Abstract
Cytosine methylation is critical in mammalian development and plays a role in diverse biologic processes such as genomic imprinting, X chromosome inactivation, and silencing of repeat elements. Several factors regulate DNA methylation in early embryogenesis, but their precise role in the establishment of DNA methylation at a given site remains unclear. We have generated a comprehensive methylation map in fibroblasts derived from the murine DNA methylation mutant Hells−/− (helicase, lymphoid specific, also known as LSH). It has been previously shown that HELLS can influence de novo methylation of retroviral sequences and endogenous genes. Here, we describe that HELLS controls cytosine methylation in a nuclear compartment that is in part defined by lamin B1 attachment regions. Despite widespread loss of cytosine methylation at regulatory sequences, including promoter regions of protein-coding genes and noncoding RNA genes, overall relative transcript abundance levels in the absence of HELLS are similar to those in wild-type cells. A subset of promoter regions shows increases of the histone modification H3K27me3, suggesting redundancy of epigenetic silencing mechanisms. Furthermore, HELLS modulates CG methylation at all classes of repeat elements and is critical for repression of a subset of repeat elements. Overall, we provide a detailed analysis of gene expression changes in relation to DNA methylation alterations, which contributes to our understanding of the biological role of cytosine methylation.
Cytosine methylation represents a crucial epigenetic modification in mammalian cells. A family of conserved DNA methyltransferases (DNMTs) performs cytosine methylation at CpG sites (Ooi et al. 2010; Jones 2012). Deletion of DNMTs in mammals is lethal, indicating their critical role in development (Ooi et al. 2010). DNA methylation patterns are highly dynamic and undergo large transitions during germ cell development and during early embryonic development (Reik 2007; Lister et al. 2009, 2011; Smith et al. 2012). Distinct methylation patterns emerge during lineage differentiation and are associated with tissue-specific gene expression (Gifford et al. 2013; Xie et al. 2013). This suggests that cytosine methylation is associated with the regulation of gene transcription; however, the precise cause and effect of events during development remain unresolved.
Several factors influence DNA methylation, including the DNMTs themselves, the TET proteins responsible for demethylation through an oxidative pathway, the hemi-methyl DNA binding protein UHRF1 involved in maintenance of methylation at the replication fork, and the two homologs HELLS and DDM1. HELLS (helicase, lymphoid specific), also called LSH (lymphoid-specific helicase) or PASG (proliferation-associated SNF2-like), is found in Mus musculus, whereas DDM1 (deficiency in DNA methylation) is present in Arabidopsis thaliana. Both proteins are members of the SNF2 family and control DNA methylation (Jeddeloh et al. 1999; Dennis et al. 2001; Bostick et al. 2007).
Several members of the SNF2 family, including DDM1 itself, can perform chromatin remodeling in vitro. This improves access of DNA binding factors to their prospective DNA sequences (Brzeski and Jerzmanowski 2003; Ryan and Owen-Hughes 2011). Nucleosomes impose a hindrance for DNMTs, and the addition of chromatin remodeling factors can facilitate methylation of nucleosomal DNA in vitro (Felle et al. 2011). Hence, it is hypothesized that the chromatin remodeling activity of HELLS or DDM1 may promote access of DNMTs to chromatin targets in vivo. DDM1 mutants display a complex pattern of cytosine methylation changes (Stroud et al. 2013). Deletion of Hells in mice alters cytosine methylation at developmental genes, repeat sequences, and many promoter regions (Zhu et al. 2006; Xi et al. 2007, 2009; Myant et al. 2011; Tao et al. 2011). Hells−/− mice die perinatally and exhibit multiple tissue deficiencies including stem cell defects. This indicates that HELLS is critical for normal development (Geiman and Muegge 2000; Geiman et al. 2001; Sun et al. 2004; De La Fuente et al. 2006; Zeng et al. 2011).
Multiple pathways regulate DNA methylation, but it remains largely unknown how specific factors control methylation at a given site. Previous studies using Hells mutants examined specific genomic loci, such as immediate promoter regions or potential enhancers, and applied techniques that are biased for regions of low CG density and that have limited resolution (Xi et al. 2007, 2009; Myant et al. 2011; Tao et al. 2011; Yu et al. 2014). In addition, fibroblast cell lines were analyzed that may have developed aberrant methylation patterns due to prolonged culture in vitro (Myant et al. 2011; Tao et al. 2011). Here, we applied whole-genome bisulfite sequencing of primary murine fibroblasts derived from Hells−/− mutants and wild-type (WT) controls. We generated high-resolution maps for cytosine methylation and provided RNA-seq analysis and ChIP-seq for two histone modifications, H3K4me3 and H3K27me3, and for Pol II association. We describe DNA methylation patterns influenced by HELLS and evaluate the relationship between DNA methylation and gene expression.
Results
Global methylation of Hells−/− (KO) MEFs, WT MEFs, and ES cells
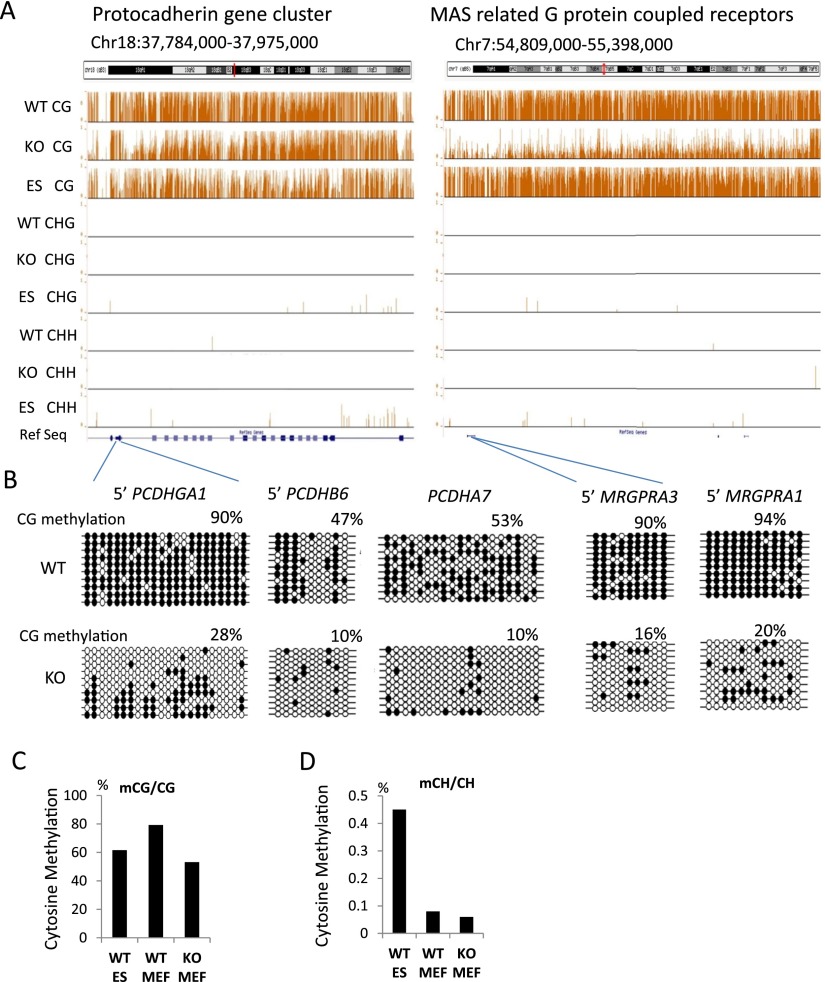
We derived genomic DNA from primary murine embryonic fibroblasts (MEFs) of day 13.5 embryos of Hells−/− (KO) or WT littermates (Geiman et al. 2001). We used an established wild-type murine embryonic stem cell line (ES) for comparison. ES cells are derived from the inner cell mass of blastocysts. Blastocysts display the lowest DNA methylation level during murine development and thus provide a reference point for somatic cells such as MEFs (Smith et al. 2012). DNA was subjected to bisulfite conversion followed by high-throughput sequencing. Using uniquely aligned reads, we obtained base-pair resolution maps of cytosine methylation for all autosomes (Lister et al. 2009). A genome browser snapshot unveils reduced cytosine methylation in the CG context in KO MEFs compared to WT MEFs and lower CG methylation in ES cells compared to WT MEFs (Fig. 1A). Using conventional bisulfite sequencing, we validated DNA methylation at 10 distinct genomic loci. These sites were chosen because they were located in large chromosomal areas with prominent CG hypomethylation. In addition, they contained gene families, such as cytochrome P450, MAS-related G protein–coupled receptor, the UDP glucuronosyltransferase 1 family, and the protocadherin family (seven sites are presented in Fig 1B; Supplemental Fig. S1). Mean methylation values of single cytosines in the CG context are 61%, 53%, and 80% for ES cells, KO MEFs, and WT MEFs, respectively (Fig. 1C). The methylation differences between samples are highly significant (Wilcoxon rank test P < 0.0001). The observation that KO MEFs display altered DNA methylation levels compared to WT MEFs is consistent with the findings of previous reports (Myant et al. 2011; Tao et al. 2011). These results indicate that HELLS is an important regulator of mammalian cytosine methylation. ES cells exhibit a low level of cytosine methylation (0.45%) in the CHG or CHH context (Fig. 1D), whereas WT MEFs and KO MEFs have extremely low amounts of non-CG methylation (0.08% and 0.06%, respectively). For this reason, only CG methylation patterns were analyzed further.
Figure 1.
Global CG methylation reduction. (A) Genome browser view presenting CG methylation values (methylated CG/CG representing methylated cytosine over the number of sequence reads covering the CG site) as a fraction of 1 (with 1 indicating 100% methylation at a given site) and CHH (methylated CHH/CHH) and CHG methylation (methylated CHG/CHG) comparing KO (Hells−/−) of primary murine embryonic fibroblasts (MEFs), wild-type (WT) MEFs, and WT embryonic stem cells (ES). (B) Bisulfite sequencing analysis of several loci comparing WT MEFs and KO MEFs to validate results of whole-genome bisulfite sequencing. Additional validations are shown in Supplemental Figure S1. The black circle represents a methylated cytosine; the open circle, an unmethylated cytosine. (C,D) Global mean CG methylation values (C) and CH methylation values (D) in KO MEFs, WT MEFs, and ES cells. First, the fraction of methylation was determined for a single cytosine in the CG or CH context (methylated cytosine/over the number of sequence reads covering the single cytosine site). Then, mean methylation values were computed based on all cytosines assessed in the genome (a value of 0.8 equals 80%). The differences of the mean methylation values are highly significant (Wilcoxon signed-rank test, P < 0.0001).
CG methylation reduction at protein-coding genes and noncoding RNA genes
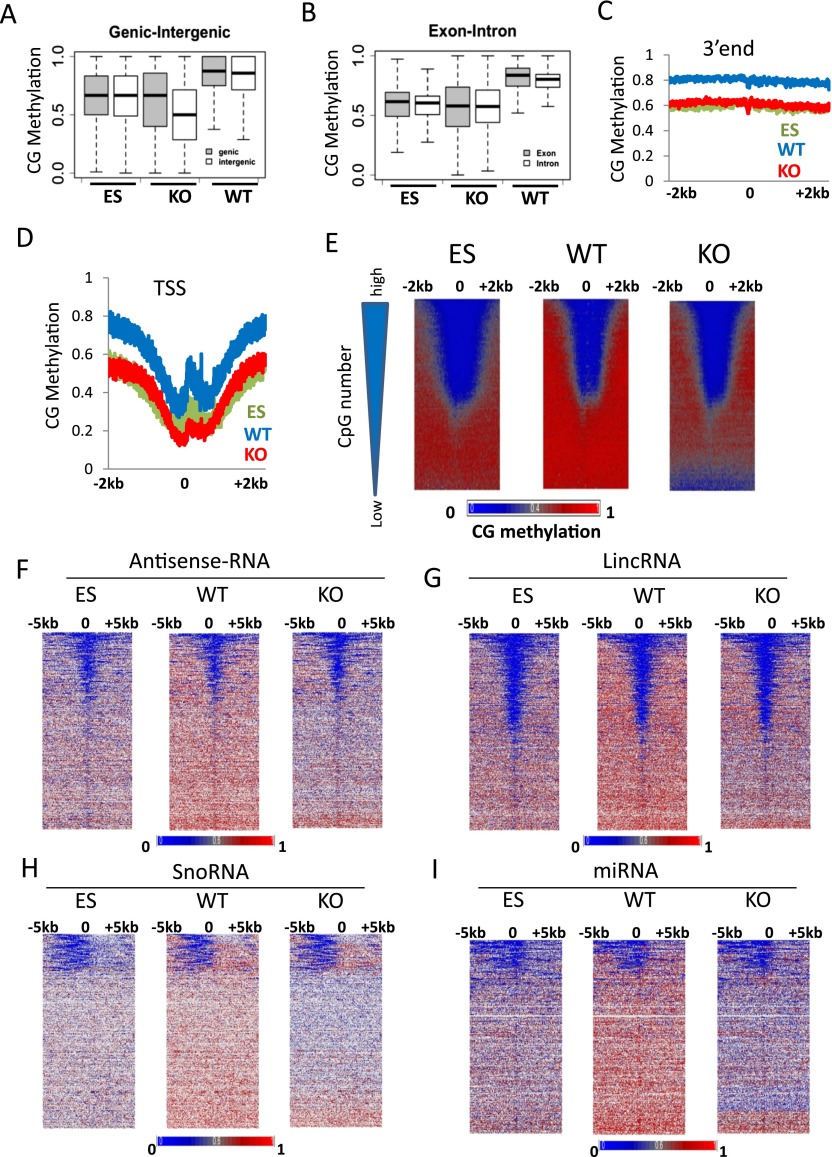
To assess CG methylation changes at distinct genomic regions, we computed mean methylation values at genic (protein-coding) and intergenic regions (Fig. 2A). CG methylation values are presented as a fraction of methylated CGs over the number of sequence reads (meCG/CG). KO MEFs display significantly reduced CG methylation at genic and intergenic regions compared to WT MEFs with a difference of 0.23 and 0.27, respectively (Wilcoxon rank test P < 0.0001). This indicates that Hells deletion has a slightly greater impact on intergenic sequences than on genic sequences. Likewise, CG methylation values at genic and intergenic regions are lower in ES cells than in WT MEFs. Furthermore, the differences in mean methylation values at exons (0.22) and introns (0.19) comparing WT MEFs and KO MEFs are significant (Wilcoxon rank test P < 0.0001) (Fig. 2B).
Figure 2.
CG methylation changes at protein-coding genes and noncoding RNA genes. (A) CG methylation values for genic (shaded) and intergenic (white) regions. First, the fraction of methylation was determined for a single cytosine in the CG context (methylated cytosine/over the number of sequence reads covering the single cytosine site). Then mean methylation values were computed for all cytosines located within a gene (shaded) or within a region between two genes (white). The plot represents the distribution of methylation values for single genes (shaded) and single intergenic regions (white). The differences of the average methylation values comparing WT and KO genic and intergenic regions are highly significant (Wilcoxon signed-rank test, P < 0.0001). (B) CG methylation values for exons (shaded) and introns (white) were assessed as in A. The differences of the mean methylation values comparing WT and KO genic and intergenic regions are highly significant (Wilcoxon signed-rank test, P < 0.0001). (C,D) Average distribution of CG methylation values at transcriptional termination sites (3′end; C) and over transcriptional start sites (TSSs; D) of protein-coding genes (n = 23,350) and 2 kb of flanking regions. (E) CG methylation at promoters of protein-coding genes. Genes (n = 23,350) are ranked based on CpG numbers (triangle at the left of the panel). Promoters with the lowest CpG density are at the bottom (low) and with the highest CpG number at the top (high). Heatmap presentation of CG methylation profiles in KO MEFs, WT MEFs, and ES cells. CG methylation distribution is shown at 1-bp resolution around TSSs and 2 kb of upstream and downstream flanking sequences (horizontal axis). Genes are arranged into 100 groups, and mean values computed (vertical axis). (F–I) Heatmap of CG methylation distribution at 1-bp resolution around transcription start sites of noncoding RNA genes comprising a region 5 kb upstream and 5 kb downstream, including antisense-RNA (F), and lincRNA (long intergenic noncoding RNA; G), and snoRNA (small nucleolar RNA; H), and miRNA (micro RNA; I). Genes are ranked based on mean CG methylation values in WT MEFs.
To analyze promoter regions in KO and WT MEFs, we averaged CG methylation levels around transcriptional start sites (TSSs) and the 3′ends of all protein-coding genes (Fig. 2C,D). A dip in CG methylation around TSSs is present in all three cell types (Fig. 2D). ES cells and KO MEFs display lower CG methylation levels around TSSs and flanking regions than do WT MEFs (Fig. 2D). Likewise, CG methylation is lower at the 3′end of protein-coding genes in KO MEFs and ES cells than in WT MEFs (Fig. 2C). Heatmap presentation of CG methylation profiles around TSSs reveals that the majority of genes have lower CG methylation levels in KO MEFs and ES cells compared to WT MEFs (Fig. 2E). Moreover, the greatest loss of CG methylation in KO MEFs is found at promoter regions with low CpG density (Fig. 2E).
To identify other genomic sequences that are influenced by HELLS, we analyzed CG methylation at noncoding RNA genes (Flicek et al. 2014). Noncoding RNA is not translated into proteins and plays multiple biological roles. Among the diverse classes of noncoding RNA are small RNA molecules (∼22 bp) known as microRNA (miRNA) which are involved in transcriptional and post-transcriptional gene regulation; small nucleolar RNA (snoRNA), playing a role in ribosome synthesis; long intergenic noncoding RNA (lincRNA) (>200 bp); and antisense RNA (asRNA). The latter RNA is complementary to mRNA and is thought to play a regulatory role in transcription. The transcriptional start sites (TSSs) of asRNA, lincRNA, and miRNA show depletion of CG methylation which is reminiscent of the profiles generated for protein-coding genes (Fig. 2F,G,I). Notably, snoRNA has a distinct CG methylation profile with reduced methylation upstream of the reported TSS, suggesting a distinct promoter organization for snoRNA (Fig. 2H). Strikingly, KO MEFs and ES cells displayed a similar distribution compared to WT MEFs, albeit at lower CG methylation levels. We noticed that a subset of miRNA loci retains DNA methylation in KO MEFs (Fig. 2I, bottom). These include several miRNAs playing a role in neuronal, hematopoietic or chondrocyte differentiation or controlling adipocyte size (mir-370, mir-181, mir-140, mir-103, respectively) (Chen et al. 2004; Fiore et al. 2009; Trajkovski et al. 2011; Karlsen et al. 2014). The persistence of DNA methylation in KO MEFs may indicate that their expression is not affected in KO MEFs.
Altogether, Hells deletion leads to widespread loss of CG methylation at uniquely mapped genomic regions. The regions that are affected by Hells deletion include TSSs of protein-coding genes and noncoding RNA genes.
Chromosomal domains are affected by Hells deletion
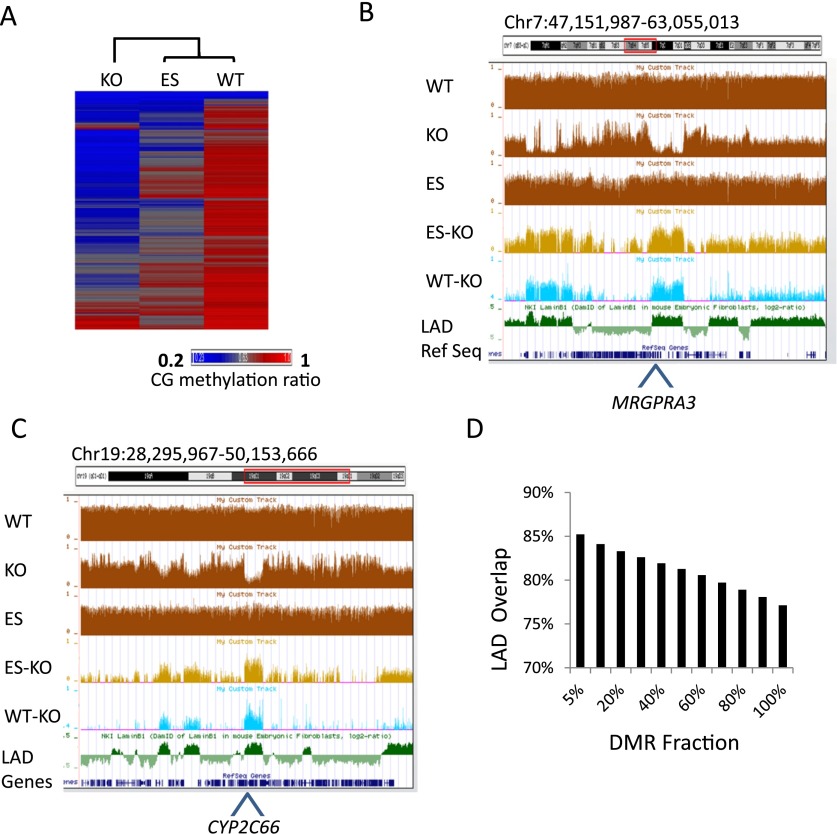
Since Hells deletion affects DNA methylation at diverse genomic features, we sought to characterize the regions with the greatest methylation difference between WT and KO MEFs. We computed mean CG methylation values at 5-kb tiles across the genome. Hierarchical clustering reveals a closer relationship between WT MEFs and ES cells than between WT and KO MEFs or between ES cells and KO MEFs (Fig. 3A). WT MEFs and ES cells display a higher correlation for CG methylation (R = 0.70) than KO MEFs and WT MEFs or KO MEFs and ES cells (R = 0.58 and R = 0.43, respectively). This indicates that CG methylation is low and greatly perturbed in KO MEFs compared to wild-type cells.
Figure 3.
Chromosomal domains with reduced CG methylation in KO MEFs. (A) Heatmap of CG methylation levels at 5-kb tiles for chromosome 1 of KO MEFs, WT MEFs, and ES cells. The rows were sorted by complete linkage hierarchical clustering with Euclidian distance as a distance measure. (B,C) Genome browser views illustrating CG methylation (mean values of 5-kb tiles) at the Cyp2 (B) and Mrgpra (C) gene clusters, comparing WT MEFs, KO MEFs, and ES cells (brown). The lower panels display differentially methylated regions (DMRs) at 5-kb tiles of ES minus KO (light brown) and WT minus KO (difference >0.4; light blue) compared to lamin B1 (LMNB1) attachment regions (green). (D) Bar graph displaying percent overlap of differentially methylated regions (DMR) with lamin B1 (LMNB1) attachment regions (P < 0.0001). The DMR is ranked based on the degree of CG methylation difference with the top 5% displaying the greatest CG methylation difference between the WT and KO MEFs.
The genome browser view reveals large chromosomal domains with reduced CG methylation in KO MEFs (Fig. 3B,C). These domains comprise gene families including olfactory receptor genes, pheromone receptor, cytochrome P450, MAS-related G protein coupled receptor, and UDP glucuronosyltransferase 1 family (Fig. 1A,B; Supplemental Fig. S1). To estimate the extent of CG methylation difference, we defined differentially methylated regions (DMRs) based on CG methylation differences between WT MEFs and KO MEFs greater than 0.4 (using 5-kb windows). About 26% of genome-wide uniquely mapped sequences are classified as DMRs.
The extent of methylation changes in KO MEFs, the diversity of genomic loci, and the fact that large chromosomal domains are affected suggest that HELLS targets a nuclear compartment rather than specific genomic sequences. This prompted a search for overlap of HELLS affected regions with known chromosomal domains. Several characteristics have been used to divide the genome into entities, for example, active and repressed compartments A and B (Zhang et al. 2012), topologically associated domains (TADs) (Dixon et al. 2012), and lamin B1 (LMNB1) attachment domains (LADs) (Peric-Hupkes et al. 2010). The lamin B1 protein belongs to the nuclear matrix and is located close to the inner nuclear membrane. High-resolution maps of LADs have been generated based on close interactions of lamin B1 protein and DNA sequences (Guelen et al. 2008; Peric-Hupkes et al. 2010). The murine genome comprises about 1200 domains ranging from 50 kb to 10 Mb. Visualization in the UCSC Genome Browser revealed a consistent overlap of DMRs with LADs (Fig. 3B,C), including Mrgpra1, Mrgpra3, Cyp2c66, and Ugt2a1 genes. We confirmed CG methylation differences at several loci located within LADs using conventional bisulfite sequencing (Fig. 3B,C; Supplemental Fig. S1). For further evaluation, we computed the overlap between DMRs and LADs. While the entire DMR overlapped 77% with LADs, the overlap steadily increases with the degree of differential methylation (P < 0.0001; Methods) (Fig. 3D). Thus the top 5% DMRs representing ∼1% of the genome have an 85% overlap with LADs (P < 0.0001), indicating a link between HELLS-dependent CG methylation changes and LADs.
These results indicate that HELLS is important for the establishment of CG methylation at a nuclear compartment that is in part defined by lamin B1 attachment regions.
Gene expression and CG hypomethylation
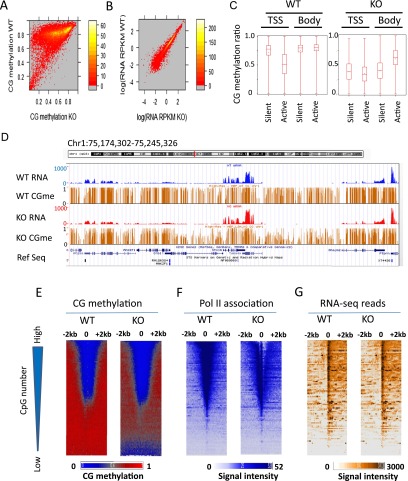
To examine the relationship between CG methylation and gene expression, we computed mean CG methylation levels at promoter regions of protein-coding genes. About 60% of promoters display a difference of CG methylation that is greater than 0.3 (WT CG methylation minus KO CG methylation). This indicates that Hells deletion has widespread effects on promoter regions (Fig. 4A). RNA-seq analysis detects similar relative transcript steady-state levels in WT and KO samples. The Pearson correlation value of RPKM values is R = 0.97 (Fig. 4B and biological replicates in Supplemental Fig. S2). The same result is obtained when protein-coding genes are grouped into CG-rich promoter regions (Supplemental Fig. S3A,B). A snapshot of the UCSC Genome Browser exhibits examples of similar relative transcript abundance levels at high-, intermediate-, and low-expressing genes (Fig. 4D). There is no evident correlation between CG methylation changes and alterations in relative RNA steady-state levels (Supplemental Fig. S3C). Hence, despite dramatic CG methylation losses at promoter regions, relative transcript steady-state levels at most low-expression genes remain unchanged and silent regions are not de-repressed.
Figure 4.
Relationship between relative transcript abundance levels and Pol II at CG hypomethylated promoters in KO MEFs. (A) Heatmap scatterplot analysis presenting the mean values of promoter CG methylation (2 kb upstream of and downstream from the TSS) in WT MEFs and KO MEFs. (B) Heatmap scatterplot analysis representing relative transcript abundance level as read coverage (RPKM values) of RNA-seq analysis comparing WT and KO MEFs. Results of biological replicates are shown in Supplemental Figure S2. (C) Box plot representing mean CG methylation values at gene bodies or TSSs (2 kb upstream of and 2 kb downstream from the TSSs) classified in groups of genes with transcripts (active) and without any detectable transcripts (silent) (Wilcoxon rank test, P < 0.0001). (D) Genome browser view illustrating CG methylation (brown) and transcript abundance level (RPKM values) at representative genes comparing WT (blue) and KO MEFs (red). (E–G) Promoters of protein-coding genes (n = 23,350) are ranked based on the presence of CpG numbers (represented by the triangle to the left). (Bottom) Genes with low CpG density; (top) genes with high CpG density. Heatmap presentation for CG methylation values at 1-bp resolution (E), Pol II ChIP signal intensities at 5-bp resolution (F), and RNA-seq signal intensities at 5-bp resolution (G) around the TSSs and 2 kb of flanking regions for WT MEFs and KO MEFs. Genes are arranged into 100 groups and mean values were computed for each genomic position.
Only 1% of genes (n = 243) are down-regulated in KO MEFs and 1.1% (n = 262) of genes are up-regulated (Supplemental Fig. S4). Using gene ontology analysis, we found that genes that are up-regulated in KO MEFs are enriched in cell adhesion and the defense response. Genes that are down-regulated are involved in the inflammatory response, cell adhesion, chemotaxis, and leukocyte differentiation (Supplemental Table S1). This indicates that primarily genes participating in host defense are deregulated in Hells-deleted MEFs.
To examine whether the extent of methylation changes is linked to gene activity, protein-coding genes were grouped into active and silent genes based on the presence of transcript abundance levels in WT MEFs. CG methylation is significantly reduced at TSS and the gene body comparing WT and KO MEFs (Wilcoxon rank test P < 0.0001) (Fig. 4C). The differences between WT and KO methylation values are greater at silent genes than at active genes (Student’s t-test P < 0.0001). These results suggest the possibility that distinct methylation mechanisms are present at active versus repressed genes. Gene body methylation is higher at active genes than at silent genes in KO MEFs (Fig. 4C). Previous studies described specific interaction of DNMT3A with H3K36me3, a histone mark preferentially found at gene bodies, and reported that DNMTs are enriched at gene bodies, suggesting the presence of distinct DNA methylation pathways at gene bodies (Dhayalan et al. 2010; Jin et al. 2012).
Finally, 14 lincRNA genes were selected that displayed CG methylation reduction in KO MEFs and that are involved in transcriptional regulation (Guttman et al. 2011). Real-time PCR analysis did not reveal any significant transcript level changes (Supplemental Fig. S5), giving no evidence that CG hypomethylation results in de-repression of this class of noncoding genes.
Pol II association at promoter regions in KO MEFs
To determine the relationship between RNA Pol II binding and CG methylation reduction in KO MEFs, Pol II ChIP-seq was performed. Promoters of protein-coding genes were ranked according to CpG numbers (Fig. 4E–G). Promoters with the greatest loss of CG methylation show low CG density. Those CG poor promoters are highly methylated in WT MEFs (Fig. 4E) and show low Pol II association (Fig. 4F) and low RNA-seq read numbers (Fig. 4G). This is consistent with the notion that HELLS guards preferentially repressed chromatin or heterochromatin, containing CG poor genes with low expression level. Pol II association is inversely related to the number of CpG sites within the promoter regions (Fig. 4F). There is no increase of Pol II association at promoter regions with reduced CG methylation. Instead, KO MEFs display slightly lower Pol II occupancy. Despite a decrease in Pol II engagement at the TSS regions in KO MEFs, relative transcript abundance levels as measured by RNA-seq reads are very similar in WT and KO MEFs. (Fig. 4G). Since we used an RNA Pol II antibody directed against the N terminus of the protein (POLR2A), we cannot discern the phosphorylation states at the C terminus. Therefore, it is possible that WT MEFs have a higher proportion of unphosphorylated Pol II (part of a preinitiation complex) or stalled Pol II compared to KO MEFs (Sims et al. 2004; Nechaev and Adelman 2008). The distinct phosphorylation states and activity of Pol II can be only determined by additional analysis.
In short, there is no evidence of increased Pol II occupancy at promoters with CG hypomethylation in KO MEFs.
Perturbation of histone modification H3K27me3 in KO MEFs
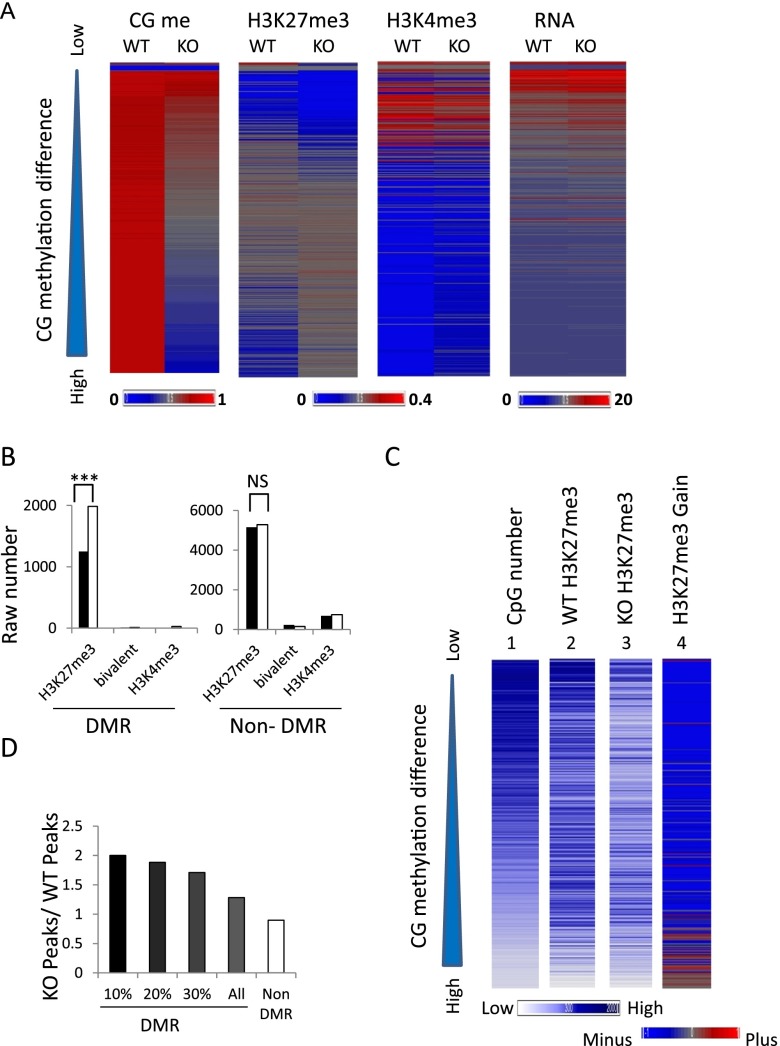
To explore other molecular mechanisms that may preserve repression at CG hypomethylated promoters, we examined H3K27me3 and H3K4me3 modifications by ChIP-seq. Genome-wide computation of histone modifications at 5-kb tiles shows no global increase of H3K27me3 level in KO MEFs. When we ranked 5-kb tiles based on CG methylation differences between WT and KO, we observed alterations in H3K27me3 distribution, while the pattern of H3K4me3 modification is unremarkable (Fig. 5A). Regions with moderate CG methylation reduction exhibit concomitant decreases in H3K27me3. These regions include immune response genes, genes involved in cell adhesion, and Hox genes; the latter had been previously reported to display reduced Polycomb association and CG methylation (Xi et al. 2007). On the other hand, regions with the greatest decrease of CG methylation are enriched for H3K27me3 in KO MEFs (Fig. 5A and three independent biological replicates in Supplemental Fig. S6). We considered the possibility that CG methylation reduction may alter accessibility to the H3K27me3 antibody. However, for a dozen regions that were validated for an H3K27me3 increase using conventional ChIPs followed by real-time PCR analysis, we did not find a change in H3 occupancy using a pan-H3 antibody (Supplemental Fig. S7). These results suggest an actual increase in the H3K27me3 mark at those loci.
Figure 5.
Increased H3K27me3 at CG hypomethylated regions. (A) Heatmap of CG methylation, H3K27me3, H3K4me3, and RNA-seq signal intensities at 5-kb tiles for chromosome 1 comparing WT and KO MEFs. The tiles (n = 38,840) are ranked based on CG methylation differences (WT values minus KO values). (Left) The ranking is presented from low (top) to high (bottom) (triangle). Biological replicates of H3K27me3 signals are presented in Supplemental Figure S6. (B) Bar graph presenting the raw number of 5-kb tiles with H3K27me3 modifications, H3K4me3, or bivalent modification in WT (black) and KO (white) MEFs in differential methylated regions (DMRs) and nondifferential methylated regions (non-DMR) on chr1. (***) The exact Pearson χ2 tests were applied. P < 0.001. (C) Heatmap presentation of mean signal intensities at promoter regions (2 kb upstream of and 2 kb downstream from TSSs) of protein-coding genes (n = 23,350). Genes are ranked based on CG methylation differences (WT minus KO MEFs) from low to high (top to bottom) (triangle). (1) Number of CpG sites located within promoter regions, (2) H3K27me3 signal intensities in WT MEFs, (3) H3K27me3 signal intensities in KO MEFs, and (4) difference in H3K27me3 modifications (KO values minus WT values). (Bottom left) The blue color scale represents signal intensities for panels 1, 2, and 3. (Bottom right) The blue/red color scale represents the difference of H3K27me3 values (KO values minus WT values) in panel 4. (D) Bar graph presenting the ratio of promoters marked by H3K27me3 in KO MEFs over the number of promoters marked by H3K27me3 in WT MEFs. The promoters have been grouped based on their CG methylation levels and fall into DMRs (n = 3794) or non-DMRs (n = 19,202). In addition, they are grouped as the top 10% DMR promoters (n = 379), 20% DMR (n = 758), and 30% (n = 1137).
Overall, DMRs are associated with a 158% increase of H3K27me3 level in KO MEFs over WT MEFs, while the rest of the genome (non-DMR) has no significant increase in H3K27me3 level (Pearson χ2 test P < 0.001) (Fig. 5B). Heatmap presentation reveals that CG methylation differences (WT minus KO) and CpG numbers at promoter regions are inversely correlated. The greatest CG methylation difference occurs at promoters with low CG density, while H3K27me3 marks are more enriched at promoters with high CG density (Fig. 5C). Nevertheless, KO MEFs display a slight increase in H3K27me3 signal intensity at promoters with the largest CG methylation difference. In addition, the number of H3K27me3 peaks in KO MEFs compared to WT MEFs is higher for promoters located in the top DMRs, whereas non-DMR promoters exhibit reduced numbers of H3K27me3 peaks (Fig. 5D). Thus, an increase of H3K27me3 at a subset of promoters could be responsible for persistent gene repression. About 200 promoters located within DMRs gain H3K27me3 peaks in KO MEFs (KO n = 717, WT n = 510). Most of those genes are silent (e.g., 20% are olfactory receptor genes) or show very little expression in WT MEFs. They stay repressed in KO MEFs, as might be expected due to the increase of H3K27me3. Only 5% show a de-repression in KO MEFs, including genes of the protocadherin family and the cytochrome P450 family. However, the vast majority of promoters located within DMRs (about 3600 genes) do not show a compensatory H3K27me3 increase.
Altogether, the distribution of H3K27me3 marks is altered in KO MEFS. Genomic regions with the greatest loss of CG methylation are linked to increases in H3K27me3 marks.
Reactivation of specific subclasses of repeat families in KO MEFs
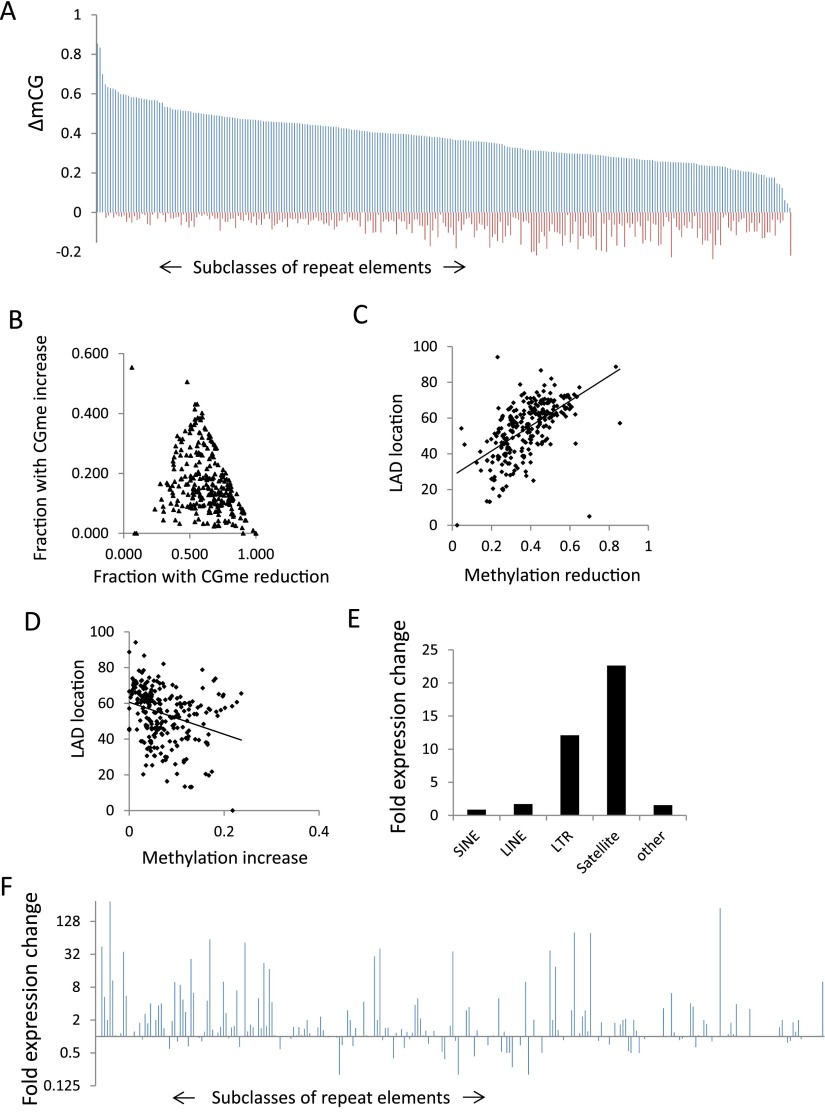
To assess the effect of Hells deletion on DNA methylation at repeat elements, bisulfite converted sequences were reevaluated and specifically aligned to repeat sequences. There are 270 distinct murine repeat elements in the RepeatMasker library (AFA Smit, R Hubley, and P Green, unpubl.), comprising about 2.3 million distinct sequences (Supplemental Table S2). Over 60% of repeat sequences are located within LADs (Supplemental Table S2). Most of the 270 subclasses fall into four superfamilies of repeats, including satellites, endogenous retroviral elements (ERVs) containing long terminal repeats (LTRs), rudiments of retrotransposons known as LINEs (long interspersed elements), or SINEs (short interspersed elements). Mean CG methylation values at repeat sequences show a 60% decrease in KO MEFs compared to WT MEFs (0.8 versus 0.49). Non-CG methylation accounts for <0.1% methylation at repeats and is decreased in KO MEFs compared to WT MEFs (CHG methylation 54% and CHH 57% decrease). Nearly all 270 subfamilies show reduced CG methylation in the absence of HELLS (Fig. 6A; Supplemental Table S2). In addition, KO MEFs display an increase in CG methylation at many subclasses (Fig. 6A). Each of the 270 subclasses consists of hundreds to thousands of distinct sequences. The majority of repeats within a specific subclass shows CG methylation reduction, and only a small subset displays an increase in CG methylation (Fig. 6B).
Figure 6.
De-repression of specific repeat subclasses in KO MEFs. (A) Bar graph representing CG methylation changes (WT methylation values minus KO methylation values) as ΔmCG for 270 repeat subclasses. The repeat classes are sorted based on CG methylation decreases (blue) in KO MEFs compared to WT MEFs. The CG methylation increases in KO MEFs are represented as minus values (red). A list of ranked repeat sequences is compiled in Supplemental Table S2. A bar graph of distinct subfamilies is presented in Supplemental Figure S8. (B) Scatterplot representing the fraction of repeat sequences within a subclass that shows methylation reduction (x-axis) and CG methylation increase (y-axis). All 270 subclasses are presented. (C,D) Scatterplot showing the DNA methylation reduction at repeat classes (C) or the DNA methylation increases (D) and the percentage of sequences within the subclass that is located in LADs. A list of repeat sequences and their frequency of LAD location is compiled in Supplemental Table S2. (E) Bar graph representing repeat element gene expression as fold changes of KO over WT levels (log2) of different repeat superfamilies (Supplemental Table S2). (F) Bar graph representing gene expression changes in KO MEFs over WT MEFs for all 270 repeat subclasses. The repeat classes are sorted as in A. A bar graph of distinct subfamilies is presented in Supplemental Figure S8.
Repeat elements with the greatest CG methylation loss show the highest proportion of LAD location (R = 0.57) (Fig. 6C,D). Conversely, repeat classes with an increase in CG methylation are less frequently localized within LADs (R = −0.3) (Fig. 6C,D). This suggests a link between CG reduction and LADs for repeat sequences.
To determine repeat gene expression RNA-seq, reads were specifically aligned to repeat sequences. Satellite sequences and endogenous retroviral elements (ERVs) showed the highest increase of transcript abundance levels in KO MEFs above wild type, whereas LINE and SINE elements had modest changes (Fig. 6E). About 60 (out of 270) subclasses have an increase greater than twofold above wild type (Fig. 6F), including IAP and MMERVK sequences (Supplemental Table S2; Supplemental Fig. S8). Thus Hells deletion affects the steady-state level of specific subclasses of repeat elements. Overall, repeat de-repression does not correlate with the extent of CG methylation changes (Supplemental Fig. S9). The length of the repeat sequence does not correlate with the expression changes (Supplemental Fig. S9). In addition, repeat sequences that are de-repressed in KO MEFs display the same percentage of LAD location as other repeat sequences that are not affected by Hells deletion (Supplemental Table S2). Since deletion of the histone methyltransferases EHMT2 and SETDB1, which mediate H3K9 methylation, leads to activation of specific repeat elements (Matsui et al. 2010; Maksakova et al. 2013), we performed ChIP analysis for selected repeat sequences. H3K9me2 is not significantly altered at MERVL and MaLR-MTA elements in KO MEFs compared to WT MEFs (Supplemental Fig. S10A). These results are consistent with a previous report demonstrating that H3K9me2 depletion in EHMT2−/− cells results in reactivation of these repeat elements (Maksakova et al. 2013). On the other hand, SETDB1 mediates H3K9me3 modifications and controls Etn1, MMEKVRC10, and IAP expression (Matsui et al. 2010). For the latter two repeats, we found reduced H3K9me3 marks and reactivation in KO MEFs (Supplemental Fig. S10B), suggesting a connection of HELLS and H3K9me3 modification at specific subclasses.
In summary, all types of repeat sequences showed substantial DNA methylation reduction in the absence of HELLS, but only subclasses of repeat elements are de-repressed.
Discussion
Our results provide a comprehensive view of genome-wide DNA methylation patterns in cells of the DNA methylation mutant Hells−/− mouse. Previous studies that reported CG methylation changes in Hells−/− tissues applied methods that were not quantitative, restricted to specific genomic loci, or biased. For example, Southern analysis was performed to report the original finding that HELLS is critical for establishment of DNA methylation, but the analysis was restricted to a limited number of restriction enzyme sites (Dennis et al. 2001). Subsequent reports used conventional bisulfite sequencing for a few selected loci, or applied biased techniques and examined only the immediate promoter region, or enriched for methylated DNA with anti-methylcytosine antibodies that are partial toward genomic regions of low CG density (Xi et al. 2007, 2009; Myant et al. 2011; Tao et al. 2011). The approach in the current study is an advance in methylation assessment because it uses an unbiased, quantitative, high-resolution, and genome-wide approach to determine CG methylation at uniquely mapped sequences and repeats. We report here that the methylome is greatly diminished in Hells−/− primary fibroblasts and, hence, HELLS is a critical factor in the establishment of CG methylation levels in murine development. A similar profound loss of cytosine methylation in a DNA methylation mutant, apart from deletion of Dnmts themselves, has been so far reported for UHRF1 (Bostick et al. 2007; Sharif et al. 2007; Ooi et al. 2009; Feng et al. 2010a,b).
CG methylation reduction in KO MEFs comprises promoter regions of protein-coding and noncoding RNA genes. The extent of methylation loss is greatest at intergenic regions and promoter regions that are CG poor, and maximum methylation decrease is observed at silent genes. This implies that HELLS is primarily required for DNA methylation at genomic regions that are embedded in repressed chromatin. Our observation that relative transcript abundance is stable in KO MEFs (despite CG methylation loss) suggests redundancies in epigenetic silencing pathways. Interestingly, several genes that are embedded in repressed chromatin (and still silenced upon CG hypomethylation) are in part marked by de novo H3K4me1 modification in KO MEFs and belong to the neuronal lineage (Yu et al. 2014). H3K4me1 can mark potential enhancers, and the lack of histone acetylation indicated that these sites are not active yet in KO MEFs but represent putative enhancers. Reprogramming of KO MEFs into induced pluripotent stem cells and challenging the cells to the neuronal pathway activated those enhancers and de-repressed neuronal lineage genes. This suggests that CG hypomethylation may in part influence cellular plasticity via alterations of potential enhancer sites.
DDM1, the HELLS homolog in A. thaliana, primarily methylates regions enriched for the linker histone H1 (Zemach et al. 2013). It is thought that the chromatin remodeling activity of DDM1 is specifically required to enhance nucleosome mobility in densely packaged H1-rich heterochromatin (Zemach et al. 2013). Consistent with the hypothesis that HELLS is mainly required for gene-poor heterochromatin is our observation that CG methylation loss is predominantly at lamin B1 attachment sites. Lamin B1, a scaffolding protein adjacent to the inner nuclear membrane, participates in the spatial organization of the nucleus. Lamin B1 is important for murine development (Kim et al. 2011). Lamin B1 attachment domains (LADs) have been identified based on physical interaction of lamin B1 with DNA sequences (Guelen et al. 2008; Peric-Hupkes et al. 2010). LADs vary in different cell types, and genes that are located within LADs show low transcriptional activity. Lamin B1–associated regions are in general gene poor, and many repeat elements are enriched in LADs (Guelen et al. 2008). We found that >60% of repeats are located in LADs, and for some types of satellite sequences and endogenous retroviral elements the frequency surpasses 80% (Supplemental Table S2). It is currently unknown whether HELLS-mediated DNA methylation alters lamin B1 association with the nuclear envelope or changes nuclear compartmentalization, or whether lamin B1 in turn is functionally interactive with the DNA methylation pathway. It is possible that the location of some genes within LADs contributes to their silenced state despite CG methylation loss, since lamin B1 is thought to influence gene expression (Kind et al. 2013). It is noteworthy that DNA hypomethylated domains in tumor cells, during transition of ES cells to somatic cells, overlap in part with LADs, supporting the link between DNA methylation and these nuclear compartments (Lister et al. 2009, 2011; Hansen et al. 2011).
We found a redistribution of H3K27me3 in the genome, and similar H3K27me3 alterations have been previously reported (Lister et al. 2009; Brinkman et al. 2012; Hagarman et al. 2013; Reddington et al. 2013). Cells that are deficient in DNA methyltransferase activity show a global increase of H3K27me3 level (Hagarman et al. 2013; Reddington et al. 2013) or a reduction of strong peaks concomitant with redistribution to broad domains (Brinkman et al. 2012). The change of H3K27me3 is more subtle in Hells−/− MEFs compared to DNMT1-depleted cells. One reason may be that DNMT1-deficient cells show a greater loss of CG methylation than do Hells−/− MEFs (down to 1.3%) (Brinkman et al. 2012; Hagarman et al. 2013; Reddington et al. 2013). Another difference is that DNMT1 deficiency reduces CG methylation rather evenly across the genome, since DNMT1 is a major maintenance methyltransferase. In Hells−/− cells we observed CG methylation changes in distinct chromosomal regions, and H3K27me3 increases were confined to regions with the greatest CG methylation loss. Only a small number of promoter regions gained H3K27me3 in DNMT1-deficient MEFs (Reddington et al. 2013), consistent with our own observation that only a subset of promoters with DNA hypomethylation is associated with H3K27me3 gains. Thus, alternate epigenetic silencing signals may grant gene repression in the absence of HELLS.
A role of DNA methylation in proviral silencing at specific genomic locations had been known, but systematic analysis of all repeat classes using whole-genome bisulfite sequencing was lacking (Jaenisch et al. 1985; Walsh et al. 1998; Howard et al. 2008). We report here that HELLS is essential for DNA methylation at nearly all subclasses of repeat sequences, supporting the notion that HELLS is a master regulator of repeat sequence methylation. Since HELLS localizes at satellites, LINE elements, and IAP sequences (Huang et al. 2004), this suggests a direct role for HELLS in CG methylation at those sites. Alternatively, HELLS may support a histone methyltransferase that influences H3K9me3 modifications, which in turn alters gene expression and CG methylation (Myant et al. 2011). Remarkably, several repeat elements also show ectopic gain of CG methylation. Cytosine hypermethylation in the absence of HELLS occurs at many loci in spontaneously immortalized cell lines derived from Hells−/− embryos and appears in the CG and non-CG context in DDM1 mutants (Tao et al. 2011; Stroud et al. 2013).
A recent study found DNA methylation reduction at repeat sequences in Hells−/− cells and reactivation of specific repeat elements (Dunican et al. 2013). While a specific role of HELLS in transcriptional silencing of satellite sequences and IAP elements is consistent with our results, there are also important differences between the two studies. For example, Dunican et al. (2013) report a DNA methylation defect at specific subfamilies, whereas we found all subfamilies affected. Another difference is the appearance of significant ectopic DNA methylation, which we report here. There may be several reasons that are responsible for the differences in the two studies: The HELP-seq technique that examines methylation patterns at HpaII restriction enzymes sites (Dunican et al. 2013), as opposed to using an unbiased whole-genome bisulfite sequencing technique as used in this study, and the use of Large T antigen for immortalization of cells (Dunican et al. 2013), as opposed to primary fibroblast cultures as used in this study and the use of a different Hells mutant mouse model that expresses a small amount of HELLS mutant protein (Sun et al. 2004; Dunican et al. 2013). Nevertheless, an important role of HELLS in silencing of some repeat elements is confirmed in either study.
Although each repeat class shows DNA hypomethylation, only some subclasses are de-repressed, and we did not detect any correlation between CG methylation and relative transcript steady-state levels. This suggests that additional pathways control repeat element silencing. For example, SETDB1 deletion results in de-repression of class I and class II elements, whereas EHMT2 and EHMT1 control silencing of some class III endogenous retroviral elements (e.g., MaLR-MTA) (Matsui et al. 2010; Maksakova et al. 2013). We found that HELLS plays a role in silencing of class I and II elements (e.g., IAP and MMEKVRC10), but not in class III (one out of 10 subclasses). We demonstrate a decrease in H3K9me3 modification concomitant with CG methylation reduction at class I/II elements (IAP and MMEKVRC10), and a similar reduction of H3K9me3 was detected at IAP elements in Hells-mutant cells (Dunican et al. 2013). It remains to be shown if HELLS can directly interact with a SETDB1-mediated silencing pathway.
Altogether, Hells−/− MEFs show profound methylation reduction at uniquely mapped sequences as well as repeat sequences. The CG methylation losses are predominantly in a nuclear compartment, in part defined by LADs. The relative transcript abundance level of protein-coding genes and the silencing state of most repeat elements are largely preserved despite DNA methylation reduction, indicating redundant pathways of epigenetic silencing.
Methods
CG methylation analysis
The protocol was followed as described previously (Lister et al. 2008, 2009). Five micrograms of genomic DNA was extracted from frozen cell pellets of WT ES cells (CCE), WT primary murine embryonic fibroblasts (MEFs) and KO (Hells−/−) MEFs. Primary MEFs were derived from day 13.5 embryos of Hells+/− heterozygotic matings and represent littermates. Samples were prepared between passages 1 and 2. The DNeasy blood & tissue kit (Qiagen) was used for DNA extraction, and the DNA was spiked with 25 ng unmethylated cI857 Sam7 Lambda DNA (Promega), serving as control for successful bisulfite treatment. MethylC-seq libraries were sequenced using the Illumina Genome Analyzer II (GAIIx) as per the manufacturer’s instructions. Final sequence coverage was obtained by sequencing all libraries for a sample separately, thus reducing the incidence of “clonal” reads that share the same alignment position and probably originate from the same template molecule in each PCR. The coverage and sequencing depth of CpG sites for each sample is as follows: The total number of CpG sites covered is 34177715 (ES), 37422208 (KO MEF), and 34700701 (WT MEF); the average coverage at covered CpG sites is 6.5 (ES), 8.4 (KO MEF), and 7.6 (WT MEF). The sodium bisulfite nonconversion rate was calculated as the percentage of cytosines sequenced at cytosine reference positions in the lambda genome. All sequence alignments were performed against the reference mouse genome mm9. For CG methylation data extraction, methylated cytosines with greater to or equal to five sequence reads (with the exception of data presentation in Fig. 1) were used. Methylation values are expressed as a fraction (number of methylated cytosine/number of reads for a specific site) or as a percentage with total methylation equal to 100% (equal to a fraction of 1.0). DMRs were determined in 5-kb tiles throughout the genome and showed a difference of mean CG methylation values (WT MEF CG methylation value minus KO MEF CG methylation value) greater than 0.4 (Stroud et al. 2013) with an estimated FDR of <0.1% using the R function qvalue. The complete set of RefSeq genes was downloaded from the UCSC website (http://hgdownload.cse.ucsc.edu/goldenPath/mm9/database/refGene.txt.gz). Complete sets of non-protein-coding sequences were downloaded from Ensembl BioMart GRCm38 (p2 antisense, lincRNA, miRNA, antisense-RNA, and snoRNA [http://useast.ensembl.org/index.html]). Mean promoter methylation was computed from TSS and flanking regions 2 kb upstream of and 2 kb downstream from TSS of protein-coding genes (n = 23,350) located on autosomes.
ChIP-seq
ChIP (chromatin immunoprecipitation) was performed as previously described (Yu et al. 2014). Two micrograms of the following antibodies were used in each reaction: anti-Pol II (Santa Cruz, sc-899x), anti-trimethyl K4 of H3 (Abcam, ab8580), anti-trimethyl K27 of H3 (Millipore, 07-449), and normal rabbit IgG (Millipore, 12-370). We fixed 107 cells at room temperature in 1% formaldehyde (v/v) for 10 min with gentle agitation. Fixation was stopped by the addition of glycine (125 mM final concentration) and agitation for 5 min on ice. Chromatin was sheared by sonication to ∼100- to 300-bp fragments. ChIP DNA libraries were made following Illumina ChIP-seq library preparation kit and subjected to Illumina sequencing (GAIIx) at the CCR-Sequencing Facility, National Cancer Institute.
RNA-seq library generation and analysis
Total RNA was isolated from cell pellets of WT (Hells+/+) MEFs and KO (Hells−/−) MEFs and their biological replicates using the RNeasy mini kit (Qiagen) and treated with DNase I (Roche) for 10 min at room temperature. rRNAs were removed from 5 μg of total RNA by RiboMinus (Life Technologies) as per the manufacturer’s instructions. The poly-A–containing mRNA molecules were purified using poly-T oligo-attached magnetic beads. After purification and PCR amplification, the final cDNA library was generated based on the RNA-seq Library Preparation Protocol from Illumina. RNA-seq libraries were sequenced with HiSeq 2000 Analyzer. Reads (102 bp each) were paired-end (PE) and mapped against the murine reference genome mm9 using RTA 1.12.4.2 software tools.
Data access
ChIP-seq, RNA-seq, and bisulfite sequencing data have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE57419 and GSE56151.
Supplementary Material
Acknowledgments
We thank Lino Tessarolo and Michael Bustin for helpful suggestions on the manuscript. We thank Karen Saylor for excellent animal technical assistance. We thank Matthew Fivash and Xiangtao Liu for help on the use of the R program, Jyoti Shetty, Yongmei Zhao, and Bao Tran from the NCI Sequencing Facility, FNLCR, and the NIH Fellows Editorial Board for suggestions on the manuscript. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and the National Library of Medicine, NIH. Frederick National Laboratory for Cancer Research is accredited by AAALAC International and follows the Public Health Service Policy for the Care and Use of Laboratory Animals. Animal care was provided in accordance with the procedures outlined in the “Guide for Care and Use of Laboratory Animals” (National Research Council, 1996, National Academy Press, Washington, DC).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.172015.114.
References
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764 [DOI] [PubMed] [Google Scholar]
- Brinkman AB, Gu H, Bartels SJ, Zhang Y, Matarese F, Simmer F, Marks H, Bock C, Gnirke A, Meissner A, et al. 2012. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res 22: 1128–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeski J, Jerzmanowski A. 2003. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J Biol Chem 278: 823–828 [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86 [DOI] [PubMed] [Google Scholar]
- De La Fuente R, Baumann C, Fan T, Schmidtmann A, Dobrinski I, Muegge K. 2006. LSH is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat Cell Biol 8: 1448–1454 [DOI] [PubMed] [Google Scholar]
- Dennis K, Fan T, Geiman T, Yan Q, Muegge K. 2001. LSH, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev 15: 2940–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. 2010. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem 285: 26114–26120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunican DS, Cruickshanks HA, Suzuki M, Semple CA, Davey T, Arceci RJ, Greally J, Adams IR, Meehan RR. 2013. LSH regulates LTR retrotransposon repression independently of Dnmt3b function. Genome Biol 14: R146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle M, Hoffmeister H, Rothammer J, Fuchs A, Exler JH, Langst G. 2011. Nucleosomes protect DNA from DNA methylation in vivo and in vitro. Nucleic Acids Res 39: 6956–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. 2010a. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci 13: 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. 2010b. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci 107: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G. 2009. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J 28: 697–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. 2014. Ensembl 2014. Nucleic Acids Res 42: D749–D755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman TM, Muegge K. 2000. LSH, an SNF2/helicase family member, is required for proliferation of mature T lymphocytes. Proc Natl Acad Sci 97: 4772–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman TM, Tessarollo L, Anver MR, Kopp JB, Ward JM, Muegge K. 2001. Lsh, a SNF2 family member, is required for normal murine development. Biochim Biophys Acta 1526: 211–220 [DOI] [PubMed] [Google Scholar]
- Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, et al. 2013. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153: 1149–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951 [DOI] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagarman JA, Motley MP, Kristjansdottir K, Soloway PD. 2013. Coordinate regulation of DNA methylation and H3K27me3 in mouse embryonic stem cells. PLoS ONE 8: e53880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. 2011. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 43: 768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. 2008. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene 27: 404–408 [DOI] [PubMed] [Google Scholar]
- Huang J, Fan T, Yan Q, Zhu H, Fox S, Issaq HJ, Best L, Gangi L, Munroe D, Muegge K. 2004. LSH, an epigenetic guardian of repetitive elements. Nucleic Acids Res 32: 5019–5028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Schnieke A, Harbers K. 1985. Treatment of mice with 5-azacytidine efficiently activates silent retroviral genomes in different tissues. Proc Natl Acad Sci 82: 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ. 1999. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet 22: 94–97 [DOI] [PubMed] [Google Scholar]
- Jin B, Ernst J, Tiedemann RL, Xu H, Sureshchandra S, Kellis M, Dalton S, Liu C, Choi JH, Robertson KD. 2012. Linking DNA methyltransferases to epigenetic marks and nucleosome structure genome-wide in human tumor cells. Cell Reports 2: 1411–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492 [DOI] [PubMed] [Google Scholar]
- Karlsen TA, Jakobsen RB, Mikkelsen TS, Brinchmann JE. 2014. microRNA-140 targets RALA and regulates chondrogenic differentiation of human mesenchymal stem cells by translational enhancement of SOX9 and ACAN. Stem Cells Dev 23: 290–304 [DOI] [PubMed] [Google Scholar]
- Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan CM, Gaiano N, Ko MS, Zheng Y. 2011. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science 334: 1706–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. 2013. Single-cell dynamics of genome-nuclear lamina interactions. Cell 153: 178–192 [DOI] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. 2011. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471: 68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Thompson PJ, Goyal P, Jones SJ, Singh PB, Karimi MM, Lorincz MC. 2013. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenetics Chromatin 6: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- Myant K, Termanis A, Sundaram AY, Boe T, Li C, Merusi C, Burrage J, de Las Heras JI, Stancheva I. 2011. LSH and G9a/GLP complex are required for developmentally programmed DNA methylation. Genome Res 21: 83–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. 2008. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle 7: 1539–1544 [DOI] [PubMed] [Google Scholar]
- Ooi SK, O’Donnell AH, Bestor TH. 2009. Mammalian cytosine methylation at a glance. J Cell Sci 122: 2787–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Wolf D, Hartung O, Agarwal S, Daley GQ, Goff SP, Bestor TH. 2010. Dynamic instability of genomic methylation patterns in pluripotent stem cells. Epigenetics Chromatin 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddington JP, Perricone SM, Nestor CE, Reichmann J, Youngson NA, Suzuki M, Reinhardt D, Dunican DS, Prendergast JG, Mjoseng H, et al. 2013. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol 14: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W. 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447: 425–432 [DOI] [PubMed] [Google Scholar]
- Ryan DP, Owen-Hughes T. 2011. Snf2-family proteins: chromatin remodellers for any occasion. Curr Opin Chem Biol 15: 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912 [DOI] [PubMed] [Google Scholar]
- Sims RJ 3rd, Belotserkovskaya R, Reinberg D. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev 18: 2437–2468 [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. 2012. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. 2013. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LQ, Lee DW, Zhang Q, Xiao W, Raabe EH, Meeker A, Miao D, Huso DL, Arceci RJ. 2004. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev 18: 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Xi S, Shan J, Maunakea A, Che A, Briones V, Lee EY, Geiman T, Huang J, Stephens R, et al. 2011. LSH, chromatin remodeling family member, modulates genome-wide cytosine methylation patterns at nonrepeat sequences. Proc Natl Acad Sci 108: 5626–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. 2011. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474: 649–653 [DOI] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20: 116–117 [DOI] [PubMed] [Google Scholar]
- Xi S, Zhu H, Xu H, Schmidtmann A, Geiman TM, Muegge K. 2007. LSH controls Hox gene silencing during development. Proc Natl Acad Sci 104: 14366–14371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi S, Geiman TM, Briones V, Guang Tao Y, Xu H, Muegge K. 2009. LSH participates in DNA methylation and silencing of stem cell genes. Stem Cells 27: 2691–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. 2013. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153: 1134–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Briones V, Lister R, McIntosh C, Han Y, Lee EY, Ren J, Terashima M, Leighty RM, Ecker JR, et al. 2014. CG hypomethylation in LSH−/− mouse embryonic fibroblasts is associated with de novo H3K4me1 formation and altered cellular plasticity. Proc Natl Acad Sci 111: 5890–5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D. 2013. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Baumann C, Schmidtmann A, Honaramooz A, Tang L, Bondareva A, Dores C, Fan T, Xi S, Geiman T, et al. 2011. Lymphoid-specific helicase (HELLS) is essential for meiotic progression in mouse spermatocytes. Biol Reprod 84: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. 2012. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148: 908–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Geiman TM, Xi S, Jiang Q, Schmidtmann A, Chen T, Li E, Muegge K. 2006. LSH is involved in de novo methylation of DNA. EMBO J 25: 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.