 34–38. In other cases, e.g. in β-adrenergic receptors, agonist binding alone is insufficient for stabilization of an active state, and therefore in the structures of these receptors obtained in the absence of G-proteins or their mimetics the intracellular side maintains the inactive conformation 39,40. In the SAG1.5 bound SMO structure, we did not observe the remodeling of the intracellular side which has been demonstrated in the active state class A GPCR structures. This could be due to the following reasons: (i) although SAG1.5 is a potent agonist for the canonical Hh signaling pathway, it is not an effective agonist for G protein activation; (ii) the insertion of BRIL at ICL3 prevented intracellular helical movement; and (iii) conformational changes at the intracellular part of the receptor require binding of G proteins as observed for β-adrenergic receptors. Due to the divergence of the canonical Hh signaling pathway from the traditional G protein pathway, it is possible that the activated SMO couples to as yet unidentified intracellular effector(s), which could eventually elicit conformational changes responsible for an active state. Alternatively, activation by agonist might induce only small conformational changes in the 7TM domain, which modify oligomerization states or lead to differential association with membrane compartments and intracellular trafficking machineries.
This long and narrow cavity responsible for the binding of small molecule ligands investigated in this study is not the only modulation site for SMO. Lipid molecules have been shown to be able to modulate SMO; for example, cholesterol depletion severely impairs SMO activation 26. It has been suggested that the endogenous modulation mechanism of SMO by Patched might also be mediated through a lipidic molecule 4. Oxysterols, for instance, have been shown to activate SMO and had been suspected to be native ligands for SMO. However, recent studies showed that oxysterols bind to the N terminal CRD domain (Fig. 5), the structure of which has been solved by crystallography27 and NMR41 methods, and that the CRD domain is important, but not essential, for the Hh-induced activation of SMO as well as for cholesterol dependence of Hh signaling26–28. Thus, there could be additional sites in the 7TM domain that serve as the binding sites of native modulators or lipidic molecules. A previous study showed that mutations within the ligand binding site that disrupt binding of synthetic ligands, such as cyclopamine, fail to impair the normal basal and Hh-induced activity, as well as the cholesterol dependence of Hh signaling 26, indicating an alternative site might be used to modulate SMO activity by Patched or lipidic molecules. W3654.50 has been shown to be a conserved lipid binding site at class A GPCRs 42–44, which locates at the interface between the 7TM domain and membrane environment, and is thus accessible to lipidic molecules. In the agonist bound structure, we observed conformational changes in a hydrogen bond network involving W3654.50 (Supplementary Figure 6), which occur in the vicinity of W5357.55, a critical residue for SMO activation. Further studies will be needed to fully investigate the role of this region in the modulation of SMO by lipidic molecules.
In summary, we provide new insights into the conformational plasticity of SMO-ligand interactions by crystallographic and biochemical characterization of several SMO complexes with diverse ligand chemotypes. Importantly, these ligands target distinct sites in the elongated cavity, have different interactions with known resistance mutants and include both antagonists and agonists of Hh signaling. The details of these interactions and ligand-dependent conformational changes, correlated with specific functional features of the ligands will help to design new clinical candidates targeting SMO with attenuated side effects and chemoresistance.
Methods
Generation of BRIL-SMO fusion constructs
The BRIL-ΔCRD-SMO-ΔC construct has been reported previously 14. For ΔCRD-SMO-BRIL(ICL3)-ΔC, BRIL was fused to the human SMO receptor 7TM domain (S190-Q555) by replacing ICL3 residues P434 to K440 using overlapping PCR. The resulting receptor chimera sequence was subcloned into a modified pFastBac1 vector (Invitrogen), designated as pFastBac1-833100, which contained an expression cassette with a haemagglutinin (HA) signal sequence followed by a Flag tag, a 10× His tag, and a TEV protease recognition site at the N terminus before the receptor sequence. Subcloning into the pFastBac1-833100 was achieved using PCR with primer pairs encoding restriction sites KpnI at the 5′ and HindIII at the 3′ termini with subsequent ligation into the corresponding restriction sites in the vector.
Expression and purification of SMO constructs
The SMO constructs were expressed in Spodoptera frugiperda (Sf9) insect cells using the Bac-to-Bac Baculovirus Expression System (Invitrogen). Sf9 cells at cell density of 2–3 × 106 cells/ml were infected with baculovirus at 27 °C. Cells were harvested by centrifugation at 48 hr post infection and stored at −80 °C until use.
Insect cell membranes were lysed by thawing frozen cell pellets in a hypotonic buffer containing 10 mM HEPES, pH 7.5, 10 mM MgCl2, 20 mM KCl and EDTA-free complete protease inhibitor cocktail tablets (Roche). Extensive washing of the raw membranes was performed by repeated centrifugation two-three times in a high osmotic buffer comprised of 1.0 M NaCl in the hypotonic buffer described above.
The washed membranes were resuspended into buffer containing 30 µM ligand, 2 mg/ml iodoacetamide (Sigma), and EDTA-free complete protease inhibitor cocktail tablets, and incubated at 4 °C for 1 hr prior to solubilization. The membranes were then solubilized in buffer containing 50 mM HEPES, pH 7.5, 200 mM NaCl, 1% (w/v) n-dodecyl-β-D-maltopyranoside (DDM, Anatrace), 0.2% (w/v) cholesteryl hemisuccinate (CHS, Sigma), 15 µM ligand, for 3–4 hours at 4 °C. The supernatant containing solubilized SMO protein was isolated from the cell debris by high-speed centrifugation, and subsequently incubated with TALON IMAC resin (Clontech) overnight at 4 °C in the presence of 20 mM imidazole and 1 M NaCl. After binding, the resin was washed with 10 column volumes of Wash I Buffer comprised of 50 mM HEPES, pH 7.5, 800 mM NaCl, 10% (v/v) glycerol, 0.1% (w/v) DDM, 0.02% (w/v) CHS, 8 mM ATP, 20 mM imidazole, 10 mM MgCl2 and 15 µM ligand, followed by 6 column volumes of Wash II Buffer comprised of 50 mM HEPES, pH 7.5, 500 mM NaCl, 10% (v/v) glycerol, 0.05% (w/v) DDM, 0.01% (w/v) CHS, 50 mM imidazole and 20 µM ligand. The protein was then eluted by 3 column volumes of Elution Buffer containing 50 mM HEPES, pH 7.5, 300 mM NaCl, 10% (v/v) glycerol, 0.03% (w/v) DDM, 0.006% (w/v) CHS, 250 mM imidazole and 50 µM ligand. PD MiniTrap G-25 column (GE Healthcare) was used to remove imidazole. The protein was then treated overnight with TEV protease (His-tagged) to cleave the N-terminal His-tag and FLAG-tag. TEV protease and cleaved N-terminal fragment were removed by TALON IMAC resin incubation at 4 °C for 2 hr. The tag-less protein was collected as the TALON IMAC column flow-through. The protein was then concentrated to 50–60 mg/ml with a 100 kDa cut-off Vivaspin concentrator. Protein monodispersity was tested by analytical size-exclusion chromatography (aSEC). Typically, the aSEC profile showed a monodisperse peak.
SANT1 ligand was purchased from Tocris Biosciences (purity > 99%); Anta XV (originally named Hh Signaling Antagonist XV) ligand was purchased from Calbiochem (purity 98.94%); and SAG1.5 ligand was purchased from Xcess Biosciences Inc. (purity > 98%).
Lipidic cubic phase crystallization
Protein samples of SMO in a complex with certain ligand were reconstituted into lipidic cubic phase (LCP) by mixing with molten lipid (monoolein and cholesterol mixture in a ratio of 9:1 (w/w)) in a mechanical syringe mixer 45. LCP crystallization trials were performed using an NT8-LCP crystallization robot (Formulatrix) as previously described 46. 96-well glass sandwich plates (Marienfeld) were incubated and imaged at 20 °C using an automated incubator/imager (RockImager 1000, Formulatrix). Crystals of SMO_SANT1 complex were grown in the condition: 150 mM NH4F, 100 mM HEPES pH 6.9, 27% PEG 400, 2.5% Jeffamine; crystals of SMO_Anta XV complex were grown in the condition: 100–115 mM NH4Cl, 100 mM HEPES pH 7.2, 36% PEG 400; and crystals of SMO_SAG1.5 complex were grown in the condition: 100 mM MgSO4, 100 mM MES pH 6.0, 30% PEG 400, 2%–3% Polypropylene glycol P 400. Crystals were harvested using MiTeGen micromounts and flash frozen in liquid nitrogen for data collection.
Crystallographic data collection and processing
X-ray data were collected at the 23ID-D beamline (GM/CA CAT) at the Advanced Photon Source, Argonne, IL using a 20 μm minibeam at a wavelength of 1.0330 Å and a MarMosaic 300 CCD detector. Crystals were aligned and data were collected using strategy similar to other GPCR structures 47. Typically 20 frames at 1° oscillation and 1 s exposure with non-attenuated beam followed by a translation of the crystal to a non-exposed position or changing the crystal to minimize the effect of radiation damage. A complete data set was obtained by indexing, integrating, scaling, and merging data using HKL2000 48. The SMO_SAG1.5 data set was highly anisotropic. The merged data were submitted to the UCLA anisotropy server (http://services.mbi.ucla.edu/anisoscale) and anisotropically truncated at 2.9 Å, 2.5 Å and 3.3 Å along a*, b* and c* axes, respectively.
Structure determination and Refinement
Initial phase information was obtained by molecular replacement with the program PHASER49 using two independent search models of SMO 7TM domain and BRIL from SMO_LY2940680 complex structure (PDB id: 4JKV). All refinements were performed with REFMAC550 and autoBUSTER51 followed by manual examination and rebuilding of the refined coordinates in the program COOT52 using both |2FO|-|FC| and |FO|-|FC| maps, as well as omit maps calculated using the Bhat’s procedure53,54. The structures have been deposited in the PDB with the accession codes: 4N4W for SMO_SANT1 complex; 4QIM for SMO_AntaXV complex; and 4QIN for SMO_SAG1.5 complex.
Radioligand competition
3H-Cyclopamine binding to WT and mutant SMO was done as previously described 9.
Molecular modeling and KAAD-cyclopamine docking
Conformational modeling and evaluation of the mutation effects in SMO complexes with different ligands was performed using an all-atom global energy optimization algorithm in ICM-Pro 3.7 (MolSoft LLC) molecular modeling package 55. Point mutations of interest (L3253.36F, V3293.40F, I4085.47F, T4666.48Q) in each of the co-crystal structures were introduced by modifying the residue side chain, followed by energy optimization that included the ligand and side chains within 6 Å distance of the mutated residue. Other side chains and the backbone of the protein were kept as in the crystal structure. The effect of the mutation was evaluated by comparing conformational energy of the ligand before and after mutation. Docking of KAAD-cyclopamine was performed with all atom energy-based docking procedure with side chain flexibility in the binding pocket 56 using the SMO_cyclopamine crystal structure. An initial conformation of KAAD-cyclopamine was generated by Cartesian optimization of the ligand model in Merck Molecular Force Field. All calculations were repeated at least in 3 independent runs, performed on a 12-core Intel Xeon 2.67 GHz Linux workstation.
Supplementary Material
Acknowledgements
This work was supported by NIH Roadmap grant P50 GM073197 for technology development (V.C. and R.C.S.), PSI:Biology grant U54 GM094618 for biological studies and structure production (target GPCR-131) (V.K., V.C. and R.C.S.); R01 MH61887, U19 MH82441, R01 DA27170; the NIMH Psychoactive Drug Screening Program (X.-P.H. and B.L.R.) and the Michael Hooker Chair of Pharmacology (B.L.R.) and DA-029925 and MH- 073853 (M.G.C.). We thank D. Stout and A. Sather for help in solution of SANT1 bound structure; J. Velasquez for help on molecular biology; T. Trinh and M. Chu for help on baculovirus expression; K. Kadyshevskaya for assistance with figure preparation; A. Walker for assistance with manuscript preparation; J. Smith, R. Fischetti, and N. Sanishvili for assistance in development and use of the minibeam and beamtime at GM/CA-CAT beamline 23-ID at the Advanced Photon Source, which is supported by National Cancer Institute grant Y1-CO-1020 and National Institute of General Medical Sciences grant Y1-GM-1104.
Footnotes
Author contributions
C.W. designed and performed crystallization experiments, solved the structures and prepared the manuscript. H.W. assisted protein expression and prepared the manuscript. T.E., E.V., X.H., S.J.H, T.J.M., and D.J.U. performed biochemical experiments. G.W.H. performed structural determination and assisted structure refinement. V.K. performed molecular docking, assisted with structural analysis and manuscript preparation. V.C. assisted with structure refinement and manuscript preparation. M.G.C. and B.L.R. supervised biochemical experiments and revised the manuscript. R.C.S. directed the study and revised the manuscript. The authors declare no competing financial interests.
References
- 1.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 2.Robbins DJ, Fei DL, Riobo NA. The Hedgehog signal transduction network. Sci Signal. 2012;5:re6. doi: 10.1126/scisignal.2002906. doi: 10.1126/scisignal.2002906 scisignal.2002906 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. doi: 10.1038/nrc1229 nrc1229 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Taipale J, et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 5.Mahindroo N, Punchihewa C, Fujii N. Hedgehog-Gli signaling pathway inhibitors as anticancer agents. J Med Chem. 2009;52:3829–3845. doi: 10.1021/jm801420y. doi: 10.1021/jm801420y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robarge KD, et al. GDC-0449-a potent inhibitor of the hedgehog pathway. Bioorg Med Chem Lett. 2009;19:5576–5581. doi: 10.1016/j.bmcl.2009.08.049. doi: 10.1016/j.bmcl.2009.08.049 S0960-894X(09)01180-9 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Von Hoff DD, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med. 2009;361:1164–1172. doi: 10.1056/NEJMoa0905360. doi: 10.1056/NEJMoa0905360 NEJMoa0905360 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Wang C, et al. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013;497:338–343. doi: 10.1038/nature12167. doi: 10.1038/nature12167 nature12167 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballesteros JA, Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure–function relations in G-protein coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 11.Yauch RL, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. doi: 10.1126/science.1179386 1179386 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dijkgraaf GJ, et al. Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res. 2011;71:435–444. doi: 10.1158/0008-5472.CAN-10-2876. doi: 10.1158/0008-5472.CAN-10-2876 0008-5472.CAN-10-2876 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Tao H, et al. Small molecule antagonists in distinct binding modes inhibit drug-resistant mutant of smoothened. Chem Biol. 2011;18:432–437. doi: 10.1016/j.chembiol.2011.01.018. doi: 10.1016/j.chembiol.2011.01.018 S1074-5521(11)00083-4 [pii] [DOI] [PubMed] [Google Scholar]
- 14.Peukert S, et al. Discovery of NVP-LEQ506, a second-generation inhibitor of smoothened. ChemMedChem. 2013;8:1261–1265. doi: 10.1002/cmdc.201300217. doi: 10.1002/cmdc.201300217. [DOI] [PubMed] [Google Scholar]
- 15.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci U S A. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. doi: 10.1073/pnas.182542899 182542899 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank-Kamenetsky M, et al. Small-molecule modulators of Hedgehog signaling: identification and characterization of Smoothened agonists and antagonists. J Biol. 2002;1:10. doi: 10.1186/1475-4924-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohatgi R, Milenkovic L, Corcoran RB, Scott MP. Hedgehog signal transduction by Smoothened: pharmacologic evidence for a 2-step activation process. Proc Natl Acad Sci U S A. 2009;106:3196–3201. doi: 10.1073/pnas.0813373106. doi: 10.1073/pnas.0813373106 0813373106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teperino R, et al. Hedgehog partial agonism drives Warburg-like metabolism in muscle and brown fat. Cell. 2012;151:414–426. doi: 10.1016/j.cell.2012.09.021. doi: 10.1016/j.cell.2012.09.021 S0092-8674(12)01128-2 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Miller-Moslin K, et al. 1-amino-4-benzylphthalazines as orally bioavailable smoothened antagonists with antitumor activity. J Med Chem. 2009;52:3954–3968. doi: 10.1021/jm900309j. doi: 10.1021/jm900309j. [DOI] [PubMed] [Google Scholar]
- 20.Weierstall U, et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nature communications. 2014;5:3309. doi: 10.1038/ncomms4309. doi: 10.1038/ncomms4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rominger CM, et al. Evidence for allosteric interactions of antagonist binding to the smoothened receptor. J Pharmacol Exp Ther. 2009;329:995–1005. doi: 10.1124/jpet.109.152090. doi: 10.1124/jpet.109.152090 jpet.109.152090 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Beachy PA. Regulators of the Hedgehog Pathway, Compostitions and Uses Related Thereto. 2001 [Google Scholar]
- 23.Carballeira L, Perez-Juste I. Influence of calculation level and effect of methylation on axial/equatorial equilibria in piperidines. J Comput Chem. 1998;19:961–976. doi: :Doi 10.1002/(Sici)1096-987x(199806)19:8<961::Aid-Jcc14>3.0.Co;2-A. [Google Scholar]
- 24.Bender MH, et al. Abstract 2819: Identification and characterization of a novel smoothened antagonist for the treatment of cancer with deregulated hedgehog signaling. Cancer Res. 2011;71:A2819. doi: 10.1158/1538-7445.AM2011-2819. [Google Scholar]
- 25.Yang H, et al. Converse conformational control of smoothened activity by structurally related small molecules. J Biol Chem. 2009;284:20876–20884. doi: 10.1074/jbc.M807648200. doi: 10.1074/jbc.M807648200 M807648200 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers BR, et al. Hedgehog pathway modulation by multiple lipid binding sites on the smoothened effector of signal response. Dev Cell. 2013;26:346–357. doi: 10.1016/j.devcel.2013.07.015. doi: 10.1016/j.devcel.2013.07.015 S1534-5807(13)00423-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nachtergaele S, et al. Structure and function of the Smoothened extracellular domain in vertebrate Hedgehog signaling. Elife. 2013;2:e01340. doi: 10.7554/eLife.01340. doi: 10.7554/eLife.01340 01340 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nedelcu D, Liu J, Xu Y, Jao C, Salic A. Oxysterol binding to the extracellular domain of Smoothened in Hedgehog signaling. Nat Chem Biol. 2013;9:557–564. doi: 10.1038/nchembio.1290. doi: 10.1038/nchembio.1290 nchembio.1290 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie J, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 30.Corbit KC, et al. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. doi: :nature04117 [pii] 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 31.Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. doi: 10.1016/j.tips.2011.09.003 S0165-6147(11)00172-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riobo NA, Saucy B, Dilizio C, Manning DR. Activation of heterotrimeric G proteins by Smoothened. Proc Natl Acad Sci U S A. 2006;103:12607–12612. doi: 10.1073/pnas.0600880103. doi: 0600880103 [pii] 10.1073/pnas.0600880103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. doi: 10.1038/nature10361 nature10361 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katritch V, Cherezov V, Stevens RC. Structure-Function of the G Protein-Coupled Receptor Superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, et al. Serotonin inhibits apoptosis of pulmonary artery smooth muscle cell by pERK1/2 and PDK through 5-HT1B receptors and 5-HT transporters. Cardiovasc Pathol. 2013;22:451–457. doi: 10.1016/j.carpath.2013.03.003. doi: 10.1016/j.carpath.2013.03.003 S1054-8807(13)00106-3 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Wacker D, et al. Structural features for functional selectivity at serotonin receptors. Science. 2013;340:615–619. doi: 10.1126/science.1232808. doi: 10.1126/science.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White JF, et al. Structure of the agonist-bound neurotensin receptor. Nature. 2012;490:508–513. doi: 10.1038/nature11558. doi: 10.1038/nature11558 nature11558 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu F, et al. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. doi: :science.1202793 [pii] 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao XJ, et al. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc Natl Acad Sci U S A. 2009;106:9501–9506. doi: 10.1073/pnas.0811437106. doi: 0811437106 [pii] 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warne T, et al. The structural basis for agonist and partial agonist action on a beta(1)-adrenergic receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. doi: 10.1038/nature09746 nature09746 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rana R, et al. Structural insights into the role of the Smoothened cysteine-rich domain in Hedgehog signalling. Nat Commun. 2013;4:2965. doi: 10.1038/ncomms3965. doi: 10.1038/ncomms3965 ncomms3965 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanson MA, et al. A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. doi: 10.1016/j.str.2008.05.001 S0969-2126(08)00174-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manglik A, et al. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. doi: 10.1038/nature10954 nature10954 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. doi: 10.1038/nprot.2009.31 nprot.2009.31 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cherezov V, Peddi A, Muthusubramaniam L, Zheng YF, Caffrey M. A robotic system for crystallizing membrane and soluble proteins in lipidic mesophases. Acta Crystallogr D Biol Crystallogr. 2004;60:1795–1807. doi: 10.1107/S0907444904019109. doi: 10.1107/S0907444904019109 S0907444904019109 [pii] [DOI] [PubMed] [Google Scholar]
- 47.Cherezov V, et al. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 microm size X-ray synchrotron beam. J R Soc Interface. 2009;6(Suppl 5):S587–S597. doi: 10.1098/rsif.2009.0142.focus. doi: 10.1098/rsif.2009.0142.focus rsif.2009.0142.focus [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. doi: :Doi 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 49.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. doi: 10.1107/S0907444996012255 S0907444996012255 [pii] [DOI] [PubMed] [Google Scholar]
- 51.BUSTER v. 2.8.0. Cambridge, UK: Global Phasing Ltd.; 2009. [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. doi: 10.1107/S0907444910007493 S0907444910007493 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhat TN, Cohen GH. Omitmap - an Electron-Density Map Suitable for the Examination of Errors in a Macromolecular Model. Journal of Applied Crystallography. 1984;17:244–248. [Google Scholar]
- 54.Bhat TN. Calculation of an Omit Map. Journal of Applied Crystallography. 1988;21:279–281. [Google Scholar]
- 55.ICM Manual v. 3.0. La Jolla, CA: MolSoft LLC; 2012. [Google Scholar]
- 56.Totrov M, Abagyan R. Flexible protein-ligand docking by global energy optimization in internal coordinates. Proteins. 1997;Suppl 1:215–220. doi: 10.1002/(sici)1097-0134(1997)1+<215::aid-prot29>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.