Abstract
Staphylococcal protein A (Spa) is an important virulence factor of Staphylococcus aureus. Transcription of the spa determinant occurs during the exponential growth phase and is repressed when the cells enter the postexponential growth phase. Regulation of spa expression has been found to be complicated, with regulation involving multiple factors, including Agr, SarA, SarS, SarT, Rot, and MgrA. Our understanding of how these factors work on the spa promoter to regulate spa expression is incomplete. To identify regulatory sites within the spa promoter, analysis of deletion derivatives of the promoter in host strains deficient in one or more of the regulatory factors was undertaken, and several critical features of spa regulation were revealed. The transcriptional start sites of spa were determined by primer extension. The spa promoter sequences were subcloned in front of a promoterless chloramphenicol acetyltransferase reporter gene. Various lengths of spa truncations with the same 3′ end were constructed, and the resultant plasmids were transduced into strains with different regulatory genetic backgrounds. Our results identified upstream promoter sequences necessary for Agr system regulation of spa expression. The cis elements for SarS activity, an activator of spa expression, and for SarA activity, a repressor of spa expression, were identified. The well-characterized SarA consensus sequence on the spa promoter was found to be insufficient for SarA repression of the spa promoter. Full repression required the presence of a second consensus site adjacent to the SarS binding site. Sequences directly upstream of the core promoter sequence were found to stimulate transcription.
Staphylococcus aureus is a significant human pathogen that causes a wide range of infections, including skin and wound infections, toxic shock syndrome, arthritis, endocarditis, osteomyelitis, and food poisoning. The bacterium produces in a regulated fashion a number of potential virulence factors, including a variety of exotoxins and cell surface-associated proteins (12, 20, 22, 30). One of the major surface proteins is staphylococcal protein A (Spa), which has been shown to comprise 7% of the cell wall (13). Spa binds the Fc fragment of immunoglobulins from several mammalian species and may be important in phagocytosis avoidance (11). Studies utilizing spa mutants have shown that protein A is an important virulence factor in a murine septic arthritis model and has a modest effect on virulence with subcutaneous infections in mice (32, 34).
Spa, like many of the surface matrix binding proteins of S. aureus, is expressed during the exponential phase of growth and then is transcriptionally down-regulated during the postexponential phase of growth. This process involves the Agr (accessory gene regulator) quorum-sensing global regulatory system of S. aureus (2, 21, 22). The Agr locus is comprised of two divergent transcriptional units, the first being a two-component regulatory system (agrBDCA). AgrB is a transmembrane protein that is responsible for the transport and processing of AgrD (44). AgrD is exported as a cyclic peptide, termed the autoinducing peptide (AIP) (21). When AIP accumulates to a threshold concentration, it binds to AgrC, the sensor of the two-component system. AIP binding induces autophosphorylation of AgrC, with the phosphate moiety subsequently transferred to AgrA (26). Activated AgrA is a transcription factor that stimulates transcription from both of the Agr locus promoters. The second Agr locus transcript is a 514-nucleotide RNA, called RNAIII. Although RNAIII encodes delta toxin, it is the RNA species itself that is the effector of the Agr system (20, 31). Accumulation of RNAIII results in the postexponential-phase activation of transcription of many exoprotein genes and inhibition of transcription of many cell wall-associated protein genes, such as spa.
Recent studies have shown that several additional regulatory factors are involved in the regulation of spa expression, such as SarA (staphylococcal accessory regulator), SarS (initially designated SarH1), Rot (repressor of toxins), SarT, and the ArlR-ArlS two-component system (1, 5-8, 10, 14, 37, 38, 42). SarA is a pleiotropic regulator for multiple genes (10). Gene chip analysis has shown that SarA represses transcription of a number of genes, including spa, and stimulates transcription of other genes, including the agr promoters (9, 12). Regulation by SarA is thought to be accomplished by both Agr-dependent and Agr-independent mechanisms (9). SarA was demonstrated to bind multiple gene promoters in vitro, and SarA binding sequences have been identified (10, 41). There is a SarA recognition sequence immediately upstream of the −35 promoter element sequence in the spa determinant (10). The mechanism(s) by which SarA differentially regulates promoter activity is unknown.
SarS, initially designated SarH1, is one of the Sar family of transcriptional regulators. The sarS determinant is located immediately upstream of spa and is a positive regulator of spa expression (8, 42). SarS was shown to be a DNA binding protein and, thus, may up-regulate spa expression by direct binding to the spa promoter. Another member of the Sar family of proteins, SarT, has been shown to be a positive regulator of SarS (38). SarT was shown to bind to the sarS promoter. Expression of sarT results in sarS, and subsequently spa, expression (38).
An additional member of the Sar family, Rot, was also shown to be a positive regulator of sarS expression and thus, indirectly, spa expression (37). A gene chip analysis indicated a 15.6-fold enhancement of spa transcription by Rot. It has not been determined if the activity of Rot on the sarS promoter is a direct or an indirect effect.
The ArlRS two-component regulatory system has been reported to affect spa expression (14). Inactivation of either the arlR or arlS determinant resulted in elevation of spa transcription. The arl mutations did not change spa transcription in an agrA or sarA mutant background, suggesting that the effect of the Arl proteins on spa expression is indirect, mediated through these global regulators (14). The mgrA-encoded protein has been shown to be responsible for the regulation of a number of genes in S. aureus, including acting as a repressor of protein A production (28). MgrA inactivation results in elevation of both protein A and alpha toxin, products whose genes are oppositely regulated by the Agr system.
A model has been proposed to explain the regulated expression of spa (38). In this model, the SarA protein binds to the spa promoter to repress transcription. Production of SarS activates transcription of spa. SarS production is indirectly regulated by the SarA and Agr systems. When the Agr system is activated, sarT transcription is repressed, with the reduction in SarT production leading to a reduction in SarS levels and consequently a loss of Spa production. The SarA protein also represses sarT transcription (38).
Although several factors have been reported to influence spa expression, it is unclear how these factors work on the spa promoter. Here we report our studies on characterization of the spa promoter and its regulation. The spa promoter has two transcriptional start sites and contains at least three upstream cis elements that are required for its regulation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The agr, sarA, and sarS mutant alleles were moved by transduction into KSI2054 to create the isogenic strains. Trypticase soy broth (Difco) and Trypticase soy agar (Difco) were used for culturing S. aureus. Luria-Bertani agar and broth were used to culture Escherichia coli. All incubations were performed at 37°C. Antibiotic concentrations used were as follows: tetracycline, 20 μg/ml; kanamycin, 20 μg/ml; chloramphenicol, 10 μg/ml; erythromycin, 5 μg/ml; ampicillin, 100 μg/ml; zeocin, 20 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK−) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Gibco-BRL |
| TOP10F′ | F′[lacIq Tn10 (Tetr)] mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| S. aureus strains | ||
| KSI2054 | 8325-4 agr+ | 45 |
| KSS5186 | Δagr rot::tetM | 43 |
| KSS5130 | agr+sarA::kan | This study |
| KSS5523 | Δagr sarA::kan | 43 |
| KSS5527 | agr+sarA::kan sarS::erm | This study |
| KSS5528 | Δagr sarA::kan sarS::erm | This study |
| KT201 | sarS::pKT200 (erm) | 42 |
| PM466 | Δagr | 29 |
| PM783 | rot::tetM | 29 |
| RN4220 | Accepts foreign DNA (r−) | 24 |
| UAMS-957 | cna sarA::kan | 4 |
| Plasmids | ||
| pJG2984 | spa −146 to +7 in pMH109 | This study |
| pJG2993 | spa −259 to +7 in pMH109 | This study |
| pJG3020 | spa −89 to +7 in pMH109 | This study |
| pJG3045 | spa −137 to +7 in pMH109 | This study |
| pJG3046 | spa −125 to +7 in pMH109 | This study |
| pJG3047 | spa −110 to +7 in pMH109 | This study |
| pJG3048 | spa −97 to +7 in pMH109 | This study |
| pJG3049 | spa −76 to +7 in pMH109 | This study |
| pJG3123 | spa −61 to +7 in pMH109 | This study |
| pJG3234 | spa −52 to +7 in pMH109 | This study |
| pJG3314 | spa −42 to +7 in pMH109 | This study |
| pJG3317 | spa −38 to +7 in pMH109 | This study |
| pJG3318 | spa −32 to +7 in pMH109 | This study |
| pJG3394 | spa −259 to +7 with −91 to −100 deletion in pMH109 | This study |
| pJG3480 | spa −259 to +7 with SarA box replaced by lacC sequences in pMH109 | This study |
| pJG3482 | spa −89 to +7 with SarA box replaced by lacC sequences in pMH109 | This study |
| pJG3483 | spa −110 to +7 with −91 to −100 deletion in pMH109 | This study |
| pMH109 | Shuttle expression plasmid | 19 |
Primer extension.
Total RNA was isolated from S. aureus cultures (A540 = 2.0) by the RNAzol B (Tel-Test, Inc.) procedure described elsewhere (16). Two primers were used. One primer, Spa1 (5′-CCTACACCTAGTTTACGAATTG-3′), is complementary to the spa structural gene (bases +42 to +63, based on the numbering scheme of Fig. 1). The other primer, cat3 (5′-GGTTATACTAAAAGTCGTTTGTTGGTTC-3′), is complementary to the cat open reading frame (ORF; 81 to 109 bp downstream of the SacI site) on the pMH109 vector. Each primer was labeled with [32P]ATP (Amersham Biosciences) with T4 polynucleotide kinase (Promega). Primer extension was performed using avian myeloblastosis virus reverse transcriptase (Primer Extension system; Promega). Briefly, 30 μg of RNA was coprecipitated with the 32P-end-labeled primer. The RNA-DNA pellet was resuspended in 7 μl of 250 mM KCl, heated to 94°C for 2 min, and then incubated at 53°C for 20 min. Avian myeloblastosis virus reverse transcriptase was then added to the reaction mixture and incubated at 42°C for 30 min. The reaction was stopped by addition of 20 μl of loading dye. A sequencing reaction using the same primer and the fmol DNA cycle sequencing system (Promega) was conducted simultaneously to provide the molecular size markers. The reaction mixtures were resolved on a 6% (7 M urea) polyacrylamide gel. After electrophoresis, the gels were transferred onto 3MM filters, dried, and subjected to radioautography.
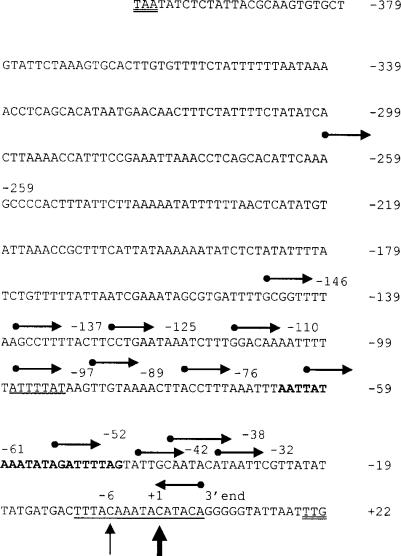
FIG. 1.
The spa promoter region. The filled circles of the rightward-pointing arrows indicate the start point of the subcloned promoter fragments (all clones end at +7). The fragments are identified by the 5′ end location of the fragments, which are listed to the right of the arrows. The spa start codon and the stop codon of the upstream sarS determinant are double underlined, the SarA binding sequence of Chien et al. (10) is in bold, and the range of fragments identified by Patel et al. (33) by an S1 protection assay is indicated by the underlining. The double wavy underlined sequence represents a SarA consensus binding site identified by SELEX (41). The upward-facing arrows indicate the start sites of transcription indicated by primer extension.
DNA manipulations.
S. aureus chromosomal DNA was isolated from 5-ml overnight cultures as follows. The cells were harvested by centrifugation (2,000 × g, 10 min), washed with 5 ml of TE buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA), and resuspended in 0.1 ml of TE. Fifty micrograms of lysostaphin was added, and the sample was incubated at 37°C for 30 min. Following this incubation, equal volumes of 1.6% Sarkosyl and 25 μg of proteinase K were added and the samples were incubated at 60°C for 60 min. The samples were then placed on ice, and the DNA was extracted with TE-saturated phenol followed by extraction with chloroform. The DNA samples were then precipitated with ethanol and resuspended in distilled water.
Plasmid DNA was isolated from E. coli by the procedure of Birnboim and Doly (3) and from S. aureus as described elsewhere (46).
Electroporation and transduction.
Introduction of plasmid DNA into S. aureus RN4220 by electroporation was conducted as described previously (23), except that a 0.1-cm electrode gap cuvette was utilized. Transduction of S. aureus strains was performed as described previously, utilizing phage 80α (36). Transformants and transductants were selected by growth at 37°C on Trypticase soy agar containing 20 μg of kanamycin/ml.
Cloning of the spa promoter and deletion derivatives.
The spa promoter (−259 to +7) and the 5′ series promoter deletion fragments were amplified by PCR (46). An XbaI recognition sequence was incorporated at the 5′ end of the upstream primers, and a SacI recognition sequence was incorporated into the downstream primers. The PCR products were cloned into PCR2.1 TOPO (Invitrogen) and then subcloned into the relatively low-copy-number promoter cloning shuttle vector pMH109 (19) following XbaI and SacI digestion. All constructs were sequenced to confirm they had the correct inserts. The plasmids were isolated from E. coli and electroporated into RN4220 (23). Plasmids in RN4220 were transduced into strains of various genetic backgrounds by transduction. Replacement mutagenesis was carried out to substitute an S. aureus lacC sequence internal to the ORF (5′-TTCGCTAAGCACAATCATACAT-3′ [35]) for the promoter-proximal SarA box sequence (5′-AATTATAAATATAGATTTTAGTA-3′ [10]).
Internal promoter deletion fragments were constructed with splicing by overlap extension (17, 18). PCR products were manipulated as described above.
CAT assays.
The activities of various spa promoter regions were measured with a chloramphenicol acetyltransferase (CAT) assay (40). The CAT assay was modified to a microformula suitable for an enzyme-linked immunosorbent assay plate reader (43). Overnight cultures were diluted to an A540 of 0.1 in Trypticase soy broth (for E. coli, Luria-Bertani broth and A600) and incubated until the A540 reached 2.0. Cells were harvested and washed once with WL buffer (25 mM Tris-HCl [pH 8], 25 mM EDTA). Pellets were resuspended in 1.0 ml of WL buffer, and 1.5 g of 0.1-mm glass beads was added. Cells were lysed using a Beadbeater 8 (Biospec Products) twice for 1 min each at 4°C. The lysed bacterial samples were centrifuged (2,500 × g, 10 min) at 4°C, and the supernatants were saved and stored at −80°C. Three microliters of the cell lysate was mixed with 37.5 μl of 10 mM 5,5′-dithiobis-2-nitrobenzoic acid (Sigma), 7.5 μl of 5 mM acetyl coenzyme A (Pharmacia), 206 μl of distilled deionized H2O, and 6.5 μl of 5 mM chloramphenicol (Sigma). Reactions were carried out in wells of a 96-well plate (Corning), and absorbance at 412 nm was determined with a Spectramax 190 microplate spectrophotometer (Molecular Devices). All strains were cultured in triplicate. The data analyses were done with Microsoft Excel. CAT activity was expressed as nanomoles of chloramphenicol acetylated per milligram of cells (dry weight) per minute at 37°C.
RESULTS
Determination of the spa transcriptional start sites.
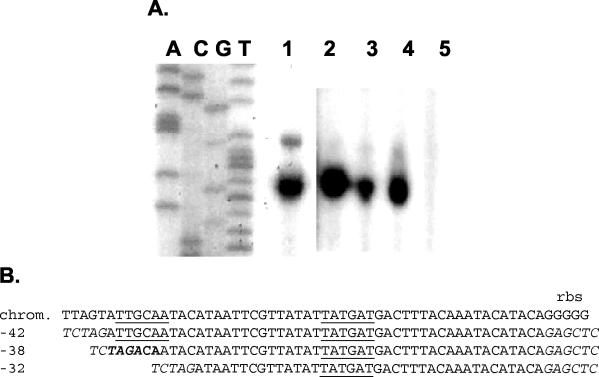
The transcriptional start site for spa was identified by an S1 nuclease assay described by Patel et al. (33). The assay yielded a series of DNA bands that corresponded to potential transcription start sites 13 to 28 bases upstream of the TTG initiation codon (Fig. 1). The multiple bands could have resulted from nibbling by S1 nuclease, although the data did not rule out the possibility of there being >1 actual transcription start sites. To more precisely define the start site of transcription, primer extension analysis was carried out. Utilizing the Spa1 primer internal to the spa ORF resulted in the appearance of two distinct bands, 6 nucleotides apart (Fig. 2A). The lower band was more intense than the upper band, and this position was designated +1. These two start sites correspond to nucleotides 19 and 25 bases upstream of the start codon, within the range of fragment sizes obtained by S1 mapping by Patel et al. (33). Examination of the sequence upstream of the two start sites revealed typical consensus −35 and −10 sequences, TTGCAA for −35 (−41 to −36) and TATGAT for −10 (−18 to −13), separated by 17 bp. To determine if these sequences constituted the spa promoter and if both transcripts arose from a single promoter element, primer extension was carried out using cloned promoter sequences containing the intact putative promoter (−42 to +7; pJG3314) and a construct lacking the first 3 nucleotides of the putative −35 element (−38 to +7; pJG3317). A positive control plasmid bearing the sequences from −259 to +7 (pJG2993) and a negative control containing an incomplete promoter (−32 to +7; pJG3318) were also tested (Fig. 2B). The DNA fragments were positioned upstream of a CAT reporter gene, and primer extension was carried out using RNA isolated from the plasmid-bearing cells (Fig. 2A). The plasmids bearing the intact putative promoter element (pJG3314 and pJG2993) resulted in the same start sites of transcription as identified with the spa ORF-specific primer, demonstrating that transcription from the plasmids faithfully reproduced the transcript pattern of the chromosomal protein A gene promoter. Deletion of the first three bases of the putative −35 element did not affect the production or location of the two transcripts. The DNA fragment with a deletion to the −32 position (pJG3318) lacked promoter activity, as expected.
FIG. 2.
Primer extension localization of the spa transcription start sites. (A) The Spa1 primer used was complementary to the 5′ end of the spa ORF, and RNA was isolated from an agr-negative mutant of S. aureus. The primer was used to generate the sequencing ladder size standards (lanes A, C, G, and T). Lane 1, primer extension with the Spa1 primer and RNA from strain PM466; lanes 2 to 5, primer extension with the cat3 primer and RNA isolated from strain PM466 bearing the −259 plasmid (lane 2), the −42 plasmid (lane 3), the −38 plasmid (lane 4), and the −32 plasmid (lane 5). The film for lanes 2 to 5 was positioned to align the bands with those of lane 1. (B) Sequence alignments of the core (−42) and truncated forms of the spa promoter used in the primer extension reactions. The sequences incorporated into the primers to provide recognition sequences for the restriction endonucleases XbaI and SacI are indicated in italics. The putative −35 and −10 promoter elements are underlined, and the putative −35 element contributed in part by the inclusion of the XbaI linker sequence in the −38 promoter construct is denoted in bold font.
The results indicated that critical features of the spa promoter, either as a −35 element or a cis element for a required transcriptional activator, lie between positions −42 and −32. It is possible that the putative −35 element is correct and that the deletion and introduction of the XbaI recognition sequence restored a functional −35 element, tcTAGACAaa, which would be an imperfect match to the consensus −35 element sequence and is shifted one nucleotide further upstream relative to the −10 element sequence.
Upstream region sequences of the spa promoter required for Agr-mediated regulation.
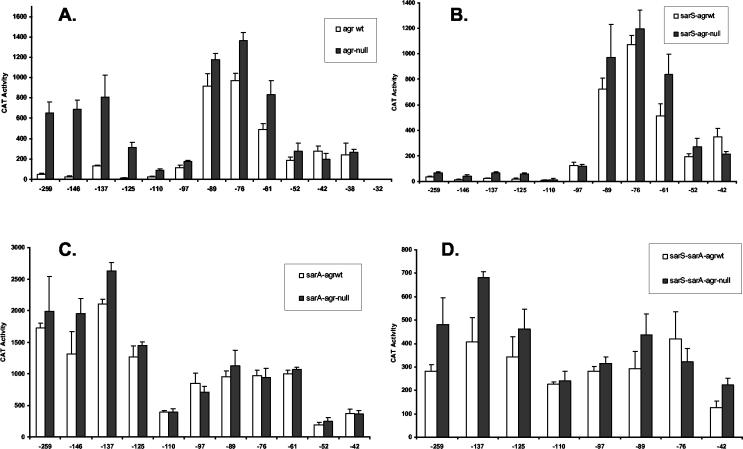
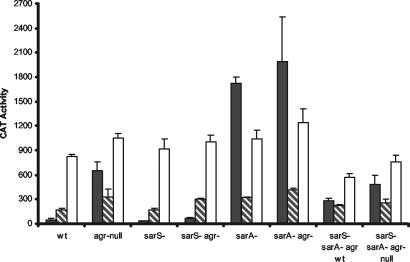
A series of nested fragments with a common downstream end (immediately upstream of the ribosome binding site) and variable upstream ends (indicated in Fig. 1) were cloned into the promoter cloning shuttle vector, pMH109, which bears a cat gene that lacks its own promoter but possesses a GGAGG ribosome binding site sequence. The reporter plasmids were transduced into agr-positive (KSI2054) and agr-null (PM466) strains. The promoter activity of each construct was then determined (Fig. 3A). Results are shown for samples taken at the postexponential phase of growth (A540 = 2), a time at which the Agr/Sar system represses spa transcription. With the largest promoter fragment (−259), there was approximately a 14-fold difference between the promoter activity from the agr-null host relative to that of the agr-positive host. This is consistent with the reported Agr effect on protein A production in 8325-derived strains (33) and indicates that this system accurately reflects the regulatory pattern of the spa promoter. The results of the deletion studies revealed the presence of three distinct regulatory sites within the spa promoter.
FIG. 3.
Effect of agr, SarS, and SarA on spa promoter activities. The x axes indicate the upstream boundary of the promoter fragments. Samples were from postexponential-phase culture lysates. CAT activity is expressed as nanomoles of chloramphenicol acetylated per minute per milligram of cells (dry weight). Values plotted are the means of at least three determinations with the standard deviations. Host strains were agr+ (white bars) and agr-negative (black bars) (A); agr+/sarS-negative (white bars) and Δagr/sarS-negative (black bars) (B); agr+/sarA-negative (white bars) and Δagr/sarA-negative (black bars) (C); and agr+/sarA-negative/sarS-negative (white bars) and Δagr/sarA-negative/sarS-negative (black bars) (D).
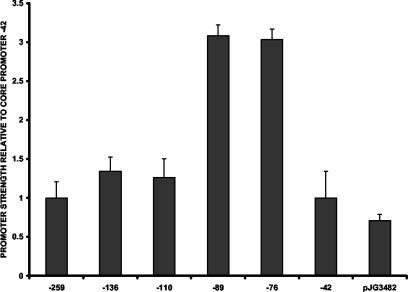
The minimal promoter element (clone −42) displays unregulated expression. The promoter strength does not change with extension to −52. However, extension of the upstream sequence to −61 resulted in an approximately twofold stimulation of the transcriptional activity, and inclusion of sequences to −76 gave an additional twofold increase. However, these clones were not regulated by the Agr system. The sequence between −52 and −76 is very AT rich and may act as a UP element to increase transcriptional efficiency rather than represent a cis element for a transcriptional factor (15). To provide an indication of the nature of the elevated transcription associated with the −76 construct, CAT assays were performed on E. coli strain DH5α cells bearing the spa promoter plasmids (Fig. 4). The spa core promoter was found to be active in E. coli, and extension of the upstream sequences to −76 resulted in an approximately threefold increase in activity. It is likely that the elevated transcription results from an UP element-like sequence, rather than from a specific transcription factor. It is improbable that E. coli and the evolutionarily distinct S. aureus would share such a transcriptional factor to up-regulate expression of this promoter.
FIG. 4.
spa promoter activity in E. coli, as shown by relative expression of the spa promoter fragments. The CAT activity of the −42 core promoter was set as 1.0, and the activities of the other promoter fragments were divided by the core promoter activity. The x axis gives the upstream boundary of the promoter fragments cloned into pMH109. The pJG3482 plasmid contains the spa −89 to +7 fragment, with the SarA box replaced by lacC sequences. Values plotted are the means of at least three determinations with the standard deviations.
Inclusion of the sequence to −97 resulted in a marked (eightfold) reduction in promoter activity in S. aureus, giving a promoter strength lower than that of the core promoter (Fig. 3A). This is suggestive of a cis element for binding a negative transcription factor. Because an additional degree of repression was observed with the −110 promoter fragment, the functional upstream boundary of this putative element is likely to be between −110 and −97.
The constructs with an upstream boundary greater than or equal to −110 had substantially higher CAT values in the agr mutant host relative to those in the agr+ host, reflecting the presence of Agr regulation. Specifically, the host ratios of CAT values of the constructs shorter than −110 were closer to 1, implying this region lacks an Agr-responsive cis element. Constructs longer than −110 gave rise to Agr−/Agr+ ratio values ranging from 10 to approximately 27, in contrast to the values of 0.7 to 1.8 for the constructs shorter than −110. The −110 construct, although displaying substantially reduced overall CAT values in both the agr mutant and in agr+ hosts, gave a ratio of approximately 4. Using a ratio of 4 as the cutoff value for determination of the presence of Agr regulation allowed us to conclude that the constructs from −110 and larger contain a cis element that is involved in Agr regulation. Full Agr system regulation became evident with the promoters that contained sequences to −125, but promoter strength with Agr regulation increased with the construct extending to −137. Therefore, the functional upstream boundary of this element lies between positions −137 and −125. With all of these spa promoter constructs, the promoter activity in the agr-positive host was less than that observed with the unregulated core promoter, suggesting that in a wild-type strain repression of promoter activity, and not simply loss of a positive regulator such as SarS, occurs.
The CAT value was reduced in the agr-deficient host with the −110 promoter fragment, relative to that in the larger constructs. This implies that the −110 construct lacks the transcriptional up-regulating element that is present on the longer constructs. The correlation between reduced promoter strength in agr-negative hosts and the appearance of an Agr effect implies that the high promoter activity with the constructs larger than the −110 promoter fragment is due to an Agr system-down-regulated transcriptional activator. The CAT values of the −110 and −97 constructs were lower than that of the core promoter construct (−42). This finding is inconsistent with a simple loss of a transcriptional activator binding site. The results suggest that the region upstream of −89 contains a regulatory element associated with reduced transcriptional activity. The evidence that this is a sequence-inherent effect, rather than representing the binding site for a negative transcriptional factor, is presented below.
SarS regulatory domain of the spa promoter.
The sarS determinant encodes a positive regulator of spa transcription (8, 42). The nested series of spa promoter fragments were introduced into sarS-negative agr+ and mutant agr isogenic strains of S. aureus and the promoter activities were determined from postexponential-phase cultures. The results are shown in Fig. 3B. Comparison of these results with the values presented in Fig. 3A with the sarS+ strains allows for a determination of the SarS element(s) within the spa promoter. With the agr+ host, there was minimal overall CAT activity with promoter fragments of −110 and greater, and there was no significant difference between the activities observed in the presence or absence of functional SarS in these postexponential-phase cultures. The results obtained with the mutant agr host strain indicated that the −110 clone was the shortest promoter fragment to be substantially affected by SarS. The −97 clone had a very modest reduction in activity, with 68% of the activity seen in the mutant agr/sarS+ host. The −110 and the larger clones in the mutant sarS hosts had activities that ranged from 6 to 18% of those seen when functional SarS protein was produced. In the absence of a functional SarS protein, the promoter activities obtained for the −97 and larger clones were less than the activity obtained with the core promoter, again indicative of active repression occurring in the sarS-negative strains. Furthermore, an Agr effect was still demonstrable in the sarS-negative strains. For example, there was a two- to threefold reduction in promoter strength with the promoter fragments −259 through −125 in the agr+/sarS-negative host relative to those in the agr-negative/sarS-negative host. Therefore, the Agr effect on spa transcription is not entirely mediated by the Agr system's effects on sarS expression, in agreement with the findings of Tegmark et al. (42).
SarS has been reported to be a DNA binding protein, although its binding site on the spa promoter has not been specifically identified (8, 25, 42). When the results of the study of the effect of Agr on the promoter constructs (Fig. 3A) and the effect of loss of SarS production (Fig. 3B) were compared, it was concluded that the upstream boundary for the fully functional SarS cis element is between −137 and −125. The smaller fragments (−97, −110, and −125) displayed reduced responses to SarS and consequently to the Agr system. The SarS protein was first identified through its binding to a fragment corresponding to −15 to −110 of the spa and other promoters (42). Our results are consistent with their finding that the −110 fragment would bind SarS. Electrophoretic mobility shift results suggest that multiple copies of SarS may be bound to the spa promoter, and this may be important for the proper function of this protein as a transcriptional regulator (8, 25).
The cis element required for SarA repression of spa expression.
SarA has been reported to be a negative regulator of spa transcription. DNase I footprinting revealed that SarA binding protected the sequence between −188 and −44, which included a 21-bp SarA box sequence 2 bp upstream of the −35 element of the spa promoter (10). Using a plasmid-based reporter system, it was determined that deletion of the SarA box sequence resulted in increased promoter activity (10). The SarA binding site identified by Cheung and coworkers is the −64 to −44 sequence shown in Fig. 1. Within this DNase I footprinting-protected sequence is the ATTTTAG (−50 to −44) imperfect SarA binding sequence (ATTTTAT) identified by Stebra et al. (41) using a SELEX procedure.
The nested series of spa promoter fragments was introduced into sarA-negative agr+ and agr-negative isogenic strains of S. aureus, and the promoter activities were determined from postexponential-phase cultures. The results are shown in Fig. 3C. Comparison of these results with the values presented in Fig. 3A with the sarA+ strains allows for a demonstration of SarA repression on the spa promoter. The previously identified SarA binding sequence is present within the −76 promoter fragment. However, this promoter fusion construct does not show a reduction in promoter expression, which would be expected if SarA were bound (Fig. 3A, compare the −76 and −61 activities). When the values of the promoter strengths in the sarA+ and sarA-negative hosts were compared, no SarA effects were observed until the upstream sequence was extended to −97. The activities of the −97 and larger clones were markedly elevated (i.e., approximately fourfold for the −97 clone) in the sarA-negative host (Fig. 3C; note that the graphs in panels C and A have different y-axis scales). The −97 promoter clone contains an ATTTTAT sequence (−97 to −91), which is a perfect match to the SarA consensus sequence identified by Stebra et al. (41) using the SELEX technique. These results suggest that this SarA site is functional. The promoter constructs from −97 and larger had elevated CAT values in sarA-negative hosts, regardless of agr status. The values were approximately 40-fold higher than those of the sarA+ agr+ host bearing the same constructs and 2- to 3-fold higher than those in the sarA+ agr-negative host. It should be noted, however, that the −110 promoter fusion construct has a lower promoter strength than the slightly larger and smaller constructs. The increased promoter strength of the larger constructs can be explained by SarS activity. The lower value relative to the smaller constructs is suggestive of an additional repression that is SarA independent. It is important to note that the promoter activity of the −110 and larger spa promoter fragments, relative to the activities of the −89 promoter fragment, were reduced in the E. coli hosts as well (Fig. 4). This again is suggestive of a sequence-inherent effect rather than a binding site for a transcriptional factor. This regulatory sequence lies between −89 and −110. Thus, two distinct mechanisms to reduce the activity of the spa promoter are found with the −97 and larger promoter fragments, one being SarA dependent while the other is SarA independent.
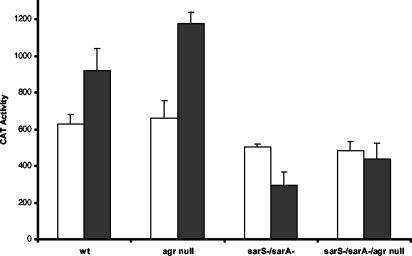
Although deletion of the promoter-proximal SarA box sequence gave rise to an increase in promoter strength (10), our studies where this sequence was included with the core promoter did not result in the expected reduction in promoter strength. Resolution of these conflicting results may involve the contribution of the upstream putative SarA box sequence. To evaluate the contributions of the upstream and downstream putative SarA boxes on SarA-mediated repression of spa expression, specific deletions of these elements were created. A 10-bp deletion was made on pJG2993 (the −259 promoter fragment) from −91 to −100, to produce pJG3394. A 10-bp deletion was utilized to remove one turn of the helix and thus minimize potential sidedness concerns. To determine if the −64-to-−44 sequence contributes to SarA-mediated repression of the spa promoter, the −259 promoter fragment was mutated such that the spa promoter sequence from −64 to −42, containing the promoter-proximal SarA binding sequence, was replaced with the same number of bases from a sequence internal to the S. aureus lacC ORF. This replacement approach was taken to avoid altering the spacing of elements within the spa promoter, and the lacC sequences are unlikely to contain regulatory sequences. These constructs, and the parental plasmid pJG2993, were introduced into the agr+ and agr-negative host strains. The CAT values were determined and are shown in Fig. 5. In the agr-positive wild-type host, removal of the promoter-proximal SarA box sequence gave rise to a substantial increase in promoter activity, whereas loss of the upstream element had a more modest stimulatory effect. In the agr-null host strain, the promoter activity of the mutant lacking the promoter-proximal element was elevated, but the activity of the wild-type promoter was actually higher than that of the mutant without the upstream element. Both mutants were impaired in Agr-mediated repression of transcription. The activities of the mutant promoters were independent of SarS, unlike the situation with the wild-type promoter. Similarly, the activities of the mutant promoters were only modestly influenced by the absence of SarA, with only slight increases in activity observed with the isogenic sarA-negative strains. Transcription of the wild-type promoter was markedly enhanced in the absence of SarA. With the sarA-negative/sarS-negative double mutant hosts, the mutant promoter fragments displayed a pattern of expression comparable to that of the wild-type promoter.
FIG. 5.
Effect of the putative SarA binding sites on spa promoter activity. Shown are the CAT activities promoted by the −259 promoter fragment (black bars), the −259 promoter fragment with the promoter-proximal SarA box (10) replaced by lacC sequences (white bars), and the −259 fragment with the upstream putative SarA binding site deleted (hatched bars). Host strains are indicated along the y axis. CAT activity is expressed as nanomoles of chloramphenicol acetylated per minute per milligram of cells (dry weight). Values plotted are the means of at least three determinations with the standard deviations.
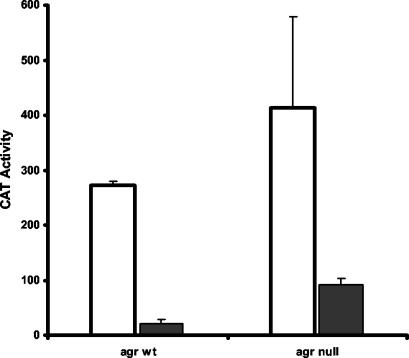
The promoter-proximal SarA box replacement mutation was introduced into the −89 promoter fragment to produce pJG3482. The −89 promoter lacks the putative upstream SarA box and was not affected by SarA (Fig. 3C). We examined the effect of loss of the remaining SarA box sequence on the activity of this promoter fragment. The results are shown in Fig. 6. Removal of the SarA box sequence actually resulted in a reduction of promoter activity in the agr+ and agr-negative hosts. Expression of these promoters in the sarS-negative/sarA-negative hosts reduced promoter activity with both the wild-type and mutant promoters, although the effect was smaller with the mutant promoter and the result was higher promoter activity for the mutant promoter in sarS-negative/sarA-negative/agr+ host cells.
FIG. 6.
Effect of the SarA binding site on spa promoter activity in the absence of the putative upstream site. Results shown are CAT activities promoted by the −89 promoter fragment (black bars) and the −89 promoter fragment with the promoter proximal SarA box (10) replaced by lacC sequences (white bars). Host strains are indicated along the y axis. CAT activity is expressed as nanomoles of chloramphenicol acetylated per minute per milligram of cells (dry weight). Values plotted are the means of at least three determinations with the standard deviations.
Extension of the upstream promoter sequences from −89 to −110 resulted in a substantial reduction in promoter activity (Fig. 3A). The −110 fragment contains the upstream SarA box consensus sequence, raising the possibility that SarA binding to this site is responsible for the decrease. However, although the activity of the −97 promoter was elevated in the sarA-deficient host (Fig. 3C), the activity remained much lower than that of the −89 promoter clone. To determine if the sequence containing the upstream SarA box were associated with the reduced transcriptional activity, perhaps in a SarA-independent fashion, the 10-bp deletion of the SarA box consensus sequence was introduced into the −110 promoter fragment. The resulting plasmid, pJG3483, was then introduced into the agr+ and agr-null strains, and promoter activity was measured (Fig. 7). Deletion of the SarA box sequence resulted in a four- to sixfold elevation of promoter activity, although the activity of the mutant was only about one-third of the activity seen with the −89 wild-type promoter fragment (compare Fig. 6 and 7). The −110 fragment, therefore, contains sequences responsible for reducing spa promoter strength that are independent of SarA-SarA box interactions. This sequence is able to exert its dampening effect on transcription in the heterologous host (E. coli) as well (Fig. 4).
FIG. 7.
Effect of the upstream SarA box sequence on expression of the −110 promoter fragment. Results shown are CAT activities promoted by the −110 promoter fragment (black bars) and the −110 promoter fragment with the −91 to −100 deletion (white bars). Host strains are indicated along the y axis. CAT activity is expressed as nanomoles of chloramphenicol acetylated per minute per milligram of cells (dry weight). Values plotted are the means of at least three determinations with the standard deviations.
The above results indicate that loss of either putative SarA binding site results in a loss of SarA responsiveness. The slight increase in promoter activity of pJG3480 in the sarA-negative host is consistent with a minimal SarA effect with only the upstream SarA box element present. The promoter strength remains high with the mutant promoter in a sarS-minus host, indicating that the requirement of SarS to promote spa transcription is lost when SarA binding is prevented. Interestingly, however, there was a modest decline in promoter activity in the sarA-negative/sarS-negative double mutant hosts. A slight, residual, Agr effect could still be observed with each of the mutant promoters, an activity that is independent of SarS for the upstream deletion mutant (pJG3394) but SarS dependent for the downstream replacement mutation promoter (pJG3480).
The lack of responsiveness to SarS with the SarA box deletion mutants suggests that the primary function of SarS in promoting transcription of spa is to relieve SarA-mediated repression, rather than stimulating transcription directly.
SarS-dependent and SarS-independent effects of SarA on spa expression.
SarA has been shown to affect the expression of spa directly, but it also has been shown to be a repressor of SarS expression (8, 42). Therefore, the elevated CAT values observed in the sarA-deficient host with the longer spa promoter constructs may have been an indirect result from derepression of sarS expression. In order to determine if the effects observed with the sarA-negative strain of S. aureus were due to a direct effect on the spa promoter or were due to an indirect effect resulting from elevated SarS levels, we introduced the deletions of the spa promoter region into the sarS-negative/sarA-negative/agr+ (KSS5527) and sarS-negative/sarA-negative/agr-negative (KSS5528) hosts. The resulting CAT values are shown in Fig. 3D. The promoter constructs longer than −110 produced an approximately three- to fourfold reduction in CAT values relative to those obtained in the isogenic sarS+ host (compare Fig. 3C and D). Thus, sarS derepression contributes substantially to the spa promoter activity in sarA-negative host cells. However, the constructs longer than the −110 still produced approximately fivefold higher values in cells lacking both SarA and SarS than from those hosts in which SarS was absent while SarA was present. The SarA-mediated repression of spa expression thus occurs through two pathways, SarS dependent and SarS independent.
An interesting finding was that the promoters containing the promoter activity enhancing sequences (putative UP element) upstream of the core promoter showed a reduced (approximately twofold) promoter strength in the sarS-negative/sarA-negative double mutant host cells relative to the values obtained with the single mutant host cells (compare the values for the −76 and −89 fragments in Fig. 3D with the corresponding values in B and C). This suggests that SarS can bind to this core promoter-proximal sequence and act directly as a transcriptional activator or better position the UP element.
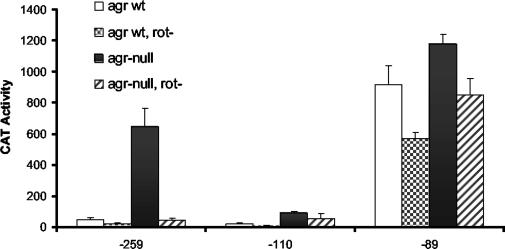
Rot is involved in up-regulation of spa expression.
The transcriptional regulator, Rot, has been reported to regulate spa expression, likely indirectly through control of SarS expression (37). We determined the effect of Rot on our various spa promoter constructs. The −259, −110, and −89 constructs were chosen as representatives. If Rot exerts its effect through SarS production, the expectation was that Rot would affect the transcription of the −110 and −259 promoters, because these constructs possessed the SarS binding site. The data (Fig. 8) were consistent with this expectation. The loss of Rot had no appreciable effect on the promoter activity observed with the −89 promoter fragment, but the activities of the −259 and −110 promoters were comparable to those observed with the sarS-deficient strains. These results are consistent with Rot acting indirectly on the spa promoter through its effects on SarS expression.
FIG. 8.
Effect of Rot on spa promoter activities. The x axis indicates the upstream boundary of the promoter fragments. Host strains were agr+ (white bars), agr+/rot-negative (stippled bars), agr-null (black bars), and agr-null/rot-negative (hatched bars). Samples were from postexponential-phase culture lysates. CAT activity is expressed as nanomoles of chloramphenicol acetylated per minute per milligram of cells (dry weight). Values plotted are the means of at least three determinations with the standard deviations.
DISCUSSION
Protein A is an important virulence determinant in S. aureus and is thought to be an important component of the immune evasion machinery of this pathogen. Protein A is expressed during the logarithmic phase of growth in vitro, and it is transcriptionally down-regulated as the cells progress into the postexponential phase of growth. A number of transcriptional regulatory systems impact expression of spa, including the Agr and ArlR-ArlS two-component systems, MgrA, SarA, SarS, and SarT. Only SarS and SarA have been shown to directly bind to the spa promoter (8, 10, 41, 42). In this study, we have more precisely defined the start site of transcription and characterized the regulatory elements within the spa promoter.
We have confirmed the SarA box findings of Chien et al. (10), but we have shown that this site is insufficient for complete repression of spa expression by SarA. A second, upstream SarA element (−97 to −91) is required for full repression. The SarS protein, a positive regulator of spa transcription, was initially shown to bind to a spa promoter-containing fragment extending from −110 to −15 (42). Our functional studies did reveal an effect on spa expression on the −110 promoter fragment, but full enhancement of spa transcription required additional upstream sequences to −137, indicating that the −110 fragment possesses an incomplete SarS cis element.
The requirement for the two SarA boxes in order to demonstrate SarS-mediated activation of transcription indicates that the primary role of SarS is to displace the SarA repressor complex from the promoter. The reduced spa promoter strength in the absence of both SarS and SarA argues for an additional role of SarS, namely, directly enhancing transcription from the spa promoter, perhaps through a direct interaction with RNA polymerase to facilitate transcription. The activity of SarS to remove SarA from the promoter and the capacity of SarA to suppress SarS production (8) make for a tightly controlled regulatory loop for spa expression.
There appear to be two sequence-inherent regulatory sites in the spa promoter. The first is a site whose upstream boundary is between −76 and −61 that increases promoter strength. The second site, whose upstream boundary is between −110 and −97, reduces promoter strength. Both elements possess the same activity in E. coli host cells. Interestingly, the promoter strength of the fragments possessing the former element displayed reduced activity when the host cells lacked both SarA and SarS, but not with either single mutant host. Binding of Sar proteins can alter the shape of the DNA, inducing bending or overwinding of the DNA (27, 39). The altered DNA topology may have a substantial impact on the functional positioning of the UP element.
The activity of the unregulated core promoter, relative to that of the larger constructs, indicated that the spa promoter is under constant regulation, regardless of whether protein A is being maximally produced during exponential growth or maximally repressed during the postexponential phase of growth. The existing model for growth phase regulation of the spa promoter suggests that the Agr system acts through a regulation of SarS expression. SarS production is thought to be regulated by the SarA and Agr systems. When the Agr system is activated, sarT transcription is repressed, leading to a reduction in SarT levels (38). SarT is a positive regulator of sarS transcription. As a consequence of the reduction in SarT activity when the Agr system is activated, SarS levels fall, which leads to a reduction in spa transcription. The SarA protein, which is constitutively expressed in S. aureus, is thought to affect spa transcription directly by binding to the spa promoter at the SarA box sequence and indirectly by acting as a repressor of sarT transcription (10, 38). The Rot protein is thought to affect spa transcription indirectly through its action as a stimulator of sarS expression (37).
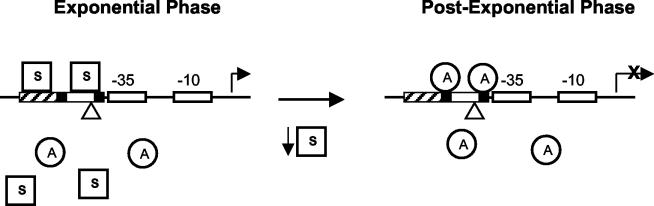
The work described herein provides for a modification of the spa regulatory model (Fig. 9). During the exponential phase of growth, the Agr system is off and SarT and Rot are active. These conditions lead to optimal expression of SarS, which then binds to the SarS binding site in the spa promoter. SarS-mediated activation of spa transcription is accomplished by preventing the binding of the SarA protein, a repressor of spa transcription. The requirement for SarA to occupy two binding sites for repression to occur fits this model. The upstream SarA box, adjacent to the SarS binding region, would be unavailable for SarA binding when SarS was present. SarS may bind in multiple copies to the spa promoter, further occluding the SarA binding sites (8). Multiple bound copies of SarS may then lead to either a direct interaction with RNA polymerase to further enhance transcription or a change in the curvature of the DNA, leading to a more active promoter.
FIG. 9.
Model for spa promoter regulation. During the exponential phase of growth when the protein A gene is maximally expressed, SarS (squares) is bound to its site (upstream boundary, approximately −137). More than one copy of SarS is likely bound to the promoter (8, 25), and one site of binding appears from this study to be immediately upstream of the core promoter. SarA (circles) is present in the cell (cytoplasmic proteins are depicted below the promoter diagram), but it is displaced from the spa promoter by SarS. As the cells enter the postexponential growth phase, activation of the Agr system results in a reduction in SarS production. This frees up the SarA binding sites. SarA binding to the two recognition sites forms the active repression complex. When the cells transition back into the exponential growth phase, SarS is produced and displaces SarA on the spa promoter to again form the transcriptionally active complex. The triangle represents the putative UP element sequence. The open boxes represent the spa −35 and −10 promoter elements (the core promoter), the black boxes denote the SarA box consensus sequences (−64 to −44 and −97 to −91), and the hatched box represents the SarS binding domain.
When the cells progress into the postexponential phase of growth, activation of the Agr system leads to a reduction in SarT levels and an inactivation of the Rot protein (29, 38). This results in a reduction in SarS production. As the SarS levels in the cell fall, SarA more successfully competes for binding to the spa promoter. When SarA occupies both binding sites, spa transcription is inhibited and protein A production declines. When favorable growth conditions return, exponential growth is accompanied by the reduction of the Agr RNAIII levels and the increase in SarS production. SarS displaces SarA, and transcription of spa ensues.
The use of an interplay between positive and negative regulatory proteins to control spa transcription allows for a finer control of the expression of this important protein. Despite what we have learned, gaps in our understanding of spa regulation still remain. It is not known how the ArlRS two-component regulatory system or MgrA affects protein A production. Furthermore, the residual Agr effect seen in SarS-negative hosts suggests an additional layer of regulation exists with this promoter.
Acknowledgments
We thank David George for technical assistance with the DNA sequencer, Staffan Arvidson for providing the sarS-negative strain KT201, Mark Smeltzer for the sarA-negative strain UAMS-979, and Peter McNamara for providing the agr deletion strain PM466 and the rot mutant PM783.
This work was supported by Public Health Services grant AI45778 from the National Institutes of Health.
Footnotes
Contribution number 04-257-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Bayer, M. G., J. H. Heinrichs, and A. L. Cheung. 1996. The molecular architecture of the sar locus in Staphylococcus aureus. J. Bacteriol. 178:4563-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benito, Y., F. A. Kolb, P. Romby, G. Lina, J. Etienne, and F. Vandenesch. 2000. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA 6:668-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung, A. L., Y.-T. Chien, and A. S. Bayer. 1999. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect. Immun. 67:1331-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y.-Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1-9. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, A. L., K. Eberhardt, and J. H. Henrichs. 1997. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect. Immun. 65:2243-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. L., K. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect. Immun. 69:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien, Y., and A. L. Cheung. 1998. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J. Biol. Chem. 273:2645-2652. [DOI] [PubMed] [Google Scholar]
- 10.Chien, Y., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 11.Dossett, J. H., G. Kronvall, R. C. Williams, Jr., and P. G. Quie. 1969. Antiphagocytic effects of staphylococcal protein A. J. Immunol. 103:1405-1410. [PubMed] [Google Scholar]
- 12.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcriptional profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsgren, A. 1969. Protein A from Staphylococcus aureus. Production of protein A by bacterial and L forms of S. aureus. Acta Pathol. Microbiol. Scand. 75:481-490. [PubMed] [Google Scholar]
- 14.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 15.Gourse, R. L., W. Ross, and T. Gaal. 2000. Ups and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 37:687-695. [DOI] [PubMed] [Google Scholar]
- 16.Hart, M. E., M. S. Smeltzer, and J. J. Iandolo. 1993. The extracellular protein regulator (xpr) affects exoprotein and agr mRNA levels in Staphylococcus aureus. J. Bacteriol. 175:7875-7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlapping extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 18.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 19.Hudson, M. C., and G. C. Stewart. 1986. Differential utilization of Staphylococcus aureus promoter sequences by Escherichia coli and Bacillus subtilis. Gene 48:93-100. [DOI] [PubMed] [Google Scholar]
- 20.Janzon, L., and S. Arvidson. 1990. The role of the δ-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 9:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji, G., R. C. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92:12055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornblum, J., B. N. Kreiswirth, S. J. Projan, H. Ross, and R. P. Novick. 1990. Agr: a polycistronic locus regulating exoproteins synthesis in Staphylococcus aureus, p. 370-402. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 23.Kraemer, G. R., and J. J. Iandolo. 1990. High-frequency transformation of Staphylococcus aureus by electroporation. Curr. Microbiol. 21:373-376. [Google Scholar]
- 24.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 25.Li, R., A. C. Manna, S. Dai, A. L. Cheung, and G. Zhang. 2003. Crystal structure of the SarS protein from Staphylococcus aureus. J. Bacteriol. 185:4219-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lina, G., S. Jarraud, G. Ji, T. Greenland, A. Pedraza, J. Etienne, R. P. Novick, and F. Vandenesch. 1998. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 28:655-662. [DOI] [PubMed] [Google Scholar]
- 27.Liu, Y., A. Manna, R. Li, W. E. Martin, R. C. Murphy, A. L. Cheung, and G. Zhang. 2001. Crystal structure of the SarR protein from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 98:6877-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luong, T. T., S. W. Newell, and C. Y. Lee. 2003. mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185:3703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNamara, P., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulatory of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182:3197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 31.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. L. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmqvist, N., T. Foster, A. Tarkowski, and E. Josefsson. 2002. Protein A is a virulence factor in Staphylococcus aureus arthritis and septic death. Microb. Pathol. 33:239-249. [DOI] [PubMed] [Google Scholar]
- 33.Patel, A. H., J. Kornblum, B. Kreiswirth, R. Novick, and T. J. Foster. 1992. Regulation of the protein A-encoding gene in Staphylococcus aureus. Gene 114:25-34. [DOI] [PubMed] [Google Scholar]
- 34.Patel, A. H., P. Nowlan, E. D. Weavers, and T. Foster. 1987. Virulence of protein A-deficient and alpha-toxin-deficient mutants of Staphylococcus aureus isolated by allele replacement. Infect. Immun. 55:3103-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosey, E. L., and G. C. Stewart. 1989. The nucleotide sequence of the lacC and lacD genes of Staphylococcus aureus. Nucleic Acids Res. 17:3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubin, S. J., and E. D. Rosenblum. 1971. Effects of recipient strain and ultraviolet irradiation on transduction kinetics of the penicillinase plasmid of Staphylococcus aureus. J. Bacteriol. 108:1192-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saïd-Salim, B., P. M. Duman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt, K. A., A. C. Manna, and A. L. Cheung. 2003. SarT influences sarS expression in Staphylococcus aureus. Infect. Immun. 71:5139-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schumacher, M. A., B. K. Hurlburt, and R. G. Brennan. 2001. Crystal structures of SarA, a pleiotropic regulator of virulence genes in S. aureus. Nature 409:215-219. [DOI] [PubMed] [Google Scholar]
- 40.Shaw, W. V. 1975. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 43:737-755. [DOI] [PubMed] [Google Scholar]
- 41.Stebra, K. M., S. G. Mackintosh, J. S. Blevins, B. K. Hurlburt, and M. S. Smeltzer. 2003. Characterization of Staphylococcus aureus SarA binding sites. J. Bacteriol. 185:4410-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398-409. [DOI] [PubMed] [Google Scholar]
- 43.Tseng, C.-W., S. Zhang, and G. C. Stewart. 2004. Accessory gene regulator (Agr) control of staphylococcal enterotoxin D gene expression. J. Bacteriol. 186:1793-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, L., L. Gray, R. P. Novick, and G. Ji. 2002. Transmembrane topology of AgrB, the protein involved in the post-translational modification of AgrD in Staphylococcus aureus. J. Biol. Chem. 277:34736-34742. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, S., J. J. Iandolo, and G. C. Stewart. 1998. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol. Lett. 168:227-233. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, S., and G. C. Stewart. 2000. Characterization of the promoter elements for the staphylococcal enterotoxin D gene. J. Bacteriol. 182:2321-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]