Abstract
Mycoplasma alligatoris causes lethal invasive disease of alligators and caimans. A homolog of the nagH gene, encoding a hyaluronidase secreted by Clostridium perfringens, and a C. perfringens hyaluronidase nagI or nagK pseudogene were discovered in the M. alligatoris genome. The nagH gene was detected by PCR in the closest relative of M. alligatoris, Mycoplasma crocodyli, but not in 40 other species representing the Mycoplasma hominis, Mycoplasma pneumoniae, and Spiroplasma phylogenetic clusters. The hyaluronidase activity in the cellular fraction of M. alligatoris and M. crocodyli SP4 broth cultures was equivalent to 10−16 U of Streptomyces hyalurolyticus hyaluronidase CFU−1. Negligible activity was present in the cell-free supernatant fraction. No chondroitinase activity was detected. There is also a novel homolog of the nanI gene, which encodes a sialidase secreted by C. perfringens, in the M. alligatoris genome. The signature YRIP and SXDXGXTW motifs and catalytic residues of the clostridial sialidase are conserved in the mycoplasmal gene, but the leader sequence necessary for its secretion by C. perfringens is absent. The gene was not detected by PCR in any other mycoplasma. Potent cell-associated sialidase activity was present in M. alligatoris colonies on agar but not in the cell-free supernatants of broth cultures or in M. crocodyli. The presence of hyaluronidase and sialidase in M. alligatoris is consistent with the rapid invasiveness and necrotizing effects of this organism, and the lack of sialidase in M. crocodyli is consistent with its comparatively attenuated virulence. This genetic and biochemical evidence suggests that the spreading factors hyaluronidase and sialidase, a combination unprecedented in mycoplasmas, are the basis of the virulence of M. alligatoris.
Mycoplasma alligatoris causes a lethal invasive disease in adult alligators (Alligator mississippiensis) and closely related caimans (Caiman latirostris). The pathology observed as early as 1 week after infection by instillation via the glottis includes necrotizing pneumonia, severe pericarditis, necrotizing myocarditis, lymphocytic interstitial nephritis, lymphocytic periportal hepatitis, splenic hyperplasia, pyogranulomatous meningitis, and necrotizing synovitis (9, 10, 11, 13, 18, 33). Hatchling alligators also rapidly developed disseminated M. alligatoris mycoplasmosis after intratracheal instillation, and the lesions are similar to those of adults and include acute multifocal brainstem hemorrhage (35). Mycoplasma crocodyli (23), the closest known relative of M. alligatoris (98% 16S rRNA gene similarity [9]), causes a similar necrotizing synovitis and occasionally subacute pneumonia in Nile crocodiles (Crocodylus niloticus) (27, 28). The severity of the lesions correlates with the numbers of M. alligatoris cells in affected tissues, suggesting that a spreading factor(s) contributes to the virulence. The extracellular matrix (ECM)-degrading enzymes that act as bacterial spreading factors include hyaluronidases, sialidases, and mucinases (26). In this report we describe the use of comparative genomics to guide the discovery of a hyaluronidase and a novel sialidase which are potential virulence factors of M. alligatoris.
MATERIALS AND METHODS
Genomics.
A whole-genome shotgun library of third-passage M. alligatoris A21JP2T (= ATCC 700619T) was constructed by random shearing, selection of 2-kb inserts, blunt end ligation into pUC18, and electroporation into Escherichia coli. High-throughput sequencing with robotic workstations, the MegaBACE capillary electrophoresis system (Pharmacia, New York, N.Y.), and the Finch-Suite relational database management system (Geospiza, Inc., Seattle, Wash.) was performed by the University of Florida Genome Sequencing Services Laboratory. More than 4× of a genome equivalent of sequence was obtained, assembled by using the PHRAP alignment engine, and used for BLASTX queries of GenBank. Sequence analyses were performed by using the Wisconsin Package of SeqWeb 2.1 (Accelrys, Inc., Burlington, Mass.) and PSORT (30) software.
PCR and reverse transcriptase PCR (RT-PCR).
PCR primers nagH-F (5′-CGTTTGAACAATTCACCCATT-3′) and nagH-R (5′-TGAACCAACATCATCGGCTA-3′) were developed by using Primer3 software (Whitehead Institute, Cambridge, Mass. [www.broad.mit.edu/genome_software/other/primer3.html]). Purified M. alligatoris nucleic acid was obtained by cell lysis with proteinase K followed by phenol-chloroform extraction. The optimum MgCl2 concentration for the PCR was 3 mM, and the 50-μl PCR mixture contained each primer at a concentration of 1 μM, each deoxynucleoside triphosphate at a concentration of 200 μM, and 0.5 U of Taq DNA polymerase (Promega, Madison, Wis.). The program consisted of 35 cycles of denaturation for 60 s at 94°C, primer annealing for 60 s at 58°C, and extension for 30 s at 72°C. The identity of the 153-bp amplification product was confirmed by digestion with restriction endonuclease AluI, which was predicted to generate 109- and 44-bp cleavage products. When PCR primers nanI-F (5′-TGGAAGAACTTGATCAGAAGCA-3′) and nanI-R (5′-AATGGCATCAATCCAGATCC-3′) were used, the reaction conditions were 35 cycles of denaturation for 60 s at 94°C, primer annealing for 60 s at 55°C, and extension for 30 s at 72°C. The identity of the 159-bp amplification product was confirmed by digestion with restriction endonuclease RsaI, which was predicted to generate 87- and 72-bp cleavage products. A crude cell lysate was used as the PCR template for M. crocodyli MP145T (= ATCC 51981T) and 40 other species that were representative of the Mycoplasma hominis, Mycoplasma pneumoniae, and Spiroplasma phylogenetic clusters. The following organisms were included in the analysis: Mycoplasma adleri ATCC 27948, Mycoplasma agalactiae PG2 (Mollicutes Reference Collection), Mycoplasma agassizii ATCC 700616, Mycoplasma alkalescens ATCC 29103, Mycoplasma alligatoris ATCC 700619, Mycoplasma arginini ATCC 25228, Mycoplasma arthritidis ATCC 14124, Mycoplasma auris UIA (Mollicutes Reference Collection), Mycoplasma bovigenitalium (a gift from Mary B. Brown), Mycoplasma bovirhinis (a gift from Mary B. Brown), Mycoplasma bovis ATCC 27368, Mycoplasma buccae ATCC 23636, Mycoplasma californicum (a gift from Mary B. Brown), Mycoplasma canadense (a gift from Mary B. Brown), Mycoplasma capricolum ATCC 23205, Mycoplasma crocodyli ATCC 51981, Mycoplasma dispar (a gift from Mary B. Brown), Mycoplasma felifaucium ATCC 43428, Mycoplasma feliminutum ATCC 25749, Mycoplasma felis ATCC 23391, Mycoplasma fermentans ATCC 19989, Mycoplasma gallisepticum ATCC 15306, Mycoplasma gatae ATCC 23392, Mycoplasma genitalium ATCC 33530, Mycoplasma hominis (a gift from Mary B. Brown), Mycoplasma hyopneumoniae J (Mollicutes Reference Collection), Mycoplasma hyorhinis (a gift from Mary B. Brown), Mycoplasma leocaptivus ATCC 49890, Mycoplasma leopharyngis ATCC 49889, Mycoplasma microti ATCC 700935, Mycoplasma mycoides subsp. capri PG3 (Mollicutes Reference Collection), Mycoplasma mycoides subsp. mycoides LC strain GM684-13 (Mollicutes Reference Collection), Mycoplasma orale ATCC 23714, Mycoplasma pneumoniae PI1428 (Mollicutes Reference Collection), Mycoplasma pulmonis X1048 (a gift from Maureen K. Davidson), Mycoplasma putrifaciens ATCC 15718, Mycoplasma salivarium ATCC 23064, Mycoplasma simbae ATCC 49888, Mycoplasma synoviae WVU1853 (Mollicutes Reference Collection), Mycoplasma testudineum ATCC 700618, Mycoplasma testudinis ATCC 43263, and Mycoplasma yeatsii G1H (Mollicutes Reference Collection).The absence of factors that might inhibit the PCR in the crude lysates was confirmed by successful amplification of the 16S rRNA gene of each species under conditions described previously (8).
High-molecular-weight M. alligatoris RNA was purified from pelleted cells in the early log phase of growth in ATCC medium 988 (SP4) (46) by lysis with guanidinium thiocyanate, followed by selective RNA adsorption to glass fibers and treatment of the final eluate with 6 U of DNase I for 60 min at 37°C (RNAqueous, DNA-free; Ambion, Inc., Austin, Tex.). For detection of mRNA by RT-PCR, cDNA synthesis was initiated in a reaction by using 300 ng of M. alligatoris total RNA, 5 U of avian myeloblastosis virus (AMV) RT, 50 pmol of primer nagH-R2 (5′-ACAACCCCTTCAGAAACAGC-3′) or nanI-R, each deoxynucleoside triphosphate at a concentration of 200 μM, and the optimum MgSO4 concentration (3 mM) in a 50-μl mixture at 48°C for 45 min. This was followed in a one-tube, two-enzyme system by second-strand cDNA synthesis and 30 cycles of PCR performed with Tfl DNA polymerase, which lacks RT activity (Access RT-PCR system; Promega), and primers nagH-F3 (5′-GGTGCTCCAAGAAAAGGTGA-3′) and nagH-R2 (the 509-bp amplification product was confirmed by the presence of 271- and 238-bp AgeI cleavage products) or primers nanI-F and nanI-R. AMV RT was omitted from the negative controls.
Mycoplasma culture.
Third-passage M. alligatoris that was previously frozen at −70°C in SP4 medium was cultured to the early log phase, as detected by acidification of the medium, at 30°C in ambient air by 1:10 dilution in SP4 broth containing 0.5% (wt/vol) glucose supplemented with 10−5 U of penicillin G liter−1, 10−5 U of polymyxin B liter−1, 65 mg of cefoperazone liter−1, and 20% (vol/vol) fetal bovine serum. Culture densities were determined by colony counting by using serial 10-fold dilutions of aliquots in SP4 broth inoculated onto SP4 agar. M. crocodyli was cultured and quantified similarly. M. pulmonis ATCC 19612 was cultured in Frey's medium (46).
To determine if glycosaminoglycanase activity was inducible with exogenous substrate, M. alligatoris cells in SP4 broth were harvested in the early log phase by centrifugation and washed three times in minimal medium CMRL 1066 without glutamine (Gibco, Carlsbad, Calif.). The cells were resuspended in an equal volume of minimal medium and incubated at 30°C in ambient air without additional supplements. After 24 h of starvation in minimal medium, a potential glycosaminoglycanase substrate (100 mg of autoclaved minced alligator tracheal cartilage ml−1) was added, and the preparation was incubated for another 72 h. Aliquots were withdrawn every 24 h for enzyme activity analysis and for quantitative subculturing in serial dilutions of SP4 broth and on SP4 agar to monitor the culture density in the minimal medium.
Glycosaminoglycanase assay.
Glycosaminoglycanase activity was measured by using a quantitative substrate precipitation assay (19). The substrates tested individually included hyaluronic acid (sodium salts) from rooster comb and from human umbilical cord, as well as bovine chondroitin sulfate A and porcine chondroitin sulfate B (sodium salts; Sigma). A 2-mg ml−1 aqueous stock solution of substrate was diluted to obtain a final concentration of 400 μg ml−1 in 1% (wt/vol) low-melting-temperature agarose (SeaPlaque; FMC, Philadelphia, Pa.) melted in phosphate-buffered saline (PBS) containing 1% (wt/vol) fatty acid-free bovine serum albumin. The optimum pH was determined by comparing the results obtained with PBS at pH 4, 6, 7.5, 8, and 10. Each well of a 96-well flat-bottom plate (no. 3072; Falcon, San Jose, Calif.), except the perimeter wells, contained 80 μg of substrate in 200 μl of solidified agarose. A specimen (100 μl) was added to each well and incubated at 30°C. After 72 h of incubation, the specimen was removed by aspiration, and 2 N glacial acetic acid (100 μl well−1) was added to precipitate any undigested substrate, which resulted in turbidity which was inversely proportional to the glycosaminoglycanase activity. The A405 of each well was measured after an additional 4 h of incubation at 30°C (Thermomix microplate reader with Softmax Pro, version 1.2.0, software; Molecular Devices, Sunnyvale, Calif.).
Glycosaminoglycanase activity was determined by using aliquots of the initial SP4 cultures, cell-free SP4 culture supernatant fluid, CMRL 1066 cultures after starvation and serially after subsequent substrate addition, and the subcultures from CMRL 1066 tested for viability with SP4 as described above. Cell-free supernatant fluid was prepared by centrifugation of a broth culture in the early log phase, and the absence of viable mycoplasmas from this fraction was demonstrated by incubation of an aliquot of the fraction diluted in fresh SP4 medium. The negative controls included PBS, sterile SP4 medium, M. pulmonis cultured to the log phase in Frey's medium, and sterile Frey's medium. The positive controls included Streptomyces hyalurolyticus hyaluronate lyase (Sigma) that was serially diluted in PBS (pH 7.5) to obtain concentrations ranging from 1 × 10−2 to 2 × 10−5 U 100 μl−1 and Proteus vulgaris chondroitinase ABC (Sigma) that was serially diluted in PBS (pH 7.5) to obtain concentrations ranging from 2 × 10−1 to 4 × 10−4 U 100 μl−1.
Sialidase assay.
Sialidase activity was assayed qualitatively by using the fluorogenic substrate 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUAN) (Sigma). Twenty-microliter aliquots of M. alligatoris and M. crocodyli in the early log phase in SP4 broth were spotted onto SP4 agar without phenol red in six-well plates and incubated for 24 h until a lawn of colonies became visible. Twenty-microliter aliquots of cell-free culture supernatant liquid were prepared similarly, and then each well was overlaid with 1 ml of top agar (2.5 g of NaCl liter−1, 10 g of tryptone liter−1, 7.5 g of Noble agar liter−1) containing 200 μl of 0.35% (wt/vol) MUAN and 30 μl of 1 M sodium acetate (pH 4) and incubated for 30 min at 25°C after the top agar solidified. Cyan fluorescence under 254-nm UV light illumination indicated a positive result (21). The negative controls were M. pulmonis colonies and cell-free culture supernatant liquid prepared similarly, as well as PBS (pH 7.5). The positive control was 0.25 U of type VI Clostridium perfringens neuraminidase (Sigma) in 20 μl of PBS (pH 7.5).
RESULTS
Genomics.
A gene (GenBank accession number AY515698) with unambiguous similarity (BLASTX E value, 1e−106) to nagH, encoding a hyaluronate lyase (GenBank protein accession number NP_561107) secreted by C. perfringens (44), was present in the M. alligatoris genome. The 1,628-amino-acid C. perfringens enzyme NagH consists of a putative 30-amino-acid N-terminal signal sequence, an approximately 700-amino-acid N-terminal domain that includes the active site binding cleft and catalytic residues, an approximately 90-amino-acid linker domain, and a C-terminal domain containing three candidate substrate-binding motifs (12). A candidate ribosome binding site (RBS) (5′-TAAAGGAGTTGA-3′) was present at position −14 with respect to the putative start codon, but no transcription promoter sequences were readily discernible. There were three in-frame putative UAA stop codons upstream of the gene, but no BLAST similarities were detected in the adjacent 5′ sequence. A 32-amino-acid N-terminal signal sequence was predicted by PSORT analysis (signal score, 0.38). The predicted M. alligatoris NagH homolog exhibited approximately 42% amino acid identity and 59% amino acid similarity to C. perfringens NagH in the N-terminal domain, 21% amino acid identity and 42% amino acid similarity in the linker domain, and 31% amino acid identity and 51% amino acid similarity in the C-terminal domain. The active site binding cleft amino acid sequence, including the presence and spacing of the catalytic N-171, H-228, and Y-236 residues, was the most highly conserved region (25, 32). The gene was interspersed with at least 24 putative UGA W codons. In addition to nagH, a pseudogene with similarities to the C. perfringens hyaluronate lyase genes nagI (GenBank protein accession number NP_561797) and nagK (GenBank protein accession number NP_562195) (BLASTX E values, 3e−24 and 2e−40, respectively) was identified in the M. alligatoris genome, but this sequence was interspersed with multiple putative UAA and UAG stop codons in addition to at least seven putative UGA W codons. No transcription promoter, RBS, or start codon sequences were discerned by alignment of the pseudogene with the C. perfringens sequences, but the remnants of the active site binding cleft region, including the catalytic residues, remained obvious.
A gene (GenBank accession number AY515695) with unambiguous similarity (BLASTX E value, 5e−20) to nanI, encoding the large exo-α-sialidase (GenBank protein accession number NP_561641) secreted by C. perfringens (44), was also present in the M. alligatoris genome. The clostridial enzyme NanI is a 694-amino-acid polypeptide that includes a 41-amino-acid hydrophobic N-terminal leader sequence. A candidate RBS, 5′-GAAAGTGTTTAA-3′, was present at position −13 with respect to the putative start codon, but no transcription promoter sequences were readily discernible. This RBS overlapped the stop codon of the upstream open reading frame (ORF), which encoded a putative sugar kinase. The predicted M. alligatoris NanI homolog was 511 amino acids long and exhibited 30% amino acid identity and 47% amino acid similarity to C. perfringens NanI excluding the C. perfringens leader sequence, which sequence alignment and PSORT analyses showed was absent in M. alligatoris (signal score, −4.29). The signature YRIP motif and four SXDXGXTW Asp box motifs, as well as the catalytic R-37, D-62, D-100, and E-230 residues of C. perfringens NanI (26, 40), were conserved in M. alligatoris. The M. alligatoris NanI homolog was interspersed with eight putative UGA W codons but no other W codons. The nanI ORF overlapped candidate transcription promoter sequences 5′-TTGA-3′ and 5′-TATAA-3′ of the downstream ORF, which encoded a putative sodium-coupled permease. The candidate RBS of this ORF, 5′-AGAAAGGATACAAT-3′, was located 2 bases 3′ of the nanI stop codon.
Mycoplasma culture.
M. alligatoris cultures in SP4 broth reached the remarkably high density of 4 × 1012 CFU ml−1 in the early log phase. The density decreased to 9 × 107 CFU ml−1 after 24 h of incubation in minimal medium and then to 2.5 × 107 and 6 × 106 CFU ml−1 in the subsequent 24 and 48 h, respectively, either with or without cartilage supplementation. Supplementation with up to 1% (wt/vol) fatty acid-free bovine serum albumin in factorial combinations with up to 0.5% (vol/vol) polyoxyethylene sorbitan (Tween 80) did not affect M. alligatoris survival in minimal medium.
PCR and RT-PCR.
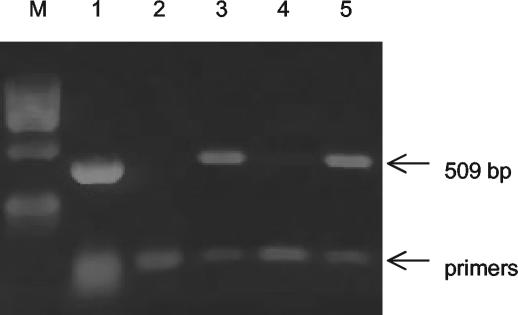
The PCR with primers nagH-F and nagH-R and an M. alligatoris template generated a 153-bp product which was cleaved by AluI as expected. The PCR with primers nanI-F and nanI-R generated a 159-bp product which was cleaved by RsaI as expected. The nagH gene was also detected by PCR in M. crocodyli, the closest known relative of M. alligatoris, but not in any of 40 other species of mollicutes tested. The nanI gene was detected by PCR only in M. alligatoris. Expression of nagH and nanI mRNA was readily detected in M. alligatoris SP4 broth cultures by RT-PCR (Fig. 1). The primer nagH-F3—primer nagH-R2 combination, which amplified a 509-bp product that was cleaved by AgeI as expected, had better specificity in the RT-PCR than the primer nagH-F—primer nagH-R combination had. No change in nagH mRNA expression following M. alligatoris passage in minimal medium, either with or without cartilage, was detected by RT-PCR.
FIG. 1.
Agarose gel electrophoresis of PCR and RT-PCR amplification products of the M. alligatoris nagH gene and mRNA, showing that the gene is expressed during culture in SP4 complex medium and after 48 h of starvation in CMRL 1066 minimal medium. Lane M, marker; lane 1, PCR with DNA template; lane 2, RT-PCR with no template (negative control); lane 3, RT-PCR of mRNA from SP4 culture; lane 4, RT-PCR of mRNA from SP4 culture with no AMV RT (negative control); lane 5, RT-PCR of mRNA from CMRL 1066 culture.
Glycosaminoglycanase assay.
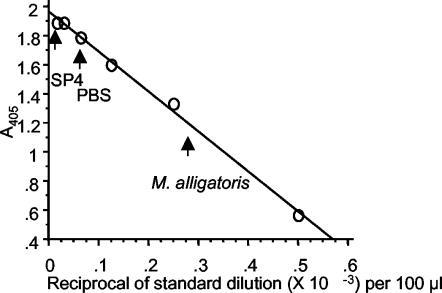
Hyaluronidase activity was found in cell suspensions of M. alligatoris and M. crocodyli in the early log phase in SP4 medium and in suspensions of M. alligatoris following 24 h of starvation in minimal medium, but not at any subsequent time in minimal medium either with or without cartilage. However, hyaluronidase activity was present in a cell suspension of M. alligatoris subcultured in SP4 following up to 48 h of growth in minimal medium. The activity in 100-μl aliquots of replicate suspensions in SP4 broth was equivalent to 2 × 10−3 to 3 × 10−3 U of purified S. hyalurolyticus hyaluronate lyase or approximately 10−16 U CFU−1 (Fig. 2). The activities were approximately equal over the pH range from 6 to 10, but the activity was 30% lower at pH 4 (data not shown). Negligible hyaluronidase activity was detected in M. alligatoris and M. crocodyli cell-free culture supernatants, and no chondroitinase activity was detected. The activity was not inducible. No hyaluronidase or chondroitinase activity was detected in either fraction of M. pulmonis cultures, indicating that none of the putative proteins encoded by the hypothetical genes of this organism is a glycosaminoglycanase.
FIG. 2.
Hyaluronidase standard curve and estimation of M. alligatoris hyaluronidase activity. S. hyalurolyticus hyaluronate lyase (Sigma) was serially diluted in PBS (pH 7.5) to obtain concentrations ranging from 1 × 10−2 to 2 × 10−5 U 100 μl−1 (open circles). The representative activity of a 100-μl aliquot of M. alligatoris in the early log phase in SP4 broth was compared to the activities of PBS and sterile SP4 negative controls (arrows).
Sialidase assay.
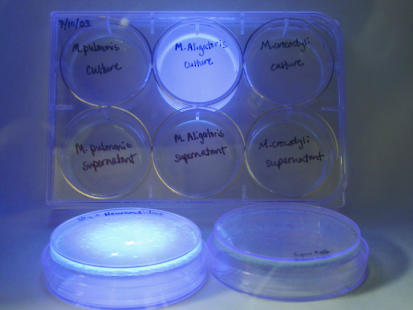
Potent sialidase activity was demonstrated by intense cyan fluorescence of M. alligatoris colonies on SP4 agar overlaid with top agar containing MUAN and illuminated with short-wave UV light (Fig. 3). No activity was detected in the cell-free M. alligatoris culture supernatant liquid or in either fraction of the M. crocodyli cultures. No sialidase activity was detected in either fraction of M. pulmonis cultures, indicating that none of the putative proteins encoded by the hypothetical genes of this organism is a sialidase.
FIG. 3.
M. alligatoris cell-associated sialidase activity. Colonies (upper center plate) and aliquots of cell-free culture supernatant (lower center plate) were overlaid with top agar containing the fluorogenic sialidase substrate MUAN (Sigma) and illuminated with short-wave UV light for comparison to M. crocodyli (right plates) and the negative control M. pulmonis (left plates). The plates in the foreground were positive (type VI C. perfringens neuraminidase [Sigma]) (left plate) and negative (PBS) (right plate) sialidase controls.
DISCUSSION
Mycoplasmas probably evolved from clostridial ancestors about 400 million years ago by genome reduction that led to obligate commensalism or parasitism of host cells (7, 47; J. Maniloff, Abstr. 13th Int. Congr. Int. Org. Mycoplasmol., seminar I-1, 2000). The best-studied mycoplasmas of vertebrate hosts are pathogens of humans and domesticated animals. These pathogens are usually associated with subclinical or subtle, slowly progressive or chronic diseases (3). Pathogenicity in all hosts is poorly understood beyond the generalization that host responses to mycoplasmal cytadherence exacerbate disease, and variation in cell surface antigens can occur with high frequency in some mycoplasmas (34). Many harmful effects on host cells have been associated with mycoplasmosis, including ciliostasis, oxidative damage to membranes, dysregulation and decreased abundance of membrane ion channels, clastogenic and oncogenic effects, chromosomal abnormalities, and cell cycle dysregulation (34, 45), but specific mechanisms of mycoplasmal pathogenicity are not well understood. Because the major mycoplasmal pathogens of humans and domesticated animals lack endotoxin or classical exotoxins but are cytadherent, current mycoplasmology studies are focused on the roles of adhesin molecules and adherence-mediated modulation of colonized hosts to explain mycoplasmal pathogenicity. Probably because it is not a usual feature of the common mycoplasmoses of humans or domesticated animals, the possibility that spreading factors like sialidases, hyaluronidases, or mucinases, the opposites of adhesins, may exist in mycoplasmas and be important contributors to virulence in other mycoplasmoses seems to have been virtually ignored, even in otherwise exhaustive reviews of mycoplasmal pathogenicity (34, 41, 45).
Hyaluronan, a polymer of N-acetylglucosamine and glucuronic acid disaccharide repeats found in many animal tissues and fluids, is a major component of the vertebrate ECM. Hyaluronan degradation increases the permeability of connective tissues and decreases the viscosity of body fluids, facilitating the spread of bacteria and their toxins, and it may promote direct contact between bacteria and host cell surfaces (32). In addition, depolymerization disrupts the role of the ECM as a reservoir of cytokines and enzymes involved in signal transduction (15). These effects are all consistent with the pathological results of M. alligatoris and M. crocodyli infections. The chemical composition of glycosaminoglycan polymers, such as hyaluronan, chondroitin, dermatan, keratin, and heparin, is highly variable even among tissues within a vertebrate species, including heterogeneities such as isomerization, acetylation, phosphorylation, and sulfation (15). Some glycosaminoglycanases have corresponding substrate specificities (44) that make them valuable as therapeutic agents or targets in management of wound healing (5, 6), tumor metastasis (14), and infection (32).
Bacterial hyaluronan lyases are endo-N-acetylhexosaminidases whose unsaturated disaccharide products are potential bacterial nutrients. As predicted from the presence of nagH, the M. alligatoris genome also encodes a glucuronyl hydrolase (GenBank accession number AY515697) that has not been found in any other mycoplasma characterized to date, which could catalyze the release of glucuronic acid from the disaccharides cleaved from hyaluronan, liberating N-acetylglucosamine for conversion to fructose-6-phosphate and entry into glycolysis by a pathway found in other mycoplasmas. The fate of the glucuronate has not been predicted. Therefore, both spreading and nutrient acquisition capabilities are probably mediated by the M. alligatoris hyaluronidase. The only other currently confirmed mycoplasmal proteins as large as NagH, besides the nucleic acid polymerases and helicases, are the adhesin P1 (1,628 amino acids) and cytoskeletal protein HMW2 (1,819 amino acids) involved in cytadherence and pathogenicity of M. pneumoniae (24, 45).
In vertebrate animals, including reptiles (16), diverse sialic acid derivatives protect against hydrolysis of the glycosidic or peptide bonds of oligosaccharides, glycoproteins, glycolipids, and gangliosides located on outer cell surfaces. They are involved in recognition processes, connection with ECM components, and intercellular interactions (1). Most sialidases occur in bacteria that live in close contact with vertebrate host cells as commensals or facultative pathogens, which allows more direct bacterial contact with the host cell surface (38, 39). However, experimental treatment with neuraminidase has been shown variously to promote, eliminate, or have no effect on glycoprotein binding or cytadherence of different species of mycoplasmas (2, 36, 42, 48). Deprotected polymers promote further enzymatic degradation of the ECM to release potential bacterial nutrients (26, 38). As predicted from the presence of nanI, the M. alligatoris genome also encodes a sialic acid lyase (nanA; GenBank accession number AY515696), which has not been found in any other mycoplasma characterized to date, that could catalyze the release of pyruvate from sialic acid, liberating N-acetylglucosamine for entry into glycolysis as described above. Therefore, both spreading and nutrient acquisition capabilities are probably also mediated by the M. alligatoris sialidase. It has also been proposed that bacterial desialylation of host glycoconjugates could expose or form new host antigens and thus play a role in autoimmune complications of infection (4, 22).
Bacterial sialidases occur irregularly even among strains within a species, and there is no correlation with the bacterial phylogenetic relationships, which suggests that there has been gene distribution via horizontal transfer (39). For example, an extracellular neuraminidase-like activity was present in an unidentified strain of M. gallisepticum (37), and potent cell surface sialidase activity of the TT strain of M. gallisepticum was characterized in detail (29, 43), but neuraminidase activity was absent from other pathogenic strains of M. gallisepticum tested (17, 22) and there is no corresponding confirmed gene in the recently annotated genome of M. gallisepticum strain Rlow (31). The sialidase of M. alligatoris is novel in that its length and amino acid sequence, including the signature YRIP motif, are most similar to the length and amino acid sequence of the large extracellular clostridial sialidase NanI, but it also resembles the small (382-amino-acid) intracellular clostridial sialidase NanH because it lacks a typical hydrophobic leader sequence (40). The absence of a sialidase in M. crocodyli may in part explain the attenuated virulence of this organism, in contrast to the virulence of M. alligatoris; this hypothesis could be tested by gene knockout and complementation experiments.
Since all mycoplasmas are thought to be descendants of clostridial ancestors (47), the absence of nag homologs in the other mycoplasmas examined to date suggests either that M. alligatoris and M. crocodyli were derived from a different ancestral lineage than other mycoplasmas, which contradicts prior 16S rRNA gene comparisons (9), or that a recent common ancestor acquired the gene by horizontal transfer (20), which might reflect the effects of mycoplasma-host interactions and a limited host range as isolating mechanisms that affect the evolution of mycoplasmas. Evidence against a single horizontal cotransfer of more than one spreading factor from C. perfringens to a recent common ancestor of M. alligatoris and M. crocodyli includes the long physical distance (>178 kb) between the nagH, nagI, or nagK gene and the nanI gene in the C. perfringens strain 13 genome (44), the many putative UAA or UAG stop codons interspersed throughout the nagI or nagK pseudogene, and the absence of nanI from M. crocodyli. Also, any selective advantage based on nutrient catabolism would require the glucuronyl hydrolase and/or sialic acid lyase activities. Only 5 kb separates nanA from nagH in the C. perfringens strain 13 genome, but the BLASTX similarity E value for M. alligatoris nanA was e7 closer to Pasteurella multocida than to C. perfringens, and the M. alligatoris glucuronyl hydrolase had no clostridial homolog. In summary, the genetic and biochemical evidence described here suggests that hyaluronidase and sialidase, a combination unprecedented in the mycoplasmas characterized to date, are candidate virulence factors of M. alligatoris.
Acknowledgments
This work was supported by Public Health Service grant 1R15HG02389-01A1 from the National Human Genome Research Institute.
REFERENCES
- 1.Achyuthan, K. E., and A. M. Achyuthan. 2001. Comparative enzymology, biochemistry and pathophysiology of human exo-α-sialidases (neuraminidases). Comp. Biochem. Physiol. B 129:29-64. [DOI] [PubMed] [Google Scholar]
- 2.Araake, M., M. Yayoshi, and M. Yoshioka. 1985. Attachment of Mycoplasma pulmonis to rat and mouse synovial cells cultured in vitro. Microbiol. Immunol. 29:601-607. [DOI] [PubMed] [Google Scholar]
- 3.Baseman, J. B., and J. G. Tully. 1997. Mycoplasmas: sophisticated, reemerging, and burdened by their notoriety. Emerg. Infect. Dis. 3:21-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biberfeld, G. 1979. Autoimmune reactions associated with Mycoplasma pneumoniae infection. Zentralbl. Bakteriol. Orig. A 245:144-149. [PubMed] [Google Scholar]
- 5.Bos, P. K., J. DeGroot, M. Budde, J. A. Verhaar, and G. J. van Osch. 2002. Specific enzymatic treatment of bovine and human articular cartilage: implications for integrative cartilage repair. Arthritis Rheum. 46:976-985. [DOI] [PubMed] [Google Scholar]
- 6.Bradbury, E. J., L. D. F. Moon, R. J. Popat, V. R. King, G. S. Bennett, P. N. Patel, J. W. Fawcett, and S. B. McMahon. 2002. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature 416:636-640. [DOI] [PubMed] [Google Scholar]
- 7.Brown, D. R. 2002. Mycoplasmosis and immunity of fish and reptiles. Front. Biosci. 7:1338-1346. (Online.) [DOI] [PubMed] [Google Scholar]
- 8.Brown, D. R., G. S. McLaughlin, and M. B. Brown. 1995. Taxonomy of the feline mycoplasmas Mycoplasma felifaucium, Mycoplasma feliminutum, Mycoplasma felis, Mycoplasma gateae, Mycoplasma leocaptivus, Mycoplasma leopharyngis, and Mycoplasma simbae by 16S rRNA gene sequence comparisons. Int. J. Syst. Bacteriol. 45:560-564. [DOI] [PubMed] [Google Scholar]
- 9.Brown, D. R., J. M. Farley, L. A. Zacher, J. M.-R. Carleton, T. L. Clippinger, J. G. Tully, and M. B. Brown. 2001. Mycoplasma alligatoris sp. nov., a new species from American alligators. Int. J. Syst. Evol. Microbiol. 51:419-424. [DOI] [PubMed] [Google Scholar]
- 10.Brown, D. R., M. F. Nogueira, T. R. Schoeb, K. A. Vliet, R. A. Bennett, G. W. Pye, and E. R. Jacobson. 2001. Pathology of experimental mycoplasmosis in American alligators. J. Wildl. Dis. 37:671-679. [DOI] [PubMed] [Google Scholar]
- 11.Brown, D. R., I. M. Schumacher, M. F. Nogueira, L. J. Richey, L. A. Zacher, T. R. Schoeb, K. A. Vliet, R. A. Bennett, E. R. Jacobson, and M. B. Brown. 2001. Detection of antibodies to a pathogenic mycoplasma in American alligators (Alligator mississippiensis), broad-nosed caimans (Caiman latirostris), and Siamese crocodiles (Crocodylus siamensis). J. Clin. Microbiol. 39:285-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canard, B., T. Garnier, B. Saint-Joanis, and S. T. Cole. 1994. Molecular genetic analysis of the nagH gene encoding a hyaluronidase of Clostridium perfringens. Mol. Gen. Genet. 243:215-224. [DOI] [PubMed] [Google Scholar]
- 13.Clippinger, T. L., R. A. Bennett, C. M. Johnson, K. A. Vliet, S. L. Deem, J. Oros, E. R. Jacobson, I. M. Schumacher, D. R. Brown, and M. B. Brown. 2000. Morbidity and mortality associated with a new mycoplasma species from captive American alligators (Alligator mississippiensis). J. Zoo Wildl. Med. 31:303-314. [DOI] [PubMed] [Google Scholar]
- 14.Delpech, B., A. Laquerriere, C. Maingonnat, P. Bertrand, and P. Freger. 2002. Hyaluronidase is more elevated in human brain metastases than in primary brain tumours. Anticancer Res. 22:2423-2427. [PubMed] [Google Scholar]
- 15.Ernst, S., R. Langer, C. L. Cooney, and R. Sasisekharan. 1995. Enzymatic degradation of glycosaminoglycans. Crit. Rev. Biochem. Mol. Biol. 30:387-444. [DOI] [PubMed] [Google Scholar]
- 16.Ferri, D., and G. E. Liquori. 1989. Lectin histochemistry of secretory cell glycoconjugates in the nasal mucosa of Podarcis sicula campestris De Betta (Reptilia, Lacertidae). Basic Appl. Histochem. 33:197-207. [PubMed] [Google Scholar]
- 17.Glasgow, L. R., and R. L. Hill. 1980. Interactions of Mycoplasma gallisepticum with sialyl glycoproteins. Infect. Immun. 30:353-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helmick, K. E., D. R. Brown, E. R. Jacobson, and M. B. Brown. 2002. In vitro antibiotic susceptibility pattern of a newly identified pathogen Mycoplasma alligatoris sp. nov. isolated from symptomatic American alligators (Alligator mississippiensis). J. Zoo Wildl. Med. 33:124-136. [DOI] [PubMed] [Google Scholar]
- 19.Hynes, W. L., and J. J. Ferretti. 1994. Assays for hyaluronidase activity. Methods Enzymol. 235:606-616. [DOI] [PubMed] [Google Scholar]
- 20.Jain, R., M. C. Rivera, and J. A. Lake. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl. Acad. Sci. USA 96:3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jost, B. H., J. G. Songer, and S. J. Billington. 2002. Identification of a second Arcanobacterium pyogenes neuraminidase and involvement of neuraminidase activity in host cell adhesion. Infect. Immun. 70:1106-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kahane, I., A. Reisch-Saada, M. Almagor, P. Abeliuck, and S. Yatziv. 1990. Glycosidase activities of mycoplasmas. Zentralbl. Bakteriol. Orig. B 273:300-305. [DOI] [PubMed] [Google Scholar]
- 23.Kirchhoff, H., K. Mohan, R. Schmidt, M. Runge, D. R. Brown, M. B. Brown, C. M. Foggin, P. Muvavarirwa, H. Lehmann, and J. Flossdorf. 1997. Mycoplasma crocodyli sp. nov., a new species from crocodiles. Int. J. Syst. Bacteriol. 47:742-746. [DOI] [PubMed] [Google Scholar]
- 24.Krause, D. C., T. Proft, C. T. Hedreyda, H. Hilbert, H. Plagens, and R. Herrmann. 1997. Transposon mutagenesis reinforces the correlation between Mycoplasma pneumoniae cytoskeletal protein HMW2 and cytadherence. J. Bacteriol. 179:2668-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, S., S. J. Kelly, E. Lamani, M. Ferraroni, and M. J. Jedrzejas. 2000. Structural basis of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. EMBO J. 19:1228-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsushita, O., and A. Okabe. 2001. Clostridial hydrolytic enzymes degrading extracellular components. Toxicon 39:1769-1780. [DOI] [PubMed] [Google Scholar]
- 27.Mohan, K., C. M. Foggin, P. Muvavarirwa, J. Honeywill, and A. Pawandiwa. 1995. Mycoplasma-associated polyarthritis in farmed crocodiles (Crocodylus niloticus) in Zimbabwe. Onderstepoort J. Vet. Res. 62:45-49. [PubMed] [Google Scholar]
- 28.Mohan, K., C. M. Foggin, P. Muvavarirwa, and J. Honywill. 1997. Vaccination of farmed crocodiles (Crocodilus niloticus) against Mycoplasma crocodyli infection. Vet. Rec. 141:476. [DOI] [PubMed] [Google Scholar]
- 29.Muller, H. E., and K. K. Sethi. 1972. Occurrence of neuraminidase in Mycoplasma gallisepticum. Med. Microbiol. Immunol. 157:160-168. [DOI] [PubMed] [Google Scholar]
- 30.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34-35. [DOI] [PubMed] [Google Scholar]
- 31.Papazisi, L., T. Gorton, G. Kutish, P. Markham, G. Browning, D. Nguyen, S. Swartzell, A. Madan, G. Mahairas, and S. J. Geary. 2003. The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain Rlow. Microbiology 149:2307-2316. [DOI] [PubMed] [Google Scholar]
- 32.Ponnuraj, K., and M. J. Jedrzejas. 2000. Mechanism of hyaluronan binding and degradation: structure of Streptococcus pneumoniae hyaluronate lyase in complex with hyaluronic acid disaccharide at 1.7 Å resolution. J. Mol. Biol. 299:885-895. [DOI] [PubMed] [Google Scholar]
- 33.Pye, G. W., D. R. Brown, M. F. Nogueira, K. A. Vliet, T. R. Schoeb, E. R. Jacobson, and R. A. Bennett. 2001. Experimental inoculation of broad-nosed caimans (Caiman latirostris) and Siamese crocodiles (Crocodylus siamensis) with Mycoplasma alligatoris. J. Zoo Wildl. Med. 32:196-201. [DOI] [PubMed] [Google Scholar]
- 34.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richey, L. J. 2001. Effects of endocrine-disrupting contaminants on the immune system of hatchling American alligators. Ph.D. dissertation. University of Florida, Gainesville.
- 36.Roberts, D. D., L. D. Olson, M. F. Barile, V. Ginsburgh, and H. C. Krivan. 1989. Sialic acid-dependent adhesion of Mycoplasma pneumoniae to purified glycoproteins. J. Biol. Chem. 264:9289-9293. [PubMed] [Google Scholar]
- 37.Roberts, D. H. 1967. Neuraminidase-like enzyme present in Mycoplasma gallisepticum. Nature 213:87-88. [Google Scholar]
- 38.Roggentin, P., B. Rothe, F. Lottspeich, and R. Schauer. 1988. Cloning and sequencing of a Clostridium perfringens sialidase gene. FEBS Lett. 238:31-34. [DOI] [PubMed] [Google Scholar]
- 39.Roggentin, P., R. Schauer, L. L. Hoyer, and E. R. Vimr. 1993. The sialidase superfamily and its spread by horizontal gene transfer. Mol. Microbiol. 9:915-921. [DOI] [PubMed] [Google Scholar]
- 40.Rood, J. R. 1998. Virulence genes of Clostridium perfringens. Annu. Rev. Microbiol. 52:333-360. [DOI] [PubMed] [Google Scholar]
- 41.Rottem, S. 2003. Interaction of mycoplasmas with host cells. Physiol. Rev. 83:417-432. [DOI] [PubMed] [Google Scholar]
- 42.Salih, B. A., and R. F. Rosenbusch. 1999. Interactions of Mycoplasma bovoculi with erythrocyte: role of p94 surface protein. Zentralbl. Veterinaermed. Reihe B 46:323-329. [DOI] [PubMed] [Google Scholar]
- 43.Sethi, K. K., and H. E. Muller. 1972. Neuraminidase activity in Mycoplasma gallisepticum. Infect. Immun. 5:260-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tryon, V. V., and J. B. Baseman. 1992. Pathogenic determinants and mechanisms, p. 457-472. In J. Maniloff (ed.), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington D.C.
- 46.Tully, J. G. 1995. Culture medium formulation for primary isolation and maintenance of mollicutes, p. 33-39. In S. Razin and J. G. Tully (ed.), Molecular and diagnostic procedures in mycoplasmology, vol. 1. Academic Press, San Diego, Calif.
- 47.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, Q., T. F. Young, and R. F. Ross. 1994. Microtiter plate adherence assay and receptor analogs for Mycoplasma hyopneumoniae. Infect. Immun. 62:1616-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]