Abstract
End-to-end fusion of critically shortened telomeres in higher eucaryotes is presumed to be mediated by nonhomologous end-joining (NHEJ). Here we describe two PCR-based methods to monitor telomere length and examine the fate of dysfunctional telomeres in Arabidopsis lacking the catalytic subunit of telomerase (TERT) and the DNA repair proteins Ku70 and Mre11. Primer extension telomere repeat amplification relies on the presence of an intact G-overhang, and thus measures functional telomere length. The minimum functional telomere length detected was 300–400 bp. PCR amplification and sequence analysis of chromosome fusion junctions revealed exonucleolytic digestion of dysfunctional ends prior to fusion. In ku70 tert mutants, there was a greater incidence of microhomology at the fusion junction than in tert mutants. In triple ku70 tert mre11 mutants, chromosome fusions were still detected, but microhomology at the junction was no longer favored. These data indicate that both Ku70 and Mre11 contribute to fusion of critically shortened telomeres in higher eucaryotes. Furthermore, Arabidopsis processes critically shortened telomeres as double-strand breaks, using a variety of end-joining pathways.
Keywords: Ku70, Mre11, NHEJ, telomerase, telomeres
Introduction
It is essential that cells have an effective mechanism to differentiate telomeres from DNA double-strand breaks. Proper telomere maintenance requires the functional elements of the telomere itself, as well as recognition by systems that would otherwise signal the presence of DNA damage. Telomeres have a distinctive nucleoprotein structure characterized by a G-rich repetitive DNA sequence ending in a 3′ single-strand overhang, which is thought to assume a complex secondary structure (t-loop) to sequester the chromosome terminus in a sheltered position (Wei and Price, 2003). Specific DNA-binding proteins recognize and associate with either double-strand or single-strand telomeric repeat sequences providing further protection for the terminus.
One function of the telomere is to prevent end-to-end chromosome fusion. Fusions can be induced through perturbation of the telomeric DNA sequence or depletion of telomere-associated proteins, such that the structure of the telomere is fundamentally altered (Ferreira et al, 2004). Telomerase activity is necessary for telomere maintenance. In addition, the appropriate complement of telomere-associated proteins must be present to assure that the telomere assumes a fully capped configuration, and is excluded from processing by DNA damage repair systems.
End-to-end chromosome fusion is a common outcome in cells with dysfunctional telomeres, suggesting that uncapped chromosome ends are often recognized and processed as DNA double-strand breaks (Ferreira et al, 2004). In both mammals and yeast, fusion of chromosomes with critically shortened telomeres requires components of the nonhomologous end-joining (NHEJ) pathway. Moreover, several DNA repair proteins, including Ku and the MRX complex (Mre11, Rad50 and Xrs2/Nbs1), localize to telomeres in wild-type cells, and are essential for recognition or maintenance of the telomere (Ferreira et al, 2004).
The precise role of DNA repair proteins at the telomere is unclear, and varies between different species. For example, loss of Ku function via disruption of the KU70 or KU80 genes leads to telomere tract extension in Arabidopsis, but chromosome fusion is not observed (Bundock et al, 2002; Riha et al, 2002; Gallego et al, 2003a). In contrast, telomere length is not dramatically altered in mammalian cells in the absence of Ku, but end-to-end chromosome fusions are formed, consistent with uncapping of the telomere (Bailey et al, 1999; Samper et al, 2000). Fusion of critically shortened telomeres proceeds efficiently without Ku70 in Arabidopsis (Riha and Shippen, 2003), but requires Ku function in mammalian cells (Espejel et al, 2002). Fusion of uncapped telomeres can also occur in the absence of conventional Ku-dependent NHEJ in yeast (Baumann and Cech, 2000; Mieczkowski et al, 2003). Together, these observations imply that dysfunctional telomeres can be joined by alternative NHEJ pathways. Examination of DNA double-strand break repair in yeast lacking Ku demonstrated a distinct pathway of microhomology-mediated end-joining (MMEJ) that requires the MRX complex (Ma et al, 2003). Although the biochemical properties of the mammalian Mre11 protein indicate that it has the potential to function in NHEJ (Paull and Gellert, 2000; D'Amours and Jackson, 2002), this hypothesis has not been tested in vivo, as null mutations involving Mre11 are lethal (Yamaguchi-Iwai et al, 1999).
Arabidopsis is emerging as an attractive higher eucaryotic model for telomere biology because it displays a remarkable tolerance to telomere dysfunction and genome instability. Progressive telomere shortening in telomerase mutants ultimately leads to sterility and massive genome instability. However, terminal generation plants are viable despite end-to-end fusion of up to half of the chromosomes (Riha et al, 2001; Riha and Shippen, 2003). Thus, Arabidopsis provides a rich source of material to examine the outcome of chromosome fusion at the molecular level. In addition, the DNA sequences immediately adjacent to the telomeric repeats in Arabidopsis are not highly repetitive, and regions of unique sequence are present near most chromosome ends. Hence, the Arabidopsis genome is well suited to the independent analysis of telomeres from various chromosome arms, and offers the potential to examine the fate of individual telomeres in different genetic settings.
Here we report the development of two PCR-based methods that exploit the unique subtelomeric DNA sequences at Arabidopsis telomeres to measure precisely the length of the telomeric tract of individual chromosomes and to amplify and characterize chromosome fusion junctions arising from telomere dysfunction. We show that when Ku is functional, chromosome fusion junctions formed between critically shortened telomeres display hallmarks of conventional NHEJ with evidence of exonucleolytic processing and frequent small insertions (Feldmann et al, 2000; Kirik et al, 2000). However, when critically shortened telomeres associate in the absence of Ku70 the architecture of the fusion joint is altered, and a more homology-driven mechanism mediates fusion. Remarkably, fusion of critically shortened telomeres still occurs in triple ku70 tert mre11 mutants, but the junctions display a significant decrease in microhomology. These data not only provide in vivo evidence that Mre11 contributes to MMEJ reactions in higher eucaryotes, but also illustrate the complexity of NHEJ pathways.
Results
Unique subtelomeric sequences in Arabidopsis
A search of the Arabidopsis genomic sequence database yielded terminal DNA sequence information for the eight non-rDNA bearing Arabidopsis chromosome arms (Richards et al, 1992; Kotani et al, 1999; Arabidopsis Genome Initiative, 2000). To confirm the sequence in wild-type plant stocks segregated from the tert mutant line (ecotype Columbia), subtelomeric sequences from 1R, 1L, 2R, 3L, 3R 4R, 5L and 5R (where R corresponds to South and L to North; Arabidopsis Genome Initiative, 2000) were amplified via PCR using a telomeric repeat primer in combination with a chromosome-specific primer. The PCR products were cloned and sequenced. Only minor deviation from the published sequence was noted, specifically involving chromosomes 2R and 4R. Although a subtelomeric primer was designed for chromosome 3R, we were unable to isolate the most distal subtelomeric sequence for that chromosome. Pairwise BLAST analysis showed limited regions of homology within the 2 kb of subtelomeric DNA immediately proximal to the start of each telomere repeat sequence, with the exception of chromosomes 1R and 4R, which share an extensive region of homology. No highly repetitive sequences were identified near the telomeric repeats (Supplementary Figure 1).
A novel PCR method to monitor individual telomere size
The presence of unique subtelomeric sequences in Arabidopsis allowed us to examine telomere length for individual chromosome ends. We devised a PCR-based technique called primer extension telomere repeat amplification (PETRA) that requires the G-overhang, a key structural element necessary for telomere function, and can accurately determine telomere length at multiple chromosome ends from a single plant.
The principle of PETRA is outlined in Figure 1A. An adaptor primer (PETRA-T) is hybridized to the 3′ G-rich overhang at the chromosome terminus. PETRA-T consists of 12 nucleotides complementary to the telomeric G-strand at its 3′ end, with a 5′ end that bears a unique sequence tag. Once annealed, the PETRA-T primer is extended by DNA PolI; the formation of PETRA products is dependent upon the action of DNA Pol1 (Figure 1C). We previously showed that this primer extension reaction requires the presence of an intact G-overhang (Riha et al, 2000). In the next step of PETRA, telomeres of specific chromosome arms are amplified by PCR using a unique subtelomeric primer and PETRA-A, a primer whose sequence is identical to the tag on the 5′ terminus of PETRA-T. PCR products are detected by Southern hybridization using a telomeric repeat probe.
Figure 1.
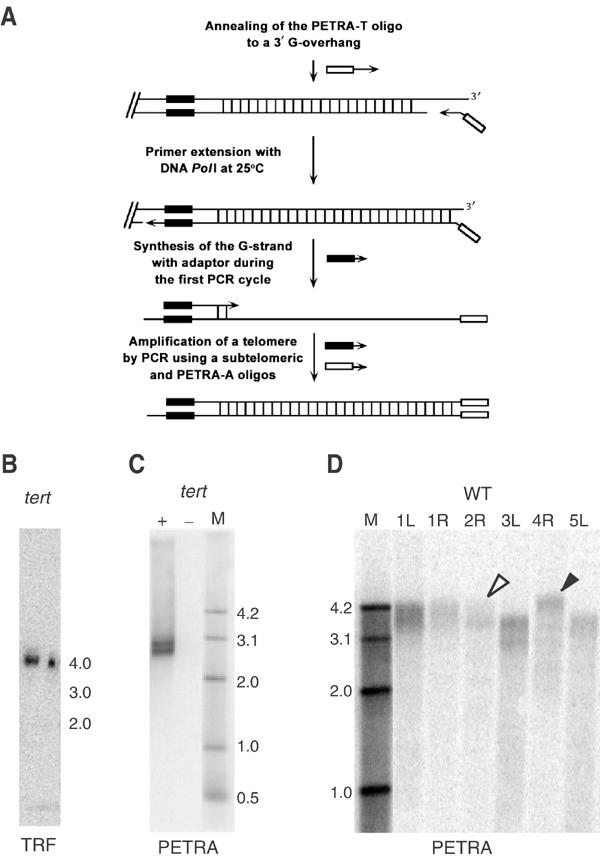
Measurement of telomere length by PETRA. (A) Diagram of the PETRA method. (B) TRF analysis for the right arm of chromosome 2 (2R) from a G4 tert mutant. The calculated telomere length is ∼2.2 kb. (C) PETRA for 2R using the same DNA sample carried out in the presence (+) or absence (−) of DNA PolI. A doublet of bands is resolved, yielding a calculated telomere length of 2.2 and 2.4 kb. (D) PETRA for different chromosome arms using a wild-type DNA template. A single diffuse band is detected for most chromosomes; calculated telomere length ranges from 3030 to 4300 bp. Open arrowhead, shortest telomere; closed arrowhead, longest telomere.
We tested the PETRA assay by evaluating the 2R telomere in a fourth-generation (G4) tert mutant. Terminal restriction fragment (TRF) analysis of genomic DNA digested with HindIII detected a single 4 kb band (Figure 1B). Since the HindIII site is located 1.8 kb proximal to the telomeric repeats, the size of the telomere at 2R in this plant is approximately 2.2 kb. PETRA reactions were performed with the same DNA, using a 2R primer that binds 500 bp upstream from the telomeric repeats. A doublet of bands (2.7 and 2.9 kb) was resolved (Figure 1C) indicating that the 2R telomere in this plant is comprised of two subpopulations of 2.2 and 2.4 kb in length. PETRA analysis of telomere length for other individual plants typically revealed only one or two bands for each chromosome arm, including 2R. Sequence analysis confirmed that PETRA products represent the predicted subtelomeric and telomeric sequences for the targeted chromosome (data not shown). These findings correlate well with previous TRF analysis of tert mutants using subtelomeric probes (Riha et al, 2001), and demonstrate that PETRA provides a sensitive and accurate measure of telomere length on individual chromosome arms.
Telomere size in telomerase-deficient mutants
In mammalian cells lacking telomerase, the shortest telomere is the one most often involved in fusions (Hemann et al, 2001b), implying the existence of a minimum functional telomere length below which the telomere is uncapped and available to participate in fusion events. To examine the relationship between telomere length, loss of telomere function and the formation of chromosome fusions in Arabidopsis, we determined telomere length for individual chromosome arms in different generations of tert mutants (Table I).
Table 1.
Summary of PETRA results for tert and ku70 tert mutants
| P | 1L | 1R | 2R | 3L | 4R | 5L | 5R | Ave | Range | |
|---|---|---|---|---|---|---|---|---|---|---|
| tert | ||||||||||
| G5, line 69 | ||||||||||
| 1 | WT | 1530 | 1280 | 1600 2150 | 1490 | 1250 1530 | 1640 | 880 | 1480 | 1270 |
| 2 | WT | 1470 | 1260 | 2200 | 1420 | 1250 | ND | 890 | 1415 | 1310 |
| 3 | WT | 1490 1230 | 1330 | 1940 | 1220 | 1020 | 1600 1380 | 580 | 1310 | 1360 |
| 4 | WT | 1460 | 1180 | 2280 2090 1840 | 1520 | 1190 | 1770 | 990 | 1590 | 1290 |
| G9, line 69 | ||||||||||
| 1 | II-T | 920 | ND | 410 | 510 | ND | 810 580 | 500 | 620 | 510 |
| 2 | II-T | 600 500 | 360 | 300 | 440 | ND | 780 | 540 | 500 | 480 |
| 3 | II-T | 950 | ND | 520 | 620 | ND | 790 | 890 320 | 680 | 630 |
| 4 | II-T | 890 | 390 | 520 | 730 | ND | 930 | ND | 690 | 540 |
| 5 | II-T | 660 | 300 | 310 | 440 | ND | 700 | 610 | 500 | 400 |
| 6 | II-T | 900 | ND | 350 | 460 | ND | 460 | 710 550 | 570 | 550 |
| 7 | II-T | 1040 | 460 | 1150 | 750 | ND | 880 | 710 | 830 | 690 |
| Pool | T | 710 530 | 360 | 1140 | 590 | ND | 880 | 760 260 | 650 | 880 |
| ku70 tert | ||||||||||
| G4, line 52 | ||||||||||
| 1 | I | ND | 1160 700 | ND | 910 | 1600 | ND | 1100 | 1090 | 900 |
| 2 | I | ND | 470 | 1100 | 970 | 1620 | ND | 630 | 960 | 1150 |
| 3 | I | ND | 640 | 1180 | 980 800 | 2670 2290 2020 1460 | ND | 1010 | 1450 | 2030 |
| 4 | I | ND | 1400 | 2570 1470 | 2010 1550 | 3200 | ND | 1610 820 | 1830 | 2380 |
| 5 | I | ND | 1310 | 2210 1410 | 1660 | 3070 | ND | 1390 | 1840 | 1760 |
| 6 | I | ND | 960 | 1100 1340 | 720 | 1900 | ND | 770 | 1130 | 1180 |
| 7 | I | ND | 810 | 840 1150 | 670 | 1530 | ND | 720 | 950 | 860 |
| 8 | T | ND | 730 | 1030 | 740 | 2280 2050 | ND | 1170 | 1330 | 1550 |
| 9 | T | 640 420 | 360 | 1040 | 1390 970 | ND | ND | 440 | 750 | 1030 |
| Pool |
T |
ND |
740 540 480 |
1080 |
1030 700 |
1690 1560 |
ND |
650 590 |
910 |
1210 |
| Values in bold indicate chromosomes for which fusion PCR products were cloned. Phenotypes (P) for the mutants are indicated. ND=not determined. | ||||||||||
PETRA detected a single diffuse band for most chromosome ends in wild type (Figure 1D) and a much sharper band in telomerase-deficient mutants (Figure 2A and B). Telomere length varied considerably between the different chromosome ends and no single telomere was consistently found to be the shortest or the longest in different generations (Table I). As expected, bulk telomere length was significantly reduced in G5 tert mutants (Figure 2A) relative to wild type and diminished further by G9 (P=0.0003) (Figure 2B). However, the size distribution of the telomere tracts (range between longest and shortest telomere) in an individual plant was dramatically compressed in G9 mutants relative to G5 (Figure 2C). On average, G5 telomeres covered a 1310 bp range (n=4), while in G9 telomeres occupied a much narrower distribution of 630 bp (n=7).
Figure 2.

Telomere length analysis for tert and ku70 tert mutants. (A, B) PETRA results for single tert plants (line 69) at G5 and G9, respectively. Telomere length in G5 ranges from 880 to 2150 bp, and in G9 from 300 to 700 bp. The shortest (open arrowhead) and longest (closed arrowhead) telomeres are indicated. (C) Bar graph illustrates the range of telomere length measured for individual G9 and G5 tert plants (line 69). See Table I for a summary of telomere length measurements. (D) PETRA results for a single ku70 tert plant at G4 (line 52). The range of telomere length is from 360 to 1390 bp.
In tert plants, cytogenetic evidence for chromosome fusion, manifest by the presence of anaphase bridges, correlates with the onset of developmental and growth defects that are either mild (Type I) or moderate (Type II). By G9, most plants exhibit abundant anaphase bridges with severe growth defects and sterility, hence the designation terminal (T) (Riha et al, 2001). To determine the size of the shortest telomeres in Arabidopsis that still retain a G-overhang, PETRA analysis was performed for seven individual G9 tert mutants displaying either a Type II or T phenotype (Table I). The shortest telomeres detected were 300–460 bp (mean=360 bp). The minimum telomere length found for a pool of terminal G9 plants was even shorter at 260 bp.
Due to an accelerated rate of telomere shortening, anaphase bridges are first detected cytogenetically in ku70 tert mutants (line 52) in G2, with many plants reaching the terminal phenotype by G4 (Riha and Shippen, 2003). To determine the minimal telomere length in this setting, we used PETRA to evaluate two G4 ku70 tert mutants that displayed a T phenotype (Figure 2D; Table I). In these plants, the shortest telomere measured 360 and 730 bp, respectively. The minimal telomere measured for a pool of terminal ku70 tert mutants was 480 bp (n=4), a value slightly higher than for tert mutants.
Molecular analysis of chromosome fusion junctions in tert mutants
To further examine the relationship between telomere length and loss of function, we developed a PCR assay to detect chromosome fusions that exploits the existence of chromosome-specific PCR primers to amplify fusion junctions between chromosome ends (Figure 3A). The primers used for fusion PCR were positioned approximately 500 bp from the telomere tract and were directed toward the chromosome terminus. PCR products were detected by Southern blot, using either a telomeric repeat or a specific subtelomeric probe. PCR products were abundant in reactions carried out with a G9 tert DNA template (Figure 3B), and typically displayed a heterogeneous pattern of hybridization, with products of varied size that sometimes appeared as a broad smear. PCR products could be obtained for all chromosome arms surveyed, although some specific primer combinations gave negative results. PCR products were occasionally generated in wild-type DNA samples (Figure 3B), but sequence analysis indicated that these rare products resulted from fortuitous amplification of interstitial sequences (data not shown).
Figure 3.
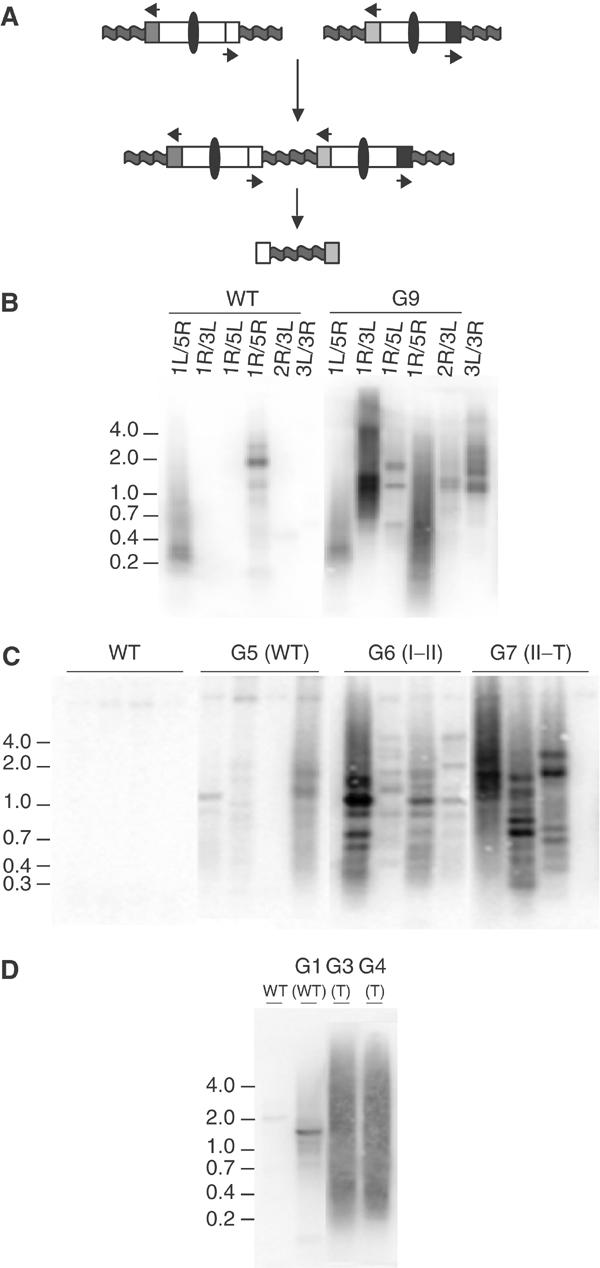
PCR amplification of chromosome fusion junctions. (A) PCR strategy for amplification of fusion junctions. Arrows denote unique subtelomeric primers directed toward the chromosome terminus. PCR amplification occurs only when two subtelomeric regions are joined end-to-end (oval: centromere; wavy line: telomere). (B–D) Southern analysis of fusion PCR products using a telomeric probe. Panel B shows fusion PCR results for different subtelomeric primer combinations, using wild-type template DNA and DNA from a pool of terminal G9 tert plants. Panels C and D show fusion PCR results using the primers for chromosomes 1R and 3L, with template DNA prepared from different generations of tert (line 20) (panel C) and ku70 tert (line 52) (panel D) mutants. Results for a single plant are shown in each lane. Plant phenotypes are indicated in parentheses.
To test whether the generation of fusion PCR products paralleled the onset of cytogenetic defects, DNA was prepared from wild-type and successive generations of tert mutant plants (line 20; G4–G7), and PCR was performed using primers specific for chromosomes 1R and 3L. PCR products were not obtained in wild-type or G4 DNA samples (Figure 3C; data not shown). In G5 faint bands were detected, and in G6 and G7 products of greater complexity and abundance were observed (Figure 3C). A similar result was obtained with DNA from ku70 tert mutants (line 52) (Figure 3D). A faint band was detected in G1 plants, products of varied size were evident in G2 (data not shown), and abundant products resulting in a broad smear of hybridization were obtained using DNA from G3 and G4. These findings correlate well with the onset of cytogenetic abnormalities (Riha et al, 2001; Riha and Shippen, 2003), and indicate that our PCR approach is a sensitive method for detecting chromosome fusions.
To further characterize the structure of the fusion junctions, and to correlate these structures with telomere length, we cloned fusion PCR products generated using tert and ku70 tert DNA samples that had previously been subjected to PETRA analysis. Three different primer combinations (1R–3L, 3R–3L, 2R–3L) were used to amplify chromosome fusion junctions. In all, 39 clones from tert mutants (Type II or T phenotype) and 38 from ku70 tert mutants (Type I or T phenotype) were evaluated. Southern blot analysis showed that the majority of the clones contained telomeric DNA and sequences from both chromosome arms, consistent with fusion of heterologous chromosomes (data not shown).
DNA sequence analysis revealed that 74 of the clones had a structure consistent with end-to-end chromosome fusion, with two chromosome arms present in a head-to-head orientation (Figure 4). Three basic configurations were observed: telomere–telomere, telomere–subtelomere and subtelomere–subtelomere. Telomere–telomere fusions made up only 11% of the clones for tert, but at 43% represented a significantly larger proportion for ku70 tert (P=0.002). Similar to what has been described for telomere–telomere fusions in yeast (McEachern et al, 2000; Mieczkowski et al, 2003), we were unable to sequence completely through the fusion junction in these clones. Telomere–subtelomere fusions were the most abundant isolates obtained from both tert (78%) and ku70 tert (51%) mutants. In contrast, subtelomere–subtelomere fusions represented only 11% of the total for tert and 5% for ku70 tert. In three additional clones, a more complex fusion junction was observed (Figure 4). In this example, two copies of 1R were fused in a telomere–subtelomere configuration; the copy of 1R with an intact terminal sequence was truncated 69 bp proximal to the telomere, and the 3L telomere was fused at that point.
Figure 4.
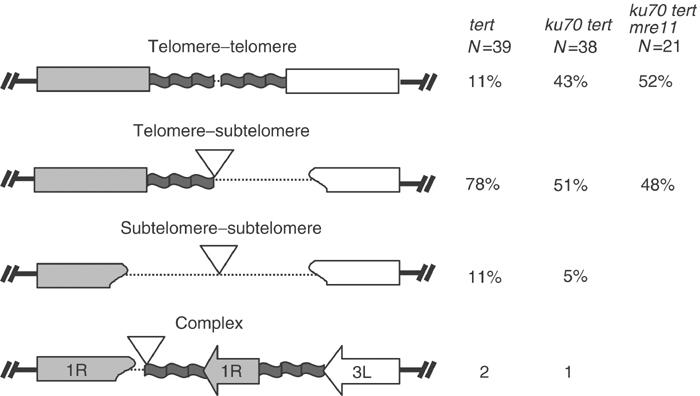
Structure of chromosome fusion junctions isolated from tert, ku70 tert and ku70 tert mre11 mutants. Only heterologous chromosome fusion events are depicted. Extent of sequence deletion varied. Occasionally, short regions of new sequence were inserted at the fusion junction (Supplementary Tables I, II andIII). Dotted lines indicate truncation of telomeric and/or subtelomeric DNA. Inverted triangles denote inserted sequences. The percentage showing a particular fusion structure for each genotype is indicated. Three examples of complex fusion junctions were isolated, but not included in the comparative analysis of junction sequences.
Telomeric DNA captured in the fusion junctions must arise from an uncapped telomere. Since PETRA suggested that the shortest functional telomeres were approximately 300–400 bp, it was of interest to determine how much telomeric DNA remained at the fusion junctions. The total length of the telomere tract for telomere–telomere fusions was determined by restriction mapping (Supplementary Tables I and II), but because we could not sequence completely across the fusion junction in these clones, the relative contribution of telomeric DNA from each chromosome end is unknown. Telomere tracts could be precisely measured in the telomere–subtelomere fusions by DNA sequence analysis. For the clones obtained from G9 tert mutants, telomeric DNA ranged from 122 to 457 bp (mean=265 bp) (Figure 5A; Supplementary Table I). A similar value was obtained for clones derived from G4 ku70 tert mutants, where telomere tracts ranged from 52 to 400 bp (mean=241 bp; Supplementary Table II). While the lengths of the telomere tracts in the fusion junctions roughly correlated with the shortest functional telomeres detected by PETRA, it is striking that these domains were often fused directly to subtelomeric DNA. Hence, dysfunctional telomeres generated by a telomerase deficiency often appear to be substrates for exonuclease attack prior to end-joining.
Figure 5.
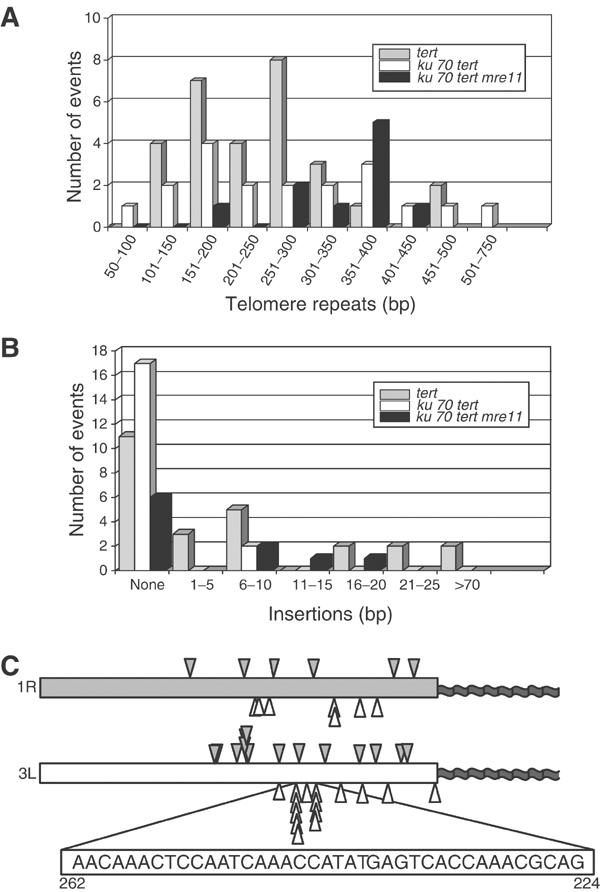
Chromosome fusion junction sequences from tert, ku70 tert and ku70 tert mre11 mutants. (A) Histogram showing the amount of telomeric DNA at the fusion junction in subtelomere–telomere clones: tert (gray bars), ku70 tert (white bars) and ku70 tert mre11 (black bars). (B) Histogram showing the size and abundance of DNA insertion sequences at the fusion junctions. (C) Distribution of the point of fusion within the deleted subtelomere for telomere–subtelomere fusion events involving 1R and 3L in tert and ku70 tert mutants. Closed arrowheads, tert fusions; open arrowheads, ku70 tert fusions. A 39 bp segment of subtelomeric sequence from 3L is expanded; this sequence was a preferred substrate for end-joining in the ku70 tert background.
Structural differences at the fusion junctions of tert and ku70 tert mutants
Sequence analysis revealed intriguing differences at fusion junctions formed in the presence or absence of Ku. In all, 54% of the tert fusion junctions evaluated contained an insertion of a short region (1–145 bp) of filler DNA (Figure 5B; Supplementary Table I). In previous studies on NHEJ, small insertions arise from both genomic and extrachromosomal DNA (Kirik et al, 2000). The origin of insertion sequences in our clones could not be determined. Significantly fewer of the ku70 tert clones harbored insertion sequences (10%; P=0.001) and these were much shorter (6–9 bp) (Figure 5B; Supplementary Table II). Overall, most insertions were less than 20 bp, and all of the longer insertions were found in the tert fusion junctions.
Deletion of sequences at the fusion junction was common for both tert and ku70 tert mutants. For tert clones, the extent of erosion of subtelomeric DNA sequences varied substantially, spanning 44–459 bp (Supplementary Table I). The fusion points were distributed fairly evenly throughout the region of the subtelomeric DNA, although a 6 bp region of chromosome 3L exhibited a slight preference for the fusion site (Figure 5C). Erosion of subtelomeric sequences was also variable in clones derived from ku70 tert mutants, extending from 3 bp to greater than 365 bp (Figure 5C; Supplementary Table II). However, in contrast to tert mutants, ku70 tert mutants displayed a stronger bias in the choice of the fusion substrate. For 10 of the 15 fusion junctions involving the subtelomeric region of chromosome 3L, joining occurred within a 39 bp region located 224–262 nucleotides from the beginning of the telomere tract (Figure 5C). This region has a similar sequence and C+A content to that of telomeric DNA, suggesting that in the absence of Ku, homology between the ends plays a more significant role in the end-joining reaction. In support of this notion, nearly all of the ku70 tert telomere–subtelomere fusions (83%) included a small region of perfect overlapping homology at the fusion junction, extending for up to 12 bp (Supplementary Table II). In contrast, only 39% of the tert telomere–subtelomere clones displayed overlapping homology (Supplementary Table I), indicating a significant increase in microhomology (P=0.006). This analysis confirms that Arabidopsis has the capacity to use both Ku-dependent and Ku-independent mechanisms for fusion of critically shortened telomeres. The data further indicate that the Ku-independent mechanism has a greater reliance on DNA homology, with a concomitant decrease in the frequency of insertions at the fusion junction.
Role of Mre11 in fusion of critically shortened telomeres
In yeast, microhomology-mediated end-joining (MMEJ) is a Ku-independent mechanism for repair of DNA double-strand breaks that requires Mre11 (Ma et al, 2003). Therefore, we asked whether Mre11 contributes to Ku-independent fusion of critically shortened telomeres. An Mre11 homolog has been characterized in Arabidopsis (Bundock and Hooykaas, 2002). As predicted, mutations in this gene (mre11-1 and mre11-2) result in increased sensitivity to DNA damage. In addition, telomeres were elongated in the mutants, suggesting that Mre11 contributes to telomere length maintenance. The mre11-1 allele resulted in severe developmental defects and sterility.
Our study focused on the DNA repair function of Mre11 in joining critically shortened telomeres. We used an mre11 allele, mre11-3, which harbors a T-DNA insertion within the conserved phosphoesterase domain IV (Puizina et al, in press). mre11-3 mutants showed vegetative growth defects and sterility, consistent with the phenotypes described for the mre11-1 mutant (Bundock and Hooykaas, 2002). However, in contrast to the previous study, we did not detect telomere elongation in the mre11-3 mutant (data not shown). A detailed description of the mre11-3 mutation, including an assessment of its impact on meiosis and genome stability, will be presented elsewhere (Puizina et al, submitted).
To determine whether Mre11 plays a role in MMEJ of critically shortened telomeres in ku70 tert mutants, we generated ku70 tert mre11 triple mutants by genetic crossing (Figure 6A). Because disruption of Mre11 results in complete sterility, sustained propagation of a triple ku70 tert mre11 mutant was impossible. However, since end-to-end chromosome fusions can be detected as early as G1 of ku70 tert double mutants, we reasoned that it would be possible to assess the impact of Mre11 dysfunction on chromosome fusion within a single generation of plant growth. Triple mutant plants (ku70 tert mre11) and ku70 tert siblings heterozygous for the insertion in Mre11 (ku70 tert Mre11+/−) were obtained by self-pollination of Ku70+/−Tert+/− Mre11+/− plants. These plants were designated as G1, representing the first generation of growth without active telomerase. Self-pollination of G1 ku70 tert Mre11+/− plants yielded G2 triple mutants (ku70 tert mre11), representing two generations of growth without active telomerase. TRF analysis showed that bulk telomere length in ku70 tert mre11 G2 plants was comparable to that of sibling plants that retained Mre11 function (Figure 6B and data not shown). Thus, Mre11 dysfunction does not appear to affect significantly telomere length in a ku70 tert background.
Figure 6.
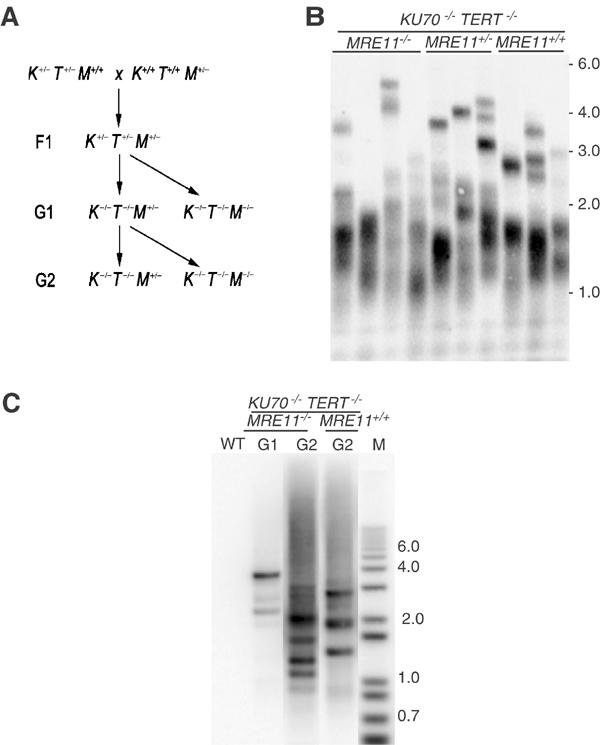
Telomere length and chromosome fusions in ku70 tert mre11 mutants. (A) Triple ku70 tert mre11 mutants were obtained by crossing a plant heterozygous for T-DNA insertions in the TERT and KU70 genes with a plant heterozygous for a T-DNA insertion in MRE11. F1 plants heterozygous for all three mutations were self-pollinated and F2 plants homozygous for the tert and ku70 mutations, and either heterozygous or homozygous for the insertion at MRE11 were identified by PCR. This population, designated G1, represents the first generation of plants without active telomerase. Second-generation (G2) tert ku70 mre11 triple mutants were derived from self-pollination of a single G1 tert ku70 MRE11+/− plant. (B) TRF analysis in G2 tert ku70 mre11 triple mutants was performed using a telomere probe. No consistent difference in telomere size was detected in tert ku70 mre11 triple mutants compared to Mre11-proficient siblings. (C) Fusion PCR products are present in ku70 tert mre11 G1, and are more abundant in G2. Fusion PCR products were generated using 1R and 3L primers.
Fusion PCR was performed using DNA prepared from G1 and G2 ku70 tert mre11 plants to investigate the contribution of Mre11 to the fusion of short telomeres. PCR products consistent with chromosome fusion were obtained from DNA samples prepared from G1 plants and were more abundant in G2 plants (Figure 6C). The increased abundance of PCR products in G2 demonstrates that the frequency of chromosome end-to-end fusions correlates with telomere shortening and not with Mre11 inactivation. Sequence analysis of cloned PCR products supports the idea that Mre11 contributes to the formation of fusion junctions. Although the frequency of telomere–telomere and telomere–subtelomere fusions was similar in ku70 tert and ku70 tert mre11 mutants (Figure 4), sequence analysis revealed striking differences in the structure of telomere–subtelomere fusion junctions. Only 50% of the ku70 tert mre11 clones displayed short regions of microhomology (1–4 bp) compared to 81% in ku70 tert mutants (Supplementary Tables II and III). Moreover, small insertions were observed in 10% of the ku70 tert clones (6–18 bp), while 40% of the ku70 tert mre11 clones harbored insertions (Supplementary Tables II and III). Overall, the fusion junctions obtained from ku70 tert mre11 mutants closely resembled junctions isolated from tert single mutants. These findings provide strong evidence that Mre11 contributes to the MMEJ reaction at critically shortened telomeres. Moreover, the detection of chromosome end-to-end fusions in plants lacking both Ku70 and Mre11 demonstrates that Arabidopsis can process dysfunctional telomeres by a variety of distinct NHEJ mechanisms, only a subset of which have been previously characterized.
Discussion
The primary function of the telomere is to define and protect the ends of chromosomes, allowing those termini to be distinguished from DNA double-strand breaks. In this study, we used two different PCR strategies to follow the fate of individual chromosome ends in Arabidopsis mutants experiencing telomere dysfunction through loss of telomerase. Arabidopsis is particularly amenable to such studies since most of its chromosome ends contain unique subtelomeric sequences. Recently, a method comparable to our PETRA approach, termed STELA, was employed to measure telomere length on the short arms of the human sex chromosomes (Baird et al, 2003). However, application of STELA to other telomeres awaits the identification of additional unique subtelomeric sequences. PETRA not only allows us to evaluate simultaneously telomere length for multiple chromosome ends, but because it relies on the presence of an intact G-overhang, it is designed to detect the shortest functional telomeres in a population. PCR amplification of fusion junctions complements PETRA, since telomere fusion is a definitive indicator of loss of function.
Analysis of critically shortened telomeres
While PETRA confirmed that telomere length decreased progressively through successive generations of telomerase mutants, we unexpectedly found that the telomere length distribution narrows sharply in later generations. It is conceivable that shorter telomeres that have lost their G-overhang exist in the population but are rapidly recruited into fusions. Removal of the G-overhang is necessary for chromosome fusion in human cells with dysfunctional TRF2 (Zhu et al, 2003). Alternatively, there may be selective pressure applied to germline cells, such that only progeny harboring telomeres within the functional range are produced. A telomere surveillance mechanism has been reported for mice wherein cells bearing dysfunctional telomeres are eliminated from the germ cell precursor pool by apoptosis (Hemann et al, 2001a).
The shortest telomeres detected by PETRA are found in plants displaying the terminal phenotype. This value is ∼300 bp in tert mutants, and is slightly longer in terminal ku70 tert mutants. By contrast, the shortest telomeres detected using a similar PCR technique in telomerase-deficient yeast were only ∼50 bp long (Forstemann et al, 2000), six times smaller than the shortest telomeres in Arabidopsis. This difference may reflect alternative modes of telomere capping in these species. Whereas telomere protection in yeast is mediated through the binding of Cdc13 protein to the G-overhang (Nugent et al, 1996), chromosome termini in plants are apparently sheltered in t-loops (Cesare et al, 2003). The length of the shortest telomeres in Arabidopsis may correspond to the minimal size required for efficient t-loop formation in vivo.
Nucleolytic processing of dysfunctional telomeres
Little is known about the molecular mechanisms that process exposed chromosome termini and their role in triggering chromosomal aberrations. The initial stages of double-strand break repair typically involve nucleolytic degradation to create suitable substrates for downstream reactions (Lieber et al, 2003). Studies in yeast reveal that deprotected telomeres are also subject to exonucleolytic resection (Baumann and Cech, 2001; Hackett and Greider, 2003). In Ku70-deficient budding yeast, chromosome ends are degradated by exonuclease 1 (Maringele and Lydall, 2002). Furthermore, in telomerase-deficient yeast, chromosome fusion is preceded by extensive telomere shortening (Chan and Blackburn, 2003; Hackett and Greider, 2003). Although relatively little information is available concerning the processing of critically shortened telomeres in higher eucaryotes, sequence analysis of a small number of chromosome fusion junctions formed in late-generation telomerase-deficient mice revealed a complete loss of telomeric DNA (Hemann et al, 2001b).
Our results in Arabidopsis support the notion that exonucleolytic processing of critically shortened telomeres occurs prior to fusion. Among the 98 clones we sequenced, the large majority involved telomere–subtelomere fusions. The extent of nucleotide deletion in these junctions is consistent with what occurs prior to repair of endonuclease-induced double-strand breaks in plants (Kirik et al, 2000). We considered the possibility that these clones represent secondary fusion events resulting from initial telomere–telomere fusion followed by one round of the BFB cycle. Since our PCR primers are targeted to sequences very close to the telomere tract, breakage of the dicentric chromosome in the next mitosis would have occurred in the immediate vicinity of the original fusion point, followed by a second round of fusion with another nonfunctional telomere. While three of our clones did display a complex structure consistent with such a secondary fusion (Figure 4), the majority appear to reflect a primary fusion event.
We suspect that telomere–telomere and sister chromatid fusions are under-represented in our study due to the inherent difficulty in generating PCR products across large palindromic regions. Indeed, in situ hybridization studies demonstrate that fusion of homologous chromosomes occurs frequently in late-generation telomerase-deficient Arabidopsis (Siroky et al, 2003), yet we identified only five examples of such junctions in our PCR survey. It is striking that the incidence of telomere–telomere fusions was significantly higher in ku70 tert double mutants than in tert singles. Analysis of telomere length by PETRA revealed that the telomeres of chromosomes 1R and 3L, which form frequent end-to-end fusions, are slightly longer in terminal ku70 tert plants than in tert single mutants. These findings, coupled with the observation that telomere loss is markedly accelerated in ku70 tert mutants (Riha and Shippen, 2003), argue that Ku70 contributes to telomere protection in Arabidopsis. Unlike mammals (Bailey et al, 1999; Samper et al, 2000), this capping function is revealed only in the context of a telomerase deficiency (Riha et al, 2002).
Role of NHEJ in fusing critically shortened telomeres in Arabidopsis
Our data strongly support the contention that critically shortened telomeres are processed as double-strand breaks and are subject to NHEJ reactions. Both Ku and Lig4 have been implicated in the fusion of aberrant telomeres in yeast and mammals (Ferreira and Cooper, 2001; Espejel et al, 2002; Smogorzewska et al, 2002; Chan and Blackburn, 2003; Mieczkowski et al, 2003). However, end-to-end chromosome fusions can also occur by other pathways, as telomere associations can form in the absence of Ku in mammalian cells (Bailey et al, 1999), fission yeast (Baumann and Cech, 2000) and Arabidopsis (Riha and Shippen, 2003). We investigated the mechanism of chromosome fusion in Arabidopsis by comparing the nucleotide sequences of fusion junctions formed in tert and ku70 tert mutants. In tert mutants, the fusion junctions are consistent with conventional Ku-dependent NHEJ, as they harbor small insertions, deletions and microhomology. In the absence of Ku, chromosome fusion appears to proceed via a more homology-driven process. We noted a marked increase in the incidence of overlapping microhomology, with a strong bias for joining of canonical telomere tracts to telomere-related sequences in the subtelomeric region. This finding supports the view that Ku acts as an alignment factor that holds DNA ends in apposition to facilitate accurate repair (Feldmann et al, 2000). Ku binding inhibits exonucleolytic attack (Tomita et al, 2003) and in its absence sequence homology between the DNA ends plays a more prominent role in proper alignment and efficient synapsis (Ma et al, 2003). The shift in the structure of the chromosome junctions formed in tert versus ku70 tert mutants demonstrates that Ku contributes to the fusion of dysfunctional telomeres in Arabidopsis. However, in its absence, an alternative and efficient NHEJ pathway operates.
The outstanding feature of this Ku-independent NHEJ pathway is increased microhomology at the fusion junction. Since studies in yeast demonstrated a Ku-independent MMEJ pathway that requires the MRX complex, we asked whether Mre11 contributes to the fusion of critically shortened telomeres in a higher eucaryote by creating a triple mutant deficient in Tert, Ku70 and Mre11. Remarkably, Arabidopsis retained the capacity to mediate chromosome fusions in this setting, but microhomology at the junction was reduced relative to that seen in double mutants lacking Ku70 and Tert.
These findings demonstrate an in vivo role for Mre11 in NHEJ in higher eucaryotes. The discovery that inactivation of both Ku-dependent NHEJ and Mre11-dependent MMEJ does not abolish end-to-end chromosome fusions in plants with critically shortened telomeres is unexpected, as nearly all end-joining activities in yeast are attributed to these two pathways (Ma et al, 2003). We conclude that plants possess at least three genetically distinct end-joining mechanisms that can efficiently substitute for each other and may directly compete in DNA repair. The robust and redundant nature of end-joining pathways is further illustrated by the capacity of Arabidopsis ku70, ku80 and lig4 mutants for T-DNA integration (Friesner and Britt, 2003; Gallego et al, 2003b; van Attikum et al, 2003). Since higher eucaryotes rely primarily on NHEJ to repair double-strand breaks, redundant end-joining activities may have evolved to ensure genome stability.
Materials and methods
Plant growth and DNA preparation
The generation of Arabidopsis thaliana tert and ku70 tert lines was described previously (Riha et al, 2001; Riha and Shippen, 2003a). Plants with a T-DNA insertion in the MRE11 gene were obtained from the Salk collection (line 054418) (Alonso et al, 2003). Triple tert ku70 mre11 lines were created as shown in Figure 6A. A. thaliana plants were grown as described (Riha et al, 2002). DNA was extracted from whole plants 4–5 weeks after germination as previously described (Riha et al, 2002). DNA concentration was determined by agarose gel electrophoresis, using lambda DNA digested with HindIII as a standard.
Subtelomeric DNA sequences and chromosome-specific PCR primers
Terminal DNA sequences for eight Arabidopsis chromosome arms were identified in GenBank (1R, AC074299; 1L, AC007323 and AC074298; 2R, AC006072; 3L, AC067753; 3R, AL732522; 4R, AL035708 and Z12169; 5L, AB033277; 5R, AB033278). Unique subtelomeric primers directed 5′ to 3′ toward the telomeric repeat were designed for each, as indicated below.
Determination of telomere length
For PETRA, primer extension was performed using a primer bound to the telomeric G-overhang (PETRA-T 5′-CTCTAGACTGTGAGACTTGGACTACCCTAAACCCT-3′), followed by PCR amplification using a chromosome-specific subtelomeric primer (1R 5′-CTATTGCCAGAACCTTGATATTCAT-3′; 1L 5′-AGGACCATCCCATATCATTGAGAGA-3′; 2R 5′-CAACATGGCCCATTTAAGATTGAACGGG-3′; 3R 5′-CTGTTCTTGGAGCAAGTGACTGTGA-3′; 3L 5′-CATAATTCTCACAGCAGCACCGTAGA-3′; 4R 5′-TGGGTGATTGTCATGCTACATGGTA-3′; 5R 5′-CAGGACGTGTGAAACAGAAACTACA-3′; 5L 5′-AGGTAGAGTGAACCTAACACTTGGA-3′) and second primer (PETRA-A 5′-CTCTAGACTGTGAGACTTGGACTAC-3′) that recognizes sequence complementary to the 5′ non-telomeric sequence present on PETRA-T. Primer extension reactions (50 μl) included 1 × DNA PolI buffer (Promega), 250 μM dNTPs, 0.2 μM PETRA-T primer, 500 ng genomic DNA and 9 U DNA PolI (Promega). The reaction was incubated at 16°C for 1 h, and then overnight at 25°C. The next day, DNA was recovered by ethanol precipitation and suspended in 20 μl of water. Each chromosome-specific PCR reaction (50 μl) included 1 × ExTaq buffer (TaKaRa), 200 μM dNTPs, 0.4 μM PETRA-A primer, 0.4 μM subtelomeric primer, 1/10 of the DNA recovered from the primer extension and 2 U ExTaq (TaKaRa). Samples were incubated at 96°C for 5 min, followed by 20–25 cycles of 94°C for 15 s, 60°C for 30 s and 72°C for 2 min, with a final incubation at 72°C for 10 min. A 15 μl aliquot of the PCR products was separated by electrophoresis through a 0.8–1% agarose gel, and transferred to a nylon membrane. Membranes were probed with a 32P end-labeled telomeric oligonucleotide (T3AG3)4. Hybridization was performed at 55°C overnight, in a buffer consisting of 0.25 M sodium phosphate buffer (pH 7.5), 7% SDS and 1 mg/ml BSA. Filters were washed in 2 × SSC/0.1% SDS and 0.2 × SSC/0.1% SDS, twice each for 10 min at 55°C. Hybridization signals were detected using a STORM PhosphorImager (Molecular Dynamics) and the data were analyzed using IMAGEQUANT software (Molecular Dynamics). TRF analysis was performed as described (Riha et al, 2001).
PCR amplification, cloning and sequence analysis of fusion junctions
Chromosome fusion junctions were amplified by PCR using subtelomeric primers in various pairwise combinations. Primer sequences were as indicated above, with the following exceptions: 1L 5′-ACAAGGATAGAAATAGAGCATCGTC-3′; 3L 5′-AGACGAGGAGACTAGGAACG-3′; 3R 5′-GTATGGATGCCGGGAAAGTTGCAGACAA-3′; 5L 5′-CGACAACGAC GACGAATGACAC-3′; 5R 5′-TCGGTTGTCGTCTTCAAG-3′. PCR reactions (20 μl) contained 1 × ExTaq reaction buffer (TaKaRa), 125 μM dNTPs, 0.5 μM each primer, 0.5 U ExTaq (TaKaRa) and 100 ng genomic DNA. Samples were incubated at 96°C for 2 min, followed by 34 cycles of 96°C for 30 s, 55°C for 1 min and 72°C for 1 min, with a final incubation at 72°C for 5 min. A 10 μl aliquot of the PCR products was separated by electrophoresis through a 1% agarose gel, and transferred to a nylon membrane. Membranes were hybridized with a 32P end-labeled (T3AG3)4 or a specific subtelomeric oligonucleotide located distal to the primer used for PCR (3L 5′-CATAATTCTCACAGCAGCACCGTAGA-3′; 1R 5′-ACAAACACAGTCAATCCTGC-3′) as probes. Hybridization and signal detection were performed as above.
For cloning of PCR products, unincorporated primers were removed using the QIAquick PCR Purification Kit (Qiagen). Products were then ligated into the pDRIVE vector (Qiagen) and transformed into SURE® competent cells (Stratagene). Fusion clones were detected by colony hybridization, using probes as described above. DNA was prepared from clones of interest using a QIAprep Spin Miniprep Kit (Qiagen). DNA sequence reactions were performed using the BigDye Reaction Mix (Perkin Elmer-ABI), and products were evaluated using an ABI PRISM 377 DNA Sequencer.
Statistical methods
An unpaired t-test was used to compare telomere lengths between mutant plants at different generations of propagation. Nonparametric data were compared using a Wilcoxon's rank sum test. Comparison between categorical variables was completed using the χ2 test. A P-value ⩽0.05 was considered significant.
Supplementary Material
Supplementary Figure 1
Supplementary Table I
Supplementary Table II
Supplementary Tables III
Acknowledgments
We thank members of the Shippen lab for helpful discussions and Jeff Kapler for critically reading the manuscript. This work was supported by NIH grant (GM65383) to DES and FWF grant (P16405) to KR.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genomic sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bailey SM, Meyne J, Chen DJ, Kurimasa A, Li GC, Lehnert BE, Goodwin EH (1999) DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc Natl Acad Sci USA 96: 14899–14904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird DM, Rowson J, Wynford-Thomas D, Kipling D (2003) Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat Genet 33: 203–207 [DOI] [PubMed] [Google Scholar]
- Baumann P, Cech TR (2000) Protection of telomeres by the Ku protein in fission yeast. Mol Biol Cell 11: 3265–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Cech TR (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Bundock P, van Attikum H, Hooykaas P (2002) Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant. Nucleic Acids Res 30: 3395–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P, Hooykaas P (2002) Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell 14: 2451–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Quinney N, Willcox S, Subramanian D, Griffith JD (2003) Telomere looping in P. sativum (common garden pea). Plant J 36: 271–279 [DOI] [PubMed] [Google Scholar]
- Chan SW-L, Blackburn EH (2003) Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Mol Cell 11: 1379–1387 [DOI] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP (2002) The Mre11 complex: at the crossroads of DNA repair and checkpoint signaling. Nat Rev Mol Cell Biol 3: 317–327 [DOI] [PubMed] [Google Scholar]
- Espejel S, Franco S, Rodriguez-Perales S, Bouffler SD, Cigudosa JC, Blasco MA (2002) Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J 21: 2207–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P (2000) DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res 28: 2585–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MG, Cooper JP (2001) The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol Cell 7: 55–63 [DOI] [PubMed] [Google Scholar]
- Ferreira MG, Miller KM, Cooper JP (2004) Indecent exposure: when telomeres become uncapped. Mol Cell 13: 7–18 [DOI] [PubMed] [Google Scholar]
- Forstemann K, Moss M, Lingner J (2000) Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res 28: 2690–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner J, Britt AB (2003) Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J 34: 427–440 [DOI] [PubMed] [Google Scholar]
- Gallego ME, Blenyard JY, Daoudal-Cotterall S, Jalut N, White CI (2003b) Ku80 plays a role in non-homologous recombination but is not required for T-DNA integration in Arabidopsis. Plant J 35: 557–565 [DOI] [PubMed] [Google Scholar]
- Gallego ME, Jalut N, White CI (2003a) Telomerase dependence of telomere length in Ku80 mutant Arabidopsis. Plant Cell 14: 782–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Greider CW (2003) End resection initiates genomic instability in the absence of telomerase. Mol Cell Biol 23: 8450–8461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Rudolph KL, Strong MA, DePinho RA, Chin L, Greider CW (2001a) Telomere dysfunction triggers developmentally regulated germ cell apoptosis. Mol Biol Cell 12: 2023–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Strong MA, Hao L-Y, Greider CW (2001b) The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107: 67–77 [DOI] [PubMed] [Google Scholar]
- Kirik A, Salomon S, Puchta H (2000) Species-specific double-strand break repair and genome evolution in plants. EMBO J 19: 5562–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani H, Hosouchi T, Tsuruoka H (1999) Structural analysis and complete physical map of Arabidopsis thaliana chromosome 5 including centromeric and telomeric regions. DNA Res 6: 381–386 [DOI] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K (2003) Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol 4: 712–720 [DOI] [PubMed] [Google Scholar]
- Ma J-L, Kim EM, Haber JE, Lee SE (2003) Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol 23: 8820–8828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L, Lydall D (2002) EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev 16: 1919–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Iyer S, Boswell Fulton T, Blackburn EH (2000) Telomere fusions caused by mutating the terminal region of telomeric DNA. Proc Natl Acad Sci USA 97: 11409–11414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski PA, Mieczkowska JO, Dominaska M, Petes TD (2003) Genetic regulation of telomere–telomere fusions in the yeast Saccharomyces cerevisae. Proc Natl Acad Sci USA 100: 10854–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V (1996) Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M (2000) A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc Natl Acad Sci USA 97: 6409–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puizina J, Siroky J, Mokros P, Schweizer D, Riha K. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. The Plant Cell, In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Chao S, Vongs A, Yang J (1992) Characterization of Arabidopsis thaliana telomeres isolated in yeast. Nucleic Acids Res 20: 4039–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, McKnight TD, Fajkus J, Vyskot B, Shippen DE (2000) Analysis of the G-overhang structures on plant telomeres: evidence for two distinct telomere architectures. Plant J 23: 633–641 [DOI] [PubMed] [Google Scholar]
- Riha K, McKnight TD, Griffing LR, Shippen DE (2001) Living with genome instability: plant responses to telomere dysfunction. Science 291: 1797–1800 [DOI] [PubMed] [Google Scholar]
- Riha K, Shippen DE (2003) Ku is required for telomeric C-rich strand maintenance, but not for end-to-end chromosome fusions in Arabidopsis. Proc Natl Acad Sci USA 100: 611–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, Shippen DE (2003a) Telomere structure, function and maintenance in Arabidopsis. Chromosome Res 11: 263–275 [DOI] [PubMed] [Google Scholar]
- Riha K, Watson J, Parkey J, Shippen DE (2002) Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku. EMBO J 21: 2819–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper E, Goytisolo FA, Slijepcevic P, van Buul PP, Blasco MA (2000) Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep 1: 244–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siroky J, Zluvova J, Riha K, Shippen DE, Vyskot B (2003) Rearrangements of ribosomal DNA clusters in late generation telomerase-deficient Arabidopsis. Chromosoma 112: 116–123 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T (2002) DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol 12: 1635–1644 [DOI] [PubMed] [Google Scholar]
- Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K-I, Ohta K, Uritani M, Ushimaru T, Yoshinaga K, Ueno M (2003) Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol 23: 5186–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Attikum H, Bundock P, Overmeer RM, Lee LY, Gelvin SB, Hooykaas PJ (2003) The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Res 31: 4247–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Price M (2003) Protecting the terminus: t-loops and telomere end-binding proteins. Cell Mol Life Sci 60: 2283–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Sonoda E, Sasaki MS, Morrison C, Haraguchi T, Hiraoka Y, Yamashita YM, Yagi T, Takata M, Price C, Kakazu N, Takeda S (1999) Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J 18: 6619–6629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X-D, Niederhofer L, Kuster B, Mann M, Hoeijmaker JHJ, de Lange T (2003) ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell 12: 1489–1498 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Table I
Supplementary Table II
Supplementary Tables III


