Abstract
In Escherichia coli, assembly of the FtsZ ring (Z ring) at the cell division site is negatively regulated by the nucleoid in a phenomenon called nucleoid occlusion (NO). Previous studies have indicated that chromosome packing plays a role in NO, as mukB mutants grown in rich medium often exhibit FtsZ rings on top of diffuse, unsegregated nucleoids. To address the potential role of overall nucleoid structure on NO, we investigated the effects of disrupting chromosome structure on Z-ring positioning. We found that NO was mostly normal in cells with inactivated DNA gyrase or in mukB-null mutants lacking topA, although some suppression of NO was evident in the latter case. Previous reports suggesting that transcription, translation, and membrane insertion of proteins (“transertion”) influence nucleoid structure prompted us to investigate whether disruption of these activities had effects on NO. Blocking transcription caused nucleoids to become diffuse, and FtsZ relocalized to multiple bands on top of these nucleoids, biased towards midcell. This suggested that these diffuse nucleoids were defective in NO. Blocking translation with chloramphenicol caused characteristic nucleoid compaction, but FtsZ rarely assembled on top of these centrally positioned nucleoids. This suggested that NO remained active upon translation inhibition. Blocking protein secretion by thermoinduction of a secA(Ts) strain caused a chromosome segregation defect similar to that in parC mutants, and NO was active. Although indirect effects are certainly possible with these experiments, the above data suggest that optimum NO activity may require specific organization and structure of the nucleoid.
When Escherichia coli cells divide by binary fission, they assemble a septal protein complex at midcell. The first known protein to localize, FtsZ, forms a ring (the Z ring) between the two segregated nucleoids at about the time of replication termination (7). The Z ring then recruits the rest of the septal complex. The selection of the site at midcell is remarkably accurate (36, 47), and two global systems have important topological roles in identifying this site (21, 47). One of these, the Min system, prevents unwanted division events at nucleoid-free cell poles. This is achieved by the continuous oscillation of an inhibitor of Z-ring formation (MinC), a membrane-associated ATPase (MinD), and an enhancer of the MinD ATPase (MinE) between the two cell poles, keeping MinC away from the midcell site (6, 12, 15-17, 30, 31). The Min system may be important for the accuracy of midcell Z-ring placement (18), or it may simply function to prevent polar divisions, as seems to be the case in Bacillus subtilis (22).
The other topological system that regulates Z-ring placement is the nucleoid itself. By an unknown mechanism, Z-ring assembly on the membrane is inhibited by the presence of the nucleoid at the same site. This negative spatial regulation is called “nucleoid occlusion” (NO) (41, 44). Under the combined negative control of both the Min system and NO, the most favorable part of the cell for assembly of the Z rings occurs at midcell at the time of chromosome segregation (21). The nucleoid and Min system appear to be independent of each other. In mutant cells lacking a nucleoid, Z rings localize near midcell, presumably because the Min system remains functional in such anucleate cells (35). Conversely, without the Min system, nucleoids can still inhibit septum formation in their vicinity (34, 47). NO was still active after arresting chromosomal DNA replication by thermoinactivation of dnaA, indicating that chromosome replication is not required for NO (34).
The molecular mechanism of NO is not yet known. One possible obstacle for division is the physical structure or local density of the nucleoid and/or nucleoid-associated proteins. To maintain the proper nucleoid structure, E. coli chromosomes need to be condensed. Disruption of MukB, a homolog of SMC that is involved in condensation of the nucleoid (32), results in thermosensitive growth and partial loss of NO (5, 25, 35). The suppression of NO in turn is thought to result in guillotining of misplaced nucleoids by septa, producing a 1,000-fold increase in the percentage of anucleate cells (25, 45).
Another important factor that governs nucleoid structure is DNA superhelicity. DNA gyrase introduces negative supercoils into DNA, while topoisomerase I (encoded by topA) removes them (8, 29). Interestingly, the defects of mukB mutants can be largely suppressed by inactivating topA, which increases negative superhelicity and thus local DNA density (32, 33). This increase in local DNA density appears to compensate for the lower levels of chromosome packing in mukB mutants. The suppression of the mukB-associated defects by a topA mutation suggests that local DNA concentration, and not some special function of MukB, is a critical factor for NO.
In addition to supercoiling and condensation, proper nucleoid structure and function also appear to require the process of transertion, which is defined as the ongoing processes of transcription, translation, and insertion of membrane and export (42). Perturbing transcription or translation causes significant changes in nucleoid density; blocking transcription expands the nucleoid, while blocking translation compacts it (26, 43). To learn more about the molecular basis of nucleoid occlusion, in this study we examine the formation and positioning of Z rings in cells with abnormal nucleoid structure caused by perturbations in superhelicity and the transertion process. By comparing the relative positions of FtsZ assembly and nucleoids, the effects of these various factors on NO were investigated.
MATERIALS AND METHODS
Strains and growth conditions.
All strains used in this study are derivatives of E. coli K-12 and are shown in Table 1. Cells were grown either in Luria-Bertani (LB) medium or M9 glucose medium supplemented with 0.5% Casamino Acids. Overnight cultures were diluted 200× in fresh medium and grown at 30°C until the early logarithmic phase. For thermosensitive mutants, cells were then shifted to the appropriate nonpermissive temperature. To study the effects of drugs on nucleoid occlusion or septation, we used final concentrations of 100-μg/ml rifampin (dissolved in dimethyl sulfoxide), 100-μg/ml chloramphenicol, 200-μg/ml cephalexin, or 5-μg/ml coumermycin A1. Drugs were obtained from Sigma Chemical Co., St. Louis, Mo.
TABLE 1.
Strains and plasmids used in this study
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| TX3772 | MG1655 ΔlacU169 | 37 |
| CAG19209 | rpoD800(Ts) | 14 |
| CGSC7141 | secA51(Ts) | 27 |
| CC4207 | ΔmukB::kan topA10 | 32 |
| WM1032 | TX3772 ΔminCDE::kan | Lab collection |
| WM1255 | pWM1255 (GFP-MinD) in JM105 | 4 |
| WM1263 | gyrB+ parent of WM1263 | LE234 (28) |
| WM1262 | F−argE gyrB(Ts) ilv metB tna | LE316 (28) |
Immunostaining and microscopy.
Immunofluorescence in combination with phase-contrast imaging and nucleoid staining with 4′, 6-diamidino-2-phenylindole (DAPI) was carried out with methanol-fixed cells as described previously (34) with a few modifications. For FtsZ staining, we used a 1:200 dilution of affinity-purified anti-FtsZ, followed by goat anti-rabbit secondary antibody conjugated with an Alexa 488 (Molecular Probes, Eugene, Oreg.). An Olympus BX60 microscope and an Optronics DEI-750 camera were used for photographing images. To overlay images of green FtsZ and DAPI, the blue DAPI signal was pseudocolored red by switching the color cables on the Scion CG-7 grabber card. The images were edited in Adobe Photoshop. FtsZ positions and cell lengths were measured by pixels in Photoshop and plotted in Microsoft Excel.
RESULTS
Effects of DNA supercoiling on nucleoid occlusion.
It was demonstrated previously that Z rings were almost always excluded from the nucleoid after inactivation of DnaA and thus replication initiation (34). To test the idea that other aspects of nucleoid structure and dynamics might also be important for nucleoid occlusion, we first examined Z-ring positioning in mutants with disrupted DNA supercoiling. DNA gyrase has two subunits, gyrA and gyrB, and the latter encodes an ATPase that binds DNA (23). Disruption of gyrB by mutation or with drugs causes trapping of gyrase on DNA (2). We first used a gyrB(Ts) mutant that inactivates gyrase at 42°C (28). This mutant is also defective in DNA replication initiation, probably because of the reduced level of negative supercoiling (11).
To avoid potential complications caused by multiple replication cycles in rapidly growing cells, we grew the gyrB mutant (WM1263) in M9 glucose medium supplemented with Casamino Acids. Under such growth conditions, the generation time at 30°C was about 2 h (data not shown). After cells were shifted to the nonpermissive temperature of 42°C for 45 min, fixed, and stained with DAPI, most cells had only one detectable nucleoid and significant nucleoid-free zones at the poles (Fig. 1E). This contrasts with the presence of two nucleoids in many wild-type MG1655 cells or the isogenic parent cells (WM1262) grown under the same conditions (Fig. 1D) (data not shown). This staining pattern is consistent with arrest of replication and segregation but continuation of some growth, similar to dnaA mutants grown at the nonpermissive temperature (34).
FIG. 1.
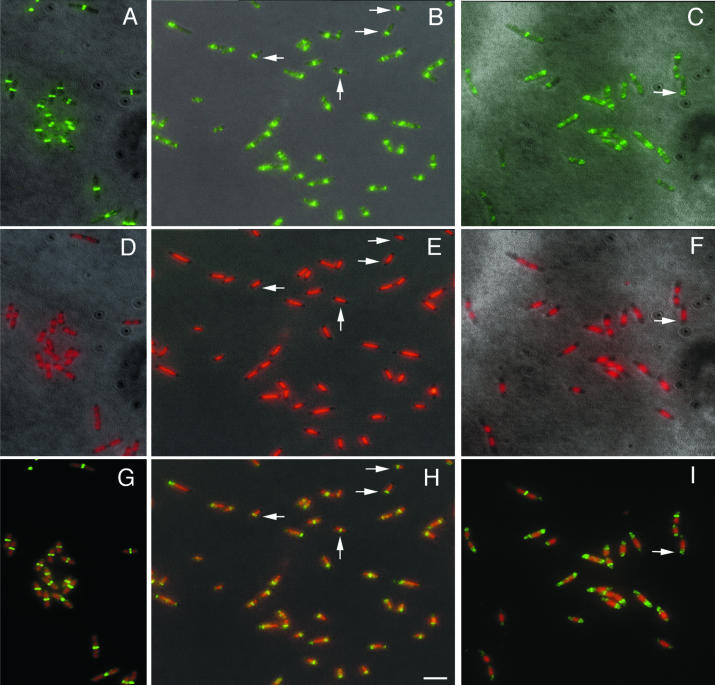
Effect of DNA supercoiling on FtsZ localization. (A to C) FtsZ staining. (D to F) DNA stained with DAPI. (G to I) Overlay of DNA and FtsZ. Panels A, D, and G show wild-type TX3772 cells. Panels B, E, and H show gyrB mutant cells grown at 28°C in M9 glucose plus Casamino Acids until the early log phase and then grown at 42°C for another 45 min. Panels C, F, and I show TX3772 cells grown in LB medium at 28°C and treated with 5-μg/ml coumermycin A1 for 40 min. Vertical arrows highlight cells containing central Z rings apparently over an unsegregated nucleoid. Horizontal arrows highlight cells containing acentral Z rings in anucleate segments or at nucleoid edges. Scale bar, 5 μm.
We then examined FtsZ positioning in these cells by immunofluorescence. It was clear that while Z rings were always at midcell in wild-type cells (Fig. 1A and G), in the gyrB mutant they tended to assemble away from midcell, often appearing at nucleoid edges. Such single acentral rings were observed in 50 out of 61 cells counted (82%) that contained a Z ring (Fig. 1B, E, and H) (data not shown), reminiscent of the patterns observed with dnaA mutants (34). In addition, some cells had more than one acentral Z ring (Fig. 1B, E, and H). In 12 of the 61 cells with Z rings (18%), a Z ring was localized at midcell, and some of these had no apparent gaps in DAPI staining (Fig. 1) (data not shown). One potential explanation for this last class of cells is that they had already initiated chromosome replication before gyrB was inactivated completely by the temperature shift and were in the process of chromosome segregation when samples were prepared. In addition, the more relaxed state of the nucleoid might make it more difficult to detect nucleoid gaps by DAPI staining. Yet another potential explanation is that NO is suppressed in some cells under these conditions for reasons that are not yet clear. Nevertheless, it is clear that Z rings often formed at acentral locations in the gyrB mutants at the nonpermissive temperature, usually at nucleoid edges. Some of these rings must have been active, because samples taken at 60 to 80 min after temperature shift exhibited a 15 to 20% frequency of anucleate cells (data not shown), indicating that the acentral rings at nucleoid edges had divided the cells into nucleate and anucleate daughters. As observed previously (35), nearly all the anucleate cells had Z rings.
To confirm these results, we examined the effects of coumermycin A1, a drug that inhibits gyrB ATPase activity (13, 23, 29), on NO and Z-ring placement. Because prolonged drug treatment leads to cell lysis, cells were grown rapidly in LB medium. Wild-type TX3772 cells were treated with 5-μg/ml coumermycin A1, and Z-ring positions were visualized by immunofluorescence (Fig. 1C, F, and I). The DAPI staining patterns and FtsZ localization were similar to those of the gyrB mutant at 42°C, with FtsZ staining in most cells to the edge of the nucleoid, away from midcell. These results support the suggestion that relaxation of the chromosome by reducing its superhelicity does not significantly suppress NO activity.
FtsZ localization in a mukB topA mutant.
Z rings often localize on top of decondensed nucleoids in a mukB mutant, indicating that nucleoid occlusion is partially suppressed (35). Cells with a defective mukB gene are thermosensitive, forming colonies in rich medium only at or below 28°C. The reason for this is not known, but perhaps slowing down the cell cycle helps to compensate for insufficient chromosomal condensation during replication (1). The suppression of anucleate cell production in a mukB mutant by inactivation of topA indicates that the nucleoids are properly positioned and condensed and probably are not cut by Z rings or division septa at a significant frequency. However, we wondered whether there might still be subtle differences in Z-ring positioning as compared with wild-type cells, and therefore we examined Z rings in a topA mukB double mutant (CC4207).
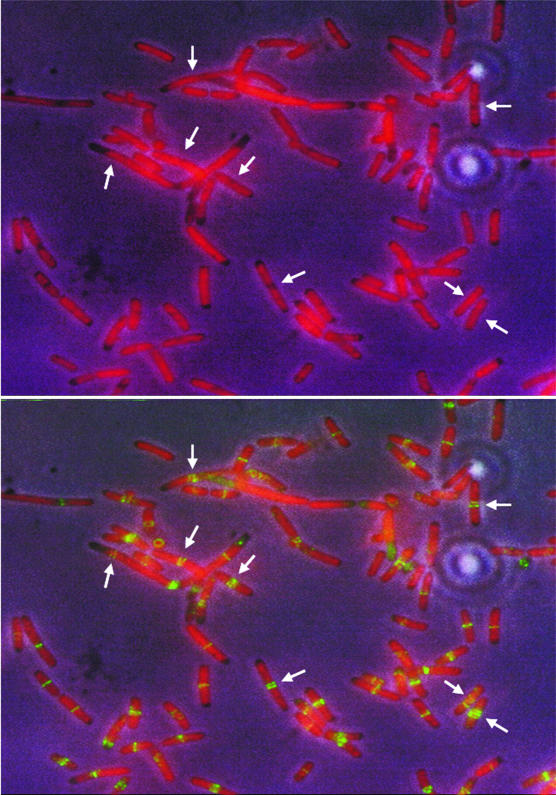
CC4207 cells were grown at various temperatures (28, 32, 37, and 42°C) before being fixed for immunofluorescence. At all temperatures examined, Z rings were present in gaps between nucleoids and lone rings were not present on top of nucleoids (data not shown). Interestingly, however, closely spaced double FtsZ bands were present in gaps between nucleoids at high frequency in cells grown at 28°C but not at higher temperatures (Fig. 2B). It was not possible to determine whether these multiple FtsZ bands represented multiple Z rings or turns of a continuous FtsZ helix. The distances between nucleoids were similar to those in wild-type cells (Fig. 2A), ruling out the possibility that the unusual FtsZ structures resulted from larger regions that lacked NO activity. In fact, it appeared that the nucleoid edges were not as potent at excluding Z rings, because the outer rings in a cluster appeared to be spread to the DAPI-stained nucleoid region (Fig. 2B). This pattern of Z-ring clusters on top of nucleoids but centered at a nucleoid-free gap is reminiscent of the Z-ring staining in ΔmukB Δmin double mutants (46). Overall, these results suggest that the topA mutation, while allowing survival of mukB mutants and suppression of most of the mukB phenotypes, does not completely suppress the NO defect. We cannot explain why we were not able to see a significant frequency of ring clusters at other growth temperatures.
FIG. 2.
Multiple FtsZ bands formed in a mukB topA double mutant. CC4207 cells were grown at 28°C and fixed for staining. Arrows highlight clusters of multiple Z rings, sometimes extending to regions occupied by nucleoids. (A) DAPI plus phase-contrast. (B) FtsZ immunostaining plus DAPI staining.
Transcription inhibition suppresses nucleoid occlusion.
It has been proposed that the structure of the E. coli nucleoid is determined by DNA binding proteins and DNA supercoiling, representing a compaction force, and by the coupled transcription/translation/translocation of plasma membrane and cell wall proteins, representing an opposing expansion force (26, 43). We were interested to study the influence of each expansion factor on NO.
To examine the role of transcription, we treated wild-type TX3772 cells with rifampin, which interferes with the β subunit of prokaryotic RNA polymerase and blocks transcription initiation (40, 48). After 10 min of rifampin treatment, nucleoids already showed some expansion as judged by DAPI staining of fixed cells compared to untreated cells (data not shown). After 30 min of rifampin treatment, the DAPI staining was uniform throughout the cells, with few detectable nucleoid-free gaps (Fig. 3F) compared to untreated cells (Fig. 3C). This nucleoid expansion effect was much easier to see in filamentous Δmin mutant cells, because of the multiple spaces between nucleoids (see below).
FIG. 3.
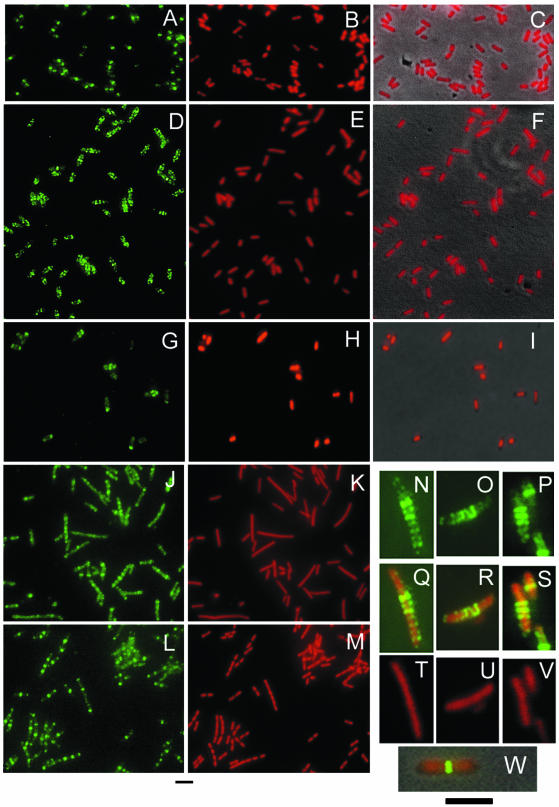
Effects of inhibiting transcription and translation on the nucleoid and FtsZ positioning. Shown are representative fields of untreated wild-type TX3772 cells (A to C), TX3772 cells 30 min after addition of 100-μg/ml rifampin (D to F), TX3772 cells 2 h after addition of 200-μg/ml chloramphenicol (G to I), WM1032 (Δmin) cells 30 min after addition of 100-μg/ml rifampin (J to K), or untreated (L and M). All cells were grown in LB medium except for those treated with chloramphenicol, which were grown in M9 glucose plus Casamino Acids. Panels A, D, G, J, and L show FtsZ immunostaining; panels B, E, H, K, and M show DAPI staining; and panels C, F, and I show DAPI staining overlaid with the phase-contrast image. Representative individual cells of CAG19209 (rpoD800) grown at 45°C for 1 h are shown either as FtsZ stained (N to P), DAPI stained (T toV), or merged (Q to S). Panel W shows a merged image of TX3722 cells for comparison. Scale bars are 5 μm for panels A to M (under panels L and M) and N to W (under panel W).
FtsZ staining patterns changed along with the change in DAPI staining during the time course. Intense FtsZ staining occurred at a nucleoid-free gap at midcell in 90% of untreated cells, with little FtsZ staining elsewhere in the cell (Fig. 3A). After 10 min of rifampin treatment, FtsZ staining was still predominantly at midcell, coincident with nucleoid-free gaps, but a significant amount of staining could be observed in other areas of the cell (data not shown). After 30 min of rifampin treatment, FtsZ staining was now predominantly at locations other than midcell. In more than 35% of these cells, multiple bands of FtsZ staining (two to five) were detected (Fig. 3D). As the sizes of most cells were in the range of 2 to 5 μm, the distances between FtsZ bands were shorter than normal, averaging 0.8 μm. This compares to 1.3 μm in Δmin mutant cells, in which Z rings are present at all nucleoid-free gaps but not on top of nucleoids (47). Because the nucleoid appeared to be distributed throughout the cell (Fig. 3E to F), by definition all of the FtsZ bands were located on top of nucleoids.
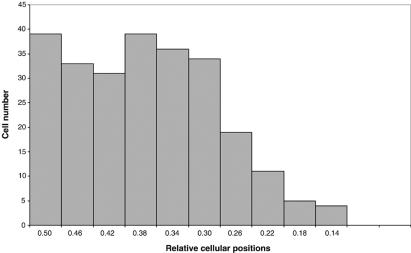
The intensities of different FtsZ bands in each cell varied. By analysis of the positions of 122 intensely staining bands, we found a tendency to localize near the cell center, but not always at the exact midcell site (Fig. 4). Bright bands at other acentral positions were also observed. Although we do not know when these bright bands formed, their presence at regions far from midcell indicates that most of these structures were not present prior to drug treatment. Furthermore, premature FtsZ assembly was detected as fluorescent foci in most cells, even in some cells that did not contain a clear Z ring. The patterns suggest that more nucleation sites are available for FtsZ assembly in these cells. Such patterns are not a result of failure of the Min system to oscillate, because we observed typical green fluorescent protein (GFP)-MinD oscillation in rifampin-treated WM1255 cells (data not shown). Perhaps the weaker FtsZ accumulations in such cells resulted from ongoing division inhibition by MinCD.
FIG. 4.
Distribution of FtsZ bands in rifampin-treated cells. A total of 122 cells, treated with rifampin as in Fig. 3D to F, were measured. The x axis represents positions of FtsZ bands relative to cell length, with 0.5 representing midcell and 0.0 representing a cell pole.
We next asked whether removing the Min system would alter the pattern of FtsZ staining in the presence of rifampin. We treated ΔminCDE::kan cells (WM1032) with rifampin and stained them for FtsZ and nucleoids. As expected, after only 30 min of rifampin treatment, nearly all cells exhibited a diffuse nucleoid stain in the short filamentous cells that are typical of a Δmin mutant (Fig. 3K). This is in contrast to the distinct multiple nucleoids visible in untreated Δmin cells (Fig. 3M). While clear Z rings formed only between well-distributed nucleoids in the untreated cells (Fig. 3L), bands of FtsZ staining were located apparently randomly in Δmin cells treated with rifampin, with the spacing between bands sometimes as short as 0.5 μm (Fig. 3J). Moreover, unlike Min-positive cells, Min-negative cells treated with rifampin exhibited FtsZ bands throughout the cell length with no bias towards the cell center, as well as greater uniformity of band intensities (Fig. 4) (data not shown). This bias toward the cell center would be consistent with the Min system being fully functional in the presence of rifampin.
To confirm independently that blocking transcription relieves nucleoid occlusion, we examined FtsZ localization in an rpoD800 mutant, in which the major transcription initiation factor, σ70, can be thermoinactivated. After a 1-h inactivation of σ70 in cells containing rpoD800 at 45°C, nucleoids stained with DAPI appeared to be diffuse throughout the whole cell, similar to those observed after rifampin treatment (Fig. 3T to V). Multiple FtsZ bands were positioned on top of nucleoids in a pattern similar to that in rifampin-treated cells (Fig. 3N to S). Wild-type cells grown at the same temperature (Fig. 3W) or rpoD800 cells grown at 30°C demonstrated normal FtsZ localization (data not shown). This result indicates that transcription inactivation itself interrupts division inhibition by the nucleoid, possibly by altering local density of the DNA.
Nucleoid occlusion remains active after blocking translation.
To test if preventing protein translation also leads to disrupted cell division, we treated wild-type TX3772 cells with chloramphenicol, which inhibits peptidyl transferase activity but leaves mRNA synthesis unchanged (19, 24). TX3772 cells growing exponentially in either M9 glucose or LB medium at 28°C were treated with chloramphenicol. After treatment for 0.5 to 2 h, nucleoids appeared to be fused and compact (Fig. 3H and I) (data not shown) as reported previously (38, 39). Z rings were often acentral, with a high proportion of FtsZ staining at the cell poles (Fig. 3G) (data not shown). Frequently abnormal FtsZ structures (foci and tilted bands) were visualized at the cell poles. However, we did not detect FtsZ staining on top of the condensed nucleoids in these cells, and rarely were there multiple FtsZ bands or helices visible. These results indicate that after chloramphenicol treatment, polar division sites are available for FtsZ assembly and compacted nucleoids retain their function to inhibit Z-ring assembly. As the Min proteins oscillate normally in the presence of chloramphenicol (31), the availability of polar sites for FtsZ assembly must be either a result of an unstable component of the Min system, such as MinC, or some other unknown factor.
Effects of inhibiting protein translocation on NO.
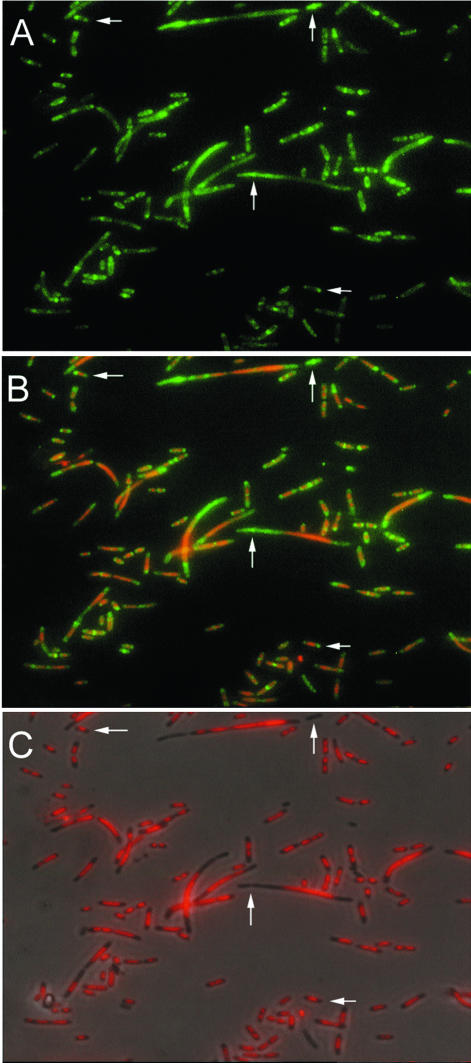
The major route for protein export or insertion into the membrane in bacteria occurs via the Sec-dependent transport apparatus. The core complex of this apparatus, consisting of SecYEG, forms a protein-conducting channel in the inner membrane, while the ATPase SecA drives translocation of substrate across the membrane (3). We tested whether SecA-mediated protein translocation and export had any effects on nucleoid occlusion. A mutant with a thermosensitivity mutation in secA (secA51) (27) was grown at the nonpermissive temperature of 42°C for 2 h and stained for FtsZ. These mutant cells appear to have a defect in chromosome segregation as well as cell division (Fig. 5C). Large nucleoids, presumably carrying multiple chromosomes, were visible in short filaments. Z rings localized between segregated nucleoids in many cells, but in cells with one central nucleoid or in filaments with large unsegregated nucleoids, FtsZ almost always localized away from the nucleoids at nucleoid edges. Septation from these rings occasionally resulted in anucleate cells, some of which had a Z ring at midcell (vertical arrows). This phenotype is very similar to that of chromosome segregation mutants like parC or parE (35). These results suggest that unpartitioned nucleoids produced in a secA mutant still exclude FtsZ from assembling in their vicinity and that disruption of Sec-mediated translocation does not abolish NO activity. The reasons for the nucleoid segregation defect after inactivation of secA are not apparent and will require further study.
FIG. 5.
Nucleoid and FtsZ distribution in a protein secretion mutant. The secA51 mutant was grown at 42°C for 2 h to inhibit sec-mediated protein translocation, and cells were fixed and stained. (A) FtsZ immunofluorescence staining. (B) Overlay of DAPI and FtsZ staining. (C) Overlay of DAPI staining and phase-contrast image. Vertical arrows highlight anucleate regions or anucleate cells containing Z rings, while horizontal arrows highlight acentral FtsZ staining in cells with a central nucleoid.
DISCUSSION
Many factors influence nucleoid structure, directly or indirectly. To understand how nucleoid structure might influence NO, we investigated the effects of altering DNA supercoiling levels or blocking transcription, translation, or protein translocation on NO activity. Our results demonstrated that superhelical relaxation of nucleoids does not affect NO, resulting in abnormal placement of Z rings at nucleoid edges, distal from midcell. This supports earlier work that showed abnormal placement of septa when gyrase is thermoinactivated (28). The active NO in gyrB mutants stands in contrast to that in mukB mutants, in which Z rings often form on top of the decondensed nucleoids. One possible explanation is that in the gyrB mutant at the nonpermissive temperature of 42°C, the activity of MukB maintains a sufficiently high level of local nucleoid density that is capable of blocking most Z rings from assembling over the nucleoid.
Our results also showed that in the absence of MukB, the nucleoid regains its ability to inhibit Z-ring assembly when topoisomerase I is inactivated in the topA mukB double mutant. This was not surprising, given that the double mutant cells grow and divide fairly normally (32), and supports the notion that increased local DNA concentration is sufficient to activate NO. However, many topA mukB cells at 28°C displayed multiple Z rings, with the ring cluster centered about a gap between nucleoids but often extending into the nucleoid region. This suggests that NO is partially suppressed under these conditions near the nucleoid edges. Nucleoid edges were also less active in NO than more central parts of the nucleoid in ΔmukB Δmin double mutants (46). This effect might reflect a gradient of NO, which might be in excess throughout normal nucleoids but is unmasked in mukB mutants under certain conditions. Another possibility is that nucleoids in a mukB mutant are not decondensed uniformly and have a higher local density towards their centers, resulting in more complete NO there than near nucleoid edges.
Importantly, we found that division inhibition by nucleoids was compromised in cells in which transcription was inhibited. Multiple FtsZ structures, possibly Z rings and possibly a continuous helix of FtsZ, assemble on top of the nucleoid. This is suggestive of relief of NO such that more potential division sites are available for FtsZ assembly. This FtsZ localization phenotype was also observed in rifampin-treated cells when grown in M9 glucose medium (data not shown). This was not seen in mukB cells, possibly because there is some residual NO in mukB cells that tends to shunt most unassembled FtsZ to cooperative assembly at a few Z rings. When transcription was inhibited by thermoinactivation of σ70, the nucleoid and FtsZ staining patterns were similar to those with rifampin, indicating that the effects were not artifacts specific to the drug.
In contrast, chloramphenicol treatment resulted in a clearly different effect on FtsZ localization. While FtsZ often still localized at midcell after rifampin treatment, midcell Z rings were extremely rare after chloramphenicol treatment. Although many indirect effects could be involved, one possible explanation for this difference is the distinct difference in nucleoid structure after treatment with rifampin versus chloramphenicol. It is generally accepted that chloramphenicol treatment leads to nucleoid fusion and compaction, while the rifampin does not (9, 10, 20, 38, 39). The diffusion of the nucleoid throughout the cell after rifampin treatment is very clear, particularly in the Δmin mutant because of the ease of visualizing multiple nucleoids in the filamentous cells characteristic of this mutant. The mechanism behind this apparent decompaction of nucleoids after transcription inhibition is unknown, but the effect is similar to that of inhibiting MukB. Likewise, the nucleoid compaction by chloramphenicol treatment might mimic the topA suppression of the mukB defect.
The effects of blocking either transcription or translation are likely to be highly complex. However, one possible explanation for the absence of Z rings at midcell after chloramphenicol treatment is that the resulting compact nucleoid occludes assembly of new Z rings. If so, this would be surprising given the assumption that the nucleoid retracts from the membrane when transertion is blocked. The disappearance of midcell Z rings in the first place may be caused by the need for continuous protein synthesis to maintain Z rings or perhaps by some type of interference by the nucleoid undergoing compaction. We favor the latter idea, only because chloramphenicol treatment does not prevent FtsZ from assembling into structures; they are present, but just not at midcell. Another possible explanation for the different effects of blocking translation versus transcription is that NO might be mediated by a specific RNA.
What might cause FtsZ to localize preferentially away from midcell after chloramphenicol treatment? One possibility is that the Min system is inactivated, allowing Z rings to form in nucleoid-free spaces, which are larger because of nucleoid compaction. As the Min proteins continue to oscillate after chloramphenicol treatment, perhaps some other aspect of Min protein function is inactivated. It is also possible that another unknown factor that is important for negatively regulating Z-ring assembly is destabilized during chloramphenicol treatment, allowing extra Z rings to form in nucleoid-free spaces. It is clear, however, that after extended blocking of translation, FtsZ protein remains sufficiently abundant and stable to be able to assemble into structures.
A model that summarizes the results in this study is shown in Fig. 6. The results are all consistent with the idea that a compact unsegregated nucleoid, either in normal cells (Fig. 6A) or in cells treated with chloramphenicol (Fig. 6B), will exclude Z-ring assembly via NO, while a diffuse nucleoid, either because of transcription inhibition (Fig. 6C) or decompaction resulting from the loss of MukB or SMC, will suppress NO and allow FtsZ to assemble in areas occupied by the nucleoid. An important caveat to the present study is that it is possible that the effects we attribute to NO are actually direct effects of the drugs or mutations on FtsZ assembly and positioning, independent of NO or at least not completely dependent on NO. Unfortunately, this caveat will persist until a more specific mechanism for NO is found, such that more specific alterations in NO can be constructed that will minimize the potential for indirect and nonspecific effects.
FIG. 6.
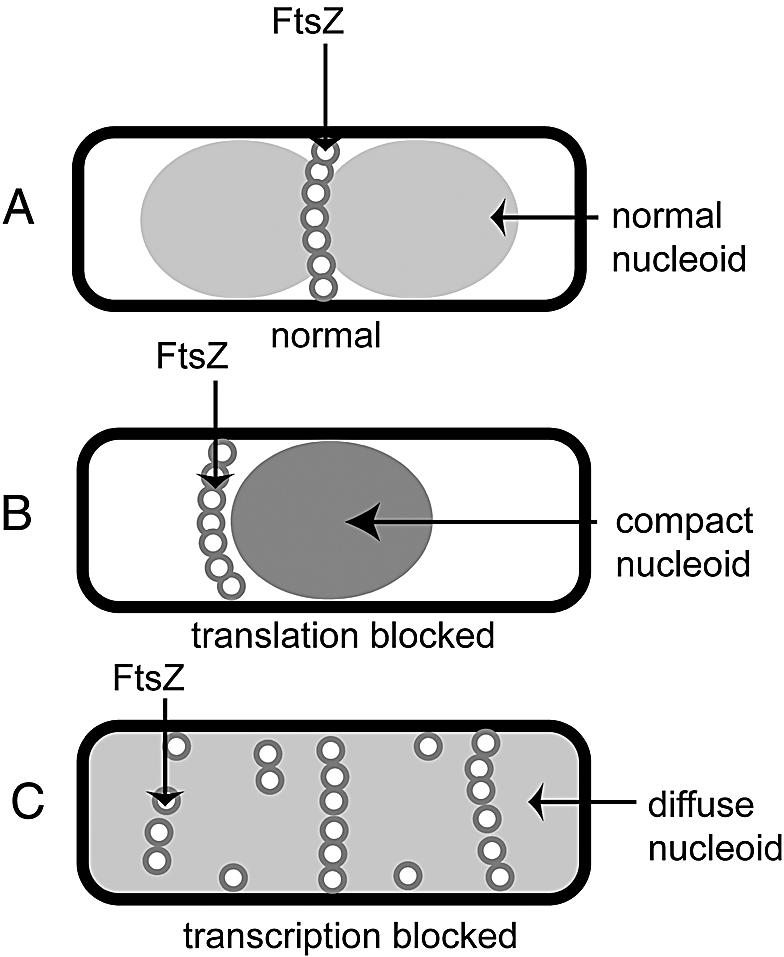
Summary diagram of the observed effects of transcription and translation inhibition on NO.
Acknowledgments
We thank Lynn Zechiedrich for the gyrB mutant strain, Alicia Dombroski for the rpoD800 strain, Jim Sawitzke for CC4207, and the E. coli Genetic Stock Center for the secA51 strain.
This work was supported by a grant (GM61074) from the National Institutes of Health.
REFERENCES
- 1.Britton, R. A., B. S. Powell, S. Dasgupta, Q. Sun, W. Margolin, J. R. Lupski, and D. L. Court. 1998. Cell cycle arrest in Era GTPase mutants: a potential growth rate regulated checkpoint in Escherichia coli. Mol. Microbiol. 27:739-750. [DOI] [PubMed] [Google Scholar]
- 2.Chen, C. R., M. Malik, M. Snyder, and K. Drlica. 1996. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J. Mol. Biol. 258:627-637. [DOI] [PubMed] [Google Scholar]
- 3.Collinson, I., C. Breyton, F. Duong, C. Tziatzios, D. Schubert, E. Or, T. Rapoport, and W. Kuhlbrandt. 2001. Projection structure and oligomeric properties of a bacterial core protein translocase. EMBO J. 20:2462-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbin, B. D., X.-C. Yu, and W. Margolin. 2002. Exploring intracellular space: function of the Min system in round-shaped Escherichia coli. EMBO J. 21:1988-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dasgupta, S., S. Maisnier-Patin, and K. Nordstrom. 2000. New genes with old modus operandi. The connection between supercoiling and partitioning of DNA in Escherichia coli. EMBO Rep. 1:323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine the proper placement of the division site in Escherichia coli. Cell 56:641-649. [DOI] [PubMed] [Google Scholar]
- 7.Den Blaauwen, T., N. Buddelmeijer, M. E. G. Aarsman, C. M. Hameete, and N. Nanninga. 1999. Timing of FtsZ assembly in Escherichia coli. J. Bacteriol. 181:5167-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiNardo, S., K. A. Voelkel, R. Sternglanz, A. E. Reynolds, and A. Wright. 1982. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell 31:43-51. [DOI] [PubMed] [Google Scholar]
- 9.Dworsky, P. 1975. Unfolding of the chromosome of Escherichia coli after treatment with rifampicin. Z. Allg. Mikrobiol. 15:243-247. [DOI] [PubMed] [Google Scholar]
- 10.Dworsky, P., and M. Schaechter. 1973. Effect of rifampin on the structure and membrane attachment of the nucleoid of Escherichia coli. J. Bacteriol. 116:1364-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filutowicz, M. 1980. Requirement of DNA gyrase for the initiation of chromosome replication in Escherichia coli K-12. Mol. Gen. Genet. 177:301-309. [DOI] [PubMed] [Google Scholar]
- 12.Fu, X., Y. L. Shih, Y. Zhang, and L. I. Rothfield. 2001. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl. Acad. Sci. USA 98:980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gellert, M., M. H. O'Dea, T. Itoh, and J. Tomizawa. 1976. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc. Natl. Acad. Sci. USA 73:4474-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross, C. A., A. D. Grossman, H. Liebke, W. Walter, and R. R. Burgess. 1984. Effects of the mutant sigma allele rpoD800 on the synthesis of specific macromolecular components of the Escherichia coli K12 cell. J. Mol. Biol. 172:283-300. [DOI] [PubMed] [Google Scholar]
- 15.Hale, C. A., H. Meinhardt, and P. A. de Boer. 2001. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 20:1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, Z., and J. Lutkenhaus. 2001. Topological regulation of cell division in E. coli. spatiotemporal oscillation of MinD requires stimulation of Its ATPase by MinE and phospholipid. Mol. Cell 7:1337-1343. [DOI] [PubMed] [Google Scholar]
- 17.Hu, Z., A. Mukherjee, S. Pichoff, and J. Lutkenhaus. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, K. C., Y. Meir, and N. S. Wingreen. 2003. Dynamic structures in Escherichia coli: spontaneous formation of MinE rings and MinD polar zones. Proc. Natl. Acad. Sci. USA 100:12724-12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imamoto, F. 1973. Diversity of regulation of genetic transcription. I. Effect of antibiotics which inhibit the process of translation on RNA metabolism in Escherichia coli. J. Mol. Biol. 74:113-136. [DOI] [PubMed] [Google Scholar]
- 20.Lin, C. G., O. Kovalsky, and L. Grossman. 1998. Transcription coupled nucleotide excision repair by isolated Escherichia coli membrane-associated nucleoids. Nucleic Acids Res. 26:1466-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margolin, W. 2001. Spatial regulation of cytokinesis in bacteria. Curr. Opin. Microbiol. 4:647-652. [DOI] [PubMed] [Google Scholar]
- 22.Migocki, M. D., M. K. Freeman, R. G. Wake, and E. J. Harry. 2002. The Min system is not required for precise placement of the midcell Z ring in Bacillus subtilis. EMBO Rep. 3:1163-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizuuchi, K., M. H. O'Dea, and M. Gellert. 1978. DNA gyrase: subunit structure and ATPase activity of the purified enzyme. Proc. Natl. Acad. Sci. USA 75:5960-5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moazed, D., and H. F. Noller. 1987. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie 69:879-884. [DOI] [PubMed] [Google Scholar]
- 25.Niki, H., A. Jaffé, R. Imamura, T. Ogura, and S. Hiraga. 1991. The new gene mukB codes for a 177-kD protein with coiled-coil domains involved in chromosome partitioning of Escherichia coli. EMBO J. 10:183-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norris, V. 1995. Hypothesis: chromosome separation in Escherichia coli involves autocatalytic gene expression, transertion and membrane-domain formation. Mol. Microbiol. 16:1051-1057. [DOI] [PubMed] [Google Scholar]
- 27.Oliver, D. B., and J. Beckwith. 1981. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell 25:765-772. [DOI] [PubMed] [Google Scholar]
- 28.Orr, E., N. F. Fairweather, I. B. Holland, and R. H. Pritchard. 1979. Isolation and characterisation of a strain carrying a conditional lethal mutation in the cou gene of Escherichia coli K12. Mol. Gen. Genet. 177:103-112. [DOI] [PubMed] [Google Scholar]
- 29.Pruss, G. J., S. H. Manes, and K. Drlica. 1982. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell 31:35-42. [DOI] [PubMed] [Google Scholar]
- 30.Raskin, D. M., and P. A. J. de Boer. 1999. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raskin, D. M., and P. A. de Boer. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawitzke, J. A., and S. Austin. 2000. Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc. Natl. Acad. Sci. USA 97:1671-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sternglanz, R., S. DiNardo, K. A. Voelkel, Y. Nishimura, Y. Hirota, K. Becherer, L. Zumstein, and J. C. Wang. 1981. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. Proc. Natl. Acad. Sci. USA 78:2747-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun, Q., and W. Margolin. 2001. Influence of the nucleoid on placement of FtsZ and MinE rings in Escherichia coli. J. Bacteriol. 183:1413-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun, Q., X.-C. Yu, and W. Margolin. 1998. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol. Microbiol. 29:491-504. [DOI] [PubMed] [Google Scholar]
- 36.Trueba, F. J. 1982. On the precision and accuracy achieved by Escherichia coli cells at fission about their middle. Arch. Microbiol. 131:55-59. [DOI] [PubMed] [Google Scholar]
- 37.Tsui, H.-C. T., G. Feng, and M. E. Winkler. 1997. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 179:7476-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Helvoort, J. M., P. G. Huls, N. O. Vischer, and C. L. Woldringh. 1998. Fused nucleoids resegregate faster than cell elongation in Escherichia coli pbpB(Ts) filaments after release from chloramphenicol inhibition. Microbiology 144:1309-1317. [DOI] [PubMed] [Google Scholar]
- 39.van Helvoort, J. M. L. M., J. Kool, and C. L. Woldringh. 1996. Chloramphenicol causes fusion of separated nucleoids in Escherichia coli K-12 cells and filaments. J. Bacteriol. 178:4289-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wehrli, W., F. Knusel, K. Schmid, and M. Staehelin. 1968. Interaction of rifamycin with bacterial RNA polymerase. Proc. Natl. Acad. Sci. USA 61:667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woldringh, C., E. Mulder, J. Valkenburg, F. Wientjes, A. Zaritsky, and N. Nanninga. 1990. Role of the nucleoid in the toporegulation of division. Res. Microbiol. 141:39-49. [DOI] [PubMed] [Google Scholar]
- 42.Woldringh, C. L. 2002. The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol. Microbiol. 45:17-29. [DOI] [PubMed] [Google Scholar]
- 43.Woldringh, C. L., P. R. Jensen, and H. V. Westerhoff. 1995. Structure and partitioning of bacterial DNA: determined by a balance of compaction and expansion forces? FEMS Microbiol. Lett. 131:235-242. [DOI] [PubMed] [Google Scholar]
- 44.Woldringh, C. L., E. Mulder, P. G. Huls, and N. Vischer. 1991. Toporegulation of bacterial division according to the nucleoid occlusion model. Res. Microbiol. 142:309-320. [DOI] [PubMed] [Google Scholar]
- 45.Yamanaka, K., T. Mitani, J. Feng, T. Ogura, H. Niki, and S. Hiraga. 1994. Two mutant alleles of mukB, a gene essential for chromosome partition in Escherichia coli. FEMS Microbiol. Lett. 123:27-31. [DOI] [PubMed] [Google Scholar]
- 46.Yu, X., Q. Sun, and W. Margolin. 2001. FtsZ rings in mukB mutants with or without the Min system. Biochimie 83:125-129. [DOI] [PubMed] [Google Scholar]
- 47.Yu, X.-C., and W. Margolin. 1999. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol. Microbiol. 32:315-326. [DOI] [PubMed] [Google Scholar]
- 48.Yura, T., and A. Ishihama. 1979. Genetics of bacterial RNA polymerases. Annu. Rev. Genet. 13:59-97. [DOI] [PubMed] [Google Scholar]