Abstract
We have shown previously that an internal ribosome entry segment (IRES) directs the synthesis of the p36 isoform of Bag-1 and that polypyrimidine tract binding protein 1 (PTB-1) and poly(rC) binding protein 1 (PCBP1) stimulate IRES-mediated translation initiation in vitro and in vivo. Here, a secondary structural model of the Bag-1 IRES has been derived by using chemical and enzymatic probing data as constraints on the RNA folding algorithm Mfold. The ribosome entry window has been identified within this structural model and is located in a region in which many residues are involved in base-pairing interactions. The interactions of PTB-1 and PCBP1 with their cognate binding sites on the IRES disrupt many of the RNA-RNA interactions, and this creates a largely unstructured region of approximately 40 nucleotides that could permit ribosome binding. Mutational analysis of the PTB-1 and PCBP1 binding sites suggests that PCBP1 acts as an RNA chaperone to open the RNA in the vicinity of the ribosome entry window while PTB-1 is probably an essential part of the preinitiation complex.
Bag-1 has been shown to regulate various cellular pathways including apoptosis, cell survival, signal transduction, proliferation, transcription, and cell motility (reviewed in reference 27). In addition, Bag-1 expression is often altered in malignant cells, suggesting that it may play a role in tumorigenesis. It is thought that the pleiotropic effects of Bag-1 are due to its interaction with diverse cellular targets. Included among its interacting partners are Hsc70 and Hsp70, BCL-2, RAF-1 kinase, nuclear hormone receptors, and subunits of the ubiquitinylation-proteosome apparatus. Bag-1 interacts with the 70-kDa heat shock proteins via a C-terminal Bag domain and thereby facilitates nucleotide exchange on the heat shock protein (3, 23). Many of the functions of Bag-1, such as its effects on cell survival and nuclear hormone receptors, are dependent on this Bag domain. Indeed, the regulation of chaperone activity by Bag-1 may prove to be a common theme linking the disparate functions of Bag-1 (27).
Multiple Bag-1 proteins are generated from a single mRNA through the use of alternative translation initiation codons. In human cells, the major polypeptides produced are Bag-1S (36 kDa), Bag-1M (46 kDa) and Bag-1L (50 kDa) (20, 26). Synthesis of Bag-1L occurs from an upstream CUG, whereas Bag-1M and Bag-1S are initiated from the first and second in-frame AUG codons, respectively. Thus, all Bag-1 isoforms share a common C terminus, but Bag-1M and Bag-1L have extended N termini. Evidence has emerged indicating that the three Bag-1 isoforms can perform distinct functions. First, these isoforms are subject to differential subcellular localization. In general, Bag-1L is found chiefly in the nucleus, whereas Bag-1S is predominantly cytoplasmic and Bag-1M is distributed between both compartments (20, 26). In addition, there are examples of interactions involving a particular Bag-1 isoform. For instance, only Bag-1L interacts with the androgen receptor and as a result enhances androgen receptor trans activation in vivo. The specific recognition of this target by Bag-1L appears to be a function of both subcellular localization and residues present in the N-terminal extension of Bag-1L (27). Hence, the use of alternative initiation codons in Bag-1 greatly expands the capabilities of this gene.
Of the three isoforms, Bag-1S is generally expressed at the highest level in cells, followed by Bag-1L and finally Bag-1M (20, 26). However, in some tumor cells, Bag-1L can also be relatively abundant. An out-of-frame AUG codon and both the Bag-1L and Bag-1M initiation codons lie upstream of the Bag-1S initiation codon; hence the high level of Bag-1S expression indicates that the synthesis of this isoform must be subject to regulation. Rather than using the classic cap-dependent mechanism of translation initiation, the synthesis of Bag-1S involves the less common mechanism of internal initiation (4). In this alternative mechanism of translation initiation, the ribosome is recruited to an internal site on the mRNA that can be some considerable distance from the cap structure (reviewed in reference 6). A complex RNA structural element present in the 5′ untranslated region (UTR) of the mRNA, known as an internal ribosome entry segment (IRES), directs the ribosome to the mRNA. The Bag-1 5′ UTR has been shown to contain an IRES that drives the synthesis of Bag-1S. Moreover, the IRES is also essential for the expression of Bag-1S during the recovery period of cells that have been stressed by heat shock (4). Indeed, maintaining the production of polypeptides during a period when cap-dependent translation is compromised appears to be a common function of cellular IRESs (6).
It has been estimated that 5% of all mRNAs are translated by internal initiation (10). Despite the numerous examples of cellular IRESs identified thus far, little is known of their mechanism of action (6). Moreover, to date few secondary structural models have been derived for cellular IRESs (2, 11, 14, 21, 29).
In the Apaf-1 IRES, the ribosome entry window is located within a structured domain. However, interaction of the RNA binding proteins upstream of N-ras (unr) and polypyrimidine tract binding protein 1 (PTB-1) with their cognate binding sites on the IRES disrupts numerous RNA-RNA interactions within this region. As a result, the ribosome entry window attains an unstructured conformation and in doing so facilitates ribosome recruitment (17). A conformational change has also been implicated in the function of the Cat-1 IRES. Translation of a small upstream open reading frame appears to unfold inhibitory structures within the Cat-1 IRES. As a consequence, the IRES undergoes major structural remodeling and assumes an active conformation (29). Thus, it is clear from these examples that deriving an RNA structural model is a key step in any mechanistic study of a cellular IRES.
It has been demonstrated that the RNA binding proteins PTB-1 and poly(rC) binding protein 1 (PCBP1) interact with the Bag-1 IRES. Furthermore, the two proteins act in concert to stimulate the activity of the IRES in vitro and in vivo (22). PTB and PCBP1 have both been shown to affect the secondary structure of viral and cellular IRESs (1, 5, 17). As such, it was of interest to determine the effect of these proteins on the secondary structure of the Bag-1 IRES. By deletion analysis, a minimal active IRES element within the 5′ UTR of Bag-1 of 186 nucleotides (nt), which has the same activity as that of the full-length IRES, was determined(22). Here, we show that within the minimal active element, PTB-1 and PCBP1 disrupt several RNA-RNA interactions in a region juxtaposed to the ribosome entry site of the Bag-1 IRES. Mutational analysis of the PCBP1 and PTB-1 binding sites has shown that while PCBP1 is required to open the RNA in the region containing the ribosome entry window, PTB-1 may be required for ribosome recruitment.
MATERIALS AND METHODS
Cell culture and transient transfections.
HeLa cells were cultured under conditions described at the American Type Culture Collection website (http://www.atcc.org). Cells were transfected with FuGene 6 (Roche) according the supplier's instructions or with calcium phosphate (12).
The activities of firefly and Renilla luciferases in in vitro translation reactions or lysates prepared from transfected cells were measured by using a dual-luciferase reporter assay system (Promega), and light emission was measured over 10 s with an OPTOCOMP I luminometer. The activity of β-galactosidase (β-Gal) in lysates prepared from cells transfected with pCDNA3.1/HisB/lacZ was measured with a Galactolight plus assay system (Tropix). IRES activity in vivo was calculated as the average of (IRES-driven firefly luciferase expression/β-Gal expression) and efficiency expressed as (mutant IRES activity/wild-type IRES activity) × 100%. Errors were calculated as the standard deviation of the three calculated IRES activities and expressed as a percentage of the average activity.
Protein expression.
PTB-1 and PCBP1 were overexpressed in Escherichia coli from a pET28a vector by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to the growth medium. The proteins that contained a His tag were purified by using a nickel affinity column according to the supplier's instructions (QIAGEN).
In vitro transcription and in vitro translation.
Vector DNA pSKBL, pSKδBL (Fig. 1), or pRBF (Fig. 2) was linearized by restriction digestion using a site downstream of the sequence of interest (NcoI or HpaI, respectively). Transcripts were synthesized as described previously (4). For radiolabeled RNAs, 50 μCi of [α-32P]CTP was included in the reaction mixtures. After incubation of the reaction mixture for 1 h at 37°C, the RNA was isolated. This RNA (5 ng/μl) was used to prime a Promega rabbit reticulocyte Flexi lysate in vitro translation system according to manufacturer's instructions. The final volume of the reaction mixture was 12.5 μl. A total of 0.2 μg of either PCBP1 or PTB-1 was added to the translation reaction mixtures where indicated. Luciferase activities were determined (as described above), and the firefly and Renilla values are expressed relative to that of the control plasmid pRF, which was assigned a value of 1. All experiments were performed in triplicate on at least three independent occasions.
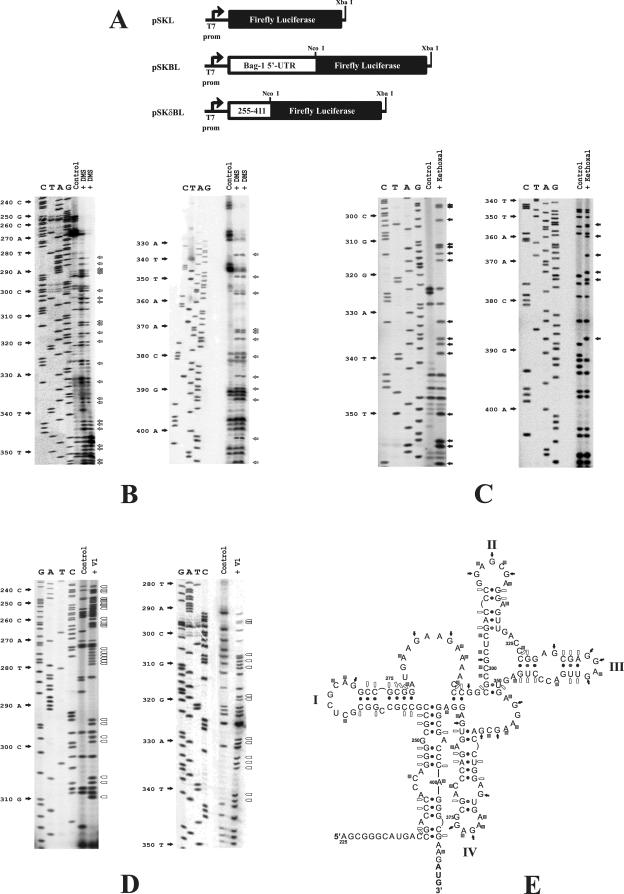
FIG. 1.
Chemical and enzymatic probing of the Bag-1 IRES. (A) Schematic diagram of the constructs used to generate the full-length version (pSKBL) or the deletion version (pSKδBL) of the Bag-1 IRES that still retains 100% of the activity of the wild-type IRES (22). (B to D) Renatured in vitro-transcribed Bag-1 IRES RNA was treated with DMS (two separate experiments are shown) (B), with kethoxal (C), or with RNaseV1 (D) as described in Materials and Methods. Primer extension was then performed with these samples and a mock-treated Bag-1 IRES RNA sample (control) as described above. Gray arrows denote bases modified by DMS, black arrows denote residues that are modified by kethoxal, and open arrows indicate residues that are sensitive to RNase V1 (V1) cleavage. (E) The secondary structural model of the Bag-1 IRES was derived by using the DMS, kethoxal, and RNase V1 accessibility data to constrain the Mfold algorithm (15, 31) Nucleotides involved in Watson-Crick pairing interactions and G-U base pairs are indicated with dots. Squares denote residues that are accessible to DMS modification, filled arrows denote residues that are sensitive to kethoxal modification, and open arrows indicate those residues that are sensitive to RNase V1 cleavage.
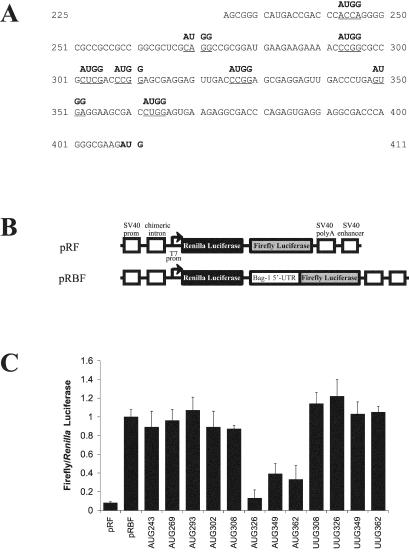
FIG. 2.
Locating the ribosome entry window in the Bag-1 IRES. (A) The sequence of the Bag-1 5′ UTR indicating the positions at which initiation codons were introduced. Site-directed mutagenesis was used to alter the underlined sequence to AUGG by using the oligonucleotides described in Materials and Methods. The position of each mutated sequence is numbered from the A of the AUGG. Mutated Bag-1 5′ UTR PCR fragments were then inserted into the dicistronic construct pRF, thereby creating the constructs pRB(AUG243)F, pRB(AUG269)F, pRB(AUG293)F, pRB(AUG302)F, pRB(AUG308)F, pRB(AUG326)F, pRB(AUG349)F, and pRB(AUG362)F. In addition, site-directed mutagenesis was used to introduce UUG codons into the Bag-1 5′ UTR. The sequence of the Bag-1 5′ UTR was altered to UUGG at nucleotides 308, 326, 349, and 362. (B) Schematic representation of the dicistronic reporter vector pRF which contains Renilla and firefly genes as the upstream and downstream cistrons, respectively. The 5′ UTR of Bag-1 was inserted into this vector in frame with the start codon to generate pRBF. (C) HeLa cells were transfected with the dicistronic construct containing the unaltered Bag-1 5′ UTR (pRBF) or the dicistronic constructs containing the mutations detailed above, in conjunction with pcDNA3.1/HisB/lacZ. Luciferase activities were then determined and normalized to the corresponding β-Gal activity. The data are represented as the ratio of firefly or Renilla luciferase activity from the mutant constructs compared to the activity from pRBF.
PCR mutagenesis.
The introduction of each upstream AUG or UUG and the production of the loop opening mutants was performed by PCR mutagenesis. First, PCR amplification of a pair of overlapping products derived from pRBF was performed by using the primer pairs and mutant primer. The products of these reactions separated on agarose Tris-borate-EDTA gels and used as a template for a second PCR with primers and PCR was performed with Pfu Turbo polymerase (Stratagene) according to the manufacturer's recommendations. The primers used for the AUG mutants were AUG 243 (5′-CGACCCAATGGGGGCGCCGCCGCCGGCGCT-3′), AUG 243R (5′-AGCGCCGGCGGCGGCGCCCCCATTGGGTCG-3′), AUG 269 (5′-GCTCGATGGCCGCGGATGAAGAAGAAAACC-3′), AUG 269R (5′-GGTTTTCTTCTTCATCCGCGGCCATCGAGC-3′), AUG 293 (5′-GGATGAAGAAAACATGGCGCCGCTCGACCC-3′), AUG 293R (5′-GGGTCGAGCGGCGCCATGTTTTCTTCATCC-3′), AUG 302 (5′-AAAACCCGGCGGCGATGGACCCGGAGCGAG-3′), AUG 302R (5′-CTCGCTCCGGGTCCATCGCCGCCGGGTTTT-3′), AUG 308 (5′-GGCGCCGCTCGACATGGAGCGAGGAGTTGA-3′), AUG 308R (5′-TCAAGTCCTCGCTCCATGTCGAGCGGCGCC-3′), AUG 326 (5′-CGAGGAGTTGACATGGAGCGAGGAGTTGAC-3′), AUG 326R (5′-GTCAACTCCTCGCTCCATGTCAACTCCTCG-3′), AUG 349 (5′-AGGAGTTGACCCGGAATGGGGAAGCGACCT-3′), AUG 349R (5′-AGGTCGCTTCCCCATTCCGGGTCAACTCCA-3′), AUG 362 (5′-AGCGACATGGAGTGAAGAGGCGACCCAGAG-3′), and AUG 362R (5′-CTCTGGGTCGCCATTTCACTCCATGTCGCT-3′). The mutant PCR product was digested with SpeI and NcoI and ligated into pRF.
Chemical structure probing.
The chemical probing protocol was adapted from a method described previously (24) and modified as described by Le Quesne et al. (14). RNA (5 μg) was combined with 5 μl of 10× standard structure probing buffer (100 mM Tris-HCl [pH 7.0], 1 M KCl), and the mixture was brought to 50 μl with the addition of water. The mixture was heated to 80°C for 3 min and cooled to 4°C over 1 h in a PCR machine and then chilled at 0°C for 10 min to permit structural equilibration. All chemical treatments were carried out at 0°C for 1 h. Dimethylsulfate (DMS) was diluted 1:12 in ethanol, and 5 μl was typically used per reaction. Kethoxal was diluted 1:20 in water, and 5 μl was used in each reaction. Mock-treated samples were prepared in parallel and were treated identically but with the omission of a chemical modifying agent. Chemical treatments were halted by ethanol precipitation after the addition of 50 μg of carrier tRNA.
For the treatment of RNA with RNase V1, the protocol was adapted from that given by the manufacturer (Ambion) for RNA structure analysis. Unlabeled RNA (2 μg) was combined with 1 μl of 10× structure buffer, 1 μg of tRNA, and 9 μl of water and RNase V1. After 10 min at room temperature, the enzyme was removed by phenol-chloroform extraction, and the cleaved RNA was precipitated with ethanol and resuspended in 5 μl of water.
Primer extension.
The procedure for primer extension was also adapted from the method described by Stern et al. (24). Primer (2 pmol/μl) was combined with 1 μl of hybridization buffer (250 mM K-HEPES [pH 7.0], 500 mM KCl) and 2.5 μl of RNA (i.e., in molar excess relative to the primer). All cDNA oligonucleotides along the length of the Bag-1 IRES were designed for primer extension analysis of RNA modification. The mixture was incubated at 85°C for 1 min and allowed to cool at room temperature for 10 to 15 min. Extension mix (3 μl) was added to the cooled hybrid consisting of 0.5 μl of C. Therm reverse transcriptase (Roche) and 1 μl of reverse transcriptase-PCR buffer (Roche), 0.33 μl of deoxynucleoside triphosphate stock (110 μM [each] dGTP, dATP, and dTTP and 6 μM dCTP), 0.66 μl of extension buffer (1.3 M Tris HCl [pH 7.4], 100 mM MgCl2, 100 mM dithiothreitol), and 0.5 μCi of [α-32P]dCTP. The reaction mixture was incubated at 37°C for 30 min, at which time 1 μl of chase mix (1 mM [each] dGTP, dCTP, dTTP, and dATP) was added, and incubation continued for a further 15 min. The reaction was stopped by precipitation of the DNA with 3 μl of 3 M NaAc (pH 5.4) and 90 μl of ethanol resuspended into 10 μl of gel loading buffer (7 M urea, 0.03% bromophenol blue dye), and the reaction products were separated on a 7 M urea-6% polyacrylamide sequencing gel.
Secondary structure prediction.
Secondary structure predictions were generated by using the web implementation of the Mfold algorithm (31) incorporating version 3.0 of the Turner rules (15).
UV cross-linking assay.
UV cross-linking was performed as described previously (16). Experiments were performed with 2.4 pmol (5 × 105 cpm) of radiolabeled RNA transcript that was incubated with 0.2 μg of PCBP1 and 0.2 μg of PTB-1. Samples were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis. Gels were then dried, and the results were visualized with a Molecular Dynamics phosphorimager.
RESULTS
Derivation of a secondary structural model for the Bag-1 IRES.
To derive a secondary structure model of the Bag-1 IRES, the accessibility of bases to chemical and enzymatic probes was determined. Briefly, Bag-1 IRES RNA was synthesized in vitro from both the full-length 5′ UTR and from the minimal active element (22) (Fig. 1A) and renatured as described previously (17). Since identical RNA modification data were obtained from both the full-length IRES (data not shown) and the minimal active element, this shorter RNA fragment was used in all subsequent experiments. Thus, this RNA was incubated with DMS (Fig. 1B), kethoxal (Fig. 1C), or RNase V1 (Fig. 1D). The effects of these reagents on the RNA are sensitive to the structural context of the individual residues; DMS will only modify unpaired A and C bases and kethoxal will modify unpaired G bases, whereas RNase V1 specifically cleaves the RNA prior to a paired residue. RNAs treated in this manner were then used as a template for primer extension analysis. In addition, renatured mock-treated RNA was subjected to the same analysis and serves as the control. The presence of an extension product that is relatively abundant in the treated reaction, compared to the control reaction, indicates a site that is accessible to modification or cleavage. The identity of these residues was determined by comparison with the corresponding DNA sequencing reaction (Fig. 1B to D).
It is clear that the sensitivity of many positions to these reagents cannot be resolved due to the presence of background in the control lanes (Table 1 and Fig. 1B to D). Such products have been attributed to robust structural features and particular sequence motifs that cause the premature termination of reverse transcription. Nevertheless, numerous residues were susceptible to modification or cleavage, and repeat experiments enabled us to generate a comprehensive set of data (Table 1). Strongly modified positions were used to constrain the RNA folding algorithm Mfold (15) and thereby generate several putative structures. The weakly modified positions were then used to further restrict the pairing interactions, and as a consequence, these structures were refined to derive a secondary structure model (Fig. 1E). The region from nt 225 to 411 of the Bag-1 IRES appears to adopt a complex structure containing four stem-loops. Interestingly, the structure suggests that a pseudoknot forms by base pairing of the G and the C at positions 297 and 298, respectively, with the U and the G at 350 and 351, a feature that has been found in other cellular IRESs, including C- and L-myc (11, 14).
TABLE 1.
Accessibility of nucleotides in the Bag-1 IRES to DMS and RNase VIa
| Position | Base | Background | RNase VI | DMS | Kethoxal |
|---|---|---|---|---|---|
| 237 | C | ++ | |||
| 238 | G | ++ | |||
| 240 | C | ++ | |||
| 241 | C | ++ | |||
| 242 | C | ++ | |||
| 243 | A | + | |||
| 245 | C | ++ | |||
| 246 | A | ++ | |||
| 247 | G | ++ | |||
| 248 | G | + | ++ | ||
| 249 | G | ++ | |||
| 251 | C | ++ | |||
| 252 | G | + | ++ | ||
| 253 | C | + | |||
| 256 | C | ++ | |||
| 257 | C | ++ | |||
| 261 | G | + | ++ | ||
| 263 | C | + | |||
| 265 | C | ++ | |||
| 268 | G | + | + | ||
| 269 | C | ++ | |||
| 270 | A | ++ | |||
| 271 | G | ++ | |||
| 272 | G | ++ | |||
| 273 | C | + | ++ | ||
| 274 | C | + | ++ | ||
| 275 | G | + | ++ | ||
| 276 | C | + | |||
| 277 | G | + | |||
| 278 | G | ++ | |||
| 279 | A | ++ | |||
| 280 | U | ++ | |||
| 282 | A | + | ++ | ||
| 283 | A | + | + | ||
| 284 | G | ++ | |||
| 285 | A | + | + | ||
| 287 | G | ++ | |||
| 288 | A | ++ | |||
| 289 | A | ++ | |||
| 290 | A | + | + | ||
| 292 | C | ++ | |||
| 293 | C | + | ++ | ||
| 294 | C | ++ | |||
| 295 | G | + | ++ | ||
| 296 | G | ++ | ++ | ||
| 297 | C | + | ++ | ||
| 298 | G | + | ++ | ||
| 299 | C | + | ++ | ||
| 300 | C | ++ | |||
| 301 | G | ++ | |||
| 302 | C | ++ | |||
| 304 | C | ++ | |||
| 305 | G | + | ++ | ||
| 306 | A | ++ | ++ | ||
| 307 | C | ++ | |||
| 308 | C | + | + | ||
| 309 | C | ++ | |||
| 310 | G | ++ | ++ | ||
| 311 | G | ++ | |||
| 312 | A | ++ | |||
| 313 | G | + | + | ++ | ++ |
| 314 | C | + | |||
| 315 | G | + | ++ | ||
| 316 | A | ++ | |||
| 317 | G | + | ++ | ||
| 318 | G | ++ | |||
| 319 | A | ++ | |||
| 320 | G | ++ | ++ | ||
| 321 | U | ++ | ++ | ||
| 322 | U | ++ | ++ | ||
| 325 | C | + | + | ||
| 326 | C | ++ | |||
| 327 | C | ++ | |||
| 328 | G | ++ | |||
| 329 | G | ++ | ++ | ||
| 330 | A | ++ | ++ | ||
| 331 | G | + | ++ | ++ | |
| 332 | C | ++ | |||
| 333 | G | ++ | |||
| 334 | A | + | ++ | + | |
| 335 | G | ++ | |||
| 336 | G | + | ++ | ++ | |
| 337 | A | ++ | |||
| 338 | G | ++ | |||
| 339 | U | + | ++ | ||
| 340 | U | ++ | |||
| 341 | G | ++ | ++ | ||
| 342 | A | + | |||
| 343 | C | + | + | ++ | |
| 344 | C | ++ | ++ | ||
| 345 | C | + | ++ | ||
| 346 | U | + | ++ | ||
| 347 | G | + | ++ | ||
| 348 | A | ++ | |||
| 349 | G | + | ++ | ||
| 350 | U | ++ | |||
| 351 | G | ++ | |||
| 352 | A | + | ++ | ||
| 353 | G | + | ++ | ++ | |
| 354 | G | ++ | ++ | ||
| 355 | A | + | |||
| 356 | A | + | ++ | ||
| 357 | G | ++ | |||
| 358 | C | + | ++ | ||
| 359 | G | + | ++ | ||
| 360 | A | ++ | |||
| 361 | C | ++ | |||
| 362 | C | ++ | ++ | ||
| 363 | U | ++ | |||
| 365 | G | ++ | |||
| 367 | G | ++ | |||
| 370 | A | ++ | |||
| 371 | A | ++ | |||
| 372 | G | + | |||
| 373 | A | ++ | |||
| 374 | G | + | ++ | ||
| 377 | G | + | |||
| 379 | C | ++ | |||
| 380 | C | ++ | |||
| 382 | A | ++ | |||
| 383 | G | + | |||
| 384 | A | ++ | |||
| 386 | U | ++ | |||
| 387 | G | + | ++ | ||
| 388 | A | + | ++ | ||
| 389 | G | ++ | |||
| 390 | G | ++ | |||
| 391 | A | ++ | |||
| 392 | G | + | + | ||
| 394 | C | ++ | + | ||
| 395 | G | ++ | |||
| 396 | A | ++ | |||
| 397 | C | ++ | |||
| 398 | C | ++ | |||
| 399 | C | ++ | |||
| 400 | A | ++ | |||
| 402 | G | ++ | |||
| 404 | C | ++ | ++ | ||
| 405 | G | ++ | |||
| 407 | A | ++ |
RNA derived from both pSKBL and pSKδBL was probed with DMS, kethoxal, and RNase V1, and the positions of modified bases that were consistently obtained in 10 separate experiments are shown. Plus signs indicate the strength of modification seen.
Locating the ribosome entry window on the Bag-1 IRES.
Having derived a structural model for the Bag-1 IRES, experiments were then performed to locate the region of the IRES to which the ribosome is recruited. For the cellular IRESs analyzed thus far, the data suggest that the 40S subunit associates with the IRES some distance upstream of the initiation codon, after which it migrates in a 5′-to-3′ direction until start codon recognition occurs (11, 14, 17). This “land and scan” mechanism is also found in the IRESs of the entero- and rhinoviruses (1).
To identify the ribosome entry window within the Bag-1 IRES, AUG codons were introduced into the 5′ UTR sequence at suitably spaced intervals by using site-directed mutagenesis (Fig. 2A). Furthermore, the sequence context of these AUG codons was altered to enhance initiation codon recognition (Fig. 2A). When these mutant 5′ UTR sequences were inserted into the intercistronic region of the dicistronic construct pRF (Fig. 2B) (25), the AUG codons were both upstream of and out of frame with the firefly luciferase initiation codon. A prior study showed that the Bag-1 5′ UTR promotes efficient internal initiation when this sequence is introduced into pRF (4). However, if an out-of-frame AUG codon is introduced into the 5′ UTR downstream of the ribosome entry window, polypeptide synthesis will occur from this upstream initiation codon, and consequently, firefly luciferase synthesis will be reduced. Conversely, an out-of-frame AUG codon placed upstream of the ribosome entry window will have no effect on firefly luciferase synthesis (17).
Dicistronic constructs (Fig. 2B) containing either the unaltered Bag-1 5′ UTR sequence or the mutant 5′ UTR sequences were transfected into HeLa cells. Comparing the expression of firefly luciferase from each mutant construct to that from the wild-type construct reveals that AUG codons introduced into the 5′ UTR at nucleotides 243, 269, 293, 302, and 308 had little or no effect (Fig. 2C). In contrast, the out-of-frame AUG codons positioned at nucleotides 326, 349, and 362 all substantially reduced the amount of firefly luciferase synthesis (Fig. 2C). To ensure that the effect of these mutations was due to the creation of an upstream open reading frame within the 5′ UTR and was not a consequence of a change in IRES structure, the sequence at these positions was altered to UUG. None of these UUG mutations decreased the synthesis of firefly luciferase (Fig. 2C). Hence, an initiation codon must be introduced at these sites to affect translation initiation at the firefly luciferase start codon. These data suggest that the ribosome engages with the Bag-1 5′ UTR between positions 308 and 326 (Fig. 2). Moreover, since the ribosome entry window is situated approximately 100 nt upstream of the authentic Bag-1 initiation codon, the data suggest that Bag-1 internal initiation occurs by a land and scan type of mechanism.
PTB-1 and PCBP1 disrupt RNA-RNA interactions in a region close to the ribosome entry window.
The mRNA binding track of the 40S ribosome can be divided into three sections: the region 5′ of the P site (11 nt), the P site itself (3 nt), and the region 3′ of the P site (12 to 17 nt) (13). Thus, in order to associate with the mRNA, the 40S subunit requires a tract consisting of 26 to 31 unpaired nucleotides. We have shown that the ribosome entry window of the Bag-1 IRES is located to a region between nt 308 and 326 (Fig. 3A), and it seems likely that structural remodeling must take place in this region prior to ribosome recruitment.
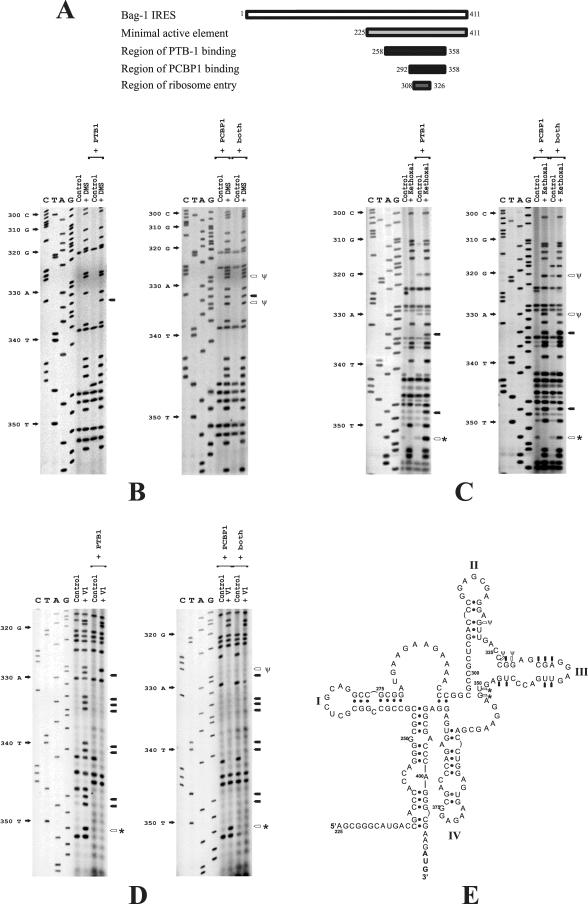
FIG. 3.
PTB-1 or PCBP1 binding to the Bag-1 IRES changes the accessibility of residues to chemical and enzymatic probes. (A) Schematic diagram showing the previously defined minimal active element of the Bag-1 IRES, the regions to which the proteins bind, and the ribosome landing site. Renatured in vitro-transcribed Bag-1 IRES RNA was incubated with a twofold molar excess of PTB-1 or PCBP1 and then treated with DMS (B), kethoxal (C), or RNase V1 (D) as described in Materials and Methods. Primer extension was then performed with these samples and a mock-treated Bag-1 IRES RNA sample (control) as described above. Arrows indicate bases that are newly modified by DMS (B) or kethoxal (C) or which are no longer available for RNase V1 cleavage (D) and are therefore predicted to be single stranded in the presence of protein. Open arrows and an asterisk denote residues that are modified by PTB alone, and open arrows and a ψ denote residues that are modified by PCBP1 alone. (E) Structure model of the Bag-1 IRES in the presence of PTB-1 or PCBP1. Black arrows indicate the bases on loop III which were formerly paired but which have become single stranded in the presence of both proteins, open arrows and an asterisk denote residues that are modified by PTB alone, and open arrows and a ψ denote residues that are modified by PCBP1 alone.
It has recently been demonstrated that the RNA binding proteins PCBP1 and PTB-1 act in concert to stimulate the activity of the Bag-1 IRES in vitro and in vivo (22). PCBP1 binds to a 66-nt region from positions 292 to 358, while PTB-1 binds to more than one site in the region that spans nt 258 to 358 (22) (Fig. 3A). Interestingly, the ribosome entry window is juxtaposed to both the PTB-1 and the PCBP1 binding regions on the Bag-1 IRES (Fig. 2 and 3A). Since PTB-1 and PCBP1 can have profound effects on RNA secondary structure, it was determined whether the interaction of these proteins with the IRES produced any changes to the RNA structure, in particular within the region around the ribosome entry window.
RNA-protein complexes were formed between renatured Bag-1 IRES RNA and either PCBP1 or PTB-1 singly or both proteins together, and the accessibility of residues within the IRES to chemical and enzymatic probes was determined (Fig. 3B to D). Primer extension analysis was performed to highlight the residues that were sensitive to modification or cleavage, and these positions were identified by comparison with the corresponding DNA sequencing reaction (Fig. 3B to D). It is clear that the binding of both PTB-1 and PCBP1 to the IRES elicits major changes in the RNA-RNA interactions. Thus, many residues within the region of positions 328 to 347 that were previously insensitive to DMS or kethoxal are modified by this reagent when the IRES is in a complex with PTB-1 and PCBP1 (Fig. 3B). Conversely, sites that were cleaved by RNase V1 in the unbound IRES were no longer accessible to this enzyme when either PTB or PCBP1 interacted with the IRES (Fig. 3C). Hence, in the absence of either PTB-1 or PCBP1, these residues are involved in base-pairing interactions, but when PTB-1 or PCBP1 bind to their cognate sites on the IRES, these residues become unpaired. However, while both proteins bind to stem-loop III, there are clear differences in the nucleotides to which the proteins bind. Thus, PTB-1 binds from nt 328 to 351, with strong interactions from 332 to 335 and 346 to 351, suggesting that this protein may either bind primarily diagonally across the stem-loop or that the loop contains a turn such that the two binding regions are adjacent. PCBP1 binds a larger section of RNA from nt 320 to 347 (Fig. 3E).
Further analysis of the Bag-1 IRES structure in the presence of PTB-1 and PCBP1 revealed that RNA-RNA interactions in other regions of the IRES are unaffected by these proteins (data not shown).
Mapping these alterations onto our secondary structural models shows that on binding of PTB-1 or PCBP1, the majority of RNA-RNA interactions from nt 320 to 355 are opened (Fig. 3E). It is noticeable that the region in which these changes occur is juxtaposed to the ribosome entry site. Consequently, the binding of PTB-1 or PCBP1 would create a largely single-stranded region of approximately 40 nt partially encompassing the ribosome entry site. Hence, ribosome recruitment would no longer be hindered by secondary structure in this region. Thus, we propose that the interactions of PTB-1 or PCBP1 with the IRES cause a conformational change in the vicinity of the ribosome entry window and thereby stimulate ribosome association with the IRES.
Mutational analysis.
To test whether protein binding was necessary for internal ribosome entry or, alternatively, if entry was due solely to the opening of the structure in the region from nt 320 to 355, four mutants were generated (named A, B, C, and D) (Fig. 4A and B). The PCBP1 consensus binding site has not been determined; however, a consensus site has been described for the α-complex which includes PCBP1 and PCBP2 (9). The α-complex consensus is (C/U)CCANXCCC(U/A)PyxUC(C/U)CC, and a C/U region fitting this general consensus is present in the Bag-1 IRES from nt 315 to 350. PTB-1 is known to bind polypyrimidine-rich tracts, and there is a 4-nt stretch of pyrimidines within the region which contains the putative PCBP1 binding site (from nt 343 to 346), although previous data suggest that PTB binds to additional sites in the region from nt 258 to 358 (22). The four mutants A, B, C, and D that were generated would both open stretches of stem-loop III and disrupt the PTB-1 and PCBP1 binding sites (Fig. 4A and B). Mutant A (Fig. 4A and B, panels ii) mimics the opening of the first half of stem-loop III by PCBP1 and PTB but at the same time alters the potential PTB1 binding region and part of the PCBP1 binding region. Mutant B disrupts a region to which both proteins bind and opens the second half of the stem-loop (Fig. 4A and B, panels iii). Mutant C mimics the opening of stem-loop III by PCBP1 and would also affect the PCBP1 binding site. However, this mutation would not be predicted to have much effect on PTB-1 binding since PTB interacts more strongly from residues 332 to 351 (Fig. 3 and 4A and B, panels iv). Finally, mutant D opens the second half of stem-loop III and may have a small effect on the binding of both PTB-1 and PCBP1 (Fig. 4A and B, panels v).
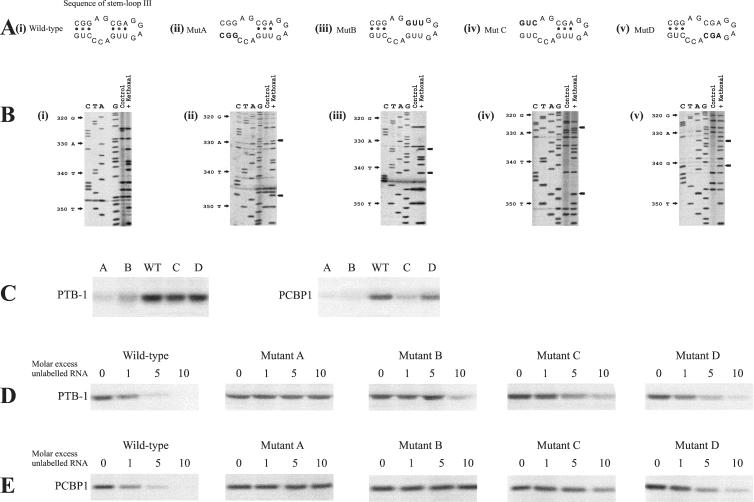
FIG. 4.
Mutation of PCBP1 and PTB-1 binding sites inhibits interaction of the Bag-1 IRES RNA with these proteins and affects the activity of the IRES in vitro. (A) Stem-loop III of the Bag-1 IRES (panel i) was mutated to create mutants A, B, C, and D (panels ii, iii, iv, and v, respectively) to alter the PCBP1 and PTB-1 binding sites and generate a structure where the RNA would be in a more open conformation. (B) Chemical probing of the Bag-1 IRES (panel i) and of the mutants A, B, C, and D (panels ii, iii, iv, and v, respectively) to identify residues which are differentially modified in the response to the loop-opening mutations. No other residues outside of this region showed any alterations (data not shown). (C) Radiolabeled RNA was generated from DNA encoding wild-type Bag-1 IRES or mutant A, B, C, or D by in vitro transcription. This RNA was then incubated with 0.2 μg of either PTB-1 or PCBP1, exposed to UV light, and then treated with RNases. The products of these reactions were separated on sodium dodecyl sulfate-10% polyacrylamide gels. The data show that the mutant versions of the IRES exhibit reduced binding of both PCBP1 and PTB-1 with mutants A and B, PCBP1 did not bind to mutant C, and mutant D had a much smaller effect. (D) UV cross-linking competition analysis was performed by incubating PTB-1 with radiolabeled wild-type Bag-1 IRES RNA in the presence of increasing amounts of unlabeled mutant A, B, C, or D IRES RNA. The data show that mutant A does not compete for binding with PTB-1 and that mutant B affects the binding of PTB-1 to the wild-type sequence only at a 10-fold molar excess. However, mutants C and D compete only slightly less well than does the wild type. (E) UV cross-linking competition analysis was performed by incubating PCPB1 with radiolabeled wild-type Bag-1 IRES RNA in the presence of increasing amounts of unlabeled mutant A, B, C, or D IRES RNA. The data show that mutants A, B, and C do not compete for binding with PCBP1. However, mutant D competes only slightly less well for binding than does the wild type.
Although the entire region from nt 225 to 441 was remapped, no additional changes other than those shown (Fig. 4B) were detected in the structure of the mutant versions of the Bag-1 IRES (data not shown).
The ability of PTB-1 and PCBP1 to bind to the mutated sequences was tested by UV cross-linking analysis. Thus, radiolabeled RNA was generated from the wild-type and mutated versions of the Bag-1 IRES and incubated with either PTB-1 or PCBP1 (Fig. 4C and D). As expected, PCBP1 interacted strongly with wild-type Bag-1; however, the binding of PCBP1 was very much reduced with mutants A, B, and C and to a smaller degree with mutant D (Fig. 4C). Again, PTB-1 interacted strongly with the wild-type sequence, and there was very much reduced binding to mutants A and B, albeit to a lesser extent (Fig. 4C). Mutants C and D did not have much effect on PTB-1 binding. The interactions of PCBP1 and PTB-1 with the mutant sequences were examined further by performing UV cross-linking analysis with radiolabeled wild-type Bag-1 IRES RNA and competing for binding with unlabeled RNA derived from the wild-type or mutant versions of the IRES (Fig. 4D and E). RNA derived from the mutant sequences A and B competed poorly for binding of PTB-1 with the wild-type IRES compared to wild-type unlabeled RNA, while the competition with mutants C and D was less affected. Thus, mutant A was unable to compete for binding of PTB-1 to the wild-type sequence, and the RNA derived from mutant B affected the binding of PTB-1 only at a 10-fold molar excess (Fig. 4D). In contrast, mutants C and D competed for binding at a 5- and 10-fold molar excess, but both of these competed less well than did wild-type RNA (Fig. 4D). The mutant sequences A, B and C were not able to compete well for binding with PCBP1; no competition at a 10-fold molar excess was observed with mutant A and B and only a slight reduction in binding of the PCBP1 to radiolabeled wild-type RNA was observed in the presence of a 10-fold molar excess of mutant C (Fig. 4E). These data agree with the data shown in Fig. 3 and suggest that the binding sites for PTB-1 and PCBP1 overlap in the central region of stem-loop III but that PCBP1 interacts more over a wider region from nt 420 to 347 while PTB-1 binds further towards stem IV.
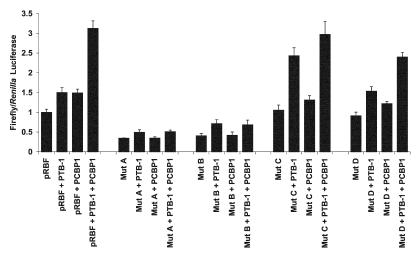
To determine whether the more open sequence generated by the mutations was sufficient to initiate translation by internal ribosome entry, in vitro translation assays were performed. Thus, RNA was generated from dicistronic constructs (Fig. 2B) from the wild-type or mutant versions of the Bag-1 IRES, and these were used to prime reticulocyte lysates in the presence or absence of PCBP1 or PTB-1. Luciferase assays were then performed to assess the effects of the mutations (Fig. 5). Mutant A and B versions of the IRES were both much less active than the wild-type IRES. This result suggests that a more open structure with mutated PTB-1 or PCBP1 binding sites was not sufficient for internal ribosome entry. Moreover, it shows that mutating the PTB-1 and PCBP1 binding sites had affected the ability of the low endogenous levels of these proteins present in these reticulocyte lysates (5) to bind and activate the IRES. The activity of the IRES was partially restored by the addition of PTB-1, albeit at a very reduced level, again suggesting that part of the PTB-1 binding site had been affected. Mutant C was as active as the wild type in the absence of proteins; however, in the presence of PTB-1, this version of the IRES was nearly twice as active as the wild-type IRES in the presence of PTB-1. There was a smaller stimulation upon the addition of PCBP1, and when assays were performed in the presence of both PTB and PCBP1, the activity of the IRES was the same as for the wild type. Mutant IRES D was less active than the wild type, yet it was stimulated by the addition of PTB-1 and PCBP1. Therefore, the opening of this part of stem-loop III is not sufficient to activate the IRES.
FIG. 5.
In vitro translation reactions using the mutant versions of the IRES in the presence of PTB-1 and PCBP1. RNA derived from pRBF or the mutated versions were used to prime rabbit reticulocyte lysates ± the addition of 200 ng of PTB-1 and/or PCBP1. Luciferase activities were determined (as described in Materials and Methods), and the firefly and Renilla values are expressed relative to that of the control plasmid pRBF, which was assigned a value of 1. All experiments were performed in triplicate on at least three independent occasions. The data show that mutants A and B exhibit a very reduced activity in this system. Mutant C is more active than the wild-type IRES in the presence of PTB-1, while mutant D shows a slight reduction in activity in all cases.
Taken together, these data suggest that PTB-1 and PCBP1 have disparate functions in activating the Bag-1 IRES with PCBP1 acting as an RNA chaperone to open part of stem-loop III to facilitate the binding of PTB-1, which would appear to be essential for Bag-1 IRES function.
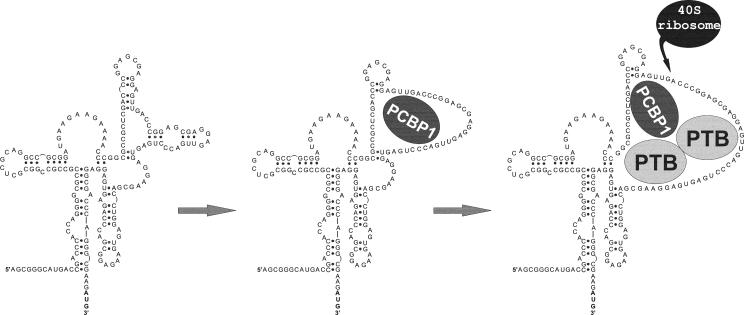
These data allow us to propose a final model (Fig. 6) whereby the binding of PCBP1 to the Bag-1 IRES RNA opens a loop region in the IRES structure that enhances the binding of PTB-1.
FIG. 6.
A model is proposed where the interactions of PCBP1 with the Bag-1 IRES in the region of nt 320 to 347 causes structural changes. As a consequence, a region of approximately 40 nt which overlaps the ribosome entry window opens to which PTB-1 can bind, and this in turn facilitates ribosome recruitment.
DISCUSSION
There is increasing evidence to show than many mRNAs (up to 10%) have the capability to initiate translation via internal ribosome entry (10). A large number of messages that contain IRESs are also subject to cap-dependent scanning, and in these cases, IRESs provide a molecular switch to initiate translation under conditions where cap-dependent scanning is compromised, including heat shock, apoptosis, and cell stress (6). Despite the growing numbers of IRESs that have been identified(http://ifr31w3.toulouse.inserm.fr/IRESdatabase/), very few structures of these RNA elements have been derived. Secondary structure models based on experimental data have to date been determined for only six cellular IRESs (2, 11, 14, 17, 21, 29). There is no obvious homology between these elements at the secondary structure level, and this is also true for the Bag-1 IRES, the structure of which bears no obvious similarity to those that have been derived previously (Fig. 1). All cellular IRESs tested to date use the land and scan mechanism, where the ribosome is recruited to a region which is upstream of the start codon and then migrates in a 5′-to-3′ direction until start codon recognition occurs (11, 14, 17) and are similar in this regard to rhino- and enteroviruses (1). The Bag-1 IRES also uses a land and scan type of mechanism, and the data show that the ribosome is recruited between nt 308 and 326 (Fig. 2).
The majority of cellular IRESs are relatively inactive in vitro and require IRES trans-acting factors (ITAFs) to function. These factors include PTB isoforms (16, 17), members of the poly(rC) protein family (5), hnRNPC (7), death-associated protein 5(19), and the autoantigen La (8). It has been proposed that a major role of ITAFs is to act as RNA chaperones either to maintain or to attain the correct three-dimensional structure of the IRES that is required for efficient assembly of the 48S complex. This has been shown for the Apaf-1 IRES, and in this case, a sequential action of two ITAFs is required. In the first instance, unr binds to the Apaf-1 IRES and opens a stem-loop structure that allows PTB-1/neuronal PTB to bind. This generates a large loop where the ribosome lands (17). For the Bag-1 IRES, the data suggest that the opening of the RNA occurs by the binding of PTB-1 and PCBP1 to a stem-loop (Fig. 3), and again, this is in a region proximal to the ribosome landing site (Fig. 2). In vitro PTB-1 and PCBP1 work in an additive manner; individually, these proteins stimulate the activity of the Bag-1 IRES by only 1.5-fold, yet together, these proteins increase the activity by 3-fold (22). Many of the same nucleotides were opened in the Bag-1 IRES by the binding of PTB-1 or PCBP1, and this can be explained by PTB-1 and PCBP1 binding to overlapping sites. However, PTB-1 binds additionally from nt 348 to 351, while PCBP1 binds a larger section of RNA from nt 320 to 347 (Fig. 3E).
To further identify the binding sites for the proteins and to test whether opening of the RNA alone was sufficient for ribosome recruitment, mutant versions of the Bag-1 IRES were created. It was possible to dissociate the functions of PTB-1 and PCBP1 by these mutations, and the data suggest that the prime role of PCBP1 is to open the structure to facilitate the binding of PTB-1 to nt 346 to 351, which is essential for activity. Therefore, mutations that affected the regions to which both PTB-1 and PCBP1 bound while opening up the structure were inactive (Fig. 4D and E and 5), but the mutant that opened up the structure and disrupted the binding of PCBP1 alone was more active in the presence of PTB-1 (Fig. 5). This finding would indeed suggest that while these proteins open a single-stranded region for ribosome entry, PTB-1 is also required for ribosome recruitment (Fig. 6). It is likely, therefore, that the activity of cellular IRESs will be regulated by changes in the intracellular levels and in subcellular localization of their trans-acting factors. Indeed, many cellular IRESs, including Bag-1 (our unpublished data), are almost completely inactive when present in dicistronic mRNAs introduced directly into the cytoplasm (by RNA transfection), suggesting that a “nuclear experience” is an essential prerequisite for internal initiation (25). In the case of Bag-1, both PCBP1 and PTB-1 are able to shuttle between the nucleus and the cytoplasm (18), and it is therefore possible that the complexes between Bag-1 and its ITAFs are formed in the nucleus. Cell signaling pathways that are activated in response to stress conditions when the Bag-1 IRES is functional (4) are probably also involved in the regulation of Bag-1 IRES function via modulating the activity or localization of the trans-acting factors. For example, it has been shown recently that nucleocytoplasmic shuttling of PTB-1 is regulated by the 3′-to-5′ cyclic AMP-dependent protein kinase A (28). In addition, PCBP1 is induced under cell stress conditions, and this induction is mediated via signaling through the mitogen-activated protein kinase pathway (30).
Further work is in progress to address how the ribosome is recruited to the IRES. This recruitment may occur by protein-protein interactions between PTB-1 and an additional factor(s), or it is possible that there is direct interaction between the 40S ribosomal subunit and single-stranded regions of Bag-1 RNA close to the ribosome landing region.
Acknowledgments
This work was supported by grants from the Wellcome Trust (S.A.M.) and the BBSRC (project grant support for K.A.S. and M.S. and an advanced fellowship held by A.E.W.). B.M.P. held an MRC studentship.
REFERENCES
- 1.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Bonnal, S., C. Schaeffer, L. Creancier, S. Clamens, H. Moine, A. C. Prats, and S. Vagner. 2003. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 278:39330-39336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briknarova, K., S. Takayama, L. Brive, M. L. Havert, D. A. Knee, J. Velasco, S. Homma, E. Cabezas, J. Stuart, D. W. Hoyt, A. C. Satterthwait, M. Llinas, J. C. Reed, and K. R. Ely. 2001. Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein. Nat. Struct. Biol. 8:349-352. [DOI] [PubMed] [Google Scholar]
- 4.Coldwell, M. J., M. L. deSchoolmeester, C. A. Fraser, B. M. Pickering, G. Packham, and A. E. Willis. 2001. The p36 isoform of BAG-1 is translated by internal ribosome entry following heat shock. Oncogene 20:4095-4100. [DOI] [PubMed] [Google Scholar]
- 5.Evans, J. R., S. A. Mitchell, K. A. Spriggs, J. Ostrowski, K. Bomsztyk, D. Ostarek, and A. E. Willis. 2003. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene 22:8012-8020. [DOI] [PubMed] [Google Scholar]
- 6.Hellen, C. U. T., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 7.Holcik, M., B. W. Gordon, and R. G. Korneluk. 2003. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol. Cell. Biol. 23:280-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holcik, M., and R. G. Korneluk. 2000. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell. Biol. 20:4648-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holcik, M., and S. A. Liebhaber. 1997. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA-protein complexes sharing cis and trans components. Proc. Natl. Acad. Sci. USA 94:2410-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johannes, G., M. S. Carter, M. B. Eisen, P. O. Brown, and P. Sarnow. 1999. Identification of eukaryotic mRNAs that are translated at reduced cap-binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. USA 96:13118-13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jopling, C. L., K. A. Spriggs, S. A. Mitchell, M. Stoneley, and A. E. Willis. 2004. L-Myc protein synthesis is initiated by internal ribosome entry. RNA 10:287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jordan, M., A. Schallhorn, and F. Wurm. 1996. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 24:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozak, M. 1997. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in position +5 and +6. EMBO J. 16:2483-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Quesne, J. P. C., M. Stoneley, G. A. Fraser, and A. E. Willis. 2001. Derivation of a structural model for the c-myc IRES. J. Mol. Biol. 310:111-126. [DOI] [PubMed] [Google Scholar]
- 15.Matthews, D., J. Sabina, M. Zuker, and D. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell, S. A., E. C. Brown, M. J. Coldwell, R. J. Jackson, and A. E. Willis. 2001. Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell. Biol. 21:3364-3374.11313462 [Google Scholar]
- 17.Mitchell, S. A., K. A. Spriggs, M. J. Coldwell, R. J. Jackson, and A. E. Willis. 2003. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 11:757-771. [DOI] [PubMed] [Google Scholar]
- 18.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 19.Nevins, T. A., Z. M. Harder, R. G. Korneluk, and M. Holcik. 2003. Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97lDAP5/NAT1. J. Biol. Chem. 278:3572-3579. [DOI] [PubMed] [Google Scholar]
- 20.Packham, G., M. Brimmell, and J. L. Cleveland. 1997. Mammalian cells express two differently localised Bag-1 isoforms generated by alternative translation initiation. Biochem. J. 328:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedersen, S. K., J. Christiansen, T. O. Hansen, M. R. Larsen, and F. C. Nielsen. 2002. Human insulin-like growth factor II leader 2 mediates internal initiation of translation. Biochem. J. 363:37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickering, B. M., S. A. Mitchell, J. R. Evans, and A. E. Willis. 2003. Polypyrimidine tract binding protein and poly r(C) binding protein 1 interact with the BAG-1 IRES and stimulate its activity in vitro and in vivo. Nucleic Acids Res. 31:639-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sondermann, H., C. Scheufler, C. Schneider, J. Hohfeld, F. U. Hartl, and I. Moarefi. 2001. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291:1553-1557. [DOI] [PubMed] [Google Scholar]
- 24.Stern, S., D. Moazed, and H. Noller. 1998. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164:481-489. [DOI] [PubMed] [Google Scholar]
- 25.Stoneley, M., T. Subkhankulova, J. P. C. Le Quesne, M. J. Coldwell, C. L. Jopling, G. J. Belsham, and A. E. Willis. 2000. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 28:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takayama, S., S. Krajewski, M. Krajewska, S. Kitada, J. M. Zapata, K. Kochel, D. Knee, D. Scudiero, G. Tudor, G. J. Miller, T. Miyashita, M. Yamada, and J. C. Reed. 1998. Expression and location of Hsp70/Hsc-binding anti-apoptotic protein BAG-1 and its variants in normal tissues and tumor cell lines. Cancer Res. 58:3116-3131. [PubMed] [Google Scholar]
- 27.Townsend, P. A., R. I. Cutress, A. Sharp, M. Brimmell, and G. Packham. 2003. BAG-1: a multi-functional regulator of cell growth and survival. Biochim. Biophys. Acta 1603:83-89. [DOI] [PubMed] [Google Scholar]
- 28.Xie, J. Y., J. A. Lee, T. L. Kress, K. L. Mowry, and D. L. Black. 2003. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 100:8776-8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yaman, I., J. Fernandez, H. Y. Liu, M. Caprara, A. A. Komar, A. E. Koromilas, L. Y. Zhou, M. D. Snider, D. Scheuner, R. J. Kaufman, and M. Hatzoglou. 2003. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell 113:519-531. [DOI] [PubMed] [Google Scholar]
- 30.Zhu, Y., Y. Sun, X. O. Mao, K. L. Jin, and D. A. Greenberg. 2002. Expression of poly(C)-binding proteins is differentially regulated by hypoxia and ischemia in cortical neurons. Neuroscience 110:191-198. [DOI] [PubMed] [Google Scholar]
- 31.Zucker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244:48-52. [DOI] [PubMed] [Google Scholar]