Abstract
Vault poly(ADP-ribose) polymerase (VPARP) was originally identified as a minor protein component of the vault ribonucleoprotein particle, which may be involved in molecular assembly or subcellular transport. In addition to the association of VPARP with the cytoplasmic vault particle, subpopulations of VPARP localize to the nucleus and the mitotic spindle, indicating that VPARP may have other cellular functions. We found that VPARP was associated with telomerase activity and interacted with exogenously expressed telomerase-associated protein 1 (TEP1) in human cells. To study the possible role of VPARP in telomerase and vault complexes in vivo, mVparp-deficient mice were generated. Mice deficient in mVparp were viable and fertile for up to five generations, with no apparent changes in telomerase activity or telomere length. Vaults purified from mVparp-deficient mouse liver appeared intact, and no defect in association with other vault components was observed. Mice deficient in mTep1, whose disruption alone does not affect telomere function but does affect the stability of vault RNA, showed no additional telomerase or telomere-related phenotypes when the mTep1 deficiency was combined with an mVparp deficiency. These data suggest that murine mTep1 and mVparp, alone or in combination, are dispensable for normal development, telomerase catalysis, telomere length maintenance, and vault structure in vivo.
Telomerase is a ribonucleoprotein (RNP) complex that replenishes telomere loss due to incomplete DNA replication in almost all eukaryotes. A loss of telomerase activity leads to an attrition of telomeric DNA which, in turn, is known to trigger end-to-end chromosome fusions, genomic instability, and cell arrest or death (31). Telomerase expression is thus critical to the prolonged viability of several human cell types and in the majority of human cancers (3). Elucidating the molecular machinery that regulates telomerase activity and its ability to maintain telomere length is critical to an understanding of malignant transformation.
Telomerase contains two core components, telomerase reverse transcriptase (TERT) and telomerase RNA, the latter serving as an integral template for the de novo synthesis of telomeric DNA. Both TERT and telomerase RNA are required for the reconstitution of telomerase activity in vitro (2, 75). In mice, the disruption of either component also abolishes telomerase activity, leading to telomere attrition, genetic instability, and eventual infertility (4, 15, 26, 44, 48, 61, 76).
Several other telomerase-associated proteins have been identified in mammals (22); these include telomerase-associated protein 1 (TEP1), which binds telomerase RNA (23, 55) and which was cloned based on its homology to ciliate Tetrahymena thermophila protein p80 (8, 20, 23, 55). T. thermophila p80 was identified as a species copurifying with telomerase, although it subsequently was found unlikely to be a core telomerase component (8, 20, 49). Although deletion of the T. thermophila p80 gene resulted in slight telomere lengthening (51), no change in telomere length was observed upon breeding of mTep1-deficient mice for up to seven generations. Telomerase activity was not affected in the absence of either the p80 gene or mTep1 (47, 51). Disruption of mTep1 did not affect the levels of telomerase RNA or its association with the telomerase RNP (34), and TEP1 appeared to associate with only a fraction of the total telomerase activity in immortalized cell extracts (Y. Liu and L. Harrington, unpublished data). Although these results suggest that TEP1 is nonessential for telomerase function in normal mouse tissues, the possibility of genetic redundancy with other telomerase-associated proteins cannot be excluded. Indeed, other telomerase-associated proteins have been identified and have been shown to interact with telomerase RNA in mammals; these include La, L22, Staufen, DKC, and several heterogeneous nuclear RNPs (11, 16-19, 29, 42, 43, 52).
TEP1 is an integral component of another RNP, the vault particle. Mammalian vaults, the largest known mammalian RNPs (13 MDa), are composed of at least four components, major vault protein (MVP), vault poly(ADP-ribose) polymerase (VPARP), TEP1, and one or more small vault RNAs (vRNAs) (71, 73). Vaults possess a distinct morphology that is highly conserved. Purified vaults display a unique eightfold barrel-like symmetry structure with caps on each end (32, 38). Although the function of the vault particle has remained elusive, its highly conserved structure, its ubiquitous distribution, and its up-regulation in several human drug-resistant cancers have led to the speculation that vaults have an important cellular function and may be carriers involved in intracellular transport (54, 71, 73). The absence of TEP1 completely disrupts the stable association of vRNA with the purified vault particle and results in decreases in the levels and stability of vRNA (34). Therefore, TEP1 is an integral vault protein and is important for the stabilization and recruitment of vRNA to the vault particle (34).
VPARP, the catalytic vault protein component, contains regions with similarity to BRCT, a poly(ADP-ribose) polymerase (PARP) catalytic domain, inter-α-trypsin and putative von Willebrand type A domains, and a C-terminal MVP-interacting domain (35, 71, 73). The putative VPARP catalytic domain shares 28% identity with the catalytic domain of PARP 1 (PARP1). Like that in PARP1, this domain is capable of catalyzing a poly(ADP-ribosyl)ation reaction, and the substrates for this vault-associated PARP activity are MVP and VPARP itself (35). Thus, VPARP is a unique member of the PARP family. In addition to its association with vaults, VPARP has also been found at other cellular locations, such as the nucleus and mitotic spindle (35), indicating that it may possess multiple roles in vivo. Here, we report that VPARP interacts with TEP1 and associates with telomerase activity in cell extracts, suggesting that VPARP and TEP1 may play roles in both cytoplasmic and nuclear RNP complexes. We generated mice deficient in mVparp or both mTep1 and mVparp and investigated telomerase function and vault structure in their absence.
MATERIALS AND METHODS
Construction of an mVparp targeting vector.
The mVparp targeting construct was designed to delete two exons (exons 3 and 4) of mVparp, including a 2-kbp region corresponding to the N terminus of the predicted PARP catalytic domain (amino acids [aa] 119 to 240). In brief, PCR primers 5′-GGT ACC TTA GGA ATC TTT GCA AAT TGT ATA TTC TGC-3′ (KpnI sense) and 5′-CTC GAG GCA GAT ATT CTG ATT GCA CTT CTG G-3′ (XhoI antisense) were used to amplify an ∼3.6-kbp long-arm fragment of mVparp from 200 ng of strain 129J genomic DNA by using an Expand long-template PCR system (Roche GmbH, Mannheim, Germany). An ∼874-bp short-arm fragment was isolated by using PCR primers 5′-ACT AGT TAG CTT AGA ACT CAT TAT GTA GAC CAG GTT AGC C-3′ (sense) and 5′-GCG GCC GCA GTA CTT CCT TCT TTG AGA CAG GGC TCC-3′ (antisense). The resulting PCR products were gel purified by using a QiaQuick gel purification system (Qiagen Inc., Chatsworth, Calif.), TA cloned into pCR2.1 (Invitrogen, San Diego, Calif.), and subcloned into pBluescript II KS (Stratagene, La Jolla, Calif.) containing a phosphoglycerate kinase-neomycin cassette. The insert sequence was obtained by using fluorescent dideoxynucleotide sequencing and automated detection (ABI/Perkin-Elmer, Forest City, Calif.).
Targeted disruption of the mVparp gene in mouse ES cells.
The targeting vector (25 μg) was linearized with restriction endonuclease NotI at the short arm and electroporated into E14 embryonic stem (ES) cells (derived from strain R129J) by using a Bio-Rad gene pulser at 0.34 kV and 0.25 mF. After G418 selection (250 μg/ml), homologous recombinants were identified by PCR and confirmed by Southern blot analysis following published protocols (21). Primer VParp Wt-1, which is specific for the deleted portion of mVparp, was used to detect the wild-type allele. Primer PGK-1, which is specific for the phosphoglycerate kinase promoter of the targeting construct, was used to detect the mutant allele. Primer VParp Wt-2 was used to detect both the wild-type and the mutant alleles of mVparp. The sequences of the above primers were as follows: VParp Wt-1, 5′-GGC CGA AGC AAC ACA GTT AGC GTC TGA G-3′; PGK-1, 5′-GCT GTC CAT CTG CAC GAG ACT AGT GAG ACG-3′; and VParp Wt-2, 5′-CTT TTG GCC CCT GTC AGT ACA CGT AAT GCA C-3′.
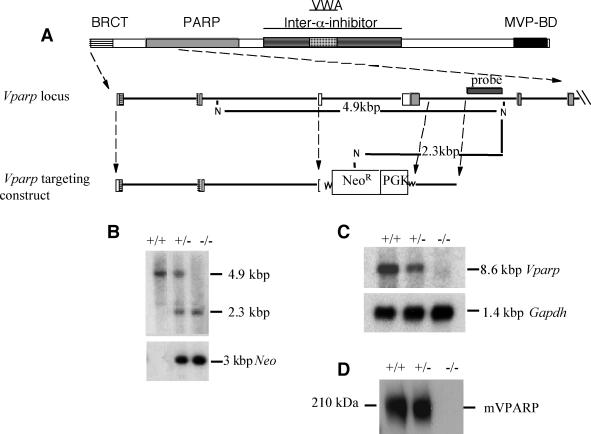
Homologous recombination of the targeting vector with the endogenous locus resulted in the insertion of a novel NcoI site into the mVparp locus, thus allowing the targeted and wild-type alleles to be distinguished by Southern analysis with a 524-bp 3′-flanking genomic probe (3′ probe). The ∼4.9-kbp NcoI fragment corresponding to the wild-type allele was decreased to ∼2.3 kbp upon disruption of the mVparp locus by integration of the neomycin phosphotransferase gene. The 3′-flanking probe (see Fig. 2A) was generated by ScaI and XhoI digestion of a genomic clone immediately 3′ of the short arm. Genomic DNA was digested with NcoI, resolved on an agarose gel, transferred to a Hybond N+ membrane, and hybridized to a probe spanning a region 3′ to the mVparp targeting construct (3′ probe).
FIG. 2.
Disruption of the mVparp gene in ES cells and mice. (A) Schematic representation of the homologous recombination of the targeting vector with the endogenous locus. Insertion of a new NcoI (N) site into the targeted locus allows the targeted and wild-type alleles to be distinguished by Southern blot analysis with the indicated 3′-flanking probe. BRCT, breast cancer gene 1; VWA, von Willebrand type A; BD, binding domain; PGK, phosphoglycerate kinase. (B) Detection of targeted and wild-type mVparp alleles by Southern blot analysis of DNA in G1 mVparp−/− mice. DNA was digested with NcoI and hybridized with the probe shown in panel A. In the upper panel, the 4.9-kbp NcoI fragment corresponding to the wild-type allele is decreased to 2.3 kbp upon disruption of the locus. The lower panel is the same blot reprobed with a probe specific for the Neor gene. (C) Detection of mVparp transcripts in wild-type (+/+), mVparp+/−, and mVparp−/− ES cells by Northern blotting. The upper panel shows no detectable mVparp transcript in mVparp−/− ES cells; in the lower panel, the blot was probed with a murine glyceraldehyde 3-phosphate dehydrogenase (Gapdh) transcript as a control. (D) Lack of detection of mVPARP in mVparp−/− MEFs by Western blot analysis. Cell lysates of MEFs derived from G1 mouse embryos of the indicated genotypes were probed with an anti-VPARP polyclonal antibody (see Materials and Methods). The position of mVPARP is shown, and a kilodalton marker is indicated at the left.
Generation of mVparp-deficient mice, ES cells, and mouse embryonic fibroblasts (MEFs).
Chimeric mice were produced by microinjection of independent mVparp heterozygous (mVparp+/−) ES cell clones into embryonic day 3.5 C57BL/6J blastocysts and transfer to ICR pseudopregnant foster mothers. Chimeric males were mated with C57BL/6J females (Jackson Laboratory). Germ line transmission of the mutant allele (mVparp+/− founder mice) was confirmed by PCR and Southern blot analysis of tail DNA from mice with an agouti coat color. The mating strategy for the mVparp-deficient (mVparp−/−) mice was the same as that used for the generation of telomerase RNA-deficient mice (4). In brief, the first generation of mVparp−/− mice was obtained through mating of mVparp+/− founder mice derived from the same ES cell line. Subsequent generations (G2 to G5) were obtained by mating two mVparp−/− mice of an equivalent generation but derived from different sets of parents (i.e., cousins). To generate mice deficient in both mVparp and mTep1, we first bred mVparp−/− with mTep1−/− mice to obtain mVparp+/− mTep1+/− mice, which were subsequently bred to generate mVparp+/− mTep1−/− or mVparp−/− mTep1+/− mice. Due to the close vicinity (<5 Mbp) of the mVparp and mTep1 genes on mouse chromosome at band 14q11.2, mVparp−/− mTep1−/− mice were obtained through mVparp−/− mTep1+/− or mVparp+/− mTep1−/− interbreeding.
mVparp−/− mouse ES cell clones were generated from G418-resistant mVparp+/− ES cell clones by incubation at an increased G418 concentration (4 mg/ml). MEFs were established from day 13.5 mouse embryos obtained from an mVparp+/− female mouse. ES cell and MEF culturing was carried out as previously described (21, 25).
Cloning of hVPARP and hTEP1.
Full-length human VPARP (hVPARP) cDNA was cloned as described previously (35) and subcloned into the pCR3 expression vector (Invitrogen). The FLAG epitope tag (DYKDDDDK) was introduced at the 5′ region of the gene by PCR and verified by sequencing as described previously (24). The full-length human TEP1 (hTEP1) gene was cloned into the pCR3.1 vector and contained two copies of an NH2-terminal MYC epitope tag as described previously (23).
Cell lines and transfections.
FLAG-tagged full-length hVPARP and/or full-length hTEP1 containing a MYC epitope (23) were transfected alone or cotranfected into human 293T cells with Lipofectamine Plus reagents (Life Technologies, Gaithersburg, Md.) following the manufacturer's instructions.
Immunoprecipitation.
A total of 500 μg of cell or tissue extract was subjected to immunoprecipitation with 10 μl of M2 affinity resin (Sigma, St. Louis, Mo.) or protein G beads conjugated with anti-MYC monoclonal antibody (Pharmingen, San Diego, Calif.) in 0.5% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)-10 mM Tris-HCl (pH 7.5)-1 mM MgCl2-1 M NaCl-5 mM β-mercaptoethanol-10% (vol/vol) glycerol. Beads were washed in the same buffer four times before they were examined by the telomere repeat amplification protocol (TRAP) or Western blot analysis (see below).
Western blot analysis.
For Western blot analysis, 40 μg of tissue or cell extract or beads from immunoprecipitation were examined for the presence of VPARP with an anti-FLAG polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.) or an anti-murine VPARP (mVPARP) rabbit polyclonal antibody (see below) or for the presence of TEP1 with an anti-MYC monoclonal antibody.
Antibody production.
The mVPARP peptide CDQNTEEFSRVRKEVLQNNRSEQ (aa 379 to 400) was used to generate a rabbit polyclonal antibody against mVPARP. The antibody was affinity purified against Affigel resin coupled to the same peptide following the manufacturer's directions (Bio-Rad). The specificity of this polyclonal antibody was confirmed by Western blot analysis with cell extracts from human 293T cells overexpressing recombinant mVPARP. This antibody was not able to efficiently precipitate either mVPARP or hVPARP (data not shown).
Cell lysate preparation and telomerase assays.
S-100 extracts from cultured cells and freshly dissected mouse tissues were prepared as described previously (58). Cells lysed in a buffer containing 0.5% (wt/vol) CHAPS were prepared as described previously (37). Cell extract or a portion of M2 beads after immunoprecipitation was assayed for the presence of telomerase activity by using the TRAP assay following the manufacturer's instructions (Intergen, Inc.) (37). Titration of a cell extract was used to demonstrate that the TRAP products were in the nearly linear range.
Telomere length measurements by FISH.
The average telomere fluorescence in populations of isolated splenocytes and thymocytes was measured by flow fluorescence in situ hybridization (FISH) (62) with minor modifications. A telomere-specific fluorescein isothiocyanate-conjugated (CCCTAA)3 peptide nucleic acid probe (0.3 μg/ml) (Perseptive Biosystems) was used. Telomere fluorescence was expressed as molecules of equivalent soluble fluorochrome.
Metaphase spread, FISH, and image analyses of activated splenocytes and MEFs were performed as described previously (4, 77). Cy-3-labeled (CCCTAA)3 peptide nucleic acid (Applied Biosystems) was used as a probe.
G overhang analysis by pulsed-field gel electrophoresis.
Telomere restriction fragment analysis of mouse genomic DNA was carried out as described previously (25). Approximately 106 ES cells or an aliquot corresponding to approximately 1 g of mouse liver was harvested and embedded in agarose plugs. The DNA was digested with 100 U of mung bean nuclease for 30 min, where indicated, and further treated with MboI (New England BioLabs) overnight. The DNA fragments were electrophoresed through 1% (wt/vol) pulsed-field-grade agarose (Bio-Rad) in Tris-acetate-EDTA. Electrophoresis was carried out with a CHEF DR-III pulsed-field apparatus (Bio-Rad) at 14°C, 3 V/cm, and a switch time of 10 s for 48 h. In-gel hybridization was carried out under native and denaturing conditions as described previously (25).
Vault particle purification and analysis.
Vaults were purified from mouse liver as described previously (34, 39). Approximately 10 g of mouse liver was used, and all gradient steps were carried out with a Sorvall AH650 rotor at 25,000 rpm. In the final purification step, vaults were purified over a single cesium chloride gradient to minimize sample loss, and the purified vaults were pelleted at 100,000 × g with a Beckman Coulter Ti80 rotor and resuspended in approximately 125 μl of 0.09 M morpholineethanesulfonic acid (pH 6.5) containing 1 mM dithiothreitol and 0.5 mM phenylmethylsulfonyl fluoride. Purified vaults were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by silver staining or immunoblot analysis. All antibodies (anti-MVP, anti-VPARP, and anti-TEP1) were described previously and were used accordingly (24, 35, 36). RNA was isolated by phenol-chloroform extraction followed by ethanol precipitation and was analyzed as previously described (34). Electron microscopy (EM) of uranyl acetate-stained vaults was carried out as described previously (33).
RESULTS
VPARP interacts with TEP1 and associates with telomerase activity in vivo.
TEP1 has been shown to associate with telomerase activity in human, mouse, and rat immortalized cell lysates (23, 55). TEP1 also has been identified as a component of the vault cap, adjacent to vRNA (34, 38). As TEP1 and VPARP have similar subcellular fractionation patterns, we decided to determine whether TEP1 and VPARP associate and/or whether VPARP, like TEP1, associates with telomerase activity in cell extracts.
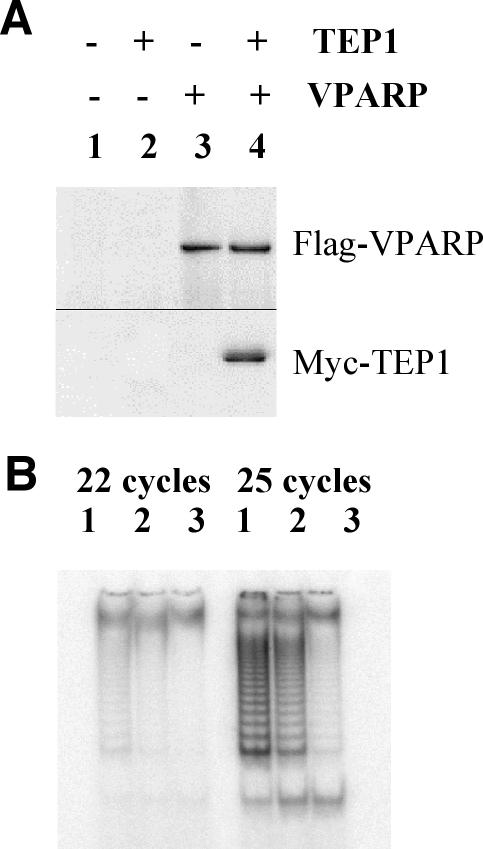
Human 293T cells were transfected with FLAG-tagged full-length hVPARP or MYC-tagged full-length hTEP1 or both. Cell extracts were subjected to immunoprecipitation with an anti-FLAG antibody followed by Western blot analysis. Coimmunoprecipitation of hTEP1 with hVPARP was observed in cells cotransfected with both hVPARP and hTEP1 (Fig. 1A, lane 4) but not in cells transfected with full-length hTEP1 or full-length hVPARP alone (Fig. 1A, lanes 2 and 3). As 293T cells do not express MVP (V. A. Kickhoefer et al., unpublished data), these results suggest that the non-vault-associated soluble forms of VPARP and TEP1 interact. Attempts to demonstrate a direct interaction of endogenous VPARP and TEP1 in 293T cells were unsuccessful (data not shown). Several explanations for these results are possible, including low levels of endogenous TEP1 in 293T cells (Liu and Harrington, unpublished), the inaccessibility of the antigens in the complex, or the potential disruption of the complex upon antibody interaction.
FIG. 1.
Overexpressed hVPARP interacts with hTEP1 and associates with telomerase activity in cell extracts. (A) FLAG-tagged full-length hVPARP and/or full-length hTEP1 containing a MYC epitope was transfected alone or cotranfected into human 293T cells. Cell extracts were subjected to immunoprecipitation with an anti-FLAG antibody and examined for the presence of FLAG-hVPARP (top panel, probed with anti-FLAG antibody) or for coimmunoprecipitation of hTEP1 (bottom panel, probed with anti-MYC antibody). (B) FLAG-tagged full-length hVPARP (lane 1), truncated hVPARP (aa 1 to 611) (lane 2), or an empty vector (lane 3) was transfected into human 293T cells. Cell extracts were subjected to immunoprecipitation with an anti-FLAG antibody and assayed for telomerase activity. A TRAP assay was performed for 22 cycles (left panel) or 25 cycles (right panel).
Since TEP1 associates with telomerase activity in human immortalized cells, we next examined whether hVPARP associates with telomerase activity in human 293T cells. Full-length FLAG-tagged hVPARP, C-terminally truncated hVPARP (aa 1 to 611), containing BRCT and PARP but lacking the von Willebrand type A domain and the MVP-interacting domain, or empty vector alone was transiently expressed in 293T cells followed by affinity purification on M2 beads. A portion of the M2 beads was further tested for the presence of telomerase activity by the TRAP assay (37). Immunoprecipitates containing either full-length hVPARP or truncated hVPARP but not mock-transfected controls possessed telomerase activity (Fig. 1B, lanes 1 and 2). This result indicates that overexpressed hVPARP associates with a fraction of telomerase activity in cell extracts. In addition, since it was previously established that the C-terminal domain of VPARP is responsible for its interaction with MVP (35), the ability of C-terminally truncated VPARP to associate with telomerase activity suggests that this interaction is independent of a VPARP association with vaults. Since we were unable to detect an association of endogenous VPARP with endogenous TERT (due to limitations of the VPARP antibody and the low levels of endogenous TERT), the robustness of the interaction at physiologically relevant levels or the stoichiometry of the association between endogenous components could not be tested.
Targeted disruption of mVparp or both mVparp and Tep1 in mice.
To study the possible role of VPARP as a telomerase-associated protein in telomerase function in mammalian cells, we generated murine ES cells containing a disruption of exons 3 and 4 of the mVparp gene (Fig. 2A). Two separate mVparp+/− ES cell clones were used to generate mVparp+/− founder mice. One of the mVparp+/− ES cell clones was used to generate mVparp−/− ES cell clones. mVparp+/− founders were bred to generate mVparp−/− mice (Fig. 2B). Northern analysis of total RNA from mVparp−/− mouse ES cells or tissues showed no detectable transcripts upon hybridization to a full-length mVparp cDNA probe (Fig. 2C and data not shown). Analysis of cell extracts prepared from mVparp−/− MEFs revealed no detectable mVPARP (Fig. 2D).
The null mice were designated generation 1 (G1), and the progeny of G1 cousins were defined as G2, and so forth, as described previously (4) (see Materials and Methods). Regardless of the generation (G1 to G5), the mVparp−/− mice appeared to be normal and fertile and exhibited no obvious phenotype.
VPARP is dispensable for telomerase activity, telomere length maintenance, telomere structure, or stability of telomerase and vault components in vivo.
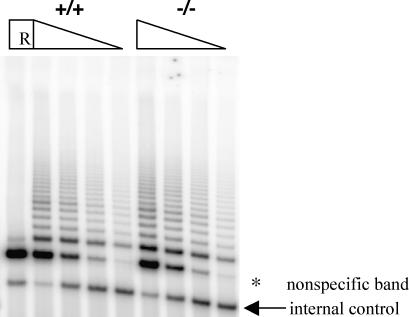
To determine the role of mVparp in telomerase catalysis, we examined mVparp−/− mouse ES cells, MEFs derived from G1 mVparp−/− embryos, and several tissues from mVparp−/− mice (up to G5) for telomerase activity by using the TRAP assay. Telomerase activity was not significantly altered in mVparp−/− mouse cells or tissues (Fig. 3 and data not shown).
FIG. 3.
Telomerase activity levels were unaltered in mVparp−/− mouse cells. TRAP was performed for 20 PCR cycles with 1.0, 0.5, 0.25, and 0.125 μg of cell extracts prepared from wild-type and mVparp−/− ES cells. An internal PCR standard for the TRAP is indicated by an arrow. R, RNase A treatment of the wild-type lysate prior to the assay (1.0 μg). The asterisk indicates a nonspecific product that was resistant to RNase A treatment.
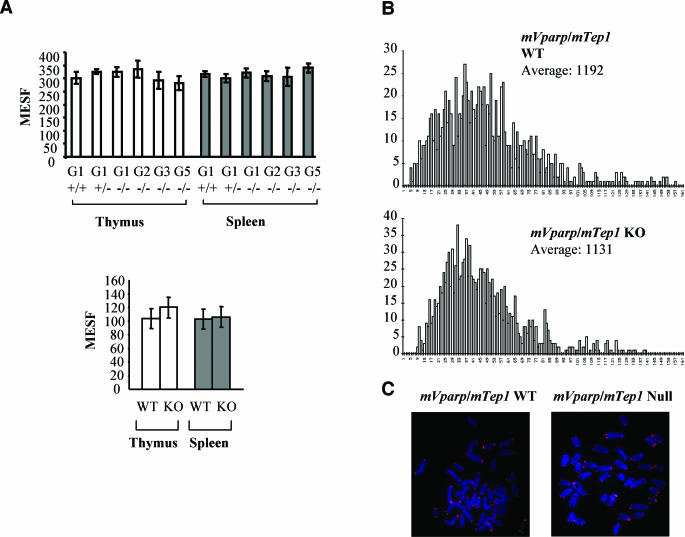
In budding yeast cells, the products of EST1 and EST3 are telomerase associated. Although they are dispensable for telomerase catalysis (7, 46), mutation or disruption of these genes results in progressive telomere loss comparable to that seen with the disruption of EST2 (also known as TERT) (45, 46). To determine whether the loss of mVparp is essential for telomere length maintenance, we analyzed telomere signal intensity in total cell populations of splenocytes and thymocytes derived from mVparp−/− mice by flow FISH. Thymocytes and splenocytes derived from different generations of mVparp−/− mice (G1 to G5) showed telomere signal intensities similar to those of G1 mVparp+/− mice or wild-type control mice (Fig. 4A, upper panel). The overall distribution of telomere fluorescence in mVparp−/− mice was also similar to that observed in G1 mVparp heterozygous mice or wild-type mice (data not shown). Using the same technique, we were able to detect telomere loss in mTert−/− mice as early as G2 (48). However, five generations of mVparp−/− mice failed to exhibit any detectable changes in overall telomere signal intensities (Fig. 4A, upper panel, and data not shown).
FIG. 4.
No detectable change in telomere signal intensity in mVparp−/− mice and mVparp−/− mTep1−/− mice. (A) Telomere signal intensity in mouse thymocytes and splenocytes was analyzed by flow FISH. (Upper panel) Average telomere fluorescence in thymocytes and splenocytes derived from different generations (G1 to G5) of mVparp−/− mice. (Lower panel) Average telomere fluorescence in thymocytes and splenocytes derived from wild-type (WT) and G1 mVparp−/− mTep1−/− (KO) mice. In each set, data were pooled from at least five individual mice (error bars represent standard deviations). MESF, molecules of equivalent soluble fluorochrome. (B) Quantitative FISH analysis of individual metaphases of activated splenocytes derived from wild-type and G1 mVparp−/− mTep1−/− mice. Data were accumulated from approximately 10 metaphases for each histogram. The number of telomeres within a given range of telomeric DNA intensities was plotted against the telomere DNA signal intensity (in arbitrary units: 0 for no telomeric DNA signal and increasing in increments of arbitrary telomere fluorescence units to 161). (C) Representative metaphase spreads of activated splenocytes derived from wild-type and G1 mVparp−/− mTep1−/− mice.
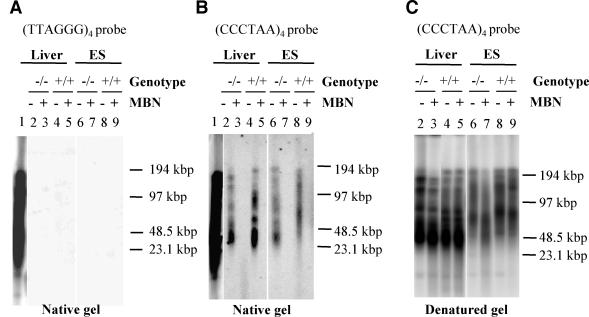
Telomere integrity depends on both telomere length and telomere structure. The telomere terminus possesses a G overhang of approximately 100 to 150 bases (74). In the event that mVPARP might affect G overhangs without affecting overall telomere length, we studied the status of G-rich overhangs in mVparp−/− mouse ES cells or tissues. Telomeric G overhangs were evident in mVparp−/− and wild-type mouse liver and ES cells (Fig. 5B, lanes 2 and 6 for mVparp−/− liver and ES cells or lanes 4 and 8 for wild-type liver and ES cells). As a control, the G overhang signal disappeared when the samples were treated with mung bean nuclease, which degrades single-stranded DNA 3′ overhangs (Fig. 5B, lanes 3, 5, 7, and 9). As expected, there was no hybridization signal when the same gel was hybridized to the G telomere probe (TTAGGG)4 (Fig. 5A). Together, these results indicate that mVparp−/− mouse ES cells or tissues possess a normal G overhang. A normal G overhang was also found in telomerase RNA-deficient mouse cells (25) or in G1 mTert−/− mice (76).
FIG. 5.
Telomeric G overhang analysis of mVparp−/− mouse ES cells and tissues. (A) Genomic DNA from wild-type and mVparp−/− ES cells and mouse liver was resolved by pulsed-field gel electrophoresis and subjected to in-gel hybridization to a radioactively labeled (TTAGGG)4 probe under native conditions. In lane 1, a denatured genomic DNA sample from wild-type mouse liver (which was excised from the same gel and subsequently denatured) was used as a positive control for hybridization. Since the telomeric G overhang consists of TTAGGG repeats, no hybridization should be evident in lanes 2 to 9. MBN, samples that were pretreated with mung bean nuclease prior to electrophoresis. (B) The same samples as those in panel A were hybridized to a radioactively labeled (CCCTAA)4 probe. (C) The same gel as that in panel B was denatured and reprobed with a radioactively labeled (CCCTAA)4 probe. The integrity of the high-molecular-weight DNA was confirmed by ethidium bromide staining of the agarose gel prior to hybridization (data not shown). DNA markers (kilobase pairs) are indicated at the right.
Previously it was demonstrated that TEP1 interacts with telomerase RNA and vRNA in a yeast three-hybrid assay (23, 36). Although the levels of telomerase RNA and its association with telomerase RNP were not affected in mTep1−/− mice, the absence of TEP1 resulted in decreases in the levels and stability of vRNA and its association with vaults in vivo (34). Deletion of mVparp did not appear to affect the steady-state levels of telomerase RNA, vRNA, or TEP1 in normal mouse cells (data not shown).
We examined cell viability, chromosome abnormalities, and the kinetics of DNA damage repair in isolated splenocytes from mVparp−/− mice exposed to ionizing radiation and found no reproducible difference compared with the results for wild-type mice. We also examined the induction of p53 and p21 in irradiated mVparp−/− MEFs and found no significant difference compared with the results for wild-type fibroblasts (data not shown).
Disruption of both mVparp and mTep1 in mice.
In the event that VPARP and TEP1 exhibit genetic redundancy in vivo, we examined mice deficient in both mVparp and mTep1. G1 double-mutant mice were viable and showed no detectable alteration in telomerase activity levels (data not shown) or telomere signal intensities in flow FISH or quantitative FISH analyses (Fig. 4A, lower panel, and Fig. 4B and C). To examine chromosome stability in mVparp−/− or mVparp−/− mTep1−/− mice, we performed FISH analysis of metaphase spreads of MEFs or mitogen-stimulated splenocytes from wild type, mVparp−/−, or mVparp−/− mTep1−/− mice. The incidences of chromosome abnormalities, including aneuploidy and chromosome breakage, were similar, regardless of genotype (Table 1, Fig. 4C, and data not shown).
TABLE 1.
Chromosomal abnormalities in MEFs and activated splenocytesa
| Gene(s) | Cellsb | No. of metaphases analyzed | % Aneuploidyc |
|---|---|---|---|
| mVparp | G1 WT splenocytesd | 41 | 17 |
| G1 KO splenocytes | 25 | 9 | |
| G2 KO splenocytes | 32 | 16 | |
| WT MEFs | 21 | 24 | |
| KO MEFs | 32 | 16 | |
| mVparp and mTep1 | G1 WT splenocytesd | 25 | 15 |
| G1 KO splenocytes | 25 | 17 |
No fusions or chromosome breakage events were detected.
WT, wild type; KO, mVparp−/− or mVparp−/− and mTep1−/−.
Percentage of metaphases with <40 or >40 chromosomes.
Activated with anti-CD3 and interleukin 2 for five days before metaphase spread preparation.
Vaults isolated from mVparp−/− mice appear to be intact.
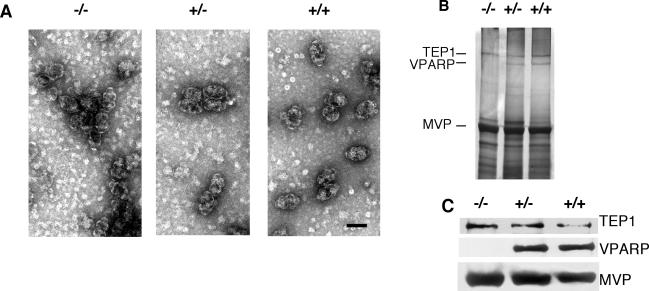
To examine the role of VPARP in vault particle formation in vivo, we purified vaults from the livers of wild-type, mVparp+/−, and mVparp−/− mice (see Materials and Methods). Vaults did not show any visible perturbation under negative EM staining (Fig. 6A). The presence of some smaller, misshapen vaults in mVparp−/− mice were comparable to representative vaults prepared from wild-type samples, and we did not detect a significant increase in misshapen vaults. Negative EM staining of vaults purified from mVparp−/− mTep1−/− mice is currently under way. Vault components purified from mVparp−/− mice through a series of gradient and centrifugation steps had properties similar to those of wild-type vaults (data not shown). Purified vault fractions from wild-type, mVparp+/−, and mVparp−/− mouse livers were analyzed by gel electrophoresis and silver staining and showed comparable levels of MVP and TEP1 (Fig. 6B). The absence of VPARP in the vault preparations was confirmed by immunoblot analysis of the purified vaults with antibodies against each vault protein component (anti-MVP, anti-TEP1, and anti-VPARP) (Fig. 6C). Finally, we examined the purified vault fractions for the presence of vRNA and found that vRNA levels were unchanged in purified vault preparations from mVparp−/− animals (data not shown). These data suggest that mVPARP is not essential for the stable interaction of the other vault components with vault particles and is not required for vault particle formation. In comparison, mTep1−/− animals showed a complete lack of vRNA in vault particles and a reduction in electron density at the vault cap in cryo-EM reconstructions (34). The exact localization of mVPARP within vaults has not yet been determined. Preliminary cryo-EM analysis of isolated mVparp−/− vault particles has revealed the presence of intact vault particles (P. L. Stewart and M. Makabi, personal communication); however, reconstruction and modeling have not yet been completed.
FIG. 6.
Vault structure and copurification of vault components in mVparp−/− mouse livers. (A) Electron micrographs show negatively stained vaults purified from livers of mVparp−/−, mVparp+/−, and wild-type (+/+) mice. Bar, 50 nm. (B) Purified vaults were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized by silver staining. (C) The identities of the vault components (TEP1, VPARP, and MVP) were confirmed by immunoblot analysis with anti-TEP1, anti-VPARP, and anti-MVP antibodies. As expected, VPARP was absent from mVparp−/− mice.
DISCUSSION
In mammalian cells, TEP1 and VPARP associate with two common RNP complexes in vivo, telomerase and vaults. Previously, it was established that TEP1 and VPARP localize to both nuclear and cytoplasmic fractions and that the vault particle itself appears to be predominantly cytoplasmic (35, 36). Therefore, it is possible that nuclear TEP1 and VPARP associate with distinct complexes in the nucleus, including telomerase. The physiological significance of this interaction is at present uncertain due to an inability to detect an association between endogenous TEP1 and VPARP and the lack of an obvious phenotype in mice deficient in mVparp and/or mTep1. Since TEP1 or VPARP appears to associate with a fraction of telomerase activity in vivo, it is possible that disruption of this association has no observable consequence. Alternatively, TEP1 and VPARP functions may be redundant with those of other telomerase-associated proteins, such as L22, Staufen, Hsp70, Hsp90, p23, DKC, hEst1A/B/C, heterogeneous nuclear RNPs, or La (11, 16-19, 27, 29, 42, 43, 52, 59, 69). Finally, the association of telomerase with TEP1 or VPARP may become important only under certain cellular stresses, a possibility currently under investigation.
Mammalian TEP1 possesses a putative RNA binding motif that is sufficient for an association with telomerase RNA and vRNA in an indirect yeast three-hybrid assay (23, 36). In support of a direct role in vRNA binding in vivo, the loss of TEP1 disrupts the stable association of vRNA with vaults and the stability of vRNA (34). VPARP, on the other hand, does not possess an obvious RNA binding motif and has no direct effect on the stability or levels of vRNA and telomerase RNA expression when disrupted. We also demonstrate that the absence of mVparp does not affect the steady-state levels of TEP1, telomerase RNA, or vRNA or the association of vRNA with the vault particle (data not shown).
Several members of mammalian PARP families are also involved in telomere protection or length regulation and thus may also share a redundant role with TEP1 or VPARP in regulating telomerase function in vivo (6, 9, 10, 67, 68). The unique feature of this family is that all proteins share a common PARP domain, which catalyzes modification of a variety of acceptor proteins (14, 65). For example, tankyrases 1 and 2 can directly bind telomere binding protein 1 (TRF1) and are capable of poly(ADP-ribosyl)ation of TRF1 (60, 68). TRF1 negatively regulates telomerase-mediated telomere length and acts in cis at chromosome ends to repress telomere elongation (1, 72). Such posttranscriptional modification of TRF1 affects its binding to telomeric DNA both in vitro and in vivo (68). Furthermore, human tumor cells expressing mutant tankyrase 1 exhibit abnormal telomere lengthening that requires the catalytic activity of tankyrase (9, 67). These findings suggest that the tankyrase-mediated removal of TRF1 from telomeres by ADP ribosylation could allow access of telomerase to chromosome termini (1, 67, 68, 72).
Another PARP family member, PARP1, is well characterized for its role in base excision repair (28). PARP1 binds to and is activated by DNA breaks and catalyzes the ribosylation of several substrates related to DNA damage and repair (65, 66). PARP1 has also been implicated in other cellular events, including the modulation of chromatin structure, DNA synthesis, gene transcription, centrosome function, cell cycle regulation, cell death, and cell survival (12, 30, 40, 41, 56). d'Adda di Fagagna et al. reported that Parp1-deficient mice exhibited telomere shortening and increased end-to-end chromosome fusions (10). In contrast, Samper et al. observed normal telomere length and G-rich overhangs and low frequencies of chromosome end-to-end fusions in Parp1-deficient mice (63). Recent evidence indicates that another PARP family member, PARP2, is also involved in base excision repair and interacts with PARP1 and TRF2 in vivo (13, 50, 64). Thus, PARP1, perhaps together with PARP2, may serve an important function at telomeres. Further experiments are required to test whether telomere integrity is further perturbed in mice deficient in both mVparp and Parp1 or Parp2.
TERT possesses poly(ADP-ribose) binding sequences (57) and thus could be a candidate poly(ADP-ribose) acceptor. Although an interaction between PARP1 and TERT was reported in a human breast cancer cell line (5), a yeast two-hybrid based assay was unable to detect such an interaction (63). Since we observed an association of VPARP with telomerase activity in vivo, we tested whether the weak poly(ADP-ribosyl)transferase activity of VPARP might modify TERT; however, the results were inconclusive (data not shown). Furthermore, a direct association between VPARP and TERT could not be demonstrated (W. Zhou and M. Robinson, unpublished data). We cannot exclude the possibility that a transient interaction occurs between these two proteins; like other PARP family members, VPARP may associate with potential protein acceptors only when activated in vivo.
MVP constitutes about 75% of the vault particle mass and was recently shown to be sufficient for vault particle formation in an insect expression system (70). Deletion of mMvp leads to a complete loss in detectable vault particles, and yet mice with this deletion appear viable (53). In contrast, VPARP and TEP1 comprise ∼20% of the vault particle mass, and vaults purified from mTep1−/− mice appear to be intact, albeit with less electron density at the vault cap (34). Taken together, these results indicate that neither TEP1 nor VPARP is essential for the formation or maintenance of vault structure in vivo.
However, TEP1 and VPARP may act as catalytic components of vaults rather than essential structural proteins. We note that TEP1 possesses a putative nucleoside triphosphate binding domain, although nucleotide binding by TEP1 has not yet been demonstrated (23, 55). Knowledge of the precise function of the vault particle in mammalian cells has remained elusive, despite compelling connections to multidrug resistance, intracellular compartmentalization, subcellular transport of biomolecules, or a cytosol or nucleoplasm detoxifier (54, 71, 73). Mice deficient in mVparp, mTep1, and mMvp may yet provide important clues to the role of the vault particle in these processes.
Acknowledgments
We thank members of the Harrington laboratory for critical comments and discussions, Denis Bouchard for assistance with FISH, and Murray Robinson for ongoing support and critical discussion and in whose laboratory the data in Fig. 1 were obtained.
Y.L. acknowledges the support of the National Cancer Institute of Canada (while a postdoctoral fellow with L.H.), with funds supplied by the Terry Fox Run and the Laboratory of Directed Research and Development Program of Oak Ridge National Laboratory (3211-2049), managed by UT-Battelle, LLD, for the U.S. Department of Energy under contract no. DE-AC05-00OR22725. L.H.R. acknowledges the support of the G. Harold and Leila Y. Mathers Charitable Foundation, and L.H. acknowledges the support of the National Institute of Health (AG16629-03) and the Canadian Institutes of Health Research (MT14340).
REFERENCES
- 1.Ancelin, K., M. Brunori, S. Bauwens, C. E. Koering, C. Brun, M. Ricoul, J. P. Pommier, L. Sabatier, and E. Gilson. 2002. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 22:3474-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 1998. Reconstitution of human telomerase activity in vitro. Curr. Biol. 8:177-180. [DOI] [PubMed] [Google Scholar]
- 3.Blasco, M. A., and W. C. Hahn. 2003. Evolving views of telomerase and cancer. Trends Cell Biol. 13:289-294. [DOI] [PubMed] [Google Scholar]
- 4.Blasco, M. A., H. W. Lee, M. P. Hande, E. Samper, P. M. Lansdorp, R. A. DePinho, and C. W. Greider. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91:25-34. [DOI] [PubMed] [Google Scholar]
- 5.Cao, Y., H. Li, S. Deb, and J. P. Liu. 2002. TERT regulates cell survival independent of telomerase enzymatic activity. Oncogene 21:3130-3138. [DOI] [PubMed] [Google Scholar]
- 6.Chang, W., J. N. Dynek, and S. Smith. 2003. TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 17:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohn, M., and E. H. Blackburn. 1995. Telomerase in yeast. Science 269:396-400. [DOI] [PubMed] [Google Scholar]
- 8.Collins, K., R. Kobayashi, and C. W. Greider. 1995. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell 81:677-686. [DOI] [PubMed] [Google Scholar]
- 9.Cook, B. D., J. N. Dynek, W. Chang, G. Shostak, and S. Smith. 2002. Role for the related poly(ADP-ribose) polymerases tankyrases 1 and 2 at human telomeres. Mol. Cell. Biol. 22:332-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.d'Adda di Fagagna, F., M. P. Hande, W. M. Tong, P. M. Lansdorp, Z. Q. Wang, and S. P. Jackson. 1999. Functions of poly(ADP-ribose) polymerase in controlling telomere length and chromosomal stability. Nat. Genet. 23:76-80. [DOI] [PubMed] [Google Scholar]
- 11.Dallaire, F., S. Dupuis, S. Fiset, and B. Chabot. 2000. Heterogeneous nuclear ribonucleoprotein A1 and UP1 protect mammalian telomeric repeats and modulate telomere replication in vitro. J. Biol. Chem. 275:14509-14516. [DOI] [PubMed] [Google Scholar]
- 12.D'Amours, D., S. Desnoyers, I. D'Silva, and G. G. Poirier. 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342:249-268. [PMC free article] [PubMed] [Google Scholar]
- 13.Dantzer, F., M. J. Giraud-Panis, I. Jaco, J. C. Ame, I. Schultz, M. Blasco, C. E. Koering, E. Gilson, J. Menissier-De Murcia, G. De Murcia, and V. Schreiber. 2004. Functional interaction between poly(ADP-ribose) polymerase 2 (PARP-2) and TRF2: PARP activity negatively regulates TRF2. Mol. Cell. Biol. 24:1595-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Murcia, G., and J. Menissier de Murcia. 1994. Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem. Sci. 19:172-176. [DOI] [PubMed] [Google Scholar]
- 15.Erdmann, N., Y. Liu, and L. Harrington. Distinct dosage requirements for the maintenance of long and short telomeres in mTert heterozygous mice. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 16.Eversole, A., and N. Maizels. 2000. In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol. Cell. Biol. 20:5425-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiset, S., and B. Chabot. 2001. hnRNP A1 may interact simultaneously with telomeric DNA and the human telomerase RNA in vitro. Nucleic Acids Res. 29:2268-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ford, L. P., J. W. Shay, and W. E. Wright. 2001. The La antigen associates with the human telomerase ribonucleoprotein and influences telomere length in vivo. RNA 7:1068-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford, L. P., J. M. Suh, W. E. Wright, and J. W. Shay. 2000. Heterogeneous nuclear ribonucleoproteins C1 and C2 associate with the RNA component of human telomerase. Mol. Cell. Biol. 20:9084-9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gandhi, L., and K. Collins. 1998. Interaction of recombinant Tetrahymena telomerase proteins p80 and p95 with telomerase RNA and telomeric DNA substrates. Genes Dev. 12:721-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakem, R., J. L. de la Pompa, C. Sirard, R. Mo, M. Woo, A. Hakem, A. Wakeham, J. Potter, A. Reitmair, F. Billia, E. Firpo, C. C. Hui, J. Roberts, J. Rossant, and T. W. Mak. 1996. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell 85:1009-1023. [DOI] [PubMed] [Google Scholar]
- 22.Harrington, L. 2003. Biochemical aspects of telomerase function. Cancer Lett. 194:139-154. [DOI] [PubMed] [Google Scholar]
- 23.Harrington, L., T. McPhail, V. Mar, W. Zhou, R. Oulton, M. B. Bass, I. Arruda, and M. O. Robinson. 1997. A mammalian telomerase-associated protein. Science 275:973-977. [DOI] [PubMed] [Google Scholar]
- 24.Harrington, L., W. Zhou, T. McPhail, R. Oulton, D. S. Yeung, V. Mar, M. B. Bass, and M. O. Robinson. 1997. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 11:3109-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemann, M. T., and C. W. Greider. 1999. G-strand overhangs on telomeres in telomerase-deficient mouse cells. Nucleic Acids Res. 27:3964-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrera, E., E. Samper, J. Martin-Caballero, J. M. Flores, H. W. Lee, and M. A. Blasco. 1999. Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J. 18:2950-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holt, S. E., D. L. Aisner, J. Baur, V. M. Tesmer, M. Dy, M. Ouellette, J. B. Trager, G. B. Morin, D. O. Toft, J. W. Shay, W. E. Wright, and M. A. White. 1999. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 13:817-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeggo, P. A. 1998. DNA repair: PARP—another guardian angel? Curr. Biol. 8:R49-R51. [DOI] [PubMed] [Google Scholar]
- 29.Kamma, H., M. Fujimoto, M. Fujiwara, M. Matsui, H. Horiguchi, M. Hamasaki, and H. Satoh. 2001. Interaction of hnRNP A2/B1 isoforms with telomeric ssDNA and the in vitro function. Biochem. Biophys. Res. Commun. 280:625-630. [DOI] [PubMed] [Google Scholar]
- 30.Kanai, M., W. M. Tong, E. Sugihara, Z. Q. Wang, K. Fukasawa, and M. Miwa. 2003. Involvement of poly(ADP-ribose) polymerase 1 and poly(ADP-ribosyl)ation in regulation of centrosome function. Mol. Cell. Biol. 23:2451-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlseder, J. 2003. Telomere repeat binding factors: keeping the ends in check. Cancer Lett. 194:189-197. [DOI] [PubMed] [Google Scholar]
- 32.Kedersha, N. L., J. E. Heuser, D. C. Chugani, and L. H. Rome. 1991. Vaults. III. Vault ribonucleoprotein particles open into flower-like structures with octagonal symmetry. J. Cell Biol. 112:225-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kedersha, N. L., D. F. Hill, K. E. Kronquist, and L. H. Rome. 1986. Subpopulations of liver coated vesicles resolved by preparative agarose gel electrophoresis. J. Cell Biol. 103:287-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kickhoefer, V. A., Y. Liu, L. B. Kong, B. E. Snow, P. L. Stewart, L. Harrington, and L. H. Rome. 2001. The telomerase/vault-associated protein TEP1 is required for vault RNA stability and its association with the vault particle. J. Cell Biol. 152:157-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kickhoefer, V. A., A. C. Siva, N. L. Kedersha, E. M. Inman, C. Ruland, M. Streuli, and L. H. Rome. 1999. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J. Cell Biol. 146:917-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kickhoefer, V. A., A. G. Stephen, L. Harrington, M. O. Robinson, and L. H. Rome. 1999. Vaults and telomerase share a common subunit, TEP1. J. Biol. Chem. 274:32712-32717. [DOI] [PubMed] [Google Scholar]
- 37.Kim, N. W., and F. Wu. 1997. Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP). Nucleic Acids Res. 25:2595-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong, L. B., A. C. Siva, V. A. Kickhoefer, L. H. Rome, and P. L. Stewart. 2000. RNA location and modeling of a WD40 repeat domain within the vault. RNA 6:890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong, L. B., A. C. Siva, L. H. Rome, and P. L. Stewart. 1999. Structure of the vault, a ubiquitous cellular component. Structure Fold Des. 7:371-379. [DOI] [PubMed] [Google Scholar]
- 40.Kraus, W. L., and J. T. Lis. 2003. PARP goes transcription. Cell 113:677-683. [DOI] [PubMed] [Google Scholar]
- 41.Kun, E., and P. I. Bauer. 2001. Cell biological functions of PARP I: an overview. Ital. J. Biochem. 50:15-18. [PubMed] [Google Scholar]
- 42.LaBranche, H., S. Dupuis, Y. Ben-David, M. R. Bani, R. J. Wellinger, and B. Chabot. 1998. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat. Genet. 19:199-202. [DOI] [PubMed] [Google Scholar]
- 43.Le, S., R. Sternglanz, and C. W. Greider. 2000. Identification of two RNA-binding proteins associated with human telomerase RNA. Mol. Biol. Cell 11:999-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, H. W., M. A. Blasco, G. J. Gottlieb, J. W. Horner II, C. W. Greider, and R. A. DePinho. 1998. Essential role of mouse telomerase in highly proliferative organs. Nature 392:569-574. [DOI] [PubMed] [Google Scholar]
- 45.Lendvay, T. S., D. K. Morris, J. Sah, B. Balasubramanian, and V. Lundblad. 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144:1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lingner, J., T. R. Cech, T. R. Hughes, and V. Lundblad. 1997. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. USA 94:11190-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu, Y., B. E. Snow, M. P. Hande, G. Baerlocher, V. A. Kickhoefer, D. Yeung, A. Wakeham, A. Itie, D. P. Siderovski, P. M. Lansdorp, M. O. Robinson, and L. Harrington. 2000. Telomerase-associated protein TEP1 is not essential for telomerase activity or telomere length maintenance in vivo. Mol. Cell. Biol. 20:8178-8184.11027287 [Google Scholar]
- 48.Liu, Y., B. E. Snow, M. P. Hande, D. Yeung, N. J. Erdmann, A. Wakeham, A. Itie, D. P. Siderovski, P. M. Lansdorp, M. O. Robinson, and L. Harrington. 2000. The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr. Biol. 10:1459-1462. [DOI] [PubMed] [Google Scholar]
- 49.Mason, D. X., C. Autexier, and C. W. Greider. 2001. Tetrahymena proteins p80 and p95 are not core telomerase components. Proc. Natl. Acad. Sci. USA 98:12368-12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Menissier de Murcia, J., M. Ricoul, L. Tartier, C. Niedergang, A. Huber, F. Dantzer, V. Schreiber, J. C. Ame, A. Dierich, M. LeMeur, L. Sabatier, P. Chambon, and G. de Murcia. 2003. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 22:2255-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller, M. C., and K. Collins. 2000. The Tetrahymena p80/p95 complex is required for proper telomere length maintenance and micronuclear genome stability. Mol. Cell 6:827-837. [DOI] [PubMed] [Google Scholar]
- 52.Mitchell, J. R., E. Wood, and K. Collins. 1999. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402:551-555. [DOI] [PubMed] [Google Scholar]
- 53.Mossink, M. H., A. van Zon, E. Franzel-Luiten, M. Schoester, V. A. Kickhoefer, G. L. Scheffer, R. J. Scheper, P. Sonneveld, and E. A. Wiemer. 2002. Disruption of the murine major vault protein (MVP/LRP) gene does not induce hypersensitivity to cytostatics. Cancer Res. 62:7298-7304. [PubMed] [Google Scholar]
- 54.Mossink, M. H., A. van Zon, R. J. Scheper, P. Sonneveld, and E. A. Wiemer. 2003. Vaults: a ribonucleoprotein particle involved in drug resistance? Oncogene 22:7458-7467. [DOI] [PubMed] [Google Scholar]
- 55.Nakayama, J., M. Saito, H. Nakamura, A. Matsuura, and F. Ishikawa. 1997. TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell 88:875-884. [DOI] [PubMed] [Google Scholar]
- 56.Nicoletti, V. G., and A. M. Stella. 2003. Role of PARP under stress conditions: cell death or protection? Neurochem. Res. 28:187-194. [DOI] [PubMed] [Google Scholar]
- 57.Pleschke, J. M., H. E. Kleczkowska, M. Strohm, and F. R. Althaus. 2000. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 275:40974-40980. [DOI] [PubMed] [Google Scholar]
- 58.Prowse, K. R., and C. W. Greider. 1995. Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc. Natl. Acad. Sci. USA 92:4818-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reichenbach, P., M. Hoss, C. M. Azzalin, M. Nabholz, P. Bucher, and J. Lingner. 2003. A human homolog of yeast est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr. Biol. 13:568-574. [DOI] [PubMed] [Google Scholar]
- 60.Rippmann, J. F., K. Damm, and A. Schnapp. 2002. Functional characterization of the poly(ADP-ribose) polymerase activity of tankyrase 1, a potential regulator of telomere length. J. Mol. Biol. 323:217-224. [DOI] [PubMed] [Google Scholar]
- 61.Rudolph, K. L., S. Chang, H. W. Lee, M. Blasco, G. J. Gottlieb, C. Greider, and R. A. DePinho. 1999. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell 96:701-712. [DOI] [PubMed] [Google Scholar]
- 62.Rufer, N., W. Dragowska, G. Thornbury, E. Roosnek, and P. M. Lansdorp. 1998. Telomere length dynamics in human lymphocyte subpopulations measured by flow cytometry. Nat. Biotechnol. 16:743-747. [DOI] [PubMed] [Google Scholar]
- 63.Samper, E., F. A. Goytisolo, J. Menissier-de Murcia, E. Gonzalez-Suarez, J. C. Cigudosa, G. de Murcia, and M. A. Blasco. 2001. Normal telomere length and chromosomal end capping in poly(ADP-ribose) polymerase-deficient mice and primary cells despite increased chromosomal instability. J. Cell Biol. 154:49-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schreiber, V., J. C. Ame, P. Dolle, I. Schultz, B. Rinaldi, V. Fraulob, J. Menissier-de Murcia, and G. de Murcia. 2002. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 277:23028-23036. [DOI] [PubMed] [Google Scholar]
- 65.Shall, S. 2002. Poly (ADP-ribosylation)—a common control process? Bioessays 24:197-201. [DOI] [PubMed] [Google Scholar]
- 66.Shall, S., and G. de Murcia. 2000. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat. Res. 460:1-15. [DOI] [PubMed] [Google Scholar]
- 67.Smith, S., and T. de Lange. 2000. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10:1299-1302. [DOI] [PubMed] [Google Scholar]
- 68.Smith, S., I. Giriat, A. Schmitt, and T. de Lange. 1998. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282:1484-1487. [DOI] [PubMed] [Google Scholar]
- 69.Snow, B. E., N. Erdmann, J. Cruickshank, H. Goldman, R. M. Gill, M. O. Robinson, and L. Harrington. 2003. Functional conservation of the telomerase protein est1p in humans. Curr. Biol. 13:698-704. [DOI] [PubMed] [Google Scholar]
- 70.Stephen, A. G., S. Raval-Fernandes, T. Huynh, M. Torres, V. A. Kickhoefer, and L. H. Rome. 2001. Assembly of vault-like particles in insect cells expressing only the major vault protein. J. Biol. Chem. 276:23217-23220. [DOI] [PubMed] [Google Scholar]
- 71.Suprenant, K. A. 2002. Vault ribonucleoprotein particles: sarcophagi, gondolas, or safety deposit boxes? Biochemistry 41:14447-14454. [DOI] [PubMed] [Google Scholar]
- 72.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385:740-743. [DOI] [PubMed] [Google Scholar]
- 73.van Zon, A., M. H. Mossink, R. J. Scheper, P. Sonneveld, and E. A. Wiemer. 2003. The vault complex. Cell Mol. Life Sci. 60:1828-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei, C., and M. Price. 2003. Protecting the terminus: t-loops and telomere end-binding proteins. Cell Mol. Life Sci. 60:2283-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weinrich, S. L., R. Pruzan, L. Ma, M. Ouellette, V. M. Tesmer, S. E. Holt, A. G. Bodnar, S. Lichtsteiner, N. W. Kim, J. B. Trager, R. D. Taylor, R. Carlos, W. H. Andrews, W. E. Wright, J. W. Shay, C. B. Harley, and G. B. Morin. 1997. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17:498-502. [DOI] [PubMed] [Google Scholar]
- 76.Yuan, X., S. Ishibashi, S. Hatakeyama, M. Saito, J. Nakayama, R. Nikaido, T. Haruyama, Y. Watanabe, H. Iwata, M. Iida, H. Sugimura, N. Yamada, and F. Ishikawa. 1999. Presence of telomeric G-strand tails in the telomerase catalytic subunit TERT knockout mice. Genes Cells 4:563-572. [DOI] [PubMed] [Google Scholar]
- 77.Zijlmans, J. M., U. M. Martens, S. S. Poon, A. K. Raap, H. J. Tanke, R. K. Ward, and P. M. Lansdorp. 1997. Telomeres in the mouse have large interchromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. USA 94:7423-7428. [DOI] [PMC free article] [PubMed] [Google Scholar]