Abstract
Background
NPM1 mutations are reported to predict a favorable prognosis in acute myeloid leukemia (AML) patients. Aberrant cytoplasmic localization of NPM protein is reported be a surrogate for NPM1 gene mutation.
Methods
Using immunohistochemical analysis (IHC), we assessed for NPM (clone 376) expression in formalin-fixed, formic acid-decalcified bone marrow biopsy specimens. DNA sequencing of the exon 12 of NPM1 gene was performed in 104 patients.
Results
The study included 252 AML patients: 192 de novo AML, 33 AML following a myelodysplastic syndrome or chronic myelomonocytic leukemia, and 27 therapy-related AML. The median age was 62 years and 115 patients were ≤ 60 years old. All patients received intensive chemotherapy. Cytoplasmic NPM was detected in 59 of 252 (23%) patients, including 48 of 192 (25%) de novo AML and 33 of 94 (35%) with a normal karyotype. DNA sequencing identified NPM1 mutations in 30 of 38 cases with cytoplasmic NPM and 10 of 66 cases with nuclear NPM. Cytoplasmic NPM was associated with young patient age (p=0.024), FLT3/ITD (p=0.005), CD34- negative blasts (p<0.001), high peripheral blood blast counts (p=0.041), and high serum albumin level (p=0.028). No statistical differences in overall or event-free survival were found on the basis of NPM localization. Similar results were obtained in patients ≤ 60 years old with normal karyotype and wild-type FLT3 (p=0.768).
Conclusions
Immunohistochemical assessment for NPM localization did not predict prognosis in this patient cohort. The discordance between immunohistochemistry and DNA sequencing results indicates that DNA sequencing cannot be replaced by IHC assessment.
Keywords: AML, NPM1 mutation, immunohistochemical analysis, prognosis, FLT3/ITD
Introduction
Nucleophosmin, encoded by the nucleophosmin (NPM1) gene on chromosome 5q35, is ubiquitously expressed in human tissues1, 2. The NPM1 gene has been shown to be commonly mutated in acute myeloid leukemia (AML)3, 4. NPM1 mutations are found in 25%–35% of all AML cases and 45%–64% of AML cases with a normal karyotype3, 4. NPM1 mutations have been associated with a favorable prognosis5 in the absence of a fms-related tyrosine kinase-3 (FLT3) gene mutation of internal tandem duplication type (ITD), a known adverse prognostic factor. In two retrospective studies, NPM1-mutated/FLT3-ITD-negative patients did not benefit from allogeneic stem cell transplantation unlike NPM1-wild-type/FLT3-ITD-negative patients5, 6. These data led to the proposal that AML patients with a normal karyotype should be tested for FLT3-ITD and NPM1 mutations to allow for risk-adapted therapy with NPM1-mutated/FLT3-ITD-negative patients being treated with chemotherapy without allogeneic stem cell transplantation, and NPM1-wild-type/FLT3-ITD-negative patients undergoing allogeneic stem cell transplantation7.
Wild type NPM is predominantly located in nuclei and nucleoli. NPM1 gene mutations typically occur in exon 12, affecting the carboxy terminus and both disrupting the localizing domain and creating a nuclear export signal. As a result, mutated NPM protein is located in the nucleus and cytoplasm of neoplastic cells8. Others have suggested that immunohistochemical analysis for NPM localization can be used in lieu of molecular analysis for NPM1 mutations9. In one study using immunohistochemical analysis of bone marrow biopsy specimens and mouse anti-NPM monoclonal antibody, the pattern of NPM expression was completely concordant with mutation status8. These data resulted in a proposed work flow to assess for NPM1 mutations using immunohistochemical analysis as an initial screening step9. Cases with cytoplasmic NPM staining subsequently need molecular confirmation of NPM1 mutations, and cases with nuclear NPM staining do not need further NPM work-up, thereby reducing unnecessary work and conserving resources9.
In this study, we investigated NPM localization using an immunohistochemical method in a large number of AML patients at our institution. The results were compared with DNA sequencing results in a subset of patients and were also correlated with clinical parameters, including overall and event-free survival.
Patients and Methods
After approval from the Institutional Review Board, we analyzed 252 bone marrow biopsy specimens from patients with AML: 70 archival and 182 that were obtained prospectively by the Department of Hematopathology at The University of Texas M. D. Anderson Cancer Center (Houston, TX) between October 1, 2006, and September 1, 2008. For all patients, we obtained clinical and demographic data as shown in Table 1.
Table 1.
Summary of clinicopathologic features
| Variables | Study groups |
|---|---|
| Age (years) | Median=62 (range: 17–88) |
| 60 year old and younger | 115 |
| Sex | |
| women | 128 |
| men | 124 |
| Race | |
| Asian | 7 |
| Black | 14 |
| Hispanic | 33 |
| White | 198 |
| Cytogenetics | |
| Favorable | 21 |
| Intermediate | 152 |
| Normal karyotype | 94 |
| Unfavorable | 79 |
| AML | |
| De novo | 192 |
| Preceding MDS or MDS/MPD | 33 |
| Therapy-related | 27 |
| FLT3 | |
| ITD | 53 |
| Wild type | 178 |
| Not tested | 21 |
| Response | |
| Complete remission | 136 |
| Relapsed | 51 |
| Failure | 116 |
| Death | |
| alive | 151 |
| dead | 101 |
| Follow-up (months) | Median=10 (range, <1–97 months) |
Formalin-fixed, paraffin-embedded tissue sections of bone marrow biopsy specimens (5 µm thick) were deparaffinized in xylene (3 times for 5 min) and then rehydrated through graded concentrations of alcohol (3 times for 3 min in 100% ethanol, then for 3 min in 80% ethanol). For antigen retrieval, tissue sections were heated in EDTA (Biocare Medical, Concord, CA) for 30 min in a Decloaking Chamber (Biocare Medical, Concord, CA). Tissue sections were incubated with Biocare Medical peroxidase blocking reagent for 5 min. Primary antibodies used were a mouse monoclonal antibody against NPM protein (clone 376) (kindly provided by Dr. Brunangelo Falini) and a mouse monoclonal antibody against nucelolin/C23 (MS-3, Santa Cruz Biotech, Santa Cruz, CA). After washing with buffer, tissue sections were incubated with the primary antibodies for 60 min at room temperature (20°C). Sections were again washed, and incubated with secondary antibodies using Mach 4 AP Universal Polymer kit (Biocare Medical) according to manufacturer’s instructions. For visualization, Vulcan Fast Red chromogen (Biocare Medical) was used for 10 min. All sections were counterstained with hematoxylin (Dako, Carpintera, CA) for 3 min. Slides were then air dried and cover-slipped.
In order to assure proper assessment of nuclear localization of NPM, all cases were stained in parallel for nucleolin (C23, Santa Cruz Biotech, Santa Cruz, CA), a protein known to be restricted to the nucleus which served as a control8. Only cases that demonstrated appropriate nuclear distribution of C23 were analyzed for the cellular distribution of NPM. The NPM staining pattern was evaluated in blasts/immature cells, which were medium-sized cells with round-to-oval nuclei, fine nuclear chromatin, and moderate-to-scant cytoplasm in immunostained sections. Cytoplasmic NPM staining in megakaryocytes and dividing cells, that always represented less than 5% of all cells, was not scored.
Conventional cytogenetic analysis using standard GTG banding was performed on bone marrow aspiration specimens according to methods described previously10. The FLT3 gene was assessed for ITD mutations using polymerase chain reaction (PCR) methods, as described previously11.
In a subset of cases we extracted genomic DNA from bone marrow aspirate specimens and used it for a PCR to amplify a 168 bp fragment of exon 12 of the NPM1 gene using Tli polymerase (Promega, Madison, WI). The forward primer (5´-GATGTCTATGAAGTGTTGTGGTTCC-3´) was labeled with 5-carboxyfluorescein (FAM) whereas the reverse primer (5´-GGACAGCCAGATATCAACTG-3´) was unlabeled. PCR fragments were resolved according to size by gel electrophoresis (3100 Genetic Analyzer; Applied Biosystems, Foster City, CA). Wildtype fragments migrated at the predicted size of 168; shifts to a larger size indicated insertion of additional nucleotides.
The Kaplan-Meier method was used to estimate overall survival (OS) and event-free survival (EFS). All patients were included in the EFS assessment. For patients who did not experience a complete remission, EFS was designated as 0. We fitted univariate and multivariate Cox proportional hazards models to assess the effects of the following patient and tumor characteristics on OS and EFS: age (> 60 vs ≤ 60 years), sex, ethnicity (black, Asian, Hispanic, or white), antecedent hematologic disorders as defined previously12, peripheral blood blast count, bone marrow blast count, hemoglobin level, platelet count, and serum bilirubin and creatinine levels. For the multivariate analysis, we fitted a full Cox proportional hazards model on OS or EFS, including all patient characteristics listed above. A reduced model was built by stepwise variable selection such that all variables remaining in the model were significant at a p value of < 0.05. All analyses were performed using SAS software (SAS Institute, Cary, NC) and S-plus software (Insightful, Seattle, WA).
Results
There were 128 women and 124 men, with a median age of 62 years (range, 17–88 years); 115 patients were ≤ 60 year old (Table 1).The median duration of clinical follow-up was 10 months (range, < 1–96 months), with the most recent follow-up in January 2009. One hundred ninety-two patients had de novo AML, 33 had AML preceded by either a myelodysplastic syndrome (n=27) or chronic myelomonocytic leukemia (n=6) (designated here as secondary AML), and 27 developed AML after chemotherapy for other malignancies (i.e. therapy-related AML). Numerous blasts were found in all BM biopsy specimens, constituting a median of 62% all cells (range, 21%–97%). All patients underwent intensive therapy according to institutional protocols. The therapeutic combinations included idarubicin and cytarabine (n=85), fludarabine and cytarabine (n=49), idarubicin, cytarabine, and sorafenib (n=14), daunorubicin and cytarabine (n=13), decitabine (n=13), clofarabine (n=11), vidaza and decitabine (n=11), daunorubicin, cytarabine, etoposide, dexamethasone and thioguanine (n=7), decitabine, cytarabine, and clofarabine (n=6), high dose cytarabine (n=5), cytarabine, cytoxan, and topotecan (n=4), decitabine cytarabine (n=4), decitabine and mylotarg (n=4), fludarabine, clofarabine (n=3), idarubicin and cytarabine decitabine (n=3), idarubicin, cytarabine, and mylotarg (n=3), idarubicin, clofarabine (n=2), clofarabine and cytarabine (n=2), decitabine and valproic acid (n=2), decitabine plus valproic acid (n=2), idarubicin, cytarabine, and SAHA (n=2), mitoxantrone, cytarabine, and gemtuzumab (n=2), idarubicin, etoposide, and daunorubicin (n=1). Four patients with t(15;17) received all trans retinoid acid (ATRA) including 3 patients who received ATRA in combination with arcenic trioxide and one patient who received a liposomal form of ATRA as a monotherapy. Fifty-one patients underwent hematopoietic stem cell transplantation (allogeneic related, n=30; matched unrelated, n=18; cord blood, n=3). One hundred thirty-six patients (54%) patients experienced a complete remission of which 51 had recurrence; 116 (56%) did not experience complete remission.
After a median follow-up of 10 months (range, < 1–97 months), 151 patients died, including 19 with no evidence of disease. The 3-year OS rate was 30% (95% CI, 26%–34%), and the 5-year OS rate was 22% (95% CI, 18%–26%). There was no difference in age, sex, and survival between patients in the prospective and retrospective groups, which allowed us to combine patients into a single group for analysis.
The conventional cytogenetic analysis showed that 21 patients had favorable-risk, 152 intermediate-risk, and 77 unfavorable-risk cytogenetic results. The 21 patients with favorable results included 9 patients with inv(16)(p13.1q22), 8 patients with t(8;21)(q22;q22), and 4 patients with t(15;17)(q22;q12). Ninety-four of 152 patients with intermediate cytogenetics had a normal karyotype. FLT3/ITD was detected in 53 of 231 (23%) patients tested.
Immunohistochemical analysis showed cytoplasmic NPM in 59 of 252 (23%) AML specimens (Fig. 1), including 48 of 192 (25%) de novo, 6 of 33 (18%) secondary, and 5 of 27 (19%) therapy-related AML. No statistically significant difference was detected in the prevalence of cytoplasmic NPM in de novo versus other AML cases (p=0.382). Cytoplasmic NPM was observed in 33 of 94 (35%) patients with a normal karyotype. No patients with inv(16), t(8;21), or t(15;17) had cytoplasmic NPM. Nuclear NPM was found in 193 cases, 172 of whom had no cytoplasmic NPM expression and 21 that had rare (<5%) immature cells with cytoplasmic NPM.
Figure 1.
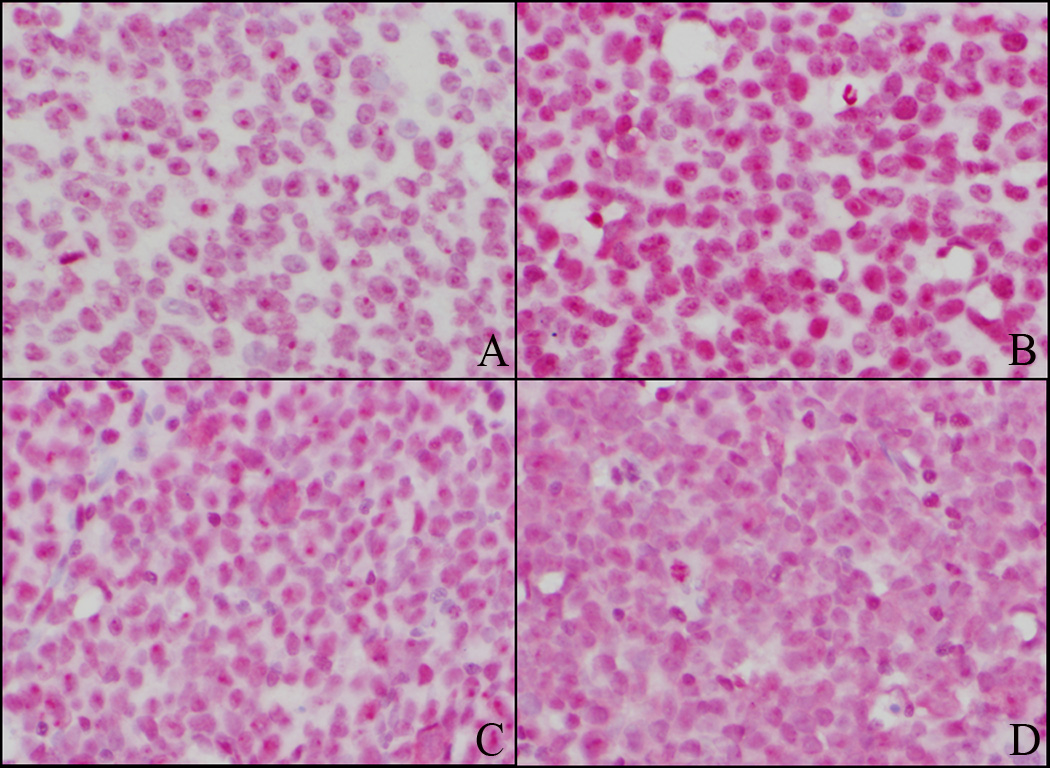
Examples of acute myeloid leukemia (AML) cases with nuclear (A&B) and cytoplasmic (C&D) NPM expression. Anti-C23 antibody is used as a control and demonstrates nuclear staining in both cases (A and C). In a case of AML with nuclear NPM, anti-NPM antibody is similar to anti-C23 staining pattern (B). In a case of AML with cytoplasmic NPM, anti-NPM antibody demonstrates predominantly cytoplasmic staining (D). (A–D, immunohistochemistry, × 400)
Cytoplasmic NPM was associated with young age (p=0.024), FLT3/ITD (p=0.005), lack of CD34 expression (p<0.001), high blast counts in bone marrow (p=0.043) and peripheral blood (p=0.041), and high albumin level (p=0.028). There was no association between cytoplasmic NPM and race, sex, leukocytosis, hemoglobin level, platelet count, or serum LDH, bilirubin, and creatinine levels.
In the univariate Cox model, prolonged OS was associated with young age (p<0.001), de novo AML (p<0.001), presence of antecedent hematologic disorders (p<0.001), favorable cytogenetics (p<0.001), hematopoietic stem cell transplantation (p<0.001), lower bone marrow blast count (p=0.014), lower peripheral blood blast count (p=0.003), lower white blood count (p=0.021), high platelet count (p=0.045), and high serum albumin level (p<0.001). No significant difference in OS was detected between patients with AML with cytoplasmic versus nuclear NPM expression, p=0.822 (Fig. 2). No significant difference in OS for cytoplasmic versus nuclear NPM was shown for the subset of patients with a normal karyotype and wild-type FLT3, p=0.987 (Fig. 3). Similar results were obtained when only patients ≤ 60 years old with a normal karyotype and wild-type FLT3 were analyzed (p=0.768).
Figure 2.
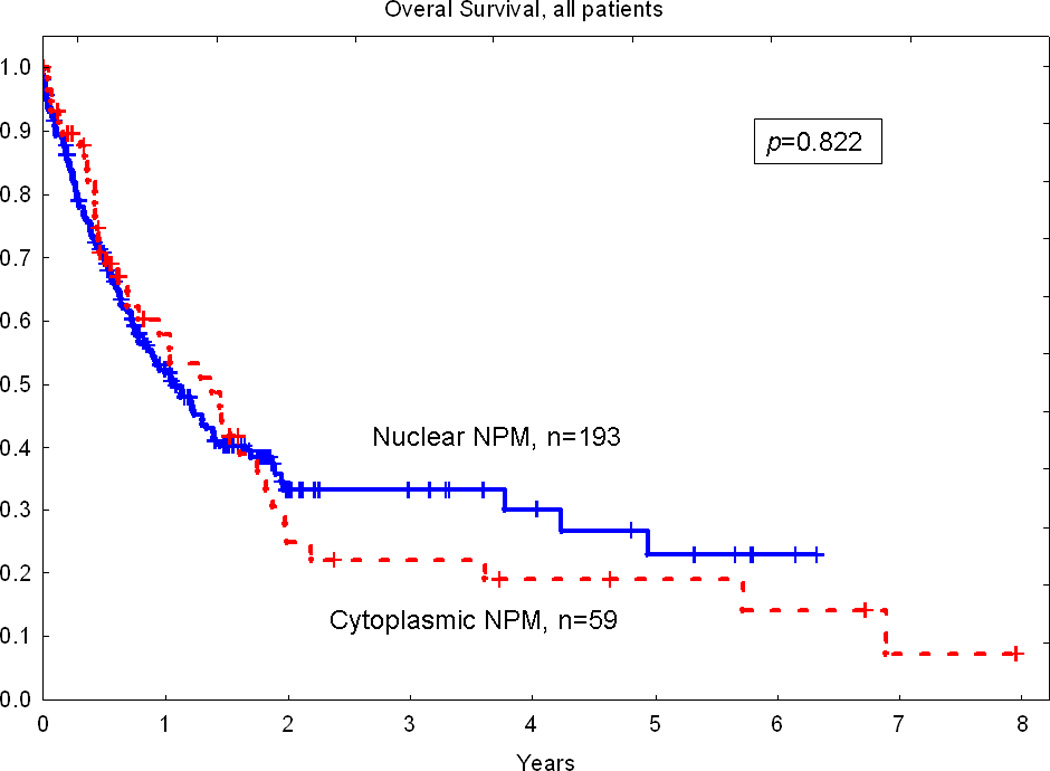
No significant difference in overall survival between patients with cytoplasmic and nuclear NPM in total study group
Figure 3.
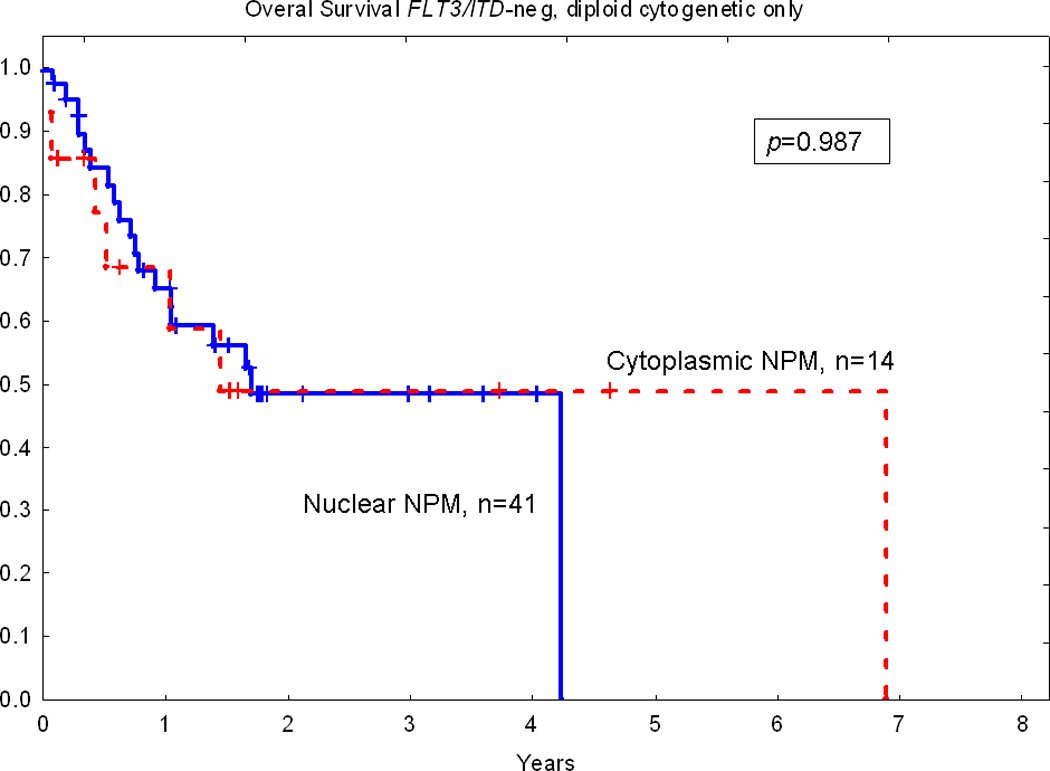
No significant difference in overall survival between patients with cytoplasmic and nuclear NPM in a subset of patients with diploid cytogenetic and wild type FLT3.
In the univariate Cox model, long EFS duration was associated with young age (p=0.0002), de novo AML (p=0.008), presence of antecedent hematologic disorders (p=0.0008), favorable cytogenetics (p=0.002), hematopoietic stem cell transplantation (p=0.0009), and high serum albumin level (p=0.006). No significant difference in EFS was detected between patients with AML with cytoplasmic versus nuclear NPM expression (p=0.534). No significant difference in EFS for cytoplasmic versus nuclear NPM was shown for the subset of patients with a normal karyotype and wild-type FLT3 (p=0.562). Similar results were obtained when only younger patients (≤ 60 years old) with a normal karyotype and wild-type FLT3 were analyzed (p=0.635).
In the multivariate Cox model, prolonged OS was associated with young age (p=0.047), a favorable cytogenetic profile (p<0.001), and hematopoietic stem cell transplantation (p<0.001). Prolonged EFS was associated with young age (p=0.014), a favorable cytogenetic profile (p<0.001), and hematopoietic stem cell transplantation (p=0.002).
We analyzed the NPM1 gene for mutations in 104 patients with AML. There were 53 women and 51 men, with a median age of 61 years (range, 17–86 years). In this subset of patients, 78 patients had de novo AML, 17 had secondary AML, and 9 had therapy-related AML. Nine patients had favorable-risk, 68 intermediate-risk, and 27 unfavorable-risk cytogenetic results. Forty of the 68 patients with intermediate cytogenetics had a normal karyotype. FLT3/ITD was detected in 28 of 90 (31%) patients tested. There was no statistical difference in sex, age, type of AML, cytogenetic findings, or frequency of FLT3/ITD between the whole group of patients and this subset of 104 patients.
NPM1 mutations were detected in 40 (38%) cases. Immunohistochemical analysis showed cytoplasmic NPM in 30 of these 40 (75%). Cytoplasmic NPM was also identified in 8 of 64 (13%) AML cases with wild type NPM1. The positive predictive value of cytoplasmic NPM was 79%, and the negative predictive value was 85%. The sensitivity of immunohistochemical analysis was 75%, with a specificity of 88%. For the subset of 104 patients with molecular data, NPM1 mutations as detected by sequencing did not impact OS (p=0.867) or EFS (p=0.561).
Discussion
NPM1 gene mutation status in AML is a subject of intense interest because several studies have shown that the presence of NPM1 mutations has a prognostic effect in FLT3-ITD-negative AML patients5, 6. The proposal to assign patients with a normal karyotype and wild-type FLT3 to different therapeutic modalities (chemotherapy with or without allogeneic stem cell transplantation)7 has resonated among clinicians leading to the need for all new AML patients to undergo assessment for NPM1 gene mutations. However, it is unknown whether the association between NPM1 mutation and clinical variables shown in European studies will be reproduced in a patient population from the United States. Furthermore, NPM1 mutational analysis which often involves relatively time-consuming sequencing methods may not be available in smaller diagnostic laboratories.
NPM1 mutations alter the carboxy terminus of NPM, leading to its cytoplasmic localization. Falini and colleagues proposed that immunohistochemical staining for NPM can be used as a screening tool for NPM mutational status8. In their study, cytoplasmic NPM had a positive predictive value of 100% for NPM1 mutations. Conversely, nuclear localization of NPM has a negative predictive value of 100% for NPM1 mutations8. These results support the notion that immunohistochemical analysis for NPM expression can be used as a screening tool, or even as a replacement for molecular assessment of NPM1 gene mutations. For these reasons, we assessed the cellular localization of NPM in a large number of AML patients at our institution. We believe that our patient population is representative because standard prognostic indicators such as age, cytogenetic profile, and de novo AML vs. therapy-related or secondary AML were predictive of OS and EFS.
NPM1 gene mutations have been reported in 25%–35% of all adult AML patients and 45%–64% of adult AML patients with a normal karyotype. In this study, cytoplasmic NPM was detected in 23% of all patients, which is a lower frequency than that reported. Surprisingly, cytoplasmic NPM was detected in only 35% of patients with a normal karyotype, which is substantially lower than what has been previously reported. As the prevalence of NPM1 gene mutations in pediatric AML cases was reported to be substantially lower in Taiwan13 than in European countries,14 it is possible that geographic differences affect the incidence of NPM1 gene mutations in adults in a similar manner.
In several European studies, NPM1 mutations in AML were associated with female sex, de novo AML, lack of CD34 expression, high blast count, high serum LDH level, and presence of FLT3-ITD5, 15. In this study, we found an association between cytoplasmic NPM and young age (p=0.024), FLT3-ITD (p=0.005), lack of CD34 expression (p<0.001), high bone marrow blast count (p=0.043), high peripheral blood blast count (p=0.041), and high serum albumin level (p=0.028). We did not show a correlation between NPM expression and sex. Of interest, no association was found between NPM1 mutation and gender in another study from the United States reported in abstract form16. These findings support the hypothesis that geographic differences affect the prevalence of NPM1 mutations as well as clinical associations.
Previously, NPM1 mutations were reported to be closely associated with de novo AML, as AML arising after a myelodysplastic syndrome, myelodysplastic/myeloproliferative neoplasm or therapy-related AML rarely show cytoplasmic NPM8. However, in our study, the prevalence of cytoplasmic NPM was similar in de novo, secondary, and therapy-related AML cases. Our findings have been recently confirmed indirectly in a large study focusing on multilineage dysplasia in AML patients17. This study found a similar frequency of multilineage dysplasia (approximately 30% of cases) among patients with NPM1 mutations and patients with wild type NPM17. Taking into consideration the reported frequency of multilineage dysplasia in secondary AML compared with de novo AML (45.4% vs. 25.2%, p<0.001)17, we suspect that this study also found a high frequency of NPM1 mutations among patients with secondary AML, although this was not specifically reported in the published data.
NPM status had no effect on the OS or EFS duration in our study, either in the total patient population or in the subset of patients with a normal karyotype and wild type FLT3. The inability of NPM to predict prognosis cannot be attributed solely to the use of immunohistochemical assessment as a surrogate for NPM1 mutations because we obtained similar results when we analyzed DNA sequencing results, although in a smaller subset of 104 patients. Although our data contradict initial reports stressing the prognostic importance of NPM1 mutations5, 6, more recent studies have failed to show a prognostic impact of NPM1 mutations on OS and EFS in AML patient populations including FLT3-ITD negative patients4, 17, 18. Although some of these studies have had relatively small numbers of patients,4, 18 a study reported by Wandt et al17 analyzed 1766 patients. The explanation for the discordance of recent studies is unknown, but possible explanations include geographic variation and differences in study design.
We compared DNA sequencing results with those of immunohistochemical analysis and found that immunohistochemical analysis had good sensitivity and excellent specificity. However, both the positive and negative predictive values of cytoplasmic and nuclear NPM, respectively, were substantially lower than that reported by others8. NPM immunostaining is nuclear in approximately 10% of molecularly proven NPM1-mutated cases (i.e. “missed” mutations). In addition, NPM cytoplasmic expression occurs in approximately 13% of AMLs without NPM1 mutation (i.e. “overcalled” mutations). The explanation for the discordance between our results and those of Falini et al are unknown. One possible explanation is that NPM1 mutations other than those occurring at exon 12 may disrupt nuclear cytoplasmic shuttling of NPM. Albiero and colleagues have reported an AML patient carrying an exon 11 mutation that encoded a truncated form of nucleophosmin with aberrant cytoplasmic localization.19,20 Another possibility, however, is that cytoplasmic localization of NPM is not entirely specific for the presence of NPM1 mutations. Technical issues also complicate our understanding of the problem as bone marrow biopsy specimens can be processed in a myriad of ways. Falini and colleagues used BM biopsy specimens that were fixed in B5 fixative and decalcified by EDTA8. We used BM biopsy specimens that were routinely processed at our institution, fixed in formalin and decalcified by formic acid. It is unclear whether formalin fixation or our decalcification process compromises the sensitivity of immunohistochemical assessment of NPM cellular localization. Nevertheless, formalin is the most popular fixative to assess tissue samples in the United States. Being conservative, we conclude that immunohistochemical analysis for NPM cannot be used as a substitute for molecular assessment using our methods.
In summary, we found no correlation between cellular NPM localization detected by immunohistochemistry and patient prognosis in this group of AML patients. The differences between our study and European studies may be related to geographical variation or other differences. In addition, our results suggest illustrate a lack of complete concordance between cytoplasmic NPM detected by immunohistochemistry and direct sequencing of the NPM1 gene. Therefore immunohistochemistry for cytoplasmic NPM appears not to be a completely reliable screening tool for detecting NPM1 mutations. The differences between our results and those of others remain to be explained. NPM1 mutations at sites other than exon 12 or technical differences related to methods of fixation, decalcification and staining are possible explanations.
Table 2.
Correlation of NPM immunohistochemistry staining and DNA Sequencing
| DNA Sequencing |
NPM IHC | |
|---|---|---|
| Cytoplasmic | Nuclear | |
| Mutant | 30 | 10 |
| Wild type | 8 | 56 |
Acknowledgments
We would like to express our gratitude to Dr. Brunangelo Falini, Institute of Hematology, University of Perugia, Perugia, Italy, who kindly provided a mouse monoclonal antibody against NPM protein (clone 376). We would like to acknowledge hematopathologists of the MD Anderson Cancer Center, Drs. Joan H. Admirand, Hesham M. Amin, Yang O. Huh, Jeffrey L. Jorgensen, Pei Lin, Ellen Schlette, Francisco Vega-Vazquez, C. Cameron Yin, and M. James You for contributing diagnostic materials for this study. We also thank LaKisha Rodgers and Geneva Williams for invaluable help in manuscript preparation.
References
- 1.Cordell JL, Pulford KA, Bigerna B, et al. Detection of normal and chimeric nucleophosmin in human cells. Blood. 1999 Jan 15;93(2):632–642. [PubMed] [Google Scholar]
- 2.Chang JH, Olson MO. Structure of the gene for rat nucleolar protein B23. J Biol Chem. 1990 Oct 25;265(30):18227–18233. [PubMed] [Google Scholar]
- 3.Schnittger S, Schoch C, Kern W, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005 Dec 1;106(12):3733–3739. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 4.Boissel N, Renneville A, Biggio V, et al. Prevalence, clinical profile, and prognosis of NPM mutations in AML with normal karyotype. Blood. 2005 Nov 15;106(10):3618–3620. doi: 10.1182/blood-2005-05-2174. [DOI] [PubMed] [Google Scholar]
- 5.Dohner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005 Dec 1;106(12):3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 6.Schlenk RF, Dohner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008 May 1;358(18):1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 7.Mrozek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood. 2007 Jan 15;109(2):431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005 Jan 20;352(3):254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 9.Falini B, Martelli MP, Bolli N, et al. Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia. Blood. 2006 Sep 15;108(6):1999–2005. doi: 10.1182/blood-2006-03-007013. [DOI] [PubMed] [Google Scholar]
- 10.Onciu M, Schlette E, Medeiros LJ, Abruzzo LV, Keating M, Lai R. Cytogenetic findings in mantle cell lymphoma cases with a high level of peripheral blood involvement have a distinct pattern of abnormalities. Am J Clin Pathol. 2001 Dec;116(6):886–892. doi: 10.1309/JQMR-323G-71Y9-M7MB. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Jones D, Medeiros LJ, Luthra R, Lin P. Acute myeloid leukaemia with FLT3 gene mutations of both internal tandem duplication and point mutation type. Br J Haematol. 2005 Sep;130(5):726–728. doi: 10.1111/j.1365-2141.2005.05666.x. [DOI] [PubMed] [Google Scholar]
- 12.Estey EH, Pierce S, Keating MJ. Identification of a group of AML/MDS patients with a relatively favorable prognosis who have chromosome 5 and/or 7 abnormalities. Haematologica. 2000 Mar;85(3):246–249. [PubMed] [Google Scholar]
- 13.Chou WC, Tang JL, Lin LI, et al. Nucleophosmin mutations in de novo acute myeloid leukemia: the age-dependent incidences and the stability during disease evolution. Cancer Res. 2006 Mar 15;66(6):3310–3316. doi: 10.1158/0008-5472.CAN-05-4316. [DOI] [PubMed] [Google Scholar]
- 14.Cazzaniga G, Dell'Oro MG, Mecucci C, et al. Nucleophosmin mutations in childhood acute myelogenous leukemia with normal karyotype. Blood. 2005 Aug 15;106(4):1419–1422. doi: 10.1182/blood-2005-03-0899. [DOI] [PubMed] [Google Scholar]
- 15.Thiede C, Koch S, Creutzig E, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006 May 15;107(10):4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 16.Weinberg JM, Beck A, Seetharam M, Gotlib J, Zehnder J, Arber DA. Significance of NPM1 and FLT3 Mutations in Acute Myeloid Leukemia with Multilineage Dysplasia: Does NPM1 Identify a Lower Risk Group? Modern Pathology. 2008;21(supplement 1s):281A. [Google Scholar]
- 17.Wandt H, Schakel U, Kroschinsky F, et al. MLD according to the WHO classification in AML has no correlation with age and no independent prognostic relevance as analyzed in 1766 patients. Blood. 2008 Feb 15;111(4):1855–1861. doi: 10.1182/blood-2007-08-101162. [DOI] [PubMed] [Google Scholar]
- 18.Bardet V, Costa LD, Elie C, et al. Nucleophosmin status may influence the therapeutic decision in de novo acute myeloid leukemia with normal karyotype. Leukemia. 2006 Sep;20(9):1644–1646. doi: 10.1038/sj.leu.2404294. [DOI] [PubMed] [Google Scholar]
- 19.Albiero E, Madeo D, Bolli N, et al. Identification and functional characterization of a cytoplasmic nucleophosmin leukaemic mutant generated by a novel exon-11 NPM1 mutation. Leukemia. 2007 May;21(5):1099–1103. doi: 10.1038/sj.leu.2404597. [DOI] [PubMed] [Google Scholar]
- 20.Falini B, Albiero E, Bolli N, et al. Aberrant cytoplasmic expression of C-terminal-truncated NPM leukaemic mutant is dictated by tryptophans loss and a new NES motif. Leukemia. 2007 Sep;21(9):2052–2054. doi: 10.1038/sj.leu.2404839. author reply 2054; discussion 2055–2056. [DOI] [PubMed] [Google Scholar]


