Abstract
ylxL was found to be the last gene of the fla/che operon in Bacillus subtilis and is cotranscribed with the gene for the flagellum-specific alternate sigma factor, σD. The ylxL gene was disrupted by insertional mutagenesis, and the resultant mutant strain was found to be compromised for σD-dependent functions.
In Bacillus subtilis, the structural genes that encode the hook-basal body complex (HBB), several genes controlling chemotaxis, and the gene for the alternate sigma factor, σD, are found adjacent to one another in a 26-kb region of the bacterial chromosome called the fla/che region (15). Several studies have demonstrated that these genes comprise a single operon (2, 10, 16, 18), while genes encoding these functions in the enteric bacterium are found in at least seven operons located throughout the bacterial chromosome (7). Many of the genes within the fla/che operon have been cloned, sequenced, and characterized (1, 15); however, the 3′ end of the operon has yet to be mapped, and the function of the ylxL gene product has not been analyzed.
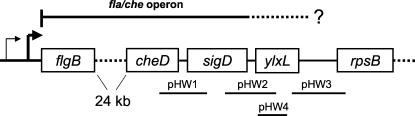
Genetic organization of ylxL.
An open reading frame, originally referred to as orfC and later renamed ylxL as a result of the B. subtilis genome project (3), was identified immediately downstream of the structural gene for σD, sigD (Fig. 1). While sigD, located 26 kb from the major promoter for this operon, has been shown to be part of fla/che, (2, 16), it was not known if ylxL is part of the fla/che transcription unit. Previous studies demonstrated that insertions between sigD and ylxL resulted in defects in chemotaxis (9, 18), swimming (9), and motility (10). Additionally, sequence analysis of this intergenic region failed to identify a transcriptional terminator, leading to the speculation that ylxL is part of the fla/che operon. Furthermore, the rpsB gene immediately downstream of ylxL is known to encode a ribosomal protein (5) and appears to be monocistronic with its own promoter (http://genolist.pasteur.fr/SubtiList), suggesting that ylxL may be the final gene of the fla/che operon.
FIG. 1.
Schematic diagram of the fla/che operon that starts with dual promoter elements dependent on σD and σA holoenzymes. The σD-dependent promoter (PD-3) lies 133 bp upstream of the σA-dependent promoter (fla/che PA), which lies 70 bp upstream of the translational start codon for the first gene in the fla/che operon, flgB. The sigD gene is found 26 kb downstream of these promoters, between the cheD and ylxL genes. DNA sequences amplified by PCR and cloned into pGEM-Zf7(+) for riboprobe synthesis or pJM102 for insertional mutagenesis are indicated by a bold line below the region that was amplified. Plasmids pHW1, pHW2, and pHW3 contain inserts that span the intergenic regions indicated, while the insertional plasmid pHW4 contains a fragment internal to the ylxL gene.
RPA of ylxL.
To determine whether ylxL is the last gene of the fla/che operon, RNase protection assays (RPA) were performed using the RPA II kit from Ambion. Riboprobes were synthesized for this analysis by first cloning the intergenic regions highlighted in Fig. 1 into plasmid pGEM-Zf7(+) from Stratagene. These intergenic regions were amplified by PCR and cloned into the plasmid in the orientation that allows for production of antisense RNA in an in vitro transcription reaction. T7 RNA polymerase and [α-32P]UTP were used in such a reaction to yield body-labeled riboprobes. Full-length protection of the riboprobes generated from plasmids pHW1 and pHW2 was obtained (data not shown), confirming that sigD is part of the fla/che operon and demonstrating that ylxL is cotranscribed with sigD. The data obtained using the riboprobe generated from pHW3 were less straightforward.
The 416-base riboprobes synthesized from pHW3 include nonhomologous sequence from the vector and 368 bases of sequence complementary to the region spanning the ylxL and rpsB genes (Fig. 1). These riboprobes were used to identify the 3′ end of the fla/che operon. The rpsB gene encodes the 30S ribosomal protein S2 (5) and is not considered to be part of the fla/che operon. Total RNA was extracted from wild-type B. subtilis cells (LMB7) grown in 2XSG sporulation medium and harvested at T0 and T0.5, when sigD expression is known to be maximal (13). Riboprobes were incubated with isolated RNA to allow annealing of complementary strands. Single-stranded portions of the riboprobes were digested with RNase provided with the RPA II kit, while regions of riboprobes complexed in the RNA duplex with ylxL-rpsB mRNA were protected from digestion and resolved by electrophoresis on a 5% denaturing gel. The resulting autoradiograph of the dried gel showed three primary products of 368, 190, and 119 nucleotides (Fig. 2A).
FIG. 2.
Results of RPA for the intergenic region between ylxL and rpsB using a body-labeled riboprobe (A) or an end-labeled DNA probe (B). The intact probe is 416 bases, and when fully hybridized to B. subtilis mRNA it gives rise to a protected fragment of 368 nucleotides upon digestion. (A) Results of RPA using body-labeled riboprobe and RNase digestion. Lane 1, RNA molecular weight marker; lane 2, intact riboprobe, no RNase added; lane 3, riboprobe incubated with RNase; lane 4, RPA using mRNA from the wild-type strain at T0 when flagellin gene expression is induced (13); lane 5, RPA using mRNA from wild-type strain at T0.5 when flagellin expression is maximal (13). (B) Results of RPA using end-labeled probe and S1 nuclease. Lane 1, DNA molecular weight marker; lane 2, empty; lane 3, intact probe, no S1 added; lane 4, RPA with yeast RNA; lane 5, RPA using mRNA from wild-type strain at T0.5.
The 368-base fragment is consistent with the lack of transcription termination by RNA polymerase at the end of ylxL and read-through into the rpsB gene: this is the predicted size for protection of homologous sequences contained within the riboprobes synthesized from pHW3. The significantly more abundant 119-base fragment is of the expected length for rpsB transcripts initiating at its own promoter, according to the SubtiList website (http://genolist.pasteur.fr/SubtiList). The protected 190-base fragment, however, suggests the termination of transcription at the end of the ylxL coding sequence prior to this promoter. In order to confirm the last two assignments, an end-labeled probe of the pHW3 region used for riboprobe synthesis was generated. The appropriate region of pHW3 was amplified by PCR and 5′ end labeled in a kinase reaction using [γ-32P]dATP. The resulting DNA probe was incubated with B. subtilis RNA and subjected to S1 nuclease digestion. As with the riboprobe assay, protection of the DNA probe from digestion requires that it anneal to complementary mRNA emanating from the ylxL-rpsB intergenic region, but it would be detected by autoradiography only if the 5′ end of the DNA probe were found within the duplex. While the 368- and 119-base fragments were still detected following S1 digestion, the 190-base fragment was no longer evident (Fig. 2B). These results demonstrate that the 5′ end of the probe is not found within the 190-base protected fragment, and this is likely due to protection of the 3′ end of the probe by ylxL mRNA. Taken together, the data obtained from the RPA show that ylxL is the last gene of the fla/che operon, although some read-through into the downstream rpsB gene is evident.
Studies of ylxL function.
ylxL, a 501-bp gene found at the 3′ end of the fla/che operon in this study, encodes a protein of unknown function. BLAST analysis showed that the predicted protein product of the ylxL gene has low similarity to other proteins, also of unknown function. However, due to previous genetic studies of the ylxL region (9, 18) and its determined location within the fla/che operon, we postulated that it might play a role in flagellar and chemotaxis functions. To study its predicted function, an insertional disruption of ylxL was generated by transformation with the disruptional plasmid pHW4 into the wild-type strain LMB7, yielding LMB231 (Table 1). pHW4 contains a 169-bp insert complementary to the 5′ end of the ylxL open reading frame immediately downstream of its translational start codon. Insertion of this plasmid disrupts ylxL without affecting expression of sigD as determined by anti-σD immunoblot analysis, described previously (16). σD protein levels in the ylxL mutant LMB231 were found to be comparable to those found in the wild-type strain (Fig. 3A) and were undetectable in the σD null mutant, LMB10. Furthermore, the phenotype of the ylxL mutant was found to be the result of the insertional disruption in the ylxL gene and not to an unlikely polar effect on rpsB expression. Insertion of the identical plasmid sequences that are found in the ylxL disruption between ylxL and rpsB in strain LMB281 did not produce the mutant ylxL phenotype (data not shown).
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype | Derivation (source or reference)a |
|---|---|---|
| LMB4 | his leu fliM1 | OI2553b (18) |
| LMB7 | trpC2 pheA1 | JH642b (J. Hoch) |
| LMB10 | trpC2 pheA1 sigD | CB100b (9) |
| LMB25 | trpC2 pheA1 hag-lacZ (Eryr) | Transform [LMB7:pDM632Ery, Eryr] (13) |
| LMB231 | trpC2 pheA1 ylxL::cat | Transform [LMB7:pHW4, Cmr] (this study) |
| LMB237 | trpC2 pheA1 ylxL::cat hag-lacZ (Eryr) | Transform [LMB231:LMB25, Eryr] (this study) |
| LMB281 | trpC2 pheA1 ylxL-rpsB::catc | Transform [LMB7:pLO3d, Cmr] (this study) |
Transformations are shown as follows: [recipient strain:plasmid DNA (pHW4) or chromosomal DNA from listed strain used for transformation, and selection for resistance].
Previous name of strain.
The insertion is between ylxL and rpsB and does not disrupt either gene.
The same insert found in pHW3 (Fig. 1) was cloned into the integrational plasmid, pJM102, to yield pLO3.
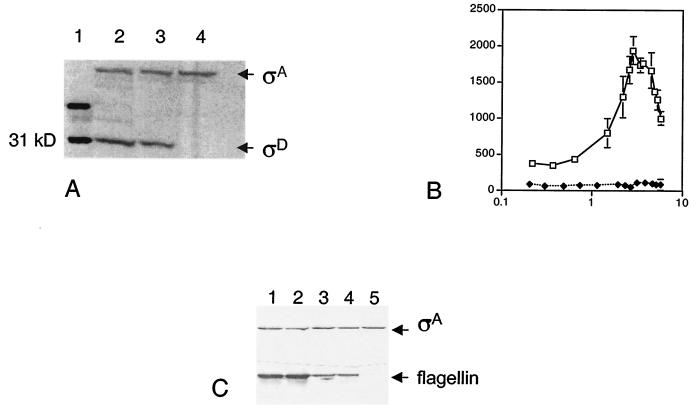
FIG. 3.
σD and flagellin gene expression in the ylxL mutant. (A) σD protein levels were measured in 50 μg of total protein extract by immunoblot analysis, using σA expression as an internal control. Lane 1, ladder; lane 2, wild-type strain (LMB7); lane 3, ylxL strain (LMB231); lane 4, sigD null mutant (LMB10). (B) β-Galactosidase activity was monitored in a strain bearing a hag-lacZ reporter construct throughout growth in complex, sporulation medium. The y axis is β-galactosidase activity expressed in Miller units, and the x axis is absorbance at 600 nm. Symbols: open squares, wild type (LMB25); filled diamonds, ylxL (LMB237). The data presented are the results of several experiments. (C) Flagellin protein levels were measured in total protein extract by immunoblot analysis, using σA expression as an internal control. Lane 1, wild-type strain (LMB7) at T0; lane 2, wild-type strain (LMB7) at T0.5; lane 3, ylxL strain (LMB231) at T0; lane 4, ylxL strain (LMB231) at T0.5; lane 5, sigD null mutant (LMB10) at T0.5.
Flagellin expression.
To determine the biological importance of ylxL in flagellin gene expression, strain LMB237 bearing both the ylxL disruption and a hag-lacZ translational fusion (Table 1) was grown in 2XSG sporulation medium, and β-galactosidase activity was measured as described previously (13). Under these conditions, flagellin gene expression increases throughout exponential growth and peaks shortly thereafter (13), as found for hag-lacZ expression in the wild-type strain, LMB25 (Fig. 3B). However, the reporter strain bearing the ylxL mutation, LMB237, displayed a drastic decrease in β-galactosidase activity (Fig. 3B). In this strain, hag-lacZ expression ranged from 50 to 100 Miller units throughout growth, indicating that ylxL is required for high-level flagellin gene expression that is developmentally regulated.
These results were confirmed and extended by antiflagellin immunoblot analysis performed as described previously (16). The level of flagellin protein was significantly reduced in the ylxL mutant LMB231 when compared to levels found in the wild-type strain (Fig. 3C). Nonetheless, flagellin protein was easily detectable in this strain and absent in the sigD null mutant. Therefore, while the alternate sigma factor encoded by sigD, σD, is absolutely required for expression of the flagellin gene as previously determined (11), ylxL appears to play a critical role in up-regulating its expression.
Motility and chemotaxis function.
Having demonstrated that the ylxL mutant is capable of synthesizing flagellin protein, we sought to test whether it is motile and able to demonstrate chemotactic behavior. Semisolid agar (0.3% agar) plates were used as described previously (9) to monitor the ability of the mutant to swim away from the site of inoculation towards areas of greater nutrient concentration. Disruption of the ylxL gene resulted in impaired swimming motility and/or chemotaxis, as demonstrated by a smaller diameter of growth found for this strain when compared to the wild-type strain (Fig. 4A and B). The ylxL mutant, LMB231, exhibited 40% of the diameter of growth found for the wild-type strain, LMB7, but was clearly capable of swimming motility when compared to two nonmotile strains bearing null mutations in the flagellum-specific alternate sigma factor, sigD, and the flagellar switch protein, fliM.
FIG. 4.
Results of chemotaxis assays for the ylxL mutant. (A) Bar graph of the average data obtained for several experiments in the plate assay shown in panel B. Results are given as a percentage of the diameter of growth obtained for the wild-type strain (LMB7). (B) Plate assay for chemotaxis and motility in the ylxL strain (LMB231), the wild-type strain (LMB7), and two nonmotile strains, the sigD strain (LMB10) and the fliM strain (LMB4). (C) Results of capillary assay for ylxL mutant (LMB231) compared to wild-type strain (LMB7) for several attractants (asparagine, proline, and mannitol) used at a concentration of 10−5 M. The data given are the average of two experiments for the number of bacteria per capillary (minus the blank).
The chemotactic efficiency of the ylxL mutant towards three attractants (asparagine, proline, and mannitol) was monitored by capillary assays that were performed as previously described (14, 17). Several concentrations of each attractant were assayed (data not shown), and the results obtained by use of 10 μM attractant are presented, since they are consistent with the data obtained for all concentrations tested (Fig. 4C). Accumulation of ylxL mutant cells in capillaries containing the three attractants tested ranged from 37 to 52% of the number of wild-type cells that accumulated under identical conditions. While this could be interpreted as compromised chemotactic behavior, a similar difference in the level of accumulation (47%) was found for the blank that contained no attractant (Fig. 4C). Therefore, our results suggest that the reduced diameter of growth found on the semisolid agar plates (Fig. 4A and B) was due to compromised swimming motility and not a defect in chemotaxis. This result is consistent with a role of the ylxL gene product in enhancing σD function. The genes for flagellin and the motility proteins (11, 12) are σD dependent, while most of the chemotaxis genes are found in the fla/che operon (1) and are not dependent on the alternate sigma factor for expression (16).
Autolysin activity.
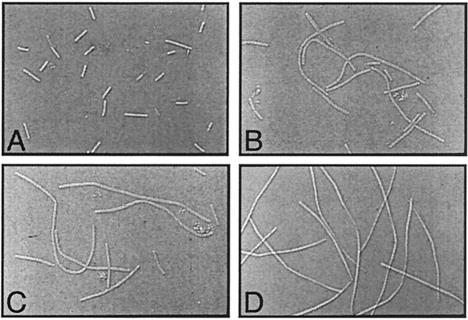
The genes for several autolysins contain σD-dependent promoters (4, 8), and cells lacking σD function grow as long filaments due to decreased autolytic activity necessary for cell separation (9). Microscopic observation of the ylxL mutant compared to the wild-type strain demonstrated filamentous growth, but the filaments were significantly shorter than those found for the sigD null mutant and included a mixture of short and long filaments (Fig. 5). These data further support the postulate that the ylxL gene product is required for maximal σD activity in the cell.
FIG. 5.
Microscopic observation of the ylxL mutant using Nomarski optics (×1,250). (A) Wild-type strain LMB7; (B and C) ylxL mutant LMB231; (D) sigD null LMB10.
Summary.
In this work we have demonstrated that the ylxL gene is the last gene of the fla/che operon that apparently encodes a protein required for maximal σD function. Sequence analysis of the intergenic region between ylxL and rpsB has failed to detect a factor-independent transcriptional terminator, suggesting that the termination of transcription identified in this study is factor dependent. The location of ylxL at the end of the fla/che transcription unit, coupled with its demonstrated role in enhancing σD function and read-through into rpsB, provides for the intriguing possibility that it acts as a sensor for complete fla/che transcription and/or couples flagellin transcription and translation.
While BLAST analysis using the predicted protein product of ylxL as the query sequence showed only weak matches to proteins of unknown function, a search of the B. subtilis genome using products of several enteric flhC genes as the query identified a single match, the ylxL gene product (2). The flhC gene is part of the master operon (7) that is required for expression of the flagellar regulon in enteric bacteria. This gene encodes part of a transcriptional activator complex absolutely required for expression of the genes that form the HBB and the flagellum-specific sigma factor in that system (6). However, ylxL does not appear to be required for sigD expression, since a strain disrupted for ylxL in this study was able to display limited σD functions. Interestingly, the similarity between the ylxL gene product and the enteric FlhC proteins is limited to a third of the protein and suggests a different domain structure. Nonetheless, the ylxL gene product may function as a transcriptional activator of σD-dependent genes. Further, its unique location at the end of the fla/che transcription unit may allow this activity to be coupled to complete transcription of the operon. Finally, read-through from ylxL to rpsB may suggest a link between σD-dependent transcription of the hag gene and its subsequent translation.
Recent work from the Hughes laboratory (P. Aldridge, H. R. Bonifield, J. Gnerer, J. E. Karlinsey, and K. T. Hughes, Abstr. BLAST VII Bact. Locomotion Signal Transduction, p. 42, 2003) suggests that hag mRNA in Salmonella enterica serovar Typhimurium may be targeted to the HBB complex for translation. The product of the rpsB gene encodes a component of the ribosome found to be localized to the poles of cells (5). The genetic organization of this gene immediately downstream of ylxL and the low level of read-through found in this study suggest such a link, but much greater investigation is required to support this prediction.
Acknowledgments
This research was supported by NSF-RUI grant MCB-0112949 to L.M.-M., DOE-GAANN P200A80128 support to H.W., NIH-MBRS RISE grant GM5-59298 to P.L., and RO1 GM54365 to G.W.O.
We are grateful to Peter Ingmire for critical reading and editing of the manuscript.
REFERENCES
- 1.Aizawa, S.-I., I. B. Zhulin, L. Márquez-Magaña, and G. W. Ordal. 2002. Chemotaxis and motility, p. 437-452. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 2.Estacio, W. E., S. Santa Anna-Arriola, M. Adedipe, and L. M. Márquez-Magaña. 1998. Dual promoters are responsible for transcription initiation of the fla/che operon of Bacillus subtilis. J. Bacteriol. 180:3548-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 4.Lazarevic, V., P. Margot, B. Soldo, and D. Karamata. 1992. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J. Gen. Microbiol. 138:1949-1961. [DOI] [PubMed] [Google Scholar]
- 5.Lewis, P. J., S. D. Thaker, and J. Errington. 2000. Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J. 19:710-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu, X., and P. Matsumura. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345-7349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macnab, R. 1992. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 26:131-158. [DOI] [PubMed] [Google Scholar]
- 8.Margot, P., M. Pagni, and D. Karamata. 1999. Bacillus subtilis 168 gene lytF encodes a gamma-d-glutamate-meso-diaminopimelate muropeptidase expressed by the alternative vegetative sigma factor, sigma-D. Microbiology 145:57-65. [DOI] [PubMed] [Google Scholar]
- 9.Márquez, L. M., J. Helmann, E. Ferrari, H. Parker, G. W. Ordal, and M. J. Chamberlin. 1990. Studies of σD-dependent functions in Bacillus subtilis. J. Bacteriol. 172:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Márquez-Magaña, L. M., and M. J. Chamberlin. 1994. Characterization of the sigD transcription unit of Bacillus subtilis. J. Bacteriol. 176:2427-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mirel, D. B., and M. J. Chamberlin. 1989. The Bacillus subtilis flagellin (hag) gene is transcribed by the σ28 form of RNA polymerase. J. Bacteriol. 174:3095-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirel, D. B., V. M. Lustre, and M. J. Chamberlin. 1992. An operon of motility genes in Bacillus subtilis is transcribed by the σD form of RNA polymerase. J. Bacteriol. 174:4197-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirel, D. B., W. F. Estacio, M. Mathieu, E. Olmsted, J. Ramirez, and L. M. Márquez-Magaña. 2000. Environmental regulation of Bacillus subtilis σD-dependent gene expression. J. Bacteriol. 182:3055-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ordal, G. W., and D. J. Goldman. 1975. Chemotaxis away from uncouplers of oxidative phosphorylation in Bacillus subtilis. Science 189:802-805. [DOI] [PubMed] [Google Scholar]
- 15.Ordal, G. W., L. Márquez-Magaña, and M. J. Chamberlin. 1993. Motility and chemotaxis, p. 765-784. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press, Washington, D.C.
- 16.West, J. T., W. Estacio, and L. M. Márquez-Magaña. 2000. Relative roles of the fla/che PA, PD-3, and PsigD promoters in regulating motility and sigD expression in Bacillus subtilis. J. Bacteriol. 182:4841-4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmer, M. A., H. Szurmant, M. M. Saulmon, M. A. Collins, J. S. Bant, and G. W. Ordal. 2002. The role of heterologous receptors in McpB-mediated signaling in Bacillus subtilis chemotaxis. Mol. Microbiol. 45:555-568. [DOI] [PubMed] [Google Scholar]
- 18.Zuberi, A. R., C. Ying, M. R. Weinrich, and G. W. Ordal. 1990. Transcriptional organization of a cloned chemotaxis locus of Bacillus subtilis. J. Bacteriol. 172:1870-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]