Abstract
Secretory proteins perform a variety of important “remote-control” functions for bacterial survival in the environment. The availability of complete genome sequences has allowed us to make predictions about the composition of bacterial machinery for protein secretion as well as the extracellular complement of bacterial proteomes. Recently, the power of proteomics was successfully employed to evaluate genome-based models of these so-called secretomes. Progress in this field is well illustrated by the proteomic analysis of protein secretion by the gram-positive bacterium Bacillus subtilis, for which ∼90 extracellular proteins were identified. Analysis of these proteins disclosed various “secrets of the secretome,” such as the residence of cytoplasmic and predicted cell envelope proteins in the extracellular proteome. This showed that genome-based predictions reflect only ∼50% of the actual composition of the extracellular proteome of B. subtilis. Importantly, proteomics allowed the first verification of the impact of individual secretion machinery components on the total flow of proteins from the cytoplasm to the extracellular environment. In conclusion, proteomics has yielded a variety of novel leads for the analysis of protein traffic in B. subtilis and other gram-positive bacteria. Ultimately, such leads will serve to increase our understanding of virulence factor biogenesis in gram-positive pathogens, which is likely to be of high medical relevance.
GENERAL INTRODUCTION
Protein export from the cytoplasm to destinations outside the cell is a phenomenon that takes place in all domains of life. Most bacterial proteins destined to leave the cytoplasm are exported via the highly conserved SecA-YEG (Sec) pathway. In addition, more specialized bacterial export pathways are used for the export of specific subsets of extracellular proteins. Most exported proteins are synthesized as precursors with an N-terminal signal peptide (151, 152). These preproteins are first recognized by soluble targeting factors for their transport to the translocation machinery in the cell membrane. Next, the polypeptide chain is transported through a proteinacious channel in the membrane, a process driven by a translocation motor that binds and hydrolyzes nucleotide triphosphates. Finally, the signal peptide is removed, resulting in the release of the mature protein from the membrane. The mature protein folds into its native conformation shortly after the release from the translocase, unless it is translocated in a folded state. These basic principles of protein transport across membranes apply to most eukaryotic and prokaryotic organisms (35, 93, 102, 111, 129).
SCOPE OF THIS REVIEW: THE PROTEOMICS OF PROTEIN SECRETION BY B. SUBTILIS
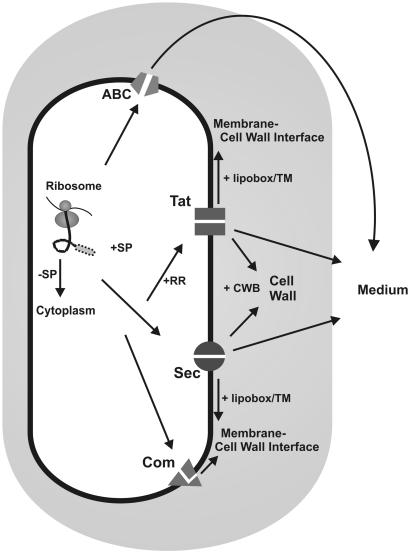
Bacterial secretory proteins are known to perform several very important “remote-control” functions, such as the provision of nutrients, cell-to-cell communication, detoxification of the environment, and killing of potential competitors. More specifically, extracellular proteins of pathogenic bacteria seem to play critical roles in virulence (53, 59, 105). The fact that exported Bacillus subtilis proteins are not retained by an outer membrane, as encountered in gram-negative bacteria, makes this gram-positive bacterium a preferred organism for the proteomic analysis of protein secretion. In addition, the availability of the complete genome sequence (58) and about 3,000 “y”-mutants constructed within the Bacillus subtilis Functional Analysis program (54, 115) make B. subtilis an ideal model organism for research on gram-positive bacteria. Furthermore, previous studies have predicted the composition of the so-called secretome of B. subtilis, which, by our definition, includes both the secreted proteins and the protein secretion machinery (129). These predictions showed that at least four distinct pathways for protein export from the cytoplasm and approximately 300 proteins with the potential to be exported could be distinguished. By far the largest number of exported proteins was predicted to follow the major Sec pathway for protein secretion. In contrast, the recently identified twin-arginine translocation Tat pathway (51, 52), a pseudopilin export pathway for competence development, and pathways using ATP-binding cassette (ABC) transporters, can be regarded as special-purpose pathways through which only few proteins appear to be transported (Fig. 1) (129). In this review, we discuss the latest views of protein secretion by B. subtilis as obtained from recent proteomic studies that were aimed at defining the extracellular complement of the B. subtilis secretome. Using different growth conditions and mutant strains, about 200 extracellular proteins could be visualized by two-dimensional (2D) gel electrophoresis, of which almost 50% could be identified by mass spectrometry (3-6, 46, 51, 52). In summary, these studies showed that in addition to the known mechanisms for protein export, B. subtilis also makes use of alternative mechanisms to release proteins into the external environment. Furthermore, the proteomic data could be used to verify genome-wide predictions concerning the secretome. Even though the process of protein secretion by B. subtilis had been documented fairly well by more classical genetic and biochemical approaches (129, 145), various secretome secrets were unveiled by proteomic approaches. These include the apparent export of cytoplasmic proteins, processing of native membrane proteins by type I signal peptidases (SPases), and the release of normally cell-associated lipoproteins and cell wall proteins into the growth medium.
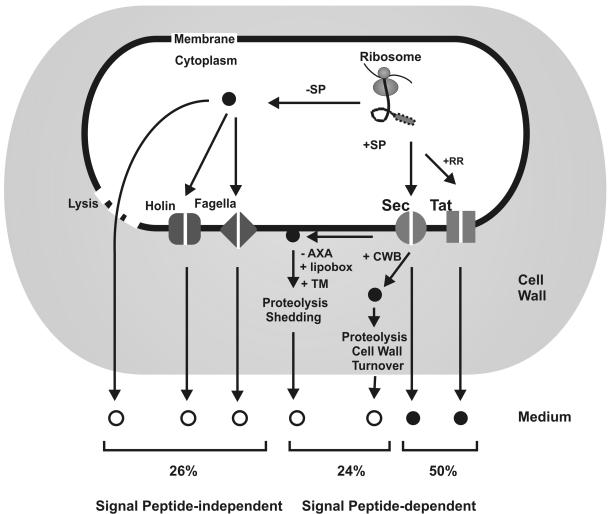
FIG. 1.
Protein export pathways in B. subtilis. Ribosomally synthesized proteins can be sorted to various destinations depending on the presence (+SP) or absence (−SP) of an N-terminal signal peptide and specific retention signals. Proteins devoid of a signal peptide remain in the cytoplasm. Proteins that have to be retained at the extracytoplasmic side of the membrane can contain either a transmembrane segment (TM) or a lipid modification (+lipobox). They are exported via the Sec or Tat pathway. Pseudopilins are exported by the Com system. Proteins that need to be retained in the cell wall can be exported via either the Sec or Tat pathway. To be retained in the cell wall, the mature parts of these proteins contain cell wall-binding repeats (+CWB). Proteins can be secreted into the medium via the Sec or Tat pathway or by ABC transporters.
PROTEIN SORTING IN B. SUBTILIS
Although the soil bacterium B. subtilis has a relatively simple cell structure, proteins can at least be delivered to, or retained at, five (sub)cellular locations: the cytoplasm, the cytoplasmic membrane, the membrane/cell wall interface, the cell wall, and the growth medium (129). The final destination of a protein is governed by the presence or absence of signal peptides and/or retention signals. Nearly all proteins of B. subtilis lacking transport signals are retained in the cytoplasm and fold, with or without the aid of chaperones, into their native conformation. Other proteins contain membrane-spanning domains that are required for their insertion into the cytoplasmic membrane. Most proteins that are completely transported across the cytoplasmic membrane are synthesized with an N-terminal signal peptide. Since B. subtilis lacks an outer membrane, many of these proteins are secreted directly into the growth medium. Other exported proteins involved in processes, such as cell wall turnover, substrate binding, and the folding and modification of translocated secretory proteins, have to be retained at the membrane/cell wall interface to fulfill their function. In the following sections, signal peptides, export routes, and retention mechanisms that are known to be involved in protein sorting in B. subtilis are discussed in the light of recent findings from proteomic analyses.
Signal Peptides
Three distinct domains, N, H, and C, are generally present in signal peptides (148-151). The N-domain contains at least one arginine or lysine residue, which has been suggested to interact with the translocation machinery and the negatively charged phospholipids in the lipid bilayer of the membrane (1, 32). The H-region, following the N-region, is formed by a stretch of hydrophobic residues that can adopt an α-helical conformation in the membrane (21). In the middle of this hydrophobic core, helix-breaking glycine or proline residues are often present to allow the formation of a hairpin-like structure that can insert into the membrane. It was proposed that unlooping of this hairpin results in insertion of the complete signal peptide into the membrane (32). Helix-breaking residues at the end of the H-domain are thought to facilitate cleavage by a specific SPase (88). The C-domain, following the H-domain, contains the cleavage site for specific SPases that remove signal peptides from the mature part of the exported protein during or shortly after translocation. The signal peptide is degraded by signal peptide peptidases and removed from the membrane. Finally, the mature part of the protein is released from the membrane and can fold into its native conformation. Despite the similar structure of signal peptides, apparently small variations can result in transport to different destinations and/or export via different pathways, as described below.
Signal Peptide Prediction and Classification
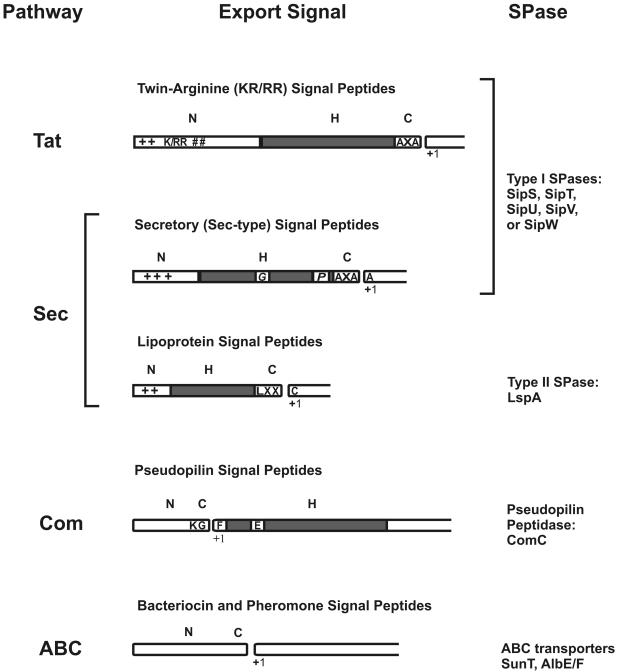
Predictions showed that 300 proteins with the potential to be exported could be distinguished in B. subtilis (129). On the basis of SPase cleavage sites and the export pathways by which these preproteins are (predicted to be) exported, signal peptides can be divided into five distinct classes: (i) twin-arginine (RR/KR) signal peptides, (ii) secretory (Sec-type) signal peptides, (iii) lipoprotein signal peptides, (iv) pseudopilin-like signal peptides, and (v) bacteriocin and pheromone signal peptides. The first group of signal peptides contains a so-called twin-arginine (RR/KR) motif, which serves to direct proteins into the Tat pathway (51). The second, and most abundant, class is composed of typical secretory signal peptides (lacking an RR/KR-motif) that direct proteins into the Sec pathway. Both the twin-arginine and secretory signal peptides appear to be cleaved by one of the various type I SPases of B. subtilis (130). The third class of signal peptides is present at the N terminus of prelipoproteins that are exported via the Sec pathway, lipid modified, and cleaved by the type II SPase (Lsp) (136). The fourth class is formed by signal peptides of pseudopilins which, in B. subtilis, are cleaved by the SPase ComC (64). Finally, the fifth class of signal peptides is found on ribosomally synthesized pheromones and lantibiotics that are exported and cleaved by ABC transporters (80). It should be noted that this specific class of signal peptides is often referred to as “leader peptides.”
Twin-arginine (RR/KR-type) signal peptides.
Signal peptide predictions resulted in the identification of ∼180 potential substrates for type I SPases. A twin-arginine motif, containing at least three residues of the consensus sequence R/K-R-X-#-# (where # is a hydrophobic residue) was found in 44 of these signals (12 RR and 32 KR signal peptides; [51]). The presence of such twin-arginine motifs was initially interpreted as an indication that the corresponding preproteins could be directed into the Tat pathway for protein export, possibly in a Sec-independent manner. The predicted twin-arginine signal peptides with a consensus R-R-X-#-# motif have an average length of 36 amino acid residues. Thus, they are significantly longer than typical Sec-type signal peptides. This is mainly because the N-domains of these R-R-X-#-# containing signal peptides have an average length of 14 amino acid residues, almost twice as long as the N-domain of the regular (Sec-type) signals (Fig. 2). Furthermore, these N-domains contain, on average, more positively charged residues than do those of Sec-type signal peptides (129). In contrast, the average features of predicted twin-arginine signal peptides with a K-R-X-#-# motif are similar to those of Sec-type signal peptides (51, 52, 129).
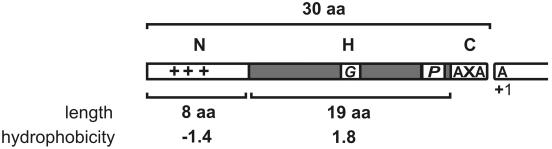
FIG. 2.
Classification of cleavable N-terminal signal peptides. On the basis of SPase cleavage sites and the export pathways via which the preproteins are exported, predicted signal peptides (129) were divided into five distinct classes: twin-arginine (RR/KR) signal peptides, secretory (Sec-type) signal peptides, lipoprotein signal peptides, pseudopilin-like signal peptides, and bacteriocin and pheromone signal peptides. The export pathways via which the preproteins are exported and the SPases responsible for their cleavage are indicated. Most signal peptides have a tripartite structure: a positively charged N-domain (N), containing lysine and/or arginine residues (indicated by +), a hydrophobic H-domain (H, indicated by a gray box), and a C-domain (C) that specifies the cleavage site for their specific SPase. The length of the signal peptides and their subdomains is drawn to the same scale. Furthermore, helix-breaking residues, mostly glycine or proline (G/P), in the H-domain of Sec-type signal peptides are indicated. These residues are, respectively, thought to facilitate loopwise membrane insertion and cleavage by SPase I (129). Finally, where appropriate, the most frequently occurring first amino acid of the mature protein (+1) is indicated.
Secretory (Sec-type) signal peptides.
The 135 predicted signal peptides lacking RR/KR motifs have an average length of 28 residues and contain two or three positively charged lysine (L) or arginine (R) residues in their N-domain. The hydrophobic core (H-domain) has an average length of 19 residues, and about 60% of the predicted Sec-type signal peptides contains a helix-breaking residue (mostly glycine) in the middle of this domain. The C-domain of the predicted signal peptides carries a type I SPase cleavage site, with the consensus sequence A-S-A at positions −3 to −1 relative to the cleavage site. About 50% of these signal peptides contain a helix-breaking residue (proline or glycine) at positions −7 to −4 relative to the predicted processing site for SPase I (129).
Lipoprotein signal peptides.
Lipoprotein signal peptide predictions resulted in the identification of 114 potential substrates for the lipoprotein-specific (type II) SPase (Lsp) (133). Signal peptides from lipoproteins have an average length of 19 residues. These are therefore considerably shorter than RR/KR- and Sec-type signal peptides. This is because both the N-domain (average of 4 residues) and the H-domain (average of 12 residues) are shorter than the corresponding domains in RR/KR- and Sec-type signal peptides. Furthermore, helix-breaking residues are not conserved in the H-region of lipoprotein signal peptides. The C-domain contains a so-called lipobox with the consensus sequence L-(A/S)-(A/G)-C. The invariable cysteine residue of the lipobox is the target for lipid modification and the first residue of the mature lipoprotein after cleavage by SPase II (Fig. 2) (129). In fact, this lipid modification is indispensable for signal peptide cleavage by SPase II. Finally, although some lipoprotein signal peptides contain an RR/KR motif (51), so far export of lipoproteins via the Tat pathway has not been reported.
Pseudopilin-like signal peptides.
Only four proteins (ComGC, ComGD, ComGE, and ComGG) with pseudopilin signal peptides have been identified in B. subtilis (129). These pseudopilin signal peptides have an average length of 33 residues. Strikingly, the C-domain of pseudopilin signal peptides, with the consensus sequence K-G-F at positions −2 to +1 relative to the SPase cleavage site, is located between the N- and H-domains (Fig. 2). This is in line with the observation that the pseudopilin signal peptidase (ComC) acts at the cytoplasmic side of the membrane (64). In addition to processing, ComC is responsible for aminomethylation of the phenylalanine at position +1 relative to the cleavage site. Although pseudopilin signal peptides show structural similarity to the previously described signal peptides, pseudopilin precursors bypass the Tat and Sec pathways and are transported via the specific Com pathway (26, 27, 129).
Signal peptides of pheromones and bacteriocins.
Pheromones and antimicrobial peptides form a distinct group of exported proteins with cleavable N-terminal signal peptides, often called leader peptides. These leader peptides consist of only N- and C-domains and completely lack a hydrophobic H-domain (Fig. 2). It has been described that parts of the mature protein are also required for export by a dedicated ABC transporter. Moreover, the leader peptide has an important function in the prevention of premature antimicrobial activity and is required for the posttranslational modification of lantibiotics (141, 144). The two known leader peptides of this type in B. subtilis 168 direct the secretion of sublancin 168 (89) and ComX (67). Like leader peptides of other lantibiotics (23, 84), the sublancin 168 leader peptide contains a double-glycine motif (GS) N-terminally of the SPase cleavage site. Interestingly, the ABC transporter SunT is likely to play a dual role in the secretion of sublancin 168 since it belongs to a class of ABC transporters that are responsible for both the removal of the leader peptide and the translocation of the mature lantibiotic across the cytoplasmic membrane (33). Although not documented, it seems likely that an ABC transporter is also responsible for the processing and secretion of the ComX pheromone. This pheromone is involved in the density-controlled onset of competence development, and, similar to sublancin 168, it is ribosomally synthesized as a precursor and modified before secretion (119).
Retention Signals
In gram-negative bacteria, the outer membrane confines numerous proteins to the periplasm. The membrane/cell wall interface of B. subtilis defines a cellular area that is analogous to the gram-negative periplasm and contains many proteins that fulfill important functions (72, 94). Proteins retained at the membrane/cell wall interface include substrate-binding proteins, chaperones for protein secretion, RNases, DNases, enzymes involved in the synthesis of peptidoglycan (penicillin-binding proteins), and cell wall hydrolases, which are involved in cell wall turnover during cell growth, cell division, sporulation, and germination (10, 14, 39, 77, 95, 129). To prevent the loss of these proteins, various retention mechanisms are employed by the cell.
Transmembrane domains.
Membrane proteins with large extracytoplasmic domains are translocated across the membrane by the Sec or Tat machinery. Due to the presence of one or more transmembrane domains and the absence of an SPase cleavage site, such proteins remain anchored to the membrane. The N-terminal transmembrane domain with an Nin-Cout topology is regarded as an uncleaved signal peptide, and the absence of a proper SPase I cleavage site is regarded as a determinant for retention in the membrane. Furthermore, certain proteins containing cleavable N-terminal signal peptides contain additional transmembrane domains in their C terminus that can function as membrane anchors (5, 51, 129). It should be noted that proteins with predicted putative transmembrane domains were regarded as nonsecretory proteins in previous secretome predictions (129, 143).
Lipid modification.
In Gram-positive bacteria, lipid modification of exported proteins can serve to retain these proteins at the extracytoplasmic membrane surface. This may explain why 32 lipoproteins of B. subtilis are homologues of periplasmic high-affinity substrate-binding proteins from gram-negative bacteria (136). Lipid-modified proteins are synthesized as prelipoproteins and have to be modified by the diacylglyceryl transferase (Lgt) (62) before the lipoprotein precursor can be processed by SPase II. The diacylglyceryl group, attached to the cysteine residue at position +1 of the mature lipoprotein, inserts into the lipid bilayer of the cytoplasmic membrane, preventing release of the protein into the environment. It is noteworthy that some lipoproteins, such as CtaC (12) and QoxA (5), contain transmembrane segments in addition to a lipoprotein signal peptide. In these cases, lipid modification seems to be required for optimal functionality rather than for cell retention.
Pseudopilin assembly.
A specific class of exported B. subtilis proteins that remain attached to the cytoplasmic membrane consists of the above-mentioned pseudopilins ComGC, ComGD, ComGE, and ComGG. These proteins are required for the binding and uptake of exogenous DNA during genetic competence (34). These resemble type IV pilins of various gram-negative bacteria that are synthesized as precursors with cleavable signal peptides. After cleavage and modification, the hydrophobic H-domains represent the N termini of mature pseudopilins, which are thought to form pilin-like structures that are attached to the cytoplasmic membrane (98).
Cell wall-binding repeats.
Several B. subtilis enzymes involved in cell wall turnover contain a variable number of repeated domains (129) in their noncatalytic C termini, which have affinity for components of the cell wall (41, 69, 100). These repeats are thought to direct enzymes for cell wall assembly and turnover to specific sites, where cell wall synthesis and/or hydrolysis take place, as was shown for Staphylococcus aureus (8, 9). The targeting to a specific location is most probably promoted by certain components of the cell wall, such as choline, which is a receptor for several cell wall proteins of Streptococcus pneumoniae (106, 109, 110).
Covalent attachment to the cell wall.
A specific group of surface proteins from gram-positive organisms is covalently anchored to the cell wall via the C terminus (112, 113). Cell wall anchoring of a variety of surface proteins in S. aureus requires, in addition to an N-terminal signal peptide, a C-terminal cell wall sorting signal consisting of the so-called LPxTG motif, a C-terminal hydrophobic domain, and a positively charged tail (82, 83, 114). A specific transpeptidase, the sortase A (SrtA), is responsible for both cleavage of the cell wall sorting signal (between the Thr and Gly residues of the LPxTG motif) and covalent attachment of the carboxyl group of the Thr residue to the cell wall (137, 138). A second, and structurally related, C-terminal cell wall sorting signal in S. aureus, Bacillus halodurans, and Bacillus anthracis contains the NPQTN motif. This sorting signal is most probably cleaved between the Thr and Asn residues by sortase B (SrtB), a paralogue of SrtA (70). Two sortase homologues, YhcS and YwpE, were identified in B. subtilis, suggesting that sortase-like enzymes for the cleavage and cell wall linkage of surface proteins are present in B. subtilis. However, no exported B. subtilis proteins with LPxTG or NPQTN motifs were identified (129). This indicates either that B. subtilis does not use this cell wall retention mechanism or that YhcS and YwpE recognize a cell wall sorting signal with a different amino acid sequence.
PROTEOMICS OF PROTEIN SECRETION BY B. SUBTILIS
The first proteomic approaches to define the extracellular complement of the secretome of B. subtilis 168 were made by Hirose et al. (46). In their study, cells were grown in minimal media with glucose, maltose, cellobiose, or starch. Extracellular proteins were separated by 2D polyacrylamide gel electrophoresis (2D PAGE) and identified by N-terminal sequencing. In subsequent studies by Antelmann et al. (3) and Jongbloed et al. (52), B. subtilis was grown under conditions of phosphate starvation, or in Luria-Bertani (LB) broth (4, 5, 6). Extracellular proteins separated by 2D PAGE were identified by matrix-assisted laser desorption ionization/time-of-flight (MALDI-TOF) mass spectrometry. The highest levels of protein secretion are usually observed when cells of B. subtilis are grown in rich media, in particular during the postexponential growth phase (see Fig. 3). Moreover, the relative amounts of most identified extracellular proteins were significantly increased during the postexponential growth phase (5). The importance of protein secretion during postexponential growth was highlighted by the fact that the extracellular levels of a subset of 13 degradative enzymes are strongly increased in the extracellular proteome of a B. subtilis degU32(hy) mutant (5, 61). Recent transcript profiling experiments (66) have confirmed that the genes encoding these degradative enzymes are indeed under the positive control of DegU-phosphate, causing their increased expression after the end of the exponential growth phase.
FIG. 3.
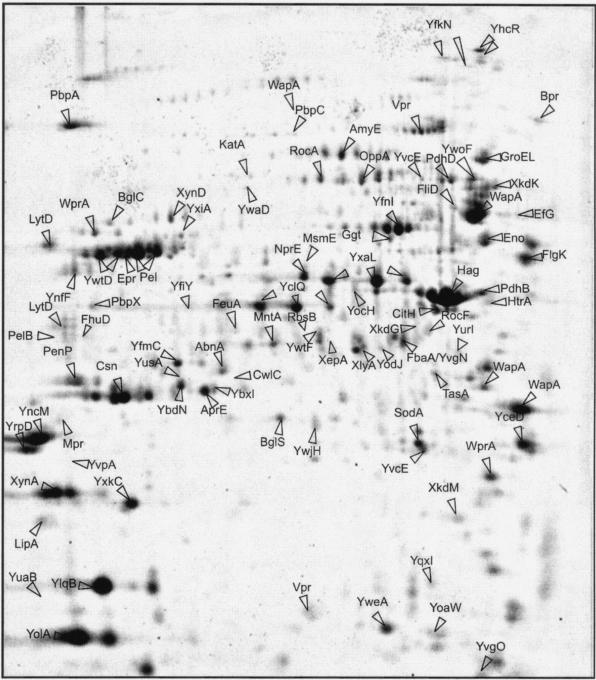
Master gel for the extracellular proteome of B. subtilis 168. Cells of B. subtilis 168 were grown in LB broth, and extracellular proteins were harvested 1 h after entry into the stationary growth phase. After precipitation with trichloroacetic acid, the extracellular proteins were separated by 2D PAGE and stained with Sypro Ruby as described by Jongbloed et al. (51). The proteins identified by mass spectrometry and/or N-terminal amino acid sequencing are indicated on the gel and listed in Table 1. The extracellular proteins found specifically during growth in minimal media (i.e., YdhT, YflE, and GapA) (46) and those specifically found during growth in phosphate starvation medium (i.e., GlpQ, PhoA, PhoB, PhoD, PstS, YdhF, YcdH, and YrpE) (5, 6, 51, 52) cannot be seen on this gel.
Extracellular Proteome of B. subtilis 168
From the approximately 200 visible extracellular protein spots, 75 different proteins could be identified as marked in the 2D master gel for the extracellular proteome (Fig. 3; Tables 1 and 2). Therefore, B. subtilis 168 cells were grown in Luria-Bertani broth and extracellular proteins were harvested from the medium 2 h after entry into the postexponential growth phase (5). In the medium of phosphate-starved cells, eight additional extracellular proteins were identified (3, 5, 52). When B. subtilis cells were grown in minimal media, much lower levels of extracellular proteins were detected (46). Nevertheless, these studies resulted in the identification of three additional extracellular proteins. In total, 90 extracellular proteins were identified, including 53 proteins to which a function has been assigned previously and 37 “Y-proteins” of unknown function (Tables 1 and 2). A possible function could be attributed to 20 Y-proteins based on their amino acid sequence similarity to proteins with a known function. In summary, the identified extracellular proteins of B. subtilis 168 include enzymes related to the metabolism of carbohydrates, proteases, or peptidases, enzymes involved in the metabolism of amino acids, enzymes involved in the decay of DNA or RNA, lipases, alkaline phosphatases, phosphodiesterases, enzymes involved in cell wall biogenesis, lipoproteins (many of which are substrate-binding components of various transport systems), proteins involved in detoxification, flagellum-related proteins, putative transcriptional regulators, proteins involved in protein synthesis and folding, prophage-related proteins, sporulation-specific proteins, and proteins of unknown function. In addition, Chu et al. (25) identified five extracellular proteins of B. subtilis strain K-1, which were specifically induced by growth in xylan-containing medium. These are a xylose isomerase homologous to XylA of B. subtilis 168, two endoxylanases homologous to XynA and XynD of B. subtilis 168, a dehydroquinate dehydratase homologous to AroC of B. subtilis 168, and a regulatory protein homologous to GltC of B. subtilis 168. The latter proteins are not included in Tables 1 and 2, which list only the extracellular proteins of the B. subtilis 168 strain.
TABLE 1.
Extracellular proteins of B. subtilis 168 with export signalsa
| Protein | Function or similarity | Export signalb | SPase | Retention signalc | Mediumd |
|---|---|---|---|---|---|
| AbnA | Arabinan-endo-1,5-α-l-arabinase | KRe | SPase I | − | LB |
| AmyE | α-Amylase | Sec | SPase I | − | LB, PS, |
| AprE | Serine alkaline protease (subtilisin E) | Sec | SPase I | − | LB, PS |
| BglC | Endo-1,4-β-glucanase, cellulase | KRe | SPase I | − | LB |
| BglS | Endo-β-1,3-1,4 glucanase | KRe | SPase I | − | LB, PS |
| Bpr | Bacillopeptidase F | Sec | SPase I | − | LB, PS |
| Csn | Chitosanase | Sec | SPase I | − | LB, MG |
| Epr | Minor extracellular serine protease | Sec | SPase I | − | LB |
| Ggt | γ-Glutamyltranspeptidase | Sec | SPase I | − | LB |
| GlpQ | Glycerophosphoryl diester | Sec | SPase I | − | PS |
| HtrA | Serine protease | TM | Unknownf | − | LB |
| LipA | Lipase | RRe | SPase I | − | LB |
| LytD | N-Acetylglucosaminidase (major autolysin) | KRe | SPase I | CWB | LB, PS |
| MntA | Manganese-binding protein | Lipo | LspA | Lipid | LB |
| Mpr | Extracellular metalloprotease | Sec | SPase I | − | LB |
| NprE | Extracellular neutral metalloprotease | Sec | SPase I | − | LB, PS |
| OppA | Oligopeptide-binding protein | Lipo/KRe | LspA | Lipid | LB, PS |
| PbpA | Penicillin-binding protein 2A | TM | Unknownf | − | LB |
| PbpX | Penicillin-binding protein | RRe | SPase I | − | LB |
| Pel | Pectate lyase | Sec | SPase I | − | LB, PS, MG |
| PelB | Pectate lyase | Sec | SPase I | − | LB |
| PenP | β-Lactamase precursor | Sec | SPase I | − | LB, MG |
| PhoA | Alkaline phosphatase A | Sec | SPase I | − | PS |
| PhoB | Alkaline phosphatase III | Sec | SPase I | − | PS |
| PhoD | Phosphodiesterase/alkaline phosphatase D | RR | SPase I | − | PS |
| PstS | Phosphate-binding protein | Lipo | LspA | Lipid | PS |
| TasA | Antimicrobial spore component | Sec | SipWg | − | LB, MG |
| Vpr | Extracellular serine protease | Sec | SPase I | − | LB, PS, |
| WapA | Cell wall-associated protein precursor | RRe | SPase I | CWB | LB, PS, MG |
| WprA | Cell wall-associated protein precursor | RRe | SPase I | CWBh | LB, PS, MG |
| XynA | Endo-1,4-β-xylanase | Sec | SPase I | − | LB, PS |
| XynD | Endo-1,4-β-xylanase | Lipo | LspA | Lipid | LB, PS, MG |
| YbdN | Unknown | Sec | SPase I | − | LB |
| YbxI | Similar to β-lactamase | Sec | SPase I | − | LB |
| YcdH | Zinc-binding protein | Lipo | LspA | Lipid | PS |
| YclQ | Ferrichrome-binding protein | Lipo | LspA | Lipid | LB, PS, MG |
| YdhF | Similar to unknown proteins from B. subtilis | Lipo/RRe | LspA | Lipid | PS |
| YdhT | Mannan endo-1,4-β-mannosidase | Sec | SPase I | − | MC |
| YfkN | 2′,3′-Cyclic-nucleotide 2′-phosphodiesterase | RRe | SPase I | TM | LB, PS |
| YfIE | Similar to anion-binding protein | TM | SPase I | − | MG |
| YfmC | Ferrichrome-binding protein | Lipo | LspA | Lipid | LB |
| YfnI | Probable transmembrane glycoprotein | TM | SipT/SipVi | − | LB, PS, MG |
| YhcR | 5′-Nucleotidase | RRe | SPase I | TM | LB |
| YlqB | Unknown | Sec | SPase I | − | LB, PS, MG |
| YncM | Similar to unknown proteins from B. subtilis | Sec | SPase I | − | LB, PS, MG |
| YnfF | Endo-xylanase | Sec | SPase I | − | LB, PS |
| YoaW | Unknown | Sec | SPase I | − | LB |
| YocH | Cell wall-binding protein | Sec | SPase I | CWBj | LB |
| YolA | Unknown | KRe | SPase I | − | LB |
| YqiX | Amino acid-binding protein | Lipo | LspA | Lipid | LB |
| YqxI | Unknown | Sec | SPase I | − | LB |
| YrpD | Similar to unknown proteins from B. subtilis | Sec | SPase I | − | LB, PS |
| YrpE | Similar to unknown proteins | Lipo | LspA | Lipid | PS |
| YuaB | Unknown | Sec | SPase I | − | LB |
| YurI | RNase | Sec | SPase I | − | LB |
| YvcE | Cell wall-binding protein | Sec | SPase I | CWB10 | LB |
| YvgO | Unknown | Sec | SPase I | − | LB |
| YvpA | Pectate lyase | Sec | SPase I | − | LB |
| YwaD | Aminopeptidase | Sec | SPase I | − | LB |
| YweA | Similar to unknown proteins from B. subtilis | Sec | SPase I | − | LB, PS |
| YwoF | Unknown | Sec | SPase I | − | LB, PS |
| YwtD | γ-dl-Glutamyl hydrolase | Sec | SPase I | CWB10 | LB, PS, MG |
| YwtF | Transcriptional regulator | Sec | SPase I | − | LB |
| YxaLk | Similar to serine/threonine protein kinase | Sec | SPase I | − | LB, PS, MG |
| YxiA | Arabinan-endo-1,5-α-l-arabinase | Sec | SPase I | − | LB |
| YxkC | Unknown | Sec | SPase I | − | LB, PS, MG |
All listed proteins were identified by 2D PAGE and subsequent MALDI-TOF mass spectrometry and/or N-terminal amino acid sequencing as described by Hirose et al. (46), Jongbloed et al. (51, 52), and Antelmann et al. (3-6). Putative signal peptides. SPase I or SPase II cleavage sites, transmembrane domains, and cell wall-binding domains were predicted as described by Tjalsma et al. (129) and Jongbloed et al. (51).
Identified transient export signals are Sec-type signal peptides (Sec), twin-arginine signal peptides (RR/KR), lipoprotein signal peptides (Lipo), and transmembrane domains (TM).
Identified retention signals present in the mature part of the protein after processing by specific SPases are lipid modifications (Lipid), transmembrane domains (TM), and cell wall-binding domains (CWB). −, absence of known retention signals.
Proteins found in the extracellular proteomes of cells were grown in LB broth (rich medium) (4, 5, 6, 51), phosphate starvation medium (PS) (3, 5, 52), or minimal medium with glucose (MG) or cellobiose (MC) (46).
Despite the presence of putative RR/KR-type signal peptides, release of AbnA, BglC, BglS, LipA, LytD, OppA, PbpX, WapA, WprA, YdhF, YfkN, YhcR, and YolA into the growth medium is not Tat dependent (51).
As pre-PbpA and pre-HtrA lack putative SPase I cleavage sites, it is unknown which protease is responsible for their cleavage and subsequent release into the medium (4, 5).
pre-TasA processing and release of mature TasA into the medium is strictly dependent on the ER-type SPase SipW (126, 131).
WprA is known to be a major cell wall protein (6, 68), but it lacks a typical cell wall-binding motif.
Release of the C-terminal part of YfnI into the medium was shown to be dependent on the presence of SipT or SipV (5).
Despite the presence of putative cell wall-binding domains, YocH, YvcE, and YwtD are not detected in the cell wall proteome of B. subtilis 168 (6).
The protein YxaL was previously annotated as YxaK (5).
TABLE 2.
Extracellular proteins of B. subtilis 168 without typical export signalsa
| Protein | Function or similarity | % Cytoplasmic abundanceb | Mediumc |
|---|---|---|---|
| CitH | Malate dehydrogenase | 1.20 | LB |
| CwlCw ex | N-Acetylmuramoyl-l-alanine amidase | − | LB |
| Ef-G | Elongation factor G | 1.91 | LB |
| Eno | Enolase | 1.23 | LB, PS |
| FlgKex | Flagellar hook-associated protein 1 | − | LB |
| FliDex | Flagellar hook-associated protein 2 | − | LB |
| GapA | Glyceraldehyde-3-phosphate | 1.20 | MG,C,M,S |
| GroEL | Class I heat shock protein (chaperonin) | 1.30 | LB, PS |
| Hagdual | Flagellin protein | 1.27 | LB, PS, MG,C,M,S |
| KatAex | Vegetative catalase 1 | − | LB, PS, MM |
| PdhA | Pyruvate dehydrogenase (E1 α subunit) | 0.71 | LB |
| PdhB | Pyruvate dehydrogenase (E1 β subunit) | 0.57 | LB |
| PdhD | Pyruvate dehydrogenase (E3 subunit) | 0.76 | LB, PS |
| RocA | Pyrroline-5 carboxylate dehydrogenase | − | LB |
| RocF | Arginase | − | LB |
| SodA | Superoxide dismutase | 1.07 | LB, PS, MG,M,S |
| XepAex | PBSX prophage lytic exoenzyme | − | LB |
| XkdGex | PBSX prophage gene | − | LB, MG |
| XkdKex | PBSX prophage gene | − | LB, PS |
| XkdMex | PBSX prophage gene | − | LB, PS |
| XlyAw ex | N-Acetylmuramoyl-l-alanine amidase | − | LB |
| YceD | Similar to tellurium resistance protein | − | LB |
| YvgNd | Similar to plant metabolite | − | LB |
| YwjH | Similar to transaldolase (pentose) | 0.37 | LB |
All listed proteins were identified by 2D PAGE and subsequent MALDI-TOF mass spectrometry and/or N-terminal amino acid sequencing as described by Hirose et al. (46), Antelmann et al. (5), and Vitikainen et al. (147). Proteins with a known dual localization (cellular and extracellular) are labeleddual, proteins that lack a typical signal peptide but have a known extracytoplasmic localization are labeledex, and proteins containing cell wall-binding repeats are marked withW.
Relative protein levels are expressed as the percentage of total protein content. Protein levels were determined by 2D PAGE of cytoplasmic protein extracts of B. subtilis cells grown in minimal medium with glucose (22). A minus (−) means that the cytoplasmic abundance is lower than 0.37%.
Proteins found in the extracellular proteomes of cells grown in LB broth (rich medium) 5), phosphate-starvation medium (PS) (4), or minimal medium with glucose (MG), cellobiose (MC), maltose (MM), or starch (MS) (46).
The YvgN protein was recently renamed FbaA (147).
Toward an Extracellular Zymoproteome of B. subtilis 168
In addition to the identification of extracellular proteins, proteomics can be used to attribute functions to extracellular proteins. An early exploration in this area was performed by Park et al. (92), who used a proteomic approach to detect fibrinolytic enzymes in the medium of B. subtilis 168. For this purpose, images of 2D PAGE gels were superimposed to detect extracellular protein spots that coincided with clearing zones on a 2D zymogram gel containing bovine fibrinogen. In this way, four protein spots with fibrinolytic activity were identified. These spots were shown to correspond to differently processed forms of the serine proteases WprA and Vpr. The processed form of WprA with proteolytic activity is usually referred to as CWBP52.
Cell Wall Proteome of B. subtilis 168
The unexpected identification of relatively large quantities of several proteins with cell wall-binding domains (e.g., LytD, WapA, YocH, YvcE, and YwtD [Tables 1 and 2]) in the extracellular proteome may relate to the well-known fact that B. subtilis undergoes cell wall turnover (7). This finding prompted Antelmann et al. (6) to define the protein composition of the cell wall, which revealed that seven LiCl-extractable proteins are present in this compartment of the B. subtilis cell (Table 3). These proteins include the known cell wall-bound proteins LytB and LytC (both involved in cell wall biogenesis), the CWBP23- and CWBP52-processing products of WprA (cell wall-located protease), and processed forms of WapA (structural cell wall-binding protein). Furthermore, the flagellum-related protein Hag and two proteins with unknown functions, YwsB and YqgA, are present in the cell wall proteome. It should be noted that although processed forms of WprA are known as cell wall proteins, they lack the typical cell wall-binding repeats that are present in LytB, LytC, and WapA (6, 68, 129). Surprisingly, two additional cell wall-located proteins, YwsB and YqgA, also lack known cell wall retention motifs. Finally, it was remarkable that extracellular proteins with cell wall-binding motifs, such as LytD, YocH, YvcE, and YwtD, were apparently absent from the cell wall proteome. It has to be emphasized that LytD, YvcE, and YwtD are abundantly present in the extracellular proteome under the growth conditions used to identify cell wall-associated proteins (5), showing that the absence of these proteins from the wall proteome is not due to a lack of expression. Probably, the same is true for YocH, but this is less clearly evident from the published data (6).
TABLE 3.
Cell wall-located proteins of B. subtilis 168a
| Protein | Function or similarity | Export signalb | SPase | Retention Signalc | Found in mediumd |
|---|---|---|---|---|---|
| Hag | Flagellin protein | − | − | − | Y |
| LytB | Modifier protein of autolysin LytC | Sec | SPase I | CWB | N |
| LytC | N-Acetylmuramoyl-l-alanine amidase | Sec | SPase I | CWB | N |
| WapA | Cell wall-associated protein precursor | RR | SPase I | CWB | Y |
| WprA | Cell wall-associated protein precursor | RR | SPase I | − | Y |
| YqgA | Similar to unknown proteins of B. subtilis | Sec | SPase I | − | N |
| YwsB | Similar to unknown proteins of B. subtilis | Sec | SPase I | − | N |
All listed cell wall-located proteins were identified by 2D PAGE and subsequent MALDI-TOF mass spectrometry and/or N-terminal amino acid sequencing as described by Antelmann et al. (6). Putative signal peptides, SPase I cleavage sites, and cell wall-binding domains were predicted as described by Tjalsma et al. (129). −, absence of known signal peptides, SPase I cleavage sites, or cell wall-binding proteins.
Identified transient export signals are Sec-type signal peptides (Sec) and RR-type signal peptides (RR).
Identified retention signals present in the mature part of the protein after processing by specific SPases are cell wall-binding domains (CWB).
The presence (Y) or absence (N) of a particular protein of the cell wall proteome in the growth medium is indicated.
Verification of Secretome Predictions
The availability of accumulating proteomic data concerning the extracellular complement of the secretome of B. subtilis 168 has allowed proteomic verification of the genome-based predictions of the composition of the secretome as previously performed (129). Intriguingly, only 48 (53%) of the 90 identified extracellular proteins are expected to be released into the medium, as judged by the presence of predicted signal peptides and a lack of retention signals (Tables 1 and 2). A potential RR/KR motif is present in the N-domains of 14 signal peptides of the latter group of proteins, suggesting their potential transport via the Tat pathway. The remaining 34 proteins contain a Sec-type signal peptide and are most probably exported by the Sec pathway of B. subtilis. Strikingly, 47% of the extracellular proteome currently cannot be predicted to end up at this location (129). This unpredicted fraction consists of proteins which have an N-terminal lipoprotein signal peptide (cleaved by SPase II) or potential transmembrane segments according to the TMHMM algorithm (28). Both groups of proteins are supposed to be retained in or at the cytoplasmic membrane. In addition, some predicted preproteins with a type I SPase cleavage site contain typical cell wall-binding repeats and therefore have a predicted cell wall localization. As listed in Table 2, 24 proteins found in the medium of B. subtilis are in fact proteins that lack a typical export signal. The latter include flagellum-related proteins, prophage-related proteins, and proteins with known or predicted enzymatic activities in the cytoplasm. The possible mechanisms by which these proteins are released from the cell are discussed in “Mechanisms for extracellular accumulation of proteins” (below).
Similarly, only about half of the identified cell wall proteins are predicted to be retained at this subcellular location, since Hag, the WprA-processing products CWBP23 and CWBP52, YqgA, and YwsB lack known cell wall-binding motifs. The last two proteins are, in fact, found exclusively in the cell wall, like the known cell wall-bound proteins LytB and LytC. This suggests that an as yet undefined cell wall retention signal is present in YqgA and YwsB. Conversely, the remarkable observation that four proteins with typical cell wall-binding domains (i.e., LytD, YocH, YvcE, and YwtD) are found extracellularly, but not cell wall bound, might indicate that the presence of a cell wall-binding repeat is not a guarantee for retention at this location. In this respect, it may be relevant that YwtD exclusively cleaves extracellular γ-polyglutamic acid whereas it cannot use cell wall peptidoglycan as a substrate (127). However, the possibilities that LytD, YocH, YvcE, and YwtD are not properly extracted from the wall with LiCl, or that these proteins are degraded during the extraction procedure, cannot be excluded.
CONTRIBUTION OF THE Sec MACHINERY TO THE EXTRACELLULAR PROTEOME
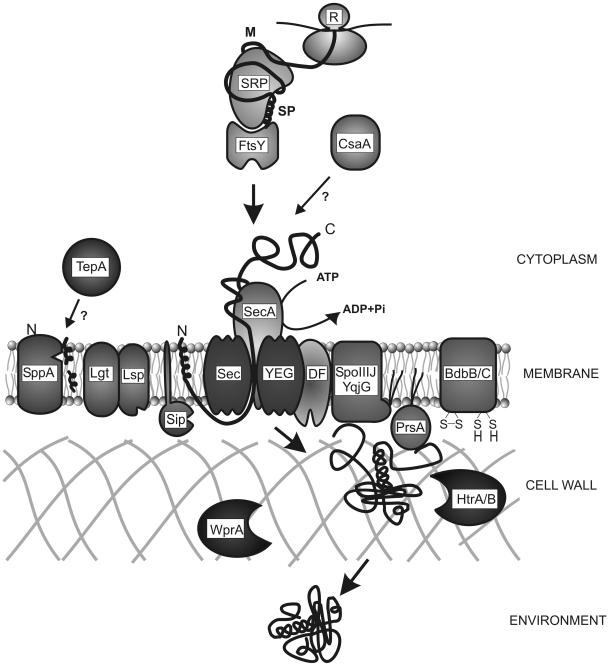
Protein secretion via the Sec pathway in B. subtilis can be divided into three functional stages: targeting, translocation, and folding and release. The following components have known or predicted functions in these stages. Cytoplasmic chaperones, such as SRP/FtsY (47) and CsaA (75, 76), keep the precursors in a translocation competent state and facilitate their targeting to the translocase in the membrane. The translocation machinery consists of SecA (motor), SecYEG (pore), and SecDF. Possibly, YrbF and SpoIIIJ/YqjG are also part of this machinery (17, 129, 135, 145). During or shortly after translocation, the preprotein is cleaved by one of the five type I signal peptidases (SipS to SipW)(130) or lipid-modified by the diacylglyceryl transferase (Lgt) (62) and cleaved by the lipoprotein-specific signal peptidase (Lsp; 133, 136). SppA and TepA may be involved in the degradation of cleaved signal peptides (16). The folding of several secreted proteins depends on the activities of PrsA (55), BdbBCD (18, 71), and/or SpoIIIJ/YqjG (135). HtrA and HtrB (85), as well as WprA (68), are involved in the quality control of secretory proteins. Importantly, HtrA and HtrB have the potential to assist in the folding or, if folding is impossible, degradation of malfolded secretory proteins. A model for the function of these main components of the Sec machinery of B. subtilis is depicted in Fig. 4. Using proteomic approaches, the extracellular proteomes of B. subtilis mutants that are affected in different stages in protein secretion have been analyzed. In the following sections, we review the currently available proteome data concerning B. subtilis strains lacking, or depleted of, various components involved in Sec-dependent protein export. These data are summarized in Table 4.
FIG. 4.
Components involved in Sec-dependent protein export in B. subtilis. Secretory proteins are ribosomally synthesized as precursor proteins with an N-terminal signal peptide (SP). Cytoplasmic chaperones, such as SRP/FtsY (47) and CsaA (75, 76), keep the precursors in a translocation-competent state and facilitate their targeting to the translocase in the membrane, consisting of SecA, SecY, SecE, SecG, and SecDF (17, 129). During or shortly after translocation, the preprotein is cleaved by one of the type I signal peptidases (SipS-W) (130) or lipid modified by the diacylglyceryl-transferase (Lgt) (62) and cleaved by the lipoprotein-specific signal peptidase (Lsp) (136). SppA and TepA may be involved in the degradation of cleaved signal peptides (16), whereas the folding of several secreted proteins depends on the activities of PrsA (55), BdbBC (18), and/or SpoIIIJ/YqjG (135). HtrA, HtrB (85), and WprA (68, 124) are involved in the quality control of secretory proteins. It should be noted that for reasons of simplicity, HtrAB are depicted in the cell wall, although HtrA is detected in both the membrane and the medium (5). On passage through the cell wall, the mature protein is released into the environment.
TABLE 4.
Impact of B. subtilis 168 secretion machinery components on extracellular proteome composition
| Limiting component | Extracellular
proteins whose levels
werea:
|
Mediumb | |
|---|---|---|---|
| Reduced | Increased | ||
| Ffh depletion | Csn, Pel, PenP, TasA, WapA, WprA, XkdG, XynD, YclQ, YflE, YfnI, YlqB, YncM, YwtD, YxaK, YxkC | SodA | MG |
| SecA depletion | Csn, Pel, PenP, TasA, WapA, WprA, XkdG, XynD, YclQ, YflE, YfnI, YlqB, YncM, YwtD, YxaK, YxkC | − | MG |
| SecA inhibition by sodium azide | Csn, LipA, WapA, XynA, YolA, YvcE, YweA, YxaL | MntA, OppA, YclQ | LB |
| SecDF deletion | − | − | LB |
| SpoIIIJ/YqjG deletion | − | − | LB |
| SipS-V deletionc | YfnI | − | LB |
| SipW deletion | TasA | − | |
| Lsp deletion | AmyE, Csn, Epr, LipA, GlpQ, LytD, PenP, XepA, XkdK, XkdM, XlyA, YncM, YolA, YrpD, YwoF, YxaK, YxkC | MntA, WprA, YxeB | LB |
| Lgt deletion | − | FeuA, FhuD, LytD, MntA, MsmE, OppA, PbpC, RbsB, XepA, XlyA, YfiY, YodJ, YusA, YvcE, YwtF, YxeB | LB |
| − | OppA, OpuAC, PbpC, PstS, YcdH, YdhF, YfiY, YqiX, YusA, YrpE, YxeB | PS | |
| PrsA depletion | AbnA, AmyE, AprE, BglC, Bpr, FlgK, LytD, MntA, Mpr, NprE, OppA, Pel, PenP, Vpr, WapA, WprA, XkdK, XlyA, XynA, XynD, YbdN, Ybxl, YclQ, YhcR, YlqB, YncM, YnfF, YvcE, YwoF, YwtD, YxiA, YxkC | CitH, Ef-G, Eno, Ggt, GroEL, LipA, PdhA, PdhB, PdhD, RocF, SodA, YlqB, YvgN, YweA, YwjH | LB |
| YacD deletion | − | − | LB |
| BdbA-D deletion | − | − | LB |
| HtrA deletion | HtrA, YqxI | − | LB |
| HtrB deletion | − | HtrA, YqxI | |
| CssS deletion | HtrA, YqxI | − | |
| WprA deletion | AbnA, AprE, Csn, WprA, YncM, YxaL, YweA | BglS, Epr, FlgK, Vpr, WapA, YclQ, YwsB | LB |
| AprE, Bpr, Epr, NprB, NprE, Mpr, Vpr deletion | AbnA, AprE, BglS, Bpr, NprE, Mpr, Vpr | HtrA, WapA, YqiX, YvcE | LB |
| Total-Tat deletion | − | − | LB |
| PhoD | − | PS | |
The relative amounts of all proteins listed were reduced or increased in the media of modified B. subtilis strains. Proteins that completely disappeared from or appeared in (see Table 5) the extracellular proteome are printed in bold. Proteins were identified by 2D PAGE and subsequent MALDI-TOF mass spectrometry and/or N-terminal peptide sequencing as described by Antelmann et al. (3-6). −, no protein(s) was reduced or increased in the medium of modified B. subtilis strains.
Proteins found in the extracellular proteomes of cells grown in minimal medium with glucose (MG) (46) LB broth (rich medium) (4, 5, 6, 51, 147), or phosphate starvation medium (PS) (3, 52).
The C-terminal part of YfnI was absent only from the medium of strains lacking at least SipT and SipV (5).
Cytoplasmic Targeting Factors
Since B. subtilis lacks a secretion-specific targeting factor similar to the SecB protein of Escherichia coli (99), an important role in this process has been attributed to the highly conserved signal recognition particle (SRP) pathway (129). An important component of this pathway is the Ffh protein (for “fifty-four homologue”), a GTPase that is homologous to the 54-kDa subunit of the eukaryotic signal recognition particle (SRP54) (47). This protein forms a complex (denoted SRP) with the small cytoplasmic RNA that is functionally related to the eukaryotic 7S RNA (78) and HBsu, a histone-like protein of B. subtilis (79). This SRP complex of B. subtilis binds to the signal peptides of nascent chains emerging from the ribosome and is targeted to the membrane with the aid of the FtsY protein, a homologue of the eukaryotic SRP receptor α-subunit (SRα) (87). Both Ffh and FtsY are essential for SRP-dependent protein secretion and cell viability (54).
Ffh depletion.
The effect of Ffh depletion on the composition of the extracellular proteome was studied by Hirose et al. (46), using a strain in which cellular Ffh levels were controlled by the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter. When this strain was grown in minimal medium without IPTG, 31 protein spots were missing and 5 spots were significantly reduced in intensity in the extracellular proteome compared to the case for the same strain grown with IPTG. Only the level of the cytoplasmic protein SodA was increased under these conditions. Of the proteins that were unaffected or only mildly affected by Ffh depletion, three were identified as Hag and GapA (both unaffected) and XkdG (mildly affected). The fact that the extracellular accumulation of the flagellin Hag is not affected by Ffh depletion is understandable since this protein is exported by a specific flagellin assembly pathway (48). In addition, GapA is a cytoplasmic protein (44) that is released into the medium by an unknown mechanism. Furthermore, XkdG is a prophage-related protein that is probably exported by a specific prophage PBSX-encoded holin system. These data suggest that flagellin assembly, release of certain cytoplasmic proteins, and export of certain phage-related proteins are not (strictly) SRP dependent. In contrast, all identified extracellular proteins with cleavable Sec-type signal peptides, Csn, Pel, PenP, TasA, WapA, WprA, XynD, YclQ, YlqB, YncM, YwtD, YxaK, and YxkC, were completely absent from the medium of Ffh-depleted cells. These data strongly suggest that signal peptide-dependent protein secretion via the Sec pathway is, directly or indirectly, SRP dependent. In this respect, it is important to consider the possibility that membrane proteins critical for protein translocation, such as certain components of the Sec translocase, could be inserted SRP dependently into the membrane. If so, SRP depletion would indirectly have a negative impact on the secretion of proteins with Sec-type signal peptides. Finally, YfnI and YflE are two paralogous transmembrane proteins whose C-terminal domain is released into the medium (5, 46). The release of YfnI was mildly affected on Ffh depletion, whereas the release of YflE was completely inhibited under these conditions. This might indicate that compared to protein secretion, lower levels of SRP are required for the insertion of certain transmembrane proteins into the cytoplasmic membrane.
Sec Translocase
The preprotein translocation machinery of the B. subtilis Sec pathway consists of at least four proteins: SecA, which is the translocation motor, and the integral membrane proteins SecE, SecG, and SecY. In the current model for preprotein translocation in B. subtilis, which has many similarities to that of E. coli, several successive steps in the translocation of proteins occur (36, 38, 73, 129, 145). First, SecA binds to the SecYEG translocase in the cytoplasmic membrane. Next, preproteins are transferred from a targeting factor (i.e., SRP or CsaA) to SecA dimers that are bound to the SecYEG complex. The binding of ATP by SecA leads to insertion of the C terminus of SecA through the pore of a SecYEG complex in the membrane, causing the translocation of a short stretch of the preprotein. Next, ATP is hydrolyzed by SecA, leading to the release of the preprotein and deinsertion of SecA. The latter step can be specifically inhibited by low concentrations of the ATPase inhibitor sodium azide. Further translocation is driven by both repeated cycling of SecA through ATP binding and hydrolysis and the proton motive force. Two proteomic approaches were performed to determine the effects of SecA limitation on the composition of the extracellular complement of the secretome. Hirose et al. (46) used a strain that contains a temperature-sensitive SecA (SecAts) protein, while Jongbloed et al. (51) used sodium azide to inhibit SecA activity. It should be emphasized that SecA limitation by inactivation of SecAts at elevated temperatures and SecA inhibition by azide represent two distinct approaches, both of which have their limitations. When SecA activity is inhibited by sodium azide, the initial targeting and translocation steps can possibly still take place, whereas these initial stages in protein transport as well as later SecA-dependent steps are affected significantly on SecA limitation. If so, initial stages in the translocation of “azide-resistant” secretory proteins may be driven by SecA while later stages in the translocation of these proteins could be more strongly dependent on the proton motive force than on SecA activity.
SecA limitation.
The effect of SecA limitation was studied by growing a strain with a temperature-sensitive SecA protein at 30°C (permissive temperature) or 42°C (nonpermissive temperature) in minimal medium (46). Of the 39 detected proteins in the medium of this strain grown at 30°C, 36 were completely absent from the medium of cells grown at 42°C (SecA limitation). Only three proteins were not affected by SecA limitation: the cytoplasmic proteins GapA and SodA, and the flagellin Hag. The latter finding suggests that flagellum assembly is both SRP (see the previous section) and SecA independent. Proteins that were completely absent from the medium of SecA-depleted cells included the signal peptide-containing proteins Csn, Pel, PenP, TasA, XynD, YclQ, YlqB, YncM, YwtD, YxaK, and YxkC, as well as the processing products of WapA and WprA. These proteins were also absent from the medium of Ffh-depleted cells (see the previous section). Furthermore, SecA limitation completely inhibited not only the release of the C-terminal domain of the transmembrane protein YflE but, unlike Ffh depletion, also that of the C-terminal domain of its paralogue YfnI. As expected, these data confirm that SecA is indispensable for protein secretion via the Sec pathway. Notably, the secretion of all identified secretory proteins with cleavable signal peptides depends on both SRP (see the previous section) and SecA, confirming the general view that SRP and the Sec machinery of B. subtilis cooperate in this process. Furthermore, these data indicate that the insertion and/or proteolytic processing of at least two transmembrane proteins does require the Sec pathway. Likewise, the provoked export inhibition by SecA limitation of the prophage-encoded protein XkdG, which lacks a typical signal peptide, may be caused by an impaired membrane insertion of certain components of the XkdG export pathway (e.g., holins [see Mechanisms for extracellular accumulation of proteins below). Thus, the fact that the export of YfnI and XkdG is unaffected or only mildly affected by Ffh depletion might indicate that at least some transmembrane proteins that are Sec-dependently inserted into the membrane can bypass the SRP pathway.
For E. coli, it was proposed that the SRP route facilitates primarily the cotranslational targeting of inner membrane proteins, which contain longer and more hydrophobic (uncleaved) signal peptides than do secretory proteins (11, 118, 140). Subsequent initial transmembrane domain insertion steps seem to be independent of SecA in E. coli (118). The fact that signal peptides of B. subtilis are, on average, longer and more hydrophobic than those of E. coli (129) might explain why the majority of secretory B. subtilis proteins are secreted in an SRP-dependent manner. Remarkably, the studies by Hirose et al. (46) suggest that the Sec-dependent insertion of transmembrane segments of some integral membrane proteins of B. subtilis could be rather SRP independent. Possibly, nascent chain-ribosome complexes can, in certain cases, dock directly onto the Sec translocase of B. subtilis without the aid of SRP. Thus, the process of membrane protein biogenesis in B. subtilis may be organized somewhat differently from the equivalent process in E. coli.
SecA inhibition by sodium azide.
The proteomic studies by Hirose et al. (46) suggested that the secretion of the majority of extracellular proteins by B. subtilis is strongly SecA dependent. However, the use of a temperature-sensitive secA mutant strain, which stops growing and dies after a temperature upshift, might influence the results. Furthermore, only a limited number of extracellular proteins were detected, since this strain was grown in minimal medium. Therefore, Jongbloed et al. (51) used a different approach, which was based on the inhibition of SecA activity by sodium azide in cells grown in a rich medium. For this purpose, it was essential to study the secretion of de novo-synthesized proteins because, otherwise, the kinetic effects of sodium azide on protein secretion would be overshadowed by the large amounts of extracellular proteins that accumulate in the growth medium of the azide-treated strain. Thus, postexponentially growing B. subtilis cells were separated from the growth medium, and grown for 20 min in fresh medium with or without sodium azide. This procedure resulted in the visualization of extracellular proteins that normally accumulate in the growth medium at relatively high levels (51). Of the 26 identified de novo-synthesized proteins in the medium of untreated cells, protein spots belonging to LipA, WapA, YolA, YvcE, YweA, and YxaL were almost completely absent from the medium of cells grown in the presence of sodium azide. Furthermore, Csn and XynA were secreted at significantly reduced levels. Notably, all eight extracellular proteins that were affected by SecA inhibition contained N-terminal signal peptides (Table 1). In contrast, no significant effect of SecA inhibition was observed on the extracellular appearance of 18 other de novo-synthesized proteins. This group of proteins consisted of the flagellum-related proteins FliD and Hag; the cytoplasmic proteins RocF, YwjH, and KatA; the membrane proteins YflE and YfnI; the lipoproteins MntA, OppA, and YclQ; and the AbnA, AprE, YbdN, YlqB, YncM, YrpD, YwtD, and YxkC proteins, which are synthesized with typical Sec-type signal peptides. These data, obtained by proteomics, support the view that the secretion of different secretory proteins depends to different extents on the activity of SecA. Accordingly, previous research has shown a difference in the SecA requirements of the α-amylase AmyE (requiring low levels of SecA activity) and the levansucrase SacB (requiring high levels of SecA activity) for secretion into the medium of B. subtilis (60). Notably, the signal peptides of SacB and AmyE are quite different (129, 143). The H-domain of the signal peptide of AmyE is longer than that of SacB (23 and 17 residues, respectively), and its overall hydrophobicity is higher (1.8 and 1.1 residues, respectively). It was therefore proposed that these differences in the H-domains could be responsible for the difference in SecA requirement of pre-AmyE and pre-SacB (60). Although AmyE and SacB were not detected in the proteomic analysis of SecA-dependent (i.e., azide-sensitive) protein secretion (51), these studies provided a good opportunity to evaluate the above-mentioned hypothesis. Primary amino acid sequence analysis showed, however, that the H-domains of the signal peptides of both azide-sensitive and azide-resistant secretory proteins have an average length of 22 residues and an average hydrophobicity between 1.5 and 1.6. Thus, it seems unlikely that the H-domain is the main determinant for the SecA requirement of a preprotein. In contrast, the N-domains of the signal peptides of the eight highly azide-sensitive secretory proteins are on average shorter (7 versus 11 residues) and more hydrophilic (−1.4 versus −1.1) than those of the eight azide-resistant secretory proteins. Nevertheless, the number of positively charged residues in the N-regions of signal peptides of azide-sensitive and azide-resistant secretory proteins did not significantly differ (3.6 on average). Although this finding might indicate that the N-domain of signal peptides is a determinant for the SecA requirement of a preprotein, a larger data set and site-directed mutagenesis approaches are needed to pinpoint the relevant features of signal peptides in relation to the extent of SecA requirement of the corresponding preprotein. It is conceivable, however, that specific properties of the mature parts of particular secretory preproteins are more important in determining their SecA requirement than are the properties of their signal peptides.
Another interesting observation from the azide inhibition experiments is that all three lipoproteins that are detectable in the extracellular proteome of untreated cells are present in equal or even larger amounts in the medium of cells treated with sodium azide. This suggests that the transport of lipoproteins via the Sec pathway requires less SecA activity than does the transport of secretory proteins. Furthermore, the insertion and release of the C-terminal domains of the transmembrane proteins YflE and YfnI do not seem to be affected by SecA inhibition with sodium azide. This is in marked contrast to the results obtained by Hirose et al. (46), who showed that the release of YflE, YfnI, and the lipoprotein YclQ into the medium of a temperature-sensitive secA mutant strain was completely blocked by SecA limitation. Similarly, the extracellular appearance of secretory proteins whose export was not (completely) inhibited by sodium azide (e.g., Csn, YwtD, YxkC, YncM and YlqB [51]) was completely blocked by SecA depletion (46). This shows that the secA mutation employed by Hirose et al. (46) is more effective in reducing the SecA translocation motor activity than is sodium azide.
SecDF deletion and SpoIIIJ and YqjG depletion.
In addition to the heterotrimeric SecYEG subcomplex, the E. coli Sec machinery contains a second heterotrimeric subcomplex that is composed of the SecD, SecF, and YajC proteins. This second subcomplex is likely to form a part of the B. subtilis Sec machinery as well, although this has not been demonstrated experimentally. In B. subtilis, this complex would be composed of the SecDF protein (a natural fusion protein of SecD and SecF) and YrbF (a homologue of E. coli YajC). The precise role of SecDF-YajC in protein export is presently not clear, but a variety of possible functions have been proposed. These include (i) removal of cleaved signal peptides or transmembrane segments from the SecYEG translocation channel; (ii) release of translocated proteins from the translocation channel; (iii) regulation of SecA cycling; and (iv) prevention of preprotein backsliding (86). Unlike SecD and SecF of E. coli, SecDF of B. subtilis 168 was shown to have little impact on cell viability and protein export, at least under standard laboratory conditions (17). A secretion defect in a secDF mutant strain was observed only under conditions of high-level expression of secretory proteins, such as AmyQ of Bacillus amyloliquefaciens. Accordingly, the disruption of secDF had no detectable influence on the composition of the extracellular proteome (H. Antelmann, unpublished observations).
A final component that can associate with the Sec translocase of E. coli is the YidC protein, which is involved in the membrane insertion of newly synthesized membrane proteins (65, 108, 116). Interestingly, YidC seems to be linked to the SecYEG subcomplex of the translocase through the SecDF-YajC subcomplex (86). B. subtilis contains two homologues of YidC, known as SpollIJ and YqjG. Remarkably, the biogenesis of a variety of integral membrane proteins in B. subtilis is only mildly affected in cells depleted of both SpoIIIJ and YqjG (135). In contrast, the simultaneous removal of SpoIIIJ and YqjG has a severe impact on (as yet undefined) posttranslocational stages in the secretion of proteins, such as AmyQ, LipA, and E. coli PhoA (135). Unfortunately, proteomic studies with SpoIIIJ/YqjG-depleted cells turned out to be difficult, since the combined activities of these proteins are essential for cell viability. The extracellular proteomes of spoIIIJ and yqjG single mutants, which display no growth defects, revealed no significant changes compared to that of the parental strain (H. Antelmann and H. Tjalsma, unpublished observations).
Type I Signal Peptidases
SPases remove signal peptides from secretory preproteins when the C-domain of the signal peptide emerges at the extracytoplasmic side of the membrane. This enzymatic reaction is a prerequisite for the release of the mature secretory protein from the membrane (29, 30, 129). One of the most remarkable features of the B. subtilis protein secretion machinery is the presence of multiple, paralogous, type I SPases. This is in contrast to many other bacteria and archaea and the endoplasmic reticulum (ER) of yeast, in which just one type I SPase seems to be sufficient for the processing of secretory preproteins (129, 130). In B. subtilis five sip genes for type I SPases are located on the chromosome (denoted sipS, sipT, sipU, sipV, and sipW [130, 134]). Interestingly, SipW is homologous to SPases found in sporulating gram-positive bacteria, archaea, and the ER membrane of eukaryotes, which, together, form the subfamily of ER-type SPases. The uniqueness of SipW was further underscored by the observation that this SPase is solely required for the processing of the spore-associated protein TasA (126, 131). In contrast, all other B. subtilis SPases are of the prokaryotic type (P-type). Such P-type SPases are typically present in eubacteria, mitochondria, and chloroplasts (130). Although all chromosomally encoded SPases in B. subtilis can process secretory preproteins, only SipS and SipT are of major importance for preprotein processing and cell viability. In contrast, SipU, SipV, and SipW play a minor role in protein secretion and have substrate specificities that differ at least in part from those of SipS and SipT (130, 134).
SPase I deletions.
The availability of proteomic techniques created a new opportunity to further investigate possible differences in the substrate specificities of the type I SPases of B. subtilis. Therefore, Antelmann et al. (5) analyzed the extracellular proteomes of single, double, triple, and quadruple SPase I mutants lacking sipS, sipT, sipU, sipV, or sipW or combinations thereof. Surprisingly, apart from the expected absence of TasA in the medium of a sipW mutant strain, no major differences in the extracellular protein patterns of these mutants were observed. This observation confirms the view that the presence of either SipS or SipT is sufficient for efficient precursor processing and that the type I SPases of B. subtilis have largely overlapping specificities (130). The only notable exception was the SipTV-dependent cleavage of the membrane protein YfnI. This observation was remarkable not only because YfnI is a polytopic membrane protein but also because the cleavage site is located 44 residues C-terminally of the fifth transmembrane segment of this protein. This suggests that despite its distant position relative to the transmembrane segment, the SPase I cleavage site of YfnI, and possibly that of the paralogous proteins YflE, YqgS, and YvgJ (46), is accessible to the catalytic sites of SipT and SipV at the extracytoplasmic membrane surface (5).
Lipoprotein Modification and Processing
Although lipoproteins are transported via the general Sec pathway, specific enzymes for their modification (Lgt) and processing (SPase II) are required. In contrast to the type I SPases, B. subtilis contains only one gene for a type II SPase (lspA) (97, 136), which is specifically required for the processing of lipid-modified preproteins. Interestingly, B. subtilis cells lacking SPase II are viable under standard laboratory conditions. This indicates that processing of lipoproteins by SPase II is not strictly required for lipoprotein function, since at least one lipoprotein, PrsA, is essential for viability (55). The fact that processing of lipoproteins by SPase II is not strictly required for lipoprotein function is probably due to activity of uncleaved lipoproteins, as was recently shown to be the case for lipoprotein precursors in Lactococcus lactis (146). In B. subtilis cells lacking SPase II, lipoprotein precursors are subject to alternative N-terminal processing by as yet unidentified proteases (132, 136). The cumulative activity of unprocessed and alternatively processed (mature-like) lipoproteins is in many cases strongly reduced compared to that of their corresponding mature form (12, 136). In B. subtilis cells lacking SPase II, the secretion of the nonlipoprotein AmyQ was strongly impaired, which could be attributed to malfunctioning of the precursor and mature forms of the lipoprotein PrsA, an extracytoplasmic folding catalyst for many secretory proteins (136).
SPase II deletion.
To explore the full impact of the absence of SPase II on the extracellular proteome, Antelmann et al. (5) analyzed the extracellular proteome of an lspA mutant strain. These studies showed that two abundant extracellular proteins of the parental strain, AmyE and YolA, were completely absent from the extracellular proteome of the lspA mutant (5). Furthermore, the relative amounts of a variety of other extracellular proteins were strongly reduced, as exemplified by the secretory proteins Csn, Epr, LipA, GlpQ, LytD, PenP, YncM, YrpD, YwoF, YxaK, and YxkC (Table 1) and the phage-related proteins XepA, XkdK, XkdM, and XlyA (Table 2). Unexpectedly, significantly increased levels of the two processed forms (CWBP23 and CWBP52) of the cell wall protease WprA were found in the medium of the lspA mutant strain. Similarly, the extracellular levels of two typical lipoproteins, MntA and YxeB, were strongly increased, showing that these proteins were not effectively retained in the membrane of the lspA mutant. In contrast, the extracellular levels of the lipoproteins OppA and YclQ were not affected by the lspA mutation. Taken together, these findings show that the absence of SPase II has rather pleiotropic effects on the composition of the extracellular proteome.
Lgt deletion.
Analysis of the extracellular proteome of B. subtilis 168 showed that at least nine different potential lipoproteins are released into the medium (Table 1); six of these can be observed when cells are grown in LB medium (MntA, OppA, YclQ, YfmC, YqiX, and YrpE), and five are present when cells are grown in phosphate starvation medium (PstS, YcdH, YdhF, YqiX, and YrpE). Moreover, elevated levels of MntA and YxeB are found in the extracellular proteome of the lspA mutant strain (see the previous section). To further investigate the factors required for lipoprotein processing and retention in the cell, the composition of the extracellular proteome of an Igt mutant, defective in the lipid modification of prelipoproteins (62), was analyzed by Antelmann et al. (5). Unexpectedly, the extracellular protein pattern of the Igt mutant grown in LB medium was completely different not only from that of the parental strain but also from the extracellular proteome of the lspA mutant. In fact, the extracellular proteome of the Igt mutant exhibited about 35 additional spots that were absent or only very weakly present in the medium of the parental strain. Furthermore, the extracellular levels of the predicted lipoproteins OppA and MntA, along with several proteins related to autolytic activities, such as the (predicted) enzymes LytD, YvcE, XepA, and XlyA and the autolysin regulator YwtF (Tables 1 and 2), were significantly increased by the Igt mutation. Of the additional extracellular proteins appearing in the medium of the Igt mutant, nine were identified as (putative) lipoproteins. These were FeuA, FhuD, MsmE, PbpC, RbsB, YfiY, YodJ, YusA, and YxeB (Table 5). By growing the Igt mutant strain in phosphate starvation medium, it was shown that the extracellular levels of the phosphate starvation-induced lipoproteins, OpuAC, PstS, YcdH, YdhF, YqiX, and YrpE, were also significantly increased in the absence of Lgt. Of the latter lipoproteins, OpuAC is the only one not detected in the extracellular proteome of the parental strain, 168 (Table 5). Finally, as shown for LB medium, the extracellular levels of the putative lipoproteins OppA, PbpC, YfiY, YusA, and YxeB were also significantly increased when the Igt mutant was grown in phosphate starvation medium (5). Taken together, these studies showed that cells lacking Lgt shed lipoproteins into their growth medium. Since these lipoproteins are, by and large, retained at the cytoplasmic membrane of the parental strain 168, these observations demonstrate that lipid modification by Lgt is the key determinant for lipoprotein retention in B. subtilis. In contrast, the cleavage by SPase II seems to be required mainly to fully activate the lipid-modified proteins of this organism. In this respect, it should be kept in mind that in the absence of Lgt, unmodified lipoprotein precursors cannot be cleaved by SPase II. Therefore, lipoprotein shedding by B. subtilis Igt can be envisaged to take place in at least two ways. First, unmodified translocated prelipoproteins, as demonstrated for OpuAC and PrsA (5), could either leak from the membrane or be actively released into the growth medium by a (hypothetical) release factor. The released prelipoproteins could form micelle-like structures or could be subject to amino-terminal proteolysis. The latter would result in the presence of mature forms in the growth medium, as observed for MntA, OppA, YclQ, YfiY, YfmC, and YxeB (5). Alternatively, these unmodified prelipoproteins could first be retained in the membrane by their uncleaved signal peptide and subsequently released from the membrane by amino-terminal proteolysis.
TABLE 5.
Additional extracellular proteins in mutant B. subtilis strainsa
| Protein | Function or similarity | Export signalb | SPase | Retention signalc | Mediumd |
|---|---|---|---|---|---|
| B. subtilis Δlgt | LB | ||||
| FeuA | Iron-binding protein | Lipo | SPase II | (lipid) | LB |
| FhuD | Ferrichrome-binding protein | Lipo | SPase II | (lipid) | LB |
| MsmE | Manganese-binding protein | Lipo | SPase II | (lipid) | LB |
| OpuAC | Glycine-betaine-binding protein | Lipo | SPase II | (lipid) | PS |
| PbpC | Penicillin-binding protein 3 | Lipo | SPase II | (lipid) | LB, PS |
| RbsB | Ribose-binding protein | Lipo | SPase II | (lipid) | LB |
| YfiY | Iron(III)-binding protein | Lipo | SPase II | (lipid) | LB, PS |
| YodJ | d-Alanyl-d-alanine carboxypeptidase | Lipo | SPase II | (lipid) | LB |
| YusA | Putative part of the S-box regulon | Lipo | SPase II | (lipid) | LB |
| YxeB | Putative binding protein | Lipo | SPase II | (lipid) | LB, PS |
| B. subtilis ΔwprA | |||||
| YwsB | Similar to unknown proteins of B. subtilis | Sec | SPase I | − | LB |
All proteins listed were identified in the medium of mutant B. subtilis strains but not seen in the medium of the parental strain, 168 (as listed in Tables 1 and 2). Proteins were identified by 2D PAGE and subsequent MALDI-TOF mass spectrometry and/or N-terminal peptide sequencing as described by Antelmann et al. (5, 6). Putative signal peptides, SPase I or SPase II cleavage sites, transmembrane domains, and cell wall-binding domains were predicted as described by Tjalsma et al. (129) and Jongbloed et al. (51).
Identified transient export signals are Sec-type signal peptides (Sec) and lipoprotein signal peptides (Lipo).
Identified retention signals present in the mature part of the protein after processing by specific SPases are only lipid modifications (lipid). It should be noted that in the lgt mutant strain, the lipid retention signal is not attached to lipoprotein precursors. Furthermore, the YwsB protein, despite the absence of known cell wall-binding repeats (−), is retained in the cell wall of parental B. subtilis cells.
Folding Catalysts
After translocation in an unfolded state, Sec-dependent secretory proteins have to fold into their native conformation. Even though proteins can fold spontaneously in vitro, their folding in vivo is frequently assisted by folding catalysts. An important folding catalyst involved in protein secretion is the lipoprotein PrsA, which shows homology to peptidyl-prolyl cis/trans-isomerases (107) and is essential for protein secretion and cell viability of B. subtilis (55, 56). Strains containing mutant forms of PrsA show impaired secretion of degradative enzymes (50, 55). It has been suggested that PrsA is required to prevent unproductive interactions of unfolded secretory proteins with the cell wall shortly after translocation (153). A similar role in posttranslocational protein folding was recently postulated for SpoIIIJ and YqjG since depletion of both of these proteins affected the stability of at least three secretory proteins during the posttranslocational stage in protein secretion (135). However, the action of SpoIIIJ and YqjG in protein folding is likely to be indirect, in view of the well-documented role of the homologues of these proteins (e.g., YidC of E. coli) in membrane protein assembly. The importance of extracytoplasmic folding catalysts is underscored by the fact that the membrane/cell wall interface and extracellular environment of B. subtilis are highly proteolytic (129) (see “Quality control factors” below). This results in a rapid degradation of exported proteins of homologous or heterologous origin that fold too slowly, or incorrectly, after translocation (19, 20).
PrsA depletion.
It seems likely that some of the phenotypes of an lspA mutant strain can be attributed to reduced levels of PrsA activity. This idea is supported by the observation that the posttranslocational folding of the PrsA-dependent nonlipoprotein AmyQ in B. subtilis cells lacking SPase II was strongly impaired (136). Most probably, the activity of pre-PrsA and/or of alternatively processed non-lipid-modified forms of PrsA is sufficient to sustain a viable cell but lower than that of properly processed PrsA. Thus, the finding that overall protein secretion in an lsp mutant strain was reduced about fourfold could very well be an effect of PrsA limitation (see “SPase II deletion” above). Very recently, studies to monitor the effects of PrsA depletion on the composition of the extracellular proteome have been documented by Vitikainen et al. (147). In these studies, a strain was used in which prsA expression was controlled by the IPTG-dependent Pspac promoter and the extracellular proteomes of cells grown in LB medium with or without IPTG were compared. Growth in the absence of IPTG resulted in PrsA depletion, and, compared to cells grown in the presence of IPTG, the relative amounts of 32 extracellular proteins were significantly reduced. Remarkably, the relative amounts of 15 other extracellular proteins were significantly increased, while 6 extracellular proteins remained unaffected by PrsA depletion. Of the proteins present in increased amounts, 11 corresponded to cytoplasmic proteins (CitH, Ef-G, Eno, GroEL, PdhA, PdhB, PdhD, RocF, SodA, YvgN, and YwjH), which is consistent with increased cell lysis in the absence of PrsA. In contrast, 29 of the 32 extracellular proteins present in decreased amounts were synthesized with an export signal (Table 4). The three remaining proteins present in decreased amounts lack a typical export signal (FlgK, XkdK, and XlyA) (Table 2). These observations support the view that PrsA is of general importance for the folding of secretory proteins (50, 55). Interestingly, eight proteins (Csn, Ggt, LipA, YfnI, YlqB, YweA, YxaK, and YolA) whose extracellular levels remained unaffected or were even increased on PrsA-depletion are synthesized with (putative) export signals (Table 1). This indicates that a specific subset of secretory proteins does not depend on PrsA for proper folding and secretion. Thus, the secretion of these proteins may depend on folding factors other than PrsA. Some of these other folding factors could be lipoproteins, as judged by the fact that the secretion of Csn, LipA, and YxaK is strongly reduced in the absence of SPase II (see above). The PrsA paralogue YacD is dispensable for the folding of PrsA-independent extracellular proteins, because the extracellular proteome of a yacD mutant strain was shown to be very similar to that of the parental strain 168 (H. Antelmann and H. Tjalsma, unpublished). This implies either that YacD plays no role in the biogenesis of extracellular proteins, that the specific substrates of YacD are not detectable in the extracellular proteome, or that YacD substrates were not expressed under the conditions tested.
Bdb mutations.
Four extracytoplasmic thiol-disulfide oxidoreductases of B. subtilis have been implicated in the formation of disulfide bonds in exported proteins. Two of these, BdbC and BdbD, are required for the biogenesis of the pseudopilin ComGC (essential for DNA binding and uptake during competence development), which contains an intramolecular disulfide bond (71). Furthermore, BdbC and BdbD are of major importance for the posttranslocational folding of a disulfide bond-containing heterologous protein, the alkaline phosphatase PhoA of E. coli (18, 71). A third thiol-disulfide oxidoreductase, BdbB, is involved in the production of the lantibiotic sublancin 168, which contains two disulfide bonds (33). Although BdbB and BdbC are highly similar paralogues, their substrate specificities overlap only partly. Thus, BdbC plays a minor role in the production of sublancin 168 (33) while BdbB is only of minor importance for the secretion of E. coli PhoA by B. subtilis, and this protein is dispensable for competence development. Thus far, no specific function has been identified for the fourth thiol-disulfide oxidoreductase of B. subtilis, which is known as BdbA. As judged by their function in the folding of exported proteins with disulfide bonds, it is believed that BdbB, BdbC, and BdbD are members of an oxidation pathway. This idea is supported by the fact that BdbB and BdbC have a high degree of similarity to DsbB of E. coli while BdbD has some similarity with DsbA of E. coli (18, 71). Remarkably, neither bdbA, bdbB, bdbC, nor bdbD single-mutant strains, nor a quadruple mutant lacking all four of these genes, displayed detectable changes of the extracellular proteome (H. Antelmann and R. Dorenbos, unpublished observations). This suggests that very few, if any, of the native secreted proteins of B. subtilis contain disulfide bonds that are critical for their stability and protease resistance and that (in some cases) Bdb-independent folding and oxidation occur.
Quality Control Factors
One important feature of B. subtilis that underscores the importance of efficient folding of secretory proteins into their native, protease-resistant conformation, is the presence of at least 27 proteases in the membrane, cell wall, and medium that can cleave (partially) unfolded polypeptide chains (129). Proteins with a known role in the quality control of secretory proteins are two HtrA-like proteases/chaperones, HtrA and HtrB (85), which are thought to have proofreading capabilities for the folding state of secretory proteins, as demonstrated for HtrA of E. coli (122). When a secretory protein is not properly folded, HtrAB can either assist in folding or, if that is impossible, degrade the malfolded secretory protein. Interestingly, transcription of the corresponding genes is induced by secretion stress, which is sensed and controlled by the CssR-CssS two-component regulatory system. This system is essential for cell viability under conditions of severe secretion stress (31, 49). Another protease, which seems to be involved in the quality control of secretory proteins, is WprA. For this protein, a chaperone domain (CWBP23) in addition to a protease domain (CWBP52) has been proposed (10, 68). Notably, HtrA and WprA have a dual localization, being present both in the cell wall proteome and in the extracellular proteome (4, 6). Like HtrB, the HtrA protein has a (predicted) N-terminal membrane anchor. As shown by N-terminal sequencing, HtrA in the medium lacks this membrane anchor domain (3). The localization of HtrB has not been documented yet. The importance of HtrAB and WprA in quality control was investigated by proteomic analyses by Antelmann et al. (4, 6).
Modulation of HtrA and HtrB levels.
To study the importance of HtrA-like proteases for the composition of the extracellular proteome, cellular levels of HtrA and HtrB were modulated in several ways (4). First, mutant strains were constructed in which htrA or htrB or both, were disrupted. Notably, disruption of one of these genes causes a secretion stress that strongly induces the activity of the promoters of both genes, and this response is mediated by the CssR-CssS two-component system (31, 49, 85). Furthermore, the transcription of both htrA and htrB can be strongly reduced by a disruption of the cssS gene, while transcription of both htr genes is significantly increased by overproducing the secretory protein AmyQ, which causes secretion stress. To monitor the impact of HtrA and HtrB on protein secretion, Antelmann et al. analyzed the extracellular proteomes of htrA and/or htrB mutant strains (4). These analyses showed that apart from the HtrA protein, only one other protein disappeared completely from the medium of an htrA mutant strain. This was YqxI, a protein of unknown function. As expected, the HtrA spot in the medium was increased on deletion of the htrB gene or overproduction of AmyQ (secretion stress) but significantly decreased in a cssS mutant strain. This pattern of HtrA appearance in the media of different mutant strains closely paralleled that of YqxI. However, in contrast to htrA, the transcription of yqxI was independent of CssRS and was not secretion stress responsive (4). Thus, HtrA seems to be specifically required for the stabilization of YqxI, a role that cannot be taken over by HtrB. The fact that the protease-active site of HtrA is not required for the appearance of YqxI in the medium (4) and that yqxI gene transcription is not secretion stress responsive (4) suggests a chaperone-like activity of HtrA involved in YqxI stabilization. Remarkably, alterations in the cellular HtrAB levels caused no other detectable changes in the extracellular proteome. These findings suggest that HtrA or HtrB are not of general importance for protein secretion under standard laboratory conditions. However, htrA htrB double mutants are very sick, and both genes are essential under conditions of severe secretion stress (49).
WprA deletion.
The cell wall-bound protease WprA degrades unstable and/or heterologous proteins at the membrane-cell wall interface (20, 124, 158). Interestingly, WprA itself is processed into two cell wall-bound products: CWBP52, which has serine protease activity, and CWBP23, which may have a chaperone-like activity (10, 68). Studies of the extracellular proteome of a wprA mutant strain showed that increased levels of BglS, Epr, FlgK, Vpr, and YclQ were present in the medium of cells lacking WprA (6). Furthermore, YwsB is released into the medium of the wprA mutant, whereas this protein is exclusively cell wall localized in the parental strain, 168. In contrast, the extracellular levels of the AbnA, AprE, Csn, YncM, YxaL, and YweA proteins were decreased under these conditions. Although the amounts of a large WapA-processing product that is released into the medium were not changed, small WapA degradation products were present at increased amounts in the medium of the wprA mutant strain. Together, these data suggest that WprA has multiple functions. Clearly, WprA can degrade various proteins prior to their release into the medium. On the other hand, this protein may also assist in the folding and cell wall binding of certain other proteins. It cannot be excluded that the observed alterations in the extracellular proteome are caused by indirect effects of the wprA mutation, such as possible alterations in the composition and structure of the cell wall.
Extracellular Proteases
Secretory proteins that have missed their last chance to be folded correctly by membrane or cell wall-attached folding catalysts and quality control factors are potential substrates for one of the many proteases in the membrane, cell wall, or medium. This can be concluded from the observation that extracellular and cell wall-associated proteases are responsible for the degradation of various heterologous proteins secreted by B. subtilis (19, 159). It should be kept in mind, however, that even correctly folded heterologous proteins can be prone to degradation and that malfolded proteins that are released into the medium might get some folding assistance from the released HtrA protein (see the previous section). To investigate the impact of secreted proteases, the extracellular proteome of a B. subtilis strain (WB700) lacking seven extracellular proteases (AprE, Epr, NprB, NprE, Mpr, Bpr, and Vpr) was investigated (4, 6). As expected, the five proteases that were visible in the extracellular proteome of the parental strain (AprE, Bpr, NprE, Mpr, and Vpr) were lacking from that of the protease mutant strain. Furthermore, consistent with the idea that homologous secretory proteins must be largely resistant to the extracellular proteases of B. subtilis, the levels of most extracellular proteins were not affected by the seven protease mutations. Nevertheless, the levels of AbnA and BglS were decreased whereas large WapA- and YvcE-processing products were present at elevated levels in the extracellular proteome of this mutant strain (6). These results indicate that released forms of the cell wall-associated proteins WapA and YvcE are degraded by extracellular proteases, a mechanism that might enable B. subtilis to recycle spoiled cell wall proteins. Accordingly, the cell wall proteome of the multiple protease deletion strain contained elevated levels of a large WapA fragment whereas the level of a smaller WapA-processing product was reduced compared to the cell wall proteome of the parental strain. Specifically, proteomic studies with the medium of a wprA epr double-mutant strain suggested that Epr is responsible for the degradation of WapA but not for the degradation of YvcE (6). It is difficult to explain why the extracellular accumulation of certain secretory proteins is reduced in the protease mutant. Possibly, extracellular proteases are important for clearing the cell wall of proteinacious waste products, a process that might be important for the secretion of certain proteins, such as AbnA and BglS. Interestingly, the levels of both HtrA and YqxI in the extracellular proteome of the multifold protease mutant B. subtilis WB700 were increased (4). This finding indicates that both proteins are subject to proteolysis after their export from the cytoplasm and is in line with the idea that YqxI needs folding assistance from HtrA in order to appear in the extracellular proteome.
Finally, it was shown that certain typical cytoplasmic proteins are present at increased levels in the medium of protease-deficient strains, which indicates that the extracellular accumulation of these proteins is inversely correlated with the activity of secreted proteases (4, 6). However, it has been reported previously that protease-deficient strains are subject to higher levels of cell lysis than is the parental strain, 168 (125). The latter finding complicates, at least to some extent, the interpretation of proteomic data obtained with (multiple) protease mutant strains.
CONTRIBUTION OF Sec-INDEPENDENT PROTEIN EXPORT TO THE EXTRACELLULAR PROTEOME
Although the Sec pathway is responsible for the secretion of most secretory proteins of B. subtilis, at least three signal peptide-dependent special-purpose pathways are present for the export of a relatively small number of proteins: the Tat pathway (51, 52), the pseudopilin export pathway for competence development, and pathways involving ABC transporters (129). The last two pathways cannot be studied by standard proteomic analysis, since the substrates of these pathways are too small to detect by 2D PAGE or, in the case of pilin export, these proteins are not released into the medium. Thus, the Tat pathway was the only special-purpose pathway that could be studied by extracellular proteome analysis.
Twin-Arginine Translocation Machinery
The known Tat components of E. coli are TatA, TatB, TatC, and TatE. Of these proteins, TatA, TatB, and TatE are structurally related. Since TatA and TatE are functionally redundant, the presence of one of these components is required for the translocation of proteins with twin-arginine signal peptides. TatB and TatC are indispensable for translocation activity (103, 142). Recent data indicate that TatB and TatC are involved in twin-arginine (RR) signal peptide reception. Furthermore, TatB and TatC, in complex with multiple copies of TatA, form a protein-conducting channel (2). In contrast to E. coli and most other bacteria, which contain only one tatC gene, B. subtilis contains two tatC genes, denoted tatCd and tatCy. Each of these genes is preceded by a tatA gene, denoted tatAd and tatAy, respectively. A third tatA gene of B. subtilis, tatAc, is not genetically linked to the tatC genes. It is not known whether the B. subtilis TatA proteins are the functional equivalents of E. coli TatA, TatB, or both TatA and TatB.
TatC and total-Tat deletions.
To identify proteins that are secreted via the Tat pathway of B. subtilis, Jongbloed et al. (51, 52) analyzed the extracellular proteomes of a tatCd tatCy double-mutant strain, and a “total-tat” mutant that lacks all known tat genes. To this end, the strains were grown in LB or phosphate starvation media. The results showed that the phosphate starvation-induced protein PhoD, containing a twin-arginine motif in its signal peptide, is the only protein whose secretion was completely blocked by the tatC or total-tat mutations. As expected, the secretion of other detectable proteins lacking a twin-arginine signal peptide, such as GlpQ, PeI, PhoA PhoB, PstS, Vpr, and YncM, was not significantly affected by the tatC or total-tat mutations. Surprisingly, however, the secretion of LipA, PbpX, WprA, WapA, YdhF, YfkN, and YhcR, all synthesized with potential RR signal peptides (Table 1), was not affected by the tatC or total-tat mutations. The same was the case for the AbnA, BglC, BglS, LytD, OppA, and YolA proteins, which contain a KR motif in their predicted twin-arginine signal peptides. Thus, not all B. subtilis precursors with RR/KR motifs in their signal peptides are transported via Tat, at least under the conditions used, suggesting that the Tat pathway of this organism is highly selective. Notably, the secretion of LipA, WapA, and YolA was shown to be inhibited by sodium azide, confirming the view that these proteins are secreted via the Sec pathway instead of the Tat pathway. Consistent with the Tat-dependent secretion of PhoD, the signal peptide of this protein conforms to the most stringent criteria for the prediction of Tat dependency, as defined for RR-signal peptides from other organisms (i.e. hydrophobic residues at the +2 and +3 positions relative to the two arginine residues, and an H-region with a hydrophobicity of less than 2.1 [51, 52, 129]). Strikingly, however, these stringent criteria also apply to LipA and LytD, which display a Tat-independent extracellular accumulation. This implies that the present criteria for the prediction of RR/KR signal peptides need to be refined, at least for B. subtilis.
MECHANISMS FOR EXTRACELLULAR ACCUMULATION OF PROTEINS
With the aid of all extracellular proteome data presently available, a first inventory of the mechanisms applied by B. subtilis to “secrete” proteins into its environment can be made. The results of this inventory are summarized in Fig. 5 (5).
FIG. 5.
Mechanisms of extracellular accumulation of B. subtilis proteins. Ribosomally synthesized proteins can be sorted to various destinations, depending on the presence (+SP) or absence (−SP) of an N-terminal signal peptide and specific retention signals. Based on the results obtained by proteomic studies, about 50% of the extracellular proteome is directly secreted into the medium via the Sec and Tat pathways. Notably, only one protein, PhoD, is (so far) known to be secreted via the Tat pathway. Proteins which have to be retained at the extracytoplasmatic side of the membrane can either lack an SPase cleavage site (−AXA), be lipid-modified (+lipobox), contain transmembrane (+TM) domains, or contain cell wall-binding repeats (+CWB). Such retained proteins are exported from the cytoplasm via the Sec or Tat pathways. About 24% of the proteins found on the extracellular proteome are predicted to have retention signals. These proteins are released into the medium by proteolysis, shedding, or cell wall turnover. Finally, about 26% of the extracellular proteome lacks typical signal peptides and can escape from the cytoplasm by cell lysis or via the flagellar export machinery, the holin systems, or other unidentified export systems. Expected locations of identified proteins, as based on previous genome-based predictions (129), are indicated by solid circles, whereas unexpected locations of identified proteins are indicated by open circles.
Protein Secretion via the Sec and Tat Pathways
A total number of 52 identified proteins of the extracellular proteome are synthesized with signal peptides that contain a cleavage site for SPase I. Eight of these proteins reach the growth medium even though they contain typical membrane or cell wall retention signals (Table 1). The latter proteins are most probably liberated from the cell through a combination of processing by SPase I and secondary processing events (see the next sections). Most signal peptides of the 52 above-mentioned proteins are likely to direct the corresponding proteins into the Sec pathway for protein translocation. However, in it was shown that Bacillus thuringiensis, FlhA, an Ffh paralogue required for flagella-assembly, is also required for the secretion of certain signal peptide-bearing preproteins (40). Thus, it cannot be excluded that, although not documented, a fraction of the secretory proteins with Sec-type signal peptides is transported via the flagellar assembly pathway in B. subtilis. Of all extracellular proteins so far identified, 14 are synthesized with signal peptides containing a potential RR/KR motif, suggesting that these proteins could be secreted via the Tat pathway. However, of these 14 proteins, only PhoD was secreted in a strictly Tat-dependent manner (52). Interestingly, a comparison of the general features of Sec-type signal peptides of identified extracellular proteins (Table 1) with the genome-based predictions for general features of Sec-type signal peptides revealed only very minor differences (Fig. 6) (129). However, the analysis of all potential SPase I cleavage sites in extracellular proteins identified by proteomics showed that the consensus A-X-A site is more frequently present than was previously predicted (129). Most importantly, based on proteomic studies, an alanine residue at the −1 position relative to the cleavage site seems to be a major determinant for SPase-I mediated cleavage of preproteins in B. subtilis (Table 6) (5).
FIG. 6.
Average features of 56 identified Sec-type signal peptides. Average signal peptide length and the length and hydrophobicity of the N- and H domains were determined on the basis of the Sec-type signal peptides from 52 identified extracellular proteins (Table 1) (5, 6, 129) and 4 cell wall-located proteins (Table 3); (6) with (putative) SPase I cleavage sites. The YfnI and YflE proteins are excluded from the analysis because they contain N-terminal transmembrane domains rather than typical signal peptides. We have included all cleavable signal peptides with RR/KR motifs, exept that of PhoD, since the export of the corresponding proteins was shown to be Tat independent (51, 52). aa, amino acids.
TABLE 6.
Amino acid residues around (putative) SPase I cleavage sitesa
| −3
|
−2
|
−1
|
+1
|
||||
|---|---|---|---|---|---|---|---|
| Residue | Frequency | Residue | Frequency | Residue | Frequency | Residue | Frequency |
| A | 0.71 | S | 0.22 | A | 1.00 | A | 0.45 |
| V | 0.18 | K, E | 0.25 | Q | 0.15 | ||
| T, I, S, G, W | 0.11 | H, Y | 0.19 | E | 0.10 | ||
| Q, G | 0.10 | K, S, V | 0.18 | ||||
| F, L, A, D, N | 0.22 | F, N, L, D, T | 0.12 | ||||
| W, P | 0.02 | ||||||
The frequency of a particular amino acid at the indicated positions around SPase I cleavage sites in signal peptides of 54 identified extracellular (Table 1) (4, 6, 129), and 4 cell wall-located (Table 3) (6) (pre)proteins with (putative) SPase I cleavage sites. Residues with similar frequencies of appearance at a certain position are grouped.
Release of Membrane Proteins by Proteolysis
The proteins Yfnl and YflE, detected in the extracellular proteome of B. subtilis 168, contain five transmembrane segments followed by a predicted SPase I cleavage site that is located about 40 residues C-terminally of the fifth transmembrane segment of this protein (5, 46). This SPase I cleavage site of Yfnl, and possibly that of YflE, is accessible to the catalytic sites of SipT and SipV. Similarly, the extracytoplasmic domains of four other membrane proteins are most probably liberated from the membrane by proteolysis. For YfkN and YhcR, this seems to be due to N-terminal processing by SPase I and C-terminal processing by an unknown protease at the membrane/cell wall interface. In contrast, the release of PbpA and HtrA requires N-terminal processing, which is probably not catalyzed by a known SPase, since PbpA and HtrA lack a typical SPase I cleavage site (4, 5).
Release of Lipoproteins by Proteolytic Shaving and/or Shedding
Although early studies by Eymann et al. (37) provided the first evidence that some lipoproteins of B. subtilis may end up in the growth medium (37), it is highly surprising that at least nine predicted lipoproteins are present in the extracellular proteome of B. subtilis 168. These lipoproteins are supposed to be retained at the cytoplasmic membrane. Since some of these extracellular lipoproteins were shown to lack the N-terminal, lipid-modified cysteine residue, these are most probably liberated from the cell by proteolytic “shaving” after their processing by SPase II (5). However, the alternative possibility that prior to N-terminal proteolysis, these lipoproteins are released by leakage from the membrane or even by a hypothetical release factor, as has been demonstrated for certain lipoproteins of gram-negative bacteria (160), cannot be excluded. Remarkably, 10 additional lipoproteins were detectable in the extracellular proteome of cells depleted of Lgt (5). The latter phenomenon suggests that unmodified prelipoproteins are actively or passively released from the membrane, which could be explained by the fact that the hydrophobic H-regions of lipoprotein signal peptides are generally too short to span the membrane completely (129). Indeed, the release of unmodified pre-PrsA and pre-OpuAC into the growth medium of an lgt mutant could be demonstrated (5). Alternatively, prelipoproteins might be released from the lgt mutant strain by proteolytic shaving, as evidenced by the fact that at least six extracellular lipoproteins were alternatively processed, lacking the cysteine residue at the +1 position of the mature lipoprotein. However, in view of the fact that some unmodified prelipoproteins can be detected in the medium of the lgt mutant, the latter proteolytic event might as well occur after the release of unmodified prelipoproteins from the membrane (5).
Release of Cell Wall Proteins by Proteolytic Shaving and Cell Wall Turnover
The fact that cell wall-bound proteins are stabilized in a multiple protease mutant strain indicates that extracellular proteases contribute to the release of cell wall-bound proteins into the medium by proteolytic shaving. However, the proteome of a σD mutant strain of B. subtilis, which displays an impaired cell wall turnover, showed a similar increase in stability of cell wall proteins (6). This suggests that in addition to proteolytic shaving, cell wall-bound proteins are simply released by cell wall turnover and subsequently degraded by extracellular proteases (6).
Release of Extracellular Proteins without Typical Export Signals
The 23 identified extracellular proteins lacking a typical secretion signal (Table 2) can potentially reach the medium via several routes. First, prophage-related proteins have the potential to be secreted via the PBSX prophage-encoded holin XhlB (57, 63), the SPβ prophage-encoded holins BhlA and BhlB (101), or the holin homologue YqxH, encoded by the SKIN prophage-like sequences. Such holins can form pores in the membrane through which the lytic enzymes of bacteriophages, which usually lack a signal peptide, gain access to the cell wall (161). Second, the flagellin Hag and two flagellar hook-associated proteins are most probably exported via a dedicated machinery for the assembly of flagella, which is related to the type III secretion machineries of gram-negative bacteria (15, 48, 81). Subsequently, these proteins could be released from the (damaged) flagella. Third, proteins that lack a signal peptide could be released by cell lysis. Nine such “extracellular” cytoplasmic proteins were shown to be highly abundant in the cytoplasmic proteome of B. subtilis (22) (Table 2), which makes it very likely that these proteins are detected in the medium due to cell lysis. Conceivably, the extracellularly encountered “cytoplasmic” proteins are significantly more resistant to degradation by extracellular proteases than are other highly abundant proteins of the cytoplasm. However, it should be noted that no additional cytoplasmic proteins were detected in the medium of a sevenfold extracellular protease mutant strain (6). Interestingly, the appearance of cytoplasmic proteins in the extracellular proteome of a strain lacking several prophages, including PBSX, SPβ, and SKIN, was not detectably affected (157). Thus, cytoplasmic proteins do not seem to leave the cytoplasm of B. subtilis via prophage-encoded holins, as proposed for L. lactis (154). Finally, the possibility that the extracellular localization of cytoplasmic proteins is due to the activity of as yet unidentified export pathways of B. subtilis cannot be excluded. For example, it is presently not clear whether B. subtilis contains an active export system homologous to the export system for the virulence factor ESAT-6 of Mycobacterium tuberculosis (90, 123).
EXTRACELLULAR PROTEOMES OF OTHER GRAM-POSITIVE BACTERIA
Secretory proteins of gram-positive pathogenic bacteria are known to perform critical roles in virulence. This knowledge has triggered many research groups to identify exported proteins of these bacteria by proteomic approaches. In the following sections, we discuss the outcomes of these studies in the light of the proteomics of protein secretion in B. subtilis (an overview is given in Fig. 7). Although B. subtilis is generally regarded as a nonpathogenic bacterium, many secreted proteins of this organism have the potential to be virulence factors. For instance, proteases can be involved in the degradation of antibacterial peptides and flagellins can be involved in adherence to host tissues (42).
FIG. 7.
Relative contributions of different mechanisms for extracellular protein accumulation in gram-positive bacteria. The relative contribution of different mechanisms for the extracellular accumulation of proteins from gram-positive bacteria was deduced from extracellular proteome studies with B. subtilis, B. cereus, C. difficile, S. aureus, GAS, and M. tuberculosis (see the text for details). For this overview, the number of proteins released by a certain export mechanism was devided by the total number of identified extracellular proteins. Sec, signal peptide- and Sec-dependent protein secretion; Lys, typical cytoplasmic proteins released by lysis or an unidentified signal peptide-independent mechanism; Hol, release of proteins by specific holin systems, Fla, release of flagellum-related proteins. It should be noted that the Sec portion of the extracellular proteomes includes all proteins with Sec-type signals, lipoprotein signals, and transmembrane domains that have the potential to direct transport across the membrane. Because only one protein of B. subtilis (PhoD) is known to be secreted in a Tat-dependent manner, this group of proteins was not included in this comparison.
Bacillus cereus
The gram-positive, spore-forming bacterium Bacillus cereus is a close relative of B. subtilis. However, this bacterium is a food-borne pathogen, causing severe food poisoning as it secretes many virulence factors and toxins into its (host) environment (45). The production of many of these factors is regulated by the transcriptional activator PlcR, which is maximally expressed at the beginning of the stationary phase. To study the impact of the PlcR regulon on the secreted proteins of B. cereus, Gohar et al. (42) compared the extracellular proteome of a plcR mutant strain with that of the parental B. cereus strain. These studies showed that most of the proteins secreted at the onset of the stationary growth phase were regulated, directly or indirectly, by PlcR.
Interestingly, the extracellular proteome of B. cereus contains about 500 proteins, which is more than twice the amount found for B. subtilis (5). This might reflect the importance of protein secretion for the pathogenesis of B. cereus. Moreover, 12 proteins, most of which seem to be completely absent in the proteome of B. subtilis, contributed to more than 80% of the total amount of extracellular proteins. Many of these highly abundant proteins were identified as collagenases, phospholipases, hemolysins, proteases, enterotoxins, and flagellins, all of which are potential virulence factors similar to those of Clostridium difficile (104). In total, 23 extracellular proteins of B. cereus were identified by this proteomic approach. Of these, 14 are synthesized with an N-terminal signal peptide (60%; B. subtilis, 50%), 1 has a lipoprotein signal peptide (4%; B. subtilis, 8%), 1 has a transmembrane anchor (4%; B. subtilis, 4%), 3 are flagellum-related proteins (14%; B. subtilis, 3%), and 4 are predicted cytoplasmic proteins (18%; B. subtilis, 13%). Thus, the relative contributions of different export mechanisms to the extracellular proteomes of B. cereus and B. subtilis are rather similar (Fig. 7). Only prophage-related proteins, responsible for about 6% of the extracellular proteins of B. subtilis, were not detected in the extracellular proteome of B. cereus. Ever though these findings suggest that the majority of extracellular proteins of B. subtilis and B. cereus are secreted via the Sec pathway, one should bear in mind that certain identified proteins with a signal peptide, such as hemolysin, seem to be secreted by the flagellar assembly pathway in B. thuringiensis (40). Despite the above-mentioned similarities, the exact composition of the extracellular proteome of B. cereus seems to be totally different from that of B. subtilis. The only homologous proteins identified in the two extracellular proteomes seem to be the lipoprotein OppA and the cytoplasmic protein Eno (Tables 1 and 2) (42).
Clostridium difficile
C. difficile-associated diarrhoea is a major problem in hospitals (96). Although it is well established that the major virulence factors of C. difficile are the two toxins A and B, this organism is thought to also secrete other virulence factors that are important for host infection. Notably, toxins A and B contribute to as much as 50% of the total protein of the extracellular proteome. The mechanism by which these toxins, which lack signal peptides of a known type, are secreted by C. difficile is unknown, and information about the other extracellular proteins of this bacterium is limited. Therefore, Mukherjee et al. (74) performed a proteomic study to identify exported proteins from a C. difficile strain during high-toxin-production conditions. Surprisingly, only 15 protein spots were detectable in the extracellular proteome of C. difficile. In addition to toxins A and B, 10 proteins could be identified, seven of which are synthesized with an N-terminal signal peptide (70%; B. subtilis, 50%) and were annotated as S-layer proteins with cell wall-binding properties, 1 of which is homologous to prophage-related proteins exported by specific holin systems (10%; B. subtilis, 6%), and 2 of which are typical cytoplasmic proteins (20%; B. subtilis, 13%). No extracellular proteins with (predicted) lipoprotein signal peptides, transmembrane anchors, or flagellum-related proteins were detected. These differences in the extracellular proteomes of B. subtilis and C. difficile (Fig. 7) may reflect the different ecological niches of the two organisms. Importantly, the fact that the relative amounts of the two cytoplasmic proteins released into the medium are less than 1% of the total protein content of the extracellular proteome argues against previous ideas that toxins A and B (lacking signal peptides) are released by cell lysis (74). In conclusion, C. difficile seems to make use of Sec-dependent and, most probably, holin(-like) pathways to release a relatively small number of proteins into its environment.
Staphylococcus aureus
S. aureus is widely recognized as a pathogen even though it is usually only a colonizer of the human host. Unfortunately, it can switch from a commensal to a lethal pathogen (24). Pathogenesis of S. aureus involves the synthesis of cell wall-associated virulence factors, a large number of extracellular proteins, and secreted toxins with damaging effects on the host cells. To identify extracellular proteins of S. aureus that are potential virulence factors, Ziebandt et al. (162) used a proteomic approach to analyze the pattern of extracellular proteins of different S. aureus strains. In total, 26 proteins of the approximately 100 protein spots could be identified. Among the newly identified proteins were enterotoxins, a leukotoxin, serine proteases, a thermonuclease, and an immunoglobulin G-binding protein, all being potential virulence factors. Strikingly, in contrast to B. subtilis (50%), about 90% of the identified extracellular proteins of S. aureus are synthesized with Sec-type signal peptides. The remaining proteins are most probably cytoplasmic proteins released by cell lysis. No extracellular proteins with (predicted) lipoprotein signal peptides, transmembrane anchors, flagellum-related proteins, or phage-related proteins were detected which, together, form a significant portion of the B. subtilis secretome (5) (Fig. 7). Interestingly, one extracellular protein was found to be homologous to the transmembrane protein YfnI of B. subtilis, whose C-terminal part is released by SPase I-mediated processing (5). However, the YfnI protein of S. aureus seems to have a potential signal peptide instead of the five membrane-spanning domains in the N terminus of YfnI from B. subtilis. Remarkably, the SPase I-processing sites are both located about 40 residues C-terminally of the fifth transmembrane segment of B. subtilis YfnI (AYA) and the potential signal peptide of S. aureus YfnI (ALA).
Bernardo et al. (13) performed a proteomic study aimed at the characterization of virulence of different S. aureus strains. Extracellular protein spots that were present in the medium of all tested strains were identified as protein A, hemolysins, lipases, and autolysins. The fact that protein A is a cell wall-anchored protein of S. aureus shows that, similarly to B. subtilis, certain cell wall-bound proteins are released into the growth medium. However, it is not clear whether this release of protein A is important for the pathogenesis of S. aureus strains.
Group A Streptococcus
Strains of group A Streptococcus (GAS) species are a common cause of severe invasive infections with unusually high rates of morbidity and mortality (155). Certain extracellular proteins from GAS strains play critical roles in human infections caused by these organisms. To perform a systematic analysis of these extracellular proteins, the proteins present in the media of different GAS strains were analyzed by Lei et al. using 2D PAGE (59). Of the about 80 protein spots that were observed, 43 distinct proteins were identified. Strikingly, only 16 of these had typical signal peptides whereas 27 proteins did not. Among the extracellular proteins with a signal peptide were the (putative) virulence factors streptolysin O, the M1 and M3 proteins, mitogenic factor, streptococcal pyrogenic exotoxin A, streptococcal inhibitor of complement, and homologues of class B acid phosphatase and serine proteases. The extracellular proteins without a typical signal peptide are presumably cytoplasmic proteins, including proteins involved in glycolytic metabolism, translation, the urea cycle, and chaperonins like GroEL. It should be noted that typical cytoplasmic proteins, such as enolases, influence pathogen-host interactions (91). Together, these data show that about half of the extracellular proteins from GAS strains are secreted in a signal peptide- and Sec-dependent manner. The remaining 50% of the proteins are released by cell lysis or by other, as yet unidentified, Sec-independent export mechanisms (Fig. 7).
Mycobacterium tuberculosis
Mycobacterium tuberculosis causes about 8 million cases of tuberculosis worldwide each year (128). To facilitate the design of novel measures for the prevention and therapy of this health threat, the proteomes of nonvirulent Mycobacterium bovis strains and virulent M. tuberculosis strains were compared by Jungblut et al. (53). Furthermore, Rosenkrands et al. (105) extended these proteome studies specifically for M. tuberculosis. The extracellular proteome of M. tuberculosis cultures contained between 600 and 800 protein spots, of which, in total, 84 proteins could be identified (105). Of these 84 proteins, only 31 were unique to the extracellular proteome since these proteins were absent from the cytoplasmic or cell wall proteomes. This suggests that a large portion of the extracellular proteins are released by cell lysis (139). The latter view is supported by the identification of typical cytoplasmic proteins, such as aldolases, enolases, elongation factor G, GroEL, superoxide dismutase, and various dehydrogenases, which were also detected in the extracellular proteome of B. subtilis (5). Strikingly, of the 31 proteins that were found exclusively in the medium fraction, only 5 are synthesized with a putative N-terminal signal peptide as previously predicted (43). However, it should be noted that proteins that contain transmembrane domains in addition to a signal peptide were excluded from the latter predictions. At least two such proteins were previously identified in the culture medium of M. tuberculosis, which shows that certain proteins are proteolytically released from the cell envelope (156), similar to the YfkN and YhcR proteins of B. subtilis (5). In addition, at least one protein with a putative lipoprotein signal peptide was identified in both the cell wall and medium fractions. Taken together, these data strongly suggest that although a small repertoire of extracellular proteins from M. tuberculosis are secreted in a signal peptide- and Sec-dependent manner, most proteins are released into the medium by cell lysis or via specific Sec-independent export mechanisms, such as the Snm system (117, 123).
PERSPECTIVES
The power of high-resolution proteomic techniques has been effectively used to gain novel insights in the general flow of proteins into the environment of B. subtilis. Specifically, these studies shed new light on signal peptide function, the role of signal peptide processing, the importance of signal peptide-independent protein export pathways, and the function of extracellular proteins in general. This boost of information will most certainly provide major leads for future research on protein transport in B. subtilis and other gram-positive bacteria. Importantly, several extracellular and surface-exposed proteins of gram-positive pathogens have been implicated as important virulence factors and mediators in the inflammatory response in human hosts during bacterial infections. Vaccines or drugs that inhibit export pathways for such proteins can therefore have broad applications in human and animal health care. This idea is especially attractive for export pathways that are absent from humans and other higher eukaryotes, such as the flagellum, holin, or Snm pathways, which have been implicated in the virulence of several gram-positive bacteria. Interestingly, the recent proteomic data imply that B. subtilis employs most of the protein export pathways or mechanisms that have been described for pathogenic gram-positive organisms. The fact that B. subtilis is genetically very amenable and nonpathogenic, combined with the availability of a large B. subtilis strain collection that contains ∼3,000 different isogenic mutants with single-gene disruptions (54) and the vast amount of readily available knowledge concerning the molecular biology of B. subtilis (120), makes this organism an ideal model to study gram-positive protein secretion in a proteome-wide context. With respect to medical applications, in particular, the elucidation of all cellular mechanisms for the export and/or release of B. subtilis extracellular proteins is an important challenge for future research.
Acknowledgments
We thank members of the Groningen and European Bacillus Secretion Groups and the ExporteRRs consortium for stimulating discussions.
H.T. was supported by Genencor International (Leiden, The Netherlands). H.A, J.D.H.J., P.G.B, J.-Y.F.D., R.D., H.W., W.J.Q., O.P.K., S.B., and J.M.V.D. were supported by grants BIO4-CT98-0250, QLK3-CT-1999-00413, and QLK3-CT-1999-00917 from the European Union. E.D. was supported by the Ubbo Emmius foundation of the University of Groningen. G.Z. was supported by grant STW VBI.1837 from the Stichting Technische Wetenschappen. M.H. was supported from grants from the Deutsche Forschungsgemeinschaft, the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Akita, M., S. Sasaki, S. Matsuyama, and S. Mizushima. 1990. SecA interacts with secretory proteins by recognizing the positive charge at the amino terminus of the signal peptide in Escherichia coli. J. Biol. Chem. 265:8162-8169. [PubMed] [Google Scholar]
- 2.Alami, M., I. Lüke, S. Deitermann, J. Brunner, and M. Müller. 2003. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell 12:937-946. [DOI] [PubMed] [Google Scholar]
- 3.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antelmann, H., E. Darmon, D. Noone, J. W. Veening, H. Westers, S. Bron, O. P. Kuipers, K. M. Devine, M. Hecker, and J. M. van Dijl. 2003. The extracellular proteome of Bacillus subtilis under secretion stress conditions. Mol. Microbiol. 49:143-156. [DOI] [PubMed] [Google Scholar]
- 5.Antelmann, H., H. Tjalsma, B. Voigt, S. Ohlmeier, S. Bron, J. M. van Dijl, and M. Hecker. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11:14984-14502. [DOI] [PubMed] [Google Scholar]
- 6.Antelmann, H., H. Yamamoto, J. Sekiguchi, and M. Hecker. 2002. Stabilization of cell wall proteins in Bacillus subtilis: a proteomic approach. Proteomics 2:591-602. [DOI] [PubMed] [Google Scholar]
- 7.Archibald, A. R., I. C. Hancock, and C. R. Harwood. 1993. Cell wall structure, synthesis and turnover, p. 381-410. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria, ASM Press, Washington, D.C.
- 8.Baba, T., and O. Schneewind. 1996. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 15:4789-4797. [PMC free article] [PubMed] [Google Scholar]
- 9.Baba, T., and O. Schneewind. 1998. Targeting of muralytic enzymes to the cell division site of Gram-positive bacteria: repeat domains direct autolysin to the equatorial surface ring of Staphylococcus aureus. EMBO J. 17:4639-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babe, L. M., and B. Schmidt. 1998. Purification and biochemical analysis of WprA, a 52-kDa serine protease secreted by Bacillus subtilis as an active complex with its 23- kDa propeptide. Biochim. Biophys. Acta 1386:211-219. [DOI] [PubMed] [Google Scholar]
- 11.Beck, K., L. F. Wu, J. Brunner, and M. Müller. 2000. Discrimination between SRP- and SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor. EMBO J. 19:134-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bengtsson, J., H. Tjalsma, C. Rivolta, and L. Hederstedt. 1999. Subunit II of Bacillus subtilis cytochrome c oxidase is a lipoprotein. J. Bacteriol. 181:685-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernardo, K., S. Fleer, N. Pakulat, O. Krut, F. Hunger, and M. Kronke. 2002. Identification of Staphylococcus aureus exotoxins by combined sodium dodecyl sulfate gel electrophoresis and matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics 2:740-746. [DOI] [PubMed] [Google Scholar]
- 14.Blackman, S. A., T. J. Smith, and S. J. Foster. 1998. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology 144:73-82. [DOI] [PubMed] [Google Scholar]
- 15.Blocker, A., K. Komoriya, and S. I. Aizawa. 2003. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl. Acad. Sci. USA 100:3027-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolhuis, A., A. Matzen, H. L. Hyyrylaïnen, V. P. Kontinen, R. Meima, J. Chapuis, G. Venema, S. Bron, R. Freudl, and J. M. van Dijl. 1999. Signal peptide peptidase- and ClpP-like proteins of Bacillus subtilis are required for efficient translocation and processing of secretory proteins. J. Biol. Chem. 274:24585-24592. [DOI] [PubMed] [Google Scholar]
- 17.Bolhuis, A., C. P. Broekhuizen, A. Sorokin, M. L. van Roosmalen, G. Venema, S. Bron, W. J. Quax, and J. M. van Dijl. 1998. SecDF of Bacillus subtilis, a molecular Siamese twin required for the efficient secretion of proteins. J. Biol. Chem. 273:21217-21224. [DOI] [PubMed] [Google Scholar]
- 18.Bolhuis, A., G. Venema, W. J. Quax, S. Bron, and J. M. van Dijl. 1999. Functional analysis of paralogous thiol-disulfide oxidoreductases in Bacillus subtilis. J. Biol. Chem. 274: 24531-24538. [DOI] [PubMed] [Google Scholar]
- 19.Bolhuis, A., H. Tjalsma, H. E. Smith, A. de Jong, R. Meima, G. Venema, S. Bron, and J. M. van Dijl. 1999. Evaluation of bottlenecks in the late stages of protein secretion in Bacillus subtilis. Appl. Environ. Microbiol. 65:2934-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolhuis, A., H. Tjalsma, K. Stephenson, C. R. Harwood, G. Venema, S. Bron, and J. M. van Dijl. 1999. Different mechanisms for thermal inactivation of Bacillus subtilis signal peptidase mutants. J. Biol. Chem. 274:15865-15868. [DOI] [PubMed] [Google Scholar]
- 21.Briggs, M. S., D. G. Cornell, R. A. Dluhy, and L. M. Gierasch. 1986. Conformations of signal peptides induced by lipids suggest initial steps in protein export. Science 233:206-208. [DOI] [PubMed] [Google Scholar]
- 22.Buttner, K., J. Bernhardt, C. Scharf, R. Schmid, U. Mader, C. Eymann, H. Antelmann, A. Völker, U. Völker, and M. Hecker. 2001. A comprehensive two- dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22:2908-2935. [DOI] [PubMed] [Google Scholar]
- 23.Chakicherla, A., and J. N. Hansen. 1995. Role of the leader and structural regions of prelantibiotic peptides as assessed by expressing nisin-subtilin chimeras in Bacillus subtilis 168, and characterization of their physical, chemical, and antimicrobial properties. J. Biol. Chem. 270:23533-23539. [DOI] [PubMed] [Google Scholar]
- 24.Cheung, A. L., S. J. Projan, and H. Gresham. 2002. The genomic aspect of virulence, sepsis, and resistance to killing mechanisms in Staphylococcus aureus. Curr. Infect. Dis. Rep. 4:400-410. [DOI] [PubMed] [Google Scholar]
- 25.Chu, P. W., M. N. Yap, C. Y. Wu, C. M. Huang, F. M. Pan, M. J. Tseng, and S. T. Chen. 2000. A proteomic analysis of secreted proteins from xylan-induced Bacillus sp. strain K-1. Electrophoresis 21:1740-1745. [DOI] [PubMed] [Google Scholar]
- 26.Chung, Y. S., and D. Dubnau. 1995. ComC is required for the processing and translocation of ComGC, a pilin-like competence protein of Bacillus subtilis. Mol. Microbiol. 15:543-551. [DOI] [PubMed] [Google Scholar]
- 27.Chung, Y. S., F. Breidt, and D. Dubnau. 1998. Cell surface localization and processing of the ComG proteins, required for DNA binding during transformation of Bacillus subtilis. Mol. Microbiol. 29:905-913. [DOI] [PubMed] [Google Scholar]
- 28.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 6:673-676. [DOI] [PubMed] [Google Scholar]
- 29.Dalbey, R. E., and G. von Heijne. 1992. Signal peptidases in prokaryotes and eukaryotes: a new protease family. Trends Biochem. Sci. 17:474-478. [DOI] [PubMed] [Google Scholar]
- 30.Dalbey, R. E., M. O. Lively, S. Bron, and J. M. van Dijl. 1997. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 6:1129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darmon, E., D. Noone, A. Masson, S. Bron, O. P. Kuipers, K. M. Devine, and J. M. van Dijl. 2002. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J. Bacteriol. 184:5661-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Vrije, T., A. M. Batenburg, J. A. Killian, and B. De Kruijff. 1990. Lipid involvement in protein translocation in Escherichia coli. Mol. Microbiol. 4:143-150. [DOI] [PubMed] [Google Scholar]
- 33.Dorenbos, R., T. Stein, J. Kabel, C. Bruand, A. Bolhuis, S. Bron, W. J. Quax, and J. M. van Dijl. 2002. Thiol-disulfide oxidoreductases are essential for the production of the lantibiotic sublancin 168. J. Biol. Chem. 277:16682-16688. [DOI] [PubMed] [Google Scholar]
- 34.Dubnau, D. 1997. Binding and transport of transforming DNA by Bacillus subtilis: the role of type-IV pilin-like proteins-a review. Gene 192:191-198. [DOI] [PubMed] [Google Scholar]
- 35.Economou, A. 1998. Bacterial pre-protein translocase: mechanism and conformational dynamics of a processive enzyme. Mol. Microbiol. 27:511-518. [DOI] [PubMed] [Google Scholar]
- 36.Economou, E. 2002. Bacterial secretome: the assembly manual and operating instructions. Mol. Membr. Biol. 19:159-169. [DOI] [PubMed] [Google Scholar]
- 37.Eymann, C., H. Mach, C. R. Harwood, and M. Hecker. 1996. Phosphate-starvation- inducible proteins in Bacillus subtilis: a two-dimensional gel electrophoresis study Microbiology 142:3163-3170. [DOI] [PubMed] [Google Scholar]
- 38.Fekkes, P., and A. J. M. Driessen. 1999. Protein targeting to the bacterial cytoplasmic membrane. Microbiol. Mol. Biol. Rev. 63:161-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foster, S. J. 1993. Molecular analysis of three major wall-associated proteins of Bacillus subtilis 168: evidence for processing of the product of a gene encoding a 258 kDa precursor two-domain ligand-binding protein. Mol. Microbiol. 8:299-310. [DOI] [PubMed] [Google Scholar]
- 40.Ghelardi, E., F. Celandroni, S. Salvetti, D. J. Beecher, M. Gominet, D. Lereclus, A. C. Wong, and S. Senesi. 2002. Requirement of flhA for swarming differentiation, flagellin export, and secretion of virulence-associated proteins in Bacillus thuringiensis. J. Bacteriol. 184:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghuysen, J. M., J. Lamotte-Brasseur, B. Joris, and G. D. Shockman. 1994. Binding site-shaped repeated sequences of bacterial wall peptidoglycan hydrolases. FEBS Lett. 342:23-28. [DOI] [PubMed] [Google Scholar]
- 42.Gohar, M., O. A. Okstad, N. Gilois, V. Sanchis, A. B. Kolsto, and D. Lereclus. 2002. Two-dimensional electrophoresis analysis of the extracellular proteome of Bacillus cereus reveals the importance of the PlcR regulon. Proteomics 2:784-791. [DOI] [PubMed] [Google Scholar]
- 43.Gomez, M., S. Johnson, and M. L. Gennaro. 2000. Identification of secreted proteins of Mycobacterium tuberculosis by a bioinformatic approach. Infect. Immun. 68:2323-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graumann, P., K. Schröder, R. Schmid, and M. A. Marahiel. 1996. Cold shock stress-induced proteins in Bacillus subtilis. J. Bacteriol. 178:4611-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guinebretiere, M. H., V. Broussolle, and C. Nguyen-The. 2002. Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J. Clin. Microbiol. 40:3053-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirose, I., K. Sano, I. Shiosa, M. Kumano, K. Nakamura, and K. Yamane. 2000. Proteome analysis of Bacillus subtilis extracellular proteins: a two-dimensional protein electrophoretic study. Microbiology 146:65-75. [DOI] [PubMed] [Google Scholar]
- 47.Honda, K., K. Nakamura, M. Nishiguchi, and K. Yamane. 1993. Cloning and characterization of a Bacillus subtilis gene encoding a homolog of the 54-kilodalton subunit of mammalian signal recognition particle and Escherichia coli Ffh. J. Bacteriol. 175:4885-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hyyrylaïnen, H. L., A. Bolhuis, E. Darmon, L. Muukkonen, P. Koski, M. Vitikainen, M. Sarvas, Z. Prágai, S. Bron, J. M. van Dijl, and V. P. Kontinen. 2001. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41:1159-1172. [DOI] [PubMed] [Google Scholar]
- 50.Jacobs, M., J. B. Andersen, V. P. Kontinen, and M. Sarvas. 1993. Bacillus subtilis PrsA is required in vivo as an extracytoplasmic chaperone for secretion of active enzymes synthesized either with or without pro-sequences. Mol. Microbiol. 8:957-966. [DOI] [PubMed] [Google Scholar]
- 51.Jongbloed, J. D. H., H. Antelmann, M. Hecker, R. Nijland, F. Pries, P. Koski, W. J. Quax, S. Bron, J. M. van Dijl, and P. G. Braun. 2002. Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis. J. Biol. Chem. 277:44068-44078. [DOI] [PubMed] [Google Scholar]
- 52.Jongbloed, J. D. H., U. Martin, H. Antelmann, M. Hecker, H. Tjalsma, G. Venema, S. Bron, J. M. van Dijl, and J. Müller. 2000. TatC is a specificity determinant for protein secretion via the twin-arginine translocation pathway. J. Biol. Chem. 275:41350-41357. [DOI] [PubMed] [Google Scholar]
- 53.Jungblut, P. R., U. E. Schaible, H. J. Mollenkopf, U. Zimny-Arndt, B. Raupach, J. Mattow, P. Halada, S. Lamer, K. Hagens, and S. H. Kaufmann. 1999. Comparative proteome analysis of Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol. Microbiol. 33:1103-1117. [DOI] [PubMed] [Google Scholar]
- 54.Kobayashi K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessières, et al. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kontinen, V. P, and M. Sarvas. 1993. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol. Microbiol. 8:727-737. [DOI] [PubMed] [Google Scholar]
- 56.Kontinen, V. P., P. Saris, and M. Sarvas. 1991. A gene (prsA) of Bacillus subtilis involved in a novel, late stage of protein export. Mol. Microbiol. 5:1273-1283. [DOI] [PubMed] [Google Scholar]
- 57.Krogh, S., S. T. Jørgensen, and K. M. Devine. 1998. Lysis genes of the Bacillus subtilis defective prophage PBSX. J. Bacteriol. 180:2110-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessières, A. Bolotin, S. Borchert, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 59.Lei, B., S. Mackie, S. Lukomski, and J. M. Musser. 2000. Identification and immunogenicity of group A Streptococcus culture supernatant proteins. Infect. Immun. 68:6807-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leloup, L., A. J. M. Driessen, R. Freudl, R. Chambert, and M. F. Petit-Glatron. 1999. Differential dependence of levansucrase and alpha-amylase secretion on SecA (Div) during the exponential phase of growth of Bacillus subtilis. J. Bacteriol. 181:1820-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lepesant, J. A., F. Kunst, J. Lepesant-Kejzlarova, and R. Dedonder. 1972. Chromosomal location of mutations affecting sucrose metabolism in Bacillus subtilis Marburg. Mol. Gen. Genet. 118:135-160. [DOI] [PubMed] [Google Scholar]
- 62.Leskelä, S., E. Wahlström, V. P. Kontinen, and M. Sarvas. 1999. Lipid modification of prelipoproteins is dispensable for growth but essential for efficient protein secretion in Bacillus subtilis: characterization of the lgt gene. Mol. Microbiol. 31:1075-1085. [DOI] [PubMed] [Google Scholar]
- 63.Longchamp, P. F., C. Mauel, and D. Karamata. 1994. Lytic enzymes associated with defective prophages of Bacillus subtilis: sequencing and characterization of the region comprising the N-acetylmuramoyl-l-alanine amidase gene of prophage PBSX. Microbiology 140:1855-1867. [DOI] [PubMed] [Google Scholar]
- 64.Lory, S. 1994. Leader peptidase of type IV prepilins and related proteins, pp. 17-29. In G. von Heijne (ed.), Signal Peptidases. R. G. Landes Co., Austin, Tex.
- 65.Luirink, J., T. Samuelsson, and J. W. de Gier. 2001. YidC/Oxa1p/Alb3: evolutionarily conserved mediators of membrane protein assembly. FEBS Lett. 501:1-5. [DOI] [PubMed] [Google Scholar]
- 66.Mader, U., H. Antelmann, T. Buder, M. K. Dahl, M. Hecker, and G. Homuth. 2002. Bacillus subtilis functional genomics: genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics. Mol. Genet. Genomics 268:455-467. [DOI] [PubMed] [Google Scholar]
- 67.Magnuson, R., J. Solomon, and A. D. Grossman. 1994. Biochemical and genetic characterization of a competence pheromone from Bacillus subtilis. Cell 77:207-216. [DOI] [PubMed] [Google Scholar]
- 68.Margot, P., and D. Karamata. 1996. The wprA gene of Bacillus subtilis 168, expressed during exponential growth, encodes a cell-wall-associated protease. Microbiology 142:3437-3444. [DOI] [PubMed] [Google Scholar]
- 69.Margot, P., M. Wahlen, A. Gholamhoseinian, P. Piggot, and D. Karamata. 1998. The lytE gene of Bacillus subtilis 168 encodes a cell wall hydrolase. J. Bacteriol. 180:749-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mazmanian, S. K., H. Ton-That, K. Su, and O. Schneewind. 2002. An iron- regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA 99:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meima, R., C. Eschevins, S. Fillinger, A. Bolhuis, L. W. Hamoen, R. Dorenbos, W. J. Quax, J. M. van Dijl, R. Provvedi, I. Chen, D. Dubnau, and S. Bron. 2002. The bdbDC operon of Bacillus subtilis encodes thiol-disulphide oxidoreductases required for competence development. J. Biol. Chem. 277:6994-7001. [DOI] [PubMed] [Google Scholar]
- 72.Merchante, R., H. M. Pooley, and D. Karamata. 1995. A periplasm in Bacillus subtilis. J. Bacteriol. 177:6176-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mori, H., and K. Ito. 2001. The Sec protein-translocation pathway. Trends Microbiol. 9:494-500. [DOI] [PubMed] [Google Scholar]
- 74.Mukherjee, K., S. Karlsson, L. G. Burman, and T. Akerlund. 2002. Proteins released during high toxin production in Clostridium difficile. Microbiology 148:2245-2253. [DOI] [PubMed] [Google Scholar]
- 75.Müller, J., S. Bron, G. Venema, and J. M. van Dijl. 2000. Chaperone-like activities of the CsaA protein of Bacillus subtilis. Microbiology 146:77-88. [DOI] [PubMed] [Google Scholar]
- 76.Müller, J., J. Ozegowski, S. Vettermann, J. Swaving, K. H. van Wely, and A. J. M. Driessen. 2000. Interaction of Bacillus subtilis CsaA with SecA and precursor proteins. Biochem. J. 348:367-373. [PMC free article] [PubMed] [Google Scholar]
- 77.Murray, T., D. L. Popham, and P. Setlow. 1997. Identification and characterization of pbpA encoding Bacillus subtilis penicillin-binding protein 2A. J. Bacteriol. 179:3021-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nakamura, K., M. Nishiguchi, K. Honda, and K. Yamane. 1994. The Bacillus subtilis SRP54 homologue, Ffh, has an intrinsic GTPase activity and forms a ribonucleoprotein complex with small cytoplasmic RNA in vivo. Biochem. Biophys. Res. Commun. 199:1394-1399. [DOI] [PubMed] [Google Scholar]
- 79.Nakamura, K., S. Yahagi, T. Yamazaki, and K. Yamane. 1999. Bacillus subtilis histone-like protein, HBsu, is an integral component of a SRP-like particle that can bind the Alu domain of small cytoplasmic RNA. J. Biol. Chem. 274:13569-13576. [DOI] [PubMed] [Google Scholar]
- 80.Nakano, M. M., and P. Zuber. 1990. Molecular biology of antibiotic production in Bacillus. Crit. Rev. Biotechnol. 10:223-240. [DOI] [PubMed] [Google Scholar]
- 81.Namba, K., I. Yamashita, and F. Vonderviszt. 1989. Structure of the core and central channel of bacterial flagella. Nature 342:648-654. [DOI] [PubMed] [Google Scholar]
- 82.Navarre, W. W., and O. Schneewind. 1994. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in Gram-positive bacteria. Mol. Microbiol. 14:115-121. [DOI] [PubMed] [Google Scholar]
- 83.Navarre, W. W., S. Daefler, and O. Schneewind. 1996. Cell wall sorting of lipoproteins in Staphylococcus aureus. J. Bacteriol. 178:441-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nes, I. F., and J. R. Tagg. 1996. Novel lantibiotics and their pre-peptides. Antonie Leeuwenhoek 69:89-97. [DOI] [PubMed] [Google Scholar]
- 85.Noone, D., A. Howell, R. Collery, and K. M. Devine. 2001. YkdA and YvtA, HtrA- like serine proteases in Bacillus subtilis, engage in negative autoregulation and reciprocal cross-regulation of ykdA and yvtA gene expression. J. Bacteriol. 183:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nouwen, N., and A. J. M. Driessen. 2002. SecDFyajC forms a heterotetrameric complex with YidC. Mol. Microbiol. 44:1397-1405. [DOI] [PubMed] [Google Scholar]
- 87.Ogura, A., H. Kakeshita, K. Honda, H. Takamatsu, K. Nakamura, and K. Yamane. 1995. Srb: a Bacillus subtilis gene encoding a homologue of the alpha- subunit of the mammalian signal recognition particle receptor. DNA Res. 2:95-100. [DOI] [PubMed] [Google Scholar]
- 88.Paetzel, M., R. E. Dalbey, and N. C. Strynadka. 1998. Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor. Nature 396:186-190. [DOI] [PubMed] [Google Scholar]
- 89.Paik, S. H., A. Chakicherla, and J. N. Hansen. 1998. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J. Biol. Chem. 273:23134-23142. [DOI] [PubMed] [Google Scholar]
- 90.Pallen, M. J. 2002. The ESAT-6/WXG100 superfamily: a new Gram-positive secretion system? Trends Microbiol. 10:209-212. [DOI] [PubMed] [Google Scholar]
- 91.Pancholi, V., and V. A. Fischetti. 1998. Alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 92.Park, S. G., C. W. Kho, S. Cho, D. H. Lee, S. H. Kim, and B. C. Park. 2002. A functional proteomic analysis of secreted fibrinolytic enzymes from Bacillus subtilis 168 using a combined method of two-dimensional gel electrophoresis and zymography. Proteomics 2:206-211. [DOI] [PubMed] [Google Scholar]
- 93.Pohlschröder, M., W. A. Prinz, E. Hartmann, and J. Beckwith. 1997. Protein translocation in the three domains of life: variations on a theme. Cell 91:563-566. [DOI] [PubMed] [Google Scholar]
- 94.Pooley, H. M., R. Merchante, and D. Karamata. 1996. Overall protein content and induced enzyme components of the periplasm of Bacillus subtilis. Microb. Drug. Resist. 2:9-15. [DOI] [PubMed] [Google Scholar]
- 95.Popham, D. L., J. Helin, C. E. Costello, and P. Setlow. 1996. Analysis of the peptidoglycan structure of Bacillus subtilis endospores. J. Bacteriol. 178:6451-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Poxton, I. R., J. McCoubrey, and G. Blair. 2001. The pathogenicity of Clostridium difficile. Clin. Microbiol. Infect. 7:421-427. [DOI] [PubMed] [Google Scholar]
- 97.Prágai, Z., H. Tjalsma, A. Bolhuis, J. M. van Dijl, G. Venema, and S. Bron. 1997. The signal peptidase II (Isp) gene of Bacillus subtilis. Microbiology 143:1327-1333. [DOI] [PubMed] [Google Scholar]
- 98.Pugsley, A. P. 1993. The complete general secretory pathway in Gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Randall, L. L., and S. J. Hardy. 2002. SecB, one small chaperone in the complex milieu of the cell. Cell Mol. Life Sci. 59:1617-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rashid, M. H., M. Mori, and J. Sekiguchi. 1995. Glucosaminidase of Bacillus subtilis: cloning, regulation, primary structure and biochemical characterization. Microbiology 141:2391-2404. [DOI] [PubMed] [Google Scholar]
- 101.Regamey, A., and D. Karamata. 1998. The N-acetylmuramoyl-l-alanine amidase encoded by the Bacillus subtilis 168 prophage SPβ. Microbiology 144:885-893. [DOI] [PubMed] [Google Scholar]
- 102.Riezman, H. 1997. The ins and outs of protein translocation. Science 278:1728-1729. [DOI] [PubMed] [Google Scholar]
- 103.Robinson, C., and A. Bolhuis. 2001. Protein targeting by the twin-arginine translocation pathway. Nat. Rev. Mol. Cell. Biol. 2:350-356. [DOI] [PubMed] [Google Scholar]
- 104.Rood, J. I. 1998. Virulence genes of Clostridium perfringens. Annu. Rev. Microbiol. 52:333-360. [DOI] [PubMed] [Google Scholar]
- 105.Rosenkrands, I., K. Weldingh, S. Jacobsen, C. V. Hansen, W. Florio, I. Gianetri, and P. Andersen. 2000. Mapping and identification of Mycobacterium tuberculosis proteins by two-dimensional gel electrophoresis, microsequencing and immunodetection. Electrophoresis 21:935-948. [DOI] [PubMed] [Google Scholar]
- 106.Rosenow, C., P. Ryan, J. N. Weiser, S. Johnson, P. Fontan, A. Ortqvist, and H. R. Masure. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25:819-829. [DOI] [PubMed] [Google Scholar]
- 107.Rudd, K. E., H. J. Sofia, E. V. Koonin, G. Plunkett, S. Lazar, and P. E. Rouviere. 1995. A new family of peptidyl-prolyl isomerases. Trends Biochem Sci. 20:12-14. [DOI] [PubMed] [Google Scholar]
- 108.Samuelson, J. C., M. Chen, F. Jiang, I. Moller, M. Wiedmann, A. Kuhn, G. J. Phillips, and R. E. Dalbey. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637-641. [DOI] [PubMed] [Google Scholar]
- 109.Sánchez-Puelles, J. M., C. Ronda, J. L. García, R. Lopez, and E. García. 1986. Searching for autolysin functions: characterization of a pneumococcal mutant deleted in the lytA gene. Eur. J. Biochem. 158:289-293. [DOI] [PubMed] [Google Scholar]
- 110.Sánchez-Puelles, J. M., J. M. Sanz, J. L. García, and E. García. 1990. Cloning and expression of gene fragments encoding the choline-binding domain of pneumococcal murein hydrolases. Gene 89:69-75. [DOI] [PubMed] [Google Scholar]
- 111.Schatz, G., and B. Dobberstein. 1996. Common principles of protein translocation across membranes. Science 271:1519-1526. [DOI] [PubMed] [Google Scholar]
- 112.Schneewind, O., A. Fowler, and K. F. Faull. 1995. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268:103-106. [DOI] [PubMed] [Google Scholar]
- 113.Schneewind, O., D. Mihaylova-Petkov, and P. Model. 1993. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 12:4803-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70:267-281. [DOI] [PubMed] [Google Scholar]
- 115.Schumann, W., S. D. Ehrlich, and N. Ogasawara. 2000. Functional analysis of bacterial genes: a practical manual. John Wiley & Sons, Inc., New York, N.Y.
- 116.Scotti, P. A., M. L. Urbanus, J. Brunner, J. W. de Gier, G. von Heijne, C. van der Does, A. J. M. Driessen, B. Oudega, and J. Luirink. 2000. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19:542-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scotti, P. A., Q. A. Valent, E. H. Manting, M. L. Urbanus, A. J. M. Driessen, B. Oudega, and J. Luirink. 1999. SecA is not required for signal recognition particle- mediated targeting and initial membrane insertion of a nascent inner membrane protein. J. Biol. Chem. 274:29883-29888. [DOI] [PubMed] [Google Scholar]
- 118.Seluanov, A., and E. Bibi. 1997. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J. Biol. Chem. 272:2053-2055. [DOI] [PubMed] [Google Scholar]
- 119.Solomon, J. M., R. Magnuson, A. Srivastava, and A. D. Grossman. 1995. Convergent sensing pathways mediate response to two extracellular competence factors in Bacillus subtilis. Genes Dev. 9:547-558. [DOI] [PubMed] [Google Scholar]
- 120.Sonenshein, A. L., J. A. Hoch, and R. Losick (ed.). 2001. Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 121.Sørensen, A. L., S. Nagai, G. Houen, P. Andersen, and A. B. Andersen. 1995. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect. Immun. 63:1710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 123.Stanley, S. A., S. Raghavan, W. W. Hwang, and J. S. Cox. 2003. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc. Natl. Acad. Sci. USA 100:13001-13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stephenson, K., and C. R. Harwood. 1998. Influence of a cell-wall-associated protease on production of alpha-amylase by Bacillus subtilis. Appl. Environ. Microbiol. 64:2875-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stephenson, K., S. Bron, and C. R. Harwood. 1999. Cellular lysis in Bacillus subtilis: the effect of multiple extracellular protease deficiencies. Lett. Appl. Microbiol. 29:141-145. [Google Scholar]
- 126.Stöver, A. G., and A. Driks. 1999. Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilis spore-associated protein. J. Bacteriol. 181:1664-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Suzuki, T., and Y. Tahara. 2003. Characterization of the Bacillus subtilis ywtD gene, whose product is involved in γ-polyglutamic acid degradation. J. Bacteriol. 185:2379-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tiruviluamala, P., and L. B. Reichman. 2002. Tuberculosis. Annu. Rev. Public Health 23:403-426. [DOI] [PubMed] [Google Scholar]
- 129.Tjalsma, H., A. Bolhuis, J. D. H. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tjalsma, H., A. Bolhuis, M. L. van Roosmalen, T. Wiegert, W. Schumann, C. P. Broekhuizen, W. Quax, G. Venema, S. Bron, and J. M. van Dijl. 1998. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 12:2318-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tjalsma, H., A. G. Stöver, A. Driks, G. Venema, S. Bron, and J. M. van Dijl. 2000. Conserved serine and histidine residues are critical for activity of the ER-type signal peptidase SipW of Bacillus subtilis. J. Biol. Chem. 275:25102-25108. [DOI] [PubMed] [Google Scholar]
- 132.Tjalsma, H., G. Zanen, G. Venema, S. Bron, and J. M. van Dijl. 1999. The potential active site of the lipoprotein-specific (type II) signal peptidase of Bacillus subtilis. J. Biol. Chem. 274:28191-28197. [DOI] [PubMed] [Google Scholar]
- 133.Tjalsma, H., G. Zanen, S. Bron, and J. M. van Dijl. 2001. The eubacterial lipoprotein-specific (type II) signal peptidase. 22:3-23. [DOI] [PubMed]
- 134.Tjalsma, H., M. A. Noback, S. Bron, G. Venema, K. Yamane, and J. M. van Dijl. 1997. Bacillus subtilis contains four closely related type I signal peptidases with overlapping substrate specificities: constitutive and temporally controlled expression of different sip genes. J. Biol. Chem. 272:25983-25992. [DOI] [PubMed] [Google Scholar]
- 135.Tjalsma, H., S. Bron, S., and J. M. van Dijl. 2003. Complementary impact of paralogous Oxa1-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J. Biol. Chem. 278:15622-15632. [DOI] [PubMed] [Google Scholar]
- 136.Tjalsma, H., V. P. Kontinen, Z. Prágai, H. Wu, R. Meima, G. Venema, S. Bron, M. Sarvas, and J. M. van Dijl. 1999. The role of lipoprotein processing by signal peptidase II in the Gram-positive eubacterium Bacillus subtilis: signal peptidase II is required for the efficient secretion of α-amylase, a non-lipoprotein. J. Biol. Chem. 274:1698-1707. [DOI] [PubMed] [Google Scholar]
- 137.Ton-That, H., G. Liu, S. K. Mazmanian, K. F. Faull, and O. Schneewind. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA 96:12424-12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ton-That, H., K. F. Faull, and O. Schneewind. 1997. Anchor structure of staphylococcal surface proteins. A branched peptide that links the carboxyl terminus of proteins to the cell wall. J. Biol. Chem. 272:22285-22292. [DOI] [PubMed] [Google Scholar]
- 139.Tullius, M. V., G. Harth, and M. A. Horwitz. 2001. High extracellular levels of Mycobacterium tuberculosis glutamine synthetase and superoxide dismutase in actively growing cultures are due to high expression and extracellular stability rather than to a protein-specific export mechanism. Infect. Immun. 69:6348-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ulbrandt, N. D., J. A. Newitt, and H. D. Bernstein. 1997. The Escherichia coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88:187-196. [DOI] [PubMed] [Google Scholar]
- 141.van der Meer, J. R., H. S. Rollema, R. J. Siezen, M. M. Beerthuyzen, O. P. Kuipers, and W. M. de Vos. 1994. Influence of amino acid substitutions in the nisin leader peptide on biosynthesis and secretion of nisin by Lactococcus lactis. J. Biol. Chem. 269:3555-3562. [PubMed] [Google Scholar]
- 142.van Dijl, J. M., P. G. Braun, C. Robinson, W. J. Quax, H. Antelmann, M. Hecker, J. Müller, H. Tjalsma, S. Bron, and J. D. H. Jongbloed. 2002. Functional genomic analysis of the Bacillus subtilis Tat pathway for protein secretion. J. Biotechnol. 98:243-254. [DOI] [PubMed] [Google Scholar]
- 143.van Dijl, J. M., A. Bolhuis, H. Tjalsma, J. D. H. Jongbloed, A. de Jong, and S. Bron. 2001. Protein transport pathways in Bacillus subtilis: a genome-based road map, p. 337-355. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives from genes to cells. ASM Press, Washington, D.C.
- 144.van Kraaij, C., W. M. de Vos, R. J. Siezen, and O. P. Kuipers. 1999. Lantibiotics: biosynthesis, mode of action and applications. Nat. Prod. Rep. 16:575-587. [DOI] [PubMed] [Google Scholar]
- 145.van Wely, K. H., J. Swaving, R. Freudl, and A. J. M. Driessen. 2001. Translocation of proteins across the cell envelope of Gram-positive bacteria. FEMS Microbiol. Rev. 25:437-454. [DOI] [PubMed] [Google Scholar]
- 146.Venema, R., H. Tjalsma, J. M. van Dijl, A. de Jong, K. Leenhouts, G. Buist, and G. Venema. 2003. Active lipoprotein precursors in the Gram-positive eubacterium Lactococcus lactis. J. Biol. Chem. 278:14739-14746. [DOI] [PubMed] [Google Scholar]
- 147.Vitikainen, M., I. Lappalainen, R. Seppala, H. Antelmann, H. Boer, S. Taira, H. Savilahti, M. Hecker, M. Vihinen, M. Sarvas, and V. P. Kontinen. 2004. Structure- function analysis of PrsA reveals roles for the parvulin-like and flanking N- and C- terminal domains in protein folding and secretion in Bacillus subtilis. J. Biol. Chem. doi:10.1074/jbc.M400861200. [DOI] [PubMed]
- 148.von Heijne, G. 1984. How signal sequences maintain cleavage specific. J. Mol. Biol. 173:243-251. [DOI] [PubMed] [Google Scholar]
- 149.von Heijne, G. 1989. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 2:531-534. [DOI] [PubMed] [Google Scholar]
- 150.von Heijne, G. 1990a. Protein targeting signals. Curr. Opin. Cell Biol. 2:604-608. [DOI] [PubMed] [Google Scholar]
- 151.von Heijne, G. 1990b. The signal peptide. J. Membr. Biol. 115:195-201. [DOI] [PubMed] [Google Scholar]
- 152.von Heijne, G. 1998. Life and death of a signal peptide. Nature 396:111, 113. [DOI] [PubMed]
- 153.Wahlstrom, E., M. Vitikainen, V. P. Kontinen, and M. Sarvas. 2003. The extracytoplasmic folding factor PrsA is required for protein secretion only in the presence of the cell wall in Bacillus subtilis. Microbiology 149:569-577. [DOI] [PubMed] [Google Scholar]
- 154.Walker, S. A., and T. R. Klaenhammer. 2001. Leaky Lactococcus cultures that externalize enzymes and antigens independently of culture lysis and secretion and export pathways. Appl. Environ. Microbiol. 67:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Weiss, K. A., and M. Laverdiere. 1997. Group A Streptococcus invasive infections: a review. Can. J. Surg. 40:18-25. [PMC free article] [PubMed] [Google Scholar]
- 156.Weldingh, K., I. Rosenkrands, S. Jacobsen, P. B. Rasmussen, M. J. Elhay, and P. Andersen. 1998. Two-dimensional electrophoresis for analysis of Mycobacterium tuberculosis culture filtrate and purification and characterization of six novel proteins. Infect. Immun. 66:3492-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Westers, H., R. Dorenbos, J. M. van Dijl, J. Kabel, T. Flanagan, K. M. Devine, F. Jude, S. J. Séror, A. C. Beekman, E. Darmon, C. Eschevins, A. de Jong, S. Bron, O. P. Kuipers, A. M. Albertini, H. Antelmann, M. Hecker, N. Zamboni, U. Sauer, C. Bruand, D. S. Ehrlich, J. C. Alonso, M. Salas, and W. J. Quax. 2003. Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol. Biol. Evol. 20:2076-2090. [DOI] [PubMed] [Google Scholar]
- 158.Wu, S. C., J. C. Yeung, Y. Duan, R. Ye, S. J. Szarka, H. R. Habibi, and S. L. Wong. 2002. Functional production and characterization of a fibrin-specific single-chain antibody fragment from Bacillus subtilis: effects of molecular chaperones and a wall- bound protease on antibody fragment production. Appl. Environ. Microbiol. 68:3261-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wu, X. C., W. Lee, L. Tran, and S. L. Wong. 1991. Engineering a Bacillus subtilis expression-secretion system with a strain deficient in six extracellular proteases. J. Bacteriol. 173:4952-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yakushi, T., K. Masuda, S. Narita, S. Matsuyama, and H. Tokuda. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2:212-218. [DOI] [PubMed] [Google Scholar]
- 161.Young, R., and U. Bläsi. 1995. Holins: form and function in bacteriophage lysis. FEMS Microbiol. Rev. 17:191-205. [DOI] [PubMed] [Google Scholar]
- 162.Ziebandt, A. K., H. Weber, J. Rudolph, R. Schmid, D. Hoper, S. Engelmann, and M. Hecker. 2001. Extracellular proteins of Staphylococcus aureus and the role of SarA and sigma B. Proteomics 1:480-493. [DOI] [PubMed] [Google Scholar]