Abstract
We have determined the 1.35- and 1.45-Å structures, respectively, of closed and open iron-loaded forms of Mannheimia haemolytica ferric ion-binding protein A. M. haemolytica is the causative agent in the economically important and fatal disease of cattle termed shipping fever. The periplasmic iron-binding protein of this gram-negative bacterium, which has homologous counterparts in many other pathogenic species, performs a key role in iron acquisition from mammalian host serum iron transport proteins and is essential for the survival of the pathogen within the host. The ferric (Fe3+) ion in the closed structure is bound by a novel asymmetric constellation of four ligands, including a synergistic carbonate anion. The open structure is ligated by three tyrosyl residues and a dynamically disordered solvent-exposed anion. Our results clearly implicate the synergistic anion as the primary mediator of global protein conformation and provide detailed insights into the molecular mechanisms of iron binding and release in the periplasm.
Iron is an essential nutrient for almost all living bacteria (7). By changing the coordinating ligands and protein environment around iron, its redox potential can be made to vary between −300 and +700 mV, making it uniquely suited to participate in a wide range of electron transfer reactions involved in intermediary metabolism (3). For example, iron is a key component of a number of essential metabolic enzymes, including the cytochromes and ribonucleotide reductase. Consequently, iron is essential for the virulence of many bacterial pathogens (5, 12). Although it is the fourth most abundant element in the earth's crust, iron is inaccessible as a component of insoluble hydroxides in the environment. In mammals, iron is also inaccessible to pathogens because it is sequestered by high-affinity iron-binding proteins (transferrins [Tf], lactoferrins [Lf], ferritin, hemoglobin, and hemopexin). Therefore, pathogenic bacteria have been forced to evolve elaborate strategies to engage in a tug-of-war with the mammalian host for this precious metal.
Mannheimia haemolytica (previously known as Pasteurella haemolytica) is a gram-negative coccobacillus that is an opportunistic pathogen of cattle, sheep, and other ruminants (28). It causes a life-threatening hemorrhagic pneumonia, termed shipping fever or pneumonic pasteurellosis, in cattle that is responsible for the deaths of at least 1% of North American feedlot cattle. In order to survive in the iron-limited environment present in the host, M. haemolytica and other pathogenic gram-negative bacteria from the Pasteurellaceae and Neisseriaceae families have developed high-affinity iron acquisition systems in which iron transport into the bacterial cell is initiated by outer membrane proteins specific for the Tf and Lf (14, 22, 24, 25). The heterodimeric Tf bacterial outer membrane receptor is composed of two proteins, Tf binding protein A (TbpA) and the extrinsic lipoprotein TbpB. Upon binding the host protein, the ferric ion is removed by the receptor proteins and transported through the TonB-dependent integral membrane protein TbpA. TbpA is proposed to form a β-barrel and function as a gated porin requiring the presence of a functional TonB protein for the translocation of iron into the periplasmic space (6).
After crossing the outer membrane, the iron is transported into the cell by an ATP binding cassette (ABC) pathway specific for iron. Within the periplasm, the ferric ion is complexed by ferric ion-binding protein A (FbpA) (9, 18). FbpA shuttles the iron to an inner membrane complex consisting of two proteins, the inner transmembrane FbpB and the cytoplasmic ATPase FbpC. The energy for the transport of iron across the inner membrane is provided by the hydrolysis of ATP by the inner membrane-associated proteins. Pathways analogous to the one described above are also utilized in gram-negative bacteria for the uptake of iron from siderophores and heme, as well as for the import of amino acids, sugars, and other nutrients (10, 19).
Several outer membrane and periplasmic protein components of different ABC transport pathways responsible for iron uptake have been well characterized biochemically and structurally (10, 19). However, there is still considerable debate surrounding the detailed molecular mechanisms of iron translocation across the outer membrane and the mechanics of iron acquisition and release by the periplasmic iron-binding proteins.
Recently, we were able to determine the 1.2-Å iron-free and formate-bound structure of M. haemolytica FbpA (MhFpbA) (apo-MhFbpA) (27), revealing a new class of periplasmic iron-binding proteins. In the same study, we were able to assign putative iron- and anion-binding residues and identified 21 bacterial homologs with apparently identical machinery for binding ferric ions and synergistic anions. To understand how MhFbpA binds a ferric ion, we sought to determine the crystal structure of the iron-loaded form of the protein. Here, we present two high-resolution structures of MhFbpA complexed with iron (MhFbpAFe) and iron-carbonate (MhFbpAFeCO3). The mode of ferric ion binding is novel and unexpected. Our results also reveal that MhFbpA is able to bind a single iron atom while the protein is in an open conformation and in the absence of a discretely bound synergistic anion, which is a highly unusual observation for characterized ferric ion transport proteins. The synergistic anion appears to play a key role in inducing large-scale global conformational changes in the protein. Our present results, analyzed in combination with the structure of apo-MhFbpA, allow us to propose models detailing the molecular mechanisms of iron uptake and release for this iron-binding protein class in the periplasm of gram-negative bacteria.
MATERIALS AND METHODS
Crystallization.
MhFbpA was expressed in Escherichia coli and purified as described previously (27). All protein samples were concentrated to 30 mg/ml in 10 mM Tris buffer (pH 7.5) with 10 mM sodium bicarbonate. The two crystal forms of the iron-loaded MhFbpA were grown by vapor diffusion by the hanging drop method. The new MhFbpA samples were initially screened around the crystallization conditions that had previously been determined for the apo-MhFbpA structure (15). Since these conditions had proved successful for the same protein, they served as a starting point in attempts to obtain an iron-loaded crystal form. Crystals of the closed holoprotein (MhFbpAFeCO3) were grown at 4°C by the hanging drop technique with 4-μl drops containing 15 mg of the FbpA/ml, 0.05 M HEPES (pH 7.5), 4.25% isopropanol, 8.5% polyethylene glycol (PEG) 4000, and 7.5% glycerol. The drops were equilibrated against a 1-ml reservoir containing 0.1 M HEPES (pH 7.5), 8.5% isopropanol, 17% PEG 4000, and 15% glycerol. Diffraction-quality orange crystals typically appeared within 1 week in these crystallization experiments and belonged to the orthorhombic space group C2221. Similarly, crystals of the open iron-loaded form (MhFbpAFe) were grown with hanging drops (4 μl) at 4°C. These drops contained 15 mg of the FbpA/ml, 11% PEG 3350, and 0.1 M citrate (pH 8.2). These drops were also equilibrated against a 1-ml reservoir containing 22% PEG 3350 and 0.2 M citrate (pH 8.2). Diffraction-quality orange crystals belonging to the monoclinic space group P21 appeared within 4 to 5 days in these crystallization experiments.
Crystal harvesting and data collection.
The crystals were harvested by scooping them with a nylon loop. Because the closed iron-loaded crystals were grown in a cocktail that already contained 15% glycerol in the reservoir, no further addition of cryoprotectant was made prior to cooling the crystals in liquid nitrogen. However, for the open iron-loaded form, the crystals were dipped into a cryoprotectant solution containing 30% (vol/vol) ethylene glycol, 22% PEG 3350, and 0.2 M citrate (pH 8.2) for ∼30 s before they were cooled in liquid nitrogen. All X-ray data sets were collected for single crystals at 100 K by using cryomounting procedures. The crystals were maintained at 100 K during data collection by use of a nitrogen gas stream. Diffraction data for both forms of MhFbpA crystals were collected by using a QUANTUM4 ADSC detector on Beamline 5.03 at the ALS Berkeley Laboratory, Berkeley, Calif., with a wavelength of 1.0 Å. The data for all experiments were reduced and scaled by using the HKL2000 suite (23). The individual data-processing statistics for the MhFbpA crystals are reported in Table 1.
TABLE 1.
X-ray data collection and refinement statistics
| Statistic | Value for Fe-loaded form of MhFbpAa
|
|
|---|---|---|
| Closed | Open | |
| Data collection statistics | ||
| Wavelength (Å) | 1.00 | 1.00 |
| Resolution (Å) | 50-1.35 | 50-1.45 |
| Space group | C2221 | P21 |
| Unit cell dimensions (Å) | a = 97.67, b = 192.94, c = 43.35, α = 90.0, β = 90.0, γ = 90.0 | a = 46.03, b = 48.19, c = 77.36, α = 90.0, β = 100.92, γ = 90.0 |
| Completeness (%) | 99.9 (99.7) | 98.7 (98.1) |
| I/σ(I) | 32.5 (3.4) | 19.1 (2.1) |
| Rsymb | 5.8 (52.5) | 5.4 (52.2) |
| Redundancy | 7.2 | 3.0 |
| Refinement statistics | ||
| Resolution (Å) | 50.0-1.35 | 50.0-1.45 |
| Completeness for range (%) | 99.86 | 98.73 |
| No. of reflectionsc | 85,709 | 55,606 |
| No. of reflections in working set | 81,183 | 52,651 |
| No. of reflections in free set | 4,526 | 2,955 |
| R-factord | 0.159 | 0.174 |
| R-free | 0.174 | 0.192 |
| No. of protein atoms | 5,070 | 5,038 |
| No. of water molecules | 508 | 433 |
| Fe atoms | 1 | 1 |
| Mean B-factors (Å2) | 11.40 | 15.55 |
| Root mean square deviation from ideality | ||
| Bond lengths (Å) | 0.006 | 0.006 |
| Bond angles (°) | 1.096 | 1.051 |
Numbers in parentheses are statistics for the highest-resolution shells.
Rsym = Σh (Σj |Ij,h − <Ih>|/ΣIj,h), where h is the set of Miller indices and j is the set of observations of reflection h.
F > 2.0σ.
R-factor = Σhkl |Fo − Fc|/Σhkl |Fo|.
Structure solution and refinement.
In order to determine the structures of the MhFbpA by molecular replacement, the coordinates of the 1.2-Å wild-type apo-MhFbpA (Protein Data Bank code 1Q35) were used as the search model. The formate anions and all of the water molecules were removed from the model. Since the structure of apo-MhFbpA was in a closed conformation compared with the open iron-loaded form found here, two independent search models comprising the N- and C-terminal domains of the protein had to be positioned independently to solve that structure. For both structures, the program MolRep from the CCP4 suite of programs was used to perform the molecular replacement (11). Iterative cycles of manual refitting of the models with the program XtalView/Xfit (20) made use of maps created with ARP/wARP 5.2 (21), and refinement with Refmac5 (29) was carried out to complete and correct the models. Restrained refinement with a maximum-likelihood target function and isotropic temperature factors was carried out. During the later stages of refinement, difference maps (|Fo| − |Fc| maps) were utilized to place the bound ethylene glycol, carbonate, ferric ion, and water molecules when required.
Structure analysis.
The refined coordinates of MhFbpA in complex with iron-carbonate and iron alone have been deposited in the Protein Data Bank (codes 1SI0 and 1SI1). Ramachandran plots of the two structures reveal the satisfactory location of all residues in allowed regions of conformational space. The DALI server was used to find structurally similar proteins in the fold classification based on structure-structure alignment in the Proteins database (16).
RESULTS
Overview of MhFbpA structure.
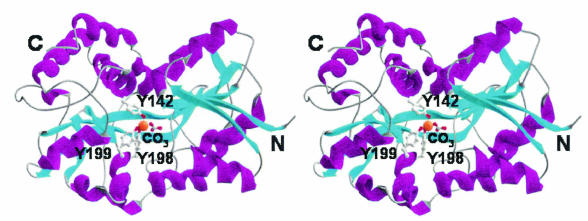
All structurally characterized bacterial periplasmic ligand-binding proteins to date adopt two basic protein folds despite low sequence homology within the protein class (10). MhFbpA belongs to the transferrin superfamily, adopting the more common of the two periplasmic binding protein folds (Fig. 1). The MhFbpA fold has also been observed in the periplasmic ferric ion-binding proteins from Neisseria (NgFbpA) (1) and Haemophilus (HiFbpA) (8). MhFbpA possesses two asymmetric globular α/β domains connected by a pair of antiparallel β-strands. The Fe3+ ion-binding site resides at the bottom of a deep cleft between the two globular domains. The domain-spanning β-strands also reside at the base of the iron binding cleft, acting as flexible hinges that allow the two domains to rotate with respect to one another. The interdomain rotations generally correspond with the opening and closing of the binding cleft in response to the presence of bound ligand in a Venus flytrap-like manner.
FIG. 1.
Stereo ribbon diagram of MhFbpA structure. MhFbpA possesses two α/β domains linked by a β-strand hinge. α-Helices are shown in purple, and β-strands are shown in cyan. The relative positions of the iron (orange sphere), carbonate, and iron-binding residues are shown.
At the topological level, MhFbpA is similar to HiFbpA and NgFbpA. The α-carbon atoms of MhFbpAFeCO3 superimpose on the α-carbons of the iron-loaded forms of HiFbpA and NgFbpA with root mean square deviations of approximately 2.3 Å. As members of the transferrin superfamily of folds, MhFbpA, HiFbpA, and NgFbpA also share a lesser but significant degree of structural and topological similarity with the N and C lobes of human Tf, which has led to the adoption of the moniker “bacterial transferrins” for HiFbpA and NgFbpA (13). However, while MhFpbA shares a similar overall fold with the other iron-chelating members of the transferrin superfamily, it differs from the other superfamily members in its iron coordination scheme and anion-binding mode. MhFbpA is also unique among the other characterized ferric ion transport proteins in that its global protein conformation appears to be mediated primarily by the anion, not the ferric ion.
Ferric ion coordination.
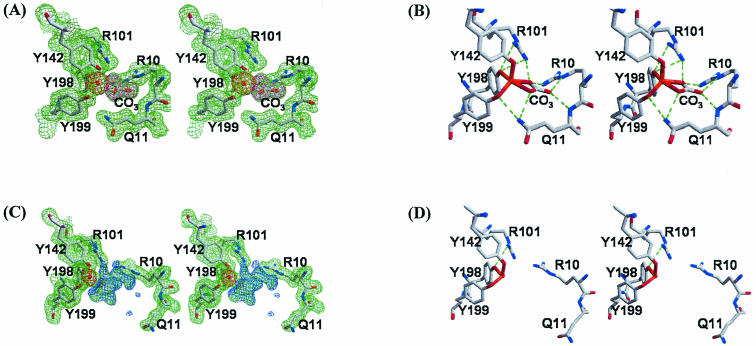
Although the global protein conformations of the MhFbpAFe and MhFbpAFeCO3 structures are dramatically different, both utilize the same protein residues in ferric ion binding. In both structures, the ferric ion is coordinated by three tyrosines, Tyr142, Tyr198, and Tyr199, from the C-terminal domain in nearly identical constellations (Fig. 2). The three tyrosinate anions act to neutralize the +3 charge on the ferric ion. The tyrosine hydroxyls form a distorted trigonal plane on the side of the ferric ion facing the C-terminal domain, approximately parallel to the C-terminal face of the binding cleft. In the MhFbpAFeCO3 structure, a carbonate anion is positioned on the opposite side of the ferric ion, where it coordinates the ferric ion in a bidentate fashion with the plane of the carbonate lying perpendicular to the plane described by the tyrosine hydroxyl groups. Bidentate coordination of iron by a carbonate anion is also observed in Lf and Tf (4). Collectively, the three tyrosyl residues and the carbonate anion surround the ferric ion in an asymmetric manner, suggesting that the interaction between the ferric ion and the protein is predominantly electrostatic in nature.
FIG. 2.
Stereoviews of the iron-binding site of MhFbpA. (A) Electron density maps of the iron-loaded closed form (green for the 2Fo-Fc map contoured at 1σ, magenta for the Fo-Fc map contoured at 5σ obtained with the reflection data after refinement of the model in which the carbonate molecule was omitted, and red for the Fo-Fc map contoured at 5σ obtained with the reflection data after refinement of the model in which the iron atom [orange sphere] was omitted). (B) Potential hydrogen bonding interactions are shown as green dotted lines. The iron ligand residues are shown with red bonds to the iron atom. (C) Electron density maps of the iron-loaded open form (green for the 2Fo-Fc map contoured at 1σ, blue for the Fo-Fc map contoured at 3σ, and red for the Fo-Fc map contoured at 5σ obtained with the reflection data after refinement of the model in which the iron atom was omitted). (D) Potential hydrogen bonding interactions are shown as green dotted lines. The iron ligand residues are shown with red bonds to the iron atom.
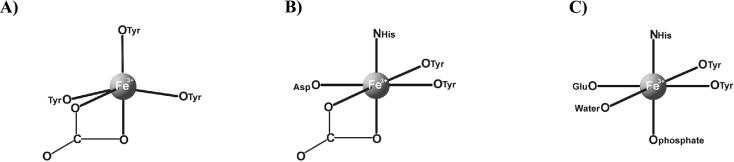
In the MhFbpAFe structure, the N- and C-terminal domains are rotated away from one another, moving the carbonate binding site away from the ferric ion and exposing one side of the ferric ion to solvent. Despite the opening of the binding cleft, the ferric ion remains coordinated by Tyr142, Tyr198, and Tyr199. On the solvent-exposed side of the ferric ion, we observed moderately strong (>3σ), continuous difference electron density within coordination distance of the iron, which extends towards the N-terminal domain and a carbonate binding Arg10 residue. However, since the density was too indistinct to reliably fit water molecules or organic ions, it was left unassigned. Since the crystals of MhFbpAFe were grown in the presence of high concentrations of citrate (see Materials and Methods), we postulate that the difference electron density around the iron arises from dynamically and/or statically disordered citrate. The distances between the ferric ion coordinating tyrosine hydroxyl groups in both structures and the ferric ion (all metal-ligand distances were not restrained during refinement) were identical within the experimental error, averaging 1.97 Å. At 1.97 Å, the tyrosyl-iron coordination distances observed in the title structures fall into the range that is typically observed in structurally characterized iron transport proteins, including Lf (2), Tf (15), HiFbpA (8), and NgFbpA (1). However, MhFbpA differs from other characterized human and bacterial ferric ion transport proteins in a number of key aspects (Fig. 2 and 3). MhFbpA utilizes only three protein residues, instead of four, to coordinate iron and, with the inclusion of the anion, coordinates iron exclusively with oxygen atoms in the inner coordination sphere. MhFbpA is the only characterized ferric transport protein to utilize three tyrosine residues in iron coordination and ferric ion charge neutralization (all others utilize two tyrosines and an aspartate or glutamate to achieve the same end). MhFbpA utilizes only five coordinating moieties, which are disposed in an asymmetric fashion around the metal atom, to ligate iron, while all other superfamily members coordinate iron with a total of six ligands and an octahedral coordination sphere. Finally, we have observed that MhFbpA is able to ligate iron without an ordered, protein-bound synergistic anion.
FIG. 3.
Coordination of ferric ion. The geometry for the coordination of a ferric ion in MhFbpA (A), Tf (B), and HiFbpA or NgFbpA (C) is illustrated.
A comparison of the UV-visible absorption properties of MhFbpA (17) to those of Tf and NgFbpA (13) also serves to highlight its uniqueness. Iron-loaded MhFbpA possesses a peak visible absorbance at 419 nm, which is significantly blue shifted compared to the same parameter from Tf and HiFbpA (∼480 nm) (13). Since the effects of anions on peak visible absorbance maxima are small (on the order of 5 to 10 nm) (13), the 61-nm blue shift in the visible absorbance maximum is most likely caused by the coordination of the ferric ion by an additional tyrosine.
The role of carbonate in mediating protein conformation.
As shown in Fig. 2 and 4, carbonate plays an important role in iron binding and global protein conformation in MhFbpA. The carbonate anion binds in a basic pocket in the N-terminal domain and has its charge compensated for by a pair of arginine residues (Arg10 and Arg101). In a classic example of molecular recognition, the full hydrogen bonding potential of the carbonate is utilized through main chain and side chain hydrogen bonding interactions with Gln11 as well as hydrogen bonding interactions with Arg10 and Arg101. The two carbonate oxygens that coordinate iron do so in a slightly asymmetric manner, with coordination distances of 2.10 and 2.20 Å, respectively. The asymmetry is likely due to a partial preference for a carbonate resonance form which places a greater partial negative charge on the carbonate oxygen positioned near the bottom of the binding cleft. The oxygen has the largest number of close contacts with positively charged groups, as it simultaneously interacts with Arg10, Arg101, and the Fe3+ ion.
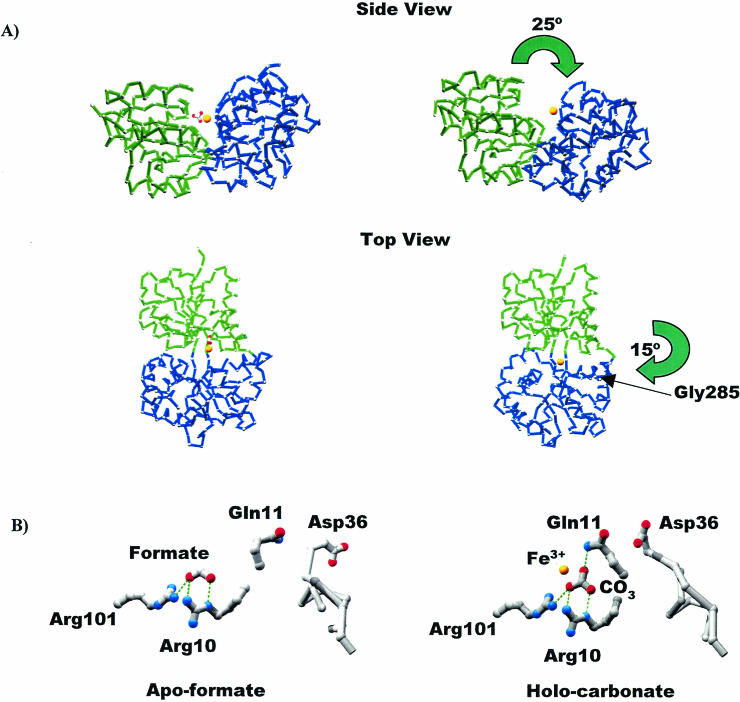
FIG. 4.
Conversion from the closed to the open form of MhFbpA. (A) Cα traces comparing the iron-loaded closed and iron-loaded open forms of the protein. The N-terminal (green) and C-terminal (blue) domains are colored differently to clarify visualization of the domain movements. The domain reorganization can be characterized by two hinge-bending movements: a 25° opening of the N and C lobes through an axis parallel to the ligand-binding cleft, and a 15° rotation through a perpendicular axis through Gly285. The iron atom (yellow sphere) and the carbonate anion are also shown. (B) Comparison of iron-apo-formate and holo-carbonate structures. H bonds are shown as green dotted lines. Formate and carbonate utilize virtually identical binding modes with respect to Arg10, Arg101, and most surrounding elements. Both structures globally adopt closed protein conformations. However, the additional oxygen atom from the carbonate anion faces the solvent-exposed side of the binding pocket, twisting the Gln11 side chain around so that it H bonds with carbonate, which in turn induces a movement in the loop containing Asp36 so that it covers the binding pocket and sequesters the ferric ion from solvent.
The carbonate modulates the conformation of the protein globally by affecting the relative orientations of the N- and C-terminal domains and also causes more localized changes in elements of the N-terminal domain in the vicinity of the ligand-binding site. The carbonate promotes a closed conformation of the protein by stitching the N- and C-terminal domains of MhFbpA together through hydrogen bonding interactions. Arg10 and Gln11 from the N-terminal domain bridge to the ferric ion through their direct interactions with carbonate. The Gln11 side chain also forms bridging hydrogen bonds between the carbonate and Tyr199, one of the ferric ion-coordinating tyrosines (Fig. 2).
A comparison of the carbonate-bound, carbonate-free and iron-apo-formate-bound (27) structures of MhFbpA reveals the exact nature of the carbonate-induced structural transitions (Fig. 4). Globally, the release of carbonate is associated with individual large-scale movements of the N- and C-terminal domains. The domain reorganization is described by rotations about a pair of orthogonal axes near the domain interface (Fig. 4). In the MhFbpAFe structure, the domains rotate away from one another compared to the MhFbpAFeCO3 structure via a 25° hinge-bending motion through an axis described by the α-carbons of Thr100 and Gly233, running along the base of the ligand-binding cleft. Thr100 and Gly233 reside in the adjacent β-strands connecting the N- and C-terminal domains. A concomitant 15° hinge bending of the N- and C-terminal domains around the Gly285 Cα also distinguishes the MhFbpAFe structure from the MhFbpAFeCO3 structure. This second rotation is through an axis that is approximately perpendicular to the floor of the ligand-binding cleft. The hinge-bending around Gly285 appears to be necessary to keep hydrophobic surfaces at the underside of the ligand-binding cleft from separating and becoming solvent exposed while allowing the ligand-binding cleft to open. Taken together, these rotations are associated with the opening of the iron-binding cleft and increased solvent exposure of the bound ferric ion. However, the domain movements were also found to maintain a closed domain interface on the underside of the ligand-binding cleft with an altered series of polar and hydrophobic domain-domain interactions when the structures of MhFbpAFeCO3 and MhFbpAFe were compared (Fig. 4). It is important to note that the underside of the ligand-binding cleft has a significant proportion of hydrophobic residues on its interface but that the faces of the ligand-binding cleft itself are almost entirely composed of polar residues. Thus, based on the analysis presented above, there is likely a minimal difference between the free energies of the open and closed states of MhFbpA, so that the protein can readily adopt either state in solution.
A comparison of the MhFbpAFeCO3 and apo-MhFbpA structures reveals that the carbonate anion not only controls the relative orientations of the N- and C-terminal domains but also plays a direct role in stabilizing conformations of loops in the solvent-exposed regions of the iron-binding pocket that sequester the ferric ion from solvent (Fig. 4). The apo-MhFbpA structure, while lacking a ferric ion, was found to contain two formate (HCO2−) anions disposed on opposite sites of the iron-binding pocket (27). Formate was present in high concentrations during crystallization. In the overlaid structures of MhFbpAFeCO3 and apo-MhFbpA, the oxygens of one of the formate anions superimpose almost exactly on the pair of carbonate oxygens that face the bottom of the binding pocket (Fig. 4). These two oxygen ligands are sufficient to promote a closed domain-domain interface without the assistance of any other ligands or iron, since the structures of MhFbpAFeCO3 and apo-MhFbpA are virtually identical and superimpose with a root mean square deviation of 0.6 Å. The structures differ significantly only in the conformations of Gln11 and the neighboring Asp36-Leu39 surface loop, which are disordered and poorly resolved in the apo-MhFbpA structure. In the apo-MhFbpA structure, the amide terminus of Gln11 points away from the iron-binding pocket and is disordered. In the MhFbpAFeCO3 structure, however, the Gln11 side chain is ordered and observed in two conformations: a major conformation in which the side chain rotates towards the ligated iron and interacts simultaneously with carbonate and Tyr199, and a minor conformation in which the Gln11 side chain hydrogen-bonds with Asp36 and stabilizes a conformation of the Asp36-Leu39 loop that partially covers the ligand-binding cleft and decreases the exposure of the iron-binding pocket to solvent. Formate lacks the third oxygen atom necessary for the simultaneous coordination of the ferric ion and tethering of Gln11.
DISCUSSION
We have carried out a detailed analysis and comparison of three high-resolution structural snapshots of MhFbpA in three unique liganded states. The MhFbpAFe form represents an open conformation, the iron-free apo-MhFbpA form represents an anion-bound closed conformation with a solvent-exposed ferric ion-binding site, and the MhFbpAFeCO3 form represents a fully closed conformation. Our analysis has allowed us to characterize the precise atomic details of ligand binding and to dissect the roles played by the ferric ion and carbonate in mediating protein conformation. The structural work also provides general insights into the possible mechanisms of iron acquisition and release utilized by MhFbpA and its homologs as part of ABC transport pathways in vivo.
It is clear from our structural analysis that the carbonate anion acts as the trigger that switches MhFbpA between its open and closed states. Surprisingly, we were able to observe iron ligation in both the open and the closed forms of MhFbpA, indicating that in the MhFbpA protein class, a specific synergistic anion may not be essential for iron ligation. However, it is likely that the affinity of MhFbpA for ferric ions is lower in the open state than in the closed state. Taken together, and focusing on the periplasmic protein component, these observations are consistent with at least two discrete mechanisms of iron transport in pathways utilizing FbpAs in the MhFbpA protein class to bind iron at the outer membrane and release it at the inner membrane (Fig. 5 and 6). A current model for iron removal from Tf and transport across the outer membrane is depicted in Fig. 5. The model proposes that binding to the receptor induces conformational changes in Tf to reduce its affinity for iron so that binding by FbpA can drive the transport process once a channel is created by interaction with TonB (26). This model is supported by recent studies with a series of HiFbpA mutants demonstrating that a relatively high binding affinity threshold is necessary for iron translocation (A. G. Khan, S. R. Shouldice, S. M. Kirby, R.-H. Yu, and A. B. Schryvers, submitted for publication). To facilitate iron translocation across the outer membrane, it is likely that MhFbpA docks against the outer membrane in a carbonate-bound state, which would have maximal affinity for iron. A comparison of the MhFbpAFeCO3 and formate-bound apo-MhFbpA structures reveals that anion-bound MhFbpA positions all of the iron-binding protein ligands within 0.5 Å of their positions in the ferric ion-bound state. Anion-bound MhFbpA would have a predefined high-affinity ferric ion-binding site available to attract a ferric ion diffusing through TbpA. Accessibility of the ferric ion-binding site to solvent is gated by the conformations of Gln11 and the Asp36-Leu39 loop, which appear highly flexible on the basis of our structural analysis.
FIG. 5.
Model for the iron uptake pathway. Binding of hTf by TbpA induces a conformational change that lowers the affinity of hTf for iron. Interaction of TbpA with TonB provides a channel for transport across the outer membrane. FbpA binds its synergistic anion (carbonate), adopting a closed conformation that has a high affinity for iron. The transport of iron across the outer membrane is driven by the high-affinity binding of iron by FbpA. Iron release occurs by two possible mechanisms. In path A, FbpA loses carbonate or swaps carbonate with a different anion, adopting an open conformation which subsequently docks to the FbpB/C inner membrane complex. In path B, carbonate removal is mediated by FbpB. In both scenarios, iron-loaded FbpA binds to the FbpB/C inner membrane complex and is released into the cytoplasm. Energy for iron translocation is provided by the ATPase activity of FbpC.
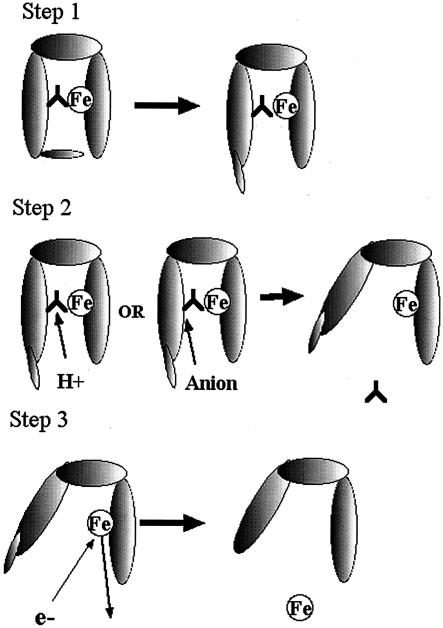
FIG. 6.
Iron removal from FbpA. In step 1, the Asp36-Leu39 loop (D36 loop) and Gln11 move to expose the carbonate anion (upside-down Y). In step 2, the carbonate anion is either protonated to facilitate release or displaced by an alternate anion (citrate). In step 3, iron is released, possibly after reduction, which lowers binding affinity.
The sequence of events surrounding iron release from MhFbpA almost certainly involves the release and/or substitution of the carbonate anion followed by the release of the bound ferric ion. As outlined in Fig. 5, the iron- and anion-free form of MhFbpA released from the inner membrane complex would complex with a readily available carbonate anion in the periplasmic space prior to accepting a ferric ion from the outer membrane receptor complex.
The first step in the iron removal process would involve the movement of the Gln11 and the Asp36-Leu39 loop to provide access to the anion binding and the iron binding site (Fig. 6). This step could occur spontaneously or be facilitated by interaction with the FbpB/C complex at the inner membrane. The removal of the carbonate anion could be facilitated by protonation through a reduction in pH in the microenvironment at the FbpA/B/C interface. The release of the carbonate anion would result in the opening of MhFbpA and the lowering of its affinity for ferric ions. A similar model for pH-mediated anion-iron release has been proposed for Tf in the endosome (4). Alternatively, the carbonate anion could be displaced by a synergistic anion, as proposed by Dhungana et al. (13). As already described, we observed difference electron density in the MhFbpAFe structure on the solvent-exposed face of the ferric ion that impinges on the carbonate-binding pocket. The difference electron density possibly arises from a disordered citrate molecule. It is also possible that the carbonate could be displaced by an acidic side chain presented by the FbpB receptor. Since domain movement in FbpA may be restricted when the protein is docked to FbpB, domain opening may be assisted by a concomitant conformational change in the FbpB receptor. Our observation of ferric ion binding in the presence of multiple anions is interesting, since it demonstrates that MhFbpA is able to transport iron in diverse chelation environments in vivo and may not require a discrete synergistic anion.
Once FbpA is opened (by any mechanism), the increased solvent exposure of the iron and the change in coordination would correspond with a significant positive shift in the ferric ion redox potential, as well as a lowered affinity of MhFbpA for the ferric ion. The positive shift in the redox potential could facilitate the reduction of the ferric ion in the opened form of MhFbpA that would further reduce the binding affinity and thus facilitate the diffusion of ferrous ions through the FbpBC channel. Our structural results will have to be coupled with additional biochemical and biophysical work to more clearly elucidate the mechanisms of iron binding and release by this novel second class of bacterial transferrins.
Acknowledgments
This work was supported by grant 49603 from the Canadian Institutes for Health Research. This work is based on diffraction experiments conducted at the Advanced Light Source (ALS). ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the U.S. Department of Energy under contract no. DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory.
We also thank the staff at ALS for their excellent support.
REFERENCES
- 1.Alexeev, D., H. Zhu, M. Guo, W. Zhong, D. J. Hunter, W. Yang, D. J. Campopiano, and P. J. Sadler. 2003. A novel protein-mineral interface. Nat. Struct. Biol. 10:297-302. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, B. F., H. M. Baker, G. E. Norris, D. W. Rice, and E. N. Baker. 1989. Structure of human lactoferrin: crystallographic structure analysis and refinement at 2.8 Å resolution. J. Mol. Biol. 209:711-734. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 4.Baker, H. M., B. F. Anderson, and E. N. Baker. 2003. Dealing with iron: common structural principles in proteins that transport iron and heme. Proc. Natl. Acad. Sci. USA 100:3579-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baltes, N., I. Hennig-Pauka, and G. F. Gerlach. 2002. Both transferrin binding proteins are virulence factors in Actinobacillus pleuropneumoniae serotype 7 infection. FEMS Microbiol. Lett. 209:283-287. [DOI] [PubMed] [Google Scholar]
- 6.Boulton, I. C., M. K. Yost, J. E. Anderson, and C. N. Cornelissen. 2000. Identification of discrete domains within gonococcal transferrin-binding protein A that are necessary for ligand binding and iron uptake functions. Infect. Immun. 68:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun, V., and H. Killmann. 1999. Bacterial solutions to the iron-supply problem. Trends Biochem. Sci. 24:104-109. [DOI] [PubMed] [Google Scholar]
- 8.Bruns, C. M., A. J. Norwalk, A. S. Avrai, M. A. McTigue, K. A. Vaughan, T. A. Mietzner, and D. E. McRee. 1997. Structure of Haemophilus influenzae Fe3+-binding protein reveals convergent evolution within a superfamily. Nat. Struct. Biol. 4:919-924. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C.-Y., S. A. Berish, S. A. Morse, and T. A. Mietzner. 1993. The ferric iron-binding protein of pathogenic Neisseria spp. functions as a periplasmic transport protein in iron acquisition from human transferrin. Mol. Microbiol. 10:311-318. [DOI] [PubMed] [Google Scholar]
- 10.Clarke, T. C., L. W. Tari, and H. J. Vogel. 2001. Structural biology of bacterial iron uptake systems. Curr. Top. Med. Chem. 1:7-30. [DOI] [PubMed] [Google Scholar]
- 11.Collaborative Computational Project Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 12.Cornelissen, C. N., M. Kelley, M. M. Hobbs, J. E. Anderson, J. G. Cannon, M. S. Cohen, and P. F. Sparling. 1998. The transferrin receptor expressed by gonococcal strain FA1090 is required for the experimental infection of human male volunteers. Mol. Microbiol. 27:611-616. [DOI] [PubMed] [Google Scholar]
- 13.Dhungana, S., C. H. Taboy, D. S. Anderson, K. G. Vaughan, P. Aisen, T. A. Mietzner, and A. L. Crumbliss. 2003. Bioinorganic chemistry special feature: the influence of the synergistic anion on iron chelation by ferric binding protein, a bacterial transferrin. Proc. Natl. Acad. Sci. USA 100:3659-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray-Owen, S. D., and A. B. Schryvers. 1996. Bacterial transferrin and lactoferrin receptors. Trends Microbiol. 4:185-191. [DOI] [PubMed] [Google Scholar]
- 15.Hall, D. R., J. M. Hadden, G. A. Leonard, S. Bailey, M. Neu, M. Winn, and P. F. Lindley. 2002. The crystal and molecular structures of diferric porcine and rabbit serum transferrins at resolutions of 2.15 and 2.60 A, respectively. Acta Crystallogr. D 58:70-80. [DOI] [PubMed] [Google Scholar]
- 16.Holm, L., and C. Sander. 1997. Dali/FSSP classification of three-dimensional protein folds. Nucleic Acids Res. 25:231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirby, S. D., F. A. Lainson, W. Donachie, A. Okabe, M. Tokuda, O. Hatase, and A. B. Schryvers. 1998. The Pasteurella haemolytica 35 kDa iron regulated protein is an FbpA homologue. Microbiology 144:3425-3436. [DOI] [PubMed] [Google Scholar]
- 18.Kirby, S. D., S. D. Gray-Owen, and A. B. Schryvers. 1997. Characterization of a ferric binding protein mutant in Haemophilus influenzae. Mol. Microbiol. 25:979-987. [DOI] [PubMed] [Google Scholar]
- 19.Koster, W. 2001. ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B12. Res. Microbiol. 152:291-301. [DOI] [PubMed] [Google Scholar]
- 20.McRee, D. E. 1999. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156-165. [DOI] [PubMed] [Google Scholar]
- 21.Morris, R. J., A. Perrakis, and V. S. Lamzin. 2002. ARP/wARP's model-building algorithms. I. The main chain. Acta Crystallogr. D 58:968-975. [DOI] [PubMed] [Google Scholar]
- 22.Ogunnariwo, J. A., and A. B. Schryvers. 1990. Iron acquisition in Pasteurella haemolytica: expression and identification of a bovine-specific transferrin receptor. Infect. Immun. 58:2091-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 267:307-326. [DOI] [PubMed] [Google Scholar]
- 24.Schryvers, A. B., and G. C. Gonzalez. 1990. Receptors for transferrin in pathogenic bacteria are specific for the host's protein. Can. J. Microbiol. 36:145-147. [DOI] [PubMed] [Google Scholar]
- 25.Schryvers, A. B., and L. J. Morris. 1988. Identification and characterization of the human lactoferrin-binding protein from Neisseria meningitidis. Infect. Immun. 56:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schryvers, A. B., and I. Stojiljkovic. 1999. Iron acquisition systems in the pathogenic neisseria. Mol. Microbiol. 32:1117-1123. [DOI] [PubMed] [Google Scholar]
- 27.Shouldice, S. R., D. R. Dougan, P. W. Williams, R. J. Skene, G. Snell, D. Scheibe, S. M. Kirby, D. J. Hosfield, D. E. McRee, A. B. Schryvers, and L. W. Tari. 2003. Crystal structure of Pasteurella haemolytica ferric ion-binding protein A reveals a novel class of bacterial iron binding proteins. J. Biol. Chem. 278:41093-41098. [DOI] [PubMed] [Google Scholar]
- 28.Whiteley, L. O., S. K. Maheswaran, D. J. Weiss, T. R. Ames, and M. S. Kannan. 1992. Pasteurella haemolytica A1 and bovine respiratory disease: pathogenesis. J. Vet. Intern. Med. 6:11-22. [DOI] [PubMed] [Google Scholar]
- 29.Winn, M. D., M. N. Isupov, and G. N. Murshudov. 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D 57:122-133. [DOI] [PubMed] [Google Scholar]