Abstract
Osmoregulated periplasmic glucans (OPGs) of Escherichia coli are anionic and highly branched oligosaccharides that accumulate in the periplasmic space in response to low osmolarity of the medium. The glucan length, ranging from 5 to 12 glucose residues, is under strict control. Two genes that form an operon, mdoGH, govern glucose backbone synthesis. The new gene mdoD, which appears to be a paralog of mdoG, was characterized in this study. Cassette inactivation of mdoD resulted in production of OPGs with a higher degree of polymerization, indicating that OpgD, the mdoD product (according to the new nomenclature), controls the glucose backbone structures. OpgD secretion depends on the Tat secretory pathway. Orthologs of the mdoG and mdoD genes are found in various proteobacteria. Most of the OpgD orthologs exhibit a Tat-dependent secretion signal, while most of the OpgG orthologs are Sec dependent.
Osmoregulated periplasmic glucans (OPGs) are a family of oligosaccharides found in the periplasm of gram-negative bacteria. Their common features are the presence of glucose as the sole constituent sugar and their increased levels in low-osmolarity media. These glucans are cyclic, branched cyclic, or branched linear, and they may be substituted by various residues in different species (3).
In Escherichia coli, OPGs are composed of 5 to 12 glucose units, and the principal species contain eight or nine glucose residues. Despite this length heterogeneity, the distribution of the various glucose backbones is strictly conserved. The structure is highly branched, and the backbone consists of linear β-1,2-linked glucose units to which the branches are attached by β-1,6 linkages; the average number of branches is three. The glucose backbone is substituted with sn-1-phosphoglycerol and phosphoethanolamine, both derived from membrane phospholipids, and with succinic acid O-ester residues from the cytoplasmic pool (15, 18).
OPGs were discovered by E. P. Kennedy and his collaborators during analysis of phosphatidylglycerol turnover in E. coli (31). sn-1-Phosphoglycerol was recovered on oligosaccharides named, for this reason, membrane-derived oligosaccharides. We now know that these compounds are found in many proteobacteria, and most of them do not contain any substituent derived from membrane lipids. The term membrane-derived oligosaccharide is confusing because these compounds are part of the envelope but may not be derived from the membrane. For these reasons the protein nomenclature was changed from Mdo to Opg in Swiss-Prot release 42 (October 2003).
Isolation of OPG-deficient mutants has been difficult because of the absence of a selectable phenotype and the absence of a reliable screening method. The first Opg− mutant (affected in glucan backbone synthesis) was isolated as a spontaneous mdoA1 mutant defective in glucosyl transferase activity (4, 19). The mdoA locus was subsequently cloned and was found to be an operon consisting of two genes, mdoGH (encoding the proteins OpgG and OpgH). The OpgH protein exhibits glucosyl transferase activity, catalyzing in vitro elongation of the linear β-1,2 glucose backbone from UDP-glucose. A nonpolar mdoG mutation was created. Although mdoG mutants are Opg−, the function of OpgG in glucan backbone synthesis remains elusive (19). Nevertheless, this protein could be involved in the formation of the β-1,6 glucose linkage and/or in the periplasmic release of newly synthesized OPGs (3). Several attempts have been made to isolate new Opg− mutants. In all cases (32; unpublished observations), the mutations were mapped in the mdoGH operon or in the galU gene necessary for UDP-glucose synthesis.
Two genes implicated in OPG substitution were found after transposon mutagenesis. The first gene, mdoB, encodes phosphoglycerol transferase I. This enzyme can transfer phosphoglycerol to an artificial acceptor, the β-glucoside arbutin. This transfer leads to the formation of diacylglycerol, which is toxic for cells unless it is immediately phosphorylated by the diacylglycerol kinase (encoded by the dgk gene), yielding phosphatidic acid. When a dgk strain was grown in the presence of arbutin, diacylglycerol accumulation increased and growth stopped. This was the basis for selection of mdoB mutants in which an increase in diacylglycerol did not occur (14). The second gene, mdoC, encodes the succinyl transferase and was isolated from an mdoB strain. In such a strain, succinyl residues are the only anionic substituents of OPGs. A thin-layer chromatographic screening protocol was based on the difference in migration between anionic and neutral OPGs (18).
OPGs are probably synthesized by a multiprotein complex embedded in the cell membrane (3). Three proteins (OpgH, OpgC, and OpgB) span the membrane; one protein is at the cytoplasmic surface of the membrane (acyl carrier protein), and two are at the periplasmic surface (OpgG, and OpgB′, a soluble version of OpgB) (Y. Lequette, E. Lanfroy, A. Bohin, J.-M. Lacroix, V. Cogez, and J.-P. Bohin, unpublished data). At least one more protein, which is necessary for phosphoethanolamine substitution, is expected to be membrane bound.
In this study, we characterized a paralog of mdoG in E. coli, ydcG, and the locus could be identified only by in silico analysis. We found that this gene encodes a protein with an unexpected function in OPG biosynthesis. Neither synthesis nor substitution was drastically affected in a ydcG mutant, but the size of the glucose backbone of OPG molecules was altered. Thus, ydcG was renamed mdoD. The OpgD protein, like OpgG, is a periplasmic protein, but, unlike OpgG, it is secreted via the twin-arginine translocation (Tat) pathway. Furthermore, phylogenetic studies showed that OpgG and OpgD belong to a family of proteins (the OpgG/D family) present in many proteobacteria. In this family, most of the OpgD orthologs exhibit a Tat signal sequence, whereas most of the OpgG orthologs appear to be secreted via the Sec system.
MATERIALS AND METHODS
Bacterial strains and media.
The E. coli K-12 strains used in this study are listed in Table 1. Bacteria were grown at 37°C with vigorous shaking in Luria broth or in minimal medium 63 supplemented with the required metabolites and glucose as the carbon source (22). Solid media were obtained by adding agar (15 g/liter). Low-osmolarity medium (LOS medium) was used for most assays (20), and NaCl was added to LOS medium to increase osmolarity.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| Strain | ||
| MC4100 | Δ(lac)U169 araD139 flbB5301 ptsF25 relA1 rpsL150 rbsR deoC1 | Laboratory collection |
| MCMTA | Δ(lac)U169 araD139 flbB5301 ptsF25 tatB::kan relA1 rpsL150 rbsR deoC1 | L.-F. Wu |
| JC7623 | argE3 hisG4 leuB6 Δ(gpt-proA)62 thr-1 thi-1 ara-14 galK2 lacY1 mtl-1 xyl-1 kdgK51 tsx33 recB21 recC22 sbcB15 sbcC201 glnV44 rpsL31 rac | A. Cohen and A. J. Clark |
| DF214 | his pgi::Mu Δ(zwf-edd)1 eda-1 rpsL | D. Fraenkel |
| NFB1967 | DF214 mdoD218::cml | This study |
| NFB1567 | NFB1967(pNF599) | This study |
| NFB1653 | NFB1967(pNF1091) | This study |
| NFB1655 | NFB1967(pNF1092) | This study |
| NFB4516 | DF214 tatB::kan | This study |
| Plasmids | ||
| pNF599 | ydcJ-mdoD in pUC18 | This study |
| pNF1091 | pNF599 with R4K and R5K mutations | This study |
| pNF1092 | pNF599 with R3K and R4K mutations | This study |
Antibiotics in media were used at the following concentrations: ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; and chloramphenicol, 15 μg/ml. In addition, isopropyl-β-d-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were used at concentrations of 1 mM and 20 μg/ml, respectively.
Transduction and transformation.
Transduction by phage P1vir and conjugation were carried out as described by Miller (22). E. coli cells were made competent by the rubidium chloride technique (22).
Recombinant DNA techniques.
Standard procedures (26) were used for genomic DNA extraction, λ DNA extraction, large-scale plasmid isolation, and rapid analysis of recombinant plasmids. Restriction endonucleases (Biolabs), the large fragment of DNA polymerase I (Klenow), and ligase of T4 phage (Gibco-BRL) were used according to the manufacturers' recommendations.
Cloning of mdoD and construction of the mdoD deletion strain.
The mdoD locus was cloned into pUC18 as a 4.7-kb SphI-HpaI fragment originating from λ phage 1A6 DNA (17) to obtain pNF599 (coordinates 1492868 to 1497608 of the E. coli MG1655 chromosome [http://asap.ahabs.wisc.edu/annotation/php/ASAP1.htm]). This plasmid harbors ydcJ upstream of mdoD. The 1.4-kb MluI-XbaI fragment of the mdoD coding sequence was replaced with the 0.9-kb SfuI Cmlr cassette of pBR328. Then the recombinant plasmid was linearized with EcoRI and transformed into the JC7623 strain (recBC sbcB sbcC). Clones resulting from homologous recombination were selected on chloramphenicol-containing plates. To generate NFB1967, a P1vir lysate of a Cmlr Amps clone was used to transduce strain DF214. Disruption of the mdoD gene was confirmed by shotgun cloning of the mutation from the chromosome into pUC18.
Site-directed mutagenesis.
Two specific mutations that changed two consecutive residues (RR to KK) were introduced into pNF599 by using a PCR-based method. DNA covering the mdoD signal peptide was amplified by using pNF599 as the template with forward primer 5′-GATTTATGCATATTCTCTCAGTTCAACAATTGG-3′ and one of two alternative mutagenic reverse primers, 5′-CACGGCGGCCATAGCCATTGAACCTTTAATAAATTTTTTACGATCCAT AC-3′ (primer 1) or 5′-CACGGCGGCCATAGCCATTGAACCTTTAATAAATCGTTTTTTATCCATACCTG-3′ (primer 2). The 200-bp reaction fragments were digested with MfeI and EaeI (present in the primers) and cloned by exchange into pNF599. Mutated plasmids were identified by screening for loss of the AccI site and were verified by sequencing to obtain pNF1091 (primer 1, R4K and R5K) and pNF1092 (primer 2, R3K and R4K).
Extraction of OPGs for structural analysis.
E. coli cultures (200 ml) were grown in LOS medium. OPGs were extracted by the charcoal adsorption procedure and were eluted with aqueous pyridine as previously described (20). The pyridine extract obtained by this procedure was chromatographed on a Bio-Gel P4 column (Bio-Rad). The column (1.5 by 65 cm) was equilibrated with 0.5% acetic acid and eluted at a rate of 15 ml/h in the same buffer. Fractions (1.5 ml) containing OPGs were pooled, concentrated by rotary evaporation, and desalted on a Bio-Gel P2 column (Bio-Rad). Fractions containing OPGs were pooled and concentrated by rotary evaporation for subsequent analysis. When necessary, the sugar content was determined colorimetrically by using the phenol-sulfuric acid reagent procedure (11).
Determination of neutral and anionic characteristics of OPGs.
Cultures (5 ml) of E. coli were grown in LOS medium with 0.24 mM d-[U-14C]glucose (125 MBq/mmol) or 0.45 mM [2-3H]glycerol (291 MBq/mmol) and 0.24 mM glucose. OPGs were prepared as described above and were desalted on a PD10 column (Pharmacia Biotech) equilibrated with 10 mM Tris-HCl (pH 7.4). Fractions containing OPGs were pooled, chromatographed on a DEAE-Sephacel column (1.5 by 38 cm; Pharmacia Biotech) equilibrated with 10 mM Tris-HCl (pH 7.4), and eluted with the same buffer containing increasing concentrations of NaCl ranging from 0 to 1 M in steps of 0.05 M. Sixty milliliters was used for each NaCl concentration, and 4-ml fractions were collected.
Removal of substituents from OPGs.
Substituents were removed from OPGs in two steps. For deesterification, OPGs were incubated in 0.05 M KOH at 37°C for 1 h. After neutralization with AG 50W-X8 (Bio-Rad) on H+ form, the samples were desalted on a Bio-Gel P2 column and lyophilized. Then phosphodiester linkages were removed from the oligosaccharides by treatment with 4 M HF for 60 h at 4°C. The HF was precipitated by adding 5 volumes of saturated LiOH, and the LiF precipitate was removed by centrifugation. The supernatant was neutralized with AG 50W-X8 on H+ form and was desalted on a Bio-Gel P2 column. The oligosaccharides were lyophilized and dissolved in water at a glucose concentration of 1 μg/ml for mass spectrometry (MS) analysis.
MALDI MS.
Matrix-assisted laser desorption ionization (MALDI) MS experiments were carried out with a Vision 2000 (Finnigan MAT, Bremen, Germany) time of flight mass spectrometer equipped with a nitrogen laser (wavelength, 337 nm; pulse width, 3 ns). After selection of the appropriate site on the target with a microscope, the laser light was focused onto the sample matrix mixture at an angle of 15° and a power level of 106 to 107 W/cm2. Positive ions were extracted with a 5- to 10-keV acceleration potential and were focused with a lens, and the masses were separated by using a Reflectron time of flight instrument. At the detector, ions were postaccelerated to 20 keV for maximum detection efficiency. The resulting signals were recorded with a fast transient digitizer with a maximum channel resolution of 2.5 ns and were transferred to a personal computer for accumulation, calibration, and storage. All MALDI mass spectra were the result of 20 single-shot accumulations.
The following matrices for carbohydrate analysis were used: 2,5-dihydrobenzoic acid (10 g/liter in water) (28) and 3-aminoquinoline (10 g/liter in water) (29). Lyophilized oligosaccharide samples were dissolved in double-distilled water and then diluted with an appropriate volume of the matrix solution (1:5, vol/vol), 1 μl of the resulting solution was deposited onto a stainless steel target, and the solvent was evaporated under a gentle stream of warm air.
Methylation analysis.
Oligosaccharides were treated first with sodium borodeuteride. Glucosidic linkage analysis was performed by methylation by using the method of Paz Parente et al. (25). The methyl ether compounds were then hydrolyzed with trifluoroacetic acid (4 N at 100°C for 4 h), reduced with sodium borodeuteride, and peracetylated. The partially methylated and acetylated alditols were analyzed by gas-liquid chromatography (GLC)-MS (12). GLC was performed by using a Delsi apparatus with a capillary column (25 m by 0.2 mm) coated with DB-1 (film thickness, 0.5-μm), a temperature gradient from 110 to 240°C in which the temperature was increased at a rate of 3°C/min, and a helium pressure of 40 kPa. The mass spectra were recorded with a 10-10B mass spectrometer (Nermag, Rueil Malmaison, France) by using an electron energy of 70 eV and an ionizing current of 0.2 mA.
Sequence analysis.
E. coli OpgG and OpgD deduced amino acid sequences were compared by using the SIM program available at the ExPASy molecular biology server (http://www.expasy.org/tools/sim-prot.html).
The DNA sequences and deduced amino acid sequences of various bacterial species were analyzed by using computer programs and sequence data available from Infobiogen (http://www.infobiogen.fr/) and the Pasteur Institute (http://bioweb.pasteur.fr/).
A preliminary alignment of the full-length sequences of MdoG homologs was generated with CLUSTAL W by using default gap penalties. The CLUSTAL W alignment was then refined by manually deleting N- and C-terminal nonconserved sequences. The 42 protein sequences, starting from a conserved phenylalanine residue, ranged from 489 to 507 amino acids long. Phylogenetic trees were constructed by using maximum-parsimony (MP) and neighbor-joining methods. For the MP analyses we used the program PROTPARS implemented in PHYLIP (Phylogeny Inference Package; Joe Felsenstein, Department of Genetics, University of Washington). The PHYLIP programs SEQBOOT, PROTPARS, and CONSENSE were used sequentially to generate an MP tree which was replicated in 100 bootstraps; on this basis bootstrap confidence levels were determined.
RESULTS
mdoD is an mdoG paralog.
A search for sequences similar to OpgG revealed a potential mdoG paralog, ydcG, in the E. coli genome (1). This gene at 32.2 min on the genetic map is not linked to the mdoC-mdoGH locus at 23.9 min. The mdoG and ydcG products exhibit 39% identity and 64% similarity in a 506-amino-acid overlapping region. The N-terminal sequence of the ydcG product (ADSDIADGQTXR; accession number P40120) has been deposited in databases by A. J. Link. As part of the work of Link et al. on E. coli proteomic analysis (21), this protein was characterized as an unknown protein located in the periplasmic space of cells at the stationary growth phase in rich medium. Since a mutation in this gene affected OPG synthesis in E. coli (see below), ydcG was renamed mdoD, and its product was designated OpgD. Both OpgG and OpgD are periplasmic proteins, but when the complete predicted translation products of mdoG and mdoD were compared, a striking difference was apparent (Fig. 1): mdoG encodes a preprotein with a 22-amino-acid signal typical of the Sec translocation system, but mdoD encodes a preprotein with a 32-amino-acid signal typical of the Tat translocation system with the XRRXFLK consensus sequence, including the characteristic twin-arginine motif and enrichment of glycine residues in the hydrophobic region (9, 10, 24). OpgD shares this distinctive feature with 22 other periplasmic proteins found in E. coli (30).
FIG. 1.
Alignment of OpgG and OpgD amino acid sequences performed with the ExPASy server by using the SIM program. Asterisks indicate amino acids conserved in both proteins. Signal sequences are underlined. N-terminal residues of mature OpgD determined by Link et al. (25) are indicated by boldface type.
The mdoD218::cml mutation increases the heterogeneity of OPG molecules.
To determine the role of OpgD in OPG biosynthesis, the mdoD gene was cloned in plasmid pNF599, and the mdoD218::cml mutation was created and introduced into strain DF214 (see Materials and Methods). Strain DF214 and its derivatives are defective for both phosphoglucose isomerase and glucose-6-phosphate dehydrogenase activities and are unable to carry out either the synthesis or the catabolism of glucose. These strains can synthesize UDP-glucose and OPGs only when exogenous glucose is present in the medium.
Cells of strains DF214 (mdo+) and NFB1967 (mdoD218::cml) were grown in the presence of radioactive glucose. When cultures were in the late stationary growth phase, cells were collected and OPGs were extracted (see Materials and Methods). The two strains produced similar amounts of OPGs, indicating that interruption of mdoD did not eliminate OPG synthesis, in contrast to the mdoG mutation.
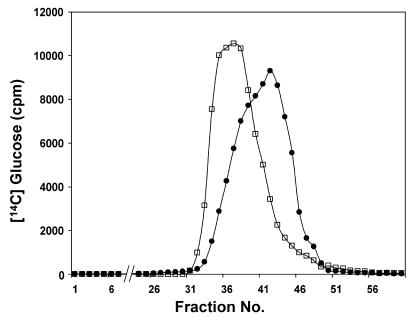
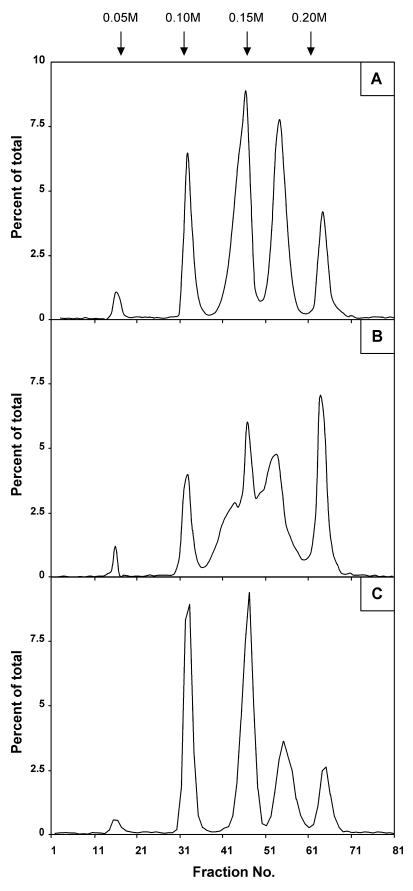
When chromatographed on a Bio-Gel P4 column, OPGs from both strains eluted in a single peak (which was missing in an mdoG or mdoH DF214 derivative strain [19, 20]), but the bulk of the OPGs of the mdoD strain reproducibly eluted seven fractions before the OPGs of wild-type strain DF214 eluted (Fig. 2). OPGs from both strains were analyzed by chromatography on a DEAE-Sephacel column (Fig. 3). By using this technique, OPGs of various strains can be separated into five well-defined subfractions (18, 23). For wild-type OPGs at pH 7.4, the anionic character is conferred by phosphoglycerol and succinyl substituents, and subfractions I to V consist of OPGs with increasing charge-to-mass ratios (Fig. 3A). This behavior originates from the limited heterogeneity in the degree of polymerization and the heterogeneity in the degree of substitution for each degree of polymerization. This complex situation appears to be under strict control since the elution pattern is very reproducible. OPGs extracted from the mdoD mutant strain exhibited a different behavior since subfractions III and IV were not well separated (Fig. 3B). When pNF599 was introduced into the mdoD strain, the wild-type phenotype was restored, OPGs eluted at the expected positions on a Bio-Gel P4 column (data not shown), and five well-defined subfractions were separated by DEAE-Sephacel chromatography (Fig. 3C). The mdoD218::cml mutation increased the heterogeneity of OPG molecules, and this phenotype could be complemented by introducing the wild-type allele on a multicopy plasmid.
FIG. 2.
Bio-Gel P-4 elution profiles of d-[U-14C]glucose-labeled OPGs from strains DF214 (mdo+) (•) and NFB1967 (mdoD218::cml) (□). The column (1.5 by 65 cm) was eluted with 0.5% acetic acid, and aliquots were counted (see Materials and Methods).
FIG. 3.
Increased heterogeneity of OPG molecules in an mdoD mutant: DEAE-Sephacel anion-exchange column chromatography profiles of d-[U-14C]glucose-labeled OPGs from strains DF214 (mdo+) (A), NFB1967 (mdoD218::cml) (B), and NFB1567 (mdoD218::cml [pNF599]) (C). The ionic strength was increased by 0.05 M NaCl steps at fractions indicated by the arrows. Fractions (4 ml) were collected, and aliquots were counted.
The mdoD218::cml mutation results in backbone structure alterations.
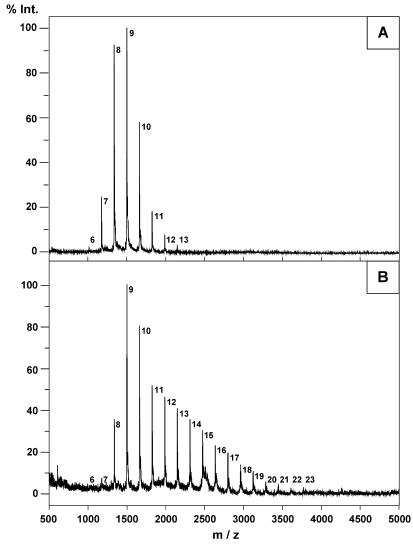
The sizes of the OPG backbones were determined by MS after removal of all the substituents (Fig. 4). As previously described (3), OPGs from the wild-type strain were composed of 6 to 13 glucose residues, and the average was between 8 and 9 glucose residues (Fig. 4A). Spectra obtained with OPGs extracted from the mdoD strain revealed the presence of 18 molecular ion species with masses corresponding to the masses of glucans composed of 6 to 23 glucose residues (the average was between 11 and 12 glucose residues) (Fig. 4B).
FIG. 4.
Positive-ion MALDI mass spectra of OPGs from strains DF214 (mdo+) (A) and NFB1967 (mdoD218::cml) (B). Prior to the analysis, all substituents were removed as described in Materials and Methods. The number to the right of each peak indicates the degree of polymerization.
OPGs were methanolized and, after acetylation, subjected to GLC-MS analysis. In both cases, the results of the methylation analysis revealed the presence of 3,4-di-, 3,4,6-tri-, and 2,3,4,6-tetra-O-methylglucoses (data not shown). This confirmed that the glucans were branched structures with branch points doubly substituted at positions 2 and 6, as previously described by Schneider et al. (27). In wild-type OPGs 33% of the glucose residues were nonreducing terminal residues, 33% were at branch points, and 25% were internal residues with no branch. In OPGs extracted from the mdoD strain, 30% of the glucose residues were nonreducing terminal residues, 24% were at branch points, and 41% were internal residues with no branch. These data indicate that mdoD inactivation results in loss of control of the backbone size, which leads to longer linear backbones with fewer branches; for example, the wild-type structure may be ⌞_ ⊥_ ⊥_ * in the wild type, and the mutant structure may be ⌞_ ⊥_ ⊥____ * or _ ⊥_ ⊥_ ⊥___ *, but there are many equivalent possibilities, as previously shown for OPGs of Erwinia chrysanthemi (8).
Therefore, OpgD appeared to be not essential for catalysis of glucosidic β-1,2 and β-1,6 linkages but to be involved in control of the structural backbone organization.
A tatB mutation mimics the effects of the mdoD mutation on OPG synthesis.
To ascertain that OpgD was translocated via the Tat pathway, as expected from its signal sequence, two kinds of approaches were used: inactivation of the Tat pathway and modification of the twin-arginine signal.
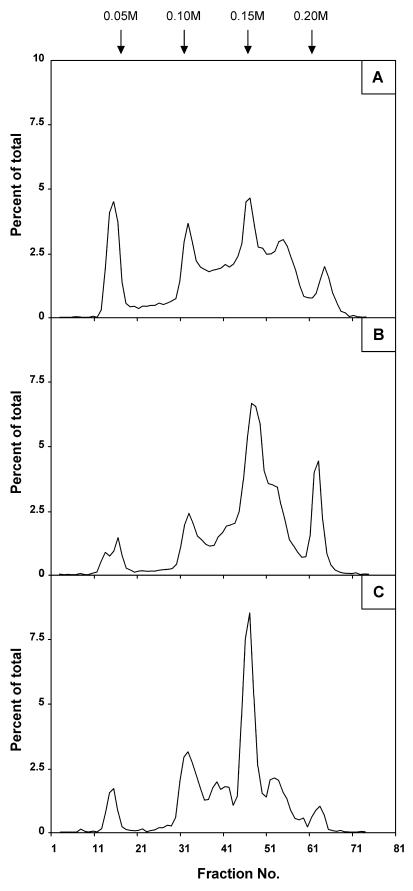
A tatB::kan mutation was introduced by transduction into strain DF214. Unlike NFB1967 (data not shown), the resulting strain, strain NFB4516, formed cell chains in the exponential growth phase. This is a typical phenotype of tat strains due to the loss of secretion of two periplasmic amidases needed for murein rearrangement, AmiA and AmiC (2, 30). The OPGs extracted from the tatB::kan strain were about 20% more abundant than the OPGs in DF214. These OPGs showed altered elution patterns, similar to those observed for the mdoD mutant, both on Bio-Gel P4 (data not shown) and on a DEAE-Sephacel column (Fig. 5A). There was a noticeable difference in the increase in subfraction I in the OPGs originating from the tat strain, indicating that there was a decrease in overall substitution. As tat mutants are known to release periplasmic proteins due to alteration of the cell envelope permeability (30), both the increase in OPG synthesis and the decrease in substitution could be explained by the release of a fraction of OPGs into the medium. Release of OPGs into the medium correlated with an increased amount of OPGs has been observed in mutant strains with increased outer membrane permeability (16). Leakage of OPGs into the growth medium partially interferes with the feedback control of OPG synthesis (3). Moreover, the fraction of OPGs released into the medium would not be subject to substitution.
FIG. 5.
Increased heterogeneity of OPG molecules in a tatB mutant or after arginine replacement in the OpgD signal sequence: DEAE-Sephacel anion-exchange column chromatography profiles of d-[U-14C]glucose-labeled OPGs from strains NFB4516 (tatB::kan) (A), NFB1655 (mdoD218::cml [pNF1092, R3K and R4K]) (B), and NFB1653 (mdoD218::cml [pNF1091, R4K and R5K]) (C). The experimental conditions were the same as those described in the legend to Fig. 3.
Mutagenesis of the OpgD twin-arginine signal peptide results in altered OPG synthesis.
Since the tatB mutation results in a pleiotropic phenotype which could indirectly affect OPG synthesis, we used a complementary approach. Three consecutive arginines are present in the OpgD signal peptide (Fig. 1), and the second arginine is thought to play a critical role in the translocation process (5). The first and second arginines or the second and third arginines were both replaced by lysines by site-directed mutagenesis of plasmid pNF599 (see Materials and Methods). Such a change was reported previously to inactivate the translocation process (2, 6). The mutant plasmids were introduced into the mdoD218::cml strain, and the OPGs produced by both strains were compared to those produced when wild-type pNF599 was present. The three strains produced normal amounts of OPGs (data not shown), indicating that overproduction of OPGs in the tatB mutant was the result of increased envelope permeability. As observed previously for OPGs extracted from the mdoD and tatB strains, the DEAE-Sephacel chromatography elution profiles appeared to be highly abnormal, even though they were not identical (Fig. 5B and C). Actually, these elution profiles were not reproducible. This is consistent with a loss of control of glucose backbone size and confirms that OpgD activity is strictly dependent on the Tat secretion apparatus. Moreover, this finding confirms that the second arginine plays a crucial role in the process (5).
Phylogenetic analysis of OpgG/OpgD homologs.
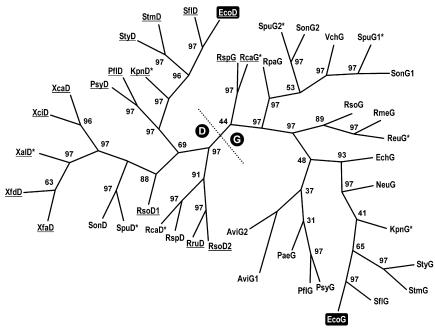
Because of the growing number of complete genomes that have been sequenced, phylogenetic analyses of OpgG/OpgD homologs is now possible. Figure 6 shows a phylogenetic tree obtained by the MP method with 42 homologs detected by BLAST in nonredundant databases and the sequencing projects in progress available at Infobiogen (http://www.infobiogen.fr/; December 2002). An opgH homolog was detected in each case analyzed. Genes which were found to be upstream of the opgH homolog were designated opgG, and the other genes were designated opgD. Several pathogens whose genomes have been completely sequenced do not possess any opgH or opgG homolog. Our analysis clearly revealed two groups of orthologs, OpgD and OpgG, which are paralogs of each other (Fig. 6). There are two exceptions to our naming rule; these are SpuG2 and SonG2, which appear to be members of the OpgG family despite their isolated genetic location (Azotobacter vinelandii possesses two copies of the opgGH operon). Interestingly, opgG is missing but opgD is present in Xanthomonas and Xylella, and opgD is missing in various unrelated bacteria. Moreover, Ralstonia solanacearum harbors an extra copy of opgD on its megaplasmid.
FIG. 6.
Unrooted phylogenetic tree for OpgG/OpgD homologs prepared by using the MP method as described in Materials and Methods. The numbers at the forks are bootstrap confidence levels. Underlined abbreviations indicate proteins with a (putative) Tat-dependent secretion signal. The asterisks indicate that data were obtained from sequencing projects in progress. Abbreviations and accession numbers: AviG1, Azotobacter vinelandii, ZP_00091295; AviG2, Azotobacter vinelandii, ZP_00091297; EcoD, Escherichia coli K-12, NP_415941 (O157:H7, NP_310056); EcoG, Escherichia coli K-12, CAA45521 (O157:H7, NP_287182); EchG, Erwinia chrysanthemi, CAC13110; NeuG, Nitrosomonas europaea, ZP_00004006; PaeG, Pseudomonas aeruginosa PAO1, NP_253765; PflD, Pseudomonas fluorescens, ZP_00084115; PflG, Pseudomonas fluorescens, ZP_00083363; PsyD, Pseudomonas syringae pv. syringae B728a, ZP_00124645; PsyG, Pseudomonas syringae pv. syringae B728a, ZP_00125042; RmeG, Ralstonia metallidurans, ZP_00023653; RpaG, Rhodopseudomonas palustris, ZP_00012410; RruD, Rhodospirillum rubrum, ZP_00014807; RsoG, Ralstonia solanacearum, NP_521029; RsoD1, Ralstonia solanacearum, NP_521178; RsoD2, Ralstonia solanacearum, NP_521823; RspD, Rhodobacter sphaeroides, ZP_00005771; RspG, Rhodobacter sphaeroides, AAG09638; SflD, Shigella flexneri 2a 301, NP_707648; SflG, Shigella flexneri 2a 301, NP_706959; SonD, Shewanella oneidensis MR-1, NP_716679; SonG1, Shewanella oneidensis MR-1, NP_717710; SonG2, Shewanella oneidensis MR-1, NP_718315; StmD, Salmonella enterica serovar Typhimurium LT2, NP_460581; StmG, Salmonella enterica serovar Typhimurium LT2, NP_460121; StyD, Salmonella enterica subsp. enterica serovar Typhi, NP_455875; StyG, Salmonella enterica subsp. enterica serovar Typhi, NP_455644; VchG, Vibrio cholerae, NP_230933; XcaD, Xanthomonas campestris pv. campestris ATCC 33913, NP_639484; XciD, Xanthomonas axonopodis pv. citri 306, NP_644583; XfdD, Xylella fastidiosa Dixon, ZP_00038586; XfaD, Xylella fastidiosa 9a5c, NP_299959. Abbreviations for data from sequencing in progress: KpnG and KpnD, Klebsiella pneumoniae MGH78578; ReuG, Ralstonia eutropha CH34; RcaG and RcaD, Rhodobacter capsulatus SB1003; SpuG1, SpuG2, and SpuD, Shewanella putrefaciens.
Upstream of the first conserved residue (a phenylalanine present in the 42 sequences) OpgG proteins have a typical Sec-dependent secretion signal, whereas most of the OpgD proteins have a Tat-dependent signal (the alignment was possible only after a search of an alternative initiation codon for several genes which have been annotated on the basis of automatic coding sequence identification). The few exceptions include the OpgD proteins in Shewanella, Rhodobacter, and Rhodopseudomonas. As mentioned previously (7), OpgG of Rhodobacter sphaeroides (which has been functionally characterized) is translated with an uncommonly long signal peptide. In fact, this signal peptide and its counterpart in Rhodobacter capsulatus exhibit a relatively high degree of similarity with the signal peptides of the OpgD proteins, whereas the signal peptide of Rhodopseudomonas palustris appears to be less closely related.
DISCUSSION
In this paper we describe initial characterization of the mdoD gene, a paralog of mdoG, and in this study we investigated the function of this gene in OPG biosynthesis. OPGs are probably synthesized by a multiprotein complex, and OpgD should be part of this complex. In such complex machinery, the structural feature of the product should depend on kinetic parameters determined by specific protein-protein interactions. We propose that OpgG and OpgD share protein domains for OPG recognition. The role of OpgD remains elusive, probably because we are still unable to reconstitute the natural conditions under which OPGs, and consequently OpgD, are needed by the bacteria to cope with the environment. The simplest explanation is that OpgD is a regulatory subunit of the OPG biosynthetic machinery. However, OpgD appears to be more abundant in the stationary growth phase than in the exponential growth phase (21). Moreover, expression of mdoD increases during the stationary phase of growth (Y. Lequette, A. Bohin, and J.-M. Lacroix, unpublished observations). However, the OPGs produced during the exponential phase of growth are indistinguishable from those recovered during the stationary phase of growth both in wild-type and mdoD mutant strains (data not shown). During the latter phase, when OPG synthesis has stopped, OpgD could function as a kind of OPG-binding protein, perhaps to favor an interaction with an unknown sensor. Since a smaller amount of OpgD is present during the biosynthetic phase, OpgD may modify the kinetic parameters of OPG synthesis because it can interact with nascent OPGs or because it shares with OpgG some surface motifs for interaction with other members of the OPG biosynthetic machinery, particularly the transmembrane glucosyltransferase OpgH. If this hypothesis is correct, OpgD would not be strictly necessary for OPG synthesis, but the other components of the multiprotein complex would have to adjust to its presence during a process of coevolution. In the absence of OpgD, the system could not control the degree of glucose polymerization as usual.
Two families of paralogous genes, opgG and opgD, have been described. When one copy of opgG is present, it is ahead of an opgH gene, forming an operon, while opgD is an isolated gene. The functions of the OpgG proteins have been established for E. coli (19), E. chrysanthemi (3), and R. sphaeroides (7), in which they are necessary for OPG synthesis. When OpgG is absent, it can be assumed that OpgH can catalyze completely backbone formation, but when OpgG is present, the two proteins are interdependent. With two exceptions, the OpgG proteins are synthesized with a Sec-dependent signal, while the OpgD proteins have a Tat-dependent signal. This is a striking example of two families of paralogous genes whose products use different routes to reach the periplasmic space. A similar situation was described recently (13) for E. coli for the three cell wall amidases encoded by the amiA, amiB, and amiC paralogous genes; AmiA and AmiC are Tat dependent, while AmiB is Sec dependent.
Acknowledgments
We thank Jérôme Lemoine for recording the MALDI MS spectra and Yves Leroy for performing the GLC-MS analyses. We thank Robert J. Kadner for critical reading of the manuscript.
Y.L. received a doctoral studentship from the Ministère de l'Enseignement Supérieur, de la Recherche et de la Technologie and from the Fondation pour la Recherche Médicale. This research was supported by a grant from the Centre National de la Recherche Scientifique (UMR8576).
REFERENCES
- 1.Aiba, H., T. Baba, K. Fujita, K. Hayashi, T. Inada, K. Isono, T. Itoh, H. Kasai, K. Kashimoto, S. Kimura, M. Kitakawa, M. Kitagawa, K. Makino, T. Miki, K. Mizobuchi, H. Mori, T. Mori, K. Motomura, S. Nakade, Y. Nakamura, H. Nashimoto, Y. Nishio, T. Oshima, N. Saito, G. Sampei, Y. Seki, S. Sivasundaram, H. Tagami, J. Takeda, K. Takemoto, Y. Takeuchi, C. Wada, Y. Yamamoto, and T. Horiuchi. 1996. A 570-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 28.0-40.1 min region on the linkage map. DNA Res. 3:363-377. [DOI] [PubMed] [Google Scholar]
- 2.Bernhardt, T. G., and P. A. J. de Boer. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohin, J.-P. 2000. Osmoregulated periplasmic glucans in Proteobacteria—a minireview. FEMS Microbiol. Lett. 186:11-19. [DOI] [PubMed] [Google Scholar]
- 4.Bohin, J.-P., and E. P. Kennedy. 1984. Mapping of a locus (mdoA) that affects the biosynthesis of membrane-derived oligosaccharides in Escherichia coli. J. Bacteriol. 157:956-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchanan, G., F. Sargent, B. C. Berks, and T. Palmer. 2001. A genetic screen for suppressors of Escherichia coli Tat signal peptide mutations establishes a critical role for the second arginine within the twin-arginine motif. Arch. Microbiol. 177:107-112. [DOI] [PubMed] [Google Scholar]
- 6.Chaddock, A. M., A. Mant, I. Karnauchov, S. Brink, R. G. Herrmann, R. B. Klosgen, and C. Robinson. 1995. A new type of signal peptide: central role of a twin-arginine motif in transfer signals for the ΔpH-dependent thylakoidal protein translocase. EMBO J. 15:2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cogez, V., E. Gak, A. Puskas, S. Kaplan, and J.-P. Bohin. 2002. The opgGIH and opgC genes of Rhodobacter sphaeroides form an operon that controls backbone synthesis and succinylation of osmoregulated periplasmic glucans. Eur. J. Biochem. 269:2473-2484. [DOI] [PubMed] [Google Scholar]
- 8.Cogez, V., P. Talaga, J. Lemoine, and J.-P. Bohin. 2001. Osmoregulated periplasmic glucans of Erwinia chrysanthemi. J. Bacteriol. 183:3127-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cristobal, S., J.-W. de Gier, H. Nielsen, and G. von Heijne. 1999. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J. 18:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dilks, K., R. W. Rose, E. Hartmann, and M. Pohlschroder. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 185:1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubois, M., K. A. Gilles, J. K. Hamilton, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Biochem. 28:350-356. [Google Scholar]
- 12.Fournet, B., G. Strecker, Y. Leroy, and J. Montreuil. 1981. Gas-liquid chromatography and mass-spectrometry of methylated and acetylated methylglycosides: application to the structural analysis of glycoprotein glycans. Anal. Biochem. 116:489-502. [DOI] [PubMed] [Google Scholar]
- 13.Ize, B., N. R. Stanley, G. Buchanan, and T. Palmer. 2003. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol. Microbiol. 48:1183-1193. [DOI] [PubMed] [Google Scholar]
- 14.Jackson, B. J., J.-P. Bohin, and E. P. Kennedy. 1984. Biosynthesis of membrane derived oligosaccharides: characterization of mdoB mutants defective in phosphoglycerol transferase I activity. J. Bacteriol. 160:976-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy, E. P. 1996. Membrane derived oligosaccharides (periplasmic beta-d-glucans) of Escherichia coli, p. 1064-1074. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Maganasik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed., American Society for Microbiology, Washington, D.C.
- 16.Kennedy, E. P., and M. K. Rumley. 1988. Osmotic regulation of biosynthesis of membrane-derived oligosaccharides in Escherichia coli. J. Bacteriol. 170:2457-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohara, Y., K. Akiyama, and K. Isono. 1987. The physical map of the whole Escherichia coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell 50:495-508. [DOI] [PubMed] [Google Scholar]
- 18.Lacroix, J.-M., E. Lanfroy, V. Cogez, Y. Lequette, A. Bohin, and J.-P. Bohin. 1999. The mdoC gene of Escherichia coli encodes a membrane protein that is required for succinylation of osmoregulated periplasmic glucans. J. Bacteriol. 181:3626-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lacroix, J.-M., I. Loubens, M. Tempête, B. Menichi, and J.-P. Bohin. 1991. The mdoA locus of Escherichia coli consist of an operon under osmotic control. Mol. Microbiol. 5:1745-1753. [DOI] [PubMed] [Google Scholar]
- 20.Lacroix, J.-M., M. Tempête, B. Menichi, and J.-P. Bohin. 1989. Molecular cloning and expression of a locus (mdoA) implicated in the biosynthesis of the membrane-derived oligosaccharides in Escherichia coli. Mol. Microbiol. 3:1173-1182. [DOI] [PubMed] [Google Scholar]
- 21.Link, A. J., K. Robison, and G. M. Church. 1997. Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12. Electrophoresis 18:1259-1313. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Miller, K. J., and E. P. Kennedy. 1987. Transfer of phosphoethanolamine residues from phosphatidylethanolamine to the membrane-derived oligosaccharides of Escherichia coli. J. Bacteriol. 169:682-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer, T., and B. C. Berks. 2003. Moving folded proteins across the bacterial cell membrane. Microbiology 149:547-556. [DOI] [PubMed] [Google Scholar]
- 25.Paz Parente, J., P. Cardon, Y. Leroy, J. Montreuil, B. Fournet, and G. Ricard. 1984. A convenient method for methylation of glycoproteins glycans in small amounts by using lithium methylsulfinyl carbanion. Carbohydr. Res. 141:41-47. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Schneider, J. E., V. Reinhold, M. K. Rumley, and E. P. Kennedy. 1979. Structural studies of the membrane-derived oligosaccharides of Escherichia coli. J. Biol. Chem. 254:10135-10138. [PubMed] [Google Scholar]
- 28.Stahl, B., M. Steup, M. Karas, and F. Hillenkamp. 1991. Analysis of neutral oligosaccharides by matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 63:1463-1466. [Google Scholar]
- 29.Stahl, B., S. Thurl, J. Zeng, M. Karas, F. Hillenkamp, M. Steup, and G. Sawatski. 1994. Oligosaccharides from human milk as revealed by matrix-assisted laser desorption/ionization mass spectrometry. Anal. Biochem. 223:218-226. [DOI] [PubMed] [Google Scholar]
- 30.Stanley, N. R., K. Findlay, B. C. Berks, and T. Palmer. 2001. Escherichia coli strains blocked in Tat-dependent protein export exhibit pleiotropic defects in the cell envelope. J. Bacteriol. 183:139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Golde, L. M. G., H. Schulman, and E. P. Kennedy. 1973. Metabolism of membrane phospholipids and its relation to a novel class of oligosaccharides in Escherichia coli. Proc. Natl. Acad. Sci. USA 70:1368-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weissborn, A. C., M. K. Rumley, and E. P. Kennedy. 1992. Isolation and characterization of Escherichia coli mutants blocked in production of membrane-derived oligosaccharides. J. Bacteriol. 174:4856-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]