Abstract
Gene expression in members of the family Bacillaceae becomes compartmentalized after the distinctive, asymmetrically located sporulation division. It involves complete compartmentalization of the activities of sporulation-specific sigma factors, σF in the prespore and then σE in the mother cell, and then later, following engulfment, σG in the prespore and then σK in the mother cell. The coupling of the activation of σF to septation and σG to engulfment is clear; the mechanisms are not. The σ factors provide the bare framework of compartment-specific gene expression. Within each σ regulon are several temporal classes of genes, and for key regulators, timing is critical. There are also complex intercompartmental regulatory signals. The determinants for σF regulation are assembled before septation, but activation follows septation. Reversal of the anti-σF activity of SpoIIAB is critical. Only the origin-proximal 30% of a chromosome is present in the prespore when first formed; it takes ≈15 min for the rest to be transferred. This transient genetic asymmetry is important for prespore-specific σF activation. Activation of σE requires σF activity and occurs by cleavage of a prosequence. It must occur rapidly to prevent the formation of a second septum. σG is formed only in the prespore. SpoIIAB can block σG activity, but SpoIIAB control does not explain why σG is activated only after engulfment. There is mother cell-specific excision of an insertion element in sigK and σE-directed transcription of sigK, which encodes pro-σK. Activation requires removal of the prosequence following a σG-directed signal from the prespore.
INTRODUCTION
Cell differentiation is a fundamental biological process. Central to it are the coordination of gene expression with morphological change and the establishment of distinct programs of gene expression in the different cell types involved. Formation of spores by Bacillus subtilis is a primitive system of cell differentiation (Fig. 1), which has become a paradigm for the study of cell differentiation in prokaryotes (59, 183, 228, 231, 281). The spores formed are dormant and show greatly increased resistance to stresses such as heat and noxious chemicals compared to what is seen with vegetative cells. It was shown 25 years ago through a study of genetic mosaics that gene expression is compartmentalized during sporulation of B. subtilis, with different genes being expressed in the prespore and the mother cell, the two cell types involved (43, 226). In the years since, the completeness of compartmentalization has been demonstrated first by immunoelectron microscopy (48, 83, 189), then by fluorescence microscopy with immunofluorescence and green fluorescent protein (105, 170, 233, 299, 314), and most recently through the use of a two-part transcriptional probe (173).
FIG. 1.
Schematic representation of the stages of spore formation. A vegetatively growing cell is defined as stage 0. It is shown as having completed DNA replication and containing two complete chromosomes (represented as disordered lines within the cells), although replication is not completed at the start of spore formation. Formation of an axial filament of chromatin, where both chromosomes (or a partially replicated chromosome) form a continuous structure that stretches across the long axis of the cell, is defined as stage I. Asymmetric division occurs at stage II, dividing the cell into the larger mother cell and smaller prespore; for clarity, the septum is indicated as a single line. At the time of division, only approximately 30% of a chromosome is trapped in the prespore, but the DNA translocase SpoIIIE will rapidly pump in the remaining 70%. Stage III is defined as completion of engulfment, and the prespore now exists as a free-floating protoplast within the mother cell enveloped by two membranes, represented by a single ellipse. Synthesis of the primordial germ cell wall and cortex, a distinctive form of peptidoglycan, between the membranes surrounding the prespore is defined as stage IV and is represented as thickening and graying of the ellipse. Deposition of the spore coat, protective layers of proteins around the prespore, is defined as stage V. The coat is represented as the black layer surrounding the engulfed prespore. Coincident with coat and cortex formation, the engulfed prespore is dehydrated, giving it a phase-bright appearance, represented here as a light grey shading. Stage VI is maturation, when the spore acquires its full resistance properties, although no obvious morphological changes occur. Stage VII represents lysis of the mother cell, which releases the mature spore into the environment.
The cell type-specific, compartmentalized programs of gene expression result from the cell type-specific activity of RNA polymerase sigma factors: σF and then σG in the prespore and σE and then σK in the mother cell (Fig. 2) (231). Compartmentalization of gene expression in each cell type is coupled to morphogenesis, with σF and σE becoming active after asymmetric division and σG and σK becoming active after engulfment of the prespore by the mother cell. Thus, the activation of σG and σK also represents compartmentalization in the sense of expression after but not before completion of engulfment. The key regulators of sporulation discussed below have been identified in all species of endospore former whose genomes have been sequenced, including the pathogens Bacillus anthracis and Clostridium difficile (277). Thus, conclusions from the study of B. subtilis are generally valid for members of the family Bacillaceae and illustrate general features of cell differentiation.
FIG. 2.
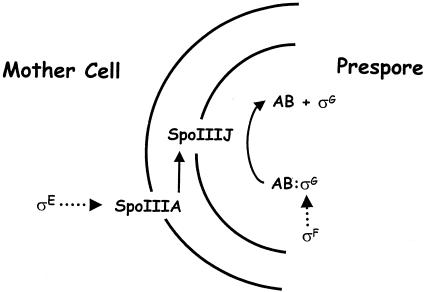
Intercompartmental communication during sporulation. The parallel vertical lines represent the two membranes separating the prespore (right) from the mother cell (left). Diagonal red lines represent pathways of intercompartmental posttranslational activation, and vertical black arrows represent intracompartmental transcriptional activation. Fluorescent micrographs represent cells stained with FM4-64 (red) to visualize the cell membranes and expressing compartment-specific gfp fusions to spoIIQ, spoIID, sspA, and gerE for σF, σE, σG, and σK, respectively. The prespore membranes are not stained in the σG and σK images because engulfment is complete and the prespore membranes are now inaccessible to the lipophilic FM4-64 stain. σF, active in the prespore, is the first compartmentalized σ factor during sporulation. It triggers expression of SpoIIR, which activates the inferred receptor protease SpoIIGA, located in the asymmetric septum. Upon receipt of the signal, SpoIIGA processes the inactive precursor pro-σE into active σE in the mother cell. RNA polymerase with σE transcribes the spoIIIA operon, whose products then signal across the prespore membrane to activate σG, expressed in the prespore under the control of σF but held inactive by SpoIIAB (and probably other factors) until this signal is received. SpoIIIJ is also required for this signaling; although only required (and therefore only represented) in the prespore, it is expressed vegetatively and is presumably present in both compartments. Although not represented here, transcription of spoIIIG (encoding σG) requires an unknown signal from the mother cell as well as the SpoIIQ protein, expressed in the prespore under the control of σF. Once σG becomes active, it causes expression of SpoIVB, which is inserted into the inner prespore membrane. SpoIVB triggers processing of pro-σK, which is synthesized in the mother cell from the σE-directed sigK gene. The processing enzyme is thought to be SpoIVFB, which also expressed in the mother cell under the control of σE but does not act upon pro-σK until it receives the SpoIVB signal from the prespore.
In this article we review the process of spore formation. We focus primarily on B. subtilis but include discussion of other species where we think it appropriate. We discuss in detail the genetic and biochemical experiments that have led to the discovery and characterization of cell-specific programs of gene expression. Since sporulation follows a distinct series of morphological and genetic stages, we detail steps in the order that they naturally occur, as though following a single cell through the entire developmental process. The cell-specific changes in gene expression that occur during sporulation are coupled to morphogenesis. The two major phases of compartmentalization are associated with two major morphological events, completion of septation and completion of engulfment. Consequently, we start with a brief description of the morphological changes during sporulation. We discuss in depth the events leading to the asymmetrically located sporulation division, which primes the organism for the compartmentalization of gene expression that follows the division. Compartmentalized gene expression is associated with the activation of the four sporulation-specific σ factors. We discuss the activation of each. We pay particular attention to regulation of σF because it is the first σ factor whose activity is compartmentalized during sporulation and because in vitro and in vivo analysis of its activation has progressed furthest.
Many of the loci discussed in this review were identified almost three decades ago as spo loci, because mutations in them blocked spore formation. Since then, our understanding of the roles of those loci has increased enormously. In general, spo loci encode proteins with unique roles in spore formation and, in some cases, in regulation of compartmentalization. They have provided the underpinning of much of our knowledge of compartmentalization. However, in the last decade it has become clear that there are also regulators, or regulatory mechanisms, which have overlapping rather than unique roles. The corresponding genes were generally missed in earlier studies because mutation in them had a comparatively mild effect on spore formation. Nevertheless, such overlapping regulatory mechanisms play an important part in the compartmentalization of gene expression, and they are an active area of research. For the main sections in the review, we use a historical approach in describing the development of our knowledge of compartmentalization. We think that this historical approach is important for appreciating much of the present and past thinking about compartmentalization.
MORPHPOLOGICAL STAGES OF SPORULATION
In order to discuss compartmentalization of gene expression, we first review the morphological changes in the developmental process, with which changes in gene expression are associated. Formation of heat-resistant spores from vegetative cells of B. subtilis takes about 7 h at 37°C. The morphological changes during sporulation were initially characterized by electron microscopy (145, 251). The basic sequence of changes is similar for all species of Bacillus and Clostridium that have been studied (79) and is illustrated in Fig. 1. Identification of successive stages by Roman numerals follows the convention introduced by Ryter (251) and now generally used. The vegetative cell is designated stage 0. Formation of an axial filament of chromatin, where two copies of the chromosome condense and elongate to form a filament that stretches across the long axis of the cell, is defined as stage I (21, 29, 308). Subsequently, the cell divides at a subpolar site, resulting in the formation of two unequally sized daughter cells.
Completion of septation is designated stage II. At the time of asymmetric division, only approximately one-third of a chromosome is present in the smaller prespore (also called the forespore), but the remaining two-thirds is rapidly pumped in by the DNA translocase SpoIIIE (17, 303), resulting in two cells with unequal volumes but identical genomes. Next, the polar septum undergoes septal thinning, followed by bulging of the prespore into the mother cell and migration of the septal membrane around both sides of the prespore. When the migration is complete, the membranes fuse at the cell pole, pinching off the prespore and releasing it within the mother cell as a free-floating protoplast that is surrounded by two membranes, one derived from each of the two cells; completion of engulfment is designated stage III (227). Engulfment is followed by the deposition of two peptidoglycan layers, the primordial germ cell wall and the cortex, in the space between the membranes surrounding the prespore (stage IV) (81). Following this deposition, a complex structure of proteins on the outside surface of the prespore, known as the coat, is constructed (stage V) (47, 109). This stage is followed by maturation of the spore (stage VI), when it gains resistance to UV radiation and high temperature (208). Lastly, the mother cell lyses (stage VII), releasing the mature spore into the environment. Germination and outgrowth, followed by a resumption of the vegetative growth cycle, occur when the spore finds itself in a nutrient-rich environment (213). Sporulation mutants are denoted by the stage in the process at which they are blocked (e.g., spoII mutants complete asymmetric septation but fail to complete engulfment). The names for sporulation loci include the stage of blockage caused by mutation and a distinguishing letter designation (e.g., spoIIA) (228, 231).
The profound morphological changes that occur during sporulation are coupled to global changes in gene expression, which are effected by activation of alternative RNA polymerase σ factors (Fig. 2) (228, 231). Activation of σH (and the response regulator Spo0A) in the predivisional cell leads to expression of factors important for axial filament formation, asymmetric division, and compartmentalization of gene expression. Immediately after asymmetric division, σF becomes active in the prespore, rapidly followed by activation of σE in the mother cell. The separate lines of gene expression drive engulfment of the prespore by the mother cell and result in synthesis of the late-compartment-specific σ factors. Upon completion of engulfment, σG becomes active in the prespore and σK becomes active in the mother cell. Coat and cortex synthesis, spore maturation, and mother cell lysis are driven by these late stages of cell-specific gene expression. Each step is dependent upon completion of all of the previous steps except axial filament formation (see below).
INITIATION OF SPORULATION
Gene expression becomes compartmentalized immediately after the spore septum has formed. To understand how this compartmentalization happens, it is important to explore the events leading to it. In this section we briefly discuss the activation of the master sporulation response regulator Spo0A and the alternative σ factor σH. For more focused reviews on the initiation of sporulation, we refer the reader to references 27 and 223.
The Phosphorelay
Spo0A is the master regulator for entry into spore formation. It is activated by the phosphorelay, a more complex version of the classic two-component system (26). In turn, Spo0A-PO4 activates transcription of genes required for axial filament formation and for asymmetric division. It also activates transcription of the genes encoding the early compartmentalized σ factors, σF and σE, as well as their regulators. Sporulation is initiated in response to a number of external and internal signals that are integrated into the phosphorelay, including signals for nutrient starvation, cell density, and cell cycle progression (27, 223, 291).
At least five kinases are involved in the phosphorelay: KinA, KinB (290), KinC (160), KinD (134), and KinE (67), of which KinA and KinB are the primary kinases for initiation of sporulation. In response to unidentified stimuli, they autophosphorylate and then donate their phosphate groups to the response regulator Spo0F. Spo0F lacks an output domain and is incapable of activating transcription; it serves only as an intermediary in the phosphorelay. The phosphotransferase Spo0B transfers the phosphate from Spo0F-PO4 to Spo0A (26). The phosphorelay is also subject to negative regulation: for example, the phosphatases Spo0E, YisI, and YnzD dephosphorylate Spo0A, thereby preventing its activation (210, 221). In order to ensure that sporulation occurs only under the appropriate conditions, the phosphorelay must integrate different intracellular and extracellular signals. The mechanisms responsible for this integration are described below.
Cell density.
Efficient sporulation requires high cell density (101). When cell density is low, the Rap (response regulator aspartyl phosphatase) proteins RapA, RapB (222), and RapE (133) dephosphorylate Spo0F-PO4, preventing Spo0A activation. The rapA and rapE genes are cotranscribed with a downstream open reading frame encoding the signaling peptide precursors PhrA and PhrE, respectively, which are processed and exported out of the cell. As cell density increases, the processed peptides are imported by the oligopeptide permease (Opp) and inhibit the activity of RapA and RapE; similarly, the processed product of PhrC (CSF [competence- and sporulation-stimulating factor])inhibits RapB. Inhibition of the Rap proteins prevents dephosphorylation of Spo0F-PO4 and allows phosphorylation of Spo0A and the initiation of sporulation when cell density is high (133, 220).
Nutrient starvation.
In addition to high cell density, nutrient starvation is also required for the initiation of sporulation. A dramatic drop in the concentration of GTP and GDP correlates with the onset of sporulation, and inhibition of GMP synthesis by decoyinine treatment induces sporulation in the absence of nutrient starvation (200). CodY has recently been identified as the key sensor of guanine nucleotide levels. Disruption of the codY gene allows sporulation to occur in the presence of excess nutrients, and the ability of the CodY repressor to bind DNA correlates with the GTP concentration. As a consequence, when GTP levels drop upon entry into stationary phase, CodY-regulated genes are derepressed (239). Microchip array analysis has identified phrA, phrE, and kinB, all positive regulators of the phosphorelay, as targets of CodY repression (204). Therefore, one way that nutrient starvation is integrated into the decision to sporulate is transcriptional regulation of phosphorelay components via CodY. In addition, sporulation is also subject to catabolite repression (117) and requires a functioning Krebs cycle (131), although the molecular basis of these dependencies is unknown.
Cell cycle.
In addition to factoring extracellular conditions such as cell density and nutrient availability into the decision to sporulate, the intracellular environment is monitored as well. Damage to DNA and blocking of either the initiation or progression of DNA replication prevent the initiation of sporulation (126, 128, 129, 132, 165). These conditions lead to the expression of Sda (suppressor of dnaA) because of the presence of binding sites in the sda promoter region for the repressors DnaA and LexA, which no longer repress when DNA replication is blocked or DNA is damaged, respectively. Sda impairs KinA autophosphorylation, blocking the phosphorelay (28). As a result, a developmental checkpoint is established that only allows cells with undamaged, replicating chromosomes to proceed into development.
An additional mechanism is thought to link chromosome partitioning status to sporulation. Spo0J and Soj (suppressor of spo0J) (also known as Spo0JB and Spo0JA, respectively) are members of the plasmid-partitioning families of proteins ParB and ParA, respectively (130). Spo0J colocalizes to cell poles with the chromosomal origin of replication (175), binds to sites near the chromosomal origin (174), and is required for optimal efficiency of chromosome segregation (130). In the absence of Spo0J, Soj binds to the promoter regions of at least four Spo0A-responsive genes (spo0A, spoIIA, spoIIE, and spoIIG) and represses their transcription (33, 191, 237, 238). This effect appears to be mediated by dynamic protein localization; when Spo0J is present, Soj oscillates between sites near the poles of the cell, presumably preventing stable DNA-protein interaction and transcriptional repression. However, in the absence of Spo0J, Soj remains static and represses developmental transcription (191, 238). Although it is tempting to speculate that Spo0J and Soj sense chromosome partitioning status and regulate the initiation of sporulation accordingly, direct evidence for this model is lacking. However, consistent with a role in monitoring the cell cycle, recent studies have linked these proteins to cell division, to initiation of DNA replication, and to axial filament formation (11, 163, 209, 308).
Spo0A Regulon
The sum of all of these interactions determines if enough Spo0A-PO4 has been generated to initiate sporulation. Spo0A-PO4 can either activate or repress transcription by binding to a 7-bp sequence, TGNCGAA, where N is any nucleotide, in or near promoters recognized by the vegetative σ factor σA and the alternative σ factor σH (223). This binding results in global changes in gene expression, altering the expression profile of over 500 genes, which represent approximately one-eighth of the total genes in B. subtilis (71). Additional genomic analysis has revealed that 121 of these genes are under the direct control of Spo0A, with approximately one-third being positively regulated and the remainder being negatively regulated; 25 of the regulated genes are themselves transcription factors, indicating that many of the transcriptional changes caused by Spo0A are indirect (203).
A number of key spo loci are directly positively regulated by Spo0A: the spoIIA and spoIIG operons, encoding the prespore- and mother cell-specific transcription factors σF and σE, respectively; and spoIIE, encoding a bifunctional protein phosphatase that is required for asymmetric division and σF activation (231). In addition, the gene encoding the effector of axial filament formation, racA, is under the control of Spo0A (21, 308). Thus, Spo0A activates the synthesis of factors required for chromosome remodeling, asymmetric division, and the compartmentalized gene expression that immediately follows. The racA gene was identified by functional analysis of the Spo0A regulon (21), and such analyses will most likely reveal additional genes involved in sporulation.
Role of σH
In addition to the phosphorelay and the main vegetative σ factor σA, the transition-state regulator σH is also required for the initiation of sporulation. Regulation of σH synthesis and activity is not well understood but involves both posttranscriptional and posttranslational mechanisms (106, 177). σH regulates a number of phosphorelay genes: σH-dependent transcription of spo0A is essential for sporulation (270), and kinA, kinE, and spo0F also have σH-dependent promoters (23, 236). In addition, transcription of several phr genes encoding peptide precursors, some of which function to reverse phosphorelay inhibition by Rap phosphatases, is also dependent upon σH (197). Other σH-regulated genes are important for later events in sporulation: σH-dependent transcription of the essential cell division operon ftsAZ is required for efficient asymmetric septation (19, 94, 98), and the spoIIA operon, which encodes the prespore-specific transcription factor σF and its regulators, is under the control of σH (302). A substantial portion of the σH and Spo0A regulons overlap: for example, spoIIA and racA are regulated by both transcription factors. The σH regulon has recently been characterized by microchip array analysis (23), and functional analysis of this regulon has resulted in the independent identification of racA (308).
AXIAL FILAMENT FORMATION
There are several substantial differences in the events associated with the sporulation division compared with those associated with the vegetative division (114, 227). Any or all may be important for priming the compartmentalization of gene expression which follows that division. The first is the formation of an axial filament of chromatin, in which both chromosomes in the predivisional cell elongate into a filament that stretches the length of the long axis of the cell. This structure was characterized by electron microscopy (78, 188, 252) and later by fluorescence microscopy (29, 300).
Genetic Control
Standard genetic analysis failed to identify effectors of axial filament formation, and only recent genomic and cell biological studies have allowed identification of the components involved. Despite their name, none of the classic spo0 mutations clearly prevented axial filament formation (228): spo0H mutants formed axial filaments (29, 94), whereas spo0A, spo0B, and spo0F mutants underwent an additional symmetric division during sporulation, and it was not clear if that was preceded by axial filament formation (29, 53). Some insight into the process was derived from analysis of the SMC (structural maintenance of chromosomes) protein, required for chromosome compaction and partitioning (25, 176). Mutants lacking this protein are able to activate Spo0A but are unable to form axial filaments (99), suggesting that an effector of axial filament formation is either missing or nonfunctional in this background. It was also found that asymmetric division did not occur in the absence of axial filament formation, suggesting the existence of a checkpoint that functioned to couple the two events (99). Such a checkpoint would ensure that asymmetric division would trap the origin-proximal region of a chromosome in the prespore, providing a template for transcription by E-σF (RNA polymerase core enzyme, E, associated with σF) and an anchor for chromosome translocation into this compartment
DivIVA.
Several lines of investigation provided insight into the genetic control of axial filament formation. The first was a study of the DivIVA protein of B. subtilis, considered the functional homologue of Escherichia coli MinE in that it restricts the division-inhibition proteins MinCD to the cell poles and so ensures mid-cell division during vegetative growth (34, 56). divIVA mutants have severe growth and sporulation defects, but the isolation of mutants specifically defective in sporulation suggested that DivIVA played a dedicated role in development (Fig. 3). These mutants frequently formed anucleate prespores (287), indicating that the proposed checkpoint coupling axial filament formation and asymmetric division (99) had been disrupted.
FIG. 3.
Chromosome partitioning and genetic asymmetry. A single cell progressing through sporulation is represented on the right. Disordered internal lines represent the nucleoids. The earliest (topmost) cell is drawn as having two complete chromosomes, although it may contain one partially replicated chromosome. Through the action of DivIVA, RacA, and Soj, the two complete chromosomes (or the partially replicated chromosome) are remodeled into an axial filament that extends across the long axis of the cell, represented in the second cell. After asymmetric division occurs, the prespore contains only the origin-proximal one-third of a chromosome, whereas the mother cell contains one complete chromosome and two-thirds of another; this partitioning results in transient genetic asymmetry between the mother cell and the prespore. For simplicity, the septum is represented as a single line. A portion of the third cell has been expanded in order to represent the asymmetry more clearly; the hatched ovals represent the DNA translocase SpoIIIE; the locations of several genetic loci are noted; and σF is depicted as being active in the prespore and σE is depicted as being active in the mother cell. Within about 15 min of the asymmetric division, SpoIIIE pumps the remaining two-thirds of the prespore chromosome into this compartment, restoring genetic symmetry.
RacA.
Genomic approaches made it possible to elucidate the role that DivIVA played in axial filament formation. One approach identified Spo0A-regulated genes by microchip array (71), systematically disrupted them, and screened the mutants for changes in chromosome segregation (21). In a separate study, σH-regulated genes were identified by microchip array, and candidate genes for partitioning function, including those predicted to have DNA-binding domains, were disrupted and characterized (308). Both approaches resulted in identification of a gene that, when disrupted, caused an anucleate prespore phenotype. The encoded protein, RacA (remodeling and anchoring of the chromosome A), was found to bind nonspecifically to the chromosome and also to the pole of the cell, acting as a bridge connecting the two. Localization of RacA to the cell pole is dependent upon DivIVA, explaining why mutations in the corresponding genes cause similar phenotypes, although those caused by mutations in divIVA are more severe (21, 287, 308). From their observations, Ben-Yehuda and colleagues (21) proposed that once the origin-proximal region of the chromosome reaches the pole of the cell, RacA binds DivIVA and displaces the division inhibitor MinCD (192, 295), triggering asymmetric division (21).
Although the prospect of a checkpoint linking axial filament formation to asymmetric division is exciting, it still requires direct testing. The original observation that smc mutants fail to form axial filaments remains unexplained (99); it may be that RacA cannot bind to the uncondensed and disorganized chromosomes of smc mutants (25, 176). racA and divIVA mutants also undergo asymmetric division even though axial filament formation is impaired (21, 287, 308), whereas the smc mutant is deficient in both processes (99). Two plausible explanations are that RacA and DivIVA prevent asymmetric division in the smc mutant, enforcing the checkpoint proposed by Graumann and Losick (99) (see above) or that defects in asymmetric division and axial filament formation are independent in this background, placing the existence of this checkpoint in doubt. Epistasis tests could help distinguish between these possibilities. For example, in the former case, racA and divIVA mutations should restore asymmetric division to an smc mutant, whereas in the latter case they should not. More work is needed to explore the relationship between axial filament formation and asymmetric division.
Soj.
The fact that racA mutations cause a less severe defect than divIVA mutations (21, 287, 308) suggested that at least one additional factor was involved in axial filament formation. Indeed, it was found that a mutant lacking RacA and the transcriptional repressor Soj (33, 191, 238) displayed a phenotype approximating the more severe phenotype caused by a divIVA mutation (308), a surprising result because no partitioning function had previously been attributed to Soj. However, such a phenotype could be mediated by its interaction partner, Spo0J, which is required for optimal efficiency of vegetative chromosome partitioning (130) and localizes to the origin region of the chromosome (174, 175). The RacA-Soj system is an example of redundancy in sporulation controls and is especially noteworthy because it led to the attribution of a novel function to Soj, one that was unlikely to be observed in a racA+ genetic background.
Polar localization region.
In a separate study of axial filament formation, the use of systematic chromosomal inversions identified a polar localization region, located ≈150 to 300 kbp from the origin of replication, that is required for efficient trapping of DNA in the prespore (307). Although an attractive hypothesis is that this region is the principal binding site for RacA, chromatin immunoprecipitation experiments suggest that a different region (60 to 80 kbp from the origin) is preferentially bound (21). As a result, the relationship of the polar localization region to the RacA-Soj-DivIVA system remains unclear.
ASYMMETRIC DIVISION
Once a cell has formed the axial filament, the next major event in sporulation is asymmetric division. Since the cell normally divides at mid-cell with remarkable accuracy (198), this switch requires a dramatic relocalization of the cell division apparatus (61). By dividing at a polar site, the cell becomes genetically and morphologically asymmetric, and the asymmetry leads to different cell fates. The asymmetric division is critical to the establishment of compartmentalized gene expression. The division has much in common with vegetative division (61, 114), but the distinguishing features are presumptively ones that might lead to compartmentalization of gene expression. It is the distinguishing features that are considered here.
FtsZ Ring Switching
Dynamic repositioning.
During vegetative growth, the essential prokaryotic tubulin homologue FtsZ forms a ring (the Z ring) at mid-cell, where division subsequently occurs (298). However, during sporulation, activation of Spo0A triggers the formation of Z rings near both poles of the cell (167). The switch has recently been characterized by deconvolution microscopy. The use of this technique revealed that, during sporulation, a Z ring initially forms at mid-cell but the FtsZ then redeploys to sites near both poles through the formation of a dynamic helical intermediate (19). This intermediate resembles the helical structures formed by two bacterial actin homologues, Mbl and MreB (137); one possibility is that FtsZ relocates by tracking along these structures.
Genetic control.
Early studies showed that the ftsAZ operon contained three promoters, one of which was activated during sporulation by σH (94, 98). However, deletion of this promoter had only a moderate effect on asymmetric division (98). Similarly, disruption of the spoIIE locus, encoding a critical activator of σF in the prespore (8, 49), also had a moderate effect on polar Z ring formation (19, 149) and asymmetric division (16, 228). However, simultaneous ablation of spoIIE and the σH-dependent promoter of ftsAZ resulted in severe impairment of polar Z ring formation and asymmetric division. Conversely, if ftsAZ overexpression was combined with expression of spoIIE, asymmetric division could be triggered during vegetative growth (19). Therefore, spoIIE induction and increased ftsAZ expression play overlapping roles during sporulation in ensuring polar Z ring formation and asymmetric division.
SpoIIE.
The discovery of a role for SpoIIE in polar Z ring formation (19, 149) reinforced previous ultrastructural studies that had implicated this protein in asymmetric division. These studies found that null mutations in spoIIE resulted in rare asymmetric septa that were aberrantly thick, whereas several point mutations blocked sporulation but allowed the formation of typical sporulation septa at normal frequency (16, 228). Consistent with this role, it was found that SpoIIE localizes to asymmetric division sites (10, 14) in an FtsZ-dependent manner (168). FtsZ and SpoIIE also interact in vitro and in yeast two-hybrid assays (186). However, how this interaction assists polar Z ring formation and asymmetric division is unknown. Some possibilities include anchoring an end of the FtsZ helix to the cell membrane, antagonizing MinCD near the pole of the cell, and recognizing a polar marker (19). In addition to SpoIIE and increased ftsAZ expression, there is evidence that MinCD and SpoVG may play minor roles in selecting the asymmetric division site (15, 195).
Abortively Disporic Phenotype
In spo+ strains, surface annular structures (cloisons) and Z rings appear at both polar sites (19, 167, 251), indicating that the cell has two potential polar division sites. However, a septum is normally formed at only one of these sites (251). In certain mutants, both potential polar division sites are utilized, resulting in a three-chambered organism consisting of two smaller prespores separated by a larger central compartment. This is the abortively disporic phenotype, which is associated with mutations in some spoII loci (228, 252). Mutants that demonstrate the abortively disporic phenotype can initiate sporulation but have defects in activating the mother cell-specific transcription factor σE (124). Three proteins expressed in the mother cell under σE control, SpoIID, SpoIIM, and SpoIIP, are required to prevent the second asymmetric division (57, 232). Although it was puzzling why B. subtilis would generate two potential division sites during sporulation, recent studies have provided some insight. Cells with anucleate prespores are frequently observed in racA mutant cells, which are deficient in axial filament formation. However, a substantial proportion of these cells undergo a second asymmetric division at the other end of the cell, and if they successfully capture DNA in the second prespore, they form spores (21, 308). This result suggests that the ability to divide at both asymmetric division sites is a failsafe mechanism to ensure successful sporulation even in the absence of axial filament formation.
Which End of the Cell?
An unanswered question is how the cell determines at which end of the cell it will divide. Although FtsZ rings form at both potential polar division sites, the next known protein to assemble, FtsA, localizes to only one of them (76), indicating that FtsA may play some role in selecting which of the two potential division sites is utilized. Chromosome segregation into the prespore or mother cell appears to be essentially random with respect to time of replication, and so chromosome age is presumably not a factor in determining which end becomes the prespore (43, 65). In most circumstances and in a range of species, spores are formed almost exclusively at the older pole of the cell, clearly suggesting that the pole is a determinant of asymmetry and compartmentalization (52, 112, 113). However, when the sporulation procedure involves centrifugation (with a force of perhaps 5,000 × g) and 25 mM Mg2+, spore position is essentially random with respect to pole age (52). Thus, polar determinism can be lost without losing asymmetric division or compartmentalization.
Differences between Sporulation Septum and Vegetative Division Septum
The asymmetrically located sporulation division is often considered the defining early morphological event in sporulation. The machinery for asymmetric division is similar to that used for vegetative division (61). However, there are several distinctive features of the sporulation division (114, 227) in addition to those described above. Since the division is critical to the compartmentalization of gene expression that follows, it is useful to summarize those distinctive features. (i) The sporulation division septum is much thinner than the vegetative division septum. (ii) The two cells that result from the sporulation division do not separate from each other, as occurs following vegetative division. Rather, the mother cell engulfs the prespore. (iii) Autolysis of the wall material (peptidoglycan) within the sporulation septum begins in the center of the septum, and ultimately there is apparently complete loss of wall material. In contrast, autolysis of the wall material of the vegetative septum begins at the periphery of the septum and proceeds inwards. Moreover, there is little loss of wall material—the split septum provides the wall for the poles of the nascent cells (227). (iv) Prior to the sporulation division, the two chromosome origins and associated proteins move to the extreme poles of the cell rather than to a subpolar location, as in vegetative division (21, 175, 300, 308). (v) The septum is asymmetrically located, with respect to the cell poles, during sporulation but not during vegetative growth (251) (vi) Several proteins become associated with the sporulation septum that are not associated with the vegetative division septum (10, 14, 72). (vii) Complete partitioning of a chromosome into the prespore occurs after septation (303, 310), so that there is genetic asymmetry between the prespore and the mother cell when they are first formed (54, 84, 305). (viii) After the sporulation division, different programs of gene expression are initiated in the two daughter cells (231). These programs are driven by activation of cell-specific σ factors that direct RNA polymerase to transcribe different genes, which encode factors responsible for establishing the very different fates of the prespore and the mother cell.
TRANSFER OF DNA INTO THE PRESPORE
At this stage of sporulation, the two chromosomes have extended into a filament stretching along the long axis of the cell, with their origin regions near the poles (21, 300, 308). The cell divides near one of the poles, trapping the origin-proximal one third of a chromosome in the smaller prespore and leaving one chromosome and two thirds of another in the larger mother cell (Fig. 3) (305). As a consequence, the two cells are now genetically asymmetric. This asymmetry has important implications for compartmentalization of gene expression (54, 84), as discussed below. The developing organism also has a major challenge in that it must ensure that the prespore compartment receives a complete chromosome The transfer of the origin-distal two-thirds of a chromosome from the mother cell into the prespore is concomitant with the activation of different transcription factors in the prespore and mother cell. Although both of these events are thought to occur simultaneously, for the sake of clarity we will first discuss the problem of DNA transfer and then turn to the compartmentalization of gene expression.
The critical locus for chromosome translocation is spoIIIE. In spoIIIE mutants, the origin-distal two-thirds of a chromosome remains trapped in the mother cell (305). In spo+ strains, the remaining DNA is actively pumped across the asymmetric septum from the mother cell into the prespore (303, 310). The DNA translocase SpoIIIE localizes to the center of the sporulation septum (264, 304), apparently by anchoring to the chromosome (20). SpoIIIE then uses ATP to transport the chromosome into the prespore (Fig. 3) (17).
The question remains why DNA is transported only from the mother cell into the prespore. Does the location of the chromosome origin in the prespore determine the direction of transfer? Or is it the orientation of SpoIIIE in the center of the spore septum, toward or away from the prespore? When discussing this question, it is important to note that SpoIIIE is widely conserved in nonsporeformers (267), is expressed vegetatively in B. subtilis (82), and is involved in chromosome partitioning in circumstances other than sporulation. For example, SpoIIIE removes trapped nucleoids from minicells, prevents chromosome bisection when DNA replication is transiently inhibited (267) and when cells lack SMC (24), and is required for efficient partitioning in mutants defective in terminating DNA replication or resolving chromosome dimers (164). These situations require either bidirectional movement or transfer out of the smaller of two cells (the minicell), suggesting that SpoIIIE lacks an inherent polarity.
Two recent studies have attempted to address the question of the polarity of SpoIIIE during spore formation by synthesizing SpoIIIE exclusively in either the prespore or mother cell (36, 265). Both laboratories agree that expression of spoIIIE only in the mother cell is sufficient to obtain DNA translocation into the prespore and substantially to restore spore formation. However, they disagree about the effect of expression only in the prespore. There were a number of technical differences between the two studies that could account for their different conclusions. As a consequence, more work may be necessary to fully understand how polarity of DNA transfer by SpoIIIE is regulated.
COMPARTMENTALIZATION OF GENE EXPRESSION
After axial filament formation and asymmetric division and concomitant with DNA transfer by SpoIIIE, different programs of gene expression are established in the prespore and the mother cell. These programs are directed by cell-specific σ factors whose activation is coupled to landmark morphological events. Asymmetric division triggers the activation of σF in the prespore, followed by activation of σE in the mother cell. Later in sporulation, the completion of engulfment of the prespore by the mother cell leads to the activation of σG in the prespore and σK in the mother cell. Therefore, in addition to complete spatial compartmentalization between prespore and mother cell, sporulation gene expression is also divided into temporal (pre- and postengulfment) phases. Throughout the intermediate and late stages of sporulation, the mother cell and prespore communicate with each other, sending and interpreting biochemical signals to ensure that their genetic programs are coordinated (Fig. 2).
In the sections that follow, we briefly review the historical development of evidence that gene expression is indeed compartmentalized. We then focus on the activation of the particular σ factors that have been shown to direct the compartmentalized gene expression. It is presumed that those activation mechanisms hold the key to why activation is compartmentalized. Whereas we now consider the evidence that gene expression directed by the different σ factors is compartmentalized to be compelling, our understanding of the mechanisms of compartmentalization is still incomplete. A variety of regulators of σ activation have been identified. Some of the regulators have an essential function in spore formation, so that their mutational inactivation eliminates spore formation. However, other regulators appear to have partially or completely overlapping functions, so that mutational inactivation of only one regulator may have little or no effect on spore formation. Regulators of both types may be critical for compartmentalization of the activity of the different σ factors.
Developing Evidence that Gene Expression Is Compartmentalized
The different fates of the prespore and the mother cell suggested that different genes are expressed in the two compartments. This suggestion was supported by biochemical characterization of extracts enriched for the contents of the prespore or the mother cell (5, 55, 88, 269). The first clear evidence that expression of spo loci was compartmentalized came from the study of genetically mosaic bacteria (43, 226). The mosaics were obtained by transforming spo mutants at the start of sporulation so that only one of the two copies of the chromosome became spo+. After division, mutant and wild-type alleles were distributed randomly into the prespore and the mother cell. Since the mother cell is destroyed and only the chromosome in the prespore is inherited upon spore germination, it was possible to infer the location of spo locus expression. For several loci, the spores obtained gave rise to spo mutant progeny, indicating that only the mother cell chromosome required a spo+ allele for sporulation to occur. Other loci that were tested yielded only spo+ spores, indicating that, for sporulation to occur, the allele on the prespore chromosome had to be transformed to spo+ (43, 226, 230). Subsequently, determination of β-galactosidase activity in prespore- and mother cell-enriched extracts from strains expressing spo-lacZ transcriptional fusions provided strong support for differential gene expression between mother cell and prespore (reviewed in reference 59).
Direct evidence that the expression of particular genes was completely compartmentalized was obtained by the use of immunoelectron microscopy with antibodies to small acid-soluble proteins (SASPs), which revealed that these proteins are found exclusively within the prespore (83). The utility of this technique was expanded by using antibodies to β-galactosidase on samples from strains expressing spo-lacZ transcriptional fusions (48, 189). The experiments demonstrated that the activities of σF and σG were confined to the prespore and those of σE and σK were confined to the mother cell (48, 83, 189).
Although informative, immunoelectron microscopy is time-consuming and difficult and suffers from low sensitivity. The study of compartmentalization took a leap forward through the use of immunofluorescence microscopy and then fluorescence microscopy of cells expressing transcriptional fusions to gfp (encoding green fluorescent protein [GFP]). These techniques provided greater sensitivity and ease of use than electron microscopy, and GFP studies have the added advantage that living cells can be analyzed. The use of these techniques demonstrated that σF and σE became active very soon after completion of septum formation and that the activities were completely compartmentalized into the prespore and mother cell, respectively, within the limits of detection. Likewise, they demonstrated that σG and σK became active very soon after completion of engulfment, also completely compartmentalized into the prespore and the mother cell, respectively (105, 169, 233, 299, 314). A two-part transcription probe provided a very different type of evidence for the completeness of compartmentalization and also provided evidence that the vegetative σ factor σA continues to be active in both prespore and mother cell throughout sporulation (173).
ACTIVATION OF σF
spoIIA Operon
Studies of the spoIIA locus have been critical to our understanding of compartmentalization of gene expression. The locus was found to be a tricistronic operon (80, 229) that was transcribed prior to asymmetric septation (93, 218) in a Spo0A- and σH-dependent manner (290, 301, 302). When the spoIIA operon was sequenced, none of the open reading frames bore obvious similarity to any genes in the limited database of the time. However, shortly thereafter, it became clear that the third gene in the operon, spoIIAC, encoded a product that was homologous to an RNA polymerase σ factor (62, 274), named σF (183). The first targets identified for E-σF action were spoIIIG and gpr (282, 285), and the site of expression of spoIIIG was shown to be the prespore (93, 142, 189), as it has been for all E-σF-directed genes analyzed subsequently (231).
Posttranslational Regulation of σF
Although the spoIIA operon is expressed prior to asymmetric division (93, 218), σF does not become active until after asymmetric division (93, 142). It seemed likely that σF was subject to some form of posttranslational regulation, and genetic analysis revealed that the other two products of the spoIIA operon, SpoIIAA and SpoIIAB, regulate its activity. Thus, overexpression of SpoIIAB inhibited σF activity, and mutation of SpoIIAB increased σF activity. Mutation of SpoIIAA blocked σF activation, but activity could be restored to these strains by mutation of SpoIIAB. Taken together, these results indicated that SpoIIAB antagonized σF, and that in turn, SpoIIAA might antagonize SpoIIAB (260). Consistent with an inhibitory role for SpoIIAB, a separate study showed that mutation of SpoIIAB caused hyperactivity of σF, blocked sporulation prior to asymmetric septation, and caused extensive lysis (38). Given that the activity of σF was confined to the prespore during sporulation (189), it was hypothesized that its regulation by SpoIIAA and SpoIIAB was responsible for compartmentalization.
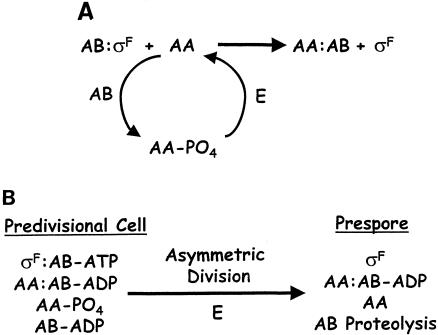
Biochemical analysis of the interactions between members of the regulatory pathway began to shed light on the mechanism of σF regulation. It was shown that the negative role of SpoIIAB was direct in that it bound σF, thus acting as an anti-sigma factor; SpoIIAA antagonized the action of SpoIIAB and so is an anti-anti-sigma factor (51, 199). SpoIIAB bore significant similarity to protein kinases, and it was found to phosphorylate SpoIIAA on a serine residue at position 58 (199, 207). Mutation of this serine residue to an alanine (mimicking dephosphorylation) resulted in constitutive σF activity, and mutation to an aspartic acid (mimicking phosphorylation) blocked σF activity in vivo (44), indicating that the phosphorylation state of SpoIIAA is critical for its ability to function as an anti-anti-sigma factor. Therefore, SpoIIAB inhibits σF both directly, as an anti-σ factor, and indirectly, by inactivating the anti-anti-σ factor SpoIIAA. SpoIIE, a membrane-bound serine phosphatase, dephosphorylates and thus activates SpoIIAA (8, 49). Therefore, it reverses the inactivation of SpoIIAA by SpoIIAB and promotes activation of σF. In addition, SpoIIE localizes to asymmetric division sites (10, 14), suggesting that it may mediate a link between asymmetric division and prespore-specific gene expression. The basic model of σF activation is illustrated in Fig. 4A.
FIG. 4.
Models of σF regulation. AA, AB, and E refer to SpoIIAA, SpoIIAB, and SpoIIE, respectively. The anti-σ factor SpoIIAB binds σF as a dimer but is represented here as a monomer for simplicity. (A)Basic model of σF regulation. The anti-σ factor SpoIIAB binds σF and holds it inactive. This inhibition can be reversed by the anti-anti-σ factor SpoIIAA. SpoIIAA is subject to regulation by its phosphorylation state; it is inactive when phosphorylated by SpoIIAB (a serine kinase as well as an anti-σ factor) and active when dephosphorylated by SpoIIE. Once dephosphorylated, SpoIIAA can bind SpoIIAB and liberate σF, activating prespore-specific transcription. In this model, the phosphorylation state of SpoIIAA is directly correlated with σF activity, and the fate of SpoIIAB after σF liberation and the nucleotide binding status of SpoIIAB are not considered. (B) Integrated model of σF regulation. In the predivisional cell, SpoIIAA and SpoIIAB are present in two forms: phosphorylation of SpoIIAA by SpoIIAB results in free phosphorylated (inactive) SpoIIAA and a SpoIIAA-SpoIIAB-ADP complex, while unreacted SpoIIAB-ATP forms an inhibitory complex with σF. As long as the level of dephosphorylated SpoIIAA remains below a certain threshold, it will be absorbed by the SpoIIAB-ADP sink. Asymmetric division triggers activation of σF in the prespore through three possible mechanisms: generation of excess dephosphorylated SpoIIAA so that the sink can no longer absorb all of it, sequestration of SpoIIAB in a long-lived complex with SpoIIAA, and proteolysis of SpoIIAB. Asymmetric division is thought to increase the level of dephosphorylated SpoIIAA either by activation of the phosphatase activity of SpoIIE or equivalent distribution of SpoIIE into both compartments, resulting in a much higher SpoIIE/SpoIIAA-PO4 ratio in the prespore. The complexes listed are not intended to reflect a stoichiometric biochemical reaction; rather, they reflect the different combinations thought to be formed by these factors and how they correlate with asymmetric division and activation of σF. The mother cell (not shown) is presumed to resemble the predivisional cell.
Mechanisms of Compartmentalization
Although the known factors involved in σF regulation have been identified, it is still not clear why σF activity is confined to the prespore. Models of compartmentalization have evolved over time; we will briefly review them in roughly the order that they were proposed. Although some of them are no longer widely accepted, we think that a discussion of their supporting data and potential flaws is useful.
ATP/ADP ratio.
Early in vitro experiments indicated that SpoIIAB bound σF in the presence of ATP, whereas it bound SpoIIAA in the presence of ADP (3, 44, 50). This result suggested that different concentrations of these nucleotides in the two compartments might be responsible for compartmentalization of σF activity to the prespore. However, it was not clear how the proposed differential ATP/ADP ratio was established in vivo, and direct evidence for the regulatory roles of these nucleotides was lacking. Subsequent in vitro studies of the interaction between these factors suggested that the ATP/ADP ratio was not a critical factor in determining the partner to which SpoIIAB bound (187).
Preferential inheritance.
As an alternative mode of regulation, it was proposed that a factor critical to σF activation was preferentially segregated to the prespore during sporulation. As evidence, it was reported that in protoplasts derived from sporulating cells expressing a SpoIIE-GFP fusion protein, the fluorescent signal was found in protoplasts derived from prespores and not from mother cells; it was inferred that SpoIIE became located predominantly in the prespore (309). However, a different study in which similar experiments were performed along with careful computerized quantitation determined that the total fluorescent signal was very similar in the prespore and in the mother cell. Therefore, it was concluded that the previous result was the consequence of similar amounts of protein being present in two compartments of dramatically different sizes. In addition, time-lapse microscopy of living cells clearly demonstrated that SpoIIE-GFP is present in both the mother cell and the prespore when first formed (150), indicating that preferential inheritance of SpoIIE into the prespore is not a primary determinant ofcompartmentalization.
Inhibitor.
One assumption that had been made in models of σF regulation was that localization of SpoIIE to the asymmetric division site was critical for its role in σF activation. When the N-terminal transmembrane domains of SpoIIE were removed, the protein failed to target to the asymmetric division site, yet sporulation was only reduced by about 50% and about half of the bacteria displaying σF activity showed prespore-specific expression (9). This result contrasts with the effect of inactivating spoIIE, where sporulation is reduced at least 107-fold (228) and σF activity is abolished (9) and indicated that when the location of SpoIIE in the cell was disturbed, some other mechanism functioned to compartmentalize σF activity. It was suggested that a cytoplasmic inhibitor of SpoIIE was able to regulate the soluble SpoIIE protein and prevent it from becoming active in the mother cell. However, the putative inhibitor has yet to be identified. It is important to note that a separate study, using a different mutant of SpoIIE that became solubilized during sporulation, found similar results (73) but interpreted them as supporting the preferential inheritance model (309).
Transient genetic asymmetry.
Studies of chromosome partitioning during sporulation helped generate a new concept for compartmentalization studies. Analysis of the effects of spoIIIE mutations revealed that at the time of asymmetric division, the prespore and mother cell contained different sets of genes in that only the origin-proximal one-third of a chromosome was present in the prespore, whereas the mother cell contained one complete chromosome and the origin-distal two-thirds of another (Fig. 3) (305). It takes about 15 min before the prespore receives a complete chromosome (148, 232). It was proposed that this transient genetic asymmetry could be the key to establishing compartmentalized gene expression (84). This proposal was tested by placing the gene encoding σF at the amyE locus very near the origin of replication, so that it was present in the prespore at the time of asymmetric division, and leaving spoIIAB near the terminus, so that it was initially absent from this compartment (Fig. 3). This arrangement of genes supported a modest level of sporulation even in the absence of the normally essential factors SpoIIAA and SpoIIE (84). These results clearly suggested that transient genetic asymmetry could play a role in compartmentalization of σF activity. Fransden and colleagues favored the existence of a gene near the terminus that encoded a cytoplasmic inhibitor of SpoIIE (84), so that transient genetic asymmetry would deplete this factor from the prespore. Although this putative factor remains unidentified, the concept of transient genetic asymmetry has had a dramatic impact on studies of compartmentalization.
A separate study addressed the role of transient genetic asymmetry by moving the entire spoIIA operon from its normal chromosomal location near the terminus to the amyE locus near the origin of replication (Fig. 3). Although this relocation, in itself, had only a mild effect on sporulation (40 to 80% of the parental strain value), when it was combined with the mutant, cytoplasmic form of SpoIIE, which also had only a mild effect in itself (9), there was a synergistic effect, and sporulation was severely reduced (<1% of that of the parental strain) (54). Therefore, targeting of SpoIIE to asymmetric division sites and the natural chromosomal location of the spoIIA operon are partially redundant factors that contribute to activation of σF in the prespore. The question remained how the chromosomal position of spoIIA could compensate for mislocalization of SpoIIE.
SpoIIAB degradation.
In a parallel line of investigation, it was discovered that a C-terminal truncation of SpoIIAB severely impaired σF activation and sporulation. Immunoblotting revealed that this mutant protein was much more stable than the wild type, indicating a key role for the instability of SpoIIAB in regulation of σF. In the absence of its binding partners SpoIIAA and σF, a unique C-terminal motif of SpoIIAB (215) targets the protein for degradation by the ClpCP protease complex (214). It was proposed that the natural chromosomal position of spoIIA near the terminus and the instability of SpoIIAB result in the anti-sigma factor's being temporarily depleted from the prespore compartment following asymmetric division. By combining this result with the transient genetic asymmetry experiments, a holistic picture began to emerge. When SpoIIE is localized to the asymmetric septum and the spoIIA operon is located near the origin, SpoIIAA is efficiently dephosphorylated that it can overcome the increased concentration of SpoIIAB that is present in the prespore. Conversely, when the phosphatase activity of SpoIIE is diminished by its mislocalization, the natural chromosomal position of spoIIA near the terminus allows SpoIIAB to be depleted from the prespore by proteolysis. In both scenarios, σF activation and sporulation occur with high efficiency. Only when the phosphatase activity of SpoIIE is reduced via impaired localization and SpoIIAB in the prespore is replenished by the presence of the spoIIA operon in this compartment is the activation of σF, and thus sporulation, severely impaired (54, 214).
Although genetic asymmetry is emerging as an exciting new concept, it is important to note that the genetic asymmetry is only transient. The DNA translocase SpoIIIE acts to export the remaining two-thirds of the chromosome from the mother cell to the prespore, a process that is estimated to take as little as 15 min (148, 232). Furthermore, the half-life of SpoIIAB is about 30 min (214). Thus, the decline in SpoIIAB relative to σF is relatively small. Nevertheless, small changes such as this are presumably sufficient to initiate prespore-specific σF activation. Not only that, σF must be activated very rapidly after septum formation and, following σF, σE must be activated in the mother cell if formation of a second septum at the other end of the organism (the abortively disporic phenotype) is to be prevented (124); the second septum may be formed as soon as 10 min after the first (77, 232). It is as if the decision to activate σF is balanced on a knife edge; just a minor reduction in the concentration of SpoIIAB in the prespore is presumably sufficient to trigger a very rapid cascade of events that ensure activation of σF in the prespore, and only in the prespore.
Cell division.
An important question is how the phosphatase activity of SpoIIE is regulated (if at all) so that the level of dephosphorylated SpoIIAA remains low enough to prevent σF activation in the predivisional cell yet can be turned on so as to rapidly activate σF after asymmetric septation. An initial study utilizing a conditional mutation (div-355) in the late-acting essential cell division protein DivIC revealed that when asymmetric division was blocked, it prevented activation of σF (166). A second study found that when cell division was blocked at a very early stage in sporulating cells by depletion of FtsZ, SpoIIAA was found largely in the phosphorylated form. In contrast, when cell division was blocked at a late stage by the div-355 mutation, there was substantial dephosphorylation of SpoIIAA. However, in neither circumstance did σF become active. These results suggested a two-step checkpoint: interaction with FtsZ triggered the phosphatase activity of SpoIIE, but the resulting dephosphorylated SpoIIAA did not activate σF until asymmetric division was complete. This checkpoint could be either enforced or bypassed by modification of SpoIIE. A mutant allele of spoIIE, spoIIE48, caused a phenotype similar to that of div-355 in that there was substantial dephosphorylation of SpoIIAA and yet σF activation was blocked. Conversely, replacement of the N-terminal transmembrane domains of SpoIIE with those of the E. coli MalF protein resulted in delocalized protein that caused hyper-σF activity even when asymmetric division was prevented (150).
SpoIIE exhibits many properties expected of a “surveillance” protein that senses asymmetric division: it localizes to asymmetric division sites (10, 14) in an FtsZ-dependent manner (168), assists in the formation of polar Z rings (19, 149), and interacts with FtsZ (186). Consistent with the checkpoint model, a number of studies have found missense mutations in SpoIIE that uncouple asymmetric division from σF activation (32, 73, 111). Indeed, one study tested the in vitro phosphatase activity of the mutant proteins and found that it was similar to that of the wild-type protein (73), further supporting the concept that SpoIIE is subject to a phosphatase-independent regulatory step that couples asymmetric division to σF activation.
SpoIIAB sink.
Separate studies have clearly established that when asymmetric division was prevented, dephosphorylated SpoIIAA accumulated but σF remained inactive (73, 150). However, how the cell prevented the dephosphorylated SpoIIAA from activating σF in the predivisional cell remained mysterious. A recent study has provided evidence that SpoIIAB, it its ADP-bound state, can serve as a “sink” absorbing SpoIIAA in the cell until a threshold level is reached (32). Precisely why asymmetric division allows the threshold level of dephosphorylated SpoIIAA to be crossed in the prespore is unknown; some speculate that the phosphatase activity of SpoIIE is stimulated by asymmetric division (73), whereas others think that equivalent inheritance of SpoIIE in the two compartments creates a favorable SpoIIE/SpoIIAA-PO4 ratio in the prespore (32). Depletion of SpoIIAB from the prespore compartment as a consequence of transient genetic asymmetry (54, 214) would logically contribute to decreasing the threshold level in this compartment. Interestingly, it was also found that in vivo, the spoIIE48 mutation substantially impairs dephosphorylation of SpoIIAA (32), rather than not affecting phosphatase activity, as previously thought (150). Indeed, a screen for suppression of a similar SpoIIE mutation identified mutations in SpoIIAA, SpoIIAB, and SpoIIE itself, all of which restored sporulation by increasing the level of dephosphorylated SpoIIAA in the cell (32). Therefore, the second step of the checkpoint model, in which SpoIIE prevents dephosphorylated SpoIIAA from liberating σF (150), appears unlikely. Rather, it appears that the SpoIIAB-ADP sink fulfills this role in the cell.
Biochemical studies.
Another area of ongoing research is the nature of the complexes that SpoIIAB forms with its binding partners. The basic model is that SpoIIAB inhibits σF activity directly by binding to it and also indirectly by acting as a serine kinase that inactivates SpoIIAA by phosphorylating it (199, 207). The phosphatase activity of SpoIIE functions to reverse this reaction (49), enabling SpoIIAA to attack the SpoIIAB-σF complex and allowing σF to become active. However, additional biochemical and genetic experiments have revealed more subtle and complex interactions between these factors.
One of the products of σF activation is the catalytically inactive SpoIIAB-ADP complex, and it has been reported that the replacement of ADP by ATP in this complex is an especially slow reaction (206), so that SpoIIAB-ADP functions to sequester SpoIIAB in an inactive form. In addition, similar experiments have shown that an even longer-lived intermediary is SpoIIAA-SpoIIAB-ADP (162). Although the experiments were performed in vitro, subsequent genetic analysis has revealed that different mutant SpoIIAA proteins incapable of forming the SpoIIAA-SpoIIAB-ADP complex in vitro cannot activate σF in vivo; the mutant strains are Spo− (161), indicating that this mechanism of sequestering SpoIIAB is physiologically relevant. However, as described above, the same complex has been found to impair σF activation in the predivisional cell (and possibly mother cell) by absorbing dephosphorylated SpoIIAA (32). Understanding how the SpoIIAA-SpoIIAB-ADP complex functions both to inhibit σF activation by absorbing dephosporylated SpoIIAA and also to promote σF activation by absorbing SpoIIAB will require further study.
Structural studies of the complex formed by SpoIIAB and σF have allowed further elucidation of the mechanism by which SpoIIAA functions. First, it was determined that the inhibitory complex had the stoichiometry SpoIIAB2:σ1F (30). Next, determination of the crystal structure of a complex of the Bacillus stearothermophilus SpoIIAB and σF revealed that only one of the two SpoIIAB molecules in the complex bound σF (31). Because of the high degree of conservation of these molecules among sporeformers (277), it is reasonable to assume that the B. subtilis homologues form a similar complex.
It had previously been thought that since the same residues of SpoIIAB contact both SpoIIAA and σF (92), SpoIIAA competed with σF to bind SpoIIAB. However, these new results required a reevaluation of how SpoIIAA interacted with the SpoIIAB-σF complex. Experiments were performed with SpoIIAB heterodimers consisting of wild-type SpoIIAB and SpoIIABR20E, a mutant deficient in binding SpoIIAA (92). The experiments revealed that SpoIIAA interacted with the molecule of SpoIIAB not bound to σF and induced release of σF from the other SpoIIAB molecule, most likely by steric displacement (115). Another interesting result from this study was that a mutant of SpoIIAB deficient in its kinase activity (SpoIIABR105A) resulted in excessive levels of σF activity. This result is notable in light of a previous study that found, counterintuitively, that the kinase activity of SpoIIAB was essential for σF activation (91). It is currently thought that the phosphorylation reaction takes place following, and therefore can be uncoupled from, σF liberation.
Summary.
A great deal of genetic and biochemical evidence has been obtained about the pathway regulating σF activity. What originated as a relatively simple biochemical model (Fig. 4A) has become far more complicated (Fig. 4B). Presently, there appear to be a number of overlapping mechanisms: localization of SpoIIE to the asymmetric septum and its regulation by interaction with division proteins (9, 10, 14, 74, 75, 111, 150, 186),transient genetic asymmetry (54, 84), proteolysis of SpoIIAB (214), and a sink to absorb dephosphorylatedSpoIIAA (32) that also sequesters SpoIIAB (161). The major challenge for the future is to understand how all of these mechanisms (and potentially others) are integrated so that activation of σF is tightly coupled to asymmetric division and completely compartmentalized to the prespore. The large number of contributing mechanism highlights how critical it is for the developing organism to efficiently regulate the earliest-acting cell-specific transcription factor.
σF Regulon
Classical genetic analysis has revealed at least four spo loci directly regulated by σF: spoIIR, required for activation of σE in the mother cell (143, 179); spoIIIG, encoding the late prespore-specific transcription factor σG (282, 283); spoIIQ, required for expression of spoIIIG and for engulfment under certain conditions (178, 284); and spoIVB, a regulator of the late mother cell-specific σ factor σK (39, 96). The gene encoding a spore catalase required for hydrogen peroxide resistance, katX, is part of the σF regulon (12), as is gpr, encoding a protease specific for SASPs (see below) (285). In addition, three other genes with roles during sporulation have been identified as being regulated by σF: bofC, an additional regulator of σK activity (97), and two regulators of σF activity, lonB (4, 263) and rsfA (306). Therefore, primary functions of σF are to couple prespore- and mother cell-specific gene expressionand to direct synthesis of the late prespore transcription factor σG.
The σF regulon has been partially defined by chip array analysis. The analysis revealed 66 genes that were active during the middle part of sporulation and whose expression depended upon both Spo0A and σF (71). However, the approach identified genes responsive to both σF as well as the mother cell-specific transcription factor σE, so more detailed analysis may be necessary to more fully define all of the genes controlled by σF.
ACTIVATION OF σE
We now turn to the next compartmentalized transcription factor, σE, which is activated specifically in the mother cell following asymmetric septation. Activation of σE depends upon receipt of a signal from the prespore, providing the first example of communication between the two compartments during development. Activation of both σE and σF is coupled to asymmetric division. However, their activation mechanisms are very different. σF is synthesized in an active state and held inactive by an anti-sigma factor, in contrast, σE is synthesized in an inactive state and activated by proteolytic cleavage.
The first sporulation-specific σ factor be purified was σE, initially known as σ29 (103). Its isolation provided the first evidence that alternative σ factors were involved in sporulation and led to the hypothesis that a cascade of σ factors drove the developmental process (181). Immunoblotting experiments revealed that σE was synthesized specifically during sporulation. The anti-σE antiserum also detected a slightly larger protein, called P31, that was synthesized earlier than and thought to be a precursor of σE (292).
A combination of genetic and protein analysis revealed that σE and P31 (now called pro-σE) were the products of the spoIIG locus (279, 292). Pulse-chase and microsequencing experiments revealed that pro-σE was processed into σE by proteolytic removal of 27 residues from its N terminus (159, 278). Subsequently, the spoIIG locus was found to be a two-gene operon: the first gene (spoIIGA) is essential for the processing reaction (136, 147, 193), and the second gene, spoIIGB, encodes pro-σE, which is processed in a SpoIIGA-dependent manner into active σE. It was proposed that SpoIIGA was the processing enzyme and that its ability to act on its substrate (pro-σE) required the appearance of the sporulation septum, thereby constituting a developmental checkpoint (278). Mutational analysis revealed that mutants of pro-σE that were poorly processed could be suppressed by mutations in SpoIIGA, providing support for a direct role for the SpoIIGA protein in processing (225). However, this role has yet to be verified by in vitro experiments. Activation of σE was also found to depend on σF activity (143, 179). The pathway regulating σE activation is illustrated in Fig. 5.
FIG. 5.
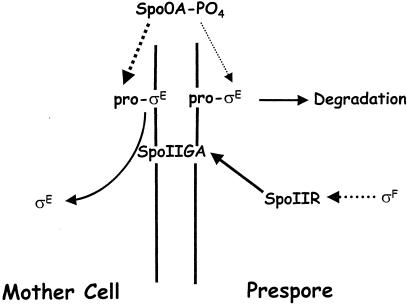
Regulation of σE activation. Parallel vertical lines separating the prespore (right) from the mother cell (left) represent the asymmetric septum. Broken arrows represent transcriptional activation, and solid arrows represent posttranslational regulation. Spo0A-PO4 is present in the predivisional cell as well as both compartments and therefore is represented above the sporulation septum. Pro-σE is synthesized in a Spo0A-PO4-dependent manner and therefore is present in both compartments; however, recent study has indicated that Spo0A-PO4-dependent transcription is largely confined to the mother cell after asymmetric division. This distinction is represented here by a thick line, indicating a high level of expression, in the mother cell and a thin line, indicating a low level of expression, in the prespore. Pro-σE, which is membrane bound, is processed into the active form, σE, by the inferred membrane-bound protease SpoIIGA. SpoIIGA becomes active in response to SpoIIR, whose expression is activated by σF. SpoIIGA is presumably present in both compartments, but σE becomes active only in the mother cell, at least in part because of the higher concentration of pro-σE in this compartment, as well as because of degradation of pro-σE in the prespore. The prespore specificity of SpoIIR expression may contribute to but is not critical for mother cell-specific activation of σE.
Compartmentalization
The spoIID locus was found to be exclusively transcribed by E-σE (246). Immunoelectron microscopy of cells containing a spoIID-lacZ fusion showed signal exclusively in the mother cell (48). This compartmentalization of σE-directed gene expression has been confirmed many times in subsequent studies (reviewed in references 231 and 281). As with σF in the prespore, a major question to be answered was how the activity of σE was confined to a specific compartment. The spoIIG operon is expressed before septation (93, 218), suggesting that posttranslational regulation played a role in its compartmentalization. Since σE activation was dependent on σF activity (135), it was suggested that a σF-controlled gene encoded a signal from the prespore that triggered processing of pro-σE to σE in the mother cell (182, 268).
Intercompartmental signaling.
The link between σF activity and σE activation was identified as the spoIIR (or csfX) locus (143, 179). Mutation of spoIIR blocked the processing of pro-σE to σE. It was found to be the only σF-directed locus that was required for σE activation, and artificial induction of spoIIR rendered σE activation independent of σF (179, 314). The SpoIIR protein contains a putative signal sequence, suggesting that it may be secreted from the prespore into the intermembrane space separating it from the mother cell. As evidence for secretion, it was found that conditioned medium from bacteria engineered to express SpoIIR could trigger processing of pro-σE to σE when added to protoplasts expressing the spoIIG operon (119). It is thought that SpoIIR acts from the prespore to trigger SpoIIGA-directed proteolysis of pro-σE to σE, although no direct biochemical evidence yet exists to support this hypothesis.
Although the signal transduction pathway linking the compartments has now been identified, the basis for mother cell-specific activation of σE remains unclear. One possibility is that the prespore-specific location of spoIIR expression, in itself, directs that σE be active only in the mother cell. To test this possibility, spoIIR was removed from σF control and expressed prior to asymmetric division from the spoIIE promoter in strains with no σF. Despite this switch in time and location of SpoIIR formation, σE was activated only in the mother cell of the organisms that underwent asymmetric division (314). Interpretation was complicated because about half of the population did not form an asymmetrically located septum, and those organisms displayed uncompartmentalized σE activity. Apparently, if an asymmetric septum was formed before σE activation, σE activity was confined to the mother cell; however, if σE activation preceded septation, it prevented septation (314), a result consistent with the σE-directed block in division that normally prevents the abortively disporic phenotype (57, 124, 232). Overall, the results indicate that compartmentalization of σE activity in the mother cell can occur independently of σF and of compartmentalized spoIIR expression.
Protein localization.
Compartmentalization studies now focused on the subcellular location of the processing reaction. It was found that fusion proteins of SpoIIGA and pro-σE to GFP localize to the asymmetric septum (72, 138). Fractionation experiments revealed that pro-σE is found primarily in the membrane, whereas processed σE is found primarily in the cytoplasm (118). These results suggested that the processing reaction took place at the membrane; consistent with this conclusion, it was found that a pro-σE mutant that failed to localize to the cell membrane was not processed (139). Immunofluorescence with anti-σE antibodies (which also interact with pro-σE) revealed a signal at the cell membrane prior to asymmetric division, a concentrated signal at the asymmetric septum upon division, and finally a signal dispersed in the cytoplasm thereafter (118). Similar immunofluorescence experiments revealed very little signal in the prespore; most of the signal was confined to the mother cell after asymmetric division (234). In addition, pro-σE artificially produced in the prespore was not processed (140) unless SpoIIGA was also expressed in this compartment (141). These studies suggested that exclusion of either SpoIIGA or pro-σE from the prespore was a mechanism of compartmentalization. In support of this model, a fusion to GFP of the N-terminal 55 residues of pro-σE, encompassing the prosequence, localized to the sporulation septum and, in protoplasts formed after asymmetric division, was found predominantly in the mother cell (138). These results suggested that pro-σE is sequestered to the mother cell face of the asymmetric septum.
However, a study assaying the expression pattern and stability of a full-length, functional pro-σE-GFP fusion (as opposed to the truncated version in the previous studies) found that the fusion protein localized nonspecifically to the cell membrane. As in the previous studies, fluorographs showed a strong signal associated with the septum (86), but a similar signal was observed with the membrane stain FM4-64, indicating that there is no specific concentration of this protein at the asymmetric septum, in contrast to previous interpretations (118, 138). Rather, the strong signal probably resulted from the two septal membranes' being viewed end on, as disks (86). In addition, it was found that the fusion protein was preferentially degraded in the prespore and the spoIIG promoter was largely active in the mother cell after asymmetric division, providing two different mechanisms to concentrate pro-σE in the mother cell (86). These results provide an alternative explanation of the earlier report of pro-σE localizing to the mother cell face of the septum (138), namely, a large increase in mother cell-specific synthesis of pro-σE and prespore-specific degradation of pro-σE following asymmetric division. Since previous studies had shown that that spoIIG transcription commenced prior to asymmetric division (93, 218), a major question to be addressed was how spoIIG transcription became largely confined to the mother cell after asymmetric division.
Role of Spo0A.
In a continuation of the study discussed above, Fujita and Losick (87) found that a substantial increase in Spo0A activity followed asymmetric division and was confined to the mother cell. This result explains the increase in transcription that they had observed (86) for the Spo0A-dependent (257, 258) spoIIG promoter. Disrupting the pattern of Spo0A activity by expression of a Spo0A inhibitor in the mother cell or a constitutively active form of Spo0A in the prespore severely reduced spore formation, indicating that preferential mother cell-specific Spo0A activity is important for development. A challenge now is to understand how the increase in Spo0A activity is coupled to asymmetric division and confined to the mother cell. Fujita and Losick propose that transient genetic asymmetry excludes important phosphorelay genes from the prespore at the time of asymmetric division. However, a recent study found that dephosphorylated SpoIIAA, thought to be found largely in the prespore (171), inhibits Spo0A-dependent transcription (6). This result suggests a supplemental or alternative mechanism to transient genetic asymmetry, that dephosphorylated SpoIIAA generates a feedback loop that prevents expression of Spo0A-dependent genes, such as spoIIG, from occurring in the prespore. Further studies will be necessary to determine the role that this feedback inhibition plays in compartmentalization of spoIIG transcription and of σE activity.
Timing of activation.
Another focus of study has been the importance of timing of σE activation during sporulation. Although it was known that pro-σE processing required asymmetric septation (278) and σF-dependent spoIIR expression (143, 179), how these events coordinated proper timing of σE activation was unknown. It was found that expression of spoIIR in the predivisional cell, rather than in the prespore, had only a mild effect on spore formation (314) and on the timing of pro-σE processing and σE activity (86). Premature spoIIG expression from a constitutive rather than its normal Spo0A-induced promoter had little if any effect on these properties. However, constitutive expression of both spoIIG and spoIIR resulted in early processing and poor sporulation (86). Therefore, correct timing of expression of both spoIIG and of spoIIR contributes to the proper timing of σE activation; they provide partly redundant mechanisms to coordinate morphogenesis and gene regulation.
Another aspect of timing to consider is that cells that fail to activate σE frequently undergo asymmetric division at both poles of the cell and subsequently fail to sporulate. This second division results in an abortively disporic phenotype in which there is a three-chambered structure consisting of two prespores separated by a large central compartment (124, 228). These two asymmetric divisions occur in rapid succession, with as little as 10 min separating them (77, 232). Genetic and cell biological experiments have revealed that three σE-directed genes that are required for engulfment of the prespore by the mother cell are also responsible for preventing the second division in the mother cell (57, 232). The rapid succession of the two divisions indicates that the σE-dependent mechanism to impair asymmetric division must be activated very rapidly after the first division to prevent the second. Moreover, the first division is needed in order to activate σF, so that spoIIR is transcribed and hence σE is activated (143, 179). Thus, some σE must become active in the mother cell very shortly after septation, probably before the large postseptation increase in Spo0A-directed spoIIG transcription can have an effect.
Regulation by gene position.
Similar to the role that the chromosomal position of the spoIIA operon plays in σF activation (54), one of the ways in which the cell ensures proper timing of σE activation is the chromosomal location of the spoIIR gene. The spoIIR gene is located very near the origin of replication, so that it is initially present in the prespore at the time of asymmetric division and will be immediately transcribed by RNA polymerase with σF (143, 179) (Fig. 3). Placing spoIIR at a location near the terminus, so that it must be imported into the prespore by the DNA translocase SpoIIIE (17, 303, 305) to become accessible to σF, curtailed its expression (148, 320). This relocation also resulted in a severe reduction in σE activity and in sporulation, and many cells in the population exhibited the abortively disporic phenotype. Therefore, the chromosomal position of spoIIR is important for the proper timing of σE activation, prevention of the second asymmetric division in the mother cell, and efficient sporulation (148, 320).
In summary, much is now known of the signal transduction pathway governing mother cell-specific gene expression (Fig. 5). The discovery that Spo0A activity is largely confined to the mother cell after septation (87) is an important recent contribution to our understanding of compartmentalization of σE activity. The mechanism for the partial compartmentalization of Spo0A remains unknown. Genetic asymmetry, already known to contribute to compartmentalization of σF activity and timing of σE activation (54, 148, 320), has been suggested as a mechanism to partially compartmentalize Spo0A activity (87). Alternatively, or additionally, differential localization of phosphorelay components could play a role; a precedent for this scenario can be found in Caulobacter crescentus development (196). In addition, the role of a novel inhibitory feedback loop involving SpoIIAA (6) has yet to be explored. The protein(s) responsible for degradation of pro-σE in the prespore remains unknown; functional analysis of the σF regulon should facilitate its identification.
σE Regulon
Members of the σE regulon that are important for sporulation include spoIID, spoIIM, and spoIIP, which are required for engulfment and to prevent a second asymmetric division from occurring in the mother cell (1, 37, 57, 85, 228, 232, 246, 271, 272); spoIVA, cotE, and spoVID, which encode scaffold proteins for spore coat assembly (18, 245, 273, 317, 318); the spoIIIA operon, which is required for activation of the late-prespore specific sigma factor σG (125, 146); sigK, the composite gene for the late mother cell-specific transcription factor σK (155, 280); and spoIVCA, the gene for the recombinase that generates sigK via a chromosomal rearrangement (155, 235, 256). In addition, σE activates transcription of spoIIID, which encodes a regulator of some σE-dependent genes (102). As a result, early mother cell-specific gene expression is divided into an initial phase, when certain genes responsive to σE alone are expressed, including spoIIID, and a later phase, when SpoIIID represses some σE-controlled genes and activates transcription of additional σE-controlled genes.
The main functions of σE are to prevent asymmetric division in the mother cell, to trigger engulfment of the prespore, to initiate spore coat assembly, and to direct synthesis of the late mother cell-specific transcription factor σK. It should be noted that functional conservation of σE has been observed in a range of species of Bacillus and Clostridium (7). In Bacillus thuringiensis, the cry1A(a) gene, encoding a protoxin crystal protein, is expressed in the mother cell under the control of σE as well as σK (2), and in Clostridium perfringens, the enterotoxin gene cpe is most likely also under the control of σE and σK (315).
The σE regulon has been defined by microchip array in two independent studies (58, 75). One study found that 253 genes (in 121 operons) are regulated by σE, and 46 of these were present in all endospore-forming bacteria whose genomes have been sequenced and absent from the genome of the related nonsporeformer Listeria monocytogenes. Null mutations in 12 of 98 previously undefined genes or operons caused substantial defects in sporulation; fusions of several of the encoded proteins to GFP showed localization to the outer prespore membrane (58). The other study found 171 genes under the control of σE, and functional analysis of this group found five novel genes required for efficient sporulation (75). The two studies agreed well and, in addition to many novel genes, identified most of the previously described σE-directed genes.
ENGULFMENT OF THE PRESPORE BY THE MOTHER CELL
The major morphological event that follows activation of σF and σE is engulfment of the prespore by the mother cell. After asymmetric division, the prespore and mother are adjacent and are in direct contact with the medium. Completion of engulfment results in the prespore's being entirely surrounded by the mother cell and so not in direct contact with the medium (Fig. 1). It should be noted that the word engulfment has been used in two senses in studies of spore formation, which at times can cause confusion. The first sense means the process of engulfment, and the second means the completion of engulfment (228). In terms of mutant designation, substages IIii and IIiii are in the process of engulfment (124), whereas stage III denotes completion of engulfment (124, 228). The activation of the late cell-specific sigma factors (σG and σK) is coupled to completion of engulfment, as activation of the early factors (σF and σE) is coupled to asymmetric division (231). Therefore, a discussion of engulfment is important for an understanding of compartmentalization, because there is compartmentalization between pre- and postengulfment (in the sense of completion of engulfment) gene expression.
Initiation of Engulfment
The process of engulfment starts with changes in the sporulation septum. The first observed change is loss of wall material from the center of the septal disk, followed by loss of much of the wall material of the septum. After this autolysis has occurred, the points of attachment of the septal membrane to the peripheral cell membrane migrate to the cell pole. Engulfment is completed by membrane fusion, resulting in the detached prespore's being entirely within the mother cell (Fig. 1) (124, 178, 227). Expression of both prespore-specific and mother cell-specific loci is required for engulfment. spoIID (37, 228), spoIIM (271), and spoIIP (85) mutants display autolysis and membrane flexibility at the center of the spore septum but not its periphery. spoIIQ mutants display a block just prior to completion of engulfment (178). Loci expressed before septation are also involved in engulfment (a spoIIB spoVG double mutant displays aberrant septal wall autolysis) (190, 224), and the spoIIIE locus has been shown to be required for the membrane fusion event (264, 266).
Transcription of the spoIID (246), spoIIM (272), and spoIIP (85) loci is directed by σE, suggesting that initiation of engulfment is driven by proteins synthesized in the mother cell. Consistent with a direct role in engulfment, GFP fusions with all three of the encoded proteins localized to the asymmetric septum and engulfing prespore membrane (1, 57). In addition, these proteins degraded partial septa and prevented a second division in the mother cell (57, 232). SpoIID bears homology with a modifier of cell wall hydrolases (157, 180). All of this indirect evidence suggests that the proteins function to degrade peptidoglycan, both to enable membrane migration around the prespore and to maintain asymmetry by preventing division in the mother cell. A recent study has shown that purified SpoIID degrades bacterial cell wall in vitro (1), providing a biochemical link with the genetic and cytological data regarding this class of proteins. In addition, this study revealed that leaky spoIID and spoIIP mutants were impaired in both autolysis and membrane migration, suggesting that these two processes are mechanistically linked (1).
The spoIIB gene, which is expressed in the predivisional cell (190), was initially thought to be required for engulfment and sporulation (37, 228). It later became clear that it is only required in strains in which a second gene expressed in the predivisional cell, spoVG (319), has been disrupted (190). Single mutations in either spoIIB and spoVG cause a very mild defect, whereas the double mutant displays little autolysis and sporulates poorly (190, 224). SpoIIB localizes to the asymmetric septum and bears weak homology with an amidase, suggesting a direct role in engulfment (190, 224). The function of SpoVG in engulfment is unclear; it appears to play some role in inhibiting asymmetric division (195, 224). The defects in double spoIIB spoVG mutants can be partially suppressed by mutation of a third gene, spoVS (241). However, the complex relationships between these genes have yet to be fully explored.
Completion of Engulfment
After autolysis and membrane migration around the prespore, the final stage of engulfment is membrane fusion. The early engulfment processes are largely driven by proteins in the mother cell. However, the SpoIIQ protein, which is expressed in the prespore from an E-σF-directed gene, is required for membrane fusion. SpoIIQ is predicted to be largely extracellular and has homology with endopeptidases, suggesting a direct role in engulfment. Consistent with this suggestion, an epitope-tagged SpoIIQ protein localized to the center of the asymmetric septum (178). Subsequently, SpoIIQ was found to be necessary for engulfment when sporulation was induced by nutrient exhaustion in rich medium but not when sporulation was induced by nutrient replacement in minimal medium (284). SpoIIQ is nevertheless required for sporulation in both conditions, one reason being that spoIIIG, encoding σG, is not transcribed in its absence. The link between SpoIIQ and spoIIIG transcription is unclear but most likely indirect (284).
A novel cytology-based screen revealed that the SpoIIIE protein, which is required for DNA transfer into the prespore (17, 303), is also required for membrane fusion (264). This result was surprising in light of earlier studies that found that spoIIIE mutants formed apparently fully engulfed, though unstable, prespores (121, 228, 275). While SpoIIIE initially localizes to the center of the asymmetric septum (304), it then travels as a focus along the engulfing membrane, finally coming to rest at the extreme pole of the cell, concomitant with membrane fusion at that location (264). In addition, a SpoIIIE mutant in which the putative ATP binding site is disrupted was found to be defective for DNA translocation but competent for membrane fusion, thereby uncoupling the two functions of the protein (264). Potentially, SpoIIIE establishes a checkpoint to ensure that chromosome segregation is completed prior to membrane fusion.
A final, puzzling finding is that mutations in the genes encoding the E1β and E2 subunits of the pyruvate dehydrogenase complex block sporulation just prior to and after the completion of engulfment, respectively. Although changes in medium composition can rescue other aspects of the defects in these mutants, they cannot restore sporulation, suggesting a sporulation-specific function independent of their enzymatic activity (90). More work will be necessary to determine what role these proteins play in engulfment.
Although several proteins involved in engulfment have been identified, biochemical analysis of the process has only recently been initiated. Several questions need to be addressed. What is the mechanism by which engulfment proteins, and the associated autolysis, are confined to septa so as not to damage the cytoplasmic cell wall? What ensures that autolysis starts in the middle of the septal disk and not its periphery, as during vegetative division? Does the chromosome's traversing the septum provide an initiating target for the autolysis? Lastly, it is likely that a major focus of study will be how the cell couples the completion of engulfment to the late stages of cell-specific gene expression, and this is discussed below.
LATE PRESPORE-SPECIFIC TRANSCRIPTION FACTOR σG
Completion of engulfment of the prespore by the mother cell signals the next phase of compartment-specific gene expression. However, synthesis of the associated σ factors commences earlier. σF directs synthesis of the late prespore transcription factor σG, and σE directs synthesis of the late mother cell transcription factor σK. Since σG is made only in the prespore and σK is made only in the mother cell, compartmentalization between prespore and mother cell is not a major focus of study. Rather, the focus of study is compartmentalization in the sense of activation after completion of engulfment but not before. σG and σK are both subject to complex regulation at different levels, and their activities are tightly coordinated with each other and with cellular morphogenesis. Activation of σK depends on σG, so it is σG activation that is most immediately tied to completion of engulfment, providing another developmental checkpoint to coordinate morphogenesis and gene regulation.
The first insight into the role of σG was provided by the study of small acid-soluble proteins (SASPs). These are present in high concentrations in the mature spore, and members of a major class, the α/β SASPs, bind to and protect DNA in the spore (213). Immunoelectron microscopy revealed that, during spore formation, several SASPs were found almost exclusively in the engulfed prespore (83). This finding suggested the existence of a prespore-specific transcription factor responsible for the compartmentalized pattern of SASP expression. Prespore-enriched fractions of sporulating cells contained a protein that supported in vitro transcription of sspE (encoding SASP-γ) and, when sequenced, was discovered to be the spoIIIG gene product and named σG (283). The spoIIIG gene had been previously identified as being located immediately downstream of spoIIGB, encoding a product with substantial homology to σ factors, and being essential for spore formation (142, 194). Consistent with the immunoelectron microscopy data, genetic analysis revealed that spoIIIG is expressed in a prespore-specific pattern (93, 142). The pathway for σG activation is illustrated in Fig. 6.
FIG. 6.
Regulation of σG activation. Two concentric semicircles represent the inner and outer membranes of the engulfed prespore. Broken arrows represent transcriptional activation, and solid arrows represent posttranslational regulation. The structural gene for σG, spoIIIG, is transcribed exclusively in the prespore under the control of σF (SpoIIAB [AB] is presumably inherited from the predivisional cell). Although not represented here, transcription also requires expression of SpoIIQ in the prespore and an unknown gene in the mother cell. Activation of σG does not occur until engulfment is complete. Activation requires release of inhibition by SpoIIAB. This release depends on the σE-directed expression of the spoIIIA operon in the mother cell. Activation of σG also requires SpoIIIJ, which is expressed vegetatively and localizes to the prespore membrane but need only be expressed in the prespore. It seems likely that some mechanism distinct from SpoIIAB inhibition keeps σG inactive prior to engulfment. The mechanism responsible for coupling activation to completion of engulfment remains unclear.
Transcriptional Regulation of spoIIIG
Transcriptional regulation of the spoIIIG locus is complex and may play an important role in coupling σG activation to engulfment. The spoIIIG locus is transcribed from the upstream spoIIG promoter during the early part of sporulation, but no detectable σG protein is produced from this transcript (194, 282), most likely because of a predicted RNA hairpin in the intergenic region (194). Moving spoIIIG to an ectopic locus does not impair spore formation, indicating that readthrough from the spoIIG promoter is not critical to σG expression (282); the role of this transcript remains obscure. The spoIIIG locus is also transcribed from its own promoter, recognized by RNA polymerase with either σF or σG; this transcript is translated, leading to active σG during the later stages of sporulation (282). Since the activity of σF is confined to the prespore (189), the presence of a σF-dependent promoter ensures that σG is present, and therefore active, only in the prespore (83, 142).
Transcription from the spoIIIG promoter differs from that of other σF-directed promoters in a number of ways. First, its expression depends upon σE activation in the mother cell (218), suggestive of a signaling pathway linking the two compartments. Transcription of spoIIIG also depends upon expression of SpoIIQ in the prespore, suggesting a potential link between σG activation and engulfment (284). Lastly, consistent with dependence upon these two events, expression occurs later than that of other known σF-dependent promoters (144). The importance of this complex transcriptional regulation in sporulation is unclear but most likely assists in proper timing of σG activation relative to early compartmentalized gene expression and morphogenesis. Because σG can direct transcription of its own structural gene, tight regulation is presumably required to prevent inappropriate initiation of an autocatalytic loop.
Activation of σG
Similar to other sporulation σ factors, σG is subject to posttranslational regulation. The first evidence was that spoIIIJ, a vegetatively expressed gene (60), and the spoIIIA operon, expressed in the mother cell (43, 125), were both required for σG activation but not spoIIIG transcription (60, 146). The anti-sigma factor for σF, SpoIIAB (38, 51, 260), has emerged as also acting on σG and indeed mediating the effects of spoIIIJ and spoIIIA. Fractionation studies revealed that SpoIIAB disappeared from the prespore at the time of σG activation. In addition, overexpression of σG resulted in toxicity that could be suppressed by simultaneous expression of SpoIIAB (151), and disruption of spoIIAB led to excessive levels of σG activity (38). These results suggested that SpoIIAB inhibited σG as well as σF during sporulation. As direct evidence for this hypothesis, it was shown that SpoIIAB bound σG in vitro, and a mutant form of σG that SpoIIAB bound poorly in vitro, SpoIIIGE155K, bypassed the requirement for spoIIIA for σG activation in vivo but not for sporulation (146). A recent study has shown that this mutant form of σG bypasses the requirement for spoIIIJ for σG activation but, once again, not for sporulation (262). The ability of this mutant to restore σG activity but not sporulation in these backgrounds indicates that either the spoIIIA and spoIIIJ locus plays an additional role in sporulation or that the restored σG activity is not properly regulated.
These results suggested a model in which SpoIIAB holds σG inactive until a signal is received from SpoIIIJ and at least one product of the spoIIIA operon. The question then is how SpoIIIJ and the products of the spoIIIA operon function to activate σG. Analysis has been hampered by the fact that the eight products of the spoIIIA operon, all predicted to be membrane bound, are not homologous to any known proteins (281). However, it was found that although SpoIIIJ is expressed vegetatively (60), expression only in the prespore, not in the mother cell, was sufficient for spore formation. In contrast, expression of the spoIIIA operon in the prespore did not support sporulation (262). SpoIIIJ is a homologue of the E. coli YidC protein (205), which is required for insertion of proteins into the lipid bilayer (254, 261). Consistent with its expression during vegetative growth (60), spoIIIJ is essential if its paralogue, yqjG, is disrupted (205), and depletion of both encoded proteins decreases the stability of membrane-bound and secreted proteins (288). In addition, SpoIIIJ localizes to the asymmetric septum and engulfing prespore membrane (205, 262). One possible role in σG activation is that SpoIIIJ is required for proper insertion and/or maintenance of a receptor in the inner prespore membrane that recognizes a signal from the mother cell.
The SpoIIAB pathway in part explains postengulfment σG activation, but several observations suggest that there must be some additional σG regulator that, minimally, prevents activation prior to engulfment (66, 262). Thus, the spoIIIGE155K allele that renders σG activation independent of SpoIIIJ and the products of the spoIIIA operon do not affect the timing of σG activation (146, 262). Furthermore, σF and σG are very similar and are both subject to regulation by SpoIIAB, with σF having much higher affinity for SpoIIAB in vitro; for both, binding is disrupted by SpoIIAA (66). It is difficult to see why σG is held inactive when σF is active if SpoIIAB is the only direct regulator (66). A major feature of σG regulation is that it only becomes active upon completion of engulfment (275, 276). The molecular basis for this linkage is unknown.
The complex transcriptional and posttranslational regulation presumably ensures that σG becomes active only in the prespore and only following engulfment. One possible reason for the complexity is that because the spoIIIG promoter is transcribed by RNA polymerase with σG (282), even a small amount of σG activity can lead to an autocatalytic loop that causes excessive levels of σG-dependent transcription. In support of this hypothesis, a mutation in lonA, encoding a putative Lon protease, resulted in high levels of σG activity under nonsporulating conditions, in which σF-directed transcription of spoIIIG does not occur (259). The complexity of the control is indicated by the observation that expression was postexponential and not constitutive in the lonA mutant; furthermore, neither expression in sporulating conditions nor spore formation was affected by the lonA mutation (259). Another reason for a multitude of controls may be the presence of the transcript of spoIIIG that originates from the upstream spoIIG promoter (194, 282). The spoIIG promoter is strongly activated in the mother cell by Spo0A (86, 87), and the cell must inactivate any σG that is produced from this transcript in order to prevent a feedback loop (282) and σG activation in this compartment.
In summary, the mechanism of coupling σG activation to the completion of engulfment is largely unknown. The mechanism underlying the dependence of spoIIIG transcription on σE activation (218) is also unknown. Likewise, the specific role of the eight products of the spoIIIA operon, all of which are thought to be membrane bound and are not obviously homologous to any known proteins, is unclear. The role of SpoIIQ in both engulfment (178) and spoIIIG transcription (284) is intriguing. One reason for our ignorance may be that genetic redundancy, as seen in σF and σE regulation, is preventing the isolation of strains with a single mutation that are blocked in σG activation or that have uncoupled activity from engulfment. Functional analysis of the σF (71) and σE (58, 75) regulons may help to overcome this problem and identify additional factors involved in σG regulation.
σG Regulon
At least three classes of genes are present in the σG regulon, those involved in sporulation, in germination, and in protecting the spore from DNA damage. The genes important for sporulation include spoIIIG, leading to an autocatalytic loop (142, 282); spoIVB, a serine peptidase that signals from the prespore to the mother cell to activate σK (39); the spoVA operon, which is required for dipicolinic acid uptake into the prespore from the mother cell (63, 202, 289); spoVT, a regulator of σG-dependent gene expression (13); and bofC, a regulator of pro-σK processing (97, 296). The germination genes include the gerA operon, involved with germination in response to alanine; the homologous gerB operon, involved with germination in response to a panel of germinants (l-asparagine, glucose, fructose, and potassium ions) (213); and pdaA, which is required for the formation of muramic δ-lactam, a unique component of spore cortex peptidoglycan (89). The genes for protection from DNA damage include splB, encoding spore-photoproduct lyase, which helps protect spore DNA from UV damage (68, 219); yqfS, encoding a type IV apurinic/apyrimidinic endonuclease (253, 293); and ssp genes, which encode SASPs, the predominant proteins of the spore core (107). Some of the SASPs bind the DNA in the spore and help protect it from exposure to heat, UV radiation, desiccation, and several other adverse conditions and also provide a source of amino acids upon germination. Transcription of all the ssp genes (sspA through sspP), except sspF and sspG, is directed by σG (107). Therefore, three main functions of σG are to couple late prespore and mother cell gene expression, to protect the spore from hazardous conditions, and to prepare the spore for germination. For detailed discussions of spore cortex formation and germination, we refer the reader to references 201, 213, and 286.
LATE MOTHER CELL-SPECIFIC TRANSCRIPTION FACTOR σK
Following the completion of engulfment and activation of σG in the prespore, σK becomes active in the mother cell. σK is only synthesized in the mother cell, so morphological coupling and compartmentalization into the postengulfment mother cell are the main focus of discussion. As is the case for σE and σG, activation of σK depends upon intercompartmental signaling (182, 231). Regulation of σK is similar to that of σE in that σK is synthesized as an inactive precursor, pro-σK, that is processed into its active form upon receipt of a signal from the prespore. However, the mechanisms of regulation are very different.
Developmental Chromosome Rearrangement
σK was first identified as a σ factor that could direct transcription in vitro of the late sporulation genes spoIVCB and cotD (152). However, the gene encoding σK was unknown. Genetic studies focused on the spoIIIC locus, which appeared to encode a small protein with high similarity to the C-terminal region of bacterial σ factors but lacking any corresponding N-terminal region (64). Interestingly, a different gene, spoIVCB, was found to encode a protein similar to the N-terminal region of a σ factor. These unusual findings were explained when it was shown that the region separating the two genes, approximately 48 kb in size, was excised during sporulation, resulting in the formation of a single, composite gene, sigK, encoding a full-length σ factor, σK (280). SpoIVCA appears to be the protein responsible for removal of the skin (sigma K intervening) element. It has substantial similarity to the Hin family of site-specific recombinases, binds to the processing site in vitro (235), and is the only sporulation-specific protein required for excision to occur in vivo (155). Transcription of both spoIVCA and the sigK gene is confined to the mother cell, initiated from a σE-dependent promoter requiring SpoIIID (102, 156, 255, 256).
Since the rearrangement generating sigK occurs only in the mother cell chromosome, which is not inherited, it could potentially be a novel mechanism of compartmentalizing gene expression. However, the presence of an intact sigK gene during growth and sporulation had no effect on development (155). Consistent with the dispensability of skin excision, other strains of B. subtilis and other species of sporeformers lack any inserted element in their sigK gene (2, 255, 277) with the notable exception of Clostridium difficile (see below) (104). However, similar to some other sporulation-regulatory elements, the role of skin excision was obscured by a genetic redundancy.
σK is synthesized as an inactive precursor, pro-σK (40, 152, 185). Removal of the prosequence (resulting in constitutively active σK) combined with artificial removal of the skin element resulted in some σK activity during vegetative growth and impaired sporulation (211). The vegetative expression is most likely because, like that of spoIIIG in the prespore (282), expression of the sigK gene is autocatalytic, being driven by a σK-dependent promoter (211). Interestingly, the sigK gene in Clostridium difficile also contains an insertion element, but it differs from the skin element in size, orientation, sequence, and site of insertion. The sigK gene of C. difficile differs from its B. subtilis counterpart in that it does not encode a prosequence and σK appears to be synthesized in an active state. Consistent with the skin element and prosequence playing redundant roles in regulation, it was found that expression of an intact sigK gene in trans in C. difficile resulted in poor sporulation (104). Presumably, regulation of σK occurs primarily via chromosomal rearrangement in this organism, whereas in B. subtilis there is redundancy in regulation; either prosequence processing or skin excision suffices to ensure the proper time and location of σK activation. Consistent with this reasoning, the chromosome of C. difficile lacks obvious homologues to known pro-σK processing components (277).
Pro-σK Processing
Based on the nucleotide sequence of sigK and the N-terminal sequence of σK, it was inferred that that the N-terminal 20 amino acid residues are removed from the sigK gene product of B. subtilis to generate active σK (152, 280). Immunoblotting revealed that pro-σK appeared approximately an hour earlier than processed σK, and processing depended upon gene expression in the prespore (40, 185). Therefore, similar to regulation of σE (159, 278, 279), it appeared that intercompartmental signaling resulted in an inactive proprotein's being processed into an active σ factor. This signaling pathway has been identified and characterized in detail (Fig. 7).
FIG. 7.
Regulation of σK activation. Two concentric semicircles indicate the inner and outer prespore membranes surrounding the engulfed prespore. Broken arrows represent transcriptional activation, and solid arrows represent posttranslational regulation. SpoIVB is expressed in the prespore under the control of σG and is thought to be inserted into the inner prespore membrane, where it undergoes autoproteolysis. σG also directs expression of BofC, which is an inhibitor of SpoIVB. In the mother cell, BofA, SpoIVFA, SpoIVFB, and pro-σK are all produced under the control of σE. SpoIVFB is thought to be the processing enzyme that acts upon pro-σK to generate active σK. BofA inhibits SpoIVFB. This inhibition is mediated by SpoIVFA, which acts to bring these proteins in contact with one another. Signaling by SpoIVB relieves inhibition of SpoIVFB, possibly by proteolysis, and so triggers pro-σK processing. Pro-σK is tethered to the outer prespore membrane by its N terminus, and it is thought that the processing reaction with SpoIVFB occurs within the membrane.
Mother cell processing components.
Genetic analysis revealed two mother cell-expressed loci important for regulation of pro-σK processing (41, 127). The first, spoIVF, is a bicistronic operon. spoIVFA encodes an inhibitor of pro-σK processing, whereas spoIVFB encodes a factor critical for processing (40, 41, 185). The second locus, bofA, also encodes an inhibitor of pro-σK processing (243). The three encoded proteins, SpoIVFA, SpoIVFB, and BofA, identified in these early studies remain the three main mother cell-expressed proteins known to be involved in pro-σK processing. SpoIVFB is considered the enzyme that cleaves the prosequence from pro-σK, and BofA and SpoIVFA act to negatively regulate this process (248).
There are multiple lines of evidence suggesting that SpoIVFB is the processing enzyme. Most compelling among these is the finding that expression of SpoIVFB is sufficient to trigger a low level of pro-σK processing during vegetative growth in B. subtilis as well as in E. coli (184, 242). SpoIVFB has been shown to share two motifs with mammalian site 2 protease, a zinc metalloprotease whose catalytic center is most likely membrane embedded. Mutations in conserved residues in these motifs in SpoIVFB abolished pro-σK processing, suggesting that pro-σK processing occurs within the membrane (247, 311). Consistent with this conclusion, biochemical and cytological experiments have indicated that pro-σK, SpoIVFB, SpoIVFA, and BofA all localize to the cell membrane (100, 240, 249, 294, 312). Models of how BofA and SpoIVFA effect the negative regulation of SpoIVFB action have undergone a series of changes, perhaps reflecting the complexity of the process (41, 100, 153, 242, 249). The most recent study suggests that SpoIVFA acts to bring BofA and SpoIVFB together in a heteromultimeric complex localized to the outer prespore membrane and that BofA is the direct inhibitor of SpoIVFB processing activity (249) (Fig. 7).
Prespore signaling.
Processing of pro-σK into mature σK was blocked in a strain lacking the late prespore transcription factor σG (40, 185), suggestive of signaling between the compartments. Several lines of evidence indicate that the signal is encoded by the spoIVB locus. Disruption of spoIVB blocks pro-σK processing, and transcription of spoIVB is largely dependent upon σG (39, 96). Transcription of spoIVB is the only function of σG that is required for pro-σK processing (95). The SpoIVB protein has a serine peptidase domain and a PDZ domain for protein-protein interactions; both domains are important for pro-σK processing and sporulation (116, 297).
It is thought that SpoIVB is inserted into the inner prespore membrane, where it undergoes autoproteolysis (297) to release signaling fragments that can diffuse across the intermembrane space and interact with the SpoIVFA-SpoIVFB-BofA complex inserted in the outer prespore membrane (249). A recent study found that SpoIVB degrades SpoIVFA in vitro (45), providing a possible mechanism of action. If this reaction occurs in vivo, it would release SpoIVFB from inhibition by BofA and trigger pro-σK processing (Fig. 7) (249). Interestingly, spoIVB mutants that show pro-σK processing but are Spo− have been obtained, suggesting that SpoIVB has an additional, unknown function in sporulation (212). Two other factors that play accessory roles in regulating pro-σK processing have been identified. The BofC protein, expressed in the prespore, inhibits SpoIVB autoproteolysis and thus pro-σK processing (296). The CtpB protein is similar to SpoIVB in that it contains PDZ and serine peptidase domains, although it is expressed in the mother cell under the control of σE. CptB is required for optimal efficiency of pro-σK processing (216).
In summary, the signaling pathway regulating σK activation is now well understood, and yet a number of issues remain. Processing of pro-σK has not been achieved in vitro. The proposal that SpoIVFA is proteolyzed by SpoIVB (45) needs to be tested in vivo. In addition, it is not known how the mother cell processing components localize specifically to the outer prespore membrane. It has been proposed that SpoIVFB is initially inserted into both the cell and prespore membranes and subsequently diffuses to and is retained by SpoIVFA only in the latter (250). However, the mechanism underlying SpoIVFA targeting to the outer prespore membrane remains unknown.
σK Regulon
The known genes in the B. subtilis σK regulon are involved in formation of the spore coat, spore maturation, and regulation of σK-dependent transcription. The regulon includes 14 cot genes, which encode proteins that make up the protective spore coat (47, 109); spoVD and spoVK, required for spore maturation (42, 69); and the structural gene for σK itself, leading to an autocatalytic loop (155). Expression of the transcriptional regulator gerE is directed by σK, dividing σK-dependent promoters into three classes: those solely regulated by σK are expressed throughout the late mother cell-specific stage, those that are repressed by GerE and are expressed early after σK activation, and those that require GerE and σK for activation and are expressed later (316). Detailed reviews of coat formation have appeared elsewhere (46, 47, 109). As mentioned previously, in other species expression of toxin genes is directed by σK: in B. thuringiensis, the cry1A(a) gene, encoding a protoxin crystal protein, is expressed in the mother cell under the control of σK as well as σE (2), and in C. perfringens, the enterotoxin-encoding gene cpe is most likely under the control of σK as well as σE (315).
TEMPORAL CONTROL AND COMPARTMENTALIZATION
The primary emphasis of our discussion has been on the spatial compartmentalization of gene expression associated with two key morphological events, completion of septation and completion of engulfment. However, an important aspect of development is temporal regulation of gene expression. Temporal control is exercised in part by coupling activation of σ factors to morphological events: σF, and hence σE, to septation and σG, and hence σK, to engulfment, as discussed above. The two prespore σ factors, σF and σG, have overlapping promoter specificities (4, 107), suggesting three temporal promoter classes in the prespore: those recognized by σF only, those recognized by σF and σG, and those recognized by σG only. A similar argument can be made for the mother cell, where σE and σK also have overlapping promoter specificities (107).
Additional layers of temporal control are exercised by activator and repressor proteins under the control of and associated with these σ factors. Such regulators have been more fully characterized for the mother cell. The SpoIIID protein acts as an activator of some and a repressor of other genes in the σE regulon and is itself a member of that regulon. Thus, SpoIIID divides the regulon into an early phase, when some genes are expressed, and a late phase, when other genes are expressed, and some genes of the first phase are turned down or off (102, 125, 152, 154). GerE effects a similar division of the σK regulon (122, 316). Further temporal division of the σK regulon was shown to result from the combined action of GerE and SpoIIID: SpoIIID repressed and so delayed the expression of some GerE-activated genes but not others (123). GerE also appears to be regulated, in part, through the action of the spore coat protein SpoVIF (158).
Different temporal classes are also found in the σF and σG regulons, but the mechanisms do not seem so clearcut (107). For example, the spoIIIG locus is expressed about 40 min later than other members of the σF regulon (144) and requires expression of spoIIQ (284) and some component(s) of the σE regulon (218). The gene for a repressor-activator, RsfA, has been identified in the σF regulon (306), but its role is less well defined than that of SpoIIID or GerE. Finally, in the σG regulon, the sspF locus is transcribed about 1 h after other members of the regulon (217). Two transcription regulators of σG-directed transcription have been identified, SplA and SpoVT (13, 68); they do not appear to have the substantial roles exercised by SpoIIID or GerE in their regulons and are not known to regulate sspF. In summary, there is a complex temporal progression of gene expression in both the prespore and the mother cell.
Analysis of expression of the σF-directed spoIIR locus indicated a very short period of transcription (148, 320), suggesting that transcription of some σF-directed genes stops before σG becomes active. This supposition was confirmed by using a two-part probe to test for compartmentalization of gene expression within individual cells (173). The result also provides clear evidence for spo gene expression being switched off during spore formation. It is not known if the temporal compartmentalization applies to other σF-directed promoters, or if it is tied to a morphological event such as the completion of engulfment, and the mechanism remains unknown. A similar switch was detected in the mother cell between the σE-directed cotEP1 promoter and σK-directed gerE expression. The mechanism is also unknown but is distinct from SpoIIID-directed repression (173). In general, the mechanisms of turning gene expression off during spore formation are much less well understood than those for turning expression on; their importance remains to be established.
SPORULATION OF COCCI
Sporulation has been studied almost exclusively in rod-shaped bacteria. However, one way to study the relationship between morphogenesis and gene expression is to extend studies to organisms with different shapes, notably cocci. Although coccal mutants of B. subtilis have been too sick to form spores (108), the coccal species Sporosarcina ureae is a naturally occurring sporeformer. S. ureae fits, by most criteria, into the genus Bacillus (70). The spores formed by S. ureae are structurally very similar to those of bacilli and clostridia (120). The sporulation septa of S. ureae resemble the sporulation septa of bacilli and clostridia in their ultrastructure and are very different from vegetative septa (244). However, the sporulation division in S. ureae is medially located with respect to cell poles, in contrast to the gross asymmetry of its location for bacilli and clostridia (244, 313). Nevertheless, compartmentalization of gene expression is observed in S. ureae, and it is inferred that spatial asymmetry is not required for compartmentalization of gene expression in this species (313). Thus, one plausible factor in the establishment of compartmentalized gene expression, gross volume asymmetry, is eliminated.
Despite the symmetry of location of the sporulation division, S. ureae has a homologue of the spoIIIE gene (35, 36) that encodes a DNA translocase critical for sporulation in B. subtilis (17, 303). This homologue is also essential for sporulation in S. ureae and can complement B. subtilis spoIIIE mutants (35, 36). Presumably the DNA translocase function of SpoIIIE is still required in S. ureae. Although the cells formed at division are similar in volume, successive division planes are at right angles to each other (22, 313). This change in direction requires that the chromosome reorient at each division. This reorientation may set the stage for a distinction in chromosome movement between the prespore and the mother cell. Conjecturally, transient genetic asymmetry (54) is still a factor in the establishment of compartmentalized gene expression in S. ureae. This conjecture raises related questions. Does trapping of the chromosome terminus region in a particular cell at septation determine that the cell will be the mother cell? Is that location predetermined by division history, or is the choice random? One intriguing possibility is that in the absence of volume asymmetry, genetic asymmetry becomes the predominant determinant of compartmentalization, so that its disruption would have a much more dramatic effect than is observed in B. subtilis (54).
DISRUPTION OF COMPARTMENTALIZATION
The spatial compartmentalization between prespore and mother cell of the activities of σF, σE, σG, and σK is, when first established and for the duration of spore formation, essentially complete. However, there are several mutant backgrounds in which this compartmentalization is disrupted. In all known examples, the mutants are impaired in their ability to form spores. This result is strong circumstantial evidence for the conclusion that complete compartmentalization of the activities of the four σ factors between prespore and mother cell is essential for spore formation, although it does not establish the conclusion unequivocally. The barrier that ordinarily prevents activation of σK before completion of engulfment appears to be important but not essential; mutants that show activation before completion of engulfment show reduced rather than no spore formation (40, 211). Below we discuss different mutations that disrupt the prespore-mother cell compartmentalization. The analysis of spoIIAB mutations (38, 260) was an important landmark in the elucidation of the regulation of σF activity. Otherwise, mutations disrupting prespore-mother cell compartmentalization have not been studied extensively; it may be that additional types of mutation can be identified and substantially more can be learned through their study.
The components for σF activation are present before septum formation, but σF normally only becomes active following septation. However, as discussed earlier, control of σF activation appears to be on a knife edge. Thus, mutations that inactivate SpoIIAB and cause hyper-σF activity (38, 260) result in uncompartmentalized σF activity. Indeed, the sporulation septum is not formed (38). Similarly, certain mutations in spoIIE result in hyper-uncompartmentalized σF activity and also prevent septation and spore formation (73, 111). Clearly, in these mutants σF becomes active before septation, and indeed a σF-directed gene appears to prevent formation of the sporulation septum (111).
Mutations that disrupt the spoIIIE locus result in loss of compartmentalization (171, 173, 234, 303-305). In contrast, many missense mutations in spoIIIE do not disrupt compartmentalization but rather result in hyper-prespore-specific expression of σF-directed genes located in the origin-proximal third of the chromosome and no expression of such genes located elsewhere in the chromosome (303-305). Both classes display a similar block in DNA translocation. However the mutant SpoIIIE protein of the latter class (class I) is located in the middle of the spore septum, as is wild-type SpoIIIE; no localization, and often no SpoIIIE protein, is detected with the former class (class II) of mutant (304). These observations led to the suggestion that SpoIIIE might form an effective seal around the DNA as it traversed the septum. In the absence of this seal, in class II mutants, small molecules, such as σF or SpoIIAA, could traverse the septum, resulting in a loss of compartmentalization (304). However, a separate study found that β-galactosidase could not diffuse across the septa of class II mutants and proposed that persistence of SpoIIE in the mother cell rather than a hole in the septum was responsible for the loss of compartmentalization (234). Our recent studies support the existence of a small hole permeable to σ factors or their regulators and to GFP (110).
A further puzzling observation was that whereas σF-directed gene expression was uncompartmentalized in class II spoIIIE mutants, it was again compartmentalized when a spoIIG mutation was introduced into such mutants (171, 234, 309), blocking σE activity. Given that σE-directed genes are required to cause septal wall autolysis during engulfment (1, 37, 57, 85, 228, 232, 246, 271, 272), it seemed plausible that similar activity might enlarge a breach in the septum resulting from loss of SpoIIIE. Indeed, σE-directed transcription of spoIID and spoIIP is required to disrupt compartmentalization in spoIIIE class II mutants (110). Presumably, autolytic activity associated with SpoIID and SpoIIP enlarges the putative hole sufficiently to allow passage of σF or its regulators and of GFP and possibly of σE.
Mutations in the spoIIIA and spoIIIJ loci have been known for some time to prevent activation of σG and hence σK (60, 146, 281). These mutations have recently been found to also cause a loss of compartmentalization of σF and σE activities (172). The activities of these σ factors are initially compartmentalized in the mutants, and compartmentalization is only lost aftercompletion of engulfment of the prespore by the mother cell. Thus, it appears to be a secondary consequence of the sporulation defect rather than the cause of the stage III blockage. It is likely that the loss of compartmentalization is a consequence of instability of the engulfed mutant prespores (172).
CONCLUSION AND FUTURE DIRECTIONS
Spore formation in B. subtilis is a simple developmental process (Fig. 1) that lends itself to genetic and cell biological analysis. B. subtilis monitors its intracellular and extracellular environment, integrating this information into a phosphorelay that regulates the initiation of sporulation. When conditions are appropriate, the action of the phosphorelay results in the phosphorylation and so activation of the master response regulator Spo0A. Active Spo0A and the transition regulator σH trigger global changes in gene regulation, setting the stage for formation of the axial filament and the asymmetrically located sporulation division. Formation of the axial filament requires the DNA-binding proteins RacA and Soj as well as the division protein DivIVA (Fig. 3). The asymmetric division requires greatly increased expression of ftsZ and induction of spoIIE, and as a consequence the cytokinetic protein FtsZ spirals from mid-cell to sites near the poles; division occurs at one of these sites, yielding the smaller prespore and the larger mother cell.
Compartmentalized activity of two sporulation-specific sigma factors commences very soon after the sporulation division, and it is now well established that compartmentalization is, within the limits of detection, complete: σF in the prespore and σE in the mother cell (Fig. 2). The first to become active is σF. A complex regulatory pathway has been elucidated for σF, centered on the anti-sigma factor SpoIIAB, the anti-anti-sigma factor SpoIIAA, and the protein phosphatase SpoIIE, which activates SpoIIAA by dephosphorylating it (Fig. 4A). Activation is associated with completion of the sporulation division septum and is confined to the prespore. When the sporulation septum is first formed, only the origin-proximal one-third of a chromosome is present in the prespore. This genetic asymmetry is resolved by SpoIIIE, which transfers the remaining portion of the trapped chromosome into the prespore (Fig. 3). It takes perhaps 15 min for the origin-distal two-thirds, including the genes for SpoIIAA, SpoIIAB, and σF, to be transferred. The transient genetic asymmetry during this time, the instability of SpoIIAB, the long-lived SpoIIAA-SpoIIAB-ADP complex sequestering dephosphorylated SpoIIAA, and the localization of the SpoIIE phosphatase to the septum have been shown to be important contributors to the prespore specificity of σF activation (Fig. 4B), although there is considerable redundancy in their contributions. What is not yet clear is why activation occurs so rapidly after septation and why σF activity is absolutely confined to the prespore. It is as if the system is very delicately balanced with respect to the prespore, and a slight push ensures prespore-specific activation, yet the system is robust in that normally there is no activation before septation and none in the mother cell.
Activation of the next factor, σE in the mother cell, requires prior activation of σF. Activation is by cleavage of a prosequence from pro-σE, probably through the action of the putative protease SpoIIGA. The activation of σE must occur rapidly (<10 min?) after formation of the sporulation septum in order to prevent the formation of a second asymmetrically located septum. Minimally, in that time σF becomes active and directs transcription of spoIIR; SpoIIR triggers the processing of pro-σE; σE directs transcription of spoIID, spoIIM, and spoIIP, whose products act to prevent the formation of the second septum. It is not clear how σE activation is achieved so rapidly and exclusively in the mother cell, although prespore-specific proteolysis of pro-σE is probably a contributory factor. Postseptation enhancement in the mother cell of Spo0A-directed transcription of spoIIG (encoding pro-σE) is an important contributing factor to the production of σE in the mother cell (Fig. 5), but it is not clear why Spo0A activity (and perhaps phosphorelay activity) is greatly enhanced in the mother cell and curtailed in the prespore, nor is it clear if this increased transcription could account for the rapidly induced, σE-directed suppression of septation.
Activation of pro-σE requires expression of the σF-directed spoIIR locus. This exemplifies a recurring theme, that compartment-specific gene expression is controlled by intercompartmental regulatory signals, a theme often referred to as crisscross regulation. Mother cell-specific activation of σE does not require that spoIIR be expressed in the prespore. Thus, the mother cell specificity of σE activity can be independent of the prespore specificity of σF action. These sigma factors direct compartmentalized gene expression but not the process of compartmentalization.
Completion of engulfment of the prespore by the mother cell starts the next phase of compartmentalized gene expression. It is associated with the action of a second pair of sporulation-specific sigma factors, σG and σK (Fig. 2) The first of these to become active, σG, does so exclusively in the prespore. Its prespore specificity can be explained, at least in part, by σF-directed transcription of spoIIIG, the structural gene for σG. However, it is not clear why activation occurs only after completion of engulfment. A chain of regulators involves the products of spoIIIA (made in the mother cell) and spoIIIJ (needed only in the prespore) antagonizing the anti-σG activity of SpoIIAB (Fig. 6), and yet the action of SpoIIAB is inadequate to explain why σG is not activated before completion of engulfment and why its activation is tied to that event. It is thought that a critical regulator remains to be identified. The morphological coupling of the activation of σF and σG to completion of septation and engulfment, respectively, is clear; the mechanisms are not.
The final σ factor to be activated during sporulation, σK, is also subject to complex regulation. First, in B. subtilis strain 168, though not in other strains or species (with the exception of C. difficile), there is mother cell-specific excision of an insertion element in sigK, the structural gene for pro-σK. There is then σE-directed transcription of sigK, also ensuring the mother cell specificity of σK. Finally, as with its predecessor in the mother cell, σE, σK is activated from an inactive prosequence form. It requires a complex of proteins synthesized in the mother cell together with a σG-directed signal from the prespore (Fig. 7). Mechanistically, the activation of pro-σK is very different from that of pro-σE. The final processing enzyme is thought to be a membrane-embedded metalloprotease, SpoIVFB.
The compartment-specific σ factors provide the bare framework of compartment-specific gene expression. However, within each σ regulon are several temporal classes of genes, some requiring additional activators and some subject to repressors. Some genes are clearly turned off before others become active. For key regulators, timing is critical. For example, accelerating the expression of σK or delaying the expression of spoIIR greatly impairs spore formation. The concept of transient genetic asymmetry emphasizes the importance of timing in the establishment of compartment-specific gene expression.
Compartmentalization of gene expression during bacterial spore formation is but part of the process of forming a heat-resistant, dormant spore. In our discussion of each of the sigma factors, we have only briefly mentioned the roles that their regulons play in building the mature spore. There is more to be done to understand compartmentalized gene expression during spore formation. There is very much more to be done to fully understand how this “simple” cell differentiation is achieved.
Acknowledgments
Work by the authors was supported in part by Public Health Service grants GM43577 (to P.J.P.) and T32AI07101 (to D.W.H.).
REFERENCES
- 1.Abanes-De Mello, A., Y. L. Sun, S. Aung, and K. Pogliano. 2002. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 16:3253-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, L. F., K. L. Brown, and H. R. Whiteley. 1991. Molecular cloning and characterization of two genes encoding σ factors that direct transcription from a Bacillus thuringiensis crystal protein gene promoter. J. Bacteriol. 173:3846-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alper, S., L. Duncan, and R. Losick. 1994. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell 77:195-205. [DOI] [PubMed] [Google Scholar]
- 4.Amaya, E., A. Khvorova, and P. J. Piggot. 2001. Analysis of promoter recognition in vivo directed by σF of Bacillus subtilis by using random-sequence oligonucleotides. J. Bacteriol. 183:3623-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreoli, A. J., S. Suehiro, D. Sakiyama, J. Takemoto, E. Vivanco, J. C. Lara, and M. C. Klute. 1973. Release and recovery of forespores from Bacillus cereus. J. Bacteriol. 115:1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arabolaza, A. L., A. Nakamura, M. E. Pedrido, L. Martelotto, L. Orsaria, and R. R. Grau. 2003. Characterization of a novel inhibitory feedback of the anti-anti-sigma SpoIIAA on Spo0A activation during development in Bacillus subtilis. Mol. Microbiol. 47:1251-1263. [DOI] [PubMed] [Google Scholar]
- 7.Arcuri, E. F., M. Wiedmann, and K. J. Boor. 2000. Phylogeny and functional conservation of σE in endospore-forming bacteria. Microbiology 146:1593-1603. [DOI] [PubMed] [Google Scholar]
- 8.Arigoni, F., L. Duncan, S. Alper, R. Losick, and P. Stragier. 1996. SpoIIE governs the phosphorylation state of a protein regulating transcription factor σF during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 93:3238-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arigoni, F., A. M. Guérout-Fleury, I. Barák, and P. Stragier. 1999. The SpoIIE phosphatase, the sporulation septum and the establishment of forespore-specific transcription in Bacillus subtilis: a reassessment. Mol. Microbiol. 31:1407-1415. [DOI] [PubMed] [Google Scholar]
- 10.Arigoni, F., K. Pogliano, C. D. Webb, P. Stragier, and R. Losick. 1995. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science 270:637-640. [DOI] [PubMed] [Google Scholar]
- 11.Autret, S., and J. Errington. 2003. A role for division-site-selection protein MinD in regulation of internucleoid jumping of Soj (ParA) protein in Bacillus subtilis. Mol. Microbiol. 47:159-169. [DOI] [PubMed] [Google Scholar]
- 12.Bagyan, I., L. Casillas-Martinez, and P. Setlow. 1998. The katX gene, which codes for the catalase in spores of Bacillus subtilis, is a forespore-specific gene controlled by σF, and KatX is essential for hydrogen peroxide resistance of the germinating spore. J. Bacteriol. 180:2057-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagyan, I., J. Hobot, and S. Cutting. 1996. A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J. Bacteriol. 178:4500-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barák, I., J. Behari, G. Olmedo, P. Guzman, D. P. Brown, E. Castro, D. Walker, J. Westpheling, and P. Youngman. 1996. Structure and function of the Bacillus SpoIIE protein and its localization to sites of sporulation septum assembly. Mol. Microbiol. 19:1047-1060. [DOI] [PubMed] [Google Scholar]
- 15.Barák, I., P. Prepiak, and F. Schmeisser. 1998. MinCD proteins control the septation process during sporulation of Bacillus subtilis. J. Bacteriol. 180:5327-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barák, I., and P. Youngman. 1996. SpoIIE mutants of Bacillus subtilis comprise two distinct phenotypic classes consistent with a dual functional role for the SpoIIE protein. J. Bacteriol. 178:4984-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bath, J., L. J. Wu, J. Errington, and J. C. Wang. 2000. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science 290:995-997. [DOI] [PubMed] [Google Scholar]
- 18.Beall, B., A. Driks, R. Losick, and C. P. Moran, Jr. 1993. Cloning and characterization of a gene required for assembly of the Bacillus subtilis spore coat. J. Bacteriol. 175:1705-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Yehuda, S., and R. Losick. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257-266. [DOI] [PubMed] [Google Scholar]
- 20.Ben-Yehuda, S., D. Z. Rudner, and R. Losick. 2003. Assembly of the SpoIIIE DNA translocase depends on chromosome trapping in Bacillus subtilis. Curr. Biol. 13:2196-2200. [PubMed] [Google Scholar]
- 21.Ben-Yehuda, S., D. Z. Rudner, and R. Losick. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532-536. [DOI] [PubMed] [Google Scholar]
- 22.Beveridge, T. J. 1980. Cell division in Sporosarcina ureae. Can. J. Microbiol. 26:235-242. [DOI] [PubMed] [Google Scholar]
- 23.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase σ factor σH regulon of Bacillus subtilis. J. Bacteriol. 184:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Britton, R. A., and A. D. Grossman. 1999. Synthetic lethal phenotypes caused by mutations affecting chromosome partitioning in Bacillus subtilis. J. Bacteriol. 181:5860-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Britton, R. A., D. C. Lin, and A. D. Grossman. 1998. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 12:1254-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burbulys, D., K. A. Trach, and J. A. Hoch. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545-552. [DOI] [PubMed] [Google Scholar]
- 27.Burkholder, W. F., and A. D. Grossman. 2000. Regulation of the initiation of endospore formation in Bacillus subtilis, p. 151-166. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. American Society for Microbiology, Washington, D.C.
- 28.Burkholder, W. F., I. Kurtser, and A. D. Grossman. 2001. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104:269-279. [DOI] [PubMed] [Google Scholar]
- 29.Bylund, J. E., M. A. Haines, P. J. Piggot, and M. L. Higgins. 1993. Axial filament formation in Bacillus subtilis: induction of nucleoids of increasing length after addition of chloramphenicol to exponential-phase cultures approaching stationary phase. J. Bacteriol. 175:1886-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell, E. A., and S. A. Darst. 2000. The anti-σ factor SpoIIAB forms a 2:1 complex with σF, contacting multiple conserved regions of the σ factor. J. Mol. Biol. 300:17-28. [DOI] [PubMed] [Google Scholar]
- 31.Campbell, E. A., S. Masuda, J. L. Sun, O. Muzzin, C. A. Olson, S. Wang, and S. A. Darst. 2002. Crystal structure of the Bacillus stearothermophilus anti-σ factor SpoIIAB with the sporulation σ factor σF. Cell 108:795-807. [DOI] [PubMed] [Google Scholar]
- 32.Carniol, K., P. Eichenberger, and R. Losick. 2004. A threshold mechanism governing activation of the developmental regulatory protein σF in Bacillus subtilis. J. Biol. Chem. 279:14860-14870. [DOI] [PubMed]
- 33.Cervin, M. A., G. B. Spiegelman, B. Raether, K. Ohlsen, M. Perego, and J. A. Hoch. 1998. A negative regulator linking chromosome segregation to developmental transcription in Bacillus subtilis. Mol. Microbiol. 29:85-95. [DOI] [PubMed] [Google Scholar]
- 34.Cha, J. H., and G. C. Stewart. 1997. The divIVA minicell locus of Bacillus subtilis. J. Bacteriol. 179:1671-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chary, V. K., D. W. Hilbert, M. L. Higgins, and P. J. Piggot. 2000. The putative DNA translocase SpoIIIE is required for sporulation of the symmetrically dividing coccal species Sporosarcina ureae. Mol. Microbiol. 35:612-622. [DOI] [PubMed] [Google Scholar]
- 36.Chary, V. K., and P. J. Piggot. 2003. Postdivisional synthesis of the Sporosarcina ureae DNA translocase SpoIIIE either in the mother cell or in the prespore enables Bacillus subtilis to translocate DNA from the mother cell to the prespore. J. Bacteriol. 185:879-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coote, J. G. 1972. Sporulation in Bacillus subtilis. Characterization of oligosporogenous mutants and comparison of their phenotypes with those of asporogenous mutants. J. Gen. Microbiol. 71:1-15. [DOI] [PubMed] [Google Scholar]
- 38.Coppolecchia, R., H. DeGrazia, and C. P. Moran, Jr. 1991. Deletion of spoIIAB blocks endospore formation in Bacillus subtilis at an early stage. J. Bacteriol. 173:6678-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cutting, S., A. Driks, R. Schmidt, B. Kunkel, and R. Losick. 1991. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-σK processing in Bacillus subtilis. Genes Dev. 5:456-466. [DOI] [PubMed] [Google Scholar]
- 40.Cutting, S., V. Oke, A. Driks, R. Losick, S. Lu, and L. Kroos. 1990. A forespore checkpoint for mother cell gene expression during development in B. subtilis. Cell 62:239-250. [DOI] [PubMed] [Google Scholar]
- 41.Cutting, S., S. Roels, and R. Losick. 1991. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis. J. Mol. Biol. 221:1237-1256. [DOI] [PubMed] [Google Scholar]
- 42.Daniel, R. A., S. Drake, C. E. Buchanan, R. Scholle, and J. Errington. 1994. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J. Mol. Biol. 235:209-220. [DOI] [PubMed] [Google Scholar]
- 43.de Lencastre, H., and P. J. Piggot. 1979. Identification of different sites of expression for spo loci by transformation of Bacillus subtilis. J. Gen. Microbiol. 114:377-389. [DOI] [PubMed] [Google Scholar]
- 44.Diederich, B., J. F. Wilkinson, T. Magnin, M. Najafi, J. Errington, and M. D. Yudkin. 1994. Role of interactions between SpoIIAA and SpoIIAB in regulating cell-specific transcription factor σF of Bacillus subtilis. Genes Dev. 8:2653-2663. [DOI] [PubMed] [Google Scholar]
- 45.Dong, T. C., and S. M. Cutting. 2003. SpoIVB-mediated cleavage of SpoIVFA could provide the intercellular signal to activate processing of pro-σF in Bacillus subtilis. Mol. Microbiol. 49:1425-1434. [DOI] [PubMed] [Google Scholar]
- 46.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Driks, A. 2002. Proteins of the spore core and coat, p. 527-535. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 48.Driks, A., and R. Losick. 1991. Compartmentalized expression of a gene under the control of sporulation transcription factor σE in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 88:9934-9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duncan, L., S. Alper, F. Arigoni, R. Losick, and P. Stragier. 1995. Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division. Science 270:641-644. [DOI] [PubMed] [Google Scholar]
- 50.Duncan, L., S. Alper, and R. Losick. 1996. SpoIIAA governs the release of the cell-type specific transcription factor σF from its anti-σ factor SpoIIAB. J. Mol. Biol. 260:147-164. [DOI] [PubMed] [Google Scholar]
- 51.Duncan, L., and R. Losick. 1993. SpoIIAB is an anti-σ factor that binds to and inhibits transcription by regulatory protein σF from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 90:2325-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunn, G., and J. Mandelstam. 1977. Cell polarity in Bacillus subtilis: effect of growth conditions on spore positions in sister cells. J. Gen. Microbiol. 103:201-205. [DOI] [PubMed] [Google Scholar]
- 53.Dunn, G., D. M. Torgersen, and J. Mandelstam. 1976. Order of expression of genes affecting septum location during sporulation of Bacillus subtilis. J. Bacteriol. 125:776-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dworkin, J., and R. Losick. 2001. Differential gene expression governed by chromosomal spatial asymmetry. Cell 107:339-346. [DOI] [PubMed] [Google Scholar]
- 55.Eaton, M. W., and D. J. Ellar. 1974. Protein synthesis and breakdown in the mother-cell and forespore compartments during spore morphogenesis in Bacillus megaterium. Biochem. J. 144:327-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edwards, D. H., and J. Errington. 1997. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol. 24:905-915. [DOI] [PubMed] [Google Scholar]
- 57.Eichenberger, P., P. Fawcett, and R. Losick. 2001. A three-protein inhibitor of polar septation during sporulation in Bacillus subtilis. Mol. Microbiol. 42:1147-1162. [DOI] [PubMed] [Google Scholar]
- 58.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 59.Errington, J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 57:1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Errington, J., L. Appleby, R. A. Daniel, H. Goodfellow, S. R. Partridge, and M. D. Yudkin. 1992. Structure and function of the spoIIIJ gene of Bacillus subtilis: a vegetatively expressed gene that is essential for σG activity at an intermediate stage of sporulation. J. Gen. Microbiol. 138:2609-2618. [DOI] [PubMed] [Google Scholar]
- 61.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Errington, J., P. Fort, and J. Mandelstam. 1985. Duplicated sporulation genes in bacteria: implication for simple developmental systems. FEBS Lett. 188:184-188. [Google Scholar]
- 63.Errington, J., and J. Mandelstam. 1984. Genetic and phenotypic characterization of a cluster of mutations in the spoVA locus of Bacillus subtilis. J. Gen. Microbiol. 130:2115-2121. [DOI] [PubMed] [Google Scholar]
- 64.Errington, J., S. Rong, M. S. Rosenkrantz, and A. L. Sonenshein. 1988. Transcriptional regulation and structure of the Bacillus subtilis sporulation locus spoIIIC. J. Bacteriol. 170:1162-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Errington, J., and R. G. Wake. 1991. Chromosome strand segregation during sporulation in Bacillus subtilis. Mol. Microbiol. 5:1145-1149. [DOI] [PubMed] [Google Scholar]
- 66.Evans, L., J. Clarkson, M. D. Yudkin, J. Errington, and A. Feucht. 2003. Analysis of the interaction between the transcription factor σG and the anti-σ factor SpoIIAB of Bacillus subtilis. J. Bacteriol. 185:4615-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fabret, C., V. A. Feher, and J. A. Hoch. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181:1975-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fajardo-Cavazos, P., and W. L. Nicholson. 2000. The TRAP-like SplA protein is a trans-acting negative regulator of spore photoproduct lyase synthesis during Bacillus subtilis sporulation. J. Bacteriol. 182:555-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fan, N., S. Cutting, and R. Losick. 1992. Characterization of the Bacillus subtilis sporulation gene spoVK. J. Bacteriol. 174:1053-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Farrow, J. A., S. Wallbanks, and M. D. Collins. 1994. Phylogenetic interrelationships of round-spore-forming bacilli containing cell walls based on lysine and the non-spore-forming genera Caryophanon, Exiguobacterium, Kurthia, and Planococcus. Int. J. Syst. Bacteriol. 44:74-82. [DOI] [PubMed] [Google Scholar]
- 71.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 97:8063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fawcett, P., A. Melnikov, and P. Youngman. 1998. The Bacillus SpoIIGA protein is targeted to sites of spore septum formation in a SpoIIE-independent manner. Mol. Microbiol. 28:931-943. [DOI] [PubMed] [Google Scholar]
- 73.Feucht, A., L. Abbotts, and J. Errington. 2002. The cell differentiation protein SpoIIE contains a regulatory site that controls its phosphatase activity in response to asymmetric septation. Mol. Microbiol. 45:1119-1130. [DOI] [PubMed] [Google Scholar]
- 74.Feucht, A., R. A. Daniel, and J. Errington. 1999. Characterization of a morphological checkpoint coupling cell-specific transcription to septation in Bacillus subtilis. Mol. Microbiol. 33:1015-1026. [DOI] [PubMed] [Google Scholar]
- 75.Feucht, A., L. Evans, and J. Errington. 2003. Identification of sporulation genes by genome-wide analysis of the σE regulon of Bacillus subtilis. Microbiology 149:3023-3034. [DOI] [PubMed] [Google Scholar]
- 76.Feucht, A., I. Lucet, M. D. Yudkin, and J. Errington. 2001. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 40:115-125. [DOI] [PubMed] [Google Scholar]
- 77.Feucht, A., T. Magnin, M. D. Yudkin, and J. Errington. 1996. Bifunctional protein required for asymmetric cell division and cell-specific transcription in Bacillus subtilis. Genes Dev. 10:794-803. [DOI] [PubMed] [Google Scholar]
- 78.Fitz-James, P. C. 1965. Spore formation in some wild and mutant strains of B. cereus and some effects of inhibitors. Colloq. Int. Centre Natl. Rech. Sci. (Paris) 124:529-544. [Google Scholar]
- 79.Fitz-James, P. C., and I. E. Young. 1969. Morphology of sporulation, p. 39-72. In G. W. Gould and A. Hurst (ed.), The bacterial spore. Academic Press, New York, N.Y.
- 80.Fort, P., and P. J. Piggot. 1984. Nucleotide sequence of sporulation locus spoIIA in Bacillus subtilis. J. Gen. Microbiol. 130:2147-2153. [DOI] [PubMed] [Google Scholar]
- 81.Foster, S. J., and D. Popham. 2002. Structure and synthesis of cell wall, spore cortex, teichoic acid, S-layers, and capsules, p. 21-41. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 82.Foulger, D., and J. Errington. 1989. The role of the sporulation gene spoIIIE in the regulation of prespore-specific gene expression in Bacillus subtilis. Mol. Microbiol. 3:1247-1255. [DOI] [PubMed] [Google Scholar]
- 83.Francesconi, S. C., T. J. MacAlister, B. Setlow, and P. Setlow. 1988. Immunoelectron microscopic localization of small, acid-soluble spore proteins in sporulating cells of Bacillus subtilis. J. Bacteriol. 170:5963-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Frandsen, N., I. Barák, C. Karmazyn-Campelli, and P. Stragier. 1999. Transient gene asymmetry during sporulation and establishment of cell specificity in Bacillus subtilis. Genes Dev. 13:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frandsen, N., and P. Stragier. 1995. Identification and characterization of the Bacillus subtilis spoIIP locus. J. Bacteriol. 177:716-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fujita, M., and R. Losick. 2002. An investigation into the compartmentalization of the sporulation transcription factor σE in Bacillus subtilis. Mol. Microbiol. 43:27-38. [DOI] [PubMed] [Google Scholar]
- 87.Fujita, M., and R. Losick. 2003. The master regulator for entry into sporulation in Bacillus subtilis becomes a cell-specific transcription factor after asymmetric division. Genes Dev. 17:1166-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fujita, Y., R. Ramaley, and E. Freese. 1977. Location and properties of glucose dehydrogenase in sporulating cells and spores of Bacillus subtilis. J. Bacteriol. 132:282-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fukushima, T., H. Yamamoto, A. Atrih, S. J. Foster, and J. Sekiguchi. 2002. A polysaccharide deacetylase gene (pdaA) is required for germination and for production of muramic δ-lactam residues in the spore cortex of Bacillus subtilis. J. Bacteriol. 184:6007-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gao, H., X. Jiang, K. Pogliano, and A. I. Aronson. 2002. The E1β and E2 subunits of the Bacillus subtilis pyruvate dehydrogenase complex are involved in regulation of sporulation. J. Bacteriol. 184:2780-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Garsin, D. A., L. Duncan, D. M. Paskowitz, and R. Losick. 1998. The kinase activity of the anti-sigma factor SpoIIAB is required for activation as well as inhibition of transcription factor σF during sporulation in Bacillus subtilis. J. Mol. Biol. 284:569-578. [DOI] [PubMed] [Google Scholar]
- 92.Garsin, D. A., D. M. Paskowitz, L. Duncan, and R. Losick. 1998. Evidence for common sites of contact between the antisigma factor SpoIIAB and its partners SpoIIAA and the developmental transcription factor σF in Bacillus subtilis. J. Mol. Biol. 284:557-568. [DOI] [PubMed] [Google Scholar]
- 93.Gholamhoseinian, A., and P. J. Piggot. 1989. Timing of spoII gene expression relative to septum formation during sporulation of Bacillus subtilis. J. Bacteriol. 171:5747-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gholamhoseinian, A., Z. Shen, J. J. Wu, and P. Piggot. 1992. Regulation of transcription of the cell division gene ftsA during sporulation of Bacillus subtilis. J. Bacteriol. 174:4647-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gomez, M., S. Cutting, and P. Stragier. 1995. Transcription of spoIVB is the only role of σG that is essential for pro-σK processing during spore formation in Bacillus subtilis. J. Bacteriol. 177:4825-4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gomez, M., and S. M. Cutting. 1996. Expression of the Bacillus subtilis spoIVB gene is under dual σF/σG control. Microbiology 142:3453-3457. [DOI] [PubMed] [Google Scholar]
- 97.Gomez, M., and S. M. Cutting. 1997. BofC encodes a putative forespore regulator of the Bacillus subtilis σK checkpoint. Microbiology 143:157-170. [DOI] [PubMed] [Google Scholar]
- 98.Gonzy-Tréboul, G., C. Karmazyn-Campelli, and P. Stragier. 1992. Developmental regulation of transcription of the Bacillus subtilis ftsAZ operon. J. Mol. Biol. 224:967-979. [DOI] [PubMed] [Google Scholar]
- 99.Graumann, P. L., and R. Losick. 2001. Coupling of asymmetric division to polar placement of replication origin regions in Bacillus subtilis. J. Bacteriol. 183:4052-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Green, D. H., and S. M. Cutting. 2000. Membrane topology of the Bacillus subtilis pro-σK processing complex. J. Bacteriol. 182:278-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grossman, A. D., and R. Losick. 1988. Extracellular control of spore formation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 85:4369-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Halberg, R., and L. Kroos. 1994. Sporulation regulatory protein SpoIIID from Bacillus subtilis activates and represses transcription by both mother-cell-specific forms of RNA polymerase. J. Mol. Biol. 243:425-436. [DOI] [PubMed] [Google Scholar]
- 103.Haldenwang, W. G., N. Lang, and R. Losick. 1981. A sporulation-induced σ-like regulatory protein from B. subtilis. Cell 23:615-624. [DOI] [PubMed] [Google Scholar]
- 104.Haraldsen, J. D., and A. L. Sonenshein. 2003. Efficient sporulation in Clostridium difficile requires disruption of the σK gene. Mol. Microbiol. 48:811-821. [DOI] [PubMed] [Google Scholar]
- 105.Harry, E. J., K. Pogliano, and R. Losick. 1995. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 177:3386-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Healy, J., J. Weir, I. Smith, and R. Losick. 1991. Post-transcriptional control of a sporulation regulatory gene encoding transcription factor σH in Bacillus subtilis. Mol. Microbiol. 5:477-487. [DOI] [PubMed] [Google Scholar]
- 107.Helmann, J. D., and C. P. Moran, Jr. 2002. RNA polymerase and σ factors, p. 289-312. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 108.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235-247. [DOI] [PubMed] [Google Scholar]
- 109.Henriques, A. O., and C. P. Moran, Jr. 2000. Structure and assembly of the bacterial endospore coat. Methods 20:95-110. [DOI] [PubMed] [Google Scholar]
- 110.Hilbert, D. W., V. K. Chary, and P. J. Piggot. 2004. Contrasting effects of σE on compartmentalization of σF activity during sporulation of Bacillus subtilis. J. Bacteriol. 186:1983-1990. [DOI] [PMC free article] [PubMed]
- 111.Hilbert, D. W., and P. J. Piggot. 2003. Novel spoIIE mutation that causes uncompartmentalized σF activation in Bacillus subtilis. J. Bacteriol. 185:1590-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hitchins, A. D. 1975. Polarized relationship of bacterial spore loci to the “old” and “new” ends of sporangia. J. Bacteriol. 121:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hitchins, A. D. 1978. Chromosome age and segregation during sporulation of Bacillus megaterium. Can. J. Microbiol. 24:1227-1235. [DOI] [PubMed] [Google Scholar]
- 114.Hitchins, A. D., and R. A. Slepecky. 1969. Bacterial sporulation as a modified procaryotic cell division. Nature 223:804-807. [DOI] [PubMed] [Google Scholar]
- 115.Ho, M. S., K. Carniol, and R. Losick. 2003. Evidence in support of a docking model for the release of the transcription factor σF from the antisigma factor SpoIIAB in Bacillus subtilis. J. Biol. Chem. 278:20898-20905. [DOI] [PubMed] [Google Scholar]
- 116.Hoa, N. T., J. A. Brannigan, and S. M. Cutting. 2001. The PDZ domain of the SpoIVB serine peptidase facilitates multiple functions. J. Bacteriol. 183:4364-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hoch, J. A., K. Trach, F. Kawamura, and H. Saito. 1985. Identification of the transcriptional suppressor sof-1 as an alteration in the Spo0A protein. J. Bacteriol. 161:552-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hofmeister, A. 1998. Activation of the proprotein transcription factor pro-σE is associated with its progression through three patterns of subcellular localization during sporulation in Bacillus subtilis. J. Bacteriol. 180:2426-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hofmeister, A. E., A. Londõno-Vallejo, E. Harry, P. Stragier, and R. Losick. 1995. Extracellular signal protein triggering the proteolytic activation of a developmental transcription factor in B. subtilis. Cell 83:219-226. [DOI] [PubMed] [Google Scholar]
- 120.Holt, S. C., and E. R. Ledbetter. 1969. Comparative ultrastructure of selective aerobic spore-forming bacteria; a freeze-etching study. Bacteriol. Rev. 33:346-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hranueli, D., P. J. Piggot, and J. Mandelstam. 1974. Statistical estimate of the total number of operons specific for Bacillus subtilis sporulation. J. Bacteriol. 119:684-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ichikawa, H., R. Halberg, and L. Kroos. 1999. Negative regulation by the Bacillus subtilis GerE protein. J. Biol. Chem. 274:8322-8327. [DOI] [PubMed] [Google Scholar]
- 123.Ichikawa, H., and L. Kroos. 2000. Combined action of two transcription factors regulates genes encoding spore coat proteins of Bacillus subtilis. J. Biol. Chem. 275:13849-13855. [DOI] [PubMed] [Google Scholar]
- 124.Illing, N., and J. Errington. 1991. Genetic regulation of morphogenesis in Bacillus subtilis: roles of σE and σF in prespore engulfment. J. Bacteriol. 173:3159-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Illing, N., and J. Errington. 1991. The spoIIIA operon of Bacillus subtilis defines a new temporal class of mother-cell-specific sporulation genes under the control of the σE form of RNA polymerase. Mol. Microbiol. 5:1927-1940. [DOI] [PubMed] [Google Scholar]
- 126.Ireton, K., and A. D. Grossman. 1992. Coupling between gene expression and DNA synthesis early during development in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 89:8808-8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ireton, K., and A. D. Grossman. 1992. Interactions among mutations that cause altered timing of gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 174:3185-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ireton, K., and A. D. Grossman. 1994. A developmental checkpoint couples the initiation of sporulation to DNA replication in Bacillus subtilis. EMBO J. 13:1566-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ireton, K., and A. D. Grossman. 1994. DNA-related conditions controlling the initiation of sporulation in Bacillus subtilis. Cell. Mol. Biol. Res. 40:193-198. [PubMed] [Google Scholar]
- 130.Ireton, K., N. W. Gunther IV, and A. D. Grossman. 1994. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 176:5320-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ireton, K., S. Jin, A. D. Grossman, and A. L. Sonenshein. 1995. Krebs cycle function is required for activation of the Spo0A transcription factor in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 92:2845-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ireton, K., D. Z. Rudner, K. J. Siranosian, and A. D. Grossman. 1993. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 7:283-294. [DOI] [PubMed] [Google Scholar]
- 133.Jiang, M., R. Grau, and M. Perego. 2000. Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis. J. Bacteriol. 182:303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jiang, M., Y. L. Tzeng, V. A. Feher, M. Perego, and J. A. Hoch. 1999. Alanine mutants of the Spo0F response regulator modifying specificity for sensor kinases in sporulation initiation. Mol. Microbiol. 33:389-395. [DOI] [PubMed] [Google Scholar]
- 135.Jonas, R. M., and W. G. Haldenwang. 1989. Influence of spo mutations on σE synthesis in Bacillus subtilis. J. Bacteriol. 171:5226-5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jonas, R. M., E. A. Weaver, T. J. Kenney, C. P. Moran, Jr., and W. G. Haldenwang. 1988. The Bacillus subtilis spoIIG operon encodes both σE and a gene necessary for σE activation. J. Bacteriol. 170:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 138.Ju, J., and W. G. Haldenwang. 1999. The “pro” sequence of the sporulation-specific σ transcription factor σE directs it to the mother cell side of the sporulation septum. J. Bacteriol. 181:6171-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ju, J., and W. G. Haldenwang. 2003. Tethering of the Bacillus subtilis σE proprotein to the cell membrane is necessary for its processing but insufficient for its stabilization. J. Bacteriol. 185:5897-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ju, J., T. Luo, and W. G. Haldenwang. 1997. Bacillus subtilis pro-σE fusion protein localizes to the forespore septum and fails to be processed when synthesized in the forespore. J. Bacteriol. 179:4888-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ju, J., T. Luo, and W. G. Haldenwang. 1998. Forespore expression and processing of the σE transcription factor in wild-type and mutant Bacillus subtilis. J. Bacteriol. 180:1673-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Karmazyn-Campelli, C., C. Bonamy, B. Savelli, and P. Stragier. 1989. Tandem genes encoding σ-factors for consecutive steps of development in Bacillus subtilis. Genes Dev. 3:150-157. [DOI] [PubMed] [Google Scholar]
- 143.Karow, M. L., P. Glaser, and P. J. Piggot. 1995. Identification of a gene, spoIIR, that links the activation of σE to the transcriptional activity of σF during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 92:2012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Karow, M. L., and P. J. Piggot. 1995. Construction of gusA transcriptional fusion vectors for Bacillus subtilis and their utilization for studies of spore formation. Gene 163:69-74. [DOI] [PubMed] [Google Scholar]
- 145.Kay, D., and S. C. Warren. 1968. Sporulation in Bacillus subtilis. Morphological changes. Biochem. J. 109:819-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kellner, E. M., A. Decatur, and C. P. Moran, Jr. 1996. Two-stage regulation of an anti-σ factor determines developmental fate during bacterial endospore formation. Mol. Microbiol. 21:913-924. [DOI] [PubMed] [Google Scholar]
- 147.Kenney, T. J., and C. P. Moran, Jr. 1987. Organization and regulation of an operon that encodes a sporulation-essential σ factor in Bacillus subtilis. J. Bacteriol. 169:3329-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Khvorova, A., V. K. Chary, D. W. Hilbert, and P. J. Piggot. 2000. The chromosomal location of the Bacillus subtilis sporulation gene spoIIR is important for its function. J. Bacteriol. 182:4425-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Khvorova, A., L. Zhang, M. L. Higgins, and P. J. Piggot. 1998. The spoIIE locus is involved in the Spo0A-dependent switch in the location of FtsZ rings in Bacillus subtilis. J. Bacteriol. 180:1256-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.King, N., O. Dreesen, P. Stragier, K. Pogliano, and R. Losick. 1999. Septation, dephosphorylation, and the activation of σF during sporulation in Bacillus subtilis. Genes Dev. 13:1156-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kirchman, P. A., H. DeGrazia, E. M. Kellner, and C. P. Moran, Jr. 1993. Forespore-specific disappearance of the σ-factor antagonist SpoIIAB: implications for its role in determination of cell fate in Bacillus subtilis. Mol. Microbiol. 8:663-671. [DOI] [PubMed] [Google Scholar]
- 152.Kroos, L., B. Kunkel, and R. Losick. 1989. Switch protein alters specificity of RNA polymerase containing a compartment-specific σ factor. Science 243:526-529. [DOI] [PubMed] [Google Scholar]
- 153.Kroos, L., Y. T. Yu, D. Mills, and S. Ferguson-Miller. 2002. Forespore signaling is necessary for pro-σK processing during Bacillus subtilis sporulation despite the loss of SpoIVFA upon translational arrest. J. Bacteriol. 184:5393-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kunkel, B., L. Kroos, H. Poth, P. Youngman, and R. Losick. 1989. Temporal and spatial control of the mother-cell regulatory gene spoIIID of Bacillus subtilis. Genes Dev. 3:1735-1744. [DOI] [PubMed] [Google Scholar]
- 155.Kunkel, B., R. Losick, and P. Stragier. 1990. The Bacillus subtilis gene for the development transcription factor σK is generated by excision of a dispensable DNA element containing a sporulation recombinase gene. Genes Dev. 4:525-535. [DOI] [PubMed] [Google Scholar]
- 156.Kunkel, B., K. Sandman, S. Panzer, P. Youngman, and R. Losick. 1988. The promoter for a sporulation gene in the spoIVC locus of Bacillus subtilis and its use in studies of temporal and spatial control of gene expression. J. Bacteriol. 170:3513-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kuroda, A., M. H. Rashid, and J. Sekiguchi. 1992. Molecular cloning and sequencing of the upstream region of the major Bacillus subtilis autolysin gene: a modifier protein exhibiting sequence homology to the major autolysin and the spoIID product. J. Gen. Microbiol. 138:1067-1076. [DOI] [PubMed] [Google Scholar]
- 158.Kuwana, R., H. Ikejiri, S. Yamamura, H. Takamatsu, and K. Watabe. 2004. Functional relationship between SpoVIF and GerE in gene regulation during sporulation of Bacillus subtilis. Microbiology 150:163-170. [DOI] [PubMed] [Google Scholar]
- 159.LaBell, T. L., J. E. Trempy, and W. G. Haldenwang. 1987. Sporulation-specific σ factor σ29 of Bacillus subtilis is synthesized from a precursor protein, P31. Proc. Natl. Acad. Sci. USA 84:1784-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.LeDeaux, J. R., and A. D. Grossman. 1995. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J. Bacteriol. 177:166-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lee, C. S., J. Clarkson, J. C. Shu, I. D. Campbell, and M. D. Yudkin. 2001. Bacillus subtilis mutations that alter the pathway of phosphorylation of the anti-anti-σF factor SpoIIAA lead to a Spo− phenotype. Mol. Microbiol. 40:9-19. [DOI] [PubMed] [Google Scholar]
- 162.Lee, C. S., I. Lucet, and M. D. Yudkin. 2000. Fate of the SpoIIAB*-ADP liberated after SpoIIAB phosphorylates SpoIIAA of Bacillus subtilis. J. Bacteriol. 182:6250-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lee, P. S., D. C. Lin, S. Moriya, and A. D. Grossman. 2003. Effects of the chromosome partitioning protein Spo0J (ParB) on oriC positioning and replication initiation in Bacillus subtilis. J. Bacteriol. 185:1326-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lemon, K. P., I. Kurtser, and A. D. Grossman. 2001. Effects of replication termination mutants on chromosome partitioning in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lemon, K. P., I. Kurtser, J. Wu, and A. D. Grossman. 2000. Control of initiation of sporulation by replication initiation genes in Bacillus subtilis. J. Bacteriol. 182:2989-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Levin, P. A., and R. Losick. 1994. Characterization of a cell division gene from Bacillus subtilis that is required for vegetative and sporulation septum formation. J. Bacteriol. 176:1451-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Levin, P. A., and R. Losick. 1996. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 10:478-488. [DOI] [PubMed] [Google Scholar]
- 168.Levin, P. A., R. Losick, P. Stragier, and F. Arigoni. 1997. Localization of the sporulation protein SpoIIE in Bacillus subtilis is dependent upon the cell division protein FtsZ. Mol. Microbiol. 25:839-846. [DOI] [PubMed] [Google Scholar]
- 169.Lewis, P. J., and J. Errington. 1996. Use of green fluorescent protein for detection of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. Microbiology 142:733-740. [DOI] [PubMed] [Google Scholar]
- 170.Lewis, P. J., T. Magnin, and J. Errington. 1996. Compartmentalized distribution of the proteins controlling the prespore-specific transcription factor σF of Bacillus subtilis. Genes Cells 1:881-894. [DOI] [PubMed] [Google Scholar]
- 171.Lewis, P. J., L. J. Wu, and J. Errington. 1998. Establishment of prespore-specific gene expression in Bacillus subtilis: localization of SpoIIE phosphatase and initiation of compartment-specific proteolysis. J. Bacteriol. 180:3276-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li, Z., F. DiDonato, and P. J. Piggot. 2004. Compartmentalization of gene expression during sporulation of Bacillus subtilis is compromised in mutants blocked at stage III of sporulation. J. Bacteriol. 186:2221-2223. [DOI] [PMC free article] [PubMed]
- 173.Li, Z., and P. J. Piggot. 2001. Development of a two-part transcription probe to determine the completeness of temporal and spatial compartmentalization of gene expression during bacterial development. Proc. Natl. Acad. Sci. USA 98:12538-12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lin, D. C., and A. D. Grossman. 1998. Identification and characterization of a bacterial chromosome partitioning site. Cell 92:675-685. [DOI] [PubMed] [Google Scholar]
- 175.Lin, D. C., P. A. Levin, and A. D. Grossman. 1997. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 94:4721-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lindow, J. C., R. A. Britton, and A. D. Grossman. 2002. Structural Maintenance of Chromosomes protein of Bacillus subtilis affects supercoiling in vivo. J. Bacteriol. 184:5317-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Liu, J., and P. Zuber. 2000. The ClpX protein of Bacillus subtilis indirectly influences RNA polymerase holoenzyme composition and directly stimulates σ-dependent transcription. Mol. Microbiol. 37:885-897. [DOI] [PubMed] [Google Scholar]
- 178.Londõno-Vallejo, J. A., C. Frehel, and P. Stragier. 1997. spoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol. Microbiol. 24:29-39. [DOI] [PubMed] [Google Scholar]
- 179.Londõno-Vallejo, J. A., and P. Stragier. 1995. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 9:503-508. [DOI] [PubMed] [Google Scholar]
- 180.Lopez-Diaz, I., S. Clarke, and J. Mandelstam. 1986. spoIID operon of Bacillus subtilis: cloning and sequence. J. Gen. Microbiol. 132:341-354. [DOI] [PubMed] [Google Scholar]
- 181.Losick, R., and J. Pero. 1981. Cascades of σ factors. Cell 25:582-584. [DOI] [PubMed] [Google Scholar]
- 182.Losick, R., and P. Stragier. 1992. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature 355:601-604. [DOI] [PubMed] [Google Scholar]
- 183.Losick, R., P. Youngman, and P. J. Piggot. 1986. Genetics of endospore formation in Bacillus subtilis. Annu. Rev. Genet. 20:625-669. [DOI] [PubMed] [Google Scholar]
- 184.Lu, S., S. Cutting, and L. Kroos. 1995. Sporulation protein SpoIVFB from Bacillus subtilis enhances processing of the σ factor precursor pro-σK in the absence of other sporulation gene products. J. Bacteriol. 177:1082-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lu, S., R. Halberg, and L. Kroos. 1990. Processing of the mother-cell σ factor, σK, may depend on events occurring in the forespore during Bacillus subtilis development. Proc. Natl. Acad. Sci. USA 87:9722-9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lucet, I., A. Feucht, M. D. Yudkin, and J. Errington. 2000. Direct interaction between the cell division protein FtsZ and the cell differentiation protein SpoIIE. EMBO J. 19:1467-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Magnin, T., M. Lord, and M. D. Yudkin. 1997. Contribution of partner switching and SpoIIAA cycling to regulation of σF activity in sporulating Bacillus subtilis. J. Bacteriol. 179:3922-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mandelstam, J., D. Kay, and D. Hranueli. 1975. Biochemistry and morphology of stage I in sporulation of Bacillus subtilis cells, p. 181-186. In P. Gerhardt, R. N. Costilow, and H. L. Sadoff (ed.), Spores VI. American Society for Microbiology, Washington, D.C.
- 189.Margolis, P., A. Driks, and R. Losick. 1991. Establishment of cell type by compartmentalized activation of a transcription factor. Science 254:562-565. [DOI] [PubMed] [Google Scholar]
- 190.Margolis, P. S., A. Driks, and R. Losick. 1993. Sporulation gene spoIIB from Bacillus subtilis. J. Bacteriol. 175:528-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Marston, A. L., and J. Errington. 1999. Dynamic movement of the ParA-like Soj protein of B. subtilis and its dual role in nucleoid organization and developmental regulation. Mol. Cell 4:673-682. [DOI] [PubMed] [Google Scholar]
- 192.Marston, A. L., H. B. Thomaides, D. H. Edwards, M. E. Sharpe, and J. Errington. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12:3419-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Masuda, E. S., H. Anaguchi, T. Sato, M. Takeuchi, and Y. Kobayashi. 1990. Nucleotide sequence of the sporulation gene spoIIGA from Bacillus subtilis. Nucleic Acids Res. 18:657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Masuda, E. S., H. Anaguchi, K. Yamada, and Y. Kobayashi. 1988. Two developmental genes encoding σ factor homologs are arranged in tandem in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 85:7637-7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Matsuno, K., and A. L. Sonenshein. 1999. Role of SpoVG in asymmetric septation in Bacillus subtilis. J. Bacteriol. 181:3392-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.McAdams, H. H., and L. Shapiro. 2003. A bacterial cell-cycle regulatory network operating in time and space. Science 301:1874-1877. [DOI] [PubMed] [Google Scholar]
- 197.McQuade, R. S., N. Comella, and A. D. Grossman. 2001. Control of a family of phosphatase regulatory genes (phr) by the alternate σ factor σH of Bacillus subtilis. J. Bacteriol. 183:4905-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Migocki, M. D., M. K. Freeman, R. G. Wake, and E. J. Harry. 2002. The Min system is not required for precise placement of the midcell Z ring in Bacillus subtilis. EMBO Rep. 3:1163-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Min, K. T., C. M. Hilditch, B. Diederich, J. Errington, and M. D. Yudkin. 1993. σF, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-σ factor that is also a protein kinase. Cell 74:735-742. [DOI] [PubMed] [Google Scholar]
- 200.Mitani, T., J. E. Heinze, and E. Freese. 1977. Induction of sporulation in Bacillus subtilis by decoyinine or hadacidin. Biochem. Biophys. Res. Commun. 77:1118-1125. [DOI] [PubMed] [Google Scholar]
- 201.Moir, A., B. M. Corfe, and J. Behravan. 2002. Spore germination. Cell. Mol. Life Sci. 59:403-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Moldover, B., P. J. Piggot, and M. D. Yudkin. 1991. Identification of the promoter and the transcriptional start site of the spoVA operon of Bacillus subtilis and Bacillus licheniformis. J. Gen. Microbiol. 137:527-531. [DOI] [PubMed] [Google Scholar]
- 203.Molle, V., M. Fujita, S. T. Jensen, P. Eichenberger, J. E. Gonzalez-Pastor, J. S. Liu, and R. Losick. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683-1701. [DOI] [PubMed] [Google Scholar]
- 204.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Murakami, T., K. Haga, M. Takeuchi, and T. Sato. 2002. Analysis of the Bacillus subtilis spoIIIJ gene and its paralogue gene, yqjG. J. Bacteriol. 184:1998-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Najafi, S. M., D. A. Harris, and M. D. Yudkin. 1997. Properties of the phosphorylation reaction catalyzed by SpoIIAB that help to regulate sporulation of Bacillus subtilis. J. Bacteriol. 179:5628-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Najafi, S. M., A. C. Willis, and M. D. Yudkin. 1995. Site of phosphorylation of SpoIIAA, the anti-anti-σ factor for sporulation-specific σF of Bacillus subtilis. J. Bacteriol. 177:2912-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ogura, Y., N. Ogasawara, E. J. Harry, and S. Moriya. 2003. Increasing the ratio of Soj to Spo0J promotes replication initiation in Bacillus subtilis. J. Bacteriol. 185:6316-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ohlsen, K. L., J. K. Grimsley, and J. A. Hoch. 1994. Deactivation of the sporulation transcription factor Spo0A by the Spo0E protein phosphatase. Proc. Natl. Acad. Sci. USA 91:1756-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Oke, V., and R. Losick. 1993. Multilevel regulation of the sporulation transcription factor σK in Bacillus subtilis. J. Bacteriol. 175:7341-7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Oke, V., M. Shchepetov, and S. Cutting. 1997. SpoIVB has two distinct functions during spore formation in Bacillus subtilis. Mol. Microbiol. 23:223-230. [DOI] [PubMed] [Google Scholar]
- 213.Paidhungat, M., and P. Setlow. 2002. Spore germination and outgrowth, p. 537-548. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 214.Pan, Q., D. A. Garsin, and R. Losick. 2001. Self-reinforcing activation of a cell-specific transcription factor by proteolysis of an anti-σ factor in B. subtilis. Mol. Cell 8:873-883. [DOI] [PubMed] [Google Scholar]
- 215.Pan, Q., and R. Losick. 2003. Unique degradation signal for ClpCP in Bacillus subtilis. J. Bacteriol. 185:5275-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Pan, Q., R. Losick, and D. Z. Rudner. 2003. A second PDZ-containing serine protease contributes to activation of the sporulation transcription factor σK in Bacillus subtilis. J. Bacteriol. 185:6051-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Panzer, S., R. Losick, D. Sun, and P. Setlow. 1989. Evidence for an additional temporal class of gene expression in the forespore compartment of sporulating Bacillus subtilis. J. Bacteriol. 171:561-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Partridge, S. R., and J. Errington. 1993. The importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol. Microbiol. 8:945-955. [DOI] [PubMed] [Google Scholar]
- 219.Pedraza-Reyes, M., F. Gutierrez-Corona, and W. L. Nicholson. 1997. Spore photoproduct lyase operon (splAB) regulation during Bacillus subtilis sporulation: modulation of splB-lacZ fusion expression by P1 promoter mutations and by an in-frame deletion of splA. Curr. Microbiol. 34:133-137. [DOI] [PubMed] [Google Scholar]
- 220.Perego, M. 1997. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc. Natl. Acad. Sci. USA 94:8612-8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Perego, M. 2001. A new family of aspartyl phosphate phosphatases targeting the sporulation transcription factor Spo0A of Bacillus subtilis. Mol. Microbiol. 42:133-143. [DOI] [PubMed] [Google Scholar]
- 222.Perego, M., C. Hanstein, K. M. Welsh, T. Djavakhishvili, P. Glaser, and J. A. Hoch. 1994. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell 79:1047-1055. [DOI] [PubMed] [Google Scholar]
- 223.Perego, M., and J. A. Hoch. 2002. Two-component systems, phosphorelays, and regulation of their activities by phosphatases, p. 473-782. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 224.Perez, A. R., A. Abanes-De Mello, and K. Pogliano. 2000. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in Bacillus subtilis. J. Bacteriol. 182:1096-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Peters, H. K., 3rd, and W. G. Haldenwang. 1994. Isolation of a Bacillus subtilis spoIIGA allele that suppresses processing-negative mutations in the pro-σE gene (sigE). J. Bacteriol. 176:7763-7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Piggot, P. J. 1978. Organization of spo locus expression during sporulation of Bacillus subtilis: evidence for different loci being expressed in the mother cell and the forespore, p. 122-126. In G. Chambliss and J. C. Vary (ed.), Spores VII. American Society for Microbiology, Washington, D.C.
- 227.Piggot, P. J., J. E. Bylund, and M. L. Higgins. 1994. Morphogenesis and gene regulation during sporulation, p. 113-117. In P. J. Piggot, C. P. Moran, Jr., and P. Youngman (ed.), Regulation of bacterial differentiation. American Society for Microbiology, Washington, D.C.
- 228.Piggot, P. J., and J. G. Coote. 1976. Genetic aspects of bacterial endospore formation. Bacteriol. Rev. 40:908-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Piggot, P. J., C. A. Curtis, and H. de Lencastre. 1984. Use of integrational plasmid vectors to demonstrate the polycistronic nature of a transcriptional unit (spoIIA) required for sporulation of Bacillus subtilis. J. Gen. Microbiol. 130:2123-2136. [DOI] [PubMed] [Google Scholar]
- 230.Piggot, P. J., and H. de Lencastre. 1978. A rapid method for constructing multiply marked strains of Bacillus subtilis. J. Gen. Microbiol. 106:191-194. [DOI] [PubMed] [Google Scholar]
- 231.Piggot, P. J., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-518. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 232.Pogliano, J., N. Osborne, M. D. Sharp, A. Abanes-De Mello, A. Perez, Y. L. Sun, and K. Pogliano. 1999. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 31:1149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Pogliano, K., E. Harry, and R. Losick. 1995. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol. Microbiol. 18:459-470. [DOI] [PubMed] [Google Scholar]
- 234.Pogliano, K., A. E. Hofmeister, and R. Losick. 1997. Disappearance of the σE transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to establishment of cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 179:3331-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Popham, D. L., and P. Stragier. 1992. Binding of the Bacillus subtilis spoIVCA product to the recombination sites of the element interrupting the σK-encoding gene. Proc. Natl. Acad. Sci. USA 89:5991-5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Predich, M., G. Nair, and I. Smith. 1992. Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing σH. J. Bacteriol. 174:2771-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Quisel, J. D., and A. D. Grossman. 2000. Control of sporulation gene expression in Bacillus subtilis by the chromosome partitioning proteins Soj (ParA) and Spo0J (ParB). J. Bacteriol. 182:3446-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Quisel, J. D., D. C. Lin, and A. D. Grossman. 1999. Control of development by altered localization of a transcription factor in B. subtilis. Mol. Cell 4:665-672. [DOI] [PubMed] [Google Scholar]
- 239.Ratnayake-Lecamwasam, M., P. Serror, K. W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Resnekov, O., S. Alper, and R. Losick. 1996. Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Genes Cells 1:529-542. [DOI] [PubMed] [Google Scholar]
- 241.Resnekov, O., A. Driks, and R. Losick. 1995. Identification and characterization of sporulation gene spoVS from Bacillus subtilis. J. Bacteriol. 177:5628-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Resnekov, O., and R. Losick. 1998. Negative regulation of the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 95:3162-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ricca, E., S. Cutting, and R. Losick. 1992. Characterization of bofA, a gene involved in intercompartmental regulation of pro-σK processing during sporulation in Bacillus subtilis. J. Bacteriol. 174:3177-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Robinson, R. W., and C. R. Spotts. 1983. The ultrastructure of sporulation in Sporosarcina ureae. Can. J. Microbiol. 29:807-814. [Google Scholar]
- 245.Roels, S., A. Driks, and R. Losick. 1992. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Rong, S., M. S. Rosenkrantz, and A. L. Sonenshein. 1986. Transcriptional control of the Bacillus subtilis spoIID gene. J. Bacteriol. 165:771-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Rudner, D. Z., P. Fawcett, and R. Losick. 1999. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc. Natl. Acad. Sci. USA 96:14765-14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Rudner, D. Z., and R. Losick. 2001. Morphological coupling in development: lessons from prokaryotes. Dev. Cell 1:733-742. [DOI] [PubMed] [Google Scholar]
- 249.Rudner, D. Z., and R. Losick. 2002. A sporulation membrane protein tethers the pro-σK processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev. 16:1007-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Rudner, D. Z., Q. Pan, and R. M. Losick. 2002. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc. Natl. Acad. Sci. USA 99:8701-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ryter, A. 1965. Étude morphologique de la sporulation de Bacillus subtilis. Ann. Inst. Pasteur Paris 108:40-60. [PubMed] [Google Scholar]
- 252.Ryter, A., H. Ionesco, and P. Schaeffer. 1966. Classification cytologique, par leur stage de blocage, des mutants de sporulation de Bacillus subtilis Marburg. Ann. Inst. Pasteur Paris 110:305-315. [PubMed] [Google Scholar]
- 253.Salas-Pacheco, J. M., N. Urtiz-Estrada, G. Martinez-Cadena, R. E. Yasbin, and M. Pedraza-Reyes. 2003. YqfS from Bacillus subtilis is a spore protein and a new functional member of the type IV apurinic/apyrimidinic-endonuclease family. J. Bacteriol. 185:5380-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Samuelson, J. C., M. Chen, F. Jiang, I. Moller, M. Wiedmann, A. Kuhn, G. J. Phillips, and R. E. Dalbey. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637-641. [DOI] [PubMed] [Google Scholar]
- 255.Sato, T., K. Harada, and Y. Kobayashi. 1996. Analysis of suppressor mutations of spoIVCA mutations: occurrence of DNA rearrangement in the absence of site-specific DNA recombinase SpoIVCA in Bacillus subtilis. J. Bacteriol. 178:3380-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Sato, T., K. Harada, Y. Ohta, and Y. Kobayashi. 1994. Expression of the Bacillus subtilis spoIVCA gene, which encodes a site-specific recombinase, depends on the spoIIGB product. J. Bacteriol. 176:935-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Satola, S., P., A. Kirchman, and C. P. Moran, Jr. 1991. Spo0A binds to a promoter used by σA RNA polymerase during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 88:4533-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Satola, S. W., J. M. Baldus, and C. P. Moran, Jr. 1992. Binding of Spo0A stimulates spoIIG promoter activity in Bacillus subtilis. J. Bacteriol. 174:1448-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Schmidt, R., A. L. Decatur, P. N. Rather, C. P. Moran, Jr., and R. Losick. 1994. Bacillus subtilis Lon protease prevents inappropriate transcription of genes under the control of the sporulation transcription factor σG. J. Bacteriol. 176:6528-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Schmidt, R., P. Margolis, L. Duncan, R. Coppolecchia, C. P. Moran, Jr., and R. Losick. 1990. Control of developmental transcription factor σF by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 87:9221-9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Scotti, P. A., M. L. Urbanus, J. Brunner, J. W. de Gier, G. von Heijne, C. van der Does, A. J. Driessen, B. Oudega, and J. Luirink. 2000. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19:542-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Serrano, M., L. Côrte, J. Opdyke, C. P. Moran, Jr., and A. O. Henriques. 2003. Expression of spoIIIJ in the prespore is sufficient for activation of σG and for sporulation in Bacillus subtilis. J. Bacteriol. 185:3905-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Serrano, M., S. Hövel, C. P. Moran, Jr., A. O. Henriques, and U. Volker. 2001. Forespore-specific transcription of the lonB gene during sporulation in Bacillus subtilis. J. Bacteriol. 183:2995-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Sharp, M. D., and K. Pogliano. 1999. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. USA 96:14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Sharp, M. D., and K. Pogliano. 2002. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science 295:137-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Sharp, M. D., and K. Pogliano. 2003. The membrane domain of SpoIIIE is required for membrane fusion during Bacillus subtilis sporulation. J. Bacteriol. 185:2005-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Sharpe, M. E., and J. Errington. 1995. Postseptational chromosome partitioning in bacteria. Proc. Natl. Acad. Sci. USA 92:8630-8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Shazand, K., N. Frandsen, and P. Stragier. 1995. Cell-type specificity during development in Bacillus subtilis: the molecular and morphological requirements for σE activation. EMBO J. 14:1439-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Singh, R. P., B. Setlow, and P. Setlow. 1977. Levels of small molecules and enzymes in the mother cell compartment and the forespore of sporulating Bacillus megaterium. J. Bacteriol. 130:1130-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Siranosian, K. J., and A. D. Grossman. 1994. Activation of spo0A transcription by σH is necessary for sporulation but not for competence in Bacillus subtilis. J. Bacteriol. 176:3812-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Smith, K., M. E. Bayer, and P. Youngman. 1993. Physical and functional characterization of the Bacillus subtilis spoIIM gene. J. Bacteriol. 175:3607-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Smith, K., and P. Youngman. 1993. Evidence that the spoIIM gene of Bacillus subtilis is transcribed by RNA polymerase associated with σE. J. Bacteriol. 175:3618-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Stevens, C. M., R. Daniel, N. Illing, and J. Errington. 1992. Characterization of a sporulation gene, spoIVA, involved in spore coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Stragier, P. 1986. Comment on “Duplicated sporulation genes in bacteria” by J. Errington, P. Fort, and J. Mandelstam. FEBS Lett. 195:9-11. [DOI] [PubMed] [Google Scholar]
- 275.Stragier, P. 1989. Temporal and spatial control of gene expression during sporulation: from facts to speculation, p. 243-254. In I. Smith, R. A. Slepecky, and P. Setlow (ed.), Regulation of prokaryotic development. American Society for Microbiology, Washington, D.C.
- 276.Stragier, P. 1992. Establishment of forespore-specific gene expression during sporulation of Bacillus subtilis, p. 297-310. In J. A. Cole, F. Mohan, and C. Dow (ed.), Prokaryotic structure and function. American Society for Microbiology, Washington, D.C.
- 277.Stragier, P. 2002. A gene odyssey: exploring the genomes of endospore-forming bacteria, p 519-525. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 278.Stragier, P., C. Bonamy, and C. Karmazyn-Campelli. 1988. Processing of a sporulation σ factor in Bacillus subtilis: how morphological structure could control gene expression. Cell 52:697-704. [DOI] [PubMed] [Google Scholar]
- 279.Stragier, P., J. Bouvier, C. Bonamy, and J. Szulmajster. 1984. A developmental gene product of Bacillus subtilis homologous to the σ factor of Escherichia coli. Nature 312:376-378. [DOI] [PubMed] [Google Scholar]
- 280.Stragier, P., B. Kunkel, L. Kroos, and R. Losick. 1989. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science 243:507-512. [DOI] [PubMed] [Google Scholar]
- 281.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 282.Sun, D. X., R. M. Cabrera-Martinez, and P. Setlow. 1991. Control of transcription of the Bacillus subtilis spoIIIG gene, which codes for the forespore-specific transcription factor σG. J. Bacteriol. 173:2977-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Sun, D. X., P. Stragier, and P. Setlow. 1989. Identification of a new σ-factor involved in compartmentalized gene expression during sporulation of Bacillus subtilis. Genes Dev. 3:141-149. [DOI] [PubMed] [Google Scholar]
- 284.Sun, Y. L., M. D. Sharp, and K. Pogliano. 2000. A dispensable role for forespore-specific gene expression in engulfment of the forespore during sporulation of Bacillus subtilis. J. Bacteriol. 182:2919-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Sussman, M. D., and P. Setlow. 1991. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis gpr gene, which codes for the protease that initiates degradation of small, acid-soluble proteins during spore germination. J. Bacteriol. 173:291-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Takamatsu, H., and K. Watabe. 2002. Assembly and genetics of spore protective structures. Cell. Mol. Life Sci. 59:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Thomaides, H. B., M. Freeman, M. El Karoui, and J. Errington. 2001. Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev. 15:1662-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Tjalsma, H., S. Bron, and J. M. van Dijl. 2003. Complementary impact of paralogous Oxa1-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J. Biol. Chem. 278:15622-15632. [DOI] [PubMed] [Google Scholar]
- 289.Tovar-Rojo, F., M. Chander, B. Setlow, and P. Setlow. 2002. The products of the spoVA operon are involved in dipicolinic acid uptake into developing spores of Bacillus subtilis. J. Bacteriol. 184:584-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Trach, K., D. Burbulys, M. Strauch, J. J. Wu, N. Dhillon, R. Jonas, C. Hanstein, P. Kallio, M. Perego, T. Bird, G. Spiegelman, C. Fogher, and J. A. Hoch. 1991. Control of the initiation of sporulation in Bacillus subtilis by a phosphorelay. Res. Microbiol. 142:815-823. [DOI] [PubMed] [Google Scholar]
- 291.Trach, K. A., and J. A. Hoch. 1993. Multisensory activation of the phosphorelay initiating sporulation in Bacillus subtilis: identification and sequence of the protein kinase of the alternate pathway. Mol. Microbiol. 8:69-79. [DOI] [PubMed] [Google Scholar]
- 292.Trempy, J. E., and W. G. Haldenwang. 1985. σ29-like protein is a common sporulation-specific element in bacteria of the genus Bacillus. J. Bacteriol. 164:1356-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Urtiz-Estrada, N., J. M. Salas-Pacheco, R. E. Yasbin, and M. Pedraza-Reyes. 2003. Forespore-specific expression of Bacillus subtilis yqfS, which encodes type IV apurinic/apyrimidinic endonuclease, a component of the base excision repair pathway. J. Bacteriol. 185:340-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Varcamonti, M., R. Marasco, M. De Felice, and M. Sacco. 1997. Membrane topology analysis of the Bacillus subtilis BofA protein involved in pro-σK processing. Microbiology 143:1053-1058. [DOI] [PubMed] [Google Scholar]
- 295.Varley, A. W., and G. C. Stewart. 1992. The divIVB region of the Bacillus subtilis chromosome encodes homologs of Escherichia coli septum placement (minCD) and cell shape (mreBCD) determinants. J. Bacteriol. 174:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Wakeley, P., N. T. Hoa, and S. Cutting. 2000. BofC negatively regulates SpoIVB-mediated signalling in the Bacillus subtilis σK-checkpoint. Mol. Microbiol. 36:1415-1424. [DOI] [PubMed] [Google Scholar]
- 297.Wakeley, P. R., R. Dorazi, N. T. Hoa, J. R. Bowyer, and S. M. Cutting. 2000. Proteolysis of SpolVB is a critical determinant in signalling of pro-σK processing in Bacillus subtilis. Mol. Microbiol. 36:1336-1348. [DOI] [PubMed] [Google Scholar]
- 298.Wang, X., and J. Lutkenhaus. 1993. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol. Microbiol. 9:435-442. [DOI] [PubMed] [Google Scholar]
- 299.Webb, C. D., A. Decatur, A. Teleman, and R. Losick. 1995. Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J. Bacteriol. 177:5906-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Webb, C. D., A. Teleman, S. Gordon, A. Straight, A. Belmont, D. C. Lin, A. D. Grossman, A. Wright, and R. Losick. 1997. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88:667-674. [DOI] [PubMed] [Google Scholar]
- 301.Wu, J. J., M. G. Howard, and P. J. Piggot. 1989. Regulation of transcription of the Bacillus subtilis spoIIA locus. J. Bacteriol. 171:692-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Wu, J. J., P. J. Piggot, K. M. Tatti, and C. P. Moran, Jr. 1991. Transcription of the Bacillus subtilis spoIIA locus. Gene 101:113-116. [DOI] [PubMed] [Google Scholar]
- 303.Wu, L. J., and J. Errington. 1994. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science 264:572-575. [DOI] [PubMed] [Google Scholar]
- 304.Wu, L. J., and J. Errington. 1997. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J. 16:2161-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Wu, L. J., and J. Errington. 1998. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol. Microbiol. 27:777-786. [DOI] [PubMed] [Google Scholar]
- 306.Wu, L. J., and J. Errington. 2000. Identification and characterization of a new prespore-specific regulatory gene, rsfA, of Bacillus subtilis. J. Bacteriol. 182:418-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Wu, L. J., and J. Errington. 2002. A large dispersed chromosomal region required for chromosome segregation in sporulating cells of Bacillus subtilis. EMBO J. 21:4001-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Wu, L. J., and J. Errington. 2003. RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol. Microbiol. 49:1463-1475. [DOI] [PubMed] [Google Scholar]
- 309.Wu, L. J., A. Feucht, and J. Errington. 1998. Prespore-specific gene expression in Bacillus subtilis is driven by sequestration of SpoIIE phosphatase to the prespore side of the asymmetric septum. Genes Dev. 12:1371-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Wu, L. J., P. J. Lewis, R. Allmansberger, P. M. Hauser, and J. Errington. 1995. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev. 9:1316-1326. [DOI] [PubMed] [Google Scholar]
- 311.Yu, Y. T., and L. Kroos. 2000. Evidence that SpoIVFB is a novel type of membrane metalloprotease governing intercompartmental communication during Bacillus subtilis sporulation. J. Bacteriol. 182:3305-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Zhang, B., A. Hofmeister, and L. Kroos. 1998. The prosequence of pro-σK promotes membrane association and inhibits RNA polymerase core binding. J. Bacteriol. 180:2434-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Zhang, L., M. L. Higgins, and P. J. Piggot. 1997. The division during bacterial sporulation is symmetrically located in Sporosarcina ureae. Mol. Microbiol. 25:1091-1098. [DOI] [PubMed] [Google Scholar]
- 314.Zhang, L., M. L. Higgins, P. J. Piggot, and M. L. Karow. 1996. Role of prespore gene expression in the compartmentalization of mother cell-specific gene expression during sporulation of Bacillus subtilis. J. Bacteriol. 178:2813-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Zhao, Y., and S. B. Melville. 1998. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J. Bacteriol. 180:136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Zheng, L., R. Halberg, S. Roels, H. Ichikawa, L. Kroos, and R. Losick. 1992. Sporulation regulatory protein GerE from Bacillus subtilis binds to and can activate or repress transcription from promoters for mother-cell-specific genes. J. Mol. Biol. 226:1037-1050. [DOI] [PubMed] [Google Scholar]
- 317.Zheng, L. B., W. P. Donovan, P. C. Fitz-James, and R. Losick. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 2:1047-1054. [DOI] [PubMed] [Google Scholar]
- 318.Zheng, L. B., and R. Losick. 1990. Cascade regulation of spore coat gene expression in Bacillus subtilis. J. Mol. Biol. 212:645-660. [DOI] [PubMed] [Google Scholar]
- 319.Zuber, P., and R. Losick. 1983. Use of a lacZ fusion to study the role of the spo0 genes of Bacillus subtilis in developmental regulation. Cell 35:275-283. [DOI] [PubMed]
- 320.Zupancic, M. L., H. Tran, and A. E. Hofmeister. 2001. Chromosomal organization governs the timing of cell type-specific gene expression required for spore formation in Bacillus subtilis. Mol. Microbiol. 39:1471-1481. [DOI] [PubMed] [Google Scholar]