Abstract
Microbial adaptation to environmental stimuli is essential for survival. While several of these stimuli have been studied in detail, recent studies have demonstrated an important role for a novel environmental parameter in which microgravity and the low fluid shear dynamics associated with microgravity globally regulate microbial gene expression, physiology, and pathogenesis. In addition to analyzing fundamental questions about microbial responses to spaceflight, these studies have demonstrated important applications for microbial responses to a ground-based, low-shear stress environment similar to that encountered during spaceflight. Moreover, the low-shear growth environment sensed by microbes during microgravity of spaceflight and during ground-based microgravity analogue culture is relevant to those encountered during their natural life cycles on Earth. While no mechanism has been clearly defined to explain how the mechanical force of fluid shear transmits intracellular signals to microbial cells at the molecular level, the fact that cross talk exists between microbial signal transduction systems holds intriguing possibilities that future studies might reveal common mechanotransduction themes between these systems and those used to sense and respond to low-shear stress and changes in gravitation forces. The study of microbial mechanotransduction may identify common conserved mechanisms used by cells to perceive changes in mechanical and/or physical forces, and it has the potential to provide valuable insight for understanding mechanosensing mechanisms in higher organisms. This review summarizes recent and future research trends aimed at understanding the dynamic effects of changes in the mechanical forces that occur in microgravity and other low-shear environments on a wide variety of important microbial parameters.
INTRODUCTION
Microbial existence and survival requires the ability to sense and respond to environmental changes, including changes in physical forces. This is because microbes inhabit an amazingly diverse range of ecological niches and therefore must constantly adapt to a wide variety of changing environmental conditions, including alterations in temperature, pH, nutrient availability, oxygen levels, and osmotic pressure gradients (2, 15, 32, 44, 46, 86). Microbes sense their environment through a variety of sensors and receptors which serve to integrate the different signals into the appropriate cellular response(s) that is optimal for survival. While numerous environmental stimuli have been examined for their effect on microorganisms, effects due to changes in mechanical and/or physical forces are also becoming increasingly apparent (6, 21, 50, 82, 97). Recently, several important studies have demonstrated a key role for microgravity and the low fluid shear dynamics associated with microgravity in the regulation of microbial gene expression, physiology, and pathogenesis (22, 54, 60, 78, 82). The mechanosensory response of microorganisms to these environmental signals, which are relevant to those encountered during microbial life cycles on Earth, may provide insight into their adaptations to physiologically relevant conditions and may ultimately lead to eludicidation of the mechanisms important for mechanosensory transduction in living cells. This review summarizes the recent and potential future research trends aimed at understanding the effect of changes in mechanical forces that occur in microgravity and other low-shear environments on different microbial parameters. The results of these studies provide an important step toward understanding how microbes integrate information from multiple mechanical stimuli to an appropriate physiological response.
Mechanosensitive Processes in Microorganisms
Microbes have the ability to sense and respond to mechanical stimuli. The response of microbes to certain mechanical stimuli has profound effects on physiology (40, 82, 97). The response of a cell to mechanical stimulation, such as stretch or shear force, is called mechanotransduction and is important for cell protection in both prokaryotes and eukaryotes (40, 48, 49). A great deal of progress has been made in understanding certain aspects of microbial mechanotransduction, for example, mechanisms used by bacteria to respond to changes in osmotic gradients (7, 40, 85). Recently, studies have also documented that microbes can sense and respond to changes in culture conditions when grown in the buoyant, low-fluid-shear environment of microgravity (21, 52, 60, 82). Moreover, it has been hypothesized that cells sense changes in mechanical forces, including shear and gravity, at their cell surface (48). While changes in the physical forces of hydrostatic pressure, gravity, and fluid shear play an important role in evolution and microbial physiology, little is known about how microbial cells convert these mechanical signals into molecular and biochemical responses. A better understanding of the mechanosensory response of microorganisms to changes in physical forces will provide important insight into the mechanisms important for mechanosensory transduction in living cells.
Microgravity and Low Fluid Shear
Mechanical culture conditions in the quiescent microgravity environment of spaceflight are characterized by significant reductions in fluid shear (41, 60). This is because convection currents are essentially absent in microgravity (60, 61). Likewise, cells cultured in specialized ground-based bioreactors designed to simulate aspects of weightlessness in the laboratory encounter a low-shear modeled microgravity environment (42, 58, 82). Thus, both microgravity and ground-based microgravity analogues represent a low-shear stress environment for cell culture. Accordingly, it is relevant that both of these mechanical culture conditions be considered together in this review, especially since recent studies have demonstrated an important role for microgravity and the low-fluid-shear dynamics associated with microgravity in the regulation of a variety of microbial parameters (60, 78, 82). Understanding how microgravity and fluid shear stress affect microorganisms may advance our understanding of the fundamental concepts of mechanotransduction, including the discovery of mechanisms which might be conserved between microbes and humans.
Why Study the Effects of Microgravity on Microorganisms?
Microgravity is a condition where the physical force of gravity is reduced and is often referred to as “weightlessness.” Because life on Earth evolved in the presence of gravity, one of the fundamental organizing forces of evolution, it is of prime interest to understand the effect of this force on the evolution of terrestrial life. By conducting studies in the reduced gravity of spaceflight, we may gain a better understanding of how the physical force of gravity shaped life on Earth. In addition, the results of spaceflight studies will lead to the discovery of other features of terrestrial life that cannot be observed on Earth. For example, how much change occurs and what form will organisms assume over time in the microgravity environment of spaceflight? Research conducted in the microgravity environment of space as well as the use of ground-based microgravity analogues, has demonstrated that microbial cells are “hardwired” to respond to changes in this physical force (22, 60, 78, 82). Although microgravity and microgravity analogues are known to have a profound effect on numerous microbial parameters, the mechanism(s) by which this occurs is unknown (22, 60, 78, 82).
Microgravity, used as a research tool, allows investigators to understand how changes in this dynamic physical force affect microbes at the molecular, physiological, and evolutionary levels. As we transition from terrestrial life to low-gravity environments, our understanding of the role of gravity in shaping evolution on Earth will increase. This is especially relevant given the ambitious new focus and vision for the future of America's space exploration program, which calls for “extending the human presence across our Solar System,” including a living base on the Moon and missions to Mars. Microgravity investigations using microbes with short generation times may reveal common characteristics shared by higher organisms, including humans, that allow adaptation and survival in low gravity and will provide important insight into how terrestrial life adapts on Earth. Moreover, since microgravity represents a profound change in physical forces encountered by cells, the use of microbes in these studies may provide clues to the underlying mechanisms responsible for the physiological adaptations that humans experience in space. Such information will lead to the advancement of our knowledge regarding cellular adaptations which may occur in outer space beyond this planet's gravitational field. In addition, just as the study of microbial life in extreme environments on Earth has led to novel biological solutions to complex medical, environmental, and agricultural problems, the investigation of microbial life in the microgravity environment of space and ground-based microgravity analogues holds significant potential for future academic and commercial applications. Indeed, the unique research environment of microgravity has already shown potential to enhance the efficiency of terrestrial fermentation processes which use microbes to produce commercial products such as antibiotics (59). The economic benefit derived from such knowledge could be substantial and might serve to initiate a new era of bioprocessing. Finally, it is important to note that the growth environment sensed during the microgravity of spaceflight and during ground-based microgravity analogue culture has distinct similarities to environments encountered by microbial cells on Earth. Specifically, the low-shear-stress, low-turbulence environment of microgravity and microgravity analogue culture is similar to that found in certain areas of the body. For example, a low-shear environment like that of microgravity is encountered in utero and in the protected environment between the brush border microvilli of epithelial cells (3, 12, 20, 39, 90). The latter environment is relevant to that encountered by numerous microbial pathogens and commensals during their normal life cycles in the gastrointestinal, respiratory, and urogenital tracts. In addition, there may be other low-shear environments that are occupied by microbes and are not presently known. Thus, the parallels between microgravity and certain Earth environments will help us understand the response of microbes to these environmental signals, which are similar to those encountered during microbial life cycles, and may provide insight into microbial adaptations to physiologically relevant conditions.
EFFECTS OF MICROGRAVITY ON MICROBIAL RESPONSES: EXPERIMENTS INVOLVING SPACEFLIGHT
Spaceflight represents a unique inhabited semiclosed environment, most notably characterized by a decreased gravitational force. Moreover, human presence in space, whether permanent or transient, is accompanied by the presence of microbes. Evaluations of the microbial ecology aboard Mir and the International Space Station suggested a predominance of common members of the environmental flora (14), although the appearance of medically significant organisms has been documented (83). Whether we are considering the true weightlessness of deep space or the microgravity observed in low-Earth orbit, it would be reasonable to predict that the microgravity-induced decrease in stress on the surface of microorganisms might affect the gene expression and physiology of both commensal and pathogenic organisms. For successful space travel, it is critical to address this issue, since studies have suggested that spaceflight negatively impacts the immune system in both humans and animals (65, 94), which would lead to an increased risk of infectious disease.
These findings, as well as the convenience of handling and short life cycle of microorganisms, mean that bacteria and yeast are excellent research tools for flight, although implementation of microbial experiments during flight can be difficult. The performance of any flight experiment is restricted by limitations in power, work area mass, and crew time (78). Safety concerns for the crew in a closed environment increase the need for multiple containment of growing microbial cultures. In addition, the lack of gravity complicates techniques requiring proper gas/liquid/solid-phase separation. These limitations often restrict the experimental design to simple, basic approaches with minimal replicates.
Brief History of Microbial Experiments Involving Spaceflight
Initial spaceflight experiments with microorganisms can be traced back to early efforts including balloon flights and Aerobee rocket payloads (22, 78). Ballooning experiments dated as early as 1935 evaluated the ability of microorganisms to survive decreased pressure and increased radiation (22, 78). The USSR was investigating microbial growth and viability characteristics during flights as early as 1957, with experiments aboard the Sputnik satellites (40). They continued their work in rocket-launched experiments in 1960 with the second Soviet satellite (Sputnik 5) (12). While the United States completed microbial experiments in satellites, such as Discoverer XVII and Biosatellite 2 (12), the National Aeronautics and Space Administration (NASA) also began experiments in the mid-1960s, utilizing the manned Gemini spacecraft (40), which permitted greater experimental interaction and complexity. The Soviet and subsequent Russian efforts continued through the Salyut and Mir programs through the 1990s (12, 40). U.S. experiments likewise continued during the Apollo, Skylab, and Spacelab programs (12, 40). Joint projects between these and other partner countries have been accomplished throughout the 1980s and 1990s and are currently ongoing aboard the International Space Station. The results of these experiments, as described below, have provided compelling evidence that spaceflight profoundly alters a variety of microbial properties.
Summary of Results from Selected Spaceflight Experiments
The overall number of microorganisms that have been investigated during spaceflight is extensive, with over 100 different experimental cultures being evaluated before 1990 (12). This review provides only a general summary of past experiments. Detailed reviews of the data and its implications have been compiled and are listed in Table 1 (17, 22, 60, 78, 95, 104). Basic microbial characteristics, such as changes in cell growth characteristics during flight, have been repeatedly evaluated. Changes in cell density were reported in early studies, including the increased Salmonella enterica serovar Typhimurium culture densities during experiments aboard Biosatillite 2 compared to ground controls (95). Escherichia coli also displayed similar increased growth during flight in several experiments (16, 57, 103). To further clarify the differences observed in bacterial growth kinetics during spaceflight compared to ground controls, a recent series of experiments was designed to examine in detail the growth of E. coli during flight (57). The results of this study indicated that the lag phase was shortened, the exponential growth phase was extended, and the bacterial cell populations were 88% greater than those of ground controls (57). A subsequent set of experiments confirmed this finding for both E. coli and Bacillus subtilis (54, 55). Potential mechanisms that could explain the differences in microbial growth parameters observed during spaceflight have not been elucidated, although several possibilities have been proposed (57). For microorganisms, most proposed mechanisms focus on physical factors, such as decreased mass diffusion or shear levels or the development of “microenvironments” (i.e., changes in the distribution of nutrients and by-products due to a lack of cell sedimentation) directly around the organism, that could alter cell growth. The difference between these indirect effects and the possible direct effects of microgravity were clarified during a flight experiment by Kacena et al. in which E. coli and B. subtilis were grown on agar to minimize any physical fluid effects (53). No difference in growth was observed compared to the ground control, suggesting that the indirect physical effects, such as changes in fluid dynamics and extracellular transport, rather than a direct gravity effect was the most likely cause of the differences seen in bacterial growth during spaceflight (53).
TABLE 1.
Detailed reviews of spaceflight experiments with microorganisms
| Description | Reference |
|---|---|
| Detailed discussion of early space through 1974, covering flight observations including lunar exploration missions and astronaut microbial flora | 95 |
| Comprehensive listing of all spaceflight experiments with microorganisms through 1990 | 22 |
| Detailed discussion of mycological studies performed during flight though 1990 | 104 |
| Discussion summarizing the organisms which have flown and a synopsis of the results through 1991 | Cioletti et al.a |
| Organized synopsis of organisms, missions, and results, as well as perspective on limitations of flight and flight experiments | 78 |
| Recent and thorough review of flight experiments, with insight into the proposed mechanisms behind the effects observed in microgravity | 60 |
L. A. Cioletti, D. L. Pierson, and S. K. Mishra, Abstr. 21st Int. Conf. Environ. Syst., abstr. 911512, 1991.
Of particular interest to NASA are changes to key microbial characteristics during flight that can directly impact the health, safety, and performance of the crew and/or affect the integrity of the spacecraft. One such series of investigations studied differences in antibiotic resistance profiles of microrganisms during space flight. In 1982, as a part of the Cytos 2 experiment, the MIC of oxacillin, chloramphenicol, and erythromycin for Staphylococcus aureus and of colistin and kanamycin for E. coli were compared to those of ground controls (98). Surprisingly, the results indicated an increased resistance of both S. aureus and E. coli to all antibiotics used in this experiment (98). Similar analyses have been performed in subsequent flight experiments to confirm this effect, and they have repeatedly shown increased antibiotic resistance in these organisms (64, 78). The mechanism for the increased resistance to antibiotics in-flight is unclear; however, this difference has been speculated to be the result of the above-mentioned differences in microbial growth kinetics or possibly a modification of cellular transport mechanisms due to changes in mass diffusion (78, 98). Some mechanistic insight has come from recent flight and MIC testing performed with E. coli grown on agar instead of in liquid culture (56). Under these conditions, no increased antibiotic resistance to gentamicin was observed, suggesting a physical rather than biological effect (56). The changes in antibiotic sensitivity observed in these microbes during spaceflight may be transient, since attempts to reproduce the resistance after return to Earth have been unsuccessful (64).
An extensive list of changes in a wide variety of microbial cell characteristics has been observed during spaceflight compared to ground controls. For example, the effect of spaceflight on genetic transfer has been investigated numerous times and has indicated increased conjugal transfer rates in E. coli (16) and changes in phage induction in E. coli and S. enterica serovar Typhimurium (1, 72). While infection studies are limited due to flight constraints concerning containment issues, some studies have been completed. For example, Saccharomyces cerevisiae cells retrieved from the Apollo 16 experiments were better able to survive in intradermal lesions in artificially infected mice compared to ground controls (104). Several studies have also indicated increased viral reactivation in astronauts in response to spaceflight; the viruses studied included Epstein-Barr virus (91), cytomegalovirus (76), and varicella-zoster virus (75). In addition, intercellular functions, such as biofilm formation by Pseudomonas aeruginosa (74) and changes in the growth and differentiation of Dictyostelium discoideum (93), have also been reported in-flight. Interestingly, the D. discoideum fruiting bodies from the flight experiments produced fewer spores that germinated later than those of ground control cultures (93). Other notable observations during cell cultivation during flight include greater cell size and enhanced swarming of Proteus vulgaris (70), a thickening of the cell wall of S. aureus (64), increased phosphate uptake of S. cerevisiae (104), impaired magnetotaxis of Magnetospirillium magnetotacticum (102), increased antibiotic production by Streptomyces plicatus and Humicola fuscoatra (by as much as 115 and 190%, respectively) (59), and morphological changes in hyphal structure, colony development, and cell shape in Trychophyton terrestre and Chaetomium globosum (104).
Numerous in-flight studies have confirmed that spaceflight has a profound effect on a variety of microbial parameters, including changes in microbial growth, morphology, metabolism, genetic transfer, and viral reactivation. However, inherent in-flight experimental limitations and some inconclusive findings have prevented our thorough understanding of the mechanisms underlying these microbial responses. While the numerous restrictions associated with in-flight experiments have limited the ability to obtain detailed data on exactly how microorganisms respond to spaceflight at the molecular and biochemical levels, future spaceflight experiments that incorporate novel technology coupled with experiments using ground-based microgravity analogues (discussed below) will provide us with important insight into the molecular mechanisms of microbial responses to microgravity and other low-shear environments that are relevant to those encountered in-flight and on Earth. This new era of in-flight experiments is exemplified by a current study by Nickerson et al., conducted onboard the International Space Station (http://science.nasa.gov/headlines/y2004/23 feb_yeastgap.htm? list691352). This experiment will be the first to use whole-genome microarrays to globally profile changes in gene expression in a microorganism (S. cerevisiae) in response to spaceflight as compared to identical ground control cultures and will also generate a fitness profile of yeast genes that convey a selective growth advantage in microgravity.
GROUND-BASED MODELS OF MICROGRAVITY: EFFECT OF MICROGRAVITY ANALOGUES ON MICROBIAL RESPONSES
As discussed in the previous section, there are a number of issues that make scientific experiments involving spaceflight particularly challenging. Such experiments require a significant amount of preflight time and planning and rely on hardware that requires extensive engineering to meet the rigorous in-flight standards that are put on any piece of equipment that is flown in space. Therefore, studies of the effect of microgravity on microorganisms during spaceflight are limited by certain constraints involved with these experiments. These constraints include: power, weight, volume, and stowage space limitations; time and labor requirements placed on the astronauts; limited flight opportunities; significant financial costs; and the need for specialized equipment necessary to perform experiments aboard spacecraft or space stations. Thus, there exists a fundamental need to assess many experiments under the conditions of ground-based modeled microgravity.
Rotating-Wall Vessel Culture Apparatus and the Low-Shear Modeled Microgravity (LSMMG) Growth Environment
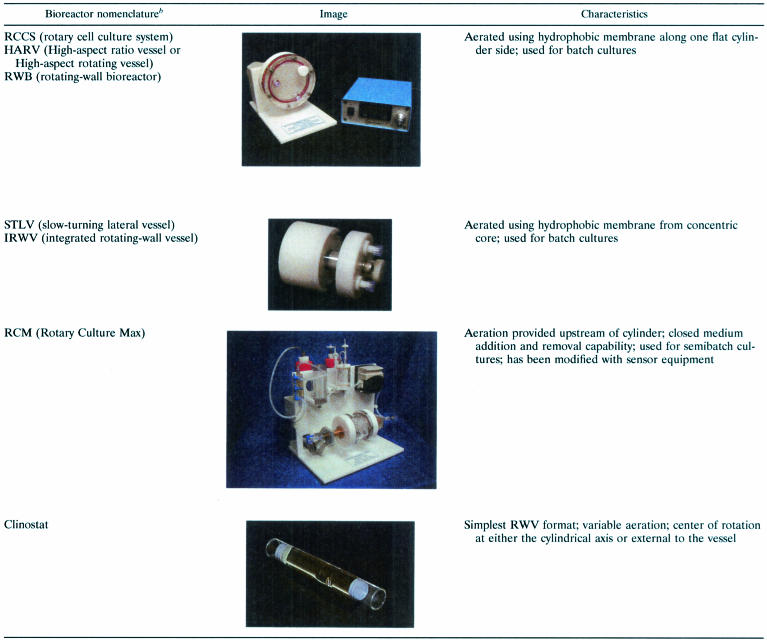
In an effort to design a novel form of low-shear, low-turbulence suspension culture that could model aspects of spaceflight (i.e., microgravity), the NASA Biotechnology Group, based at Johnson Space Center in Houston, Tex., invented the rotating-wall vessel (RWV) culture apparatus (42, 82, 89). The RWV is a powerful laboratory tool but is rather simple in concept and design. Several variations of the apparatus exist (Table 2). The RWV is essentially an optimized form of suspension culture and consists of a hollow disk or cylinder that is completely filled with medium (no bubbles, i.e., “zero headspace”) and rotates on an axis parallel to the ground (and perpendicular to the gravitational force vector) (Fig. 1) (42, 58, 82, 105). The result is solid-body rotation of the medium within and a constant rotation normal to the gravitational field that results in an environmental culture condition in which the gravitational vectors are randomized over the surface of the cells. Under these culture conditions, the cells are maintained in suspension as the RWV is rotated and a sustained low-shear environment for cell growth is achieved (42, 58, 82). Exchange of nutrients and localized “mixing” of the microenvironment is facilitated by the constant falling of the cells through the local fluid environment and the gentle rotation of the culture medium. A gas-permeable membrane on one side of the RWV allows constant air exchange during growth.
TABLE 2.
RWV bioreactors used for LSMMG cell culturea
Reprinted from reference 82 with permission.
RCCS, STLV, and RCM photographs printed with permission from Synthecon, Inc., Houston, Tex.
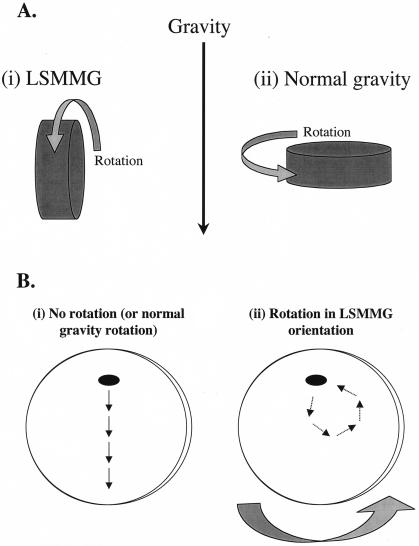
FIG. 1.
Operating orientations of the RWV and the effect of RWV rotation on particle (microbe) suspension. (A) The two operating orientations of the RWV are depicted. In the LSMMG orientation (panel i), the axis of rotation of the RWV is perpendicular to the direction of the gravity force vector. In the normal-gravity (or 1 × g) orientation (panel ii), the axis of rotation is parallel to the gravity vector. (B) Effect of RWV rotation on particle suspension. When the RWV is not rotating or is rotating in the 1 × g orientation (panel i), the force of gravity will cause particles in the apparatus to sediment and eventually settle on the bottom of the RWV. When the RWV is rotating in the LSMMG position (panel ii), particles are continually suspended in the medium. The medium within the RWV rotates as a single body, and the sedimentation of the particle due to gravity is offset by the upward forces of rotation. The result is a low-shear aqueous suspension that is strikingly similar to what would occur in true microgravity. Panel B is not drawn to scale.
The fluid mechanics that affect objects in the RWV have been evaluated in detail (42, 58, 82, 105). Basically, particles or cells in the RWV are in a state of constant fluid suspension such that hydrodynamic forces offset the gravitational sedimentation of the bacteria in the reactor, allowing the organism to fall at a constant terminal velocity without being allowed to settle on the bottom of the apparatus (Fig. 1B) (42, 58, 82). The solid-body rotation of the medium is believed to minimize turbulence and shear within the vessel based on the Stokes law for flow around spherical objects (36, 42, 99). The hydrodynamic forces involved may include centrifugal, Coriolis, and shear components (42, 58). A model developed by Gao et al. (36) explains that shear force around a spherical bead in the RWV increases primarily with either increased particle radius or increased particle density relative to the medium. Assuming that microbes react in a similar fashion to a spherical particle, their size and density would suggest a minimal shear. Thus, the low shear associated with cell growth in the RWV is most probably the result of a combination of fluidic principals and gravitational factors. However, the precise individual contribution of these forces on particles or microbes in the RWV is not clear and requires further research. We refer the reader to some excellent reviews that extensively discuss the technical aspects of the forces that affect growth in the RWV environment (33, 42, 58, 59, 61, 105).
There are two compelling reasons why the RWV growth environment is a microgravity analogue. First, cells in the RWV are maintained in a constant state of suspension in a fluid environment such that they mimic the way in which objects would be suspended in true microgravity. Indeed, suspension in a fluid environment is already used to model space walks during astronaut training at Johnson Space Center. Astronauts are submerged and suspended underwater in a pool to practice repairs to be performed during actual space walks in an effort to simulate the space environment. While these two situations differ in a number of ways, the design of the RWV takes advantage of the same concept by creating a state of low-shear suspension where sedimentation is absent and turbulent motion is greatly minimized (42, 58, 59, 61). Although it is true that the gravity force vector present in a liquid environment on Earth is very different in magnitude from that present in the space atmosphere, the low-shear buoyant sensation experienced in both environments is very similar. There is increasing evidence not only that entire organisms sense and respond physiologically to low-shear, buoyant environments but also that cells respond at the molecular level to this environment (43, 52, 81, 106, 107).
Second, cellular responses observed in space have been observed during culture in the RWV on Earth. The most dramatic of these responses is the formation of three-dimensional (3-D) tissue aggregates that structurally and functionally resemble in vivo tissues. In fact, RWV bioreactor technology was originally designed for the growth of suspension cultures of mammalian cells under conditions of extremely low turbulence, which permits the generation of these 3-D differentiated tissue-like assemblies which model many aspects of in vivo human tissues (34, 35, 101, 105). Accordingly, the 3-D aggregates are physiologically relevant in vitro tissue models and are currently being engineered for use in infectious-disease research, tissue transplantation, and other fundamental biomedical applications (34, 80, 82a, 101; A. J. Carterson, C. M. Ott, M. S. Clark, C. R. Vanderburg, C. A. Nickerson, and M. J. Schurr, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol. 2003; abstr. B-131, 2003; H. LaMarca, C. M. Ott, K. Honer zu Bentrup, C. L. LeBlanc, D. L. Pierson, C. A. Nickerson, A. Nelso, and C. Morris, Abstr. Fifteenth Annu. Tulane Health Sci. Res. Days, 2003; A, Meijer, J. Siekman, P. Roholl, and Ossewararde, Abstr. Eur. Chlamydia Congr., 2000). The same 3-D aggregates also form in space, and, in fact, this phenomenon was first observed during culture of tissue cell suspensions on space missions (23, 47, 101). There are numerous examples of different cell types and lineages forming 3-D aggregates during culture in space aboard spacecraft and space stations (23, 34, 47, 101). The fact that these novel 3-D aggregates are formed both in space and in the RWV culture apparatus points to the similarities between the two growth environments.
It is also relevant to mention that the growth environment achieved through optimized suspension culture in the RWV provides low-shear growth cues similar to those encountered in utero and in certain low-shear areas of the body such as between the brush border microvilli of epithelial cells (3, 12, 20, 39, 90). Comparisons of the in utero growth environment and the RWV environment have obvious implications in possibly explaining why tissue aggregates form in the RWV, since the low-shear suspension of cells and tissues in the two environments is strikingly similar. In relation to microbes, the low-shear environment between brush border microvilli is probably encountered by numerous bacterial pathogens during the natural course of infection of the gastrointestinal, respiratory, and urogenital tracts. This may be a niche where the reduced shear serves as a signal to microbes that reside there.
Since the RWV apparatus provides a low-shear culture environment that simulates aspects of space (and therefore “models microgravity”), we have adopted the terminology LSMMG (low-shear modeled microgravity) to refer to the RWV culture environment. Although designed to provide an environment of LSMMG, the RWV can also be used to grow cells under normal gravity by simply changing the position of the bioreactor. Figure 1A shows how the RWV bioreactors are oriented to grow cells under conditions of LSMMG (Figure 1A, i) with the vessel perpendicular to the gravitational vector; or normal gravity (i.e. 1 × g) (Fig. 1A, panel ii) with the vessel parallel to the gravitational vector. Thus, the LSMMG and normal-gravity conditions in the RWV are identical except for the physical orientation of the apparatus.
Although it is true that the RWV is used for Earth-based studies to learn how the environment of space will impact both eukaryotic and prokaryotic physiology, perhaps the most exciting aspect of RWV culture techniques is that the low-shear growth environment allows these cells to assume medically and biologically important phenotypes that cannot be observed using conventional culture methods. Although designed with spaceflight implications in mind, it is becoming increasingly evident that the RWV may find its greatest utility in ground-based applications such as (i) the formation and engineering of 3-D tissue aggregates that structurally and functionally resemble in vivo tissues (80, 82a, 101); (ii) understanding of the molecular mechanisms used by cells to sense and respond to LSMMG and the identities of the genes involved in response to this signaling pathway (43, 48, 52, 107); (iii) understanding of how microbial pathogens modulate their virulence both on Earth and in space, which could provide clues to the functioning of known virulence systems or to the identification of novel, uncharacterized bacterial virulence strategies (81, 106, 107); and (iv) studies to further understand microbial metabolism and physiology (21, 82). This section of the review focuses on the growing body of information regarding the responses of microbes to the LSMMG growth environment of the RWV.
Responses of Bacteria to LSMMG
Altered secondary-metabolite production.
Studies by Demain, Fang, and colleagues have examined the effects of growth in the RWV on the production of secondary metabolites by a variety of bacteria (21). These studies have provided substantial evidence that bacteria alter their metabolic properties during LSMMG cultivation in the RWV apparatus. They also provide a first step in examining the use of RWV technology as a way to optimize the production of microbial metabolites for biotechnological purposes. These metabolites include many important chemicals and compounds that have utility in the pharmaceutical, food, and bioprocessing industries.
In these studies, production of the peptide antibiotics cephalosporin and microcin B17 and the polyketide macrolide rapamycin by Streptomyces clavuligerus, E. coli, and Streptomyces hygroscopicus, respectively, was shown to be inhibited by LSMMG whereas production of gramicidin S by Bacillus brevis was unaffected (26-30, 37). Interestingly, the latter finding with B. brevis indicates that LSMMG does not have a universally negative effect on secondary metabolism and suggests that microbes respond to LSMMG in specific ways. The repressive effect of glycerol on gramicidin production by B. brevis normally seen in shake flasks was not observed during culture in the RWV (30). Similarly, glucose repression of microcin B17 production by E. coli was found to be dramatically inhibited in the RWV (27). These findings suggest that carbon source repression of these processes in B. brevis and E. coli are influenced by growth in the RWV, although it is unclear how LSMMG is related to this phenomenon since it was observed in both the LSMMG and 1 × g RWV orientations.
The site of microcin B17 and rapamycin accumulation was found to be markedly different when the bacteria were cultured in the RWV compared to when they were cultured in shaking flasks (28). In flasks, accumulation of these peptide antibiotics was intracellular, whereas in the RWV, the majority of the product was found in the medium (i.e., extracellular). The authors noted that the shift in localization of microcin from intracellular to extracellular was probably due to the much lower degree of shear stress in the bioreactors, since addition of a single glass bead to the RWV medium created enough shear to change the site of micrococin accumulation from the medium to the cells (28). This type of phenotype associated with growth in the RWV could be exploited to increase the secretion and extracellular accumulation of a cellular product of interest for use in bioprocessing and biotechnological applications.
Enhanced Salmonella enterica serovar Typhimurium virulence.
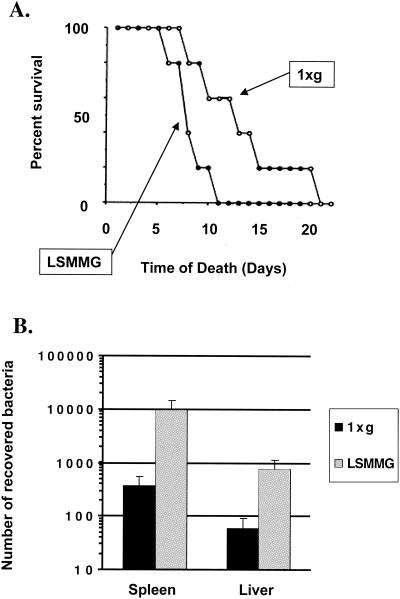
As mentioned above, spaceflight has the capacity to alter immune system function in a manner which suggests a decreased ability to mount a robust immune response to infection (65, 94). In addition, inherent in the habitation of spacecraft and space stations is exposure to microbes in a closed, self-contained environment with little to no ability to quarantine a serious infectious-disease outbreak, should one occur. Likewise, the use of regenerative life support systems will serve to increase exposure to pathogens and potential pathogens. Therefore, the response of microbes, especially pathogens, to a low-shear microgravity environment is of prime interest. Recent studies have demonstrated that the LSMMG growth environment of the RWV can enhance the virulence of a gram-negative bacterial pathogen, S. enterica serovar Typhimurium (81). In the murine model of infection, the oral lethal dose of S. enterica serovar Typhimurium grown under LSMMG for killing 50% of the infected animal subjects (LD50) was 5.2 times lower the LD50 of the same strain grown under normal gravity. Mice infected with 106 CFU of LSMMG-grown S. enterica serovar Typhimurium displayed a decreased average time to death compared to mice given the same dosage of cells grown under normal gravity (Fig. 2A). In addition, LSMMG-grown S. enterica serovar Typhimurium showed increased colonization of the murine liver and spleen following oral infection compared to the normal gravity-grown strain (Fig. 2B). This study was the first direct evidence that LSMMG could alter microbial virulence; it further demonstrates the global impact of LSMMG on microbial physiology. The molecular mechanisms responsible for the LSMMG-induced increase in Salmonella virulence are not yet known, and microarray analysis (see below) did not reveal any of the known Salmonella virulence factors to be induced during growth under LSMMG (107). Surprisingly, the expression of several genes in two major Salmonella pathogenicity islands (SPI-1 and SPI-2) was decreased under LSMMG (107). An exciting possibility is that LSMMG is inducing novel Salmonella virulence mechanisms or is “fine-tuning” the expression and/or function of known Salmonella virulence mechanisms in a new way. The results indicate that LSMMG can be added to the list of environmental signals already known to regulate the expression of virulence determinants in Salmonella, including osmolarity, pH, oxidative stress, starvation, and growth phase (32, 69). The elucidation of the molecular mechanisms responsible for enhanced Salmonella virulence by LSMMG could lead to the discovery of previously unknown virulence mechanisms or signaling pathways. It will also be of interest to see how LSMMG modulates the virulence of other microbial pathogens.
FIG. 2.
LSMMG-enhanced virulence of S. enterica serovar Typhimurium in the murine model of infection. (A) Shortened time to death of mice infected with LSMMG-grown S. enterica serovar Typhimurium cells compared to mice infected with 1 × g-grown cells. BALB/c mice (8 weeks old) were inoculated perorally with 2 × 106 CFU of LSMMG- or 1 × g-grown S. enterica serovar Typhimurium, and the survival of the animals was monitored for 20 days postinfection. The percent survival of the infected animals over this period is plotted, and the curves for LSMMG- and 1 × g-infected mice are indicated. (B) Enhanced ability of LSMMG-grown S. enterica serovar Typhimurium cells to colonize the murine spleen and liver compared to that of 1 × g-grown cells. LSMMG- and 1 × g-grown S. enterica serovar Typhimurium cells (2 × 106) were administered perorally as individual infections to 8-week-old BALB/c mice. The spleen and liver were excised 6 days after infection, and the recovered bacteria were quantitated. The standard deviation represents the statistical difference between five mice for each infection group. (Reprinted from reference 81.)
Altered stress resistance and examination of LSMMG responses in an S. enterica serovar Typhimurium rpoS mstant.
In general, the ability of a pathogen to resist environmental stresses such as extreme fluctuations in pH, osmolarity, and temperature correlates with its virulence potential (32, 69). Recent studies have shown that when cultured in the LSMMG environment of the RWV, S. enterica serovar Typhimurium demonstrated increased resistance to acid, osmotic and thermal stresses, and ability to survive within macrophages than did normal-gravity-grown cells (81, 106, 107). This may help to explain the increased virulence of S. enterica serovar Typhimurium induced by LSMMG. However, the results also showed that LSMMG increased the sensitivity of S. enterica serovar Typhimurium to oxidative stress (106). This is an interesting finding since it indicates that LSMMG does not induce a general resistance to all environmental stresses and affects resistance differentially. LSMMG has also been observed to alter the stress resistance of E. coli. LSMMG-grown E. coli cells are more resistant to the growth-inhibitory effects of ethanol than are cells grown in shaking flasks and are more resistant to osmotic and thermal shock than are cells grown at 1 × g (37; S. V. Lynch and A. Matin, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol. 2003, abstr. I-038, 2003).
RpoS is the primary sigma factor responsible for the expression of genes that are required for resistance to environmental stresses and has accordingly been described as the master regulator of the general stress response in E. coli and Salmonella (45, 67). This assertion is clearly demonstrated by the fact that RpoS-deficient strains are highly sensitive to a wide range of environmental stresses and cannot induce the stress resistance that is observed under certain culture conditions such as acid shock, osmotic shock, stationary phase, and carbon starvation (63, 73). In addition, Salmonella rpoS mutants are avirulent in the murine model of infection, most probably because of deficient expression of several genes required for full pathogenicity (18, 19, 31, 79). Because of the central role of RpoS in Salmonella stress resistance and virulence, the effects of LSMMG were recently examined in an S. enterica serovar Typhimurium rpoS mutant (106). The authors reasoned that if RpoS plays a role in the transmission of the LSMMG signal, altered responses to LSMMG would be observed in the rpoS mutant compared to the wild-type parent strain. The study showed that RpoS is not required for the LSMMG response to occur in S. enterica serovar Typhimurium, since the same physiological responses to acid, osmotic, thermal, and oxidative stresses were observed using wild-type and isogenic rpoS mutant Salmonella strains (106). In addition, the study used microarray analysis to show that 25 genes belonging to the RpoS regulon (i.e., genes regulated by RpoS) did not undergo any change in expression under LSMMG but that a separate set of genes (i.e., genes not regulated by RpoS) displayed LSMMG responsiveness in both the wild-type and rpoS mutant strains (106). This indicates that transmission of the LSMMG signal does indeed occur in the rpoS mutant strain as it does in the wild-type strain and that the altered stress resistance phenotypes associated with LSMMG in S. enterica serovar Typhimurium are occurring via an RpoS-independent pathway(s). It is also worth noting that this study also demonstrated that LSMMG represents a novel environmental culture condition that can serve to preadapt a Salmonella rpoS mutant for resistance to multiple environmental stresses. Interestingly, recent evidence indicates that in E. coli, the RpoS protein level is increased during culture at LSMMG as compared to 1 × g (Lynch and Matin, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol 2003). This indicates that RpoS may be a part of the LSMMG regulon in E. coli, but it is not known how that observation relates to LSMMG-induced stress responses.
Altered growth kinetics.
Monitoring the growth kinetics of bacteria under different conditions can reveal differences in the way in which the cells respond to the growth signals present in each environment. When the effect of LSMMG on the growth kinetics of Salmonella in broth culture was recently examined, the results showed that LSMMG shortened the generation time of S. enterica serovar Typhimurium in minimal medium by 25 to 30 min compared to the time measured under 1 × g conditions (106). This result further characterized LSMMG as a signal that has direct effects on S. enterica serovar Typhimurium metabolism and that acts to reprogram the physiological state of the bacteria. This phenotype was observed in both wild-type S. enterica serovar Typhimurium and an isogenic rpoS mutant, providing further evidence that RpoS is not required for LSMMG signals to be transmitted by the cell. Interestingly, the growth kinetics of LSMMG-grown and 1 × g-grown Salmonella appear identical over the same time course for cultures in Luria-Bertani broth, a rich medium that contains many energy-providing factors compared to minimal medium. The combination of minimal medium and LSMMG seems to induce increased Salmonella metabolic activity that drives growth, although the specific pathways and genes involved are not yet known. However, microarray analysis did reveal many genes that could be involved in altering the growth-related metabolism to be induced in Salmonella at LSMMG, but their role in this phenotype has not been determined (107). A similar growth-related phenotype has also been observed with E. coli grown at LSMMG. A study by Fang et al. demonstrated an increase in the dry-cell weight of E. coli during culture at LSMMG compared to culture at 1 × g (28). It is interesting that, similar to the S. enterica serovar Typhimurium growth experiments, the E. coli growth experiments were performed with cells in minimal media. Together, these findings indicate that bacteria can more readily proliferate in an environment of LSMMG (similar to space or certain in vivo niches) and suggest that bacteria in this environment are more readily able to initiate growth that could lead to contamination, colonization, and infection.
A possible interpretation of this result is that there are differences in mass diffusion between the LSMMG and 1 × g environments that may affect nutrient uptake and metabolism independently of any Salmonella or E. coli LSMMG responses. For example, a possible result of lowered mass diffusion that may be experienced under LSMMG is a difference in the accumulation of nutrients and cellular by-products in the local environment around the cell (58, 59). It is interesting that the altered growth kinetics observed in Salmonella and E. coli are consistent with experiments performed on various space missions that found that bacteria grew to higher densities in liquid culture during space flight than the densities achieved by equivalent ground-based controls (16, 57, 103). The altered microbial growth kinetics observed during spaceflight has been hypothesized to arise as a result of changes in extracellular mass transport of nutrients and by-products (57). Differences in mass diffusion and other chemical alterations of the cellular microenvironment could play a significant role in altering microbial metabolic responses in microgravity and LSMMG.
LSMMG regulon and evidence for Fur as a potential regulator of the S. enterica serovar Typhimurium LSMMG response.
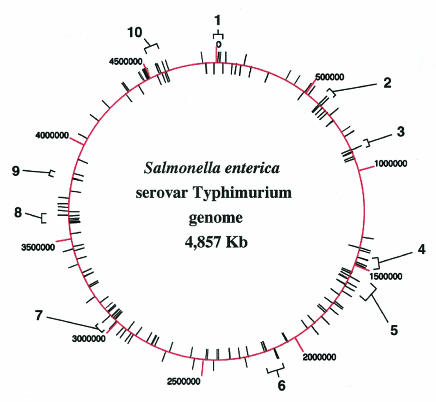
The LSMMG-induced phenotypic changes observed with numerous bacterial species suggest that LSMMG represents a global environmental regulatory signal in prokaryotes that serves to reprogram gene expression (21, 82). Elucidation of the mechanisms involved in transmitting this signal and identification of the genes that are altered in expression in response to LSMMG would significantly aid our understanding of the LSMMG culture responses and possibly lead to ways of manipulating this signal for beneficial engineering of microbes. Two-dimensional (2-D) gel analysis has shown that the expression of numerous S. enterica serovar Typhimurium proteins is altered when the cells are grown under LSMMG (81). A recent study also used 2-D gel analysis to show that several E. coli proteins are LSMMG regulated (Lynch and Matin, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol. 2003). However, these analyses did not allow identification of the LSMMG-regulated proteins. Subsequently, DNA microarrays were used to elucidate the global transcriptional response of S. enterica serovar Typhimurium to LSMMG (107). Compared to identical growth conditions under normal gravity (1 × g), LSMMG differentially regulated the expression of 163 genes distributed throughout the Salmonella chromosome, representing functionally diverse groups encoding transcriptional regulators, virulence factors, lipopolysaccharide (LPS) biosynthetic enzymes, ribosomal proteins, iron utilization enzymes, and proteins of unknown function (Fig. 3) (107). Several of the identified genes are located in the same transcriptional operon or in physically linked clusters, indicating that certain genetic loci may be targeted by the LSMMG response. It may be important to delineate these physically linked clusters since these genes may be part of a large “island” that may contain operons that are coregulated by a single common regulator protein. Two such S. enterica serovar Typhimurium islands, SPI-1 and SPI-2, contain several LSMMG-regulated genes (see Table 3). A representative list of the 163 identified genes is presented in Table 3. Reverse transcriptase PCR analysis was used to verify the results obtained from the microarray analysis. The study indicated that the expression of a large operon containing the rfb LPS biosynthetic genes was decreased in response to LSMMG, and, strikingly, Salmonella LPS levels changed accordingly, as predicted by this microarray result. This finding takes on additional significance since it has been hypothesized that cells sense changes in mechanical forces, including shear and gravity, at their cell surface (7,48, 97).
FIG. 3.
Chromosomal organization of the S. enterica serovar Typhimurium LSMMG regulon. The circular chromosome is schematically depicted, with kilobase coordinates noted and labeled. LSMMG-regulated genes as identified by microarray analysis are noted as unlabeled lines extending from the chromosome. The genes belong to diverse functional groups including transcriptional regulators, virulence factors, LPS biosynthetic enzymes, ribosomal proteins, iron utilization functions, and proteins of unknown function. The numbered brackets indicate clusters of LSMMG-regulated genes that are physically linked (within 50 kb) or part of the same operon. Identification of such physically linked gene clusters is important because they may represent genes that are part of the same “island,” which may contain operons that are coregulated by the same transcriptional regulator. This could give clues to the identity of potential LSMMG regulators. In fact, clusters 7 and 4 contain several LSMMG-responsive genes that belong to the Salmonella pathogenicity islands SPI-1 and SPI-2, respectively. (Reprinted from reference 107 with permission.)
TABLE 3.
S. enterica serovar Typhimurium genes belonging to the LSMMG regulona
| STM gene ID no. | Gene name | Expression ratiob (mean ± SD) | Gene function(s) |
|---|---|---|---|
| Up-regulated genes | |||
| STM0459 | ybaO | 6.50 ± 3.04 | Putative transcriptional regulator, AsnC family |
| STM1625 | ydcI | 5.00 ± 0 | Putative transcriptional regulator, LysR family |
| STM3014 | lysR | 5.00 ± 4.36 | Positive transcriptional regulator, LysR family |
| STM4322 | yjdC | 8.33 ± 2.89 | Putative bacterial regulatory protein, merR family |
| STM0592 | fepD | 3.17 ± 1.61 | ABC superfamily, ferric enterobactin transporter |
| STM1471 | rstB | 10.00 ± 0 | Sensory histidine kinase, two-component with RstA |
| STM3630 | dppA | 8.33 ± 2.89 | ABC superfamily, dipeptide transport protein |
| STM1327 | ydiY | 10.00 ± 0 | Putative salt-induced outer membrane protein |
| STM1517 | ydeD | 7.33 ± 4.62 | Putative permease, integral membrane protein |
| STM4591 | sthE | 6.67 ± 2.89 | Putative major fimbrial subunit |
| STM3069 | pgk | 10.00 ± 0 | Phosphoglycerate kinase |
| STM3939 | cyaA | 5.73 ± 3.96 | Adenylate cyclase |
| Down-regulated genes | |||
| STM2869 | orgA | 0.26 ± 0.04 | SPI-1 type III secretory protein |
| STM2874 | prgH | 0.35 ± 0.11 | SPI-1 type III secretion machinery |
| STM2883 | sipD | 0.37 ± 0.07 | SPI-1 type III secreted protein, regulator of secretion |
| STM2893 | invI | 0.33 ± 0.18 | SPI-1 type III secretory protein |
| STM2896 | invA | 0.35 ± 0.11 | SPI-1 type III secretion machinery |
| STM2902 | pigB | 0.41 ± 0.07 | SPI-1 pathogenicity island-associated protein |
| STM1398 | sseB | 0.42 ± 0.11 | SPI-2 type III secretion system effector |
| STM1412 | ssaL | 0.28 ± 0.11 | SPI-2 type III secretion system apparatus |
| STM1414 | ssaV | 0.35 ± 0.03 | SPI-2 type III secretion system apparatus |
| STM1631 | sseJ | 0.37 ± 0.03 | SPI-2 type III secretion system effector |
| STM0543 | fimA | 0.41 ± 0.1 | Major type I fimbrial subunit |
| STM2082 | rfbP | 0.41 ± 0.08 | LPS side chain synthesis |
| STM2084 | rfbM | 0.42 ± 0.05 | LPS side chain synthesis |
| STM2093 | rfbI | 0.32 ± 0.01 | LPS side chain synthesis |
| STM1371 | sufC | 0.46 ± 0.06 | Putative ABC superfamily transport protein |
| STM1373 | sufS | 0.34 ± 0.06 | Selenocysteine lyase |
This list is not comprehensive but is representative of the 163 LSMMG regulon genes identified by microarray analysis.
Expression ratio is the value for the LSMMG fluorescent channel divided by the value for the 1 × g channel as described in reference 107.
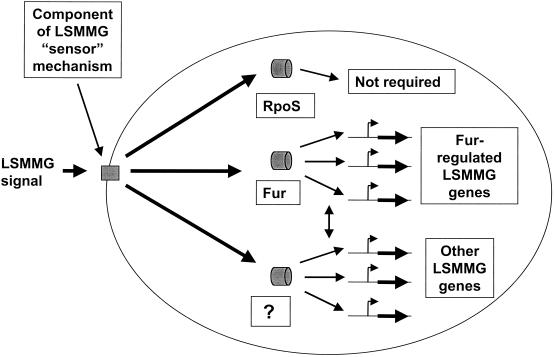
On DNA sequence analysis of the LSMMG regulon genes, it was observed that ferric uptake regulator (Fur) binding sites were associated with several of these genes. In addition, several genes that are involved in iron metabolism or that could potentially use iron for normal function were identified as LSMMG regulated. The authors investigated the LSMMG response in an S. enterica serovar Typhimurium fur mutant strain compared to that in an isogenic wild-type control. The wild-type strain displayed LSMMG-induced acid resistance as expected, but the fur mutant did not show any detectable acid resistance induced by LSMMG (107). This indicates that Fur is required for LSMMG-induced acid resistance in S. enterica serovar Typhimurium and suggests that Fur is involved in the transmission of the LSMMG signal. The role of Fur as a global regulator of a variety of cellular functions in response to an environmental signal (in addition to iron concentration) has been previously suggested (25). However, it is expected that other regulators in S. enterica serovar Typhimurium are involved in addition to Fur for LSMMG signal transmission, given the large number of functional gene groups that are affected by LSMMG. Based on studies with S. enterica serovar Typhimurium to date, Fig. 4 illustrates the current knowledge of LSMMG signal transmission in the bacterial cell. One of the most exciting aspects of future LSMMG studies will be to identify additional regulators of this signaling process. The microarray study indicates that LSMMG is a major global regulatory signal in Salmonella and provides an important step toward understanding the response of bacteria to LSMMG growth signals by forming a potential “roadmap” for future studies involving the molecular response to LSMMG.
FIG. 4.
Diagram summarizing how the LSMMG signal could be transmitted in S. enterica serovar Typhimurium. Based on current data, this diagram depicts a potential picture of LSMMG signal transmission in S. enterica serovar Typhimurium. The mechanical, physical, and/or chemical changes associated with LSMMG culture in the RWV are sensed by the bacterial cell using a hypothetical sensor mechanism. The sensor component of this mechanism (depicted as a rectangle) is probably located at the cell envelope (similar to common prokaryotic response regulator systems), but an intracellular location is possible. Suggested candidates and mechanistic models for this sensor are discussed in the text. This signal is probably transduced to intracellular regulators that regulate the expression of LSMMG-responsive genes. The potential regulators (depicted as cylinders) of the LSMMG response are labeled. Based on experimental data, it appears that the RpoS sigma factor, a logical candidate regulator, is not required for the LSMMG response in S. enterica serovar Typhimurium. Data obtained from microarray-related experiments suggest that the Fur transcriptional regulator is involved in S. enterica serovar Typhimurium LSMMG signal transmission. Given the large number of functional groups of genes regulated by LSMMG, it is likely that other regulators, indicated by the question mark, are also involved. In addition, there may be overlap in the regulation of Fur-regulated LSMMG genes and other such genes controlled by the unknown regulator(s). This potential cross talk is indicated by a double-headed arrow.
Microarray analysis is also currently being used to identify E. coli genes that are regulated in response to LSMMG (D. Tucker, C. M. Ott, D. L. Pierson, and G. E. Fox, Abstr. 103rd Gen. Meet. Am. Soc Microbiol. 2003, abstr. I-043, 2003) Thus far, the study has revealed a number of genes from different functional groups to be regulated by LSMMG including genes involved in acid tolerance, chaperone function, and cell motility. It will be very interesting to see how the full body of these results, as well as the results of similar analyses done with other bacterial species, compare to those obtained with S. enterica serovar Typhimurium. Comparison of results from such analyses with those for other bacterial species will provide knowledge about whether the known effects of LSMMG are a general phenomenon or species specific.
Responses of Yeast to LSMMG
Recently, microarray analysis was used to examine the kinetic changes in gene expression in the yeast S. cerevisiae during culture in the RWV for different periods (52). The S. cerevisiae RWV cultures incubated at LSMMG were compared to cultures grown in identical RWV bioreactors in the 1 × g orientation on a gyrorotatory shaker. The study identified LSMMG-responsive genes at different time points and used cluster analysis to group genes that displayed the same pattern of expression changes over the time course. The DNA sequences of the identified genes were scanned for a variety of promoter elements to define the possible mechanisms mediating these genetic changes. This analysis revealed candidate regulatory binding motifs similar to the Rap1p transcription factor binding site and the stress-responsive element (52). Rap1p is a transcriptional regulator of many genes including those whose expression is altered in response to changes in growth rate, including ribosomal proteins (84). Several of the S. cerevisiae LSMMG-responsive genes which contained Rap1p binding sites are involved in glycolysis and were indicated to be upregulated in response to LSMMG by comparison with the control culture. In agreement with the microarray data, increased glucose utilization and increased expression of ribosomal-protein genes were also observed in yeast cultured in the RWV compared to the control culture. Although a change in Rap1p expression was not observed, the authors suggested that the increased glucose utilization in the RWV might provide a model for increased Rap1p-mediated transcription since there is evidence that activation of Rap1p sites could be associated with this phenomenon (52). Interestingly, it has been proposed by Li et al. that plasma membrane stretching in S. cerevisiae is important in mediating the coordination of ribosome and tRNA synthesis with cell growth (66). In this regard, it is relevant to reiterate that S. enterica serovar Typhimurium exhibited altered production of genes encoding ribosomal components and tRNA synthetases in response to LSMMG culture in the RWV (107). Taken together, the results from both S. cerevisiae and S. enterica serovar Typhimurium culture in the RWV suggest that changes in genotype and phenotype by these two model microbes in response to LSMMG may be initially sensed as mechanical deformation or perturbation of the cell surface and subsequently transmitted into a molecular response.
Collectively, the results from microarray analyses using model prokaryotic and eukaryotic organisms have begun to define the molecular mechanisms of the microbial response to the LSMMG culture environment of the RWV. The results from the various studies of microbial responses to LSMMG are summarized in Table 4. Future studies hold exciting promise to provide insight into the specific microbial signaling pathways involved in sensing and responding to changes in the mechanical and physical forces of fluid shear stress and gravitational force, respectively.
TABLE 4.
Summary of LSMMG-induced effects on microbial physiology
| Physiological effect induced by LSMMG | Bacterial species | Reference(s) |
|---|---|---|
| Altered production of secondary metabolites | ||
| Production inhibited | ||
| Cephalosporin | Streptomyces clavuligerus | 29 |
| Microcin B 17 | Escherichia coli | 28 |
| Rapamycin | Streptomyces hygroscopicus | 26 |
| Glycerol or glucose repression inhibited in RWV (not specific to LSMMG) | ||
| Gramicidin S (glycerol) | Bacillus brevis | 30 |
| Microcin B17 (glucose) | Escherichia coli | 27 |
| Increased extracellular accumulation | ||
| Microcin B17 | Escherichia coli | 28 |
| Rapamycin | Streptomyces hygroscopicus | 26 |
| Increased virulence in murine model of infection (decreased LD50, shortened host time to death, increased liver and spleen colonization) | Salmonella enterica serovar Typhimurium | 81 |
| Altered stress resistance | ||
| Increased ethanol stress resistance | Escherichia coli | 37 |
| Increased acid, osmotic, and thermal stress resistance | Salmonella enterica serovar Typhimurium | 81, 106 |
| Decreased oxidative stress (H2O2) resistance | Salmonella enterica serovar Typhimurium | 106 |
| Increased survival within J774 macrophages | Salmonella enterica serovar Typhimurium | 81, 106 |
| Decreased generation time in M9 minimal medium | Salmonella enterica serovar Typhimurium | 106 |
| Global alteration of gene expression (LSMMG regulon) | Salmonella enterica serovar Typhimurium | 107 |
| Saccharomyces cerevisiae | 52 |
HOW DO CELLS RESPOND TO MICROGRAVITY, MICROGRAVITY ANALOGUES, AND OTHER LOW-SHEAR ENVIRONMENTS?
It is clear that both prokaryotic and eukaryotic microbes demonstrate profound changes in response to microgravity and microgravity-analogue environments; however, the specific mechanism(s) of responses due to gravity reduction and the resulting low-fluid shear is not well defined. Studies in numerous laboratories are under way to address this important issue. However, insight gained from the response of microbes to changes in other mechanical and physical forces may provide insight into the mechanistic effects of microgravity at the cellular level. To better understand how microbial cells transduce mechanical force into biological responses, it is relevant to revisit what is currently known about how microbes respond to changes in the mechanical forces of osmotic pressure gradients and fluid shear. This is because numerous reports suggest that the cell perceives changes in gravity, as well as changes in osmotic gradients and fluid shear, at the cell surface and transduces the resulting signals to the inside of the cell (6, 48, 49, 97). Indeed, microgravity and microgravity analogues, osmotic gradients, and fluid shear all cause changes in the cell surface of microbial cells (6, 22, 86, 96, 97, 107). Thus, local distortion in the cell surface appears to be common to mechanisms of cellular mechanotransduction. Moreover, mechanical restructuring or deformation of the cell surface results in changes in cell-signaling pathways (48). While there is no evidence that there is a shared mechanism used by cells to sense changes in osmotic gradients, fluid shear levels, and gravity, it is not uncommon for microbes to exhibit cross talk in response to different environmental stimuli (87, 88).
Influence of Fluid Shear Force on Cellular Physiology
The study of phenotypic alterations in microbes as a result of the mechanical force of flow-induced shear is a growing and exciting field of research. This is because the level of shear force experienced by microbes has important and far-reaching implications, since (i) changes in fluid shear profoundly affect microbial responses (10, 82, 97) and (ii) the level of shear force experienced by microorganisms is expected to vary greatly during the natural course of their life cycles (this is especially true for pathogens). Conventional cultivation of pathogens has utilized either static or vigorously shaken cultures, neither of which may be accurate representations of what occurs during pathogen interactions with hosts. In 1983, Brooks and Trust demonstrated that changes in fluid shear force have a profound effect on bacterial adhesion (11). This study was the first to demonstrate that enhanced shear force increased the binding of bacteria to red blood cells, although a mechanism was not identified. Recently, an elegant study by Thomas et al. provided mechanistic evidence as to how bacterial adhesion is affected by changes in fluid shear stress (97). This study used red blood cell agglutination assays in combination with flow chamber experiments to compare the ability of several structural variants of E. coli FimH adhesin to bind to red blood cells under a variety of fluid shear conditions. The authors showed that adhesion was enhanced by increased shear force, thus demonstrating direct mechanosensing of fluid shear by the FimH adhesin. In their molecular model of FimH response to fluid shear, the authors describe FimH as a two-domain protein consisting of a lectin domain (which binds host cell surface mannose residues) and a pilin domain (which incorporates FimH subunits into the body of the pilus appendage). Connecting the two domains is a flexible linker that is maintained in a compact state (due to hydrogen bonding) under conditions of lowered shear. In this conformation, the lectin domain binds mannose loosely. When shear is increased, the hydrogen bond(s) is broken, the linker region extends, and the resulting conformational change decreases the off-rate of FimH binding to mannose. Binding of FimH to mannose is predicted to be optimized under these conditions. Then when shear decreases, the FimH off-rate increases and dissociation from mannose occurs. This has been described as a “catch-bond” model and can explain how FimH modulates its binding affinity to mannose in response to changes in shear force.
The functional significance of shear-enhanced adhesion has important implications for essentially any microbe, since microorganisms encounter a wide variety of dynamic shear forces throughout their life cycles and must be able to respond appropriately. This is especially true for pathogens, which often initiate infection by colonizing host tissues via fimbrial adhesions and are subjected to both continuous and intermittent shear forces in vivo during the natural course of infection (11).
Mechanisms To Sense Deformation of the Cell Membrane
Fluctuation of external osmolarity is a common mechanical force change to which microbial cells must adapt throughout their life cycles. The response of a microbial cell to changes in extracellular solute concentration, both a decrease (hypoosmotic stress) and an increase (hyperosmotic stress), is essential for cellular metabolism and survival. Microbes sense changes in osmotic pressure gradients directly through tension in their cell membrane (6, 7, 86). For example, in response to sudden osmolarity decreases, bacteria use mechanosensitive membrane mechanisms to help maintain osmotic balance and prevent cell lysis (6). These membrane mechanisms are mechanically gated protein channels that prevent cell lysis by sensing membrane deformation induced by turgor and allowing the release of hydrostatic pressure buildup (6, 40). Since their initial discovery in E. coli, the existence of mechanically gated channels has been documented in both gram-positive and other gram-negative bacteria (4, 5, 8, 9, 71, 92, 109, 110). There are two major families of mechanosensitive channels in bacteria which regulate cellular turgor by sensing and responding to perturbations in the lipid bilayer of the cell membrane caused by osmotic swelling: the large-conductance channel (MscL) and the small-conductance channel (MscS) (7, 9, 40, 85). Both MscL and MscS form gated protein channels which directly detect stretch forces transmitted from the lipid bilayer induced during osmotic downshift (i.e., stretch-sensitive ion channels) (7, 8, 40, 85). The open conformation of these channels, which is induced by tension in the lipid bilayer, serves as a safety valve for the release of mechanosensitive ions and the integrity of the cell structure (6, 7, 40, 85). Recently, the yeast vacuolar channel protein Y vc1p was demonstrated to be a mechanosensitive ion channel which is activated in response to stretch forces on the vacuolar membrane during osmotic upshift (108). The stretch force on the vacuolar membrane was shown to induce the open conformation of Y vc1p with the concomitant release of Ca2+ from the yeast vacuole into the cytoplasm. It is interesting that Y vc1p is a member of the transient receptor potential family channels (Trp), several of which have been associated with mechanosensation in animals, including detection of touch, hearing, balance, vibration, limb location, and osmotic pressure (24). In addition, it has been recently proposed that yeast maintain balances in their protein-synthesizing machinery, i.e., tRNA and rRNA levels, by sensing changes in membrane stretching induced by turgor (66). The ability to coordinate protein synthesis with changes in membrane deformation due to turgor pressure may help to achieve metabolic economy. It may also serve to promote survival under adverse conditions by preventing protein synthesis when cells are unable to expand their plasma membranes. Thus far, the unifying theme of mechanosensitive ion-gated channels is that they are membrane-bound protein complexes that open and close in response to mechanical changes involving membrane deformation. Further analysis of mechanically gated protein channels in both bacteria and yeast will provide fundamental insight into the similarities and differences among the force-transducing mechanisms used by different types of cells. The mechanisms of how cells respond to changes in mechanical forces such as fluid shear and membrane stretching are very likely to be relevant in forming an overall picture of how cells respond to microgravity and the associated low-shear fluid mechanics.
Molecular Model for Microbial “Sensing” of Microgravity and LSMMG
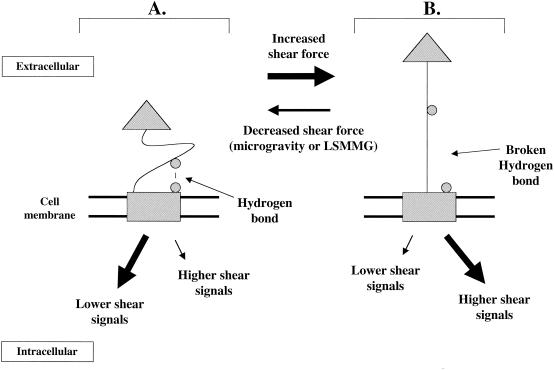
Since liquid environments in microgravity and during LSMMG culture in the RWV are associated with lowered fluid shear forces, this may provide one explanation of how microbes “sense” these environments. The results of the E. coli FimH study described above provide an excellent basis for such a model (51, 97). In this model (Fig. 5), the functioning of a sensor protein embedded in the prokaryotic cell membrane is based on the data for the FimH adhesin. This sensor protein would have two domains connected by a flexible linker region. One domain is embedded in the membrane (the rectangle in Fig. 5) and can serve to initiate signaling inside the cell in response to changes in the linker domain conformation. This domain would be analogous to the membrane-bound response protein in the well-characterized two-component or response regulator model of prokaryotic signal transduction. The other domain is extracellular (the triangle in Fig. 5) and can initiate changes in linker domain conformation in response to changes in shear stress. Under conditions of lowered shear (such as those in a fluid environment in microgravity or LSMMG), a hydrogen bond is formed between a residue on the linker domain and a residue on the membrane domain (both depicted as circles in Fig. 5) and keeps the linker in a compact conformation. (Note that this bond could also be another type of noncovalent bond. One hydrogen bond is depicted here, but several bonds could theoretically exist, involving multiple residues.) The compact conformation of the linker domain causes the membrane domain to send responses inside the cell. These responses could result in (i) activation of signaling pathways associated with lower shear stress, (ii) deactivation of signaling pathways associated with higher shear stress, or (iii) both of these events. This would result in phenotypic responses associated with lower shear. Under conditions of increased shear force, perturbation of the extracellular domain weakens and breaks the hydrogen bond(s) between the linker domain and membrane domain. This conformational change causes the membrane domain to activate signaling pathways associated with higher shear and/or deactivate signals associated with lowered shear. The result would be phenotypic responses associated with higher shear. In such a model, the possible mechanisms used by the membrane domain to transmit the signal are numerous. The membrane domain could phosphorylate regulator proteins that alter the transcription of targeted genes, similar to the two-component model. Alternatively, the membrane domain may interact with microbial cytoskeletal elements (see below) to initiate the mechanical changes in cell structure or function that may be associated with microgravity or LSMMG. Several other possibilities for transmission of this signal exist as well. However, the detailed example illustrated by the responsiveness of the FimH protein to shear stress provides an excellent overall picture of a potential molecular mechanism for the way in which cells can cause the phenotypic changes associated with an altered mechanical force.
FIG. 5.
Model for the way in which microbes may sense changes in aqueous shear force and transmit this signal at the molecular level. Since aqueous environments in microgravity and during LSMMG culture in the RWV are associated with lowered shear forces, this model may explain how microbes “sense” these environments. The functioning of the sensor molecule in this model is based on the data for the shear-responsive E. coli FimH adhesin. A membrane-bound protein with two domains connected by a flexible linker is depicted. The rectangular domain is embedded in the membrane and can also serve to initiate signaling inside the cell in response to changes in the linker domain conformation. The triangular domain is extracellular and can initiate changes in linker domain conformation in response to changes in shear stress. (A), Lowered shear. A hydrogen bond (or another type of noncovalent bond) is formed between a residue on the linker domain and a residue on the membrane domain (both depicted as circles) and keeps the linker in a compact conformation. One hydrogen bond is depicted, but several such bonds could exist, involving multiple residues. This compact linker domain conformation causes the membrane domain to respond by activating signaling pathways associated with lower shear and/or deactivating pathways associated with high shear responses. The result would be responses associated with lower shear. (B) Increased shear. The increased shear force sensed by the extracellular domain weakens and breaks the hydrogen bond(s) between the linker domain and the membrane domain. This causes the membrane domain to fire signaling pathways associated with higher shear and decrease signals associated with lowered shear. The result would be responses associated with higher shear. In this model, the microbe would not be responding to changes in gravity force directly but to the lowered aqueous shear force that occurs when a microbe is suspended in an aqueous environment at microgravity or LSMMG.
Is There a Dedicated Gravitational Sensor Mechanism in Microbes?
In the above model, the microbe would not be responding to changes in gravity force directly but would be responding to the lowered fluid shear force that occurs when a microbe is suspended in an aqueous environment at microgravity or LSMMG. This naturally leads to the question whether there is a cellular sensor that microbes use to sense gravity directly. In other words, is there a bona fide microbial gravitational sensor molecule(s)? Indeed, gravitational force is very weak compared to the other fundamental physical forces of nature, and there is debate about whether cells are able to directly sense such a weak force (48, 60). However, there is ample evidence that both eukaryotic and prokaryotic cells have molecular mechanisms to sense and respond to mechanical changes and stresses (6,48, 97). Moreover, there is no question that both eukaryotic and prokaryotic cells respond and phenotypically change when cultured in microgravity and LSMMG environments (21, 22, 82, 101). However, in relation to the changes observed during microgravity and LSMMG, are the cells responding to altered gravity (or perception of altered gravity in LSMMG) or to other mechanical or chemical changes in the local environment that result from the altered gravity? In the two different explanations, the cells are responding either directly to altered gravity or indirectly to altered gravity by sensing other changes that result from that environment. Of course, both events could be happening at the same time. However, the question whether there is a dedicated gravitational sensor molecule is unanswered at present.
In microbes, there are certainly some possible molecular candidates that may serve as sensors of the mechanical changes associated with microgravity. One such candidate is represented by the microbial cytoskeletal proteins and their polymerized structures. Elegant work by Ingber and colleagues has shown how mammalian cells probably use tensegrity-based cytoskeletal architecture (triangular, geodesic shapes) to translate changes in mechanical forces at the membrane to corresponding molecular responses inside the cell, including changes in cell signaling (48, 49). A similar type of mechanosensing mechanism could be used in microbes as well. Actin and tubulin homologues (Mbl/MreB and FtsZ, respectively) in bacteria polymerize into cytoskeleton-like structures, and mutants of these proteins are defective in cell shape (38). The microtubule and actin-like elements of yeast are well characterized and function to establish cell shape and division site location. Although these elements in bacteria and yeast have yet to be shown to form tensegrity structures (the Mbl/MreB and FtsZ proteins actually form spiral structures), they could serve to translate mechanical and physical changes into molecular responses that alter cell-signaling pathways. Another set of possible molecular candidates for sensing mechanical changes due to microgravity are the mechanosensitive channels in bacteria and yeast (85). In bacteria, the members of the MscL and MscS protein families are membrane-bound gating channels that sense changes in membrane deformation and respond by open or closing a large pore formed by multimer subunits as described above (85). A member of the Trp (transient receptor potential) family of mechanosensitive channels has recently been cloned and isolated from yeast cells and shown to be a Ca2+ channel that responds to mechanical signals (108). Although the major mechanical signal for these channels is thought to involve changes in osmotic pressure and cell turgor, interactions that occur at the membrane surface under normal gravity and/or higher-shear conditions may cause local changes in membrane fluidity and deformation, either related or unrelated to osmotic pressure. Mechanosensitive channels such as those mentioned here could translate changes in membrane deformation associated with microgravity or LSMMG into cellular responses.
However, with these potential mechanisms, it is highly likely that gravity is being sensed not directly but indirectly by the sensing of other changes associated with altered gravity. An essential requirement for a possible microbial gravity-sensing molecule or mechanism would be to respond directly to the gravity force vector that is oriented toward the Earth's surface. Thus, a major factor in such a mechanism would be one of orientation. In microgravity, the sensed orientation is altered because particles or objects can “float” in a direction opposite to that of the gravity vector. This would provide a key distinction between the two environments of the Earth's surface and of microgravity. Therefore, a key question is this: in all the positions that microbes assume relative to the gravitational vector, is there an orientation sensor that always “knows” where the true gravity vector is relative to the current position? The discovery of such a molecular sensor would be truly astounding, if one exists at all. However, the alternative to a true microbial gravitational sensor mechanism would be the combined action of other mechano- and chemosensitive mechanisms that respond to the changes associated with alterations in gravity. At present, this view best fits the available experimental information regarding cellular gravity sensing.
CONCLUSIONS AND FUTURE DIRECTIONS
Throughout their life cycles, microbes are exposed to an array of physical and mechanical forces ranging from those generated by hydrostatic pressure gradients and fluid shear stress to the constant forces exerted on cells by gravity. Alterations in these forces result in dynamic modifications in the function and phenotype of microbial cells. Recently, several important studies have demonstrated the remarkable and varied roles that mechanical forces play in microbial gene expression, physiology, and pathogenesis. However, limited information is available regarding how mechanical signals, such as pressure or mechanical force delivered to a cell, are translated to direct biological responses. It is probable that mechanical and physical signals sensed by the microbe may be integrated with other environmental signals and transduced into a biochemical response. Thus, studying the response of microbes to changes in mechanical and physical forces may provide insight into a key mechanism to orchestrate and fine-tune the cellular response to environmental stimuli. Moreover, the study of microbial mechanotransduction may identify common conserved mechanisms used by cells to perceive changes in mechanical and/or physical forces and has potential to provide valuable insight into understanding mechanosensing mechanisms in higher organisms, including plants and animals.
Acknowledgments
We are grateful to Tim Hammond and Pat Allen for helpful discussions.
Work in the Nickerson laboratory was supported by grants NAG2-1378, NAG9-1350, and NCC 2-1362 from the National Aeronautics and Space Administration (NASA) and LEQSF-1999-02-RD-A-41 from the Louisiana Board of Regents Support Foundation. Work in the Pierson laboratory was supported by grants NAS9-02078, NAG2-1378, NCC 2-1362, NAG9-1350, and PWC 111-30-40-97 from NASA.
REFERENCES
- 1.Alpatov, A., Y. Il'in, V. Antipov, and M. Tairbekov. 1988. Biological experiments on COSMOS-1887. Kosm. Biol. Aviakosm. Med. 23:26-32. [PubMed] [Google Scholar]
- 2.Audia, J. P., C. C. Webb, and J. W. Foster. 2001. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int. J. Med. Microbiol. 291:97-106. [DOI] [PubMed] [Google Scholar]
- 3.Beeson, J. G., S. J. Rogerson, B. M. Cooke, J. C. Reeder, W. Chai, A. M. Lawson, M. E. Molyneux, and G. V. Brown. 2000. Adhesion of Plasmodium falciparum-infected erythrocytes to hyaluronic acid in placental malaria. Nat. Med. 6:86-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berrier, C., M. Besnard, B. Ajouz, A. Coulombe, and A. Ghazi. 1996. Multiple mechanosensitive ion channels from Escherichia coli, activated at different thresholds of applied pressure. J. Membr. Biol. 151:175-187. [DOI] [PubMed] [Google Scholar]
- 5.Berrier, C., A. Coulombe, C. Houssin, and A. Ghazi. 1989. A patch-clamp study of ion channels of inner and outer membranes and of contact zones of E. coli, fused into giant liposomes. Pressure-activated channels are localized in the inner membrane. FEBS Lett. 259:27-32. [DOI] [PubMed] [Google Scholar]
- 6.Blount, P. 2003. Molecular mechanisms of mechanosensation: big lessons from small cells. Neuron 37:731-734. [DOI] [PubMed] [Google Scholar]
- 7.Blount, P., and P. C. Moe. 1999. Bacterial mechanosensitive channels: integrating physiology, structure and function. Trends Microbiol. 7:420-424. [DOI] [PubMed] [Google Scholar]
- 8.Blount, P., S. I. Sukharev, P. C. Moe, B. Martinac, and C. Kung. 1999. Mechanosensitive channels of bacteria. Methods Enzymol. 294:458-482. [DOI] [PubMed] [Google Scholar]
- 9.Blount, P., S. I. Sukharev, P. C. Moe, M. J. Schroeder, H. R. Guy, and C. Kung. 1996. Membrane topology and multimeric structure of a mechanosensitive channel protein of Escherichia coli. EMBO J. 15:4798-4805. [PMC free article] [PubMed] [Google Scholar]
- 10.Brooks, D. E., and T. J. Trust. 1983. Enhancement of bacterial adhesion by shear forces: characterization of the haemagglutination induced by Aeromonas salmonicida strain 438. J. Gen. Microbiol. 129:3661-3669. [DOI] [PubMed] [Google Scholar]
- 11.Brooks, D. E., and T. J. Trust. 1983. Interactions of erythrocytes with bacteria under shear. Ann. N. Y. Acad. Sci. 416:319-331. [DOI] [PubMed] [Google Scholar]
- 12.Cai, Z., J. Xin, D. M. Pollock, and J. S. Pollock. 2000. Shear stress-mediated NO production in inner medullary collecting duct cells. Am. J. Physiol. Ser. F 279:F270-F274. [DOI] [PubMed] [Google Scholar]
- 13.Reference deleted.
- 14.Castro, V. A., A. N. Thrasher, M. Healy, C. M. Ott, and D. L. Pierson. 2004. Microbial characterization during the early habitation of the International Space Station. Microb. Ecol., in press. [DOI] [PubMed]
- 15.Cavicchioli, R., T. Thomas, and P. M. Curmi. 2000. Cold stress response in Archaea. Extremophiles 4:321-331. [DOI] [PubMed] [Google Scholar]
- 16.Ciferri, O., O. Tiboni, G. Di Pasquale, A. M. Orlandoni, and M. L. Marchesi. 1986. Effects of microgravity on genetic recombination in Escherichia coli. Naturwissenschaften 73:418-421. [DOI] [PubMed] [Google Scholar]
- 17.Reference deleted.
- 18.Coynault, C., and F. Norel. 1999. Comparison of the abilities of Salmonella typhimurium rpoS, aroA and rpoS aroA strains to elicit humoral immune responses in BALB/c mice and to cause lethal infection in athymic BALB/c mice. Microb. Pathog. 26:299-305. [DOI] [PubMed] [Google Scholar]
- 19.Coynault, C., V. Robbe-Saule, and F. Norel. 1996. Virulence and vaccine potential of Salmonella typhimurium mutants deficient in the expression of the RpoS (sigma S) regulon. Mol. Microbiol. 22:149-160. [DOI] [PubMed] [Google Scholar]
- 20.Creasy, R., and R. Resnik. 1984. Maternal-fetal medicine: principles and practice. The W. B. Saunders Co., Philadelphia, Pa.
- 21.Demain, A. L., and A. Fang. 2001. Secondary metabolism in simulated microgravity. Chem. Rec. 1:333-346. [DOI] [PubMed] [Google Scholar]
- 22.Dickson, K. J. 1991. Summary of biological spaceflight experiments with cells. ASGSB Bull. 4:151-260. [PubMed] [Google Scholar]
- 23.Dintenfass, L. 1986. Execution of “ARC” experiment on space shuttle “Discovery” STS 51-C: some results on aggregation of red blood cells under zero gravity. Biorheology 23:331-347. [DOI] [PubMed] [Google Scholar]
- 24.Ernstrom, G. G., and M. Chalfle. 2002. Genetics of sensory mechanotransduction. Annu. Rev. Genet. 36:411-453. [DOI] [PubMed] [Google Scholar]
- 25.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang, A., D. Pierson, S. Mishra, and A. Demain. 2000. Growth of Streptomyces hygroscopicus in rotating-wall bioreactor under simulated microgravity inhibits rapamycin production. Appl. Microbiol. Biotechnol. 54:33-36. [DOI] [PubMed] [Google Scholar]
- 27.Fang, A., D. Pierson, S. Mishra, and A. Demain. 2000. Relief from glucose interference in microcin B-17 biosynthesis by growth in a rotating-wall bioreactor. Lett. Appl. Microbiol. 31:39-41. [PubMed] [Google Scholar]
- 28.Fang, A., D. Pierson, S. Mishra, D. Koenig, and A. Demain. 1997. Effect of simulated microgravity and shear stress on microcin B7 production by Escherichia coli and on its excretion into the medium. Appl. Environ. Microbiol. 63:4090-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang, A., D. Pierson, S. Mishra, D. Koenig, and A. Demain. 1997. Secondary metabolism in simulated microgravity: beta-lactam production by Streptomyces clavuligerus. J. Ind. Microbiol. 18:22-25. [DOI] [PubMed] [Google Scholar]
- 30.Fang, A., D. L. Pierson, S. K. Mishra, D. W. Koenig, and A. L. Demain. 1997. Gramicidin S production by Bacillus brevis in simulated microgravity. Curr. Microbiol. 34:199-204. [DOI] [PubMed] [Google Scholar]
- 31.Fang, F. C., S. J. Libby, N. A. Buchmeier, J. Switala, J. Harwood, and D. Guiney. 1992. The alternative sigma factor KatF (RpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11978-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foster, J. W., and M. P. Spector. 1995. How Salmonella survive against the odds. Annu. Rev. Microbiol. 49:145-174. [DOI] [PubMed] [Google Scholar]
- 33.Freed, L., and G. Vunjak-Novakovic. 2000. Tissue engineering bioreactors, p. 143-156. In R. P. Lanza, R. Langer, and J. Vacanti (ed.), Principles of tissue engineering. 2nd ed. Academic Press, Inc., San Diego, Calif.
- 34.Freed, L. E., R. Langer, I. Martin, N. R. Pellis, and G. Vunjak-Novakovic. 1997. Tissue engineering of cartilage in space. Proc. Natl. Acad. Sci. USA 94:13885-13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freed, L. E., and G. Vunjak-Novakovic. 1997. Microgravity tissue engineering. In Vitro Cell Dev. Biol. Anim. 33:381-385. [DOI] [PubMed] [Google Scholar]
- 36.Gao, H., P. S. Ayyaswamy, and P. Ducheyne. 1997. Dynamics of a microcarrier particle in the simulated microgravity environment of a rotating-wall vessel. Micrograv. Sci. Technol. 10:154-165. [PubMed] [Google Scholar]
- 37.Gao, Q., A. Fang, D. L. Pierson, S. K. Mishra, and A. L. Demain. 2001. Shear stress enhances microcin B17 production in a rotating wall bioreactor, but ethanol stress does not. Appl. Microbiol. Biotechnol. 56:384-387. [DOI] [PubMed] [Google Scholar]
- 38.Gitai, Z., and L. Shapiro. 2003. Bacterial cell division spirals into control. Proc. Natl. Acad. Sci. USA 100:7423-7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo, P., A. M. Weinstein, and S. Weinbaum. 2000. A hydrodynamic mechanosensory hypothesis for brush border microvilli. Am. J. Physiol. Ser. F 279:F698-F712. [DOI] [PubMed] [Google Scholar]
- 40.Hamill, O. P., and B. Martinac. 2001. Molecular basis of mechanotransduction in living cells. Physiol. Rev. 81:685-740. [DOI] [PubMed] [Google Scholar]
- 41.Hammond, T. G., E. Benes, K. C. O'Reilly, D. A. Wolf, R. M. Linnehan, A. Taher, J. H. Kaysen, P. L. Allen, and T. J. Goodwin. 2000. Mechanical culture conditions effect gene expression: gravity-induced changes on the space shuttle. Physiol. Genomics 3:163-173. [DOI] [PubMed] [Google Scholar]
- 42.Hammond, T. G., and J. M. Hammond. 2001. Optimized suspension culture: the rotating-wall vessel. Am. J. Physiol. Ser. F. 281:F12-F25. [DOI] [PubMed] [Google Scholar]
- 43.Hammond, T. G., F. C. Lewis, T. G. Goodwin, R. M. Lennehan, D. A. Wolf, K. P. Hire, W. C. Campbell, E. Benes, K. C. O'Reilly, R. K. Globus, and J. H. Kaysen. 1999. Gene expression in space. Nat. Med. 4:359. [DOI] [PubMed] [Google Scholar]
- 44.Hecker, M., and U. Volker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 45.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 46.Hengge-Aronis, R. 2002. Recent insights into the general stress response regulatory network in Escherichia coli. J. Mol. Microbiol. Biotechnol. 4:341-346. [PubMed] [Google Scholar]
- 47.Hymer, W. C., R. E. Grindeland, T. Salada, R. Cenci, K. Krishnan, C. Mukai, and S. Nagaoka. 1996. Feeding frequency affects cultured rat pituitary cells in low gravity. J. Biotechnol. 47:289-312. [DOI] [PubMed] [Google Scholar]
- 48.Ingber, D. 1999. How cells (might) sense microgravity. FASEB J. 13:S3-S15. [DOI] [PubMed] [Google Scholar]
- 49.Ingber, D. E. 1997. Integrins, tensegrity, and mechanotransduction. Grav. Space Biol. Bull. 10:49-55. [PubMed] [Google Scholar]
- 50.Ingber, D. E. 2003. Mechanosensation through integrins: cells act locally but think globally. Proc. Natl. Acad. Sci. USA 100:1472-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isberg, R. R., and P. Barnes. 2002. Dancing with the host; flow-dependent bacterial adhesion. Cell 110:1-4. [DOI] [PubMed] [Google Scholar]
- 52.Johanson, K., P. L. Allen, F. Lewis, L. A. Cubano, L. E. Hyman, and T. G. Hammond. 2002. Saccharomyces cerevisiae gene expression changes during rotating wall vessel suspension culture. J. Appl. Physiol. 93:2171-2180. [DOI] [PubMed] [Google Scholar]
- 53.Kacena, M. A., P. E. Leonard, P. Todd, and M. W. Luttges. 1997. Low gravity and inertial effects on the growth of E. coli and B. subtilis in semi-solid media. Aviat. Space Environ. Med. 68:1104-1108. [PubMed] [Google Scholar]
- 54.Kacena, M. A., G. A. Merrell, B. Manfredi, E. E. Smith, D. M. Klaus, and P. Todd. 1999. Bacterial growth in space flight: logistic growth curve parameters for Escherichia coli and Bacillus subtilis. Appl. Microbiol. Biotechnol. 51:229-234. [DOI] [PubMed] [Google Scholar]
- 55.Kacena, M. A., E. E. Smith, and P. Todd. 1999. Autolysis of Escherichia coli and Bacillus subtilis cells in low gravity. Appl. Microbiol. Biotechnol. 52:437-439. [DOI] [PubMed] [Google Scholar]
- 56.Kacena, M. A., and P. Todd. 1999. Gentamicin: effect on E. coli in space. Micrograv. Sci. Technol. 12:135-137. [PubMed] [Google Scholar]
- 57.Klaus, D., S. Simske, P. Todd, and L. Stodiek. 1997. Investigation of space flight effects on Escherichia coli and a proposed model of underlying physical mechanisms. Microbiology 143:449-455. [DOI] [PubMed] [Google Scholar]
- 58.Klaus, D. M. 2001. Clinostats and bioreactors. Gravity Space Biol. Bull. 14:55-64. [PubMed] [Google Scholar]
- 59.Klaus, D. M. 1998. Microgravity and its implications for fermentation biotechnology. Trends Biotechnol. 16:369-373. [DOI] [PubMed] [Google Scholar]
- 60.Klaus, D. M. 2002. Space microbiology: microgravity and microorganisms, p. 2996-3004. In G. Bitton (ed.), Encyclopedia of environmental microbiology. John Wiley & Sons, Inc., New York, N.Y.
- 61.Klaus, D. M., P. Todd, and A. Schatz. 1998. Functional weightlessness during clinorotation of cell suspensions. Adv. Space Res. 21:1315-1318. [DOI] [PubMed] [Google Scholar]
- 62.Reference deleted.
- 63.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 64.Lapchine, L., N. Moatti, G. Gasset, G. Richoilley, J. Templier, and R. Tixador. 1986. Antibiotic activity in space. Drugs Exp. Clin. Res. 12:933-938. [PubMed] [Google Scholar]
- 65.Lesnyak, A., G. Sonnenfeld, L. Avery, I. Konstantinova, M. Rykova, D. Meshkov, and T. Orlova. 1996. Effect of SLS-2 spaceflight on immunologic parameters of rats. J. Appl. Physiol. 81:178-182. [DOI] [PubMed] [Google Scholar]
- 66.Li, Y., R. D. Moir, I. K. Sethy-Coraci, J. R. Warner, and I. M. Willis. 2000. Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol. Cell. Biol. 20:3843-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loewen, P., and R. Hengge-Aronis. 1994. The role of the sigma factor sigma S (KatF) in bacterial global regulation. Annu. Rev. Microbiol. 48:53-80. [DOI] [PubMed] [Google Scholar]
- 68.Reference deleted.
- 69.Mahan, M., J. Slauch, and J. Mekalanos. 1996. Environmental regulation of virulence gene expression in Escherichia coli, Salmonella, and Shigella spp., p. 2803-2815. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 70.Manko, V., V. Kordyum, L. Vorob'yev, N. Konshin, and G. Nechitaylo. 1986. Changes over time in Proteus vulgaris cultures grown in the ROST-4M2 device on the Salyut-7 space station, p. 3-10. In K. Sytnik (ed.), Sapce biology and biotechnology: a collection of papers. Naukov Dumka, Kiev, Russia.
- 71.Martinac, B., M. Buechner, A. H. Delcour, J. Adler, and C. Kung. 1987. Pressure-sensitive ion channel in Escherichia coli. Proc. Natl. Acad. Sci. USA 84:2297-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mattoni, R. 1968. Space flight effects and gamma radiation interaction on growth and induction of lysogenic bacteria: a preliminary report. Bioscience 18:602-608. [Google Scholar]
- 73.McCann, M. P., J. P. Kidwell, and A. Matin. 1991. The putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J. Bacteriol. 173:4188-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McLean, R. J. C., J. M. Cassanto, M. B. Barnes, and J. H. Koo. 2001. Bacterial biofilm formation under microgravity conditions. FEMS Microbiol. Lett. 195:115-119. [DOI] [PubMed] [Google Scholar]
- 75.Mehta, S. K., R. J. Cohrs, B. Forghani, G. Zerbe, D. H. Gilden, and D. L. Pierson. 2004. Stress-induced subclinical reactivation of varicella zoster virus in astronauts. J. Med. Virol. 72:174-179. [DOI] [PubMed] [Google Scholar]
- 76.Mehta, S. K., R. P. Stowe, A. H. Feiveson, S. K. Tyring, and D. L. Pierson. 2000. Reactivation and shedding of cytomegalovirus in astronauts during spaceflight. J. Infect. Dis. 182:1761-1764. [DOI] [PubMed] [Google Scholar]
- 77.Reference deleted.
- 78.Mishra, S. K., and D. L. Pierson. 1992. Space flight: effects on microorganisms, p. 53-60. In J. Lederberg (ed.), Encyclopedia of microbiology, vol. 4. Academic Press, Inc., San Diego, Calif. [Google Scholar]
- 79.Nickerson, C. A., and R. Curtiss III. 1997. Role of sigma factor RpoS in initial stages of Salmonella typhimurium infection. Infect. Immun. 65:1814-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nickerson, C. A., T. J. Goodwin, J. Terlonge, C. M. Ott, K. L. Buchanan, W. B. Uicker, K. Emami, C. L. Cedor, R. Ramamurthy, M. S. Clarke, T. Hammond, and D. L. Pierson. 2001. Three-dimensional tissue assemblies: novel models for the study of Salmonella pathogenesis. Infect. Immun. 69:7106-7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nickerson, C. A., C. M. Ott, S. J. Mister, B. J. Morrow, L. Burns-Keliher, and D. L. Pierson. 2000. Microgravity as a novel environmental signal affecting Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 68:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nickerson, C. A., C. M. Ott, J. W. Wilson, R. Ramamurthy, C. L. LeBlanc, K. Honer zu Bentrup, T. Hammond, and D. L. Pierson. 2003. Low-shear modeled microgravity: a global environmental regulatory signal affecting bacterial gene expression, physiology, and pathogenesis. J. Microbiol. Methods 54:1-11. [DOI] [PubMed] [Google Scholar]
- 82a.Nickerson, C. A., and C. M. Ott. 2004. A new dimension in modeling infectious disease. ASM News 70:169-175.
- 83.Ott, C. M., R. J. Bruce, and D. L. Pierson. Microbial characterization aboard spacecraft: evaluation of the Mir space station. Microb. Ecol. in press. [DOI] [PubMed]
- 84.Pina, B., J. Fernandez-Larrea, N. Garcia-Reyero, and F. Z. Idrissi. 2003. The different (sur)faces of Rap1p. Mol. Genet. Genomics 268:791-798. [DOI] [PubMed] [Google Scholar]
- 85.Pivetti, C. D., M. R. Yen, S. Miller, W. Busch, Y. H. Tseng, I. R. Booth, and M. H. Saier, Jr. 2003. Two families of mechanosensitive channel proteins. Microbiol. Mol. Biol. Rev. 67:66-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Poolman, B., P. Blount, J. H. Folgering, R. H. Friesen, P. C. Moe, and T. van der Heide. 2002. How do membrane proteins sense water stress? Mol. Microbiol. 44:889-902. [DOI] [PubMed] [Google Scholar]
- 87.Rowbury, R. J. 2001. Extracellular sensing components and extracellular induction component alarmones give early warning against stress in Escherichia coli. Adv. Microb. Physiol. 44:215-257. [DOI] [PubMed] [Google Scholar]
- 88.Sansonetti, P. 2002. Host-pathogen interactions: the seduction of molecular cross talk. Gut 50(Suppl. 3):III2-III8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schwarz, R. P., D. A. Wolf, and T. Trinh. June. 1991. Rotating cell culture vessel. U.S. patent 5,026,650.
- 90.Stock, U. A., and J. P. Vacanti. 2001. Cardiovascular physiology during fetal development and implications for tissue engineering. Tissue Eng. 7:1-7. [DOI] [PubMed] [Google Scholar]
- 91.Stowe, R. P., D. L. Pierson, D. L. Feeback, and A. D. Barrett. 2000. Stress- induced reactivation of Epstein-Barr virus in astronauts. Neuroimmunomodulation 8:51-58. [DOI] [PubMed] [Google Scholar]
- 92.Sukharev, S. I., P. Blount, B. Martinac, and C. Kung. 1997. Mechanosensitive channels of Escherichia coli: the MscL gene, protein, and activities. Annu. Rev. Physiol. 59:633-657. [DOI] [PubMed] [Google Scholar]
- 93.Takahashi, A., K. Ohnishi, S. Takahashi, M. Masukawa, K. Sekikawa, T. Amano, T. Nakano, S. Nagaoka, and T. Ohnishi. 2001. Differentiation of Dictostelium discoideum vegetative cells into spores during Earth orbit in space. Adv. Space Res. 28:549-553. [DOI] [PubMed] [Google Scholar]
- 94.Taylor, G., I. Konstantinova, G. Sonnenfeld, and R. Jennings. 1997. Changes in the immune system during and after spaceflight. Adv. Space Biol. Med. 6:1-32. [DOI] [PubMed] [Google Scholar]
- 95.Taylor, G. R. 1974. Space microbiology. Annu. Rev. Microbiol. 28:121-137. [DOI] [PubMed] [Google Scholar]
- 96.Thevenet, D., R. D'Ari, and P. Bouloc. 1996. The SIGNAL experiment in BIORACK: Escherichia coli in microgravity. J. Biotechnol. 47:89-97. [DOI] [PubMed] [Google Scholar]
- 97.Thomas, W. E., E. Trintchina, M. Forero, V. Vogel, and E. V. Sokurenko. 2002. Bacterial adhesion to target cells enhanced by shear force. Cell 109:913-923. [DOI] [PubMed] [Google Scholar]
- 98.Tixador, R., G. Richoilley, G. Gasset, J. Templier, J. Bes, N. Moatti, and L. Lapchine. 1985. Study of minimum inhibitory concentration of antibiotics on bacteria cultivated in vitro in space (cytos 2 experiment) Aviat. Space Environ. Med. 56:748-751. [PubMed] [Google Scholar]
- 99.Tsao, Y., E. Boyd, D. A. Wolf, and G. F. Spaulding. 1994. Fluid dynamics within a rotating bioreactor in space and Earth environments. J. Spacecraft Rockets 31:937-943. [Google Scholar]
- 100.Reference deleted.
- 101.Unsworth, B. R., and P. I. Lelkes. 1998. Growing tissues in microgravity. Nat. Med. 4:901-907. [DOI] [PubMed] [Google Scholar]
- 102.Urban, J. E. 2000. Adverse effects of microgravity on the magnetotactic bacterium Magnetospirillium magnetotacticum. Acta Astronaut. 47:775-780. [DOI] [PubMed] [Google Scholar]
- 103.Volkmann, D. 1988. Microgravity and the organisms: results of the Spacelab mission D1. Acta Astronau. 17:267-270. [DOI] [PubMed] [Google Scholar]
- 104.Volz, P. A. 1990. Mycology studies in space. Mycopathologia 109:89-98. [DOI] [PubMed] [Google Scholar]
- 105.Vunjak-Novakovic, G., N. Searby, J. De Luis, and L. E. Freed. 2002. Microgravity studies of cells and tissues. Ann. N. Y. Acad. Sci. 974:504-517. [DOI] [PubMed] [Google Scholar]
- 106.Wilson, J. W., C. M. Ott, R. Ramamurthy, S. Porwollik, M. McClelland, D. L. Pierson, and C. A. Nickerson. 2002. Low-shear modeled microgravity alters the Salmonella enterica serovar Typhimurium stress response in an RpoS- independent manner. Appl. Environ. Microbiol. 68:5408-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wilson, J. W., R. Ramamurthy, S. Porwollik, M. McClelland, T. Hammond, P. Allen, C. M. Ott, D. L. Pierson, and C. A. Nickerson. 2002. Microarray analysis identifies Salmonella genes belonging to the low-shear modeled microgravity regulon. Proc. Natl. Acad. Sci. USA 99:13807-13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhou, X. L., A. F. Batiza, S. H. Loukin, C. P. Palmer, C. Kung, and Y. Saimi. 2003. The transient receptor potential channel on the yeast vacuole is mechanosensitive. Proc. Natl. Acad. Sci. USA 100:7105-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zoratti, M., and V. Petronilli. 1988. Ion-conducting channels in a gram-positive bacterium. FEBS Lett. 240:105-109. [DOI] [PubMed] [Google Scholar]
- 110.Zoratti, M., V. Petronilli, and I. Szabo. 1990. Stretch-activated composite ion channels in Bacillus subtilis. Biochem. Biophys. Res. Commun. 168:443-450. [DOI] [PubMed] [Google Scholar]