Abstract
Mutation is associated with developmental and hereditary disorders, aging, and cancer. While we understand some mutational processes operative in human disease, most remain mysterious. We used Caenorhabditis elegans whole-genome sequencing to model mutational signatures, analyzing 183 worm populations across 17 DNA repair-deficient backgrounds propagated for 20 generations or exposed to carcinogens. The baseline mutation rate in C. elegans was approximately one per genome per generation, not overtly altered across several DNA repair deficiencies over 20 generations. Telomere erosion led to complex chromosomal rearrangements initiated by breakage–fusion–bridge cycles and completed by simultaneously acquired, localized clusters of breakpoints. Aflatoxin B1 induced substitutions of guanines in a GpC context, as observed in aflatoxin-induced liver cancers. Mutational burden increased with impaired nucleotide excision repair. Cisplatin and mechlorethamine, DNA crosslinking agents, caused dose- and genotype-dependent signatures among indels, substitutions, and rearrangements. Strikingly, both agents induced clustered rearrangements resembling “chromoanasynthesis,” a replication-based mutational signature seen in constitutional genomic disorders, suggesting that interstrand crosslinks may play a pathogenic role in such events. Cisplatin mutagenicity was most pronounced in xpf-1 mutants, suggesting that this gene critically protects cells against platinum chemotherapy. Thus, experimental model systems combined with genome sequencing can recapture and mechanistically explain mutational signatures associated with human disease.
Maintenance of DNA fidelity in both germline and somatic cells is critical to life. Genome duplication is fraught with the risk of replication errors, and this is compounded by damage to DNA from environmental agents, ultraviolet light, ionizing radiation, and endogenous by-products of metabolism such as reactive oxygen species. Unchecked, the resulting mutations in somatic cells can lead to cancer, neurodegeneration, and premature aging, as evidenced by an increased incidence of such disorders in individuals who inherit abnormalities of DNA repair pathways. Mutations arising in germ cells can lead to hereditary disease and major developmental disorders.
Given these multifarious threats to the fundament of cellular inheritance, eukaryotes have evolved sophisticated pathways for detecting and repairing DNA damage. Our understanding of the mechanisms underpinning detection of DNA lesions, the relative contribution of different repair pathways to each type of lesion, and the regulation of pathway choice is still fragmentary. DNA repair ideally occurs by restoring the original genetic information. However, it is well established that some DNA repair pathways are prone to errors and yet necessary for cellular survival. These include nonhomologous DNA end-joining (NHEJ), a repair modality that leads to direct ligation of broken DNA ends, and translesion synthesis (TLS), a process where error-prone polymerases maintain replication opposite damaged DNA bases (for review, see Ho and Scharer 2010). Homologous recombination is overall considered to be an error-free pathway of DNA double-strand break repair, but DNA synthesis associated with gene conversion leads to increased mutation rates, as is the case for synthesis-dependent strand annealing (SDSA) and break-induced replication (BIR) repair (Smith et al. 2007; Hicks et al. 2010; Deem et al. 2011).
A “mutational process” results from the interplay of DNA damaging agents, either endogenous or exogenous, with local DNA chemistry and repair pathways. Such processes leave distinct “signatures” in the genome, which we can define using a number of variables, including type of event, local sequence context, chromatin landscape, timing, genomic clustering, and imprint of attempted DNA repair. In somatic cells, these signatures can be deciphered from cancer genomes (Stratton et al. 2009; Alexandrov et al. 2013) or single somatic cell sequencing (Welch et al. 2012); in the germline, they are best inferred from sequencing parent–child trios (Kong et al. 2012). They differ across tumor types, among individuals within a tumor type, and even among clones within an individual’s tumor. Systematic sequencing of cancer and germline genomes has enabled description of these signatures in exquisite detail, and many new mutational processes have been delineated, often with surprising properties.
The leap from describing a mutational signature to a mechanistic understanding of its etiology is challenging. For several, we have a reasonably complete picture: C>T mutations at CpG dinucleotides are due to spontaneous deamination of methylated cytosine (Waters and Swann 2000); C>T substitutions in skin cancers result from the failure to repair UV-induced cyclobutane pyrimidine dimers (Pfeifer et al. 2005; Pleasance et al. 2010a); and microsatellite instability arises in cells with loss of the mismatch repair pathway (Aaltonen et al. 1993; Ionov et al. 1993; Thibodeau et al. 1993). For the majority of mutational signatures, however, we have limited insights into their etiology, resulting in a proliferation of neologisms that describe their genomic manifestations rather than the underlying (and unknown) mechanisms. These include “kataegis”: referring to a processive clustering of cytosine mutations in a TpC context often near genomic rearrangements, thought to be caused by DNA-editing enzymes (Nik-Zainal et al. 2012a,b; Roberts et al. 2012, 2013; Burns et al. 2013; Taylor et al. 2013); “chromoplexy”: balanced chains of rearrangements across multiple chromosomes in prostate cancer (Baca et al. 2013); “chromothripsis”: clusters of tens to hundreds of localized genomic rearrangements occurring in a single catastrophic event in 2%–3% of all cancers (Stephens et al. 2011); and “chromoanasynthesis”: a phenomenon occurring in the germline, driving localized gains and losses of chromosomal material through a replication-based mechanism (Liu et al. 2011a,b).
Experimental model systems such as yeast and Caenorhabditis elegans have a tradition in the study of mutational processes, due to their convenient life cycles, compact genome, and ease of genetic manipulation. The ∼100 million base pair C. elegans genome was the first animal genome to be sequenced (The C. elegans Sequencing Consortium 1998). Mutational signatures have been investigated for ethyl methanesulfonate (EMS) and N-ethyl-N-nitrosourea (ENU), alkylating agents commonly used for forward genetic screens (Flibotte et al. 2010). EMS and ENU mutagenesis approaches have been recently used systematically, aiming for generating multiple mutations in every worm gene. The sequencing of ∼1000 strains was used to generate >800,000 unique single-nucleotide mutations and thousands of indels (Thompson et al. 2013). Using classic genetic methods, mutation frequencies have also been estimated in response to ionizing radiation (Rosenbluth et al. 1985) and as a consequence of DNA repair deficiencies in the dog-1 helicase and the mrt-2 and clk-2 checkpoint genes (Harris et al. 2006; Kruisselbrink et al. 2008; Zhao et al. 2008). Moreover, limited genome sequencing was used to estimate mutation rates in C. elegans (Cheung et al. 2002; Denver et al. 2006, 2009; Harris et al. 2006) and studies were more recently extended genome-wide to strains deficient for translesion polymerases (Koole et al. 2014; Roerink et al. 2014). An initial study focused on yeast mutant accumulation, provided a baseline mutation rate, and showed increased mutagenesis for several repair deficiencies (Serero et al. 2014). Nevertheless, in general, though, the DNA repair community largely relies on toxicity assays that score for reduced survival to study consequences of carcinogen exposure and/or loss of repair fidelity. These assays and assays that rely on single locus reporter systems lack the genomic breadth to characterize a full mutational signature. With the advent of whole-genome sequencing, however, it has become feasible to systematically exploit these experimental systems for a large-scale, comprehensive description of mutational signatures.
We established massively parallel sequencing of C. elegans genomes as a model to analyze the patterns and etiology of mutations arising with exposure to carcinogens and across different DNA repair backgrounds. In total, we sequenced whole genomes from 183 populations of worms, drawn from 17 different genetic backgrounds (wild-type and knockouts for various DNA repair and DNA damage response genes) followed for 20 generations or exposed in a single generation to one of three mutagens. We identified and analyzed 1559 base substitutions, 406 indels, and 281 genomic rearrangements. From these data, we extracted mutational signatures that resemble those seen in human cancers and in developmental genomic disorders.
Results
Mutation accumulation in wild-type and DNA repair-deficient C. elegans
In C. elegans, knockouts have been systematically generated by the research community and by international consortia using random mutagenesis protocols that cause either inactivating point mutations or deletions in the gene of interest (The C. elegans Deletion Mutant Consortium 2012). We identified a set of lines with knockout of genes involved in DNA damage response and repair and backcrossed these to our N2 wild-type reference strain to obtain lines with genomes virtually identical to wild-type, except for the gene disruption under investigation (Supplemental Material; Supplemental Table S1). Based on backcrossing for a minimum of six times against the wild-type reference, we expect >98% of the genome of wild-type and knockout worms to be identical. Stocks were frozen to safeguard against mutation accumulation between experiments.
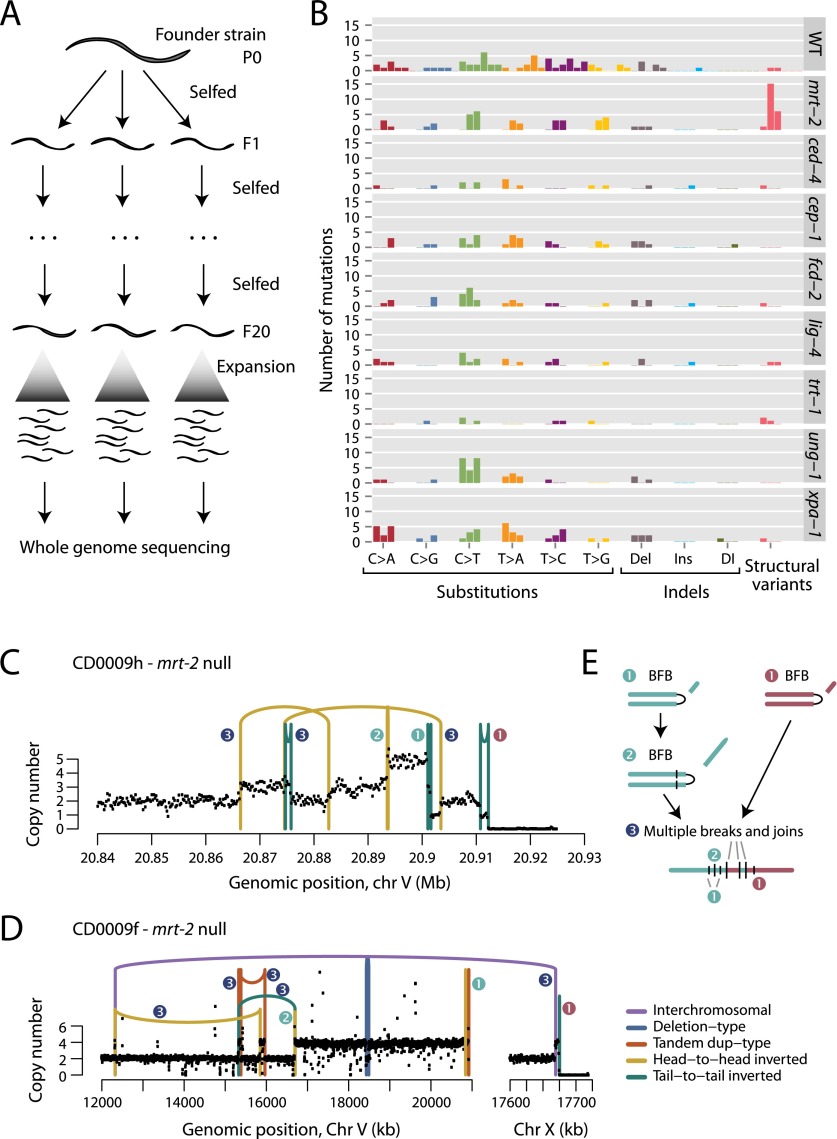
To study spontaneous mutation accumulation rates per generation, we transferred a single hermaphrodite of a given genotype onto a plate (P0 generation) (Fig. 1A). After 3 to 4 d, single F1 L4 animals were randomly chosen and propagated on a new plate, a procedure repeated for 20 generations and performed in quintuplicate for each genotype. Since propagation is by self-fertilization, each new generation effectively derives from a diploid zygote representing a single-cell bottleneck, thereby enabling mutations to be identified in shotgun sequencing data (Denver et al. 2009). Genomic DNA from three F20 animals of each genotype was sequenced to a target depth of 30-fold, and compared with the genome of the founder worm using well-established bioinformatics algorithms to call newly acquired base substitutions, small indels, and genomic rearrangements (Nik-Zainal et al. 2012a,b).
Figure 1.
Mutations in C. elegans followed for 20 generations. (A) The experimental design used three progeny of a founder or parental (P0) animal, propagated by self-fertilization for 20 generations (F1: filial 1; F20: filial 20). The 20th generation worm was expanded to generate sufficient DNA for whole-genome sequencing. (B) Numbers and distribution of acquired mutations across wild-type (WT) and different genetic knockout lines. Each replicate worm is shown as a separate bar, colored by the class of variant. (Del) Deletion; (Ins) insertion; (DI) complex indel. (C) Copy-number profile and genomic rearrangements observed on chromosome V for an mrt-2 deficient animal. Numbers next to the genomic rearrangements refer to the inferred sequence of events. (D) Copy-number profile and genomic rearrangements observed on chromosomes V and X for another mrt-2-deficient animal. Numbers next to the genomic rearrangements refer to the inferred sequence of events. (E) Proposed model for the evolution of the copy-number changes and genomic rearrangements in C and D. A telomere crisis precipitates several breakage-fusion-bridge (BFB) cycles, which are resolved by a cell cycle in which multiple DNA breaks are simultaneously joined.
As expected, base substitutions were the most frequent mutations observed in wild-type C. elegans and most of the DNA repair-deficient backgrounds analyzed (Fig. 1B; Supplemental Tables S2–S4). Mutation rates were consistent across the replicate lines in each given genetic background, indicating that the accumulation of de novo mutations per se did not significantly change the observed mutation rate, at least within the time course of our experiment. The six possible single-base substitutions (C>A, C>G, C>T, T>A, T>C, and T>G) occurred at broadly equal frequency (Fig. 1B), although C>T mutations (mean, 2.8 per 20 generations per line) were more common than other categories (range in means, 0.7–1.7 per 20 generations per line), as previously reported (Denver et al. 2009). In the ung-1 knockout line, we observed a greater proportion of C>T transitions than in other lines (P = 0.003). This signature is expected, because the function of UNG-1 is to eliminate uracil that arises from spontaneous deamination of cytosine in genomic DNA by initiating base excision repair. Without this, DNA replication can misincorporate an adenine opposite the uracil, leading to a C>T mutation. In the absence of UNG-1, redundant pathways may take over in line with no overt increase in mutation frequency.
For each round of self-fertilization, a newly acquired heterozygous mutation has a 25% chance of becoming homozygous and fixed in the line, assuming no effect on viability (Thompson et al. 2013). Also, with each generation, a variant has a 50% chance of remaining heterozygous and a 25% chance of being lost to the line. Allowing for this, we used maximum likelihood methods to estimate mutation rates per generation in each background. We observed a remarkably consistent average of approximately one mutation per genome per C. elegans generation (Supplemental Fig. S1), approximately twice that previously estimated (Denver et al. 2009). This discrepancy in estimated rates is probably due to the greater portion of the genome that we can now access with 100-bp paired end reads than was available in earlier experiments. Based on the known developmental lineage and germ cell expansion in adult animals, ∼15 cell divisions occur between successive generations (Sulston et al. 1983). We therefore estimate a spontaneous mutation rate in the C. elegans germ cell lineage of ∼6.7 × 10−10 per base pair per cell division, in line with the reported rate of ∼3.6 × 10−10 in budding yeast (Serero et al. 2014). Germ cell genome maintenance appears surprisingly robust even when the ability to initiate one of several different DNA repair pathways is compromised. The absence of increased mutation accumulation might indicate that the corresponding repair modalities might be selected to cope with exogenous DNA damaging agents. However, mutation accumulation experiments need to be done for substantially more generations to exclude increased spontaneous mutation rates. Also endogenous damage levels might be increased when worms grow in their natural environment.
C. elegans deficient for mrt-2 develop telomere crisis and breakage-fusion-bridge cycles
Both TRT-1, the catalytic subunit of telomerase, and MRT-2, the C. elegans RAD1 subunit of the conserved 9-1-1 replication factor C-like complex, are required for telomere maintenance. In addition, MRT-2 has a role in homologous recombination and DNA damage checkpoint signaling (Gartner et al. 2000). Telomeres become critically short after propagating trt-1 and mrt-2 homozygous mutant lines for 15–25 generations (Ahmed and Hodgkin 2000; Meier et al. 2006). Eventually, the entire mutant strain population succumbs to sterility but fused chromosomes have previously been isolated, suggesting the presence of genomic rearrangements near telomeres (Ahmed and Hodgkin 2000; Meier et al. 2006; Lowden et al. 2008). Sequencing late-generation mrt-2 lines, we identified several lines with stable but convoluted clusters of genomic rearrangements (Fig. 1C,D). These rearrangements clustered near the ends of chromosomes and were associated with multiple changes in copy-number state. We used a graph theory algorithm (Greenman et al. 2012) to infer the sequence of events leading to these complex genomic structures (Fig. 1E; Supplemental Fig. S2).
In each case, the initial rearrangements (marked ❶ in Fig. 1C–E) comprised inverted rearrangements that demarcated sharp changes in copy number. These rearrangements most likely result from breakage-fusion-bridge (BFB) cycles, first described by Barbara McClintock in maize (McClintock 1941). During these cycles, an existing dsDNA break is replicated in S phase and resolved in G2 by fusing the broken ends of the chromatids together (Supplemental Fig. S2). This results in chromosomes that form an anaphase bridge during cytokinesis. With completion of cell division, scission of this bridge generates new dsDNA breaks in the two daughter cells, and the breakage-fusion-bridge cycle can repeat. We see evidence for a second such cycle in the two genomes illustrated (rearrangements marked ❷).
In both mrt-2 mutant lines, the process is completed through simultaneous acquisition of several rearrangements (marked ❸) (Supplemental Fig. S2). Although the number of rearrangements in this final series of clustered events is small, the pattern bears some resemblance to chromothripsis (Stephens et al. 2011) in that the rearrangements are acquired simultaneously, demarcate oscillating copy-number changes, and form a chain of linked chromosomal segments (Korbel and Campbell 2013). This is of particular interest because the anaphase bridge associated with breakage-fusion-bridge cycles can cause micronucleus formation as the cytokinetic furrow divides daughter cells (Pampalona et al. 2010), and micronuclei may sequester chromosomal fragments for DNA repair before rejoining the main nucleus (Crasta et al. 2012; Hatch et al. 2013). Breakage-fusion-bridge cycles may therefore increase the chance of chromothripsis-type events occurring. This final event solves the initial telomere crisis through end-to-end fusion of two chromosomes.
Taken together, then, telomere crises occurring in mrt-2-deficient animals are associated with breakage-fusion-bridge cycles of repair and rearrangement. Telomere attrition occurs in many cancer types (Xu et al. 2013), but the genomic signature of breakage-fusion-bridge repair is seen rather less frequently. It is, however, especially prominent in pancreatic cancer, accounting for one-sixth of rearrangements in this cancer type (Campbell et al. 2010) compared to, for example, 1% of breast or ovarian cancer rearrangements (Stephens et al. 2009; McBride et al. 2012).
Substitutions induced by aflatoxin B1 in wild-type and DNA repair-deficient C. elegans
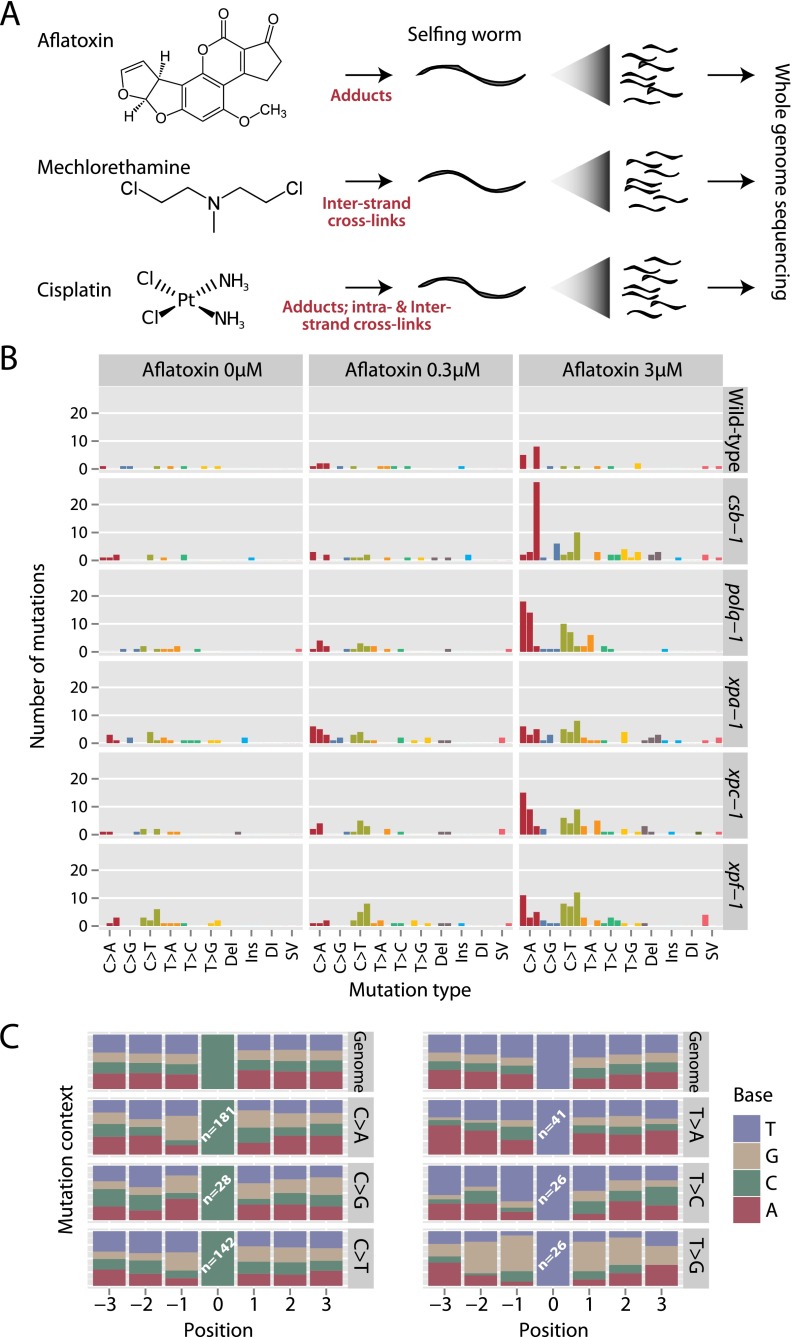
Many carcinogens drive cancer by directly modifying nucleotides, leading to errors in either DNA replication or DNA repair. The specific chemistry of the mutagen and DNA dictates a rather stereotypical interaction, which reads out as a reproducible mutational signature in the genome that is distinctive for each carcinogen (Pfeifer et al. 2002). We exploited the C. elegans model system to analyze mutation spectra associated with three carcinogens across different DNA repair deficiencies (Fig. 2A). In this protocol, we exposed individual mature hermaphrodites to the carcinogen and expanded a single offspring derived from mutagenized male and female germ cells for sequencing—each carcinogen-background combination was performed in triplicate. The carcinogens were chosen to encompass different types of DNA lesion: aflatoxin causes bulky DNA adducts on guanine residues; mechlorethamine and cisplatin are reported to cause a variety of monoadducts, intra- and interstrand crosslinks.
Figure 2.
Mutations in C. elegans exposed to carcinogens. (A) The experimental design exposed a selfing animal to a specified dose of carcinogen and expanded progeny for whole-genome sequencing. The three carcinogens used were chosen because they generate different classes of DNA lesions. (B) Numbers and distribution of acquired mutations across wild-type (WT) and different genetic knockout lines exposed to different doses of aflatoxin. Each replicate worm is shown as a separate bar, colored by the class of variant. (Del) Deletion; (Ins) insertion; (DI) complex indel; (SV) structural variant. (C) Surrounding sequence context for base substitutions in animals exposed to aflatoxin. The genomic sequence from three bases upstream to three bases downstream from each observed mutation was aggregated and presented as proportions in a stacked bar chart, compared to the reference genome.
Aflatoxin B1 is a naturally occurring mycotoxin, produced by Aspergillus flavus, a fungus linked to the development of many hepatocellular carcinoma in sub-Saharan Africa and East Asia (Kensler et al. 2011). Aflatoxin B1 is metabolized to the toxicologically relevant epoxide, which preferentially attacks guanine residues leading to bulky DNA adducts that are removed by nucleotide excision repair (NER) (Smela et al. 2001). Aflatoxin B1, a mutagen requiring metabolic activation, has been shown to be metabolized and inflict DNA damage in C. elegans (Leung et al. 2010). Exposure of C. elegans wild-type, NER mutants, and polq-1 translesion synthesis mutants to low doses of Aflatoxin B1 led to a slight reduction in viability of embryos reaching 15%–20% embryonic lethality in xpf-1 mutants (Supplemental Fig. S3A). Low doses of mutagen exposure were chosen to avoid secondary mutagenic effects relating to swamping the DNA damage response.
For the genomic analyses we sequenced the clonal progeny of three independently derived F1 animals from each dose level of Aflatoxin B1 (0 μM, 0.3 μM, and 3 μM) across six genetic backgrounds. From the genome sequencing, we called newly acquired mutations (Fig. 2B). There was a clear relationship between the dose of aflatoxin and the number of base substitutions (P < 0.0001) and a markedly uneven distribution across the six possible classes of base substitution (P < 0.0001). C>T and C>A (equivalently, G>A and G>T) changes were particularly prominent, consistent with the known property of aflatoxin to generate adducts at guanines. All NER deficiency backgrounds showed an increase in substitutions compared to wild-type (P = 0.007). However, among the different NER-deficient backgrounds, there was no significant difference in substitution rates (P = 0.3). Accumulation of mutations without concomitant reduction in embryonic progeny survival indicates that sequencing provides a more sensitive genotoxicity assay than measuring reduced survival of progeny. The aflatoxin signature matches mutation profiles reported in the TP53 gene in human aflatoxin-induced hepatocellular carcinomas, namely, a preponderance of C>T transitions and C>A transversions (equivalently, G>A and G>T) occurring with a strand bias indicative of evidence of transcription-coupled nucleotide excision repair (Petitjean et al. 2007; Besaratinia et al. 2009).
In many carcinogen-induced cancers, the surrounding sequence context influences the mutation rate at adducted bases. We therefore compared the distribution of bases around the mutated base in aflatoxin-exposed animals to the expected distribution from the reference genome (Fig. 2C). For mutations at C:G bases, we found strong evidence for an enrichment of G immediately 5′ to the mutated C (P < 0.0001) and immediately 3′ (P = 0.0002). Statistically significant biases in a sequence context were also found in mutations at T:A pairs (P < 0.0001), albeit with much lower numbers.
Substitutions induced by crosslinking agents mechlorethamine and cisplatin
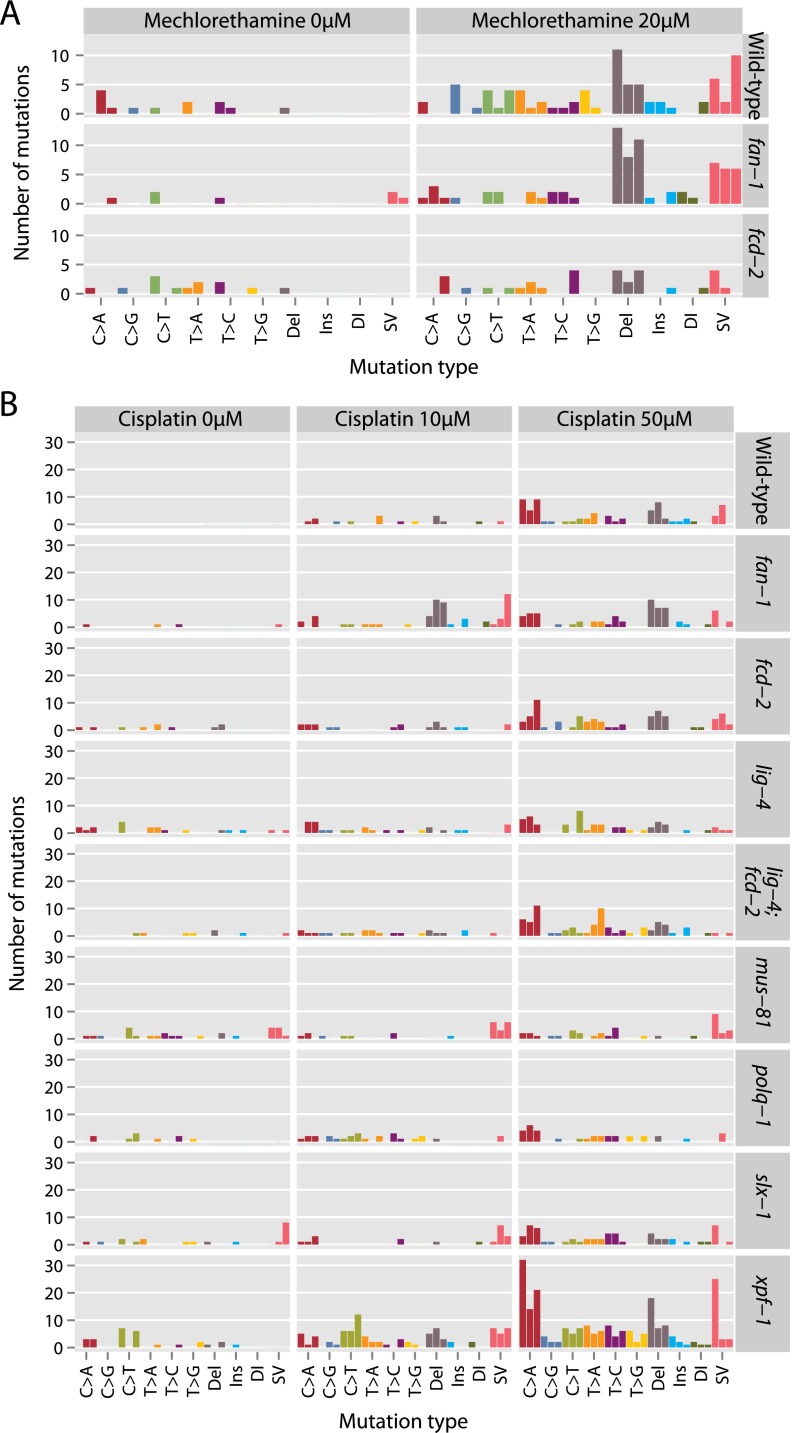
We next investigated mutational signatures induced by two widely used chemotherapeutic agents, mechlorethamine and cisplatin. Mechlorethamine, a nitrogen mustard, induces monoadducts at guanines and interstrand crosslinks, especially at CG:GC or GC:CG dinucleotides (Povirk and Shuker 1994). Cisplatin is a platinum-based drug widely used in cancer chemotherapy. The platinum atoms form covalent bonds with purines resulting in 65% GpG, 25% ApG, and 5%–10% GpNpG intrastrand crosslinks, as well as a small percentage of interstrand crosslinks and monoadducts (Lemaire et al. 1991). Monoadducts are repaired by the base excision, nucleotide excision, and mismatch repair pathways. Intrastrand crosslinks are thought to be largely repaired by the NER pathway, while the Fanconi anemia pathway, in conjunction with components of the homologous recombination pathway and translesion synthesis, are required to repair DNA interstrand crosslinks.
Treating germ cells with a moderate dose of mechlorethamine and cisplatin resulted in a reduction of progeny survival, accentuated in DNA repair mutants (Supplemental Fig. S3B,C). In the genome, mechlorethamine treatment caused a modest but statistically significant increase in single-base substitutions compared to no treatment controls (P = 0.04). Compared to wild-type, this dose-response relationship was no more pronounced in knockouts of fan-1 or fcd-2 (Fig. 3A), genes encoding components of the Fanconi anemia crosslink repair pathway. Indels and structural variants also substantially increased with mechlorethamine exposure (P < 0.0001), as discussed later.
Figure 3.
Mutations in C. elegans exposed to mechlorethamine (A) and cisplatin (B). Numbers and distribution of acquired mutations across wild-type and different genetic knockout lines exposed to the mutagen. Each replicate worm is shown as a separate bar, colored by the class of variant. (Del) Deletion; (Ins) insertion; (DI) complex indel; (SV) structural variant.
Cisplatin treatment caused a striking, dose-dependent increase in base substitutions (P < 0.0001) (Fig. 3B). This was especially pronounced for C>A mutations, but all classes of substitution increased in frequency with cisplatin exposure. There were significant differences in the mutation prevalence depending on the genetic background (P < 0.0001), largely relating to an increase in substitutions for xpf-1 knockouts (P = 0.3 for differences across the other backgrounds). This result is consistent with the expected role of XPF-1 in nucleotide excision repair and intrastrand crosslink repair.
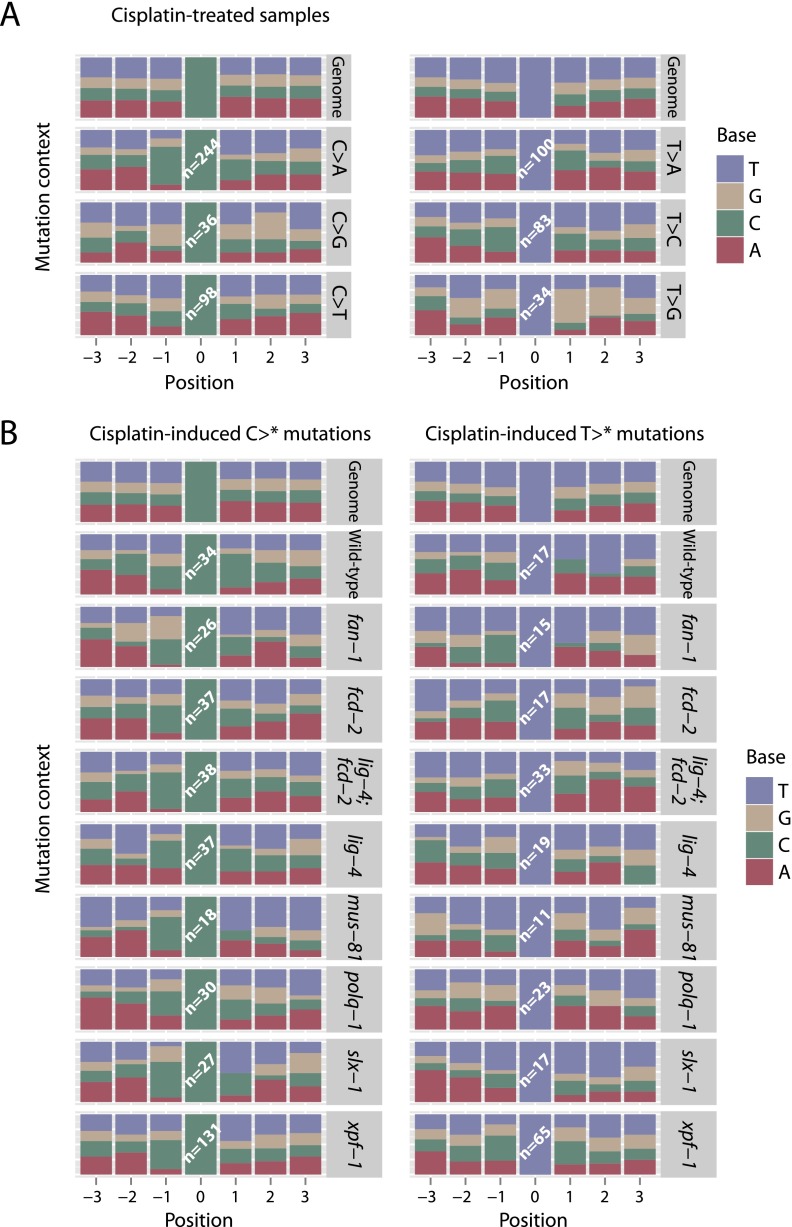
As for aflatoxin, the local sequence context strongly influences the mutagenicity of cisplatin (Fig. 4A). The base immediately 5′ to a mutated C (P < 0.0001) and 3′ (P < 0.0001) showed a different distribution compared to the genome, and there were significant differences among the three different types of cytosine mutation (P < 0.0001 for 5′ base context and P = 0.01 for 3′ base). Thus, C>A mutations were especially prominent in a CpC context (equivalently, G>T in a GpG context). However, in a TpC context, C>T mutations were more pronounced, suggesting that local sequence context influences which incorrect base is incorporated opposite a damaged base. Similar findings applied to mutations at thymine residues (P < 0.0001 for difference from genome at 5′ and 3′ bases; P < 0.0001 and P = 0.01 for differences across mutation types at 5′ and 3′ bases, respectively). Among the various genetic backgrounds, there were only minor differences in the local sequence context for cytosine mutations (P = 0.05 and P = 0.07 for 5′ and 3′ base) (Fig. 4B).
Figure 4.
Sequence context for base substitutions observed in cisplatin-treated C. elegans. (A) Surrounding sequence context for cytosine and thymine substitutions in animals exposed to cisplatin, split by type of mutation. The genomic sequence from three bases upstream to three bases downstream from each observed mutation was aggregated and presented as proportions in a stacked bar chart, compared to the reference genome. (B) Surrounding sequence context for cytosine and thymine substitutions in animals exposed to cisplatin, split by genetic background. The genomic sequence from three bases upstream to three bases downstream from each observed mutation was aggregated and presented as proportions in a stacked bar chart, compared to the reference genome.
Taken together, across all three carcinogens, the signature of mutations observed reflects the specific chemistry of a mutagen’s interaction with a single base or adjacent bases. The most frequent intrastrand crosslink induced by cisplatin is at GpG dinucleotides (Lemaire et al. 1991), and this matches the high rate of C>A mutations in a CpC context (equivalently, G>T in a GpG context). The interaction of mutagen and base is modulated by subtle changes in local DNA shape rendered by the sequence context. The acquisition of a mutation further requires a failure of DNA repair, as evidenced by the high rate of cisplatin-induced mutations in xpf-1 deficiency and aflatoxin-induced mutations in NER deficiency.
Patterns of dinucleotide substitutions in C. elegans and human tumors
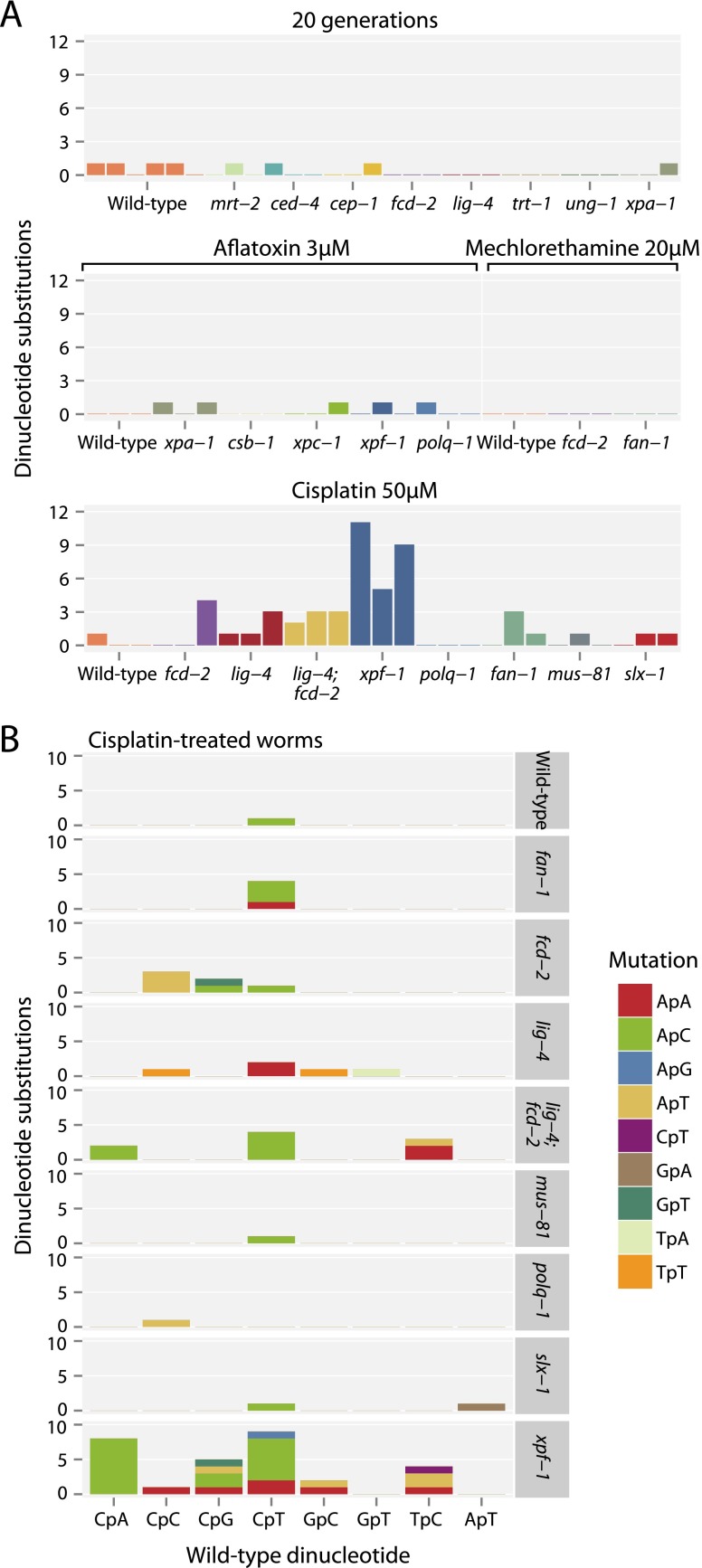
Dinucleotide substitutions, where two adjacent bases are mutated, were rare in both wild-type and DNA repair-deficient animals followed for 20 generations (Fig. 5A). Similarly, they were not seen to any great extent in worms treated with either aflatoxin or mechlorethamine (Fig. 5A). However, dinucleotide substitutions were substantially more frequent in C. elegans exposed to cisplatin, especially in the context of xpf-1, but also in fcd-2 and lig-4 (nonhomologous end-joining) deficiency. The signature of dinucleotide substitutions observed with cisplatin showed some specificity, both in which dinucleotides were mutated and what they mutated to (Fig. 5B). The most commonly mutated dinucleotide was CpT (equivalently, ApG), followed by CpA (TpG). These were most frequently transformed to ApC dinucleotides in the form of CT>AC or CA>AC mutations. The observed substitution patterns match the known chemistry of cisplatin, namely, that intrastrand crosslinks comprise 65% GpG and 25% ApG lesions (Lemaire et al. 1991). In our data, the former appear to result in G>T (equivalently, C>A) single nucleotide substitutions (that is, GG>TG or GG>GT), whereas the latter seem to cause dinucleotide substitutions (especially AG>GT).
Figure 5.
Patterns of dinucleotide substitutions observed. (A) Numbers of dinucleotide substitutions in animals propagated for 20 generations and those exposed to the different carcinogens. (B) Dinucleotide substitutions in cisplatin-exposed animals. The sequences of the dinucleotides that are mutated are on the x-axis and what they are mutated to is denoted by the color of the bar.
Overall, then, dinucleotide substitutions appear to be rather specific in their origin. They are rare occurrences spontaneously, even in the setting of the DNA repair deficiencies we analyzed. Moreover, of the three carcinogens studied, only cisplatin caused these events at appreciable rates. These data raise the hypothesis that intrastrand crosslinks, the most frequent lesion induced by cisplatin, may be especially associated with dinucleotide substitutions.
Signatures of dinucleotide substitutions have not been explicitly explored in human cancers. Given that such variants are rare spontaneous events, we explored whether any human tumors had specific signatures of dinucleotide substitution. We extracted dinucleotide substitutions observed in 7042 cancer samples, comprising 6535 exomes and 507 whole genomes across 30 tumor types (Alexandrov et al. 2013). Ranked by the overall frequency of dinucleotide mutations per Mb sequenced, the tumor types with the highest rates were melanoma, lung adenocarcinoma, lung squamous, small cell lung, and bladder cancers (Supplemental Fig. S4A). Like the C. elegans experiments, there was specificity across tumor types in both the dinucleotides mutated and what they mutated to (Supplemental Fig. S4B). In melanoma, the predominant pattern was CC>TT mutations, in keeping with ultraviolet light inducing intrastrand covalent bonds between adjacent cytosines, effectively an intrastrand crosslink. In the lung cancers, the predominant dinucleotide substitution was CC>AA (GG>TT). Acetaldehyde, one of the major chemical constituents of cigarette smoke, induces this exact dinucleotide mutation signature in vitro through the formation of intrastrand crosslinks at GpG (Matsuda et al. 1998). That it is a carcinogen acting at GpG dinucleotides in the lung cancers is further supported by a clear transcriptional strand bias—1214 GG>TT events were observed on the nontranscribed strand compared to 724 on the transcribed strand (P < 0.0001 for departure from a 50% split). This strand bias is indicative of the action of transcription-coupled repair, which utilizes the nucleotide-excision repair machinery (Bohr et al. 1985; Mellon et al. 1987; Mellon and Hanawalt 1989). Thus, human tumors with the highest rates of dinucleotide substitutions are those associated with clear mutagen exposures.
Signatures of small insertions and deletions
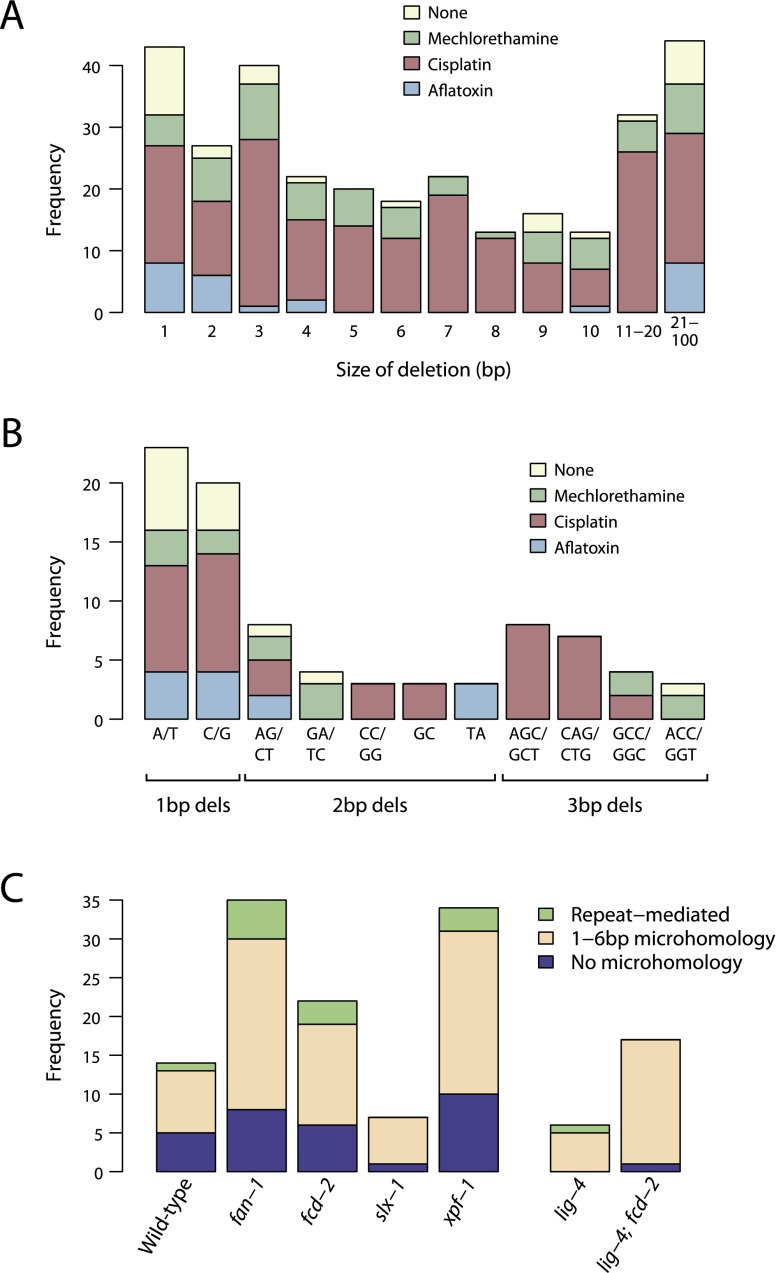
Both cisplatin and mechlorethamine treatment led to a dramatic increase in the number of small deletions compared to untreated animals (Fig. 3), and a change in their size distribution (Fig. 6A). Deletions in the absence of mutagen, or upon treatment with aflatoxin, tended to be 1–2 bp or >20 bp in size, whereas we observed a much more uniform distribution of sizes for mechlorethamine and cisplatin. As might be expected for the known chemistry of these latter two agents, the deletions tended to affect guanine-rich sequences (Fig. 6B).
Figure 6.
Patterns of indels observed. (A) Histogram of the size of deletions observed across the different experimental designs. (B) Histogram of the most frequently observed small deletions across the different experiments. (C) Distribution of repeat-mediated, microhomology-mediated, or blunt end-joining deletions by different genetic backgrounds. The difference between lig-4-deficient and other mutant backgrounds for the number of blunt end-joining deletions is significant (P = 0.01).
There were interesting differences across the various genetic backgrounds. Indel numbers were somewhat reduced in animals lacking the SLX-1 or MUS-81 nucleases involved in homologous recombination, the LIG-4 DNA end-joining ligase or the POLQ-1 translesion polymerase (Fig. 3). This suggests that these proteins might have a role in the generation of indels upon cisplatin exposure. This hypothesis is further supported by the observation that the extent of microhomology at the indel was reduced in lig-4 and lig-4; fcd-2 knockout lines (Fig. 6C). For these two lines, only one of 22 indels had flanking microhomology, compared to 30/100 for the other lines combined (P = 0.01). These data suggest that deletions with no detectable microhomology are generated via lig-4-dependent DNA end-joining. We also note that the number of complex rearrangements and indels is reduced in lig-4; fcd-2 double mutants compared to knockouts of fcd-2 alone. This finding provides molecular evidence that inappropriate end-joining of broken DNA strands occurring as a repair intermediate during interstrand crosslink excision contributes to chromosomal alterations. Our results corroborate previous findings demonstrating that the reduced survival of Fanconi anemia-deficient cells in response to crosslinking agents is suppressed by eliminating nonhomologous end-joining (Adamo et al. 2010; Pace et al. 2010). On the other hand, our results also suggest that NHEJ might help in sealing some ICL lesions, leading to a positive outcome of ICL repair.
Complex genomic rearrangements caused by interstrand crosslinks, fork stalling, and template switching
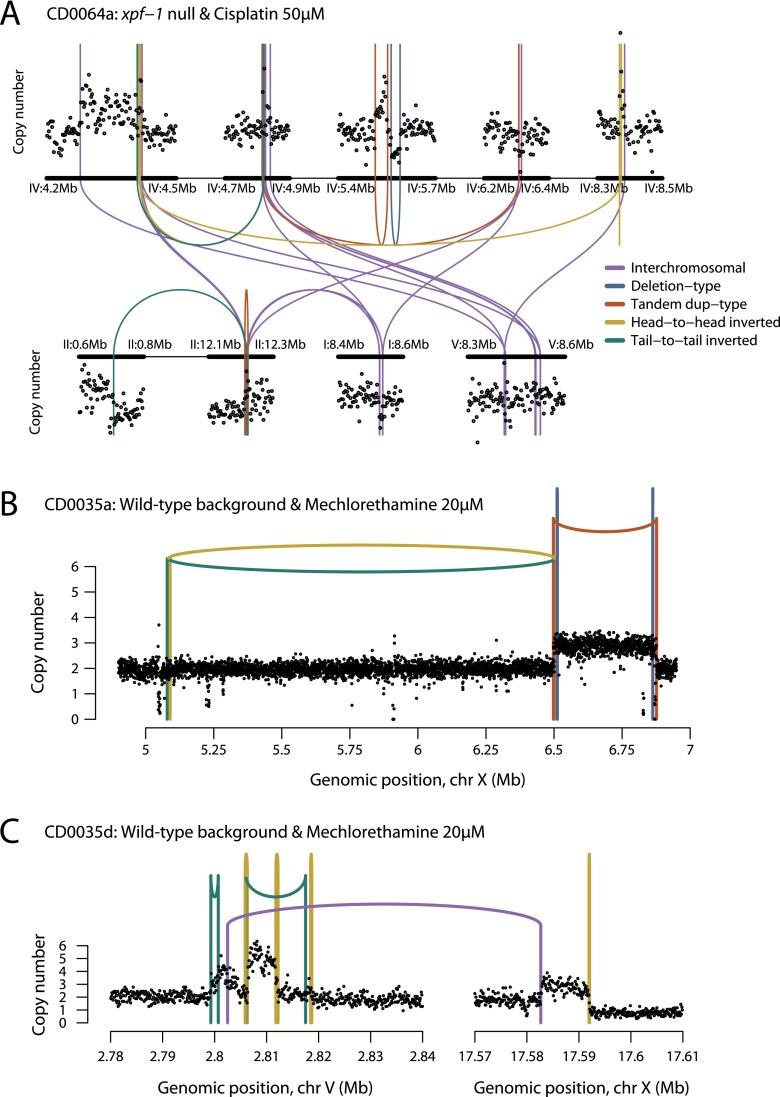
Among structural variants identified in these experiments, we found several examples of multiple rearrangements tightly clustered in one or a few genomic regions (Fig. 7; Supplemental Fig. S5). These occurred in C. elegans treated with mechlorethamine or cisplatin, but not in animals exposed to aflatoxin or followed for 20 generations without mutagen exposure. The clusters in carcinogen-treated worms differed from those described earlier in the mrt-2-deficient background (Fig. 1C–E) in that they occurred anywhere within a chromosome rather than being limited to telomeric regions. Interestingly, clusters occurred in animals with DNA repair defects such as xpf-1 (Fig. 7A) or fan-1 deficiency (Supplemental Fig. S5), but they clearly also arose in a wild-type background exposed to crosslinking agents (Fig. 7B,C). Thus, interstrand crosslinks appear to be strongly linked with this signature of clustered structural variants, possibly an outcome of blocked DNA repair intermediates, with the resulting genomic disruption worsened by defects in crosslink repair.
Figure 7.
Chromoanasynthesis in animals treated with crosslinking agents. (A) Copy-number profile and genomic rearrangements observed on chromosomes I, II, IV, and V for an xpf-1-deficient animal treated with cisplatin. (B) Copy-number profile and genomic rearrangements observed on chromosome X for a wild-type animal exposed to mechlorethamine. (C) Copy-number profile and genomic rearrangements observed on chromosomes V and X for another wild-type animal exposed to mechlorethamine.
Several genomic features of these events argue that the clustered rearrangements result from a replication-based mechanism, such as fork stalling and template switching, dubbed “chromoanasynthesis” (Liu et al. 2011b). First, by virtue of the experimental design, each cluster must have occurred in a single catastrophic event in the germ cell, since each replicate was only exposed transiently to the mutagen at a critical stage of the life cycle. Secondly, unlike chromothripsis, which is fundamentally a deletion mechanism (Korbel and Campbell 2013), the events observed here were associated with multiple regions of chromosomal gain that suggest a replicative process. For example, in one wild-type animal exposed to mechlorethamine, we saw two regions of copy number 3 and one of copy number 5 created by the rearrangements (Fig. 7C). Given that this occurred in a single germ cell, there must have been at least two rounds of over-replication in the same S phase to generate a copy number of 5. Thirdly, there is striking clustering of breakpoints within several regions even across different chromosomes, with multiple examples of rearrangements crossing back and forth between these regions (Fig. 7A). This pattern indicates that template switching can occur many times during the resolution of the chromosomal crisis, but is limited to discrete genomic regions. Fourthly, persistent interstrand DNA crosslinks conferred by mechlorethamine and cisplatin are known to impede the progress of DNA polymerases (Knipscheer et al. 2009), and hence place replication fork stalling and template switching at the center of this process.
We postulate that each region affected by clustered rearrangements might result from as little as one persistent DNA crosslink. The massive scale of genomic disruption in these regions suggests that the most cytotoxic effects of mechlorethamine and cisplatin derive from such unrepaired, persistent crosslinks. The number of complex rearrangements is highly variable across replicates in the cisplatin-treated animals (Figs. 3, 7). We expect that such rearrangements confer increased lethality. The fluctuation in progeny survival between single cisplatin-treated animals hints that complex rearrangements might be the most lethal lesions (Supplemental Fig. S6).
Replication fork stalling and template switching has been hypothesized to underpin some of the complex genomic rearrangements found in constitutional genomic disorders (Liu et al. 2011b). One study, for example, showed that 19% of individuals with chromosomal abnormalities had evidence for complexity of genomic rearrangements at the breakpoints (Chiang et al. 2012). In many cases, these complexes of structural variation are driven by clustered rearrangements with a striking resemblance to those described here, and cause severe developmental disorders (Liu et al. 2011b). Our data provide evidence from an experimental model for the proposed fork stalling and template switching mechanism, and identify persistent interstrand crosslinks as an important potential etiologic agent for these events. This is of particular interest given the recent report that endogenous crosslinking agents, such as alcohol-derived aldehydes, can cause genotoxic damage in mice (Langevin et al. 2011) and are associated with esophageal cancer (Brooks et al. 2009).
Discussion
We have used C. elegans as an experimental system to study the effects of carcinogens and DNA repair deficiencies on mutational signatures, and their interactions. Mutation profiles obtained from the three carcinogens differ in several key aspects, faithfully reflecting the known chemistry of these agents. Aflatoxin B1, associated with monoadducts on guanines, causes a mutational signature characterized by single base G>T and G>A substitutions, and few dinucleotide substitutions, indels, or structural variants. The substitutions are influenced by the local sequence context, and increase when genes critical to the nucleotide excision repair pathway are absent. Mechlorethamine, causing interstrand crosslinks, has a relatively modest effect on rates of single or double nucleotide substitutions, but is instead associated with a marked increase in small deletions and complex genomic rearrangements indicative of replication fork stalling and template switching. Finally, cisplatin, inducing a mixture of intrastrand crosslinks, interstrand crosslinks, and monoadducts, generates an equally complex mix of mutations, with increased rates of single base and dinucleotide substitutions, indels, and structural variants. Deficiencies in crosslink repair have variable consequences on the rates of these events, with XPF-1 being particularly critical to protection against the genotoxic effects of cisplatin. Intriguingly, increased expression of the XPF regulatory subunit ERCC1 is correlated with poor prognosis upon cisplatin treatment in human cancers (Dabholkar et al. 1994; Shirota et al. 2001; Lord et al. 2002; Ho and Scharer 2010), which chimes with our experimental findings.
The Environmental Protection Agency currently scores 240 chemicals for potential carcinogenicity, of which 11 and 72 are recorded as definite and probable human carcinogens, respectively (http://www.epa.gov/iris/). Of the 240 chemicals, 116 cannot be classified because available data are too limited. We believe genomics offers an effective approach to augment these analyses. Many (but not all) carcinogens act through increasing the mutation rate in exposed tissues. We have shown here and elsewhere (Pleasance et al. 2010a,b; Alexandrov et al. 2013) that mutational signatures associated with carcinogens have a high degree of specificity when assessed across the several dimensions of single base changes, dinucleotide substitutions, indels, and structural variants; effects of the local sequence context; and imprints of DNA repair such as transcriptional strand bias. In a genomics-focused approach, a candidate carcinogen could be assessed by sequencing human cancers with documented exposures coupled with parallel assessment in an experimental system such as the C. elegans model outlined here. Our model will allow the detection of specific profiles that may trigger early stages of carcinogenesis, but in tumors are later superseded by further signatures, possibly linked to acquired mutator phenotypes.
To ascertain the safety of new compounds is a major challenge of the pharmaceutical, cosmetic, and food industries. Current standard procedures for genotoxicity testing are rather limited. The widely used bacterial Ames assay is based on the reversion frequency of a single gene (Ames 1979). Alternative assays such as micronuclei counts or the comet assay only uncover large-scale genomic alterations (for review, see Fenech et al. 2011). Whole-genome sequencing of model organisms, both wild-type and DNA repair deficient, would significantly enlarge the repertoire of genotoxicity assays. C. elegans provides the advantage of being more closely related to human systems using DNA repair and apoptosis modalities not conserved in budding or fission yeast. Many carcinogens need to be metabolically activated by cytochrome P450 enzymes. A large number of these enzymes are conserved in nematodes, and members of the cytochrome P450 group I enzymes, which are missing in the nematode genome, could be expressed in C. elegans (Leung et al. 2010). Genome-wide assays will also be useful in the field of DNA repair. The direct measurement of DNA lesions has historically been restricted to locus-specific reporter assays. Defects associated with a given DNA repair pathway are conventionally assayed by showing decreased viability of the corresponding knockout when treated with genotoxic agents. However, decreased viability does not necessarily correlate with defects in genome integrity, and genome sequencing will provide a much more explicit description of the degree and nature of defects in genome maintenance. Next-generation sequencing provides an accurate measurement of fixed DNA damage; the nature of DNA damaging agents and how these are handled in intact or repair-deficient backgrounds affects mutation frequency and mutational profiles. Examining repair modalities, where DNA repair intermediates can be visualized as distinct repair foci, as can be done with the RAD-51 recombinase during homologous recombination, will allow for a more precise correlation of actual DNA damage, the efficiency of repair and the ultimate repair outcome at the level of the genome. Ultimately, though, the sequencing approach outlined here is correlative, not definitive—our hope is that coupling these large-scale screens with experimental systems such as C. elegans will generate hypotheses and guide experiments into the mechanistic basis for our observations.
With sequencing of thousands of cancer and normal genomes now complete, mutational signatures are being dissected and characterized at a pace and scale that evokes the revolution in taxonomy of species sparked by Linnaeus. Currently, however, the taxonomy of mutational signatures is too often Aristotelian, based on a genomic “morphology” rather than a deep understanding of the underlying causative mechanisms. As we develop systematic catalogs of mutation signatures associated with chemicals and DNA repair defects in experimental systems such as described here, we will increasingly be able to explain the etiology of human cancer genomes in mechanistic detail.
Methods
C. elegans strains and maintenance
Strains were grown at 20°C under standard conditions (Brenner 1974) unless indicated otherwise. The N2 Bristol reference line TG1813 is used in the Gartner laboratory as the wild-type reference strain. All mutant strains were backcrossed six times to TG1813. Strains are listed in Supplemental Table S1.
C. elegans propagation for mutation accumulation
To study the accumulation of mutations over several generations, starved F2 plates (first generation homozygous) from the sixth backcross were split onto two large plates and frozen for glycerol stocks and genomic DNA preparation (P0 generation) (Fig. 1A). In parallel, five single L4 stage animals were randomly chosen from these plates and singled each genotype (F1). After every 3–4 d, one single L4 stage animal was randomly chosen among the progeny per plate and propagated. This procedure was repeated until the F20 generation (Fig. 1A). Lines were frozen at generations P0, F5, F10, F15, and F20 for DNA isolation and glycerol stocks. Genomic DNA was sequenced from the P0 and F20 generation.
C. elegans intoxication
To assess drug sensitivity, 20–30 young adult hermaphrodites were packed into 2 mL of S basal + 5 μL concentrated OP50 bacteria in a 24-well plate. Aflatoxin B1, mechlorethamine hydrochloride, and cisplatin (all Sigma-Aldrich) solutions were freshly prepared and added to indicated concentrations. Animals were incubated under gentle shaking for 16 h at 20°C and then transferred to plates to recover. Progeny viability was scored 22–24 h post-treatment as described previously (Collis et al. 2006). Single offspring (derived from mutagenized male and female germ cells) from three individual adults of each genotype and carcinogen dose were expanded for sequencing.
Genomic DNA isolation
Animals were washed from 9-cm 3×NGM plates within 12 h post-starvation, washed three times 10 min in M9 buffer, and frozen in liquid nitrogen. Genomic DNA was isolated using ChargeSwitch gDNA Mini Tissue Kit (Invitrogen). DNA quality was determined by agarose gel electrophoresis and quantified using a Qubit Fluorometer (Invitrogen) or photometric OD260/OD280 measurement. Minimally, 5 μg DNA of each sample was used for whole-genome sequencing.
DNA sequencing
Genomic DNA was shotgun sequenced on an Illumina HiSeq 2000 platform in pools of five to six samples per lane. Library preparation followed manufacturer protocols and was sequenced at 100 bp paired-end.
Bioinformatic analysis
Raw sequencing reads were aligned to the reference C. elegans genome (build WS220/ce10) using the BWA algorithm (Li and Durbin 2009). For every “test” worm (that is, a worm exposed to carcinogen or followed for 20 generations), we identified a “control” worm (that is, one with the same genetic background, but not exposed or followed for 20 generations). Base substitutions, indels, genomic rearrangements, and copy-number changes acquired in the “test” worm, not found in the “control” worm, were called using a series of in-house algorithms described in detail elsewhere (Nik-Zainal et al. 2012a,b). From the raw output of these pipelines, we ran a series of post-processing filters developed specifically for this project, in order to produce a set of variant calls with high positive predictive value (see Supplemental Material for details).
Statistical analysis
To compare numbers of mutations across different genetic backgrounds, carcinogen doses, and types of mutations, we used generalized linear mixed effects models. Essentially, these models allow for correlations in the numbers of mutations of each different type within a given worm while enabling formal hypothesis testing (likelihood ratio tests) of questions of interest. To compare differences in sequence context, χ2 tests were used. To estimate the mutation rate per generation per genome in the 20-generation experiment, we used the maximum likelihood estimate from the observed number of mutations, assuming that with each generation a heterozygous mutation has a 25% chance of becoming fixed and homozygous, a 50% chance of remaining heterozygous, and a 25% chance of being lost.
Data access
Sequencing data generated for this study have been submitted to the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena/) under study accession number ERP000975.
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust (grant reference 077012/Z/05/Z). P.J.C. and A.G. are personally funded through Wellcome Trust Senior Clinical Research Fellowships and Senior Basic Research Fellowships, respectively, and are members of the Wellcome funded COMSIG consortium. We thank the Japanese National BioResource Project for the Nematode C. elegans, the Caenorhabditis Genetics Center for providing strains, and John Rouse and Karim Labib for comments on the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.175547.114.
Freely available online through the Genome Research Open Access option.
References
- Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR, et al. 1993. Clues to the pathogenesis of familial colorectal cancer. Science 260: 812–816 [DOI] [PubMed] [Google Scholar]
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A. 2010. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell 39: 25–35 [DOI] [PubMed] [Google Scholar]
- Ahmed S, Hodgkin J. 2000. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 403: 159–164 [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN. 1979. Identifying environmental chemicals causing mutations and cancer. Science 204: 587–593 [DOI] [PubMed] [Google Scholar]
- Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, Macdonald TY, Ghandi M, et al. 2013. Punctuated evolution of prostate cancer genomes. Cell 153: 666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besaratinia A, Kim SI, Hainaut P, Pfeifer GP. 2009. In vitro recapitulating of TP53 mutagenesis in hepatocellular carcinoma associated with dietary aflatoxin B1 exposure. Gastroenterology 137: 1127–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. 1985. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 40: 359–369 [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks PJ, Enoch MA, Goldman D, Li TK, Yokoyama A. 2009. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med 6: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Temiz NA, Harris RS. 2013. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 45: 977–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Deletion Mutant Consortium . 2012. Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda) 2: 1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium . 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018 [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, et al. 2010. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467: 1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung I, Schertzer M, Rose A, Lansdorp PM. 2002. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat Genet 31: 405–409 [DOI] [PubMed] [Google Scholar]
- Chiang C, Jacobsen JC, Ernst C, Hanscom C, Heilbut A, Blumenthal I, Mills RE, Kirby A, Lindgren AM, Rudiger SR, et al. 2012. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet 44: 390–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collis SJ, Barber LJ, Ward JD, Martin JS, Boulton SJ. 2006. C. elegans FANCD2 responds to replication stress and functions in interstrand cross-link repair. DNA Repair 5: 1398–1406 [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482: 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabholkar M, Vionnet J, Bostick-Bruton F, Yu JJ, Reed E. 1994. Messenger RNA levels of XPAC and ERCC1 in ovarian cancer tissue correlate with response to platinum-based chemotherapy. J Clin Invest 94: 703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, Chabes A, Malkova A. 2011. Break-induced replication is highly inaccurate. PLoS Biol 9: e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Feinberg S, Steding C, Durbin M, Lynch M. 2006. The relative roles of three DNA repair pathways in preventing Caenorhabditis elegans mutation accumulation. Genetics 174: 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Dolan PC, Wilhelm LJ, Sung W, Lucas-Lledo JI, Howe DK, Lewis SC, Okamoto K, Thomas WK, Lynch M, et al. 2009. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc Natl Acad Sci 106: 16310–16314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J, Norppa H, Eastmond DA, Tucker JD, Thomas P. 2011. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis 26: 125–132 [DOI] [PubMed] [Google Scholar]
- Flibotte S, Edgley ML, Chaudhry I, Taylor J, Neil SE, Rogula A, Zapf R, Hirst M, Butterfield Y, Jones SJ, et al. 2010. Whole-genome profiling of mutagenesis in Caenorhabditis elegans. Genetics 185: 431–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. 2000. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol Cell 5: 435–443 [DOI] [PubMed] [Google Scholar]
- Greenman CD, Pleasance ED, Newman S, Yang F, Fu B, Nik-Zainal S, Jones D, Lau KW, Carter N, Edwards PA, et al. 2012. Estimation of rearrangement phylogeny for cancer genomes. Genome Res 22: 346–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J, Lowden M, Clejan I, Tzoneva M, Thomas JH, Hodgkin J, Ahmed S. 2006. Mutator phenotype of Caenorhabditis elegans DNA damage checkpoint mutants. Genetics 174: 601–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. 2013. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154: 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks WM, Kim M, Haber JE. 2010. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science 329: 82–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TV, Scharer OD. 2010. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ Mol Mutagen 51: 552–566 [DOI] [PubMed] [Google Scholar]
- Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. 1993. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 363: 558–561 [DOI] [PubMed] [Google Scholar]
- Kensler TW, Roebuck BD, Wogan GN, Groopman JD. 2011. Aflatoxin: a 50-year odyssey of mechanistic and translational toxicology. Toxicol Sci 120: S28–S48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. 2009. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326: 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Wong WS, et al. 2012. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488: 471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koole W, van Schendel R, Karambelas AE, van Heteren JT, Okihara KL, Tijsterman M. 2014. A polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat Commun 5: 3216. [DOI] [PubMed] [Google Scholar]
- Korbel JO, Campbell PJ. 2013. Criteria for inference of chromothripsis in cancer genomes. Cell 152: 1226–1236 [DOI] [PubMed] [Google Scholar]
- Kruisselbrink E, Guryev V, Brouwer K, Pontier DB, Cuppen E, Tijsterman M. 2008. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr Biol 18: 900–905 [DOI] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. 2011. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475: 53–58 [DOI] [PubMed] [Google Scholar]
- Lemaire MA, Schwartz A, Rahmouni AR, Leng M. 1991. Interstrand cross-links are preferentially formed at the d(GC) sites in the reaction between cis-diamminedichloroplatinum(II) and DNA. Proc Natl Acad Sci 88: 1982–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung MC, Goldstone JV, Boyd WA, Freedman JH, Meyer JN. 2010. Caenorhabditis elegans generates biologically relevant levels of genotoxic metabolites from aflatoxin B1 but not benzo[a]pyrene in vivo. Toxicol Sci 118: 444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Bi W, Carvalho CM, Simmons AD, Wiszniewska J, Fang P, Eng PA, Cooper ML, et al. 2011a. Copy number gain at Xp22.31 includes complex duplication rearrangements and recurrent triplications. Hum Mol Genet 20: 1975–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. 2011b. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord RV, Brabender J, Gandara D, Alberola V, Camps C, Domine M, Cardenal F, Sanchez JM, Gumerlock PH, Taron M, et al. 2002. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res 8: 2286–2291 [PubMed] [Google Scholar]
- Lowden MR, Meier B, Lee TW, Hall J, Ahmed S. 2008. End joining at Caenorhabditis elegans telomeres. Genetics 180: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Kawanishi M, Yagi T, Matsui S, Takebe H. 1998. Specific tandem GG to TT base substitutions induced by acetaldehyde are due to intra-strand crosslinks between adjacent guanine bases. Nucleic Acids Res 26: 1769–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride DJ, Etemadmoghadam D, Cooke SL, Alsop K, George J, Butler A, Cho J, Galappaththige D, Greenman C, Howarth KD, et al. 2012. Tandem duplication of chromosomal segments is common in ovarian and breast cancer genomes. J Pathol 227: 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1941. The stability of broken ends of chromosomes in Zea mays. Genetics 26: 234–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier B, Clejan I, Liu Y, Lowden M, Gartner A, Hodgkin J, Ahmed S. 2006. trt-1 is the Caenorhabditis elegans catalytic subunit of telomerase. PLoS Genet 2: e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I, Hanawalt PC. 1989. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature 342: 95–98 [DOI] [PubMed] [Google Scholar]
- Mellon I, Spivak G, Hanawalt PC. 1987. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51: 241–249 [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. 2012a. Mutational processes molding the genomes of 21 breast cancers. Cell 149: 979–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, et al. 2012b. The life history of 21 breast cancers. Cell 149: 994–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. 2010. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science 329: 219–223 [DOI] [PubMed] [Google Scholar]
- Pampalona J, Soler D, Genesca A, Tusell L. 2010. Telomere dysfunction and chromosome structure modulate the contribution of individual chromosomes in abnormal nuclear morphologies. Mutat Res 683: 16–22 [DOI] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. 2007. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 28: 622–629 [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. 2002. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene 21: 7435–7451 [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, You YH, Besaratinia A. 2005. Mutations induced by ultraviolet light. Mutat Res 571: 19–31 [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. 2010a. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. 2010b. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk LF, Shuker DE. 1994. DNA damage and mutagenesis induced by nitrogen mustards. Mutat Res 318: 205–226 [DOI] [PubMed] [Google Scholar]
- Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, et al. 2012. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell 46: 424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. 2013. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 45: 970–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roerink SF, van Schendel R, Tijsterman M. 2014. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res 24: 954–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth RE, Cuddeford C, Baillie DL. 1985. Mutagenesis in Caenorhabditis elegans. II. A spectrum of mutational events induced with 1500 r of γ-radiation. Genetics 109: 493–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serero A, Jubin C, Loeillet S, Legoix-Ne P, Nicolas AG. 2014. Mutational landscape of yeast mutator strains. Proc Natl Acad Sci 111: 1897–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirota Y, Stoehlmacher J, Brabender J, Xiong YP, Uetake H, Danenberg KD, Groshen S, Tsao-Wei DD, Danenberg PV, Lenz HJ. 2001. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J Clin Oncol 19: 4298–4304 [DOI] [PubMed] [Google Scholar]
- Smela ME, Currier SS, Bailey EA, Essigmann JM. 2001. The chemistry and biology of aflatoxin B1: from mutational spectrometry to carcinogenesis. Carcinogenesis 22: 535–545 [DOI] [PubMed] [Google Scholar]
- Smith CE, Llorente B, Symington LS. 2007. Template switching during break-induced replication. Nature 447: 102–105 [DOI] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. 2009. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. 2009. The cancer genome. Nature 458: 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100: 64–119 [DOI] [PubMed] [Google Scholar]
- Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. 2013. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife 2: e00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau SN, Bren G, Schaid D. 1993. Microsatellite instability in cancer of the proximal colon. Science 260: 816–819 [DOI] [PubMed] [Google Scholar]
- Thompson O, Edgley M, Strasbourger P, Flibotte S, Ewing B, Adair R, Au V, Chaudry I, Fernando L, Hutter H, et al. 2013. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res 23: 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters TR, Swann PF. 2000. Thymine-DNA glycosylase and G to A transition mutations at CpG sites. Mutat Res 462: 137–147 [DOI] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, et al. 2012. The origin and evolution of mutations in acute myeloid leukemia. Cell 150: 264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Li S, Stohr BA. 2013. The role of telomere biology in cancer. Annu Rev Pathol 8: 49–78 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tarailo-Graovac M, O’Neil NJ, Rose AM. 2008. Spectrum of mutational events in the absence of DOG-1/FANCJ in Caenorhabditis elegans. DNA Repair 7: 1846–1854 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.