Abstract
The etiology of post-traumatic stress disorder (PTSD) likely involves the interaction of numerous genes and environmental factors. Similarly, gene-expression levels in peripheral blood are influenced by both genes and environment, and expression levels of many genes show good correspondence between peripheral blood and brain tissues. In that context, this pilot study sought to test the following hypotheses: 1) post-trauma expression levels of a gene subset in peripheral blood would differ between Marines with and without PTSD; 2) a diagnostic biomarker panel of PTSD among high-risk individuals could be developed based on gene expression in readily assessable peripheral blood cells; and 3) a diagnostic panel based on expression of individual exons would surpass the accuracy of a model based on expression of full-length gene transcripts. Gene-expression levels in peripheral blood samples from 50 U.S. Marines (25 PTSD cases and 25 non-PTSD comparison subjects) were determined by microarray following their return from deployment to war-zones in Iraq or Afghanistan. The original sample was carved into training and test subsets for construction of support vector machine classifiers. The panel of peripheral blood biomarkers achieved 80% prediction accuracy in the test subset based on the expression of just two full-length transcripts (GSTM1 and GSTM2). A biomarker panel based on 20 exons attained an improved 90% accuracy in the test subset. Though further refinement and replication of these biomarker profiles are required, these preliminary results provide proof-of-principle for the diagnostic utility of blood-based mRNA-expression in PTSD among trauma-exposed individuals.
Keywords: alternative splicing, mRNA, peripheral blood mononuclear cells, gene expression, microarray, transcriptome, trauma, diagnosis, biomarker, antioxidant, oxidative stress
Introduction
Post-traumatic stress disorder (PTSD) is a severe anxiety syndrome that is currently diagnosed based on the emergence and persistence of core clinical symptoms including hyperarousal, re-experiencing, avoidance, or emotional numbing for a period greater than one month. Early psychosocial intervention after stress exposure may help reduce some of the symptoms and prevent the development of chronic PTSD (Litz et al., 2002). However, many individuals initially presenting with PTSD-like symptoms recover spontaneously and do not develop chronic PTSD (McFarlane, 1997). Thus, identifying which individuals will benefit most from early intervention can be challenging. Despite possible benefits of early intervention and a growing knowledge of the pathophysiology accompanying PTSD, a readily assessable diagnostic biomarker for PTSD has yet to be validated.
Classical descriptions of PTSD pathophysiology have included dysregulation of the hypothalamic-pituitary-adrenal (HPA) axis, but a specific pattern of dysregulation is not consistently observed across studies. Similarly, heightened inflammatory signaling has been reported in some (but not all) contexts (Baker et al., 2012b). Some have proposed a model of insufficient regulation of inflammatory signaling (Heinzelmann and Gill, 2013). Yet, there is an apparent paradox; i.e., the observation that peripheral blood mononuclear cells (PBMCs) from PTSD patients show increased sensitivity to glucocorticoid-mediated suppression of an in-situ inflammatory response (van Zuiden et al., 2012b).
There is considerable evidence that genetic effects, environmental influences, and their interaction play a role in the development of PTSD (Afifi et al., 2010; Koenen et al., 2009; True et al., 1993). There is a well-established body of clinical literature supporting a link between early life events, previous exposure to traumatic stress, and other psychosocial factors with the development of PTSD (Brewin et al., 2000; Ozer et al., 2003; DiGangi et al., 2013). Many biological investigations of PTSD have focused on the HPA axis and glucocorticoid receptor (GR) signaling pathways. In a cross-sectional study, Binder and colleagues (Binder et al., 2008) identified an interaction between child abuse history and genetic polymorphisms in FKBP5 (a negative regulator of GR sensitivity) in predicting adult PTSD symptomology among a sample of non-psychiatric medical clinic patients. Mehta and colleagues (Mehta et al., 2011) described the association between genetic polymorphisms in FKBP5 and dysregulated neuroendocrine profiles described in PTSD. Van Zuiden and others (2012a) provided evidence that increased GR density is a pre-trauma risk factor for the development of PTSD and that dysregulation of GR density may be associated with an interaction between polymorphisms in the GR gene and concomitant early life stress. Another line of research suggests that genetic variants in corticotropin-releasing hormone type 1 receptor (CRHR1) are a risk factor for PTSD in children who were abused at an early age (Gillespie et al., 2009). PTSD is thus thought to be a disorder whose development is influenced by multiple genetic and environmental effects that establish a susceptible biological state; this vulnerability may be reflected in gene expression signatures.
In light of the less-than-absolute heritability of PTSD and the prominent role of environmental factors, the pursuit of static genetic markers alone (e.g., single nucleotide polymorphisms and copy-number variations) likely will not yield a suitable diagnostic biomarker. Gene expression (i.e., mRNA) levels, which potentially reflect the effects of both heredity and environment, may be better indicators of the aberrant biology underlying PTSD. PTSD clearly is a brain disorder, but assaying gene-expression levels—either acutely or longitudinally—in the brains of living human subjects at risk for PTSD is impossible. Yet, peripheral blood expression levels of many genes are moderately correlated with the expression levels of those genes in other tissues, including postmortem brain (Tylee et al., 2013) suggesting the possibility that peripheral blood gene expression can be harnessed to construct useful profiles of brain disorders. Previous work by our group and by others has demonstrated that peripheral blood gene expression provides a useful biomarker signal for a number of neuropsychiatric disorders, including schizophrenia, bipolar disorder, and autism spectrum disorders (Glatt et al., 2009; Glatt et al., 2005; Tsuang et al., 2005).
In the context of PTSD, several prior studies identified differences in peripheral blood gene-expression levels between individuals with PTSD and similarly exposed comparison subjects without PTSD (Neylan et al., 2011; Segman et al., 2005; Yehuda et al., 2009; Zieker et al., 2007) (see Glatt and others, 2013 for a brief review of these studies). Taken together, these studies suggest that PTSD is associated with alterations in peripheral blood gene transcripts thought to play a role in HPA axis function, glucocorticoid signaling, immune and inflammatory signaling, and metabolism of reactive oxygen species. Consolidating this evidence with the results from a large body of epidemiologic, genomic, and neurobiological studies of the disorder (e.g., (Uddin et al., 2010)) led us to recently propose a theory of PTSD predicated on dysregulation of immune and inflammatory processes in general, and cellular immunity in particular (Baker et al., 2012b). We maintain that a variety of specific genetic factors and environmental influences may play a role in producing this dysregulated immune and inflammatory phenotype within different individuals. For this reason, we propose that a blood-based diagnostic biomarker calibrated to detect commonly-dysregulated immune and inflammatory transcripts may ultimately provide the best sensitivity for detecting PTSD within a clinical sample. Our previous work in this domain supports this hypothesis and further proposes that pre-existing dysregulation of immune and inflammatory processes may dispose individuals to develop PTSD at some future time, following exposure to traumatic stress (Glatt et al., 2013). Another recent publication, examining a large group of Marine Resiliency Study subjects across multiple cohorts, provided strong evidence that pre-deployment inflammation levels, assessed via measurement of plasma C-reactive protein level, were a strong positive predictor for the development of post-deployment PTSD after controlling for other risk factors (Eraly et al., 2014).
In the context of this prior work, we report here the results of a pilot study examining transcriptome-wide expression-profiling of pre- and post-exposure peripheral blood samples from individuals with uniquely elevated rates of trauma exposure and PTSD development: participants in the Marine Resiliency Study (MRS) following return from active war zones in Iraq or Afghanistan, as part of an ongoing longitudinal investigation (Baker et al., 2012a). The objectives of this pilot study were to evaluate the following hypotheses: 1) post-trauma expression levels of some genes in peripheral blood cells would differ between Marines with PTSD and matched comparison subjects; 2) a readily assessable, predictive biomarker panel of the PTSD diagnostic status could be developed based on gene expression levels in peripheral blood cells; and 3) a diagnostic panel based on the expression of individual exons would surpass the accuracy of a model based on the expression of full-length transcripts of genes. We interpret the results of these analyses in two contexts: 1) as a means of identifying biological functions, processes, pathways, and protein domains whose genomic dysregulation may indicate or influence the development of the disorder; and 2) as an approach to the construction of classifiers that might ultimately assist in the clinical diagnosis of PTSD in such populations.
Methods
Ascertainment and Clinical Characterization of Subjects
The MRS is a prospective study of factors predictive of PTSD among approximately 2,600 Marines in four battalions deployed to Iraq or Afghanistan. The research team conducted structured clinical interviews on Marine bases and collected blood samples and data at four time points: pre-deployment, and 1-week, 3-months, and 6-months post-deployment. Measures collected, including those used in this study, have been described in detail previously (Baker et al., 2012a).
The principal exclusion criteria were identical to those used for the pre-deployment gene expression studies (Glatt et al., 2013). Subjects were excluded if they showed clinically significant PTSD prior to deployment, manifesting in: 1) a pre-deployment PTSD Checklist (PCL) score > 44; and/or 2) a pre-deployment diagnosis of PTSD based on the Clinician-Administered PTSD Scale (CAPS). PTSD cases were identified as those subjects who were issued a CAPS-based PTSD diagnosis at 3- and/or 6-months post-deployment. Unaffected comparison subjects were identified as those subjects who, at no time, attained a PCL score >44 and who were not issued a CAPS-based PTSD diagnosis at any post-deployment interview. Among subjects who were included in the full MRS sample and assigned to case or comparison groups based on these criteria, we then selected for analysis 25 male PTSD cases and 25 male comparison subjects based on similar demographics, pre-deployment clinical characteristics, deployment history, and levels of trauma exposure as determined from the Combat and Post-Battle Experiences subscales of the Deployment Risk and Resilience Inventory (DRRI). The group of subjects selected for this study largely overlapped with those featured in our previous study of pre-deployment gene expression (Glatt et al., 2013); 24 of the twenty-five PTSD cases and 23 of the twenty-five comparison subjects within the present study were also featured in the pre-deployment study. The demographic, clinical, and combat-experiential characteristics of the subjects are shown in Table 1. The two groups were comparable on all demographic and combat-experiential variables. Within both the case and comparison groups, 50% of the subjects had previously been deployed on at least one occasion and the average number of previous deployments was not significantly different between the two groups. Although no subject met diagnostic threshold for PTSD at pre-deployment as determined by either clinician ratings on the CAPS or self-ratings on the PCL, the eventual PTSD cases did have significantly higher clinician ratings on the CAPS at pre-deployment, whereas no significant difference in pre-deployment self-ratings on the PCL were observed.
Table 1. Demographic, Clinical, and Experiential Characteristics of PTSD Cases and non-PTSD Comparison Subjects.
| PTSD Cases | Comparison Subjects | p | |
|---|---|---|---|
| Sample Size: n | 25 | 25 | |
| Age: | 22.4 ± 3.1 | 21.9 ± 3.1 | 0.576 |
| Previously Deployed: n (%) | 13 (52.0) | 13 (52.0) | 1.000 |
| Ancestry: Caucasian n (%) | 17 (68.0) | 19 (76.0) | 0.754 |
| Cohort n (%): 1 | 3 (12.0) | 5 (20.0) | 0.721 |
| 2 | 8 (32.2) | 8 (32.2) | |
| 3 | 14 (56.0) | 12 (48.0) | |
| DRRI Combat Experiences | 18.5 ± 13.0 | 19.3 ± 14.8 | 0.846 |
| DRRI Post-Battle Experiences | 7.25 ± 4.5 | 8.0 ± 4.5 | 0.518 |
| CAPS Pre-Deployment | 22.4 ± 118 | 14.0 ± 8.7 | 0.006* |
| CAPS 3-Months Post-Deployment | 62.8 ± 19.0 | 11.8 ± 10.8 | <0.001* |
| PCL Pre-Deployment | 24.3 ± 6.5 | 22.8 ± 3.4 | 0.330 |
| PCL 1-Week Post-Deployment | 42.9 ± 17.2 | 23.0 ± 5.2 | <0.001* |
| PCL 3-Months Post-Deployment | 49.0 ± 12.4 | 21.6 ± 6.1 | <0.001* |
| PCL 6-Months Post-Deployment | 39.3 ± 15.0 | 19.8 ± 2.4 | <0.001* |
Notes:
1) Demographic characteristics of each sample are reported as mean + s.d. unless otherwise noted.
2) Sample means and proportions were compared using independent samples t-tests and chi-square tests, respectively.
3) P-values < .05 are indicated with *.
mRNA Sample Acquisition, Stabilization, Isolation, and Storage
Close collaboration with the Marine Corps and the Navy, which provides health support for the Marine Corps, enabled comprehensive on-site data collection. Clinical interviews and sample blood draw (10 ml) were both performed within 4 hours of each other on the same day, 3 months after return from deployment. Specific methods for stabilization, isolation and storage of mRNA samples were described previously (Glatt et al., 2013).
mRNA Quantitation, Quality Control, and Hybridization
Specific methods employed for mRNA sample quantitation and quality control assessment were also described previously (Glatt et al., 2013). The quantity and purity of mRNA in each of the 50 samples were sufficient for microarray hybridization assay. Two batches of 25 samples each (balanced with PTSD cases and controls) were then assayed on GeneChip® Human Exon 1.0 ST Arrays (Affymetrix, Inc.; Santa Clara, CA) per the “Whole Transcript Sense Target Labeling Assay” protocol (Affymetrix, 2006) using 1μg of total RNA from each sample.
Microarray Data Import, Normalization, Transformation, Summarization, and Quality Control
Partek® Genomics Suite software, version 6.6 © 2012 (Partek Incorporated; St. Louis, MO), was utilized for all analytic procedures performed on microarray scan data. Interrogating probes were imported, and corrections for background signal were applied using the robust multi-array average (RMA) method (Irizarry et al., 2003), with additional corrections applied for the GC-content of probes. The set of GeneChips was standardized using quantile normalization and expression levels of each probe underwent log-2 transformation to yield distributions of data that more closely approximated normality. As most genes were measured by multiple probe sets (typically one probe set per exon, but sometimes more), summarization of probes took place at two levels: first, probes tagging the same exon were summarized by median polish to arrive at one expression value per exon; second, exons tagging the same gene were summarized by median polish to arrive at one expression value per gene. All probesets were expressed with a signal:noise ratio ≥3; thus, no probesets were excluded from analyses of differential expression. A total of 257,106 probesets were analyzed, mapping to 28,536 whole transcripts and 253,002 exons. Unsupervised clustering of subjects revealed no evidence of batch effects based on scan date. Principal Components Analysis (PCA) of the 50 post-deployment data points identified no outliers; all 50 subjects' data were well within the four-SD ellipsoid on each of the first three PCA dimensions, and deviation among redundant probes located within the same chip was low.
Microarray Data Analyses
Four independent sets of analyses were performed on the microarray data, as described below. For analyses of covariance (ANVOCAs), nominal uncorrected p-value thresholds were selected in order to generate reasonably sized lists of differentially expressed genes and exons for biological annotation analysis and machine learning classifier construction.
1) Identification of Differentially Expressed Genes and their Associated Biological Terms
We utilized ANVOCAs to determine which full-length genetic transcripts were differentially expressed at post-deployment in peripheral blood cells between PTSD cases (n=25) and comparison subjects (n=25). We performed 28,536 ANCOVAs to assess each gene's expression level as a function of PTSD status (case or control), deployment cohort (three levels corresponding to three battalions deployed at different times), age, ancestry (dichotomized as Caucasian or not, as most subjects were Caucasian), and prior deployment status (first or subsequent deployment).
Family-wise Bonferroni-correction was applied to the ANCOVA p-values to determine whether any genes reached a genome-wide level of significance. To generate a relatively large candidate-gene list for functional profiling and construction of classifiers, we utilized an uncorrected type-I-error rate for diagnosis in these analyses at 0.01. We then reduced the dimensionality of the resulting list of candidate biomarkers through analysis of annotation-enrichment using the DAVID algorithm (Dennis et al., 2003) to determine if the gene list disproportionately represented any biological “terms”. Details of the enrichment analysis are described previously (Glatt et al., 2013). Bonferroni-correction was applied to the p-values obtained in the enrichment analyses of these annotation terms.
Pearson correlations were examined between each gene and the summed score from both DRRI subscales (Combat Experience Scale and Post-Battle Experience Scale), first within the PTSD group, and separately within the comparison group, in order to identify genes whose expression level varied with the degree of combat stress exposure. Family-wise Bonferonni and FDR q-values were used to correct for multiple observations. Among PTSD cases, the genes associated with the 200 most significant correlations were analyzed for biological annotations enrichment using DAVID.
2) Discovery and Replication of Gene-Based Diagnostic Predictors
We utilized a machine-learning technique (support vector machine, SVM) to construct, evaluate, optimize, and cross-validate classification algorithms predicting PTSD status based on gene-expression levels at post-deployment for a training subset of our full sample. Training (n=40) and validation (n=10) subsets (distinct from those utilized in Glatt et al., 2013) were carved from the full sample using pseudo-random selection in order to preserve a similar distributions of diagnostic status, demographic features (age, ancestry), and covariate values (deployment cohort, prior deployments) for both subsets. All analyses for classifier construction were carried out in the training subset and completely independent from the test subset. Using the same panel of factors and covariates described above, 28,536 ANCOVAs were performed; we generated a large list of candidate genes based on a nominally-selected uncorrected p<.01. The probes on this list were then supplied as potential predictors in an SVM, as various model parameters and predictor combinations were evaluated to identify the model with the highest accuracy in identifying cases and comparison subjects based solely on the expression levels of a minimal gene set identified by shrinking centroids after two-level nested 10-fold cross-validation. The top-performing model was then deployed on the test subset (5 cases and 5 comparison subjects) to determine its generalizability in accurately predicting case status based on gene-expression levels. (The 10 subjects used for model validation were not significantly different from those in the training set in terms of demographic, gene-expression QC, experiential, or clinical factors; data not shown).
3) Identification of Differentially Expressed Exons and their Associated Biological Terms
Within the full sample (n=50), we examined exon-expression levels utilizing 22,204 ANCOVAs to identify putative alternative splicing differences between individuals with PTSD and comparison subjects. The same factors evaluated in gene-based analyses (PTSD status, cohort, age, ancestry and prior deployment status) were assessed for their main effects and their interaction with exonID as predictors of exon-expression levels; the PTSD status x exonID interaction term was examined as an indicator of putative alternative splicing, c.f., (Glatt et al., 2009). Family-wise Bonferroni-correction was applied to the ANCOVA p-values to determine whether any interaction term reached a genome-wide level of significance. We utilized an uncorrected type-I-error rate of .0005 to obtain a candidate gene-list for functional profiling and classifier construction. Enrichment analyses were performed using the DAVID algorithm and were evaluated against a Bonferroni-corrected p-value accounting for the number of terms evaluated.
4) Discovery and Replication of Exon-Based Diagnostic Predictors
As outlined above for full-length transcripts under section 2, we used SVMs to construct, evaluate, optimize, and cross-validate classification algorithms predicting eventual PTSD status based on exon-expression levels at pre-deployment for the same training subset of our full sample. Using identical subject allocations to training and validation subsets; we first generated a large candidate list of putatively alternatively spliced genes within the training subset (nominal uncorrected p<0.0005 for the interaction of PTSD status and exonID), using 22,204 ANCOVAs and the same panel of factors, covariates, and interaction terms described above. For each gene on the list, the most significantly dysregulated exon was identified and supplied as a potential predictor in the SVM classifiers. Various model parameters and predictor combinations were then evaluated to identify the model with the highest accuracy in identifying cases and comparison subjects based solely on the expression levels of a minimal exon set identified by shrinking centroids after two-level nested 10-fold cross-validation. The top-performing model was then deployed on the test subset (5 cases and 5 comparison subjects) to determine its generalizability in accurately predicting case status based on exon-expression levels.
Validation of Gene Expression with Quantitative Real Time Polymerase Chain Reaction
A subset of nine transcripts featured in SVM classifiers were selected for validation with quantitative real time polymerase chain reaction (QRTPCR). First, total RNA was quantitatively converted with High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems, San Diego City, CA) to generate single-stranded cDNA (for a 20μL reaction). Aliquots of 20ng of cDNA were analyzed via QRTPCR using the Prism 7900 HT Fast Real-Time PCR system (Applied Biosystems). Statistical analysis was performed using the comparative ΔCT method. All reactions were run in duplicate and normalized against gyceraldehyde-3-phosphate dehydrogenase (GAPDH) and hypoxanthine phosphoribosyltransferase 1 (HPRT1). For one transcript (GSTM1), QRTPCR analysis was repeated with 75ng cDNA in order to compensate for low signal. The fold change values were compared using independent samples t-tests (p<0.05).
Results
1) Identification of Differentially Expressed Genes and their Associated Biological Terms
No gene's expression level was related to PTSD status at a Bonferroni-corrected level of significance, which is not surprising given the relatively small sample size and large number of transcripts tested. We did, however, identify 64 probes dysregulated with a nominally significant p<0.01 in Marines diagnosed with PTSD (Table 2). Thirty-three of these 64 probes were down-regulated, whereas 31 were up-regulated. Log2 fold-change (FC) of these probes in eventual PTSD cases ranged from 2.00-fold down-regulation to 1.66-fold up-regulation. Among the 64 probes, 59 were recognized pathway participants within the DAVID database; however, no significantly enriched annotations were identified. Exploratory pathway analysis of the differentially expressed genes in Table 2 using the Reactome database also revealed no significant enrichments. When examining gene expression levels significantly correlated with summed DRRI score, no correlation p-values survived genome-wide correction among comparison subjects. Among PTSD cases, 13,336 correlation p-values survived an FDR correction threshold of 5%. The probesets featured in the 200 most significant correlations were submitted to DAVID for annotation enrichment analysis. The following terms were significantly enriched, with corresponding Bonferroni-corrected p-values: regulation of actin cytoskeleton (5 of 8 probesets down-regulated in PTSD, p=.03), nucleotide-binding (17 of 30 probesets down-regulated, p=.04), host-virus interaction (6 of 10 probesets down-regulated, p=.07), and long-term depression (4 of 5 up-regulated in PTSD, p=.08).
Table 2. Genes Significantly Dysregulated (p<0.01) in Peripheral Blood Mononuclear Cells from the Full Sample of PTSD Cases at Post-Deployment and Used in Predictive SVM Classifiers*.
| Transcript Cluster ID | Gene Symbol | Gene Product | Diagnostic Group Main Effect | ||
|---|---|---|---|---|---|
|
| |||||
| Fold-Change in Cases | F | p | |||
| 7971296 | EPSTI1 | Epithelial stromal interaction 1 (breast) | 1.66 | 7.6 | 8.6E-03 |
| 7921434 | AIM2 | Absent in melanoma 2 | 1.47 | 10.4 | 2.4E-03 |
| 8056408 | GALNT3 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) | 1.32 | 17.7 | 1.3E-04 |
| 7970096 | ING1 | Inhibitor of growth family, member 1 | 1.28 | 12.3 | 1.0E-03 |
| 8046861 | ITGAV | Integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) | 1.23 | 10.3 | 2.5E-03 |
| 8102817 | ELF2 | E74-like factor 2 (ets domain transcription factor) | 1.23 | 8.7 | 5.2E-03 |
| 8124022 | DTNBP1 | Dystrobrevin binding protein 1 | 1.23 | 13.3 | 7.0E-04 |
| 8145175 | PDLIM2 | PDZ and LIM domain 2 (mystique) | 1.21 | 9.3 | 3.9E-03 |
| 7953032 | LRTM2 | Leucine-rich repeats and transmembrane domains 2 | 1.20 | 8.3 | 6.1E-03 |
| 7905131 | CA14 | Carbonic anhydrase XIV | 1.20 | 11.4 | 1.5E-03 |
| 8044613 | CBWD1 | COBW domain containing 1 | 1.20 | 10.7 | 2.1E-03 |
| 8067680 | PRIC285 | Peroxisomal proliferator-activated receptor A interacting complex 285 | 1.18 | 7.9 | 7.5E-03 |
| 8161537 | CBWD3 | COBW domain containing 3 | 1.18 | 8.3 | 6.2E-03 |
| 8155636 | CBWD3 | COBW domain containing 3 | 1.16 | 8.4 | 5.7E-03 |
| 8077099 | SCO2 | SCO cytochrome oxidase deficient homolog 2 (yeast) | 1.15 | 7.5 | 9.0E-03 |
| 8012953 | TRIM16 | Tripartite motif-containing 16 | 1.15 | 8.2 | 6.5E-03 |
| 8037355 | ZNF428 | Zinc finger protein 428 | 1.15 | 8.4 | 5.8E-03 |
| 8161587 | CBWD3 | COBW domain containing 3 | 1.15 | 7.4 | 9.3E-03 |
| 7963157 | RACGAP1 | Rac GTPase activating protein 1 | 1.15 | 9.2 | 4.0E-03 |
| 7982868 | CHAC1 | ChaC, cation transport regulator homolog 1 (E. coli) | 1.15 | 7.3 | 9.6E-03 |
| 7969638 | ENST00000459449 | Ncrna:snoRNA chromosome:GRCh37:13:95862598:95862702:1 | 1.14 | 9.1 | 4.3E-03 |
| 8053248 | C2orf65 | Chromosome 2 open reading frame 65 | 1.14 | 9.9 | 3.0E-03 |
| 8067773 | ZNF512B | Zinc finger protein 512B | 1.13 | 10.4 | 2.4E-03 |
| 8074916 | C22orf43 | Chromosome 22 open reading frame 43 | 1.12 | 10.7 | 2.1E-03 |
| 8139921 | CALN1 | Calneuron 1 | 1.11 | 11.1 | 1.7E-03 |
| 7921492 | IGSF9 | Immunoglobulin superfamily, member 9 | 1.10 | 7.3 | 9.6E-03 |
| 7975562 | PAPLN | Papilin, proteoglycan-like sulfated glycoprotein | 1.10 | 7.8 | 7.9E-03 |
| 7996219 | NDRG4 | NDRG family member 4 | 1.09 | 8.7 | 5.0E-03 |
| 7919267 | AK125616 | cDNA FLJ43628 fis, clone SPLEN2027268. | 1.09 | 8.4 | 5.8E-03 |
| 7919347 | AK125616 | cDNA FLJ43628 fis, clone SPLEN2027268 | 1.09 | 8.4 | 5.8E-03 |
| 8064242 | NCRNA00176 | Non-protein coding RNA 176 | 1.06 | 12.1 | 1.1E-03 |
|
| |||||
| 8161133 | SPAG8 | Sperm associated antigen 8 | -1.07 | 11.1 | 1.7E-03 |
| 7960434 | GENSCAN00000019809 | cDNA:Genscan chromosome:GRCh37:12:5141715:5142095:-1 | -1.07 | 8.6 | 5.4E-03 |
| 7901967 | ENST00000489463 | Ncrna_pseudogene:scRNA_pseudogene chromosome:GRCh37:1:64121426:64121718:1 | -1.08 | 8.5 | 5.7E-03 |
| 7926670 | ENST00000430957 | cDNA:known chromosome:GRCh37:10:23425901:23426107:1 | -1.08 | 8.6 | 5.2E-03 |
| 8098167 | C4orf39 | Chromosome 4 open reading frame 39 | -1.08 | 8.6 | 5.4E-03 |
| 7937474 | NS3BP | NS3BP | -1.08 | 8.2 | 6.3E-03 |
| 7935690 | ENST00000471360 | Ncrna_pseudogene:Mt_tRNA_pseudogene chromosome:GRCh37:10:101817589:101817658:-1 | -1.08 | 7.6 | 8.5E-03 |
| 8090366 | UROC1 | Urocanase domain containing 1 | -1.08 | 7.5 | 8.8E-03 |
| 8103753 | MORF4 | Mortality factor 4 | -1.09 | 8.0 | 7.2E-03 |
| 8130939 | DLL1 | Delta-like 1 (Drosophila) | -1.09 | 7.7 | 8.0E-03 |
| 8019437 | CCDC57 | Coiled-coil domain containing 57 | -1.09 | 8.0 | 7.0E-03 |
| 7966596 | IQCD | IQ motif containing D | -1.10 | 13.5 | 6.3E-04 |
| 7974695 | ENST00000480540 | cDNA:pseudogene chromosome:GRCh37:14:59261372:59261747:1 | -1.10 | 8.3 | 6.0E-03 |
| 8175815 | PNCK | Pregnancy up-regulated non-ubiquitously expressed CaM kinase | -1.10 | 9.6 | 3.3E-03 |
| 8030292 | DKKL1 | Dickkopf-like 1 | -1.10 | 8.5 | 5.6E-03 |
| 7997533 | OSGIN1 | Oxidative stress induced growth inhibitor 1 | -1.11 | 7.7 | 8.0E-03 |
| 8067671 | SRMS | Crc-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites | -1.11 | 7.6 | 8.3E-03 |
| 7998929 | ENST00000470337 | Ncrna_pseudogene:tRNA_pseudogene chromosome:GRCh37:16:3220961:3221031:-1 | -1.12 | 7.3 | 9.7E-03 |
| 8017361 | ENST00000460492 | cDNA:pseudogene chromosome:GRCh37:17:60593682:60594128:-1 | -1.12 | 15.2 | 3.2E-04 |
| 7973900 | C14orf19 | Immunoglobulin (CD79A) binding protein 1 pseudogene | -1.13 | 12.8 | 8.4E-04 |
| 7950321 | UCP3 | Uncoupling protein 3 (mitochondrial, proton carrier) | -1.13 | 11.0 | 1.8E-03 |
| 7965838 | ENST00000411000 | Ncrna:snRNA chromosome:GRCh37:12:102190188:102190280:-1 | -1.15 | 9.1 | 4.2E-03 |
| 8113413 | NUDT12 | Nudix (nucleoside diphosphate linked moiety X)-type motif 12 | -1.16 | 9.2 | 4.0E-03 |
| 8175775 | MAGEA1 | Melanoma antigen family A, 1 (directs expression of antigen MZ2-E) | -1.16 | 7.5 | 8.8E-03 |
| 7948667 | AHNAK | AHNAK nucleoprotein | -1.18 | 7.9 | 7.3E-03 |
| 7913252 | PINK1 | PTEN induced putative kinase 1 | -1.18 | 7.9 | 7.5E-03 |
| 8118974 | RPL10A | Ribosomal protein L10a | -1.19 | 7.8 | 7.7E-03 |
| 7979551 | PPP2R5E | Protein phosphatase 2, regulatory subunit B', epsilon isoform | -1.20 | 7.3 | 9.6E-03 |
| 7972021 | TBC1D4 | TBC1 domain family, member 4 | -1.21 | 8.2 | 6.4E-03 |
| 8106393 | F2R | Coagulation factor II (thrombin) receptor | -1.23 | 9.1 | 4.3E-03 |
| 7900597 | C1orf50 | Chromosome 1 open reading frame 50 | -1.24 | 10.5 | 2.3E-03 |
| 7903753 | GSTM2 | Glutathione S-transferase mu 2 (muscle) | -1.58 | 16.5 | 2.0E-04 |
| 7903765 | GSTM1 | Glutathione S-transferase mu 1 | -2.00 | 22.0 | 2.6E-05 |
Rows are sorted by decreasing fold-change in PTSD cases relative to non-PTSD comparison subjects.
2) Discovery and Replication of a Gene-Based Diagnostic Predictor
To construct a gene-based classifier and assess its generalizability, we first derived a list of potential classifier transcripts as those probes with a difference in expression between PTSD case and comparison subjects attaining p<0.01 in a training subsample of 20 cases and 20 comparison subjects while controlling for the same factors and covariates as in analysis 1. This analysis and filtering left 66 probes (Table 3) that were then used to build and optimize SVM classifiers. The optimal SVM (identified through two-level nested 10-fold cross-validation with shrinking centroids, cost=401, tolerance=0.001, kernel=radial basis function, and gamma=0.001) comprised just 2 of the 66 starting probes (Table 3, probes in bold font) and attained 78% accuracy in classifying those individuals with PTSD in the training sample. We then tested the identical 2-gene SVM (with the same parameters, but with no shrinkage or cross-validation) in the remaining test subset (5 cases and 5 comparison subjects), where it yielded 80% accuracy. Among cases, four of five were correctly classified, while four of five comparison subjects were also classified correctly. These values correspond to a sensitivity, specificity, positive predictive value and negative predictive value in the test sample of 80%, 80%, 80%, and 80%, respectively. Expression levels for GSTM1 and GSTM2 are shown for PTSD cases and comparison subjects in Figure 1. QRTPCR analysis demonstrated that GSTM2 expression was reduced among PTSD cases, but results were less consistent for GSTM1 (Table 4).
Table 3. Genes Significantly Dysregulated (p<0.01) in Peripheral Blood Mononuclear Cells from a Subset of PTSD Cases at Post-Deployment and Used in Predictive SVM Classifiers*.
| Transcript Cluster ID# | Gene Symbol | Gene Product | Diagnostic Group Main Effect | ||
|---|---|---|---|---|---|
|
| |||||
| Fold-Change in Cases | F | p | |||
| 7904853 | GPR89A | G protein-coupled receptor 89A (GPR89A), transcript variant 1, mRNA. | 1.46 | 9.7 | 3.8E-03 |
| 8095139 | SRD5A3 | Steroid 5 alpha-reductase 3 (SRD5A3), mRNA. | 1.44 | 7.7 | 8.9E-03 |
| 8056408 | GALNT3 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-Acetylgalactosaminyltransferase 3 (GalNAc-T3) (GALNT3), mRNA. | 1.40 | 21.9 | 4.4E-05 |
| 8102817 | ELF2 | E74-like factor 2 (ets domain transcription factor) (ELF2), transcript variant 1, mRNA. | 1.30 | 10.0 | 3.3E-03 |
| 7970096 | ING1 | Inhibitor of growth family, member 1 (ING1), transcript variant 4, mRNA. | 1.26 | 7.9 | 8.2E-03 |
| 7938208 | RBMXL2 | RNA binding motif protein, X-linked-like 2 (RBMXL2), mRNA. | 1.26 | 8.8 | 5.6E-03 |
| 8047784 | ZDBF2 | Zinc finger, DBF-type containing 2 (ZDBF2), mRNA. | 1.23 | 8.3 | 6.8E-03 |
| 8124022 | DTNBP1 | Dystrobrevin binding protein 1 (DTNBP1), transcript variant 2, mRNA. | 1.20 | 7.9 | 8.0E-03 |
| 7961418 | ENST00000364606 | Ncrna:rRNA chromosome:GRCh37:12:13593818:13593935:-1 | 1.18 | 11.6 | 1.7E-03 |
| 8162562 | C9orf130 | Chromosome 9 open reading frame 130 (C9orf130), transcript variant 2, non-coding RNA. | 1.16 | 9.3 | 4.5E-03 |
| 7969638 | ENST00000459449 | Ncrna:snoRNA chromosome:GRCh37:13:95862598:95862702:1 | 1.16 | 8.6 | 5.9E-03 |
| 7930561 | HABP2 | Hyaluronan binding protein 2 (HABP2), transcript variant 1, mRNA. | 1.15 | 10.0 | 3.3E-03 |
| 8067773 | ZNF512B | Zinc finger protein 512B (ZNF512B), mRNA. | 1.14 | 8.9 | 5.3E-03 |
| 7999356 | AF090898 | Clone HQ0149 PRO0149 mRNA, complete cds. | 1.13 | 7.6 | 9.3E-03 |
| 8139921 | CALN1 | Calneuron 1 (CALN1), transcript variant 2, mRNA. | 1.10 | 9.0 | 5.0E-03 |
| 7949668 | CCDC87 | Coiled-coil domain containing 87 (CCDC87), mRNA. | 1.10 | 7.9 | 8.2E-03 |
|
| |||||
| 8091239 | ENST00000516936 | Ncrna:rRNA chromosome:GRCh37:3:142310519:142310633:-1 | -1.03 | 7.5 | 9.8E-03 |
| 7910188 | ENST00000365394 | Ncrna:rRNA chromosome:GRCh37:1:227748882:227749001:1 | -1.04 | 9.7 | 3.7E-03 |
| 8161133 | SPAG8 | Sperm associated antigen 8 (SPAG8), transcript variant 2, mRNA. | -1.07 | 9.3 | 4.4E-03 |
| 7925250 | GNG4 | Guanine nucleotide binding protein (G protein), gamma 4 (GNG4), transcript variant 2, mRNA. | -1.08 | 8.7 | 5.8E-03 |
| 8141423 | MIR106B | MicroRNA 106b (MIR106B), microRNA. | -1.08 | 9.3 | 4.3E-03 |
| 7926670 | ENST00000430957 | Cdna:known chromosome:GRCh37:10:23425901:23426107:1 | -1.08 | 7.7 | 9.0E-03 |
| 7966596 | IQCD | IQ motif containing D (IQCD), mRNA. | -1.09 | 10.1 | 3.2E-03 |
| 7946977 | SAA4 | Serum amyloid A4, constitutive (SAA4), mRNA. | -1.09 | 8.7 | 5.8E-03 |
| 8017361 | ENST00000460492 | Cdna:pseudogene chromosome:GRCh37:17:60593682:60594128:-1 | -1.10 | 9.1 | 4.8E-03 |
| 8090366 | UROC1 | Urocanase domain containing 1 (UROC1), transcript variant 1, mRNA. | -1.10 | 8.5 | 6.2E-03 |
| 8018673 | QRICH2 | Glutamine rich 2 (QRICH2), mRNA. | -1.11 | 8.2 | 7.1E-03 |
| 7973900 | C14orf19 | Chromosome 14 open reading frame 19 (C14orf19), non-coding RNA. | -1.11 | 7.5 | 9.9E-03 |
| 7974695 | ENST00000480540 | Cdna:pseudogene chromosome:GRCh37:14:59261372:59261747:1 | -1.11 | 9.7 | 3.7E-03 |
| 8019437 | CCDC57 | Coiled-coil domain containing 57 (CCDC57), mRNA. | -1.11 | 8.5 | 6.1E-03 |
| 8130939 | DLL1 | Delta-like 1 (Drosophila) (DLL1), mRNA. | -1.12 | 9.0 | 5.0E-03 |
| 8146334 | ENST00000343867 | Cdna:pseudogene chromosome:GRCh37:8:48068735:48069425:1 | -1.12 | 9.2 | 4.7E-03 |
| 7988283 | LOC645212 | Hypothetical LOC645212 (LOC645212), transcript variant 1, non-coding RNA. | -1.12 | 8.6 | 5.9E-03 |
| 8029693 | FOSB | FBJ murine osteosarcoma viral oncogene homolog B (FOSB), transcript variant 1, mRNA. | -1.12 | 8.4 | 6.6E-03 |
| 8034276 | ZNF653 | Zinc finger protein 653 (ZNF653), mRNA. | -1.13 | 12.8 | 1.1E-03 |
| 8142997 | PLXNA4 | Plexin A4 (PLXNA4), transcript variant 1, mRNA. | -1.13 | 8.3 | 6.7E-03 |
| 8012126 | CLDN7 | Claudin 7 (CLDN7), transcript variant 1, mRNA. | -1.13 | 7.7 | 9.0E-03 |
| 8071382 | ZNF74 | Zinc finger protein 74 (ZNF74), transcript variant 1, mRNA. | -1.13 | 8.5 | 6.3E-03 |
| 8148607 | GLI4 | GLI family zinc finger 4 (GLI4), mRNA. | -1.14 | 8.2 | 7.2E-03 |
| 8099279 | ABLIM2 | Actin binding LIM protein family, member 2 (ABLIM2), transcript variant 1, mRNA. | -1.14 | 11.7 | 1.7E-03 |
| 8070744 | C21orf2 | Chromosome 21 open reading frame 2 (C21orf2), mRNA. | -1.14 | 8.3 | 6.8E-03 |
| 8125149 | SLC44A4 | Solute carrier family 44, member 4 (SLC44A4), transcript variant 1, mRNA. | -1.15 | 7.5 | 9.7E-03 |
| 8178653 | SLC44A4 | Solute carrier family 44, member 4 (SLC44A4), transcript variant 1, mRNA. | -1.15 | 7.5 | 9.7E-03 |
| 8179861 | SLC44A4 | Solute carrier family 44, member 4 (SLC44A4), transcript variant 1, mRNA. | -1.15 | 7.5 | 9.7E-03 |
| 7928306 | ENST00000363300 | Ncrna:misc_RNA chromosome:GRCh37:10:73980510:73980610:1 | -1.15 | 9.4 | 4.3E-03 |
| 8141795 | POLR2J3 | Polymerase (RNA) II (DNA directed) polypeptide J3 (POLR2J3), mRNA. | -1.16 | 8.0 | 7.9E-03 |
| 8010629 | CCDC137 | Coiled-coil domain containing 137 (CCDC137), mRNA. | -1.16 | 14.6 | 5.3E-04 |
| 8112159 | ANKRD55 | Ankyrin repeat domain 55 (ANKRD55), mRNA. | -1.16 | 9.4 | 4.2E-03 |
| 8064014 | SLC17A9 | Solute carrier family 17, member 9 (SLC17A9), mRNA. | -1.16 | 12.1 | 1.4E-03 |
| 8164665 | RAPGEF1 | Rap guanine nucleotide exchange factor (GEF) 1 (RAPGEF1), transcript variant 1, mRNA. | -1.16 | 8.6 | 6.0E-03 |
| 8113413 | NUDT12 | Nudix (nucleoside diphosphate linked moiety X)-type motif 12 (NUDT12), mRNA. | -1.17 | 7.9 | 8.3E-03 |
| 7948995 | ATL3 | Atlastin-3 gene:ENSG00000184743 | -1.17 | 8.0 | 7.9E-03 |
| 8130867 | THBS2 | Thrombospondin 2 (THBS2), mRNA. | -1.17 | 8.3 | 6.9E-03 |
| 8010848 | TBCD | Tubulin folding cofactor D (TBCD), mRNA. | -1.18 | 7.4 | 1.0E-02 |
| 7965838 | ENST00000363300 | Ncrna:snRNA chromosome:GRCh37:12:102190188:102190280:-1 | -1.18 | 10.9 | 2.3E-03 |
| 8157804 | OLFML2A | Olfactomedin-like 2A (OLFML2A), mRNA. | -1.19 | 8.3 | 6.8E-03 |
| 8118974 | RPL10A | Ribosomal protein L10a (RPL10A), mRNA. | -1.20 | 8.8 | 5.4E-03 |
| 7972021 | TBC1D4 | TBC1 domain family, member 4 (TBC1D4), mRNA. | -1.26 | 8.1 | 7.4E-03 |
| 7898679 | NBPF3 | Neuroblastoma breakpoint family, member 3 (NBPF3), mRNA. | -1.27 | 12.4 | 1.3E-03 |
| 8088458 | FHIT | Fragile histidine triad gene (FHIT), transcript variant 1, mRNA. | -1.27 | 7.7 | 8.8E-03 |
| 8135268 | EIF4B | Eukaryotic translation initiation factor 4B (EIF4B), mRNA. | -1.30 | 8.0 | 7.7E-03 |
| 7900597 | C1orf50 | Chromosome 1 open reading frame 50, mRNA (cDNA clone MGC:2448 IMAGE:2959109), complete cds. | -1.31 | 14.8 | 4.9E-04 |
| 8138950 | RP9 | Retinitis pigmentosa 9 (autosomal dominant) (RP9), mRNA. | -1.33 | 9.1 | 4.8E-03 |
| 8137464 | PSPH | Phosphoserine phosphatase (PSPH), mRNA. | -1.35 | 7.8 | 8.4E-03 |
| 7903753 | GSTM2 | Glutathione S-transferase mu 2 (muscle) (GSTM2), transcript variant 1, mRNA. | -1.51 | 12.9 | 1.0E-03 |
| 7903765 | GSTM1 | Glutathione S-transferase mu 1 (GSTM1), transcript variant 1, mRNA. | -2.14 | 19.7 | 9.0E-05 |
Rows are sorted by decreasing fold-change in PTSD cases relative to non-PTSD comparison subjects.
Transcripts in bold comprised the optimal 2-probe SVM classifier of PTSD status identified by training and testing in independent samples.
Figure 1.
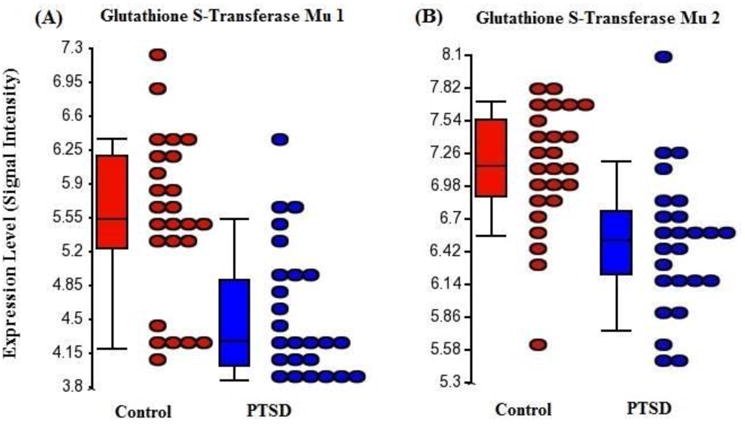
Microarray-derived expression levels (ordinate) of summarized exon probesets reflecting whole-transcript expression levels (abscissa) of glutathione s-transferase mu 1 (GSTM1) and glutathione s-transferase mu 2 (GSTM2) in peripheral blood mononuclear cells from PTSD cases (n=25, red) and comparison subjects (n=25, blue). These transcripts were notably down-regulated among PTSD cases within the full sample (fold changes -1.58 and -2.00, respectively) and were identified as the sole components of the optimal performing SVM classifier of diagnostic status, which achieved 80% accuracy in the test subset (n=10; 4 of 5 cases correctly identified).
Table 4.
QRT-PCR Validation of Gene and Exon Expression in Full Sample of PTSD Cases and Comparison Subjects.
| Assay ID | Gene | GADPH-normalized Average 2-ΔCt | GADPH-normalized p-values | HPRT1-normalized Average 2-ΔCt | HPRT1-normalized p-values | ||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| PTSD | Control | PTSD | Control | ||||
| Hs01683722_gH | GSTM1 | 0.367±0.741 | 1.138±1.529 | 0.031* | 0.685±1.367 | 1.251±1.139 | 0.128 |
| Hs03044640_gH | GSTM2 | 0.732±0.596 | 1.114±0.655 | 0.036* | 0.300±0.184 | 0.531±0.262 | 0.001* |
|
| |||||||
| Hs00180203_m1 | CUL2 | 1.000±0.323 | 1.223±0.612 | 0.114 | 0.768±0.250 | 0.872±0.350 | 0.236 |
| Hs00211676_m1 | DYNC1LI1 | 0.277±0.261 | 0.333±0.204 | 0.401 | 0.441±0.434 | 0.466±0.243 | 0.803 |
| Hs00948075_m1 | HUWE1 | 0.333±0.204 | 0.586±0.570 | 0.192 | 0.144±0.105 | 0.208±0.215 | 0.184 |
| Hs00277883_m1 | LARP7 | 0.674±0.400 | 0.711±0.272 | 0.709 | 0.689±0. 381 | 0.752±0.331 | 0.537 |
| Hs00382272_m1 | PNPLA8 | 1.115±0.240 | 1.123±0.276 | 0.910 | 1.520±0.694 | 1.254±0.478 | 0.122 |
| Hs01554570_m1 | RBM5 | 0.462±0.317 | 0.617±0.621 | 0.271 | 0.268±0.183 | 0.329±0.303 | 0.390 |
| Hs00208869_m1 | TRAPPC8 | 0.788±0.536 | 0.787±0.375 | 0.990 | 0.548±0.372 | 0.612±0.313 | 0.512 |
Values are reported as mean + standard deviation.
P-values < 0.05 are indicated with *.
3) Identification of Differentially Expressed Exons and their Associated Biological Terms
The interaction of diagnosis and exonID identified putative isoform-expression differences (p<0.0005) in 63 genes, 11 of which attained Bonferroni-corrected significance (Table 5). An example of between-group differences in exon expression for one of these eleven genes (DYNC1LI1) is illustrated in Figure 2, where the PTSD cases have significantly lower levels of expression of a single probe corresponding to the fifth exon; this region corresponds to a retained intron, which could account for this pattern of expression difference. The list of 63 genes was analyzed by the DAVID algorithm and Reactome database (Table 6). DAVID analysis revealed five significantly enriched annotations (armadillo-like helical domain, macromolecule catabolic process, acetylation, modification-dependent protein catabolic process, modification-dependent macromolecule catabolic process). Analysis using the Reactome database revealed a single enriched pathway (class 1 MHC mediated antigen processing and presentation).
Table 5.
Exons Significantly Dysregulated in Peripheral Blood Mononuclear Cells from the Full Sample of Eventual PTSD Cases at Post-Deployment.
| Transcript Cluster ID |
Gene Symbol | Gene Product | Accession Number |
F | p | Adjusted p |
q | Probesets (n) |
Dysregulated Probesets (n) |
Dysregulated Probeset ID+ |
|---|---|---|---|---|---|---|---|---|---|---|
| 7903765 | GSTM1 | Glutathione S-transferase mu 1 | NM_000561 | 7.8 | 1.1E-09 | 2.3E-05 | 2.3E-05 | 9 | 9 | 7903767 |
| 7954810 | LRRK2 | Leucine-rich repeat kinase 2 | NM_198578 | 2.6 | 2.8E-09 | 5.8E-05 | 2.9E-05 | 53 | 10 | 7954831 |
| 8158597 | GPR107 | G protein-coupled receptor 107 | NM_001136557 | 3.1 | 2.2E-07 | 4.4E-03 | 1.5E-03 | 27 | 5 | 8158617 |
| 8052443 | USP34 | Ubiquitin specific peptidase 34 | NM_014709 | 2.0 | 3.9E-07 | 7.9E-03 | 2.0E-03 | 80 | 4 | 8052505 |
| 8022767 | TRAPPC8 | Trafficking protein particle complex 8 | NM_014939 | 2.8 | 5.9E-07 | 1.2E-02 | 2.3E-03 | 31 | 2 | 8022775 |
| 8086008 | DYNC1LI1 | Dynein, cytoplasmic 1, light intermediate chain 1 | NM_016141 | 4.5 | 6.7E-07 | 1.4E-02 | 2.3E-03 | 13 | 1 | 8086013 |
| 8136662 | MGAM | Maltase-glucoamylase (alpha-glucosidase) | NM_004668 | 2.4 | 9.3E-07 | 1.9E-02 | 2.3E-03 | 46 | 1 | 8136700 |
| 8070467 | TMPRSS2 | Transmembrane protease, serine 2 | NM_001135099 | 4.2 | 9.6E-07 | 1.9E-02 | 2.3E-03 | 14 | 1 | 8070472 |
| 8059596 | TRIP12 | Thyroid hormone receptor interactor 12 | NM_004238 | 2.4 | 1.0E-06 | 2.0E-02 | 2.3E-03 | 43 | 2 | 8059600 |
| 8142307 | PNPLA8 | Patatin-like phospholipase domain containing 8 | NM_015723 | 3.9 | 1.1E-06 | 2.3E-02 | 2.3E-03 | 16 | 1 | 8142322 |
| 7978285 | ADCY4 | Adenylate cyclase 4 | NM_001198592 | 2.9 | 2.1E-06 | 4.3E-02 | 4.0E-03 | 26 | 2 | 7978294 |
| 8149986 | ZNF395 | Zinc finger protein 395 | NM_018660 | 3.9 | 2.6E-06 | 5.2E-02 | 4.3E-03 | 15 | 1 | 8149998 |
| 8058118 | KCTD18 | Potassium channel tetramerisation domain containing 18 | NM_152387 | 5.5 | 5.0E-06 | 0.10 | 7.8E-03 | 8 | 1 | 8058121 |
| 8010454 | RNF213 | Ring finger protein 213 | NM_020914 | 2.2 | 1.0E-05 | 0.21 | 1.5E-02 | 44 | 1 | 8010469 |
| 7946610 | EIF4G2 | Eukaryotic translation initiation factor 4 gamma, 2 | NM_001418 | 2.8 | 1.5E-05 | 0.31 | 2.1E-02 | 24 | 1 | 7946612 |
| 7982904 | RTF1 | Rtf1, Paf1 | NM_015138 | 2.8 | 2.6E-05 | 0.52 | 3.3E-02 | 22 | 3 | 7982911 |
| 8079869 | RBM5 | RNA binding motif protein 5 | NM_005778 | 2.6 | 2.9E-05 | 0.58 | 3.4E-02 | 26 | 1 | 8079878 |
| 8076077 | DDX17 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | NM_006386 | 3.2 | 4.1E-05 | 0.84 | 4.5E-02 | 16 | 3 | 8076093 |
| 7968128 | PABPC3 | Poly(A) binding protein, cytoplasmic 3 | NM_030979 | 11.3 | 4.2E-05 | 0.85 | 4.5E-02 | 3 | 1 | 7968129 |
| 8172914 | HUWE1 | HECT, UBA and WWE domain containing 1 | NM_031407 | 1.7 | 5.0E-05 | 1.00 | 5.0E-02 | 90 | 5 | 8172940 |
| 7903777 | GSTM5 | Glutathione S-transferase mu 5 | NM_000851 | 4.7 | 5.2E-05 | 1.00 | 5.0E-02 | 8 | 3 | 7903782 |
| 8139896 | PMS2P4 | Postmeiotic segregation increased 2 pseudogene 4 | NR 022007 | 8.0 | 6.3E-05 | 1.00 | 5.8E-02 | 4 | 1 | 8139900 |
| 8076455 | RRP7A | Ribosomal RNA processing 7 homolog A (S. cerevisiae) | NM_015703 | 6.3 | 9.0E-05 | 1.00 | 7.5E-02 | 5 | 1 | 8076458 |
| 7996677 | NUTF2 | Nuclear transport factor 2 | NM_005796 | 4.9 | 1.0E-04 | 1.00 | 7.5E-02 | 7 | 2 | 7996683 |
| 8015642 | PSMC3IP | PSMC3 interacting protein | NM_016556 | 3.5 | 1.0E-04 | 1.00 | 7.5E-02 | 12 | 1 | 8015646 |
| 8058182 | FAM126B | Family with sequence similarity 126, member B | NM_173822 | 3.5 | 1.0E-04 | 1.00 | 7.5E-02 | 12 | 0 | - |
| 8029884 | SAE1 | SUMO1 activating enzyme subunit 1 | NR 027280 | 4.1 | 1.1E-04 | 1.00 | 7.5E-02 | 9 | 1 | 8029892 |
| 8096938 | LARP7 | La ribonucleoprotein domain family, member 7 | NM_016648 | 3.0 | 1.1E-04 | 1.00 | 7.5E-02 | 16 | 2 | 8096944 |
| 7965359 | ATP2B1 | ATPase, Ca++ transporting, plasma membrane 1 | NM_001001323 | 2.5 | 1.1E-04 | 1.00 | 7.5E-02 | 24 | 1 | 7965379 |
| 8159984 | C9orf46 | Chromosome 9 open reading frame 46 | NM_018465 | 4.8 | 1.1E-04 | 1.00 | 7.5E-02 | 7 | 1 | 8159991 |
| 7965652 | CDK17 | Cyclin-dependent kinase 17 | NM_002595 | 2.8 | 1.2E-04 | 1.00 | 7.6E-02 | 18 | 1 | 7965654 |
| 8077858 | ATG7 | ATG7 autophagy related 7 homolog (S. cerevisiae) | NM_006395 | 2.8 | 1.2E-04 | 1.00 | 7.6E-02 | 18 | 1 | 8077874 |
| 7971602 | RCBTB1 | Regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 | NM018191 | 3.4 | 1.2E-04 | 1.00 | 7.6E-02 | 12 | 1 | 7971613 |
| 7971620 | KPNA3 | Karyopherin alpha 3 (importin alpha 4) | NM_002267 | 2.7 | 1.3E-04 | 1.00 | 7.8E-02 | 19 | 1 | 7971637 |
| 7956910 | CAND1 | Cullin-associated and neddylation-dissociated 1 | NM_018448 | 2.7 | 1.4E-04 | 1.00 | 7.8E-02 | 19 | 3 | 7956914 |
| 8002778 | MLKL | Mixed lineage kinase domain-like | NM_152649 | 3.3 | 1.4E-04 | 1.00 | 7.8E-02 | 13 | 1 | 8002781 |
| 8160213 | TTC39B | Tetratricopeptide repeat domain 39B | NM_152574 | 2.6 | 1.4E-04 | 1.00 | 7.8E-02 | 22 | 2 | 8160226 |
| 7962112 | CAPRIN2 | Caprin family member 2 | NM_001002259 | 2.5 | 1.5E-04 | 1.00 | 7.8E-02 | 24 | 1 | 7962123 |
| 7981068 | SERPINA1 | Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | NM_001002236 | 4.0 | 1.6E-04 | 1.00 | 8.1E-02 | 9 | 2 | 7981074 |
| 8103951 | ACSL1 | Acyl-CoA synthetase long-chain family member 1 | NM_001995 | 2.5 | 1.8E-04 | 1.00 | 9.3E-02 | 23 | 1 | 8103965 |
| 7999841 | SMG1 | SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) | NM_015092 | 2.0 | 2.0E-04 | 1.00 | 1.0E-01 | 42 | 4 | 7999869 |
| 7933047 | CUL2 | Cullin 2 | NM_003591 | 2.5 | 2.1E-04 | 1.00 | 0.10 | 23 | 2 | 7933056 |
| 8017162 | RNFT1 | Ring finger protein, transmembrane 1 | NM_016125 | 3.6 | 2.2E-04 | 1.00 | 0.10 | 10 | 3 | 8017163 |
| 7967881 | MPHOSPH8 | M-phase phosphoprotein 8 | NM_017520 | 3.0 | 2.4E-04 | 1.00 | 0.11 | 14 | 1 | 7967895 |
| 7900426 | SMAP2 | Small ArfGAP2 | NM_022733 | 3.4 | 2.5E-04 | 1.00 | 0.11 | 11 | 1 | 7900432 |
| 7903893 | CD53 | CD53 molecule | NM_000560 | 3.3 | 2.5E-04 | 1.00 | 0.11 | 12 | 2 | 7903894 |
| 8078569 | GOLGA4 | Golgin A4 | NM_002078 | 2.2 | 2.6E-04 | 1.00 | 0.11 | 30 | 2 | 8078594 |
| 7967117 | OASL | 2′-5′-oligoadenylate synthetase-like | NM_003733 | 3.8 | 2.6E-04 | 1.00 | 0.11 | 9 | 3 | 7967123 |
| 7975416 | PCNX | Pecanex homolog (Drosophila) | NM_014982 | 2.1 | 2.7E-04 | 1.00 | 0.11 | 36 | 1 | 7975429 |
| 8179298 | CSNK2B | Casein kinase 2, beta polypeptide | NM_001320 | 3.6 | 2.8E-04 | 1.00 | 0.11 | 10 | 1 | 8179308 |
| 8143327 | PARP12 | Poly (ADP-ribose) polymerase family, member 12 | NM_022750 | 3.1 | 2.9E-04 | 1.00 | 0.11 | 13 | 2 | 8143336 |
| 8021496 | KIAA1468 | KIAA1468 | NM_020854 | 2.2 | 2.9E-04 | 1.00 | 0.11 | 31 | 4 | 8021504 |
| 8141846 | FBXL13 | F-box and leucine-rich repeat protein 13 | NM_145032 | 2.3 | 3.0E-04 | 1.00 | 0.12 | 25 | 4 | 8141865 |
| 7929677 | PI4K2A | Phosphatidylinositol 4-kinase type 2 alpha | NM_018425 | 3.3 | 3.2E-04 | 1.00 | 0.12 | 11 | 0 | - |
| 8088348 | FAM116A | Family with sequence similarity 116, member A | NM_152678 | 2.5 | 3.3E-04 | 1.00 | 0.12 | 20 | 2 | 8088366 |
| 7937363 | PKP3 | Plakophilin 3 | NM_007183 | 3.0 | 3.5E-04 | 1.00 | 0.13 | 14 | 2 | 7937370 |
| 8089785 | POPDC2 | Popeye domain containing 2 | NM_022135 | 3.7 | 3.6E-04 | 1.00 | 0.13 | 9 | 2 | 8089794 |
| 7967563 | UBC | Ubiquitin C | NM_021009 | 2.9 | 3.6E-04 | 1.00 | 0.13 | 14 | 2 | 7967584 |
| 8077171 | RABL2B | RAB, member of RAS oncogene family-like 2B | NM_001130921 | 3.0 | 3.7E-04 | 1.00 | 0.13 | 13 | 1 | 8077180 |
| 8112772 | AP3B1 | Adaptor-related protein complex 3, beta 1 subunit | NM_003664 | 2.2 | 3.7E-04 | 1.00 | 0.13 | 28 | 2 | 8112794 |
| 7997626 | KLHL36 | Kelch-like 36 (Drosophila) | NM_024731 | 4.7 | 3.9E-04 | 1.00 | 0.13 | 6 | 2 | 7997629 |
| 8043251 | PTCD3 | Pentatricopeptide repeat domain 3 | NM_017952 | 2.3 | 5.0E-04 | 1.00 | 0.16 | 24 | 0 | - |
| 8072170 | KREMEN1 | Kringle containing transmembrane protein | NM_032045 | 2.9 | 5.0E-04 | 1.00 | 0.16 | 14 | 4 | 8072173 |
Rows are sorted by increasing p-value for the interaction of diagnosis and exonID.
Exon probesets listed were the most significantly differentially expressed (per gene) between diagnostic groups. Information on the identities of all dysregulated exons for each gene can be obtained from the authors upon request.
Figure 2.
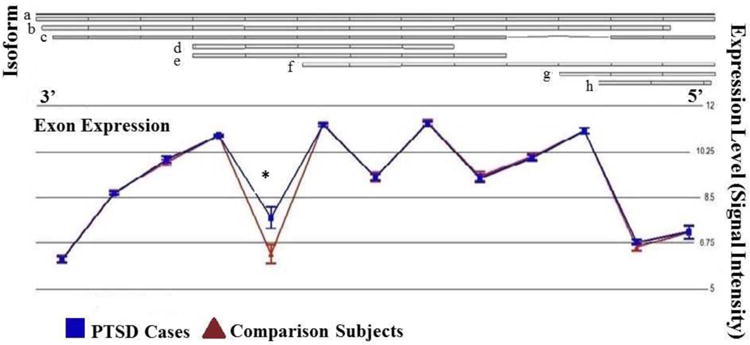
Microarray-derived expression levels (ordinate) of individual exon-probes (abscissa) of dynein, cytoplasmic 1, light intermediate chain 1 (DYNC1LI1) in peripheral blood mononuclear cells from PTSD cases (n=25, triangles) and comparison subjects (n=25, squares). The interaction of diagnosis and exon ID was highly significant (p = 6.7E-07, Bonferroni-corrected p = 1.4E-02) owing to the selective down-regulated of an exon (probeset ID 8086013; p = 0.019) in the context of comparable expression levels of all other exons.
Table 6. Annotations Enriched at Corrected Significance Levels among Differentially-Expressed Exons (p<0.0005) in Peripheral Blood Mononuclear Cells from the Full Sample of PTSD Cases at Post-Deployment*.
| DAVID Category | Term | Count (%) | Fold- Enrichment | P | Bonferroni- Corrected p | Gene Corresponding to Dysregulated Exon |
|---|---|---|---|---|---|---|
| INTERPRO | IPR011989:Armadillo-like helical | 7 (11.1%) | 14.9 | 6.5E-06 | 1.0E-03 | KIAA1468, AP3B1, PKP3, CAND1, TRIP12, KPNA3, LRRK2 |
| GOTERM_BP_FAT | GO:0009057∼macromolecule catabolic process | 12 (19.0%) | 4.5 | 4.2E-0.5 | 1.8E-0.2 | SAE1, SMG1, USP34, FBXL13, HUWE1, CAND1, ATG7, TRIP12, MGAM, CUL2, KLHL36, UBC |
| SP_PIR_KEYWORDS | Acetylation | 22 (34.9%) | 2.5 | 5.8E-05 | 7.9E-03 | EIF4G2, AP3B1, NUTF2, SMG1, HUWE1, PTCD3, CUL2, LARP7, UBC, ACSL1, KIAA1468, CSNK2B, SAE1, PABPC3, RTF1, ATG7, CAND1, KPNA3, RNF213, RBM5, MPHOSPH8, GSTM5 |
| GOTERM_BP_FAT | GO:0019941∼modification-dependent protein catabolic process | 10 (15.9%) | 5.1 | 1.0E-04 | 4.3E-02 | SAE1, USP34, FBXL13, HUWE1, CAND1, ATG7, TRIP12, CUL2, KLHL36, UBC |
| GOTERM_BP_FAT | GO:0043632∼modification-dependent macromolecule catabolic process | 10 (15.9%) | 5.1 | 1.0E-04 | 4.3E-02 | SAE1, USP34, FBXL13, HUWE1, CAND1, ATG7, TRIP12, CUL2, KLHL36, UBC |
|
| ||||||
| Reactome | Term | Count (%) | p | FDR- Corrected p | Gene Corresponding to Dysregulated Exon | |
|
| ||||||
| Class 1 MHC mediated antigen processing and presentation | 6 (9.9%) | 1.0E-04 | 1.5E-02 | SAE1,CUL2,ATG7,TRIP12,HUWE1,UBC | ||
Rows are sorted by increasing p-value for the enrichment of annotations.
4) Discovery and Replication of an Exon-Based Diagnostic Predictor
To construct an exon-based classifier and assess its generalizability we first identified potentially differentially spliced exons within our training subsample of 20 cases and 20 comparison subjects based on the diagnosis-x-exonID interaction term, using a nominal threshold of p<0.00005, while controlling for the same factors and covariates as in the analyses above. For genes displaying more than one dysregulated probe between diagnostic groups, we selected the probe with the most significant between-group difference in expression level based on the p-values from planned comparisons. This analysis and filtering yielded 56 exons with expression differences between PTSD cases and comparison subjects (Table 7) that were then used to build and optimize SVM classifiers. The optimal SVM (identified through two-level nested ten-fold cross-validation with shrinking centroids, cost=401, tolerance=0.001, kernel=radial basis function, and gamma=0.001) comprised 20 of the 56 starting probes (Table 7, probes in bold font) and attained 100% accuracy in classifying those individuals in the training sample with PTSD. We then tested the identical 20-exon SVM (with the same parameters, but with no shrinkage or cross-validation) in the remaining test subset (n=10; 5 cases and 5 comparison subjects), where it yielded a diminished but reasonable 90% accuracy (higher than the accuracy observed in gene-based analyses). All PTSD cases were correctly classified, while four of five comparison subjects were classified correctly. These values correspond to sensitivity, specificity, positive predictive and negative predictive values of 100%, 80%, 83% and 100%, respectively. QRTPCR analysis of seven exons in the classifier failed to detect significant differences in expression levels between PTSD cases and comparison subjects (Table 4).
Table 7. Exons Significantly Dysregulated in Peripheral Blood Mononuclear Cells from a Subset of PTSD Cases at Post-Deployment and Used in Predictive SVM Classifiers*.
| Transcript Cluster ID# | Gene Symbol | Gene Product | Interaction p | Exon ID | Fold-Change | F | p |
|---|---|---|---|---|---|---|---|
| 8096938 | LARP7 | La ribonucleoprotein domain family, member 7 | 7.2E-09 | 8096944 | 3.52 | 8.6 | 5.9E-03 |
| 8097148 | RNF213 | Ring finger protein 213 | 7.1E-11 | 8010469 | 3.06 | 9.6 | 3.8E-03 |
| 8086008 | DYNC1LI1 | Dynein, cytoplasmic 1, light intermediate chain 1 | 4.0E-10 | 8086013 | 3.06 | 7.9 | 8.3E-03 |
| 8134122 | PTCD3 | Pentatricopeptide repeat domain 3 | 1.6E-07 | 8043256 | 3.05 | 7.7 | 9.0E-03 |
| 8142307 | PNPLA8 | Patatin-like phospholipase domain containing 8 | 5.3E-09 | 8142322 | 2.87 | 9.3 | 4.4E-03 |
| 8172914 | DIDO1 | Death inducer-obliterator 1 | 3.7E-06 | 8067576 | 2.77 | 5.1 | 3.0E-02 |
| 8076455 | RRP7A | Ribosomal RNA processing 7 homolog A (S.cerevisiae) | 1.7E-05 | 8076458 | 2.74 | 12.8 | 1.1E-03 |
| 8158597 | GPR107 | G protein-coupled receptor 107 | 2.3E-10 | 8158617 | 2.74 | 11.5 | 1.8E-03 |
| 8054092 | CUL2 | Cullin 2 | 3.4E-06 | 7933056 | 2.71 | 11.9 | 1.5E-03 |
| 8105191 | KIAA1468 | KIAA1468 | 1.0E-07 | 8021504 | 2.71 | 11.1 | 2.1E-03 |
| 8045090 | ZC3H11A | Zinc finger CCCH-type containing 11A | 1.1E-05 | 7908985 | 2.67 | 6.5 | 1.6E-02 |
| 8056837 | TTC17 | Tetratricopeptide repeat domain 17 | 1.1E-06 | 7939453 | 2.63 | 6.3 | 1.7E-02 |
| 8079869 | NEK9 | NIMA (never in mitosis gene a)- related kinase 9 | 6.7E-06 | 7980282 | 2.62 | 8.4 | 6.4E-03 |
| 8079869 | RBM5 | RNA binding motif protein 5 | 1.5E-08 | 8079878 | 2.62 | 8.9 | 5.3E-03 |
| 8105191 | PARP8 | Poly (ADP-ribose) polymerase family, member 8 | 1.6E-05 | 8105199 | 2.57 | 4.6 | 4.0E-02 |
| 8083523 | AQR | Aquarius homolog (mouse) | 2.6E-05 | 7987328 | 2.57 | 6.3 | 1.7E-02 |
| 8136662 | MGAM | Maltase-glucoamylase (alpha-glucosidase) | 5.1E-08 | 8136700 | 2.57 | 4.4 | 4.3E-02 |
| 8107375 | TRAPPC8 | Trafficking protein particle complex 8 | 6.8E-08 | 8022775 | 2.56 | 7.1 | 1.2E-02 |
| 8136662 | UGGT1 | UDP-glucose glycoprotein glucosyltransferase 1 | 2.1E-06 | 8045104 | 2.53 | 10.7 | 2.5E-03 |
| 8169541 | XRN2 | 5′-3′ exoribonuclease 2 | 2.7E-05 | 8061333 | 2.46 | 6.4 | 1.6E-02 |
| 8103951 | ACSL1 | Acyl-CoA synthetase long-chain family member 1 | 1.2E-05 | 8103961 | 2.45 | 5.2 | 3.0E-02 |
| 8172914 | HUWE1 | HECT, UBA and WWE domain containing 1 | 1.3E-07 | 8172982 | 2.42 | 7.7 | 9.0E-03 |
| 8061324 | CAND1 | Cullin-associated and neddylation-dissociated 1 | 5.1E-06 | 7956914 | 2.42 | 8.1 | 7.4E-03 |
| 8160213 | TTC39B | Tetratricopeptide repeat domain 39B | 3.3E-06 | 8160226 | 2.37 | 10.2 | 3.0E-03 |
| 8107375 | YTHDC2 | YTH domain containing 2 | 9.5E-06 | 8107388 | 2.35 | 12.7 | 1.1E-03 |
| 8149986 | ZNF395 | Zinc finger protein 395 | 4.6E-08 | 8149998 | 2.32 | 8.1 | 7.4E-03 |
| 8076455 | CDK17 | Cyclin-dependent kinase 17 | 5.8E-06 | 7965654 | 2.25 | 8.2 | 7.2E-03 |
| 8149986 | USP34 | Ubiquitin specific peptidase 34 | 1.5E-10 | 8052505 | 2.20 | 7.7 | 9.0E-03 |
| 8092933 | SMG1 | SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. ele | 5.0E-07 | 7994025 | 2.18 | 5.0 | 3.2E-02 |
| 8156321 | TMEM131 | Transmembrane protein 131 | 2.2E-05 | 8054124 | 2.18 | 5.6 | 2.3E-02 |
| 8158597 | GPR155 | G protein-coupled receptor 155 | 3.6E-06 | 8056852 | 2.15 | 6.7 | 1.4E-02 |
| 8086008 | ADAM10 | ADAM metallopeptidase domain 10 | 7.6E-06 | 7989240 | 2.10 | 5.1 | 3.0E-02 |
| 8092933 | ACAP2 | ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 | 2.4E-05 | 8092954 | 2.10 | 5.0 | 3.1E-02 |
| 8051882 | DENND2D | DENN | 3.4E-06 | 7918493 | 2.07 | 5.7 | 2.3E-02 |
| 8156321 | SYK | Spleen tyrosine kinase | 2.0E-06 | 8156330 | 1.93 | 13.3 | 8.7E-04 |
| 8083523 | GMPS | Guanine monphosphate synthetase | 8.9E-06 | 8083535 | 1.83 | 5.9 | 2.1E-02 |
| 8134122 | AKAP9 | A kinase (PRKA) anchor protein (yotiao) 9 | 7.8E-07 | 8134144 | 1.81 | 8.3 | 6.9E-03 |
| 8160213 | TRIP12 | Thyroid hormone receptor interactor 12 | 1.2E-06 | 8059619 | 1.79 | 6.5 | 1.6E-02 |
| 8059596 | EIF4G2 | Eukaryotic translation initiation factor 4 gamma, 2 | 7.3E-07 | 7946612 | 1.76 | 5.9 | 2.1E-02 |
| 8130151 | APC2 | Adenomatosis polyposis coli 2 | 3.6E-05 | 8024308 | 1.75 | 7.0 | 1.2E-02 |
| 8130151 | RAET1E | Retinoic acid early transcript 1E | 3.7E-05 | 8130160 | 1.74 | 8.1 | 7.4E-03 |
| 8076077 | CAPRIN2 | Caprin family member 2 | 6.6E-06 | 7962123 | 1.69 | 6.4 | 1.6E-02 |
| 8103951 | NUP88 | Nucleoporin 88kDa | 4.9E-05 | 8011838 | 1.68 | 5.6 | 2.3E-02 |
| 8078569 | MPHOSPH8 | M-phase phosphoprotein 8 | 1.9E-05 | 7967895 | 1.63 | 18.3 | 1.5E-04 |
| 8052443 | FAM175B | Family with sequence similarity 175, member B | 2.7E-05 | 7931218 | 1.61 | 6.8 | 1.3E-02 |
| 8097148 | KIAA1109 | KIAA1109 | 1.7E-05 | 8097151 | 1.59 | 7.7 | 9.1E-03 |
| 8169541 | DOCK11 | Dedicator of cytokinesis 11 | 1.4E-05 | 8169559 | 1.41 | 5.5 | 2.5E-02 |
|
| |||||||
| 8022767 | SMAP2 | Small ArfGAP2 | 3.0E-05 | 7900432 | -1.12 | 6.1 | 1.9E-02 |
| 8179298 | DDX17 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | 1.1E-06 | 8076093 | -1.12 | 5.6 | 2.4E-02 |
| 8067563 | MDM2 | Mdm2 p53 binding protein homolog (mouse) | 6.3E-07 | 7957003 | -1.16 | 5.5 | 2.5E-02 |
| 8096938 | SMG1 | SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) | 2.7E-08 | 7999869 | -1.20 | 14.1 | 6.6E-04 |
| 8043251 | GSTM5 | Glutathione S-transferase mu 5 | 4.2E-05 | 7903782 | -1.57 | 18.5 | 1.4E-04 |
| 8078569 | GOLGA4 | Golgin A4 | 5.0E-06 | 8078597 | -1.62 | 5.2 | 2.9E-02 |
| 8142307 | LRPPRC | Leucine-rich PPR-motif containing | 5.6E-07 | 8051896 | -1.77 | 10.4 | 2.8E-03 |
| 8179298 | CSNK2B | Casein kinase 2, beta polypeptide | 2.3E-05 | 8179308 | -1.83 | 7.8 | 8.4E-03 |
| 8024306 | GSTM1 | Glutathione S-transferase mu 1 | 4.5E-09 | 7903772 | -2.52 | 21.4 | 5.3E-05 |
Rows are sorted by decreasing fold-change in eventual PTSD cases relative to non-PTSD comparison subjects.
Exons of Transcript Cluster IDs in bold comprised the optimal 20-probe SVM classifier of eventual PTSD status identified by training and testing in independent samples.
Discussion
There is emerging support for the hypothesis that peripheral blood transcriptomic signatures associated with PTSD involve dysregulation of genes that function in immune and inflammatory processes or their regulation To this picture we add new and compelling pilot data suggesting that dysregulation of genes whose proteins function in the management of cellular oxidative stress may also be clinically useful biomarkers for distinguishing PTSD cases from trauma-exposed subjects who are resilient to PTSD. Yet, dysregulation of genes with immune-, inflammatory- and antioxidant-activity is probably only a small piece of the biological puzzle of PTSD pathophysiology, as many of the differentially expressed genes, as well as the exons comprising the best-performing PTSD-diagnostic classifier, were apparently unrelated to these functions; these other genes had highly disparate functions. Collectively, profiles of dysregulated genes in immune, inflammatory and other pathways may serve as potent biological indicators upon which diagnosis and early intervention may ultimately be based. The present study demonstrates proof-of-principle for the construction of blood-based PTSD diagnostic biomarkers that ranged in accuracy from 80-90% in a small subset that was held completely independent from classifier construction.
It is important to note that these classifiers employed decision-rules based solely on mRNA expression levels. To our knowledge, our group is among the first to employ data-driven (SVM) modeling on a list of differentially expressed transcripts in order to identify a subset of transcripts that were most predictive of PTSD status. These two strategies may be useful for identifying exons, genes, and pathways that play a role in the etiology of PTSD, but that may have been overlooked by other approaches focusing on well-established candidate genes. If these profiles of mRNA-expression differences in PTSD cases can be further refined and replicated, and if SVM-based models are found to perform reliably in larger or more diverse populations, then this study proposes an avenue for early diagnosis among trauma-exposed individuals, potentially fostering earlier intervention. However, it is likely that a more accurate classification model can be constructed in the future by taking into account additional known risk factors for PTSD, such as family history, personality traits, pre-existing mental disorders (Koenen et al., 2003a; Koenen et al., 2003b), peri-traumatic dissociation and post-trauma social support (Brewin et al., 2000; Ozer et al., 2003), non-genomic biological markers available in the MRS dataset (Baker et al., 2012b; Eraly et al., 2014), and other factors not necessarily related to gene expression.
The present study did not account for many of these pre- and peri-traumatic risk factors, but future efforts to construct diagnostic models should seek to incorporate such data. Nevertheless, a single diagnostic classifier of PTSD (no matter how precisely constructed) may never perform with 100% accuracy, which is why it will be essential to pursue (in larger samples) the characteristics of subjects for whom such a classifier does not work. Of equal interest is the possibility that there are two or more unique biomarker profiles that are diagnostic of the same phenotypic outcome. In fact, etiologic heterogeneity may be a hallmark of complex disorders including PTSD, so it may not be possible to identify a single “one-size-fits-all” biomarker profile. In the future, methodologies that facilitate the identification of distinct biomarker profiles associated with the same phenotype may be required in order to account for etiologic heterogeneity in PTSD and other complex disorders. Another distinct possibility is that for some cases of PTSD there is no blood-based biomarker signature to be found. We are currently investigating each of these possibilities further. It is also important to acknowledge that the present study did not account for possible effects of pharmacological therapy (e.g., anti-depressants, anxiolytics, and antipsychotics) or other treatments on post-deployment gene expression profiles. Five of the 25 PTSD subjects reported using at least one psychiatric medication at the time blood samples were obtained, while none of the comparison subjects reported psychiatric medication use. It is plausible that between-group differences in medication use could account for some of the gene-expression differences observed between these groups. In order to account for this possibility, we performed a separate ANCOVA comparing non-medicated PTSD subjects (n=20) and comparison subjects (n=25). The removal of medicated subjects from the PTSD group produced only minor changes in ANCOVA fold-change values for the genes of interest; the average difference in fold-change value was < 2%. Previous genome-wide expression studies have addressed this issue by using samples from PTSD subjects who were not currently medicated (Zeiker et al., 2007; Yehuda et al., 2009; Neylan et al., 2011). Other studies have not explicitly address medication status (Mehta et al., 2011; Segmen et al., 2005). However, if the ultimate goal is to develop gene expression-based diagnostic classifiers that are robust to real-world variability, then the inclusion of medicated subjects may be valuable. Future studies should attempt to account for medication status and statistically control for its effect on gene expression in order to identify genes that are specific to PTSD pathophysiology.
Because of our relatively small sample size and the severe corrections for multiple-testing required when examining the entire transcriptome, we did not detect individual gene-expression differences in PTSD cases that surpassed stringent criteria for declaring statistical significance. As such, the focus of our efforts and interpretations has been on groups of genes, either in regard to their biological annotations or their collective ability to identify PTSD cases. Nevertheless, one gene identified here as dysregulated has been identified previously in studies seeking to identify blood-based diagnostic biomarkers for PTSD. Prior to rigorous correction for multiple observations, Neylan and others (Neylan et al., 2011) reported up-regulation of GSTM1 in PTSD cases, whereas we observed down-regulation of GSTM1 in PTSD cases. It is plausible that differences in subject characteristics or study design could account for the discrepant findings. Neylan and colleagues found increased GSTM1 expression in PTSD subjects compared to a non-trauma exposed control group. Perhaps these discrepant findings could make sense in the context of a model where increased GSTM1 expression reflects an adaptive response to traumatic stress and the attenuation of this response disposes some trauma-exposed individuals to developing PTSD., These studies also differed with respect to the time-span between disease onset and blood sample collection. Remarkably, GSTM1 and GSTM2 were identified as the lone predictors within a diagnostic classifier that achieved 80% accuracy in the test subset, and the down-regulation of GSTM2 was confirmed by QRTPCR. In previous work, we observed down-regulation of GSTM1 among these same subjects in samples taken prior to their deployment and the development of clinically significant PTSD symptoms; GSTM1 expression levels were also part of a pre-deployment predictor of subsequent PTSD diagnosis (Glatt et al., 2013). Members of this enzyme class function in the detoxification of electrophilic compounds--including carcinogens, therapeutic drugs, environmental toxins and products of oxidative stress--by conjugation with glutathione. Down-regulation of genes whose proteins are responsible for the metabolism of reactive oxygen species (ROS) was also observed in lifetime PTSD cases with current symptoms (Zieker et al., 2007). The apparent link between ROS metabolism and PTSD may make more sense in light of previous in vitro studies demonstrating redox regulation of intracellular GR signaling. Specifically, reduced expression of antioxidant protein or direct administration of ROS negatively modulated GR signaling and resulted in reduced expression of glucocorticoid-induced genes; this effect could be rescued by the administration of antioxidant compounds (Makino et al., 1996; Okamoto et al., 1999). It is also interesting to note that other groups have found associations between GSTM1 polymorphisms and other brain disorders, including schizophrenia (Gravina et al., 2011; Rodriguez-Santiago et al., 2010), bipolar disorder (Mohammadynejad et al., 2011), and alcohol withdrawal symptoms (Okubo et al., 2003).
Despite our relatively small sample size and the additional levels of correction for multiple-testing required for exon analyses, a number of differentially expressed exons surpassed stringent criteria for declaring statistical significance. Additionally, the exon-based predictive classifier appeared to perform better than the gene-based predictive classifier. Taken together, these data support our previous findings (Glatt et al., 2013), suggesting that exon expression (indexing the activity of individual splice variants) may be more reliable and biologically informative than the expression of full-length “genes” or aggregated transcript clusters. Yet, we could not successfully recapitulate these array-derived results by QRTPCR, so further validation attempts must be made. Two of the dysregulated exons we identified by array analysis are components of genes DDX17 and FAM175B, which have been identified as differentially expressed in previous PTSD biomarker studies. Yehuda and others (Yehuda et al., 2009) observed up-regulation of DDX17 among PTSD cases. Sarapas and others (Sarapas et al., 2011) observed down-regulation of FAM175B among current PTSD cases, but not lifetime PTSD cases or trauma-exposed comparison subjects. It is also curious to note that the list of alternatively spliced transcripts was enriched for acetylation-dependent protein catabolism and acetylation-regulated proteins more generally. Emerging evidence indicates that the acetylation of amino acids within non-histone proteins plays a role in regulation of cell metabolism (Choudhary et al., 2009; Yang and Seto, 2008). If the differentially expressed exons in PTSD are found to contain acetylation-dependent regulatory domains, then it is plausible that the PTSD proteome may be abnormally (hypo- or hyper-) responsive to acetylation.
It is interesting to compare the results of the present pilot study with our own previous work examining pre-deployment gene expression profiles associated with subsequent development of PTSD after return from deployment (Glatt et al., 2013), as many of the same subjects (24 of 25 cases, 23 of 25 comparison) were featured in both studies. A number of whole-gene transcripts appeared dysregulated both prior to deployment (in those who would later develop PTSD) and after deployment (in current PTSD cases). AIM2 and EPSTI1 were up-regulated in both analyses (i.e., at both pre- and post-deployment), while RPL10A and GSTM1 were down-regulated in both analyses; these may reflect stable markers for PTSD vulnerability. Alternatively, they could have been dysregulated at pre-deployment assessment because pathophysiogical changes had already begun among these subjects, many of whom had previously been deployed to war zones (or because of other unmeasured factors, such as early adversity). LRTM2, on the other hand, was down-regulated in pre-deployment samples, but up-regulated in post-deployment samples; this may reflect a maladaptive change or a compensatory yet ultimately ineffective change. Additionally, one putatively alternatively spliced transcript was identified in both pre- and post-deployment analyses. For MGAM, a similar pattern of dysregulated exon expression was observed in both analyses, with PTSD cases showing increased expression of several probes and the largest expression difference observed in a probe (8136700) spanning the 23rd and 24th exons. This may also reflect a stable marker for PTSD vulnerability.
The present pilot study broadens the search for diagnostic biomarkers for PTSD beyond that of previous work. Several studies have compared genome-wide transcriptional profiles between PTSD cases and controls, but to our knowledge, very few transcripts have been identified as dysregulated across studies using independent samples. Two studies with overlapping sample pools reported reduced expression of FKBP5 among current PTSD cases (Sarapas et al., 2011; Yehuda et al., 2009). A third study by Mehta and colleagues (Mehta et al., 2011) reported that the interaction of PTSD status and FKBP5 genotype (rs9296158) was associated with dysregulation of FKBP5 expression. Pre-deployment expression levels of FKBP5 were also shown to independently predict post-deployment PTSD symptoms (van Zuiden et al., 2012a). In the present microarray study, diagnostic status was not associated with differences in FKBP5 expression, suggesting a need to further consider heterogeneity in PTSD etiology. As discussed above, GSTM1 was originally identified as up-regulated among PTSD cases by Neylan and others (Neylan et al., 2011), but was found to be down-regulated prior to deployment among US Marines who would later develop PTSD and down-regulated among current PTSD cases within the present analysis, which utilized many of the same subjects as Glatt and others (Glatt et al., 2013). Also discussed above, the present study was the second to implicate DDX17 and FAM175B transcripts. The paucity of overlapping findings across studies may be accounted for by a number of factors, including the high risk for Type 1 error inherent when the number of subjects is small and statistical correction for multiple observations is inadequate. Other factors could also contribute to discrepant findings, including differences in trauma exposure (military combat, catastrophic event), sample type (PBMC vs. whole blood), the timing of sampling with respect to trauma exposure and disease onset, or differences in PTSD treatment effects across studies.
Comparing annotation-enrichment results across studies provided no additional consensus on the nature of gene expression dysregulation in PTSD; we performed DAVID and Reactome analyses on individual transcript lists provided by each of the reviewed studies (Mehta et al., 2011; Neylan et al., 2011; Sarapas et al., 2011; van Zuiden et al., 2012a; Yehuda et al., 2009; Zieker et al., 2007), but few studies demonstrated significant enrichment of terms and no common terms were observed across studies. However, when these transcript lists were combined with the present data to create a single list, significant enrichment was observed for a number of terms related to cytokine signaling, lysosomal activity, and other immune-cell activities (Table 8). It is apparent that genes involved in cellular immunity are reliably and disproportionately represented among those that are dysregulated in PTSD cases. This is supported by a large body of evidence for dysfunctional cellular immune processes in individuals with PTSD, which we recently reviewed in depth (Baker et al., 2012b). Our review of the collective evidence suggests that systemic inflammation and deleterious health consequences in PTSD are strongly linked. Given this evidence, treatment strategies to reduce inflammation or modulate cell-mediated immunological processes may be of value to pursue in preclinical models of PTSD.
Table 8. Annotations Enriched at Corrected Significance Levels among Dysregulated Transcripts across Studies Comparing PTSD Cases and Comparison Subjects.
| DAVID Category | Term | Count (%) | Fold- Enrichment |
p | Bonferroni- Corrected p |
Gene Corresponding to Dysregulated Exon |
|---|---|---|---|---|---|---|
| GOTERM_BP_FAT | GO:0001775∼cell activation | 15 (8.1%) | 5.1 | 1.2E-06 | 2.1E-03 | BLM, SWAP70, STAT5B, KLRK1, MINK1, PF4, IGF2, SLAMF1, TGFB1, AZU1, CCND3, C14ORF19, TREML1, F2R, CD7 |
| GOTERM_BP_FAT | GO:0009611∼response to wounding | 19 (10.2%) | 3.5 | 6.6E-06 | 1.1E-02 | LIPA, CCR1, STAT5B, CXCR1, PF4, IGF2, SOD1, CCL5, DTNBP1, TGFB1, GP9, AZU1, ORM1, CX3CR1, CTSB, PLA2G4C, TREML1, KDM6B, F2R |
| GOTERM_CC_FAT | GO:0030141∼secretory granule | 10 (5.4%) | 5.7 | 5.9E-05 | 1.3E-02 | AZU1, SPAG8, PPBP, CPA3, PF4, SPARC, TREML1, RACGAP1, TGFB1, GP9 |
| GOTERM_BP_FAT | GO:0001817∼regulation of cytokine production | 11 (5.9%) | 6.0 | 1.5E-05 | 2.5E-02 | AZU1, IL18, TIA1, STAT5B, KLRK1, IGF2, PF4, TRIM16, SOD1, TGFB1, F2R |
| GOTERM_CC_FAT | GO:0031091∼platelet alpha granule | 6 (3.2%) | 11.0 | 2.0E-04 | 4.1E-02 | PPBP, PF4, SPARC, TREML1, TGFB1, GP9 |
| GOTERM_CC_FAT | GO:0000323∼lytic vacuole | 10 (5.4%) | 4.9 | 2.0E-04 | 4.2E-02 | AZU1, LAPTM5, LIPA, MMD, IFI30, CPA3, CTSC, CTSB, ASAH1, GBA |
| GOTERM_CC_FAT | GO:0005764∼lysosome | 10 (5.4%) | 4.9 | 2.0E-04 | 4.2E-02 | AZU1, LAPTM5, LIPA, MMD, IFI30, CPA3, CTSC, CTSB, ASAH1, GBA |
| KEGG_PATHWAY | hsa04060:cytokine-cytokine receptor interaction | 13 (7.0%) | 3.2 | 5.0E-04 | 4.5E-02 | TNFRSF21, IL18R1, IL18, CCR1, CXCR1, PF4, CCL5, TGFB1, PPBP, CX3CR1, CSF2RB, IL2RG, IL3RA |
| GOTERM_MF_FAT | GO:0019955∼cytokine binding | 8 (4.3%) | 7.0 | 1.4E-04 | 4.9E-02 | IL18R1, CCR1, CX3CR1, CSF2RB, CXCR1, IL2RG, TRIM16, IL3RA |
|
| ||||||
| Reactome | Term | Count (%) | p | FDR-corrected p | Gene Corresponding to Dysregulated Exon | |
|
| ||||||
| Cytokine-cytokine receptor interaction | 13 (7.0%) | 1.0E-05 | 3.0E-03 | IL18,TGFB1,PPBP,CX3CR1,CCL5,IL3RA,CXCR1,CSF2RB,TNFRSF21, CCR1,PF4,IL2RG,IL18R1 | ||
| Lysosome | 7 (3.8%) | 3.0E-04 | 3.5E-02 | NAPSA,LAPTM5,ASAH1,CTSC,CTSB,GBA,LIPA | ||
| Interleukin signaling pathway | 5 (2.7%) | 3.0E-04 | 4.7E-02 | IL16,IL18,STAT5B,IL3RA,CXCR1 | ||
In conclusion, as the development of PTSD following initial trauma exposure remains quite variable and unpredictable, we sought to identify readily assessable biomarkers to aid in early diagnosis based on evaluations of blood-based gene expression among Marines participating in the MRS. Our analyses converged on a diverse group of genes and exons that appeared to be differentially expressed in peripheral blood cells from individuals with PTSD. Reduced expression of two genes involved in ROS metabolism were predictive of PTSD diagnostic status, while altered exon expression within a larger and more heterogeneous group of transcripts also predicted the PTSD diagnosis with apparently high accuracy. If blood-based biomarkers (such as the panels of genes and exons identified here) can be validated in additional cohorts exposed to a wider variety of traumatic stressors, then they may serve as useful adjuncts to the prevailing gold-standard behavioral diagnostic systems (Brewin et al., 2005; Ozer et al., 2003). Enabling clinicians to more confidently diagnose PTSD at earlier stages would be particularly important in groups such as these Marines, for whom it is known in advance that exposure to serious trauma is highly likely. This may also prove highly relevant for first-responders, such as police, fire, and emergency medical teams, for whom a regular part of their job is also exposure to potentially traumatic situations. Furthermore, blood-based biomarkers may help clinicians identify instances of determination of fitness for duty so that support services and limited resources can be made available to those individuals with the greatest need.
Acknowledgments
This work was supported in part by grants R21MH085240 (M.T.T.), R01MH093500 (C.M.N.), and P50MH081755-0003 (S.J.G.) from the U.S. National Institutes of Health, grant SDR 09-0128 (D.G.B.) from the VA Health Service Research and Development Service (D.G.B.), the Marine Corps and Navy Bureau of Medicine and Surgery (D.G.B), an Independent Investigator Award and the Sidney R. Baer, Jr. Prize or Schizophrenia Research (S.J.G.) from NARSAD: The Brain and Behavior Research Fund, a Research Grant from the Gerber Foundation, a grant from National Natural Science Foundation of China (81000588), and a “Young Talents Training Plan” award from the Shanghai Health Bureau (XYQ2011016). We gratefully acknowledge the assistance of Melanie Collins in the preparation of the RNA samples, the constructive feedback of Dr. Stephen V. Faraone and Brian Reardon on a previous version of this manuscript, and all of the MRS staff members, including Amela Ahmetovic, Nilima Biswas, William H. Black, Mahalah R. Buell, Teresa Carper, Andrew De La Rosa, Benjamin Dickstein, Heather Ellis-Johnson, Caitlin Fernandes, Susan Fesperman, David Fink, Summer Fitzgerald, Steven Gerard, Abigail A. Goldsmith, Gali Goldwaser, Patricia Gorman, Jorge A. Gutierrez, John A. Hall, Jr., Christian J. Hansen, Laura Harder, Pia Heppner, Alexandra Kelada, Christopher L. Lehnig, Jennifer Lemmer, Morgan LeSuer-Mandernack, Manjula Mahata, Adam X. Maihofer, Arame Motazedi, Elin Olsson, Ines Pandzic, Anjana H. Patel, Dhaval H. Patel, Sejal Patel, Shetal M. Patel, Taylor Perin-Kash, James O.E. Pittman, Stephanie Raducha, Brenda Thomas, Elisa Tsan, Maria Anna Valencerina, Chelsea Wallace, Kate Yurgil, Kuixing Zhang, and the many intermittent on-site MRS clinician-interviewers and data-collection staff.
Footnotes
The authors report no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Affymetrix I. GeneChip® Whole Transcript (WT) Sense Target Labeling Assay Manual, Version 4. Affymetrix Inc.; 2006. [Google Scholar]
- Afifi TO, Asmundson GJ, Taylor S, Jang KL. The role of genes and environment on trauma exposure and posttraumatic stress disorder symptoms: a review of twin studies. Clinical psychology review. 2010;30(1):101–12. doi: 10.1016/j.cpr.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Baker DG, Nash WP, Litz BT, Geyer MA, Risbrough VB, Nievergelt CM, O'Connor DT, Larson GE, Schork NJ, Vasterling JJ, et al. Predictors of risk and resilience for posttraumatic stress disorder among ground combat Marines: methods of the Marine Resiliency Study. Prev Chronic Dis. 2012a;9:E97. doi: 10.5888/pcd9.110134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DG, Nievergelt CM, O'Connor DT. Biomarkers of PTSD: neuropeptides and immune signaling. Neuropharmacology. 2012c;62(2):663–73. doi: 10.1016/j.neuropharm.2011.02.027. [DOI] [PubMed] [Google Scholar]
- Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB, Tang Y, Gillespie CF, Heim CM, Nemeroff CB, et al. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA : the journal of the American Medical Association. 2008;299(11):1291–305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewin CR. Risk factor effect sizes in PTSD: what this means for intervention. J Trauma Dissociation. 2005;6(2):123–30. doi: 10.1300/J229v06n02_11. [DOI] [PubMed] [Google Scholar]
- Brewin CR, Andrews B, Valentine JD. Meta-analysis of risk factors for posttraumatic stress disorder in trauma-exposed adults. Journal of consulting and clinical psychology. 2000;68(5):748–66. doi: 10.1037//0022-006x.68.5.748. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biology. 2003;4(5):P3. [PubMed] [Google Scholar]
- DiGangi JA, Gomez D, Mendoza L, Jason LA, Keys CB, Koenen KC. Pretrauma risk factors for posttraumatic stress disorder: A systematic review of the literature. Clinical Psychology Review. 2013 doi: 10.1016/j.cpr.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Eraly SA, Nievergelt CM, Maihofer AX, Barkauskas DA, Biswas N, Agorastos A, O'Connor DT, Baker DG. Assessment of plasma C-reactive protein as a biomarker of posttraumatic stress disorder risk. JAMA psychiatry. 2014;71(4):423–31. doi: 10.1001/jamapsychiatry.2013.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie CF, Phifer J, Bradley B, Ressler KJ. Risk and resilience: genetic and environmental influences on development of the stress response. Depression and anxiety. 2009;26(11):984–92. doi: 10.1002/da.20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt SJ, Chandler SD, Bousman CA, Chana G, Lucero GR, Tatro E, May T, Lohr JB, Kremen WS, Everall IE, et al. Alternatively spliced genes as biomarkers for schizophrenia, bipolar disorder and psychosis: A blood-based spliceome-profiling exploratory study. Current Pharmacogenomics and Personalized Medicine. 2009;7(3):164–188. doi: 10.2174/1875692110907030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt SJ, Everall IP, Kremen WS, Corbeil J, Sasik R, Khanlou N, Han M, Liew CC, Tsuang MT. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15533–8. doi: 10.1073/pnas.0507666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt SJ, Tylee DS, Chandler SD, Pazol J, Nievergelt CM, Woelk CH, Baker DG, Lohr JB, Kremen WS, Litz BT, et al. Blood-based gene-expression predictors of PTSD risk and resilience among deployed marines: a pilot study. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2013;162B(4):313–26. doi: 10.1002/ajmg.b.32167. [DOI] [PubMed] [Google Scholar]
- Gravina P, Spoletini I, Masini S, Valentini A, Vanni D, Paladini E, Bossu P, Caltagirone C, Federici G, Spalletta G, et al. Genetic polymorphisms of glutathione S-transferases GSTM1, GSTT1, GSTP1 and GSTA1 as risk factors for schizophrenia. Psychiatry research. 2011;187(3):454–6. doi: 10.1016/j.psychres.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Heinzelmann M, Gill J. Epigenetic Mechanisms Shape the Biological Response to Trauma and Risk for PTSD: A Critical Review. Nursing research and practice. 2013;2013:417010. doi: 10.1155/2013/417010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Koenen KC, Amstadter AB, Nugent NR. Gene-environment interaction in posttraumatic stress disorder: an update. Journal of traumatic stress. 2009;22(5):416–26. doi: 10.1002/jts.20435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenen KC, Lyons MJ, Goldberg J, Simpson J, Williams WM, Toomey R, Eisen SA, True W, Tsuang MT. Co-twin control study of relationships among combat exposure, combat-related PTSD, and other mental disorders. J Trauma Stress. 2003a;16(5):433–8. doi: 10.1023/A:1025786925483. [DOI] [PubMed] [Google Scholar]
- Koenen KC, Lyons MJ, Goldberg J, Simpson J, Williams WM, Toomey R, Eisen SA, True WR, Cloitre M, Wolfe J, et al. A high risk twin study of combat-related PTSD comorbidity. Twin Res. 2003b;6(3):218–26. doi: 10.1375/136905203765693870. [DOI] [PubMed] [Google Scholar]
- Litz BT, Gray MJM, Bryant R, Adler A. Early interventions for trauma: Current status and future directions. Clinical Psychology: Science and Practice. 2002;9:112–134. [Google Scholar]
- Makino Y, Okamoto K, Yoshikawa N, Aoshima M, Hirota K, Yodoi J, Umesono K, Makino I, Tanaka H. Thioredoxin: a redox-regulating cellular cofactor for glucocorticoid hormone action. Cross talk between endocrine control of stress response and cellular antioxidant defense system. The Journal of clinical investigation. 1996;98(11):2469–77. doi: 10.1172/JCI119065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane AC. The prevalence and longitudinal course of PTSD. Implications for the neurobiological models of PTSD. Annals of the New York Academy of Sciences. 1997;821:10–23. doi: 10.1111/j.1749-6632.1997.tb48265.x. [DOI] [PubMed] [Google Scholar]
- Mehta D, Gonik M, Klengel T, Rex-Haffner M, Menke A, Rubel J, Mercer KB, Putz B, Bradley B, Holsboer F, et al. Using polymorphisms in FKBP5 to define biologically distinct subtypes of posttraumatic stress disorder: evidence from endocrine and gene expression studies. Archives of general psychiatry. 2011;68(9):901–10. doi: 10.1001/archgenpsychiatry.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadynejad P, Saadat I, Ghanizadeh A, Saadat M. Bipolar disorder and polymorphisms of glutathione S-transferases M1 (GSTM1) and T1 (GSTT1) Psychiatry research. 2011;186(1):144–6. doi: 10.1016/j.psychres.2010.06.017. [DOI] [PubMed] [Google Scholar]
- Neylan TC, Sun B, Rempel H, Ross J, Lenoci M, O'Donovan A, Pulliam L. Suppressed monocyte gene expression profile in men versus women with PTSD. Brain, behavior, and immunity. 2011;25(3):524–31. doi: 10.1016/j.bbi.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Tanaka H, Ogawa H, Makino Y, Eguchi H, Hayashi S, Yoshikawa N, Poellinger L, Umesono K, Makino I. Redox-dependent regulation of nuclear import of the glucocorticoid receptor. The Journal of biological chemistry. 1999;274(15):10363–71. doi: 10.1074/jbc.274.15.10363. [DOI] [PubMed] [Google Scholar]
- Okubo T, Harada S, Higuchi S, Matsushita S. Association analyses between polymorphisms of the phase II detoxification enzymes (GSTM1, NQO1, NQO2) and alcohol withdrawal symptoms. Alcoholism, clinical and experimental research. 2003;27(8 Suppl):68S–71S. doi: 10.1097/01.ALC.0000078616.63296.41. [DOI] [PubMed] [Google Scholar]
- Ozer EJ, Best SR, Lipsey TL, Weiss DS. Predictors of posttraumatic stress disorder and symptoms in adults: a meta-analysis. Psychological bulletin. 2003;129(1):52–73. doi: 10.1037/0033-2909.129.1.52. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Santiago B, Brunet A, Sobrino B, Serra-Juhe C, Flores R, Armengol L, Vilella E, Gabau E, Guitart M, Guillamat R, et al. Association of common copy number variants at the glutathione S-transferase genes and rare novel genomic changes with schizophrenia. Molecular psychiatry. 2010;15(10):1023–33. doi: 10.1038/mp.2009.53. [DOI] [PubMed] [Google Scholar]
- Sarapas C, Cai G, Bierer LM, Golier JA, Galea S, Ising M, Rein T, Schmeidler J, Muller-Myhsok B, Uhr M, et al. Genetic markers for PTSD risk and resilience among survivors of the World Trade Center attacks. Disease markers. 2011;30(2-3):101–10. doi: 10.3233/DMA-2011-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segman RH, Shefi N, Goltser-Dubner T, Friedman N, Kaminski N, Shalev AY. Peripheral blood mononuclear cell gene expression profiles identify emergent post-traumatic stress disorder among trauma survivors. Molecular psychiatry. 2005;10(5):500–13. doi: 10.1038/sj.mp.4001636. 425. [DOI] [PubMed] [Google Scholar]
- True WR, Rice J, Eisen SA, Heath AC, Goldberg J, Lyons MJ, Nowak J. A twin study of genetic and environmental contributions to liability for posttraumatic stress symptoms. Archives of general psychiatry. 1993;50(4):257–64. doi: 10.1001/archpsyc.1993.01820160019002. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Nossova N, Yager T, Tsuang MM, Guo SC, Shyu KG, Glatt SJ, Liew CC. Assessing the validity of blood-based gene expression profiles for the classification of schizophrenia and bipolar disorder: a preliminary report. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2005;133B(1):1–5. doi: 10.1002/ajmg.b.30161. [DOI] [PubMed] [Google Scholar]
- Tylee DS, Kawaguchi DM, Glatt SJ. On the outside looking in: A review and evaluation of the comparability of blood and brain “-omes”. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2013;162B(7):595–603. doi: 10.1002/ajmg.b.32150. [DOI] [PubMed] [Google Scholar]
- Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, Goldmann E, Galea S. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2010;107(20):9470–5. doi: 10.1073/pnas.0910794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zuiden M, Geuze E, Willemen HL, Vermetten E, Maas M, Amarouchi K, Kavelaars A, Heijnen CJ. Glucocorticoid receptor pathway components predict posttraumatic stress disorder symptom development: a prospective study. Biological psychiatry. 2012a;71(4):309–16. doi: 10.1016/j.biopsych.2011.10.026. [DOI] [PubMed] [Google Scholar]
- van Zuiden M, Heijnen CJ, Maas M, Amarouchi K, Vermetten E, Geuze E, Kavelaars A. Glucocorticoid sensitivity of leukocytes predicts PTSD, depressive and fatigue symptoms after military deployment: A prospective study. Psychoneuroendocrinology. 2012b;37(11):1822–36. doi: 10.1016/j.psyneuen.2012.03.018. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Molecular cell. 2008;31(4):449–61. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Cai G, Golier JA, Sarapas C, Galea S, Ising M, Rein T, Schmeidler J, Muller-Myhsok B, Holsboer F, et al. Gene expression patterns associated with posttraumatic stress disorder following exposure to the World Trade Center attacks. Biological psychiatry. 2009;66(7):708–11. doi: 10.1016/j.biopsych.2009.02.034. [DOI] [PubMed] [Google Scholar]
- Zieker J, Zieker D, Jatzko A, Dietzsch J, Nieselt K, Schmitt A, Bertsch T, Fassbender K, Spanagel R, Northoff H, et al. Differential gene expression in peripheral blood of patients suffering from post-traumatic stress disorder. Molecular psychiatry. 2007;12(2):116–8. doi: 10.1038/sj.mp.4001905. [DOI] [PubMed] [Google Scholar]


