Abstract
c-Myc is a well-known oncogene frequently upregulated in different malignancies, whereas liver specific miR-122, a bona fide tumor suppressor, is downregulated in hepatocellular cancer (HCC). Here, we explored the underlying mechanism of reciprocal regulation of these two genes. Real-time RT-PCR and Northern blot analysis demonstrated reduced expression of the primary, precursor and mature miR-122 in c-MYC induced HCCs compared to the benign livers, indicating transcriptional suppression of miR-122 upon MYC overexpression. Indeed, chromatin immunoprecipitation (ChIP) assay showed significantly reduced association of RNA polymerase II and histone H3K9Ac, markers of active chromatin, with the miR-122 promoter in tumors relative to the c-Myc-uninduced livers, indicating transcriptional repression of miR-122 in c-Myc overexpressing tumors. ChIP assay also demonstrated significant increase in c-Myc association with the miR-122 promoter region that harbors a conserved noncanonical c-Myc binding site in tumors compared to the livers. Ectopic expression and knockdown studies showed that c-Myc indeed suppresses expression of primary and mature miR-122 in hepatic cells. Additionally, Hnf-3β, a liver enriched transcription factor that activates miR-122 gene, was suppressed in c-MYC-induced tumors. Notably, miR-122 also repressed c-Myc transcription by targeting transcriptional activator E2f1 and coactivator Tfdp2, as evident from ectopic expression and knockdown studies and luciferase reporter assays in mouse and human hepatic cells. Conclusion: c-Myc represses miR-122 gene expression by associating with its promoter and by downregulating Hnf-3β expression whereas miR-122 indirectly inhibits c-Myc transcription by targeting Tfdp2 and E2f1. In essence, these results suggest a double-negative feedback loop between a tumor suppressor (miR-122) and an oncogene (c-Myc).
Keywords: miR-122 knockout mice, microRNA-122, miR-122, c-Myc, Tfdp2, Pkm2, Iqgap1, Mapre1, LETF, HNF-3b
Introduction
microRNAs (miRNAs) are a class of small (~22nt), non-coding RNAs that post-transcriptionally repress target gene expression by pairing with mRNAs of protein coding genes, mainly in the 3’ untranslated regions (UTR) (1, 2). Recent studies have shown that miRNAs may also regulate gene expression through interaction with coding region or 5’UTR of target genes, as demonstrated by transcriptome-wide identification of miRNA target sites (3). Over the past few years, many studies have proved that miRNAs play important physiological role in almost every aspect of biological processes, including development and differentiation, immune response, metabolism, cell proliferation and apoptosis (4). Thus, dysregulation of some miRNAs is involved in the pathogenesis of a variety of diseases, such as vascular diseases, immunological diseases, neurological disorders, and cancers (5). Aberrations in miRNA expression have been attributed to several mechanisms, including amplification, deletion or mutation of miRNA genes, altered transcriptional regulation of miRNA genes, or epigenetic regulation, such as DNA methylation (6).
miR-122 is the most abundant liver specific microRNA that plays fundamental roles in liver (7). It has been shown to regulate cholesterol metabolism (8), hepatitis C virus replication (9). We have previously demonstrated that it is suppressed in diet induced liver cancer in rat (10) and mouse (11). Several investigators including us also showed that miR-122 is significantly repressed in hepatocellular carcinoma (HCC) patients and in HCC cell lines (12, 13). Moreover, some studies have illustrated correlation between reduced expression of miR-122 and metastasis and poor prognosis, higher tumor burden and gene expression signature of aggressive tumors in HCC patients (14, 15). Recently, we have generated liver specific knockout and germ-line miR-122 knockout mice (16), which develop steatohepatitis, fibrosis and HCC with age, further reinforcing the important physiological role and intrinsic tumor suppressor function of miR-122 in liver. Although miR-122 expression is reduced in HCCs, the mechanism of this downregulation is still unknown. Several studies have reported regulation of miR-122 expression by liver-enriched transcription factors (LETFs) during liver development and hepatocyte differentiation (17–19). Whether these LETFs are involved in miR-122 suppression in liver cancer remains to be established.
The Myc proto-oncogene encodes c-Myc transcription factor that is frequently upregulated in a variety of human cancers (20), including liver cancer. As a transcription factor, c-Myc dimerizes with Max, binds to E boxes in the promoter region of target genes and activates transcription of target genes involved in cell growth and proliferation (20). Activation of c-Myc can initiate tumorigenesis as documented in several c-Myc transgenic mouse models (21, 22). For example, tet-o-MYC; LAP-tTA bi-transgenic mice harboring a tetracycline (tet)-repressible MYC transgene (tet-o-MYC) and a transgene that produces the tet-transactivator protein (tTA) driven by the liver activator promoter (LAP) develop HCC within a few weeks after c-Myc induction (21, 23). In addition to transactivation of target gene expression, c-Myc is also known to repress some gene expression by mechanisms that may involve interaction with other transcription factors, such as Myc-interacting zinc finger protein 1 (Miz-1) (24). Interestingly, activation of c-Myc results in widespread miRNA repression by directly binding to miRNA promoter region, which facilitates tumorigenesis (25). Although c-Myc has been demonstrated to repress the expression of several miRNAs in liver cancer such as miR-100, let-7a, miR-26a and miR-125b (26), there is no study investigating whether c-Myc can regulate miR-122 expression, the most abundant and frequently suppressed miRNA in liver cancer.
Previously, we have observed dramatic repression of miR-122 in liver tumors formed by induction of c-Myc in tet-o-MYC; LAP-tTA bi-transgenic mice following doxycycline withdrawal, indicating that c-Myc may inhibit miR-122 expression in liver cancer (16). In contrast, hepatic c-Myc is significantly upregulated in miR-122 knockout mice (16), suggesting a double-negative feedback loop between miR-122 and c-Myc. In this study, we investigated the possible mechanisms underlying the inverse regulation between miR-122 and c-Myc.
Materials and Methods
Animals
miR-122 knockout mice and tet-o-MYC; LAP-tTA bi-transgenic mice were described previously (16). LAP-tTA mice were obtained from Jackson Laboratory. All animals were housed in a temperature-controlled room under a 12 h light/12 h dark cycle and under pathogen free conditions. All animal studies were reviewed and approved by the Ohio State University Institutional Laboratory Animal Care and Use Committee.
Hepatocyte isolation
Hepatocytes were isolated as described previously (27). Briefly, miR-122 knockout or wild type mice (20–30gms) were anesthetized with ketamine and xylazine injected intraperitoneally (i.p.). Livers were perfused with 25ml perfusion buffer (5ml/min) and then with 50 ml of warm (37°C) liver digestion buffer via the portal vein. The livers were aseptically removed to a sterile Petri dish containing DMEM at 4°C to stop digestion. The hepatocytes were released by peeling off hepatic capsule and dispersed by shaking the digested liver in DMEM medium at 4°C, followed by passing through 70 micron strainer and collected by centrifugation at 1000×g at 4°C. The cells were resuspended in William E medium supplemented with 10% serum. The hepatocytes were counted and viability was determined by trypan blue dye exclusion. The cells were plated on 6-well plate coated with rat tail type I collagen (BD Bioscience) at a density of 1×106 cells per well. Next day cells were transfected as described below.
Cell lines and transfection
Hepa (mouse hepatoma) and human HCC (Huh-7, Hep3B, PLC/PRF5) cell lines were obtained from ATCC and cultured as recommended by the supplier. For miR-122 overexpression and knockdown experiments, these cells and mouse hepatocytes were transfected with miR-122 or control miR (NC) mimic (50nM) (Thermo Scientific), and anti-miR-122 or anti-miR control (anti-NC) (100nM) (Thermo Scientific), respectively using Lipofectamine 2000 (Invitrogen) following manufacturer’s protocol. For gene knockdown experiments, Hepa cells and mouse hepatocytes were transfected with control (siNC: D-001206-13-05, Thermo Scientific) or gene specific siRNAs (60nM) (si-Myc: M-040813-02, si-Tfdp2: M-057863-01, si-E2f1: M-044993-03, Thermo Scientific).
RNA isolation and Northern blot analysis
The total liver and tumor RNA was isolated using TRIzol and subjected to Northern blot analysis using 32P-labeled anti-miR-122 or anti-5S-rRNA oligo as described before (10).
Real-time reverse-transcription polymerase chain reaction (qRT-PCR)
The TaqMan miRNA Assay (Applied Biosystems) was used to quantify mature and primary miR-122 expression in total liver and tumor RNA according to manufacturer's instructions. Normalization was performed with RNU6B and 18S rRNA, respectively. For gene expression assay, DNase I treated total RNA was reverse transcribed into cDNA using high capacity cDNA reverse transcription kit (Applied Biosystems) and real-time PCR was performed using SYBR Green chemistry. The expression was normalized to that of Gapdh. All real-time reactions, including controls without cDNA, were run in triplicate in a thermocycler. Relative expression was calculated using the comparative CT method. Primer sequences are provided in the Supplemental Information.
Western blot analysis
Whole cell or tissue extracts were prepared in SDS lysis buffer followed by immunoblotting with specific antibodies. The signal was developed with ECL reagent (ThermoFisher) after incubation with appropriate secondary antibodies. Western blot signals were quantified by ImageJ software (NIH) following online manual. The antibodies used are: c-Myc: sc-40X, Tfdp2: sc-1209, E2f1: sc-193, Iqgap1: sc-10792, Mapre1: sc-15347, Hnf-1: sc-6548), Hnf-1β: sc-22840, Hnf-3α: sc-22841, Hnf-3β: sc-6554, Hnf-4α: sc-8987, Hnf6: sc-13050, Pkm2: cs-3198, histone H3: ab1791 and c-MYC (human specific): cs-5605.
Plasmids construction and luciferase assay
Wild type (WT) or mutant 3’UTR (deletion of miR-122 targeting sites) of Tfdp2 and E2f1 were cloned into psiCHECK2 (Promega) luciferase reporter vector downstream of renilla luciferase coding region. For luciferase assay, HCC cells were cotransfected with 200 ng of each luciferase reporter construct and miR-122 or negative control (NC) mimic (50nM) (ThermoScientific). After 48h, luciferase activity was measured using Dual-Luciferase Reporter Assay kit (Promega) and renilla luciferase activity was normalized to that of firefly luciferase.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay in c-Myc induced tumors and control livers were performed as described (28). Briefly, liver and tumor tissues were homogenized in 10 volumes of 2mM DSG (0.5M stock in DMSO) in PBS, incubated at room temperature for 10 min and filtered through ~70um cell strainer. The cells were resuspended in 1% formaldehyde in PBS and cross-linked at room temperature for 10 min, which was stopped by adding glycine (0.125M). The cells were washed twice with cold PBS and resuspended in ice-cold cell lysis buffer (150mM NaCl, 50mM Tris.HCl pH 7.5, 5mM EDTA, 0.5% NP-40, 1% Triton X-100, 1× complete EDTA free protease inhibitor (Roche, 25X stock)). The nuclei were washed with cell lysis buffer and resuspended in nuclear lysis buffer (1% SDS, 5mM EDTA, 50mM Tris.HCl pH 8.1, protease inhibitor). The chromatin was sonicated to 300–500bp, followed by standard ChIP analysis with the following antibodies, c-Myc: sc-40x, Pol II (sc-899X), AcH3K9 (cs-9649) and H3Me3K9 (ab8898). Primer sequences are provided in the Supplemental Information.
Statistical analysis
qRT-PCR and transfection analysis was performed in triplicate. The data presented as means ± standard deviation (SD). Most of the experiments were repeated twice. Statistical significance was calculated with a Student’s t test with a P value of <0.05 considered significant. In the figures, P values ≤0.05 and ≤0.01 are represented as * and **, respectively.
Results
miR-122 expression was suppressed whereas some of its targets were upregulated in c-MYC induced liver tumors
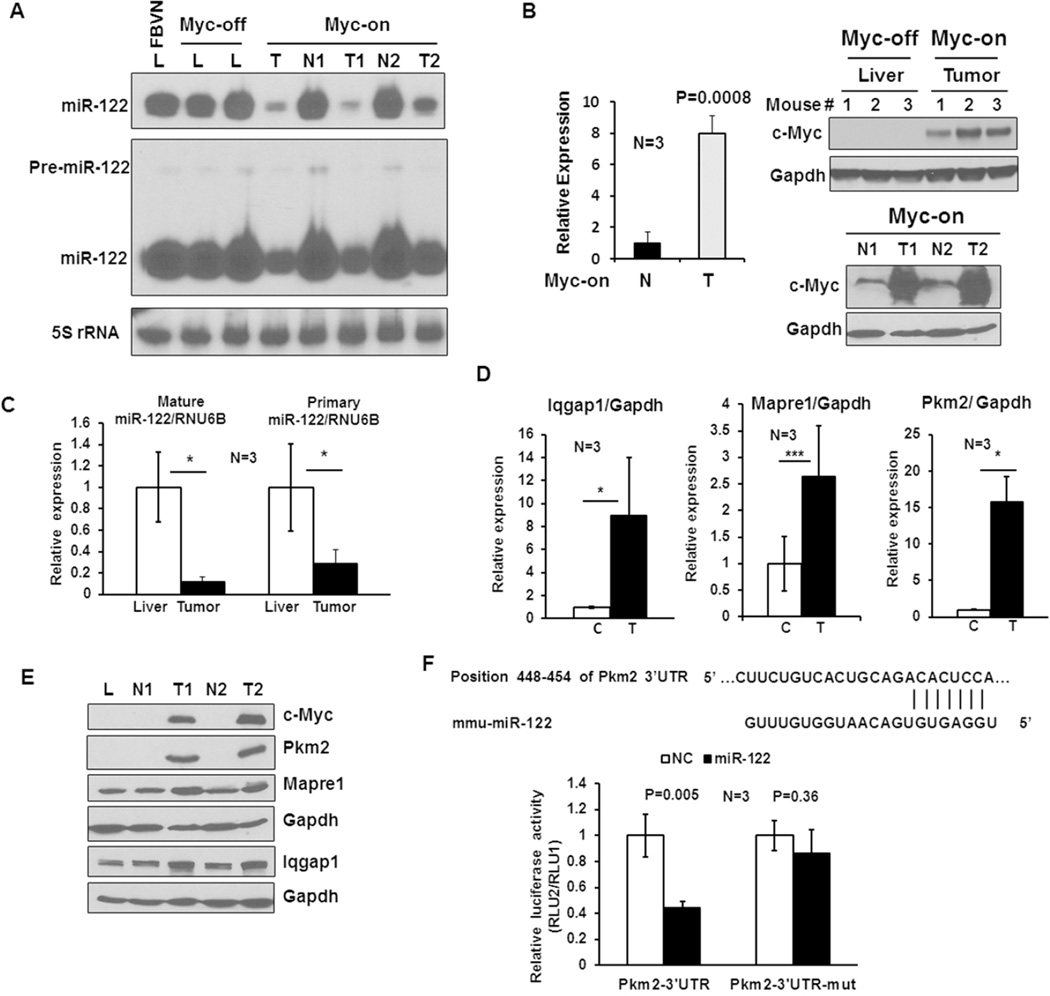
Bi-transgenic Tet-o-MYC; LAP-tTA mice develop tumors after withdrawal of doxycycline from the diet that initiates liver tumorigenesis due to induction of human MYC transgene, and all liver lobes are progressively transformed with numerous tumors within a few weeks (Supplemental Fig. 1). To determine whether miR-122 is downregulated upon c-MYC induction, tumors and benign livers were removed at early stages of tumor development after withdrawal of doxycycline after 3 weeks and its level was analyzed by Northern blotting which exhibited dramatic decrease in miR-122 level in tumors but not in benign liver tissues compared to c-MYC-uninduced (Myc off) and the parental mouse (FVBN) livers (Fig. 1A). Since miR-122 level in the benign livers was not altered, we measured ectopic MYC level in these tissues by qRT-PCR and immunoblotting with an antibody that specifically detects human c-MYC protein. The results showed that c-MYC RNA level was upregulated ~8 fold whereas protein level was increased more than 12 fold in tumors compared to the benign livers (Fig. 1B), suggesting stabilization of the protein in tumors. These results indicate that miR-122 expression is specifically suppressed in tumors expressing high levels of c-Myc (Fig. 1A). Precursor miR-122 (pre-miR-122) that could be detected after longer exposure of the Northern blot because of its very short half-life, was detectable in benign livers but was barely detectable in tumors (Fig. 1A), suggesting that the Dicer-mediated processing of pre-miR-122 does not play any major role in reducing mature miR-122 level in tumors. qRT-PCR analysis showed profound decrease (~70–90%) in both primary and mature miR-122 in tumors compared to livers (Fig. 1C), indicating that miR-122 was downregulated primarily due to transcriptional repression upon c-MYC induction. Although decrease in mature miR was more than that of pri-miR-122, it was not significant.
Figure 1. miR-122 is downregulated in c-Myc-induced liver tumors.
A. Northern blot analysis of precursor and mature miR-122 levels in the normal liver (L) from FBVN and (Myc-off) mice, as well as benign liver (N) and tumor (T) from the same (Myc-on) mouse. Total RNA (25 µg) from each sample was separated by denaturing PAGE (15% acrylamide), transferred onto Nylon membrane, cross-linked and subjected to Northern blot analysis with 32P-labeled anti-miR-122 deoxyoligonucleotide. Membrane was exposed to 4hr and 72hr at −80°C to detect mature and precursor miR-122, respectively. The blot was hybridized to anti-sense 5S rRNA probe to demonstrate equal loading of RNA. B. qRT-PCR and Western blot analysis of c-Myc expression in the liver (Myc-off), Myc-on benign liver (N) and tumor (T) tissues in the presence and absence of doxycycline, respectively. Whole tissue extracts (100 µg protein) were immunoblotted with an antibody specific for human c-Myc and reprobed with Gapdh antibody. C. qRT-PCR analysis of primary and mature miR-122 in Myc-off livers and Myc-on tumors using Taqman kit. Data was normalized to RNU6B. D. qRT-PCR analysis of mRNA levels of specified miR-122 targets in livers and tumors. C: control liver, T: tumor. E. Western blot analysis of selected miR-122 targets in liver and tumor whole tissue extracts (100 µg). L: FBVN liver, N: benign liver and T: Tumor. F. Luciferase reporter assay demonstrating Pkm2 is a target of miR-122. Upper panel: miR-122 cognate site in 3’-UTR of Pkm2 gene. Lower panel: relative luciferase activity in Hepa cells transfected with the psiCHECK2 harboring wild type (Pkm2-3’-UTR) or mutant (Pkm2-3’-UTR-mut) site along with miR-122 mimic (miR-122) or negative control RNA (NC).
To investigate the consequence of miR-122 suppression in c-Myc tumors, we searched the microarray data available from GEO database (GSE28198) (29) for miR-122 targets, which showed upregulation of several known targets such as Adam10, Agpat1, Ccng1, Ndrg3, Aldoa, Iqgap1 and Mapre1 (16). Among these, Iqgap1 and Mapre1 were significantly upregulated at the RNA and protein levels in the tumors compared to benign liver tissues (Fig. 1D and E). Notably, Pkm2, a predicted target of miR-122 upregulated in many cancer including HCC (30), was also highly elevated in these tumors both at the RNA and protein level (Fig. 1D and E). To substantiate that the mouse Pkm2 is a direct miR-122 target, we performed luciferase reporter assay. This study demonstrated that ectopic miR-122 inhibited mouse Pkm2 3’-UTR driven luciferase activity by 60%, which could be reversed by deletion of miR-122 seed match from the 3’-UTR (Fig. 1F). Collectively, these data showed that miR-122 is transcriptionally suppressed in c-Myc induced tumors correlating with upregulation of its selected targets.
c-Myc direct binds to miR-122 immediate upstream promoter region and also suppresses Hnf-3β level
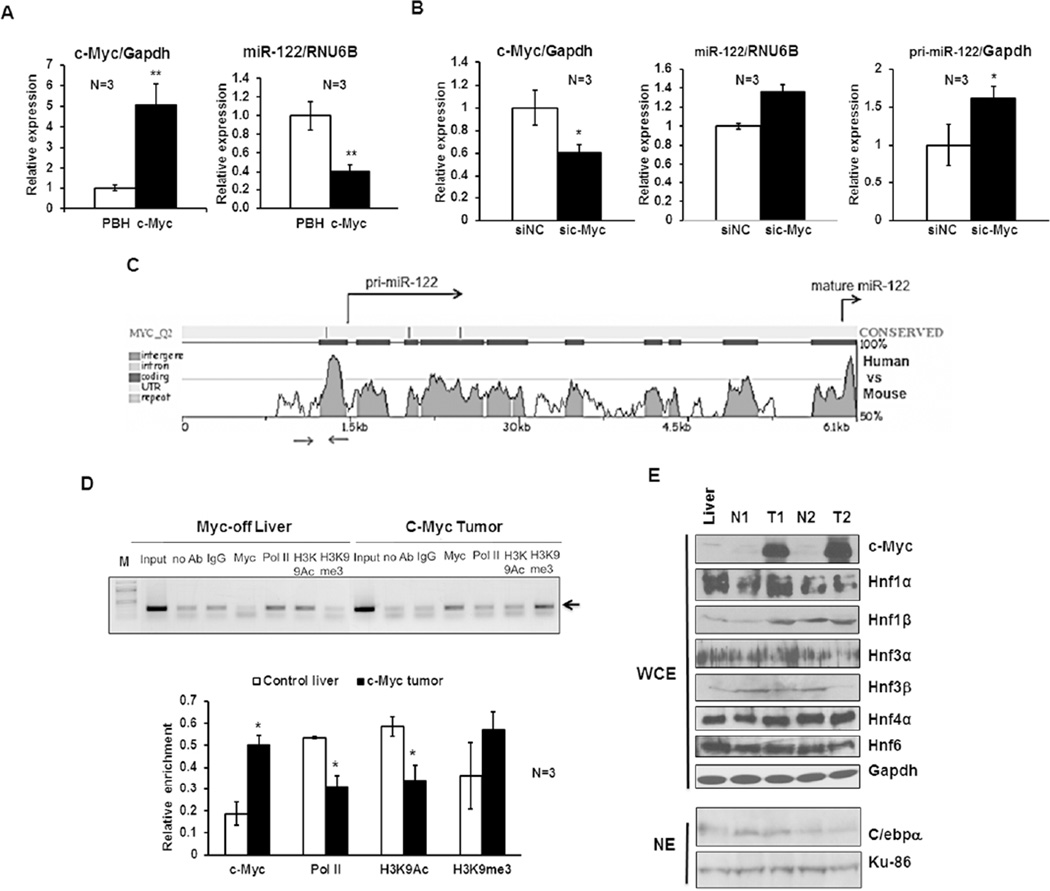
Next we sought to elucidate the mechanism by which miR-122 was repressed in c-Myc induced tumors. To this end, we first examined if c-Myc could inhibit miR-122 expression in vitro. Indeed, overexpression of c-Myc in mouse Hepa cells resulted in ~60% decrease in miR-122 level compared to vector transfected cells (Fig. 2A). In contrast, siRNA mediated depletion of c-Myc significantly increased both mature and primary miR-122 expression in these cells (Fig. 2B). These results suggested that c-Myc could negatively regulate miR-122 expression at transcriptional level.
Figure 2. c-Myc represses miR-122 expression in vitro.
A. Relative c-Myc and mature miR-122 expression in Hepa cells transfected with MYC expression vector (pBH-c-Myc) or pBH (vector control). Hepa cells were transfected with specified plasmids and RNAs isolated 36–48 hr post-transfection were subjected to qRT-PCR analysis. The data was normalized to Gapdh or RNU6B. Relative level in vector-transfected cells was assigned a value of 1. B. Relative c-Myc, mature and primary miR-122 expression in Hepa cells transfected with c-Myc siRNA (sic-Myc) or negative control siRNA (siNC). Cellular RNAs isolated after 36–48 hr were analyzed by qRT-PCR. C. Conserved miR-122 promoter region upstream of +1 site harbors a candidate noncanonical c-Myc binding site predicted by rVista program. Sequence conservation of miR-122 gene between mouse and human as analyzed by rVista. Arrows denote the chromatin immunoprecipitation (ChIP) analysis primers spanning c-Myc site. D. ChIP analysis showed increased binding of c-Myc but reduced association of RNA polymerase II (Pol II) and histone H3K9Ac (a marker of active gene) with miR-122 promoter region in c-Myc induced tumors compared to Myc-off livers. Equal amounts of chromatin (DNA) from livers and tumors were subjected to ChIP assay with specific antibodies. Rabbit IgG and protein G beads alone were used as negative controls. 1:100 dilution of input was used for amplification of miR-122 promoter. Upper panel: A representative photograph of ethidium bromide stained agarose gel containing PCR products obtained with specified antibodies in ChIP assay. Lower panel: Quantitative analysis of ChIP data from 3 different livers and tumors. E. Western blot analysis of c-Myc, LETFs and C/ebpα expression in whole cell extracts (WCE) or nuclear extracts (NE) of Myc-off liver, as well as benign liver (N) and tumor (T) from Myc-on.
To gain further insight into the mechanism of suppression, we investigate if c-Myc inhibits miR-122 expression by directly interacting with its promoter. Indeed, searching of miR-122 promoter region for evolutionarily conserved transcription factor binding sites using rVista program (http://rvista.dcode.org) identified one conserved noncanonical c-Myc binding site in its immediate upstream promoter and two downstream of +1 site (Fig. 2C). To test if c-Myc could bind to this region, chromatin immunoprecipitaiton (ChIP) analysis was performed in c-Myc induced tumor and control liver tissues using an antibody that precipitates both human and mouse c-Myc (see Methods for details). Results showed that the association of c-Myc at miR-122 promoter region was ~2.7-fold higher (P=0.02) in tumors compared to control livers (Fig. 2D). Moreover, the association of RNA polymerase II (Pol II) and histone H3K9Ac, a marker of activate chromatin, was reduced significantly in tumors compared to that of control livers, correlating with reduced transcription of primary miR-122 in c-Myc tumors (Fig. 1C).
Previous studies have shown that liver enriched transcription factors (LETFs) Hnf-1α, Hnf-3α, Hnf-3β (15, 18), Hnf-4α (19), Hnf6 (17) as well as C/ebpα (18) are involved in transcriptional activation of miR-122 during liver development and hepatocyte differentiation. It is possible that c-Myc may suppress miR-122 expression indirectly by inhibiting the expression of these LETFs. To test this possibility, we measured expression of these transcription factors in c-Myc induced tumors. Western blot analysis showed that among these transcription factors expression of only Hnf-3β was consistently reduced by at least 50% in both the tumors relative to those in the respective adjacent benign livers (Fig. 2E), suggesting that Hnf-3β downregulation may contribute to suppression of miR-122 in tumors.
Downregulation of c-Myc expression by miR-122
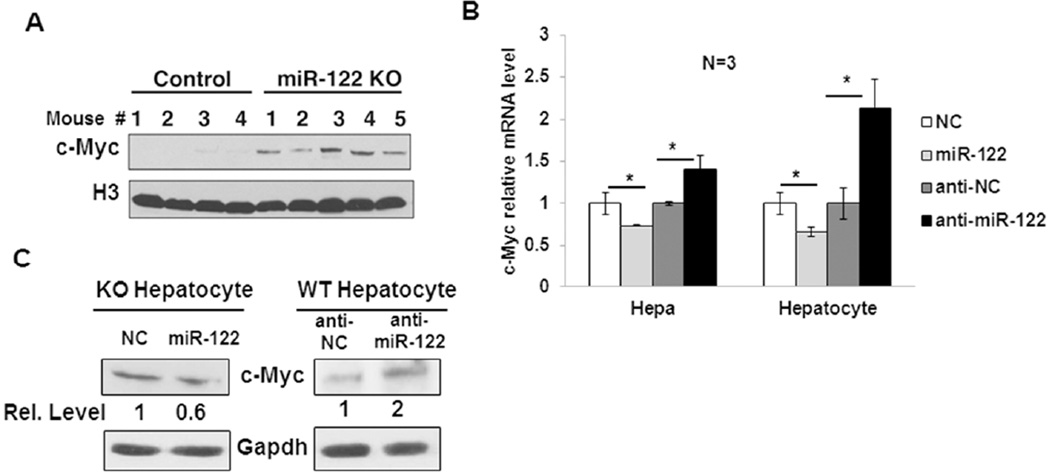
A key observation we made while characterizing miR-122 knockout mice is the upregulation of c-Myc in their livers (16) (Fig. 3A). Since c-Myc is an oncogene with multifarious functions, it was of interest to elucidate how miR-122 modulates its expression. For this purpose, we transiently overexpressed miR-122 in Hepa cells that express miR-122 but at a significantly reduced level relative to the wild type (miR-122+/+) livers (data not shown), and miR-122−/− (KO) hepatocytes by transfecting miR-122 mimic, and measured c-Myc expression. qRT-PCR analysis showed that c-Myc mRNA level was reduced by ~20% and ~40% in miR-122 mimic transfected Hepa and KO hepatocytes, respectively (Fig. 3B). In contrast, c-Myc mRNA level was significantly elevated in Hepa cells and wild type (WT) hepatocytes depleted of miR-122 by transfecting anti-miR-122 oligonucleotide (Fig. 3B). Similarly, overexpression of miR-122 in KO hepatocytes reduced c-Myc protein level (~45%), while its depletion in WT hepatocytes increased c-Myc protein level (~2 fold) (Fig. 3C), suggesting that miR-122 negatively regulates c-Myc expression in vitro. To examine if c-Myc is a direct target of miR-122, we searched for potential miR-122 binding sites in c-Myc 3’UTR. However, we could not find any miR-122 cognate site in its 3’UTR searching different databases, indicating that miR-122 may repress c-Myc expression by an alternate mechanism.
Figure 3. c-Myc is upregulated in miR-122KO mouse livers.
A. Western blot analysis of c-Myc protein levels in the control and miR-122KO liver nuclear extracts using an antibody that detects both mouse and human Myc proteins. Histone H3 was used as a loading control. B. qRT-PCR analysis of endogenous c-Myc mRNA expression in Hepa and KO hepatocytes transfected with miR-122 mimic (miR122) or negative control mimic (NC) as well as in Hepa cells and wild-type (WT) hepatocytes transfected with miR-122 inhibitor (anti-miR122) or negative control inhibitor (anti-NC). C. Western blot analysis of c-Myc in KO and WT hepatocytes transfected with miR-122 mimic and inhibitor, respectively along with respective negative controls as described in 3B.
miR-122 suppressed c-Myc expression through its targets Tfdp2 and E2f1
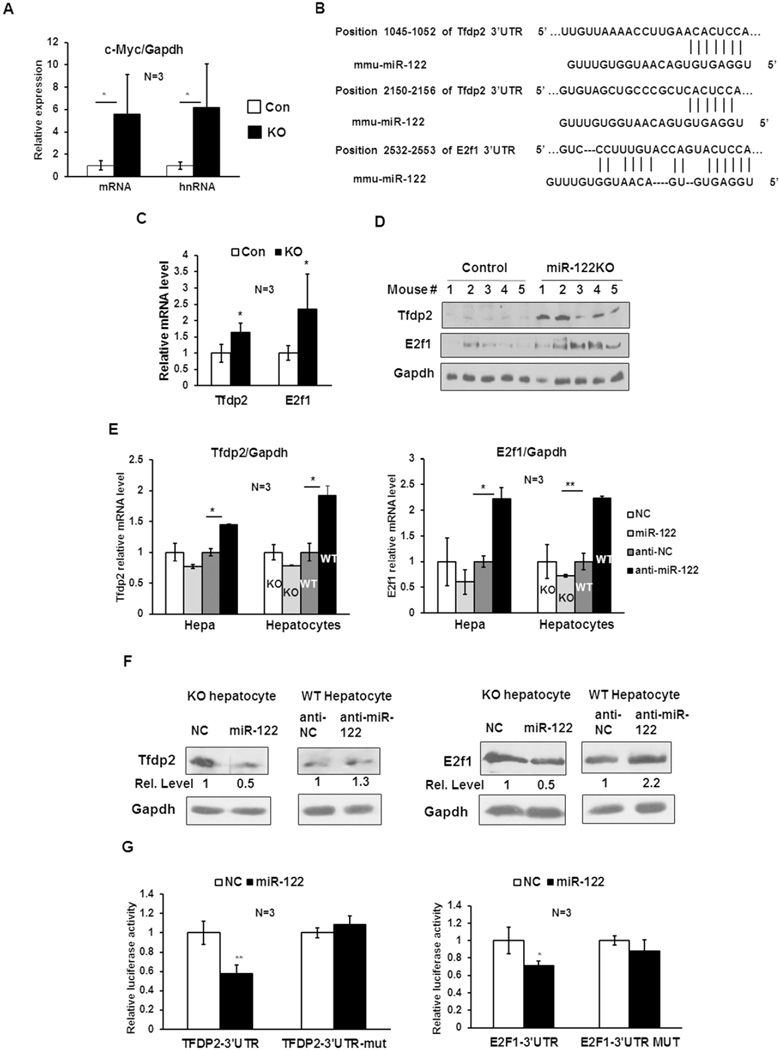
To gain insight into the molecular mechanism of c-Myc repression by miR-122, we first determined whether c-Myc was upregulated in miR-122KO liver at the transcriptional or posttranscriptional level. qRT-PCR analysis showed that hepatic c-Myc mRNA and hnRNA (primary transcript) levels were augmented at a comparable level in both 5 week old KO mice compared to those in the control (miR-122fl/fl) mice (Fig. 4A), suggesting its transcriptional regulation by miR-122. Previous studies have shown that E2f family of transcription factors together with transcription factor dimerization partner (TFDP) form a complex and trans-activate c-Myc promoter through E2f binding sites in the promoter region of c-Myc (31, 32). Tfdp2, a member of TFDP family, is predicted as a conserved miR-122 target by several databases including TargetScan (Fig. 4B). Interestingly, RNA22 program also predicted one miR-122 targeting site in E2f1 3’UTR (Fig. 4B). Based on these observations, we hypothesized that miR-122 may negatively regulate c-Myc gene expression through targeting Tfdp2 and E2f1. To this end, we first examined Tfdp2 and E2f1 expression in miR-122KO livers. qRT-PCR revealed ~1.65 and ~2.35 fold rise in Tfdp2 and E2f1 mRNA level in KO livers compared to controls (Fig. 4C). Similarly, their protein levels were increased in KO livers compared to controls (Fig. 4D). Furthermore, in miR-122 mimic transfected Hepa cells and KO hepatocytes, Tfdp2 and E2f1 mRNA level was reduced by ~30% and ~40%, respectively (Fig. 4E). In contrast, upregulation of their expression in miR-122 depleted Hepa cells and WT hepatocytes was more robust and significant (Fig. 4E). Western blot analysis confirmed that overexpression of miR-122 in KO hepatocytes decreased Tfdp2 and E2f1 protein levels by ~50%, whereas knockdown miR-122 in WT hepatocytes increased respective protein levels by ~1.3 and ~2.2 fold (Fig. 4F). Taken together, these data imply that miR-122 negatively regulates Tfdp2 and E2f1 expression.
Figure 4. Tfdp2 and E2f1, targets of miR-122, are involved in the regulation of c-Myc expression by miR-122.
A. qRT-PCR analysis of c-Myc mRNA and hnRNA (primary transcript) levels in control (floxed) and miR-122KO (KO) mouse livers demonstrated comparable increase in both in KO livers. B. Potential miR-122 seed match in Tfdp2 and E2f1 3’UTRs predicted by TargetScan and RNA22 programs, respectively. C. qRT-PCR analysis of Tfdp2 and E2f1 mRNA levels in control and KO mouse livers. D. Western blot analysis of Tfdp2, E2f1 and Gapdh in the whole liver extracts. E. qRT-PCR analysis of Tfdp2 and E2f1 in Hepa cells and KO hepatocytes transfected with miR-122 mimic (miR-122), negative control mimic (NC) as well as in Hepa cells and WT hepatocytes transfected with miR-122 inhibitor (anti-miR-122) or negative control inhibitor (anti-NC). F. Western blot analysis of Tfdp2 and E2f1 in KO and WT hepatocytes transfected with miR-122 mimic or inhibitors as described in 4E. G. Tfdp2 and E2f1 3’UTR driven luciferase assays were performed as described in 1F.
To determine further if Tfdp2 and E2f1 are bona fide targets of miR-122, we cloned their wild type and mutant (deleted of miR-122 targeting sites) 3’UTR into psiCHECK2 dual luciferase vector downstream of renilla cDNA and performed reporter assay. The relative luciferase activities (renilla to firefly) were significantly repressed by miR-122 mimic compared to negative control RNA in both wild type 3’UTR constructs (E2f1 and Tfdp2), while these activities were not affected in the mutant 3’UTR transfected cells (Fig. 4G), suggesting that miR-122 directly represses Tfdp2 and E2f1 expression by interacting with their 3’UTRs.
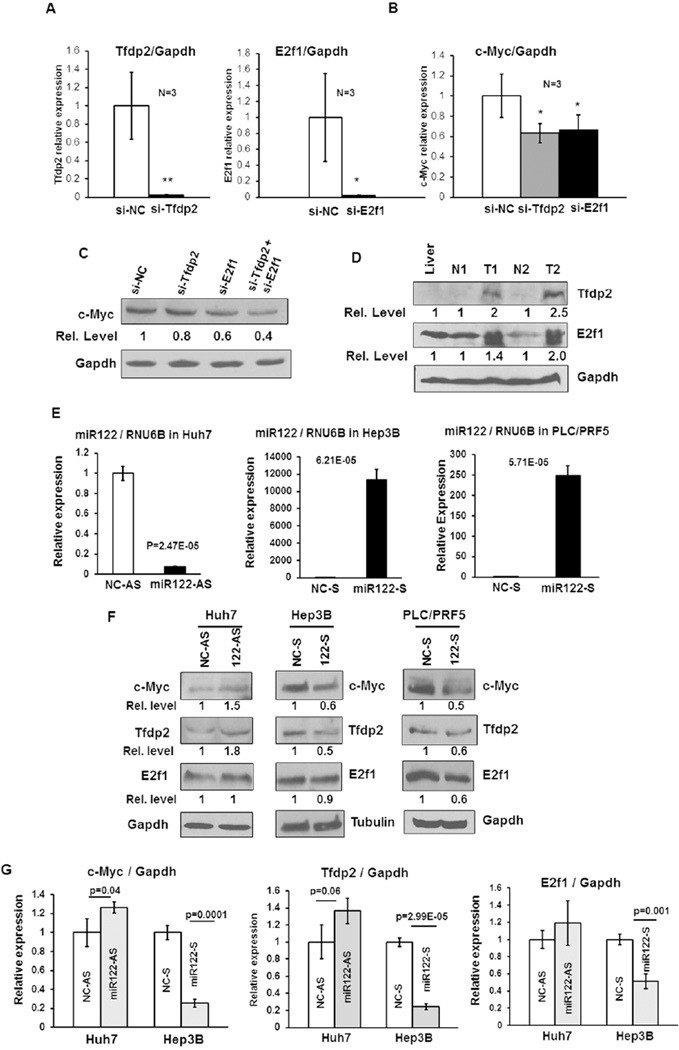
Finally, we explored the possibility that Tfdp2 and E2f1 are involved in the upregulation of hepatic c-Myc expression in KO mice by siRNA mediated knockdown of Tfdp2 and E2f1 in KO hepatocytes followed by measurements of c-Myc mRNA and protein levels. The results showed that indeed c-Myc was downregulated upon depletion of Tfdp2 or E2f1 (Fig. 5A–C), suggesting that they are involved in transcription of c-Myc in KO mouse livers. As expected, both Tfdp2 and E2f1 were upregulated in c-Myc induced tumors compared to benign livers (Fig. 5D). Extension of this study to human hepatic (Huh-7) cells depleted of endogenous miR-122 by transfecting anti-miR-122 oligo (Fig. 5E) showed elevated expression of c-MYC and TFDP2 both at the protein (Fig. 5F) and RNA (Fig. 5G) levels. In contrast, ectopic expression of miR-122 in nonexpressing Hep3B cells resulted in downregulation of these transcription factors (Fig. 5E–G). Interestingly, PLC/PRF5 cells ectopic expression of miR-122 resulted in downregulation of all three factors at the protein level (Fig. 5E and F).
Figure 5. Tfdp2 and E2f1 are involved in the regulation of c-Myc expression in miR-122KO mouse livers.
A. qRT-PCR analysis of Tfdp2 and E2f1 in respective siRNA or negative control siRNA (siNC) transfected KO hepatocytes. B. qRT-PCR analysis of c-Myc in KO hepatocytes transfected with siRNA specific for Tfdp2, E2f1 or negative control siRNA (siNC). C. Western blot analysis of c-Myc in Tfdp2 and E2f1 siRNA transfected KO hepatocytes. D. Western blot analysis of E2f1 and Tfdp2 in c-Myc-induced tumors (T), benign (N) and control livers. E, F. qRT-PCR analysis of miR-122 (E) and western blot analysis of c-MYC, E2F1 and TFDP2 (F) in Huh7 cells transfected with 50 nM of anti-miR-122 (122-AS) or anti-sense NC-RNA (NC-AS) and Hep3B and PLC/PRF5 cells transfected with 50nM miR-122 mimic (122-S) or sense NC-RNA (NC-S). G. qRT-PCR analysis of c-MYC, E2F1 and TFDP2 in Huh7 and Hep3B cells.
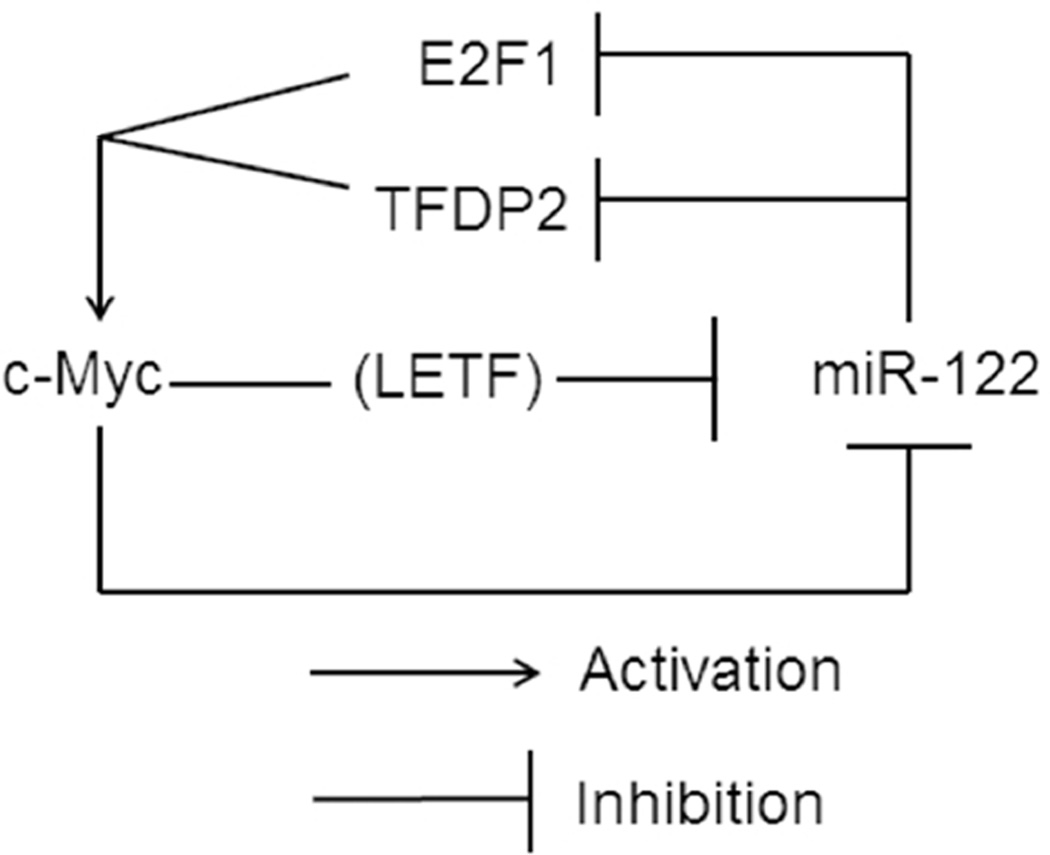
Based on all these data we propose a model delineating reciprocal regulation of miR-122 and c-Myc in the liver and tumor (Fig. 6). Intriguingly, the regulation of both appears to occur at the transcriptional level; c-Myc overexpression suppresses miR-122 by directly binding to its promoter and indirectly, by downregulating liver enriched transcription factor, Hnf-3β. In contrast, miR-122 indirectly suppresses c-Myc expression by targeting the activator (E2f1) and co-activator (Tfdp2) of c-Myc gene.
Figure 6. A model depicting reciprocal regulation between c-Myc and miR-122 based on the observations in miR-122KO and c-MYC transgenic mouse models.
Discussion
It is now well established that miR-122 functions as a tumor suppressor in the liver and its downregulation is a characteristic of HCCs with poor prognosis. However, the mechanism of its suppression in hepatocellular cancer is not well understood. In the present study, we have discovered a novel reciprocal regulation between miR-122 and the pleiotropic oncogene c-Myc in HCC. This study originated from two key observations made while characterizing miR-122KO mice generated in our laboratory (16). First, hepatic c-Myc was consistently elevated in young adult miR-122KO mice, which was further upregulated in spontaneous HCCs developed in these mice. Second, while searching for a mouse model of liver cancer to test therapeutic efficacy of miR-122, we found that c-MYC induced liver tumors exhibited dramatic decrease in miR-122 expression. Present study showed that c-MYC induction causes suppression of miR-122 gene transcription in liver tumors that correlated with association of c-MYC with miR-122 immediate upstream promoter region that harbors a conserved noncanonical c-Myc cognate site predicted by rVista, and also by downregulating Hnf3β protein level that activates miR-122 expression (15, 18). Although repression occurs predominantly at the pri-miRNA transcript level, regulation can also occur at the level of mature miR-122 stability in some tumors since it has been reported that addition of a single A residue at the 3’-end of miR-122 by a noncanonical poly(A) polymerase stabilizes miR-122 (33). It would be of interest to explore if there is any regulation at the level of precursor processing or mature miR-122 stability. Notably, several miR-122 targets such as Iqgap1, Mapre1 and Pkm2 with demonstrated role in oncogenesis, were upregulated in c-MYC induced liver tumor. Thus, transcriptional suppression of miR-122 is likely to be one of the mechanisms by which induction of c-MYC promotes HCC development.
c-Myc has been shown to both positively and negatively regulate microRNA expression (34). For example, c-Myc directly activates transcription of the polycistronic miR-17-92 cluster by interacting with canonical E box located in the promoter region. In contrast, Myc represses expression of several miR genes miR-15a/16-1, miR-26a, miR-29 family members, and miR-34a. Interestingly, these downregulated miRs exhibit tumor suppressor function. Although the mechanisms underlying Myc mediated transcriptional activation of target genes is well studied, that of transcriptional repression mediated by c-Myc is poorly understood. c-Myc interacts with its dimerization partner, Max, to promote gene activation upon occupancy of E box. In contrast, interaction of c-Myc with Miz-1 represses expression of certain genes due to displacement of the co-activator p300 (20). It would be of interest to investigate how c-Myc inhibits expression of miR genes including miR-122. Notably, c-Myc represses expression of certain miRs such as let-7 variants by interfering with their processing but not transrepression (35). However, our Northern blot and qRT-PCR data suggest that primarily transcription of miR-122 is affected in c-Myc-induced liver tumors.
c-Myc is a well-known oncogene overexpressed in various neoplasms, including hepatocellular cancer (36, 37). Amplification of c-Myc genomic locus and overexpression of c-Myc have been observed in ~70% of HCV and alcohol-induced HCCs (38) as well as in animal models of HCC (39). Additionally, c-Myc expression can be directly regulated at the post-transcriptional level by microRNAs, such as miR-34, let-7, miR-145 (20). Our study revealed that the suppression of miR-122 in c-MYC induced liver tumors correlated with direct association with the promoter and downregulation of LETF (HNF3β) (Fig. 6), whereas miR-122 can indirectly repress c-Myc expression by targeting E2f1 (in mouse) and its dimerization partner Tfdp2 (both in mouse and human). Thus, downregulation of miR-122 could be a critical mechanism of upregulation of c-Myc in HCC. Interaction of Tfdp2 with E2f1 has been shown to enhance both the DNA binding activity and the transactivation function of the heterodimer. Indeed, knockdown of E2f1 and Tfdp2 exhibited additive effects on c-Myc induction in Hepa cells.
Both miR-122 (16, 40) and c-Myc (29, 41) are major players in hepatic metabolism. Recently, it has been shown that energy metabolism is profoundly altered in c-Myc induced liver tumors compared to benign liver tissues. Since miR-122 gene delivery inhibited c-Myc induced liver tumor growth in mice, it would be of interest to determine whether miR-122 can reverse metabolic profile of these tumors and the metabolic pathways affected in this process.
Supplementary Material
Acknowledgements
We thank Drs. Michael Bishop, Gustavo Leone and Addgene for providing TET-o-Myc mice and c-Myc expression vector, respectively, and Julia Shreve and Corie Klepper for technical assistance.
Grant support: Supported, in part, by grants DK088076 and CA086978 from NIH
Abbreviations
- KO
miR-122 knockout mice
- Pol II
RNA polymerase II
- qRT-PCR
real-time RT-PCR
Footnotes
Disclosures: None
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 3.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filipowicz W, Grosshans H. The liver-specific microRNA miR-122: biology and therapeutic potential. Prog Drug Res. 2011;67:221–238. [PubMed] [Google Scholar]
- 8.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 9.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, Schmittgen TD, et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu SH, Ghoshal K. MicroRNAs in Liver Health and Disease. Curr Pathobiol Rep. 2013;1:53–62. doi: 10.1007/s40139-012-0005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, Lin CD, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 15.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laudadio I, Manfroid I, Achouri Y, Schmidt D, Wilson MD, Cordi S, Thorrez L, et al. A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology. 2012;142:119–129. doi: 10.1053/j.gastro.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, Qu LH. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology. 2010;52:1431–1442. doi: 10.1002/hep.23818. [DOI] [PubMed] [Google Scholar]
- 19.Li ZY, Xi Y, Zhu WN, Zeng C, Zhang ZQ, Guo ZC, Hao DL, et al. Positive regulation of hepatic miR-122 expression by HNF4alpha. J Hepatol. 2011;55:602–611. doi: 10.1016/j.jhep.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 20.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 22.Calvisi DF, Factor VM, Loi R, Thorgeirsson SS. Activation of beta-catenin during hepatocarcinogenesis in transgenic mouse models: relationship to phenotype and tumor grade. Cancer Res. 2001;61:2085–2091. [PubMed] [Google Scholar]
- 23.Beer S, Zetterberg A, Ihrie RA, McTaggart RA, Yang Q, Bradon N, Arvanitis C, et al. Developmental context determines latency of MYC-induced tumorigenesis. PLoS Biol. 2004;2:e332. doi: 10.1371/journal.pbio.0020332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wanzel M, Herold S, Eilers M. Transcriptional repression by Myc. Trends Cell Biol. 2003;13:146–150. doi: 10.1016/s0962-8924(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 25.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cairo S, Wang Y, de Reynies A, Duroure K, Dahan J, Redon MJ, Fabre M, et al. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci U S A. 2010;107:20471–20476. doi: 10.1073/pnas.1009009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang B, Hsu SH, Majumder S, Kutay H, Huang W, Jacob ST, Ghoshal K. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010;29:1787–1797. doi: 10.1038/onc.2009.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jonker JW, Suh JM, Atkins AR, Ahmadian M, Li P, Whyte J, He M, et al. A PPARgamma-FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis. Nature. 2012;485:391–394. doi: 10.1038/nature10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu S, Balakrishnan A, Bok RA, Anderton B, Larson PE, Nelson SJ, Kurhanewicz J, et al. 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab. 2011;14:131–142. doi: 10.1016/j.cmet.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu S, Le H. Dual roles of PKM2 in cancer metabolism. Acta Biochim Biophys Sin (Shanghai) 2013;45:27–35. doi: 10.1093/abbs/gms106. [DOI] [PubMed] [Google Scholar]
- 31.Hiebert SW, Lipp M, Nevins JR. E1A-dependent trans-activation of the human MYC promoter is mediated by the E2F factor. Proc Natl Acad Sci U S A. 1989;86:3594–3598. doi: 10.1073/pnas.86.10.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elliott MJ, Dong YB, Yang H, McMasters KM. E2F-1 up-regulates c-Myc and p14(ARF) and induces apoptosis in colon cancer cells. Clin Cancer Res. 2001;7:3590–3597. [PubMed] [Google Scholar]
- 33.Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T. Selective stabilization of mammalian microRNAs by 3' adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bui TV, Mendell JT. Myc: Maestro of MicroRNAs. Genes Cancer. 2010;1:568–575. doi: 10.1177/1947601910377491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A. 2009;106:3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaposi-Novak P, Libbrecht L, Woo HG, Lee YH, Sears NC, Coulouarn C, Conner EA, et al. Central role of c-Myc during malignant conversion in human hepatocarcinogenesis. Cancer Res. 2009;69:2775–2782. doi: 10.1158/0008-5472.CAN-08-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 38.Schlaeger C, Longerich T, Schiller C, Bewerunge P, Mehrabi A, Toedt G, Kleeff J, et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology. 2008;47:511–520. doi: 10.1002/hep.22033. [DOI] [PubMed] [Google Scholar]
- 39.Motiwala T, Ghoshal K, Das A, Majumder S, Weichenhan D, Wu YZ, Holman K, et al. Suppression of the protein tyrosine phosphatase receptor type O gene (PTPRO) by methylation in hepatocellular carcinomas. Oncogene. 2003;22:6319–6331. doi: 10.1038/sj.onc.1206750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Mates JM, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.