Summary
Imaging host-pathogen interactions in real time can provide significant insight into dynamic processes and provide information about time and space of their occurences. Here we present detailed experimental instructions on how to image the membrane penetration process of the non-enveloped adenovirus in rel time. The system is based on a cell line stably expressing the lectin galectin-3 fused to a fluorophore. Membrane-lytic events during adenovirus cell entry can be monitored by the recruitment of galectin-3 to galactose-containing membrane glycoproteins on the exo-surface of ruptured membranes. The simultaneous use of fluorescently labeled adenoviral capsids allows to image the events in unmatched temporal resolution.
Keywords: adenovirus, galectin-3, membrane damage, endosomal escape, trafficking, live cell imaging, virus entry
2. Introduction
While considerable structural and mechanistic detail of enveloped virus fusion with cell membranes has allowed the development of fusion inhibitory antiviral drugs and more effective vaccines, much less is known about analogous events in non-enveloped virus cell entry. Non-enveloped virus capsids often exist as hydrophilic particles in the extracellular environment, retaining a much higher stability and prolonged infectivity then enveloped viruses [1]. In many cases these capsids must physically penetrate and cross hydrophobic cellular membranes to initiate replication. A major limitation to our understanding of non-enveloped virus membrane penetration is in part due to the lack of experimental systems that allow a detailed study of this process.
Adenoviruses (Ad) are prototypic non-enveloped viruses with a large capsid of ~90 nm in diameter that are mild pathogens for immune competent individuals but can cause more severe infections in immune suppressed subjects. Adenovirus disease pathogenesis is thought to result from the strong proinflammatory response to the virus [2]. This response is thought to provide some advantage in the exploitation of Ads as vaccine vectors but has limited the utility of Ad vectors for gene therapy [3].
Ads enter cells by receptor-mediated endocytosis, but must rapidly exit the endosome to avoid lysosomal degradation. We and colleagues have previously shown that the membrane lytic activity of Ad resides in an internal capsid protein, protein VI, which is exposed upon entry during or shortly after endosomal uptake [4, 5]. As a consequence, the virus is thought to penetrate the endosomal membrane and gain access to the cytoplasm for subsequent microtubule-directed transport towards the nucleus. Further analysis of protein VI using in vitro studies showed that membrane fragmentation is achieved by protein VI through an amphipathic helix and the induction of positive membrane curvature [6]. This model for protein VI membrane lysis is consistent with a lack of size selectivity in adenovirus rupture of endosomal membrane [7, 8] and observations that leakage of endosomal contents during adenovirus entry is a danger signal sensed by the host [9–11].
In this chapter we describe a new method that permits the visualization of the membrane lysis and escape from endosomal structures of the non-enveloped adenoviruses using live cell fluorescence microscopy [12]. The method relies on the recruitment of the cytosolic galactose binding lectin galectin-3 (Gal-3) to ruptured membranes. Under physiological conditions galactose-containing polysaccharides are present exclusively on the extracellular or intraluminal domains of membrane glycoproteins. Upon membrane damage cytosolic Gal-3 can access and bind to exposed galactose residues, effectively concentrating on damaged membranes and thus providing a measurable fluorescent signal when fluorescently labeled Gal-3 is used. This technique was originally established to visualize vacuole lysis by invasive bacteria [13, 14]. Here we adapted this technique to show membrane lysis and penetration by adenoviruses. Using this approach we recently showed that membrane rupture and endosomal escape are mechanistically distinct in time and space with Ad particles residing and trafficking in Gal-3 positive structures followed by viral egress from these structures occurring later and often at sites distal from the site of membrane rupture [12]. In this detailed protocol we highlight the experimental set up to image the membrane lysis and escape from ruptured membranes by adenoviruses. However, provided that viruses can be fluorescently labeled without influencing infectivity this approach should be easy to adapt for the use of any viral system whether they are enveloped or non-enveloped.
2. Materials
All work with infectious adenovirus and adenovirus derived vectors should be performed in agreement with the local safety guidelines.
2.1 Cell Culture
| U-2 OS cell line | (ATCC: HTB-96) |
| 293 cells | (ATCC: CRL-1573) |
| DMEM GlutaMAX | (Life Technologies # 1966016) |
| FBS | (check batch with specific cell line) |
| Opti-MEM GlutaMAX | (Life Technologies # 51985026) |
| Penicillin/Streptomycin | (Life Technologies # 15140122) |
| D-PBS | (Life Technologies # 14190235) |
| Trypsin/EDTA | (Life Technologies # 25300054) |
| Hygromycin B | (InvivoGen # ant-hm-1) |
2.2 Virus labeling & quantification
| purified virus | (> 0.2 µg/µl, Note 1) |
| Microscale protein labeling kit | (e.g. Molecular Probes® ref. A30006) |
| Zeba Spin Desalting Columns, 7K MWCO | (ref. 89882 or 89883) |
| DMSO (1mg/ml) or milliQ water to dissolve fluorophore | |
| PBS, 10% glycerol | |
| 1M Tris/HCl pH 7.4 | |
| SDS | |
| 0.2M EDTA | |
| 6-well dish | |
| low melting agarose | |
2.3 Microscopy material
| Ibidi μ-slides VI0.4, 6-channel slides | (Ibidi, ref. 80606) |
| Live cell imaging solution | (Molecular Probes® ref. A14291DJ) |
3. Methods
3.1 Virus labeling protocol
Ads are non-enveloped viruses that can be labeled with different fluorophores using primary amine groups on the exposed surface of the capsid. If done correctly the labeling reaction does not interfere with the infectivity of the virus and allows very good signal-to-noise ratios for live cell imaging.
3.1.1 Labeling the virus
Purify Ad or Ad derived vectors using the CsCl density gradient centrifugation and dialyze extensively against PBS, 10% glycerol. Resulting virus preparations should be >0.25 mg/ml for efficient labeling. (see Notes 1) To label the virus we routinely use the Alexa Fluor® 488 Microscale Protein Labeling Kit from Invitrogen (see Note 2).
Method
Prepare labeling solution following kit procedure by diluting the dye in 10 µL DMSO. One vial of fluorophore generally allows labeling of up to three virus preparations of 100 µL each. Because labeling solution cannot be stored after reconstitution, we label different viruses/vectors in parallel.
Combine 100 µL virus in PBS/10% Glycerol, 12 µL 1M Na-bicarbonate buffer ph 8.3 and 3 µL dye in a reaction tube (e.g. a Screwcap Eppendorf tube).
Mix the reaction slowly and incubate for 30–60 min on a rotating mixer at room temperature. Cover tube using aluminium foil to protect mix from light.
Following the labeling reaction, separate the remaining dye from labeled virus/vector using the Zeba desalting column. Desalting columns allow viruses to pass the resin, while the free dye is retained. This procedure will dilute the virus/vector preparation between two or three fold but is fast and efficient. (see Note 3).
Equilibrate the column by adding 300 µl PBS, 10% glycerol on a Zeba-column and spin it at 1500 ×g in a microcentrifuge tube for 1 min, repeat at least three times.
Add the reaction mix at the top of the resin carefully without destroying the resin pellet and spin again for 2 min and collect the flow through. The resin in the column should be colored by the free dye and the flow through should also be colored by the labeled virus/vector.
Use the flow through containing the labeled virus/vector and repeat the dye separation process at least one more time with new equilibrated columns. Free dye trapped in the remaining resin should be low after second or third desalting while the virus/vector flow through should remain slightly colored.
After the last dye removal step aliquot the flow through and prepare 10 µL aliquots. Snap freeze aliquots in liquid nitrogen and store aliquots at -80°C. Each aliquot can be thawed up to three times without loss of titer.
3.1.2 Quantification of virus (physical particles)
The fastest way to quantify CsCl purified virus/vector preparation is by determining the physical particle number (pp/µL) by measuring absorbance of particle containing DNA at 260nm [15]. The same method can be applied for the labeled end product.
To release the DNA, particles are lysed in Ad-lysis buffer (10 mMTris/HCl pH7.4, 0.1% SDS, 1 mM EDTA). Mix virus preparation of purified virus (and original stock for comparison) with Ad-lysis buffer at 1:10 (or 1:20 for good preps). For example use 10 µL virus prep and complete to 100µL using Ad-lysis buffer in low binding microcentrifuge tube.
Heat the mix ten minutes at 56°C and measure DNA in a spectrophotometer at OD 260nm. Measured absorbance should be between 0.5 and 0.05 O.D. To calculate the physical particle per µL ratio multiply the measured O.D. by dilution factor and divide by extinction coefficient for adenoviruses (9.09 × 10−13). One O.D. equals 1.1 × 109 pp/µL.
Labeling reactions depend on the protein concentration. Thus labeling reactions work better and faster if the virus/vector preparation is more concentrated. We do not proceed with labeling reactions if the concentration drops below one O.D. (1.1 × 109 pp/µL).
3.1.3 Infectivity of labeled virus/vector
To retain the biological properties of labeled adenoviruses it is essential to avoid overlabeling of the capsid with dye. We routinely determine the post-labeling infectivity using plaque forming assays or measure fluorescence protein (FP) expression from Ad derived vectors and compare it to the parental preparation.
3.1.3.1 Infectivity of labeled virus using plaque forming assays (PFA)
Day1
Seed HEK293 E1 transcomplementing cells at a density of 1 × 106 cells/well in a 6-well plate. Use one 6-well plate per virus tested.
Day 2
Three different MOI in duplicate can be tested by plate. To determine the infectivity of the labeled virus assume that your labeled virus has the same ratio of physical to infectious particles as your parental preparation. For wildtype Ad5 we use a MOI of 0,1 – 0,01 – 0,001 per cell. For attenuated viruses higher MOI’s should be used. Prepare virus dilutions accordingly, adjust to 1 ml final volume and infect cells by adding 1 ml per well final volume. Incubate cells over night at 37°C in the incubator. Make independently pipetted duplicates for each MOI. (Note 4)
Day3
On day three the cells are washed with PBS to remove unbound virus and are then covered with an overlay of DMEM agar solution to avoid virus spread. Some training on non-infected cells can be beneficial for this step, because DMEM agar solution preparation and pouring has to be done fast and carefully.
Make 2% stock solution by dissolving 2g of low melting agarose into 100mL 1X PBS. Warm solution can be kept soluble at 60°C (e.g. in a waterbath) for later use. Fill a 50mL tube with 20mL pre-warmed DMEM (37°C). Cool the 2% PBS-agar until hand warm and add 10mL to the DMEM and mix 5 times by inverting the tube. If PBS-agar is to warm it will degrade thermosensitive antibiotics and kill cells (they will detach immediately or the next day). Too cold agar will solidify before pouring.
Remove PBS from the infected cells and add 2 mL of the mix to each well and keep under the hood until agar solidifies then move to 37°C incubator. From now on the agar overlay should not dry out. If gel surface appears dry add carefully 2 or 3 drops of complete DMEM on top of the well. Move plates as little as possible to allow nice plaque formation. Plaques formation will take around 10 days. If a GFP expressing vector was used, plaque formation can be followed by spread of GFP fluorescence, otherwise plaques become visible as “hole” in the cell layer.
After 8–12 days plaques should be ready for counting. Choose an MOI resulting in ~ 10–100 plaques for highest accuracy.
Quantification will lead to Plaque Forming Units (PFU)/physical particle or PFU/µL of virus prep.
3.1.3.1 Alternative infectivity assay using fluorescent protein (FP) expressing Ad derived vectors and fluorescence activated cell sorting (FACS)
A faster way to determine the infectivity of labeled virus is the use of FACS and the quantification of FP expression following a single round infection. As above this requires the quantification of the original and the labeled vector preparation in pp/µl. In addition access to a FACS machine is required. We prefer this assay because for most applications GFP expressing Ad5 derived vector particles are sufficient.
Day1
Seed U2OS at 5 × 104 cells/well of a 24 wells plate in a final volume of 500µL DMEM. Prepare enough wells for triplicates and 2wells for non-transduced control cells.
Day2
To compare transduction efficiency of labeled versus non-labeled vectors we use different dilutions such as 1000:1 – 100:1 – 10:1 – 1:1 pp/cell of e.g. a GFP expressing Ad derived vector. We dilute the corresponding amout of virus in 500µL DMEM and infect cells for 3 hours without pre-binding step. After three hours we discard the infection solution and replace with fresh 500µL complete DMEM.
Day3
Discard Medium and wash once with 500 µL PBS. Add 2 drops Trypsin (~ 50 µl), incubate for ~1 minute at 37°C until cells detach. Neutralize trypsin by addition of 450 µl complete DMEM and transfer 200 µL of the cell suspension into a 96 wells plate (for FACS devices compatible with plates, e.g. BD FACS Canto II) or the full 500µL in a tube compatible with other FACS devices.
Proceed with FACS analysis to count GFP positives cells in your samples. Typically 10 000 events (from cell population) are counted for each sample and GFP transduction levels can be compared for the labeled versus the non-labeled vector.
3.2 Imaging
3.2.2 Microscopy setup (hardware)
Imaging of adenovirus membrane rupture and endosomal escape events occur within time scales of seconds to minutes, with very fast acceleration events due to microtubule directed transport [12]. For this reason an advanced imaging system capable of rapid dual color image acquisition is recommended. To image membrane rupture and endosomal escape in real time we used a spinning disk microscope based on a Leica DMI6000 (Leica Microsystems, Wetzlar, Germany) equipped with a confocal Scanner Unit CSU-X1 (Yokogawa Electric Corporation, Tokyo, Japan) using objectives HCX PL Apo 100× oil NA 1.4 and an Evolve EMCCD camera (Photometrics, Tucson, USA). The diode lasers used were at 491 nm and 561 nm. The 37°C atmosphere was created with an incubator box and an air heating system (Life Imaging Services, Basel, Switzerland) (Note 5)
2.2.3 Microscopy setup (software)
Acquisitions were done using MetaMorph software (Molecular Devices, Sunnyvale, USA). For maximum time resolution all acquisitions were performed in streaming mode using 2 wavelengths (491 nm and 561 nm) and a dual band pass filter (BP502–552/BP615–675). Image acquisition was at 50 msec per channel without binning at 1024 × 1024 px at 10 Hz. This results in rates of 10 frames per second (fps, dual-color) of total cells with an excellent signal-to-noise ratio.
3.2.1 Filling the Ibidi slides
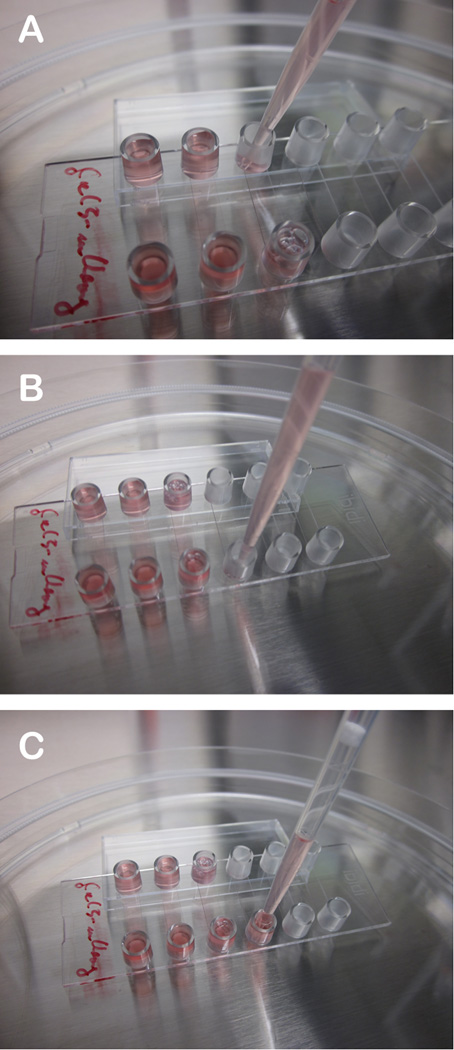
To monitor Ad membrane lysis and escape we use stably expressing U2OS-FRT-Galectin-3-mCherry cells. This cell line was essentially generated as described in chapter xyz of this book. U2OS-FRT-Galectin-3-mCherry are kept at 200 µg/ml of hygromycin in complete medium and split at regular intervals at 1:4 for maintenance. To use the cells for imaging we seed cells and perform infections in Ibidi μ-slides VI0.4 (6-channel slides, see Figure 1).
Figure 1. How to seed cells for the live cell imaging experiment.
A) Place the Ibidi slide at an angle by placing one edge of the slide on the cover. B) Start by filling cell solution into the lower reservoir. The channel between the two reservoir will fill by capillary forces. C) Finish by filling the lower reservoir first and then the upper reservoir with cell solution. The reservoirs should be filled about 2/3.
On the day prior to your experiment prepare cell solution at a density of approximately 1×105 cells per ml by trypsinization and resuspending the cells in complete medium. Position the Ibidi slide in a 15 cm dish at a slight angle by using the cover as indicated in Figure 1. Seed 150µl of this suspension by starting with the bottom opening. The liquid phase will fill the channel by capillary forces and provide a connection to the second reservoir. Finish by adding the remaining cells into the second reservoir and put the slide flat to reach a homogenous distribution of liquid between both reservoirs so that they are filled approximately by 2/3. This procedure will result in about 25–50% confluent cells the next day per channel or ~3000 cells per channel (Note 6).
3.2.2 Setting up infections in Ibidi slides
On the day of the experiment exchange the medium in the channels for live cell imaging solution just prior to start your experiment using the same technique as shown in Figure 1. This clear, HEPES buffered, physiological solution allows keeping the cells for several hours under ideal imaging conditions. Exchange the liquid 2–3 times to fully remove any residual medium and cover the slide with the lid. Transfer your slide to the preheated microscope and let the slide equilibrate for 10 minutes. Before adding the virus/vector to the cells optimize the image acquisition parameters on the microscope using the uninfected cells. Galectin-3 positive cells provide a solid red signal, with few vesicles visible. After infection severe membrane damage can be seen (see Figure 2 compare left and right panel).
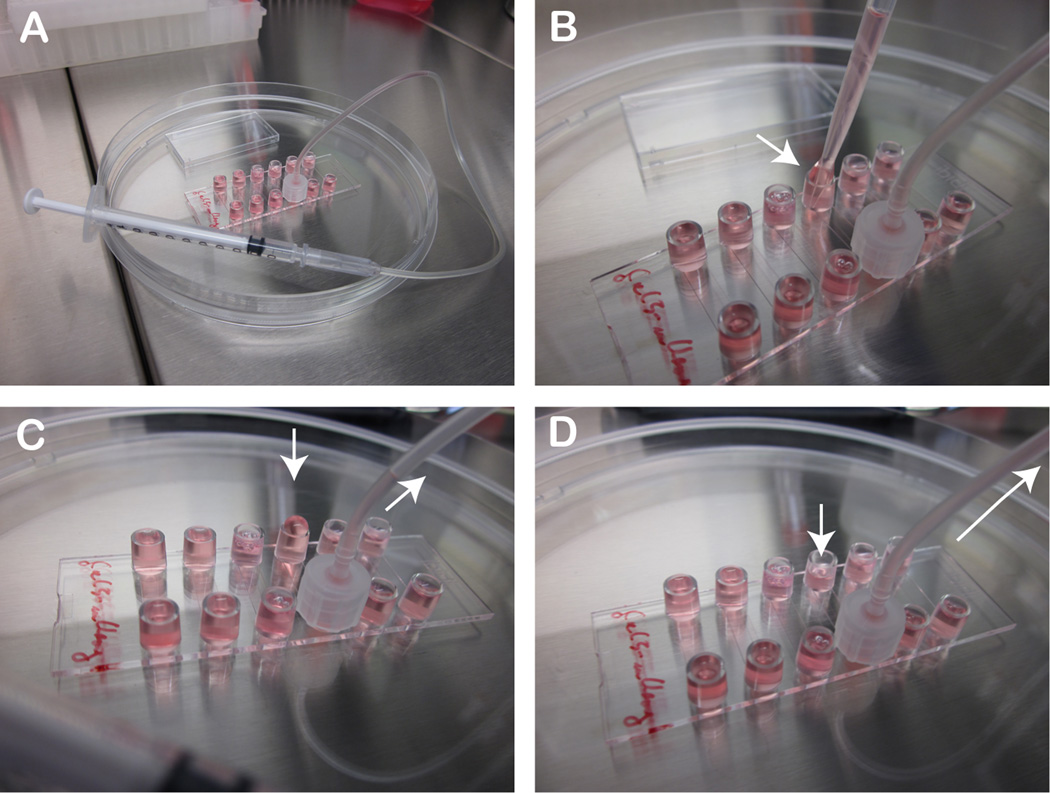
Figure 2. How to infect cells for the live cell imaging experiment.
A) Connect a female Luer adapter with plastic tubing to one reservoir and on the other end to a 1 ml syringe and carefully aspirate the liquid to empty the opposite reservoir B-C) Add fresh virus/vector containing solution to reservoir D) Aspirate the virus/vector containing solution into the channel and repeat the procedure 2–3 times.
To start the infection dilute your labeled virus/vector at 5× 10E7 to 1× 10E8 physical particles per 500 µl of live cell imaging solution (you will need ~200 µl virus solution for one channel). At this dilution you have approximately 1000 pp/cell respectively 5000 pp/cell. To add the virus we connect a female Luer-adapter to one of the reservoirs and connect it with a tube to a 1ml insulin syringe as depicted in Figure 3. Next using the syringe slowly remove the liquid from the second reservoir and replace it with your virus solution. Slowly aspirate the virus solution through the channel into the tube/syringe making sure that there is always solution covering the cells in the channel. Repeat the procedure by refilling the opposite reservoir 2–3 times as indicated in Figure 3. This setting allows to quickly and safely fill the channel with virus solution (Note 7).
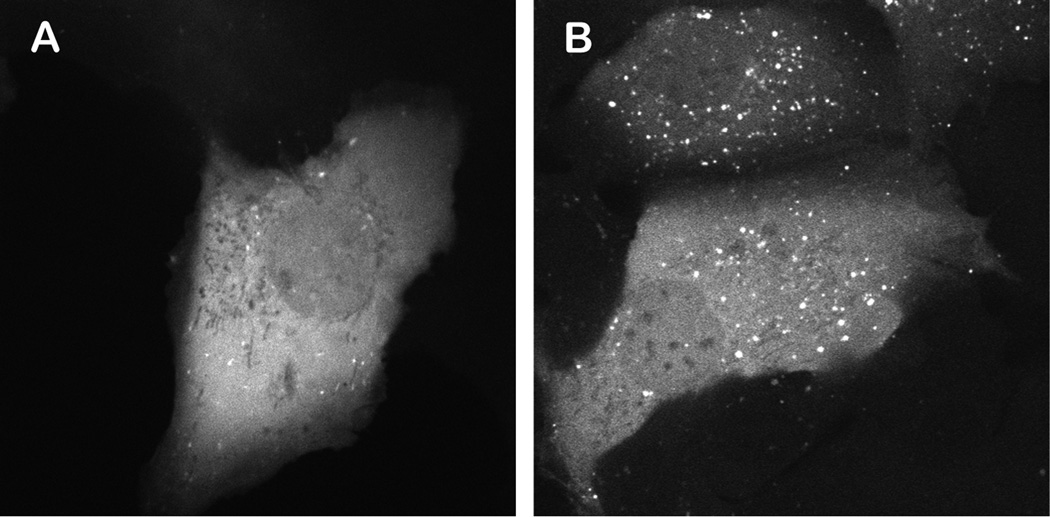
Figure 3. U2OS-FRT-Gal-3 cell line.
A) Midcellular confocal section of a single U2OS-FRT cell stably expressing Gal-3 fused to mCherry. B) As in A following infection with adenovirus. Note the strong increase in Gal-3 labeled vesicular structures.
The above protocol does no synchronize the steps for infection. An alternative protocol is to pre-bind the virus/vector and perform a synchronized infection. In this case virus solutions and imaging media should be kept on ice. Following the addition of the ice cold virus solution (as described above) place the slide on a cold metal block (e.g. kept on ice) and incubate for 30 minutes to allow virus binding. Prior to the transfer to the microscope replace the virus solution with fresh cold imaging medium. The bound virus will remain attached to the cells. However, one should be aware that incubating cells at 4°C also de-polymerizes the microtubule network [16].
3.2.3 Selecting cells and parameters for imaging
Selecting cells for imaging is somewhat subjective and requires some experience. High temporal imaging resolution, especially when involving two colors, can be achieved by avoiding time consuming steps such as Z-stack acquisition, physical filter changes and data transfer. To achieve the best time resolution (in our case 50 msec per frame per color) we restrict imaging to a single confocal slice and acquire individual frames in stream mode using a dual-bandpass filter. This setting reduces the length of the acquisition to the storage capacity of the camera but allows excellent temporal and spatial resolution. We use U2OS cells for most imaging approaches (see also chapter xyz). U2OS cells are flat with a large cytoplasm to nucleus ratio and thus a good choice for imaging requiring high temporal resolution. For optimal imaging we select cells as shown in Figure 3, where the cell periphery and the nucleus can be displayed in a single confocal slice and which have a brightness that gives good signal-to-noise ratios at the desired acquisition speed. Selecting cells with flat morphology is optimal for single confocal frame acquisitions and is best achieved by seeding cells to subconfluency as indicated above. Depending on the events (e.g. for membrane lysis by acquisition of Gal-3 stain) we select a more peripheral slice as shown in Figure 4.
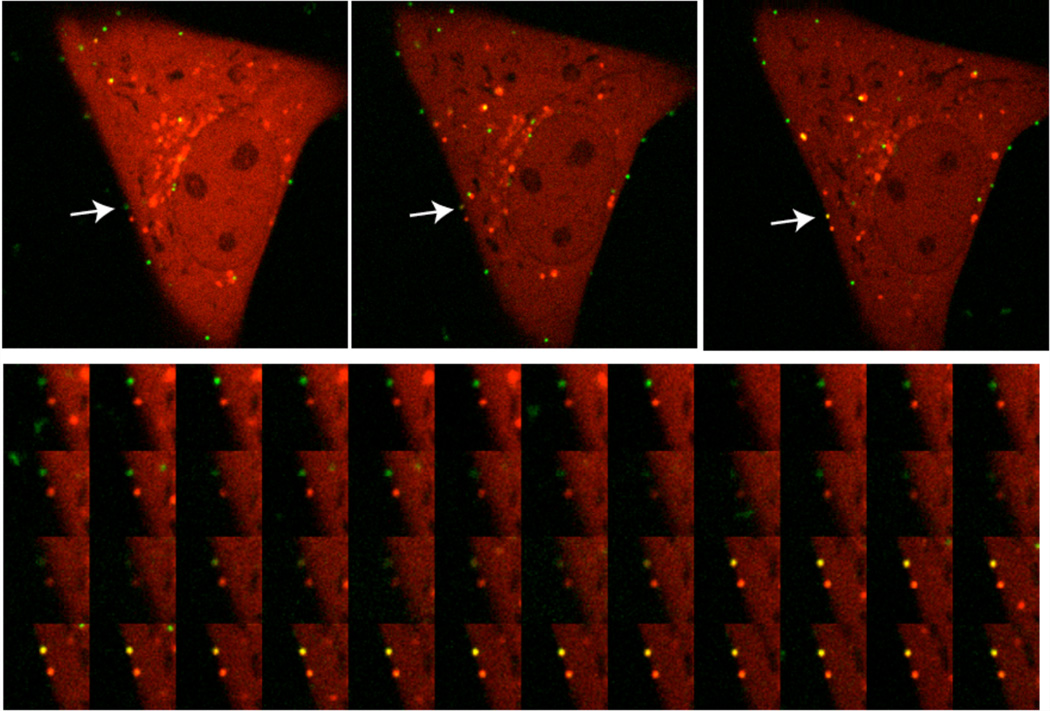
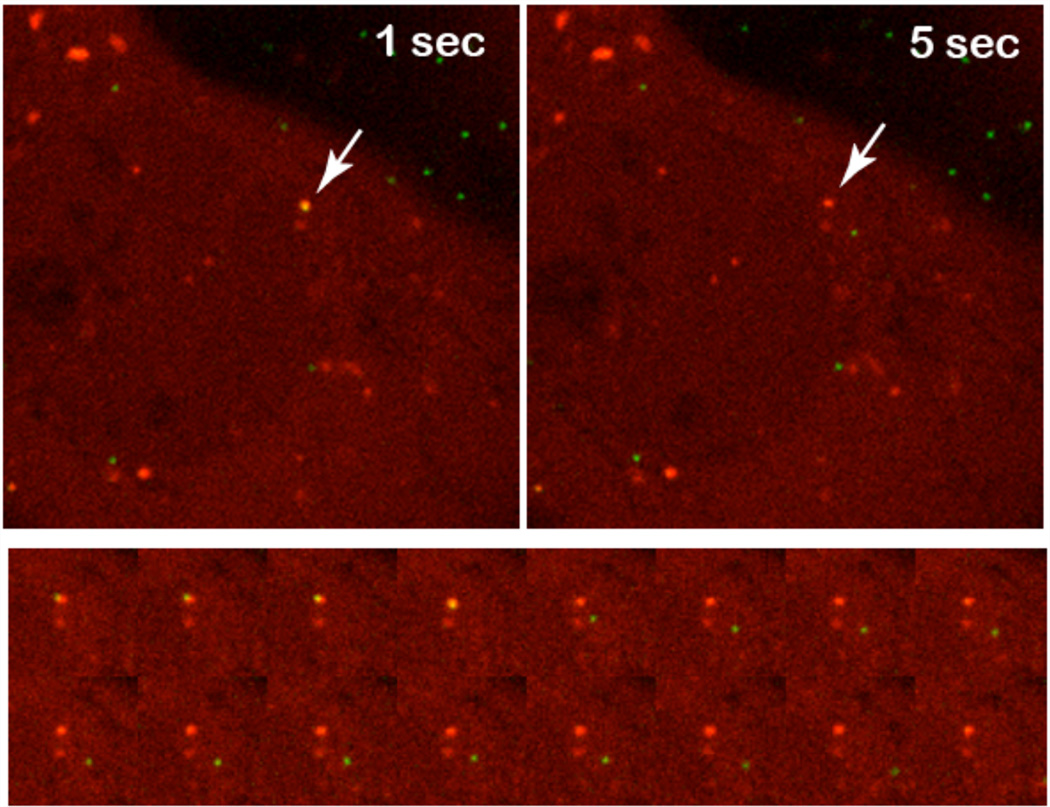
Figure 4. Adenovirus causing membrane lysis.
Top row) Overview of Alexa488 labeled adenoviruses (green signal) bound to the cell surface of a single U2OS-FRT-Gal-3-mCherry cell (red signal). From left to right the arrow points at an individual particle that acquires Gal-3 stain. Bottom row) Detailed image series of the acqusition of Gal-3 by an Alexa488 labeled particle. Note that the acquisition of the Gal-3 coat becomes visible as yellow color from the overlay of both signals.
3.2.4 Imaging membrane damage
The time delay between virus/vector addition and first cell surface binding is approximately 5 minutes. This is enough time to transfer the cells with the virus to the microscope and adjust focus and laser intensities. The membrane lysis through adenoviruses occurs at or near the cell surface but seldom immediately after binding. To image membrane lysis as shown in Figure 4 the focus should be adjusted to an area of the cell surface with several bound viruses. Membrane lysis can be seen by the appearance of Gal-3 positive signal at the site of the virus attachment (Figure 4, Note 8). This recruitment of Gal-3 occurs rapidly, in under a second. Thus image acquisition must occur with high temporal resolution (e.g. >5 fps)
3.2.5 Imaging escape from damaged membranes
Following membrane lysis transport of virus/vector particles in Gal-3 positive vesicles from the site of lysis can be observed. In most cases post-lysis transport is initiated by short rapid local movements followed by long range movements and/or separation of the virus/vector from the Gal-3 positive structure as shown in Figure 5. To image endosomal escape events it is best to focus on the perinuclear region because most escape events occur in nuclear proximity [12]. Adjusting the focal plane during acquisition can be helpful because the relative mobility of the Gal-3 positive particle containing structures and the thickness of the cell makes capturing escape events more difficult. Rapid image acquisition at >5 fps is essential to capture the event in detail (Note 9).
Figure 5. Adenovirus escaping from Gal-3 positive vesicular structures.
Top row) Detail of a single U2OS-FRT-Gal-3-mCherry cell. The arrow points to an Alexa488 labeled particle before (left) and after (right) escaping from the inside of a Gal-3 positive vesicular structure. Bottom row) Detailed image series of the endosomal escape process. Note that separation of the combined yellow signal into the green signal for the escaping particle and the red signal for the empty vesicular structure left behind.
3.2.6 Controls
The assay can be controlled by using the adenovirus temperature sensitive mutant Ad2ts (ts1)[17, 18]. The ts1 particles can be produced, labeled and used as decribed in this protocol. When grown at the permissive temperature of 33°C this virus behaves like wildtype viruses/vectors permitting the visualization of membrane lysis and escape events as described above. When grown at the non-permissive temperature above 38.5°C the ts1 mutation located in the viral protease results in the production of hyperstable particles due to unprocessed capsid proteins. When used with the Gal-3 expressing U2OS cells these particles enter cells. Because they lack subsequent disassembly steps and protein VI release they do not acquire Gal3 and end up in structures presumed to be lysosomes.
Acknowledgements
Part of this work was supported by Equipe FRM 2011 Projet DEQ 20110421299 (HW). CW acknowledges funding from the NIH (AI082430) and American Heart Association (2261306). AMB acknowledges support from the NIH (AI007508). We acknowledge the Bordeaux imaging centre (BIC) for help setting up the live cell imaging acquisition. HW is an INSERM fellow.
Footnotes
Several excellent methods for CsCl purification of Ads or Ad derived vectors have been published [19–21]. Some of these protocols use TRIS based buffers for purification. Because most labeling reactions are based on primary amines TRIS has to be eliminated from purified virus stocks prior to the labeling reaction. We dialyze purified viruses into PBS/10% Glycerol for long term storage at −80°C. The purity of CsCl purified virus should be confirmed by SDS-PAGE. Since contaminating proteins will likely be labeled as well, these contaminants could influence the background staining and may lead to misinterpretations of results.
Although the kit is designed to label antibodies and small scale proteins preparations we have used this protocol successfully to label Ads and Ad derived vectors with the protein microscale labeling kits for Alexa Fluor® 488, Alexa Fluor® 555, Alexa Fluor® 494 and Alexa Fluor® 647. However, in the literature several examples exist where groups have successfully used dyes from other suppliers to label Ads [4, 11]. In such cases, it is advised that the labeling be optimized to maximize fluorescence signal and minimize loss of virus infectivity upon labeling.
The kit contains a limited amount of columns and purification matrix. Because we split the labeling reaction we use the alternative described in the protocol. However, dye separation should be feasible using several size exclusion columns from many suppliers. The labeling reaction can also be terminated by the addition of TRIS to 40 mM to quench not reacted dye. However, in our hands efficient dye removal is sufficient.
The physical particle (pp/µl) to infectious particles per µL (multiplicity of infection; MOI) ratio should always be determined for the parental virus preparation. The reason for using different MOI’s is to obtain ~ 10–100 plaques per well at a given MOI for reliable quantification.
We prefer an environmental system that surrounds and heats up the whole microscopic setup to avoid thermal aberrations in the optical system. In our experience heated stages alone can result in focus fluctuations during acquisition. Alternatives such as infrared autofocus systems to keep the focus can be used.
Depending on the application, cell densities can be varied. The conditions provided in the protocol are optimized for single cell imaging. We sometimes also use medium with reduced serum (Opti-MEM®) to prevent cells from fast growing or to keep the slide for the next day (e.g. when not all channels have been used in one experiment).
This step is crucial because of the involved safety issues. Tubing and syringes can easily be obtained from any hospital (e.g. ask for a “butterfly” blood drawing system). To reduce the risk of contaminating the microscope set-up we use i) viral vectors instead of replication competent viruses and ii) fill the channels physically separated from the microscope hardware so that the slide can be covered with the lid before imaging. However, the syringe setup as described in the protocol can also be conveniently fixed to the microscope allowing to image while adding solutions. The long flexible tubing holds the slide in place while changing the solution. Lastly, always make sure to discard the aspirated virus/vector solution as infectious liquid and rinse the tubing/syringe system with bleach, plenty of water and ethanol before the next use.
We observed membrane lysis almost excusively at the cell surface. Viruses/vectors can sit on the cell surface for prolonged times (> 30 min) without significant movement or any sign of membrane lysis. Even following membrane lysis the virus/vector can remain at the same position for several minutes before it subsequently starts moving to the cell interior. As such imaging Ad induced membrane lysis requires patience. Alternatively we also performed synchronized infections prebinding viruses/vectors in the cold for 30 minutes (see alternative protocol). However we disfavor this approach because the temperature shifts provides focus aberrations making imaging more difficult and the results have proven to be more difficult to analyze (e.g. incubation at 4°C results in microtubule de-polymerisation).
The nature of Gal-3 positive structures that carry virus/vector particles is currently not known. It could consist of damaged or resealed membrane structures. Similar to imaging the membrane lysis event, we have not identified any visual cue to predict when a virus/vector will escape the Gal-3 positive structure. However post-escape the particles engage in fast movements resembling microtubule based transport.
Material transfer:
To obtain the U2OS-FRT-Gal3-mCherry cell line contact the corresponding author
Contributor Information
Ruben Martinez, Email: ruben.martinez@u-bordeaux2.fr, Microbiologie Fondamental et Pathogénicité, MFP CNRS UMR 5234, University of Bordeaux SEGALEN, 146 rue Leo Seignat, 33076 Bordeaux, France.
Andrew M. Burrage, Email: aburrage@lumc.edu, Loyola University Medical Center, Department of Microbiology and Immunology, 2160 S. First Ave, Maywood, IL 60153, USA.
Christopher M. Wiethoff, Email: cwiethoff@lumc.edu, Loyola University Medical Center, Department of Microbiology and Immunology, 2160 S. First Ave, Maywood, IL 60153, USA.
Harald Wodrich, Email: harald.wodrich@u-bordeaux2.fr, Microbiologie Fondamental et Pathogénicité, MFP CNRS UMR 5234, University of Bordeaux SEGALEN, 146 rue Leo Seignat, 33076 Bordeaux, France.
References
- 1.Tsai B. Penetration of nonenveloped viruses into the cytoplasm. Annu Rev Cell Dev Biol. 2007;23:23–43. doi: 10.1146/annurev.cellbio.23.090506.123454. [DOI] [PubMed] [Google Scholar]
- 2.Ginsberg HS, Horswood RL, Chanock RM, Prince GA. Role of early genes in pathogenesis of adenovirus pneumonia. Proc Natl Acad Sci U S A. 1990;87(16):6191–6195. doi: 10.1073/pnas.87.16.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldhamen YA, Seregin SS, Amalfitano A. Immune recognition of gene transfer vectors: focus on adenovirus as a paradigm. Front Immunol. 2011;2:40. doi: 10.3389/fimmu.2011.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burckhardt CJ, Suomalainen M, Schoenenberger P, Boucke K, et al. Drifting motions of the adenovirus receptor CAR and immobile integrins initiate virus uncoating and membrane lytic protein exposure. Cell Host Microbe. 2011;10(2):105–117. doi: 10.1016/j.chom.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Wodrich H, Henaff D, Jammart B, Segura-Morales C, et al. A Capsid-Encoded PPxY-motif Facilitates Adenovirus Entry. PLoS Pathog. 2010;6(3):e1000808. doi: 10.1371/journal.ppat.1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maier O, Galan DL, Wodrich H, Wiethoff CM. An N-terminal domain of adenovirus protein VI fragments membranes by inducing positive membrane curvature. Virology. 402(1):11–19. doi: 10.1016/j.virol.2010.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farr GA, Zhang LG, Tattersall P. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc Natl Acad Sci U S A. 2005;102(47):17148–17153. doi: 10.1073/pnas.0508477102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prchla E, Plank C, Wagner E, Blaas D, et al. Virus-mediated release of endosomal content in vitro: different behavior of adenovirus and rhinovirus serotype 2. J Cell Biol. 1995;131(1):111–123. doi: 10.1083/jcb.131.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGuire KA, Barlan AU, Griffin TM, Wiethoff CM. Adenovirus type 5 rupture of lysosomes leads to cathepsin B-dependent mitochondrial stress and production of reactive oxygen species. J Virol. 2011;85(20):10806–10813. doi: 10.1128/JVI.00675-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barlan AU, Griffin TM, McGuire KA, Wiethoff CM. Adenovirus membrane penetration activates the NLRP3 inflammasome. Journal of Virology. 2011;85(1):146–155. doi: 10.1128/JVI.01265-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barlan AU, Danthi P, Wiethoff CM. Lysosomal localization and mechanism of membrane penetration influence nonenveloped virus activation of the NLRP3 inflammasome. Virology. 2011;412(2):306–314. doi: 10.1016/j.virol.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maier O, Marvin SA, Wodrich H, Campbell EM, et al. Spatiotemporal dynamics of adenovirus membrane rupture and endosomal escape. J Virol. 2012;86(19):10821–10828. doi: 10.1128/JVI.01428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paz I, Sachse M, Dupont N, Mounier J, et al. Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell Microbiol. 2010;12(4):530–544. doi: 10.1111/j.1462-5822.2009.01415.x. [DOI] [PubMed] [Google Scholar]
- 14.Dupont N, Lacas-Gervais S, Bertout J, Paz I, et al. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009;6(2):137–149. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Mittereder N, March KL, Trapnell BC. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol. 1996;70(11):7498–7509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubin RW, Weiss GD. Direct biochemical measurements of microtubule assembly and disassembly in Chinese hamster ovary cells. The effect of intercellular contact, cold, D2O, and N6,O2'-dibutyryl cyclic adenosine monophosphate. J Cell Biol. 1975;64(1):42–53. doi: 10.1083/jcb.64.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greber UF, Webster P, Weber J, Helenius A. The role of the adenovirus protease on virus entry into cells. EMBO J. 1996;15(8):1766–1777. [PMC free article] [PubMed] [Google Scholar]
- 18.Weber J. Genetic analysis of adenovirus type 2 III. Temperature sensitivity of processing viral proteins. J Virol. 1976;17(2):462–471. doi: 10.1128/jvi.17.2.462-471.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jager L, Hausl MA, Rauschhuber C, Wolf NM, et al. A rapid protocol for construction and production of high-capacity adenoviral vectors. Nat Protoc. 2009;4(4):547–564. doi: 10.1038/nprot.2009.4. [DOI] [PubMed] [Google Scholar]
- 20.Tollefson AE, Kuppuswamy M, Shashkova EV, Doronin K, et al. Preparation and titration of CsCl-banded adenovirus stocks. Methods Mol Med. 2007;130:223–235. doi: 10.1385/1-59745-166-5:223. [DOI] [PubMed] [Google Scholar]
- 21.Barry MA, Weaver EA, Hofherr SE. Rescue, amplification, purification, and PEGylation of replication defective first-generation adenoviral vectors. Methods Mol Biol. 2011;651:227–239. doi: 10.1007/978-1-60761-786-0_13. [DOI] [PubMed] [Google Scholar]