Abstract
The chemotactic sensory system of Escherichia coli comprises membrane-embedded chemoreceptors and six soluble chemotaxis (Che) proteins. These components form signaling complexes that mediate sensory excitation and adaptation. Previous determinations of cellular content of individual components provided differing and apparently conflicting values. We used quantitative immunoblotting to perform comprehensive determinations of cellular amounts of all components in two E. coli strains considered wild type for chemotaxis, grown in rich and minimal media. Cellular amounts varied up to 10-fold, but ratios between proteins varied no more than 30%. Thus, cellular stoichiometries were almost constant as amounts varied substantially. Calculations using those cellular stoichiometries and values for in vivo proportions of core components in complexes yielded an in vivo stoichiometry for core complexes of 3.4 receptor dimers and 1.6 CheW monomers for each CheA dimer and 2.4 CheY, 0.5 CheZ dimers, 0.08 CheB, and 0.05 CheR per complex. The values suggest a core unit of a trimer of chemoreceptor dimers, a dimer (or two monomers) of kinase CheA, and two CheW. These components may interact in extended arrays and, thus, stoichiometries could be nonintegral. In any case, cellular stoichiometries indicate that CheY could be bound to all signaling complexes and this binding would recruit essentially the entire cellular complement of unphosphorylated CheY, and also that phosphatase CheZ, methylesterase CheB, and methyltransferase CheR would be present at 1 per 2, per 14, and per 20 core complexes, respectively. These characteristic ratios will be important in quantitative treatments of chemotaxis, both experimental and theoretical.
Motile bacterial cells respond to chemical gradients by moving toward favorable environments. The phenomenon, termed chemotaxis, is mediated by a set of modular components found across the taxonomic diversity of prokaryotes. The molecular and mechanistic basis of chemotaxis has been most extensively defined in Escherichia coli. In this species, the chemotactic sensory system consists of five related, membrane-embedded chemoreceptors and six different soluble proteins, the Che proteins (see reference 15 for an overview). These components interact, forming signaling complexes that mediate sensory excitation and adaptation. Some components interact to create a stable core complex, and others interact more weakly with this core. How many copies of the components of the signaling complex are contained in a cell? Several studies have reported values for one or a few of these proteins. However, as documented in an informative website (www.anat.cam.ac.uk/comp-cell), published values for a given protein differ, sometimes by as much as 10-fold. The origin and significance of this variation has been unknown and, thus, there has been no basis for choosing from among the different values. In addition, the stoichiometry of signaling complexes is an area of active investigation, and independent investigators have come to different conclusions (2, 13, 27, 36). Again, the basis of these differences has been unclear. Yet an understanding of the complexities of signaling, amplification, and adaptation in chemotaxis will require a clear definition of the stoichiometry of signaling complexes.
The core signaling complex consists of a ternary complex of chemoreceptors, the chemotaxis-specific, autophosphorylating histidine kinase CheA, and an SH3-related coupling protein, CheW. This core complex is stable over durations relevant to sensory excitation and adaptation (13). Other Che proteins bind to the core complex components, with apparent dissociation constants in the micromolar range. Two response regulators, CheY and CheB, bind to the P2 domain of CheA (26) and interact with the P1 domain of CheA to capture a phosphoryl group carried on a histidinyl side chain. Phospho-CheY is inherently unstable, undergoing hydrolysis to release the phosphate with a half time of a few seconds (20). Hydrolysis is substantially enhanced by CheZ (20), a protein that binds to phospho-CheY (50). CheZ also binds to CheAS (46), the “short” form of kinase CheA that results from an alternative translational start site in E. coli approximately 90 codons interior to the start site that generates the “long” form, CheAL (24). CheAS lacks the phosphorylated histidine, but truncation results in a binding site for CheZ not available in the intact protein. Receptors are methylated and demethylated at specific methyl-accepting glutamates in reactions that are crucial to sensory adaptation and catalyzed by methyltransferase CheR and methylesterase-deamidase CheB. Each enzyme interacts at multiple substrate sites on each receptor monomer, the four to six methyl-accepting glutamyl residues in their unmodified and methylester-amide forms, respectively. In addition, both enzymes interact with a specific pentapeptide sequence present at the extreme carboxyl terminus of high-abundance receptors (5, 49). In E. coli, the high-abundance chemoreceptors Tsr and Tar are present at higher dosages than the three low-abundance receptors, Trg, Tap, and Aer, a difference commonly estimated as ∼10-fold (17, 18, 45, 48).
To determine the cellular content and stoichiometry of the components of the signaling complex, we used immunoblotting to quantify amounts of chemoreceptors and Che proteins.
MATERIALS AND METHODS
Bacterial strains and plasmids.
OW1 (29) and RP437 (31, 32) are strains of E. coli K-12 wild type for chemotaxis. RP3098 (38) and RP2867 (32) are derivatives of RP437 that lack, respectively, all chemoreceptors and Che proteins or CheR and CheB. PCR mutagenesis was used to create forms of trg, tsr, and tar contained in plasmids pAL1 (12), pCT1 (12), and pNT201 (8), respectively, that coded for receptors Trg-6H, Tsr-6H, and Tar-6H carrying six histidines at their carboxyl termini. Plasmid pJB100 carries a truncated form of trg under the control of the T7 promoter that codes for Trg-pd, a methionine followed by residues 52 through 198, i.e., the periplasmic domain.
Purified proteins.
Purified CheZ was a gift from Birgit Scharf (Universität Regensburg). Purification of CheA, CheW, and CheY followed the method described by Barnakov et al. (4). CheR and CheB were purified using a pentapeptide affinity column (5). Histidine-tagged receptors were purified using a Ni2+-nitrilotriacetic acid-agarose column (QIAGEN). A. N. Barnakov provided Trg-pd purified by solubilizing washed inclusion bodies with urea and renaturing by dialysis.
Concentrations of proteins used as immunoblotting standards were determined by quantitative amino acid analysis, using means of determinations for 7 to 10 amino acids, adjusted for the yield of the specific run determined by recovery of an internal standard, ornithine. Yields were 90 to 111% of an ornithine-alone sample. Standard deviations for mean values of different quantifications of a specific protein derived from determinations of individual amino acids were 3 to 10%, with the exception of 16% for Trg-6H. Concentrations of the protein standards were adjusted for the proportion of intact protein determined by densitometry of overloaded gels. This proportion was ≥87% for Che proteins and 59 to 85% for chemoreceptors.
Antisera and antibodies.
Covance Research Products (Richmond, Calif.) raised polyclonal antisera to CheA, CheW, Tar-6H, and Trg-pd in rabbits, using purified proteins that we provided. Generous colleagues gave us mouse monoclonal anti-CheY (Birgit Scharf, Universität Regensburg), rabbit polyclonal anti-CheB and anti-CheR (Ann Stock, University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School and Howard Hughes Medical Institute), and rabbit polyclonal anti-CheZ (Philip Matsumura, University of Illinois-Chicago). To reduce nonspecific background in immunoblots, some antisera were adsorbed with acetone powder (14) of a culture of RP3098 or, for anti-Trg-pd, of CP177 (30) by diluting antiserum 1:10 in 20 mM Tris-HCl (pH 7.5), 500 mM NaCl, adding acetone powder to ∼1% (wt/vol), incubating at room temperature 20 to 30 min, and spinning for 5 min at 16,000 × g in a table-top centrifuge. Secondary antibodies were goat anti-rabbit immunoglobulin G (heavy and light) horseradish peroxidase conjugate (Bio-Rad) diluted 1:2,000 or 1:4,000 for colorimetric or chemiluminescent detection, respectively, and goat anti-mouse immunoglobulin G (heavy and light) horseradish peroxidase conjugate (Molecular Probes, Eugene, Oreg.) diluted 1:3,000 from a 1-mg/ml stock for chemiluminescence detection.
Cell growth.
Cultures were inoculated to an optical density at 560 nm (OD560) of 0.05 (∼3.75 × 107 cells/ml) with highly motile cells grown in the same medium to an OD560 of ∼0.5 and grown at 35°C with good aeration. At OD560 = 0.5, cells were harvested by adding 0.2 ml of 50% (wt/wt) ice-cold trichloroacetic acid (TCA) to 1-ml samples, incubating 15 min on ice, centrifuging for 15 min at 16,000 × g in a table-top centrifuge at 4°C, adding 1 ml of ice-cold acetone to the pellet, centrifuging as before, air drying the pellet, and storing at −20°C. TCA precipitation was important to avoid proteolysis of chemoreceptors. In a control experiment, we compared the amount of protein detected in samples harvested by TCA precipitation to parallel samples harvested by centrifugation, washing, and solubilization of cells in sodium dodecyl sulfate (SDS) electrophoresis sample buffer. For Che proteins, values determined by direct sampling were within 9% of values determined by TCA precipitation. For Tsr, Tar, and Trg, direct sampling values were ∼25% lower than for TCA precipitation, presumably because of proteolysis during sample preparation. Thus, we used TCA precipitation for all samples. Tryptone broth (TB) contained 1% Difco Bacto-Tryptone (Becton Dickinson and Company, Sparks, Md.), and 0.4% NaCl, pH 7.0 (with NaOH), and H1 minimal salts medium (19) contained 50 mM potassium phosphate (pH 7.0), 7.6 mM (NH4)2SO4, 0.5 mM MgSO4, 1.25 μM Fe2(SO4)3, 0.01% vitamin B1, and 0.4% ribose and required 1 mM amino acids (histidine, leucine, and threonine for OW1 and those plus methionine for the other strains).
For each strain and growth condition, we determined the concentration of cellular protein at the optical density of harvest, OD560 = 0.5, by the bicinchoninic acid assay (Pierce, Rockford, Ill.) with bovine serum albumin as the standard, and the concentration of cells was determined by plating dilutions on Luria-Bertani agar plates. Protein concentrations, in micrograms of protein per milliliter of culture, were as follows: RP437 in TB, 150; RP437 in H1, 120; OW1 in TB, 140; OW1 in H1, 125. Since the amount of protein is a measure of cell volume (10), these values indicated only modest differences in cell volume per milliliter as strain and growth conditions varied. However, cell concentrations expressed as 108 cells/ml were as follows: RP437 in TB, 2.9; RP437 in H1, 5.35; OW1 in TB, 4.1; OW1 in H1, 2.9. These findings implied a greater percent variation in cell size. This was confirmed under a microscope: RP437 cells were significantly smaller in H1 cultures than in TB cultures, and OW1 were smaller in TB cultures than in H1 cultures. Thus, the most informative parameter was not molecules per cell, but molecules per cell volume, measured by determining total cellular protein. Yet, expressing cellular amounts in units of molecules per total cellular protein would not have provided ready comparison to literature values of molecules per cell. Thus, we defined the TB-grown RP437 value, 0.51 pg of total protein/cell, as a standard cell volume and have expressed cellular contents as molecules per standard cell volume.
Quantitative immunoblotting.
SDS electrophoresis sample buffer was added to tubes containing TCA-precipitated cells and boiled for 5 min. Solubilized samples were submitted to SDS-polyacrylamide gel electrophoresis and immunoblotting, and the intensity of bands was converted to the protein amount by using a calibration curve of pure protein present on each blot. Each 0.5-mm gel was loaded with nine different dilutions of a protein standard (usually comprising a ninefold concentration range in equal steps), three serial dilutions of three experimental samples (usually representing independent cultures of the same strain in the same growth medium and adjusted after pilot runs for amounts that would fall within the range of the standard), and a negative control (a sample of cells lacking the protein of interest at a dilution equivalent to the most concentrated of experimental samples). Both standards and experimental samples were adjusted to contain the same amount of total cellular protein by addition of solubilized, TCA-precipitated RP3098 or CP177.
Each protein was quantified using gels with a polyacrylamide concentration that provided a well-focused band, as follows: CheA and chemoreceptors, 12%; CheB, CheR, and CheZ, 13%; CheW, 14%; CheY, a gradient from 12 to 18%. Proteins were transferred electrophoretically from gels to 0.45-μm-pore-size nitrocellulose (Hybond ECL; Amersham Pharmacia Biotech) for 2 h at 12 V. Nitrocellulose sheets were immersed in 2.5% instant nonfat powdered milk in 20 mM Tris (pH 7.0) and 0.5 M NaCl (PM-TBS) and placed on a rocking platform at room temperature for ≥1 h, immersed in a dilution of primary antiserum in PM-TBS, placed on a rocking platform at room temperature overnight, washed with TBS-Tween (0.05% [vol/vol]) three times for 5 min (colorimetric detection) or four times for 15 min (chemiluminescent detection), incubated for 2 h with a secondary antibody, washed as above, and immersed for 30 min in a solution of 4-chloro-1-naphthol created by dissolving 15 mg in 5 ml of methanol, mixing with 25 ml of TBS, and adding 15 μl of 30% H2O2 immediately before use (colorimetric detection) or treated with ECL Western blotting detection reagents (Amersham Pharmacia Biotech) according to the manufacturer's protocol and exposed to Kodak scientific imaging film (chemiluminescent detection). CheA, CheW, and chemoreceptors were quantified by colorimetric detection, CheB, CheR, and CheY were quantified by chemiluminescent detection (ECL Western blotting detection reagents; Amersham Pharmacia Biotech), and CheZ was quantified by both. Colorimetric and chemiluminescent images were captured by using a digital camera, and intensities were quantified using TotalLab software (version 2.01; Nonlinear Dynamics Ltd.). CheAL and CheAS were quantified separately, and the values were summed to determine total CheA. Antisera varied in avidity and degree of specificity, and thus the following dilutions, preabsorptions with acetone powders, and amounts of protein standards were used: anti-CheA, 1:5,000, 2 preabsorptions, 0.1 to 0.9 ng standard; anti-CheB, 1:500, 3, 0.1 to 0.9 ng; anti-CheR, 1:500, 3, 0.2 to 1.8 ng; anti-CheW, 1:50, 0, 1 to 9 ng; anti-CheY, 1:1,250 (0.01 mg/ml), 0, 0.5 to 4.5 ng; anti-CheZ, 1:250 (colorimetric) or 1:500 (chemiluminescent), 0, 0.5 to 4.5 ng (colorimetric) or 0.1 to 0.9 ng (chemiluminescent); anti-Tar, 1:2,500, 0, 0.5 to 4.5 ng (Tar-6H or Tsr-6H); anti-Trg-pd, 1:40, 5, 1 to 8 ng Trg-6H.
High-abundance receptors Tsr and Tar were quantified together (Fig. 1C), using an anti-Tar serum that reacted essentially equally with the two proteins (Fig. 1E). The low-abundance receptor Trg was quantified (Fig. 1D) with a Trg-specific antiserum raised to its periplasmic domain, the sequence of which is essentially unrelated to other receptors (7). With the aim of quantifying Tsr and Tar individually, we used RP2867, a derivative of RP437 lacking CheR and CheB (32) and thus producing only one electrophoretic form of each receptor (Fig. 1E). However, in this strain Tsr/Tar ratios were close to 1:1 (56:44 in TB and 52:48 in H1), values inconsistent with many published observations indicating ∼2:1 ratios in wild-type cells (e.g., references 11, 16, 39, and 42). Also, the cellular contents of high-abundance receptors, CheA and CheW in RP2867, were all 25 to 50% lower than in the parent RP437 (see Fig. 2). We surmise that insertion and excision of a lambda transducing phage that created RP2867 from RP437 (32) altered expression of chemosensory genes and may well have altered the cellular ratio of Tsr to Tar. Thus, we have not used Tsr/Tar ratios determined in RP2867 to deduce the ratio in wild-type cells.
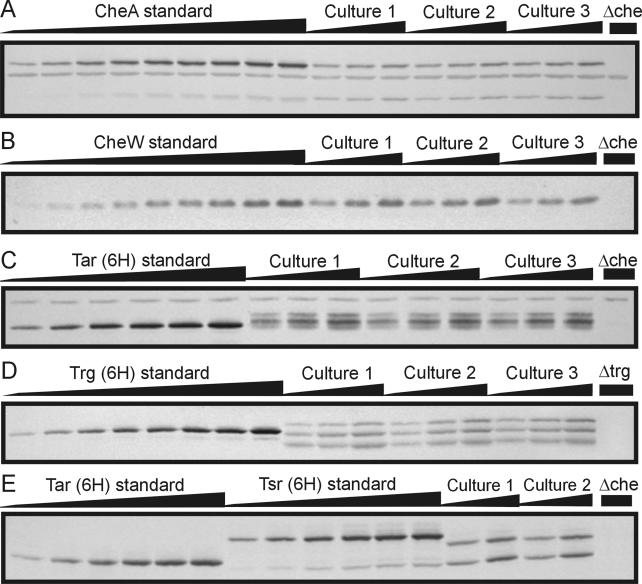
FIG. 1.
Quantitative immunoblots. The figure shows segments of immunoblots designed to quantify the cellular content of CheA (A), CheW (B), high-abundance receptors (C and E), or Trg (D). Samples were adjusted to contain the same amount of total cellular material by addition of cellular material from a strain missing all chemotaxis components or Trg (D). This accounts for nonspecific, cross-reacting bands in all lanes of panels A and C. In panel A, CheAL and CheAS are the upper and lower bands, respectively. In panels C and D, experimental samples exhibit multiple bands because of adaptational covalent modification of chemoreceptors. In panel E, Tar and Tsr standards migrated slightly slower than Tar and Tsr from experimental samples because the standards had six appended carboxyl-terminal histidines.
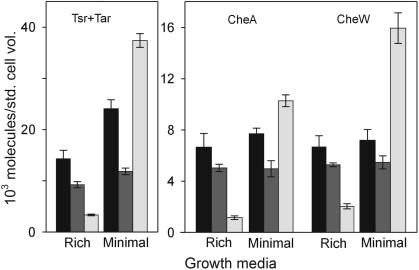
FIG. 2.
Cellular content of high-abundance receptors, CheA and CheW, in three strains grown in two media. Values are for RP437 (black bars), RP2867 (dark gray bars), and OW1 (light gray bars), grown in TB (rich) or minimal salts medium (minimal). Error bars are standard deviations for mean values from three independent cultures, tested in at least two (RP2867) or three separate assays.
RESULTS
Quantification of cellular content.
We used quantitative immunoblotting to determine the cellular content of chemoreceptors and Che proteins in two strains of E. coli K-12 considered wild type for chemotaxis (Table 1). These strains, OW1 (29) and RP437 (31, 32), have been utilized as chemotactically wild type in many studies for over 25 years. Some studies used cells grown in rich medium, most commonly TB, and others used cells grown in a minimal salts medium, often H1 salts plus a source of carbon and energy. Thus, we examined the two strains grown in two different media, TB and H1 salts plus ribose and required amino acids. For each combination of strain and growth medium, determinations were done on three independent, highly motile cultures actively growing in mid-exponential phase. Multiple samples from these cultures were harvested directly into TCA, washed, and stored at −20°C for later analysis.
TABLE 1.
Cellular contents of chemotaxis components
| Component | Content (no. of molecules/standard cell vol)a in strain:
|
|||||
|---|---|---|---|---|---|---|
| RP437
|
OW1
|
RP2867 (ΔcheR cheB)
|
||||
| Rich medium | Minimal medium | Rich medium | Minimal medium | Rich medium | Minimal medium | |
| Receptors (total)b | 15,000 ± 1,700 | 26,000 ± 1,800 | 3,600 ± 170 | 41,000 ± 1,400 | ||
| Tsr + Tar | 14,000 ± 1,700 | 24,000 ± 1,800 | 3,300 ± 170 | 37,000 ± 1,300 | 9,200 ± 600c | 12,000 ± 660c |
| Trg | 440 ± 70 | 910 ± 20 | 150 ± 20 | 1,800 ± 200 | ||
| CheA (total) | 6,700 ± 1,100 | 7,700 ± 440 | 1,100 ± 130 | 10,000 ± 460 | 5,100 ± 290 | 5,000 ± 530 |
| CheAL | 4,500 ± 940 | 5,100 ± 380 | 810 ± 110 | 6,700 ± 340 | 3,200 ± 190 | 3,300 ± 450 |
| CheAS | 2,200 ± 520 | 2,600 ± 230 | 340 ± 90 | 3,600 ± 310 | 1,900 ± 220 | 1,700 ± 280 |
| CheW | 6,700 ± 890 | 7,200 ± 830 | 2,000 ± 200 | 16,000 ± 1,200 | 5,300 ± 150 | 5,500 ± 510 |
| CheY | 8,200 ± 310 | 6,300 ± 70 | 1,400 ± 90 | 14,000 ± 1,400 | ||
| CheZ | 3,200 ± 90 | 2,700 ± 80 | 560 ± 90 | 5,700 ± 130 | ||
| CheB | 240 ± 10 | 270 ± 10 | 50 ± 10 | 440 ± 20 | ||
| CheR | 140 ± 10 | 160 ± 10 | 40 ± 10 | 320 ± 10 | ||
Means ± standard deviations for three independent cultures analyzed four to six times, except for Tsr and Tar in RP2867 (two cultures).
Sum of the experimental measurements for Tsr + Tar and Trg and an estimate for Tap and Aer. See text for details.
Sum of separate values for Tsr and Tar. Tsr rich, 5,200 ± 520; minimal, 6,100 ± 510. Tar rich, 4,000 ± 290; minimal, 5,800 ± 410.
Each quantitative immunoblot assay included a standard curve of pure protein and samples of the cultures being tested (Fig. 1). To avoid effects of differing amounts of total cell extract on intensities of immunoblot reactions (34), all samples on each particular immunoblot, both experimental and protein standards, were adjusted to contain the same amount of total cellular protein by using an extract of cells that did not contain the component being quantified. The amount of a protein was calculated using at least two and often three points that fell within the segment of the standard curve for which intensity was a linear function of protein amount. Concentration of protein standards was determined by quantitative amino acid analysis.
To quantify each Che protein, we performed separate immunoblot assays specific for that protein. To quantify the sum of Tsr and Tar, we used an antiserum that was raised to Tar but recognized the two high-abundance chemoreceptors with essentially the same efficiency (Fig. 1E). Under immunoblotting conditions that yielded a linear relation between intensity and amount of the high-abundance receptors, low-abundance receptors were not detected. However, it was possible to quantify specifically the low-abundance receptor Trg in samples containing all chemoreceptors by using a Trg-specific antiserum, raised to the periplasmic domain of that receptor, that did not react with the other receptors (Fig. 1D). Receptor-specific antisera were not available for the other low-abundance receptors, Tap and Aer. However, a measurement for Aer cited in a review (45) and the intensity on a two-dimensional gel of radiolabeled protein likely to be Tap (18) indicate that these two receptors are present at levels no higher, and likely lower, than that of Trg. To include these low-abundance receptors into our values for cellular content of chemoreceptors, we estimated that the content of the two together would be approximately the content of Trg. Values for total chemoreceptors are thus the sum of experimental values for total high-abundance receptors and Trg plus the estimate for Tap plus Aer.
Cell size varied almost twofold as a function of strain and growth condition. To make comparisons among different strains and growth conditions that would reflect the concentrations of the sensory components and be independent of differences in cell size, we expressed cellular contents in units of molecules per standard cell volume, defining TB-grown RP437 as the standard cell (see Materials and Methods).
Cellular content as a function of strain and growth condition.
The cellular content of chemoreceptors and Che proteins varied as much as 10-fold as a function of strain and growth medium (Table 1 and Fig. 2). However, these variations occurred in concert for all components (Fig. 2) and, thus, hardly changed the relative amounts of the proteins. This is illustrated by pair-wise ratios (Fig. 3). These ratios were relatively constant as a function of strain and growth condition, with standard deviations of 4 to 31%. These standard deviations were comparable to or only modestly larger than those observed for protein ratios determined for independent cultures of the same strain grown under the same condition. Thus, within the limits of our assays, ratios varied little or not at all as amounts of components varied as much as 10-fold. This indicates a rather constant cellular stoichiometry among the proteins of the chemosensory system.
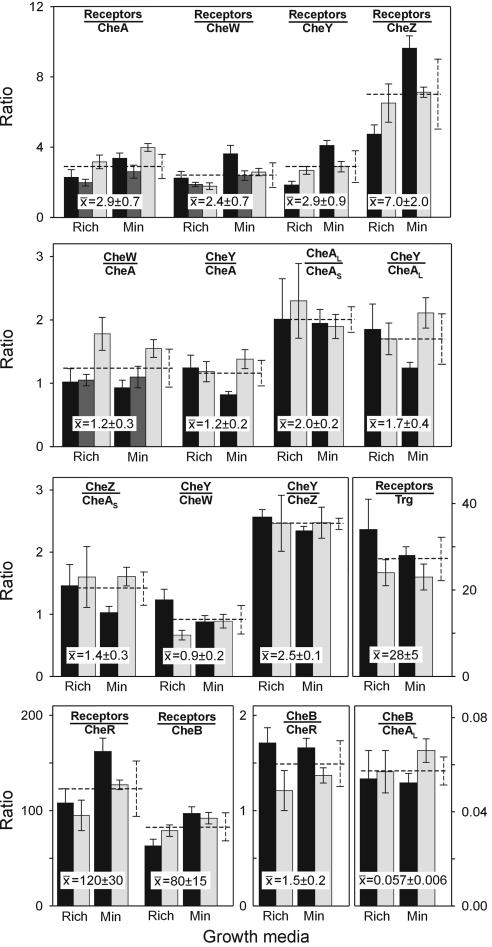
FIG. 3.
Ratios of cellular contents of components of the chemotaxis signaling complex. Values in Table 1 were used to calculate ratios for RP437 (black bars), RP2867 (dark gray bars), and OW1 (light gray bars), grown in TB (Rich) or minimal medium (Min). Error bars are standard deviations of means from three independent cultures, tested in at least two (RP2867) or three separate assays. Means and standard deviations for ratios determined for different strains and media are shown as numerical overlays and dotted lines.
In the one case we investigated, growth phase also influenced cellular content but not the relative amounts of Che proteins. This observation was the result of investigating an apparent disparity between our data and previously published work. In designing our experimental approach for quantification of chemotaxis components, we had utilized several aspects of the careful study of Scharf et al. (34), which had observed a constant ratio of CheY to CheZ in two E. coli strains considered wild type for chemotaxis even though the two strains had an ∼2.5-fold difference in content of the two proteins. Thus, we were concerned when our values for CheY and CheZ in TB-grown RP437 were approximately threefold higher than theirs, even though we observed essentially the same ratio between these two proteins. However, Scharf et al. had analyzed cells growing in TB at a density equivalent to 1.5 additional generations, near the end of the exponential phase of growth in that medium. When we performed quantifications of CheY and CheZ in RP437 at that cell density, we obtained essentially the same values as Scharf et al. Thus, modest differences in growth phase can have significant effects on cellular content of chemotaxis components, as previously documented by Wang and Matsumura for CheAL and CheAS (47).
DISCUSSION
Variable contents, constant ratios.
We aimed to determine the cellular content of each component of the chemotaxis signaling complex of E. coli. We found there was no single value for any protein. Instead, cellular amounts varied up to 10-fold as a function of bacterial strain and growth medium (and, where examined, growth phase). However, ratios between proteins varied little, with standard deviations only slightly greater than those for multiple determinations of a single measurement or for ratios between proteins in different strains and growth conditions. This provides an important insight for analysis of the chemotactic signaling system, particularly as analyses become more quantitative: the most relevant parameters are not the amounts of the components, but the ratios among them.
The two strains we characterized are wild type for chemotaxis, exhibiting effective responses to many compounds when grown in rich or minimal media. Thus, the chemosensory system functions effectively over at least a 10-fold range of cellular dosage. This situation reflects robustness in the bacterial chemosensory system (3). Importantly, it also provides a test for mathematical models of chemotaxis. In addition, maintenance of relative amounts, as strain and growth medium vary, must involve yet-unstudied mechanisms for maintaining relative expression in four different operons and for relative protein amounts that vary from 1:1 to ∼50:1.
As described in the introduction, independently determined values for cellular content of specific chemotaxis components published previously have differed by as much as 10-fold, and the origin of this variation has been unclear. Since our data demonstrate that the cellular content of each component can vary as much as 10-fold as a function of strain, growth medium, and growth phase, variation in published values could well reflect one or more of these factors.
Cellular ratios and the stoichiometry of the core signaling complex.
We observed cellular ratios of 2.9 ± 0.7 receptor monomers per CheA monomer, 2.4 ± 0.7 receptor monomers per CheW, and 1.2 ± 0.3 CheW per CheA monomer. Both chemoreceptors and CheA are native dimers, whereas CheW is considered a monomer, so these values suggest interaction of three receptor dimers, one CheA dimer and two CheW monomers. This is tantalizing, because X-ray crystallography showed that a fragment of much of the cytoplasmic domain of chemoreceptor Tsr formed trimers of dimers in vitro (23), and biochemical studies have provided evidence that intact chemoreceptors form trimers in vivo (44). In addition, in vitro reconstitution studies of core signaling complexes have observed maximal kinase activation at ratios of receptors or receptor fragments to CheA and CheW significantly greater than one receptor to one kinase (2, 25, 27). The ratio of receptors to CheA required for effective activation in those in vitro studies was higher than the stoichiometry we observed in vivo, perhaps because of reduced activities for isolated proteins that were used or because receptor participation in core complexes is not identical to kinase activation.
In any case, extrapolation of cellular stoichiometry to stoichiometry of the ternary core signaling complex in vivo requires knowing the proportion of each protein in a cell that is in such complexes. Values have been provided by immunogold studies of RP437 cells, which identified 79% of CheA and 53% of CheW, both water-soluble proteins, as membrane associated in cells containing all three components of the core complex (28). The CheW value may be artificially low because of the small number of particles in analyses using anti-CheW (J. Maddock, personal communication). CheA, CheW, and receptors appear in clusters, and effective clustering requires all three (28), implying that clustering is linked to formation of ternary complexes. An assessment of chemoreceptor clustering in wild-type cells, using original and subsequent data, indicates that at least 93% of receptors are in clusters (J. Maddock, personal communication) and, thus, participants in ternary complexes. We used the cellular stoichiometry we determined and the values for participation in signaling complexes to calculate a deduced in vivo stoichiometry of core signaling complexes. Expressed per CheA dimer, the stoichiometry was 1 CheA dimer per 1.6 ± 0.5 CheW monomers and 3.4 ± 0.8 receptor dimers. Like the cellular stoichiometry, these values are within 1 standard deviation of an integral stoichiometry of three receptor dimers, 1 CheA dimer (or two CheA monomers from different dimers [36]), and two CheW monomers. However, in the cell, stoichiometries may not be integral. For instance, if the components of the core complex form an extended array (36, 43), not every trimer of receptor dimers would necessarily associate with exactly two monomers of CheA and of CheW. A finding of 3.4 receptor dimers per CheA dimer could reflect an array in which 13% of receptor trimers interacted with one monomer of a CheA dimer instead of two, perhaps at borders of arrays in which CheA dimers bridged trimers (36). A finding of 1.6 CheW per CheA dimer could reflect an array in which 40% of CheA dimer-receptor trimer units bound 1 CheW monomer, not 2. Alternatively, multiple dynamic equilibria across a signaling array could result in nonintegral steady-state stoichiometries.
Low-abundance chemoreceptor Trg was present in cells at a ratio to total receptors of 0.036 ± 0.007, a value close to the early estimate that Trg was present at ∼1/10 the level of a high-abundance receptor (16). If different receptor dimers have equal probabilities of being in a trimer, then ∼10% of signaling complexes would contain Trg.
Loosely bound components of the signaling complex.
Most clusters of core signaling components are at cell poles (28). CheY, CheZ, and CheR, loosely bound components of signaling complexes, are also localized there, dependent on presence of intact core components (9, 37, 40). The localization of CheY is consistent with persistent interaction with core complexes in plasmon resonance studies (35). The notion of molecular brachiation (25) implies that CheB, as well as CheR, could be retained in extended receptor clusters. Thus, cellular stoichiometries are relevant to steady-state interactions of loosely bound components in the signaling complex.
In vivo ratios of 2.9 ± 0.9 receptors per CheY, 1.2 ± 0.2 CheY per CheA, and 0.9 ± 0.2 CheY per CheW indicated that essentially all 70% of cellular CheY that is not phosphorylated (1) could be bound to P2 domains of CheA contained in signaling complexes and, conversely, most signaling complexes would be occupied with at least one unphosphorylated CheY. As we varied the strain and growth condition, the ratio of CheAL to CheAS was notably constant at 2.0 ± 0.2. However, this ratio has been observed to vary fourfold as a function of growth phase (47). In any case, at the growth phase we tested, cells contained two molecules of CheAL for every molecule of CheAS, even though the absolute amounts of the two components varied close to 10-fold. If probabilities of heterologous and homologous dimerization of CheAL and CheAS were equal, as might be expected since the dimerization domain is structurally distinct (6) from the P1 region truncated in CheAS, then 45% of core signaling units would have only CheAL, 44% would have both CheAL and CheAS, and 11% would have only CheAS. Thus, 11% of core signaling units could not donate phosphoryl groups to CheY and CheB. There would be CheAS binding sites for CheZ in approximately half the core signaling units that could donate phosphoryl groups to CheY: the 44% that contained both CheAL and CheAS, but not the 45% containing only CheAL. The ratio of CheZ monomer to CheAS was 1.4 ± 0.3, corresponding to a ratio of ∼0.7 for native CheZ dimer to CheAS. Thus, there would be sufficient sites for all CheZ to interact with signaling complexes, a situation consistent with localization of a CheZ-green fluorescent protein fusion to cell poles in the presence of signaling complexes but not in their absence (9, 40).
We found that methyltransferase CheR and methylesterase-deamidase CheB were present in the cell at lower levels than their protein substrates, a common cellular relationship of enzyme to substrate. Ratios of chemoreceptor monomers to CheR and CheB were 120 ± 30 and 80 ± 15, respectively. Since there are four and six potential methylation and demethylation sites on each monomer of high-abundance receptors Tar and Tsr, respectively (21, 33), the ratio of cellular substrate to enzyme is several hundred to one, depending on the adaptational state of the receptor population. Considering stoichiometry in terms of core signaling complexes, cells would contain 1 CheR molecule for every 20 signaling complexes and 1 CheB for every 13. Previously published values for cellular content of the two enzymes, determined independently, had implied that CheB was present at substantially higher levels than CheR (www.anat.cam.ac.uk/comp-cell). This was consistently not the case for the strains and conditions we investigated.
Conclusions.
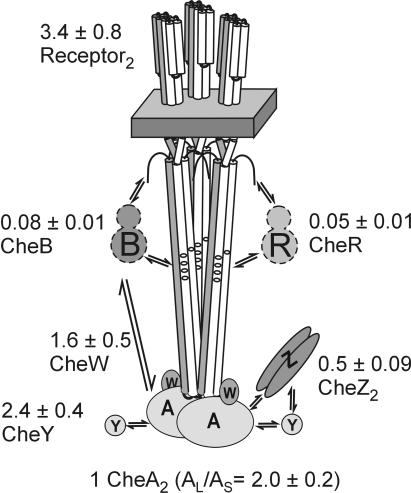
It is striking that variation as large as 10-fold in cellular content of the components of the chemotaxis signaling complex did not substantially alter cellular stoichiometry. It is equally striking that the cellular stoichiometry of the core signaling complex suggests an organization based on trimers of receptor dimers. Our data imply that signaling clusters are built from core units of three receptor dimers, two monomers of the dimeric kinase, and two monomers of CheW. Figure 4 provides a cartoon of the complete signaling complex, labeled with the specific stoichiometry deduced from our studies. The ratios of CheY and CheZ to components of the core complex mean there are sufficient copies of the two proteins to interact with all (CheY) or half (CheZ) of the core complexes. These substantial presences in complexes could be important in sensory mechanisms and ought to be considered experimentally and theoretically. One CheB for every 80 receptor monomers and approximately every 13 signaling complexes means that the role of CheB in signal amplification (22, 41), which is not yet understood, cannot involve mechanisms that require close to stoichiometric interaction with the population of receptors or signaling complexes. Although there is no unique set of cellular amounts for the chemotaxis components of E. coli, it is useful to define representative values. If a cell contained 7,000 CheA monomers, the average ratios we determined (Table 1; Fig. 3) suggest a cellular content, expressed as monomers, of 7,000 CheA, 8,400 CheW, 20,000 receptors, 8,400 CheY, 3,300 CheZ, 280 CheB, and 180 CheR.
FIG. 4.
Cartoon of the chemotaxis signaling complex with deduced in vivo stoichiometry. The basic unit of a core signaling complex is shown with interacting proteins. Values for the stoichiometry of the complex in vivo, calculated as described in the text and expressed as molecules per CheA dimer, are shown with standard deviations.
We found that E. coli maintained a notably constant cellular stoichiometry for the components of the chemotaxis signaling complex as cellular contents varied as much as 10-fold. Both cellular stoichiometry and derived values for an in vivo stoichiometry of the signaling complex suggested an organization of the core complex consistent with previous in vitro structural and biochemical studies. The stoichiometry of the signaling complex provides an important parameter for further experimental and theoretical analysis of one of the most extensively characterized biological signaling systems.
Acknowledgments
We thank A. Barnakov and L. Barnakova for protein purification, A. Lilly for plasmid construction and for coordinating production of antisera, P. Matsumura and A. Stock for antisera, G. Munske (Laboratory for Bioanalysis and Biotechnology, Washington State University) for quantitative amino acid analysis, J. Maddock for interpretations of her localization studies, and B. Scharf for pure protein, antibodies, advice, and encouragement.
This work was supported by GM29963 from NIGMS.
REFERENCES
- 1.Alon, U., L. Camarena, M. G. Surette, B. Aguera y Arcas, Y. Liu, S. Leibler, and J. B. Stock. 1998. Response regulator output in bacterial chemotaxis. EMBO J. 17:4238-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames, P., Y. A. Yu, and J. S. Parkinson. 1996. Methylation segments are not required for chemotactic signalling by cytoplasmic fragments of Tsr, the methyl-accepting serine chemoreceptor of Escherichia coli. Mol. Microbiol. 19:737-746. [DOI] [PubMed] [Google Scholar]
- 3.Barkai, N., and S. Leibler. 1997. Robustness in simple biochemical networks. Nature 387:913-917. [DOI] [PubMed] [Google Scholar]
- 4.Barnakov, A. N., L. A. Barnakova, and G. L. Hazelbauer. 1998. Comparison in vitro of a high- and a low-abundance chemoreceptor of Escherichia coli: similar kinase activation but different methyl-accepting activities. J. Bacteriol. 180:6713-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnakov, A. N., L. A. Barnakova, and G. L. Hazelbauer. 1999. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc. Natl. Acad. Sci. USA 96:10667-10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bilwes, A. M., L. A. Alex, B. R. Crane, and M. I. Simon. 1999. Structure of CheA, a signal-transducing histidine kinase. Cell 96:131-141. [DOI] [PubMed] [Google Scholar]
- 7.Bollinger, J., C. Park, S. Harayama, and G. L. Hazelbauer. 1984. Structure of the Trg protein: homologies with and differences from other sensory transducers of Escherichia coli. Proc. Natl. Acad. Sci. USA 81:3287-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borkovich, K. A., N. Kaplan, J. F. Hess, and M. I. Simon. 1989. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc. Natl. Acad. Sci. USA 86:1208-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantwell, B. J., R. R. Draheim, R. B. Weart, C. Nguyen, R. C. Steward, and M. D. Manson. 2003. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J. Bacteriol. 185:2354-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cayley, D. S., H. J. Guttman, and M. T. R. Record, Jr. 2000. Biophysical characterization of changes in amounts and activity of Escherichia coli cell and compartment water and turgor pressure in response to osmotic stress. Biophys. J. 78:1748-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke, S., and D. E. Koshland, Jr. 1979. Membrane receptors for aspartate and serine in bacterial chemotaxis. J. Biol. Chem. 254:9695-9702. [PubMed] [Google Scholar]
- 12.Feng, X., A. A. Lilly, and G. L. Hazelbauer. 1999. Enhanced function conferred on low-abundance chemoreceptor Trg by a methyltransferase-docking site. J. Bacteriol. 181:3164-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gegner, J. A., D. R. Graham, A. F. Roth, and F. W. Dahlquist. 1992. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell 70:975-982. [DOI] [PubMed] [Google Scholar]
- 14.Harlow, E., and D. Lane (ed.). 1988. Antibodies: a laboratory manual, p. 633. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 15.Hazelbauer, G. L. 2004. Bacterial chemotaxis: a model for sensory receptor systems. In G. Adelman and B. H. Smith (ed.), Encyclopedia of neuroscience, 3rd ed. Elsevier, Amsterdam, The Netherlands. http://203.200.24.140:8080/Neuroscience.
- 16.Hazelbauer, G. L., and P. Engström. 1981. Multiple forms of methyl-accepting chemotaxis proteins distinguished by a factor in addition to multiple methylation. J. Bacteriol. 145:35-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazelbauer, G. L., and P. Engström. 1980. Parallel pathways for transduction of chemotactic signals in Escherichia coli. Nature 283:98-100. [DOI] [PubMed] [Google Scholar]
- 18.Hazelbauer, G. L., P. Engström, and S. Harayama. 1981. Methyl-accepting chemotaxis protein III and transducer gene trg. J. Bacteriol. 145:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hazelbauer, G. L., R. E. Mesibov, and J. Adler. 1969. Escherichia coli mutants defective in chemotaxis toward specific chemicals. Proc. Natl. Acad. Sci. USA 64:1300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hess, J. F., K. Oosawa, N. Kaplan, and M. I. Simon. 1988. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 53:79-87. [DOI] [PubMed] [Google Scholar]
- 21.Kehry, M. R., M. W. Bond, M. W. Hunkapiller, and F. W. Dahlquist. 1983. Enzymatic deamidation of methyl-accepting chemotaxis proteins in Escherichia coli catalyzed by the cheB gene product. Proc. Natl. Acad. Sci. USA 80:3599-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, C., M. Jackson, R. Lux, and S. Khan. 2001. Determinants of chemotactic signal amplification in Escherichia coli. J. Mol. Biol. 307:119-135. [DOI] [PubMed] [Google Scholar]
- 23.Kim, K. K., H. Yokota, and S. H. Kim. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787-792. [DOI] [PubMed] [Google Scholar]
- 24.Kofoid, E. C., and J. S. Parkinson. 1991. Tandem translation starts in the cheA locus of Escherichia coli. J. Bacteriol. 173:2116-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levin, M. D., T. S. Shimizu, and D. Bray. 2002. Binding and diffusion of CheR molecules within a cluster of membrane receptors. Biophys. J. 82:1809-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, J., R. V. Swanson, M. I. Simon, and R. M. Weis. 1995. The response regulators CheB and CheY exhibit competitive binding to the kinase CheA. Biochemistry 34:14626-14636. [DOI] [PubMed] [Google Scholar]
- 27.Liu, Y., M. Levit, R. Lurz, M. G. Surette, and J. B. Stock. 1997. Receptor-mediated protein kinase activation and the mechanism of transmembrane signaling in bacterial chemotaxis. EMBO J. 16:7231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 29.Ordal, G. W., and J. Adler. 1974. Properties of mutants in galactose taxis and transport. J. Bacteriol. 117:517-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park, C., and G. L. Hazelbauer. 1986. Mutations specifically affecting ligand interaction of the Trg chemosensory transducer. J. Bacteriol. 167:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkinson, J. S. 1978. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J. Bacteriol. 135:45-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice, M. S., and F. W. Dahlquist. 1991. Sites of deamidation and methylation in Tsr, a bacterial chemotaxis sensory transducer. J. Biol. Chem. 266:9746-9753. [PubMed] [Google Scholar]
- 34.Scharf, B. E., K. A. Fahrner, and H. C. Berg. 1998. CheZ has no effect on flagellar motors activated by CheY13DK106YW. J. Bacteriol. 180:5123-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuster, S. C., R. V. Swanson, L. A. Alex, R. B. Bourret, and M. I. Simon. 1993. Assembly and function of a quaternary signal transduction complex monitored by surface plasmon resonance. Nature 365:343-347. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu, T. S., N. Le Novere, M. D. Levin, A. J. Beavil, B. J. Sutton, and D. Bray. 2000. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat. Cell Biol. 2:792-796. [DOI] [PubMed] [Google Scholar]
- 37.Shiomi, D., I. B. Zhulin, M. Homma, and I. Kawagishi. 2002. Dual recognition of the bacterial chemoreceptor by chemotaxis-specific domains of the CheR methyltransferase. J. Biol. Chem. 277:42325-42333. [DOI] [PubMed] [Google Scholar]
- 38.Smith, R. A., and J. S. Parkinson. 1980. Overlapping genes at the cheA locus of Escherichia coli. Proc. Natl. Acad. Sci. USA. 77:5370-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sourjik, V., and H. C. Berg. 2004. Cross-talk between receptors in bacterial chemotaxis. Nature 428: 437-441 [DOI] [PubMed] [Google Scholar]
- 40.Sourjik, V., and H. C. Berg. 2000. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 37:740-751. [DOI] [PubMed] [Google Scholar]
- 41.Sourjik, V., and H. C. Berg. 2002. Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 99:123-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Springer, M. S., M. F. Goy, and J. Adler. 1977. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc. Natl. Acad. Sci. USA 74:3312-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stock, J., and M. Levit. 2000. Signal transduction: hair brains in bacterial chemotaxis. Curr. Biol. 10:R11-R14. [DOI] [PubMed] [Google Scholar]
- 44.Studdert, G. A., and J. S. Parkinson. 2004. Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. USA 101:2117-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor, B. L., A. Rebbapragada, and M. S. Johnson. 2001. The FAD-PAS domain as a sensor for behavioral responses in Escherichia coli. Antioxid. Redox Signal. 3:867-879. [DOI] [PubMed] [Google Scholar]
- 46.Wang, H., and P. Matsumura. 1996. Characterization of the CheAS/CheZ complex: a specific interaction resulting in enhanced dephosphorylating activity on CheY-phosphate. Mol. Microbiol. 19:695-703. [DOI] [PubMed] [Google Scholar]
- 47.Wang, H., and P. Matsumura. 1997. Phosphorylating and dephosphorylating protein complexes in bacterial chemotaxis. J. Bacteriol. 179:287-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weerasuriya, S., B. M. Schneider, and M. D. Manson. 1998. Chimeric chemoreceptors in Escherichia coli: signaling properties of Tar-Tap and Tap-Tar hybrids. J. Bacteriol. 180:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, J., J. Li, G. Li, D. G. Long, and R. M. Weis. 1996. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry 35:4984-4993. [DOI] [PubMed] [Google Scholar]
- 50.Zhao, R., E. J. Collins, R. B. Bourret, and R. E. Silversmith. 2002. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat. Struct. Biol. 9:570-575. [DOI] [PubMed] [Google Scholar]