Abstract
A sensor histidine kinase of Synechococcus sp. strain PCC7942, designated nblS, was previously identified and shown to be critical for the acclimation of cells to high-light and nutrient limitation conditions and to influence the expression of a number of light-responsive genes. The nblS orthologue in Synechocystis sp. strain PCC6803 is designated dspA (also called hik33). We have generated a dspA null mutant and analyzed global gene expression in both the mutant and wild-type strains under high- and low-light conditions. The mutant is aberrant for the expression of many genes encoding proteins critical for photosynthesis, phosphate and carbon acquisition, and the amelioration of stress conditions. Furthermore, transcripts from a number of genes normally detected only during exposure of wild-type cells to high-light conditions become partially constitutive in the low-light-grown dspA mutant. Other genes for which transcripts decline upon exposure of wild-type cells to high light are already lower in the mutant during growth in low light. These results suggest that DspA may influence gene expression in both a positive and a negative manner and that the dspA mutant behaves as if it were experiencing stress conditions (e.g., high-light exposure) even when maintained at near-optimal growth conditions for wild-type cells. This is discussed with respect to the importance of DspA for regulating the responses of the cell to environmental cues.
Photosynthetic organisms have evolved intricate mechanisms for sensing and acclimating to environmental change. Parameters such as light quality, light intensity, and nutrient availability can modulate both the structure and the function of the photosynthetic machinery. Physiological and biochemical changes elicited by external cues include modification of light-harvesting complex (LHC) synthesis and degradation (6, 12, 27, 29, 38, 45, 65, 75), changes in absorption and excitation energy transfer properties of LHC (11, 18, 28, 59), and a modification of reaction center function (59, 80). Intracellular cues critical for controlling cellular processes during acclimation may reflect the cell's growth potential, cellular redox conditions, and/or accumulation of reactive oxygen species (52, 77).
Precise control over the fate of absorbed excitation energy is critical for cell viability during exposure of photosynthetic cells to excess excitation, since energized pigment molecules may trigger the production of damaging, reactive oxygen species (2). Over the short term, cells can dissipate excess absorbed excitation energy as heat by quenching excited pigment molecules in the LHC or by eliciting a state transition in which the LHC of photosystem II (PS II) directs its excitation energy to PS I, where quenching can occur. Over the long term, excess excitation may cause a dramatic reduction in the level of LHC. In cyanobacteria, a reduction in LHC size is reflected in reduced levels of transcripts encoding light-absorbing polypeptides (or phycobiliproteins) of the major LHC (or phycobilisomes). Several studies have shown that light and nutrient conditions modulate levels of phycobiliprotein transcripts (e.g., from the cpcBA and apcAB genes) (9, 30, 40, 41, 53, 60), enzymes required for pigment biosynthesis (e.g., from hem and chl genes), and subunits of the photosynthetic reaction centers (e.g., from psaA, psaB, psbA, and psbD genes) (36).
PS II is a primary site of damage to the photosynthetic machinery in high light (HL), causing photoinhibition. The PS II reaction center polypeptides are composed of D1 and D2, and damage to the reaction center reflects a rapid loss of function of D1. Cellular processes have evolved to facilitate rapid turnover and exchange of photodamaged D1 subunits (10). In cyanobacteria, the D1 and D2 polypeptides are encoded by two gene families, psbA and psbD, respectively. Synechococcus sp. strain PCC7942 has three distinct psbA genes and two distinct psbD genes. The three psbA genes encode two forms of D1 (7, 25); psbAI encodes D1, form I, which is abundant under low-light (LL) conditions, while psbAII and psbAIII each encode D1, form II, which becomes abundant when cells are shifted from LL to HL. The latter form of D1 is important for maintaining PS II function and the fitness of cells under HL conditions (1, 8, 31, 50, 51). Synechocystis sp. strain PCC6803 also has three psbA genes, but the expression of only psbAII and psbAIII has been observed; transcripts from both psbAII and psbAIII increase during exposure of the cells to HL (13). Synechocystis sp. strain PCC6803 also has two psbD genes (psbDI and psbDII) encoding D2 (35, 56).
Another family of cyanobacterial genes that are highly expressed during HL exposure and nutrient deprivation has been designated hli (HL induced) (15) or scp (small chlorophyll binding protein). The genes in this family have homology to light-harvesting polypeptides of vascular plants but have one membrane-spanning helix, rather than three (15). In Synechocystis sp. strain PCC6803, there are four hli genes, hliA, hliB, hliC, and hliD (also called scpB, scpC, scpD, and scpE) (21), as well as an hli sequence fused to the ferrochelatase coding region (21, 34). All of the Hli polypeptides accumulate following the exposure of cells to HL conditions, although the kinetics of accumulation may be different. These polypeptides also accumulate under conditions of nitrogen or sulfur deprivation and low temperature, suggesting that they are stress-related polypeptides that are synthesized when cells absorb excess excitation energy (34). A null mutant for all four of the hli genes (hli quadruple mutant) grows well in LL but, unlike wild-type cells, dies upon exposure to HL (34). The Hli polypeptides may be involved in protecting cells from HL damage and may mechanistically facilitate the dissipation of excess absorbed light energy (33) and/or limit the accumulation of free phototoxic intermediates in chlorophyll biosynthesis (81).
Recently, we isolated a photosynthetic mutant of Synechococcus sp. strain PCC7942 that exhibited abnormal expression of the light-regulated hli, psbA, and cpcBA genes and that died rapidly following exposure to either HL or nutrient deprivation. The gene altered (by two point mutations) in the mutant encodes a sensor histidine kinase (62, 71, 72), designated NblS (77), that has a PAS domain and may interact with the response regulator NblR (69). The PAS domain may bind a chromophore and function in monitoring light, oxygen, and/or redox conditions (76, 77). We were unable to obtain an nblS null mutant of Synechococcus sp. strain PCC7942 (77). Recently, a mutant of Synechocystis sp. strain PCC6803 that was null for dspA (also called hik33) (73) was isolated, and the mutant phenotype was rescued by the Synechococcus sp. strain PCC7942 nblS gene (42). Like the nblS mutant, the dspA strain showed aberrations in expression of the psbA and hli genes, although the phenotype of the dspA null mutant was somewhat different than that of the nblS mutant.
Microarray-based methods for high-throughput monitoring of gene expression have been developed over the last decade (14, 66, 82). Such methods are powerfully used for analyses of gene expression in organisms for which there is a complete genome sequence, such as Synechocystis sp. strain PCC6803 (46). Recent microarray studies have exploited Synechocystis sp. strain PCC6803 for identifying genes whose activities are modulated by light levels and UV irradiation (22, 36, 43). To further define the role of NblS/DspA/Hik33 (3, 54, 73) in the responses of cells to environmental change, we used microarrays to compare global transcript levels in wild-type Synechocystis sp. strain PCC6803 and the dspA mutant both in LL and following exposure of cells to HL. The results suggest that DspA has a far-reaching effect on the levels of numerous transcripts, including those encoding polypeptides directly or indirectly associated with photosynthesis and other metabolic processes, and that these effects are apparent under both HL and LL conditions.
MATERIALS AND METHODS
Strains and growth conditions.
The nonmotile strain of Synechocystis sp. strain PCC6803 and the dspA mutant (42) were cultured in BG-11 medium with or without supplementation with 10 mM glucose. Cells were maintained in LL (30 μmol of photon m−2 s−1) and bubbled with 3% CO2 in air. For HL treatment, cells were grown according to a method described previously (34). Briefly, when cultures attained mid-logarithmic growth (optical density at 750 nanometers of ∼0.6), they were diluted to an optical density at 750 nanometers of ∼0.3 and either placed back in LL or illuminated with 500 μmol of photon m−2 s−1 of white light for various periods of time. The cells were then cooled in liquid nitrogen and collected by centrifugation (5,000 × g for 5 min), and the cell pellets were stored at −80°C until they were used for RNA isolation.
Amplification of gene fragments and array construction.
The whole set of PCR-amplified gene-specific fragments used to construct the microarray was generated as previously described (63); all primers used contained an additional universal sequence (forward primer, 5′-CCAGGGGAGCTCTTCCATG-3′; reverse primer, 5′-GCACGGGTGCTCTTCCACC-3′) that allowed for the reamplification of the fragments with a single primer pair. The PCRs were performed in 384-well microtiter plates with 0.4 U of Pfx polymerase (Invitrogen, Carlsbad, Calif.), 10 to 30 ng of the original amplified DNA fragment, 5 μmol of oligonucleotides, 50 μmol of deoxynucleoside triphosphates, and 1.5 mM MgCl2 in a 20-μl reaction volume. For PCR amplification of specific gene fragments from purified Synechocystis sp. strain PCC6803 genomic DNA (74), reactions were performed in 96-well microtiter plates using 2 U of Taq polymerase (MBI Fermentas, Amherst, N.Y.), 100 ng of genomic DNA, 5 μmol of oligonucleotides, 50 μmol of deoxynucleoside triphosphates, and 1.5 mM MgCl2 in a 100-μl reaction volume. Reactions were cycled 25 times at 94°C for 2 min, 92°C for 30 s, 55°C for 30 s, 72°C for 2 min, and, after the final cycle, 72°C for 5 min. Approximately 5% of the PCR products contained multiple DNA fragments; these fragments were reamplified either at different annealing temperatures or from electrophoretically resolved fragments that were purified from agarose gels. There were still 67 genes for which products did not amplify well; 70-mer oligonucleotides specific for these genes were included on the array. The PCR products, resolved on 1.2% agarose gels and sized with respect to molecular mass markers prepared by Life Technology (Rockville, Md.), were purified by MultiScreen PCR 384-well filter plates (Millipore, Bedford, Mass.), and the nucleic acid concentration was measured with the PicoGreen double-stranded DNA quantification reagents (Molecular Probes, Eugene, Ore.). Purified PCR products were resuspended in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) to a final concentration of 250 to 300 ng/μl. Eight microliters of each PCR product was transferred from 384-well microtiter plates to the printing plates, also in the 384-well microtiter format (Genetix Limited, Hampshire, United Kingdom). The PCR products were arrayed onto CMT-GAPS slides coated with γ amino propyl silane (Corning Incorporated, New York, N.Y.) by using an Omnigrid Microarrayer (GeneMachines, San Carlos, Calif.) with ChipMaker 2 pins (TeleChem International, Sunnyvale, Calif.). Four complete cDNA sets, each consisting of 3,707 array elements, were printed onto each slide. Printed slides were allowed to dry for 12 h at room temperature (25°C) at <50% relative humidity and then cross-linked by UV irradiation in the Stratalinker 1800 (Stratagene, La Jolla, Calif.) at a total power of 300 mJ. Just prior to prehybridization, arrays were baked at 65°C for 10 min. Following prehybridization for 1 h at 50°C in 3× SSC-0.1% sodium dodecyl sulfate (SDS)-0.1 mg of bovine serum albumin/ml, the arrays were rinsed by immersion in double-distilled H2O, transferred to isopropanol for 1 min, dried by centrifugation for 5 min in a SpeedVac Plus model SC210A (Savant, Holbrook, N.Y.), and then stored in a desiccator at room temperature until they were used.
Nucleic acid isolation.
RNA was isolated from 50-ml cultures of wild-type cells and the dspA mutant; cells were collected by centrifugation (5,000 × g for 5 min), and cell pellets were resuspended in 500 μl of NAES extraction buffer (50 mM sodium acetate [pH 5.1], 10 mM EDTA, 1% SDS). An equal volume of acidic phenol (equilibrated with NAES) was added to the suspension (approximately 1.5 ml) along with 100 mg of glass beads (0.1-μm average diameter; Sigma, St. Louis, Mo.) prior to agitation (two times for 30 s each) at the maximum speed (∼5,000 rpm) in a Mini-Bead Beater (Biospec Products, Bartlesville, Okla.). The beads and unbroken cells were pelleted by a brief centrifugation (5 to 10 s at full speed in an Eppendorf centrifuge 5415C), and the supernatant was subjected to two phenol-chloroform (1:1) and one chloroform extractions. Two volumes of ethanol were added to the aqueous phase of the final chloroform extraction, and the solution was incubated for 2 h at −20°C. Precipitated RNA was collected by centrifugation (10 min at full speed in an Eppendorf Microfuge), washed with 70% ethanol, resuspended in Tris-EDTA, and treated with 25 U of RNase-free DNase I (Amersham, Piscataway, N.J.) at room temperature for 30 min. The DNase I was removed by two phenol-chloroform extractions, and the RNA was precipitated with 2 volumes of ethanol.
Microarray hybridization and data analysis.
Hybridization to the microarray was performed according to the method described by Schena et al. (66), with minor modifications. First-strand cDNA was synthesized by incubating 25 μg of total RNA with mixed gene-specific reverse primers and 30 U of Superscript II RNase H-minus reverse transcriptase (Invitrogen). The reaction was performed at 42°C for 1 h in a volume of 40 μl. After digesting total RNA with RNase H, second-strand synthesis was accomplished by using the Klenow fragment of DNA polymerase (USB, Cleveland, Ohio) and random primers; Cy3- or Cy5-dUTP fluorescent nucleotides were incorporated into the second strand (Amersham) over a labeling period of 3 h at 37°C. Labeled DNA fragments were purified by using the QIAquick spin column (QIAGEN, Valencia, Calif.), and the cDNA derived from 6.25 μg of total RNA was resuspended in hybridization buffer (3× SSC, 0.3% SDS, 0.17 μg of tRNA/μl) to a final volume of 30 μl. The fluorescent cDNA was denatured at 95°C for 90 s, and its application to the array was done by capillary delivery under a glass lifter slip (Eric Scientific, Portsmouth, N.H.). Three spots each of 20 μl of 3× SSC were positioned at the edges of the slides to maintain humidity during hybridization. Slides were sealed in custom-built hybridization chambers, and hybridizations were done at 65°C for 16 h. Following hybridization, the arrays were washed for 2 min at room temperature in low-stringency wash buffer (2× SSC, 0.03% SDS), 2 min in moderate-stringency wash buffer (1× SSC), and 2 min in high-stringency wash solution (0.05× SSC); dried in a Speedvac Plus (Savant); and scanned with a GenePix Laser Scanner 4000 (Axon, Foster city, Calif.). The TIFF images generated by the GenePix Laser Scanner 4000 were imported into the GenePix Pro 3.0 program (Axon), and spot intensities from scanned slides were quantified. Grids were predefined and manually adjusted to ensure optimal spot recognition. Spot signals that were distorted by dust or local high backgrounds were flagged and not included in the analysis. After determining the relative ratios of the fluorescent signals from the two channels (532 and 635 nm), the data files (.gpr files) were imported into the Stanford Microarray Database (http://genome-www.stanford.edu/microarray/) (26, 70) and normalized by using an internal control, rps1b (for details, see the Stanford Microarray Database website). The primary data sets may be viewed and downloaded (see above); the experiment identification numbers are 38980, 38981, 38985 to 38988, 39071, 39080, 39085, 39086, 39563, 39578, and 39583 to 39590.
Only those spots with a minimum normalized net intensity for the median (channel 1 net median intensity or normalized channel 2 net median intensities) of >350 for both wild-type cells and the dspA mutant, in at least one of the two channels, were included in the analyses. Log (base 2) channel 1/channel 2 ratios after normalization were calculated. The significance of the differences in gene expression observed (at P = 0.05) was statistically evaluated by t tests using a df of 7 with an α of 2.8, representing P = 0.05. The degree of freedom (df) value was calculated from the 8 samples used for the data analysis; for each data point, two slides were analyzed, with four duplicates for each array element on each slide. Two independently isolated RNA samples were used for generating the cDNA that was hybridized to each of the slides.
Microarray experimental design.
The effect of glucose on gene expression in wild-type cells and the HL effects on both the wild-type and the dspA mutant of the Synechocystis sp. strain PCC6803 were examined. For measuring long-term acclimation of wild-type cells to glucose, total RNA was extracted from cells cultured in BG-11 medium or BG-11 medium supplemented with 10 mM glucose. The cells were grown for 2 weeks in LL but were diluted every 2 days to maintain their logarithmic growth. Total RNA isolated from wild-type cells grown in BG-11 medium in the absence of glucose was reverse transcribed, and the second-strand cDNA was labeled with Cy3-dUTP. Total RNA isolated from the cells grown in BG-11 medium supplemented with glucose was used for labeling cDNA with Cy5-dUTP. The Cy3- and Cy5-labeled cDNA samples were mixed and applied to a microarray slide. A “dye-swap” Cy-dye-dUTP labeling was performed with RNA isolated from a duplicate, independent experiment. A similar procedure was used for analyzing transcripts in wild-type cells and the dspA mutant grown in glucose-supplemented medium following transfer from LL (zero time point reference) to HL (for 30 min, 1 h, 3 h, and 6 h). We also examined differential transcript accumulation in the dspA mutant in LL compared to that of wild-type cells grown under the same conditions. Total RNA from the LL-grown dspA mutant was reverse transcribed, and the second-strand cDNA was labeled with Cy5-dUTP and mixed with Cy3-dUTP-labeled cDNA generated from LL-grown wild-type cells and applied to a microarray slide. The experiment was duplicated, but the dyes used to label the cDNAs were swapped; the cDNA generated from LL-grown dspA mutant was labeled with Cy3-dUTP, while the cDNA from LL-grown wild-type cells was labeled with Cy5-dUTP. The dspA mutant grew extremely slowly and was difficult to maintain in the absence of glucose.
Real-time quantitative RT-PCR.
To confirm the validity of the results generated from microarray analyses, we performed real-time reverse transcription (RT)-PCR for 8 specific transcripts and compared the results to the mean of the quantified array measurements for those same transcripts. To remove trace amounts of genomic DNA contamination from RNA samples used for the real-time RT-PCR, RNA (100 ng/μl) was treated with 2 U of RNase-free DNase at 37°C for 30 min (Ambion, Austin, Tex.), and the DNase was removed by DNase removal reagent (Ambion). Levels of specific transcripts in total mRNA isolated from HL-treated cells were quantified by real-time RT-PCR with the LightCycler (Roche Applied Science, Indianapolis, Ind.). Amplifications were performed by incubating 200 ng of DNA-free RNA with specific primer pairs, as given in Table 1, and LightCycler RNA Master SYBR Green I according to the protocols of the manufacturer (Roche Applied Science). Amplifications performed according to the protocols are as follows: 1 cycle of RT reaction at 61°C for 20 min, 1 cycle at 95°C for 2 min, and 45 cycles at 95°C for 5 s, 55°C for 5 s, and 72°C for 13 s. A “no-RT” PCR control reaction was performed by incubating an equal amount of DNA-free RNA samples with a mixture of specific primer pairs and LightCycler DNA Master SYBR Green I at a final volume of 20 μl (Roche Applied Science). Amplification was done for 1 cycle at 95°C for 30 s followed by 45 cycles of 95°C for 5 s, 55°C for 5 s, and 72°C for 10 s.
TABLE 1.
Primer pairs used for quantitative real-time RT-PCRa
| Gene | Forward primer | Reverse primer |
|---|---|---|
| cpcB (sll1577) | CCGGTGGAAACGCCTACA | GCAGCAGCGGCGCGGTCGA |
| psaA (slr1834) | CGTCATCAACTCCTACGGTTC | GTAACAATACCTCCGAGGAGAT |
| psbAII (slr1311) | ACAGCCGTTCCTTGCACTTCTT | TTAACCGTTGACAGCAGGAGCG |
| pstS (sll0680) | ACTGTTGGTCTGGCAGCCTG | CCACGGACTGATAGTTAACTTC |
| groEL-1 (slr2076) | GAGTTAACCGGTGCCCTGATTG | AAGTCACCACCGGGGGCACCA |
| groEL-2 (sll0416) | GCCGACATCATCGCCAAAGCC | CCATGCCACCCATATCGGGCA |
| ccmK (sll1029) | TGACCGCTGGCATTGAAA | GGCGGATGATGGAAGGATTA |
| rps1B (slr1984) | TCCAACAGTGCAGCCTTTTC | GCAATCCTTCCACATCGC |
Forward and reverse primers specific for transcripts encoding CpcB, PsaA, PsbAII, PstS, GroEL-1, GroEL-2, CcmK, and the control polypeptide Rps1B that were used for quantitative real-time RT-PCR are shown.
A number of standards were developed as controls for these experiments. The rps1b (slr1984) and rnpB (slr0249) transcripts were used as external standards since the levels of these transcripts did not change in either wild-type cells or the dspA mutant following HL exposure (under all conditions that we used). PCR efficiency differences between target and reference transcripts were corrected by generating fit coefficients. A dilution series was used for constructing standard curves that enabled us to determine the fit coefficients for PCR amplification efficiency of target relative to the reference (rps1b and rnpB) transcripts. Quantification of the relative change (n-fold) in mRNA levels was calculated as described by Technical Note no. LC13/2001 (Roche Applied Science, 2001). Each quantitative RT-PCR experiment was performed with at least two replicates.
RESULTS
(i) Effect of glucose on expression in wild-type cells and the dspA mutant.
In LL, the dspA mutant exhibited a markedly reduced growth rate (doubling time of ∼40 h) under photoautotrophic conditions relative to wild-type Synechocystis sp. strain PCC6803 and was difficult to maintain. This slow growth rate reflected very low light-dependent O2 evolution (42), demonstrating that a strain lacking DspA is strongly affected in both growth and photosynthetic performance. In contrast, in the presence of glucose, the mutant grew, with a doubling time of 8 h, compared to a 6-h doubling time for wild-type cells (42). We examined how glucose influenced global transcript accumulation in wild-type Synechocystis sp. strain PCC6803. Cells were grown long term (2 weeks with the dilution of cultures to maintain the cells in logarithmic growth phase) in LL in either BG-11 or BG-11 supplemented with 10 mM glucose. Total RNA was purified (5) and reverse transcribed, second-strand cDNAs were labeled with Cy3- or Cy5-dUTP, and equal amounts of the two labeled cDNA samples were hybridized to the arrays (see Materials and Methods). The use of second-strand synthesis for probe preparation, compared to using just first-strand synthesis, increased the array signals generated without significantly changing the relative intensities of the signals.
Of the 3,164 distinct Synechocystis sp. strain PCC6803 genes represented on the array, ∼95% showed no statistically significant difference in transcript levels between wild-type cells grown photoautotrophically and those grown mixotrophically (supplemented with 10 mM glucose). Statistically significant changes in the levels of transcripts for 5% of the genes were relatively small; all changes were less than twofold (data not shown). These results demonstrate that during long-term growth, the pattern of transcript accumulation in wild-type cells is not dramatically influenced by the inclusion of glucose in the growth medium. Therefore, we compared the effects of HL on gene expression in wild-type cells and the dspA mutant during growth in glucose-supplemented medium.
(ii) Gene expression in wild-type cells and the dspA mutant.
Expression from 3,164 Synechocystis sp. strain PCC6803 genes was measured for wild-type cells and the dspA mutant grown in LL and then transferred to HL for 0.5, 1, 3, and 6 h. A comparison of transcript abundance in wild-type cells and the dspA mutant grown in LL demonstrated that the levels of 84 transcripts were altered by more than twofold in the mutant relative to wild-type cells (data not shown). Nearly all of these transcripts showed increased abundance in the dspA mutant relative to wild-type cells during LL growth, with the exceptions of chlL, chlN, hofG, cpcC, psaA, psaB, cpcG, sll0177, and slr0147, for which the transcripts were lower in the mutant strain.
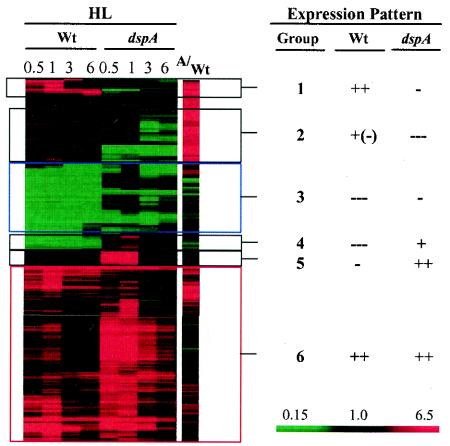
We identified a set of 143 genes for which the transcript abundance changed by at least threefold in either wild-type cells or the dspA mutant following the transfer of cells to HL (for at least one of the time points following transfer) or that were already elevated in the dspA mutant compared to wild-type cells during LL growth. A hierarchical clustering analysis was used to group the genes on the basis of similarities in their expression patterns and to allow for easy viewing of the changes that we observed; six different groups were established, as shown in Fig. 1. For each gene in the different expression categories, the change in transcript levels over the 6-h time course following the transfer of cells to HL (for both wild-type and dspA mutant cells), the difference in the initial expression levels for wild-type cells and the dspA mutant under LL conditions (0 h), and the statistical significance of the data are given in Table 2. Table 3 shows trends in expression within specific categories of genes critical for photosynthesis and acclimation of cells to phosphorus deprivation. In this table, all changes of twofold or greater are shown.
FIG. 1.
Gene expression profiles of wild-type Synechocystis sp. strain PCC6803 and the dspA mutant following HL exposure presented as hierarchical clusters. Global expression patterns of 143 genes whose transcript levels changed by at least threefold in either wild-type (Wt) cells or the dspA mutant for at least one time point following the transfer of cells to HL. The time following the transfer of wild-type cells (left) and the dspA mutant (right) are 0.5, 1, 3, and 6 h. Also presented is the ratio of transcripts in dspA relative to those of wild-type cells (A/Wt) in LL-grown cells. Expression patterns with respect to wild-type cells and the dspA mutant were divided into groups 1 to 6, and the trends within each of the groups are presented in the right columns for both wild-type cells and the dspA mutant. Increases in transcript abundance are indicated in red (+), and decreases are indicated in green (−). The scale, extending from 0.15- to 6-fold, is shown on the right.
TABLE 2.
Influence of HL on levels of transcripts in wild-type cells and the dspA mutanta
| Geneb | Function | Statistical significance of transcript levels after HL treatment (h)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wt
|
dspA
|
dspA/Wt
|
||||||||
| 0.5 | 1 | 3 | 6 | 0.5 | 1 | 3 | 6 | 0 | ||
| Group 1 | ||||||||||
| slr0228 (ftsH) | Protease | 3.69 | 3.81 | 1.36 | 0.81 | −1.40 | 0.88 | −1.53 | −2.48 | 2.61 |
| slr1604 (ftsH) | Cell division | 2.25 | 2.50 | 1.09 | 0.69 | −1.91 | −1.49 | −1.63 | −2.15 | 3.24 |
| slr0927 (psbD2) | PS II reaction center D2 protein | 1.76 | 2.79 | 1.52 | 1.02 | −2.47 | −1.79 | −1.87 | −2.39 | 3.43 |
| sll0680 (pstS) | Phosphate-binding periplasmic protein precursor | 2.66 | 4.17 | 2.32 | 2.58 | −3.37 | −3.19 | 1.21 | 0.77 | 17.48 |
| sll0681 (pstC) | Phosphate transport system permease protein | 1.77 | 2.21 | 1.24 | 1.14 | −5.48 | −4.60 | −1.56 | −2.45 | 10.35 |
| sll1515 | Unknown | 1.94 | 2.11 | 2.32 | 3.46 | −2.24 | −1.50 | −1.66 | −2.07 | 5.23 |
| ssr0692 | Unknown | 1.23 | 1.34 | 1.41 | 2.47 | −1.72 | −1.32 | −1.41 | −1.66 | 3.43 |
| Group 2 | ||||||||||
| slr1247 (pstS) | Phosphate-binding periplasmic protein | 0.92 | 1.00 | 0.95 | 0.95 | −7.30 | −6.98 | −4.70 | −5.76 | 14.79 |
| sll0683 (pstB) | Phosphate transport ATP-binding protein | 1.46 | 1.63 | 1.02 | 0.87 | −3.39 | −2.79 | −1.68 | −2.16 | 5.91 |
| sll0684 (pstB) | Phosphate transport ATP-binding protein | 1.23 | 1.40 | 0.96 | −1.78 | −3.15 | −2.70 | −1.72 | −2.13 | 4.71 |
| slr1248 (pstC) | Phosphate transport system permease protein | 0.88 | 0.90 | 0.89 | −1.42 | −4.44 | −4.38 | −4.41 | −4.78 | 4.61 |
| slr1249 (pstA) | Phosphate transport system permease protein | 0.85 | 0.85 | 0.87 | −1.36 | −2.43 | −2.27 | −2.49 | −3.14 | 2.48 |
| sll0654 | Alkaline phosphatase | 0.83 | 0.89 | 0.88 | 0.77 | −6.82 | −5.82 | −5.60 | −4.90 | 9.19 |
| slr0897 | Probable endoglucanase | −1.25 | 0.77 | 0.87 | −1.56 | −1.76 | −1.67 | −2.04 | −2.76 | 11.98 |
| sll1349 (cbbZp) | Phosphoglycolate phosphatase | 0.98 | 1.15 | 1.04 | 0.96 | −1.86 | −1.55 | −1.51 | −1.89 | 4.99 |
| sll1535 (rfbP) | Putative sugar transferase | 1.18 | 1.29 | 0.90 | −1.32 | −1.74 | 0.82 | −1.83 | −2.82 | 3.23 |
| slr1204 (htrA) | Protease | −1.33 | 0.98 | −1.44 | 0.87 | −2.13 | −2.00 | −2.83 | −2.57 | 4.70 |
| sll0656 | Unknown | −1.30 | 0.83 | 0.80 | −1.41 | −3.71 | −3.14 | −4.05 | −4.38 | 6.33 |
| slr2123 | Similar to d-3-phosphoglycerate dehydrogenase | −1.32 | 0.81 | 0.80 | −1.52 | −1.69 | 0.84 | −1.75 | −2.22 | 3.24 |
| slr2122 | Unknown | −1.28 | 0.90 | 0.91 | −1.34 | −1.42 | 0.86 | −1.71 | −2.11 | 3.21 |
| ssr2912 | Unknown | −1.22 | 0.89 | 0.87 | 0.81 | −1.50 | −1.32 | −1.66 | −2.50 | 3.38 |
| slr1116 | Unknown | −1.29 | −1.21 | 0.83 | −1.19 | −1.56 | −1.36 | −1.27 | −1.58 | 3.32 |
| slr0238 | Unknown | 0.91 | 0.93 | −1.23 | 0.81 | 0.79 | 0.82 | −1.32 | −1.91 | 3.74 |
| ssl3769 | Unknown | 0.99 | 1.30 | 1.08 | 1.01 | −2.06 | −1.49 | −3.06 | −3.36 | 6.87 |
| Group 3 | ||||||||||
| sll1471 (cpcG) | Phycobilisome rod-core linker polypeptide | −10.05 | −8.42 | −7.85 | −11.58 | −3.23 | −3.14 | −2.61 | −3.27 | −2.97 |
| ssl3093 (cpcD) | Phycobilisome small rod linker polypeptide | −8.26 | −6.77 | −5.41 | −5.13 | −2.48 | −1.69 | −2.06 | −2.26 | −1.58 |
| sll1580 (cpcC) | Phycobilisome rod linker polypeptide | −14.16 | −12.11 | −16.11 | −10.14 | −5.19 | −3.03 | −3.99 | −3.34 | −2.00 |
| sll1579 (cpcC) | Phycobilisome rod linker polypeptide | −17.20 | −14.42 | −12.19 | −10.64 | −5.87 | −3.45 | −4.31 | −3.79 | −1.93 |
| sll1578 (cpcA) | Phycocyanin alpha subunit | −14.20 | −15.06 | −22.71 | −9.60 | −8.55 | −4.06 | −4.37 | −3.14 | −1.42 |
| sll1577 (cpcB) | Phycocyanin beta subunit | −13.59 | −16.82 | −24.57 | −7.86 | −9.60 | −4.27 | −4.07 | −2.85 | −1.33 |
| ssr3383 | Phycobilisome small core linker polypeptide | −4.48 | −3.41 | −2.32 | −1.61 | −2.76 | −2.43 | −1.81 | −1.78 | 0.99 |
| slr1986 | Allophycocyanin beta subunit | −7.50 | −5.09 | −2.83 | −1.57 | −5.03 | −2.94 | −1.93 | −1.56 | 0.89 |
| slr2067 | Allophycocyanin alpha subunit | −4.36 | −3.21 | −2.60 | −1.40 | −2.85 | −2.05 | −1.36 | 0.81 | 0.98 |
| sll0819 (psaF) | PS I reaction center subunit III precursor | −5.20 | −4.77 | −4.81 | −3.91 | −1.97 | 0.68 | −1.68 | −1.55 | −1.35 |
| sml0008 (psaJ) | PS I subunit IX | −3.45 | −3.52 | −3.55 | −2.95 | −1.50 | 0.80 | −1.50 | −1.51 | 0.83 |
| slr1835 (psaB) | P700 apoprotein subunit lb | −3.83 | −5.46 | −5.65 | −7.19 | −2.09 | 0.86 | −1.86 | −1.93 | −2.53 |
| slr1834 (psaA) | P700 apoprotein subunit Ia | −4.22 | −5.62 | −5.97 | −6.98 | −2.33 | 0.91 | −1.76 | −2.33 | −2.27 |
| slr1655 (psaL) | PS I subunit XI | −5.13 | −5.02 | −6.15 | −4.18 | −2.64 | −1.55 | −1.79 | −1.74 | −1.59 |
| slr0533 | Two-component sensor histidine kinase (hik10) | −3.59 | −3.41 | −2.96 | −3.69 | 0.89 | 1.09 | −2.13 | −2.33 | 0.94 |
| slr0083 (crhR) | RNA helicase light | −2.53 | −3.05 | −3.87 | −5.29 | −3.33 | −1.50 | −3.77 | −4.35 | 1.94 |
| sll1451 (nrtB) | Nitrate/nitrite transport system permease protein | 1.09 | 1.24 | −1.74 | −3.76 | 0.88 | −1.67 | −1.47 | −2.54 | 2.07 |
| slr0750 (chlN) | Light-independent protochlorophyllide reductase subunit | −7.28 | −7.68 | −5.30 | −7.64 | 0.87 | 0.86 | −1.35 | −1.64 | −3.51 |
| slr0749 (chlL) | Light-independent protochlorophyllide reductase subunit | −6.71 | −5.98 | −5.15 | −6.86 | 0.79 | 0.85 | −1.29 | −1.49 | −3.14 |
| slr1634 | Unknown | −10.12 | −10.40 | −9.02 | −14.25 | −7.72 | −5.86 | −8.50 | −9.80 | 0.99 |
| sll1184 | Heme oxygenase | −5.92 | −4.73 | −3.10 | −1.48 | −2.79 | −2.07 | −1.66 | −1.44 | 0.85 |
| sll1330 | Two-component system response regulator OmpR subfamily | −5.42 | −4.87 | −4.83 | −4.78 | −2.43 | −1.94 | −2.72 | −2.33 | 0.86 |
| sll0756 | Unknown | −4.03 | −3.14 | −3.13 | −4.01 | −1.25 | 0.94 | −1.38 | −1.86 | −1.46 |
| sll0022 | Unknown | −3.99 | −2.92 | −3.50 | −3.53 | −1.24 | 1.32 | −2.20 | −2.82 | 1.49 |
| sll1783 | Unknown | −3.83 | −3.55 | −2.16 | −2.26 | −2.35 | −1.97 | −1.96 | −1.58 | 1.17 |
| sll1306 | Periplasmic protein, function unknown | −3.67 | −3.52 | −3.00 | −3.51 | −2.01 | −1.65 | −1.94 | −1.98 | 1.00 |
| sll1305 | Probable hydrolase | −3.30 | −3.43 | −2.72 | −3.27 | −1.95 | −1.69 | −2.09 | −2.08 | 1.05 |
| slr1535 | Unknown | −3.01 | −2.96 | −2.72 | −3.17 | −1.46 | −1.37 | −1.41 | −1.68 | 0.99 |
| slr0082 | Unknown | −2.98 | −3.04 | −3.66 | −4.57 | −2.82 | −1.37 | −3.08 | −3.66 | 2.03 |
| sll1304 | Unknown | −2.84 | −2.65 | −2.41 | −2.77 | −1.74 | −1.52 | −1.64 | −1.66 | 1.19 |
| sll1784 | Periplasmic protein, function unknown | −2.75 | −2.42 | −2.16 | −2.03 | −1.69 | −1.43 | −1.31 | −1.38 | 1.33 |
| sll1307 | Periplasmic protein, function unknown | −2.73 | −2.59 | −2.24 | −2.60 | −1.56 | 0.83 | −1.46 | −1.66 | 1.11 |
| slr0374 | Unknown | −1.62 | −2.48 | −3.36 | −4.10 | −2.42 | 0.90 | −3.74 | −4.84 | 1.43 |
| slr1974 | GTP binding protein | −1.60 | −1.59 | −1.69 | −2.15 | −1.81 | −1.45 | −1.98 | −2.80 | 2.88 |
| slr1403 | Unknown | −1.36 | −1.35 | −1.30 | −1.47 | 0.75 | 0.90 | −1.47 | −2.17 | 5.14 |
| slr0287 | Unknown | −1.33 | −2.09 | −2.88 | −3.93 | −2.44 | 0.89 | −3.83 | −5.86 | 1.08 |
| Group 4 | ||||||||||
| sll1626 (lexA) | LexA repressor | −5.42 | −4.30 | −2.55 | 0.99 | 0.89 | 1.16 | 1.64 | 1.68 | 1.09 |
| slr0737 (psaD) | PS I subunit II | −5.30 | −5.07 | −3.96 | −3.02 | 1.03 | 1.12 | 0.81 | 0.69 | 1.06 |
| sll1695 (hofG) | Pilin polypeptide PilA2 | −3.14 | −3.38 | −3.86 | −4.11 | 1.57 | 1.86 | 1.30 | 1.14 | −1.87 |
| sll1694 (hofG) | Pilin polypeptide PilA1 | −2.56 | −3.43 | −5.47 | −5.14 | 1.37 | 1.65 | 1.13 | 0.90 | −2.03 |
| sll0517 (rbpA) | Putative RNA binding protein | −3.03 | −3.72 | −2.64 | −2.14 | 0.86 | 1.74 | 0.84 | 0.70 | 1.43 |
| sll1696 | Unknown | −2.74 | −3.12 | −3.17 | −3.83 | 1.52 | 2.01 | 1.44 | 1.36 | −1.66 |
| sll0177 | Unknown | −3.68 | −3.34 | −3.88 | −4.29 | 0.99 | 1.13 | 0.89 | 0.75 | −2.11 |
| Group 5 | ||||||||||
| sll1029 (ccmK) | Carbon dioxide concentrating mechanism protein | −1.31 | −1.44 | −1.67 | −1.72 | 3.93 | 5.97 | 1.00 | −1.73 | 1.34 |
| sll1028 (ccmK) | Carbon dioxide concentrating mechanism protein | −1.36 | −1.52 | 0.70 | −1.61 | 3.26 | 4.56 | 0.89 | −1.72 | 1.29 |
| sll1031 (ccmM) | Carbon dioxide concentrating mechanism protein | −1.17 | −1.43 | −1.45 | −1.83 | 2.77 | 4.45 | −1.32 | −2.03 | 1.19 |
| sll1032 (ccmN) | Carbon dioxide concentrating mechanism protein | −1.19 | −1.38 | −1.48 | −2.00 | 2.76 | 4.51 | 0.88 | −1.78 | 1.47 |
| sll1030 (ccmL) | Carbon dioxide concentrating mechanism protein | −1.34 | −1.41 | −1.41 | −1.50 | 2.87 | 4.24 | 1.02 | −1.58 | 1.43 |
| slr0009 (rbcL) | Ribulose bisphosphate carboxylase large subunit | 0.94 | 0.73 | −1.47 | 0.91 | 2.12 | 3.92 | 0.93 | −1.55 | 1.19 |
| slr0012 (rbcS) | Ribulose bisphosphate carboxylase small subunit | −1.33 | −1.79 | 0.70 | −1.04 | 2.82 | 3.60 | 0.95 | −1.59 | 1.09 |
| sll1853 | Unknown | 0.94 | 0.98 | 0.88 | −1.41 | 2.51 | 3.32 | 0.95 | −1.37 | 1.22 |
| slr0006 | Unknown | −1.72 | −1.71 | −1.69 | −1.59 | 4.44 | 5.78 | 1.19 | 0.79 | 1.02 |
| Group 6 | ||||||||||
| ssr2595 (hliB) | HL-inducible polypeptide (scpD) | 8.85 | 10.13 | 3.47 | 2.18 | 2.17 | 3.69 | 2.60 | 1.43 | 1.86 |
| ssl1633 (hliC) | HL-inducible polypeptide (scpB) | 5.28 | 7.33 | 3.92 | 4.25 | 5.02 | 4.30 | 4.61 | 1.79 | 6.85 |
| ssl2542 (hliA) | HL-inducible polypeptide (scpC) | 1.16 | 1.73 | 1.49 | 1.55 | 3.10 | 2.81 | 2.90 | 1.65 | 1.92 |
| sll0416 (groEL-2) | 60-kDa chaperonin 2 | 4.89 | 6.04 | 2.21 | 1.89 | 5.59 | 3.93 | 3.10 | 1.38 | 1.51 |
| slr2076 (groEL) | 60-kDa chaperonin | 4.45 | 7.38 | 2.66 | 2.40 | 7.65 | 5.28 | 5.93 | 2.21 | 1.62 |
| slr2075 (groES) | 10-kDa chaperonin | 3.43 | 6.01 | 2.46 | 2.52 | 5.66 | 3.74 | 4.74 | 1.90 | 1.52 |
| sll0430 (htpG) | Heat shock protein 90 | 3.13 | 2.24 | 0.98 | 0.96 | 2.66 | 3.35 | 1.89 | 1.03 | 1.66 |
| sll1514 (hsp17) | 16.6-kDa small heat shock protein | 1.81 | 1.87 | 1.12 | 1.72 | 1.32 | 1.30 | 1.12 | 1.34 | 4.51 |
| slr1291 (ndhD2) | NADH dehydrogenase subunit 4 | 3.36 | 3.80 | 1.89 | 1.78 | 1.00 | 2.37 | −1.35 | −1.67 | 1.82 |
| sll1732 (ndhF) | NADH dehydrogenase subunit 5 | 1.05 | 1.13 | 1.51 | −1.36 | 1.47 | 3.14 | 1.12 | −1.41 | 1.40 |
| sll1733 (ndhD3) | NADH dehydrogenase subunit 4 | 1.03 | 1.04 | 1.32 | 0.80 | 1.71 | 3.16 | 1.08 | −1.42 | 1.45 |
| sll1867 (psbA3) | PS II D1 protein | 2.94 | 4.31 | 2.56 | 2.48 | 1.38 | 1.64 | 1.15 | 0.70 | 6.30 |
| slr1311 (psbA2) | PS II D1 protein | 2.81 | 4.33 | 2.42 | 2.46 | 1.47 | 1.59 | 1.20 | 0.68 | 5.80 |
| slr1908 | Probable porin, major outer membrane protein | 2.86 | 4.00 | 1.86 | 2.05 | 1.03 | 1.38 | 1.25 | −0.53 | 2.74 |
| slr1746 (murI) | Glutamate racemase | 1.61 | 1.93 | 1.72 | 1.85 | 1.06 | 1.78 | 0.83 | −1.83 | 4.10 |
| ssl0020 (petF) | Ferredoxin I | 2.58 | 2.99 | 1.72 | 1.76 | 1.77 | 1.39 | 1.54 | −1.30 | 1.32 |
| sll1363 (ilyC) | Ketol-acid reductoisomerase | 1.83 | 2.07 | 1.74 | 1.52 | 3.07 | 2.53 | 3.76 | 1.82 | 0.98 |
| slr0447 (amiC) | ABC-type urea transport system substrate-binding protein | 1.46 | 2.09 | 1.18 | 1.09 | 3.33 | 2.58 | 2.51 | 1.12 | −1.90 |
| sll0330 (fabG) | Sepiapterine reductase | 1.44 | 1.66 | 1.27 | 1.06 | 1.12 | 1.24 | 0.84 | 0.94 | 5.50 |
| slr1469 (rnpA) | Protein subunit of ribonuclease P | 1.38 | 1.50 | 1.09 | 0.97 | 3.06 | 2.71 | 2.76 | 1.39 | 1.14 |
| slr0394 (pgk) | Phosphoglycerate kinase | 1.35 | 1.42 | 1.16 | 1.01 | 1.23 | 1.65 | 1.32 | −1.45 | 9.08 |
| slr1251 (cvp) | Peptidyl-prolyl cis-trans isomerase | 1.18 | 1.43 | 1.89 | 2.22 | 1.99 | 1.59 | 3.48 | 2.03 | 0.87 |
| sll1799 (rpl3) | 50S ribosomal protein L3 | 2.31 | 2.41 | 1.51 | 1.19 | 5.76 | 6.25 | 4.58 | 2.04 | −1.60 |
| sll1800 (rpl4) | 50S ribosomal protein L4 | 2.13 | 2.07 | 1.34 | 1.08 | 5.72 | 6.18 | 3.99 | 2.06 | −1.31 |
| sll1801 (rpl23) | 50S ribosomal protein L23 | 2.13 | 2.08 | 1.33 | 1.05 | 6.51 | 6.77 | 4.35 | 1.94 | −1.33 |
| sll1802 (rpl2) | 50S ribosomal protein L2 | 1.99 | 2.07 | 1.12 | 0.87 | 5.03 | 6.20 | 3.44 | 1.71 | −1.40 |
| ssl3432 (rps19) | 30S ribosomal protein S19 | 1.90 | 1.90 | 1.05 | 0.86 | 6.33 | 6.48 | 3.94 | 1.89 | −1.38 |
| sll1804 (rps3) | 30S ribosomal protein S3 | 1.85 | 1.79 | 1.02 | 1.46 | 4.51 | 4.95 | 3.12 | 1.52 | −1.73 |
| sll1803 (rpl22) | 50S ribosomal protein L22 | 1.82 | 1.87 | 1.13 | 0.83 | 5.09 | 5.42 | 3.44 | 1.65 | −1.34 |
| sll1805 (rpl16) | 50S ribosomal protein L16 | 1.73 | 1.78 | 1.05 | −1.34 | 4.54 | 5.37 | 3.06 | 1.57 | −1.57 |
| sll1806 (rpl14) | 50S ribosomal protein L14 | 1.57 | 1.55 | 0.97 | −1.40 | 5.37 | 5.68 | 3.06 | 1.65 | −1.63 |
| ssl3436 (rpl29) | 50S ribosomal protein L29 | 1.57 | 1.58 | 1.06 | −1.19 | 5.14 | 5.70 | 3.18 | 1.55 | −1.64 |
| sll1808 (rpl5) | 50S ribosomal protein L5 | 1.52 | 1.62 | 0.95 | −1.40 | 4.63 | 5.60 | 2.59 | 1.55 | −1.68 |
| sll1807 (rpl24) | 50S ribosomal protein L24 | 1.52 | 1.53 | 0.96 | −1.37 | 5.35 | 6.11 | 3.37 | 1.62 | −1.66 |
| ssl3437 (rps17) | 30S ribosomal protein S17 | 1.50 | 1.57 | 1.10 | −1.27 | 4.73 | 5.29 | 2.85 | 1.61 | −1.59 |
| sll1260 (rps2) | 30S ribosomal protein S2 | 1.45 | 1.65 | 1.07 | 0.99 | 3.36 | 3.19 | 3.18 | 1.71 | −1.39 |
| sll1816 (rps13) | 30S ribosomal protein S13 | 1.40 | 1.49 | 0.96 | −1.30 | 3.05 | 2.74 | 2.93 | 1.43 | −1.42 |
| sll1744 (rpl1) | 50S ribosomal protein L1 | 1.38 | 1.56 | 1.00 | 0.93 | 3.85 | 3.81 | 2.94 | 1.58 | −1.31 |
| sll1812 (rps5) | 30S ribosomal protein S5 | 1.34 | 1.34 | 0.89 | −1.65 | 4.26 | 4.65 | 3.09 | 1.58 | −1.83 |
| slr0628 (rps14) | 30S ribosomal protein S14 | 1.34 | 1.78 | 1.59 | 1.85 | 2.39 | 2.09 | 3.31 | 2.02 | 1.25 |
| sll1809 (rps8) | 30S ribosomal protein S8 | 1.34 | 1.44 | 0.84 | −1.49 | 3.97 | 4.71 | 2.94 | 1.43 | −1.41 |
| sll1810 (rpl6) | 50S ribosomal protein L6 | 1.33 | 1.41 | 0.86 | −1.69 | 4.16 | 4.62 | 2.70 | 1.36 | −1.65 |
| slr0469 (rps4) | 30S ribosomal protein S4 | 1.33 | 1.54 | 1.09 | 1.15 | 3.60 | 3.05 | 2.96 | 1.52 | 0.94 |
| sll1745 (rpl10) | 50S ribosomal protein L10 | 1.28 | 1.53 | 1.05 | 0.97 | 3.21 | 3.64 | 2.54 | 1.41 | −1.38 |
| sll1811 (rpl18) | 50S ribosomal protein L18 | 1.27 | 1.36 | 0.85 | −1.57 | 4.18 | 4.82 | 2.74 | 1.55 | −1.70 |
| sll1813 (rpl15) | 50S ribosomal protein L15 | 1.24 | 1.40 | 0.95 | −1.46 | 2.96 | 3.53 | 2.25 | 1.39 | −1.56 |
| sll1743 (rpl11) | 50S ribosomal protein L11 | 1.23 | 1.46 | 0.99 | 0.92 | 3.76 | 3.38 | 2.94 | 1.49 | −1.33 |
| sll1746 (rpl12) | 50S ribosomal protein L12 | 1.22 | 1.56 | 1.04 | 1.15 | 3.31 | 3.51 | 2.54 | 1.43 | −1.31 |
| ssr2016 | Unknown | 12.06 | 16.02 | 5.58 | 2.41 | 7.90 | 10.08 | 5.37 | 2.14 | 1.67 |
| sll1483 | Similar to transforming growth factor-induced protein | 5.59 | 8.75 | 7.00 | 9.22 | 4.14 | 2.48 | 5.39 | 3.99 | 9.00 |
| slr1544 | Unknown | 4.54 | 4.70 | 2.26 | 1.45 | 1.48 | 2.27 | 1.60 | 0.91 | 1.48 |
| sll1911 | Unknown | 4.46 | 6.09 | 3.85 | 3.66 | 1.45 | 1.19 | 1.81 | 1.03 | 3.04 |
| slr1963 | Water-soluble carotenoid protein | 3.92 | 3.61 | 1.62 | 1.14 | 3.22 | 2.55 | 2.55 | 1.33 | 1.89 |
| sll0788 | Unknown | 3.26 | 3.44 | 1.49 | 1.39 | 1.32 | 1.91 | 0.87 | 0.74 | 2.56 |
| slr0075 (vcf16) | ABC transporter ATP-binding protein | 3.02 | 3.03 | 1.62 | −1.53 | 1.64 | 2.69 | 2.33 | −1.54 | 0.97 |
| slr1992 (gpx2) | Glutathione peroxidase-like NADPH peroxidase | 2.96 | 3.58 | 2.73 | 2.54 | 1.36 | 1.22 | 1.78 | 1.01 | 3.33 |
| slr0074 (ycf24) | ABC transporter subunit | 2.90 | 3.01 | 1.61 | −1.47 | 1.25 | 2.32 | 1.92 | −1.67 | 0.98 |
| sll0661 (ycf35) | Unknown | 2.51 | 3.35 | 2.55 | 2.12 | 1.27 | 1.29 | 1.32 | 0.97 | 1.76 |
| sll0185 | Unknown | 2.44 | 4.00 | 1.64 | 1.28 | 2.07 | 3.48 | 1.47 | 0.99 | 1.81 |
| slr1982 | Two-component response regulator CheY subfamily | 2.31 | 3.07 | 2.04 | 2.13 | 1.39 | 1.60 | 1.13 | 0.75 | 4.78 |
| sll0982 | Unknown | 2.00 | 3.92 | 2.43 | 1.56 | 1.22 | 1.11 | 1.07 | 0.95 | 2.11 |
| sll0905 | Unknown | 1.71 | 2.07 | 1.34 | 1.58 | 0.99 | 1.22 | 0.85 | −1.56 | 3.75 |
| slr1512 (sbtA) | Sodium-dependent bicarbonate transporter | 1.24 | 1.29 | 1.51 | 1.05 | 3.02 | 7.68 | 1.20 | −1.55 | 2.05 |
| slr1513 | Unknown | 1.19 | 1.24 | 1.38 | 1.23 | 2.47 | 3.56 | 1.51 | 0.97 | 1.97 |
| sll1219 | Unknown | 1.19 | 1.19 | 1.06 | 1.04 | 1.91 | 2.93 | 1.37 | −1.36 | 3.50 |
| ssr1853 | Unknown | 1.15 | 1.49 | 1.83 | 2.53 | 4.02 | 2.17 | 4.83 | 3.16 | 1.83 |
| slr0581 | Unknown | 1.11 | 1.31 | 1.21 | 1.91 | 1.51 | 1.34 | 1.56 | 1.41 | 3.25 |
| slr0821 | Unknown | 1.07 | 1.58 | 1.69 | 1.70 | 2.93 | 1.80 | 3.41 | 1.74 | 0.92 |
| sll0219 | Flavoprotein | 0.94 | 0.97 | 0.88 | 0.77 | 1.20 | 3.02 | 0.91 | −1.30 | 1.82 |
Transcript levels of 143 genes were changed by at least threefold in either wild-type (Wt) cells or the dspA mutant in at least one of the time points following the transfer of cells to HL and the ratio of dspA to the wild type (dspA/Wt) at time zero are presented. Expression patterns with respect to both wild-type cells and the dspA mutants were divided into six groups based on similarity of hierarchical clustering analyses. The statistical significance of the expression patterns are indicated for each time point and for the dspA/Wt time zero ratio; boldface type indicates an increase in the transcript level, underlining indicates a decrease in the transcript level, and plain type indicates no significant change in the transcript level. The significance was computed by t tests (see Materials and Methods).
For group 1, transcripts in wild-type cells increased while dspA transcripts declined; group 2, transcripts changed slightly in wild-type cells and declined in the dspA mutant; group 3, levels of transcripts in both wild-type cells and the dspA mutant strongly declined; group 4, levels of transcript markedly declined in wild-type cells but increased in dspA; group 5, transcripts declined slightly in wild-type cells but increased in the dspA mutant; group 6, transcripts increased in both wild-type cells and dspA.
TABLE 3.
Influence of light on levels of transcripts encoding phycobilisomes, PS I, PS II, and polypeptides involved in HL acclimation and phosphate acquisition in wild-type cells and the dspA mutanta
| Gene ID | Function | Statistical significance of transcript levels after HL treatment (h)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wt
|
dspA
|
dspA/Wt
|
||||||||
| 0.5 | 1 | 3 | 6 | 0.5 | 1 | 3 | 6 | 0 | ||
| Phycobilisome | ||||||||||
| slr2067 (apcA) | Allophycocyanin a chain | −4.36 | −3.21 | −2.60 | −1.40 | −2.85 | −2.05 | −1.36 | 0.81 | 0.98 |
| slr1986 (apcB) | Allophycocyanin b chain | −7.50 | −5.09 | −2.83 | 0.64 | −5.03 | −2.94 | −1.93 | −1.56 | 0.89 |
| ssr3383 (apcC) | Phycobilisome LC linker polypeptide | −4.48 | −3.41 | −2.32 | 0.62 | −2.76 | −2.43 | −1.81 | −1.78 | 0.99 |
| sll1471 (cpcG) | Phycobilisome rod-core linker polypeptide | −10.05 | −8.42 | −7.85 | −11.58 | −3.23 | −3.14 | −2.61 | −3.27 | −2.97 |
| slr2051 (cpcG) | Phycobilisome rod-core linker polypeptide | −0.50 | −2.01 | −1.86 | −1.56 | −2.19 | −1.76 | −1.73 | −1.72 | 1.43 |
| sll1578 (cpcA) | Phycocyanin a subunit | −14.20 | −15.06 | −22.71 | −9.60 | −8.55 | −4.06 | −4.37 | −3.14 | −1.42 |
| sll1577 (cpcB) | Phycocyanin b subunit | −13.59 | −16.82 | −24.57 | −7.86 | −9.60 | −4.27 | −4.07 | −2.85 | −1.33 |
| sll1579 (cpcC) | Phycocyanin associated linker protein | −17.20 | −14.42 | −12.19 | −10.64 | −5.87 | −3.45 | −4.31 | −3.79 | −1.93 |
| sll1580 (cpcC) | Phycocyanin associated linker protein | −14.16 | −12.11 | −16.11 | −10.14 | −5.19 | −3.03 | −3.99 | −3.34 | −2.00 |
| ssl3093 (cpcD) | Phycocyanin associated linker protein | −8.26 | −6.77 | −5.41 | −5.13 | −2.48 | −1.69 | −2.06 | −2.26 | −1.58 |
| HL-inducible protein | ||||||||||
| ssr2595 (hliB) | HL-inducible protein | 8.85 | 10.13 | 3.47 | 2.18 | 2.17 | 3.69 | 2.60 | 1.43 | 1.86 |
| ssl1633 (hliC) | HL-inducible protein | 5.28 | 7.33 | 3.92 | 4.25 | 5.02 | 4.30 | 4.61 | 1.79 | 6.85 |
| ssr1789 (hliD) | HL-inducible protein | 1.68 | 1.79 | 1.21 | 1.18 | 1.08 | 1.36 | 1.15 | −1.36 | 2.16 |
| ssl2542 (hliA) | HL-inducible protein | 1.16 | 1.73 | 1.49 | 1.55 | 3.10 | 2.81 | 2.90 | 1.65 | 1.92 |
| PS I | ||||||||||
| slr1834 (psaA) | P700 apoprotein subunit la | −4.22 | −5.62 | −5.97 | −6.98 | −2.33 | 0.91 | −1.76 | −2.33 | −2.27 |
| slr1835 (psaB) | P700 apoprotein subunit lb | −3.83 | −5.46 | −5.65 | −7.19 | −2.09 | 0.86 | −1.86 | −1.93 | −2.53 |
| slr0737 (psaD) | PS I subunit II | −5.30 | −5.07 | −3.96 | −3.02 | 0.97 | 1.12 | 0.81 | 0.69 | 1.06 |
| sll0819 (psaF) | PS I subunit III | −5.20 | −4.77 | −4.81 | −3.91 | −1.97 | −1.47 | −1.68 | −1.55 | −1.35 |
| sml0008 (psaJ) | PS I subunit IX | −3.45 | −3.52 | −3.55 | −2.95 | −1.50 | 0.80 | −1.50 | −1.51 | 0.83 |
| ssl0563 (psaC) | PS I subunit VII | −2.07 | −2.02 | −1.93 | −2.46 | −1.52 | 0.87 | −1.47 | −1.53 | 0.84 |
| slr1655 (psaL) | PS I subunit XI | −5.13 | −5.02 | −6.15 | −4.18 | −2.64 | 0.65 | 0.56 | 0.58 | −1.59 |
| PS II | ||||||||||
| sll1194 (psbU) | PS II 12-kDa extrinsic protein | 1.19 | 1.64 | 1.37 | 1.49 | 0.98 | 0.99 | 1.70 | 1.11 | 2.00 |
| sll0851 (psbC) | PS II CP43 protein | 0.96 | 1.30 | 0.77 | −1.80 | −2.43 | −1.77 | −1.99 | −2.29 | 1.48 |
| slr0906 (psbB) | PS II CP47 protein | −0.64 | 1.08 | 0.96 | 0.91 | −2.85 | −2.25 | −1.59 | −1.71 | 1.52 |
| sll1867 (psbA3) | PS II D1 protein | 2.94 | 4.31 | 2.56 | 2.48 | 1.38 | 1.64 | 1.15 | 0.70 | 6.30 |
| slr1311 (psbA2) | PS II D1 protein | 2.81 | 4.33 | 2.42 | 2.46 | 1.47 | 1.59 | 1.20 | 0.68 | 5.80 |
| slr0927 (psbD2) | PS II D2 protein | 1.76 | 2.79 | 1.52 | 1.02 | −2.47 | −1.79 | −1.87 | −2.39 | 3.43 |
| sll0849 (psbD) | PS II D2 protein | 1.52 | 2.20 | 1.21 | 0.85 | −2.36 | −1.63 | −1.95 | −2.40 | 2.62 |
| sml0005 (psbK) | PS II PsbK protein | 0.94 | 0.98 | 0.74 | 0.95 | 1.04 | 1.19 | −1.32 | −1.71 | 2.09 |
| smr0007 (psbL) | PS II PsbL protein | −1.22 | 0.91 | 0.96 | 1.13 | 0.84 | 0.91 | 1.01 | 0.82 | 2.18 |
| smr0001 (psbT) | PS II PsbT protein | 1.70 | 1.82 | 1.09 | 0.87 | 1.72 | 2.04 | 1.62 | 1.03 | 1.18 |
| sml0002 (psbX) | PS II PsbX protein | 1.75 | 1.76 | 1.12 | 1.01 | 1.97 | 2.08 | 1.72 | 1.13 | 1.10 |
| Pho regulon | ||||||||||
| sll0680 (pstS) | Phosphate binding periplasmic protein precursor | 2.66 | 4.17 | 2.32 | 2.58 | −3.37 | −3.19 | 1.21 | 0.77 | 17.48 |
| sll0681 (pstC) | Phosphate transport system permease protein | 1.77 | 2.21 | 1.24 | 1.14 | −5.48 | −4.60 | −1.56 | −2.45 | 10.35 |
| sll0682 (pstA) | Phosphate transport system permease protein | 1.28 | 1.39 | 1.04 | 0.84 | −1.83 | −1.59 | 0.79 | −1.69 | 2.63 |
| slr1248 (pstC) | Phosphate transport system permease protein | 0.88 | 0.90 | 0.89 | −1.42 | −4.44 | −4.38 | −4.41 | −4.78 | 4.61 |
| slr1249 (pstA) | Phosphate transport system permease protein | 0.85 | 0.85 | 0.87 | 0.74 | −2.43 | −2.27 | −2.49 | −3.14 | 2.48 |
| sll0683 (pstB) | Phosphate transport ATP-binding protein | 1.46 | 1.63 | 1.02 | 0.87 | −3.39 | −2.79 | −1.68 | −2.16 | 5.91 |
| sll0684 (pstB) | Phosphate transport ATP-binding protein | 1.23 | 1.40 | 0.96 | −1.78 | −3.15 | −2.70 | −1.72 | −2.13 | 4.71 |
| slr1250 (pstB) | Phosphate transport ATP-binding protein | 0.96 | 1.00 | 1.01 | 0.92 | −1.71 | −1.73 | −1.81 | −2.19 | 2.58 |
| slr1247 (pstS) | Periplasmic phosphate binding protein | 0.92 | 1.00 | 0.95 | 0.95 | −7.30 | −6.98 | −4.70 | −5.76 | 14.79 |
| sll0222 (phoA) | Alkaline phosphatase | 0.99 | 1.15 | −1.30 | −1.65 | −1.40 | 0.87 | −1.31 | −2.05 | 2.02 |
| sll0654 | Alkaline phosphatase | 0.83 | 0.89 | 0.88 | 0.77 | −6.82 | −5.82 | −5.60 | −4.90 | 9.19 |
For the genes included in this table, the transcripts changed by at least twofold in either wild-type (Wt) cells or the dspA mutant in at least one of the time points (0.5, 1, 3, and 6 h) following the transfer of cells to HL. The ratio of dspA to the wild-type (dspA/Wt) at time zero for each of the genes is also presented. Statistical significances of the expression patterns are indicated for each time point and for the dspA/Wt time zero ratio, boldface type indicates an increase in the transcript level, underlining indicates a decrease in the transcript level, and plain type indicates no significant change in the transcript level. The significance was computed by t tests (see Materials and Methods).
Group 1: transcripts in wild-type cells increased, while dspA transcripts declined.
Seven genes were placed in group 1. When wild-type cells were exposed to HL, the level for all of the seven genes increased to a significant extent, while they declined by 30 to 70% in the dspA mutant. The genes in this group include two genes related to cell division, ftsH (or slr0228) and slr1604. The ftsH gene encodes a metalloprotease involved in the replacement of damaged D1 polypeptides in the reaction center of PS II 46a, 64a, 70a). The other genes in this group include psbD2 (slr0927), which encodes a second reaction center polypeptide; pstS (sll0680), which encodes a phosphate binding protein; pstC (sll0681), which encodes a subunit of the phosphate permease; and two genes encoding proteins of unknown function (Table 2). Interestingly, transcript levels of genes in this category were 2.6- to 17-fold higher in the mutant than in wild-type cells during LL growth; the pstS and the pstC transcripts were 17- and 10-fold more abundant, respectively, in the mutant than in wild-type cells during LL growth.
Group 2: transcripts that changed slightly in wild-type cells and declined in the dspA mutant.
Surprisingly, most of the phosphate starvation-induced genes (pho), extensively studied in E. coli (78), were in group 2; this group includes pstA (slr1249), pstB (sll0684, sll0683), and pstS (slr1247). Some transcripts for pho regulon polypeptides exhibited a transient increase in HL, as is clearly observed for pstC and pstS (Tables 2 and 3). A similar increase in pstS mRNA in HL was observed previously (4). Interestingly, even in LL, the dspA strain showed very high levels (relative to wild-type cells) of pstS (both sll0680 and slr1247), pstC (sll0681 and slr1248, integral membrane protein), and pstB (sll0683 and slr1250, nucleotide binding protein) transcripts, as well as of transcripts encoding an alkaline phosphatase (sll0654) and the extracellular NucH nuclease (sll0656) (Table 3). Although nucleases have been associated with the pho regulon, it is not known whether the nucH transcript increases in response to phosphorus limitation. However, it is likely to be part of the pho regulon since it is immediately downstream of the alkaline phosphatase and the two genes may be part of the same operon. These data demonstrate the apparent derepression of several genes associated with phosphorus starvation in the dspA mutant in LL. There was no further increase in the levels of pho transcripts when the dspA mutant was exposed to HL, and the abundance of some of the transcripts declined significantly (Table 3).
Interestingly, while the transcript encoding phosphoglycolate phosphatase (CbbZp, sll1349) was not elevated significantly in wild-type cells during HL treatment, it was fivefold higher in the mutant relative to wild-type cells under LL growth conditions. This enzyme is part of the photorespiratory pathway, which can serve as an energy sink that prevents an overreduction of the photosynthetic electron transport chain (and consequent photoinhibition) and may be especially valuable under stress conditions that lead to reduced CO2 fixation (79). Photorespiration also provides metabolites for the production of glutathione, a tripeptide that protects cells from oxidative damage. Other genes in this category included htrA (slr1204; 4.7-fold), encoding a serine protease; rfbP (sll1535), encoding a putative sugar transferase; and six genes encoding proteins of unknown function.
Group 3: levels of transcript in both wild-type cells and the dspA mutant strongly declined.
Genes associated with photosynthetic function, especially those encoding phycobilisomes and PS I polypeptides, are in group 3. The levels of most cpc transcripts showed a marked decrease in both wild-type and dspA mutant cells following the imposition of HL conditions (Tables 2 and 3). The cpcE and cpcF transcripts were exceptions to this trend. These transcripts code for subunits of a lyase involved in chromophorylation of the α-PC subunit (16, 17). Interestingly, most cpc transcripts (cpcA, cpcB, cpcC, cpcD, and cpcG) were already 25 to 50% lower in the mutant than in wild-type cells during LL growth (Table 3). In contrast, most apc transcripts were equally abundant in LL-grown mutant and wild-type cells. There was a pronounced transient change in apc transcript levels following exposure of wild-type cells to HL, but by 6 h in HL, the transcript levels approached those observed for LL-grown cells.
The transcripts for most PS I polypeptides (including psaA, psaB, psaF, and psaL) were already 30 to 60% lower in the dspA mutant than in wild-type cells in LL. The most pronounced difference between the mutant and wild-type cells was for the psaA/psaB transcripts, which were 60% lower in LL-grown mutant cells (Tables 2 and 3). Furthermore, the levels of many transcripts declined in wild-type cells following exposure to HL and stayed low (psaA, psaB, psaC, psaD, psaF, psaJ, and psaL), while others didn't decline nearly as much or showed a transient decline (psaE, psaI, and psaK). For a number of PS I genes, the decline in the dspA mutant was not quite as severe as is it was in wild-type cells (e.g., psaD, psaF, and psaJ) (Table 3, compare the wild type and the dspA mutant).
Transcripts encoding polypeptides associated with chlorophyll biosynthesis are also in this category. Transcripts encoding two of the subunits of the light-independent prochlorophyllide reductase (chlL [slr0749] and chlN [slr0750]) were already reduced in the dspA mutant relative to wild-type cells (30 to 40%) in LL. While chlL and chlN transcripts declined markedly following the exposure of wild-type cells to HL (Table 2), they did not decline to nearly the same extent in the mutant (they were already low in the mutant strain in LL). The other genes in this category with characterized functions were nrtB (sll1451), encoding a nitrate/nitrite permease; crhR (slr0083), encoding a RNA helicase; and ho1 (sll1184), encoding a heme oxygenase.
Group 4: transcript that markedly declined in wild-type cells but increased in dspA.
A small number of genes were clustered in group 4, which included genes encoding the pilin polypeptide (hofG or pilA [sll1694 and sll1695]), psaD, rbpA (sll0517), and lexA (sll1626) (Table 2).
Group 5: transcripts that declined slightly in wild-type cells but increased in dspA.
The transcripts for most genes encoding constituents of the carbon-concentrating mechanism (CCM) (47) were slightly higher in the dspA mutant than in wild-type cells in LL conditions. Furthermore, the levels of most of these transcripts did not change much when wild-type cells were transferred to HL, although the ccmM and ccmK transcripts may have declined slightly. Interestingly, there was a relatively large transient increase in the levels of transcripts (four- to sixfold) from a number of the CCM genes (ccmK [sll1028 and sll0129], ccmL, ccmM, and ccmN) (Table 2) following exposure of the dspA mutant to HL, suggesting that the mutant cells were starved for CO2 immediately following the transfer to HL. There was essentially no difference in the levels of rbcL and rbcS transcripts (and in transcripts encoding components of the reductive pentose phosphate pathway; data not shown) between wild-type cells and the dspA mutant grown in LL, and HL did not significantly affect the abundance of these transcripts in wild-type cells, although there may have been a small increase in rbcL and rbcS transcript levels following the exposure of dspA to HL. Furthermore, small but significant increases in the prk transcript (encoding phosphoribulokinase) were observed in both wild-type and mutant cells following HL exposure.
Group 6: transcripts that increased in both wild-type cells and dspA.
There were a total of 69 genes whose transcript levels were increased significantly in both wild-type cells and the dspA mutant following the transfer from LL to HL. These genes included those with functions associated with photosynthesis (PS II, NADH dehydrogenase, and ATP synthase), HL and heat shock acclimation processes, and ribosome biosynthesis. Importantly, the transcripts from a number of these genes were already higher in the LL-grown dspA mutant relative to LL-grown wild-type cells. As presented in Tables 2 and 3, the level of the psbA transcripts (we were not able to distinguish between psbAII and psbAIII transcripts because they are nearly identical) was five- to sixfold higher in the mutant than in the wild-type strain grown under LL conditions. Similarly, the levels of psbDI and psbDII transcripts were 2.6- to 3.4-fold more in the dspA mutant than in wild-type cells grown in LL (Tables 2 and 3). Similar to the psbA genes, the psbDI and psbDII genes show strong sequence similarity, and substantial cross-hybridization of the cDNA probes to the psbD array elements almost surely occurred.
Upon the transfer of wild-type cells from LL to HL, the levels of transcripts for some PS II polypeptides showed a transient increase and then declined to near-LL levels (psbDI and psbDII) (Table 3), while others did not change very much. However, the psbA transcript levels went up sharply (approximately 5-fold) but declined to 2.5-fold of the LL level following a 6-h exposure to HL. Transient features of psbA transcript accumulation in cyanobacteria following HL exposure were previously described (24, 50). The changes in transcript levels for the psbA genes in the dspA mutant were not as pronounced as those in wild-type cells, and while there was some increase in psbA mRNA at early times following HL imposition (between 1.5- and 2-fold), the change was only transient, and the final level after 6 h in HL was similar to the level in LL (Table 3) (although, as already mentioned, the initial levels were much higher in the dspA mutant than in wild-type cells in LL).
Transcripts for all hli genes were also higher in the dspA mutant than in wild-type cells in LL, which had been previously observed from RNA blot hybridizations (42); the transcript for hliC showed the highest increase (approximately sevenfold) relative to wild-type cells (Table 3). Furthermore, there was a large transient increase in the levels of hliB and hliC transcripts in wild-type cells following 1 h of HL exposure (7- to 10-fold), with a subsequent decline; but the level still remained well above the LL level. The dspA mutant also exhibited elevated levels of hli transcripts following the exposure of cells to HL, although the relative increases were not as great as those observed for wild-type cells (Table 3).
Transcripts from many of the genes encoding subunits of the pyridine nucleotide dehydrogenase complex were elevated by 1.4- to 2.0-fold in the dspA mutant relative to wild-type cells grown in LL (Table 2). Furthermore, for most ndh genes, there was either no significant change in transcript levels or a transient change immediately following exposure of cells to HL (data not shown). The ndhD2 (slr1291) transcript exhibited a relatively large transient change in wild-type cells at 30 min (3.6-fold) and 1 h (3.8-fold) after HL exposure (the transient change was greater in the mutant than in wild-type cells), but the level dropped to 1.78-fold the LL value after 6 h in HL (Table 2). The NdhD2 protein has been associated with the transfer of electrons between NADPH and the plastoquinone pool (61), which may be important for controlling cyclic electron flow and redox poise under HL conditions.
Transcripts encoding subunits of the ATP synthase and cytochrome b6f complexes were generally slightly higher in the dspA mutant than in the wild-type strain in LL (data not shown). However, there were only small, transient effects of HL on levels of transcripts encoding polypeptides of these complexes, with the most significant influence on atpD, atpG, and petF transcripts, which exhibited approximately twofold transient increases.
There were pronounced changes in transcript levels for a number of stress-related genes following the exposure of both mutant and wild-type cells to HL; the transcripts for many of these increased in both mutant and wild-type cells exposed to HL (Table 2, group 6). The level of groEL1 (slr2076), groEL2 (sll0416), and groES (slr2075) transcripts were increased in both wild-type cells and the dspA mutant by six- to eightfold following HL imposition and were present at only slightly higher levels in the mutant strain during LL growth. The hsp17 mRNA showed a twofold increase following HL exposure but was already high in the dspA mutant in LL (4.5-fold dspA/wild-type cell mRNA). One of the transcripts encoding glutathione peroxidase (slr1992) was also already elevated by about threefold in LL in the dspA mutant relative to the wild type. The mRNA for the heat shock protein HtpG (sll0430) increased three- to fourfold in both wild-type and mutant cells following HL exposure but was only slightly more abundant in the dspA mutant in LL. Most transcripts for the other hsp genes (including the dnak, dnaJ, and clp genes) were not dramatically affected under the conditions used for these experiments (data not shown).
A number of other transcripts increased in both wild-type cells and the dspA mutant following exposure to HL (Table 2, group 6); some of these transcripts exhibited large transient increases. These included transcripts encoding a glucanase (slr0897; 12-fold), phosphoglycerate kinase; phosphoribulokinase (prk [slr0394]; 9-fold), which is involved in both photosynthetic and respiratory metabolism; FabG (sll0330; 5.5-fold), which is involved in lipid metabolism; CheY (slr1982; 4.8-fold), a chemosensory protein; HlyD (sll1181; 6-fold), which is involved in secretory processes; and a number of hypothetical proteins encoded by genes of unknown function. Analyzing the activities and functions of the genes encoding hypothetical proteins are likely to unveil novel aspects of acclimation processes.
(iii) Analysis of transcript levels by real-time RT-PCR.
We have used real-time RT-PCR to examine transcript levels in wild-type cells and the dspA mutant following the imposition of HL to help evaluate the microarray data. These analyses were performed for some genes whose transcripts increased and others whose transcripts declined following HL exposure. The results of these analyses are presented in Table 4. Similar to the microarray results, real-time RT-PCR showed that HL stimulated an increase in the levels of groEL-1 and groEL-2 transcripts; the highest levels in wild-type cells were observed 1 h after the imposition of HL. Similar trends were noted for the dspA mutant, although peak transcript levels were at 30 min (for both the microarray and real-time RT PCR analysis). Furthermore, similar to the groEL results, both microarray and real-time RT-PCR results showed that the psbAIII and pstS transcripts in wild-type cells accumulated to maximal levels after 1 h in HL, although the increase measured by real-time RT-PCR was two to three times that determined by microarray analysis. In contrast, the levels of these transcripts did not increase much in the dspA strain following the imposition of HL; this result probably reflects the fact that they are already high in the mutant strain prior to HL exposure. Finally, the cpcB and psaA transcripts generally declined in both wild-type and mutant cells (as measured by both methods), and the ccmK transcript declined in wild-type cells but showed a strong transient increase in the dspA mutant. Overall, the comparisons of real-time RT-PCR and microarray results were in excellent agreement with respect to the direction of change in transcript abundance for any given gene, while the extent of change determined by these two methods was often significantly different (note that the standard deviations for the real-time RT-PCR are relatively high).
TABLE 4.
Comparison of microarray and real-time RT-PCR dataa
| Gene | Test type | Fold change (SD) at time points following HL exposure
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild type
|
dspA mutant
|
||||||||
| 0.5 h | 1 h | 3 h | 6 h | 0.5 h | 1 h | 3 h | 6 h | ||
| cpcB (sll1577) | Array | 0.07 (0.01) | 0.06 (0.013) | 0.04 (0.07) | 0.13 (0.023) | 0.1 (0.04) | 0.23 (0.09) | 0.24 (0.06) | 0.35 (0.144) |
| PCR | 0.02 (0) | 0.017 (6E-04) | 0.012 (0.001) | 0.0431 (0.0055) | 0.03 (0) | 0.1 (0.01) | 0.08 (0.01) | 0.185 (0.063) | |
| psbAll (slr1311) | Array | 2.94 (0.44) | 4.31 (1.295) | 2.56 (0.062) | 2.48 (0.948) | 1.38 (0.58) | 1.64 (0.75) | 1.15 (0.26) | 0.7 (0.24) |
| PCR | 5.79 (0.33) | 9.278 (1.822) | 7.476 (0.39) | 3.4102 (0.6257) | 0.79 (0.04) | 1.35 (0.01) | 0.62 (0.05) | 0.388 (0.096) | |
| pstS (sll0680) | Array | 2.66 (0.03) | 4.17 (0.53) | 2.32 (0.07) | 2.58 (0.36) | 0.3 (0.04) | 0.31 (0.04) | 1.21 (0.19) | 0.77 (0.23) |
| PCR | 6.66 (0.66) | 11.04 (2.285) | 9.257 (0.382) | 6.8607 (1.9526) | 0.18 (0.03) | 0.18 (0.04) | 0.7 (0.13) | 0.794 (0.248) | |
| groEl-1 (slr2076) | Array | 4.45 (0.75) | 7.38 (1.57) | 2.66 (0.001) | 2.4 (0.68) | 7.65 (1.4) | 5.28 (2.69) | 5.93 (1.53) | 2.21 (0.8) |
| PCR | 14.7 (1.58) | 24.29 (7.31) | 3.92 (0.72) | 3.79 (0.05) | 20.6 (5.55) | 7.94 (1.58) | 15.5 (4.82) | 3.49 (0.243) | |
| groEl-2 (sll0416) | Array | 4.89 (0.15) | 6.04 (0.68) | 2.2 (0.33) | 1.89 (0.42) | 5.59 (2.17) | 3.93 (1.06) | 3.1 (1.5) | 1.38 (0.8) |
| PCR | 12.2 (0.24) | 15.96 (3.241) | 3.42 (0.134) | 4.48 (1.1331) | 13.6 (3.27) | 7.61 (0.12) | 8.49 (3.12) | 2.648 (0.293) | |
| ccmK (sll1029) | Array | 0.76 (0.02) | 0.69 (0.048) | 0.6 (0.011) | 0.58 (0.075) | 3.93 (0.94) | 5.97 (1.83) | 1.0 (0.2) | 0.58 (0.19) |
| PCR | 0.92 (0.04) | 0.378 (0.133) | 0.357 (0.081) | 0.2284 (0.0128) | 1.66 (0.64) | 5.57 (0.53) | 0.19 (0.08) | 0.103 (0.005) | |
| psaA (slr1834) | Array | 0.24 (0.02) | 0.18 (0.034) | 0.17 (0.007) | 0.14 (0.016) | 0.43 (0.03) | 0.91 (0.12) | 0.57 (0.09) | 0.43 (0.139) |
| PCR | 0.05 (0.03) | 0.005 (0.002) | 0.002 (4E-04) | 0.0054 (0.0023) | 0.02 (0.01) | 0.18 (0.07) | 0.58 (0.58) | 0.031 (0.003) | |
| rps1B (slr1984) | PCR | 0.9 (0.06) | 1.142 (0.265) | 1.149 (0.199) | 1.0596 (0.2681) | 0.96 (0.29) | 0.97 (0.25) | 1.43 (0.46) | 1.128 (0.343) |
The levels of cpcB, psbAII, pstS, groEL-1, groEL-2, ccmK, and psaA transcripts in both wild-type cells and the dspA mutant were quantified by real-time RT-PCR (PCR) at each time point following HL exposure and compared to spot intensities generated from microarray (Array) analysis. For the real-time RT-PCR quantification, the rps1b (slr1984) transcript was used as an external standard since the level of this transcript did not change in either wild-type cells or the dspA mutant following HL exposure (under all conditions that we used). Quantitative real-time RT-PCR analysis of the rps1b transcript was repeated 10 times at each of the 5 time points (0, 0.5, 1, 3, and 6 h) following HL exposure, while the target genes were analyzed two times using independently isolated RNA samples. SD indicates the standard deviation from the mean.
DISCUSSION
Analysis of wild-type cells.
Cells grow in environments that may rapidly fluctuate with respect to light, temperature, and nutrient availability. Changing environmental conditions will cause changes in the flux of electrons through the photosynthetic electron transport system and may also cause an imbalance in electron flow between the two photosystems, which either directly or indirectly stimulates a change in gene activity and the activities of specific protein complexes.
A number of groups have begun to examine the effects of light on levels of mRNAs in Synechocystis sp. strain PCC6803 (36, 43), as well as in other photosynthetic organisms such as Chlamydomonas reinhardtii (44) and Arabidopsis thaliana (49, 64), using both cDNA and oligonucleotide-based arrays. This “semiquantitative” technology has been validated with RNA blot hybridizations and real-time RT-PCR, as presented in this paper (and the companion paper [42]). We have been especially interested in how the photosynthetic machinery changes in response to changing light conditions. In cyanobacteria, the PS II/PS I ratio generally increases upon a shift from LL to HL (37, 48, 57). It was suggested that diminished PS I content would be expected to lower the susceptibility of cells to HL damage, particularly during prolonged exposure. Many of the results obtained with wild-type cells in this study are congruent with this idea as well as with results obtained in other studies with Synechocystis sp. strain PCC6803, in which levels of various transcripts were analyzed following a transfer of cells from LL to HL. For example, both we an others have demonstrated that levels of transcripts encoding psbAII/psbAIII and psbD genes increase following the exposure of Synechocystis sp. strain PCC6803 to HL (13, 36, 42). Increased transcript production may reflect the need for more rapid synthesis of these proteins, especially D1, which is known to rapidly turn over under HL conditions. Levels of the cpc, psaA, and psaB transcripts declined, and levels of apc transcripts remained relatively constant following the transfer of cultures to HL conditions. This result suggests that both light-harvesting capacity and PS I activity are declining under the light conditions used in this experiment, which agrees with physiological and biochemical experiments (19, 20) as well as with molecular experiments (58) performed by others. Transcripts from other genes such as those belonging to the pho regulon, and especially those involved in phosphate transport, were shown to increase following the exposure of cells to HL. This was first observed in studies in which differential display was used to identify genes responsive to HL conditions (4). Differences in values and precise sets of genes modulated by HL conditions are likely to reflect differences in conditions (and strains) used for the experiments. For example, such differences are observed when our data are compared with that of Hihara et al. (36), with respect to some of the ndh genes (ndhF3 and ndhD3), some of the ccm genes (ccmM and ccmK), and the rbcS and rbcL genes. The experiments presented in this paper result from a transfer of cells from 30 to 500 μmol of photon m−2 s−1, while Hihara et al. transferred cells from 50 to 200 μmol of photon m−2 s−1 (36). It should be noted that even the HL levels used in the experiments presented in this study are not detrimental to cell growth (the cells maintain an optimal doubling time of ∼6 h), which suggests that modulating components and activities of the photosynthetic apparatus (and other metabolic processes in the cell) compensates for the relatively high-intensity light conditions, limiting photodamage and its potential deleterious effects on cell growth. Higher light levels might result in more extreme changes in transcript levels and the activation of additional genes encoding chaperones and oxidative stress-related polypeptides.
Chaperones and heat shock proteins are also affected by light conditions. Heat shock proteins have been shown to bind unfolded polypeptides and to release them in an ATP-dependent reaction under stress condition such as those of high temperature and HL (32). These proteins may assist in the proper folding of nascent polypeptide chains as they emerge from ribosomes, participate in transmembrane protein transport, prevent aggregation of denatured proteins, and disassemble multiprotein complexes (32, 39). There are a number of genes in Synechocystis sp. strain PCC6803 that encode proteins homologous to heat shock proteins from other organisms (23, 46). Many of these genes were shown to be induced during both heat shock and HL exposure (36, 52a); these genes include clpB, htpG, dnaK (sll0170), groEL-1, groEL-2, groES, and hsp17. In our experiments, the genes for the chaperones groEL-1, groEL-2, and groES showed strong induction under HL stress; the transcripts for many of the other hsp genes did not increase to very high levels following the exposure of cells to HL. Although the actual function of GroEL or Hsp60 in Synechocystis sp. strain PCC6803 is not clear, recent studies have shown that Hsp70 of C. reinhardtii helps protect cells from photoinhibition; the overexpression of Hsp70 reduced photoinactivation of PS II and enhanced recovery from photoinhibition, while reduced expression caused the opposite effect (67). The role of the Hsp proteins in protection against photoinhibition had already been proposed based on the finding that the exposure of C. reinhardtii to heat shock resulted in enhanced resistance to light stress (68).
Analysis of mutant cells.
We recently identified the nblS gene (nonbleaching mutant sensor), which is important for the acclimation of cyanobacteria to both HL and nutrient limitation conditions (77). This gene encodes a sensor molecule predicted to be integral to the membranes and contains a PAS domain that might bind a redox active molecule such as a flavin (this still needs to be clearly established). The homolog to nblS in Synechocystis sp. strain PCC6803, which is dspA or hik33, is important for Synechocystis sp. strain PCC6803 to acclimate to HL, nutrient limitation, cold, and osmotic conditions (42, 55, 58, 73, 77). NblS and DspA polypeptides may be global regulators that tune cellular metabolism to the growth potential (77).
In our initial studies of a dspA null mutant, we demonstrated abnormal regulation of several genes that respond to light intensity (42), and it was therefore important to understand how wild-type Synechocystis sp. strain PCC6803 responded to HL and the relationship between the HL responses observed in the mutant and wild-type strains. In this study, we have demonstrated that numerous transcripts were markedly different in abundance in wild-type Synechocystis sp. strain PCC6803 and the dspA mutant, even under our LL conditions. Importantly, many transcripts that accumulated in wild-type cells in HL were already high in the dspA strain in LL. Furthermore, a number of transcripts that declined in wild-type cells following exposure to HL were already low in LL-grown dspA. Such changes include increases in levels of transcripts encoding PsbAII/PsbAIII, PsbD, the Hli polypeptides, the HtpG and to some extent the GroEL/GroES chaperones and decreases in transcripts encoding the PC subunits and the associated linker proteins (CpcA, CpcB, CpcC, CpcD, and CpcG), the reaction center polypeptides of PS I (PsaA and PsaB), and polypeptides involved in chlorophyll biosynthesis (ChlL and ChlN); many of these changes were subsequently verified by real-time RT-PCR. Additionally, the dspA mutant showed increased levels of transcripts for some genes encoding polypeptides associated with oxidative stress, such as glutathione peroxidase. Finally, additional increases in at least some of these transcripts (e.g., psbAII/III and hliA, hliB, hliC, and hliD) were observed upon exposure of the dspA mutant to HL. The transcript accumulation data presented in this paper, along with the phenotypes determined for the dspA mutant (42, 58, 73, 77), demonstrate that the mutant strain is constitutively derepressed for many genes associated with stress conditions. The DspA protein may be directly or indirectly involved in negative regulation of stress-responsive genes or the dspA mutant may be experiencing stress even under normal growth conditions. The responses of at least some of these genes that appear to be derepressed in the dspA mutant still show some response to HL (although the extent of the response might be reduced), suggesting that other mechanisms may also function to control the expression levels of these genes. However, the results from the studies in this paper do suggest that the DspA protein is likely to be a global regulatory element that either directly or indirectly controls numerous genes in the cell and may in fact be key for maintaining an appropriate homeostatic cellular milieu as environmental conditions fluctuate. Of course, there are transcripts whose abundances in wild-type cells and the dspA mutant cannot be easily explained by the hypothesis that the mutant is more sensitive to light levels. Therefore, it is likely that the DspA regulatory activity integrates with other regulatory systems to generate a final homeostatic output. It is important to understand how DspA might be sensing changing environmental conditions; the most obvious possibilities are that DspA monitors intracellular redox levels, the concentration of reactive oxygen species, and possibly even levels of blue or UV-A light (or a combinations of these factors) (77). Other sensors thought to participate in redox control of gene expression in Synechocystis sp. strain PCC6803 have recently been identified by Li and Sherman (52).
The dspA mutant appeared to be constitutively activated for many genes that are thought to be constituents of the pho regulon. These components include pstS, pstC, a gene encoding an alkaline phosphatase, and nucH. However, many of these genes are also activated under HL conditions. Interestingly, the sensory polypeptide SphS (sll0337) of Synechocystis sp. strain PCC6803 may contain a PAS domain (4), which has the potential to bind a chromophore and sense light and/or redox levels and coordinate light availability with the acquisition and assimilation of phosphate. Alternatively, it is possible that a major function of DspA is to manage the phosphate budget of the cell, and when this is not properly achieved in the mutant strain, the pho regulon is activated and many genes that are generally associated with stress and the absorption of excess excitation energy become active. Secondary effects may also cause the activation of some of the ccm genes when the mutant is exposed to HL (the mutant cannot take up enough for optimal CO2 fixation). All of these results raise the question, what are the primary and secondary effects of deleting dspA in Synechocystis sp. strain PCC6803? It is clear that the phenotype of the mutant and its pattern of transcript accumulation strongly suggest that it is experiencing generalized stress responses that occur even under LL and high-nutrient conditions; these responses in many cases match the responses observed for wild-type cells transferred to HL. However, it is still not clear which genes are under the direct control of DspA as a consequence of its interactions with specific response regulators and exactly what factor(s) in the intracellular environment is being monitored by this sensory polypeptide.
Acknowledgments
We thank Bi-Huei Hou, Katrina Ramonell, and Sue Thayer in Shanna Somerville's laboratory for providing technical support and useful discussion on microarray spotting and processing. We are grateful for the help provided by Janos Demeter in the Department of Genetics and the Stanford Microarray Database (SMD) for microarray data normalization, retrieval, and analysis. We specially thank Q. He for his helpful suggestions during the course of this work.
This work was supported by funds awarded to A.R.G. by the Carnegie Institution of Washington, NSF (grant MCB9727836), and USDA (grant USDA98-35301).
Footnotes
This is manuscript no. 1632 from the Carnegie Institution.
REFERENCES
- 1.Aro, E.-M., I. Virgin, and B. Andersson. 1993. Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143:113-134. [DOI] [PubMed] [Google Scholar]
- 2.Asada, K. 1994. Production and action of active oxygen species in photosynthetic tissues, p. 77-104. In C. H. Foyer and P. M. Mullineaux (ed.), Causes of photooxidative stress and amelioration of defense systems in plants. CRC Press, Boca Raton, Fla.
- 3.Bartsevich, V. V., and S. V. Shestakov. 1995. The dspA gene product of the cyanobacterium Synechocystis sp. strain PCC6803 influences sensitivity to chemically different growth inhibitors and has amino acid similarity to histidine protein kinases. Microbiology 141:2915-2920. [DOI] [PubMed] [Google Scholar]
- 4.Bhaya, D., D. Vaulot, P. Amin, A. W. Takahashi, and A. R. Grossman. 2000. Isolation of regulated genes of the cyanobacterium Synechocystis sp. strain PCC6803 by differential display. J. Bacteriol. 182:5692-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhaya, D., N. Watanabe, T. Ogawa, and A. R. Grossman. 1999. The role of an alternate sigma factor in motility and pili formation in the cyanobacterium Synechocystis sp. strain PCC6803. Proc. Natl. Acad. Sci. USA 96:3188-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogorad, L. 1975. Phycobiliproteins and complementary chromatic adaptation. Annu. Rev. Plant Physiol. 26:369-401. [Google Scholar]
- 7.Bustos, S. A., and S. S. Golden. 1992. Light-regulated expression of the psbD gene family in Synechococcus sp. strain PCC7942: evidence for the role of duplicated psbD genes in cyanobacteria. Mol. Gen. Genet. 232:221-230. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, D. 1996. D1 exchange and the photosystem II repair cycle in the cyanobacterium Synechococcus. Plant Sci. 115:183-190. [Google Scholar]
- 9.Casey, E. S., and A. R. Grossman. 1994. In vivo and in vitro characterization of the light-regulated cpcB2A2 promoter of Fremyella diplosiphon. J. Bacteriol. 176:6362-6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow, W. S. 1994. Photoprotection and photoinhibitory damage. Adv. Mol. Cell. Biol. 10:151-196. [Google Scholar]
- 11.Collier, J. L., and A. R. Grossman. 1992. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC7942: not all bleaching is the same. J. Bacteriol. 174:4718-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collier, J. L., and A. R. Grossman. 1994. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 13:1039-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Constant, S., I. Perewoska, M. Alfonso, and D. Kirilovsky. 1997. Expression of the psbA gene during photoinhibition and recovery in Synechocystis PCC6714: inhibition and damage of transcriptional and translational machinery prevent the restoration of photosystem II activity. Plant Mol. Biol. 34:1-13. [DOI] [PubMed] [Google Scholar]
- 14.Conway, T., and G. K. Schoolnik. 2003. Microarray expression profiling: capturing a genome-wide portrait of the transcriptome. Mol. Microbiol. 47:879-889. [DOI] [PubMed] [Google Scholar]
- 15.Dolganov, N. A. M., D. Bhaya, and A. R. Grossman. 1995. Cyanobacterial protein with similarity to the chlorophyll a/b-binding proteins of higher plants: evolution and regulation. Proc. Natl. Acad. Sci. USA 92:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fairchild, C. D., and A. N. Glazer. 1994. Oligomeric structure, enzyme kinetics, and substrate specificity of the phycocyanin α-subunit phycocyanobilin lyase. J. Biol. Chem. 269:8686-8694. [PubMed] [Google Scholar]
- 17.Fairchild, C. D., J. Zhao, J. Zhou, S. E. Colson, D. A. Bryant, and A. N. Glazer. 1992. Phycocyanin α-subunit phycocyanobilin lyase. Proc. Natl. Acad. Sci. USA 89:7017-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finazzi, G., F. Rappaport, A. Furia, M. Fleischmann, J. D. Rochaix, F. Zito, and G. Forti. 2002. Involvement of state transitions in the switch between linear and cyclic electron flow in Chlamydomonas reinhardtii. EMBO J. 3:280-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujita, Y. 1997. A study on the dynamic features of photosystem stoichiometry: accomplishments and problems for future studies. Photosynth. Res. 53:83-93. [Google Scholar]
- 20.Fujita, Y., A. Murakami, and K. Aizawa. 1992. Light acclimation of thylakoid system in cyanophytes: regulation of PSI formation in response to light regime, p. 301-308. In N. Murata (ed.), Research in photosynthesis, vol. IV. Kluwer, Dordrecht, The Netherlands.
- 21.Funk, C., and W. Vermaas. 1999. A cyanobacterial gene family coding for single-helix proteins resembling part of the light-harvesting proteins from higher plants. Biochemistry 38:9397-9404. [DOI] [PubMed] [Google Scholar]
- 22.Gill, R. T., E. Katsoulakis, W. Schmitt, G. Taroncher-Oldenburg, J. Misra, and G. Stephanopoulos. 2002. Genome-wide dynamic transcriptional profiling of the light-to-dark transition in Synechocystis sp. strain PCC 6803. J. Bacteriol. 184:3671-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glatz, A., I. Vass, D. A. Los, and L. Vígh. 1999. The Synechocystis model of stress: from molecular chaperones to membranes. Plant Physiol. Biochem. 37:1-12. [Google Scholar]
- 24.Golden, S. S. 1994. Light-responsive gene expression and the biochemistry of the photosystem II reaction center, p. 693-714. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer, Dordrecht, The Netherlands.
- 25.Golden, S. S., J. Brusslan, and R. Haselkorn. 1986. Expression of a family of psbA genes encoding a photosystem II polypeptide in the cyanobacterium Anacystis nidulans R2. Eur. Mol. Biol. Org. J. 5:2789-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gollub, J., C. A. Ball, G. Binkley, J. Demeter, D. B. Finkelstein, J. M. Hebert, T. Hernandez-Boussard, H. Jin, M. Kaloper, J. C. Matese, M. Schroeder, P. O. Brown, D. Botstein, and G. Sherlock. 2003. The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res. 31:94-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grossman, A. R. 1990. Chromatic adaptation and the events involved in phycobilisome biosynthesis. Plant Cell Environ. 13:651-666. [Google Scholar]
- 28.Grossman, A. R., D. Bhaya, K. E. Apt, and D. M. Kehoe. 1995. Light-harvesting complexes in oxygenic photosynthesis: diversity, control and evolution. Annu. Rev. Genet. 29:231-287. [DOI] [PubMed] [Google Scholar]
- 29.Grossman, A. R., D. Bhaya, and Q. He. 2001. Tracking the light environment by cyanobacteria and the dynamic nature of light harvesting. J. Biol. Chem. 276:11449-11452. [DOI] [PubMed] [Google Scholar]
- 30.Grossman, A. R., M. R. Schaefer, G. G. Chiang, and J. L. Collier. 1993. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol. Rev. 57:725-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han, B. P. 2002. A mechanistic model of algal photoinhibition induced by photodamage to photosystem-II. J. Theor. Biol. 214:519-527. [DOI] [PubMed] [Google Scholar]
- 32.Hartl, F. 1996. Molecular chaperones in cellular protein folding. Nature 381:571-579. [DOI] [PubMed] [Google Scholar]
- 33.Havaux, M., G. Guedeney, Q. He, and A. R. Grossman. 2003. Elimination of high-light-inducible polypeptides related to eukaryotic chlorophyll a/b-binding proteins results in aberrant photoacclimation in Synechocystis PCC6803. Biochim. Biophys. Acta 1557:21-33. [DOI] [PubMed] [Google Scholar]
- 34.He, Q., N. Dolganov, O. Bjorkman, and A. R. Grossman. 2001. The high light-inducible polypeptides in Synechocystis PCC6803. Expression and function in high light. J. Biol. Chem. 276:306-314. [DOI] [PubMed] [Google Scholar]
- 35.Herranen, M., E. M. Aro, and T. Tyystjarvi. 2001. Two distinct mechanisms regulate the transcription of photosystem II genes in Synechocystis sp. PCC 6803. Physiol. Plant. 112:531-539. [DOI] [PubMed] [Google Scholar]
- 36.Hihara, Y., A. Kamei, M. Kanehisa, A. Kaplan, and M. Ikeuchi. 2001. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13:793-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hihara, Y., K. Sonoike, M. Kanehisa, and M. Ikeuchi. 2003. DNA microarray analysis of redox-responsive genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 185:1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofmann, E., P. M. Wrench, F. P. Sharples, R. G. Hiller, W. Welte, and K. Diederichs. 1996. Structural basis of light harvesting by carotenoids: peridinin-chlorophyll-protein from Amphidinium carterae. Science 272:1788-1791. [DOI] [PubMed] [Google Scholar]
- 39.Horst, M., W. Oppliger, S. Rospert, H. J. Schonfeld, G. Schatz, and A. Azem. 1997. Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J. 16:1842-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Houmard, J., V. Capuano, T. Cousin, and N. Tandeau de Marsac. 1988. Genes encoding core components of the phycobilisome in the cyanobacterium Calothrix sp. strain 7601: occurrence of a multigene family. J. Bacteriol. 170:5512-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Houmard, J., V. Capuano, T. Cousin, and N. Tandeau de Marsac. 1988. Isolation and molecular characterization of the gene encoding allophycocyanin B, a terminal energy acceptor in cyanobacterial phycobilisomes. Mol. Microbiol. 2:101-107. [PubMed] [Google Scholar]
- 42.Hsiao, H.-Y., Q. He, L. G. van Waasbergen, and A. R. Grossman. 2004. Control of photosynthetic and high-light-responsive genes by the histidine kinase DspA: negative and positive regulation and interactions between signal transduction pathways. J. Bacteriol. 186:3882-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang, L., M. P. McCluskey, H. Ni, and R. A. LaRossa. 2002. Global gene expression profiles of the cyanobacterium Synechocystis sp. strain PCC 6803 in response to irradiation with UV-B and white light. J. Bacteriol. 184:6845-6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Im, C. S., Z. Zhang, J. Shrager, C. W. Chang, and A. R. Grossman. 2003. Analysis of light and CO2 regulation in Chlamydomonas reinhardtii using genome-wide approaches. Photosynth. Res. 75:111-125. [DOI] [PubMed] [Google Scholar]
- 45.Jansson, S. 1994. The light-harvesting chlorophyll a/b-binding proteins. Biochim. Biophys. Acta 1184:1-19. [DOI] [PubMed] [Google Scholar]
- 46.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, T. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 46a.Kanervo, E., C. Spetea, Y. Nishiyama, N. Murata, B. Andersson, and F. M. Aro. 2003. Dissecting a cyanobacterial proteolytic system: efficiency in inducing degradation of the D1 protein of photosystem II in cyanobacteria and plants. Biochim. Biophys. Acta 1607:131-140. [DOI] [PubMed] [Google Scholar]
- 47.Kaplan, A., and L. Reinhold. 1999. CO2 concentrating mechanisms in photosynthetic microorganisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:539-570. [DOI] [PubMed] [Google Scholar]
- 48.Kawamura, M., M. Mimuro, and Y. Fujita. 1979. Quantitative relationship between two reaction centers in the photosynthetic system of blue-green algae. Plant Cell Physiol. 20:697-705. [Google Scholar]
- 49.Kimura, M., Y. Y. Yamamoto, M. Seki, T. Sakurai, M. Sato, T. Abe, S. Yoshida, K. Manabe, K. Shinozaki, and M. Matsui. 2003. Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem. Photobiol. 77:226-233. [DOI] [PubMed] [Google Scholar]
- 50.Kulkarni, R. D., and S. S. Golden. 1994. Adaptation to high light intensity in Synechococcus sp. strain PCC 7942: regulation of three psbA genes and two forms of the D1 protein. J. Bacteriol. 176:959-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kulkarni, R. D., and S. S. Golden. 1995. Form II of D1 is important during transition from standard to high light intensity in Synechococcus sp. strain PCC7942. Photosynth. Res. 46:435-443. [DOI] [PubMed] [Google Scholar]
- 52.Li, H., and L. A. Sherman. 2000. A redox-responsive regulator of photosynthesis gene expression in the cyanobacterium Synechocystis sp. strain PCC6803. J. Bacteriol. 182:4268-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52a.Mary, I., C.-J. Tu., A. R. Grossman, and D. Vaulot. 2004. Effects of high light on transcripts of stress-associated genes for the cyanobacteria Synechocystis sp. PCC 6803 and Prochlorococcus MED4 and MIT9313. Microbiology 150:1271-1281. [DOI] [PubMed] [Google Scholar]
- 53.Mazel, D., J. Houmard, and N. Tandeau de Marsac. 1988. A multigene family in Calothrix sp. PCC7601 encodes phycocyanin, the major component of the cyanobacterial light-harvesting antenna. Mol. Gen. Genet. 211:296-304. [DOI] [PubMed] [Google Scholar]
- 54.Melis, A. 1989. Spectroscopic methods in photosynthesis: photosystem stoichiometry and chlorophyll antenna size. Philos. Trans. R. Soc. Lond. B Biol. Sci. 323:397-410. [Google Scholar]
- 55.Mikami, K., Y. Kanesaki, I. Suzuki, and N. Murata. 2002. The histidine kinase Hik33 perceives osmotic stress and cold stress in Synechocystis sp. PCC6803. Mol. Microbiol. 46:905-915. [DOI] [PubMed] [Google Scholar]
- 56.Mohamed, A., and C. Jansson. 1989. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol. Biol. 13:693-700. [DOI] [PubMed] [Google Scholar]
- 57.Murakami, A., and Y. Fujita. 1991. Steady state of photosynthetic electron transport in cells of the cyanophyte Synechocystis PCC6714 having different stoichiometry between PS I and PS II: analysis of flash-induced oxidation-reduction of cytochrome f and P700 under steady state of photosynthesis. Plant Cell Physiol. 32:213-222. [Google Scholar]
- 58.Muramatsu, M., and Y. Hihara. 2003. Transcriptional regulation of genes encoding subunits of photosystem I during acclimation to high-light conditions in Synechocystis sp. PCC6803. Planta 216:446-453. [DOI] [PubMed] [Google Scholar]
- 59.Niyogi, K. K. 1999. Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:333-359. [DOI] [PubMed] [Google Scholar]
- 60.Oelmüller, R., P. B. Conley, N. Federspiel, W. R. Briggs, and A. R. Grossman. 1988. Changes in accumulation and synthesis of transcripts encoding phycobilisome components during acclimation of Fremyella diplosiphon to different light qualities. Plant Physiol. 88:1077-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohkawa, H., H. B. Pakrasi, and T. Ogawa. 2000. Two types of functionally distinct NAD(P)H dehydrogenases in Synechocystis sp. strain PCC6803. J. Biol. Chem. 275:31630-31634. [DOI] [PubMed] [Google Scholar]
- 62.Parkinson, J. S. 1993. Signal transduction schemes of bacteria. Cell 73:857-871. [DOI] [PubMed] [Google Scholar]
- 63.Postier, B. L., H. L. Wang, A. Singh, L. Impson, H. L. Andrews, J. Klahn, H. Li, G. Risinger, D. Pesta, M. Deyholos, D. W. Galbraith, L. A. Sherman, and R. L. Burnap. 2003. The construction and use of bacterial DNA microarrays based on an optimized two-stage PCR strategy. BMC Genomics 4:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rossel, J. B., I. W. Wilson, and B. J. Pogson. 2002. Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol. 130:1109-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64a.Sakamoto, W., A. Zaltsman, Z. Adam, and Y. Takahashi. 2003. Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15:2843-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sauer, J., U. Schreiber, R. Schmid, U. Volker, and K. Forchhammer. 2001. Nitrogen starvation-induced chlorosis in Synechococcus PCC 7942. Low-level photosynthesis as a mechanism of long-term survival. Plant Physiol. 126:233-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 67.Schroda, M., O. Vallon, F. A. Wollman, and C. F. Beck. 1999. A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11:1165-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schuster, G., R. Timberg, and I. Ohad. 1988. Turnover of thylakoid photosystem II proteins during photoinhibition of Chlamydomonas reinhardtii. Eur. J. Biochem. 177:403-410. [DOI] [PubMed] [Google Scholar]
- 69.Schwarz, R., and A. R. Grossman. 1998. A response regulator of cyanobacteria integrates diverse environmental signals and is critical for survival under extreme conditions. Proc. Natl. Acad. Sci. USA 95:11008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sherlock, G., T. Hernandez-Boussard, A. Kasarskis, G. Binkley, J. C. Matese, S. S. Dwight, M. Kaloper, S. Weng, H. Jin, C. A. Ball, M. B. Eisen, P. T. Spellman, P. O. Brown, D. Botstein, and J. M. Cherry. 2001. The Stanford Microarray Database. Nucleic Acids Res. 29:152-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70a.P., Silva, E. Thompson, S. Bailey, O. Kruse, C. W. Mullineaux, C. Robinson, N. H. Mann, and P. J. Nixon. 2003. FtsH is involved in the early stages of repair of photosystem II in Synechocystis sp PCC 6803. Plant Cell 15:2152-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stock, J. 1999. Signal transduction: gyrating protein kinases. Curr. Biol. 9:R364-R367. [DOI] [PubMed] [Google Scholar]
- 72.Stock, J., and S. Da Re. 2000. Signal transduction: response regulators on and off. Curr. Biol. 10:R420-R424. [DOI] [PubMed] [Google Scholar]
- 73.Suzuki, I., Y. Kanesaki, K. Mikami, M. Kanehisa, and N. Murata. 2001. Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol. Microbiol. 40:235-244. [DOI] [PubMed] [Google Scholar]
- 74.Tandeau de Marsac, N., W. E. Borrias, C. J. Kuhlemeier, A. M. Castets, G. A. van Arkel, and C. A. M. J. J. van den Hondel. 1982. A new approach for molecular cloning in cyanobacteria: cloning of an Anacystis nidulans met gene using a Tn901-induced mutant. Gene 20:111-119. [DOI] [PubMed] [Google Scholar]
- 75.Tandeau de Marsac, N., and J. Houmard. 1993. Adaptation of cyanobacteria to environmental stimuli: new steps towards molecular mechanisms. FEMS Microbiol. Rev. 104:119-190. [Google Scholar]
- 76.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Waasbergen, L. G., N. Dolganov, and A. R. Grossman. 2002. nblS, a gene involved in controlling photosynthesis-related gene expression during high light and nutrient stress in Synechococcus elongatus PCC 7942. J. Bacteriol. 184:2481-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wanner, B. L. 1996. Signal transduction in the control of phosphate-regulated genes of Escherichia coli. Kidney Int. 49:964-967. [DOI] [PubMed] [Google Scholar]
- 79.Wingler, A., P. J. Lea, W. P. Quick, and R. C. Leegood. 2000. Photorespiration: metabolic pathways and their role in stress protection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:1517-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wykoff, D., J. Davies, and A. Grossman. 1998. The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 117:129-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu, H., D. Vavilin, C. Funk, and W. Vermaas. 2002. Small cab-like proteins regulating tetrapyrrole biosynthesis in the cyanobacterium Synechocystis sp. PCC6803. Plant Mol. Biol. 49:149-160. [DOI] [PubMed] [Google Scholar]
- 82.Ye, R. W., T. Wang, L. Bedzyk, and K. M. Croker. 2001. Applications of DNA microarrays in microbial systems. J. Microbiol. Methods 47:257-272. [DOI] [PubMed] [Google Scholar]