SUMMARY
Programmed DNA rearrangements in the single-celled eukaryote Oxytricha trifallax completely rewire its germline into a somatic nucleus during development. This elaborate, RNA-mediated pathway eliminates noncoding DNA sequences that interrupt gene loci and reorganizes the remaining fragments by inversions and permutations to produce functional genes. Here, we report the Oxytricha germline genome and compare it to the somatic genome to present a global view of its massive scale of genome rearrangements. The remarkably encrypted genome architecture contains >3,500 scrambled genes, as well as >800 predicted germline-limited genes expressed, and some posttranslationally modified, during genome rearrangements. Gene segments for different somatic loci often interweave with each other. Single gene segments can contribute to multiple, distinct somatic loci. Terminal precursor segments from neighboring somatic loci map extremely close to each other, often overlapping. This genome assembly provides a draft of a scrambled genome and a powerful model for studies of genome rearrangement.
INTRODUCTION
Genomes are dynamic structures. Humans possess genomic variation among tissues from the same individual (O'Huallachain et al., 2012). Furthermore, genome instability can be a common factor in cancer transformation (Stephens et al., 2011), when thousands of genome rearrangement events contribute to cancer-causing lesions. Curiously, programmed genome rearrangements occur during development in a variety of eukaryotes, with DNA elimination the most frequent type. Examples include chromatin diminution in the parasitic nematode Ascaris (Wang et al., 2012) and DNA loss in lamprey (Smith et al., 2012). In both cases, ~10%–20% of germline DNA is eliminated in somatic cells. Rejoining of flanking sequences follows DNA deletion in some cases, but sometimes whole chromosomes are discarded, as in sciarid flies (Goday and Esteban, 2001). Genome-wide DNA rearrangements are most exaggerated in ciliates, particularly in the model organism Oxytricha trifallax, which programs not only DNA deletion, but also total reorganization, through RNA-mediated events (Fang et al., 2012; Nowacki et al., 2008). Hence, Oxytricha presents a unique opportunity to study the intricate process of large-scale genome remodeling.
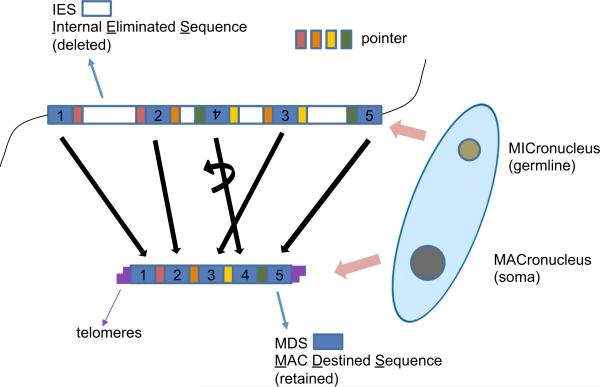
O. trifallax, like most ciliates, possesses two types of nuclei in a single cell: a germline micronucleus (MIC) and a transcriptionally-active somatic macronucleus (MAC) (Prescott, 1994). After sexual conjugation, the old MAC disintegrates and a new MAC develops from a copy of the diploid zygotic MIC through an elaborate cascade of events that delete >90% of the germline DNA and reorganize and join the remaining DNA pieces. The germline precursors of MAC gene loci are highly interrupted by short non-coding elements called internal eliminated sequences (IESs) (Figure 1) that are removed during development. The retained gene segments, called macronuclear-destined sequences (MDSs), are often disordered (scrambled) or inverted in the germline. Thus, macronuclear development requires the rearrangement of MDS segments by inversion or permutation to assemble functional genes. Pairs of short direct repeats, called pointers, are present at consecutive MDS-IES junctions, and one copy of each pointer is retained in the MAC. It is unknown whether the recombination mechanism is a cis or trans process, or a combination of both, because the polytene chromosome stage during MAC development (Spear and Lauth, 1976) could permit recombination in trans between identical copies of MIC DNA molecules. Massive chromosome fragmentation breaks long MIC chromosomes into ~16,000 gene-sized “nanochromo somes” that are bound by short telomeres, average just 3.2 kb, and are each amplified to high copy number (Swart et al., 2013). Recent studies have revealed roles of noncoding RNAs in these events, suggesting that 27 nt Piwi-associated small RNAs (piRNAs) mark MDS regions for retention (Fang et al., 2012) and long, noncoding RNAs serve as epigenetic templates to program segment order and orientation (Nowacki et al., 2008), as well as DNA copy number (Nowacki et al., 2010), in the rearranged, somatic genome.
Figure 1. Development of the Oxytricha Macronuclear Genome from the Micronuclear Genome.
In the micronucleus (MIC), macronuclear destined sequences (MDSs) are interrupted by internal eliminated sequences (IESs); MDSs may be disordered (e.g., MDS 3, 4, and 5) or inverted (e.g., MDS 4). During development after conjugation, IESs, as well as other MIC-limited DNA, are removed. MDSs are stitched together, some requiring inversion and/or unscrambling. Pointers are short identical sequences at consecutive MDS-IES junctions. One copy of the pointer is retained in the new macronucleus (MAC). The old macronuclear genome degrades. Micronuclear chromosome fragmentation produces genesized nanochromosomes (capped by telomeres) in the new macronuclear genome. DNA amplification brings nanochromosomes to a high copy number. See also Figure S1.
Previously, very little was known about the Oxytricha MIC genome, aside from a general understanding of a small number of scrambled genes and its vast excess of noncoding and repetitive DNA, including satellites and transposons. Among the latter, only the TBE elements (a group of abundant DNA transposons) were previously characterized and shown to participate in programmed deletion of themselves as well as IESs (Nowacki et al., 2009). The MIC genome has been a mysterious puzzle, harboring not only a labyrinth of hundreds of thousands of intricately organized gene segments but also other germline-limited elements that could be involved in genome rearrangement.
High quality draft somatic genome sequences have been reported for the main model ciliates Tetrahymena (Eisen et al., 2006), Paramecium (Aury et al., 2006), and Oxytricha (Swart et al., 2013). Although genome-wide IES studies have been reported for Paramecium and Tetrahymena (Arnaiz et al., 2012; Fass et al., 2011), neither of which has scrambled genes, no comprehensive germline genome has been described to date for any ciliate species, nor for any organism with a scrambled genome. Here, we present a draft assembly of the Oxytricha micronuclear genome and compare it to the somatic genome (Swart et al., 2013) to reveal an unprecedented level of programmed rearrangements and genomic complexity, arguably the most complex genome architecture of any known eukaryote. We demonstrate that the MIC genome sequence is fragmented into over 225,000 segments, tens of thousands of which are complexly scrambled and interwoven. Gene segments from neighboring loci are located in extreme proximity to each other, often overlapping. Furthermore, the discovery of more than 800 germline-restricted genes provides insights into genome rearrangement events.
RESULTS AND DISCUSSION
Genome Sequencing Reveals a Dispersed Set of Fragments that Produce a Somatic Nucleus
We isolated Oxytricha micronuclei using sucrose purification (Lauth et al., 1976) and sequenced the DNA to a coverage of ~110× using a shotgun Illumina method and ~15× with single molecule real-time (SMRT) DNA sequencing (Pacific Biosciences). De novo assembly of error-corrected PacBio reads with the Celera assembler (Miller et al., 2008) yields an ~496 Mb draft assembly (Table 1 and Table S1 available online). Additional data (BAC and Fosmid sequencing) were used to validate the assembly (Table S1; Extended Experimental Procedures). Previous studies using reassociation kinetics (Lauth et al., 1976) estimated the MIC genome as 0.3–2.3 Gb and the somatic-destined (MDS) portion to be 2.4%–18% (generally cited as ~5%) of the MIC genome (Prescott, 1994), but we infer it could be as high as 10%, based on read statistics (Figure S1; Extended Experimental Procedures). This small fraction of the germline gives rise to all functional somatic genes.
Table 1.
Assembly Statistics from Oxytricha Micronuclear and Macronuclear Genomes
| MIC Genome Assembly | MAC Genome Assemblya | |
|---|---|---|
| Estimated genome size (Mb) | ~490–500 | ~50b |
| Total assembly size (Mb) | 496.2 | 55.4 |
| Contig number | 25,720 | 19,152 |
| Number ≥ 200 bp | 25,720 | 19,078 (18,405)c |
| Number ≥ 10 kb | 15,942 | 249 |
| N50 (bp) (≥ 200 bp) | 27,807 | 3,597 |
| Longest (kb) | 381 | 66 |
| GC (%) | 28.4 | 31.0 |
| Repeat (%) | 35.9 | 0 |
| Gene number | 810d | 20,883 (~18,400)e |
See also Table S1.
The MAC genome assembly was clustered using CD-HIT (Fu et al., 2012) at 95% identity to remove redundancy before calculation of statistics. Repetitive contigs assembled from MIC contamination and bacterial contigs were also removed.
Taken from Swart et al. (2013).
Containing one or both telomeres.
Not including IES-less genes.
Estimated number (18,400) of nonredundant genes. Taken from Swart et al. (2013).
The MIC assembly contains 98.9% of all nucleotides in the MAC assembly. Of 18,405 MAC contigs with one or both telomeres (Swart et al., 2013), 18,097 (98.3%) are at least 90% covered in the MIC assembly, and 16,220 (88.1%) are at least 90% covered on single MIC contigs, suggesting completely resolved germline-somatic maps. MIC gene loci are typically interrupted by at least one IES, except for 548 IES-less nanochromosomes. Hence, most functional information is encrypted in the MIC, and macronuclear development is a process of decryption. Most IESs interrupt exons (84.7%), making their removal a strict requirement for gene expression.
The germline genome is fragmented into over 225,000 precursor DNA segments (MDSs) that massively rearrange during development to produce nanochromosomes containing approximately one gene each. Note that this number is on the same order of magnitude as the total number of exons in the human genome (Harrow et al., 2012), but these segments fuse via DNA splicing at short direct repeats (pointers) at their ends. The six tiniest of these segments (0 bp MDSs) are merely a splint joining two other MDSs. We identified six strong cases of 0 bp MDSs (four nonscrambled and two scrambled; Figure 2C). These comprise just two tandem pointers with no intervening MAC sequence, underscoring the minimalist role of these MDSs as a splint between two adjacent regions that would otherwise share no pointer repeat between them (and be misannotated as 0 bp pointers).
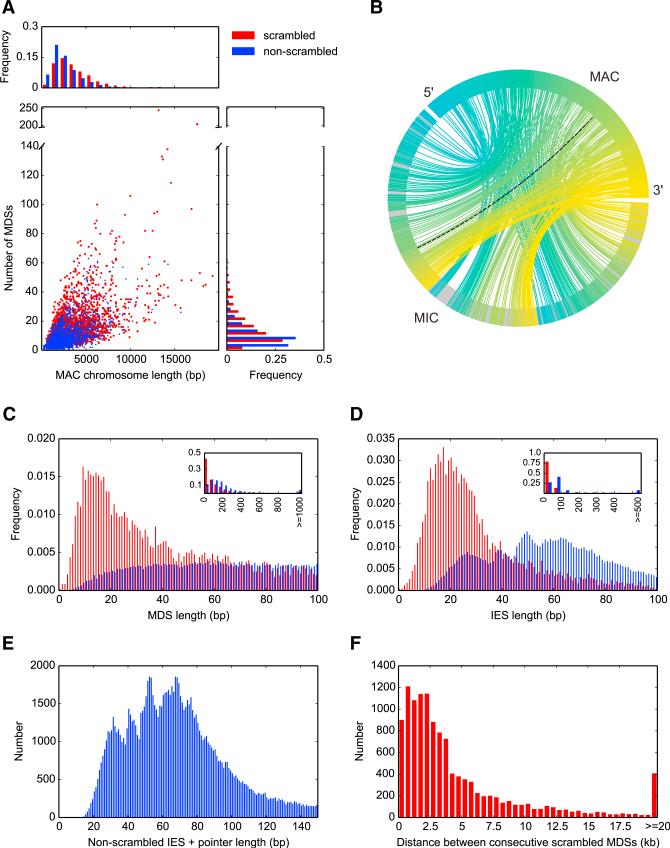
Figure 2. The MIC Genome Is Fragmented into Hundreds of Thousands of Segments with Massive Levels of Scrambling.
(A) Comparison of chromosome length and number of MDS segments between 13,910 non-scrambled (blue) and 2,310 scrambled nanochromosomes (red) completely covered on single MIC contigs.
(B) A chord diagram mapping MIC ctg7180000068801 to its rearranged form, MAC Contig17454.0. Lines connect precursor (MIC) and product (MAC) MDS locations; black dotted line, an inverted MDS; all 245 MDSs (242 are scrambled) drawn in a blue to yellow MAC color gradient; IESs, gray.
(C) Length distribution of 44,191 nonscrambled and 9,841 scrambled MDSs <100 nt (excluding pointers). Inset: 150,615 nonscrambled MDSs and 16,350 scrambled MDSs excluding pointers, showing the most typical length of scrambled MDSs is <50 nt.
(D) Length distribution of 101,345 nonscrambled and 8,333 scrambled high-confidence IESs <100 nt (excluding pointers and IESs that contain other MDSs). Inset: 147,122 nonscrambled versus 9,040 scrambled IESs excluding pointers and IESs that contain other MDSs. We identified six strong cases of 0 bp MDSs (four nonscrambled [Contig7827.0 MDS3, Contig11190.0.1 MDS18, Contig13633.0 MDS3, and Contig9208.0.0 MDS18] and two scrambled [Contig6325.0.0 MDS58 and Contig1267.1 MDS7]).
(E) Length distribution of 112,125 nonscrambled IESs (excluding those that contain other MDSs) <150 nt, with one copy of the pointer included (i.e., the total length of DNA deleted).
(F) MIC genomic distance between scrambled MDSs that are consecutive in the MAC (n = 12,197); distance calculated from the pointer flanking MDS N to its paired pointer flanking MDS N+1.
See also Figure S2.
Of all the millions of piRNA sequences (Fang et al., 2012) that map to the MIC genome assembly, 96.0% map to MDS regions. Among the remaining 4% that map to non-MDS regions, the majority (2.4% of total) map to the MAC genome assembly of another strain, JRB510, suggesting that they belong to MIC regions that are either MAC-destined in the other strain or were missed in the JRB310 MAC assembly. The remaining 311,012 (1.6%) reads could also derive from MAC-destined regions that are present on lower copy number nanochromosomes and absent from either MAC assembly. Just 0.11% (21,946) of piRNAs map to MIC-limited repeats, such as TBE transposons and satellites. Therefore, Oxytricha piRNAs rarely map to IESs or other MIC-limited sequences. These observations support the model in Fang et al. (2012) that piRNAs mark the precise regions of the MIC genome for retention during genome rearrangement.
Massive Genome Reorganization
In addition to the intense dispersal of all somatic coding information into >225,000 DNA fragments in the germline, a second unprecedented feature of the Oxytricha MIC genome is its remarkable level of scrambling (disordered or inverted MDSs). The germline maps of at least 3,593 genes, encoded on 2,818 nanochromosomes, are scrambled. No other sequenced genome bears this level of structural complexity.
Scrambled nanochromosomes are typically longer and contain more MDSs (average 4.9 per kb) than nonscrambled nanochromosomes (average 3.7 per kb; Figure 2A). Among the 2,818 scrambled MAC chromosomes, 1,676 contain at least one inverted segment and 644 contain extended regions that partition odd- and even-numbered segments, a pattern previously observed for a limited number of scrambled genes (Prescott, 1994). The most scrambled gene is a 22 kb MIC locus fragmented into 245 precursor segments that assemble to produce a 13 kb nanochromosome encoding a dynein heavy chain family protein (Figure 2B).
Scrambled MDSs are typically much shorter than non-scrambled MDSs (Figure 2C; median 81 bp for scrambled MDSs, 181 bp for nonscrambled MDSs), presumably reflecting the increased fragmentation of scrambled genes (Figure 2A). While IESs are generally short and GC poor (18% GC), scrambled IESs (median 27 nt) are also much shorter on average than nonscrambled IESs (median 68 nt, Figure 2D). Like the smallest MDS, the shortest IES is 0 nt, just two adjacent scrambled pointers. Figure 2E plots the length distribution of non-scrambled IESs plus one copy of the pointer, which is the full length deleted, since one copy of a pointer is retained. (Scrambled IESs, on the other hand, are flanked by different pointers.) Several weak peaks are present in this length distribution, with a periodicity of approximately 10 bp, similar to one turn of a DNA double helix. A stronger trend in length distribution among nonscrambled IESs in Paramecium (Arnaiz et al., 2012) suggests DNA loop formation during assembly of a transposase-containing excision complex. Despite the prevalence of small IESs, gene scrambling permits consecutive MDSs in the MAC to be far apart in the MIC, up to 208 kb (Figure 2F, median distance 2.9 kb).
Scrambled pointers are also longer and more GC-rich than those flanking nonscrambled MDSs (Figure S2A; average scrambled pointer 11 bp and 30% GC, average nonscrambled pointer 5 bp and 19% GC). These longer, more GC-rich pointers may facilitate pointer alignment and unscrambling, even over a distance.
Most 2 bp pointers are TA (Figures S2B and S2C), the only pointer sequence in Paramecium IESs and the most common among Euplotes IESs, all of which appear to be nonscrambled and have a short terminal consensus sequence resembling the ends of Tc1/mariner transposons (Jacobs and Klobutcher, 1996; Klobutcher and Herrick, 1995). This suggests that such IES may be relics from ancient transposon invasion (Klobutcher and Herrick, 1997). Sequences at the ends of TA IESs in Oxytricha do not match the Paramecium consensus (Figure S2H) but do display a complementary overall base composition that resembles an inverted repeat (Figure S2I).
Among 3 bp pointers, an A nucleotide is overrepresented at the 5′ most position and a T at the 3′ most position (Figures S2D and S2E). ANT is the target duplication site of TBE transpo-sons, present in thousands of copies in the Oxytricha MIC genome. Such IESs may also be relics of nonfunctional transpo-sons (Klobutcher and Herrick, 1997) or reflect constraints on splice sites processed by transposon-derived machinery (Nowacki et al., 2009). Their terminal sequences (Figure S2J) also bear a weak resemblance to the first few bases of the CA4C4 telomeric repeats at TBE transposon ends (Figure S2K).
Thousands of Interwoven Gene Loci
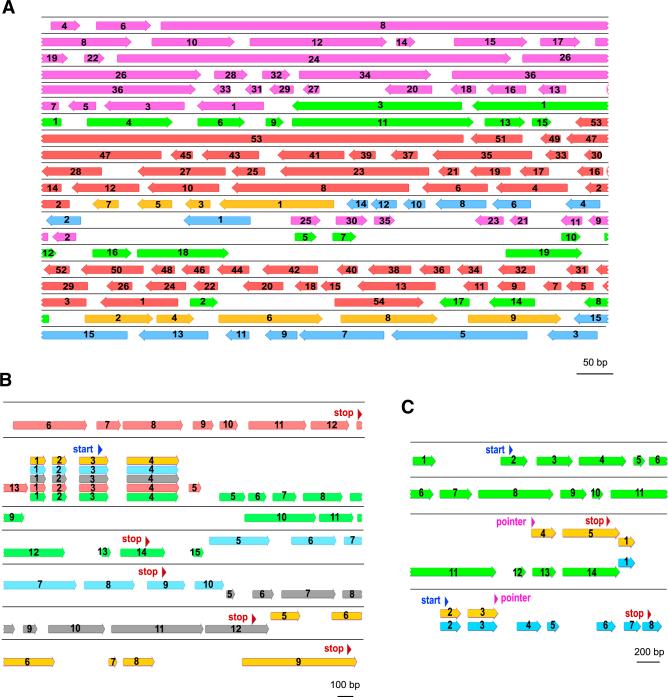
A third exceptional feature we noted is 1,537 cases (1,043 of which are scrambled) of nested genes, with the precursor MDS segments for multiple different MAC chromosomes interwoven on the same germline locus, such that IESs for one gene contain MDSs for another. Previously, in an earlier diverged genus, Uroleptus, Kuo et al. (2006) discovered that the precursor of a two-gene MAC chromosome exhibits a nested structure, with a precursor segment of one gene present among those for the other gene—but these segments descramble into only one chromosome. The finding in Oxytricha of nested structures occurring even among segments for different MAC chromosomes implicates a massive scale and coordination of genome rearrangement to assemble separate MAC chromosomes. Though simple cases of nested genes exist in other eukaryotes; for example, 158 human genes reside completely within the intron of another gene (Kumar, 2009; Yu et al., 2005), the nested gene segments in Oxytricha interweave with each other in an elaborately entangled order and orientation. Figure 3A shows a germline locus that contains precursors of 5 nanochromosomes, whose MDSs are not only heavily scrambled themselves, but also deeply interwoven with each other. This type of interwoven architecture also contributes to the large micronuclear distances between MDS segments that are consecutive in the MAC (Figure 2F).
Figure 3. Gene Segments for Multiple Distinct MAC Chromosomes Are Sometimes Interwoven or Reused.
(A) Germline map of MIC ctg7180000067411 (drawn to scale), containing precursor MDSs (bars, orientation as shown, including pointers) for five MAC chromosomes (purple, Contig1267.1; green, Contig18709.0; red, Contig20652.0; gold, Contig18297.0; blue, Contig6980.0) whose MDSs are scrambled and interwoven with each other. IES regions are gaps. MDS numbers are consecutive in the MAC.
(B) Germline map of a MIC region (ctg7180000067243) with four shared MDSs that assemble into five distinct MAC chromosomes with identical 5′ ends (red, scrambled Contig14686.0; green, Contig7507.0; blue, Contig7395.0; gray, Contig15152.0; gold, Contig4858.0); start/stop codons annotated in blue and red, respectively.
(C) Germline map (ctg7180000068430) depicting a scrambled MAC chromosome (gold, Contig19716.0) that arose by recombination between MDSs from two different gene loci (green, Contig16277.0; blue, Contig22490.0) at a new pointer (11 bp direct repeat, magenta triangles). Note that the green Contig16277.0 is an alternatively processed chromosome, itself, with two predicted stop codons; the shorter, more abundant isoform (not shown) terminates at an alternative telomere addition site between MDS 12-13, upstream of an intron 3′ splice site. This creates an earlier, in-frame stop codon within the retained portion of the unspliced intron (Swart et al., 2013).
Alternative Processing of MDSs Produces Multiple Genes and Chromosomes
A fourth notable feature arising from this radical genome architecture is that a single MDS in the MIC may contribute to multiple, distinct MAC chromosomes. Like alternative splicing, this modular mechanism of “MDS shuffling” (proposed in Prescott, 1999 and suggested in Katz and Kovner, 2010) can be a source of genetic variation, producing different nanochromosomes and even new genes and scrambled patterns (Figures 3B and 3C). At least 1,267 MDSs from 105 MIC loci are reused, contributing to 240 distinct MAC chromosomes. A single MDS can contribute to the assembly of as many as five different nanochromosomes. Figure 3B shows an example where the precursors of five single-gene nanochromosomes (one scrambled) share 4 MDSs at the 5′ end of their encoded genes but have different sets of 3′ MDSs. The shared MDSs preserve reading frame and use the same start codon.
MDS sharing can also produce new MAC chromosome architectures. For example, in Figure 3C, the gold nanochromosome fuses the first three MDSs from the 5′ end of the downstream gene (blue) to the last two MDSs from the upstream gene (green) via recombination at an 11 bp direct repeat (magenta triangle, labeled “pointer”). This single event creates a novel chimeric, scrambled nanochromosome from two precursor nonscrambled loci, supporting the ability of MDS shuffling (Prescott, 1999) to contribute to the origins of both new genes and new scrambled genetic architectures.
We examined a set of potential new genes that are produced through combinatorial assembly of both reused and unique MDSs. Of the 105 MIC loci that share some MDSs, 55 encode paralogous proteins (i.e., some of their unique MDSs also share sequence similarity at the predicted protein level). Such cases might derive from duplication and divergence of MDSs but not complete gene loci. Thirty-two cases lack paralogy outside the shared MDS, resulting in genuinely chimeric proteins that fuse identical sequence blocks to completely unique blocks. In one case, the shared MDSs do not extend into the coding regions. In 12 cases, entire reading frames reside within the shared MDSs, producing no new predicted gene structures. The remaining five cases have no predicted protein-coding genes.
Precursors of Chromosome Ends Often Overlap
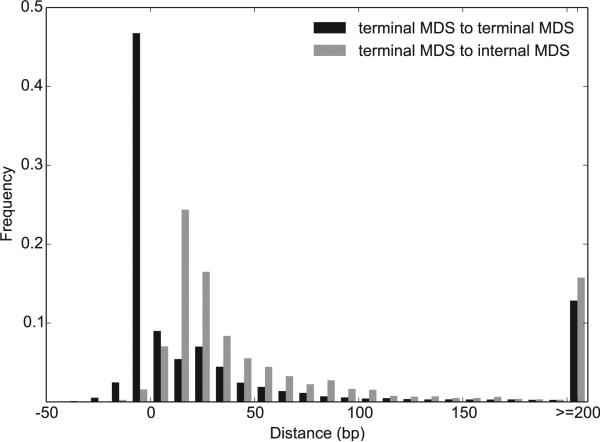
In addition to removal of MIC-limited sequences and descrambling of MDSs, macronuclear development also involves fragmentation of the long MIC chromosomes and addition of telomeres at the new termini, producing gene-sized nanochromosomes in the MAC. Among the 22,875 terminal MDSs we identified in the MIC genome, most (20,012) are adjacent to a terminal MDS of another nanochromosome, while 2,863 reside next to an MDS that is an internal segment of another nanochromo-some. A fifth remarkable feature of Oxytricha's MIC genome, deriving from the proximity between terminal segments for different nanochromosomes, is that these MDSs frequently overlap. This creates vanishingly short intergenic regions, to the point where the median distance between 10,006 pairs of terminal MDSs that are adjacent to each other in the MIC (Figure 4, black bars) is precisely 0 bp. (The range is −34 bp to 19 kb, where a negative value indicates the length of overlap.) This is in striking contrast with the absence of gene linkage on most somatic chromosomes (Swart et al., 2013). While preliminary studies of the related ciliate, O. nova, hinted that MDS-containing regions cluster in the germline (Boswell et al., 1983; Klobutcher, 1987; Klobutcher et al., 1988), this type of gene density is exaggerated to the point where nearly half of nanochromosome ends actually overlap. The variable length of overlap is consistent with a mechanism of micronuclear chromosome fragmentation that involves staggered cuts to allow production of both chromosomes from one precursor or production of two chromosomes with overlapping ends from different polytene chromosomes (Klobutcher et al., 1988).
Figure 4. The Distance between Adjacent Terminal MDSs Is Much Smaller Than that between Terminal MDSs and Adjacent Internal MDSs for a Different MAC Chromosome.
Negative values represent the length of overlapping regions, with the peak distance between terminal MDSs from −1 to −10 bp (10,006 pairs, black) and the peak between terminal MDS to internal MDS between 10–19 bp (2,863 pairs, gray).
See also Figure S3.
If IES removal and chromosome fragmentation are separate events, then during the stage of IES removal, two adjacent terminal MDSs for different nanochromosomes could sometimes be processed as a single MDS, with chromosome fragmentation and telomere addition to follow. This would economically require just a single cut when the distance between terminal MDSs is <1 bp. Terminal MDSs next to an internal MDS, on the other hand, must be processed separately, because they need to be joined to consecutive MDSs. Correspondingly, the distance between a terminal MDS and an adjacent internal MDS is generally much larger (median 30 bp, Figure 4, gray bars). Sequence features flanking chromosome fragmentation sites are discussed in the Extended Experimental Procedures and Figure S3.
Germline-Restricted Protein-Coding Genes
With TBE transposases a notable exception (Nowacki et al., 2009), the deleted portion of Oxytricha's germline has generally been considered transcriptionally inactive. However, a sixth main conclusion of this study was the discovery and confirmation of hundreds of expressed germline-limited genes, many with predicted functions that could relate to genome rearrangement. In addition to 548 IES-less nanochromosomes, some of which appear expressed from both the MIC and the MAC (see next section), we predicted 810 expressed, nonrepetitive (single copy) MIC-limited protein-coding genes (including one MT-A70 gene family; see below). Sixty-eight of these MIC-limited genes fully reside within an IES of another MAC-destined locus. Therefore, IESs, often considered to be AT-rich “junk” DNA, can not only harbor MDSs of other genes, but they also bear germline-limited genes that are discarded during genome rearrangement.
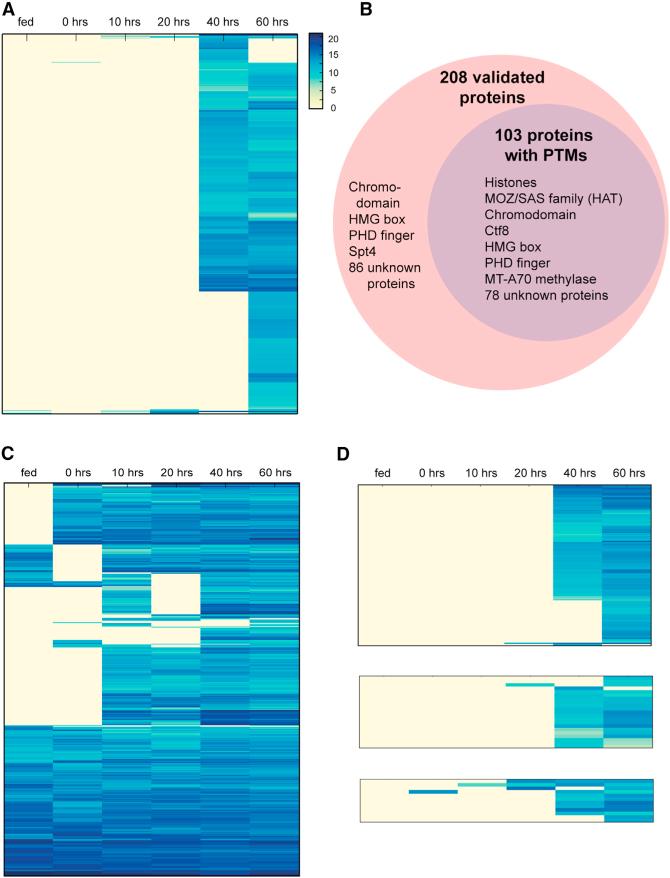
Based on RNA sequencing (RNA-seq) data (Swart et al., 2013) these 810 germline-limited genes are almost exclusively expressed during conjugation (peaking 40–60 hr after the onset of conjugation, with 98% expressed only at 40 and/or 60 hr and little transcription in asexually dividing (vegetative) cells or at the “0 hr” time point when cells of compatible mating types are mixed to initiate conjugation, Figure 5A). The developmentally-limited expression of these germline genes is naturally abrogated by DNA diminution, which has been proposed as a mechanism of germline gene regulation in lamprey (Petromyzon marinus) (Smith et al., 2012) and Ascaris suum (Wang et al., 2012). The ciliate Euplotes crassus possesses a telomerase gene that is expressed only during development, after activation by IES deletion, but the gene itself is absent from the vegetative MAC, suggesting that programmed gene elimination may shut off its expression (Karamysheva et al., 2003). In lamprey and Ascaris, the genes eliminated from somatic cells are mostly expressed during gametogenesis or early embryogenesis. They are often associated with basic cellular functions such as transcription, translation and chromatin remodeling, suggesting that these genes are likely to be involved in development or maintenance of the germline. In the unicellular Oxytricha, however, in addition to germline differentiation and maintenance, germline-limited genes could also provide functions in early somatic differentiation, specifically in macronuclear development and genome rearrangement, bridging the interval from degradation of the parental MAC through production of new macronuclei. This also suggests that, despite unicellularity, these microbial eukaryotes may harbor orthologs of genes required for the evolution of differentiated multicellularity, at least a refined germ-line-soma distinction. MIC-limited genes could technically be expressed from either the micronucleus or the developing macronucleus (before they are eliminated). Because RNA-seq data (Swart et al., 2013) derive from whole cells, we are unable to deduce at this time whether expression derives exclusively from either organelle.
Figure 5. MIC-Limited Genes and Transpo-sons Are Preferentially Expressed during Conjugation, while IES-less Genes Are More Constitutive but Show Universally High Expression during Conjugation.
(A) Clustered expression profile of 810 germline-limited nonrepetitive genes across different time points (vegetative stage (fed); 0, 10, 20, 40, and 60 hr during the conjugating time course). Gene expression levels are represented by log2 (100,000 × normalized RNA-seq counts/coding sequence length).
(B) Mass spectrometry validated 208 MIC-limited genes (outer, pink circle) and 103 were found to contain posttranslational modifications (PTMs) (inner, purple circle). Representative members of each group are shown within the circles.
(C) Clustered expression profiles of 530 IES-less genes.
(D) Clustered expression profiles of MIC-limited transposon-associated genes. Upper: 275 reverse transcriptase and endonuclease domain proteins encoded by LINEs; Middle: 21 Helitron-associated helicases; Lower: 12 DDE_Tnp_IS1595 (ISXO2-like transposase) domain proteins from insertion sequences (ISs).
See also Tables S2, S3, S4, S5, S6, S7, and S8 and Figure S4.
Table S2 compares the properties of predicted Oxytricha germline-limited genes to the categories of both IES-less genes and MAC-specific genes on completely sequenced nanochromosomes (Swart et al., 2013). Predicted genes and introns are both shorter in MIC-limited genes than in the MAC. In addition, the MIC genome is much more intron-poor (chi-square test, p value <2.2 × 10−16). A possible explanation may be that the micronucleus or the developing macronucleus could have limited access to intron splicing machinery. Among MIC genes expressed at 40 or 60 hr, their expression levels are not significantly different from expressed MAC genes (2 sample t test, p value = 0.3148, 0.1285 for 40 and 60 hr, respectively). Among 810 predicted MIC genes, only 311 are located on MIC contigs that contain MDSs for MAC loci, while the others map either to short contigs or to entirely MIC-limited regions of the genome, including contigs with repetitive elements.
Proteomic analysis unambiguously supported translation of 208 predicted germline-limited genes (26% of the 810; Figure 5B; Tables S3 and S4). High-resolution, accurate-mass ultra-high performance liquid chromatography mass spectrometry (UPLC-MS) analysis of Oxytricha 40 hr nuclear lysate with minimal upfront fractionation by SDS-PAGE detected MIC-encoded proteins, based on high-confidence MS characterization of over 100 peptides per protein in some cases (Table S3). With over one million tandem mass spectra from MAC and MIC peptides assigned to >6,900 proteins, this analysis was sufficiently deep to reveal significant stoichiometric and substoichiometric post-translational modifications (PTMs) on half (103) of the 208 validated proteins. These modifications include methylation, acetylation, and phosphorylation, consistent with functional regulation of the MIC-encoded proteins. Both phosphorylation and acetylation occur on proteins with predicted diverse cellular functions, as well as unknown proteins (Figures S4A– S4D) that account for most (74%) of the phosphorylated cases. Such candidates might participate in signaling pathways that coordinate genome rearrangements.
While 118 predicted MIC genes have paralogs in the Oxytricha MAC genome, several MIC-specific protein domains are not identified in the MAC (Table S5). An example is Ctf8 (chromosome transmission fidelity 8), a component of a DNA clamp loader involved in sister chromatid cohesion and usually associated with mitosis/meiosis, both specific to the MIC, although it could also associate with polytene chromosomes during differentiation (Spear and Lauth, 1976). Proteomic analysis confirmed Ctf8 expression, with seven unique peptides, one of which contained serine 36 phosphorylation (Table S3), suggesting regulation. Most other MIC-specific protein domains are virus-associated, although the hits are often weak, as suggested by high E-values. This could be due to viral integration into the Oxytricha MIC genome. Moreover, these virus-associated genes use the Oxytricha genetic code and some of the contigs containing them bear MDSs and/or TBEs, suggesting that they are not contaminants. In addition, mass spectrometry validated the presence of four viral proteins (Filoviridae VP35 domain and three Parvovirus nonstructural protein NS1 domain; Table S3). Mass spectrometry also identified specific phosphor-ylation or methylation of three of these proteins.
Similar to lamprey and Ascaris, the Oxytricha germline genome encodes a repertoire of chromatin-associated genes (Table S6), and these are significantly enriched in the phosphorylated and acetylated protein sets, relative to the total predicted MIC-limited genes (GO term enrichment test, p value = 7.55 × 10−5 and 1.63 × 10−3, respectively). These include a complete set of core histones (one H2A, two H2B, one H3, and one H4; H1 has not been identified in either the Oxytricha MAC or MIC) and genes associated with histone modification (three PHD, one SET, and six chromodomain proteins), suggesting a direct involvement of chromatin and chromatin remodeling proteins in genome rearrangement and germline maintenance. Histone N-terminal tails were the most heavily modified MIC-limited peptides in the proteomic analysis (Table S3). For example, Histone H3 contains both repressive marks (e.g., H3K9 and H3K27 trimethylation) and activating marks (e.g., H3K9 acetylation) plus H3K4 monomethylation, which can be either activating or repressing (Cheng et al., 2014). Histone H4 was heavily modified (61 detected PTMs) with both activating and repressive marks identified on the same residues. The most heavily modified MIC-limited gene was an H2B variant, with 83 PTMs. We note that a Euplotes development-specific histone H3 (encoded in the MAC) has conjugation-specific expression in the developing MAC (Ghosh and Klobutcher, 2000). The presence of Oxytricha MIC-specific chromatin components and modifiers could allow changes in nucleosome composition and chromatin structure to regulate genome rearrangement. Development-specific his-tones or their modifications could either mark DNA segments for genome restructuring or alter the chromatin structure to allow access to machinery for DNA deletion and rearrangement. Proteomic analysis confirmed expression of a histone acetyltransferase of the MOZ/SAS family (18 unique peptides) with its own acetylation at lysines 7 and 257 (Table S3), suggesting the possibility of self-regulation through autoacetylation. Thirty unique peptides also confirmed expression of a MIC-limited SET domain histone methyltransferase. In addition to known epigenetic modifiers, the MIC genome encodes other genes that could directly manipulate DNA during genome rearrangement, such as a DNA topoisomerase (that could regulate DNA unwinding during recombination), a helicase, and an HTH_Tnp_Tc5 (Tc5 transposase DNA-binding domain) protein.
GO terms associated with predicted MIC-limited genes are especially enriched in activities related to methylation (Table 2). The MIC encodes a large set of 61 MT-A70 domain proteins (RNA adenosine methyltransferases, four of which are confirmed by proteomic analysis) that could participate in many steps during RNA-guided DNA rearrangement. During conjugation, both long, noncoding RNAs and 27 nt piRNAs are produced from the parental MAC and transported to the developing MAC, providing essential information about which sequences to retain and their rearrangement order (Fang et al., 2012; Nowacki et al., 2008). RNA adenosine methylation is a widespread and dynamically regulated posttranscriptional RNA modification (Meyer et al., 2012). It might function in Oxytricha to regulate noncoding RNAs or to mark specific sites or sequences on noncoding RNAs that guide genome rearrangement during development.
Table 2.
GO Terms Enriched in Predicted Germline-Limited Genes
| GO Term | Description | Ratio in MIC-Limited Genes | Ratio in All Genes (MIC + MAC) | Fold Enrichment | Bonferroni-Corrected p Value |
|---|---|---|---|---|---|
| GO:0008168 | methyltransferase activity | 56/810 | 171/21,693 | 8.8 | 1.50 × 10–8 |
| GO:0016741 | transferase activity, transferring one-carbon groups | 56/810 | 173/21,693 | 8.7 | 1.99 × 10–8 |
| GO:0032259 | methylation | 56/810 | 165/21,693 | 9.1 | 3.38 × 10–8 |
| GO:0006139 | nucleobase-containing compound metabolic process | 64/810 | 760/21,693 | 2.3 | 6.79 × 10–7 |
| GO:0006725 | cellular aromatic compound metabolic process | 64/810 | 784/21,693 | 2.2 | 1.46 × 10–6 |
| GO:0046483 | heterocycle metabolic process | 64/810 | 786/21,693 | 2.2 | 2.40 × 10–6 |
| GO:1901360 | organic cyclic compound metabolic process | 64/810 | 793/21,693 | 2.2 | 2.81 × 10–6 |
| GO:0034641 | cellular nitrogen compound metabolic process | 64/810 | 798/21,693 | 2.1 | 3.26 × 10–6 |
IES-less Genes
IES-less genes are a second category of genes that can be expressed from the micronucleus or developing macronucleus, but also the MAC, itself. While they display different expression levels during a conjugating time-series, most IES-less genes are highly expressed at 40 and 60 hr (Figure 5C). Furthermore, they have significantly higher expression during conjugation than genes whose MIC precursors contain IESs (2 sample t test, p value = 2.585 × 10−9 and 0.0003037 for 40 and 60 hr, respectively). Their gene features lie between those of MAC-specific and MIC-specific genes (Table S2). In particular, they contain fewer introns per gene than genes encoded on MAC nanochromosomes with IESs (chi-square test, p value = 3.38 × 10−7). Among the 530 genes encoded on 548 IES-less nanochromosomes, 278 lack introns. It is possible that expression from the MIC contributes to their high expression levels, especially when the parental MAC is degraded. To query MIC-specific expression, we examined RNA-seq reads that mapped to MIC loci for these 530 genes and found 21 cases where reads mapped beyond all detected MAC telomere addition sites, consistent with transcription from the MIC or from incorrectly processed MAC chromosome ends.
While GO term enrichment analysis suggests that these IES-less genes are not enriched for specific functions (except carboxylesterase activity, GO:0004091), future studies can address whether the absence of IESs specifically regulates their early expression or is a requirement for function during genome rearrangement.
Repetitive Elements
Finally, we analyzed the types and percentages of various repetitive elements in the genome, with the caveat that genome assembly of repetitive regions poses a special challenge and may be an underrepresentation. The MIC genome contains four types of germline-limited transposable elements, of which only TBEs were previously described in Oxytricha (Doak et al., 1994; Herrick et al., 1985; Nowacki et al., 2009): TBE transpo-sons (DDE family cut-and-paste DNA transposons), LINEs, Helitrons (rolling-circle transposons), and insertion sequences (Table S8). Oxytricha's germline genome appears less transposon-rich than our own, which is roughly half transposon-derived (de Koning et al., 2011; Lander et al., 2001). With the caveat that transposable elements may be degenerate and individual sequences not assembled accurately, we predicted hundreds of genes associated with these transposons (reverse transcriptase and endonuclease domain genes, Helitron-associated helicases and DDE_Tnp_IS1595 [ISXO2-like transposase] domain genes). Their expression is also limited to 40–60 hr into conjugation (Figure 5D), suggesting that, like TBE transposases (Nowacki et al., 2009), they could be recruited to function during genome rearrangement. Mass spectrometry confirmed expression at 40 hr of Helitron-associated helicases and LINE-associated genes, both of which are often posttranslationally modified, suggesting regulated functions (Table S7).
The largest class of repetitive elements, TBE transposons frequently map near MDSs and interrupt at least 6,776 nanochromosome gene loci. They encode three genes: a 42 kDa transposase, implicated in genome rearrangement (Nowacki et al., 2009), a 22 kDa unknown ORF, and a 57 kDa gene with a zinc finger/kinase domain (Witherspoon et al., 1997) and fall into three classes: TBE1, TBE2, and TBE3, with two subfamilies that we identified within TBE2 (see Extended Experimental Procedures). Long PacBio reads allowed us to successfully assemble as many as 16 complete TBE transposons on a single MIC contig (three TBE1, seven TBE2, and six TBE3), demonstrating the power of this approach to resolve repetitive regions.
LINE elements, also present in the Tetrahymena germline (Fillingham et al., 2004), interrupt at least nine Oxytricha precursor gene loci. In the Oxytricha MAC genome, telomerase is the only protein containing an RVT_1 (reverse transcriptase) domain, commonly associated with telomerases or retrotransposons. Curiously, in the MAC genomes of both Paramecium and Tetrahymena, the RVT_1 domain is present in other genes besides telomerase. Paramecium gene model GSPATP00023049001 matches the RVT_1 domain with a HMMER E-value of 1.3 × 10−4 and does not show significant differential expression during conjugation (http://Paramecium.cgm.cnrs-gif.fr/db/feature/217802). In Tetrahymena, gene model TTHERM_00129610 matches RVT_1 domain with a HMMER E-value of 8.9 × 10−30 and is upregulated during conjugation (http://tfgd.ihb.ac.cn/search/detail/ gene/TTHERM_00129610). It is possible that their LINE-associated reverse transcriptases were domesticated in the MAC, unless those genome assemblies are contaminated by MIC sequences (that is less likely because the two RVT_1 domain genes are located on long MAC contigs [>300 kb]). Helitrons, on the other hand, do not appear to interrupt any Oxytricha precursor gene loci. Seven Helitron-associated genes are also present in the Tetrahymena MIC, suggesting a possible deeper evolutionary origin.
Swart et al. (2013) previously predicted 21 proteins containing Phage_integrase, DDE_Tnp_IS1595, or MULE transposase domains in the MAC proteome of Oxytricha but not Tetrahymena or Paramecium. All these Oxytricha transposase domain genes show highly conjugation-specific expression. Their MIC precursors all contain IESs, however, and none are full-length transpo-sons. We did not find any germline transposons bearing Phage_integrase or MULE domain genes, but we did identify hundreds of MIC insertion sequences (0.12% – 0.2% of the MIC genome; Table S8) that carry DDE_Tnp_IS1595 domain genes. These sequences interrupt at least 24 precursor gene loci in the MIC. Although usually rare in eukaryotes, this protein domain is present in Stylonychia's MAC genome and Perkinsus (Swart et al., 2013), but absent from Paramecium's MAC genome and also appears absent from both the MAC and MIC genomes of Tetrahymena. Full-length insertion sequences are also rare in eukaryotes. Hence, the discovery of insertion sequences bearing these transposase genes and with conjugation-specific expression suggests they could be another class of domesticated transposase recruited to genome rearrangement.
Two major classes of satellite repeats that were previously identified by hybridization, but not sequencing (Dawson et al., 1984) are also present in the germline, with repeat units of 170 and 380 bp, respectively (Table S8; Extended Experimental Procedures). They rarely interrupt MDSs (just 11 cases) and some fosmid clones have the same repeat sequence present at both ends, suggesting that large, satellite repeat-dense regions may cluster independently of MDS-rich clusters. This satellite organization may facilitate their complete elimination during development.
Conclusions
The assembled Oxytricha micronuclear genome greatly expands our perspective on the limits of genome complexity, displaying an unprecedentedly fragmented and scrambled genome architecture, with thousands of scrambled genes. We provide complete germline-somatic maps for the majority of genes and a window into nuclear development at a whole-genome level.
The correct interpretation of complex MDS-IES structures relies on the accuracy of genome assembly. Because macronu-clear contamination was nearly absent from our micronuclear DNA preparations (see Extended Experimental Procedures), interference of macronuclear sequences in the assembly was kept to a minimum. We also validated portions of the assembly via several different approaches (see Extended Experimental Procedures). The assembly agrees with the validation data in all cases in nonrepetitive regions. Therefore, we conclude that this assembly is an accurate representation of the Oxytricha germline genome. Repetitive regions offer a significant challenge for the assembly of any complex genome. Despite the fragmentary nature of the assembly at repetitive regions, we were able to achieve a relatively continuous assembly of the nonrepetitive regions, especially the MDS-containing regions, from which we derive most of our biological conclusions.
The discovery of hundreds of germline-limited nonrepetitive genes is unique among unicellular eukaryotes, so far, and elegantly suggests a cache of functional genes that support somatic differentiation and genome rearrangement when the maternal somatic genome is destroyed, consistent with the proposed use of chromatin diminution for germline gene regulation in lamprey (Smith et al., 2012) and Ascaris (Wang et al., 2012). Validation of 26% of these germline-limited genes by MS-based proteomics, plus identification of posttranslational modifications affecting half of the validated genes, hints at a complex protein regulatory network during somatic differentiation. The Oxytricha germline genome assembly presented here provides a valuable resource for comparative genomics, even within a single cell, a window into the extreme limits of eukaryotic genome architecture, and a platform for future studies of genome remodeling.
EXPERIMENTAL PROCEDURES
See the Extended Experimental Procedures for detailed protocols.
Nuclei Isolation, DNA Extraction, and Genome Sequencing
We grew vegetative cultures of Oxytricha strain JRB310 and isolated micronuclei using sucrose gradient centrifugation, as described in Lauth et al. (1976). Different libraries were prepared from extracted DNA and sequenced with Illumina HiSeq 2000 and PacBio platforms.
Genome Assembly
Illumina unitigs assembled from MaSuRCA (Zimin et al., 2013) were used to error correct PacBio reads with ectools (https://github.com/jgurtowski/ectools). Corrected PacBio reads were assembled using the Celera assembler (Miller et al., 2008).
Identifying Genome Rearrangement Junctions and MDS, IES, and Pointer Designations
We mapped the MAC genome assembly (excluding telomeres) onto the MIC assembly using BLASTN (BLAST+ [Camacho et al., 2009] parameters: -ungapped -word_size 20 -outfmt 6). Paralogous MDS regions were filtered out if they had poor sequence similarity (<98%) to the MAC and no pointers between consecutive matches. Custom Python scripts were used to extract MDSs, IESs, and pointers from the BLAST output. MDSs, IESs, and pointers are defined below. Complete MDS-IES maps are available at http://trifallax. princeton.edu/cms/raw-data/genome/mic/Oxytricha_trifallax_micronuclear_genome_MDS_IES_maps.gff.
Pointers
Pointers are short sequences of microhomology repeated at MDS-IES junctions and present as one copy at consecutive MDS-MDS junctions in the MAC. No mismatch is allowed between the two copies of each pointer.
MDSs
MDSs are sequence blocks retained in the MAC, excluding the pointers.
IESs
IESs are sequence blocks that separate MDSs in the MIC and are absent from the MAC, excluding the pointers.
Nonscrambled MDSs
Nonscrambled MDSs are MDSs that are in the same orientation and order in the MIC relative to the MAC.
Scrambled MDSs
Scrambled MDSs are MDSs that are not in the same orientation or order in the MIC relative to the MAC.
Nonscrambled IESs
Nonscrambled IESs are IESs between two MDSs that are in the same orientation and order in the MIC relative to the MAC. They are flanked by identical pointer repeats.
Scrambled IESs
Scrambled IESs are IESs between two MDSs that are not in the same order or orientation in the MIC relative to the MAC. They are flanked by different pointer sequences.
Gene Prediction
We gathered RNA-seq reads from Swart et al. (2013) and assembled a transcriptome using only reads that do not map to the MAC genome assembly with SOAPdenovo-Trans (Li et al., 2010), Trinity (Grabherr et al., 2011) and PASA (Haas et al., 2003). We predicted gene models with AUGUSTUS (version 2.5.5) (Stanke and Morgenstern, 2005) using assembled transcripts as hints.
Supplementary Material
ACKNOWLEDGMENTS
We thank the late David Prescott for suggesting sucrose purification of the Oxytricha micronuclei, Jingmei Wang for laboratory assistance, Jessica Wiggins, Wei Wang, and Donna Storton of the Princeton Sequencing Core Facility for assistance with Illumina library preparation and sequencing, Gintaras Deikus for assistance with PacBio sequencing, Klaas Schotanus, Mariusz Nowacki, Wenwen Fang, and all current laboratory members for discussion. We thank Jue Ruan at Beijing Institute of Genomics for advice on the assembly strategy. We are grateful to National Center for Genome Analysis Support (NCGAS) computing resources (supported by National Science Foundation [NSF] grant ABI-1062432 to Indiana University). This study was supported by NIH grant GM59708 and GM109459 and NSF grants 0900544 and 0923810 to L.F.L.. J.R.B. was supported by NIH fellowship 1F32GM099462 and A.D.G. was a National Aeronautics and Space Administration (NASA) postdoctoral fellow.
Footnotes
AUTHOR CONTRIBUTIONS
J.R.B. optimized experimental procedures and isolated micronuclei and extracted genomic DNA. X.C. assembled the genome, conducted the bioinformatic analyses in the paper, and drafted the manuscript. T.G.D., J.R.B., A.D.G., and L.F.L. conceived the project, which L.F.L. supervised. R.P.S. performed PacBio library preparation and sequencing. D.H.P performed mass spectrometry experiments. J.R.B. and D.H.P. analyzed the proteomic data. J.R.B., A.D.G., A.S., and C.T.A. prepared and sequenced the fosmid and BAC clones. D.M.C. performed assembly validation by PCR. E.D. produced chord diagrams for visualization of germline-soma maps. E.C.S. provided advice on genome analysis and edited the manuscript. J.R.B., D.H.P., and R.P.S. contributed to the writing of the manuscript, which X.C., J.R.B., and L.F.L. extensively edited.
ACCESSION NUMBERS
The GenBank accession number for the genome assembly and the raw sequencing data reported in this paper is ARYC00000000.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, four figures, and eight tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2014.07.034.
REFERENCES
- Arnaiz O, Mathy N, Baudry C, Malinsky S, Aury J-M, Wilkes CD, Garnier O, Labadie K, Lauderdale BE, Le Mouël A, et al. The Paramecium germline genome provides a niche for intragenic parasitic DNA: evolutionary dynamics of internal eliminated sequences. PLoS Genet. 2012;8:e1002984. doi: 10.1371/journal.pgen.1002984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury J-M, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Ségurens B, Daubin V, Anthouard V, Aiach N, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Boswell RE, Jahn CL, Greslin AF, Prescott DM. Organization of gene and non-gene sequences in micronuclear DNA of Oxytricha nova. Nucleic Acids Res. 1983;11:3651–3663. doi: 10.1093/nar/11.11.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Blum R, Bowman C, Hu D, Shilatifard A, Shen S, Dynlacht BD. A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol. Cell. 2014;53:979–992. doi: 10.1016/j.molcel.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson D, Buckley B, Cartinhour S, Myers R, Herrick G. Elimination of germ-line tandemly repeated sequences from the somatic genome of the ciliate Oxytricha fallax. Chromosoma. 1984;90:289–294. doi: 10.1007/BF00287037. [DOI] [PubMed] [Google Scholar]
- de Koning APJ, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doak TG, Doerder FP, Jahn CL, Herrick G. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc. Natl. Acad. Sci. USA. 1994;91:942–946. doi: 10.1073/pnas.91.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Wang X, Bracht JR, Nowacki M, Landweber LF. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell. 2012;151:1243–1255. doi: 10.1016/j.cell.2012.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass JN, Joshi NA, Couvillion MT, Bowen J, Gorovsky MA, Hamilton EP, Orias E, Hong K, Coyne RS, Eisen JA, et al. Genome-scale analysis of programmed DNA elimination sites in Tetrahymena thermophila. G3 (Bethesda) 2011;1:515–522. doi: 10.1534/g3.111.000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham JS, Thing TA, Vythilingum N, Keuroghlian A, Bruno D, Golding GB, Pearlman RE. A non-long terminal repeat retrotransposon family is restricted to the germ line micronucleus of the ciliated protozoan Tetrahymena thermophila. Eukaryot. Cell. 2004;3:157–169. doi: 10.1128/EC.3.1.157-169.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Klobutcher LA. A development-specific histone H3 localizes to the developing macronucleus of Euplotes. Genesis. 2000;26:179–188. doi: 10.1002/(sici)1526-968x(200003)26:3<179::aid-gene3>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Goday C, Esteban MR. Chromosome elimination in sciarid flies. BioEssays. 2001;23:242–250. doi: 10.1002/1521-1878(200103)23:3<242::AID-BIES1034>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Jr., Smith RK, Jr., Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick G, Cartinhour S, Dawson D, Ang D, Sheets R, Lee A, Williams K. Mobile elements bounded by C4A4 telomeric repeats in Oxytricha fallax. Cell. 1985;43:759–768. doi: 10.1016/0092-8674(85)90249-1. [DOI] [PubMed] [Google Scholar]
- Jacobs ME, Klobutcher LA. The long and the short of developmental DNA deletion in Euplotes crassus. J. Eukaryot. Microbiol. 1996;43:442–452. doi: 10.1111/j.1550-7408.1996.tb04503.x. [DOI] [PubMed] [Google Scholar]
- Karamysheva Z, Wang L, Shrode T, Bednenko J, Hurley LA, Ship-pen DE. Developmentally programmed gene elimination in Euplotes crassus facilitates a switch in the telomerase catalytic subunit. Cell. 2003;113:565–576. doi: 10.1016/s0092-8674(03)00363-5. [DOI] [PubMed] [Google Scholar]
- Katz LA, Kovner AM. Alternative processing of scrambled genes generates protein diversity in the ciliate Chilodonella uncinata. J. Exp. Zoolog. B Mol. Dev. Evol. 2010;314:480–488. doi: 10.1002/jez.b.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA. Micronuclear organization of macronuclear genes in the hypotrichous ciliate Oxytricha nova. J. Protozool. 1987;34:424–428. doi: 10.1111/j.1550-7408.1987.tb03206.x. [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Herrick G. Consensus inverted terminal repeat sequence of Paramecium IESs: resemblance to termini of Tc1-related and Euplotes Tec transposons. Nucleic Acids Res. 1995;23:2006–2013. doi: 10.1093/nar/23.11.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA, Herrick G. Developmental genome reorganization in ciliated protozoa: the transposon link. In: Cohn WE, Moldave K, editors. Progress in Nucleic Acid Research and Molecular Biology. Academic Press; Waltham, MA: 1997. pp. 1–62. [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Huff ME, Gonye GE. Alternative use of chromosome fragmentation sites in the ciliated protozoan Oxytricha nova. Nucleic Acids Res. 1988;16:251–264. doi: 10.1093/nar/16.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. An overview of nested genes in eukaryotic genomes. Eukaryot. Cell. 2009;8:1321–1329. doi: 10.1128/EC.00143-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S, Chang W-J, Landweber LF. Complex germline architecture: two genes intertwined on two loci. Mol. Biol. Evol. 2006;23:4–6. doi: 10.1093/molbev/msj017. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lauth MR, Spear BB, Heumann J, Prescott DM. DNA of ciliated protozoa: DNA sequence diminution during macronuclear development of Oxytricha. Cell. 1976;7:67–74. doi: 10.1016/0092-8674(76)90256-7. [DOI] [PubMed] [Google Scholar]
- Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 30 UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Delcher AL, Koren S, Venter E, Walenz BP, Brownley A, Johnson J, Li K, Mobarry C, Sutton G. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics. 2008;24:2818–2824. doi: 10.1093/bioinformatics/btn548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Higgins BP, Maquilan GM, Swart EC, Doak TG, Landweber LF. A functional role for transposases in a large eukaryotic genome. Science. 2009;324:935–938. doi: 10.1126/science.1170023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Haye JE, Fang W, Vijayan V, Landweber LF. RNA-mediated epigenetic regulation of DNA copy number. Proc. Natl. Acad. Sci. USA. 2010;107:22140–22144. doi: 10.1073/pnas.1012236107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Huallachain M, Karczewski KJ, Weissman SM, Urban AE, Snyder MP. Extensive genetic variation in somatic human tissues. Proc. Natl. Acad. Sci. USA. 2012;109:18018–18023. doi: 10.1073/pnas.1213736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM. The DNA of ciliated protozoa. Microbiol. Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM. The evolutionary scrambling and developmental un-scrambling of germline genes in hypotrichous ciliates. Nucleic Acids Res. 1999;27:1243–1250. doi: 10.1093/nar/27.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Baker C, Eichler EE, Amemiya CT. Genetic consequences of programmed genome rearrangement. Curr. Biol. 2012;22:1524–1529. doi: 10.1016/j.cub.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear BB, Lauth MR. Polytene chromosomes of Oxytricha: biochemical and morphological changes during macronuclear development in a ciliated protozoan. Chromosoma. 1976;54:1–13. doi: 10.1007/BF00331828. [DOI] [PubMed] [Google Scholar]
- Stanke M, Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–W467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart EC, Bracht JR, Magrini V, Minx P, Chen X, Zhou Y, Khurana JS, Goldman AD, Nowacki M, Schotanus K, et al. The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol. 2013;11:e1001473. doi: 10.1371/journal.pbio.1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mitreva M, Berriman M, Thorne A, Magrini V, Koutsovoulos G, Kumar S, Blaxter ML, Davis RE. Silencing of germline-expressed genes by DNA elimination in somatic cells. Dev. Cell. 2012;23:1072–1080. doi: 10.1016/j.devcel.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherspoon DJ, Doak TG, Williams KR, Seegmiller A, Seger J, Herrick G. Selection on the protein-coding genes of the TBE1 family of transposable elements in the ciliates Oxytricha fallax and O. trifallax. Mol. Biol. Evol. 1997;14:696–706. doi: 10.1093/oxfordjournals.molbev.a025809. [DOI] [PubMed] [Google Scholar]
- Yu P, Ma D, Xu M. Nested genes in the human genome. Genomics. 2005;86:414–422. doi: 10.1016/j.ygeno.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Zimin AV, Marçais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. The MaSuRCA genome assembler. Bioinformatics. 2013;29:2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.