Abstract
A systematic survey for the presence of plasmids in 17 different xenobiotic-degrading Sphingomonas strains was performed. In almost all analyzed strains, two to five plasmids with sizes of about 50 to 500 kb were detected by using pulsed-field gel electrophoresis. A comparison of plasmid preparations untreated or treated with S1 nuclease suggested that, in general, Sphingomonas plasmids are circular. Hybridization experiments with labeled gene probes suggested that large plasmids are involved in the degradation of dibenzo-p-dioxin, dibenzofuran, and naphthalenesulfonates in S. wittichii RW1, Sphingomonas sp. HH69, and S. xenophaga BN6, respectively. The plasmids which are responsible for the degradation of naphthalene, biphenyl, and toluene by S. aromaticivorans F199 (pNL1) and of naphthalenesulfonates by S. xenophaga BN6 (pBN6) were site-specifically labeled with a kanamycin resistance cassette. The conjugative transfer of these labeled plasmids was attempted with various bacterial strains as putative recipient strains. Thus, a conjugative transfer of plasmid pBN6 from S. xenophaga BN6 to a cured mutant of strain BN6 and to Sphingomonas sp. SS3 was observed. The conjugation experiments with plasmid pNL1 suggested a broader host range of this plasmid, because it was transferred without any obvious structural changes to S. yanoikuyae B1, Sphingomonas sp. SS3, and S. herbicidovorans. In contrast, major plasmid rearrangements were observed in the transconjugants after the transfer of plasmid pNL1 to Sphingomonas sp. HH69 and of pBN6 to Sphingomonas sp. SS3. No indications for the transfer of a Sphingomonas plasmid to bacteria outside of the Sphingomonadaceae were obtained.
The genus Sphingomonas accommodates strictly aerobic, chemoheterotrophic, gram-negative, rod-shaped, usually yellow-pigmented bacteria that contain glycosphingolipids as cell envelope components and belong to the α-4-subgroup of the Proteobacteria (62). Sphingomonas strains appear to be widely distributed in various aquatic and terrestrial environments. They have been isolated from anthropogenic polluted river water and sediments (17, 45, 67) and medical material (69), and they constitute an important part of the marine bacterial plankton (11). Recently, sphingomonads have also been detected in rather high cell densities on the surfaces of various plants (31) and in biofilms found in drinking water supplies (32). Furthermore, they have been repeatedly isolated from extreme environments such as arctic and antarctic soils and deeply buried (>200 m) sediments (2, 3, 13, 56). The genus Sphingomonas is becoming increasingly interesting in environmental microbiology because various xenobiotic-degrading organisms belong to this group. Previously, Sphingomonas strains have been described that degrade compounds such as biphenyl, (substituted) naphthalene(s), fluorene, (substituted) phenanthrene(s), pyrene, (chlorinated) diphenylether(s), (chlorinated) furan(s), (chlorinated) dibenzo-p-dioxin(s), carbazole, polyethylene glycols, chlorinated phenols, and different herbicides and pesticides (e.g., references 10, 13, 20, 28, 34, 39, 40, 41, 42, 45, 55, 59, 60, 61, 64, 66, 71, 72, and 73).
The isolation of various Sphingomonas strains which harbor different metabolic pathways for the degradation of a wide range of xenobiotic compounds suggests that the members of this genus have the ability to adapt more quickly or more efficiently to the degradation of new compounds in the environment than members of other bacterial genera. This ability does not seem to be related to different general strategies in the degradative pathways used for the mineralization of xenobiotic compounds, because previous studies about the physiology and enzymology of a number of degradative pathways [e.g., those for (substituted) naphthalene(s) or biphenyl] did not demonstrate any significant differences between sphingomonads and other bacteria (30, 59, 61). However, a comparison of the genes encoding the degradation of 2,4-dichlorophenoxyacetate or biphenyl suggested that usually only a low degree of sequence similarity is found between the sphingomonads and other Proteobacteria (e.g., authentic pseudomonads) (15, 16). This indicated that in the evolution of degradative pathways, some kind of barriers must have existed between sphingomonads and other Proteobacteria.
Currently, the major known difference between Sphingomonas strains and other gram-negative bacteria is a frequently observed unusual organization of the degradative genes in the sphingomonads. Previous studies with authentic pseudomonads suggested that the genes which encode catabolic enzymes are often organized in operons and are coordinately regulated. Classical examples for such a genetic organization are meta-cleavage pathways from the TOL or NAH plasmids, modified ortho-cleavage pathways for chlorocatechols from various plasmids, or the chromosomally encoded β-ketoadipate pathway (19, 21, 70). In contrast, it is becoming increasingly evident that the genes for catabolic pathways in Sphingomonas strains are often localized separately from each other or are at least not organized in coordinately regulated operons. This has been described, e.g., for the genes involved in the degradation of γ-hexachlorocyclohexane (lindane) by S. paucimobilis UT26 (39, 41), pentachlorophenol by S. chlorophenolica (5), protocatechuate by S. paucimobilis SYK-6 (37), naphthalene, biphenyl, and toluene by S. yanoikuyae B1 and S. aromaticivorans F199 (52, 73), and dibenzo-p-dioxin by S. wittichii RW1 (1). (For a current review see reference 50.)
There are several reports which indicate that large plasmids may be important for the degradation of xenobiotic compounds by Sphingomonas strains. Thus, in S. aromaticivorans F199 and some other sphingomonads isolated from the same location, the genes encoding the degradative pathways for biphenyl, naphthalene, m-xylene, and p-cresol were detected on large plasmids (29, 52). There is also some evidence that the genes for the degradation of carbazole by some other sphingomonads (e.g., Sphingomonas CF06) are also (at least in part) encoded on plasmids (12, 47).
A detailed analysis of the naphthalenesulfonate-degrading strain S. xenophaga BN6 has previously been performed, and in the course of these investigations, various tools for the molecular analysis of sphingomonads were established (e.g., references 27, 33, 51, and 59). In the present study it was therefore attempted to localize the genes for the degradation of naphthalenesulfonates in the genome of strain BN6 and to obtain more general information about the presence, importance, and host range of plasmids in xenobiotic-degrading sphingomonads.
MATERIALS AND METHODS
Bacterial strains and media.
The sphingomonads studied and some characteristic compounds which are degraded by the respective strains are shown in Table 1. The members of the former genus Sphingomonas sensu lato were classified in Table 1 according to the suggestion of Takeuchi et al. (62) as members of the newly created genera Sphingomonas, Sphingobium, Novosphingobium, or Sphingopyxis. For some strains indicated in Table 1, no valid new descriptions have been performed, but according to their 16S rDNA, most of them probably belong to the genera Sphingobium or Novosphingobium and do not belong to the newly defined genus Sphingomonas sensu stricto. In order to circumvent these taxonomical problems, the older nomenclature is used and all of the strains are referred to as Sphingomonas sensu lato throughout this report.
TABLE 1.
Sphingomonas strains analyzed during the present study
| Straina | Compounds degraded | Reference(s) |
|---|---|---|
| Sphingomonas (Sphingobium) yanoikuyae B1 DSM 6900 | Toluene, biphenyl, naphthalene, anthracene, phenanthrene | 69 |
| Sphingomonas (Sphingobium) herbicidovorans DSM 11019 | 2-(2,4-Dichlorophenoxy) propioniate, 2,4-dichlorophenoxy propionic acid, mecoprop | 72 |
| Sphingomonas (Sphingobium) chlorophenolica ATCC 33790 | Pentachlorophenol, 2,4,6-trichlorophenol | 44 |
| “Sphingomonas paucimobilis” Q1 | Toluene, xylene, naphthalene, biphenyl, anthracene | 17 |
| “Sphingomonas” wittichii RW1 DSM 6014 | (Chlorinated) dibenzo-p-dioxin(s), dibenzofuran(s) | 67, 68 |
| “Sphingomonas” sp. HH69 DSM 7135 | (Acetoxy-, hydroxy-) dibenzofuran(s) | 20 |
| “Sphingomonas” sp. SS3 DSM 6432 | (4-Chloro-, 4-fluoro-) diphenylether | 55 |
| “Sphingomonas” sp. EPA505 DSM 7526 | Fluoranthene, (substituted) naphthalene(s), phenanthrene, anthracene | 40 |
| “Sphingomonas” sp. A175 DSM 13477 | Benzene, 1,4-dichlorobenzene | 43 |
| “Sphingomonas” sp. K39 | 2,3,4,6-Tetrachlorophenol | 64 |
| “Sphingomonas” xenophaga BN6 DSM 6383 | (Substituted) naphthalene-2-sulfonate(s) | 45, 58 |
| Sphingomonas (Sphingopyxis) macrogoltabidus DSM 8826 | Polyethylene glycol 4000 | 62, 63 |
| Sphingomonas (Novosphingobium) subterranea DSM 12447 | Naphthalene, toluene, biphenyl, dibenzothiophene, fluorene | 2, 62 |
| Sphingomonas (Novosphingobium) aromaticivorans F199 DSM 12444 | Naphthalene, toluene, cresoles, biphenyl, dibenzothiophene, fluorene | 2, 62 |
| Sphingomonas (Novosphingobium) aromaticivorans B0695 | 2-Methylnaphthalene, acenaphthene, anthracene, fluoranthene, phenanthrene | 56 |
| Sphingomonas (Novosphingobium) subarctica KF1 DSM 10700 | 2,3,4,6-Tetrachlorophenol, 2,4,6-trichlorophenol | 43 |
| 62 | ||
| Sphingomonas (Novosphingobium) stygia DSM 12445 | Toluene, biphenyl, dibenzothiophene, fluorene | 62 |
| Sphingomonas paucimobilis DSM 1098 | Cannot degrade aromatic hydrocarbons | 69 |
Quotation marks indicate strains for which no valid new descriptions have been performed.
The non-Sphingomonas strains which were used for the conjugation experiments were obtained from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany). These bacteria were cultivated in the media suggested by the DSMZ.
For the plasmid detection experiments, the bacterial strains were routinely subcultured on nutrient broth (NB) or R2A medium (Difco, Becton Dickinson, Sparks, Md.). A mineral medium described by Dorn et al. (9) was used as selection medium for the curing and conjugation experiments. The following carbon and energy sources were added for the selection of transconjugants or cured mutants: malate (5 mM) and glucose (0.5%, wt/vol) were from aqueous stock solutions, 2(2,4-dichlorophenoxy)propionate was supplied from a stock solution in acetone (363 mg/ml) to a final concentration of 640 mg/liter, and dibenzofuran and diphenylether were added as solid particles in the lid of mineral agar plates.
Escherichia coli strains DH5α (Gibco, Eggenstein, Germany) and S17.1 (57) were used for recombinant DNA work.
Molecular techniques for the manipulation of recombinant E. coli strains.
Plasmid DNA was isolated from E. coli DH5α with a Gfx-Micro plasmid prep kit from Amersham Biosciences (Freiburg, Germany). Digestion of DNA with restriction endonucleases (MBI Fermentas, St. Leon-Rot, Germany), electrophoresis, DNA purification, alkaline phosphatase treatment, and ligation with T4 DNA ligase were performed according to the standard procedures (54). Transformation of E. coli was done by the method of Chung et al. (7).
Preparation of genomic DNA from sphingomonads for the detection of megaplasmids.
The different Sphingomonas strains were usually grown overnight at 30°C in 20 ml of NB, and the plasmids were prepared basically as previously described by Barton et al. (4). Approximately 4 × 109 cells were harvested by centrifugation, washed with 1 ml of 1 M NaCl in 10 mM Tris-HCl (pH 7.6), and resuspended in 500 μl of EC buffer (1 M NaCl, 100 mM EDTA, 6 mM Tris-HCl [pH 7.6], 0.5% [wt/vol] Brij 58, 0.2% [wt/vol] deoxycholate, 0.5% [wt/vol] N-lauroylsarcosine). The cell suspensions were thoroughly mixed with an equal volume of InCert agarose (10 mg/ml in EC buffer; BMA, Rockland, Maine) which was melted and maintained at 85°C in a thermomixer (Eppendorf, Hamburg, Germany). Two hundred microliters of each of the preparations was immediately transferred into the plug molds. The molder was put on ice for 15 min to allow the plugs to solidify. The solidified plugs were transferred into 2-ml Eppendorf tubes and incubated at 37°C in 1 ml of EC buffer, which additionally contained 20 μg of RNase/ml and 1 mg of lysozyme/ml until the cells were lysed. The complete clearance of plugs could be observed usually after 45 to 60 min of incubation. Subsequently, the plugs were incubated overnight at 50°C in ES buffer (1% [wt/vol] N-lauroylsarcosine, 0.5 M EDTA [pH 8.0]) supplemented with 1 mg of proteinase K/ml (Gerbu, Gaiberg, Germany). The ES buffer and the proteinase K were incubated together at 37°C for 2 h before usage. Finally, the proteinase K was inactivated by incubating the plugs in 1 ml of 1 mM phenylmethylsulfonyl fluoride in TE buffer (10 mM Tris [pH 7.5], 1 mM EDTA) for 45 min at 37°C. The plugs were then incubated twice in 1 ml of TE buffer (45 min each) to remove traces of phenylmethylsulfonyl fluoride. The agarose plugs prepared in this way could be stored at least for 2 weeks at 4°C.
In order to linearize circular megaplasmids, slices of 2 to 3 mm were cut out of each plug, and the incorporated DNA was digested for 10 min at 37°C with 1 U of Aspergillus oryzae S1 nuclease (MBI Fermentas) in 200 μl of S1 buffer (50 mM NaCl, 30 mM sodium acetate [pH 4.5], 5 mM ZnSO4). The reactions were stopped by transferring the slices into 100 μl of ice-cold ES buffer. After 15 min of incubation on ice, the slices were loaded separately on the teeth of a gel comb, and the comb with the attached gel slices was transferred to the horizontal gel mounting chamber (14 by 14 by 1 cm). The comb with the attached gel slices was embedded in 100 ml of 1% agarose (molecular biology grade; Eurogentec, Herstal, Belgium) prepared in 0.5× TBE buffer (45 mM Tris-HCl, 45 mM boric acid, 1 mM EDTA [pH 8.0]). The comb was removed from the gel after solidification of the agarose, leaving the DNA-containing plugs within the agarose. Finally, the pulsed-field gel electrophoresis (PFGE) was run in a clamped homogenous electric field apparatus (CHEF Mapper; Bio-Rad Laboratories, Richmond, Calif.) in 0.5× TBE buffer at 14°C at 6 V/cm with linearly increasing pulse times from 7.23 to 24 s for 26 h. λ-DNA concatemers (1 μg) were used as size standards (New England Biolabs, Beverly, Mass.). The gels were subsequently stained with ethidium bromide and documented by using an image documentation system (Raytest, Straubenhardt, Germany).
PCR experiments.
PCR experiments were performed in a Genius thermal cycler (Techne, Cambridge, United Kingdom). The PCR mixtures contained, in a volume of 30 μl, 50 to 200 ng of DNA, 0.3 μM of each forward and reverse primer (Eurogentec), 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM magnesium acetate, 0.2 mM dNTP (Eppendorf), and 0.5 to 1 U of Taq DNA polymerase (Eppendorf) under the conditions indicated below.
The primers for the preparation of the pcpB and pcpC probes (Table 2) were derived from the corresponding nucleotide sequences from S. chlorophenolica ATCC 33790 and ATCC 39723 deposited at the NCBI databases. The following PCR program was used for the amplification of the indicated regions of pcpB and pcpC: an initial denaturation (3 min, 94°C) was followed by 30 cycles consisting of annealing at 60°C (30 s), polymerization at 72°C (40 s), and denaturation at 94°C (30 s). The last polymerization step was extended to 5 min.
TABLE 2.
Oligonucleotides used for the amplification of specific gene probes and construction of hybrid DNA molecules
| Primer | Sequencea | Location in the relevant gene | NCBI no. |
|---|---|---|---|
| pcpB-UP | ACGATCAACCCGCGCTGG | bp 475-492 | U60175 |
| pcpB-LOW | ACGGCGAATGGCAATGCG | bp 916-899 | U60175 |
| pcpC-UP | GCGCCGTTTCGAAGCTGC | bp 21171-21187 | AF512952 |
| pcpC-LOW | CGATGAAGACCCGTCTGGCG | bp 21842-21823 | AF512952 |
| dxnA1A2-UP | CTCTAAGGGACGAAAATGCTGTTG | bp 8783-8806 | X72850 |
| dxnA1A2-LOW | GCCTGGCGAGTCTGAATCCTA | bp 10714-10694 | X72850 |
| PrimerA-Km | tatatctagaGTAATGCCCTCCCGGTGCTC | 5′-Flanking sequence of ORF363 | |
| PrimerB-Km | cttgctgtGTTGATCGGCGGCGTG | Fusion of 5′-flanking sequence ORF363 and 5′ neo end | |
| PrimerC-Km | cgatcaacACAGCAAGCGAACCGG | Fusion of 5′ neo end and 5′-flanking sequence ORF363 | |
| PrimerD-Km | tcccgcgcTCAGAAGAACTCGTCAAGAAGG | Fusion of 3′ neo end and 3′-flanking sequence ORF363 | |
| PrimerE-Km | tcttctgaGCGCGGGACAACGACT | Fusion of 3′-flanking sequence ORF363 and 3′ neo end | |
| PrimerF-Km | tatagtttaaacAACCGGCCTCCTCTCCCT | 3′-Flanking sequence of ORF363 | |
| S2051 | tatatctagaACATCAATCTGACGCTCGCG | 5′-Flanking sequence of ORF6 | |
| S2052 | cttgctgtTACCTGTCTCTCCCGC | Fusion of 5′ neo end and 5′-flanking sequence of ORF6 | |
| S2053 | gacaggtaACAGCAAGCGAACCGG | Fusion of 5′-flanking sequence ORF6 and 5′ neo end | |
| S2054 | gacgtcatTCAGAAGAACTCGTCA | Fusion of 3′-flanking sequence ORF6 and 3′ neo end | |
| S2055 | tcttctgaATGACGTCGATGCAAA | Fusion of 3′ neo end and 3′-flanking sequence ORF6 | |
| S2056 | aataattcatatgAAGCACAACCGTGTTACCCG | 3′-Flanking sequence of ORF6 |
Recognition sequences for restriction endonucleases NdeI, XbaI, and PmeI are underlined; lowercase characters indicate the sequences which are not homologous to the template.
The primers for the amplification of dxnA1A2 (large and small subunit of the dioxin dioxygenase) were derived from published nucleotide sequences obtained from S. wittichii RW1 (Table 2). The PCR program applied consisted of an initial denaturation (3 min, 94°C), followed by 30 cycles of annealing at 60°C (30 s), polymerization at 72°C (90 s), and denaturation at 94°C (30 s). The last polymerization step was extended to 5 min.
Hybridization procedures.
A digoxigenin (DIG) DNA labeling and detection kit was used according to the instructions of the supplier (Roche, Mannheim, Germany). The pcpB, pcpC, and dxnA1A2 probes were prepared by PCR using the known gene sequences as described above.
Plasmid pAKE3/5 (26) was used for the localization of the genes participating in the degradation of naphthalenesulfonates in S. xenophaga BN6. This recombinant plasmid contained a 12-kb insert encoding several genes of the naphthalenesulfonate degradative pathway (e.g., 1,2-dihydroxynaphthalene dioxygenase and 2′-hydroxybenzalpyruvate aldolase-hydratase). The insert in pAKE3/5 corresponded to the 16.2-kb fragment deposited at the NCBI database as U65001, without the terminal ∼1.5-kb PstI fragment at the 3′ end and a terminal ∼3-kb HindIII fragment at the 5′ end.
The hybridization temperatures in the experiments with the dxnA1A2, pcpB, and pcpC probes and the 12-kb fragment from pAKE3/5 were set to 60, 62, 58, and 68°C, respectively.
Introduction of a kanamycin resistance cassette into plasmid pNL1.
The suicide vector pTB200 which carried a fusion between the 5′- and 3′-flanking region of an open reading frame (ORF) of unknown function (ORF363) on plasmid pNL1 and a kanamycin resistance (neo) gene was constructed by inserting the neo gene into the respective ORF using splicing by overlap extension (SOE) (24). The oligonucleotides PrimerA-Km and PrimerB-Km (for the nucleotide sequences of all oligonucleotides, see Table 2) were used to amplify an 845-bp fragment of the 5′-flanking region of ORF363 by PCR, and the oligonucleotides PrimerE-Km and PrimerF-Km were used to amplify an 857-bp fragment of the 3′-flanking region of ORF363 with genomic DNA of S. aromaticivorans F199 as template. To facilitate the further cloning of the SOE product, XbaI and PmeI restriction sites were added to the primers PrimerA-Km and PrimerF-Km, respectively. The neo gene was amplified together with its own promoter region using the oligonucleotides PrimerC-Km and PrimerD-Km and pUTminiTn5 (22) as template. This resulted in the amplification of a 960-bp fragment. The three PCR fragments were fused in two subsequent PCRs. In the first reaction, the PrimerA-Km and PrimerD-Km were used, and the fragment obtained in the course of this PCR was finally fused to the third fragment using PrimerA-Km and PrimerF-Km (for the primer sequences, see Table 2).
The final 2,662-bp SOE product was first introduced into pBluescriptII SK+ prepared as T vector (36) to give pTB100. The SOE product was cut out of pTB100 using XbaI and PmeI and subsequently inserted into XbaI/PmeI-cleaved plasmid pLO3 (35) to give pTB200. The correct insertion of the SOE product into pTB200 was verified by sequencing with primers B-Seq (5′ ACACGCCGGGAGCCAGAT 3′) and E-Seq (5′ CGGGTAACGCTTCAGGCC 3′).
Plasmid pTB200 was conjugatively transferred to S. aromaticivorans F199 in a plate-mating experiment using E. coli S17.1 λ pir as donor strain (8). Kanamycin (50 μg/ml)-resistant transconjugants were selected and further investigated. In order to obtain clones that had integrated the disrupted form of ORF363 by a double crossover event, approximately 100 clones were transferred to agar plates with NB plus kanamycin (50 μg/ml) and with kanamycin (50 μg/ml) plus tetracycline (10 μg/ml). Those clones that were kanamycin resistant and tetracycline sensitive were further studied. In order to verify the correct insertion of the neo gene into ORF363 on plasmid pNL1, a PCR experiment was performed with primers vor363 (5′ CGGCGACGACGTTGTGGC 3′), which binds upstream of ORF363, and PrimerD-Km and primers hinter363 (5′ GGGTGGGATCGGGACATGG 3′), which binds downstream of ORF363, and PrimerC-Km. The sizes of the obtained PCR products corresponded to the expected sizes, which implicated the disruption of ORF363 by the neo gene. In an additional control experiment, the wild-type and the mutated form of pNL1 were separated from the chromosomal DNA of the wild-type strain or the presumed mutant of S. aromaticivorans F199 by PFGE. Genomic DNA of both strains was transferred by Southern blotting on a nylon membrane, which was hybridized against a DIG-labeled (Roche) neo probe. As expected, the neo probe hybridized only with the mutated form of plasmid pNL1.
Introduction of the kanamycin resistance cassette to plasmid pBN6.
The kanamycin resistance gene was inserted into the coding region (ORF6) for a putative α-subunit of a ring-hydroxylating dioxygenase located on the 180-kb plasmid of S. xenophaga BN6 by using the same SOE strategy previously described for the disruption of the 1,2-dihydroxynaphthalene dioxygenase gene (27). For this construction, the 5′-flanking sequence of ORF6 (1,034 bp) was amplified by use of the primer pair S2051 and S2052. The neo gene (960 bp) and the 3′-flanking fragment (955 bp) were amplified with the primer pairs S2053-S2054 and S2055-S2056. In the second PCR step, the 5′-flanking sequence was fused to the neo gene (primers S2051 and S2054; 1,978 bp). In the third PCR (primers S2051 and S2056), the 5′-flanking sequence neo fusion was joined to the 3′-flanking sequence (2,917 bp). This PCR fragment was cut with NdeI and XbaI and inserted into conjugative plasmid pAKE35.1 (also cut with NdeI and XbaI), which contains the sacB gene and allows gene replacements to get pAKE36 (26). E. coli S17.1 (57) was transformed with plasmid pAKE36, and the plasmid was conjugatively transferred to S. xenophaga BN6. Plasmid integration mutants (cointegrates) were isolated on mineral medium agar plates with glucose which were supplemented with kanamycin and tetracycline. To isolate integration mutants which had lost the vector fragment by double crossover events, the mutants obtained were transferred to agar plates with glucose plus kanamycin and sucrose (4%, wt/vol).
Conjugative transfer of plasmids.
The recipient and donor strains were grown overnight in 125-ml flasks with 20 ml of NB medium which was supplemented with 50 μg of kanamycin/ml for the donor strains. The cells were harvested by centrifugation (1 min, 20,000 × g), washed with 1 ml of a saline solution (0.85% [wt/vol] NaCl, 1 mM MgSO4), and resuspended in 50 μl of the saline to yield an optical density at 546 nm of approximately 40. The donor and recipient cells were mixed and subsequently transferred to sterile filters (cellulose acetate filters, pore size 0.2 μm, 25-mm diameter; Sartorius, Göttingen, Germany) which were placed on NB agar plates. In control experiments, the donor and recipient cells were transferred individually to two separate filters. The cells were incubated at 30°C for 8 to 16 h on the filters and resuspended afterwards in 500 μl of saline. From these suspensions, serial dilutions (100 to 10−3) were plated on the selective media. Furthermore, serial dilutions of the donor and recipient cells were plated on both selective and nonselective agar plates, which served as negative and positive controls, respectively. To determine the cell numbers of the donor and recipients, dilutions of approximately 10−7 to 10−9 of donor and recipient cells were plated on NB agar plates. The colonies of the transconjugants appeared usually after 2 to 7 days. Some of the colonies were purified first on nonselective and subsequently on selective media and were then subjected to PCR and PFGE analysis.
The media to detect a transfer of the 2NS+ (conveying the ability to convert naphthalene-2-sulfonate) 180-kb plasmid from S. xenophaga BN6 AKE2/5 to the recipient strains contained 50 μg of kanamycin/ml plus the following carbon sources: biphenyl (for S. aromaticivorans F199), malate (for S. yanoikuyae B1 and S. subarctica KF1), and dibenzofuran (for Sphingomonas sp. HH69).
The transfer of plasmid RP1 from S. xenophaga BN6(RP1) to Pseudomonas putida was demonstrated by using Simmons agar plus 40 μg of kanamycin/ml plus 10 μg of tetracycline/ml.
Enzyme assays.
Naphthalenesulfonate dioxygenase, 1,2-dihydroxynaphthalene dioxygenase, 2′-hydroxybenzalpyruvate aldolase-hydratase, and salicylaldehyde dehydrogenase were determined as described previously (33).
Chemicals.
Naphthalene-2-sulfonic acid was obtained from Bayer AG (Leverkusen, Germany). All other chemicals were obtained from Sigma-Aldrich Chemie (Deisenhofen, Germany) or Merck (Darmstadt, Germany). Biochemicals were from Roche Diagnostics, and restriction enzymes, RNase, and DNA ligase for molecular biology were from MBI Fermentas.
RESULTS
Detection of megaplasmids in different sphingomonads.
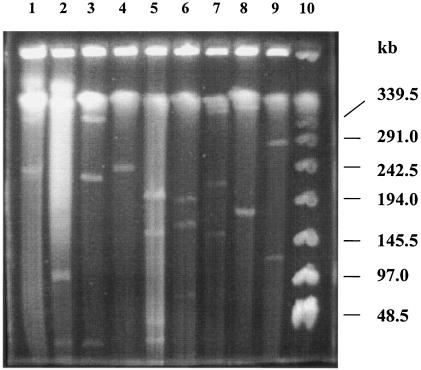
A collection of sphingomonads was analyzed which degraded a wide range of different xenobiotic compounds, such as polycyclic aromatic hydrocarbons, chlorinated and sulfonated aromatics, herbicides, aromatic ethers, and polyethylene glycol (Table 1). The genomic DNA of the strains was analyzed using PFGE in order to detect plasmids in these strains (Fig. 1). The PFGE analysis demonstrated that almost all tested Sphingomonas strains harbored two to five plasmids with sizes of about 50 to 500 kb. Furthermore, in some strains plasmids with sizes smaller than 50 kb were also detected (Table 3).
FIG. 1.
Detection of megaplasmids in different Sphingomonas strains by PFGE. Lane 1, S. wittichii RW1; lane 2, Sphingomonas sp. A175; lane 3, Sphingomonas sp. SS3; lane 4, S. yanoikuyae B1; lane 5, S. paucimobilis EPA505; lane 6, S. chlorophenolica; lane 7, S. subterranea; lane 8, S. aromaticivorans F199; lane 9, S. stygia; lane 10, λ-DNA standard.
TABLE 3.
Numbers and sizes of plasmids detected in various Sphingomonas strains
| Strain | Compounds degraded | No. of plasmids | Approx size(s) of plasmids (kb) |
|---|---|---|---|
| S. yanoikuyae B1 | Toluene, biphenyl, naphthalene, anthracene, phenanthrene | 1 | 240 |
| S. herbicidovorans | 2-(2,4-Dichlorophenoxy) propioniate, 2,4-dichlorophenoxypropionic acid, mecoprop | 2 | 300, 160 |
| S. chlorophenolica ATCC 33790 | Pentachlorophenol, 2,4,6-trichlorophenol | 4 | 200, 160, 50, <50 |
| S. paucimobilis Q1 | Toluene, xylene, naphthalene, biphenyl, anthracene | 3 | 240, 80, <50 |
| S. wittichii RW1 | (Chlorinated) dibenzo-p-dioxin(s), dibenzofuran(s) | 2 | 340, 240 |
| Sphingomonas sp. HH69 | (Acetoxy-, hydroxy-) dibenzofuran(s) | 5 | 230, 150, 70, 50, <50 |
| Sphingomonas sp. SS3 | (4-Chloro-, 4-fluoro-) diphenylether | 3 | 340, 230, <50 |
| S. paucimobilis EPA 505 | Fluoranthene, (substituted) naphthalene(s), phenanthrene, anthracene | 3 | 200, 160, 50 |
| Sphingomonas sp. A175 | Benzene, 1,4-dichlorobenzene | 2 | 100, <50 |
| Sphingomonas sp. K39 | 2,3,4,6-Tetrachlorophenol | 4 | 290, 280, 180, 120 |
| S. xenophaga BN6 | (Substituted) naphthalene-2-sulfonate(s) | 4 | 260, 180, 100, 50 |
| S. macrogoltabidus | Polyethylene glycol 4000 | 2 | 450, 150 |
| S. subterranea | Naphthalene, toluene, biphenyl, dibenzothiophene, fluorene | 3 | ∼450, 220, 150 |
| S. aromaticivorans F199 | Naphthalene, toluene, cresoles, biphenyl, dibenzothiophene, fluorene | 2 | ∼500, 180 |
| S. aromaticivorans B0695 | 2-Methylnaphthalene, acenaphthene, anthracene, fluoranthene, phenanthrene | 2 | 195, <50 |
| S. subarctica KF1 | 2,4,6-Trichlorophenol, 2,3,4,6-tetrachlorophenol | 2 | 300, 220 |
| S. stygia | Toluene, biphenyl, dibenzothiophene, fluorene | 2 | 290, 120 |
| S. paucimobilis | Cannot degrade aromatic hydrocarbons | 2 | 340, 150 |
Sphingomonas plasmids are generally circular.
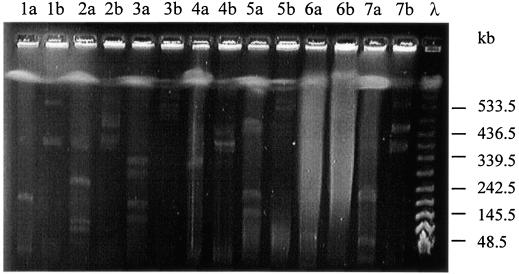
In order to allow a reliable size determination, the plasmid preparations for the PFGE analysis were routinely treated with S1 nuclease in order to linearize circular plasmids (see Materials and Methods). In the PFGE, the circular and linear forms of plasmids can be readily distinguished because the supercoiled forms move slower than the linear forms. Because there were several recent reports about the presence of linear plasmids in various bacterial species (38), it was determined whether the plasmids detected during the PFGE were circular or linear in vivo. Therefore, for all of the sphingomonads analyzed, the plasmid isolation procedures were performed in duplicate with or without the S1 nuclease treatment (for an example, see Fig. 2). Thus, it was found that all detected Sphingomonas plasmids were presumably circular.
FIG. 2.
PFGE analysis of the plasmids from different Sphingomonas strains with (a) or without (b) S1 nuclease treatment. Lane 1, S. aromaticivorans B0695; lane 2, S. xenophaga BN6; lane 3, Sphingomonas sp. K39; lane 4, S. herbicidovorans; lane 5, S. subterranea; lane 6, Sphingomonas sp. A175; lane 7, S. paucimobilis Q1; λ, λ-DNA standard.
Detection of genes encoding enzymes for the degradation of naphthalenesulfonates and dibenzo-p-dioxin on megaplasmids from S. xenophaga BN6 and S. wittichii RW1.
The PFGE analysis demonstrated the presence of several plasmids in S. xenophaga BN6, S. wittichii RW1, and S. chlorophenolica ATCC 33790 (Table 3). From these three organisms, different genes coding for enzymes involved in the degradation of naphthalenesulfonates, dibenzo-p-dioxin, and pentachlorophenol, respectively, have been described previously by different groups (1, 10, 27). However, no further investigations regarding the localization of the respective genes in the genomes of these three strains were conducted.
The PFGE analysis of the genomic DNA of S. xenophaga BN6 showed four prominent bands which corresponded to plasmids with sizes of about 50, 100, 180, and 260 kb (Table 3). In order to localize the genes encoding the degradative pathway for naphthalenesulfonates, a 12-kb fragment was cloned and labeled which contained the genes encoding the 1,2-dihydroxynaphthalene dioxygenase and 2′-hydroxybenzalpyruvate aldolase-hydratase from the naphthalenesulfonate pathway. The subsequent hybridization experiment demonstrated that the genes were localized on the 180-kb plasmid.
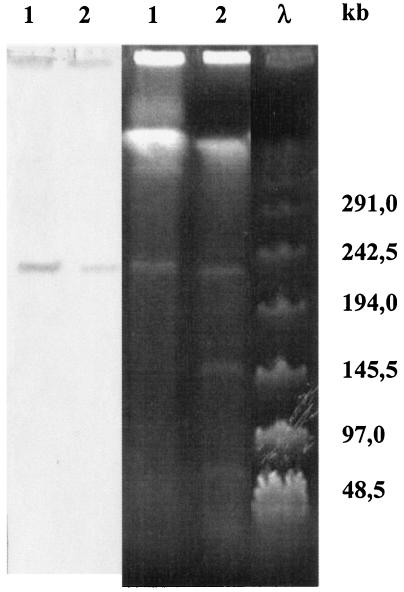
In the following experiments, dxnA1A2 (coding for the large and small subunits of the dibenzo-p-dioxin dioxygenase from S. wittichii RW1) and pcpB and pcpC (coding for the pentachlorophenol-4-monooxygenase and tetrachlorohydroquinone dehalogenase, respectively, from S. chlorophenolica ATCC 33790) were amplified by PCR and labeled with DIG, and these probes were hybridized against the genomes of the respective strains after PFGE. Thus, it was found that dxnA1A2 was localized on an approximately 240-kb plasmid in S. wittichii RW1 (Fig. 3). A positive hybridization signal was also observed between the labeled dxnA1A2 probe and a plasmid with a similar size from the dibenzofuran-degrading strain Sphingomonas sp. HH69. This suggested that dibenzo-p-dioxin and dibenzofuran are oxidized by rather similar dioxygenases by S. wittichii RW1 and Sphingomonas sp. HH69, respectively. A positive hybridization signal between another dibenzofuran-degrading sphingomonad and a dxnA1A2 probe obtained from S. wittichii RW1 has recently also been described by Fukuda et al. (14).
FIG. 3.
PFGE analysis of the plasmid profile of S. wittichii RW1 and Sphingomonas sp. HH69 (right) and hybridization of the genomic DNAs with a labeled dxnA1A2 probe (left).
In contrast to the situation with S. xenophaga BN6 and S. wittichii RW1, the hybridization experiments with genomic DNA from S. chlorophenolica ATCC 33790 demonstrated that pcpB and pcpC were chromosomally located.
Curing of plasmid pBN6 from S. xenophaga BN6.
It was previously demonstrated that salicylate concentrations of >1 mM almost completely inhibited growth of S. xenophaga BN6 on glucose (46). This caused a strong selection pressure for mutant strains of S. xenophaga which had lost the ability to convert 2NS to salicylate using a mineral medium with glucose plus 2NS, because under these conditions the wild-type strains converted 2NS to salicylate and are thus inhibited in growth. Therefore, a liquid culture of strain BN6 was grown on NB medium for approximately 100 generations, and the culture was then plated on mineral agar plates with glucose (0.5%, wt/vol) which were supplemented with 2NS (1 mM). This resulted in two different colony phenotypes: small brown colonies, which demonstrated the characteristic growth retardation of strain BN6 in the presence of salicylate, and large colonies (>50% of the total number of colonies), which showed the characteristic yellow pigmentation of S. xenophaga BN6. Four random clones which showed the new phenotype (designated as JK0.1, JK0.2, JK0.3, and JK0.4) were isolated, purified, and maintained for further biochemical investigations. In subsequent resting-cell experiments, it was demonstrated that the mutant strains did not convert 2NS. Furthermore, no activities for different key enzymes of the naphthalenesulfonate degradative pathway (1,2-dihydroxynaphthalene dioxygenase, 2′-hydroxybenzalpyruvate aldolase-hydratase, and salicylaldehyde dehydrogenase) were detected in the mutant strains with the enzyme assays previously established (33) (data not shown), and no revertants to the wild-type phenotype could be detected.
The mutant strains were analyzed by PFGE, and it was found that the 260-, 100-, and 50-kb plasmids were still present, but the mutant strains did not contain the 180-kb plasmid. Furthermore, it was demonstrated in hybridization experiments with the 12-kb DNA fragment containing the genes encoding the dihydroxynaphthalene dioxygenase and 2′-hydroxybenzalpyruvate aldolase-hydratase that the mutant strains indeed showed no hybridization signal.
Introduction of an antibiotic resistance cassette on plasmids pNL1 from S. aromaticivorans F199 and pBN6 from S. xenophaga BN6.
The plasmids from S. aromaticivorans F199 and S. xenophaga BN6 were tagged with a kanamycin resistance gene in order to test their conjugatability using a strong selective pressure and to avoid any possible problems connected to the expression of the respective plasmid-encoded degradative pathways in different genetic backgrounds. Therefore, suicide vectors were constructed carrying the levan sucrase gene (sacB) and a fusion between the 5′- and 3′-flanking regions of an ORF (ORF363) with unknown function from plasmid pNL1 and a ferredoxin reductase gene from plasmid pBN6 and the neo gene from Tn5. The suicide vectors were conjugatively transferred to S. aromaticivorans F199 and S. xenophaga BN6, respectively, and transconjugants were selected on agar plates with kanamycin and tetracycline. The loss of the integrated vector DNA from plasmid pBN6 was enforced by the addition of sucrose, as described previously by Keck et al. (27).
Transfer of plasmid pBN6 to a plasmid-free variant of S. xenophaga BN6 and to Sphingomonas sp. SS3.
In order to demonstrate the principle feasibility of the attempted conjugation experiments, a spontaneous nalidixinic acid-resistant derivative of the cured 2NS− mutant of S. xenophaga BN6 described above was produced. This strain was used as a potential recipient in a conjugation experiment with the S. xenophaga BN6 derivative, which carried the kanamycin resistance gene, as the donor strain (BN6 AKE2/5). The kanamycin resistance was conjugatively transferred by a plate mating to the plasmid-free nalidixin-resistant 2NS− mutant of strain BN6 (JK0.1) with a conjugation rate of 1.1 × 10−6 transconjugants per recipient.
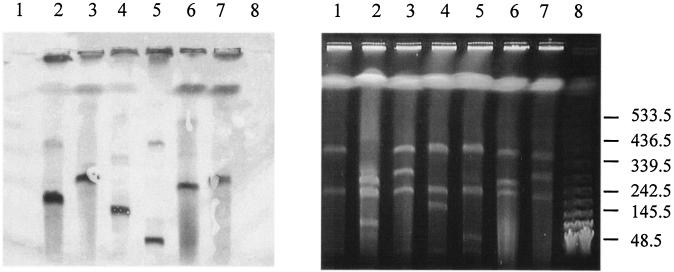
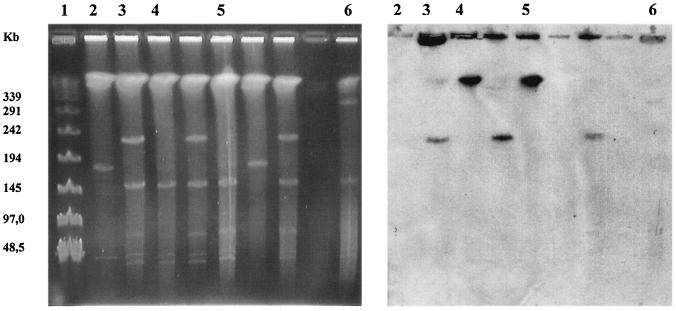
In the following experiments, S. xenophaga BN6 AKE2/5 was used as the donor in different conjugation experiments with different sphingomonads as putative recipient strains. The selection for transconjugants in these experiments was performed using carbon sources which were only utilized by the presumed plasmid recipient strains and the antibiotic resistance encoded on the recombinant derivative of plasmid pBN6. A plasmid transfer was only observed in a mating of S. xenophaga BN6 AKE2/5 with Sphingomonas sp. SS3 with a conjugation rate of 5 × 10−7 transconjugants per recipient. Transconjugants were selected for their ability to grow with diphenylether in the presence of kanamycin, and the transfer of the neo gene was additionally confirmed by PCR. S. xenophaga BN6 and Sphingomonas sp. SS3 could be easily differentiated by the colors of their respective colonies (bright yellow versus dark yellow). As expected, all transconjugant colonies demonstrated the typical appearance of Sphingomonas sp. SS3. The subsequent PFGE analysis revealed unexpected plasmid patterns in all of the investigated transconjugants (Fig. 4). These PFGEs suggested that the endogenous plasmids of strain Sphingomonas sp. SS3 seemed not to have undergone any obvious changes but that in all transconjugants the “incoming” plasmid from strain BN6 had undergone significant alterations in size. In order to localize the neo gene which had been introduced by the plasmid from strain BN6, a hybridization experiment was performed using a neo gene probe. The probe hybridized with plasmids of significantly different sizes, ranging from approximately 50 kb up to approximately 290 kb (Fig. 4). This suggested that the plasmid pBN6 in its original structure is possibly not stable in Sphingomonas sp. SS3. Furthermore, out of five investigated transconjugants, four carried different plasmid patterns, suggesting that there are different ways to stabilize the plasmid from strain BN6 after transfer to strain SS3.
FIG. 4.
PFGE of the total DNA of different transconjugants, which were obtained after the conjugation of S. xenophaga BN6 AKE2/5 Kmr with Sphingomonas sp. SS3 (right), and Southern hybridization of the gel using a neo probe (left). Lanes: 1, Sphingomonas sp. SS3; 2, S. xenophaga AKE2/5; 3 to 7. transconjugants; 8, λ DNA standard.
In further conjugation experiments with S. aromaticivorans F199, S. yanoikuyae B1, S. subarctica KF1, and Sphingomonas sp. HH69, no indications for transfer of the 2NS+ 180-kb plasmid from S. xenophaga BN6 AKE2/5 to the recipient strains were detected.
Transfer of plasmid pNL1 from S. aromaticivorans F199 to different Sphingomonas strains.
A comparison of the carbon sources utilized by S. yanoikuyae B1 and S. aromaticivorans F199 demonstrated that malate supported growth of S. yanoikuyae B1 but not of S. aromaticivorans F199. Therefore, putative transconjugants were selected on malate (5 mM) plus kanamycin (50 μg/ml). After 24 h, this selection already resulted in the formation of visible colonies on the selective medium. In contrast, no colonies were formed on the control plates with the strains plated separately on the same medium. The conjugation frequency was estimated to be 2 × 10−4 transconjugants per recipient. The color and the morphology of the colonies formed by the transconjugants (whitish yellow) clearly demonstrated that S. yanoikuyae B1 and not S. aromaticivorans F199 served as recipient, because the latter strain formed brighter yellow colonies on all media used. In order to prove the conjugative transfer of plasmid pNL1 to S. yanoikuyae B1, several of the presumed transconjugants were analyzed by PFGE, and the presence of a plasmid with the expected size was observed for all transconjugants. It was therefore deduced that plasmid pNL1 was indeed conjugatively transferable to S. yanoikuyae B1. In similar experiments, the transfer of the antibiotic resistance genes was demonstrated between S. aromaticivorans F199 and Sphingomonas sp. SS3 (selection for growth on diphenylether plus 50 μg of kanamycin/ml) and S. aromaticivorans F199 and S. herbicidovorans [selection for growth on 2-(2,4-dichlorophenoxy)propionate plus 50 μg of kanamycin/ml]. Also in these experiments, a transfer of plasmid pNL1 could be confirmed by the size and color of the respective colonies formed by the transconjugants. The analysis of the plasmid pattern in the transconjugants clearly demonstrated the presence of an additional plasmid with the size of pNL1. No transfer of pNL1 could be detected to S. wittichii RW1, S. subarctica KF1, and S. chlorophenolica ATCC 33790.
Plasmid rearrangements after the transfer of plasmid pNL1 to Sphingomonas sp. HH69.
A transfer of the kanamycin resistance gene was also detected in a conjugation experiment with S. aromaticivorans F199 and Sphingomonas sp. HH69 (selecting for growth on dibenzofuran and 50 μg of kanamycin/ml). In contrast to the experiments with S. yanoikuyae B1, Sphingomonas sp. SS3, and S. herbicidovorans, in this case massive alterations were observed in the plasmid pattern of the transconjugants (Fig. 5). An analysis of the different transconjugants suggested that all transconjugants had lost the 240-kb plasmid, which hybridized with the dxnA1A2 gene probe (see above) and which therefore presumably coded for at least part of the dibenzofuran degradation pathway in Sphingomonas sp. HH69. It was therefore investigated, in hybridization experiments, if the transconjugants harbored the dxnA1A2 genes on different loci. These experiments demonstrated that in some of the transconjugants (e.g., the transconjugants shown in Fig. 5, lanes 4 and 5), the dxnA1A2 genes were found on the chromosome. In contrast, in some other transconjugants (e.g., the one shown in Fig. 5, lane 6), the gene(s) hybridizing with dxnA1A2 was found on a newly formed 320-kb plasmid.
FIG. 5.
PFGE of the total DNA of different transconjugants, which were obtained after the conjugation of S. aromaticivorans F199 Kmr with Sphingomonas sp. HH69 (left) and Southern hybridization of DNA from the gel using a dxnA1A2 probe (right). Lane 1, λ-DNA standard; lane 2, S. aromaticivorans F199 Kmr; lane 3, Sphingomonas sp. HH69; lane 4, Sphingomonas sp. HH69-1 transconjugant; lane 5, Sphingomonas sp. HH69-2 transconjugant; lane 6, Sphingomonas sp. HH69-3 transconjugant.
Attempts to transfer plasmid pNL1 from S. aromaticivorans F199 to bacteria not belonging to the genus Sphingomonas.
The ability of a conjugative transfer of pNL1 was also tested with different bacteria belonging to various genera within the α-, β-, and γ-subgroups of Proteobacteria (Table 4). No indications for a transfer of plasmid pNL1 to bacteria belonging to the β- or γ-subgroup of Proteobacteria could be detected. The only example for a transfer of plasmid pNL1 to a strain not belonging to the genus Sphingomonas was obtained with Porphyrobacter sanguineus DSM 11032, which is also a member of the family Sphingomonadaceae and which has been described as an aerobic bacteriochlorophyll a-containing organism with the ability to degrade biphenyl and dibenzofuran (23).
TABLE 4.
Bacterial strains and selection conditions used in order to demonstrate the transfer of plasmid pNL1 from S. aromaticivorans F199 to nonsphingomonadsa
| Strain | DSM no. | Taxonomical classification | Selection medium | Transfer frequency of pNL1 |
|---|---|---|---|---|
| Porphyrobacter sanguineus | 11032 | α-Proteobacteria; Sphingomonadales; Sphingomonadaceae | MBM + Km (200 μg/ml) | 1.9 × 10−4 |
| Rhizobium radiobacter | 30147 | α-Proteobacteria; Rhizobiales; Rhizobiaceae | Malate (5 mM) + Tc (10 μg/ml) | No transfer |
| Brevundimonas subvibrioides | 4375 | α-Proteobacteria; Caulobacteriales; Caulobacteriaceae | MBM + Km (200 μg/ml) | No transfer |
| Pigmentiphaga kullae | 13608 | β-Proteobacteria; Burkholderiales; Alcaligenaceae | Simmons agar + Km (50 μg/ml) | No transfer |
| Burkholderia glathei | 50014 | β-Proteobacteria; Burkholderiales; Burkholderiaceae | Simmons agar + Km (50 μg/ml) | No transfer |
| Serratia proteamaculans | 4597 | γ-Proteobacteria; Enterobacteriales; Enterobacteriaceae | Simmons agar + Km (50 μg/ml) | No transfer |
| Pseudomonas putida | 291 | γ-Proteobacteria; Pseudomonadales; Pseudomonadaceae | Simmons agar + Km (50 μg/ml) | No transfer |
MBM, marine broth medium (DSMZ medium 514); Km, kanamycin; Tc, tetracycline.
Transfer of plasmid RP1 from S. xenophaga BN6 to other bacterial genera.
The outer membranes of sphingomonads contain huge amounts of sphingoglycolipids instead of lipid A, which is almost ubiquitously found among other Proteobacteria (25). This was a possible reason for the restrictions in plasmid transfer from Sphingomonas strains to other Proteobacteria, because these differences could influence cell aggregation or change the functionality of pili and other structures involved in the conjugative transfer of DNA. The transfer of broad-host-range plasmids of the incompatibility group P-1 from E. coli to S. xenophaga BN6 had been previously demonstrated (53). The transfer of plasmid RP1 (49) from S. xenophaga BN6(RP1) to P. putida was achieved in a plate-mating experiment with a conjugation frequency of 2.3 × 10−6. Plasmid RP1 was also successfully transferred in similar conjugation experiments from S. xenophaga BN6(RP1) to most of the strains tested in the previous experiments as possible recipient strains for the transfer of plasmid pNL1 (Table 5). This demonstrated that, in principle, the transfer of plasmids from Sphingomonas strains to nonsphingomonads is possible.
TABLE 5.
Transfer of plasmid RP1 from S. xenophaga BN6 to different bacterial strainsa
| Recipient strain | Selective medium | Transfer frequency of RP1 (per recipient cell) |
|---|---|---|
| Rhizobium radiobacter | MM + malate (5 mM) + Km (40 μg/ml) + Tc (10 μg/ml)b | 7.2 × 10−7 |
| Pigmentiphaga kullae | Simmons agar + Km (40 μg/ml) + Tc (10 μg/ml) | 1.1 × 10−4 |
| Burkholderia glathei | Simmons agar + Km (40 μg/ml) + Tc (10 μg/ml) | 4 × 10−8 |
| Serratia proteamaculans | Simmons agar + Km (40 μg/ml) + Tc (10 μg/ml) | 2.4 × 10−4 |
| Pseudomonas putida | Simmons agar + Km (40 μg/ml) + Tc (10 μg/ml) | 2.3 × 10−6 |
MBM, marine broth medium (DSMZ medium 514); Km, kanamycin; Tc, tetracycline; MM, mineral medium.
MM containing malate as a single source of carbon and energy did not support the growth of S. xenophaga BN6(RP1).
DISCUSSION
The present study clearly demonstrates that large plasmids are ubiquitous in sphingomonads and are very important for the degradation of various harmful and/or xenobiotic compounds by this group of organisms. In previous work, it was reported that plasmids are presumably involved in the degradation of naphthalene, biphenyl, toluene, and carbazole by sphingomonads (6, 12, 47, 52). We have obtained evidence that in different sphingomonads the degradative pathways for naphthalenesulfonates, dibenzo-p-dioxin, and dibenzofuran are also encoded on plasmids. Therefore, it is very probable that with the progress in the detection and sequencing of genes involved in the degradation of xenobiotics by sphingomonads, the majority of the genes will be detected on large plasmids similar to those described here. The degradative plasmids in sphingomonads generally appear to be larger (160 to 240 kb) than the previously studied degradative plasmids from pseudomonads, such as the TOL (117 kb) or NAH (usually 80 to 120 kb) plasmid(s) (18, 70). This explains why Sphingomonas plasmids are only reproducibly detected by PFGE and not by previously established plasmid isolation procedures, such as agarose gel electrophoresis or CsCl gradient centrifugation.
It was clearly demonstrated in the course of the present study that plasmid pNL1 could be conjugatively transferred from S. aromaticivorans F199 to S. yanoikuyae B1, Sphingomonas sp. SS3, and S. herbicidovorans. These four strains belong to different subgroups within the genus Sphingomonas sensu lato and would be classified in the scheme suggested by Takeuchi et al. (62) as members of the genera Novosphingobium (S. aromaticivorans) and Sphingobium (S. yanoikuyae and S. herbicidovorans). The conjugatability of plasmid pNL1 was also suggested by the previously performed sequence analysis of the plasmid, which demonstrated the presence of three gene clusters encoding homologs of the E. coli F plasmid required for conjugative sex pilus formation and mating-pair stabilization (52). In contrast, it appears that the host range of the plasmid pBN6 from S. xenophaga BN6 is much more restricted, because for this plasmid only a transfer to Sphingomonas sp. SS3 was observed.
The generally observed presence of several plasmids within single isolates suggested that there are plasmids belonging to several incompatibility groups present in Sphingomonas strains. The major rearrangements observed in the plasmid pattern after the attempted transfer of pNL1 to Sphingomonas sp. HH69 might be an indication that a plasmid from the same incompatibility group is present in strain HH69 and thus may prevent the establishment of plasmid pNL1 in this host.
According to the results of the present study, the plasmids found within the Sphingomonas strains are transferable by conjugation among different Sphingomonas strains (and presumably some closely related genera) but are presumably only rarely transferred to other Proteobacteria. Thus, this will result in a reduced gene flow from the sphingomonads to other bacteria. This is probably one of the reasons for the comparable low degree of sequence homology between Sphingomonas genes and those from other Proteobacteria which have been detected during sequence analysis and hybridization experiments with different 2,4-dichlorophenoxyacetate- or biphenyl-degrading bacteria (15, 16, 29). A similar conclusion has recently also been obtained from the molecular analysis of the distribution of the pcpB gene within a Finnish groundwater supply heavily polluted with chlorinated phenols. In this study, it was demonstrated that highly similar or identical copies of the pcpB gene could be found among different sphingomonads but that homologous genes were totally absent from nonsphingomonads which also degraded chlorophenols (65). A possible reason for the reduced (or maybe abolished) plasmid transfer rates from Sphingomonas strains (and closely related genera) to other Proteobacteria could be that specific Sphingomonas plasmids exist which are only able to transfer among or to replicate within Sphingomonas strains.
It was initially assumed that a further possible reason for the restrictions in plasmid transfer from Sphingomonas strains to other Proteobacteria could be due to the composition of the outer membranes of sphingomonads. These outermost cellular structures of the sphingomonads differ from those of other Proteobacteria because they contain huge amounts of sphingoglycolipids instead of lipid A, which is almost ubiquitously found among other Proteobacteria (25). We could exclude a principle impediment of plasmid transfer by these differences because it was possible to transfer the broad-host-range plasmid RP1 from S. xenophaga BN6 to various Proteobacteria. Although the observed conjugation frequencies in the range of 10−4 to 10−8 transconjugants per recipient appear to be rather low, similar conjugation frequencies have also been observed for various intergeneric conjugation experiments with plasmid RP1 (48).
Although we could clearly demonstrate in our present study the importance of plasmids for the degradation of xenobiotic compounds by Sphingomonas strains, it is also evident that degradative genes may in some cases also be found encoded on the bacterial chromosome. Thus, it was previously found that the genes coding for the degradative pathways for naphthalene, biphenyl, and toluene are encoded on plasmid pNL1 in S. aromaticivorans F199 but that the homologous pathways are chromosomally encoded in S. yanoikuyae B1 and S. paucimobilis Q1 (29). A similar situation might exist for the location of the genes involved in the degradation of pentachlorophenol. Although we could demonstrate in the present study that the genes coding for pentachlorophenol degradation by S. chlorophenolica ATCC 33790 are chromosomally located, a very recent study presented strong evidence for a natural horizontal transfer of the pcpB gene among different sphingomonads within a Finnish groundwater treatment plant (65). This may indicate that even in the case of currently chromosomally encoded pathways in sphingomonads, some recent plasmid exchange events have taken place.
REFERENCES
- 1.Armengaud, J., B. Happe, and K. N. Timmis. 1998. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. RW1: catabolic genes dispersed on the genome. J. Bacteriol. 180:3954-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balkwill, D. L., G. R. Drake, R. H. Reeves, J. K. Fredrickson, D. C. White, D. B. Ringelberg, D. P. Chandler, M. F. Romine, D. W. Kennedy, and C. M. Spadoni. 1997. Taxonomic study of aromatic-degrading bacteria from deep-terrestrial-subsurface sediments and description of Sphingomonas aromaticivorans sp. nov., Sphingomonas subterranea sp. nov., and Sphingomonas stygia sp. nov. Int. J. Syst. Bacteriol. 47:191-201. [DOI] [PubMed] [Google Scholar]
- 3.Baraniecki, C. A., J. Aislabie, and J. M. Foght. 2002. Characterization of Sphingomonas sp. Ant 17, an aromatic hydrocarbon-degrading bacterium isolated from Antarctic soil. Microb. Ecol. 43:44-54. [DOI] [PubMed] [Google Scholar]
- 4.Barton, B. M., G. P. Hardening, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:234-240. [DOI] [PubMed] [Google Scholar]
- 5.Cai, M., and L. Xun. 2002. Organization and regulation of pentachlorophenol-degrading genes in Sphingobium chlorophenolicum ATCC 39723. J. Bacteriol. 184:4672-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho, J. C., and S. J. Kim. 2001. Detection of mega plasmid from polycyclic aromatic hydrocarbon-degrading Sphingomonas sp. strain KS14. J. Mol. Microbiol. Biotechnol. 3:503-506. [PubMed] [Google Scholar]
- 7.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5 and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 9.Dorn, E., M. Hellwig, W. Reineke, and H.-J. Knackmuss. 1974. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol. 99:61-70. [DOI] [PubMed] [Google Scholar]
- 10.Ederer, M. M., R. L. Crawford, R. P. Herwig, and C. S. Orser. 1997. PCP degradation is mediated by closely related strains of the genus Sphingomonas. Mol. Ecol. 6:39-49. [DOI] [PubMed] [Google Scholar]
- 11.Eguchi, M., M. Ostrowski, F. Fegatella, J. Bowman, D. Nichols, T. Nishino, and R. Cavicchioli. 2001. Sphingomonas alaskensis strain AFO1, an abundant oligotrophic ultramicrobacterium from the North Pacific. Appl. Environ. Microbiol. 67:4945-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng, X., L.-T. Ou, and A. Ogram. 1997. Plasmid-mediated mineralization of carbofuran by Sphingomonas sp. CF-06. Appl. Environ. Microbiol. 63:1332-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fredrickson, J. K., D. L. Balkwill, G. R. Drake, M. F. Romine, D. B. Ringelberg, and D. C. White. 1995. Aromatic-degrading Sphingomonas isolates from the deep surface. Appl. Environ. Microbiol. 61:1917-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuda, K., S. Nagata, and H. Taniguchi. 2002. Isolation and characterization of dibenzofuran-degrading bacteria. FEMS Microbiol. Lett. 208:179-185. [DOI] [PubMed] [Google Scholar]
- 15.Fulthorpe, J. J., C. McGowan, O. V. Maltseva, W. E. Holben, and J. M. Tiedje. 1995. 2,4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaic of catabolic genes. Appl. Environ. Microbiol. 61:3274-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furukawa, K., N. Hayase, K. Taira, and N. Tomizuka. 1989. Molecular relationship of chromosomal genes encoding biphenyl/polychlorinated biphenyl catabolism: some soil bacteria possess highly conserved bph operons. J. Bacteriol. 171:5467-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furukawa, K., J. R. Simon, and A. M. Chakrabarty. 1983. Common induction and regulation of biphenyl, xylene/toluene, and salicylate catabolism in Pseudomonas paucimobilis. J. Bacteriol. 154:1356-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greated, A., L. Lambertsen, P. A. Williams, and C. M. Thomas. 2002. Complete sequence of the IncP-9 TOL plasmid pWW0 from Pseudomonas putida. Environ. Microbiol. 4:856-871. [DOI] [PubMed] [Google Scholar]
- 19.Harayama, S., and M. Rekik. 1990. The meta cleavage operon of TOL degradative plasmid pWW0 comprises 13 genes. Mol. Gen. Genet. 221:113-120. [DOI] [PubMed] [Google Scholar]
- 20.Harms, H., H. Wilkes, R.-M. Wittich, and P. Fortnagel. 1995. Metabolism of hydroxydibenzofurans, methoxybenzofurans, acetoxydibenzofurans, and nitrodibenzofurans by Sphingomonas sp. HH69. Appl. Environ. Microbiol. 61:2499-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 22.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiraishi, A., Y. Yonemitsu, M. Matsushita, Y. K. Shin, H. Kuraishi, and K. Kawahara. 2002. Characterization of Porphyrobacter sanguineus sp. nov., an aerobic bacteriophyll-containing bacterium capable of degrading biphenyl and dibenzofuran. Arch. Microbiol. 178:45-52. [DOI] [PubMed] [Google Scholar]
- 24.Horton, R. M., D. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 25.Kawasaki, S., R. Moriguchi, K. Sekiya, T. Nakai, E. Ono, K. Kume, and K. Kawahara. 1994. The cell envelope structure of the lipopolysaccharide-lacking gram-negative bacterium Sphingomonas paucimobilis. J. Bacteriol. 176:284-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keck, A. 2000. Conversion of azo dyes by a redox mediator dependent mechanism which is linked to the naphthalenesulfonate degradation of Sphingomonas sp. strain BN6. Ph.D. thesis. Universität Stuttgart, Stuttgart, Germany.
- 27.Keck, A., J. Rau, T. Reemstma, R. Mattes, A. Stolz, and J. Klein. 2002. Identification of quinoide redox mediators that are formed during the degradation of naphthalene-2-sulfonate by Sphingomonas xenophaga BN6. Appl. Environ. Microbiol. 68:4341-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan, A. A., R.-F. Wang, W.-W. Cao, W. Franklin, and C. E. Franklin. 1996. Reclassification of a polycyclic aromatic hydrocarbon-metabolizing bacterium, Beijerinckia sp. strain B1, as Sphingomonas yanoikuyae by fatty acid analysis, protein pattern analysis, DNA-DNA hybridization, and 16S ribosomal DNA sequencing. Int. J. Syst. Bacteriol. 46:466-469. [DOI] [PubMed] [Google Scholar]
- 29.Kim, E., P. J. Aversano, M. F. Romine, R. P. Schneider, and G. J. Zylstra. 1996. Homology between genes for aromatic hydrocarbon degradation in surface and deep-subsurface Sphingomonas strains. Appl. Environ. Microbiol. 62:1467-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, E., and G. J. Zylstra. 1995. Molecular and biochemical characterization of two meta-cleavage dioxygenases involved in biphenyl and m-xylene degradation by Beijerinckia sp. strain B1. J. Bacteriol. 177:3095-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, H., M. Nishiyama, T. Kunito, K. Senoo, K. Kawahara, K. Murakami, and H. Oyaizu. 1998. High population of Sphingomonas species on plant surface. J. Appl. Microbiol. 85:731-736. [Google Scholar]
- 32.Koskinen, R., T. Ali-Vehmas, P. Kämpfer, M. Laurikkala, I. Tsitko, E. Kostyal, F. Atroshi, and M. Salkinoja-Salkonen. 2000. Characterization of Sphingomonas isolates from Finnish and Swedish drinking water distribution systems. J. Appl. Microbiol. 89:687-696. [DOI] [PubMed] [Google Scholar]
- 33.Kuhm, A. E., A. Stolz, K.-L. Ngai, and H.-J. Knackmuss. 1991. Purification and characterization of a 1,2-dihydroxynaphthalene dioxygenase from a bacterium that degrades naphthalenesulfonic acids. J. Bacteriol. 173:3795-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumari, R., S. Subudhi, M. Suar, G. Dhingra, V. Raina, C. Dogra, S. Lal, J. R. van der Meer, C. Holliger, and R. Lal. 2002. Cloning and characterization of lin genes responsible for the degradation of hexachlorocyclohexane isomers by Sphingomonas paucimobilis strain B90. Appl. Environ. Microbiol. 68:6021-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lenz, O., E. Schwartz, J. Dernedde, M. Eitinger, and B. Friedrich. 1994. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J. Bacteriol. 176:4385-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchuk, D., M. Drumm, A. Saulino, and F. S. Collins. 1991. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 19:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masai, E., S. Shinohara, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 1999. Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J. Bacteriol. 181:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meinhardt, F., R. Schaffrath, and M. Larsen. 1997. Microbial linear plasmids. Appl. Microbiol. Biotechnol. 47:329-336. [DOI] [PubMed] [Google Scholar]
- 39.Miyauchi, K., S.-K. Suh, Y. Nagata, and M. Takagi. 1998. Cloning and sequencing of a 2,5-dichlorohydroquinone reductive dehalogenase gene whose product is involved in degradation of γ-hexachlorocyclohexane by Sphingomonas paucimobilis. J. Bacteriol. 180:1354-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mueller, J. G., R. Devereux, D. L. Santavy, S. E. Lantz, S. G. Willis, and P. H. Pritchard. 1997. Phylogenetic and physiological comparison of PAH-degrading bacteria from geographically diverse soils. Antonie Leeuwenhoek 71:329-343. [DOI] [PubMed] [Google Scholar]
- 41.Nagata, Y., M. Miyauchi, and M. Takagi. 1999. Complete analysis of genes and enzymes for γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26. J. Ind. Microbiol. 23:380-390. [DOI] [PubMed] [Google Scholar]
- 42.Nagata, Y., R. Ohtomo, K. Miyauchi, M. Fukuda, K. Yano, and M. Takagi. 1994. Cloning and sequencing of a 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase gene involved in the degradation of γ-hexachlorocyclohexane in Pseudomonas paucimobilis. J. Bacteriol. 176:3117-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nohynek, L. J., E.-L. Nurmiaho-Lassila, E. L. Suhonen, H.-J. Busse, M. Mohammadi, J. Hantula, F. Rainey, and M. Salkinoja-Salonen. 1996. Description of chlorophenol-degrading Pseudomonas sp. strains KF1, KF3, and NKF1 as a new species of the genus Sphingomonas, Sphingomonas subarctica sp. nov. Int. J. Syst. Bacteriol. 46:1042-1055. [DOI] [PubMed] [Google Scholar]
- 44.Nohynek, L. J., E. L. Suhonen, E.-L. Nurmiaho-Lassila, J. Hantula, and M. Salkinoja-Salonen. 1995. Description of four pentachlorophenol-degrading bacterial strains as Sphingomonas chlorophenolica sp. nov. Syst. Appl. Microbiol. 18:527-538. [Google Scholar]
- 45.Nörtemann, B., J. Baumgarten, H. G. Rast, and H.-J. Knackmuss. 1986. Bacterial communities degrading amino- and hydroxynaphthalenesulfonates. Appl. Environ. Microbiol. 52:1195-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nörtemann, B., A. E. Kuhm, H.-J. Knackmuss, and A. Stolz. 1994. Conversion of substituted naphthalenesulfonates by Pseudomonas sp. BN6. Arch. Microbiol. 161:320-327. [Google Scholar]
- 47.Ogram, A. V., Y.-P. Duan, S. L. Trabue, X. Feng, H. Castro, and L.-T. Ou. 2000. Carbofuran degradation mediated by three related plasmid systems. FEMS Microbiol. Ecol. 32:197-203. [DOI] [PubMed] [Google Scholar]
- 48.Olsen, R. H., and P. Shipley. 1973. Host range and properties of the Pseudomonas aeruginosa R factor R1822. J. Bacteriol. 113:772-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncPα plasmids—compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 50.Pinyakong, O., H. Habe, and T. Omori. 2003. The unique aromatic catabolic genes in sphingomonads degrading polycyclic aromatic hydrocarbons. J. Gen. Appl. Microbiol. 49:1-9. [DOI] [PubMed] [Google Scholar]
- 51.Riegert, U., S. Bürger, and A. Stolz. 2001. Altering catalytical properties of 3-chlorocatechol-oxidizing extradiol dioxygenase from Sphingomonas xenophaga BN6 by random mutagenesis. J. Bacteriol. 183:2322-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romine, M. F., L. C. Stillwell, K.-K. Wong, S. J. Thurston, E. C. Sisk, C. Sensen, T. Gaasterland, J. K. Fredrickson, and J. D. Saffer. 1999. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruβ, R. 1996. Verbesserung der Abbauleistung des Bakterienstammes BN6 gegenüber sulfonierten aromatischen Verbindungen-Vergleich zwischen Hybridorganismus und Mischkultur. Ph.D. thesis. Universität Stuttgart, Stuttgart, Germany.
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 55.Schmidt, S., R.-M. Wittich, D. Erdmann, H. Wilkes, W. Francke, and P. Fortnagel. 1992. Biodegradation of diphenyl ether and its monohalogenated derivatives by Sphingomonas sp. strain SS3. Appl. Environ. Microbiol. 58:2744-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi, T., J. K. Fredrickson, and D. L. Balkwill. 2001. Biodegradation of polycyclic aromatic hydrocarbons by Sphingomonas strains isolated from the terrestrial subsurface. J. Ind. Microbiol. Biotechnol. 26:283-289. [DOI] [PubMed] [Google Scholar]
- 57.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 58.Stolz, A., C. Schmidt-Maag, E. B. M. Denner, H.-J. Busse, T. Egli, and P. Kämpfer. 2000. Description of Sphingomonas xenophaga sp. nov. for strains BN6T and N,N which degrade xenobiotic aromatic compounds. Int. J. Syst. Bacteriol. 50:35-41. [DOI] [PubMed] [Google Scholar]
- 59.Stolz, A. 1999. Degradation of substituted naphthalenesulfonic acids by Sphingomonas xenophaga BN6. J. Ind. Microbiol. Biotechnol. 23:391-399. [DOI] [PubMed] [Google Scholar]
- 60.Sugimoto, M., M. Tanabe, M. Hataya, S. Enokibara, J. A. Duine, and F. Kawai. 2001. The first step in polyethylene glycol degradation by sphingomonads proceeds via a flavoprotein alcohol dehydrogenase containing flavin adenine dinucleotide. J. Bacteriol. 183:6694-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taira, K., N. Hayase, N. Arimura, S. Yamashita, T. Miyazaki, and K. Furukawa. 1988. Cloning and nucleotide sequence of the 2,3-dihydroxybiphenyl dioxygenase gene from the PCB-degrading strain of Pseudomonas paucimobilis Q1.0 Biochemistry 27:3990-3996. [DOI] [PubMed] [Google Scholar]
- 62.Takeuchi, M., K. Hamana, and A. Hiraishi. 2001. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int. J. Syst. E vol. Microbiol. 51:1405-1417. [DOI] [PubMed] [Google Scholar]
- 63.Takeuchi, M., F. Kawai, Y. Shimada, and A. Yokota. 1993. Taxonomic study of polyethylene glycol-utilizing bacteria: emended description of the genus Sphingomonas and new descriptions of Sphingomonas macrogoltabidus sp. nov., Sphingomonas sanguis sp. nov. and Sphingomonas terrae sp. nov. Syst. Appl. Bacteriol. 16:2227-2238. [Google Scholar]
- 64.Tiirola, M. A., M. K. Mannisto, J. A. Puhakka, and M. S. Kulomaa. 2002. Isolation and characterization of Novosphingobium sp. strain MT1, a dominant polychlorophenol-degrading strain in a groundwater bioremediation system. Appl. Environ. Microbiol. 68:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tiirola, M. A., H. Wang, L. Paulin, and M. S. Kulomaa. 2002. Evidence for natural horizontal transfer of the pcpB gene in the evolution of pentachlorophenol-degrading sphingomonads. Appl. Environ. Microbiol. 68:4495-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wattiau, P., L. Bastiaens, R. van Herwijnen, L. Daal, J. R. Parsons, M.-E. Renard, D. Springael, and G. R. Cornelis. 2001. Flourene degradation by Sphingomonas sp. LB126 proceeds through protocatechuic acid: a genetic analysis. Res. Microbiol. 152:861-872. [DOI] [PubMed] [Google Scholar]
- 67.Wittich, R.-M., H. Wilkes, V. Sinnwell, W. Francke, and P. Fortnagel. 1992. Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl. Environ. Microbiol. 58:1005-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yabuuchi, E., H. Yamamoto, S. Terakubo, N. Okamura, T. Naka, N. Fujiwara, K. Kobayashi, Y. Kosako, and A. Hiraishi. 2001. Proposal of Sphingomonas wittichii sp. nov. for strain RW1T, known as a dibenzo-p-dioxin metabolizer. Int. J. Syst. E vol. Microbiol. 51:281-292. [DOI] [PubMed] [Google Scholar]
- 69.Yabuuchi, E., I. Yano, H. Oyaizu, Y. Hashimoto, T. Ezaki, and H. Yamamoto. 1990. Proposals of Sphingomonas paucimobilis gen. nov. and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata comb. nov., and two genospecies of the genus Sphingomonas. Microbiol. Immunol. 34:99-119. [DOI] [PubMed] [Google Scholar]
- 70.Yen, K.-M., and C. M. Serdar. 1988. Genetics of naphthalene catabolism in pseudomonads. Crit. Rev. Microbiol. 15:247-268. [DOI] [PubMed] [Google Scholar]
- 71.Yrjälä, K., S. Suomalainen, E. L. Suhonen, S. Kilpi, L. Paulin, and M. Romantschuk. 1998. Characterization and reclassification of an aromatic- and chloroaromatic-degrading Pseudomonas sp., strain HV3, as Sphingomonas sp. HV3. Int. J. Syst. Bacteriol. 48:1057-1062. [DOI] [PubMed] [Google Scholar]
- 72.Zipper, C., K. Nickel, W. Angst, and H.-P. Kohler. 1996. Complete microbial degradation of both enantiomers of the chiral herbicide mecoprop [(RS)-2-(4-chloro-2-methylphenoxy)propionic acid] in an enantioselective manner by Sphingomonas herbicidovorans sp. nov. Appl. Environ. Microbiol. 62:4318-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zylstra, G. J., and E. Kim. 1997. Aromatic hydrocarbon degradation by Sphingomonas yanoikuyae B1. J. Ind. Microbiol. Biotechnol. 19:408-414. [DOI] [PubMed] [Google Scholar]