Abstract
Vibrio fischeri possesses two acyl-homoserine lactone quorum-sensing systems, ain and lux, both of which are involved in the regulation of luminescence gene expression and are required for persistent colonization of the squid host, Euprymna scolopes. We have previously demonstrated that the ain system induces luminescence at cell densities that precede lux system activation. Our data suggested that the ain system both relieves repression and initially induces the lux system, thereby achieving sequential induction of gene expression by these two systems. Analysis of the V. fischeri genome revealed the presence of a putative third system based on the enzyme LuxS, which catalyzes the synthesis of the Vibrio harveyi autoinducer 2 (AI-2). In this study, we investigated the impact of V. fischeri LuxS on luminescence and colonization competence in comparison to that of the ain system. Similar to the ain system, inactivation of the AI-2 system decreased light production in culture, but not in the squid host. However, while an ainS mutant produces no detectable light in culture, a luxS mutant expressed approximately 70% of wild-type luminescence levels. A mutation in luxS alone did not compromise symbiotic competence of V. fischeri; however, levels of colonization of an ainS luxS double mutant were reduced to 50% of the already diminished level of ainS mutant colonization, suggesting that these two systems regulate colonization gene expression synergistically through a common pathway. Introduction of a luxO mutation into the luxS and ainS luxS background could relieve both luminescence and colonization defects, consistent with a model in which LuxS, like AinS, regulates gene expression through LuxO. Furthermore, while luxS transcription appeared to be constitutive and the AI-2 signal concentration did not change dramatically, our data suggest that ainS transcription is autoregulated, resulting in an over 2,000-fold increase in signal concentration as culture density increased. Taken together, these data indicate that V. fischeri LuxS affects both luminescence regulation and colonization competence; however, its quantitative contribution is small when compared to that of the AinS signal.
Vibrio fischeri, the bacterial light organ symbiont of the Hawaiian squid Euprymna scolopes, utilizes at least two quorum-sensing systems whose relative importance is dependent on cell density: (i) the well-known lux system, consisting of LuxI, which synthesizes N-(3-oxo-hexanoyl) homoserine lactone (3-oxo-C6-HSL), and the transcriptional regulator LuxR (14); and (ii) the ain system, which includes the second V. fischeri acyl-HSL synthase AinS, generating N-octanoyl HSL (C8-HSL) (16, 23, 24). We have previously demonstrated that these two systems are important at different cell densities: i.e., the ain system is the predominant inducer of luminescence in culture (24), whereas the lux system is necessary for luminescence expression at the higher cell densities present in the squid light organ (44). This sequential induction of luminescence gene expression is accomplished by a hierarchically regulated cascade in which the activated ain system exerts two effects: (i) relief of a negative regulation of gene expression of the lux system and (ii) low-level induction of the lux system through binding of the AinS signal to LuxR, which results in initial lux expression (24). The relief of gene repression is carried out through inactivation of the transcriptional regulator protein LuxO. LuxO inactivation results in an increased transcription of litR (32), which encodes a positive regulator of luxR transcription (10), thereby linking the ain and lux quorum-sensing systems in V. fischeri (24). Mutation of either the ain or the lux system results in a light organ colonization defect of V. fischeri, demonstrating the importance of quorum-sensing-dependent luminescence and possibly other colonization gene regulation in the Vibrio-squid symbiosis (24).
The recently sequenced genome of V. fischeri (http://www.ergo-light.com/ERGO/) revealed the presence of a putative third system, consisting of LuxS, the enzyme synthesizing a molecule named autoinducer 2 (AI-2), a furanosyl borate diester (7), and the putative AI-2 receptor proteins LuxP and LuxQ. LuxS was originally discovered in Vibrio harveyi as part of the quorum-sensing circuit that regulates luminescence gene expression (43). This signal synthase is hypothesized to serve in interspecies communication, as it can be found in many gram-negative and gram-positive bacterial species (29, 50). Luminescence gene expression in V. harveyi is coordinately controlled by both the LuxS-derived AI-2 and N-(3-hydroxybutanoyl) HSL (3-OH-C4-HSL), synthesized by the AinS homolog LuxM (33). Both signals interact with cognate hybrid two-component sensor-kinase proteins, LuxPQ and LuxN, respectively (2, 13), which transduce information through a phosphorelay cascade involving a protein designated LuxU (12), leading to inactivation of the transcriptional regulator LuxO (3, 11). Inactivation of LuxO results in increased transcription of V. harveyi luxR (a V. fischeri litR homolog), which positively regulates luminescence gene expression (32). Besides our own study of V. fischeri (24), other recent studies have demonstrated the involvement of homologous proteins of this regulatory cascade in colonization factor expression in Vibrio anguillarum (8, 31), Vibrio cholerae (30, 52), and Vibrio vulnificus (22, 26, 39), and both Vibrio alginolyticus and Vibrio parahaemolyticus have been reported to possess AI-2 activity (1). While the complete regulatory system remains to be determined for most of these species, the accumulating data are suggesting a common mechanism in the genus Vibrio (27, 32).
These homologies within the genus Vibrio, as well as the fact that the ain system is apparently conserved between V. fischeri and V. harveyi, led us to speculate that an AI-2 system might also operate in V. fischeri. Besides the potential impact of AI-2 on the expression of luminescence, we were curious to learn whether this signal is also important in light organ colonization by V. fischeri, as AI-2 has been implicated in colonization factor regulation in a variety of bacterial species (50). Furthermore, the V. fischeri ainS mutant is defective in colonizing its squid host, suggesting that ainS regulates the expression of genes important in symbiosis (24). If the AI-2 system functions in V. fischeri as it does in V. harveyi (33), those factors might also be coordinately regulated by luxS and ainS.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Medium components were purchased from Difco Laboratories, Inc. (Sparks, Md.), and Sigma Chemical Co. (St. Louis, Mo.). Strains of V. fischeri were grown at 28°C in either a seawater-based nutrient medium (SWT) containing Bacto-Tryptone (Difco), yeast extract, and glycerol (5) or Luria-Bertani salt (LBS) medium (17). Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) medium (37). Media were solidified with 1.5% (wt/vol) agar as needed. Antibiotics were added to the media at the following concentrations when appropriate: chloramphenicol at 2.5 μg/ml for V. fischeri and 20 μg/ml for E. coli, kanamycin at 100 μg/ml for V. fischeri and E. coli, and erythromycin at 5 μg/ml for V. fischeri and 150 μg/ml for E. coli. 3-Oxo-C6-HSL was obtained from Sigma Chemical Co., and C8-HSL was obtained from Aurora Biosciences (Coralville, Iowa).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| V. fischeri | ||
| ES114 | Wild-type isolate from E. scolopes light organ | 5 |
| ainS mutant CL21 | ainS gene partially deleted and replaced by chloramphenicol resistance (cat) marker | 24 |
| litR mutant PMF8 | litR gene inactivated by insertion of kanamycin resistance marker (kanR) | 10 |
| luxI mutant VCW2G7 | luxI gene inactivated by a frameshift mutation | 24 |
| luxO mutant CL42 | luxO gene inactivated by insertion of kanamycin resistance marker (kanR) | 24 |
| luxR mutant CL53 | luxR gene inactivated by insertion of erythromycin resistance marker (ermR) | This study |
| luxS mutant CL39 | luxS gene inactivated by insertion of kanamycin resistance marker (kanR) | This study |
| ainS luxS mutant CL41 | Double mutant carrying mutations as described above for CL21 and CL39 | This study |
| luxS luxO mutant CL90 | Double mutant carrying mutations as described above for CL39 and CL42 | This study |
| ainS luxS luxO mutant CL91 | Triple mutant carrying mutations as described above for CL21, CL39 and CL42 | This study |
| V. harveyi | ||
| BB120 | Wild type | 1 |
| BB152 | AI-2 producer | 1 |
| BB170 | AI-2 sensor | 1 |
| BB886 | AI-2 sensor mutant | 1 |
| Plasmids | ||
| pCL112 | 2.1-kb HaeIII fragment carrying ainS gene in pVO8 | 24 |
| pCL115 | 2.7-kb V. fischeri ES114 DNA with luxS gene cloned into pEVS79 | This study |
| pCL122 | pCL115 with random TnKan insertion 70 bp downstream of the luxS transcriptional start site | This study |
| pCL125 | 1.1-kb V. fischeri ES114 DNA with luxS gene cloned into pVO8 | This study |
| pCL145 | 1.8-kb V. fischeri ES114 DNA with luxO gene cloned into pEVS79 | 24 |
| pCL149 | Allelic exchange vector pEVS79 carrying partially deleted luxR gene replaced by erythromycin resistance (erm) marker | This study |
| pCL152 | pCL112 with lacZ gene from pKV124 cloned into EcoRV site approximately 200 bp downstream of translational start site | This study |
| pCL154 | pCL125 with the lacZ gene from pKV124 cloned into the EcoRV site approximately 200 bp downstream of translational start site | This study |
| pCL155 | pCL145 with luxO gene inactivated by insertion of ermR-marker into NsiI site | This study |
| pEVS79 | Allelic exchange vector, camR | 41 |
| pEVS94 | Source of the ermR cassette | 41 |
| pKV30 | Cloning vector carrying partially deleted luxR gene replaced by erythromycin resistance (erm) marker | 44 |
| pKV124 | Source of lacZ gene | 47 |
| pMU106 | Allelic exchange vector carrying partially deleted ainS gene replaced by chloramphenicol resistance (cat) marker | 24 |
| pVO8 | V. fischeri cloning vector, ermR | 45 |
Genetic techniques.
Genomic and plasmid DNAs were extracted by using the Qiagen DNeasy and Qiaprep Miniprep systems (Qiagen, Inc., Valencia, Calif.), respectively. PCR was performed according to standard protocols (37) with AmpliTaq DNA polymerase (Perkin-Elmer, Inc., Branchburg, N.Y.). For plasmid constructions, restriction enzymes and DNA ligase were obtained from New England Biolabs, Inc. (Beverly, Mass.), and used according to the manufacturer's protocol. Transfer of plasmids into E. coli host strains was accomplished by standard techniques (37). Triparental conjugation was used to transfer plasmids into V. fischeri strains (40). Sequencing was carried out on a Perkin-Elmer ABI Prism automated sequencer at the University of Hawaii Biotechnology/Molecular Biology Instrumentation and Training Facility.
Construction of V. fischeri mutants.
To generate the luxS and the ainS-luxS mutant strains, a 2.7-kb fragment carrying the luxS gene was PCR amplified from V. fischeri strain ES114 based on the genomic sequence provided by Integrated Genomics, Inc. (Chicago, Ill.), at http://www.ergo-light.com/ERGO/. The fragment was cloned into the mobilizable vector pEVS79 (Table 1), generating pCL115. Plasmid pCL115 was randomly mutagenized with the EZ::TN <Kan-2> insertion kit (Epicentre, Madison, Wis.). One clone, designated pCL122, was identified that carried the <Kan-2> cassette insertion 70 bp downstream of the translational start site of the luxS gene. Plasmid pCL122 was transferred into both the wild-type (ES114) and the ainS mutant (CL21) strains of V. fischeri by triparental mating. Single and double recombinants were selected as previously described (40), generating the V. fischeri luxS and the ainS-luxS mutant strains (CL39 and CL41, respectively). Introduction of the luxS mutation into the genome of V. fischeri was confirmed by PCR analysis. The luxS-complementing plasmid pCL125 was generated by subcloning a 1.1-kb BamHI-EcoRI fragment carrying the intact luxS gene from pCL115 into the V. fischeri cloning vector pVO8 (Table 1).
To generate the luxS luxO and the ainS luxS luxO mutants, the luxO gene carried on pCL145 (Table 1) was mutagenized by inserting a 1.2-kb erythromycin resistance (ermR) gene from pEVS94 (Table 1) into the NsiI site located approximately 300 bp downstream of the luxO gene start site. The resulting plasmid, pCL155, was transferred into V. fischeri luxS by triparental mating, and single and double recombinants were selected as previously described (40), generating the V. fischeri luxS luxO mutant CL90. Similarly, plasmid pMU106, carrying a partially deleted ainS gene replaced by a chloramphenicol resistance marker, was introduced into the luxS luxO mutant to create the ainS luxS luxO mutant strain CL91. Using a previously described bioassay (38), we saw no evidence that strain CL91 produces C8-HSL activity, confirming that the ainS mutation was introduced into the genome.
The luxR mutant was constructed by subcloning a 4.1-kb HaeIII fragment from pKV30 into the mobilizable vector pEVS79 (Table 1). This fragment carries the luxR gene from V. fischeri ES114, which has been inactivated by partial deletion and insertion of an erythromycin resistance cassette (44). The resulting plasmid, pCL149, was transferred into the chromosome of V. fischeri ES114 by triparental mating. Single and double recombinants were selected as previously described (40), generating the V. fischeri luxR mutant strain CL53. The introduction of the mutation into the V. fischeri genome was confirmed by PCR analysis. Furthermore, luminescence characteristics of strain CL53 were determined and found to be similar to the luminescence characteristics of a previously described V. fischeri luxR mutant strain (44). Specifically, luminescence in culture was decreased to approximately 10 to 20% of wild-type levels. Similarly, whereas wild-type luminescence was induced several 100-fold upon exogenous addition of 120 nM 3-oxo-C6-HSL, as expected, this addition produced no induction of the luxR mutant.
Luminescence in culture.
To determine their luminescence characteristics, V. fischeri wild-type and mutant strains were inoculated to an optical density at 600 nm (OD600) of about 0.05 into 10 ml of SWT containing either (i) no addition, (ii) 120 nM C8-HSL, or (iii) 120 nM 3-oxo-C6-HSL. Cultures were kept shaking at 28°C, samples were taken at various times during growth, and luminescence and OD were measured. Maximum specific luminescence (luminescence/OD) values were averaged for measurements made between ODs of 1.0 and 5.0. Growth rate and growth yield were also determined during these experiments by measuring OD as a function of time. Experiments were repeated at least once.
Colonization assays.
Three colonization phenotypes of V. fischeri wild-type and mutant strains were assessed.
(i) Symbiotic bioluminescence.
The progress of early colonization events was monitored as described previously (35). Briefly, newly hatched squids were placed into vials with 4 ml of filter-sterilized seawater (FSSW) containing an inoculum of approximately 1,000 CFU of the indicated strain per ml. Twenty-four individual animals were infected per treatment group; 6 animals served as uninoculated controls and were placed into FSSW without added bacteria. Animal bioluminescence, an indirect measure of the degree of squid colonization, was monitored periodically over 24 h using a modified Packard Tri-Carb 2100TR scintillation counter (Packard Instruments Inc., Meriden, Conn.) as a photometer.
(ii) Colonization level in the squid light organ.
The number of CFU per squid was determined at 24 and 48 h postinoculation following a previously described method (34). Newly hatched squid were placed into 50 ml of FSSW containing about 1,000 CFU of the indicated strain per ml, incubated for 12 h, and then transferred into FSSW. Some animals were placed into FSSW without added bacteria. At subsequent times, 15 animals per treatment group and 2 uninoculated animals were homogenized, and the homogenate was diluted and spread onto SWT agar. The colony number was counted after overnight incubation, and the mean number of CFU per squid was calculated.
(iii) Competitive phenotype.
The ability of bacterial symbionts to compete during host colonization under conditions of coinoculation was tested for the V. fischeri wild-type and luxS mutant strains, using a previously described approach (46) with the following modifications. About 15 newly hatched squid were placed into 50 ml of FSSW containing approximately 1,000 CFU of each of the competing strains per ml and were incubated for 12 h. An aliquot of the inoculated seawater was spread onto LBS agar to determine the total number and exact ratio of the two strains in the inoculum. At 48 h postinoculation, squids were homogenized and a dilution of the homogenate spread onto LBS agar. Approximately 100 CFU from the inoculum and each of the homogenates was patched onto antibiotic-containing or antibiotic-free LBS agar to determine the ratio of V. fischeri wild-type (Kans) to luxS mutant (Kanr) cells.
LuxS and AinS signal activities of V. fischeri cultures.
A modification of a previously described assay (1) was used to determine the ability of V. fischeri to produce LuxS signal activity. Briefly, V. fischeri strains were grown to the indicated OD in SWT. The medium was separated from the cells by centrifugation and subsequent filtration through a 0.2-μm-pore-size filter. Overnight cultures of V. harveyi strains BB170 (AI-2 sensor) and BB886 (AI-2 sensor mutant) (Table 1), grown in autoinducer bioassay (AB) medium (18), were diluted 1:5,000 into fresh AB medium. Cell-free culture supernatants of V. fischeri strains grown in SWT were added to these bioassay strains to a final concentration of 10% (vol/vol). The luminescence of 1-ml aliquots of the V. harveyi bioassay cultures was periodically measured with a TD20/20 luminometer (Turner Design, Sunnyvale, Calif.). SWT served as a negative control, and cell-free AB broths of an overnight culture of either the V. harveyi wild type (BB120) or the acyl-HSL-defective V. harveyi BB152 strain were used as the AI-2-producing positive controls. Results are reported as a percentage of the induction level produced by the positive control.
The concentration of C8-HSL produced by V. fischeri strains grown in SWT was determined with the same cultures assayed above. Supernatants of cultures at different ODs were extracted with acidified ethyl acetate and concentrated by evaporation. The amount of C8-HSL present in the concentrated samples was determined by using a previously described β-galactosidase-linked bioassay (38).
LuxS signal and AinS signal were extracted from V. fischeri cultures in three independent experiments. From each extraction, both the induction of luminescence expression of the AI-2 reporter strain and the induction of β-galactosidase expression in the C8-HSL reporter strain were determined in triplicate. It should be noted that while the concentration of C8-HSL can be determined directly by using synthetic C8-HSL standard, determination of AI-2 activity is indirect and assumes a linear relationship between signal concentration and induction of luminescence in the V. harveyi reporter strain.
Transcriptional activity of luxS and ainS.
Two plasmids were generated to estimate the transcriptional activity of the luxS and ainS genes in V. fischeri. The lacZ gene was isolated from plasmid pKV124 (Table 1) by BamHI restriction digestion and was blunt ended with Klenow fragment. To obtain the transcriptional fusions ainS::lacZ and luxS::lacZ, the lacZ gene was ligated either (i) into the EcoRV site located approximately 200 bp downstream of the translational start site of the ainS gene carried on pCL112 (Table 1), creating pCL152, or (ii) into the EcoRV site located approximately 200 bp downstream of the translational start site of the luxS gene carried on pCL125 (Table 1), creating pCL154. Each of these low-copy-number plasmids was introduced into V. fischeri wild-type and mutant strains by triparental mating.
V. fischeri strains carrying in trans either pCL152 or pCL154 were cultured in either SWT plus erythromycin or SWT plus chloramphenicol, and the relative transcriptional activity of luxS and ainS was assayed in terms of the β-galactosidase activity of triplicate cultures at different times during growth with a standard o-nitrophenyl-β-d-thiogalactopyranoside (ONPG) method (37). Because these are in trans fusions, their expression levels are used only to estimate the level of native transcriptional activity.
RESULTS
LuxS signal activity in V. fischeri.
To investigate whether V. fischeri produces a LuxS-dependent signal activity, we tested the ability of culture supernatants of the wild type and the luxS mutant to induce luminescence in the V. harveyi reporter strains BB170 (AI-2 sensor) and BB886 (AI-2 sensor mutant). Cell supernatants harvested from mid-log-phase cultures of the V. fischeri wild type, but not of the luxS mutant, induced luminescence in the AI-2 sensor strain to approximately 50% of the level of culture supernatants from either wild-type V. harveyi BB120 or the acyl-HSL mutant V. harveyi BB152 (Table 2). Complementation of the V. fischeri luxS mutation by providing a functional luxS gene in trans on pCL125 restored the ability to induce luminescence (Table 2). These results establish that V. fischeri possesses an activity capable of inducing V. harveyi luminescence and that this activity is dependent on a functional luxS gene. Neither the V. fischeri wild type nor the luxS mutant induced luminescence in the AI-2 sensor mutant, whereas medium conditioned by wild-type V. harveyi BB120 did (Table 2), demonstrating that the V. fischeri LuxS activity is dependent on the presence of the AI-2 receptor protein. Taken together, these data indicate that V. fischeri cells produce an activity in culture that is indistinguishable from that of V. harveyi AI-2.
TABLE 2.
LuxS activity of V. fischeria
| Strainb | % LuxS activity in V. harveyi reporter strain:
|
|
|---|---|---|
| BB170 (AI-2 sensor) | BB886 (AI-2 sensor mutant) | |
| V. harveyi | ||
| Wild-type BB120 | 147 (14) | 100 (37) |
| AI-1 mutant BB152 | 100 (18) | 1 (12) |
| V. fischeri | ||
| Wild type (vector) | 49 (3) | 0 (1) |
| luxS (vector) | 0 (3) | 1 (2) |
| Wild type (pCL125) | 44 (8) | 2 (2) |
| luxS (pCL125) | 41 (16) | 2 (2) |
The V. harveyi reporter strains BB170 (AI-2 sensor) and BB886 (AI-2 sensor mutant) were incubated in medium containing 10% (vol/vol) culture supernatants of either the V. fischeri wild type or the luxS mutant carrying in trans either a vector control (pVO8) or a functional copy of the luxS gene on pCL125 (Table 1). Shown is the normalized response of a representative experiment. Standard errors are indicated in parentheses.
The addition of culture supernatants from either wild-type V. harveyi BB120 (which produces both V. harveyi AI-1, an acyl-HSL, and AI-2) or V. harveyi BB152 (which produces only AI-2) served as positive controls.
Luminescence of V. fischeri luxS mutants in culture.
Having shown that V. fischeri possesses LuxS signal activity, we investigated whether this signal contributes to light production in V. fischeri as it does in V. harveyi. The luminescence levels of the V. fischeri wild type, as well as the luxS, ainS, and ainS luxS mutants, were determined in culture either with or without the addition of C8-HSL or 3-oxo-C6-HSL (Table 3). Because V. fischeri strain ES114 produces only very low levels of 3-oxo-C6-HSL activity in culture (5), exogenous supplementation with this compound triggers a dramatic response in light production (24, 44). In contrast, C8-HSL has been previously demonstrated to affect luminescence gene expression of V. fischeri in culture by two means: (i) relief of lux repression through inactivation of LuxO and (ii) direct induction of the lux system, apparently through low-affinity binding to and activation of LuxR (24). Thus, a mutation in ainS results in a dark phenotype due to repression and absence of induction of lux. Exogenous addition of C8-HSL to wild-type V. fischeri stimulates light production in a LuxR-dependent manner, although to a lesser extent than 3-oxo-C6-HSL addition (24, 44).
TABLE 3.
Luminescence of V. fischeri luxS mutantsa
| Strain type | Luminescence values (10−2 quanta s−1 cell−1) with addition ofb:
|
||
|---|---|---|---|
| None | C8-HSL | 3-Oxo-C6-HSL | |
| Wild type | 5.7 (0.1) | 8.2 (1.1) | 17,000 (3,900) |
| luxS | 4.2 (0.2) | 5.1 (0.4) | 22,000 (3,600) |
| ainS | BDc | 4.3 (0.2) | 5 (1) |
| ainS luxS | BD | 2.6 (0.1) | 4 (1) |
| luxO | 6.8 (0.8) | 12.2 (2.9) | 24,000 (4,000) |
| luxS luxO | 8.3 (1.1) | 17.2 (4.0) | 37,000 (1,500) |
Specific luminescence values are the means of cultures with an OD600 of between 1.0 and 5.0; standard errors of the means are indicated in parentheses. The results shown are from a representative experiment.
Cultures were grown in SWT either without additions or with the addition of either 120 nM C8-HSL or 120 nM 3-oxo-C6-HSL.
BD, below detection (<0.02 quanta s−1 cell−1).
Light production by the luxS mutant was decreased to between 60 and 80% of the wild-type level (Table 3). Addition of C8-HSL to the luxS mutant increased light production, but not to wild-type levels, indicating that C8-HSL cannot fully complement the AI-2 deficiency. In contrast, when the LuxI-derived 3-oxo-C6-HSL was supplied exogenously, the luminescence levels of the luxS mutant and the wild type were both increased dramatically to levels that were approximately the same (Table 3), suggesting that the response of V. fischeri to 3-oxo-C6-HSL is not dependent on a functional AI-2 system. Both the ainS and ainS luxS mutants were not detectably luminous in culture (Table 3). The addition of C8-HSL could restore light production of the ainS and the ainS luxS mutants to nearly wild-type and luxS mutant levels, respectively. Similarly, the luminescence levels of luxS and ainS luxS were indistinguishable from those of their parent strains (wild type and ainS, respectively) when 3-oxo-C6-HSL was supplied exogenously. Because complementation of both the luxS and ainS mutations with functional genes in trans could restore luminescence to wild-type levels (data not shown), these mutations are unlikely to exert a downstream effect.
In V. harveyi, AI-2 activates a phosphorylation cascade resulting in the inactivation of LuxO, a negative regulator of luminescence (3, 11). To determine whether this pathway is conserved in V. fischeri, we compared the luminescence of a luxS luxO double mutant to that of a luxO mutant and found that the levels of light production by these two strains were similar, either with or without the addition of acyl-HSLs (Table 3). These data are consistent with a model in which V. fischeri LuxS produces a signal that regulates luminescence through inactivation of LuxO, as has been reported for the AinS signal, C8-HSL (24).
Symbiotic luminescence and colonization competence of V. fischeri luxS mutants.
To determine whether symbiotic competence of V. fischeri is compromised by a mutation in luxS, we monitored the luminescence of juvenile animals colonized by the V. fischeri wild type or the luxS, ainS, or ainS luxS mutants during the first 24 h of colonization (Fig. 1). As previously shown (24), ainS mutant-colonized animals expressed between 10 and 20% of the luminescence level of those colonized by the wild type. In contrast, the luminescence of luxS mutant-colonized animals was indistinguishable from that of wild-type-colonized animals (Fig. 1). However, the luminescence levels of animals colonized by the ainS luxS mutant were consistently 50% lower than those of animals colonized by the ainS single mutant, suggesting that either their specific luminescence or their colonization levels were decreased (Fig. 1).
FIG. 1.
Luminescence of luxS mutants in the symbiotic light organ. Animal luminescence was monitored during the initial stages of E. scolopes colonization by V. fischeri wild-type (solid diamonds), ainS mutant (solid squares), luxS mutant (solid triangles), and ainS luxS mutant (solid circles) strains. For each time point, mean values of 24 animals were calculated and standard errors of the mean are indicated. The experiment was repeated with the same outcome.
To differentiate between these two possibilities, we compared the number of bacterial cells that were present in the light organ populations of animals colonized by either wild-type V. fischeri or the luxS, ainS, or ainS luxS mutants at 24 and 48 h (Fig. 2). While no significant difference in colonization ability was detected between the luxS mutant and wild-type V. fischeri, the ainS luxS double mutant colonized the juvenile squid to only 50 to 75% of the level reached by the ainS single mutant (Fig. 2), indicating a synergistic effect of these two mutations, similar to what has been shown for the effect of homologous systems on light emission by V. harveyi (33). Because squid luminescence levels were similarly reduced (Fig. 1), the levels of symbiotic light production per cell of the ainS and the ainS luxS mutants are likely to be the same, suggesting that colonization factors other than luminescence are cooperatively regulated by these signals. Complementation of the ainS luxS mutant with an intact luxS gene in trans restored luminescence and colonization to the levels of an ainS mutant, indicating that the combined effect is due to a mutation in luxS and not to an unrelated defect of this strain (data not shown). When squid were colonized with an ainS luxS luxO triple mutant, colonization levels were similar to wild type (Fig. 2). The latter result might be solely due to the reversal of the ainS phenotype by the luxO mutation, thus restoring the essentially wild-type phenotype of a luxS mutant. However, because the effect of a luxS mutation can only be detected in the absence of AinS, suggesting a synergistic effect, and AinS has been shown to operate through LuxO (24), it is possible that LuxS regulates colonization gene expression through LuxO as well.
FIG. 2.
Colonization competence of quorum-sensing mutants. Colonization levels of the ainS mutant (A), luxS mutant (S), ainS luxS mutant (A S), and ainS-luxS-luxO (A S O) mutant relative to V. fischeri wild type (wt) were measured at 24 and 48 h postinoculation. Each bar represents the mean value of 15 animals with the associated standard errors. The experiment was conducted twice with the same outcome.
To ensure that the observed limitation in colonization level was not a result of a general metabolic defect, we determined the growth characteristics of the luxS mutant strains. The luxS mutant exhibited both wild-type growth rate and yield in rich medium. In contrast, the ainS luxS mutant displayed a wild-type growth rate but was decreased in its final growth yield; however, this decrease was comparable to that previously reported for the ainS mutant (24). Thus, the absence of a functional luxS gene did not significantly impact the growth characteristics of V. fischeri.
Mixed-inoculum colonization experiments were performed to determine whether the luxS mutation would reduce the competitive competence of V. fischeri. We found no detectable defect in the competitive colonization ability of either the luxS mutant or the ainS luxS mutant relative to their parent strains (data not shown). Because the ainS luxS mutant displayed a colonization defect (Fig. 2) one might expect to also observe this defect in a mixed-inoculum experiment; however, because both LuxS and AinS synthesize small molecules that will diffuse into the surrounding environment (48), it is likely that the presence of wild-type cells can complement the deficiency of the luxS mutation.
LuxS and AinS signal synthesis during growth.
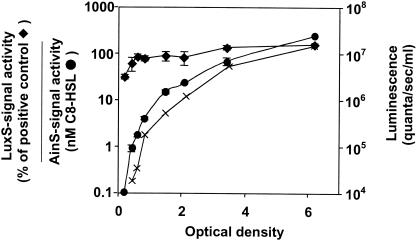
To regulate gene expression in a cell density-dependent manner, quorum-sensing signals accumulate in the growing bacterial population until a threshold concentration is reached that is sufficient for gene induction (48). To determine whether the synthesis of either the V. fischeri LuxS signal or the AinS-derived C8-HSL signal, which both positively influence luminescence (Table 3), correlates with light production, the levels of both signal activities were determined during growth (Fig. 3). While the concentration of the C8-HSL signal increased over 2,000-fold during the course of the experiment, mirroring the expression of luminescence in culture, AI-2 activity increased less than 4-fold (from 40 to 150% that of the positive control) (Fig. 3).
FIG. 3.
Relationship between luminescence and quorum-sensing signal activity during growth of V. fischeri in culture. The relative level of LuxS signal activity (solid diamonds) in the culture supernatant is given as a percentage of the level produced by the positive control, an overnight V. harveyi BB152 culture (see Materials and Methods). The concentration of the AinS signal, C8-HSL (solid circles), in the V. fischeri culture was determined with synthetically produced C8-HSL as standard. Luminescence of the culture (crosses) is presented for comparison. Shown is a representative experiment; standard deviation bars are indicated.
Transcriptional activity of luxS and ainS.
The transcriptional autoregulation of a quorum-sensing signal synthase gene, as occurs in the V. fischeri lux system (15), provides a mechanism to rapidly induce gene expression once a threshold concentration of the signal is reached. To investigate whether the expression of either ainS or luxS is similarly autoregulated, we determined the relative transcriptional activities of these two genes throughout culture growth by using lacZ transcriptional fusions. The rate of ainS transcription changed significantly during growth, increasing exponentially until the culture reached an OD of approximately 2.0 (Fig. 4). This pattern coincided with the increase in both C8-HSL signal production and luminescence (Fig. 3). In contrast, the rate of increase in luxS gene transcription was much lower and did not change throughout the experiment (Fig. 4), correlating with the relatively constant production of the signal (Fig. 3) and consistent with a constitutive expression of this gene.
FIG. 4.
Transcriptional activity of luxS and ainS in culture. β-Galactosidase activity of V. fischeri wild-type cells carrying either a luxS::lacZ fusion (solid diamonds) or an ainS::lacZ fusion (open diamonds) in trans on low-copy-number plasmids pCL152 and pCL154, respectively, was measured during growth. Shown are the cumulative data of three cultures; standard deviation bars were smaller than the symbols.
To determine whether proteins involved in the three V. fischeri quorum-sensing systems: lux (i.e., LuxR and LuxI), ain (i.e., AinS, LuxO, and LitR), and/or LuxS, regulate transcription of luxS and ainS, we measured the relative transcriptional activity of these two genes in a variety of quorum-sensing mutants. Because in several cases we were interested in the effect of the presence of the genes on their own transcription, the lacZ fusions had to be carried in trans. The inactivation of neither of these genes influenced luxS transcription (Table 4), providing further evidence for a constitutive transcription of this gene. In contrast, ainS gene transcription was decreased almost 10-fold in an ainS mutant, and transcription levels could be restored to wild-type levels either by supplying C8-HSL exogenously or by introducing a luxO mutation into the ainS mutant background (Table 4). These data are consistent with a model in which the ainS gene induces its own transcription through the synthesis of the AinS signal and this regulation involves inactivation of LuxO (Fig. 5). A similarly decreased level of ainS transcription was also observed in a litR mutant, but not in either a luxR or a luxI mutant, implying that the ain quorum-sensing system is autoregulated independently of the lux system. Mutations in neither luxS nor luxO changed transcription levels of ainS significantly (Table 4).
TABLE 4.
Transcriptional activity of V. fischeri luxS and ainS
| Strain type | β-Galactosidase activity (Miller units) from fusion toa:
|
|
|---|---|---|
| luxS | ainS | |
| Wild type | 205 (46) | 17.8 (4.4) |
| ainS | 189 (15) | 2.0 (0.1) |
| ainS (+ C8-HSL)b | NDc | 17.2 (4.1) |
| luxS | 204 (23) | 16.0 (4.1) |
| ainS luxS | 216 (35) | 1.9 (0.1) |
| luxO | 225 (46) | 18.9 (3.9) |
| ainS luxO | ND | 15.1 (6.0) |
| litR | 181 (30) | 2.7 (0.4) |
| luxR | 167 (24) | 17.4 (4.3) |
| luxI | 206 (36) | 17.4 (3.8) |
Shown is transcriptional activity of luxS and ainS in different genetic backgrounds. β-Galactosidase activity was measured during growth and averaged for several measurements taken at ODs of between 1 and 5. Shown is a representative experiment, with standard errors in parentheses.
The ainS mutant was grown in the presence of 120 nM C8-HSL.
ND, not determined.
FIG. 5.
Conceptual model for the regulation of luminescence and colonization genes by V. fischeri LuxS and AinS. Arrows indicate positive, inducing effects, and bars indicate negative, inhibitory ones. (See Discussion for explanation.)
DISCUSSION
Previous studies have identified two acyl-HSL-based signaling systems in V. fischeri, designated lux and ain (14, 24). The activation of the lux system is dependent on ain induction, thereby providing a genetic mechanism to sequentially induce gene expression with increasing cell density (24). In this study, we investigated the impact of a putative third system, based on the enzyme LuxS, on luminescence and colonization ability in comparison to AinS. Our results suggests that (i) V. fischeri LuxS synthesizes a compound that is indistinguishable from V. harveyi AI-2; (ii) both V. fischeri AinS and LuxS signals exert their regulatory effects on gene expression through the same pathway, which is likely to operate through LuxO (Fig. 5); and (iii) ainS transcription and C8-HSL signal concentration are regulated by a positive feedback loop, whereas rates of luxS transcription and LuxS signal production appear to be relatively constant (Fig. 5).
Regulation of luminescence and colonization gene expression by LuxS.
In V. harveyi, both the LuxM-derived 3-OH-C4-HSL and the LuxS-derived AI-2 regulate the expression of luminescence and other gene products through the inactivation of the transcriptional regulator LuxO (3, 11, 33). Similarly, our results demonstrate that the inactivation of the V. fischeri luxS gene causes a defect in luminescence expression in culture and that this defect could be relieved by the mutational inactivation of LuxO (Table 3). Furthermore, while a mutation in luxS did not affect colonization competence of V. fischeri, introduction of a luxS mutation into an ainS mutant background further decreased colonization levels (Fig. 2). These data, together with our previous finding that the signal synthesized by the LuxM homolog, AinS, operates through LuxO (24), suggest that these two systems regulate gene expression synergistically through the same downstream LuxO-mediated cascade (Fig. 5) that has been described in V. harveyi (3).
In contrast to V. harveyi (33), however, the effect of the V. fischeri LuxS-synthesized signal on luminescence and other colonization gene expression (Table 3) appears to be considerably smaller than that of the AinS signal (24). These findings are similar to the results of a recent study on the V. cholerae quorum-sensing circuit (30). V. cholerae possesses a regulatory cascade resembling that of V. harveyi (and now V. fischeri), with at least two signal inputs: the LuxS-derived AI-2 and a yet unidentified signal synthesized by an enzyme designated CqsA. While a mutation in cqsA abolished the ability of V. cholerae to express luminescence from V. harveyi lux genes carried in trans, a mutation in luxS did not have a significant effect. Similarly, the LuxO-regulated phenotype of biofilm formation by V. cholerae is under the control of CqsA, but the LuxS signal is largely dispensable (51). Taken together, these data suggest that although a regulatory circuit involving LuxO appears to be conserved in Vibrio species (32), the relative importance of each of the involved signals may vary widely among bacterial species.
Regulation of LuxS and AinS signal production.
We determined the activity of the LuxS- and AinS-derived signals and estimated the transcriptional activity of the signal synthase genes, luxS and ainS, during growth in rich medium. Although AI-2 is produced in a growth phase-dependent manner in several bacterial species (6, 9, 21, 22, 42), the nutrient composition of the medium and other physiological factors appear to impact AI-2 production more than cell density (6, 19, 22, 43). We did not investigate the environmental conditions under which V. fischeri produces AI-2, but instead chose growth conditions that resulted in a >1,000-fold difference in luminescence output, a phenotype regulated by luxS (Table 3). Under these conditions, LuxS signal concentration and luxS transcription did not reflect the magnitude of change that occurred in bacterial luminescence (Figs. 3 and 4). Significantly, even at cell densities that preceded luminescence induction (i.e., OD of <0.5), both measures of LuxS activity were comparably high (Fig. 3 and 4), suggesting the absence of a characteristic signal dose response. As expected by the relatively constant transcription levels throughout growth (Fig. 4), luxS transcription apparently was not affected by mutation of any of the known V. fischeri quorum-sensing proteins (Table 4).
In contrast, the patterns of both C8-HSL concentration and ainS transcriptional activity correlated well with light emission; i.e., both were highest at cell densities at which luminescence is induced and reached saturation thereafter (Fig. 3 and 4). Because the changes in both the rate of C8-HSL signal production and the apparent transcriptional activity of ainS correlated, it is likely that ainS transcription rate directly determines signal concentration. The observation that ainS transcription was decreased considerably in the absence of C8-HSL (Table 4) suggests that the cell density-dependent increase in C8-HSL concentration is accomplished by an autoregulatory mechanism of the ainS gene. The inactivation of the transcriptional regulator LitR decreased the activity of the ainS transcriptional reporter as well, supporting the hypothesis that LitR positively regulates ainS gene transcription either directly or through another transcriptional protein (Fig. 5). The notion that C8-HSL activates a phosphorelay cascade resulting in the inactivation of LuxO, a negative regulator of litR transcription (24, 32), predicts that mutation of luxO should result in increased levels of litR and, therefore, increased levels of ainS. However, ainS reporter levels were indistinguishable between the wild type and the luxO mutant (Table 4). While this result might suggest an effect of ainS on litR that is independent of LuxO, a simpler explanation is that the positive effect of LuxO on ainS in this assay is below our detection levels. This idea is supported by the observation that ainS reporter levels were indistinguishable from those of the wild type when measured in an ainS luxO mutant background (Table 4). Mutations in neither luxR nor luxI affected ainS gene transcription, indicating that the regulation is independent of the lux quorum-sensing system (Fig. 5). Because our data suggest that LuxS and AinS operate through the same pathway, one would expect decreased ainS transcription levels in a luxS mutant as well. However, ainS transcription levels were indistinguishable in the luxS mutant and wild type (Table 4), providing additional evidence that the relative effect of the LuxS signal in this regulatory cascade is much lower than that of the AinS signal (Fig. 5).
LuxS signal synthase or a metabolic enzyme?
Since the discovery of AI-2 activity in V. harveyi, the question of why a bacterium would utilize two inputs into the same regulatory cascade has led to much speculation. It was originally proposed that the acyl-HSL signal senses an individual species' cell density, whereas the nonspecific AI-2 senses the cumulative cell density of the bacterial community (1). The discovery that AI-2 signal production is dependent on the growth conditions rather than cell density per se has led to the alternative hypothesis that AI-2 reflects a change in environmental conditions through a change in the metabolic activity of the bacterial community (4, 43). These findings, together with the fact that LuxS has a role in central metabolism, led to the proposal that in most bacteria AI-2 is not a specific signal but a metabolic by-product of a common detoxifying pathway (49). On the other hand, the regulation by LuxS of niche-specific genes has been demonstrated for several pathogens (50), suggesting that this signal synthase has a specific role in host colonization. However, colonization competence of luxS mutants has been investigated in only a few bacterial species, and the observed defects were in many cases either small or not detectable (28), implying that other regulatory factors might be more important than the LuxS signal.
Our study does not definitively determine the role of LuxS in V. fischeri. The inactivation of luxS did not result in a growth defect either in culture (data not shown) or in symbiosis (Fig. 2), suggesting that the absence of LuxS activity does not significantly affect metabolic processes, at least under our experimental conditions. Instead, our data demonstrate that a luxS mutation has an effect on luminescence and that this effect can be relieved by a luxO mutation (Table 3 and Fig. 2), thereby providing evidence for a signal function exerting its effect through a transduction cascade common in Vibrio species. Although this putative signal alone did not appear to be important for the symbiotic lifestyle of V. fischeri cells in the environment of the squid light organ, inactivation of luxS had a significant effect on colonization competence in an ainS mutant background (Fig. 2). Thus, one might speculate that in environments in which the ain system is not active, the AI-2 system becomes an important player in the regulation of colonization factors. In addition to its monospecific light organ associations, V. fischeri is part of the multispecies bacterial community present in the enteric tracts of fishes and marine invertebrates (20, 25, 36). It is possible that the V. fischeri AI-2 system in these associations is important and/or that AI-2 produced by other bacteria might have an impact on colonization competence.
Acknowledgments
B. L. Bassler kindly provided V. harveyi reporter strains. D. S. Millikan, P. M. Fidopiastis, and J. R. Graber provided helpful comments on the manuscript. Integrated Genomics Inc., Chicago, made V. fischeri ES114 genomic sequence information available.
This work was supported by the National Institutes of Health grant RR12294 to E. G. Ruby and M. J. McFall-Ngai; National Science Foundation grant IBN0211673 to M. J. McFall-Ngai and E. G. Ruby; and a W. M. Keck Foundation grant to E. P. Greenberg, E. G. Ruby, M. J. McFall-Ngai, and others.
REFERENCES
- 1.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol. Microbiol. 12:403-412. [DOI] [PubMed] [Google Scholar]
- 4.Beeston, A. L., and M. G. Surette. 2002. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3450-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgess, N. A., D. F. Kirke, P. Williams, K. Winzer, K. R. Hardie, N. L. Meyers, J. Aduse-Opoku, M. A. Curtis, and M. Camara. 2002. LuxS-dependent quorum sensing in Porphyromonas gingivalis modulates protease and haemagglutinin activities but is not essential for virulence. Microbiology 148:763-772. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X., S. Schauder, N. Potier, A. van Dorsselaer, I. Pelczer, B. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 8.Croxatto, A., V. J. Chalker, J. Lauritz, J. Jass, A. Hardman, P. Williams, M. Cámara, and D. L. Milton. 2002. VanT, a homologue of Vibrio harveyi LuxR, regulates serine metalloprotease, pigment, and biofilm production in Vibrio anguillarum. J. Bacteriol. 184:1617-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dove, J. E., K. Yasukawa, C. R. Tinsley, and X. Nassif. 2003. Production of the signalling molecule, autoinducer-2, by Neisseria meningitidis: lack of evidence for a concerted transcriptional response. Microbiology 149:1859-1869. [DOI] [PubMed] [Google Scholar]
- 10.Fidopiastis, P. M., C. M. Miyamato, M. G. Jobling, E. A. Meighen, and E. G. Ruby. 2002. LitR, a newly described transcriptional activator homologue in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol. Microbiol. 45:131-143. [DOI] [PubMed] [Google Scholar]
- 11.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31:665-677. [DOI] [PubMed] [Google Scholar]
- 12.Freeman, J. A., and B. L. Bassler. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J. Bacteriol. 181:899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freeman, J. A., B. N. Lilley, and B. L. Bassler. 2000. A genetic analysis of the functions of LuxN: a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol. Microbiol. 35:139-149. [DOI] [PubMed] [Google Scholar]
- 14.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Gene 35:439-468. [DOI] [PubMed] [Google Scholar]
- 15.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 16.Gilson, L., A. Kuo, and P. V. Dunlap. 1995. AinS and a new family of autoinducer synthesis proteins. J. Bacteriol. 177:6946-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graf, J., P. V. Dunlap, and E. G. Ruby. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 176:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg, E. P., J. W. Hastings, and S. Ulitzer. 1979. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch. Microbiol. 120:87-91. [Google Scholar]
- 19.Hardie, K. R., C. Cooksley, A. D. Green, and K. Winzer. 2003. Autoinducer-2 activity in Escherichia coli culture supernatants can be actively reduced despite maintenance of an active synthase, LuxS. Microbiology 149:715-728. [DOI] [PubMed] [Google Scholar]
- 20.Haygood, M. G. 1993. Light organ symbioses of fishes. Crit. Rev. Microbiol. 19:191-216. [DOI] [PubMed] [Google Scholar]
- 21.Joyce, E. A., B. L. Bassler, and A. Wright. 2000. Evidence for a signaling system in Helicobacter pylori: detection of a luxS-encoded autoinducer. J. Bacteriol. 182:3638-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, S. Y., S. E. Lee, Y. R. Kim, C. M. Kim, P. Y. Ryu, H. E. Choy, S. S. Chung, and J. H. Rhee. 2003. Regulation of Vibrio vulnificus virulence by the LuxS quorum-sensing system. Mol. Microbiol. 48:1647-1664. [DOI] [PubMed] [Google Scholar]
- 23.Kuo, A., S. M. Callahan, and P. V. Dunlap. 1996. Modulation of luminescence operon expression by N-octanoyl-l-homoserine lactone in ainS mutants of Vibrio fischeri. J. Bacteriol. 178:971-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupp, C., M. Urbanowski, E. P. Greenberg, and E. G. Ruby. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 50:319-331. [DOI] [PubMed] [Google Scholar]
- 25.Makemson, J., and G. V. Hermosa, Jr. 1999. Luminous bacteria cultured from fish guts in the Gulf of Oman. Luminescence 14:161-168. [DOI] [PubMed] [Google Scholar]
- 26.McDougald, D., S. A. Rice, and S. Kjelleberg. 2001. SmcR-dependent regulation of adaptive phenotypes in Vibrio vulnificus. J. Bacteriol. 183:758-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDougald, D., S. Srinivasan, S. A. Rice, and S. Kjelleberg. 2003. Signal-mediated cross-talk regulates stress adaptation in Vibrio species. Microbiology 149:1923-1933. [DOI] [PubMed] [Google Scholar]
- 28.McNab, R., and R. J. Lamont. 2003. Microbial dinner-party conversations: the role of LuxS in interspecies communication. J. Med. Microbiol. 52:541-545. [DOI] [PubMed] [Google Scholar]
- 29.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 30.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 31.Milton, D. L., V. J. Chalker, D. Kirke, A. Hardman, M. Camara, and P. Williams. 2001. The LuxM homologue VanM from Vibrio anguillarum directs the synthesis of N-(3-hydroxyhexanoyl)homoserine lactone and N-hexanoylhomoserine lactone. J. Bacteriol. 183:3537-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyamoto, C. M., P. V. Dunlap, E. G. Ruby, and E. A. Meighen. 2003. LuxO controls luxR expression in Vibrio harveyi: evidence for a common regulatory mechanism in Vibrio. Mol. Microbiol. 48:537-548. [DOI] [PubMed] [Google Scholar]
- 33.Mok, K. C., N. S. Wingreen, and B. L. Bassler. 2003. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 22:870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruby, E. G. 1996. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 50:591-624. [DOI] [PubMed] [Google Scholar]
- 35.Ruby, E. G., and L. M. Asato. 1993. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch. Microbiol. 159:160-167. [DOI] [PubMed] [Google Scholar]
- 36.Ruby, E. G., and K.-H. Lee. 1998. The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. Appl. Environ. Microbiol. 64:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schaefer, A. L., B. L. Hanzelka, M. R. Parsek, and E. P. Greenberg. 2000. Detection, purification and structural elucidation of the acylhomoserine lactone inducer of Vibrio fischeri luminescence and other related molecules. Methods Enzymol. 305:288-301. [DOI] [PubMed] [Google Scholar]
- 39.Shao, C.-P., and L.-I. Hor. 2001. Regulation of metalloprotease gene expression in Vibrio vulnificus by a Vibrio harveyi LuxR homologue. J. Bacteriol. 183:1369-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stabb, E. V., K. A. Reich, and E. G. Ruby. 2001. Vibrio fischeri genes hvnA and hvnB encode secreted NAD+-glycohydrolases. J. Bacteriol. 183:309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stabb, E. V., and E. G. Ruby. 2002. New RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413-426. [DOI] [PubMed] [Google Scholar]
- 42.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visick, K. L., J. Foster, J. Doino, M. McFall-Ngai, and E. G. Ruby. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Visick, K. L., and E. G. Ruby. 1997. New genetic tools for use in the marine bioluminescent bacterium Vibrio fischeri, p. 119-122. In J. W. Hastings, L. J. Kricka, and P. E. Stanley (ed.), Bioluminescence and chemiluminescence. John Wiley & Sons, New York, N.Y.
- 46.Visick, K. L., and E. G. Ruby. 1998. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J. Bacteriol. 180:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visick, K. L., and L. M. Skoufos. 2001. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J. Bacteriol. 183:835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitehead, N. A., A. M. L. Barnard, H. Slater, N. J. L. Simpson, and G. P. C. Salmond. 2001. Quorum sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 49.Winzer, K., K. R. Hardie, and P. Williams. 2002. Bacterial cell-to-cell communication: sorry, can't talk now—gone to lunch! Curr. Opin. Microbiol. 5:216-222. [DOI] [PubMed] [Google Scholar]
- 50.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 51.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647-656. [DOI] [PubMed] [Google Scholar]
- 52.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]