Abstract
We describe 2127 new human leukocyte antigen (HLA) class I alleles found in registered stem cell donors. These alleles represent 28.9% of the currently known class I alleles. Comparing new allele sequences to homologous sequences, we found 68.1% nonsynonymous nucleotide substitutions, 28.9% silent mutations and 3.0% nonsense mutations. Many substitutions occurred at positions that have not been known to be polymorphic before. A large number of HLA alleles and nucleotide variations underline the extreme diversity of the HLA system. Strikingly, 156 new alleles were found not only multiple times, but also in carriers of various parentage, suggesting that some new alleles are not necessarily rare. Moreover, new alleles were found especially often in minority donors. This emphasizes the benefits of specifically recruiting such groups of individuals.
Keywords: genetic diversity, hematopoietic stem cell transplantation, human leukocyte antigens, new alleles, sequencing-based typing
More than 9500 human leukocyte antigen (HLA) alleles have been described so far 1. Most new alleles are identified in the context of unrelated hematopoietic stem cell transplantation. The number of known HLA alleles has grown considerably in the last years, as donor centers and registries increasingly carry out comprehensive HLA typing at donor recruitment. The resulting DNA sequences typically include the antigen recognition sites of HLA class I loci A, B and C, and HLA class II loci DRB1, DQB1 and sometimes DPB1.
Here, we describe 2127 new HLA class I alleles, accounting for 28.9% of all HLA class I alleles described so far (see Table1; Table S1, Supporting Information) 2. Most of the new alleles are HLA-C (n = 774) and HLA-B alleles (n = 755), followed by 598 HLA-A alleles (Figure 1). These alleles were found in potential hematopoietic stem cell donors registered with DKMS donor centers in Germany, the United States and Poland.
Table 1.
Number of new HLA class I alleles found in individuals registered with different DKMS donor centers located in Germany, the United States and Poland. These alleles were included in the monthly HLA nomenclature updates between February 2009 3 and March 2013 4
| Allele group | Germanya | The United Statesa | Polanda | ≥2 countriesb | Total |
|---|---|---|---|---|---|
| A*01 | 41 | 17 | 5 | 4 | 67 |
| A*02 | 92 | 27 | 10 | 15 | 144 |
| A*03 | 60 | 10 | 6 | 3 | 79 |
| A*11 | 26 | 14 | 3 | — | 43 |
| A*23 | 11 | 5 | 3 | 1 | 20 |
| A*24 | 39 | 10 | 9 | 9 | 67 |
| A*25 | 7 | 1 | 3 | — | 11 |
| A*26 | 21 | 5 | 3 | 3 | 32 |
| A*29 | 8 | 4 | 1 | 2 | 15 |
| A*30 | 11 | 5 | 2 | 2 | 20 |
| A*31 | 9 | 1 | 2 | 2 | 14 |
| A*32 | 16 | 7 | 1 | 3 | 27 |
| A*33 | 10 | 8 | 2 | 1 | 21 |
| A*66 | 2 | — | 1 | 1 | 4 |
| A*68 | 20 | 10 | 3 | 1 | 34 |
| B*07 | 55 | 7 | 12 | 5 | 79 |
| B*08 | 29 | 9 | 6 | 1 | 45 |
| B*13 | 13 | 4 | 4 | — | 21 |
| B*14 | 15 | 4 | 1 | — | 20 |
| B*15 | 35 | 13 | 3 | 3 | 54 |
| B*18 | 23 | 12 | 2 | 3 | 40 |
| B*27 | 23 | 5 | 7 | 5 | 40 |
| B*35 | 49 | 17 | 8 | 8 | 82 |
| B*37 | 9 | — | 2 | 1 | 12 |
| B*38 | 10 | 2 | 3 | — | 15 |
| B*39 | 15 | 2 | 4 | 1 | 22 |
| B*40 | 32 | 19 | 8 | 3 | 62 |
| B*41 | 9 | 1 | 3 | — | 13 |
| B*44 | 58 | 17 | 4 | 4 | 83 |
| B*45 | 2 | — | — | — | 2 |
| B*46 | 1 | 3 | — | — | 4 |
| B*47 | 2 | — | — | — | 2 |
| B*48 | — | 1 | — | — | 1 |
| B*49 | 8 | 2 | — | — | 10 |
| B*50 | 8 | 2 | — | 2 | 12 |
| B*51 | 37 | 7 | 8 | 8 | 60 |
| B*52 | 5 | 6 | — | — | 11 |
| B*53 | 3 | 3 | — | — | 6 |
| B*55 | 8 | 3 | 1 | 1 | 13 |
| B*56 | 7 | — | 3 | 2 | 12 |
| B*57 | 19 | 4 | 3 | 2 | 28 |
| B*58 | 5 | — | — | — | 5 |
| B*78 | — | 1 | — | — | 1 |
| C*01 | 15 | 5 | 2 | 1 | 23 |
| C*02 | 32 | 3 | 6 | 5 | 46 |
| C*03 | 74 | 17 | 13 | 8 | 112 |
| C*04 | 49 | 26 | 6 | 10 | 91 |
| C*05 | 41 | 12 | 2 | 7 | 62 |
| C*06 | 37 | 12 | 7 | 4 | 60 |
| C*07 | 125 | 30 | 23 | 14 | 192 |
| C*08 | 14 | 7 | 1 | 1 | 23 |
| C*12 | 36 | 12 | 9 | 4 | 61 |
| C*14 | 12 | 3 | 2 | — | 17 |
| C*15 | 17 | 10 | 2 | 2 | 31 |
| C*16 | 23 | 12 | 1 | 2 | 38 |
| C*17 | 8 | 5 | 3 | 1 | 17 |
| C*18 | — | — | — | 1 | 1 |
| Total | 1336 | 422 | 213 | 156 | 2127 |
Location of DKMS donor centers.
New alleles found in more than one DKMS donor center.
Figure 1.
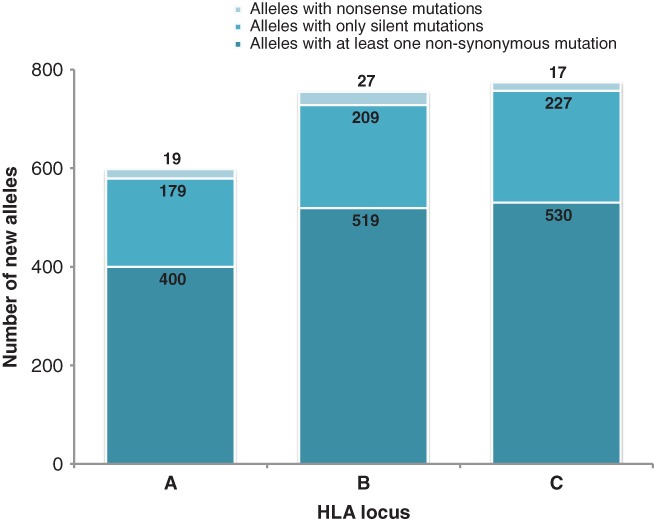
Total new HLA class I alleles per locus. Type of mutation (i.e. nonsynonymous, silent and nonsense mutations) is color coded.
All new sequences were reported to the IMGT/HLA Database and named by the World Health Organization (WHO) Nomenclature Committee. These newly named alleles were then included in the monthly HLA nomenclature updates between February 2009 3 and March 2013 4.
HLA class I (HLA-A, -B, -C) alleles were genotyped at the ASHI-accredited laboratory HistoGenetics (Ossining, NY) using sequencing-based typing (SBT). Sequencing templates were produced by locus- or group-specific pairs of oligonucleotide primers from genomic DNA by polymerase chain reaction to amplify exons 2 and 3 5–7. A total of 25 locus- and group-specific primers were used to amplify the target sequences. Sanger cycle sequencing was carried out using BigDye V3.1 (Applied Biosystems, Foster City, CA) chemistry and ABI 3730xl capillary sequencer for base calling. In order to sequence the entire exons, class I sequencing primers were designed for each HLA locus using locus-specific sequences located in the intron/exon boundary regions. Furthermore, when new substitutions of generic amplification were found, the strand carrying the new allele was sequenced in isolation by sequencing group-specific amplification products, whenever possible. Otherwise, sequence-specific primers were used.
Subsequently, DNA sequences of all known HLA alleles cataloged in Release 3.12.0 of the IMGT/HLA Database 8 were aligned by locus to determine each new allele's most homologous equivalent. The most homologous equivalent was defined as follows:
First, we identified alleles whose DNA sequences showed highest similarity to the new allele's DNA sequence (i.e. having a minimum amount of nucleotide substitutions). For example, if a new allele's sequence differed by two or more nucleotides from any other allele, the minimum amount of nucleotide substitutions was two. Hence, all alleles whose sequences differed by exactly two nucleotides from the new allele's sequence fulfilled the first criterion.
Among those alleles that met criterion 1, alleles potentially encoding the most similar polypeptide were selected, namely this criterion includes those alleles with a maximum number of silent substitutions.
Finally, if more than one allele met criteria 1 and 2, the earliest allele reported was chosen (avoiding null alleles).
Only exons 2 and 3 were considered during the definition of homologous alleles.
Comparing each new allele to its most homologous equivalent, we were able to detect and describe variations in DNA sequences (Table2; Table S1). This comparison showed that most new alleles (1995; 93.8%) were single nucleotide variants of their most homologous equivalents (Figure 1). Yet, some alleles differed by several nucleotides from their most homologous equivalents. These alleles included HLA-A*01:95 that varied by seven nucleotides as well as alleles HLA-B*44:90, HLA-B*40:166, HLA-B*46:32 and HLA-C*07:242 that differed by six nucleotides from their respective most homologous counterparts. Further examination of the nucleotide variations demonstrated that 68.1% of the new alleles comprised nonsynonymous nucleotide substitutions, while 28.9% comprised only silent mutations. The remaining 3.0% of the new alleles showed nonsense mutations (Figure 1). These alleles were further classified as null alleles, due to the presence of a premature stop codon in their sequences.
Table 2.
Description of nucleotide substitutions for all new HLA class I alleles that were found in at least 10 individuals
| HLA | New allele | Most homologous allele | NVa | CAb | Codon changec | AA changed | Type of mutation | IRe | Accession no. |
|---|---|---|---|---|---|---|---|---|---|
| A | A*01:40 | A*01:01:01:01 | 1 | 15 | GCG≫GTG | A136V | Nonsynonymous | 15 | FJ940763 |
| A | A*02:01:23 | A*02:01:01:01 | 1 | 36 | TCC≫TCT | S13S | Silent | 11 | FJ224141 |
| A | A*02:01:26 | A*02:01:01:01 | 1 | 36 | CCG≫CCA | P50P | Silent | 14 | FJ224211 |
| A | A*02:01:34 | A*02:01:01:01 | 1 | 37 | CGC≫CGT | R111R | Silent | 10 | FJ619451 |
| A | A*02:01:37 | A*02:01:01:01 | 1 | 36 | GAC≫GAT | D129D | Silent | 10 | FJ875539 |
| A | A*02:05:02 | A*02:05:01 | 1 | 3 | GAG≫GAA | E173E | Silent | 19 | FJ224147 |
| A | A*02:158 | A*02:01:01:01 | 1 | 36 | ACT≫GCT | T73A | Nonsynonymous | 21 | FJ224142 |
| A | A*03:01:11 | A*03:01:01:01 | 1 | 18 | CTG≫CTC | L81L | Silent | 12 | FJ765913 |
| A | A*03:01:14 | A*03:01:01:01 | 1 | 17 | GCG≫GCA | A153A | Silent | 22 | FJ976868 |
| A | A*03:01:17 | A*03:01:01:01 | 1 | 17 | TCG≫TCT | S105S | Silent | 12 | FJ619442 |
| A | A*03:48 | A*03:01:01:01 | 1 | 17 | TAC≫TCC | Y27S | Nonsynonymous | 10 | FJ358629 |
| A | A*11:47 | A*11:01:01 | 1 | 14 | TAC≫CAC | Y9H | Nonsynonymous | 10 | FJ222571 |
| A | A*23:01:02 | A*23:01:01 | 1 | 7 | CAC≫CAT | H3H | Silent | 17 | FJ224143 |
| A | A*24:02:19 | A*24:02:01:01 | 1 | 26 | CCG≫CCC | P57P | Silent | 14 | FJ619421 |
| A | A*24:106 | A*24:66 | 2 | 4 | GAA≫GAC | E114D | Nonsynonymous | 10 | FJ619413 |
| CAC≫GAC | H116D | Nonsynonymous | |||||||
| A | A*26:39 | A*26:01:01 | 1 | 7 | GGG≫CGG | G107R | Nonsynonymous | 12 | FJ224158 |
| A | A*29:21 | A*29:02:01:01 | 1 | 5 | CGC≫CTC | R21L | Nonsynonymous | 12 | FJ875544 |
| A | A*30:29 | A*30:04:01 | 1 | 1 | TGG≫TCG | W167S | Nonsynonymous | 13 | FJ976741 |
| A | A*31:26 | A*31:01:02 | 1 | 11 | GCC≫CCC | A125P | Nonsynonymous | 23 | FJ224188 |
| A | A*31:27 | A*31:01:02 | 1 | 11 | CGG≫TGG | R48W | Nonsynonymous | 18 | FJ765932 |
| A | A*32:01:04 | A*32:01:01 | 1 | 4 | ATC≫ATA | I124I | Silent | 13 | FJ976750 |
| A | A*33:27 | A*33:01:01 | 1 | 1 | GAC≫GAG | D106E | Nonsynonymous | 14 | FJ594710 |
| B | B*07:02:10 | B*07:02:01 | 1 | 15 | CTC≫CTT | L110L | Silent | 11 | FJ346326 |
| B | B*07:87 | B*07:02:01 | 1 | 15 | GCG≫GTG | A139V | Nonsynonymous | 16 | FJ875564 |
| B | B*08:01:08 | B*08:01:01 | 2 | 4 | CCG≫CCC | P47P | Silent | 10 | FJ875561 |
| CCG≫CCA | P50P | Silent | |||||||
| B | B*51:69 | B*51:01:01 | 1 | 11 | GCG≫ACG | A135T | Nonsynonymous | 11 | FJ392179 |
| C | C*01:02:07 | C*01:02:01 | 1 | 8 | GCC≫GCG | A135A | Silent | 16 | FJ594542 |
| C | C*01:02:08 | C*01:02:01 | 1 | 9 | ACC≫ACG | T143T | Silent | 17 | FJ614613 |
| C | C*01:32 | C*01:02:01 | 1 | 8 | CGC≫AGC | R131S | Nonsynonymous | 19 | FJ976875 |
| C | C*02:02:07 | C*02:02:02 | 1 | 3 | GAG≫GAA | E58E | Silent | 21 | FJ594540 |
| C | C*02:02:09 | C*02:02:02 | 1 | 3 | ACC≫ACT | T94T | Silent | 10 | FJ976839 |
| C | C*03:03:07 | C*03:03:01 | 1 | 5 | ACC≫ACG | T143T | Silent | 11 | FJ554597 |
| C | C*04:01:08 | C*04:01:01:01 | 1 | 15 | ACG≫ACC | T138T | Silent | 18 | FJ594538 |
| C | C*04:01:13 | C*04:01:01:01 | 1 | 14 | CGC≫CGT | R17R | Silent | 13 | FJ619434 |
| C | C*04:56 | C*04:01:01:01 | 1 | 14 | CCG≫CGG | P47R | Nonsynonymous | 15 | FJ875616 |
| C | C*05:01:08 | C*05:01:01:01 | 1 | 8 | CTC≫CTT | L168L | Silent | 16 | FJ969926 |
| C | C*05:34 | C*05:01:01:01 | 1 | 8 | GCC≫ACC | A135T | Nonsynonymous | 10 | FJ875589 |
| C | C*07:02:08 | C*07:02:01:01 | 1 | 14 | GCT≫GCA | A73A | Silent | 17 | FJ976872 |
| C | C*07:02:10 | C*07:02:01:01 | 1 | 14 | GCC≫GCT | A11A | Silent | 12 | FJ792489 |
| C | C*07:91 | C*07:01:01:01 | 1 | 10 | GCC≫ACC | A150T | Nonsynonymous | 13 | FJ618927 |
| C | C*08:28 | C*08:02:01 | 1 | 3 | GCC≫GAC | A90D | Nonsynonymous | 15 | FJ976837 |
| C | C*12:30 | C*12:02:01 | 1 | 2 | GTG≫ATG | V52M | Nonsynonymous | 20 | FJ976876 |
| C | C*15:24 | C*15:06:01 | 1 | 1 | GAC≫AAC | D114N | Nonsynonymous | 12 | FJ976807 |
NV, number of nucleotide variations between new and homologous alleles.
CA, number of complementary alleles, i.e. those alleles that fulfill criterion 1 (alleles whose DNA sequences showed highest similarity to the new allele's DNA sequence) and criterion 2 (allele with a maximum number of silent substitutions).
The altered codon sequence of the most homologous allele is listed first and the compared codon sequence of the respective new allele is listed second. The nucleotide change(s) are given in bold.
AA change, amino acid change. Numbering from the first codon of the mature protein. The reference amino acid from the homologous allele (listed first) is followed by the codon number where the mutation was found and then the compared amino acid in the respective new allele (listed second). Stop codons are designated by X.
IR, number of individuals reported, i.e. the number of individuals carrying the new allele within the current sample.
The observed nucleotide substitutions distributed along all codons in exons 2 and 3: there were no apparent conserved regions among the new sequences (Figure 2). Many of these nucleotide substitutions were new nucleotide variations that are unique among class I alleles. Moreover, of those substitutions, many occurred at sequence positions that had not even been known as polymorphic so far (Table3). Altogether, 39.8% (i.e. 847 alleles) of the new HLA class I alleles described here contain unique nucleotide variations. This reflects the extreme polymorphism that typically characterizes HLA alleles 9. Figure 2 depicts a histogram of the polymorphic codon positions in HLA-A compared to the respective most homologous alleles, as an example. Equivalent information for HLA loci B and C can be found in Figure S1.
Figure 2.

Histogram of nucleotide variations along codon positions in exons 2 and 3 of HLA-A alleles. The x-axis represents the codon position. The y-axis represents the number of pairs (i.e. new allele and homologous allele) that have nucleotide substitutions at the respective codon positions.
Table 3.
Amount of DNA sequence positions of new HLA class I alleles (exons 2 and 3) with new nucleotide variations
| HLA locus | Number and percent of new polymorphic DNA positionsa | Number and percent of DNA positions with new nucleotide variationsa |
|---|---|---|
| A | 59 (10.8%) | 173 (31.7%) |
| B | 69 (12.6%) | 199 (36.4%) |
| C | 112 (20.5%) | 244 (41.0%) |
Percent indicates the number of the DNA positions in comparison with the total number of nucleotides in exon 2 and exon 3 (546 nucleotides).
Typically, new alleles are likely to occur on a particular haplotype. Therefore, we analyzed the haplotypes of all new alleles that differed by at least one nonsynonymous nucleotide variation from their homologous allele, and that were found in at least 10 different individuals. In particular, each individual's phenotype was decomposed into all its possible haplotypes. Among these haplotypes, we then selected those that agreed with most individuals' phenotypes. This procedure was carried out for each of the 20 considered new alleles separately. Results demonstrated that new alleles may occur on a particular haplotype: for each new allele, a particular haplotype could be identified, which agreed with 73% ± 20 of the respective phenotypes on average (Table4).
Table 4.
Most common haplotypes for new alleles that have been found in at least 10 different individuals and that show at least one nonsynonymous nucleotide variationa
| HLA-A | HLA-B | HLA-C | HLA-DRB1 | HLA-DQB1 | Number of donors showing the haplotype listed |
|---|---|---|---|---|---|
| 31:27 | 56:01 | 01:02 | 11:01 | 03:01G | 12 of 24 donors |
| 11:47 | 55:01 | 03:03G | 15:01 | 06:02 | 6 of 10 donors |
| 02:158 | 40:01 | 03:04 | 04:01 | — | 14 of 21 donors |
| 03:48 | 35:01 | 04:01 | 03:04 | 03:01 | 9 of 10 donors |
| 33:27 | 35:01G | 04:01G | 13:05 | 03:01 | 8 of 14 donors |
| 01:40 | 08:01G | 07:01G | 03:01 | 02:01 | 15 of 15 donors |
| 30:29 | 07:02 | 07:02G | 01:01 | — | 4 of 13 donors |
| 26:39 | 38:01 | 12:03 | 13:01 | 06:03 | 5 of 12 donors |
| 31:26 | 39:01 | 12:03 | 09:01 | — | 21 of 23 donors |
| 24:106 | 44:03 | 16:01 | 07:01 | 02:01 | 5 of 10 donors |
| 29:21 | 44:03 | 16:01 | 07:01 | 02:01 | 9 of 12 donors |
| 02:01G | 07:87 | 07:02 | 15:01 | 06:11 | 14 of 16 donors |
| 11:01G | 51:69 | 15:02G | 04:02 | 03:02 | 9 of 11 donors |
| 24:02G | 51:01G | 01:32 | 11:01G | 03:01 | 10 of 19 donors |
| 23:01 | 44:03 | 04:56 | 07:01 | 02:01 | 15 of 15 donors |
| 02:01G | 44:02G | 05:34 | 10:01 | 05:01 | 10 of 10 donors |
| 01:01G | 08:01 | 07:91 | 03:01 | 02:01 | 10 of 13 donors |
| 11:01 | 44:02 | 08:28 | 12:01 | 03:01 | 11 of 15 donors |
| 11:01G | 52:01G | 12:30 | 14:01G | 05:03 | 13 of 20 donors |
| 11:01 | 51:01G | 15:24 | 14:01 | — | 11 of 12 donors |
New alleles are printed in bold. G stands for G-codes (i.e. summarizing those alleles that have identical nucleotide sequences across exons 2 and 3).
In order to shed a light on the origin of alleles, we examined the self-assessed parentage records of individuals carrying new HLA class I alleles [Table S2; 10] registered with either one of the three DKMS donor centers located in Germany, the United States and Poland. Each of these donor centers assesses individual's origin differently. In Germany, the recorded parentage refers to nationalities. By contrast, in the United States, origin refers to ethnic groups (such as African American, Western European or South Asian). Moreover, only those individuals registered in the United States could specify more than one ethnic group. These cases were labeled as ‘mixed’ throughout this report. Lastly, no information about individuals' parentage was available for Poland. Due to the different classification schemes, the origin of donors carrying new alleles was assessed separately for the various donor centers.
Of the 2127 new alleles described in this report, 1336 (62.8%) were observed in donors from Germany, 422 (19.8%) in donors from the United States, 213 (10.0%) in donors from Poland, and 156 (7.3%) new alleles were found in donors from at least two different donor centers (Table1). Figure 3 presents the origin of new HLA class I alleles that were found in donors registered in Germany (Figure 3A) and the United States (Figure 3B). Alleles that were found in several individuals in more than one donor center are excluded, as these alleles' origin cannot be assigned unequivocally due to the different classification schemes.
Figure 3.
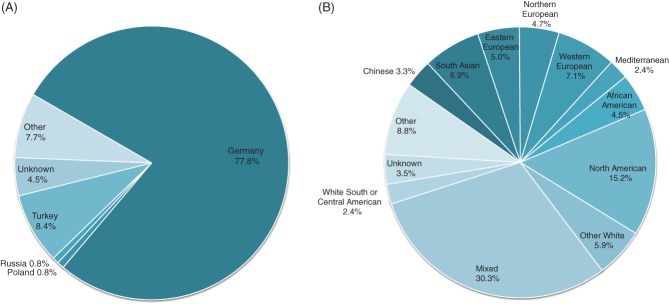
New alleles' origin based on self-assessed parentage records. (A) Individuals registered in Germany: parentage based on nationality of all individuals carrying new alleles is shown. (B) Individuals registered in the United States: ethnic groups of all individuals carrying new alleles are shown. The category ‘Mixed’ refers to new alleles that were observed in individuals who specified two ethnic groups. For visualization purposes, origins that appeared <10 times were summarized as ‘Other’ in both plots.
Most new alleles (1040; 77.8%) that were identified in donors from Germany were found in individuals with self-reported German parentage, followed by 112 (8.4%) new alleles identified in individuals of Turkish origin. Carriers of new alleles from the United States showed a higher diversity: out of 422 alleles, almost one third (30.3%) were found in individuals with ‘mixed’ origin. Furthermore, 19.2% of the alleles were identified in Europeans (Western, Eastern and Northern European and Mediterranean), 15.2% in North Americans, 10.2% in Asians (South Asian and Chinese) and 4.5% in African-Americans.
As expected, new alleles were found disproportionally more often in minority donors. For example, while 8.4% of new alleles identified in donors from Germany were found in individuals of Turkish origin, this ethnic group represents only 3.4% of all donors of this donor center. Similarly, 6.9% of the new alleles identified in US donors were found in South Asian individuals, while South Asians represent only 1.8% of all registered donors of the respective donor center.
Interestingly, 527 new alleles (i.e. 24.8%) were found in several individuals. The frequency an allele was identified and reported to the WHO Nomenclature Committee for Factors of the HLA System 1 is listed in Table S1 as ‘number of individuals reported (IR)’. Note, however, that allele sequences that have been found more than 20 times were usually no longer reported. Table2 shows alleles that have been reported at least 10 times. Strikingly, many new alleles (156; 7.3%) were found not only multiple times but also in more than one donor center (in Table2 referred to as ≥2 countries), thus indicating that carriers of these alleles are of various parentages.
Detailed quantitative analyses or future projections regarding the frequency of the occurrence of new alleles are confounded by several difficult-to-quantify effects, including many registered stem cell donors of the global donor pool have been typed with methods that do not allow the identification of new alleles. Some of these methods are still in use for HLA typing of new registrants. Moreover, a substantial fraction of new HLA alleles that has been identified in a laboratory has never been reported to the Nomenclature Committee or only with a significant time delay. In many cases, the delay probably resulted in the fact that the new allele had been reported elsewhere, maybe in an individual from another ethnicity. These practical obstacles affect both the number of observed new alleles and the underlying population-specific sample sizes, thus preventing detailed quantitative analyses regarding the occurrences of new alleles.
In conclusion, we described 2127 new HLA class I alleles that have been identified in high throughput HLA typing of newly registered DKMS stem cell donors in Germany, the United States and Poland. Importantly, these alleles also disclosed novel polymorphic positions among HLA class I alleles so far considered as conserved sequence positions.
The large number of HLA class I alleles presented in this work and the ongoing identification of novel alleles in new registrants of our donor centers underline the extreme diversity of the HLA system. Many new alleles were observed in several individuals who originated partly from different populations. It follows that alleles that are newly identified nowadays are not necessarily rare and may thus be of practical relevance for actual stem cell donor searches 11. Therefore, laboratories should use methods that allow the identification of new HLA alleles and report these alleles to the WHO Nomenclature Committee. Finally, the overrepresentation of minority donors among donors with new alleles confirms earlier results that emphasize the benefits of specific minority donor recruitment efforts 12,13.
Conflict of interest
The authors have declared no conflicting interests.
Supporting Information
The following supporting information is available for this article:
(A) Histogram of nucleotide variations along codon positions in exons 2 and 3 of HLA-B alleles. (B) Histogram of nucleotide variations along codon positions in exons 2 and 3 of HLA-C alleles. The x-axis represents the codon position. The y-axis represents the number of pairs (i.e. new allele and homologous allele) that have nucleotide substitutions at the respective codon positions.
Description of all new HLA class I alleles included in this work.
Origin and HLA phenotypes of individuals carrying new HLA class I alleles described in this work.
References
- Marsh SGE, Albert E, Bodmer W, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SGE. The IMGT/HLA Database. Nucleic Acids Res. 2011;39:D1171–6. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh SGE. Nomenclature for factors of the HLA system, update February 2009. Tissue Antigens. 2009;74:180–2. doi: 10.1111/j.1399-0039.2009.01285.x. [DOI] [PubMed] [Google Scholar]
- Marsh SGE. Nomenclature for factors of the HLA system, update March 2013. Tissue Antigens. 2013;81:480–4. doi: 10.1111/tan.12132. [DOI] [PubMed] [Google Scholar]
- Cereb N, Maye P, Lee S, Kong Y, Yang SY. Locus-specific amplification of HLA class I genes from genomic DNA: locus-specific sequences in the first and third introns of HLA-A, -B, and -C alleles. Tissue Antigens. 1995;45:1–11. doi: 10.1111/j.1399-0039.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- Cereb N, Yang SY. Dimorphic primers derived from intron 1 for use in the molecular typing of HLA-B alleles. Tissue Antigens. 1997;50:74–6. doi: 10.1111/j.1399-0039.1997.tb02839.x. [DOI] [PubMed] [Google Scholar]
- Isobe N, Gourraud PA, Harbo HF, et al. Genetic risk variants in African Americans with multiple sclerosis. Neurology. 2013;81:219–27. doi: 10.1212/WNL.0b013e31829bfe2f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Halliwell JA, McWilliam H, Lopez R, Parham P, Marsh SGE. The IMGT/HLA Database. Nucleic Acids Res. 2013;41:D1222–7. doi: 10.1093/nar/gks949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P, Adams EJ, Arnett KL. The origins of HLA-A, B.C polymorphism. Immunol Rev. 1995;143:141–80. doi: 10.1111/j.1600-065x.1995.tb00674.x. [DOI] [PubMed] [Google Scholar]
- WMDA reference website. List of letter codes for encoding of allelic ambiguities. http://bioinformatics.nmdp.org/HLA/Allele_Codes/Allele_Codes.aspx.
- Cano P, Klitz W, Mack SJ. Common and well-documented HLA alleles. Hum Immunol. 2007;68:392–417. doi: 10.1016/j.humimm.2007.01.014. Report of the Ad-Hoc Committee of the American Society for Histocompatiblity and Immunogenetics. [DOI] [PubMed] [Google Scholar]
- Schmidt AH, Solloch UV, Baier D, et al. Criteria for initiation and evaluation of minority donor programs and application to the example of donors of Turkish descent in Germany. Bone Marrow Transplant. 2009;44:405–12. doi: 10.1038/bmt.2009.55. [DOI] [PubMed] [Google Scholar]
- Pingel J, Solloch UV, Hofmann JA, Lange V, Ehninger G, Schmidt AH. High-resolution HLA haplotype frequencies of stem cell donors in Germany with foreign parentage: how can they be used to improve unrelated donor searches? Hum Immunol. 2013;74:330–40. doi: 10.1016/j.humimm.2012.10.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Histogram of nucleotide variations along codon positions in exons 2 and 3 of HLA-B alleles. (B) Histogram of nucleotide variations along codon positions in exons 2 and 3 of HLA-C alleles. The x-axis represents the codon position. The y-axis represents the number of pairs (i.e. new allele and homologous allele) that have nucleotide substitutions at the respective codon positions.
Description of all new HLA class I alleles included in this work.
Origin and HLA phenotypes of individuals carrying new HLA class I alleles described in this work.


