Abstract
An allele of rpoD (rpoD1181) that results in increased synthesis of the pyrimidine moiety of thiamine in Salmonella enterica was identified. The S508Y substitution caused by rpoD1181 is analogous to the S506F derivative of the Escherichia coli protein. The properties of this E. coli mutant protein have been well characterized in vitro. Identification of a metabolic phenotype caused by the rpoD1181 allele of S. enterica allows past in vitro results to be incorporated in continuing efforts to understand cellular processes that are integrated with the thiamine biosynthetic pathway.
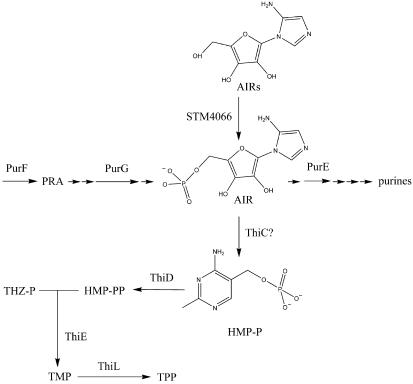
The essential cofactor thiamine pyrophosphate is generated by the condensation of two independently synthesized molecules, 4-methyl-5(β-hydroxyethyl) thiazole phosphate and 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate(HMP-PP) (Fig. 1) (3). The precursor of the pyrimidine moiety is 5-aminoimidazole ribotide (AIR), an intermediate in the purine biosynthetic pathway. HMP-P synthesis involves a complex intramolecular rearrangement and requires at least the product of the thiC gene (24, 26). Reconstitution of HMP-P synthesis in vitro has not yet been reported.
FIG. 1.
Purine mononucleotide and thiamine pyrophosphate biosynthetic pathways in S. enterica. Relevant intermediates are indicated. The documented phosphorylation of AIRs to AIR by STM4066 (8) is represented. Abbreviations: THZ-P, 4-methyl-5(β-hydroxyethyl) thiazole phosphate; TMP, thiamine phosphate; TPP, thiamine pyrophosphate.
Past work has identified loci other than thi biosynthetic enzymes that are required for efficient HMP-P synthesis. Characterization of these loci has implicated the oxidative pentose phosphate pathway, pantothenate (and/or coenzyme A [CoA]), and iron-sulfur cluster metabolism in the synthesis of HMP (10, 13, 14, 21, 23).
Work described here was initiated to probe the integration of distinct metabolic processes with thiamine synthesis. Genetic analysis identified an allele of rpoD (rpoD1181) that allowed several HMP-requiring mutant strains to grow in the absence of thiamine. Our working model suggests that the rpoD1181 (S508Y) allele restores thiamine-independent growth by altering expression of an unidentified gene(s) whose product is required for efficient HMP-P synthesis.
Thiamine synthesis in a purF iscA strain is restored by an rpoD allele.
Salmonella enterica strain DM6176 (purF iscA) is unable to grow in the absence of thiamine (23). The iscA allele used is an insertion that is polar on hscAB (23). Mutations that suppressed the thiamine synthetic defect caused by the iscA lesion were identified. A 0.1-ml aliquot of an overnight culture of DM6176 was spread on a plate containing minimal medium plus gluconate and adenine. After overnight incubation at 37°C, more than 300 colonies that no longer required thiamine for growth were found. Ten random colonies were chosen for further analysis. In nine of the mutants, an insertion near the gltA locus, zbg-6391::Tn10d(Tc), was genetically linked to the causative lesion. On the basis of the proximity of gltA to the sdh genes and the knowledge that null mutations in sdh spared cellular thiamine pools (11), succinate dehydrogenase activity was measured in the nine mutants. Each of the nine mutants lacked detectable succinate dehydrogenase activity (data not shown), and they were not analyzed further.
Multiple transposon insertions (MudJ, Tn10d) linked to the suppressor mutation in the remaining mutant (S. enterica DM6229) were identified. The chromosomal location of each transposon insertion was determined by a PCR-based protocol using degenerate primers (5, 25). Linkage analyses and three-factor cross data focused attention on a small region of the chromosome (∼10 kb) at approximately 70 min that contained five open reading frames. Each of the open reading frames was amplified from wild-type (DM6176) and mutant (DM6229) strains of S. enterica and sequenced. The causative lesion was found to be a CG-to-AT transversion in rpoD, resulting in the substitution of a tyrosine for serine at residue 508. Subsequently, rpoD was amplified and the PCR product was sequenced from the chromosome of DM6229 eight independent times to confirm the causative lesion.
Residue 508 is in conserved region 3.1 of RpoD and is analogous to S506 in the Escherichia coli protein. In E. coli, the similar rpoD (S506F) allele was isolated as a suppressor of the multiple amino acid auxotrophy of a relA spoT double mutant (15) and subsequently shown to reduce abortive transcription at several promoters in vitro (6, 16, 22). This substitution belonged to a class of mutants that responded as if ppGpp were always present. Most characterized members of this mutant class contained lesions in rpoBC. An rpoB (T563P) allele that belongs to this class (16) was isolated as a suppressor of the nutritional requirements of an E. coli dksA mutant (4). One interpretation of these results was that reduction in abortive transcription at a global set of promoters suppressed the nutritional requirements of the dksA mutant. To complete the correlation between this defined class of polymerase mutants and the Salmonella rpoD1181 allele, an isogenic pair of strains DM7088 (dksA3::MudJ) and DM7087 (dksA3::MudJ rpoD1181) was constructed. Doubling times on minimal medium containing glucose (240 and 100 min, respectively) determined that the rpoD1181 allele substantially relieved the nutritional requirements of the dksA mutant.
The rpoD (S508Y) allele alters transcription on a global level.
β-Galactosidase activity was measured from several MudJ transcriptional fusions in the presence or absence of the rpoD1181 allele (Table 1). Fusions to several loci involved in thiamine biosynthesis and CoA biosynthesis were used, as well as fusions to loci not expected to affect these pathways directly. The data show that expression from several promoters was slightly, but significantly, increased by the rpoD1181 allele. In particular, it was noted that expression from the panBCD fusion (1.6-fold), ilvC fusion (1.5-fold), and thiC fusion (1.6-fold) was increased (Table 1).
TABLE 1.
Expression from transcriptional fusions is affected by rpoD1181
| Strain | rpoD allele | Locusa | β-Galactosidase activity (Miller units)b | Relative activityc |
|---|---|---|---|---|
| DM7358 | rpoD1181 | ilvCd | 352 ± 5 | 1.5 |
| DM7359 | wte | ilvCd | 235 ± 6 | |
| DM7081 | rpoD1181 | panCd | 38.9 ± 1.3 | 1.6 |
| DM7082 | wt | panCd | 23.6 ± 0.4 | |
| DM7114 | rpoD1181 | thiC | 104 ± 4 | 1.6 |
| DM7115 | wt | thiC | 63.0 ± 1.4 | |
| DM7106 | rpoD1181 | dksA | 144 ± 2 | 1.3 |
| DM7107 | wt | dksA | 107 ± 4 | |
| DM7108 | rpoD1181 | stm4065 | 3.7 ± 0.0 | 0.8 |
| DM7109 | wt | stm4065 | 4.5 ± 0.1 | |
| DM7201 | rpoD1181 | gnd | 27.3 ± 0.9 | 1.0 |
| DM7202 | wt | gnd | 26.1 ± 1.9 | |
| DM7203 | rpoD1181 | yigF | 48.4 ± 0.2 | 1.0 |
| DM7204 | wt | yjgF | 48.9 ± 1.9 | |
| DM7205 | rpoD1181 | hisD | 22.6 ± 3.0 | 1.5 |
| DM7206 | wt | hisD | 15.3 ± 1.0 | |
| DM7207 | rpoD1181 | ilvD | 26.3 ± 0.3 | 1.1 |
| DM7208 | wt | ilvD | 24.5 ± 1.5 |
Strains carried a MudJ fusion (7) inserted in the indicated locus.
Cells from a full-density nutrient broth (NB) culture were inoculated into NB and grown to an optical density at 650 nm of ≈0.3. β-Galactosidase activity was determined as previously described (12, 17). A Miller unit is defined as 1 nmol of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per min.
β-Galactosidase activity in the rpoD1181 strain divided by activity of the wild-type strain.
Gene products from the panBCD operon are required for synthesis of pantothenate. IlvC function is required for pantothenate synthesis when panE is absent (20).
wt, wild type.
The rpoD1181 allele suppresses several conditional thiamine auxotrophs.
Multiple loci that prevent PurF-independent thiamine synthesis have been identified (9, 10, 18). In general, mutations in these loci are thought to reduce either (i) PurF-independent formation of phosphoribosylamine (PRA) or (ii) efficiency of conversion of AIR to HMP (M. J. Dougherty and D. M. Downs, unpublished data). The rpoD1181 allele was transduced into mutants representing both classes, and thiamine-independent growth was monitored. Results from these experiments are shown in Table 2. With the exception of the purF gnd mutant, each of the strains that contained the rpoD1181 allele regained thiamine-independent growth. Significantly, the purF gnd mutant has a defect in PRA formation (10), while the other mutants have been shown genetically to have defective conversion of AIR to HMP (Dougherty and Downs, unpublished).
TABLE 2.
The rpoD1181 allele alters thiamine-independent growth and CoA levels
| Strain | Relevant genotype | Doubling time (min)a | Total CoA thioesterb |
|---|---|---|---|
| DM6293 | purF iscAc rpoD1181 | 97 ± 6 | 0.115 ± 0.035 |
| DM6294 | purF iscA | NGd | 0.082 ± 0.004 |
| DM6324 | purF rpoD1181 | 80 ± 1 | 0.072 ± 0.002 |
| DM6325 | purF | NG | 0.155 ± 0.007 |
| DM6326 | purF panE rpoD1181 | 82 ± 5 | 0.024 ± 0.002 |
| DM6327 | purF panE | NG | 0.003 ± 0.005 |
| DM6328 | panE rpoD1181 | 95 ± 2 | NDe |
| DM6329 | panE | NG | ND |
| DM7291 | purF apbC rpoD1181 | 68 ± 4 | 0.058 ± 0.000 |
| DM7292 | purF apbC | NG | 0.090 ± 0.010 |
| DM7360 | purF apbE rpoD1181 | 74 ± 1 | 0.038 ± 0.004 |
| DM7361 | purF apbE | NG | 0.079 ± 0.001 |
| DM7362 | purF gshA rpoD1181 | 103 ± 2 | 0.033 ± 0.004 |
| DM7363 | purF gshA | NG | 0.044 ± 0.003 |
| DM7289 | purF gnd rpoD1181 | NG | 0.079 ± 0.003 |
| DM7290 | purF gnd | NG | 0.136 ± 0.006 |
Growth in liquid medium was assessed as described previously (18) using NCE (no carbon E medium) with 11 mM glucose or gluconate as the carbon source and 0.4 mM adenine.
Total amount of CoA thioester is given in nanomoles of CoA per milligram (dry weight). CoA levels were determined by the method of Allred and Guy (2).
iscA mutation is a polar insertion affecting iscA and hscAB (23).
NG, no growth. The absorbance at 650 nm did not increase during the course of the experiment.
ND, not done.
The rpoD1181 allele specifically affects the conversion of AIR to HMP-P.
The above results suggested a model in which altered gene expression caused by the rpoD1181 allele resulted in increased conversion of AIR to HMP, thus suppressing strains that are compromised in that step. The effect of the rpoD1181 allele on the efficiency of AIR to HMP conversion was monitored in vivo. Isogenic strains, DM7256 (purG purE stm4068 rpoD1181) and DM7257 (purG purE stm4068) were constructed. These strains are blocked in the de novo purine biosynthetic pathway before and after the thiamine precursor AIR (Fig. 1). An insertion in stm4068 allows efficient utilization of aminoimidazole riboside (AIRs) as a source of thiamine by increasing the expression of a kinase that converts AIRs to AIR (8).
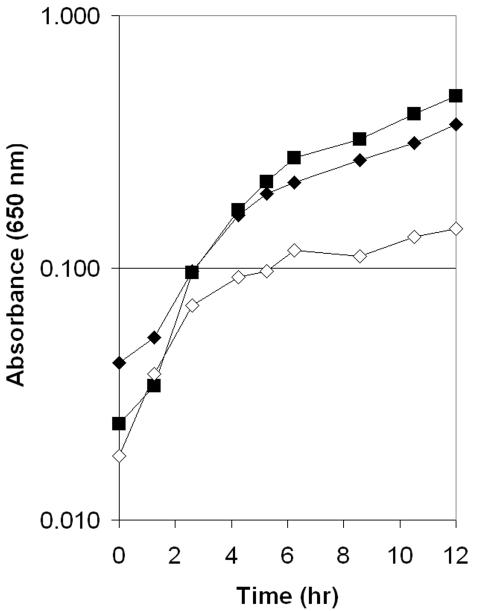
The two relevant strains were tested for growth on minimal medium containing glucose and adenine supplemented with variable amounts of AIRs. A 10 nM concentration of AIRs was determined empirically to be limiting for thiamine synthesis in strain DM7257, and this concentration was used in growth analyses. Results of the growth analyses (Fig. 2) showed that when 10 nM AIRs was provided as the source of HMP, the rpoD1181 allele slightly increased the growth rate and significantly increased the final yield of the strain. This result was interpreted to reflect increased conversion efficiency of AIR to HMP. In a similar assay, pantothenate was shown to stimulate the conversion of AIR to HMP (1). As shown in Fig. 2, addition of pantothenate to the rpoD mutant strain failed to further stimulate growth, suggesting that the same cellular process was being targeted by each. Although the rpoD1181 allele increased the transcription of the thi operon (Table 1), increasing the level of ThiC fails to result in thiamine-independent growth in the mutant backgrounds suppressed by the rpoD1181 allele (Table 2) (S. Allen and D. M. Downs, unpublished data).
FIG. 2.
The rpoD1181 allele increases the efficiency of the conversion of AIR to HMP-P. Growth in liquid medium was assessed as described previously (19) using NCE (no carbon E medium) with 11 mM glucose and 0.4 mM adenine; 10 nM AIRs and 100 μM pantothenate were added as indicated. The stm4068-6::Tn10d(Tc) insertion results in an ∼100-fold increase in expression of stm4066 (8). Representative growth for strain DM7256 [purG3111 purE3043 stm4068-6::Tn10d(Tc) zxx-9149::MudJ rpoD1181] grown with 10 nM AIRs (filled diamonds) or with 10 nM AIRs plus 100 μM pantothenate (filled squares) and strain DM7257 [purG3111 purE3043 stm4068-6::Tn10d(Tc) zxx-9149::MudJ] grown with 10 nM AIRs (open diamonds) is depicted.
The rpoD allele alters the level of total CoA thioesters.
The thiamine requirement of each strain suppressed by the rpoD1181 allele could be satisfied by pantothenate (D. M. Downs, unpublished data). Past work showed that in some cases, the effect of pantothenate on thiamine synthesis was via an increase in CoA levels (21). Taken together, these results suggested that the rpoD mutation could be altering endogenous CoA levels to allow growth, possibly by increasing expression of the panBCD operon (Table 1). Total CoA levels were determined in multiple isogenic strains differing in the allele of rpoD present (Table 2). In the majority of strain backgrounds, the mutant rpoD allele decreased CoA levels approximately twofold. This result was not consistent with increased CoA levels being responsible for restoring thiamine-independent growth.
In contrast to other strains, the rpoD1181 allele in the panE mutants increased total CoA levels approximately eightfold. However, even with this increase, the total CoA level in the panE strains was well below that found in the strains unable to grow independent of thiamine (Table 2). Strains lacking panE are proficient in pantothenate synthesis due to the ability of the IlvC enzyme to catalyze the conversion of α-ketopantoate to pantoate at a low efficiency (20). Thus, increased expression of the panBCD operon and the ilvC gene by the rpoD1181 allele (Table 1) could be responsible for increased pantothenate (and thus CoA) levels in this strain.
The working model implicates an additional factor(s) in the conversion of AIR to HMP.
Taken together, the above data do not support a model in which the rpoD1181 allele restores thiamine synthesis simply by elevating the cellular level of CoA thioesters. In considering data presented here and elsewhere, a working model was developed. This model suggests that the expression of a gene required for optimal conversion of AIR to HMP is increased by the rpoD1181 allele. In this scenario, the resulting gene product is involved (directly or indirectly) in the conversion of AIR to HMP and its function requires a Fe-S cluster and involves a CoA thioester.
Conclusions.
This work has identified a metabolic phenotype caused by the rpoD1181 allele. Results presented herein, in addition to previous work with the analogous E. coli allele, emphasize that small changes in global gene expression can result in metabolic flux changes significant enough to generate clear growth phenotypes. The suppressor allele described herein (rpoD1181) mediates its effect by stimulating the conversion of AIR to HMP by an as yet undefined mechanism. On the basis of the small magnitude of the transcriptional effect, the growth phenotype defined here provides the best means to identify cellular factors involved in HMP-P synthesis in this genetic background that may have eluded more standard genetic approaches.
Acknowledgments
We thank M. Cashel for helpful discussions.
This work was supported in part by competitive grant GM47296 from NIH. Funds were also provided from a 21st Century Scientist Scholars Award from the J.S. McDonnell Foundation. M.J.D. was supported by a Biotechnology Traineeship from NIH (T32 GM08349), a Louis and Elsa Thomsen Wisconsin Distinguished Fellowship Award, and the William H. Peterson Predoctoral Fellowship from the Department of Bacteriology.
REFERENCES
- 1.Allen, S., J. L. Zilles, and D. M. Downs. 2002. Metabolic flux in both the purine mononucleotide and histidine biosynthetic pathways can influence synthesis of the hydroxymethyl pyrimidine moiety of thiamine in Salmonella enterica. J. Bacteriol. 184:6130-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allred, J. B., and D. G. Guy. 1969. Determination of coenzyme A and acetyl CoA in tissue extracts. Anal. Biochem. 29:293-299. [DOI] [PubMed] [Google Scholar]
- 3.Begley, T. P., D. M. Downs, S. E. Ealick, F. W. McLafferty, A. P. Van Loon, S. Taylor, N. Campobasso, H. J. Chiu, C. Kinsland, J. J. Reddick, and J. Xi. 1999. Thiamine biosynthesis in prokaryotes. Arch. Microbiol. 171:293-300. [DOI] [PubMed] [Google Scholar]
- 4.Brown, L., D. Gentry, T. Elliott, and M. Cashel. 2002. DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol. 184:4455-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caetano-Annoles, G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 3:85-92. [DOI] [PubMed] [Google Scholar]
- 6.Cashel, M., L. M. Hsu, and V. J. Hernandez. 2003. Changes in conserved region 3 of Escherichia coli σ70 reduce abortive transcription and enhance promoter escape. J. Biol. Chem. 278:5539-5547. [DOI] [PubMed] [Google Scholar]
- 7.Castilho, B. A., P. Olfson, and M. J. Casadaban. 1984. Plasmid insertion mutagenesis and lac gene fusion with mini-Mu bacteriophage transposons. J. Bacteriol. 158:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dougherty, M., and D. M. Downs. 2003. The stm4066 gene product of Salmonella enterica serovar Typhimurium has aminoimidazole riboside (AIRs) kinase activity and allows AIRs to satisfy the thiamine requirement of pur mutant strains. J. Bacteriol. 185:332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Downs, D. M., and L. Petersen. 1994. apbA, a new genetic locus involved in thiamine biosynthesis in Salmonella typhimurium. J. Bacteriol. 176:4858-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enos-Berlage, J. L., and D. M. Downs. 1996. Involvement of the oxidative pentose phosphate pathway in thiamine biosynthesis in Salmonella typhimurium. J. Bacteriol. 178:1476-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enos-Berlage, J. L., and D. M. Downs. 1997. Mutations in sdh (succinate dehydrogenase genes) alter the thiamine requirement of Salmonella typhimurium. J. Bacteriol. 179:3989-3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escalante-Semerena, J. C., and J. R. Roth. 1987. Regulation of cobalamin biosynthetic operons in Salmonella typhimurium. J. Bacteriol. 169:2251-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frodyma, M., A. Rubio, and D. M. Downs. 2000. Reduced flux through the purine biosynthetic pathway results in an increased requirement for coenzyme A in thiamine synthesis in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:236-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gralnick, J., E. Webb, B. Beck, and D. Downs. 2000. Lesions in gshA (encoding γ-l-glutamyl-l-cysteine synthetase) prevent aerobic synthesis of thiamine in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 182:5180-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernandez, V. J., and M. Cashel. 1995. Changes in conserved region 3 of Escherichia coli sigma 70 mediate ppGpp-dependent functions in vivo. J. Mol. Biol. 252:536-549. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez, V. J., L. M. Hsu, and M. Cashel. 1996. Conserved region 3 of Escherichia coli final σ70 is implicated in the process of abortive transcription. J. Biol. Chem. 271:18775-18779. [DOI] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Petersen, L., and D. M. Downs. 1996. Mutations in apbC (mrp) prevent function of the alternative pyrimidine biosynthetic pathway in Salmonella typhimurium. J. Bacteriol. 178:5676-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen, L., J. Enos-Berlage, and D. M. Downs. 1996. Genetic analysis of metabolic crosstalk and its impact on thiamine synthesis in Salmonella typhimurium. Genetics 143:37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Primerano, D. A., and R. O. Burns. 1983. Role of acetohydroxyacid isomeroreductase in biosynthesis of pantothenic acid in Salmonella typhimurium. J. Bacteriol. 153:259-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubio, A., and D. M. Downs. 2002. Elevated levels of ketopantoate hydroxymethyltransferase (PanB) lead to a physiologically significant coenzyme A elevation in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:2827-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sen, R., H. Nagai, V. J. Hernandez, and N. Shimamoto. 1998. Reduction in abortive transcription from the λPR promoter by mutations in region 3 of the σ70 subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 273:9872-9877. [DOI] [PubMed] [Google Scholar]
- 23.Skovran, E., and D. M. Downs. 2000. Metabolic defects caused by mutations in the isc gene cluster in Salmonella enterica serovar Typhimurium: implications for thiamine synthesis. J. Bacteriol. 182:3896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vander Horn, P. B., A. D. Backstrom, V. Stewart, and T. P. Begley. 1993. Structural genes for thiamine biosynthetic enzymes (thiCEFGH) in Escherichia coli K-12. J. Bacteriol. 175:982-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webb, E., K. Claas, and D. Downs. 1998. thiBPQ encodes an ABC transporter required for transport of thiamine and thiamine pyrophosphate in Salmonella typhimurium. J. Biol. Chem. 273:8946-8950. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, Y., and T. P. Begley. 1997. Cloning, sequencing and regulation of thiA, a thiamine biosynthesis gene from Bacillus subtilis. Gene 198:73-82. [DOI] [PubMed] [Google Scholar]