Abstract
We have deleted a gene for a sensor histidine kinase, dspA (or hik33), in the cyanobacterium Synechocystis sp. strain PCC6803. In low and moderate light, the mutant grew slowly under photoautotrophic conditions, with a doubling time of ∼40 h, and had severely reduced photosynthetic oxygen evolution. When the mutant was maintained in low or moderate light in the presence of glucose, its growth rate was only somewhat lower than that of wild-type cells. However, the mutant was light sensitive and rapidly died in high light. Furthermore, levels of many transcripts encoding genes associated with photosynthesis were altered in the mutant relative to wild-type Synechocystis sp. strain PCC6803 both in low light and following exposure to high light. There was constitutive expression of several high-light-inducible genes, including hli, psbAIII, and gpx2; there was little increased accumulation of sodB mRNA in high light; and the cells failed to accumulate cpcBA and psaAB mRNAs in low light in the presence of glucose, although a normal decline in the levels of these mRNAs was observed during exposure to high light. These results suggest that DspA is involved in controlling sets of photosynthetic and high-light-responsive genes, either directly or indirectly. These and other results, some of which are presented in a companion paper (C.-J. Tu, J. Shrager, R. Burnap, B. L. Postier, and A. R. Grossman, J. Bacteriol. 186:3889-3902, 2004), suggest that DspA acts as a global regulator that helps coordinate cellular metabolism with growth limitations imposed by environmental conditions.
Photosynthetic organisms adjust their photosynthetic activity to balance the absorption of light energy with the energy and growth requirements of the cell. This coordination reduces the accumulation of potentially damaging reactive oxygen species that may result from the excitation of pigment molecules; reactive oxygen species may interact with proteins, lipids, and nucleic acids, ultimately causing a loss of cell viability, but they may also have a signaling role (3, 10, 20, 21, 25).
Both microalgae and vascular plants have evolved mechanisms for photoacclimation that enable them to tolerate the absorption of excess excitation energy (10, 24, 25, 31, 32). Acclimation mechanisms include, but are not limited to, changes in the composition of light-harvesting and/or reaction center pigment-protein complexes (4, 6, 7, 38), dissipation of excess absorbed excitation energy as heat, and synthesis of enzymes with antioxidant function, such as superoxide dismutase (26, 35), catalase (27, 29, 42), and peroxidases (11, 16, 40). Efficient degradation and repair of photodamaged polypeptides also occur when photosynthetic organisms are exposed to high light (HL). The D1 polypeptide of photosystem II (PS II), encoded by members of the psbA gene family, carries the reaction center P680 chlorophyll molecules. This protein represents the primary site of photodamage during photoinhibition (2, 18), and its turnover rate increases in HL. In Synechocystis sp. strain PCC6803, there are two active psbA genes, termed psbAII and psbAIII. The psbAII mRNA predominates in low light (LL), but there is a dramatic increase in the level of psbAIII mRNA and a slight increase in psbAII mRNA when Synechocystis sp. strain PCC6803 is exposed to HL (8).
Other cyanobacterial genes have been shown to be sensitive to light conditions and important for acclimation of cells to HL. The HliA protein of Synechococcus sp. strain PCC7942 is a small thylakoid membrane-associated polypeptide (72 amino acids) with sequence similarity to members of the chlorophyll a/b-binding family of proteins (9; N. A. Dolganov, Q. He, and A. R. Grossman, unpublished data). There are four hli genes present on the Synechocystis sp. strain PCC6803 genome, with a fifth sequence fused to the ferrochelatase gene (12, 15). The levels of Hli polypeptides increase in response to HL, low temperature, and nutrient limitation and are necessary for the survival of cells in HL. Strains unable to synthesize specific Hli polypeptides could not cope with HL as effectively as wild-type cells, and a mutant in which all four of the hli genes were deleted rapidly dies in HL (15). The Hli polypeptides may enable cyanobacterial cells to cope with excess absorbed excitation energy, possibly by promoting its dissipation as heat (14), or they may be important in controlling tetrapyrrole biosynthesis (and perhaps binding intermediates in this pathway) (39).
In photosynthetic microbes, acclimation responses are generally mediated by signaling systems comprised of transmembrane sensory kinases that sense extra- or intracellular cues and transmit the information to response regulators that may directly control the transcriptional activity of specific sets of genes. Recently, a sensory histidine kinase termed NblS, identified in Synechococcus sp. strain PCC7942, was shown to be critical for both acclimation to HL and nutrient limitation. This sensory kinase controls HL-regulated and blue or UV-A light-regulated expression from a number of genes whose polypeptide products are involved in photosynthetic function. NblS appears to influence light-dependent modulation of hliA gene expression, HL regulation of the psbA genes, and the biosynthesis and degradation of light-harvesting phycobilisome polypeptides. The deduced polypeptide sequence of NblS revealed the presence of a PAS domain that may bind a flavin (37). The association of NblS with a pigmented electron carrier (the flavin) may allow for direct monitoring of both light and intracellular redox conditions.
NblS of Synechococcus sp. strain PCC7942 has strong sequence similarity to DspA (also called Hik33) of Synechocystis sp. strain PCC 6803 (5, 34). Studies of the nonhomoplasmic hik33 mutant showed that this polypeptide is involved in sensing and controlling gene expression in response to low-temperature conditions (33). Low-temperature treatment reduces the anabolic activity of cells, resulting in the absorption of excess excitation energy and elevated cellular redox, even under moderate light conditions. Recent studies suggest that Hik33 also controls the expression of osmostress-regulated genes (19). These findings add support to the hypothesized role of NblS as a global regulator that integrates redox and light signals and suggest that this regulatory polypeptide may influence other signaling pathways involved in acclimation responses (37).
Recently, under heterotrophic growth conditions, we have been able to obtain a homoplasmic strain disrupted for dspA or hik33. The data in this report suggest that DspA acts as a global regulator that helps coordinate cellular metabolism with growth limitations imposed by environmental conditions. Additional support for a global regulatory role for NblS/DspA/Hik33 comes from microarray analyses (36), which demonstrate pronounced changes in the levels of numerous transcripts in the dspA mutant, even under LL conditions.
MATERIALS AND METHODS
Culture conditions.
Synechocystis sp. strain PCC6803 was cultivated in BG-11 medium (1) buffered with 10 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid], pH 8.2, at 30°C. The cultures were bubbled with 3% CO2 in air under both LL (40 μmol of photon m−2 s−1) and HL (500 μmol of photon m−2 s−1) conditions. For HL treatments, cells grown to mid-logarithmic growth phase (optical density at 730 nm [OD730] of ∼0.8) were diluted with fresh medium to an OD730 of ∼0.3, and then the cultures (∼35 ml in 50-ml culture tubes) were placed in a temperature-controlled glass chamber (maintained at 30°C) and exposed to HL (incandescent bulbs) for various lengths of time, as indicated in the text.
DNA manipulation and mutant construction.
To inactivate the dspA (hik33) gene sll0698 in CyanoBase (34), the entire coding region was amplified by PCR to yield a fragment of 1992 bp. The two primers used for the amplification were 5′-gc(TT)ATGc(G)GGACTTCTGTGTCCAATCCA-3′ and 5′-gg(CT)At(G)CCCACCACCATCAACATGGATTG-3′. Lowercase letters indicate mutations introduced into the sequence, with the original nucleotides given in parentheses to the left of those nucleotides that were modified. The PCR product was cloned into pGEM-T vector (TA ligation), and a 0.85-kbp DNA fragment with the cat gene (chloramphenicol acetyltransferase), under the control of the Synechocystis sp. strain PCC6803 psbAII promoter, was exchanged with a 29-bp fragment (335 to 364 bp downstream of the dspA start codon, from a BglII site to an XmnI site) of the dspA gene, generating the plasmid pDsp-cat. The direction of transcription of the cat gene was opposite of that of dspA. The plasmid was partially sequenced to determine the relative orientation of the dspA and cat genes and to ensure that no modifications in the nucleotide sequence occurred during cloning.
The pDsp-cat plasmid was used to transform Synechocystis sp. strain PCC6803, and transformants were selected by screening for resistance to 20 μg of chloramphenicol/ml in BG-11 medium supplemented with 10 mM glucose. Transformants were restreaked into the same medium, and segregation of the inactivated dspA gene was monitored by PCR using genomic DNA of transformants used as the template and primers that recognize sequences upstream or downstream of the inserted cat gene. The homoplasmic mutant obtained was designated dspA-cat.
To test complementation of the Synechocystis sp. strain PCC6803 dspA-cat mutant with the nblS gene from Synechococcus sp. strain PCC7942, the cat cartridge of the plasmid pDsp-cat was replaced by the nblS gene (under the control of its own promoter) linked to a kanamycin resistance cassette, generating a plasmid that we designated pNblSrDspA. The nblS gene was in the same reading direction as that of dspA. pNblSrDspA was transformed into the dspA-cat mutant, and transformants were repeatedly streaked onto BG-11 medium containing 25 μg of kanamycin/ml until the chloramphenicol resistance phenotype was lost. In the new strain (nblS+ dspA mutant), the nblS gene was nested within the dspA gene (Fig. 1).
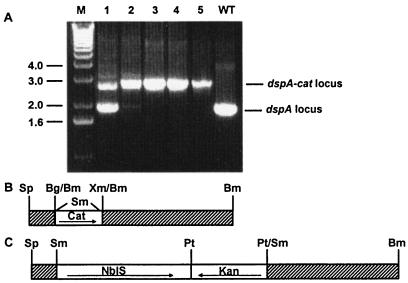
FIG. 1.
Disruption of the dspA gene. (A) PCR analysis of the dspA gene in potential mutants in which the dspA gene was insertionally inactivated with the cat gene. WT, wild type; M, DNA size marker. (B and C) Depiction of the constructs used to generate the dspA-cat strain (B) and the nblS+ dspA mutant strain (C). To generate the dspA-cat disruption, an internal 29-bp dspA fragment from the dspA gene (from Sp to Bm; shaded area) was deleted and replaced by a 0.85-kbp chloramphenicol resistance cartridge (Cat); in panel C, the cat gene shown in panel B was replaced by a 2.2-kb nblS gene linked with a 1.2-kb kanamycin resistance cartridge (Kan); the nblS and kanr genes are transcribed in opposite directions. In panel A, both the intact dspA (∼2.0 kbp) and the dspA-cat (∼2.8 kbp) loci were efficiently amplified from genomic DNA preparations by PCR. A total of 5 chloramphenicol-resistant transformants were isolated (lanes 1 to 5) and analyzed for a disruption of the dspA gene; transformants 3 and 4 showed no detectable wild-type dspA PCR product, and transformant 4 was used in all subsequent analyses. Sp, SphI; Bg, BglII; Bm, BamHI; Xm, XmnI; Sm, SmaI; Pt, PstI.
For PCR analysis, Synechocystis sp. strain PCC6803 genomic DNA was prepared by a mini preparation procedure. Briefly, a loopful of cyanobacterial cells was suspended in 200 μl of Tris-EDTA buffer (pH 8.0) and transferred to a microcentrifuge tube with 200 μl of glass beads (0.1-mm diameter; Sigma). The cells were broken in a MiniBead Beater (Biospec Products, Bartlesville, Okla.) by two cycles of agitation at the low speed setting for 30 s; between each cycle, the cells were cooled on ice for 2 to 3 min. Lysates were extracted with phenol-chloroform, and the DNA was precipitated, washed, and dried according to standard procedures (28). The dry DNA pellet was dissolved in 40 μl of H2O, of which 1 μl was used for each PCR.
Photosynthetic O2 evolution.
O2 exchange in cell suspensions was measured at 30°C with a Clark-type O2 electrode (Hansatech DW2/2). Cells were illuminated with white light produced by a Schott KL1500 lamp equipped with a flexible light guide. The intensity of illumination was adjusted with combinations of neutral density filters. Cell viability was monitored by using the vital stain TO-PRO-1 iodine (see the legend to Fig. 2).
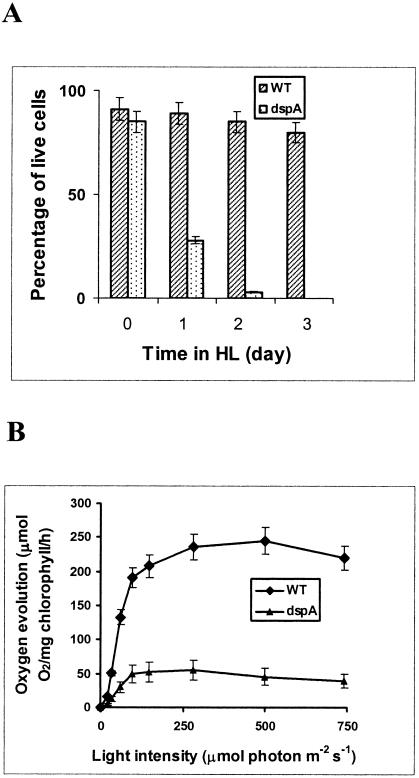
FIG. 2.
Light sensitivity of the dspA mutant. (A) Cells were grown in BG-11 medium with 10 mM glucose in LL to an OD730 of ∼0.3 (day 0) before the cultures were transferred and incubated in HL (days 1 to 3). Cell viability was determined by staining with TO-PRO-1 iodine, a nonpermeable fluorescence dye that stains nucleic acid (only in dead cells). Curves were generated by averaging the data obtained from three representative experiments. (B) Light-response curves of photosynthetic O2 evolution in wild-type Synechocystis sp. strain PCC6803 (WT) and dspA-cat mutant (dspA). Cells were grown in LL in the presence of 10 mM glucose to an OD730 of ∼0.6. Cultures were concentrated to a OD730 of 0.9 for O2 evolution measurements under various actinic light intensities.
RNA isolation and Northern hybridizations.
RNA was isolated from cells as previously described (22). For RNA blot hybridizations, equal amounts of RNA (determined spectroscopically) were resolved by electrophoresis in formaldehyde gels; ethidium bromide was included in the loading buffer, allowing for visualization of the rRNA bands and confirmation of equal loading of RNA samples. The DNA probes used for hybridization were prepared by PCR in a 15-μl reaction mixture containing PCR buffer, 2 U of Taq DNA polymerase (Roche, Palo Alto, Calif.), 0.1 μg each of the two PCR primers, 30 μCi of [32P]dCTP (3,000 Ci/mmol, 10 μCi/μl; NEN-DuPont, Boston, Mass.), and ∼10 ng of gene-specific PCR products as a template. The gene-specific PCR products were originally generated from plasmid clones or from genomic DNA by two cycles of PCR. In the first cycle, a larger fragment was amplified, which was purified from agarose gels and used as the template for a second PCR to generate gene-specific probes. The second PCR cycle employed either one or two new internal primers, and the product was purified from agarose gels. The different probes generated and primers used for amplification of these probes are given in Table 1.
TABLE 1.
Sequences of primers used for generating gene-specific probesa
| Gene | Template | Primer | Position on genome |
|---|---|---|---|
| hliA | Plasmid | TAATCCAATTATGACCACCCG | 701360-701340 |
| CTCGGCCTATTCTACAGGCTA | 701127-701147 | ||
| hliB | Plasmid | CAACACTATGACTAGCCGC | 982961-982979 |
| ACCCAGCCAATTAGAGAGAG | 983190-983171 | ||
| hliC | Plasmid | ACAGACTTGCCATGGGCGCAA | 1142026-1142006 |
| CTATGGAAAAATTACAGAATGC | 1141792-1141813 | ||
| hliD | Plasmid | GGAAATCCCATGAGTGAAGAAC | 398179-398200 |
| ACTCCCTAGCGCAGTCCCAACC | 398366-398345 | ||
| psbA | Plasmid | ACCATTGCCGTTGCCGGTAAAG | 943222-943243 |
| GGTGGTAACAATACCTCCGAGG | 943902-943881 | ||
| gpx2 | PCR | ATGCCATTACCCACTTCCCTGAC | 1436699-1436721 |
| TTAGCCCAAGGCTTTTTCGATCG | 1437163-1437141 | ||
| CGGCTTAACCCCCCAATAC | 1436800-1436818 | ||
| TTAGCCCAAGGCTTTTTCGATCG | 1437163-1437141 | ||
| sodB | PCR | ATGGCTTACGCACTAC | 1607353-1607373 |
| CTAACCTAGGCCGCTGCTAAGTTAGC | 1607952-1607932 | ||
| CAAACACCATGCCGCCTAC | 1607439-1607457 | ||
| AGCATGCTCCCATACGTCC | 1607850-1607832 | ||
| cpcBA | Plasmid | GTTTCCACCGCTGACTCTCAAG | 726813-726792 |
| CTAGCTCAGAGCATTGATGGCG | 726349-726370 | ||
| psaAB | Plasmid | CCATTGCCGTTGCCGGTAAAGTC | 943223-943245 |
| GTGGTAACAATACCTCCGAGGAG | 943901-943879 |
The nucleotide sequences of the primers used for amplification of the gene-specific probes are given, and the positions of these primer sequences on the Synechocystis sp. strain PCC6803 genome are defined by the base numbers provided by CyanoBase (http://www.kazusa.or.jp/cyanobase/) (right-hand column). The templates used for amplification were either plasmids containing full-length genes or second-round PCR products purified from agarose gels. The hliA and hliB DNA fragments used as probes are 95% identical. Transcripts from these two genes were distinguishable based on their size difference. The psbA probe was prepared by using a plasmid containing the full-length psbAII gene; it could hybridize to all three of the potential psbA transcripts of Synechocystis sp. strain PCC6803.
RESULTS
Impacts on cell viability and photosynthetic function.
To investigate the functions of DspA, we inactivated the dspA gene in Synechocystis sp. strain PCC6803 (see Materials and Methods). The construct (pDsp-cat) used to generate the dspA-disrupted strain is shown in Fig. 1B. A homoplasmic mutant was obtained, as evaluated by PCR analysis (Fig. 1A, lanes 3 to 5). The growth of the dspA mutant under photoautotrophic and photomixotrophic conditions was monitored in LL and HL (data not shown). The dspA mutant grew very slowly, with a doubling time of ∼40 h under photoautotrophic growth conditions (in contrast to ∼8 h for wild-type cells); however, it grew well, although not as well as wild-type cells, in medium supplemented with glucose; the doubling time of wild-type cells under our photomixotrophic growth conditions was 6 h, while that of the mutant was 8 h. In contrast, when the dspA mutant was placed in HL for more than 1 day in either the presence (Fig. 2A) or absence (data not shown) of glucose, it stopped dividing and lost viability, as measured by both the replating of cells onto solid BG-11 medium with 10 mM glucose (unpublished data) and cell viability assays with vital stains (Fig. 2A); by 48 h in HL, nearly all of the cells were dead. These results demonstrated that glucose cannot sustain the dspA mutant in HL and that photodamage in HL resulted in a loss of cell viability. The cells also died much more rapidly than wild-type cells following exposure to nutrient deprivation (unpublished data).
Photosynthetic O2 evolution in the wild-type and mutant strains was measured as a function of light intensity (Fig. 2B). For wild-type cells, O2 evolution saturated at ∼250 μmol of photon m−2 s−1 with a peak activity of ∼250 μmol of O2/mg of chlorophyll/h. In contrast, the photosynthetic efficiency in LL was reduced in the mutant relative to the wild-type strain, and the maximum rate of O2 evolution attained by the mutant was 50 μmol of O2/mg of chlorophyll/h. These results demonstrate that the dspA lesion caused a marked decline in photosynthetic performance and explain why a homoplasmic strain disrupted for dspA was difficult to obtain under photoautotrophic growth conditions, even using antibiotic selection (34). In our study, complete segregation was made possible by the supplementation of the antibiotic-containing growth medium with 10 mM glucose.
Analyses of transcripts for photosynthetic and stress-responsive genes.
The growth data and measurements of O2 evolution demonstrated that the dspA mutant was severely compromised in photosynthetic function and was unable to acclimate to HL conditions. To further characterize responses of the mutant to HL, we measured levels of transcripts from the various genes involved in photosynthesis and in the amelioration of oxidative stress. Total RNA from cells grown in LL or 30 min following the transfer of cells to HL was hybridized to specific fragments of genes encoding polypeptides of the photosynthetic apparatus or associated with oxidative stress responses. As shown in Fig. 3 (columns 1 and 2), in wild-type cells, the transcripts for all four of the hli genes and for one of the two gpx genes (encoding glutathione peroxidase) accumulated to high levels following the transfer of cells to HL. Furthermore, total psbA mRNA increased by more than threefold following 30 min of HL treatment, consistent with results previously reported (13). The transcript for the sodB gene, encoding superoxide dismutase, increased by two- to threefold following HL treatment, with the generation of a high-molecular- mass transcript (the ends of these transcripts have not been characterized). In contrast, levels of cpcBA and psaAB transcripts declined, in agreement with the observations of Muramatsu and Hihara (23).
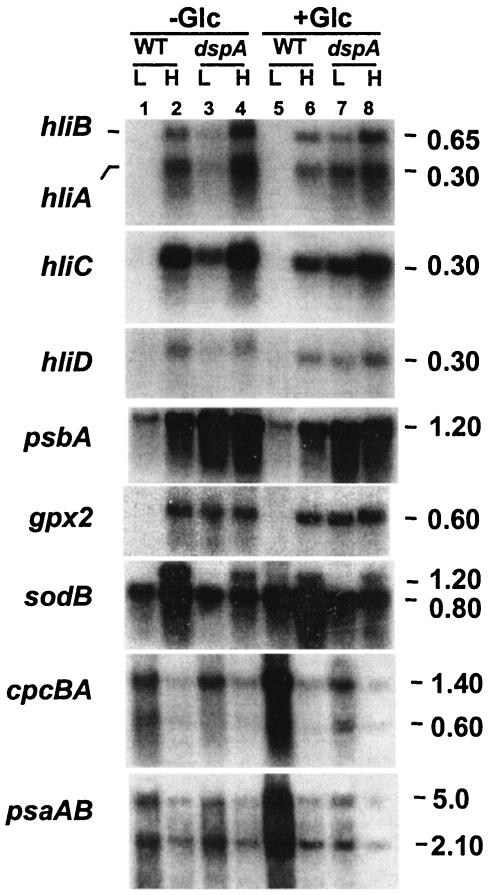
FIG. 3.
RNA blot hybridizations with specific probes for genes involved in photosynthesis and stress responses. Total RNA was isolated from cells grown in LL (L) or exposed to HL (H) for 30 min. The RNA was resolved by denaturing electrophoresis on 1.3% agarose gels, transferred to nylon membranes, and hybridized with the labeled probes indicated. The three psbA genes were not distinguished in these analyses. The sodB gene fragment consistently hybridized to two transcripts (bands), the lower band having the size predicted from the CyanoBase gene sequence. The upper band may have a different transcription start site or represent a read-through product, but this possibility was not investigated. The psaAB probe hybridized to two transcripts as previously reported (30). The cpcBA fragment also hybridized to two transcripts; the signal from the 1.4-kb transcript represents the full-length transcript of cpcBA. The size of each transcript is indicated. gpx2 (slr1992), glutathione peroxidase; −Glc, without glucose in the growth medium; +Glc, with 10 mM glucose in the growth medium; WT, wild-type cells.
In the dspA mutant, as in wild-type cells, transcripts from hli, psbA, and gpx2 genes accumulated to high levels following 30 min of HL treatment. However, levels of these mRNAs were already relatively high under LL conditions. In addition, the sodB transcript in the mutant strain no longer accumulated to very high levels in HL, while the HL-induced decline of cpcBA and psaAB transcripts was similar to that observed for wild-type cells.
Interestingly, wild-type cells grown for 1 to 2 days in LL in medium supplemented with 10 mM glucose showed a marked accumulation of cpcBA and psaAB transcripts; this elevated accumulation was not observed in the dspA mutant. The transcript levels of other genes shown in Fig. 3 were not strongly affected by the addition of glucose to the growth medium.
Complementation of the dspA mutant with the nblS gene from Synechococcus sp. strain PCC7942.
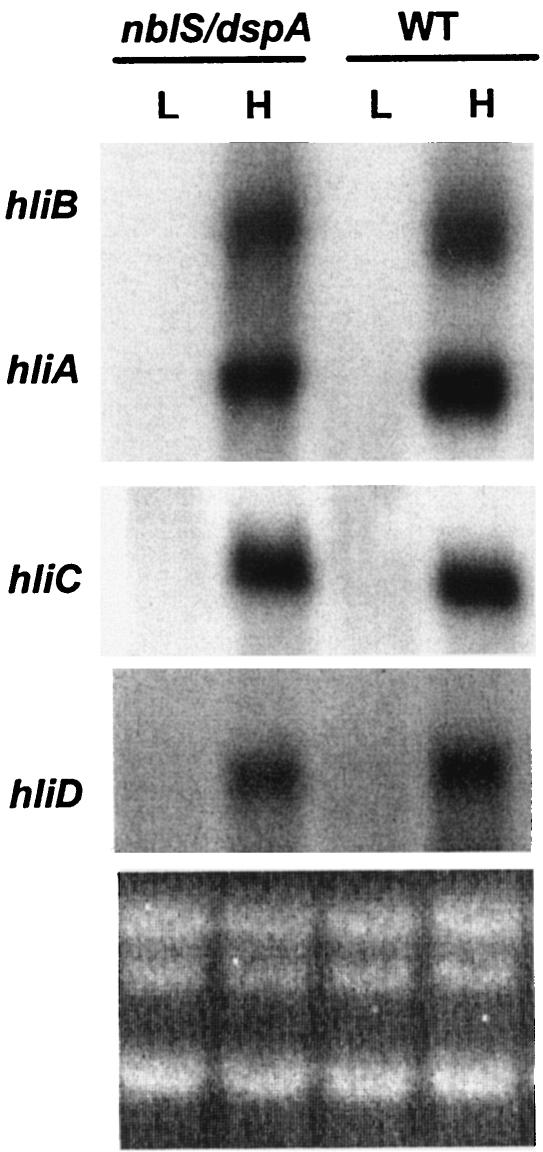
Expression of hli genes in the dspA mutant (as shown in Fig. 3) was different from that observed in the nblS1-1 mutant of Synechococcus sp. strain PCC7942, although it should be remembered that the latter strain contains two point mutations in nblS, and we were never able to generate a null mutation. To investigate whether DspA is functionally related to NblS, the full-length nblS gene of Synechococcus sp. strain PCC7942 was transformed into the dspA mutant of Synechocystis sp. strain PCC6803 (see Materials and Methods for details); in this strain, the nblS gene was nested within the dspA gene containing an internal deletion of 29 bp (Fig. 1C). As shown in Fig. 4, the nblS-harboring transformant regained the ability to suppress accumulation of hli transcripts under LL conditions (just like wild-type cells), although the levels of mRNA accumulation in HL appeared to be slightly lower than those observed for wild-type cells. Furthermore, the rescued mutant survived HL conditions and grew at a rate similar to that of wild-type cells in both LL and HL (data not shown). These results strongly suggest that DspA and NblS are functional orthologs.
FIG. 4.
Expression of hli genes in the dspA-cat mutant complemented with the nblS gene (nblS/dspA). Total RNA was isolated from cells grown in LL (L) or exposed to HL (H) for 30 min. The RNA was hybridized with the fragments from the four hli genes, as indicated. WT, wild type.
Stability of transcripts.
Since the absolute levels of specific transcripts are governed by a combination of the rates of transcription and transcript degradation, we investigated whether mRNA stability contributed to differential accumulation of transcripts observed in the dspA mutant under LL and HL conditions. LL-grown cells were exposed to HL for 30 min before rifampin (150 μg/ml) was added to cultures to block transcription. Cultures were then divided and incubated in either LL or HL for an additional 60 min. Aliquots were removed at various time points following the administration of rifampin, and RNA levels were monitored by RNA blot hybridizations. As shown in Fig. 5, a disruption of the dspA gene did not strongly influence the rate of transcript degradation under LL or HL conditions. The hliA messages appeared to be slightly more stable in the dspA mutant than in wild-type cells, but the differences were not very large. Furthermore, glucose did not significantly influence the stability of any of the transcripts examined (data not shown). The half-lives of the transcripts deduced from Fig. 5, under both LL and HL conditions, are presented in parentheses in the figure.
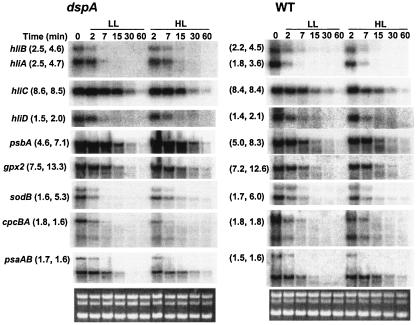
FIG. 5.
Transcript stability in wild-type cells (WT) and the dspA-cat mutant (dspA). The cells were exposed to HL for 30 min followed by the addition of 150 μg of rifampin/ml (transcription inhibitor) immediately after placing half of the culture in LL and the other half in HL; the addition of rifampin occurred at time zero. Aliquots of the cultures were withdrawn at the times indicated at the top of the figure, and the mRNA levels were determined by RNA blot hybridizations. The number pairs in the parentheses indicate the estimated mRNA half-life of each gene under LL (numbers at left within each set of parentheses) and HL (numbers at right within each set of parentheses) in the dspA-cat mutant (parentheses at the left side of the figure) and in the wild type (parentheses in the center of the figure). To estimate the mRNA half-life of a messenger, the intensities of the specific hybridization signals on the autoradiograms were measured by densitometry, and the half-life of each transcript was calculated. The half-lives of the cpcBA and psaAB transcripts were determined from the loss of full-length transcripts following the addition of rifampin, while the half-life of sodB was estimated from the loss of the transcript, represented by the lower hybridization signal (which is the expected size of the transcript based on the gene sequences; the upper transcript might be a consequence of a second transcription initiation or termination site).
DISCUSSION
Our results demonstrate that the loss of DspA function strongly impacts photosynthetic performance and survival of the cells in HL. This finding may be a consequence of aberrant assembly of the photosynthetic apparatus in the mutant strain, which leads to increased photodamage and the production of reactive oxygen species, especially under HL conditions. Furthermore, there was a high level of accumulation of the hli, psbA, and gpx2 transcripts in the dspA mutant but not in wild-type cells grown in LL, regardless of glucose supplementation. Constitutive expression of hli, psbA, and gpx2 genes suggests that these genes are repressed by DspA activity in LL and that HL activation involves removing the repression imposed by DspA (either by direct or indirect effects on gene activity). While this is the first report that suggests that the HL activation of bacterial genes may involve, at least in part, derepression, negative regulation of stress-responsive genes has been reported for both prokaryotic and eukaryotic systems. For example, the catalase gene (katG) of Mycobacterium tuberculosis is negatively regulated by the global ferric uptake regulator FurA (41), and ethylene responses in vascular plants are negatively regulated by a family of receptors encoded by ETR genes (17).
Interestingly, in the dspA mutant, the hli transcripts still accumulated to a significantly higher level in HL than in LL (Fig. 3). This enhanced accumulation may be due to differential mRNA stability, as the hli transcripts are slightly more stable in HL than in LL, regardless of whether or not dspA is functional (Fig. 5). However, it is also possible that the hli genes are subject to a positive regulation in addition to the negative control imposed by DspA. The convergence of a potential negative and a positive regulation in controlling hli genes resembles what has been observed in the regulation of the katG gene in M. tuberculosis (41). Furthermore, in contrast to the negative regulation of the hli and psbA genes by DspA, sodB expression appears to be positively regulated by DspA. These results demonstrate that DspA has both a positive and a negative impact on the accumulation of different transcripts.
Another interesting finding of this study is that the transcript levels of cpcBA and psaAB dramatically increase if glucose is added to the growth medium of wild-type cells growing in LL and that this effect is dependent on DspA. The increase was sustained for 1 to 2 days after adding glucose to the medium and resulted in a ∼25% increase in the corresponding polypeptides (data not shown) over the time period. These findings suggest an interaction (either direct or indirect) between the sugar and DspA signaling pathways.
In sum, the results presented in this paper demonstrate that DspA profoundly influences the growth characteristics and photosynthetic capacity of Synechocystis sp. strain PCC6803, that it is required for survival of cyanobacterial cells in HL, and that it appears to be functionally equivalent to NblS of Synechococcus sp. strain PCC7942 (which is critical for the acclimation of cells to both HL and nutrient limitation conditions). The results also suggest that DspA or NblS polypeptides may elicit both negative and positive regulatory responses under a variety of conditions and that they may also influence glucose signaling and, potentially, the integration of respiratory and photosynthetic function. Combined with previous results (34, 37), the data reported here suggest that DspA or NblS plays a central role in integrating cellular metabolism with environmental cues (and the potential for cell growth). While a direct regulatory relationship between DspA and potential target genes cannot be clearly established, the inability of the nblS1-1 allele to derepress (or activate) hli or psbA genes suggests a primary role for DspA in controlling the expression of at least some of the HL-activated genes. The precise environmental cues that NblS or DspA senses are not clear; however, it is likely that these polypeptides are able to perceive light, redox, and/or reactive oxygen species and that their signaling outputs are integrated into a regulatory web that controls the activities of numerous cyanobacterial genes (36).
Acknowledgments
We thank D. Bhaya, J. Christie, C.-J. Tu, G. Thompson, and W. Briggs for their helpful suggestions during the course of this work. We specially thank G. Thompson for generously making the resources in his laboratory available to us.
This work was supported by NSF grant MCB9727836 and USDA 98-35301 awarded to A.R.G. and by startup funds provided by the University of Arkansas at Little Rock to Q.H.
REFERENCES
- 1.Allen, M. M. 1969. Simple conditions for the growth of unicellular blue-green algae in plates. J. Phycol. 4:1-3. [DOI] [PubMed] [Google Scholar]
- 2.Aro, E.-M., I. Virgin, and B. Andersson. 1993. Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143:113-134. [DOI] [PubMed] [Google Scholar]
- 3.Asada, K. 1994. Production and action of active oxygen species in photosynthetic tissues, p. 77-104. In C. H. Foyer and P. M. Mullineaux (ed.), Causes of photooxidative stress and amelioration of defense systems in plants. CRC Press, Boca Raton, Fla.
- 4.Bailey, S., R. G. Walters, S. Jansson, and P. Horton. 2001. Acclimation of Arabidopsis thaliana to the light environment: the existence of separate low light and high light responses. Planta 213:794-801. [DOI] [PubMed] [Google Scholar]
- 5.Bartsevich, V. V., and S. V. Shestakov. 1995. The dspA gene product of the cyanobacterium Synechocystis sp. strain PCC 6803 influences sensitivity to chemically different growth inhibitors and has amino acid similarity to histidine protein kinases. Microbiology 141:2915-2920. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, D., J. Houmard, and N. Tandeau de Marsac. 1993. Electron transport regulates cellular differentiation in the filamentous cyanobacterium Calothrix. Plant Cell 5:451-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casey, E. S., D. M. Kehoe, and A. R. Grossman. 1997. Suppression of mutants aberrant in light intensity responses of complementary chromatic adaptation. J. Bacteriol. 179:4599-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Constant, S., I. Perewoska, M. Alfonso, and D. Kirilovsky. 1997. Expression of the psbA gene during photoinhibition and recovery in Synechocystis PCC 6714: inhibition and damage of transcriptional and translational machinery prevent the restoration of photosystem II activity. Plant Mol. Biol. 34:1-13. [DOI] [PubMed] [Google Scholar]
- 9.Dolganov, N. A. M., D. Bhaya, and A. R. Grossman. 1995. Cyanobacterial protein with similarity to the chlorophyll a/b-binding proteins of higher plants: evolution and regulation. Proc. Natl. Acad. Sci. USA 92:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foyer, C. H., and J. M. Fletcher. 2001. Plant antioxidants: colour me healthy. Biologist (London) 48:115-120. [PubMed] [Google Scholar]
- 11.Fryer, M. J., L. Ball, K. Oxborough, S. Karpinski, P. M. Mullineaux, and N. R. Baker. 2003. Control of ascorbate peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J. 33:691-705. [DOI] [PubMed] [Google Scholar]
- 12.Funk, C., and W. Vermaas. 1999. A cyanobacterial gene family coding for single-helix proteins resembling part of the light-harvesting proteins from higher plants. Biochemistry 38:9397-9404. [DOI] [PubMed] [Google Scholar]
- 13.Golden, S. S. 1995. Light-responsive gene expression in cyanobacteria. J. Bacteriol. 177:1651-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Havaux, M., G. Guedeney, Q. He, and A. R. Grossman. 2003. Elimination of high-light-inducible polypeptides related to eukaryotic chlorophyll a/b-binding proteins results in aberrant photoacclimation in Synechocystis PCC6803. Biochim. Biophys. Acta 1557:21-33. [DOI] [PubMed] [Google Scholar]
- 15.He, Q., N. Dolganov, O. Bjorkman, and A. R. Grossman. 2001. The high light-inducible polypeptides in Synechocystis PCC6803. Expression and function in high light. J. Biol. Chem. 276:306-314. [DOI] [PubMed] [Google Scholar]
- 16.Horling, F., P. Lamkemeyer, J. Konig, I. Finkemeier, A. Kandlbinder, M. Baier, and K. J. Dietz. 2003. Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis. Plant Physiol. 131:317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua, J., H. Sakai, S. Nourizadeh, Q. G. Chen, A. B. Bleecker, J. R. Ecker, and E. M. Meyerowitz. 1998. EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10:1321-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyle, D. J., C. B. Osmond, and C. J. Arntzen. 1987. The biochemical basis for photoinhibition of photosystem II, p. 197-226. In D. J. Kyle, C. B. Osmond, and C. J. Arntzen (ed.), Photoinhibition. Elsevier, Amsterdam, The Netherlands.
- 19.Mikami, K., Y. Kanesaki, I. Suzuki, and N. Murata. 2002. The histidine kinase Hik33 perceives osmotic stress and cold stress in Synechocystis sp. PCC 6803. Mol. Microbiol. 46:905-915. [DOI] [PubMed] [Google Scholar]
- 20.Mittler, R. 2002. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7:405-410. [DOI] [PubMed] [Google Scholar]
- 21.Mittler, R., and E. Tel-Or. 1991. Oxidative stress responses in the unicellular cyanobacterium Synechococcus PCC 7942. Free Radic. Res. Commun. 12-13(Pt. 2):845-850. [PubMed] [Google Scholar]
- 22.Mohamed, A., and C. Jansson. 1989. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol. Biol. 13:693-700. [DOI] [PubMed] [Google Scholar]
- 23.Muramatsu, M., and Y. Hihara. 2003. Transcriptional regulation of genes encoding subunits of photosystem I during acclimation to high-light conditions in Synechocystis sp. PCC 6803. Planta 216:446-453. [DOI] [PubMed] [Google Scholar]
- 24.Nayak, L., M. K. Raval, B. Biswal, and U. C. Biswal. 2002. Topology and photoprotective role of carotenoids in photosystem II of chloroplast: a hypothesis. Photochem. Photobiol. Sci. 1:629-631. [DOI] [PubMed] [Google Scholar]
- 25.Niyogi, K. K. 1999. Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:333-359. [DOI] [PubMed] [Google Scholar]
- 26.Perl-Treves, R., and E. Galun. 1991. The tomato Cu,Zn superoxide dismutase genes are developmentally regulated and respond to light and stress. Plant Mol. Biol. 17:745-760. [DOI] [PubMed] [Google Scholar]
- 27.Redinbaugh, M. G., M. Sabre, and J. G. Scandalios. 1990. Expression of the maize Cat3 catalase gene is under the influence of a circadian rhythm. Proc. Natl. Acad. Sci. USA 87:6853-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Skadsen, R. W., and J. G. Scandalios. 1987. Translational control of photo-induced expression of the Cat2 catalase gene during leaf development in maize. Proc. Natl. Acad. Sci. USA 84:2785-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smart, L. B., and L. McIntosh. 1991. Expression of photosynthesis genes in the cyanobacterium Synechocystis PCC6803. psaA-psaB and psbA transcripts accumulate in dark-grown cells. Plant Mol. Biol. 17:959-971. [DOI] [PubMed] [Google Scholar]
- 31.Smirnoff, N. 2000. Ascorbate biosynthesis and function in photoprotection. Philos. Trans. R. Soc. Lond. B 355:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smirnoff, N., and G. L. Wheeler. 2000. Ascorbic acid in plants: biosynthesis and function. Crit. Rev. Biochem. Mol. Biol. 35:291-314. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki, I., Y. Kanesaki, K. Mikami, M. Kanehisa, and N. Murata. 2001. Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol. Microbiol. 40:235-244. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki, I., D. A. Los, Y. Kanesaki, K. Mikami, and N. Murata. 2000. The pathway for perception and transduction of low-temperature signals in Synechocystis. EMBO J. 19:1327-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsang, E. W., C. Bowler, D. Herouart, W. Van Camp, R. Villarroel, C. Genetello, M. Van Montagu, and D. Inze. 1991. Differential regulation of superoxide dismutases in plants exposed to environmental stress. Plant Cell 3:783-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tu, C.-J., J. Shrager, R. Burnap, B. L. Postier, and A. R. Grossman. 2003. Consequences of a deletion in dspA on transcript accumulation in Synechocystis sp. strain PCC6803. J. Bacteriol. 186:3501-3514. [DOI] [PMC free article] [PubMed]
- 37.van Waasbergen, L. G., N. Dolganov, and A. R. Grossman. 2002. nblS, a gene involved in controlling photosynthesis-related gene expression during high light and nutrient stress in Synechococcus elongatus PCC 7942. J. Bacteriol. 184:2481-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walters, R. G., J. J. Rogers, F. Shephard, and P. Horton. 1999. Acclimation of Arabidopsis thaliana to the light environment: the role of photoreceptors. Planta 209:517-527. [DOI] [PubMed] [Google Scholar]
- 39.Xu, H., D. Vavilin, C. Funk, and W. Vermaas. 2002. Small Cab-like proteins regulating tetrapyrrole biosynthesis in the cyanobacterium Synechocystis sp. PCC6803. Plant Mol. Biol. 49:149-160. [DOI] [PubMed] [Google Scholar]
- 40.Yabuta, Y., T. Motoki, K. Yoshimura, T. Takeda, T. Ishikawa, and S. Shigeoka. 2002. Thylakoid membrane-bound ascorbate peroxidase is a limiting factor of antioxidative systems under photo-oxidative stress. Plant J. 32:915-925. [DOI] [PubMed] [Google Scholar]
- 41.Zahrt, T. C., J. Song, J. Siple, and V. Deretic. 2001. Mycobacterial FurA is a negative regulator of catalase-peroxidase gene katG. Mol. Microbiol. 39:1174-1185. [DOI] [PubMed] [Google Scholar]
- 42.Zhong, H. H., J. C. Young, E. A. Pease, R. P. Hangarter, and C. R. McClung. 1994. Interactions between light and the circadian clock in the regulation of CAT2 expression in Arabidopsis. Plant Physiol. 104:889-898. [DOI] [PMC free article] [PubMed] [Google Scholar]